Abstract
Background
A comprehensive understanding of host factors modulated by the antiviral cytokine interferon-α (IFNα) is imperative for harnessing its beneficial effects while avoiding its detrimental side-effects during HIV infection. Cytokines modulate host glycosylation which plays a critical role in mediating immunological functions. However, the impact of IFNα on host glycosylation has never been characterized.
Methods
We assessed the impact of pegylated IFNα2a on IgG glycome, as well as CD8+ T and NK cell-surface glycomes, of 18 HIV-infected individuals on suppressive antiretroviral therapy. We linked these glycomic signatures to changes in inflammation, CD8+ T and NK cell phenotypes, and HIV DNA.
Findings
We identified significant interactions that support a model in which a) IFNα increases the proportion of pro-inflammatory, bisecting GlcNAc glycans (known to enhance FcγR binding) within the IgG glycome, which in turn b) increases inflammation, which c) leads to poor CD8+ T cell phenotypes and poor IFNα-mediated reduction of HIV DNA. Examining cell-surface glycomes, IFNα increases levels of the immunosuppressive GalNAc-containing glycans (T/Tn antigens) on CD8+ T cells. This induction is associated with lower HIV-gag-specific CD8+ T cell functions. Last, IFNα increases levels of fucose on NK cells. This induction is associated with higher NK functions upon K562 stimulation.
Interpretation
IFNα causes host glycomic alterations that are known to modulate immunological responses. These alterations are associated with both detrimental and beneficial consequences of IFNα. Manipulating host glycomic interactions may represent a strategy for enhancing the positive effects of IFNα while avoiding its detrimental side-effects.
Funding
NIH grants R21AI143385, U01AI110434.
Keywords: HIV, Interferon, Glycosylation, IgG, CD8+T cells, NK cells
Research in context.
Evidence before this study
Interferon-α (IFNα) is a key modulator of immunological functions during acute viral infections. However, its role is paradoxical during chronic diseases, such as cancer and chronic viral infections. During chronic diseases, IFNα can impose both beneficial effects and detrimental side-effects on immunological functions [1], [2], [3], [4], [5], [6]. To take full advantage of IFNα antiviral potential, there is a need for a comprehensive understanding of the host factors that modulate its beneficial effects and detrimental side-effects. Some proinflammatory cytokines are known to regulate cell signaling and progression of inflammatory diseases by modulating host glycosylation machinery. However, the effects of IFNα on the host glycosylation machinery in vivohave remained an open question. Almost four decades ago, there was a debate on whether IFN alters host glycosylation [7], [8], [9] or not [10,11]. However, these studies were performed using in vitro cell lines and with limited glycomic analysis. A comprehensive understanding of the impact of IFN on the host glycans in vivo may allow us to enhance the beneficial impact of IFN while avoiding its detrimental influence, during chronic viral infections, such as HIV infection.
Added value of this study
We performed the first-of-its-kind in vivo longitudinal analysis on the impact of IFNα on the host glycosylation machinery in humans. We identified specific glycomic alterations caused by IFNα on IgG, CD8+ T cell, and NK cell glycomes, that are linked to both beneficial and detrimental consequences of this cytokine on innate and adaptive immune functions during antiretroviral therapy (ART)-suppressed HIV infection.
Implications of all the available evidence
Given the documented functional significance of host glycomic alterations on immunological functions, our results could have significant implications in solving the interferon paradox during viral infections and chronic diseases. Resolving this paradox could allow for novel interventions to manipulate glycomic interactions as a strategy to improve the beneficial impact of IFNs (both endogenous and exogenous) while avoiding its detrimental side-effects.
Alt-text: Unlabelled box
1. Introduction
Interferons are cytokines that work as a first-line defense against viral infection by interfering with viral replication and modulating host immune responses. During acute infection, type I IFNs are important for limiting early replication and activating immune cells. However, prolonged and sustained exposure to IFN during chronic viral infections, and possibly other chronic diseases, can drive a persistent inflammatory state that is detrimental to immunological functions [1], [2], [3]. Among type I IFNs, IFNα is a family of controversial molecules that has been described to cause both potent antiviral effects but also detrimental immunomodulatory effects during chronic viral infections, such as HIV infection [2], [3], [4], [5]. The balance between the antiviral and the pro-inflammatory effects may determine the overall beneficial or detrimental impact of IFN during chronic diseases [1,2,6]. Therefore, understanding the potential host determinants behind this delicate balance could deepen our knowledge about endogenous IFNs and improve the therapeutic efficacy of exogenous IFNs by biasing them towards an efficient immunological effect rather than inflammation and immune exhaustion.
Several cytokines have been shown to modulate host glycosylation [12]. Glycans on cell-surface proteins and lipids, as well as on circulating glycoproteins such as antibodies, play a critical role in mediating several cellular processes and immunological functions. The specific structure of a glycan allows it to bind to glycan-binding proteins called lectins, leading to the modulation of essential signaling pathways. Antibody glycans, for example, directly impacts its features and functions. The glycomic structures on the antibody can alter antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), complement-dependent cytotoxicity (CDC), as well as several pro- and anti-inflammatory activities [13], [14], [15], [16], [17], [18]. In addition, glycans and lectins on the cell surface are involved in modulating molecular interactions such as cell adhesion, cell trafficking, receptor activation, among others. For example, β-galactosides binding to galectins, sialic acid binding to siglecs, and fucosylated glycans binding to selectins can modulate immunological and inflammatory functions as well as cell death and migration [19], [20], [21], [22], [23], [24]. Some pro-inflammatory cytokines are known to regulate cell signaling and progression of inflammatory diseases by modulating cell surface glycosylation [12]. However, the effects of IFN on the host glycosylation machinery in vivo have remained an open question. Almost four decades ago, there was a debate on whether IFN alters host glycosylation [7], [8], [9] or not [10,11]. However, these studies were performed using in vitro cell lines and limited glycomic analysis. A comprehensive understanding of the impact of IFN on the host glycans in vivo may allow us to enhance the beneficial impact of IFN while avoiding its detrimental influence, during chronic viral infections, such as HIV infection.
Currently, people living with HIV still require lifelong treatment to maintain viral suppression and avoid viral rebound [25]. Exogenous IFN has been proposed as a potential immuno-based therapy for HIV latency. In a clinical trial, a combination of antiretroviral therapy (ART) and pegylated (Peg) interferon alfa-2a (Peg-IFNα2a), followed by Peg-IFNα2a monotherapy, was associated with significant suppression of HIV viremia and a reduction in integrated HIV DNA [26]. Other studies confirmed this observation [27]. Furthermore, therapeutics that enhance endogenous IFN production, such as TLR9 agonists, have gained attention as promising tools to reduce the size of the HIV reservoir and achieve functional HIV cure [28], [29], [30]. However, the lack of a comprehensive understanding of the impact of IFN on host functions, including the impact on host glycosylation machinery, has hampered the efforts to take full advantage of endogenous and exogenous IFN antiviral potential. We hypothesized that IFNα alters the host glycomic machinery in a way that modulates host immunological functions during ART-suppressed HIV infection. Here, using in vivo human samples, we show that IFNα treatment causes specific host glycomic alterations, to both the IgG glycome and the CD8+ T and NK cell-surface glycomes, that are known to variously mediate immunological responses. Indeed, these alterations associated with both detrimental and beneficial consequences of IFNα on innate and adaptive immune functions in vivo, in a manner consistent with their documented anti- or pro-immunological functions.
2. Materials and methods
2.1. Study cohort
We profiled the circulating and cell-surface glycomic signatures from banked plasma and peripheral blood mononuclear cell (PBMC) samples of 18 HIV-infected individuals on suppressive ART who participated in previous clinical trial (registered at Clinical-Trials.gov (http://www.clinicaltrials.gov/; NCT00594880) [26] in which Peg-IFNα2a was added to their ART regimen. All individuals participating in NCT00594880 had CD4+ T cell count for > 6 months at ≥ 400 cells/mm3 (nadir ≥ 200 cells/mm3) and undetectable HIV RNA <50 copies/ml at screening. The plasma and PBMC samples from NCT00594880 that were analyzed in the current study correspond to weeks 8, 13, and 25 of the clinical trial (Fig. 1A).
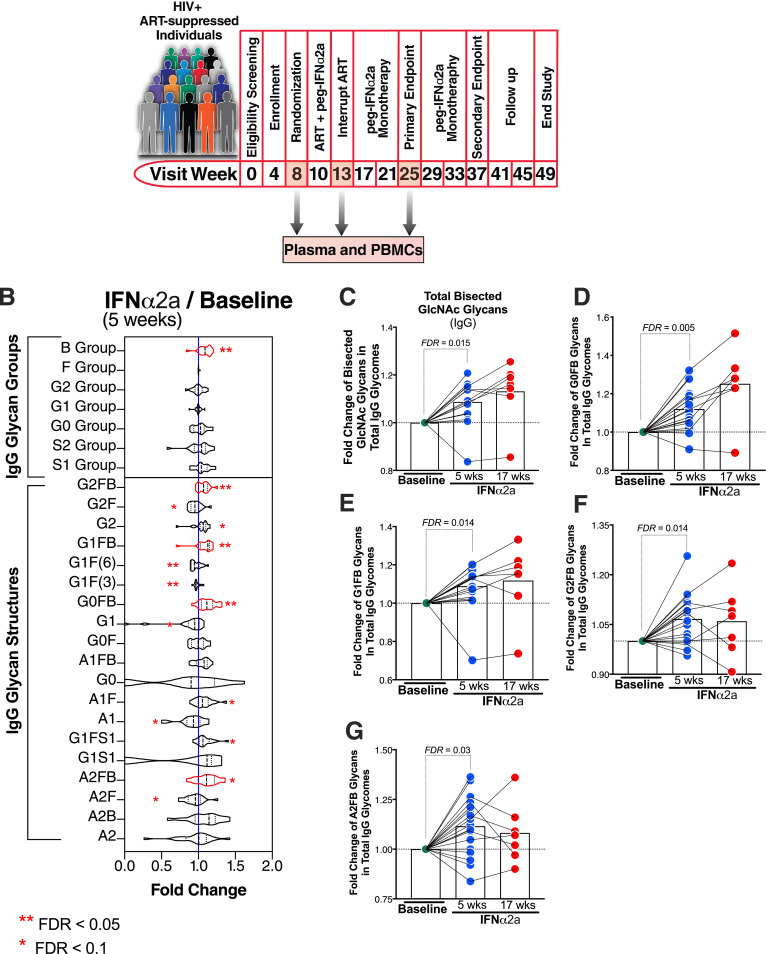
Fig. 1.
Peg-IFNα2a treatment induces the proportion of the pro-inflammatory, bisecting GlcNAc glycans on plasma IgG. (A) A schematic of the clinical study from which samples had been obtained. (B) Violin plots of the IgG glycan structures that were modulated by IFNα. Nineteen glycan structures (displayed on the bottom half of the graph) are grouped into seven groups (displayed in the top half of the graph), depending on the presence or absence of four monosaccharides: bisecting GlcNAc (B group), sialic acid (S1 and S2 groups), galactose (G0, G1, and G2 groups), or fucose (F group). Wilcoxon signed-rank test. (C-G) Bar graphs of selected B group IgG glycan structures modulated by IFNα. Wilcoxon signed-rank test. Lines represent medians. FDR values were calculated using the Benjamini–Hochberg method. N = 18 for study visits 8 and 13, and n = 7 for study visit 25.
2.2. Ethics
Written informed consent was obtained from all participants of NCT00594880 according to the directives of the institutional review boards (IRB) at the Wistar Institute, University of Pennsylvania, Philadelphia FIGHT, and Drexel University. All research protocols of the study were approved by The Wistar Institute (IRB approval # 2709237), University of Pennsylvania (IRB approval # 806138), Philadelphia FIGHT (protocol ID. DAIDS ES 10401), and Drexel University (IRB approval # 17175). All human experimentation was conducted in accordance with the guidelines of the US Department of Health and Human Services and those of the authors’ institutions.
2.3. IgG isolation and IgG subclass quantification
Bulk IgG was purified from plasma using Pierce™ Protein G Spin Plate (ThermoFisher Scientific, catalogue# 45204). IgG purity was confirmed by SDS gel. IgG subclasses were quantified by commercial ELISA kit (ThermoFisher Scientific, catalogue# 991000).
2.4. N-glycan analysis using capillary electrophoresis
To profile the circulating (plasma and bulk IgG) glycome, N-glycans were released using peptide-N-glycosidase F (PNGase F) and labeled with 8-aminopyrene-1,3,6-trisulfonic acid (APTS) using the GlycanAssure APTS Kit (ThermoFisher Scientific, catalogue# A33952) following the manufacturer's protocol. Labeled N-glycans were analysed using the 3500 Genetic Analyzer capillary electrophoresis system. IgG N-glycan samples were separated into 19 peaks and total plasma N-glycans were separated into 24 peaks (structures and names are in Supplementary Figs. 1 and 2, respectively). The relative abundance of N-glycan structures was quantified by calculating the area under the curve of each glycan structure divided by the total glycans.
2.5. Membrane protein purification from CD8+ T cells and NK cells
To profile cell surface glycomes from CD8+ T cells and NK cells, we first isolated CD8+ T cells and NK cells from cryopreserved PBMCs by negative selection using a Stem Cell Human CD8+ T Cell Isolation Kit (STEMCELL Technologies, catalogue# 17953) and Human NK Cell Isolation Kit (STEMCELL Technologies, catalogue# 17955) according to the manufacturer's instructions. Membrane proteins were then extracted using Membrane Protein Extraction Kit (ThermoFisher Scientific, catalogue# 89842), adding protease and phosphatase inhibitors (Halt™ Protease Inhibitor Cocktail, ThermoFisher Scientific, catalogue# 78425) to extraction buffers. proteins were quantified using Micro BCA™ Protein Assay Kit (ThermoFisher Scientific, catalogue# 23235)
2.6. Glycan analysis of cell-surface proteins using the lectin array
Membrane proteins extracted from CD8+ and NK cells were labeled with Cy3 dye (GE Healthcare, catalogue# GEPA23001) and hybridized to the lectin microarrays. The resulting chips were scanned for fluorescence intensity on each lectin-coated spot using an evanescent-field fluorescence scanner GlycoStation Reader (GlycoTechnica Ltd.), and data were normalized using the global normalization method.
2.7. Measurement of cytokines and soluble markers in plasma
Plasma levels of galectin-3, galectin-3B, galectin-1, galectin-9, l-Selectin, E-Selectin, P-selectin, IFN-gamma, IL-10, IL-12p40, IL-15, IL-17A, IL-18, IL-22, IL-4, IP-10, RANTES, and TNF-alpha, were measured using custom multiplex Luminex assays (catalogue# LXSAHM-16 and LXSAHM-02). Plasma levels of soluble CD14 and soluble CD163 were measured using Quantikine Elisa kits (R&D Systems, catalogue# DC140 and CD1630, respectively) according to the manufacturer's instructions. Optical density was measured at 450 nm and 540 nm using Versa Max microplate reader. Plasma levels of TGF-β1, TGF-β2, and TGF-β3 were determined using MSD U-PLEX TGF-β Combo assay (Meso Scale Diagnostic catalogue# K15241K-1) according to manufacturer instructions.
2.8. CD8+ T cell and NK cell phenotyping
Cryopreserved PBMC were thawed, counted, examined for viability, treated with DNAse (Roche Diagnostics, catalogue# 11513500) for 2 h, and rested overnight at 1 × 106 cells per tube at 37 °C and 5% CO2 in complete medium (RPMI supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin). The next day, cells were either co-cultured with K562 cells (2 × 105 cells), treated with aCD28/CD49d (BD Biosciences, catalogue# 347690) together with DMSO 0.5% (Fisher BioReagents/ ThermoFisher Scientific, catalogue# BP231–1), or HIV-1 gag consensus subtype B peptide set (pool of JPT peptides, NIH AIDS Reagent Program catalogue# 8117). CD107a [Fluorescein isothiocyanate (FITC); clone H4A3; BD Biosciences, catalogue# 555800, RRID:AB_396134] was added at this stage and cells were incubated for 2 h at 37 °C and 5% CO2 in complete medium. Subsequently, Golgi stop (monensin, BD Biosciences, catalogue# 554724) and Brefeldin A (Millipore Sigma, catalogue# B7651) were added, and the incubation was continued for an additional 4 h. Cells were washed in PBS and stained for viability for 10 min at room temperature using the Live/Dead Fixable Aqua Dead Cell Stain Kit (Invitrogen/ThermoFisher Scientific, catalogue# L34957). Cells were then surface stained for 20 min at room temperature with CD3 [allophycocyanin (APC)Cy7; clone UCHT1; Biolegend catalogue# 300458, RRID:AB_2564151], CD8 [phycoerythrin (PE) Texas red; clone 3B5; Life Technologies/ThermoFisher Scientific catalogue# MHCD0804, RRID:AB_10372952], CD4 (PECy5.5; clone S3.5; Life Technologies/ThermoFisher Scientific, catalogue# MHCD0418, RRID:AB_10376013), Dump [a) CD14 Brilliant Violet (BV)510; clone M5E2 (Biolegend, catalogue# 301842, RRID:AB_2561946), b) CD19 BV510; clone HIB19 (Biolegend, catalogue# 302242, RRID:AB_2561668), c) CD20 BV510; clone 2H7; (Biolegend, catalogue# 302340, RRID:AB_2561941)], CD56 (BV605; clone HCD56; Biolegend, catalogue# 318334, RRID:AB_2561912), CD16 (PECy5; clone 3G8; Biolegend, catalogue# 302010, RRID:AB_314210), PD1 (BV711; clone EH12.2H7; Biolegend, catalogue# 329928, RRID:AB_2562911). Subsequently, cells were washed with FACS buffer, permeabilized with eBio FoxP3 Fixation/Permeabilization working solution (eBioscience/ThermoFisher Scientific, catalogue# 00–5523–00) for 45 min at room temperature, washed with eBio permeabilization buffer (eBioscience/ThermoFisher Scientific, catalogue# 00–5523–00) and stained intracellularly for 1 h at room temperature with Tbet (PE; clone 4B10; Biolegend, catalogue# 644810, RRID:AB_2200542), Eomes (eFluor (ef)660; clone WD1928; eBioscience/ThermoFisher Scientific, catalogue# 50–4877–42, RRID:AB_2574229), Perforin (BV421; clone B-D48; Biolegend, catalogue# 353307, RRID:AB_11149688), IFNγ (Alexa Fluor (AF700); clone B27; BD Biosciences, catalogue# 557995, RRID:AB_396977), and TNFα (PECy7; clone MAb11; eBioscience/ThermoFisher Scientific, catalogue# 25–7349–41, RRID:AB_1257208). Finally, cells were washed with eBio 1x permeabilization buffer, fixed with 1% paraformaldehyde, and analyzed on a BD FACSymphony A5 flow cytometer (BD Biosciences) by collecting > 200,000 events. Data were analyzed using FlowJo software (Version 8.8.4; Tree Star, Ashland, OR). Results were expressed as mean fluorescence intensity (MFI), and percentages, and cells/mm3. HIV gag-stimulation was expressed after taking into account the background (i.e. DMSO stimulation). CD8+ T and NK cell gating are described in Supplementary Figs. 3 and 4, respectively.
2.9. Integrated HIV DNA quantification
Levels of integrated HIV DNA in total PBMCs were measured using an Alu-gag–based polymerase chain reaction, as described [31]. Values were converted to copies of integrated HIV DNA per number of CD4+ T cells, following the equation: [number of copies of integrated HIV DNA per PBMC] × [(monocyte count + lymphocyte count)/(CD4+ T-cell count)] [26].
2.10. Statistical analysis
Wilcoxon signed-rank test was used for matched-pairs comparisons in Fig. 1 (N = 18 for study visits 8 and 13, and n = 7 for study visit 25). FDR was calculated using the Benjamini–Hochberg approach (* FDR < 0.1, ** FDR < 0.05). Wilcoxon signed-rank test was used for matched-pairs comparisons in Fig. 2 (N = 18 for study visits 8 and 13, and n = 7 for study visit 25). P values <0.05 considered statistically significant. Wilcoxon signed-rank test was used for matched-pairs comparisons in Figs. 5 and 7A (N = 8). FDR was calculated using the Benjamini–Hochberg approach. P values <0.05 considered statistically significant. Spearman's rank correlation was used for bivariate correlation analysis in Fig. 2 (n = 18), Fig. 3 (n = 8), Fig. 4 (n = 8), Fig. 6 (N = 8, * p < 0.05, ** p < 0.01), and Fig. 7B (N = 8, * p < 0.05, ** p < 0.01). P values <0.05 considered statistically significant. Shapiro-Wilk test was used to examine normality in all figures. Statistical analyses were performed in R and Prism 7.0 (GraphPad).
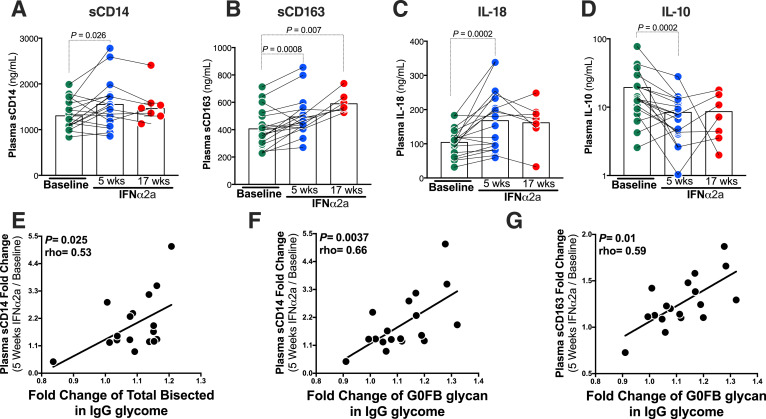
Fig. 2.
Peg-IFNα2a-mediated induction of the pro-inflammatory bisecting GlcNAc glycans on IgG associates with higher monocyte and macrophage activation/inflammation. (A-D) IFNα treatment induces the levels of monocyte activation/inflammation marker (sCD14), macrophage activation/inflammation marker (sCD163), pro-inflammatory cytokine (IL-18), and reduces the levels of the anti-inflammatory marker (IL-10). Wilcoxon signed-rank test. Lines represent medians. (E-G) IFNα2-mediated induction of pro-inflammatory bisecting GlcNAc glycans on IgG correlates with higher levels of IFNα2-mediated induction of markers of monocyte and macrophage activation/inflammation (comparing visit week 13 to visit week 8). (E) IFNα-mediated fold change of total bisecting GlcNAc (i.e. B group) glycans correlates with IFNα-mediated fold change of sCD14. Fold change of the specific B group G0FB trait also correlated positively with fold change of sCD14 (F), and with fold change of sCD163 (G). Correlations were evaluated using Spearman's rank correlation tests. N = 18 for study visits 8 and 13, and n = 7 for study visit 25.
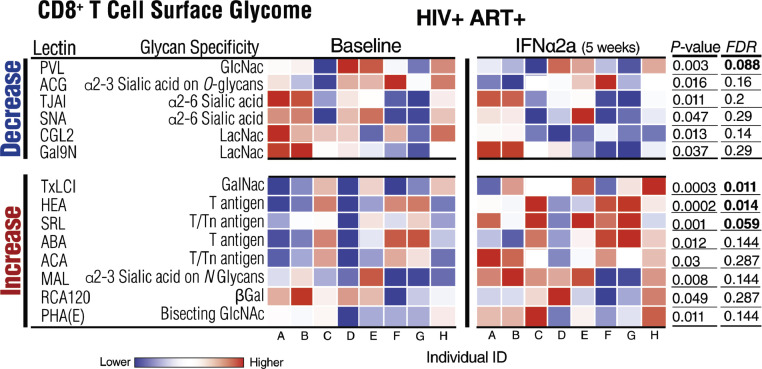
Fig. 5.
IFNα modulates the CD8+ T cell-surface glycome. Heat-map depicting normalized lectin-binding intensity of CD8+ T cell-surface proteins isolated from CD8+ T cells before and five weeks during Peg-IFNα2a treatment. Heat colors show standardized Z-scores; red indicates higher binding, and blue indicates lower binding. Top half lists glycan structures whose levels decreased by IFNα treatment; bottom half lists glycan structures whose levels increased by IFNα treatment. Wilcoxon signed-rank tests. Nominal p values and corrected FDR are displayed. N = 8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
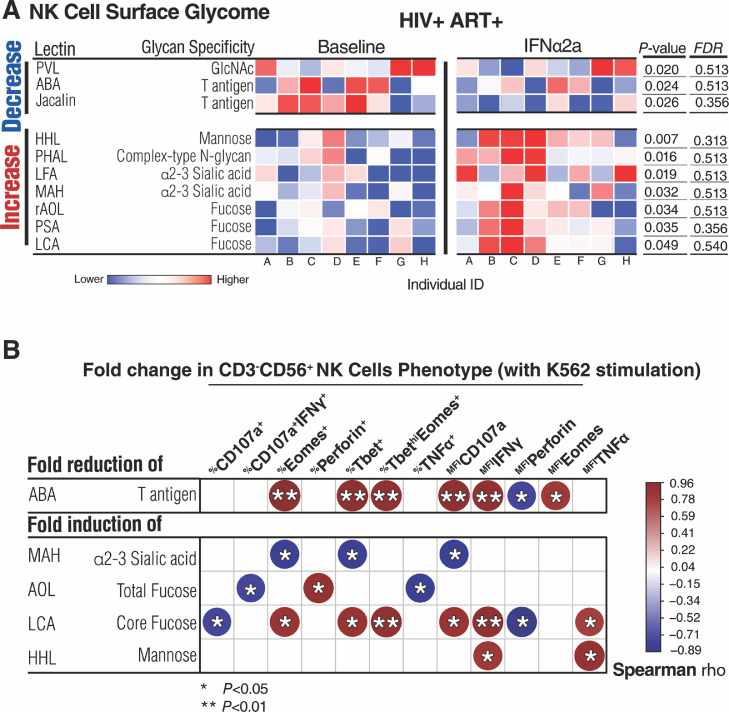
Fig. 7.
IFNα modulates the NK cell-surface glycome. (A) Heat-map depicting normalized lectin-binding intensity of NK cell-surface proteins isolated from NK cells before and five weeks during Peg-IFNα2a treatment. Heat colors show standardized Z-scores; red indicates higher binding, and blue indicates lower binding. Top half lists glycan structures whose levels decreased by IFNα treatment, bottom half lists glycan structures whose levels increased by IFNα treatment. Wilcoxon signed-rank tests. Nominal p values and corrected FDR are displayed. (B) Correlation heat-map showing associations between IFNα2a-mediated changes on the NK cell glycomes and K652-stimulated NK cell phenotypes. The size and colour of circles represent the strength of correlation, with blue shades representing negative correlations, and red shades representing positive correlations. Correlations were evaluated using Spearman's rank correlation tests. *=P < 0.05, and **=P < 0.01. N = 8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
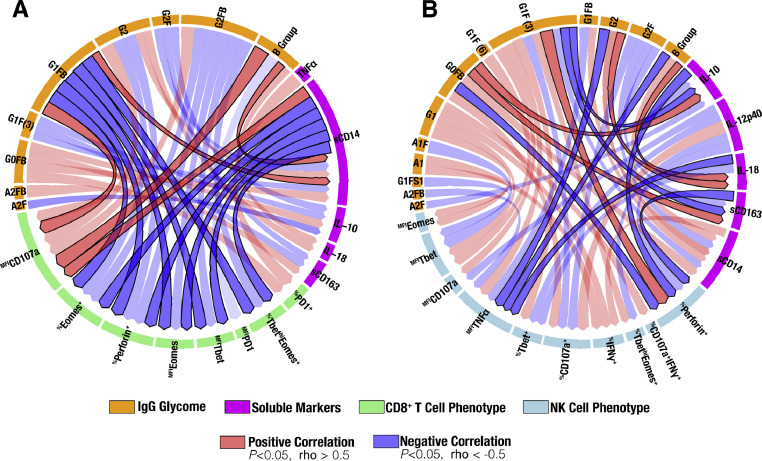
Fig. 3.
Interaction network of IFNα-mediated modulation of the IgG glycomes, plasma inflammatory markers, and CD8+ T and NK cell phenotypes. Circos plots describe the correlations between Peg-IFNα2a-mediated changes in the IgG glycomes (orange), plasma markers of inflammation and immune activation (fuchsia), and (A) CD8+ T cell (green) and (B) K652-stimulated NK cell (light teal) phenotypes. Interactions in which all three domains (IgG glycomes, soluble plasma markers of inflammation, and immune cell phenotype) significantly correlate with each other are indicated by the darker red or blue connections. Correlations were evaluated using Spearman's rank correlation tests. N = 8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
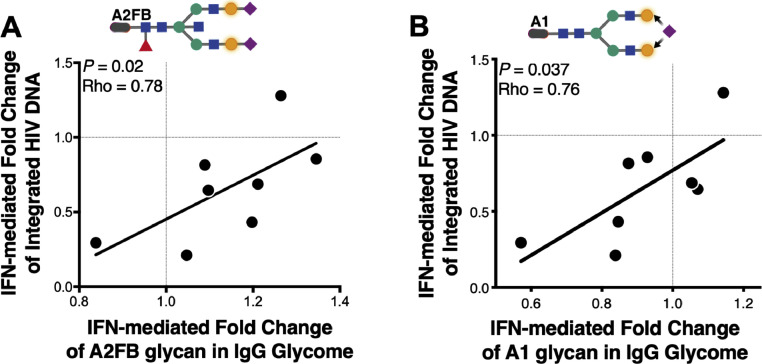
Fig. 4.
IFNα-mediated modulation of the IgG glycome correlates with the IFNα-mediated reduction of integrated HIV DNA. (A) Induction in levels of the bisecting A2FB glycan trait in IgG glycome correlates with poor reduction of integrated HIV DNA during IFNα treatment (comparing visit week 13 to visit week 8). (B) Reduction in levels of the mono-sialylated A1 glycan trait in IgG glycome correlates with a better reduction of integrated HIV DNA during IFNα treatment (comparing visit week 13 to visit week 8). Correlations were evaluated using Spearman's rank correlation tests. N = 8.
Fig. 6.
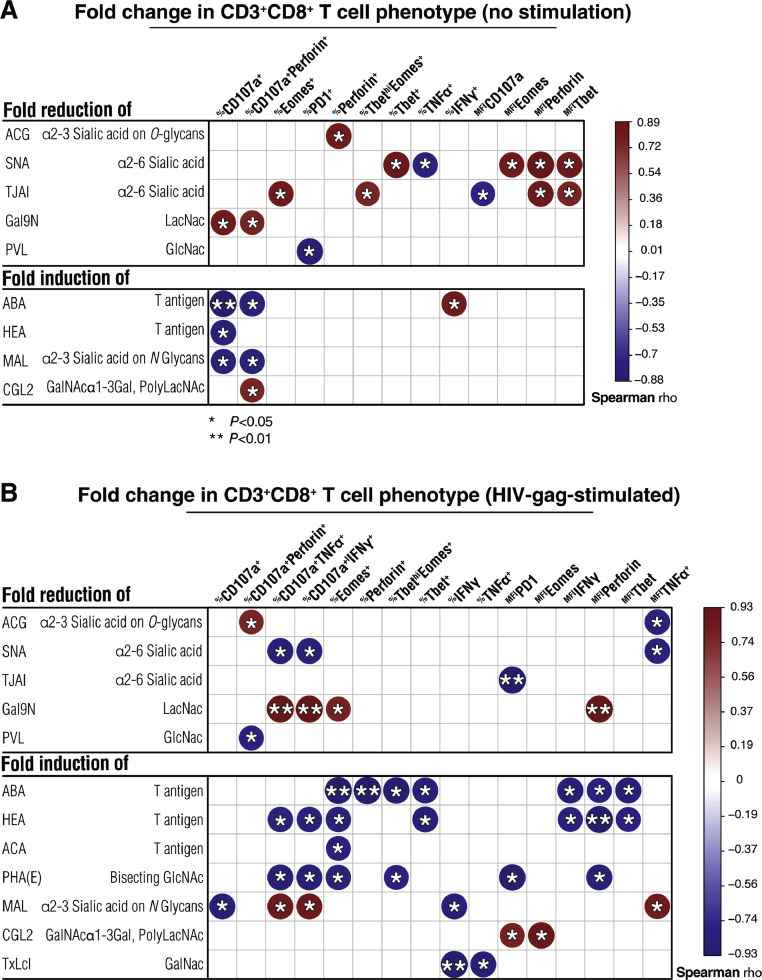
IFNα-mediated changes CD8+ T cell-surface glycome correlate with constitutive and HIV-specific CD8+ T functions. Correlation heat-maps showing associations between IFNα2a-mediated changes in CD8+ T cell-surface glycomes and IFNα2a-mediated changes in CD8+ T cell phenotypes measured without stimulation (A) or after HIV gag stimulation (B). The size and color of circles represent the strength of correlation, with blue shades representing negative correlations and red shades representing positive correlations. Correlations were evaluated using Spearman's rank correlation tests. *=P < 0.05, and **=P < 0.01. N = 8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.11. Role of the funding source
Funders had no role in the study design; no role in the collection, analysis, and interpretation of data; no role in the writing of the report; and no role in the decision to submit the paper for publication.
3. Results
3.1. Peg-IFNα2a treatment alters the IgG glycome during ART-suppressed HIV infection
We used capillary electrophoresis to profile the N-glycomic signatures of total plasma and isolated IgG samples from 18 HIV-infected individuals on suppressive ART who participated in a clinical trial in which Peg-IFNα2a was added to their ART regimen (NCT00594880) [26]. In this clinical trial, Peg-IFNα2a resulted in control of HIV replication and decreased levels of integrated HIV DNA. Plasma samples analyzed correspond to weeks 8 (before Peg-IFNα2a treatment; on ART alone with plasma HIV RNA load of <50 copies/mL), and 13 (five weeks after starting Peg-IFNα2a treatment; on both Peg-IFNα2a + ART with plasma HIV RNA load of <50 copies/mL) (Fig. 1A). Depends on sample availability, we also analyzed a subset of samples from study visit 25 (12 weeks after stopping ART; on Peg-IFNα2a treatment alone) of this study (Fig. 1A).
Peg-IFNα2a treatment was associated with significant alterations in 13 of the 19 glycan structures identified within the IgG glycome (Fig. 1B, false discovery rate (FDR) < 0.1). These 19 glycan structures can be grouped into seven groups, depending on the presence or absence of four key monosaccharides: bisecting GlcNAc (B group), sialic acid (S1 and S2 groups), galactose (G0, G1, and G2 groups), and fucose (F group). Following Peg-IFNα2a treatment there was a significant increase in the proportion of pro-inflammatory, bisecting GlcNAc (Group B) glycan structures (Fig. 1C, FDR=0.015; Wilcoxon signed-rank test). Specific increases were observed in specific B group glycan structures including G0FB, G1FB, G2FB, and A2FB (Fig. 1D–G, FDR < 0.05; Wilcoxon signed-rank test). Bisecting GlcNAc glycans on IgG are known to enhance FcγR binding to induce both innate immune function and inflammation [32], [33], [34]. In the total plasma glycome, B group was also higher following Peg-IFNα2a (data not shown); however, no changes in specific glycan structures were statistically significant, suggesting that the IFN-induced changes on plasma glycans depend on the IFN impact on IgG-associated glycans.
In addition, Peg-IFNα2a treatment was associated with a reduction in the levels of non-bisected fucosylated glycan structures G2F, G1F(3), G1F(6), and A2F, and an induction in the levels of the non-bisected non-fucosylated digalactosylated glycan structure, G2. Non-fucosylated galactosylated glycomic traits in IgG glycomes have been associated with higher ADCC/ADCP activities [17,18,35], and their levels associate positively with longer time-to-viral-rebound upon ART-cessation [36]. The magnitude of IFN modulation on IgG glycomes was relatively limited; however, recent publications showed that even subtle glycan modulations and minor glycan species can cause significant differences in antibody effector functions and activities [15,37].
To ensure that the observed IFN-mediated glycomic alterations in IgG glycomes were not a result of changes in total levels of IgG, we measured total levels of IgG before and during IFNα treatment. Five weeks of Peg-IFNα2a treatment did not impact the total levels of plasma IgG (Supplementary Fig. 5). We also measured levels of IgG subclasses IgG1, IgG2, IgG3, and IgG4. Peg-IFNα2a treatment did not impact levels of IgG1 or IgG3 but caused minimal induction in the levels of IgG2 and IgG4 (Supplementary Fig. 5). Together these data suggest that IFNα treatment causes alterations in the IgG glycome that are compatible with activating immune mechanisms associated with both higher inflammation and higher innate immune functions.
3.2. Peg-IFNα2a-mediated induction of the pro-inflammatory, bisecting GlcNAc on IgG associates with higher myeloid activation/inflammation
To assess if the increased levels of the pro-inflammatory bisecting GlcNac glycan structures caused by IFNα associate with inflammation, we evaluated levels of 23 pro- and anti-inflammatory plasma markers and cytokines (including soluble CD14 (sCD14) and soluble CD163 (sCD163) as markers of monocyte and macrophage inflammation). First, as expected, we found that levels of pro- and anti-inflammatory cytokines, and of monocyte and macrophage activation markers, changed after IFNα treatment. Comparison between baseline and five weeks on Peg-IFNα2a treatment showed a significant increase in levels of monocyte activation marker sCD14 (Fig. 2A, P = 0.026; Wilcoxon signed-rank test), macrophage activation marker sCD163 (Fig. 2B, P = 0.0008; Wilcoxon signed-rank test), and pro-inflammatory cytokine IL-18 (Fig. 2C, P = 0.0002; Wilcoxon signed-rank test). Also increased were levels of E-Selectin (P = 0.008), l-Selectin (P = 0.03), Galectin-3BP (P = 0.002), Galectin-9 (P<0.0001), and RANTES (P = 0.002) (Supplementary Table 1; Wilcoxon signed-rank tests). Consistently, IFNα2a treatment reduced the levels of the anti-inflammatory cytokine IL-10 (Fig. 2D, P = 0.0002; Wilcoxon signed-rank test). However, certain pro-inflammatory markers were reduced by IFNα treatment such as IL-12p40 (P = 0.04), IL-17A (P = 0.0003), and TNF-α (P = 0.0002) (Supplementary Table 1; Wilcoxon signed-rank tests).
We then evaluated the hypothesis that IFNα-mediated induction of the pro-inflammatory IgG glycomic features, mainly bisecting GlcNac glycans, is associated with IFNα-mediated induction of this inflammatory phenotype. Indeed, we found a positive correlation between IFNα-mediated fold change of B group (bisecting GlcNAc) glycans and IFNα-mediated fold change of sCD14 (Fig. 2E, P = 0.025, rho=0.53; Spearman's rank correlation). The change of the individual B group trait G0FB also correlated positively with fold change of sCD14 (Fig. 2F, P = 0.0037, rho=0.66; Spearman's rank correlation) and fold change of sCD163 (Fig. 2G, P = 0.01, rho=0.59; Spearman's rank correlation). Together these data suggest that IFNα treatment causes mostly a pro-inflammatory phenotype that is compatible with its induction of pro-inflammatory alterations in IgG glycomes that are known to enhance FcγR binding on monocytes, macrophages, and NK cells.
3.3. Peg-IFNα2a-mediated inductions in pro-inflammatory IgG glycans and plasma inflammatory markers correlate with differential CD8+ T and NK cell phenotypes
CD8+ T and NK cell functions are important for the antiviral effect of IFNα treatment [38] and the impact of IFNα on levels of integrated HIV DNA was associated with NK activation and cytotoxic response to K562 target cells [39]. To examine the links between IFNα-mediated alterations in IgG glycosylation, inflammatory markers, and immune functions, we evaluated CD8+ T cell (without stimulation) and NK cell (upon K562 stimulation) phenotypes using flow cytometry (green and blue arcs in Fig. 3A and B, respectively). We focused on identifying interactions in which all three domains evaluated (IgG glycome, soluble plasma markers of inflammation, and immune cell phenotype) significantly correlated with each other, after IFNα treatment (indicated by the darker-colored correlations in Fig. 3 (P<0.05, rho >0.5 or rho <−0.5; Spearman's rank correlation).
As a number of 3-way correlations reached significance, we highlight here two examples. IFNα-mediated induction of the pro-inflammatory IgG B group glycan G1FB correlates with higher induction of sCD14, lower levels with several markers of T cell function (including Eomes, Perforin, and Tbet), and higher levels of CD107a. Concurrently, levels of sCD14 correlate with lower levels of CD8+ T Eomes, Perforin, and Tbet but higher CD107a (Fig. 3A), which is compatible with results showing that sCD14 negatively impacts CD8+ T cell function by directly interacting with them [40]. As another example, the pro-inflammatory IgG B group glycan G0FB correlates negatively with the percentage of Perforin on NK cells, which correlates negatively with levels of sCD163 (Fig. 3B). This interactome analysis highlighted significant interactions that support a potential model in which a) IFNα increases the proportion of pro-inflammatory, bisecting GlcNAc (B group) glycans within the IgG glycome (Fig. 1), which in turn b) may increase levels of pro-inflammatory cytokine and markers of monocyte/macrophage inflammation (Fig. 2), which c) are linked to differential levels, mostly lower, of markers of CD8+ T and NK cell functions (Fig. 3).
3.4. IFN-mediated changes in the IgG glycome correlate with IFNα-mediated reduction of integrated HIV DNA
The prior study by Azzoni et al. observed that IFNα treatment reduces levels of integrated HIV DNA in CD4+ T cells, but with different efficiency (in some HIV+ individuals Peg-IFNα2a failed to reduce levels of integrated HIV DNA while in the majority of HIV+ individuals it did) [26]. Our results in Figs. 1–3 of the current study suggest that IFNα treatment mediates pro-inflammatory changes in antibody glycomes, which are linked to both higher systemic inflammation and to (in most cases) lower markers of CD8+ T and NK cell functions. Next, we examined if these links extended to the overall antiviral ability of IFNα to reduce levels of HIV integrated DNA in CD4+ T cells (comparing visit week 13 to visit week 8). Indeed, we found that IFNα-mediated induction of the pro-inflammatory B group glycan A2FB associates with poor IFNα-mediated fold reduction in levels of integrated HIV DNA (Fig. 4A, P = 0.02, rho=0.78; Spearman's rank correlation). In addition to bisecting GlcNac glycan traits we also found that the IFNα-mediated reduction of the mono-sialylated non-bisecting glycomic trait, A1, associates with better IFNα-mediated fold reduction in levels of integrated HIV DNA (Fig. 4B, P = 0.037, rho=0.76; Spearman's rank correlation). These data further highlight the potential detrimental impact that IFNα-mediated induction of pro-inflammatory B group glycans can have on IFNα antiviral capacity, possibly through mediating a systemic inflammatory state. However, these data also suggest that some effects of IFNα on IgG glycome are associated with favorable outcomes. It is likely that a fine balance between the detrimental and beneficial impacts of IFNα on the IgG glycome determines the overall effect of IFNα on cellular viral load.
3.5. IFNα modulates CD8+ T cell-surface glycomes in a way linked to their constitutive and HIV-gag-specific functions
Glycosylation of the cell surface shapes several CD8+ T [41] and NK [42] cell processes and functions. Therefore, in addition to evaluating the effect of IFNα on circulating glycomes, we sought to determine whether Peg-IFNα2a treatment also alters cell surface glycosylation of CD8+ T and NK cells, and thereby their functions. We negatively isolated CD8+ T and NK cells from peripheral blood mononuclear cells (PBMCs) from eight individuals from the same clinical trial (individuals with available PBMCs before and during Peg-IFNα2a treatment). We then profiled their cell membrane glycosylation using lectin arrays. The lectin microarrays enable an analysis of multiple cell-surface glycan structures by employing a panel of immobilized lectins (glycan-binding proteins) with known glycan-binding specificity [43,44]. In this study, we used two versions of the lectin array, one with 45 lectins and one with 96 lectins (Supplementary Tables 2 and 3 detail the names of the lectins and their glycan-binding specificity in the 45-plex and 96-plex lectin arrays, respectively).
Focusing on CD8+ T cells, after five weeks of IFNα treatment, there were several alterations in CD8+ T cell surface glycomes, including a decrease in the levels of GlcNac (measured by binding to PVL lectin), α2–3 sialic acid on O-glycans (measured by binding to ACG lectin), and α2–6 sialic acid (measured by binding to TJAI and SNA lectins) (Fig. 5, P<0.05; Wilcoxon signed-rank test). There also was an increase in the levels of the immunosuppressive GalNAc-containing glycans, such as T and Tn antigens (measured by binding to HEA, SRL, ABA, and ACA lectins) (Fig. 5, P<0.05; Wilcoxon signed-rank test). This glycomic profile (i.e. a decrease in sialic acid and an increase in GalNAc-containing glycans) is compatible with a higher activation status of CD8+ T cells [45], [46], [47], [48], [49].
We then examined the links between IFNα-mediated changes of CD8+ T cell surface glycomes and CD8+ T cell phenotypes (without stimulation and after HIV-gag stimulation) (Fig. 6). Compatible with the known suppressive impact of cell-surface sialic acid on T cell functions [41,[50], [51], [52], [53], the IFNα-mediated fold reduction of α2–6 sialylated glycans correlated positively with IFNα-mediated fold change of CD8+ T cell functional markers such as %Eomes+, %Tbet+ and %TbethiEomes+. These data suggest that levels of T cell surface sialylation change by IFNα to possibly enhance CD8+ T cell functions (Fig. 6A, P<0.05, rho>0.5; Spearman's rank correlation). In contrast, the fold induction of GalNAc-containing glycans correlated negatively with 1) fold change in %CD107a+ and %CD107a+Perforin+ (without stimulation; Fig. 6A, P<0.05, rho <−0.5; Spearman's rank correlation), and 2) fold change in %CD107a+TNFα+, %CD107a+IFNγ+, %Perforin+, %Tbet+, %TbethiEomes+, Tbet mean fluorescence intensity (MFI), IFNγ MFI, and Perforin MFI (after HIV-gag stimulation; Fig. 6B, P<0.05, rho <−0.5; Spearman's rank correlation). These data suggest that lower functions of CD8+ T cells (constitutive and HIV-specific) are linked to IFNα-mediated increase of this immunosuppressive glycomic trait (T and Tn antigens). These results are compatible with work showing that CD8+ T cell functions are down-regulated by GalNAc-containing glycans (T and Tn antigens) on T cells by binding to C-type lectin macrophage galactose type lectin (MGL) on antigen-presenting cells [54]. Supplementary Tables 4 and 5 detail the P and rho values of the correlations displayed in Fig. 6A and B, respectively. Together, these data suggest that CD8+ T cell surface glycomic profiles change after IFNα treatment in such a way that may impact CD8+ T cell functionality (constitutive and HIV-specific), in particular, a decrease in sialic acid-containing glycans may enhance CD8+ T cell functionality, whereas an increase in GalNAc-containing glycans (T or Tn antigen) may reduce constitutive and HIV-specific CD8+ T cell functionality.
3.6. IFNα modulates NK cell-surface glycomes in a way linked to their cellular functions
Focusing on NK cells, after five weeks of IFNα treatment, there were several changes in NK cell surface glycosylation. Similar to its impact on CD8+ T cells, Peg-IFNα2a treatment decreased the levels of GlcNac (measured by binding to PVL lectin), but different than its impact on CD8+ T cells, IFNα treatment decreased levels of T and Tn antigens on NK cells (measured by binding to ABA and Jacalin lectins) (Fig. 7A, P<0.05; Wilcoxon signed-rank test). The decrease in T/Tn antigens correlated positively with IFNα-mediated changes in several NK cell functional markers (upon K562 stimulation) such as %Eomes+,%Tbet+, and %TbethiEomes+, suggesting that reducing NK cell-surface T/Tn antigens improves functioning of NK cells (Fig. 7B, P<0.05, rho>0.5; Spearman's rank correlation). The treatment also led to an increase in mannose (binding to HHL lectin), α2–3 sialic acid (binding to LFA and MAH lectins), and fucose (binding to AOL, LCA, and PSA lectins) (Fig. 7A, P<0.05; Wilcoxon signed-rank test). The increase in fucose correlated positively with the IFNα-mediated changes in the MFI of CD107a, IFNγ, and TNFα, and percentage of Eomes+, Perforin+, Tbet+, and TbethiEomes+, also suggesting that a better function of NK cells is associated with this IFNα-mediated impact on NK cell-surface fucosylation (Fig. 7B, P<0.05, rho>0.5; Spearman's rank correlation). Finally, the increase in sialic acid levels on NK cells by IFNα associated with lower NK functionality, compatible with the immunosuppressive effects of sialic acid on NK cells [55] (Fig. 7B, P<0.05, rho <−0.5; Spearman's rank correlation). Supplementary Table 6 details the P and rho values of the correlations displayed in Fig. 7B. These combined data suggest that NK cell surface glycomic profiles change after IFNα treatment in such a way as to favor better NK functionality, in particular, by increasing fucose and decreasing T antigen. However, the IFNα-mediated induction of the immunosuppressive sialic acid may impose a negative impact on NK functionality.
Together, data from Fig. 5, Fig. 6, Fig. 7 suggest that IFNα treatment impacts immune cell-surface glycosylation in a cell-type-dependent manner and that specific alterations (namely levels of sialic acid, T/Tn antigens, and fucose) are associated with both detrimental and beneficial consequences of IFNα on CD8+ T and NK cell functions.
4. Discussion
Here we describe the first comprehensive longitudinal analysis of the impact of IFNα treatment on host glycosylation machinery in vivo. We found that IFNα treatment causes several alterations to both antibody and immune cell-surface glycomes. We also examined the links between these IFNα-mediated glycomic alterations and the IFNα impact on inflammatory, immunological, and antiviral responses during ART-suppressed HIV-infection. In particular, IFNα alters the IgG glycome by inducing the pro-inflammatory, bisecting GlcNac glycans that are known to enhance FcγR binding between IgG and immune cells. This induction of bisecting IgG glycans was associated with IFNα-mediated induction of inflammation, lower markers of CD8+ T and NK functions, and poor ability of IFNα to reduce levels of integrated HIV DNA. In addition, IFNα treatment alters CD8+ T and NK cell-surface glycomes in a cell-type-dependent manner. These alterations, namely modulation of sialic acid, T/Tn antigens, and fucose, are associated with both detrimental and beneficial consequences of IFNα on the functionality of these immune cells, in a manner consistent with their documented anti- or pro-immunological functions.
Our findings represent the first evidence that alterations in the host glycome may contribute to the IFNα paradox during chronic infections. While our longitudinal in vivo data do not unequivocally demonstrate a causal relationship between IFNα-mediated glycomic alterations and both inflammation and immune functions, the robust literature on the role of these specific glycomic alterations is relevant to and consistent with our findings and hypotheses. This consistency suggests that our observed glycomic alterations and associated changes in immunological and antiviral responses should be further explored for their potential functional significance as mediators of both beneficial and detrimental effects of IFNα during chronic HIV infection and other chronic diseases. Recently, a number of glycan-based strategies have been tested as novel immunotherapy agents, e.g., glycan-lectin interaction blockers, glycan-specific antibodies, glycan-coated nanoparticles, and metabolic inhibitors for certain glycans [56], [57], [58], [59], [60], [61]. Our current findings can lay the groundwork for future studies to investigate using some of these glycan-based strategies to enhance the antiviral effects of IFNα (both endogenous and exogenous) while avoiding its detrimental side-effects.
IFNα treatment significantly alters antibody glycomes. The glycosylation of IgG significantly impacts its ability to bind to Fcγ receptors and therefore impacts IgG functionality [13], [14], [15], [16], [17], [18]. Our finding that exogenous IFNα treatment significantly induces bisecting GlcNac glycans on IgG is consistent with our recent findings that TLR9 agonist treatment in vivo induces the same glycan structures on IgG [29]. TLR9 agonists induce the production of endogenous type I IFNs in vivo [29]. This consistency suggests that the impact of endogenous IFN production on the host glycome may be similar to the impact of exogenous IFN, expanding the utility of our findings to possibly advise strategies for enhancing the impact of both endogenously induced or exogenously added IFNs. The IFNα-mediated induction of bisecting GlcNac glycans on IgG is associated both with production of soluble markers of monocyte/macrophage inflammation (sCD14 and sCD163) and with poor CD8+ T cell phenotypes. These results are consistent with studies showing that bisecting GlcNac glycans enhance FcγR binding and induce inflammation [32], [33], [34],62,63]. They are also consistent with reports showing that IFNα treatment induces levels of sCD14 in hepatitis B infected individuals [64]. Soluble CD14 negatively impacts CD8+ T cells activation and function by directly interacting with T cells [40]. The robust literature on the direct impact of IgG glycomic alterations (including bisecting glycans) on inflammation [16,32,34,[65], [66], [67], [68], [69], supports our model in which IFNα increases the proportion of pro-inflammatory, bisecting GlcNAc glycans in the IgG glycome, which in turn increases levels of monocyte/macrophage inflammation/activation, leading to poor CD8+ T functions. This model warrants further investigation as it may help explain some of the detrimental impact of IFNα on host immune functions, especially given the known link between sCD14/sCD163 levels and both mortality and morbidity during chronic HIV infection [70,71]. While IgG glycomic dysregulations can induce inflammation directly [16,32,34,[65], [66], [67], [68], [69], IFNs are known to induce hundreds of interferon-stimulated genes (ISGs) and modulate the production of other cytokines. Therefore, future studies will be needed to investigate the direct versus indirect consequences of IFNα on the host glycosylation machinery as well as on monocyte/macrophage inflammatory and glycomic interactions.
IFNα also reduces the levels of several non-bisected fucosylated glycans, known to inhibit ADCC activity, and induces the non-fucosylated galactosylated glycan trait called G2. Non-fucosylated galactosylated glycomic structures in IgG glycomes are known to be associated with higher ADCC/ADCP activities and lower levels of HIV persistence during suppressive ART [13,14,17,18,35,72]. Levels of G2 on bulk plasma IgG are associated with longer time-to-viral-rebound upon ART-cessation [36]. Whether these IFNα-mediated alterations to the IgG glycomes contribute to any beneficial antiviral impact of IFNα on the whole-body burden of HIV reservoirs or time-to-viral-rebound upon ART-cessation warrants further investigation. Future strategies aimed at preventing or normalizing the detrimental effects of inducing pro-inflammatory bisecting GlcNac glycans, while amplifying the beneficial effects of reducing anti-ADCC fucosylated glycans, may maximize the antiviral effects of IFNα.
IFNα causes several alterations to CD8+ T and NK cell surface glycomes. Altered immune cell surface glycosylation affects cellular functions and immune responses, mainly by modulating the interaction between cell surface glycoproteins and lectins expressed on the same cells 'cis' or on other cells ‘trans’ [73]. The glycomic signature induced by IFNα on the CD8+T cell surface – increasing T/Tn antigens and decreasing sialic acid – is consistent with an activation status of these T cells [47], [48], [49],74]. This glycomic activation signature is known to change CD8+ T cell functions significantly. First, the truncated O GalNAc-containing glycans such as T/Tn antigens are ligands of the MGL lectin that is expressed on activated antigen presenting cells (APCs). MGL interaction with GalNAc-containing glycans on the T cell results in reduction of pro-inflammatory cytokines secretion, reduction of T cell proliferation, and induction of T cell apoptosis [54]. This is consistent with our results that the IFNα-mediated induction of T/Tn antigens on CD8+ T cells correlates with worse constitutive and HIV-specific CD8+ T cell functions. On the other hand, the decrease of sialylated glycans correlates with better T cell functionality. This is also consistent with studies showing that sialylated glycans exert immunosuppressive effects on T cells by binding to the negative regulators of cell signaling, Siglecs (sialic acid recognizing Ig-superfamily lectins) [50]. The binding of sialylated antigens on T cells to siglecs on APCs (or on T cells themselves) can lower the T effector cell responses [51,52]; conversely, loss of sialic acid can enhance T cell reactivity against antigens [53]. Understanding the upstream modulators of the IFNα-mediated CD8+ T cell surface glycomic alterations, as well as the downstream consequences of these alterations, should be the subject of future studies. In addition, it will be important to examine the impact of these IFN-mediated T cell glycomic alterations on antigen presentation to CD8+ T cells by APCs, given that the glycan-lectin interactions described above are mainly interactions between glycans on CD8+ T cells and lectins on APCs.
The functions of NK cells can be influenced by their cell-surface glycosylation and the glycosylation of their target cells. NK cells express two cell-surface siglecs, Siglec-7 and Siglec-9. Siglecs bind to sialic acid on the surface of NK cells (cis) or the surface of target cells (trans), leading to the inhibition of NK activity [59]. Our observations that IFNα induces sialic acid on the NK surface and that this induction correlates negatively with NK functions warrant further investigation. IFNα is an activator of NK cell functions [75,76], and the positive impact of IFNα on levels of integrated HIV DNA was associated with NK activation and cytotoxic response to K562 target cells [39]. However, IFNα-mediated induction of sialic acid may exert a negative effect that hampers taking full advantage of the positive impact of IFNα on NK cells. Targeting sialic acid/siglec binding [59], as proposed in the cancer field [59], may represent a useful strategy to enhance NK functionality during exogenous IFNα treatment or treatments aimed to enhance endogenous IFN production. Examining the impact of IFN on the cell-surface glycosylation (and cell-surface lectin expression) of other immune cells, as well as identifying the exact proteins backbones of these IFNα-mediated alterations to CD8+ T and NK cell surface glycomes, should be studied as it will likely allow for a better understanding of the role of cell-surface glycosylation in modulating immune functions during HIV infection.
Our study has limitations, including the small sample size. While we were able to apply a correction for multiple comparisons in our investigation on the IFNα-mediated alterations to IgG and plasma glycans (n = 18), the limited availability of CD8+ T and NK cells (n = 8) from the same study limited our ability to fully apply multiple comparisons correction to that exploratory section of our study. Validating these results using larger cohorts should be the subject of future studies. Second, gender, genetics, diet, and age may impact host glycosylation. Larger sample size and controlled animal studies will be needed to explore the full extent of the link between Type I IFNs and the host glycosylation machinery. Third, our study was performed on peripheral blood. Investigating the impact of IFN on the glycosylation of immune cells and other cells in different tissues and how this impact may influence the tissue microenvironment and HIV persistence will be needed. In summary, our study, using longitudinal in vivo human samples, provides the first insight into a potential role of host glycomic alterations as a mediator of the IFN paradox during HIV infection. The role of IFN-mediated alterations to host glycomes in mediating immunological and antiviral responses during chronic diseases, such as HIV infection, warrants further investigation, in order to advise novel glycomic-based interventions to manipulate glycan-lectin interactions to maximize the antiviral effects of both endogenous and exogenous IFNs while avoiding their detrimental side-effects.
Funding sources
M.A-M is supported by NIH grants (R01 DK123733, R01 AG062383, R01 NS117458, R21 AI143385, R21 AI129636, and R21 NS106970), The Foundation for AIDS Research (amfAR) impact grant # 109840–65-RGRL, and W.W. Smith Charitable Trust grant # A1901. L.J.M. supported by the NIH-funded BEAT-HIV Martin Delaney Collaboratory to cure HIV-1 infection (1UM1Al126620), U01 AI110434, R01 DA048728, R01 DA049666, Kean Family Professorship, and the Philadelphia Foundation (Roberts I. Jacobs Fund).
Author contributions
L.B.G., F.C., E.P., L.A., M.F., A.A., M.D., U.O., H.T., M.R.B., L.J.M., and M.A-M designed and carried out experiments. E.P., L.A., K.M., J.R.K., P.T., and L.J.M selected study participants and provided samples. L.B.G, F.C., E.P., X.Y., Q.L., and M.A-M analyzed and interpreted data. L.B.G, F.C., L.J.M., and M.A.M. wrote the manuscript, and all authors edited it.
Declaration of Interests
Dr. Mounzer reports grants and personal fees from Janssen Therapeutics, grants and personal fees from ViiV healthcare, grants and personal fees from Gilead Sciences, outside the submitted work. All other authors have nothing to disclose.
Acknowledgments
We would like to thank all donor participants. We would also like to thank Rachel E. Locke, Ph.D., for providing comments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102945.
Contributor Information
Luis J. Montaner, Email: montaner@wistar.org.
Mohamed Abdel-Mohsen, Email: mmohsen@wistar.org.
Appendix. Supplementary materials
Supplementary Fig. 1. The structures and names of the N-glycans identified in IgG by capillary electrophoresis.
Supplementary Fig. 2. The structures and names of the N-glycans identified in plasma by capillary electrophoresis.
Supplementary Fig. 3. A representative example of the gating strategy used for analyses of CD8\elsamp #x002B;T cells. FSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;forward scatter, and SSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;side scatter.
Supplementary Fig. 4. A representative example of the gating strategy used for analyses of NK cells. FSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;forward scatter, and SSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;side scatter.
Supplementary Fig. 5. Impact of Peg-IFN\elsamp #x03B1;2a treatment on levels of plasma total IgG, IgG1, IgG2, IgG3, and IgG4. Five weeks of Peg-IFN\elsamp #x03B1;2a treatment did not impact total levels of plasma total IgG, IgG1, or IgG3 and caused minimal induction of IgG2 and IgG4. Wilcoxon signed-rank test. Lines represent medians.
References
- 1.Scagnolari C., Antonelli G. Type I interferon and HIV: subtle balance between antiviral activity, immunopathogenesis and the microbiome. Cytokine Growth Factor Rev. 2018;40:19–31. doi: 10.1016/j.cytogfr.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Utay N.S., Douek D.C. Interferons and HIV Infection: the good, the bad, and the ugly. Pathog Immun. 2016;1(1):107–116. doi: 10.20411/pai.v1i1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng L., Ma J., Li J. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J Clin Invest. 2017;127(1):269–279. doi: 10.1172/JCI90745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugawara S., Thomas D.L., Balagopal A. HIV-1 Infection and Type 1 interferon: navigating through uncertain waters. AIDS Res Hum Retroviruses. 2019;35(1):25–32. doi: 10.1089/aid.2018.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van der Sluis R.M., Zerbato J.M., Rhodes J.W. Diverse effects of interferon alpha on the establishment and reversal of HIV latency. PLoS Pathog. 2020;16(2) doi: 10.1371/journal.ppat.1008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207(10):2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhlbrigge R.C., Edwards B.S., Borden E.C. Effects of interferon alpha on the sheep erythrocyte "receptor" of human lymphocytes. J Interferon Res. 1984;4(4):499–505. doi: 10.1089/jir.1984.4.499. [DOI] [PubMed] [Google Scholar]
- 8.Maheshwari R.K., Banerjee D.K., Waechter C.J., Olden K., Friedman R.M. Interferon treatment inhibits glycosylation of a viral protein. Nature. 1980;287(5781):454–456. doi: 10.1038/287454a0. [DOI] [PubMed] [Google Scholar]
- 9.Maheshwari R.K., Sreevalsan T., Silverman R.H., Hay J., Friedman R.M. Tunicamycin enhances the antiviral and anticellular activity of interferon. Science. 1983;219(4590):1339–1341. doi: 10.1126/science.6187067. [DOI] [PubMed] [Google Scholar]
- 10.Faltynek C.R., Baglioni C. Treatment of human cells and interferon has no effect on the glycosylation of viral and cellular proteins. Virology. 1983;127(1):225–229. doi: 10.1016/0042-6822(83)90386-0. [DOI] [PubMed] [Google Scholar]
- 11.Olden K., Bernard B.A., Turner W., White S.L. Effect of interferon on protein glycosylation and comparison with tunicamycin. Nature. 1982;300(5889):290–292. doi: 10.1038/300290a0. [DOI] [PubMed] [Google Scholar]
- 12.Dewald J.H., Colomb F., Bobowski-Gerard M., Groux-Degroote S., Delannoy P. Role of cytokine-induced glycosylation changes in regulating cell interactions and cell signaling in inflammatory diseases and cancer. Cells. 2016;5(4) doi: 10.3390/cells5040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colomb F., Giron L.B., Trbojevic-Akmacic I., Lauc G., Abdel-Mohsen M. Breaking the glyco-code of HIV persistence and immunopathogenesis. Curr HIV AIDS Rep. 2019;16(2):151–168. doi: 10.1007/s11904-019-00433-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. 2018;18(1):46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko Y., Nimmerjahn F., Ravetch J.V. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313(5787):670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 16.Karsten C.M., Pandey M.K., Figge J. Anti-inflammatory activity of IgG1 mediated by Fc galactosylation and association of FcgammaRIIB and dectin-1. Nat Med. 2012;18(9):1401–1406. doi: 10.1038/nm.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomann M., Reckermann K., Reusch D., Prasser J., Tejada M.L. Fc-galactosylation modulates antibody-dependent cellular cytotoxicity of therapeutic antibodies. Mol Immunol. 2016;73:69–75. doi: 10.1016/j.molimm.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Shields R.L., Lai J., Keck R. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277(30):26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 19.Mendez-Huergo S.P., Blidner A.G., Rabinovich G.A. Galectins: emerging regulatory checkpoints linking tumor immunity and angiogenesis. Curr Opin Immunol. 2017;45:8–15. doi: 10.1016/j.coi.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Smith L.K., Boukhaled G.M., Condotta S.A. Interleukin-10 directly inhibits CD8(+) T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity. 2018;48(2):299–312. doi: 10.1016/j.immuni.2018.01.006. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrne B., Donohoe G.G., O'Kennedy R. Sialic acids: carbohydrate moieties that influence the biological and physical properties of biopharmaceutical proteins and living cells. Drug Discov Today. 2007;12(7–8):319–326. doi: 10.1016/j.drudis.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Rabinovich G.A., Rubinstein N., Toscano M.A. Role of galectins in inflammatory and immunomodulatory processes. Biochim Biophys Acta. 2002;1572(2–3):274–284. doi: 10.1016/s0304-4165(02)00314-8. [DOI] [PubMed] [Google Scholar]
- 23.Zhuo Y., Bellis S.L. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem. 2011;286(8):5935–5941. doi: 10.1074/jbc.R110.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley K. The role of selectins in inflammation and disease. Trends Mol Med. 2003;9(6):263–268. doi: 10.1016/s1471-4914(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 25.Deeks S.G. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azzoni L., Foulkes A.S., Papasavvas E. Pegylated Interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis. 2013;207(2):213–222. doi: 10.1093/infdis/jis663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H., Buzon M.J., Shaw A. Hepatitis C therapy with interferon-alpha and ribavirin reduces CD4 T-cell-associated HIV-1 DNA in HIV-1/hepatitis C virus-coinfected patients. J Infect Dis. 2014;209(9):1315–1320. doi: 10.1093/infdis/jit628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krarup A.R., Abdel-Mohsen M., Schleimann M.H. The TLR9 agonist MGN1703 triggers a potent type I interferon response in the sigmoid colon. Mucosal Immunol. 2018;11(2):449–461. doi: 10.1038/mi.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schleimann M.H., Kobbero M.L., Vibholm L.K. TLR9 agonist MGN1703 enhances B cell differentiation and function in lymph nodes. EBioMedicine. 2019;45:328–340. doi: 10.1016/j.ebiom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vibholm L., Schleimann M.H., Hojen J.F. Short-course toll-like receptor 9 Agonist treatment impacts innate immunity and plasma viremia in individuals with human immunodeficiency virus infection. Clin Infect Dis. 2017;64(12):1686–1695. doi: 10.1093/cid/cix201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graf E.H., Mexas A.M., Yu J.J. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies J., Jiang L., Pan L.Z., LaBarre M.J., Anderson D., Reff M. Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FC gamma RIII. Biotechnol Bioeng. 2001;74(4):288–294. [PubMed] [Google Scholar]
- 33.Takahashi M., Kuroki Y., Ohtsubo K., Taniguchi N. Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydr Res. 2009;344(12):1387–1390. doi: 10.1016/j.carres.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Umana P., Jean-Mairet J., Moudry R., Amstutz H., Bailey J.E. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 1999;17(2):176–180. doi: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- 35.Chung A.W., Crispin M., Pritchard L. Identification of antibody glycosylation structures that predict monoclonal antibody Fc-effector function. AIDS. 2014;28(17):2523–2530. doi: 10.1097/QAD.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giron L.B., Papasavvas E., Azzoni L. Plasma and antibody glycomic biomarkers of time to HIV rebound and viral setpoint. AIDS. 2020;34(5):681–686. doi: 10.1097/QAD.0000000000002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ackerman M.E., Crispin M., Yu X. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J Clin Invest. 2013;123(5):2183–2192. doi: 10.1172/JCI65708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manion M., Rodriguez B., Medvik K. Interferon-alpha administration enhances CD8+ T cell activation in HIV infection. PLoS ONE. 2012;7(1):e30306. doi: 10.1371/journal.pone.0030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papasavvas E., Azzoni L., Kossenkov A.V. NK response correlates with HIV decrease in pegylated IFN-alpha2a-treated antiretroviral therapy-suppressed subjects. J Immunol. 2019;203(3):705–717. doi: 10.4049/jimmunol.1801511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey Nores J.E., Bensussan A., Vita N. Soluble CD14 acts as a negative regulator of human T cell activation and function. Eur J Immunol. 1999;29(1):265–276. doi: 10.1002/(SICI)1521-4141(199901)29:01<265::AID-IMMU265>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 41.Pereira M.S., Alves I., Vicente M. Glycans as Key Checkpoints of T Cell Activity and Function. Front Immunol. 2018;9:2754. doi: 10.3389/fimmu.2018.02754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson V., Grobarova V., Richter J., Fiserova A. Glycosylation regulates NK cell-mediated effector function through PI3K pathway. Int Immunol. 2010;22(3):167–177. doi: 10.1093/intimm/dxp123. [DOI] [PubMed] [Google Scholar]
- 43.Tateno H., Kuno A., Itakura Y., Hirabayashi J. A versatile technology for cellular glycomics using lectin microarray. Meth Enzymol. 2010;478:181–195. doi: 10.1016/S0076-6879(10)78008-3. [DOI] [PubMed] [Google Scholar]
- 44.Hirabayashi J., Kuno A., Tateno H. Development and applications of the lectin microarray. Top Curr Chem. 2015;367:105–124. doi: 10.1007/128_2014_612. [DOI] [PubMed] [Google Scholar]
- 45.Priatel J.J., Chui D., Hiraoka N. The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity. 2000;12(3):273–283. doi: 10.1016/s1074-7613(00)80180-6. [DOI] [PubMed] [Google Scholar]
- 46.Chervenak R., Cohen J.J. Peanut lectin binding as a marker for activated T-lineage lymphocytes. Thymus. 1982;4(2):61–67. [PubMed] [Google Scholar]
- 47.Galvan M., Murali-Krishna K., Ming L.L., Baum L., Ahmed R. Alterations in cell surface carbohydrates on T cells from virally infected mice can distinguish effector/memory CD8+ T cells from naive cells. J Immunol. 1998;161(2):641–648. [PubMed] [Google Scholar]
- 48.Jenner J., Kerst G., Handgretinger R., Muller I. Increased alpha2,6-sialylation of surface proteins on tolerogenic, immature dendritic cells and regulatory T cells. Exp Hematol. 2006;34(9):1212–1218. doi: 10.1016/j.exphem.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 49.Piller F., Piller V., Fox R.I., Fukuda M. Human T-lymphocyte activation is associated with changes in O-glycan biosynthesis. J Biol Chem. 1988;263(29):15146–15150. [PubMed] [Google Scholar]
- 50.Crocker P.R., Redelinghuys P. Siglecs as positive and negative regulators of the immune system. Biochem Soc Trans. 2008;36(Pt 6):1467–1471. doi: 10.1042/BST0361467. [DOI] [PubMed] [Google Scholar]
- 51.Perdicchio M., Ilarregui J.M., Verstege M.I. Sialic acid-modified antigens impose tolerance via inhibition of T-cell proliferation and de novo induction of regulatory T cells. Proc Natl Acad Sci U S A. 2016;113(12):3329–3334. doi: 10.1073/pnas.1507706113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanczak M.A., Siddiqui S.S., Trefny M.P. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J Clin Invest. 2018;128(11):4912–4923. doi: 10.1172/JCI120612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pappu B.P., Shrikant P.A. Alteration of cell surface sialylation regulates antigen-induced naive CD8+ T cell responses. J Immunol. 2004;173(1):275–284. doi: 10.4049/jimmunol.173.1.275. [DOI] [PubMed] [Google Scholar]
- 54.van Vliet S.J., Gringhuis S.I., Geijtenbeek T.B., van Kooyk Y. Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat Immunol. 2006;7(11):1200–1208. doi: 10.1038/ni1390. [DOI] [PubMed] [Google Scholar]
- 55.Daly J., Carlsten M., O'Dwyer M. Sugar Free: novel Immunotherapeutic Approaches Targeting Siglecs and Sialic Acids to Enhance Natural Killer Cell Cytotoxicity Against Cancer. Front Immunol. 2019;10:1047. doi: 10.3389/fimmu.2019.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bull C., Boltje T.J., Wassink M. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Mol Cancer Ther. 2013;12(10):1935–1946. doi: 10.1158/1535-7163.MCT-13-0279. [DOI] [PubMed] [Google Scholar]
- 57.Dube D.H., Bertozzi C.R. Glycans in cancer and inflammation–potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 58.Posey A.D., Jr., Schwab R.D., Boesteanu A.C. Engineered CAR T cells targeting the cancer-associated Tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity. 2016;44(6):1444–1454. doi: 10.1016/j.immuni.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao H., Woods E.C., Vukojicic P., Bertozzi C.R. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci U S A. 2016;113(37):10304–10309. doi: 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q., Chung C.Y., Chough S., Betenbaugh M.J. Antibody glycoengineering strategies in mammalian cells. Biotechnol Bioeng. 2018;115(6):1378–1393. doi: 10.1002/bit.26567. [DOI] [PubMed] [Google Scholar]
- 61.Spence S., Greene M.K., Fay F. Targeting Siglecs with a sialic acid-decorated nanoparticle abrogates inflammation. Sci Transl Med. 2015;7(303):303ra140. doi: 10.1126/scitranslmed.aab3459. [DOI] [PubMed] [Google Scholar]
- 62.Liu D., Zhao Z., Wang A. Ischemic stroke is associated with the pro-inflammatory potential of N-glycosylated immunoglobulin G. J Neuroinflamm. 2018;15(1):123. doi: 10.1186/s12974-018-1161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plomp R., Ruhaak L.R., Uh H.W. Subclass-specific IgG glycosylation is associated with markers of inflammation and metabolic health. Sci Rep. 2017;7(1):12325. doi: 10.1038/s41598-017-12495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carotenuto P., van Riel D., Artsen A. Antiviral treatment with alpha interferon up-regulates CD14 on liver macrophages and its soluble form in patients with chronic hepatitis B. Antimicrob Agents Chemother. 2005;49(2):590–599. doi: 10.1128/AAC.49.2.590-599.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bohm S., Kao D., Nimmerjahn F. Sweet and sour: the role of glycosylation for the anti-inflammatory activity of immunoglobulin G. Curr Top Microbiol Immunol. 2014;382:393–417. doi: 10.1007/978-3-319-07911-0_18. [DOI] [PubMed] [Google Scholar]
- 66.Maverakis E., Kim K., Shimoda M. Glycans in the immune system and the altered glycan theory of autoimmunity: a critical review. J Autoimmun. 2015;57:1–13. doi: 10.1016/j.jaut.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Washburn N., Schwab I., Ortiz D. Proceedings of the national academy of sciences of the United States of America. Vol. 112. 2015. Controlled tetra-Fc sialylation of IVIg results in a drug candidate with consistent enhanced anti-inflammatory activity; pp. E1297–E1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Epp A., Hobusch J., Bartsch Y.C. Sialylation of IgG antibodies inhibits IgG-mediated allergic reactions. J Allergy Clin Immunol. 2018;141(1):399–402. doi: 10.1016/j.jaci.2017.06.021. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng J., Vongpatanasin W., Sacharidou A. Supplementation with the sialic acid precursor N-acetyl-d-mannosamine breaks the link between obesity and hypertension. Circulation. 2019;140(24):2005–2018. doi: 10.1161/CIRCULATIONAHA.119.043490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knudsen T.B., Ertner G., Petersen J. Plasma soluble CD163 level independently predicts all-cause mortality in HIV-1-infected individuals. J Infect Dis. 2016;214(8):1198–1204. doi: 10.1093/infdis/jiw263. [DOI] [PubMed] [Google Scholar]
- 71.Sandler N.G., Wand H., Roque A. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vadrevu S.K., Trbojevic-Akmacic I., Kossenkov A.V. Frontline Science: plasma and immunoglobulin G galactosylation associate with HIV persistence during antiretroviral therapy. J Leukoc Biol. 2018;104(3):461–471. doi: 10.1002/JLB.3HI1217-500R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Reilly M.K., Paulson J.C. Multivalent ligands for siglecs. Meth Enzymol. 2010;478:343–363. doi: 10.1016/S0076-6879(10)78017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Vliet S.J., Vuist I.M., Lenos K. Human T cell activation results in extracellular signal-regulated kinase (ERK)-calcineurin-dependent exposure of Tn antigen on the cell surface and binding of the macrophage galactose-type lectin (MGL) J Biol Chem. 2013;288(38):27519–27532. doi: 10.1074/jbc.M113.471045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomescu C., Mavilio D., Montaner L.J. Lysis of HIV-1-infected autologous CD4+ primary T cells by interferon-alpha-activated NK cells requires NKp46 and NKG2D. AIDS. 2015;29(14):1767–1773. doi: 10.1097/QAD.0000000000000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomescu C., Tebas P., Montaner L.J. IFN-alpha augments natural killer-mediated antibody-dependent cellular cytotoxicity of HIV-1-infected autologous CD4+ T cells regardless of major histocompatibility complex class 1 downregulation. AIDS. 2017;31(5):613–622. doi: 10.1097/QAD.0000000000001380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. The structures and names of the N-glycans identified in IgG by capillary electrophoresis.
Supplementary Fig. 2. The structures and names of the N-glycans identified in plasma by capillary electrophoresis.
Supplementary Fig. 3. A representative example of the gating strategy used for analyses of CD8\elsamp #x002B;T cells. FSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;forward scatter, and SSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;side scatter.
Supplementary Fig. 4. A representative example of the gating strategy used for analyses of NK cells. FSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;forward scatter, and SSC\elsamp #x00A0;\elsamp #x003D;\elsamp #x00A0;side scatter.
Supplementary Fig. 5. Impact of Peg-IFN\elsamp #x03B1;2a treatment on levels of plasma total IgG, IgG1, IgG2, IgG3, and IgG4. Five weeks of Peg-IFN\elsamp #x03B1;2a treatment did not impact total levels of plasma total IgG, IgG1, or IgG3 and caused minimal induction of IgG2 and IgG4. Wilcoxon signed-rank test. Lines represent medians.