Abstract
Group 2 innate lymphoid cells (ILC2s) in mouse lungs are activated by the epithelium-derived alarmin IL-33. Activated ILC2s proliferate and produce IL-5 and IL-13 that drive allergic responses. In neonatal lungs, the occurrence of spontaneous activation of lung ILC2s is dependent on endogenous IL-33. Here, we report that neonatal lung ILC2 activation by endogenous IL-33 has significant effects on ILC2 functions in adulthood. Most neonatal lung ILC2s incorporated 5-bromo-2′-deoxyuridine (BrdU) and persisted into adulthood. BrdU+ ILC2s in adult lungs responded more intensely to IL-33 treatment compared with BrdU– ILC2s. In IL-33–deficient (KO) mice, lung ILC2s develop normally, but they are not activated in the neonatal period. Lung ILC2s in KO mice responded less intensely to IL-33 in adulthood compared with WT ILC2s. While there was no difference in the number of lung ILC2s, there were fewer IL-13+ ILC2s in KO mice compared with those in WT mice. The impaired responsiveness of ILC2s in KO mice was reversed by i.n. administrations of IL-33 in the neonatal period. These results suggest that activation of lung ILC2s by endogenous IL-33 in the neonatal period may “train” ILC2s seeding the lung after birth to become long-lasting resident cells that respond more efficiently to challenges later in life.
Keywords: Immunology
Keywords: Allergy, Asthma, Innate immunity
Endogenous IL-33 trains neonatal mouse lung ILC2s to respond strongly to IL-33 administration in adulthood, establishing the neonatal period as important for lung ILC2 function.
Introduction
Group 2 innate lymphoid cells (ILC2s) are key drivers of type 2 immunity (1). Lung ILC2s respond to the alarmin IL-33 released by epithelial cells upon exposure to inhaled allergens and secrete large amounts of IL-5 and IL-13. IL-5 promotes eosinophil differentiation, recruitment, activation, and survival. IL-13 is responsible for goblet cell hyperplasia, mucus secretion, and collagen secretion from fibroblasts and drives the development of alternatively activated macrophages (2). Additionally, IL-13 promotes Th2 cell differentiation (3–6). Through the production of those type 2 cytokines, lung ILC2s play a key role in allergic lung diseases, tissue repair, lung fibrosis, and antiparasite immunity (7, 8).
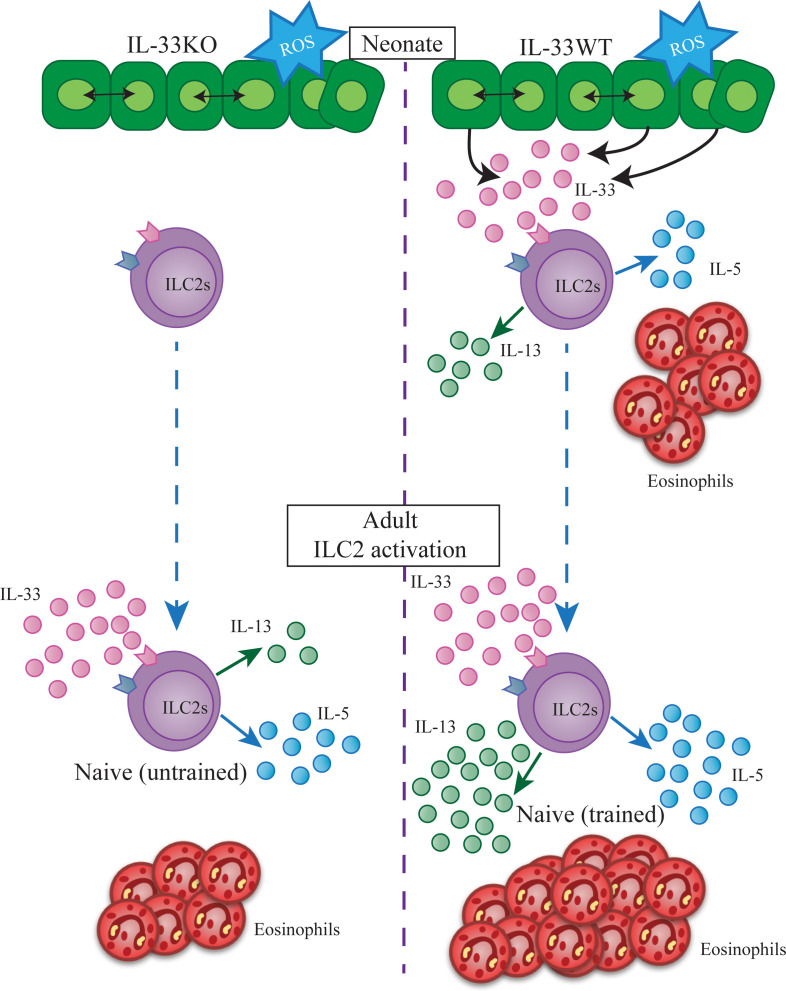
Analyses of parabiosis mice showed that ILCs are tissue-resident lymphocytes and very few ILCs circulate in adult mice (9). Recently, Schneider et al. have also shown that most ILC2s in adult mouse lungs develop in the neonatal period and only a minor fraction is slowly replaced by newly developing ILC2s in adulthood (10). Therefore, the neonatal period is critical for ILC2 development. We (11) and others (12–14) have found that lung epithelial and stromal cells express IL-33 at around 10–14 days after birth. This may be due to early life signals, including the mechanical stretch of the first few breathes of life, oxidative stress, or even lung developmental processes. The wave of IL-33 causes transient activation and proliferation of neonatal lung ILC2s and eosinophil infiltration. It is thought that the activation of ILC2s is responsible for the Th2-biased response to antigens in the neonatal period (11).
We have previously shown that activation of adult mouse lung ILC2s by inhaled allergen or recombinant IL-33 has long-lasting effects on ILC2 functions (15). After the resolution of inflammation, some activated ILC2s persist for several months as resting “allergen-experienced” memory cells that are more responsive to inhaled allergens or IL-33 than naive ILC2s. Upon challenge with an unrelated allergen or IL-33, months after the first allergen exposure, memory ILC2s vigorously respond and induce enhanced airway inflammation characterized by increased numbers of eosinophils in the lung and higher mucus accumulation in the airways compared with naive mice receiving the same challenge.
While the exact mechanism behind ILC2 memory has yet to be elucidated, our transcriptome analyses of purified naive and memory ILC2s identified a small set of differentially expressed genes that included Il17rb, encoding the IL-25 receptor (15). The expression of Il17rb was very low in naive lung ILC2s but was rapidly upregulated by activation, and the higher expression of Il17rb was maintained in memory ILC2s for more than 4 months after the initial activation. Flow cytometric analyses confirmed higher expression of the IL-25R on memory ILC2s compared with naive lung ILC2s. Furthermore, memory, but not naive, lung ILC2s were activated by i.n. administration of IL-25. These results suggested that IL-25R upregulation is an ILC2-intrinsic mechanism by which lung ILC2s acquire memory functions.
We now have investigated whether the activation of neonatal lung ILC2s by endogenous IL-33 has a similar effect as adult ILC2 activation, which results in the generation of memory ILC2s. ILC2s in IL-33–deficient (KO) mice are not activated in the neonatal period (11). Therefore, we compared lung ILC2s from KO and WT mice. Our results show that activation of lung ILC2s in the neonatal period made them more functionally competent in adulthood. This process is different from the generation of memory ILC2s in adult mice. The former does not involve acquisition of new functions, whereas memory ILC2s become responsive to IL-25 (15). Therefore, we termed the process “ILC2 training.”
Results
Characterization of neonatal lung ILC2s.
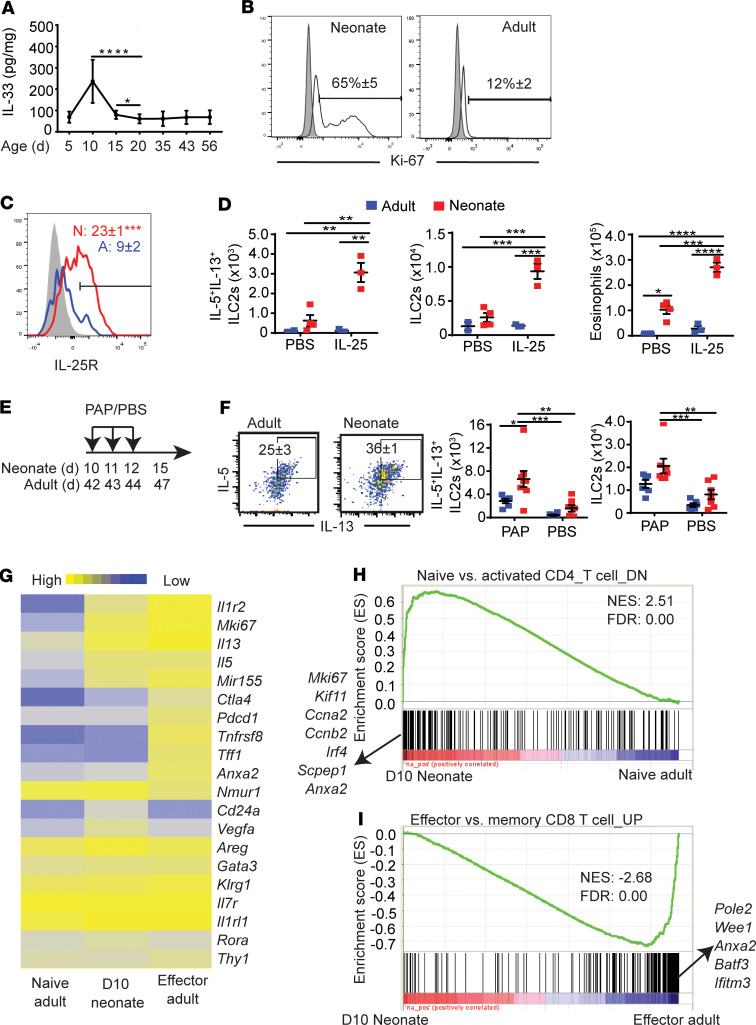
We previously found that the ILC2 population in the lung expands at around 10 days after birth, and this was not observed in other tissues (11). Immunohistochemical analysis of neonatal lung suggested that endogenous IL-33 is elevated at the time of ILC2 expansion (11). To further investigate the activation of neonatal lung ILC2s, we first analyzed the amounts of IL-33 in neonatal mouse lung homogenates. ELISA analysis of lung homogenates showed that the amounts of IL-33 were elevated at around 10 days after birth (Figure 1A). Lung ILC2s in this period highly expressed the proliferation marker Ki-67 (Figure 1B). Neonatal ILC2s expressed the IL-25R (Figure 1C) and responded to i.n. administration of IL-25 (Figure 1D). In contrast, naive adult lung ILC2s expressed very low amounts of IL-25R and did not respond to i.n. IL-25. The protease allergen papain, along with other allergens, induces IL-33–mediated allergic inflammation but also induces the release of IL-25 (16). As expected, exposure to papain in the neonatal period led to more ILC2 proliferation and intracellular IL-5 and IL-13 expression compared with those treated in adulthood (Figure 1, E and F).
Figure 1. Neonatal ILC2s are activated by endogenous IL-33.
(A) The amount of IL-33 in whole lung homogenates of naive mice at indicated ages (n > 4). (B) Ki-67 expression of neonatal (day 10) and adult (4–6 weeks) lung ILC2s (gated as in Supplemental Figure 1A) (n = 4). Shading indicates isotype control; white indicates anti-Ki-67. (C) Neonatal and adult ILC2s were examined for IL-25R/IL-17RB expression (n = 5). Shading indicates isotype control. Numbers in the plot indicate the mean percentage ± SEM. (D) B6 neonates (red squares) and adults (blue squares) were treated with 1 administration of i.n. IL-25 on postnatal day 10 and 42, respectively. Three days later, numbers of IL-5+IL-13+ ILC2s, total ILC2s, and eosinophils in the lungs were analyzed by flow cytometry (n = 3–4, 2 independent experiments). (E) Scheme of i.n. papain (PAP)/PBS injections (n = 5–8, 2 independent experiments). d, days after birth. (F) Flow cytometry plots show intracellular IL-5 and IL-13 expression in ILC2s in adult and neonatal papain-treated mice. Numbers in flow plots represent the percentage of double-positive ILC2s ± SEM. Scatterplots show numbers of IL-5+IL-13+ ILC2s and total ILC2s. Blue squares represent adults; red squares represent neonates. (G) Heatmap shows the relative expression levels of genes in lung ILC2s from naive adults (8 weeks of age), day 10 neonatal pups, and effector adults (8 weeks of age) that have been given three 0.25 μg IL-33 i.n. injections and then harvested 1 day after the last injection (n = 3 per condition, each replicate represents sorted ILC2s from 8 pooled mice). (H) Gene set enrichment analysis (GSEA) of neonatal ILC2s vs. naive adult ILC2s. The log2-fold change for each comparison was ranked in descending order and compared with the immunological signatures gene set (C7). The x axis shows where the members of the gene set appear in the ranked list of genes. The y axis represents the enrichment score. The highest ranked genes are listed on the x axis. The genes that were downregulated in naive CD4 T cells compared with activated CD4 T cells were significantly enriched in neonatal day 10 ILC2s. (I) GSEA of neonatal ILC2s and effector adult ILC2s. Genes that were upregulated in effector CD8 T cells compared with memory CD8 T cells were enriched in effector adult ILC2s. NES, normalized enrichment score; FDR, false discovery rate. *P < 0.05; **P < 0.01; ***P < 0.005 (1-tailed unpaired Student’s t test for C, 2-way ANOVA with Bonferroni’s post hoc test for D and F).
We then compared the global gene expression profile of neonatal lung ILC2s with those of naive adult lung ILC2s and after activation by recombinant IL-33 (effector adult) (Figure 1G). As expected, based on the protein expression, neonatal lung ILC2s had higher expression levels of Mki67, Il5, and Il13 compared with naive adult ILC2s. Neonatal lung ILC2s also had a higher expression of activation-associated genes than naive adult ILC2s, including the gene encoding the decoy IL-1 receptor, Il1r2 (15, 17), and ILC2 activation–associated micro-RNA Mir155 (18, 19). However, neonatal ILC2s differed from effector adult ILC2s in the expression of several genes. The former had a slightly lower expression of Il1r2 (Supplemental Figure 2A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.135961DS1) as well as Tnfrsf8 and Anxa2, genes that have been previously shown to be overexpressed in asthmatics (Figure 1G) (20–22). Neonatal and naive adult ILC2s had a higher expression of Cd24a than effector adult ILC2s, which was confirmed by flow cytometry (Figure 1G and Supplemental Figure 2B). Effector adult ILC2s also had higher expression of the checkpoint molecule genes Pdcd1 and Ctla4, which are upregulated in activated ILC2s (23, 24), although protein expression of intracellular CTLA4 was not different between neonatal and adult effector ILC2s (Supplemental Figure 2C). Effector ILC2s expressed lower levels of Nmur1 than naive adult and neonatal ILC2s. Nmur1, which encodes neuromedin-U receptor 1, has been known to be downregulated in response to IL-33 treatment (25) (Figure 1G and Supplemental Figure 2D). Neonatal ILC2s also had a higher expression of Vegfa, encoding VEGFA, than adult naive and effector ILC2s, suggesting a role in lung development, as has been proposed previously (12). However, this was not recapitulated with qPCR analysis (Supplemental Figure 2E). Additionally, periodic acid–Schiff staining (PAS) of 10-day-old mouse lungs confirmed that the amount of cytokine being released by activated neonatal ILC2s was not sufficient to induce mucus secretion, a hallmark of type 2 inflammation (data not shown).
Gene set enrichment analysis (GSEA) showed that the differences in gene expression between naive adult and neonatal lung ILC2s were very similar to those between naive and activated CD4+ T cells (Figure 1H), whereas the differences between neonatal ILC2s and effector adult lung ILC2s were similar to those between memory and effector CD8+ T cells (Figure 1I). These results suggested that activation of neonatal lung ILC2s by endogenous IL-33 is not as strong as activation of adult ILC2s by exogenous recombinant IL-33.
Neonatal lung ILC2s persist and respond more vigorously than those that develop in adulthood.
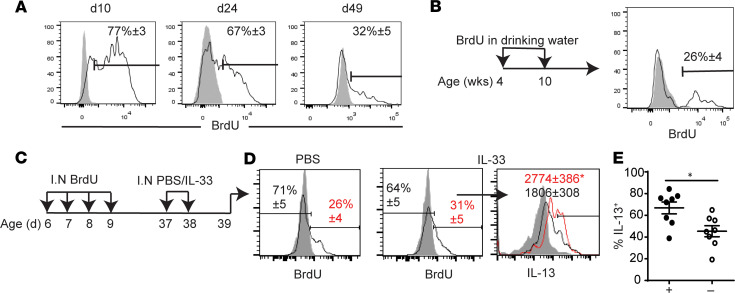
5-Bromo-2′-deoxyuridine (BrdU) i.n. administration from postnatal day 6 to 9 resulted in 77% of lung ILC2s labeled with BrdU at postnatal day 10 (Figure 2A). Two weeks and forty-nine days after the BrdU treatment, 67% and 32% of the adult ILC2 population, respectively, was BrdU+, indicating that neonatal ILC2s persisted into adulthood, as previously reported (11, 26). ILC2s are tissue resident at steady state (9), and it is unlikely that the decrease in the BrdU-labeled ILC2s was due to their migration out of the lung. Adult mice given BrdU in their drinking water for 6 weeks had approximately 26% of their ILC2s labeled with BrdU, indicating a very slow turnover of lung ILC2s in adulthood (Figure 2B). These findings are consistent with the recent report showing that most adult mouse ILC2s develop in the neonatal period, and they are very slowly replaced by newly developing ILC2s in adulthood (10).
Figure 2. ILC2s labeled with BrdU in the neonatal period respond more intensely than BrdU– ILC2s to IL-33 treatment later in life.
(A) BrdU was administered i.n. at postnatal day (d) 6–9. Lung ILC2s were analyzed at indicated ages for BrdU (n > 4). Shading indicates isotype control; white indicates anti-BrdU. (B) Adult mice were given BrdU in their drinking water for 6 weeks, as indicated by the scheme, and analyzed for ILC2 incorporation of BrdU. Numbers in the plot represent mean percentage ± SEM (n = 9, 2 independent experiments). (C) Mice received BrdU and IL-33 i.n. administrations as shown. (D) Lung ILC2s in PBS-injected and IL-33–injected mice were stained by anti-BrdU (white) or isotype control (shaded). Numbers in plots represent mean percentage ± SEM. The histogram on the right shows intracellular IL-13 in BrdU+ (red line) and BrdU– (black line) ILC2s from IL-33–injected mice with isotype control (shaded). Numbers in the plot show mean fluorescence intensity ± SEM. (E) Scatter plot shows the percentages of IL-13+ cells among BrdU+ (black) and BrdU– (white) ILC2s (n = 8, 2 independent experiments). *P < 0.05 (1-tailed unpaired Student’s t test).
We next investigated whether there is a difference in responsiveness of neonatal (BrdU+) ILC2s and BrdU– ILC2s in the adult period. Two i.n. injections of IL-33 (0.5 μg) were given to adult mice that had been given i.n. BrdU in the neonatal period (Figure 2C). BrdU+ ILC2s more intensely expressed intracellular IL-13 compared with the BrdU– ILC2s (Figure 2D, right). A higher proportion of BrdU+ ILC2s were positive for IL-13 (Figure 2E). Therefore, neonatal ILC2s that persist in adult lungs are more responsive to IL-33 stimulation than ILC2s that develop in adulthood.
ILC2s from KO adult mice respond to IL-33 treatment more weakly than WT mouse ILC2s.
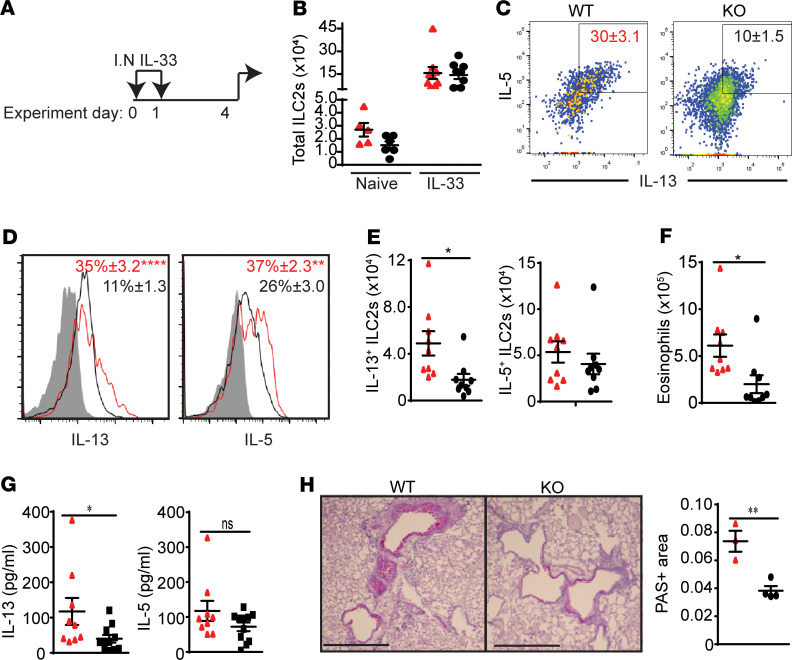
To test the effects of neonatal exposure to endogenous IL-33 on ILC2 functions in adulthood, we compared lung ILC2s from adult WT and KO mice. KO mice are not exposed to endogenous IL-33 and not activated in the neonatal period (11). We gave 2 daily i.n. administrations of IL-33 (0.5 μg) to KO and WT littermates at 8 weeks of age and analyzed the lung 3 days later (Figure 3A). IL-33 deficiency had no significant effect on the number of lung ILC2s in adulthood at steady state and after IL-33 treatment, the number of ILC2s expanded similar to that of WT ILC2s (Figure 3B). However, intracellular cytokine staining showed that the intensity of IL-13 and IL-5 was significantly lower in lung ILC2s from KO mice compared with that in WT mice (Figure 3, C and D). Interestingly, the numbers of IL-13+ ILC2s from KO mice were significantly lower than those from WT mice, while there was no significant difference in the number of intracellular IL-5+ ILC2s (Figure 3E). Nevertheless, eosinophil numbers in KO mouse lungs were lower than those in WT lungs (Figure 3F). Analysis of bronchoalveolar lavage also showed less IL-13, but not IL-5, in KO than WT mice (Figure 3G). IL-33 (i.n.) induced less mucus production in KO mouse lungs than WT mouse lungs (Figure 3H). These results suggest that lung ILC2s in KO mice respond more weakly than those in WT mice to i.n. IL-33 administration in adulthood.
Figure 3. ILC2s from IL-33–deficient mice respond more weakly than WT mice to IL-33 treatment.
(A) Adult WT (red) and KO (black) mice at 8 weeks of age received 2 daily i.n. IL-33 administrations and were analyzed 3 days later (n = 9, 3 independent experiments). (B) Total number of lung ILC2s in naive and IL-33–treated WT and KO mice. (C) Plots show intracellular IL-5 and IL-13 expression in WT and KO ILC2s. Numbers in plots represent mean percentage ± SEM. (D) Histograms of intracellular IL-13 and IL-5 staining in WT and KO ILC2s. Numbers in plots represent mean percentage ± SEM. Shading indicates isotype control. (E) Numbers of IL-13+ and IL-5+ ILC2s. (F) Eosinophil numbers in the lung. (G) The amount of IL-13 and IL-5 in the bronchoalveolar lavage. (H) Mucus production by PAS staining of lung tissue sections and PAS+ area (mm2 per mm of basement membrane) (n = 4). Scale bar: 0.05 mm. *P < 0.05; **P < 0.01; ****P < 0.0001 (1-tailed unpaired Student’s t test).
Gene expression analysis demonstrates intrinsic differences between KO and WT ILC2s.
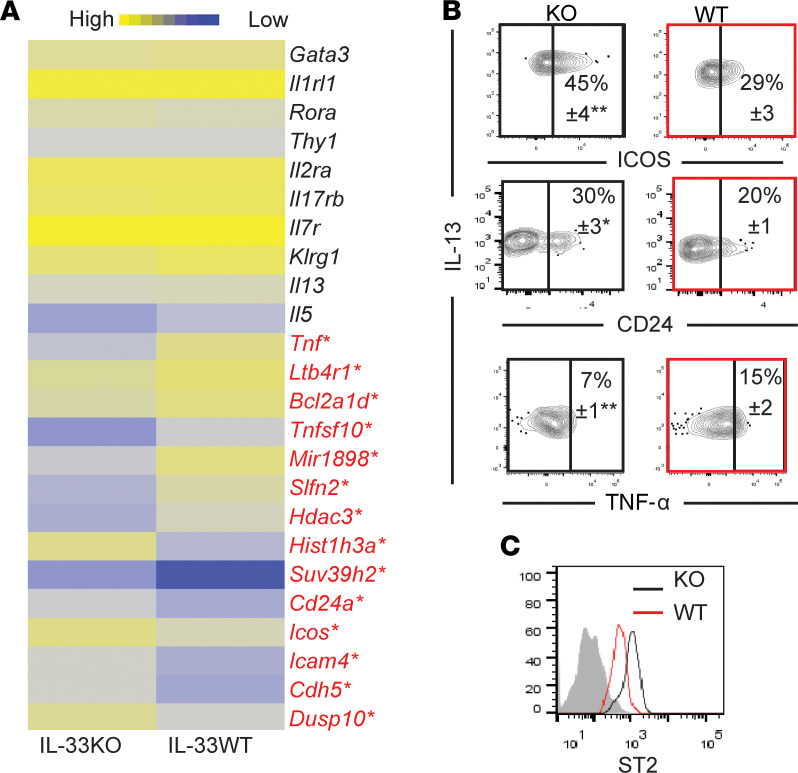
We compared the gene expression profiles of lung ILC2s from adult KO and WT mice. There was no difference in the expression of Il13, Il5, Gata3, and Rora (Figure 4A). WT ILC2s had a higher expression of the proinflammatory gene Tnf, the antiapoptotic-related gene Bcl2a1d, Tnfsf10 (encoding TRAIL), Ltb4r1 (encoding the activating receptor of leukotriene B4), and Hdac3 (encoding the histone deacetylase). KO ILC2s have a higher expression of Icam4, Icos, Cd24a, and Cdh5, genes associated with cell adhesion and activation, as well as Dusp10, a regulatory enzyme (27). We confirmed protein expression of CD24, ICOS, and intracellular TNF-α (Figure 4B). It should also be noted that WT ILC2s had a lower protein expression of ST2 than KO ILC2s, likely due to ligand-induced internalization (28), reflecting an environment of constitutive expression of IL-33 in adulthood in WT mice, albeit at low levels (Figure 1A and Figure 4C). These results highlight intrinsic differences between WT and KO ILC2s.
Figure 4. Comparison of IL-33–deficient and WT adult ILC2s.
(A) Heatmap shows the relative expression levels of genes that are differentially expressed in lung ILC2s from adult IL-33–deficient (KO) (n = 2) and WT mice (n = 3). Each replicate represents sorted ILC2s from 8 pooled mice *Genes in red indicate those that are differentially expressed more than 2-fold between KO and WT ILC2s. (B) Flow cytometry plots show expression of CD24, ICOS, and intracellular TNF-α on KO (black) and WT (red) adult mouse lung ILC2s (n = 6). Numbers in the plots indicate the mean percentage ± SEM. (C) Histogram shows ST2 expression on KO and WT ILC2s. Shading indicates isotype control. *P < 0.05; **P < 0.01. (1-tailed unpaired Student’s t test for B and C).
Neonatal IL-33 administration into KO mice reverses impaired ILC2 functions in adulthood.
To test whether the weak responsiveness of KO ILC2s (shown in Figure 3) was due to the lack of activation in the neonatal period, we investigated if administrations of recombinant IL-33 into KO neonates were able to rescue the weak responses to IL-33 later in life. In this experiment, IL-33 was administered only once in adulthood (Figure 5A). This weakly activated ILC2s and highlighted the differences between WT and KO adult ILC2 responses. While there were slightly more ILC2s in WT mice than KO mice, the difference was not statistically significant (Figure 5B, left). As in Figure 3, WT mice had significantly more eosinophil recruitment than KO mice (Figure 5B, right) and higher frequency of IL-13+ ILC2s (Figure 5C) but no difference in IL-5+ ILC2s (Figure 5D). The frequency of IL-13+IL-5+ ILC2s was also higher in WT mice compared with KO mice (Figure 5E).
Figure 5. i.n. IL-33 administration in the neonatal period rescues impaired KO ILC2 responses in adulthood.
(A) WT (n = 9) (red triangles) and KO (n = 11) (black circles) mice were treated with i.n. IL-33 in adulthood. (B) Total numbers of ILC2s and eosinophils. (C) Frequency of IL-13+ and (D) IL-5+ ILC2s. (E) Frequency of IL-5+IL-13+ ILC2s. (F) KO mice (white circles) were treated with 3 daily i.n. IL-33 administrations as neonates and challenged with IL-33 in adulthood (n = 13). WT and KO control mice did not receive IL-33 as neonates. (G) Total numbers of ILC2s and eosinophils. (H) Flow cytometry plots show intracellular IL-5 and IL-13 expression, and the bar graph shows the frequency of IL-13+IL-5+ ILC2s. (I) Frequency of IL-13+ ILC2s. Data are representative of 4 independent experiments. *P < 0.05; **P < 0.01. (1-tailed unpaired Student’s t test for B–E; 1-way ANOVA with Bonferroni’s post hoc test for G–I).
To rescue the KO response to a single IL-33 treatment, 3 daily i.n. administrations of recombinant IL-33 (0.1 μg) in KO pups at 9–11 days after birth were given, and 6 weeks later all groups were challenged with IL-33 (Figure 5F). Mouse lungs were analyzed for ILC2 proliferation, eosinophil recruitment, and intracellular cytokine expression of ILC2s. Neonatal IL-33 treatment significantly enhanced ILC2 proliferation beyond WT ILC2 numbers and increased eosinophil recruitment to the level of WT mice (Figure 5G). There were no differences in the frequencies of IL-5/IL-13 double-positive ILC2s between WT and neonatally IL-33–treated KO mouse lungs (Figure 5H). The frequency of IL-13+ ILC2s increased with neonatal IL-33 treatment in KO mice in comparison with KO mice with no neonatal treatment, and there was no significant difference compared with the WT mice (Figure 5I). There was no difference between the groups in terms of IL-5+ ILC2 frequency. as expected (data not shown). These results suggest that the impaired responsiveness of lung ILC2s in adult KO mice is due to the lack of IL-33 in the neonatal period and can be rescued by neonatal IL-33 administration.
Discussion
ILC2s are tissue-resident lymphocytes that develop early and persist throughout life (9, 10). We have previously shown that there are very few lymphocytes in newborn mouse lungs (11). While T, B, and NK cell numbers steadily increase over 3 weeks to reach the adult levels, ILC2 numbers increase more rapidly, reaching the adult level (about 4 × 103 cells per lung) by postnatal day 8 and then further increasing by 4-fold (to about 1.5 × 104 cells per lung) at between 10 and 14 days of age. Previous studies have suggested that the expansion of the ILC2 population is due to IL-33 derived from lung epithelial and stromal cells (13, 14). Our current study has confirmed that the amount of IL-33 in the lung peaks in 10-day-old pups. Day 10 neonatal lung ILC2s express activation-associated genes and proteins, similar to adult lung ILC2s after i.n. administration of recombinant IL-33. We then investigated the effects of the activation of neonatal lung ILC2s by endogenous IL-33 on adult lung ILC2s in 2 ways. First, we labeled neonatal ILC2s with BrdU and compared BrdU+ and BrdU– ILC2s in adult lungs. The former were activated in the neonatal period and persisted in adults, whereas the latter were likely a mixture of neonatal ILC2s that were not activated by endogenous IL-33 and did not proliferate in the neonatal period (~25%, Figure 2A), those that lost BrdU due to cell proliferation, and ILC2s that newly developed in the adult period (~26% in 6 weeks, Figure 2B). BrdU+ ILC2s in adult lungs responded more than BrdU– ILC2s to i.n. administration of recombinant IL-33. We also compared lung ILC2s in adult WT and IL-33 KO mice. ILC2s in KO mouse lungs are not activated in the neonatal period; nevertheless, ILC2 development is not impaired by IL-33 deficiency, as the number of lung ILC2s in adult KO mice did not significantly differ from that in WT mice. However, i.n. administration of recombinant IL-33 induced less production of type 2 cytokines, less mucus production, and less eosinophil infiltration in KO than WT mice. Moreover, lung ILC2s in adult KO mice that received i.n. administration of recombinant IL-33 in the neonatal period were no less responsive to IL-33 than WT mouse ILC2s with or without neonatal IL-33 treatment. These results showed that the activation of lung ILC2s by endogenous IL-33 in the neonatal period has significant effects on the functions of lung ILC2s in adulthood.
We have previously shown that adult mouse lung ILC2s activated by i.n. administration of allergens or recombinant IL-33 persist for many months as memory ILC2s, which are more responsive to allergen/IL-33 challenges than naive ILC2s (15). Although neonatal lung ILC2 activation appears to have similar effects as the generation of memory ILC2s, there are important differences between the two. Memory ILC2s in adult mice maintain the expression of the IL-25R for many months and acquire the ability to respond to IL-25. Neonatal ILC2s also express the IL-25R and are responsive to IL-25; however, they do not maintain the expression of the IL-25R into adulthood and become unresponsive to IL-25. Therefore, ILC2s do not acquire new functions by exposure to endogenous neonatal IL-33. Instead, they become more fit in their responses to IL-33, hence the term “training.”
It is unknown why the activation of lung ILC2s by endogenous IL-33 in the neonatal period results in IL-25R– “trained” ILC2s, but not IL-25R+ memory ILC2s similar to those generated in adult mice by i.n. administration of recombinant IL-33. It is likely that neonatal ILC2 activation is weak, as demonstrated by the lack of mucus hyperproduction (data not shown). Based on the gene expression data in the current study, the neonatal activation is weak, as neonatal ILC2s do not upregulate some genes, including Il1r2, Tnfrsf8, Anxa2, Pdcd1, Ctla4, and Nmur1 to the same extent as IL-33–treated adult effector ILC2s. This may be due to the difference in the amount of bioactive IL-33 in the lung. In neonatal lungs, we detected approximately 250 pg/mg IL-33, whereas in adult lungs given i.n. recombinant IL-33, we measured around 150 pg/mg IL-33 (29). However, in contrast to recombinant IL-33, it is difficult to determine how much neonatal IL-33 is nuclear versus cytoplasmic or extracellular and whether it is bioactive and available to stimulate ILC2s (30). It should also be noted that IL-33 alone is not sufficient to activate purified ILC2s in vitro (16), and other stimuli are required, including TSLP, IL-7, IL-2, neuropeptides (NMU, CGRP, VIP), or prostaglandins (PGD2) (31). Whether neonatal and adult lungs differ in the amount of these costimulatory molecules remains to be elucidated.
The concept of neonatal ILC2 training implies that naive adult lung ILC2s in WT mice are trained. Untrained ILC2s in IL-33–KO and trained ILC2s in WT mice have a small number of differentially expressed genes. For example, Hdac3 is downregulated in KO ILC2s. Interestingly, the HDAC inhibitor, trichostatin A, which targets class I HDACs, such as HDAC3, has been shown to downregulate the number of ILC2s expressing IL-5 and IL-13 upon Alternaria alternata extract challenge, suggesting an important role of HDACs in activation (32). Additionally, WT ILC2s express more Tnfsf10, a proapoptotic gene, consistent with a cytoprotective function that occurs upon lymphocyte activation (33). DUSP10 encoded by the Dusp10 gene is normally expressed at low levels in ILC2s. Forced expression of DUSP10 in ILC2s has been shown to inhibit IL-5 and IL-13 production upon IL-33 treatment, attenuating allergic responses (27). The differential expression of Icos may imply differences in cell-to-cell contact that may be important for activation of ILC2s. While ICOS/ICOSL interactions between ILC2s have been found to be important for function and survival (34), they also have been shown to inhibit ILC2 responses via interactions with Tregs (35). Additionally, CD24, which regulates cell surface receptor signaling (36), is expressed more on KO ILC2s than WT ILC2s. Tnf is a proinflammatory-associated gene, and it has recently been found to be increased in peripheral tissue ILC2s at steady state compared with bone marrow ILC2s (37). It remains to be determined whether, in addition to these ILC2-intrinsic differences, there might be differences in lung microenvironments between WT and KO mouse lungs that contribute to ILC2 activation in adulthood.
The term “trained immunity” has been used to describe the phenomenon of memory-like myeloid cells (38). Activation of macrophages by pathogen-derived molecules, including β-glucan, results in their differentiation into highly functional macrophages that live up to 4 weeks and mediate antigen-nonspecific innate protection against subsequent infections. Naive macrophages are also trained by helminth infection and acquire the ability to kill larvae in a later infection. Trained immunity in myeloid cells has been shown to be mediated by epigenetic modifications. It remains to be determined whether similar mechanisms are involved in neonatal ILC2 training.
The neonatal period is critical for the development of lung immunity and establishing within the lung the ILC2 population, which plays a critical role in allergic lung diseases. Modulating ILC2 training during childhood could dampen the propensity of ILC2s to develop into effective mediators of allergic disease.
Methods
Mice.
C57BL/6 (B6) mice were purchased from The Jackson Laboratory and bred and maintained in the British Columbia Cancer Research Centre (BCCRC) animal facility. An equal number of males and females were used for each experiment. B6.Il33−/− mice (obtained from the Knockout Mouse Project Repository) were used and maintained at the BCCRC. Mice were used at 10 days after birth for neonatal experiments and at 4–8 weeks of age for adults.
Primary leukocyte preparation.
Cell suspensions were prepared from the lungs as described previously (39). The protocols were modified for neonatal lungs, which used 2.5 mL of the appropriate digestion buffer for each lung. Single cells from all tissues were counted using a hemocytometer, incubated with 2.4G2 monoclonal antibody to block Fc receptors, and stained with fluorochrome-conjugated antibodies (Table 1) and phenotypically analyzed by flow cytometry.
Table 1. List of antibodies used for flow cytometry sorting and analysis.
Antibodies and flow cytometry.
Antibodies used are listed in Table 1. BD Fortessa was used for phenotypic analysis. FlowJo (Tree Star, versions 8.6 and 10) was used for data analysis. Lung mouse cells were first gated as singlets, live, CD45+ lymphocytes, and ILC2s were identified as lineage– (Gr-1, Ter119, CD11b, CD11c, CD19, NK1.1, CD3ε, CD4, TCRβ, TCRγδ), Thy1.2+, CD127+, CD25+, ST2+ (Supplemental Figure 1A). Lung mouse eosinophils were identified as lymph– (CD3ε, NK1.1, CD19), 7/4–, CD11c–, SiglecF+. Total cell numbers were calculated based on their frequency out of live, CD45+ cells determined by FlowJo multiplied by the hemocytometer total cell count.
Bronchoalveolar lavage.
Mice were euthanized and a catheter attached to a syringe was inserted into the trachea. 1 mL plain PBS was instilled into the lungs and then collected. The bronchoalveolar lavage was centrifuged at 400 RCF for 10 minutes, and the supernatant was collected. IL-5 and IL-13 were quantified by ELISA Ready-SET-Go! (Thermo Fisher Scientific) according to the manufacturer’s protocols.
In vivo stimulation.
Injections (i.n.) were given after mice were anesthetized by isoflurane inhalation. The concentration of proteins used i.n. is shown in Table 2.
Table 2. Concentrations of i.n. administered treatments.
Intracellular staining.
All intracellular staining for IL-5 and IL-13 was performed with the Cytofix/Cytoperm kit (BD Biosciences) after a 3-hour incubation of cells in 500 μL Dulbecco modified Eagle medium containing 10% FBS, penicillin/streptomycin, 2-mercaptoethanol, Brefeldin A (GolgiPlug, BD Biosciences), PMA (30 ng/mL), and ionomycin (500 ng/mL) (MilliporeSigma) at 37°C. Dead cells were excluded with eFluor780 (Thermo Fisher Scientific) fixable viability dye. Ki-67 staining was performed without incubation using the Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) according to the manufacturer’s protocol.
BrdU.
Neonates were given 0.8 mg BrdU (MilliporeSigma) in 10 μl preheated PBS for 4 days starting at day 6–9 after birth. Adult mice were given BrdU at the concentration of 0.8 mg/mL in sterilized drinking water. To ensure that mice drank the water, 20% dextrose was added. Light-sensitive bottles were used to protect BrdU. Water was changed every 2 days. One mouse was euthanized per week to check the incorporation of BrdU. After harvesting the lungs from BrdU-treated adults or neonates, single-cell suspensions were prepared from lung tissues, and cells were stained with viability dye and antibodies for surface markers to identify ILC2s. Cells were then fixed, followed by staining of BrdU using the APC BrdU Flow Kit (BD Biosciences). For BrdU and IL-13 staining, cells were first incubated with PMA, ionomycin, and GolgiPlug for 3 hours at 37°C. After, the cells were harvested and fixed using the APC BrdU Flow Kit and then stained for both IL-13 and BrdU.
ILC2 enrichment and sorting.
Individual mouse lungs were prepared as described previously and, after lysing, were pooled, and the suspension was then enriched for ILC2s using EasySep Mouse ILC2 Enrichment Kit (STEMCELL Technologies) according to the manufacturer’s protocols. BD Aria was used for purity sort of ILC2s identified as lineage–, Thy1.2+, ST2+, CD127+ (Supplemental Figure 1B).
Microarray.
Total RNA was isolated using TRIzol (Life Technologies). RNA was quantified and checked for quality with an Agilent Bioanalyzer 2100. RNA amplification and microarray services were performed by the Centre for Applied Genomics, Toronto, Ontario, using the Affymetrix GeneChip Mouse Gene 2.0ST Array. The expression profile was normalized using robust multiarray algorithm. All data analysis was performed with FlexArray 1.6 (Genome Quebec). The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO GSE148539 for Figure 1G and GSE148545 for Figure 4A; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148539 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148545).
GSEA.
GSEA was performed using GSEA software. Log2-fold expression values for each comparison were ranked in descending order and then run through GSEA (V2.2.2, http://software.broadinstitute.org/gsea/index.jsp) (40). The gene set collections that were used were C7 immunologic signatures (41), with the following parameters: number of permutations, 1000; number of genes plotted per graph, 200. High normalized enrichment scores and low false discovery rates indicated that the similarity between a priori gene set and the 2 states analyzed was highly significant.
cDNA synthesis.
cDNA was prepared from 1.5 μg RNA using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) according to the manufacturer’s protocols.
RT-qPCR.
RT-qPCR was carried out with the StepOnePlus system (Thermo Fisher Scientific) using TaqMan fast reagents. 75 ng cDNA was used per reaction, and the following probe/primers sets were used for detection: Il1r2 (IDT assay ID Mm.PT.58.30874994), probe, -5′-/56-FAM/CCCATTACA/ZEN/TCGGAGAAGCCCACA/3IABkFQ/-3′; primer 1, 5′-TGCTTTCACCACTCCAACAG-3′, primer 2, 5′-CCTTCCAGCCTCAATTCAGAT-3′; Nmur1 (IDT assay ID Mm.PT.58.32232111), probe, 5′-/56-FAM/ACCACAACC/ZEN/AGTGCAAACAGCATC/3IABkFQ/-3′; primer 1, -5′-AGGATTCAGCTGCAAGATAGG-3′, primer 2, 5′-ATACGGTCAGCATGGAATGG-3′; Vegfa (IDT assay ID Mm.PT.58.14200306), probe, 5′-/56-FAM/TGCTGTACC/ZEN/TCCACCATGCCAAG/3IABkFQ/-3′; primer 1, -5′-CCGAAACCATGAACTTTCTGC-3′, primer 2, 5′-GACTTCTGCTCTCCTTCTGTC-3′; and Tbp (IDT Assay ID Mm.PT.39a.22214839), probe, 5′-/56-FAM/ACTTGACCT/ZEN/AAAGACCATTGCACTTCGT/3IABkFQ/-3′; primer 1, 5′-TGTATCTACCGTGAATCTTGGC-3′; primer 2, 5′-CCAGAACTGAAAATCAACGCAG-3′.
Acquired data were analyzed by the 2-ΔΔCt method (42). Threshold cycles were normalized to the expression of Tbp and then to adult naive ILC2s (controls).
Lung homogenate preparation and analyses.
Lungs were collected from naive mice at various days after birth and homogenized in HBSS with EDTA and Halt protease inhibitor cocktail (Thermo Fisher Scientific) at 200 mg per lung tissue per ml. After centrifugation at 800 RCF for 20 minutes, supernatant was analyzed using IL-33 ELISA (Thermo Fisher Scientific) according to the manufacturer’s protocols. Total protein was quantified using the Protein Quantification Kit-Rapid (MilliporeSigma). The amount of IL-33 per sample (pg) was normalized by the total amount of protein within the lung homogenate (mg).
PAS staining.
Lungs were perfused with 1 mL (adults) 4% paraformaldehyde and harvested. Fixed lungs were embedded in paraffin by the Centre for Translational and Applied Genomics (Vancouver, British Columbia, Canada), and 4-μm sections were placed on glass. Slides were PAS stained by the Department of Pathology at Vancouver General Hospital (Vancouver, British Columbia, Canada). To quantify the PAS+ area, ImageJ (NIH) was used as previously described (with a magnification of ×4) (43, 44). Results were expressed as PAS+ area (square millimeters) per mm of basement membrane length.
Statistics.
Data were analyzed with GraphPad Prism 6. Unpaired 1-tailed Student’s t test was used to determine statistical significance between groups; for multiple comparisons 2-way (Figure 1, D and E) and 1-way (Figure 5, G–I) ANOVA with Bonferroni’s post hoc test were used. n refers to an individual biological replicate unless otherwise specified. Data in plots represent the mean ± SEM. A P value less than 0.05 was considered significant.
Study approval.
All animal use was approved by the animal care committee of the University of British Columbia in accordance with the guidelines of the Canadian Council on Animal Care. Genotyping was done by the Biomedical Research Centre Genotyping Facility.
Author contributions
CAS designed and performed experiments and wrote the manuscript. LM and HS performed experiments. FT designed experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
CAS is the recipient of a University of British Columbia Four-Year Fellowship and a Canadian Institutes of Health Research studentship. LM is a recipient of a University of British Columbia Four-Year Fellowship and a Vanier Canada Graduate Scholarship. This work was supported by grants from the Canadian Institute of Health Research (PJT-153304, MOP-126194).
Version 1. 06/23/2020
In-Press Preview
Version 2. 07/23/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(14):e135961.https://doi.org/10.1172/jci.insight.135961.
Contributor Information
Catherine A. Steer, Email: catherine.steer@ucsf.edu.
Laura Mathä, Email: lmatha@bccrc.ca.
Hanjoo Shim, Email: hj.bshim@gmail.com.
Fumio Takei, Email: ftakei@bccrc.ca.
References
- 1.Martinez-Gonzalez I, Steer CA, Takei F. Lung ILC2s link innate and adaptive responses in allergic inflammation. Trends Immunol. 2015;36(3):189–195. doi: 10.1016/j.it.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Gieseck RL, Wilson MS, Wynn TA. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol. 2018;18(1):62–76. doi: 10.1038/nri.2017.90. [DOI] [PubMed] [Google Scholar]
- 3.Halim TY, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40(3):425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliphant CJ, et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 2014;41(2):283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halim TYF, et al. Tissue-restricted adaptive type 2 immunity is orchestrated by expression of the costimulatory molecule OX40L on group 2 innate lymphoid cells. Immunity. 2018;48(6):1195–1207.e6. doi: 10.1016/j.immuni.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halim TY, et al. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat Immunol. 2016;17(1):57–64. doi: 10.1038/ni.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahlgren MW, Molofsky AB. All along the watchtower: group 2 innate lymphoid cells in allergic responses. Curr Opin Immunol. 2018;54:13–19. doi: 10.1016/j.coi.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuijs MJ, Halim TYF. Group 2 innate lymphocytes at the interface between innate and adaptive immunity. Ann N Y Acad Sci. 2018;1417(1):87–103. doi: 10.1111/nyas.13604. [DOI] [PubMed] [Google Scholar]
- 9.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350(6263):981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider C, et al. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity. 2019;50(6):1425–1438.e5. doi: 10.1016/j.immuni.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steer CA, Martinez-Gonzalez I, Ghaedi M, Allinger P, Mathä L, Takei F. Group 2 innate lymphoid cell activation in the neonatal lung drives type 2 immunity and allergen sensitization. J Allergy Clin Immunol. 2017;140(2):593–595.e3. doi: 10.1016/j.jaci.2016.12.984. [DOI] [PubMed] [Google Scholar]
- 12.de Kleer IM, et al. Perinatal activation of the interleukin-33 pathway promotes type 2 immunity in the developing lung. Immunity. 2016;45(6):1285–1298. doi: 10.1016/j.immuni.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 13.Saluzzo S, et al. First-breath-induced type 2 pathways shape the lung immune environment. Cell Rep. 2017;18(8):1893–1905. doi: 10.1016/j.celrep.2017.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlgren MW, et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity. 2019;50(3):707–722.e6. doi: 10.1016/j.immuni.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Gonzalez I, Mathä L, Steer CA, Ghaedi M, Poon GF, Takei F. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity. 2016;45(1):198–208. doi: 10.1016/j.immuni.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012;36(3):451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 17.Lang D, et al. The type II IL-1 receptor interacts with the IL-1 receptor accessory protein: a novel mechanism of regulation of IL-1 responsiveness. J Immunol. 1998;161(12):6871–6877. [PubMed] [Google Scholar]
- 18.Knolle MD, et al. MicroRNA-155 protects group 2 innate lymphoid cells from apoptosis to promote type-2 immunity. Front Immunol. 2018;9:2232. doi: 10.3389/fimmu.2018.02232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson K, Malmhäll C, Ramos-Ramírez P, Rådinger M. MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation. J Allergy Clin Immunol. 2017;139(3):1007–1016.e9. doi: 10.1016/j.jaci.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 20.Heshmat NM, El-Hadidi ES. Soluble CD30 serum levels in atopic dermatitis and bronchial asthma and its relationship with disease severity in pediatric age. Pediatr Allergy Immunol. 2006;17(4):297–303. doi: 10.1111/j.1399-3038.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 21.Katsunuma T, et al. Analysis of gene expressions of T cells from children with acute exacerbations of asthma. Int Arch Allergy Immunol. 2004;134(1):29–33. doi: 10.1159/000077530. [DOI] [PubMed] [Google Scholar]
- 22.Sekigawa T, et al. Gene-expression profiles in human nasal polyp tissues and identification of genetic susceptibility in aspirin-intolerant asthma. Clin Exp Allergy. 2009;39(7):972–981. doi: 10.1111/j.1365-2222.2009.03229.x. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz C, et al. ILC2s regulate adaptive Th2 cell functions via PD-L1 checkpoint control. J Exp Med. 2017;214(9):2507–2521. doi: 10.1084/jem.20170051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salimi M, et al. Activated innate lymphoid cell populations accumulate in human tumour tissues. BMC Cancer. 2018;18(1):341. doi: 10.1186/s12885-018-4262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallrapp A, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 2017;549(7672):351–356. doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghaedi M, Steer CA, Martinez-Gonzalez I, Halim TYF, Abraham N, Takei F. Common-lymphoid-progenitor-independent pathways of innate and t lymphocyte development. Cell Rep. 2016;15(3):471–480. doi: 10.1016/j.celrep.2016.03.039. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T, et al. DUSP10 constrains innate IL-33-mediated cytokine production in ST2hi memory-type pathogenic Th2 cells. Nat Commun. 2018;9(1):4231. doi: 10.1038/s41467-018-06468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Wei J, Bowser RK, Traister RS, Fan MH, Zhao Y. Focal adhesion kinase-mediated activation of glycogen synthase kinase 3β regulates IL-33 receptor internalization and IL-33 signaling. J Immunol. 2015;194(2):795–802. doi: 10.4049/jimmunol.1401414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathä L, Shim H, Steer CA, Yin YH, Martinez-Gonzalez I, Takei F. Female and male mouse lung group 2 innate lymphoid cells differ in gene expression profiles and cytokine production. PLoS One. 2019;14(3):e0214286. doi: 10.1371/journal.pone.0214286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cayrol C, Girard JP. Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol Rev. 2018;281(1):154–168. doi: 10.1111/imr.12619. [DOI] [PubMed] [Google Scholar]
- 31.Hurrell BP, Shafiei Jahani P, Akbari O. Social networking of group two innate lymphoid cells in allergy and asthma. Front Immunol. 2018;9:2694. doi: 10.3389/fimmu.2018.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toki S, et al. The histone deacetylase inhibitor trichostatin A suppresses murine innate allergic inflammation by blocking group 2 innate lymphoid cell (ILC2) activation. Thorax. 2016;71(7):633–645. doi: 10.1136/thoraxjnl-2015-207728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falschlehner C, Ganten TM, Koschny R, Schaefer U, Walczak H. TRAIL and other TRAIL receptor agonists as novel cancer therapeutics. Adv Exp Med Biol. 2009;647:195–206. doi: 10.1007/978-0-387-89520-8_14. [DOI] [PubMed] [Google Scholar]
- 34.Maazi H, et al. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 2015;42(3):538–551. doi: 10.1016/j.immuni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigas D, et al. Type 2 innate lymphoid cell suppression by regulatory T cells attenuates airway hyperreactivity and requires inducible T-cell costimulator-inducible T-cell costimulator ligand interaction. J Allergy Clin Immunol. 2017;139(5):1468–1477.e2. doi: 10.1016/j.jaci.2016.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayre DC, Christian SL. CD24: a rheostat that modulates cell surface receptor signaling of diverse receptors. Front Cell Dev Biol. 2016;4:146. doi: 10.3389/fcell.2016.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricardo-Gonzalez RR, et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol. 2018;19(10):1093–1099. doi: 10.1038/s41590-018-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netea MG, van der Meer JW. Trained immunity: an ancient way of remembering. Cell Host Microbe. 2017;21(3):297–300. doi: 10.1016/j.chom.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Romera-Hernández M, Mathä L, Steer CA, Ghaedi M, Takei F. Identification of group 2 innate lymphoid cells in mouse lung, liver, small intestine, bone marrow, and mediastinal and mesenteric lymph nodes. Curr Protoc Immunol. 2019;125(1):e73. doi: 10.1002/cpim.73. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godec J, et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44(1):194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Zuberi RI, et al. Deficiency of endothelial heparan sulfates attenuates allergic airway inflammation. J Immunol. 2009;183(6):3971–3979. doi: 10.4049/jimmunol.0901604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge XN, et al. Allergen-induced airway remodeling is impaired in galectin-3-deficient mice. J Immunol. 2010;185(2):1205–1214. doi: 10.4049/jimmunol.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.