Abstract
In the present study, we hypothesized that hypoxia-inducible factor 1α (HIF-1α)-mediated mitophagy plays a protective role in ischemia/reperfusion (I/R)-induced acute kidney injury (AKI). Mitophagy was evaluated by measuring the changes of mitophagy flux, mitochondria DNA copy number, and the changes of mitophagy-related proteins including translocase of outer mitochondrial membrane 20 (TOMM20), cytochrome c oxidase IV (COX IV), microtubule-associated protein 1 light chain 3B (LC3B), and mitochondria adaptor nucleoporin p62 in HK2 cells, a human tubular cell line. Results show that HIF-1α knockout significantly attenuated hypoxia/reoxygenation (H/R)-induced mitophagy, aggravated H/R-induced apoptosis, and increased the production of reactive oxygen species (ROS). Similarly, H/R induced significantly increase in Bcl-2 19-kDa interacting protein 3 (BNIP3), a downstream regulator of HIF-1α. Notably, BNIP3 overexpression reversed the inhibitory effect of HIF-1α knockout on H/R-induced mitophagy, and prevented the enhancing effect of HIF-1α knockout on H/R-induced apoptosis and ROS production. For in vivo study, we established HIF-1αflox/flox; cadherin-16-cre mice in which tubular HIF-1α was specifically knockout. It was found that tubular HIF-1α knockout significantly inhibited I/R-induced mitophagy, and aggravated I/R-induced tubular apoptosis and kidney damage. In contrast, adenovirus-mediated BNIP3 overexpression significantly reversed the decreased mitophagy, and prevented enhanced kidney damage in tubular HIF-1α knockout mice with I/R injury. In summary, our study demonstrated that HIF-1α-BNIP3-mediated mitophagy in tubular cells plays a protective role through inhibition of apoptosis and ROS production in acute kidney damage.
Keywords: Tubular cells, Hypoxia-inducible factor 1α, Bcl-2 19-kDa interacting protein 3, Mitophagy, Acute kidney injury
1. Introduction
Acute kidney injury (AKI) is a major kidney disease which is often caused by renal ischemia/reperfusion (I/R), nephrotoxins, and sepsis. In clinical practice, AKI remains a difficult problem for clinicians in diagnosis and treatment in spite of the high attention to this common clinical disease, till now, the morbidity and mortality rates of AKI in intensive care unit patients could reach 50%–70% [1]. In molecular level, the commonest form of renal AKI is acute tubular necrosis, characterized by tubular epithelial cell death and dysfunction in one or several tubular segments [2,3]._ENREF_3.
Ischemia/reperfusion injury (IRI) is caused by a sudden temporary impairment of the blood flow to the particular organ [4,5]. Of note, IRI due to hypotension or sepsis is the most common cause of human AKI [4,5]. In particular, it has been demonstrated that an increase of hypoxia-inducible factor 1 (HIF-1) is a hallmark change in I/R [6]. HIF-1 is a heterodimer composed of a constitutively expressed HIF-1β subunit and an O2-regulated HIF-1α subunit.4 Under normoxic conditions, HIF-1α is continuously produced and degraded proteasomally; whereas under hypoxic conditions, HIF-1α protein stabilizes and transactivates genes involved in adaptation to hypoxic conditions. It is generally believed that HIF-1α exerts a protective role in IRI at least partially through increased expression of HIF-1α-targeted genes involved in the shift of glucose metabolism to glycolysis [5,7], scavenging oxygen free radicals [7], and cell survive [8]. For example, It has been shown that miR-30c-5p stabilizes HIF-1α expression by targeting suppressor of cytokine signaling-3 (SOCS3), thereby protecting hypoxia/reoxygenation (H/R)-induced apoptosis and inducing cell proliferation [[9], [10], [11]]._ENREF_19 _ENREF_8However, the mechanism underlying the protective effect of HIF-1α in I/R-induced AKI remains largely unknown.
The mitochondrial homeostasis is maintained by two interlinked processes, mitochondrial dynamics and mitophagy [12]. Mitochondria are highly dynamic organelles that undergo constant fission and fusion. The elimination of damaged mitochondria, named mitophagy, helps to maintain the adenosine triphosphate (ATP) production required by the cells [13]. There are accumulating data showing that Bcl-2 19-kDa interacting protein 3 (BNIP3), a mitochondrial protein, plays a critical role in mitophagy. For example, Chourasia et al., reported that loss of BNIP3 leads to mitophagy defects in mammary tumor cells [14], and that tumor suppressor p53 inhibits the transcription and expression of BNIP3, resulting in mitophagy stagnation [15]._ENREF_14 Furthermore, it was found that BNIP3-mediated mitophagy plays a protective role in IRI-induced AKI [23], and in the generation of natural killer cell memory [16].
Studies show that mitophagy plays a protective role in liver [17] and lung [18,19]. In the brain and heart, however, the role of mitophagy remains elusive, it can be protective or detrimental [20,21]. Recently, Liu et al., found that the loss of autophagy in proximal tubules worsens tubular injury and renal function, and causes the accumulation of damaged mitochondria in the autophagy-deficient kidneys subjected to IRI in AKI model [22]. However, the precise mechanism for the regulation of mitophagy and its potential role in the kidney, especially in renal tubular cells, remains largely unknown. _ENREF_5In the present study, we performed in vitro and in vivo study to investigate whether HIF-1α exerts a protective role by inducing BNIP3-mediated mitophagy in IRI-induced AKI.
2. Materials and methods
2.1. Cell culture and H/R model
Human proximal tubular cells (HK2, CRL-2190), from American Type Culture Collection (ATCC, Manassas, VA, USA), were cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12, Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS, Gibco, Waltham, MA, USA) in the incubator with 5% CO2 at 37 °C. For the H/R model, cells were firstly cultured in serum-free medium for 24h, and they were transferred into a humidified hypoxic incubator with 1% O2, 5% CO2 and 94% N2 for another 24h. After the hypoxia treatment, cells were transferred into a normoxic incubator (21% O2, 5% CO2 and 74% N2) with fresh culture medium supplemented with 10% FBS, and cultured for 2, 6, 12, 24 or 72h as indicated. Cells cultured in culture medium with 10% FBS without H/R were used as no H/R cells.
2.2. Transfection of HIF-1α siRNA and BNIP3-overexpressing plasmid
The HIF-1α siRNA duplex was designed to target nucleotides 1521–1541 of the HIF-1α mRNA sequence (NM001530), which comprised of sense 5′-CUGAUGACCAGCAACUUGAdTdT-3′ and antisense 5′-UCAAGUUGCUGGUCAUCAGdTdT-3’. The BNIP3 siRNA duplex was from Santa Cruz Biotechnology (NM_004052, Dallas, TX, USA). The inverted HIF-1α control duplex comprised of sense 5′-AGUUCAACGACCAGUAGUCdTdT-3′ and antisense 5′-GACUACUGGUCGUUGAdTdT-3’. BNIP3-overexpression plasmid was purchased from LncBio (Shanghai, China). Briefly, human BNIP3 cDNA was cloned from NM_004052, the resultant fragments were inserted into pCDH-CMV-MCS-Flag-EF1α-Puro lentivector between NheI and EcoRI sites. The construction details of BNIP3-overexpression plasmid were shown in part one of supplementary Materials and Methods.
After HK2 cells were transfected with HIF-1α siRNA or scramble RNA under normoxia for 12h, they were transfected into HK2 cells by using Lipofectamine 3000 reagents (Thermo Fisher, Waltham, MA, USA). In particular, some cells were transfected with red fluorescence protein-microtubule-associated protein 1 light chain 3B (RFP-LC3B) plasmid (p36236, Thermo Fisher, Waltham, MA, USA) for autophagy, treated with MitoTracker Green (a specific mitochondria marker, 50 nM, 15min, 37 °C; C1048, Beyotime, Shanghai, China), and then treated with Hoechst 33342 (a fluorescent dye to stain nuclei in live or fixed cells, 1 μg/mL, 5min, 37 °C, Sigma-Aldrich, St. Louis, MO, USA). Some cells were transfected with BNIP3-overexpression plasmid or vector under normoxia for 12h before the staining with MitoTracker Green and Hoechst. The cells were observed with confocal microscopy (LSCM, LSM800, ZEISS, Oberkochen, Germany), and the formation of mitophagosomes was evaluated by counting the amounts of cells with co-localized RFP-LC3B and MitoTracker Green per 0.25 mm [24].
2.3. Quantitative PCR
Total RNA from the cells was extracted by using Trizol reagent (Invitrogen, Waltham, MA, USA) according to the manufacture manual. Complementary DNA (cDNA) was synthesized by using oligo (dT) primer and PrimeScript™ RT Reagent Kit (Takara, Shiga, Japan). Quantitative PCR (qPCR) was performed to amplify the cDNA using the SYBR Premix Ex Tag Kit (Takara, Shiga, Japan) and an ABI 7500 Sequencing Detection System (Applied Biosystems, Foster City, CA, USA). The gene primers were as follows: BNIP3 (access No. NM_004052.3), forward 5′-TTGGATGCACAACATGAATCAGG-3′, reverse 5′-TCTTCTGACTGAGAGCTATGGTC-3’; β-actin (access No. NM_001101.4), forward 5′-CATGTACGTTGCTATCCAGGC-3′, reverse 5′-CTCCTTAATGTCACGCACGAT-3’. The relative gene expressions were calculated in accordance with the ΔΔCt method. Relative mRNA levels were expressed by the values of 2−ΔΔCt as described previously [25].
2.4. Western blot
The protein of HK2 cells and renal cortex tissues were prepared following a routine procedure in the lab [26]. After the protein samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and moved to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA), they were blocked with 5% skim milk for 60min and then incubated with primary antibodies overnight at 4 °C. The primary antibodies used were at dilutions of 1:1000 for HIF-1α (A11945, Abclonal, Wuhan, China), 1:1000 for LC3B (L7543, Sigma-Aldrich, St. Louis, MO, USA), 1:500 for COX IV (A10098, Abclonal, Wuhan, China), 1:1000 for translocase of outer mitochondrial membrane 20 (TOMM20) (11802-1-AP, Proteintech, Chicago, IL, USA), 1:500 for p62 (A7758, Abclonal, Wuhan, China), 1:1000 for BNIP3 (ab38621, Abcam, Cambridge, FL, USA), 1:1000 for cleaved caspase 3 (CC3) (9664, Cell Signaling Test, Danvers, MA, USA), and 1:3000 for β-actin (A5316, Sigma-Aldrich, St. Louis, MO, USA). After incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA), the membranes were developed by using an enhanced chemiluminescence detection system. β-actin was set as internal control with anti-β-actin antibody (Sigma-Aldrich, St. Louis, MO, USA). The expression of the proteins was quantified according to the gray value by using Image-J software (Version 1.51).
2.5. mtDNA copy number
Total DNA was extracted from cell samples. To quantify mitochondria DNA (mtDNA) copy number, real-time PCR was performed using an ABI 7500 Sequencing Detection System (Applied Biosystems, Foster City, CA, USA) as described previously [27]. The mtDNA primers were: forward 5′-CGAAAGGACAAGAGAAATAAGG-3′, reverse 5′-CTGTAAAGTTTTAAGTTTTATGCG-3’. β-glubin was used as genomic DNA control, and the primers were: forward 5′-CAACTTCATCCACGTTCACC-3′, reverse 5′-GAAGAGCCAAGGACAGGTAC-3’.
2.6. Mitophagy flux
Mitophagy flux was evaluated as described previously [28]. Briefly, cells were stained with MitoTracker Green FM (500 nM, C1048, Beyotime, Shanghai, China), and analyzed by using CytoFlex platform (Beckman Co., Ltd, South Kraemer Boulevard Brea, CA, USA). Mitophagy flux was defined as the ratio of MitoTracker Green FM fluorescence in the presence of mitophagy and lysosomal inhibitor (bafilomycin A1, Baf, 10 nM, Sigma-Aldrich, St. Louis, MO, USA) to that in the absence of inhibitor, normalized to the corresponding value in control cells.
2.7. TUNEL assay
For apoptosis detection, cell sections and paraffin-embedded renal sections were stained by using a TdT-mediated dUTP Nick-End Labeling (TUNEL) kit (Roche, Basel, Switzerland), and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for the nuclei. Cells were fixed in 4% paraformaldehyde (10min) and 0.1% Triton X-100 (15 min, X10010, Abcone, Shanghai, China) before staining. The apoptotic cells were determined and quantified by counting the positive cells in 3 fields with at least 300 cells per field in each group. For the tissue sections, the quantification was performed according to the number of TUNEL-positive cell per mm2 in 5 random selected fields.
2.8. Determination of reactive oxygen species
The ROS in HK2 cells was measured using dichloro-dihydro-fluorescein diacetate (DCFH-DA) according to the manufacture instruction [29]. Briefly, cells incubated with 10 μM DCFH-DA (S0033, Beyotime, Shanghai, China) for 10 min, then the cells were stained with Hoechst (1X, 5min, 37 °C) for nucleus. Then images were obtained by using a laser scanning confocal microscopy (LSCM, LSM800, ZEISS, Oberkochen, Germany). And the relative intensity of fluorescence expression was analyzed by using the Image J software (Version 1.51).
2.9. Animal model
All animal experiments were approved by the Ethics Committee for Animal Care and Use of Ruijin Hospital. Male C57BL/6 mice (10 weeks, 20–25g) were purchased from SLAC Laboratory Animal Corporation (Shanghai, China). Renal tubular specific HIF-1α knockout mice were generated by crossing HIF-1α-floxed mice (B6.129-HIF-1αtm3Rsjo, The Jackson Laboratory, Bar Harbor, ME, USA) with cadherin-16-cre mice. Cadherin 16 is expressed exclusively in the basolateral membrane of tubular epithelial cells in the kidney [30]. HIF-1αflox+/+: cadherin16-cre+ mice were used as HIF-1α−/-, and HIF-1αflox+/-: cadherin16-cre mice born in the same litter were used as HIF-1α+/+. The genotyping method and representative results were included as s part two of supplementary Materials and Methods.
Bilateral ischemia-reperfusion injury was induced in mice under inhalation anesthesia as we described previously [26]. Briefly, the bilateral renal pedicles of mice were exposed and clamped for 30min to induce ischemia. Then the clamps were released, and the animals were sacrificed 2, 6, 12, 24 or 72h later as indicated. Mice without renal pedicle clamping were used as no I/R mice.
2.10. Adenovirus-mediated BNIP3 expression in mice
Mouse BNIP3-overexpression adenovirus was from Vector Biolabs (Malvern, PA, USA). Transfection of adenovirus into rat kidneys was performed as we described previously [25]. In brief, mice were anesthetized with 2% isoflurane, and 10 μg of plasmids mixed with 2 μL of in vivo jetPEI (Polyplus Transfection, New York, NY) in 10% glucose (600 μL) were injected into the kidneys via the left renal artery when the renal artery and vein were temporarily blocked (<5 min). After injection, an ultrasound transducer (Sonitron 2000, Rich-Mar) was applied directly onto the kidney with an output of 1 MHz at 10% power for a total of 60 s with 30-s intervals, and then the renal artery and vein were unblocked to recover renal blood flow. This technique has been shown to effectively deliver DNA into the renal cells without toxicity to the kidney [31].
2.11. Immunohistochemistry and immunofluorescence
For tissue immunohistochemistry, hematoxylin-eosin (H&E) staining was performed and the kidney damage was evaluated using the criterion as reported previously [32]. Tissue damage was examined in a blinded manner and scored by the percentage of damaged tubules: 0, no damage; 1, <25%; 2, 25–50%; 3, 50–75%; 4, >75%.
For cell and tissue immunofluorescence, paraffin-embedded kidney sections were deparaffinized in dimethylbenzene, dehydrated using graded ethanol, and incubated with proteinase K (P78893, Abcone, Shanghai, China) for antigen retrieval. The incubation condition was as follows: anti-LC3B antibody, anti-TOMM20 antibody, or anti–HIF–1α antibody overnight at 4 °C, Alexa Fluor555-or Alexa Fluor488-coupled secondary antibodies at room temperature for 1h. To specifically stain tubules, some slides were stained with lotus tetragonolobus lectin (LTL, 1:200 dilution; Vector Laboratories Inc, Burlingame, CA, USA) for 20 min at room temperature. 4, 6-Diamidino-2-phenyindole dilactate (DAPI, Life Technologies, Rockville, MD, USA) was used to counterstain cellular nuclei. Images were obtained by using a laser scanning confocal microscopy (LSCM, LSM800, ZEISS, Oberkochen, Germany). Mitophagy was evaluated by measuring the relative fluorescence intensity of yellow dots (LC3B and TOMM20 overlay) per field with the Image J software (Version 1.51).
2.12. Transmission electron microscopy
For transmission electron microscopy (TEM) analysis, the renal tissues (1 mm3) were quickly fixed in 2.5% glutaraldehyde. Then they were post fixed in 1% osmium tetroxide, dehydrated using graded ethanol, and embedded in hard resin. Next, they were cut into ultra-thin slices (80 nm) by using the ultra-microtome instrument (Leica, Germany). Then the slices were stained with uranyl acetate and lead citrate before the TEM observation, and the mitophagosome or autophagosome was determined as previously reported [33]. Autophagosomes are defined as early autophagic compartments containing morphologically intact cytosol or organelles, and autolysosomes are defined as degradative autophagic structures containing partially degraded cytoplasmic as well as organelle material. In particular, mitophagosomes are defined as autophagosomes containing mitochondria and no more than a small amount of other cytoplasmic components, as observed during selective macromitophagy. Apoptotic bodies are a number of membrane-bound domains characterized of pyknotic, shrunken nucleus and the very condensed cytoplasm. For quantification of abnormal mitochondria that were swollen with severely disrupted cristae, at least 300 individual mitochondria from three different kidneys per group were counted. The percentage of abnormal mitochondrial over all mitochondria was analyzed to indicate the degree of mitochondrial damage [34].
2.13. Serum creatinine measurement
The level of serum creatinine (SCr) was measured by using a commercial kit (700460, Cayman Chemical, Ann Arbor, MI, USA) according to manufacture instruction.
2.14. Statistics
All data were expressed as mean ± SD, and all assays were performed at least in triplicate and experiments were repeated three times to verify the results for cell study. The statistics was carried out by using the GraphPad Prism software (Version 5.0, USA). Comparison between groups was made using one-way analysis of variance followed by Turkey post-test. P < 0.05 was considered statistically significant.
3. Results
3.1. Effect of H/R on HIF-1α and mitophagy-related proteins
Firstly, we evaluated the effect of H/R on HIF-1α and mitophagy using the protocol described in Fig. 1A. As shown in Fig. 1B&C, HIF-1α level was significantly increased and peaked at 24h after reoxygenation in HK2 cells. Meanwhile, the protein level of BNIP3, a downstream regulator of HIF-1α, was also significantly increased in response to H/R (Fig. 1B&D). Fig. 1B&E shows that H/R induced significant increase in LC3B-II, an autophagosome marker, whereas COX IV and TOMM20, two mitochondria proteins, were significantly decreased at 24h after reoxygenation (Fig. 1B&F-G). During mitophagy, p62, a mitophagy adaptor, would translocate to damaged mitochdondria and is digested. Therefore the decrease of p62 indicates mitophagy. Then we evaluated the changes of p62. As expected, H/R induced significant decrease of p62 protein (Fig. 1B&H). Accordingly, mtDNA copy number, a mitochondria function indicator, was also significantly decreased (sFig. 1). These results suggest that H/R elicits HIF-1α increase, autophagy, mitophagy, and mitochondrial dysfunction in renal tubular cells.
Fig. 1.
H/R induced mitophagy in HK2 cells. A, A timeline scheme showing the procedures of H/R. After HK2 cells were cultured in serum-free culture medium for 24h, they were cultured in hypoxic (24h) followed by culture in reoxygenation condition for the time indicated. Cells cultured in culture medium with 10% FBS without H/R were used as control. Representative Western blot images (B) and summarized data (C, D, E, F, G, H) showing the changes of protein level of HIF-1α, BNIP3, autophagy-related protein LC3B and mitophagy-related proteins including COX IV, TOMM20, and p62 in HK2 cells with hypoxia (24h)/reoxygenation (0.5, 1.5, 3, 6, 12, 24, 48h). n = 9. *P < 0.05, ***P < 0.001 vs control for HIF-1α, BNIP3, LC3B-II, COX IV, TOMM20, and p62 respectively.
3.2. HIF-1α-BNIP3 signaling pathway mediates H/R-induced mitophagy
We then evaluated whether the increased HIF-1α was involved in H/R-induced mitophagy. 24h reoxygenation was chosen in the following experiments due to the decreased mitophagy-related proteins at this time point (Fig. 1B&E-H). As shown in supplementary sFig. 2, HIF-1α siRNA effectively inhibited H/R-induced HIF-1α expression. It was found that HIF-1α knockdown significantly prevented H/R-induced increase in autophagy marker LC3B-II (Fig. 2A&B). Importantly, HIF-1α knockdown significantly prevented H/R-induced decrease in mitophagy-related proteins including COX IV, p62 and TOMM20 (Fig. 2A&C&D&E). In contrast, H/R significantly decreased mtDNA copy number, and this decrease was further enhanced by HIF-1α knockdown (Fig. 2F). These results suggest that H/R induced mitophagy and mitochondrial dysfunction in renal tubular cells.
Fig. 2.
HIF-1α knockout reduced H/R-induced mitophagy in HK2 cells. Representative Western blot images (A) and summarized data (B, C, D, E) showing the changes of LC3B-II, COX IV, TOMM20, and p62 in H/R-treated cells with or without HIF-1α siRNA. F, Summarized data showing the changes of mtDNA copy numberin H/R-treated cells with or without HIF-1α siRNA. n = 9. **P < 0.01, ***P < 0.001 vs scramble group without H/R; #P < 0.05, ###P < 0.001 vs scramble group with H/R.
Next, we evaluated whether BNIP3 is a downstream regulator in HIF-1α-mediated mitophagy. As shown in Fig. 3A&B, BNIP3 protein level was significantly increased in H/R-treated cells, and this increase was significantly inhibited by HIF-1α siRNA. We then investigated whether BNIP3 is a downstream regulator in HIF-1α-mediated mitophagy. As shown in Fig. 3C, BNIP3 overexpression significantly reversed the inhibitory effect of HIF-1α knockdown on the changes of LC3B-II and mitophagy-related proteins including COX IV, TOMM20 and p62 (Fig. 3A&E-G). In contrast, BNIP3 overexpression significantly reversed the inhibitory effect of HIF-1α knockdown on mtDNA copy number (Fig. 3H).
Fig. 3.
BNIP3 overexpression reversed the decreased H/R-induced mitophagy. Representative Western blot images (A) and summarized data (B) showing the inhibitory effect of HIF-1α siRNA on H/R-induced BNIP3 expression. C, Representative Western blot images (C) and summarized data (D, E, F, G) showing that BNIP3 overexpression reversed the changes of LC3B, COX IV, TOMM20 and p62 in H/R-treated cells. H, Summarized data showing that BNIP3 overexpression reversed the changes of mtDNA copy number in H/R-treated cells. n = 9. **P < 0.01, ***P < 0.001 vs scram group without H/R; ###P < 0.001 vs scramble group with H/R in A&B. **P < 0.01 vs vector group without H/R; #P < 0.05, ##P < 0.01, ###P < 0.001 vs vector group with H/R in C&E.
3.3. HIF-1α-BNIP3 signaling pathway mediates H/R-induced mitophagosome formation
The involvement of HIF-1α was further evaluated by evaluating mitophagosome formation by the co-localization of autophagy marker LC3B and mitochondria marker Mito-Tracker Green. As shown in Fig. 4A&C, mitophagy was significantly increased in H/R-treated cells, as indicated by increased colocalization of RFP-LC3B and MitoTracker, and this increase was significantly decreased in H/R-treated cells with HIF-1α siRNA pretreatment (Fig. 4A&C). As expected, Fig. 4B&D shows that BNIP3 overexpression significantly reversed the inhibitory effect of HIF-1α siRNA on H/R-induced increase in mitophagy formation as indicated by the increased co-localization of LC3B and MitoTracker Green. These results suggest that HIF-1α-BNIP3 mediates H/R-induced mitophagosome formation in HK2 cells.
Fig. 4.
HIF-1α-BNIP3 signaling pathway contributes to mitophagosome formation in renal tubular cells in confocal assay. Representative immunofluorescence images (A) and summarized data (C) showing the inhibitory effect of HIF-1α siRNA on H/R-induced mitophagosome formation as indicated by the colocalization of LC3B with Mitotracker-Green. Representative immunofluorescence images (B) and summarized data (D) showing that BNIP3 overexpression reversed the inhibitory effect HIF-1α siRNA on H/R-induced mitophagosome formation as indicated by the colocalization of LC3B with Mitotracker-Green. Bar = 10 μm n = 9. *P < 0.05, **P < 0.01 vs scramble group without H/R; #P < 0.05, ###P < 0.001 vs scramble group with H/R. **P < 0.01, ***P < 0.001 vs scram group without H/R; ###P < 0.001 vs scramble group with H/R. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. HIF-1α mediates to H/R-induced mitophagy flux
We evaluated whether HIF-1α signaling pathway contributes to H/R-induced mitophagy flux in tubular cells. Mitophagy flux was evaluated by measuring the changes of mitochondria that is inhibited by lysosome function inhibitor Baf. As shown in Fig. 5A&B, H/R significantly increased mitophagy flux, which was significantly inhibited by HIF-1α siRNA. In addition, the mitochondria mass was also evaluated by MitoTracker Green fluorescence. As shown in Fig. 5C, H/R significantly decreased mitochondria mass, which was significantly inhibited by HIF-1α siRNA. These results suggest that HIF-1α was involved in H/R-induced mitophagy flux.
Fig. 5.
HIF-1α-BNIP3 signaling pathway mediated H/R-induced mitophagy flux in HK2 cells. A, Representative immunofluorescence images showing the inhibitory effect of HIF-1α siRNA on H/R-induced mitophagy flux in HK2 cells. Cells were stained with mito-Tracker Green, and mitophagy flux was defined as the inhibitory portion of Baf. B, Summarized data showing the inhibitory effect of HIF-1α siRNA on H/R-induced mitophagy flux in HK2 cells. C, Summarized data showing the effect of HIF-1α siRNA on mitochondria mass as indicated by the changes of.mito-Tracker Green fluorescence. n = 5–6. ***P < 0.001 vs control without H/R group; ###P < 0.001 vs siHIF-1α+BNIP3 overexpression with H/R group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. HIF-1α-BNIP3-mediated mitophagy prevented H/R-induced apoptosis and ROS production
Next, we evaluated whether HIF-1α-BNIP3-mediated mitophagy protects cells from H/R-induced apoptosis and ROS production. As shown in Fig. 6A, H/R significantly increased apoptosis in HK2 cells, which was further enhanced by HIF-1α siRNA. Consistently, in Fig. 6B, the protein level of CC3, a cell apoptosis marker, was significantly increased in H/R-treated cells, which was enhanced by HIF-1α siRNA. Similarly, H/R significantly increased ROS production, and the increase was exacerbated by HIF-1α siRNA (Fig. 6C). Notably, the stimulatory effect of HIF-1α siRNA on apoptosis, CC3 expression, and ROS production was significantly reversed by BNIP3 overexpression (Fig. 6D–F). These results suggest that HIF-1α-BNIP3-mediated mitophagy may exert its protective effect by decreasing H/R-induced apoptosis and ROS production.
Fig. 6.
HIF-1α-BNIP3 signaling pathway attenuated H/R-induced apoptosis and ROS production in HK2 cells. A, Representative immunofluorescence images (upper panel) and summarized data (lower panel) showing the effect of HIF-1α siRNA on H/R-induced tubular apoptosis in TUNEL assay. B, Representative Western blot images (upper panel) and summarized data (lower panel) showing the effect of HIF-1α siRNA on H/R-induced CC3 increase. C, Representative immunofluorescence images (upper panel) and summarized data (lower panel) showing the effect of HIF-1α siRNA on H/R-induced ROS production. D, Representative immunofluorescence images (upper panel) and summarized data (lower panel) showing the effect of BNIP3 overexpression on H/R-induced tubular apoptosis in TUNEL assay. E, Representative Western blot images (upper panel) and summarized data (lower panel) showing the effect of BNIP3 overexpression on H/R-induced CC3 increase. F, Representative immunofluorescence images (upper panel) and summarized data (lower panel) showing the effect of BNIP3 overexpression on H/R-induced ROS production. Bar = 50 μm in A&D, Bar = 100 μm in C&F. n = 9–10. **P < 0.01vs scramble group without H/R; ##P < 0.01, ###P < 0.001 vs scramble group with H/R in A-C. ***P < 0.001 vs vector group without H/R; ###P < 0.001 vs vector group with H/R in D-F.
3.6. Tubular HIF-1α knockout aggravated I/R-induced kidney damage and decreased kidney function
Next, we evaluated whether specific tubular HIF-1α knockdown would aggravate kidney damage in mice. Fig. 7A shows the protocol for establishment of I/R mice. As shown in supplementary sFigure 3A&C, there was high HIF-1α expression in tubule in I/R mice, as stained with LTL, a specific tubule marker. In tubular HIF-1α knockout mice, however, I/R-induced HIF-1α expression was significantly inhibited (sFigure 3A&C). In contrast, there was also obvious HIF-1α expression in outer medullary collecting duct (OMCD) in I/R mice, but this HIF-1α was not decreased in tubular HIF-1α knockout mice (sFigure 3A&C). Similarly, immunohistochemistry staining in sFigure 3B shows that I/R significantly increased HIF-1α in tubule, which was significantly inhibited in tubular HIF-1α knockout mice. Furthermore, Western blot in sFigure 3D shows that I/R increased HIF-1α protein level in isolated tubules, which was significantly inhibited in tubular HIF-1α knockout mice. As expected, I/R induced significantly increase of BNIP3 in renal tissue, which was significantly inhibited in tubular HIF-1α knockout mice (sFig. 4). Based on these findings, tubular HIF-1α was specifically knocked out in HIF-1α+/+; cadherin16-cre mice, and that this knockout also significantly attenuated I/R-induced BNIP3 expression.
Fig. 7.
Specific tubular HIF-1α knockout aggravated I/R-induced kidney damage. A, A timeline scheme showing the procedures of I/R. Mice were subjected to 30 min of renal ischemia (clapping bilateral renal pedicles) followed by reperfusion for different periods (2, 6, 12, 24 or 72h). Mice received the same treatment without renal pedicle clamping were used as control. Representative renal H&E staining images (B) (upper panel, 10x, lower panel 40x) and summarized tubular damage score (C) in HIF-1α+/+ and HIF-1α−/- mice with or without ischemia (30min)/reperfusion (2, 6, 12, 24 or 72h). D, Summarized data showing the effect of tubular HIF-1α knockout on the changes of SCr in mice with ischemia (30min)/reperfusion (2, 6, 12, 24 or 72h). Bar = 100 μm n = 6. **P < 0.01, ***P < 0.001 vs HIF-1α+/+ group without I/R; #P < 0.05, ##P < 0.01vs HIF-1α−/- group with the same reperfusion duration in C. *P < 0.05, **P < 0.01, ***P < 0.001 vs other groups with the same reperfusion duration in D.
H&E staining in Fig. 7B shows that cell swelling was observed after 2h reperfusion and exfoliated cells were observed after 24h reperfusion in HIF-1α+/+ mice. There were more swollen cells in HIF-1α −/− mice compared with HIF-1α+/+ mice with I/R (Fig. 7B). Notably, the brush edge of the lumen is detached, the nucleus is dissolved and the cells are detached from the lumen after 24h reperfusion in HIF-1α−/- mice, but not in HIF-1α+/+ mice (Fig. 7B). In contrast, the regenerated cells, characterized by two nuclei vertically arranged on cell wall below the exfoliated cells, appeared after 72h reperfusion in HIF-1α+/+ mice, but not in HIF-1α−/- mice (Fig. 7B). Fig. 7C shows that tubular damage score, as calculated from H&E staining, was significantly increased in I/R-treated mice, and the increase was further aggravated in tubular HIF-1α knockout mice. The kidney function was evaluated by measuring SCr. As shown in Fig. 7D, SCr was significantly increased after I/R operation, and this increase was further aggravated in tubular HIF-1α knockout mice. These results suggest that tubular HIF-1α knockout aggravated I/R-induced tubular apoptosis and kidney damage.
3.7. Tubular HIF-1α knockout inhibited mitophagy, and aggravated renal tubular apoptosis
We then evaluated the effect of tubular HIF-1α knockout on mitophagy by TEM. As shown in Fig. 8A, double membrane was wrapped around the cell after 2h reperfusion, and autophagosomes/mitophagosomes were observed after 6–24h reperfusion in HIF-1α+/+ mice. Summarized data in Fig. 8B shows that mitophagosome was significantly increased in kidney in response to I/R, which was significantly inhibited in kidney from HIF-1α−/- mice. Accordingly, the number of damaged mitochondria was significantly increased in kidney from HIF-1α+/+ mice, which was further increased in HIF-1α−/- mice (Fig. 8C). These results suggest that HIF-1α-mediated mitophagy was involved in the clearance of damaged mitochondria in IRI.
Fig. 8.
Specific tubular HIF-1α knockout decreased mitophagosome formation in TEM. A, Representative TEM images of kidney tissues in mice with or without ischemia (30min)/reperfusion (2, 6, 12, 24 or 72h). Summarized data showing the effect of tubular HIF-1α knockout on mitophagosome formation (B) and the content of damaged mitochondria (C) in kidney tissue in mice with ischemia (30min)/reperfusion (2, 6, 12, 24 or 72h). Black arrows, normal mitochondria; green arrows, damaged mitochondria; red arrows, cup-like bilayer inclusion body; white arrows, autophagosome; yellow arrows, mitophagosome; purple arrows, apoptotic bodies; blue arrows, remaining vacuoles after digestion by autophagosome. Bar = 1 μm n = 6. ***P < 0.001 vs HIF-1α+/+ group without I/R; ##P < 0.01, ###P < 0.001vs HIF-1α−/- group with the same reperfusion duration. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We further evaluated whether I/R induces the changes of HIF-1α-BNIP3 signaling pathway and mitophagy by Western blot assay. Fig. 9A–C shows that protein level HIF-1α and BNIP3 was significantly increased in renal cortex post I/R operation. Accordingly, LC3B-II was also significantly increased (Fig. 9A&D). In contrast, mitophagy-related proteins including COX IV, TOMM20 and p62 were significantly decreased in renal cortex in I/R mice (Fig. 9A&E-G). Similarly, mtDNA copy number was also significantly decreased in renal cortex in I/R mice (Fig. 9H). These results suggest that I/R stimulated HIF-1α-BNIP3 signaling pathway and induced mitophagy in vivo.
Fig. 9.
I/R induced significant autophagy and mitophagy in mice in Western blot assay. Representative Western blot images (A) and summarized data (B, C, D, E, F, G) showing the time-dependent effect of reperfusion on the expression of HIF-1α, BNIP3, LC3B-II, COX IV and TOMM20, and p62 in the kidney. I, Summarized data showing the time-dependent effect of reperfusion on mtDNA copy number in the kidney. n = 6. *P < 0.05, **P < 0.01, ***P < 0.001 vs control for HIF-1α, BNIP3, LC3B-II, COX IV, TOMM20, and p62.
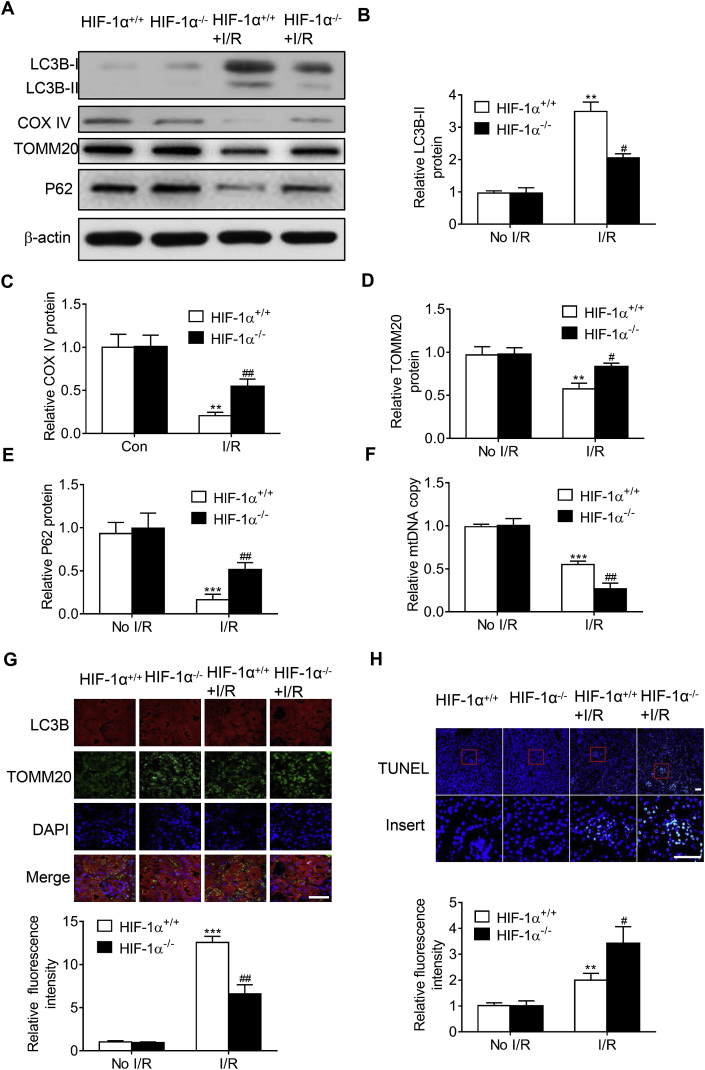
Then the involvement of HIF-1α-BNIP3 signaling pathway in mitophagy and apoptosis was further investigated. Fig. 10A–F shows that I/R-induced mitophagy was significantly inhibited in kidney from tubular HIF-1α knockout mice compared with control mice, as indicated by the decrease of LC3B-II, and the increase of mitophagy-related proteins including COX IV, TOMM20, p62, and mtDNA copy number. Fig. 10G shows that mitophagosome formation, as indicated by the yellow fluorescence of LC3B-II and TOMM20 overlay, was significantly increased in kidney from I/R mice, and this increase was significantly inhibited in kidney from tubular HIF-1α knockout mice. In addition, TUNEL assay shows that I/R-induced cell apoptosis was further aggravated in HIF-1α−/- mice (Fig. 10H). These results suggest that HIF-1α knockout inhibited I/R-induced mitophagy, and aggravated I/R-induced tubular apoptosis.
Fig. 10.
Specific renal tubular HIF-1α knockout decreased mitophagy and aggravated I/R-induced apoptosis. Representative Western blot images (A) and summarized data (B, C, D, E) showing the inhibitory effect of tubular HIF-1α knockout on I/R-induced changes in LC3B, TOMM20 and BNIP3 in the kidney. F, Summarized data showing the inhibitory effect of tubular HIF-1α knockout on I/R-induced changes in mtDNA copy number in the kidney. G, Representative fluorescence images (upper panel) and summarized data (lower panel) showing the inhibitory effect of tubular HIF-1α knockout on I/R-induced mitophagosome formation. H, Representative fluorescence images (upper panel) and summarized data (lower panel) showing the inhibitory effect of tubular HIF-1α knockout on I/R-induced TUNEL in kidney. Bar = 50 μm in G, bar = 100 μm in H. n = 6. **P < 0.01, ***P < 0.001 vs HIF-1α+/+ group without I/R; #P < 0.05, ##P < 0.01vs HIF-1α+/+ group with I/R.
3.8. BNIP3 overexpression improved kidney damage and increased mitophagy in tubular HIF-1α knockout mice with I/R
Finally, we evaluated whether BNIP3 overexpression could reverse the inhibitory effect of tubular HIF-1α on mitophagy and improve kidney function. Supplementary sFigure 5A&B shows that adenovirus-mediated transfection of BNIP3 worked efficiently in the kidney as indicated by the increased green fluorescence in the kidney from adenovirus-treated mice. As shown in Fig. 11A&B, BNIP3 overexpression significantly alleviated acute kidney damage and improved kidney function in tubular HIF-1α knockout mice with I/R. Fig. 11C shows that BNIP3 overexpression significantly increased mitophagosome formation, and decreased the accumulation of damaged mitochondria in kidney from tubular HIF-1α knockout mice with I/R. The potential effect of BNIP3 overexpression on mitophagy was also evaluated by Western blot. As shown in Fig. 11D&E, BNIP3 overexpression significantly increased mitophagy as indicated by the increase of LC3B, and decrease of mitophagy-related proteins including COX IV, TOMM20, and p62 in kidney from tubular HIF-1α knockout mice with I/R. Furthermore, BNIP3 overexpression significantly decreased the accumulation of mtDNA in tubular HIF-1α knockout mice with I/R (Fig. 11F). Finally, BNIP3 overexpression significantly attenuated I/R-induced apoptosis in tubular HIF-1α knockout mice with I/R (Fig. 11G&H). These results suggest that BNIP3 works as a downstream regulator in HIF-1α-mediated mitophagy in vivo.
Fig. 11.
BNIP3 overexpression improved renal function and increased mitophagy in tubular HIF-1α knockout mice with I/R. A, Summarized data showing the effect of BNIP3 overexpression on the changes of SCr in tubular HIF-1α knockout mice with I/R. B, Representative H&E images (upper panel) and summarized tubular damage score (lower panel) showing that BNIP3 overexpression reversed the aggravated tubular damage in tubular HIF-1α knockout mice with I/R. C, Representative TEM images (upper panel) and summarized data (middle and lower panels) showing that BNIP3 overexpression increased the mitophagosome formation and decreased the accumulation of damaged mitochondria in kidney from tubular HIF-1α knockout mice with I/R. Representative Western blot images (D) and summarized data (E) showing that BNIP3 overexpression reversed the effect of tubular HIF-1α knockout on the expression of LC3B, COX IV, TOMM20, p62 and CC3 in the kidney from tubular HIF-1α knockout mice with I/R. F, Summarized data showing that BNIP3 overexpression inhibited the enhancing effect of tubular HIF-1α knockout on I/R-induced mtDNA accumulation in the kidney. Representative fluorescence images (G) and summarized data (H) showing that BNIP3 overexpression reversed the enhancing effect of tubular HIF-1α knockout on I/R-induced tubular apoptosis in TUNEL assay. Bar = 100 μm in B, bar = 1 μm in C, bar = 100 μm in J. n = 6. *P < 0.05, **P < 0.01 vs HIF-1α+/+ group without I/R; #P < 0.05 vs HIF-1α+/+ group with I/R.
4. Discussion
In the present study, we evaluated whether tubular HIF-1α-BNIP3 signaling pathway is involved in I/R-induced mitophagy and plays a protective role in acute kidney damage. Our results shows that HIF-1α knockout significantly attenuated H/R-induced mitophagy, aggravated H/R-induced apoptosis, and increased the production of ROS in tubular cells. Notably, overexpression of BNIP3, a downstream regulator of HIF-1α, reversed the inhibitory effect of HIF-1α knockout on H/R-induced mitophagy, and prevented the enhancing effect of HIF-1α knockout on H/R-induced apoptosis and ROS production. Further in vivo study show that specific tubular HIF-1α knockout decreased mitophagy and aggravated I/R-induced apoptosis. In contrast, BNIP3 overexpression significantly reversed the inhibitory effect of tubular HIF-1α knockout on I/R-induced mitophagy, and improved I/R-induced AKD in tubular HIF-1α knockout mice.
Firstly, we performed in vivo and in vitro study demonstrating that HIF-1α in renal tubular cells plays a protective role in acute kidney damage. Indeed, there are accumulating data showing that HIF-1α protects renal I/R-induced injury [6,9]. For example, preconditional activation of HIF-1α with specific prolyl-hydroxylase inhibitors FG-4487 and FG-4497 attenuated I/R-induced AKI [11,35]._ENREF_26 Further study suggests that HIF-1α-induced downstream genes have a protective effect on ischemia reperfusion renal injury, these genes include vascular endothelial growth factor (VEGF), erythropoietin, heme oxygenase 1 (HO1), and glucose transporter type 1 (Glut1) [36]. Subsequent studies confirmed that HIF-1α is associated with the protection of kidney injury after ischemia reperfusion [35,37,38]. In consistent these findings, the present study demonstrated that renal tubular cell plays a protective role in AKD, since tubular HIF-1α knockout significantly aggravated IRI.
There are accumulating data showing that mitophagy plays an important role in eliminating damaged or dysfunctional mitochondria and maintaining mitochondria homeostasis [39]. Mitochondria contain less cytochrome c and more pro-apoptotic protein Bax expression; it tends to swell, break, and have a lower membrane potential [40]. These changes may result in the failure of renal tubular epithelial cells to maintain their functional and structural integrity [41,42]. In consistent with these findings, the present study shows that mitochondria swollen and mitochondria damage occurs in renal tubular epithelial cells in AKI. Furthermore, our study demonstrated that HIF-1α-BNIP3-mediated mitophagy protects against AKI by reducing apoptosis of renal tubular cells. The present finding that HIF-1α directly increased the transcription and expression of BNIP3 is consistent with previous studies demonstrating that HIF-1α plays a critical role in hypoxia-induced mitophagy [43], and that BNIP3 acts as a direct downstream regulator of HIF-1α in hypoxia [44].
Mitochondrial autophagy is generally regarded as an adaptive metabolic response which is necessary to prevent increased levels of ROS and cell death [45]. In dysfunctional mitochondria, the formation of superoxide anion free radicals and ROS production are increased due to the disrupted electron transport along the respiratory chain and decreased oxygen formation [14,46]. Indeed, it has been demonstrated that BNIP3-dependent mitophagy reduces mitochondrial mass and promotes the integrity of the mitochondrial pool thereby limiting generation of ROS in tumor cells [47,48]. In particular, Tang et al., reported that BNIP3 was increased in tubules in IRI, and that inhibition of BNIP3 with specific short hairpin RNAs reduced mitophagy and worsen IR injury [23]. In consistent with these studies, _ENREF_40_ENREF_14the present study demonstrated that HIF-1α-BNIP3-mediated mitophagy plays a role in reducing the increased apoptosis and increased ROS production. However, the relationship between mitophagy and ROS may be complex since a previous study demonstrating that excessive ROS accumulation induces redox stress and hence up-regulated the expression of HIF-1α and its target genes [14,48].
In consistent with the protective role of mitophagy in the kidney in the present study, it has been documented that mitophagy plays a protective role in I/R-related damage in liver and lung. For example, it has been shown that pharmacological inhibition of autophagy by 3-methyladenine or chloroquine exacerbated acetaminophen-induced hepatotoxicity [49]. Mannam et al. demonstrated that mitogen-activated protein kinase (MAPK) kinase 3 (MKK3) deficiency simultaneously increase mitochondrial biogenesis and mitophagy, which led to a more robust mitochondrial network and provides protection against sepsis [50]. In addition, it has been shown that propofol (2,6-diisopropylphenol) exerts protective effects partly through attenuating hypoxia-induced apoptosis in alveolar epithelial type II [51]. In particular, studies suggest that HIF-1α-BNIP3-mediated autophagy/mitophagy plays a protective role in experimental retinal detachment [52], and in mouse granulosa cells in response to follicle-stimulating hormone stimulation [53].
In cerebral and cardiac ischemia injury, however, autophagy can be protective or destructive [20,21]._ENREF_17 For example, it has been reported that hydrogen, heat shock protein B8 (HSPB8) exert neuroprotective effects by protecting mitochondrial function via enhanced mitophagy in oxygen-glucose deprivation/reoxygenation-induced neuron damage in rat hippocampal [39,54]._ENREF_37 In contrast, it has been reported that knockout of programmed cell death 5 (PCD5) has a protective role in middle cerebral artery occlusion model by promoting apoptosis and autophagy through the activation of HIF-1α-BNIP3 signaling pathway [55]. This inconsistency is similar with cardiomyocyte. For example, it has been reported that hypoxia trigger mitochondria-dependent cardiomyocyte apoptosis through HIF-1α-BNIP3-dependent signaling pathway [56], and that 17β-estradiol, ellagic acid, and dual-specificity protein phosphatase1 prevents cardiomyocyte hypertrophy, autophagy and apoptosis by inhibiting HIF-1α-BNIP3 signaling pathway [57,58]_ENREF_43 [59]. In contrast, it has been reported that loss of peroxisome proliferator-activated receptor γ (PPARγ) results in FUN14 domain containing 1-dependent mitophagy during I/R-induced cardiac injury, the increased mitophagy then enhanced mitochondrial electron transport chain complex activity, mitochondrial respiratory function, and elevated ATP production, which eventually results in myocardial dysfunction [60]. In addition, another study demonstrated that activation of G protein-coupled estrogen receptor 1 (GPER1) at the onset of reperfusion protects the myocardium against I/R-induced injury by reducing mitochondrial dysfunction and mitophagy [61]. These studies suggest that mitophagy may have various roles in different tissues.
In summary, the present study demonstrates that HIF-1α protects AKI through BNIP3-mediated mitophagy in ischemia-reperfusion injury (Fig. 12). This finding may provide pharmacological target for the prevention and treatment of acute kidney damage.
Fig. 12.
Scheme showing that HIF-1α-BNIP3-mediated mitophagy in tubular cells protects I/R-induced AKD by inhibiting cell apoptosis and ROS production.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81470967, 81670613, 81100814), Shanghai Nature Science Foundation (17ZR1423700, 18ZR1423700), the Shanghai Medical Bureau Fund (201540037), the Scientific Fund of Shanghai Jiao Tong University School of Medicine (14XJ10042), and the Scientific Research Foundation for the Returned Overseas Chinese Scholars of the State Education Ministry.
Declaration of competing interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101671.
Contributor Information
Wei-Qing Han, Email: hanweiqing2000@163.com.
Wen Zhang, Email: zhangwen255@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bellomo R., Kellum J.A., Ronco C. Acute kidney injury. Lancet. 2012;380(9843):756–766. doi: 10.1016/S0140-6736(11)61454-2. [DOI] [PubMed] [Google Scholar]
- 2.Sancho-Martinez S.M., Lopez-Novoa J.M., Lopez-Hernandez F.J. Pathophysiological role of different tubular epithelial cell death modes in acute kidney injury. Clin. kidney J. 2015;8(5):548–559. doi: 10.1093/ckj/sfv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duann P., Lianos E.A., Ma J., Lin P.H. Autophagy, innate immunity and tissue repair in acute kidney injury. Int. J. Mol. Sci. 2016;17(5) doi: 10.3390/ijms17050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malek M., Nematbakhsh M. Renal ischemia/reperfusion injury; from pathophysiology to treatment. J. Ren. Inj. Prev. 2015;4(2):20–27. doi: 10.12861/jrip.2015.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastor-Soler N.M., Sutton T.A., Mang H.E., Kinlough C.L., Gendler S.J., Madsen C.S. Muc1 is protective during kidney ischemia-reperfusion injury. Am. J. Physiol. Ren. Physiol. 2015;308(12):F1452–F1462. doi: 10.1152/ajprenal.00066.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z., Haimovich B., Kwon Y.S., Lu T., Fyfe-Kirschner B., Olweny E.O. Unilateral partial nephrectomy with warm ischemia results in acute hypoxia inducible factor 1-alpha (HIF-1alpha) and toll-like receptor 4 (TLR4) overexpression in a porcine model. PloS One. 2016;11(5) doi: 10.1371/journal.pone.0154708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahling M., Mathia S., Paliege A., Koesters R., Mrowka R., Peters H. Tubular von Hippel-Lindau knockout protects against rhabdomyolysis-induced AKI. J. Am. Soc. Nephrol. : JASN (J. Am. Soc. Nephrol.) 2013;24(11):1806–1819. doi: 10.1681/ASN.2013030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conde E., Alegre L., Blanco-Sanchez I., Saenz-Morales D., Aguado-Fraile E., Ponte B. Hypoxia inducible factor 1-alpha (HIF-1 alpha) is induced during reperfusion after renal ischemia and is critical for proximal tubule cell survival. PloS One. 2012;7(3) doi: 10.1371/journal.pone.0033258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou Y.F., Liao W.T., Fu Z.J., Zhao Q., Chen Y.X., Zhang W. MicroRNA-30c-5p ameliorates hypoxia-reoxygenation-induced tubular epithelial cell injury via HIF1alpha stabilization by targeting SOCS3. Oncotarget. 2017;8(54):92801–92814. doi: 10.18632/oncotarget.21582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaushal G.P., Shah S.V. Autophagy in acute kidney injury. Kidney Int. 2016;89(4):779–791. doi: 10.1016/j.kint.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernhardt W.M., Campean V., Kany S., Jurgensen J.S., Weidemann A., Warnecke C. Preconditional activation of hypoxia-inducible factors ameliorates ischemic acute renal failure. J. Am. Soc. Nephrol. : JASN (J. Am. Soc. Nephrol.) 2006;17(7):1970–1978. doi: 10.1681/ASN.2005121302. [DOI] [PubMed] [Google Scholar]
- 12.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxidants Redox Signal. 2011;14(10):1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W., Chen C., Wang J., Liu L., He Y., Chen Q. Mitophagy in cardiomyocytes and in platelets: a major mechanism of cardioprotection against ischemia/reperfusion injury. Physiology. 2018;33(2):86–98. doi: 10.1152/physiol.00030.2017. [DOI] [PubMed] [Google Scholar]
- 14.Chourasia A.H., Tracy K., Frankenberger C., Boland M.L., Sharifi M.N., Drake L.E. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H., Du W., Li Y., Shi C., Hu N., Ma S. Effects of melatonin on fatty liver disease: the role of NR4A1/DNA-PKcs/p53 pathway, mitochondrial fission, and mitophagy. J. Pineal Res. 2018;64(1) doi: 10.1111/jpi.12450. [DOI] [PubMed] [Google Scholar]
- 16.O'Sullivan T.E., Johnson L.R., Kang H.H., Sun J.C. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015;43(2):331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S., Kim J.S. Mitophagy: therapeutic potentials for liver disease and beyond. Toxicol. Res. 2014;30(4):243–250. doi: 10.5487/TR.2014.30.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sureshbabu A., Bhandari V. Targeting mitochondrial dysfunction in lung diseases: emphasis on mitophagy. Front. Physiol. 2013;4:384. doi: 10.3389/fphys.2013.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H., Huang H., Li R., Bi W., Feng L., E L Mitophagy protects SH-SY5Y neuroblastoma cells against the TNFalpha-induced inflammatory injury: involvement of microRNA-145 and Bnip3. Biomed. Pharmacother. Biomed. Pharmacother. 2019;109:957–968. doi: 10.1016/j.biopha.2018.10.123. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y.C., Tian H.X., Yi T., Chen H.B. The critical roles of mitophagy in cerebral ischemia. Protein Cell. 2016;7(10):699–713. doi: 10.1007/s13238-016-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shires S.E., Gustafsson A.B. Mitophagy and heart failure. J. Mol. Med. 2015;93(3):253–262. doi: 10.1007/s00109-015-1254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., Hartleben B., Kretz O., Wiech T., Igarashi P., Mizushima N. Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy. 2012;8(5):826–837. doi: 10.4161/auto.19419. [DOI] [PubMed] [Google Scholar]
- 23.Tang C., Han H., Liu Z., Liu Y., Yin L., Cai J. Activation of BNIP3-mediated mitophagy protects against renal ischemia-reperfusion injury. Cell Death Dis. 2019;10(9):677. doi: 10.1038/s41419-019-1899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Q., Li S., Jiang N., Shao X., Zhang M., Jin H. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han W.Q., Xu L., Tang X.F., Chen W.D., Wu Y.J., Gao P.J. Membrane rafts-redox signalling pathway contributes to renal fibrosis via modulation of the renal tubular epithelial-mesenchymal transition. J. Physiol. 2018;596(16):3603–3616. doi: 10.1113/JP275952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen D., Zou Y.F., Gao Y.H., Zhao Q., Xie Y.Y., Shen P.Y. Inhibitor of DNA binding 1 is induced during kidney ischemia-reperfusion and is critical for the induction of hypoxia-inducible factor-1alpha. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/4634386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaral A., Ramalho-Santos J., St John J.C. The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm. Hum. Reprod. 2007;22(6):1585–1596. doi: 10.1093/humrep/dem030. [DOI] [PubMed] [Google Scholar]
- 28.Mauro-Lizcano M., Esteban-Martinez L., Seco E., Serrano-Puebla A., Garcia-Ledo L., Figueiredo-Pereira C. New method to assess mitophagy flux by flow cytometry. Autophagy. 2015;11(5):833–843. doi: 10.1080/15548627.2015.1034403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao W., Fu Z., Zou Y., Wen D., Ma H., Zhou F. MicroRNA-140-5p attenuated oxidative stress in Cisplatin induced acute kidney injury by activating Nrf2/ARE pathway through a Keap1-independent mechanism. Exp. Cell Res. 2017;360(2):292–302. doi: 10.1016/j.yexcr.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Igarashi P., Shashikant C.S., Thomson R.B., Whyte D.A., Liu-Chen S., Ruddle F.H. Ksp-cadherin gene promoter. II. Kidney-specific activity in transgenic mice. Am. J. Physiol. 1999;277(4):F599–F610. doi: 10.1152/ajprenal.1999.277.4.F599. [DOI] [PubMed] [Google Scholar]
- 31.Newman C.M., Bettinger T. Gene therapy progress and prospects: ultrasound for gene transfer. Gene Ther. 2007;14(6):465–475. doi: 10.1038/sj.gt.3302925. [DOI] [PubMed] [Google Scholar]
- 32.Tang C., Han H., Yan M., Zhu S., Liu J., Liu Z. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy. 2018;14(5):880–897. doi: 10.1080/15548627.2017.1405880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizumura K., Cloonan S.M., Nakahira K., Bhashyam A.R., Cervo M., Kitada T. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Invest. 2014;124(9):3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberger C., Rosen S., Shina A., Frei U., Eckardt K.U., Flippin L.A. Activation of hypoxia-inducible factors ameliorates hypoxic distal tubular injury in the isolated perfused rat kidney. Nephrol. Dial. Transplant. : Off. Publ. European Dialysis and Transplant Assoc. European Renal Association. 2008;23(11):3472–3478. doi: 10.1093/ndt/gfn276. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto M., Makino Y., Tanaka T., Tanaka H., Ishizaka N., Noiri E. Induction of renoprotective gene expression by cobalt ameliorates ischemic injury of the kidney in rats. J. Am. Soc. Nephrol. : JASN (J. Am. Soc. Nephrol.) 2003;14(7):1825–1832. doi: 10.1097/01.asn.0000074239.22357.06. [DOI] [PubMed] [Google Scholar]
- 37.Sutton T.A., Wilkinson J., Mang H.E., Knipe N.L., Plotkin Z., Hosein M. p53 regulates renal expression of HIF-1{alpha} and pVHL under physiological conditions and after ischemia-reperfusion injury. Am. J. Physiol. Ren. Physiol. 2008;295(6):F1666–F1677. doi: 10.1152/ajprenal.90304.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Hoermann C., Ruther J., Reibe S., Madea B., Ayasse M. The importance of carcass volatiles as attractants for the hide beetle Dermestes maculatus (De Geer) Forensic Sci. Int. 2011;212(1–3):173–179. doi: 10.1016/j.forsciint.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Wu X., Li X., Liu Y., Yuan N., Li C., Kang Z. Hydrogen exerts neuroprotective effects on OGD/R damaged neurons in rat hippocampal by protecting mitochondrial function via regulating mitophagy mediated by PINK1/Parkin signaling pathway. Brain Res. 2018;1698:89–98. doi: 10.1016/j.brainres.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Plotnikov E.Y., Kazachenko A.V., Vyssokikh M.Y., Vasileva A.K., Tcvirkun D.V., Isaev N.K. The role of mitochondria in oxidative and nitrosative stress during ischemia/reperfusion in the rat kidney. Kidney Int. 2007;72(12):1493–1502. doi: 10.1038/sj.ki.5002568. [DOI] [PubMed] [Google Scholar]
- 41.Molitoris B.A. Actin cytoskeleton in ischemic acute renal failure. Kidney Int. 2004;66(2):871–883. doi: 10.1111/j.1523-1755.2004.00818.x. [DOI] [PubMed] [Google Scholar]
- 42.Schrier R.W., Wang W., Poole B., Mitra A. Acute renal failure: definitions, diagnosis, pathogenesis, and therapy. J. Clin. Invest. 2004;114(1):5–14. doi: 10.1172/JCI22353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazure N.M., Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol. 2010;22(2):177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z., Zhang W., Kang Y.J. Copper affects the binding of HIF-1alpha to the critical motifs of its target genes. Metall : Integrated Biometal Sci. 2019;11(2):429–438. doi: 10.1039/c8mt00280k. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H., Bosch-Marce M., Shimoda L.A., Tan Y.S., Baek J.H., Wesley J.B. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94(3):909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chourasia A.H., Macleod K.F. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy. 2015;11(10):1937–1938. doi: 10.1080/15548627.2015.1085136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sathiyaseelan P., Rothe K., Yang K.C., Xu J., Chow N.S., Bortnik S. Diverse mechanisms of autophagy dysregulation and their therapeutic implications: does the shoe fit? Autophagy. 2018:1–4. doi: 10.1080/15548627.2018.1509609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni H.M., Bockus A., Boggess N., Jaeschke H., Ding W.X. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55(1):222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mannam P., Shinn A.S., Srivastava A., Neamu R.F., Walker W.E., Bohanon M. MKK3 regulates mitochondrial biogenesis and mitophagy in sepsis-induced lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306(7):L604–L619. doi: 10.1152/ajplung.00272.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ning H.J., Yuan H.B., Xu H.T., He X.Y. Propofol reduces hypoxiainduced autophagic cell death through downregulating HIF 1alpha in alveolar epithelial type II cells of rats. Mol. Med. Rep. 2017;16(2):1509–1515. doi: 10.3892/mmr.2017.6697. [DOI] [PubMed] [Google Scholar]
- 52.Liu H., Zhu H., Li T., Zhang P., Wang N., Sun X. Prolyl-4-Hydroxylases inhibitor stabilizes HIF-1alpha and increases mitophagy to reduce cell death after experimental retinal detachment. Invest. Ophthalmol. Vis. Sci. 2016;57(4):1807–1815. doi: 10.1167/iovs.15-18066. [DOI] [PubMed] [Google Scholar]
- 53.Zhou J., Yao W., Li C., Wu W., Li Q., Liu H. Administration of follicle-stimulating hormone induces autophagy via upregulation of HIF-1alpha in mouse granulosa cells. Cell Death Dis. 2017;8(8) doi: 10.1038/cddis.2017.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F., Tan J., Zhou F., Hu Z., Yang B. Heat shock protein B8 (HSPB8) reduces oxygen-glucose deprivation/reperfusion injury via the induction of mitophagy. Cell. Physiol. Biochem. : Int. J. Exp. Cell. Physiol. Biochem. and Pharmacol. 2018;48(4):1492–1504. doi: 10.1159/000492259. [DOI] [PubMed] [Google Scholar]
- 55.Lu J., Jiang Z., Chen Y., Zhou C., Chen C. Knockout of programmed cell death 5 (PDCD5) gene attenuates neuron injury after middle cerebral artery occlusion in mice. Brain Res. 2016;1650:152–161. doi: 10.1016/j.brainres.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Feng C.C., Lin C.C., Lai Y.P., Chen T.S., Marthandam Asokan S., Lin J.Y. Hypoxia suppresses myocardial survival pathway through HIF-1alpha-IGFBP-3-dependent signaling and enhances cardiomyocyte autophagic and apoptotic effects mainly via FoxO3a-induced BNIP3 expression. Growth Factors. 2016;34(3–4):73–86. doi: 10.1080/08977194.2016.1191480. [DOI] [PubMed] [Google Scholar]
- 57.Hsieh D.J., Kuo W.W., Lai Y.P., Shibu M.A., Shen C.Y., Pai P. 17beta-Estradiol and/or estrogen receptor beta attenuate the autophagic and apoptotic effects induced by prolonged hypoxia through HIF-1alpha-Mediated BNIP3 and IGFBP-3 signaling blockage. Cell. Physiol. Biochem. : Int. J. Exp. Cell. Physiol. Biochem. and Pharmacol. 2015;36(1):274–284. doi: 10.1159/000374070. [DOI] [PubMed] [Google Scholar]
- 58.Dhingra A., Jayas R., Afshar P., Guberman M., Maddaford G., Gerstein J. Ellagic acid antagonizes Bnip3-mediated mitochondrial injury and necrotic cell death of cardiac myocytes. Free Radic. Biol. Med. 2017;112:411–422. doi: 10.1016/j.freeradbiomed.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Jin Q., Li R., Hu N., Xin T., Zhu P., Hu S. DUSP1 alleviates cardiac ischemia/reperfusion injury by suppressing the Mff-required mitochondrial fission and Bnip3-related mitophagy via the JNK pathways. Redox Biol. 2018;14:576–587. doi: 10.1016/j.redox.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H., Li D., Zhu P., Hu S., Hu N., Ma S. Melatonin suppresses platelet activation and function against cardiac ischemia/reperfusion injury via PPARgamma/FUNDC1/mitophagy pathways. J. Pineal Res. 2017;63(4) doi: 10.1111/jpi.12438. [DOI] [PubMed] [Google Scholar]
- 61.Feng Y., Madungwe N.B., da Cruz Junho C.V., Bopassa J.C. Activation of G protein-coupled oestrogen receptor 1 at the onset of reperfusion protects the myocardium against ischemia/reperfusion injury by reducing mitochondrial dysfunction and mitophagy. Br. J. Pharmacol. 2017;174(23):4329–4344. doi: 10.1111/bph.14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.