Abstract
Tissue regeneration capacity declines with aging in association with heightened oxidative stress. Expression of the oxidant-generating enzyme, NADPH oxidase 4 (Nox4), is elevated in aged mice with diminished capacity for fibrosis resolution. Bromodomain-containing protein 4 (Brd4) is a member of the bromodomain and extraterminal (BET) family of proteins that function as epigenetic “readers” of acetylated lysine groups on histones. In this study, we explored the role of Brd4 and its interaction with the p300 acetyltransferase in the regulation of Nox4 and the in vivo efficacy of a BET inhibitor to reverse established age-associated lung fibrosis. BET inhibition interferes with the association of Brd4, p300, and acetylated histone H4K16 with the Nox4 promoter in lung fibroblasts stimulated with the profibrotic cytokine, TGF-β1. A number of BET inhibitors, including I-BET-762, JQ1, and OTX015, downregulate Nox4 gene expression and activity. Aged mice with established and persistent lung fibrosis recover capacity for fibrosis resolution with OTX015 treatment. This study implicates epigenetic regulation of Nox4 by Brd4 and p300 and supports BET/Brd4 inhibition as an effective strategy for the treatment of age-related fibrotic lung disease.
Keywords: Aging, Cell Biology
Keywords: Epigenetics, Fibrosis
BET inhibition reduces Brd4, p300, and H4K16ac at the Nox4 promoter region to decrease Nox4 expression and restore lung fibrosis resolution capacity in aged mice.
Introduction
Idiopathic pulmonary fibrosis (IPF) is an age-related, progressive, and usually fatal lung disease with limited treatment options (1). IPF is characterized by the aberrant tissue remodeling associated with declining lung function and progressive respiratory failure (1). Currently approved drug therapies merely slow the decline in lung function, without arresting/reversing disease progression or improving quality of life (2, 3). The pathogenesis of IPF is incompletely understood, although our group and others have demonstrated that accumulation/persistence of activated fibroblasts/myofibroblasts is a hallmark of nonresolving fibrosis (4, 5).
Based on the observation that contractile, activated myofibroblasts are key effector cells in fibrosis, they have been deemed to represent a potentially effective therapeutic target (6, 7). Myofibroblasts express α-smooth muscle actin (α-SMA) and myosin stress filaments as well as the ROS-generating enzyme, NADPH oxidase 4 (Nox4) (4, 5). Increased levels of the profibrotic cytokine, TGF-β1, have been implicated in persistent myofibroblast differentiation/activation that leads to this aberrant wound-healing process (8). TGF-β1 differentiates lung fibroblasts into myofibroblasts (9) and markedly upregulates Nox4 expression (5). Nox4 is constitutively upregulated in activated myofibroblasts and in lung tissues of humans with IPF (5). Nox4 expression is also constitutively upregulated in senescent (myo)fibroblasts, in part, through epigenetic regulation involving histone acetylation (10). However, epigenetic mechanisms that control Nox4 expression in IPF fibroblasts and in response to TGF-β1 remain unclear.
Nox4 has been reported to be involved in many fibrotic diseases, including those of the heart (11), lung (5), liver (12), kidney (13), and skin (14); however, the development of Nox4-specific inhibitors has been challenging (15). Among the new epigenetic modifiers, the bromodomain and extraterminal (BET) inhibitors have emerged as a transcriptional regulator and are being developed as novel therapeutics for cancer (16). However, the mechanisms of action of this class of epigenetic inhibitors on Nox4 expression and their efficacy in treatment of established lung fibrosis in animal models are yet to be defined.
Epigenetic mechanisms, including histone acetylation, have been implicated in the pathogenesis of lung fibrosis (17–21). Histone modifications control gene expression, for example, by acetylation of lysine on histones that most often confers an open chromatin conformation to facilitate active gene transcription (22). Lysine acetylation is usually carried out by histone acetyltransferases, such as p300, which may serve as a transcriptional coactivators (23). Acetylated lysines are a binding site for bromodomain (Brd) proteins, which were originally identified as chromatin-associated proteins and functional components of histone acetyltransferase complexes (24). The Brd protein family includes BET proteins, which are epigenetic “readers” that bind to acetylated histone lysine and recruit additional coactivators to regulate gene transcription (25). Brd-containing protein 4 (Brd4), a member of the BET family, has been implicated in many fibrotic disorders, such as liver (26), lung (27), renal (28), pancreatic (29), and cardiac (30) fibrosis. However, the epigenetic regulation of Nox4 expression by Brd4 and other epigenetic modifiers in the fibrotic process has not been well defined. In this study, we investigated the effects of a BET/Brd4 inhibition on p300 and H4K16ac in regulating Nox4 expression in response to TGF-β1 and in the constitutive activation of IPF fibroblasts; we also examined the efficacy of the BET inhibitor, OTX015, in the treatment of established and persistent lung fibrosis in aged mice.
Results
Nox4 is downregulated in IPF myofibroblasts by Brd4 inhibition.
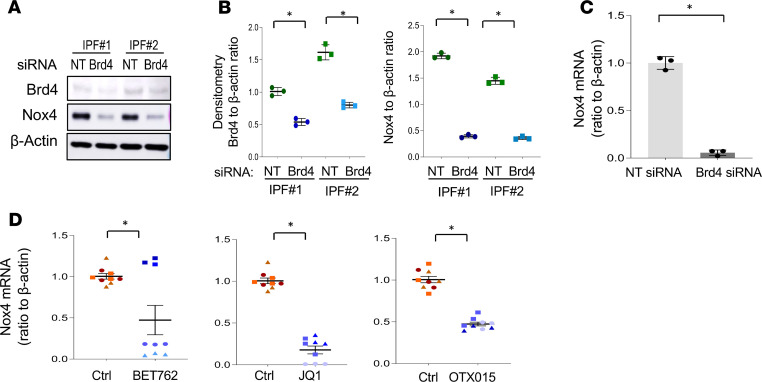
First, we investigated whether Brd4 regulates Nox4 gene expression. Primary IPF lung fibroblasts were transfected with Brd4 siRNA or nontargeting (NT) control siRNA, and gene and protein expression were analyzed after 48 hours. Decreased Nox4 expression, at both the protein and mRNA levels, were observed in Brd4 siRNA–transfected cells (Figure 1, A–C). We then tested the effects of pharmacological BET inhibitors to determine if they mediate similar effects. Although these drugs are potent inhibitors of Brd4, they may also inhibit other BET proteins, such as Brd2 and Brd3. We examined the effects of I-BET-762, JQ1, and OTX015; both I-BET-762 and OTX015 have undergone early-stage clinical trials for cancer (31, 32). Primary IPF fibroblasts from at least 3 different humans were examined with each inhibitor. JQ1 and OTX015 decreased Nox4 mRNA levels in all 3 IPF cell lines, while I-BET-762 was effective in 2 of the 3 samples tested (Figure 1D). Similar decreases in Nox4 expression at the protein level were observed in these individuals (Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137127DS1). These data indicate that genetic or pharmacologic BET/Brd4 inhibition decreases Nox4 expression at both the mRNA and protein levels in primary IPF lung fibroblasts.
Figure 1. Brd4 inhibition downregulates Nox4 expression in IPF lung fibroblasts.
(A–C) Primary IPF lung fibroblasts were transfected with Brd4 siRNA for 48 hours. (A) Cells were collected and subjected to Western blots for Nox4, Brd4, and β-actin. (B) Densitometry of Brd4 and Nox4 protein expression relative to β-actin, as in A. *P < 0.05, Brd4 siRNA compared with NT control, by 2-tailed t test. (C) RNA from NT and Brd4 siRNA–treated cells was analyzed for Nox4 mRNA by real-time PCR. *P < 0.05, compared with the NT control of the same cell line, by 2-tailed t test. (D) BET inhibitors, BET-762 (0.5 μM), JQ1 (1 μM), and OTX015 (0.5 μM) were added to primary IPF lung fibroblasts at 70% confluence for 48 hours, and RNA was collected and Nox4 mRNA expression analyzed by real-time PCR. Triangles, squares, or circles indicate 3 different IPF individuals from whom primary cells were derived. Expressed values represent mean ± SD; n = 3 experimental replicates of each cell line. *P < 0.05, treated group vs. control (vehicle) group, by 2-tailed t test.
Brd4 inhibition blocks TGF-β1–induced Nox4 expression.
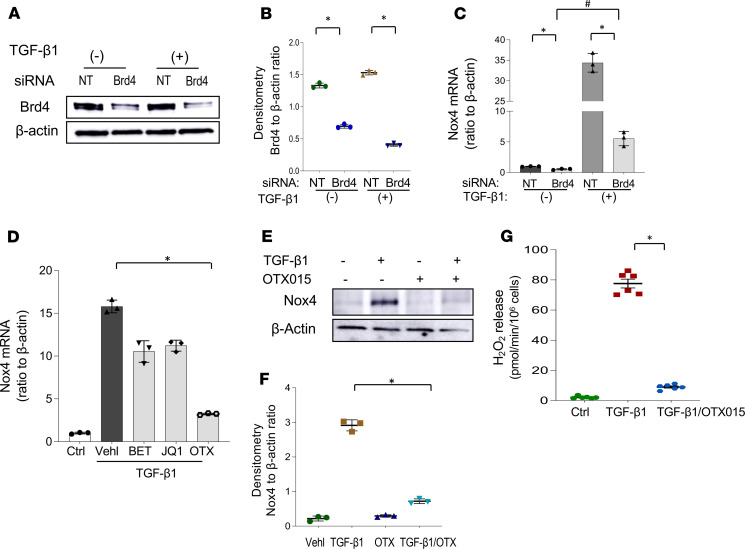
TGF-β1 is a cardinal profibrotic cytokine (8) that induces Nox4 expression in fibroblasts (5); we further examined the effects of BET inhibitors on TGF-β1–induced Nox4 expression. Normal human lung fibroblasts (IMR90) were transfected with Brd4 siRNA or NT siRNA control, followed by treatment with TGF-β1 (2 ng/mL) for 48 hours. Fibroblasts transfected with Brd4 siRNA failed to upregulate Nox4 expression (Figure 2, A–C). We then examined the effect of BET inhibitors on Nox4 expression in response to TGF-β1. IMR90 fibroblasts were pretreated with BET inhibitors for 2 hours before TGF-β1 (2 ng/mL) treatment for 48 hours; the upregulation of Nox4 mRNA was suppressed by all 3 Brd4 inhibitors, although OTX015 was the most potent with >95% inhibition at 0.5 μM (Figure 2D; corresponding changes at the protein levels were also observed, Supplemental Figure 2B). In subsequent experiments, we focused on the effects of OTX015 for both in vitro and in vivo studies. The effects of OTX015 on Nox4 expression were confirmed at the protein level (Figure 2, E and F) and at the level of enzymatic activity, as assessed by extracellular H2O2 release (Figure 2G). Although Nox4 has been reported to promote myofibroblast differentiation and profibrotic responses, it is not known whether putative antifibrotic effects of BET inhibition can be fully accounted for by Nox4 inhibition. We tested the effect of OTX015 treatment on Nox4-silenced cells and observed a small, but appreciable, additive inhibitory effect on TGF-β1–induced expression of α-SMA and collagen. The effect was shown in Supplemental Figure 3. Together, our data indicate that Brd4 inhibition, either by siRNA-mediated gene silencing or by pharmacologic BET inhibitors, downregulates not only the constitutive but also TGF-β1–inducible Nox4 expression/activity in lung fibroblasts.
Figure 2. Brd4 inhibition blocks TGF-β1–induced Nox4 gene upregulation in human lung fibroblasts.
(A–C) Normal human lung fibroblasts (IMR90) were transfected with siRNA Brd4 or NT and then treated with vehicle or TGF-β1 (2 ng/mL) for 48 hours. (A) The whole cell lysate were collected to examine Brd4 expression by Western blots. (B) Densitometry of Brd4-associated signals detected (ratio to β-actin) in A. *P < 0.05, Brd4 siRNA–transfected cells compared with NT control of the same cell line, by 2-tailed t test. (C) Treated as in A, cells were analyzed for Nox4 mRNA by real-time PCR (mean ± SD; n = 3 in each group). *P < 0.05, each group compared with vehicle only; #P < 0.05, TGF-β1–treated siRNA Brd4 vs. NT cells, by 2-tailed t test. (D) IMR90 fibroblasts were incubated overnight with 1% fetal bovine serum at 70% confluence and then treated with vehicle or various Brd4 inhibitors with the same concentration as in Figure 1 for 2 hours before stimulation with TGF-β1 (2 ng/mL) for 48 hours. Cells were analyzed for Nox4 mRNA by real-time PCR (mean ± SD; n = 3 in each group). *P < 0.05, each group compared with TGF-β1 with vehicle only (Vehl/TGF-β1), by 2-tailed t test. (E) IMR90 fibroblasts were pretreated with or without OTX015 for 2 hours and then with or without TGF-β1 for 48 hours. Cells were collected and subjected to SDS-PAGE and Western blot analysis for Nox4 and β-actin (loading control). (F) The densitometry of Nox4-associated signals detected (ratio to β-actin) in E. *P < 0.05, OTX015 pretreated cells with TGF-β1 compared with TGF-β1, by 2-tailed t test. (G) IMR90 fibroblasts stimulated with/without TGF-β1 (2 ng/mL for 24 hours) in the presence/absence of OTX015 (0.5 μM) were analyzed for extracellular H2O2 production, as a marker of Nox4 activity (mean ± SD; n = 6 in each group). *P < 0.05, compared with TGF-β1/OTX015, by 2-tailed t test.
BET inhibition blocks the enrichment of transcriptional components (H4K16ac, Brd4, and p300) associated with the Nox4 promoter.
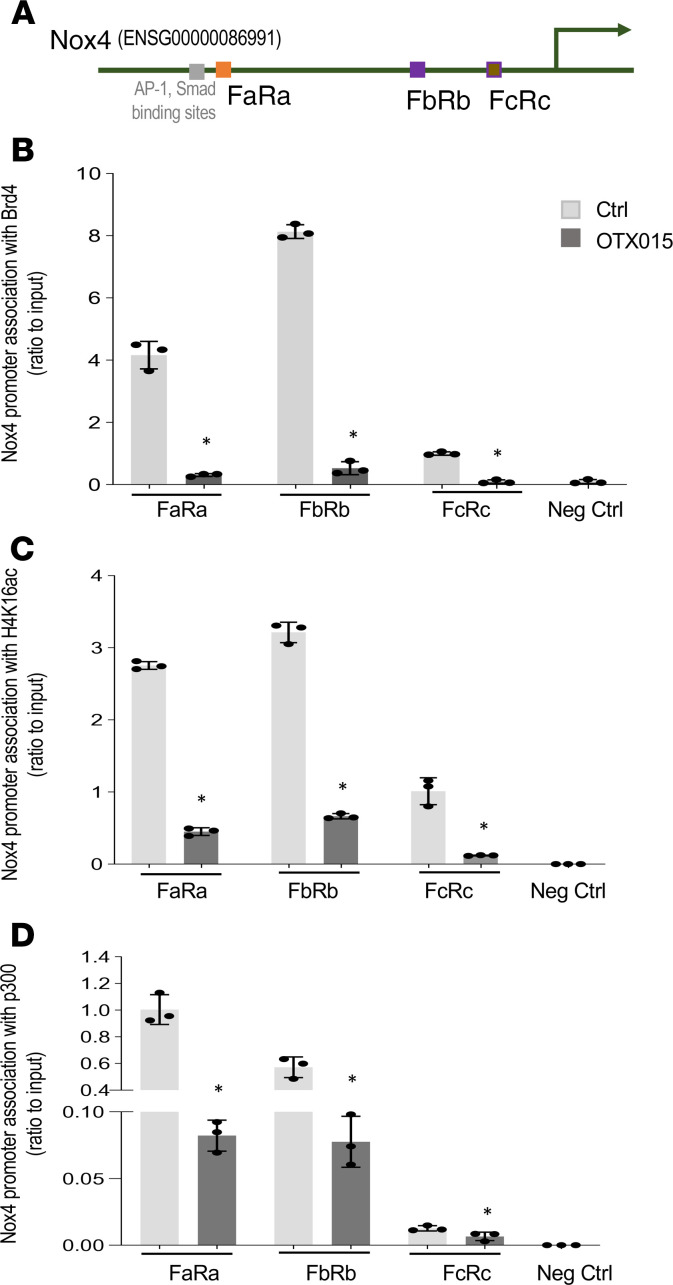
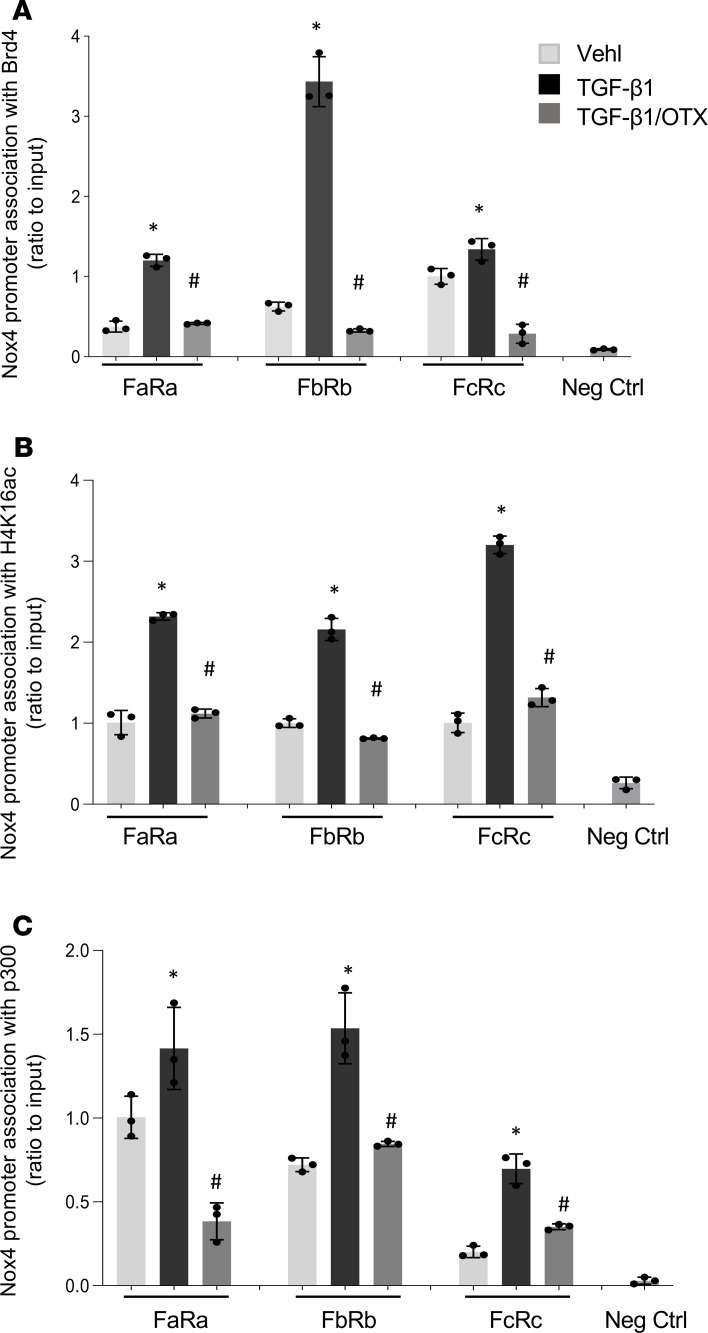
Previously, we demonstrated that Nox4 transcription is associated with an active histone mark, H4K16ac (10), whereas inactivated p300 has been reported to reduce Nox4 expression in cancer cells (33). We examined whether decreased Nox4 gene expression by the BET inhibitor, OTX015, was due to the reduction of these transcriptional components at the Nox4 promoter. We used 3 sets of PCR primers to examine regions within the Nox4 promoter that associate with these components (Figure 3A). We first examined the association of Brd4 with the Nox4 promoter region in OTX015-treated IPF fibroblasts. Brd4 association was significantly inhibited at the Nox4 promoter by OTX015, suggesting that this inhibitor is effective at preventing the association of Brd4 protein at this promoter (Figure 3B). In addition to decreased association of Brd4 with the Nox4 promoter, OTX015 treatment reduced the association of H4K16ac (Figure 3C) and p300 (Figure 3D) with the Nox4 promoter in IPF fibroblasts. This indicates that, in parallel with the observed reduction in constitutive expression of Nox4 gene expression in IPF fibroblasts (Figure 1D), OTX015 treatment depletes the recruitment of Brd4, H4K16ac, and p300 at the Nox4 promoter.
Figure 3. BET inhibitor OTX015 blocks the Nox4 promoter association with Brd4, H4K16ac, and p300 in IPF fibroblasts.
(A) Schematic of the relative locations of 3 PCR primer sets used to examine the association of transcriptional complex proteins with Nox4 (ENSG00000086991) by ChIP assays. (B–D) Primary IPF lung fibroblasts were treated with vehicle or OTX015 (0.5 μM) for 48 hours before the cells were subjected to ChIP assays. The Nox4 promoter region association with Brd4, H4K16Ac, and p300 was analyzed by ChIP assays, with primer sets indicated in A. DNA was immunoprecipitated with specific antibodies against Brd4 (B), H4K16Ac (C), and p300 (D). The relative levels of the PCR product are represented in the bar graphs as Nox4 promoter region association with Brd4, H4K16ac, or p300. Negative control represents IgG pull-down. qPCR data were analyzed by the 2-ΔΔCt method and were normalized to input DNA, expressed as fold change relative to primer set FcRc or FaRa. The values are expressed as mean ± SD from average of 3 independent experiments of 1 representative cell line. *P < 0.05, OTX015-treated compared with control (vehicle only), by 2-tailed t test.
Next, we tested the effects of OTX015 on TGF-β1–induced responses of these transcriptional components at the Nox4 promoter in IMR90 fibroblasts (Figure 4 and Supplemental Figure 4). TGF-β1 induced an enrichment of Brd4 association with the Nox4 promoter, an effect that is blocked by OTX015 pretreatment (Figure 4A). Concurrently, the enhanced associations of H4K16ac and p300 at the Nox4 promoter region induced by TGF-β1 are also blocked by this BET inhibitor (Figure 4, B and C). Together with the finding that OTX015 is a potent inhibitor of Nox4 upregulation by TGF-β1 (Figure 2), these data indicate that BET inhibition not only blocks the association of Brd4 with the Nox4 promoter, but also its association with the open chromatin mark, H4K16ac, and the transcriptional cofactor/acetyltransferase p300, which together regulate both constitutive and inducible Nox4 gene expression.
Figure 4. Association of Brd4, H4K16ac, and p300 with the Nox4 promoter region by TGF-β1 with or without BET inhibitor OTX015.
Normal lung fibroblasts (IMR90) were pretreated with vehicle or OTX015 (0.5 μM) 2 hours before stimulation with TGF-β1 (2ng/mL) for 48 hours. Then, the cells were collected for ChIP assays. The association of the Nox4 promoter region with Brd4, H4K16Ac, and p300 was analyzed by ChIP assays, with primer sets indicated in Figure 3A. DNA was immunoprecipitated with specific antibodies against Brd4 (A), H4K16ac (B), and p300 (C). Bars represent the relative levels of the PCR product of the Nox4 promoter region association with Brd4, H4K16ac, or p300. Negative control represents IgG pull-down. qPCR data were analyzed by using the 2-ΔΔCt method, and results were normalized to input DNA, expressed as fold changes relative to primer set FcRc or FaRa. The values are expressed as mean ± SD from average of 3 independent experiments of 1 representative cell line. *P < 0.05, compared with control (vehicle only), #P < 0.05, compared with TGF-β1, by 2-tailed t test.
p300 regulates Nox4 gene expression in lung myofibroblasts.
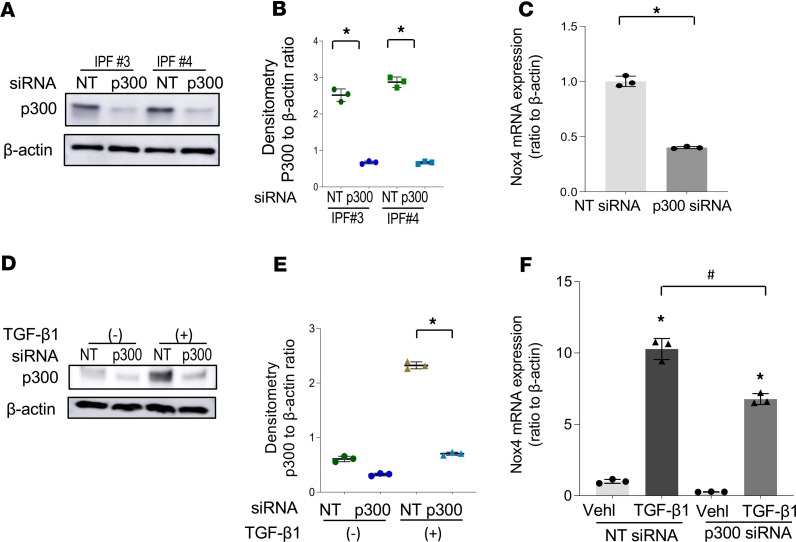
Based on our findings of p300 association with the Nox4 promoter under constitutive (Figure 3D) or TGF-β1–inducible (Figure 4C) conditions, we tested the requirement of p300 for Nox4 gene regulation under both settings. In primary IPF fibroblasts, siRNA silencing of p300 resulted in constitutive downregulation of Nox4 at mRNA levels (Figure 5, A–C, and Supplemental Figure 5A for Nox4 at protein levels). Silencing of p300 in IMR90 fibroblasts significantly reduced TGF-β1–induced Nox4 at the mRNA level (Figure 5, D–F, and Supplemental Figure 5B for Nox4 protein). These data indicate that p300 regulates both constitutive and inducible Nox4 gene expression in lung fibroblasts.
Figure 5. p300 mediates Nox4 gene expression in lung fibroblasts.
(A–C) Primary IPF fibroblasts were transfected with nontargeting (NT) siRNA or p300 siRNA. (A) Representative Western blots of fibroblasts from 2 different individuals with IPF transfected with siRNA NT or p300 demonstrated the levels of p300 and β-actin. (B) The densitometry of p300-associated signals detected (ratio to β-actin) in A. *P < 0.05, compared with NT control of the same cell line, by 2-tailed t test. (C) The expression of Nox4 mRNA was analyzed by real-time quantitative PCR of the IPF cells transfected with siRNA NT or p300 (mean ± SD, n = 3 experimental replicates of a representative cell line). *P < 0.05 compared with NT control, by 2-tailed t test. (D–F) IMR90 cells were transfected with siRNA NT or p300, and then the cells were treated with TGF-β1 (2 ng/mL) for 48 hours and analyzed for p300 expression by Western blot and Nox4 mRNA expression by real-time PCR. (D) Representative Western blots of p300 and β-actin. (E) Densitometric analysis of p300-associated signals detected (ratio to β-actin) in D. *P < 0.05, TGF-β1–treated cells with p300 siRNA vs. NT siRNA, by 2-tailed t test. (F) The mRNA expression of Nox4 by real-time PCR (mean ± SD, n = 3 experimental replicates). *P < 0.05, compared with vehicle control; #P < 0.05, the transfected cells with TGF-β1 treatment, siRNA p300 vs. NT, by 2-tailed t test.
BET inhibitor OTX015 interferes with p300 induction/recruitment by TGF-β1.
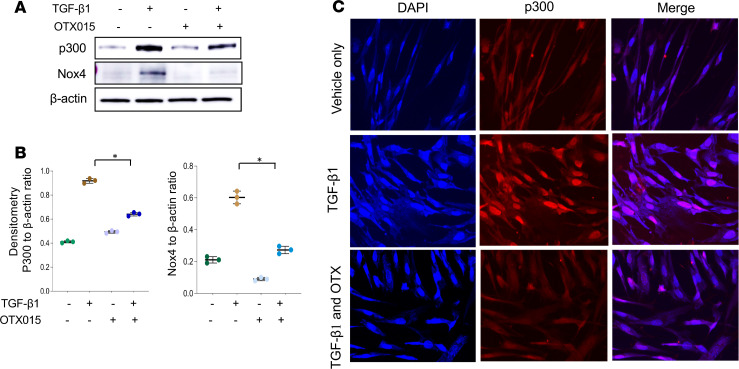
We then tested whether the BET inhibitor, OTX015, regulates the induction and/or recruitment of p300 by TGF-β1. OTX015 inhibited TGF-β1–induced p300 expression in parallel with Nox4 downregulation in IMR90 fibroblasts (Figure 6, A and B) and in non-IPF primary human lung fibroblasts (Supplemental Figure 6). OTX015 pretreatment also inhibited TGF-β1–induced nuclear localization of p300, as shown by immunofluorescence staining (Figure 6C). Together, these data indicate that p300 mediates TGF-β1–induced Nox4 gene upregulation and that Brd4 may function to recruit/stabilize p300 at the transcriptional complex.
Figure 6. The BET inhibitor OTX015 blocks p300-mediated Nox4 upregulation by TGF-β1.
Lung fibroblasts IMR90 were treated either with vehicle control or OTX015 (0.5 μM) or pretreated with vehicle or OTX015 for 2 hours, before adding TGF-β1 (2 ng/mL) for 48 hours. (A) Representative Western blots showing p300, Nox4, and β-actin levels in the whole cell lysate with the above treatment. (B) Densitometric analysis of p300- or Nox4-associated signals detected (ratio to β-actin) in A. *P < 0.05, TGF-β1–treated cells without OTX015 vs. with OTX015 (mean ± SD from 3 independent experiments), by 2-tailed t test. (C) Representative images of immunofluorescence of IMR90 cells treated as in A. Cells are stained with antibodies of p300 (red) and DAPI (blue) or merged for stains of both. The images were taken with a Keyence microscope (original magnification, ×20).
Pharmacologic BET inhibition with OTX015 reverses established lung fibrosis in aged mice.
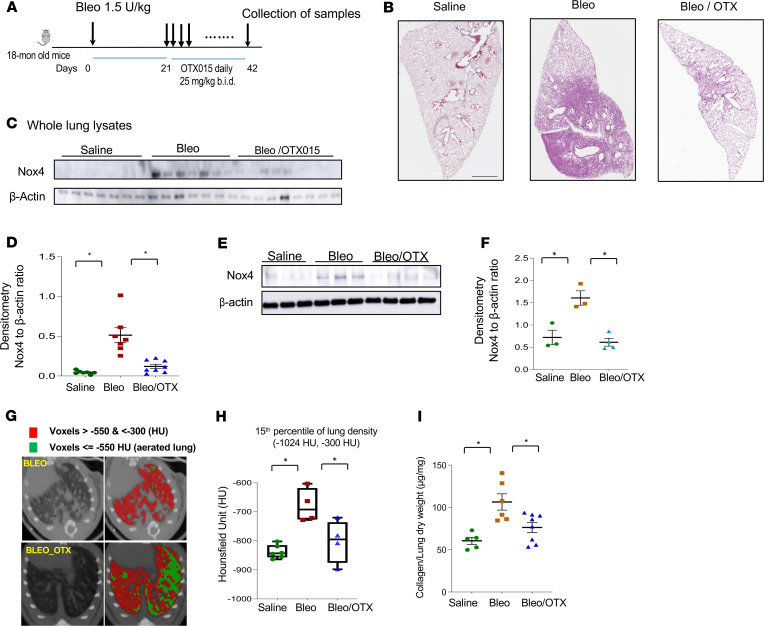
We have established a model of persistent lung fibrosis in aged mice subjected to airway epithelial injury by administration of intratracheal bleomycin (34). In 18-month-old mice, we induced lung fibrosis with bleomycin through oropharyngeal installation (1.25 units/kg). OTX015 was administered twice daily by oral gavage at a dose of 25 mg/kg, which has previously been shown to be effective in a murine model of cancer (35). Dosing was initiated 21 days after bleomycin injury when lung fibrosis is well established and continued for the ensuing 3 weeks (at 42 days) before sacrifice and analyses (Figure 7A). In mice receiving OTX015 treatment, marked improvement in fibrosis resolution was observed by histopathology (Figure 7B). Whole lung lysates showed decreased Nox4 protein levels by Western blot analyses in the OTX015-treated group in comparison with induced expression in bleomycin-injured mice (Figure 7, C and D); this effect was also observed in isolated primary lung fibroblasts from lung tissues (Figure 7, E and F). Lungs of mice were analyzed by micro-CT scans. Using –550 Hounsfield units (HU) as a discriminator of aerated versus dense lung (36), mice receiving OTX015 treatment showed a marked increase in aerated lung and a relative decrease in nonaerated lung (Figure 7G). Additionally, we used the 15th percentile of lung density as a sensitive cutoff to discriminate air content from lung parenchyma, as previously described (37); this analysis demonstrated that lung tissue density was markedly reduced in OTX015-treated mice in comparison with the bleomycin group (Figure 7H). Accumulation of total lung collagen, measured by hydroxyproline levels, demonstrated that the OTX015-treated bleomycin group had lower steady-state levels of collagen in comparison with the bleomycin-only group (Figure 7I). These studies indicate that OTX015, a drug that inhibits Brd4, H4K16ac, and p300 recruitment to the Nox4 promoter region and downregulates Nox4 in lung tissues and in isolated lung fibroblasts, is effective in mediating resolution of established/persistent lung fibrosis in aged mice.
Figure 7. The inhibitor OTX015 facilitates resolution of established lung fibrosis in aged mice.
(A) The schedule of lung fibrosis induced by bleomycin injury in 18-month-old mice. OTX015 was given twice daily orally from day 21–42 after bleomycin injury. All the mice were sacrificed at day 42 after bleomycin injury. (B) Whole lung histology by immunohistochemistry with H&E staining of mice with saline, bleomycin injury (bleo), or bleomycin injury with OTX015 treatment (Bleo/OTX). (C) Mouse whole lung tissue lysates from saline (n = 7), bleomycin (n = 7), and bleomycin with OTX015 treatment (n = 9) groups were subjected to SDS-PAGE and Western blots analysis for Nox4 and β-actin. (D) Densitometry of the Western blots in C for the ratio of Nox4 to β-actin (mean ± SD). (E) Primary lung fibroblasts were cultured from mouse lungs. Whole cell lysate from saline (n = 3), bleomycin (n = 3), and bleomycin with OTX015 treatment (n = 4) groups were collected at passage 1 and subjected to SDS-PAGE and Western blots analysis for Nox4 and β-actin. (F) Densitometry of the Western blots in E for the ratio of Nox4 to β-actin in the primary murine lung fibroblasts (mean ± SD). (G) Representative axial micro-CT images of mouse lungs 6 weeks after they were subjected to bleomycin injury with/without OTX015 treatment. Voxels of the lung field that are below –550 Hounsfield units are in green (representing aerated lung) and those between –550 and –350 Hounsfield units are in red (representing nonaerated lung). (H) Quantitation of lung density in uninjured (saline) and injured (Bleo) mice treated with/without OTX015. Whisker plots represent mean ± SD; n = 4–6 per group (saline, n = 6; bleo, n = 4; bleo/OTX015, n = 4). *P < 0.05, by 2-tailed t test. (I) Hydroxyproline content in lungs of mice after saline (n = 5), bleomycin (n = 6), or bleomycin with OTX015 treatment (n = 8) (mean ± SD). *P < 0.05, for comparisons of indicted groups as compared with the bleomycin group, by 2-tailed t test.
Discussion
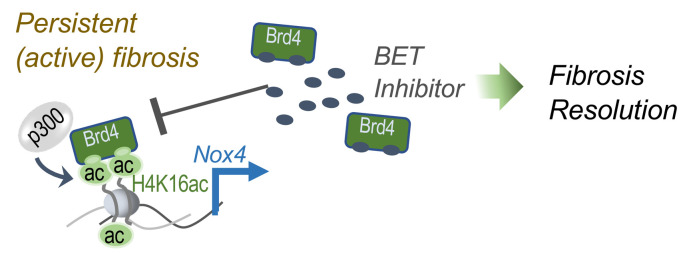
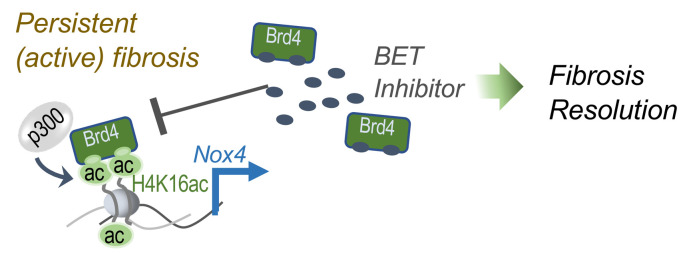
Epigenetic regulation of gene expression by dynamic changes in chromatin structure are essential for physiological adaptive responses and contribute to pathological states. Posttranslational acetylation of histone tails are recognized by epigenetic “readers,” in particular Brd proteins, that typically facilitate active transcription of target genes (38). Brd proteins have emerged as attractive epigenetic targets for cancer and other diseases (39). Epigenetic mechanisms contribute to the pathogenesis of IPF, including histone acetylation (7, 18, 19, 40). In this study, we demonstrate that inhibition of the epigenetic reader, Brd4, promotes resolution of established and persistent lung fibrosis in aged mice. This restored capacity for fibrosis resolution is associated with a marked reduction in the expression of Nox4, a ROS-generating enzyme implicated in age-associated pulmonary fibrosis (34). Both constitutive and TGF-β1–inducible Nox4 expression are downregulated in lung fibroblasts with genetic or pharmacologic inhibition of Brd4. Gene repression of Nox4 by Brd4 inhibition is coordinated by reduced associations of Brd4, H4K16ac, and p300 at the Nox4 promoter (Figure 8).
Figure 8. Schematic diagram of proposed mechanisms from this study’s findings.
BET inhibitor inhibits epigenetic reader proteins, such as Brd4, thus blocking the association of Brd4, H4K16ac, and p300 with Nox4 promoter region, which would downregulate the expression of Nox4 (and other genes) and, ultimately, restore lung capacity and improve lung fibrosis resolution.
Targeting BET proteins is emerging as a potentially effective strategy for treatment of organ fibrosis in preclinical models (41). We investigated a BET inhibitor that is already in cancer clinical trials, OTX015 (16), while other drugs in this class may mediate similar activities. In an in vitro study of human lung fibroblasts, the BET inhibitor JQ1 downregulated Nox4 by decreasing the association of its promoter with Brd3 and Brd4 but not Brd2 (42); in this study, JQ1 was proposed to counterregulate the Nox4/SOD2 imbalance by decreasing Nox4 gene expression and increasing Nrf2 activity. Previous studies from our group showed that Nox4 silencing itself was sufficient to induce Nrf2 activation in human lung fibroblasts (43). Brd4 inhibition with JQ1 was reported to protect against renal fibrosis by downregulating Nox4 expression and interrupting upstream Smad3 and ERK1/2 signaling (28). In human dermal fibroblasts, JQ1 was shown to inhibit the upregulation of Nox4 and differentiation of skin fibroblasts to myofibroblasts induced by TGF-β1 by interruption of the interaction of Smad3 with Brd4 (44). However, our studies implicate a more direct role for Brd4 in regulating the association of acetylated histone H4 and the transcriptional coactivator/acetyltransferase p300 with the Nox4 promoter, which in turn downregulates Nox4 gene expression in fibrotic lung fibroblasts. We demonstrate that Brd4 inhibition partially blocks p300 recruitment/stabilization, and its nuclear translocation/expression, in lung fibroblasts to downregulate Nox4 gene expression. This is similar to findings in adipose tissue that showed that Brd4 binding to p300 at active enhancers regulates genes that confer cellular identity (45); additionally, in embryonic stem cells, Brd4 interacts with p300 to maintain the expression and chromatin patterns of pluripotency-associated genes (46). Together, these studies suggest that Brd4 enhances p300-mediated histone acetylation. Our studies reveal that active Nox4 transcription-dependent gene expression is associated with enriched Brd4, p300, and acetylated H4K16 at the Nox4 promoter, while BET/Brd4 inhibition depletes p300 and acetylated H4K16 at this promoter site, resulting in Nox4 gene downregulation.
By virtue of its role as a protein that coordinates and integrates acetylated histones and transcription factor recruitment/binding (47), Brd4 is involved in many normal and disease cellular processes (39, 48). While BET inhibitors have been investigated in fibrotic disease models with promising results (49), previous studies have not tested antifibrotic efficacy of these drugs in therapeutic approach with established fibrosis but rather in a preventative model. Additionally, the influence of age and capacity for fibrosis resolution, which may be more relevant in preclinical models, has not been addressed. Previous studies of BET inhibition in lung fibrosis models were conducted with drug administration at early stages of inflammation following injury and in young mice that have the capacity to resolve fibrosis spontaneously (27, 50). For example, the inhibitor JQ1 was administrated on day 5 after bleomycin injury (27) or 1 day before bleomycin challenge (50); thus, it is not clear if the beneficial effects were related to antiinflammatory (vs. antifibrotic) effects. On the other hand, we examined the therapeutic effects of the inhibitor OTX015 in aged mice with established and persistent fibrosis, specifically at 3 weeks following bleomycin injury in aged mice that manifest nonresolving fibrosis in the absence of drug treatment (34, 51). Importantly, our studies support a direct effect of BET/Brd4 inhibition on the fibrosis resolution process. In OTX015-treated animals, Nox4 expression in the lungs was markedly decreased with appreciable restoration of lung structure. Although Nox4 is a well-recognized profibrotic gene in lung fibrosis (5, 34), the observed restitution of resolution capacity may be attributable, not only to decreased Nox4 expression, but to a skewing of gene expression that favors tissue regeneration over fibrosis. Other antifibrotic effects of BET inhibitors may include their antiproliferative and antiinflammatory effects as well as potential Nox4-independent effects on fibroblast activation (26, 27, 42).
The rapid expansion of novel BET inhibitors offers opportunities for more targeted approaches and precision in heterogeneous diseases, such as IPF. While our studies showed that the pan-BET inhibitors, OTX015, I-BET-762, and JQ1, consistently downregulate Nox4 expression, we also observed variable responses/sensitivities to individual drugs in primary cells derived from different patients with IPF, supporting a need for a more “personalized/precision” approach to drug selection (52). Despite the promising results of BET inhibitors, off-target effects of these drugs are emerging. For example, cardiomyopathy developed in animals receiving the I-BET-151 inhibitor (53); additionally, acquired resistance to BET inhibitors has been reported (54). More specific BET inhibitors are currently under development, in addition to combination therapies to combat resistance (55). An understanding of the full spectrum of profibrotic effects of BET proteins may allow for more precise and targeted approaches. Despite these limitations, our studies support the future development of specific BET/Brd4 inhibitors for the treatment of fibrotic disorders such as IPF.
Methods
Cell culture, reagents, and treatments.
The diagnosis of IPF was made by a multidisciplinary approach according to the ATS/ERS guidelines (56). All primary cell lines were used before passage 5. Normal lung fibroblasts (IMR90) were from the Coriell Institute for Medical Research and were used at early passages. All cells were collected for various assays at about 85% confluent. TGF-β1 was from R&D Systems. I-BET-762 was from Cayman Chemical and JQ1 was a gift from James Bradner, Dana-Farber Cancer Institute Inc. (Boston, Massachusetts, USA). The BET inhibitor, OTX015, was purchased from Chemietek. For IPF fibroblasts, I-BET-762 was added at 0.5 μM, JQ1 at 1 μM, and OTX015 at 0.5 μM for 48 hours. At this dose of OTX015, no cellular toxicity was noted and no effect on growth of lung fibroblasts were observed (Supplemental Figure 7). For normal lung fibroblasts (IMR90 or non-IPF fibroblasts), cells were kept in 1%FBS medium overnight and then pretreated with vehicle control or the above inhibitors at the indicated concentrations for 2 hours before adding TGF-β1 at 2 ng/mL for 48 hours; cells were then collected for various assays.
siRNA-mediated gene silencing.
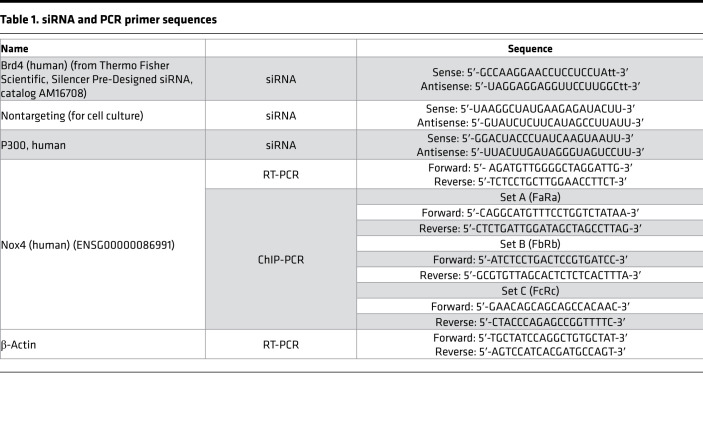
siRNA was transfected with Opti-MEM (Thermo Fisher Scientific) using lipofectamine RNAi/MAX (Invitrogen). The sequences for Brd4, p300 siRNA and for NT are listed in Table 1. IPF or normal lung fibroblasts were kept in full medium with 10% FBS for 48 hours after transfection and then were subjected to various assays.
Table 1. siRNA and PCR primer sequences.
Real-time PCR.
mRNA expression levels were determined by real-time PCR with SYBR Green PCR Master Mix from Life Technologies. RNA was extracted with the RNeasy Mini Kit (QIAGEN) and transcribed into cDNA with a cDNA synthesis kit (Takara Bio). Real-time PCR was performed in triplicate and normalized to β-actin using the ΔΔCt method (17). The primer sequences are listed in Table 1.
H2O2 production measurements.
The measurement of extracellular H2O2 production from the cultured cells was according to previously published protocols (57).
Immunoblotting.
Western immunoblotting was performed as previously described (58). Whole cell lysates were collected and quantified with a Micro BCA Protein Assay kit (Thermo Fisher Scientific). Anti-Nox4 (AF8158) was from R&D Systems, and validation of the antibody was showed in Supplemental Figure 1; anti-p300 (catalog 61903) was from Active Motif; anti-Brd4 (A301-985A100) was from Bethyl Laboratories; and anti–β-actin (catalog 2128) was from Cell Signaling.
Immunofluorescence.
Immunofluorescence staining was performed as descried previously (59). Mouse anti-p300 was purchased from Active Motif (catalog 61903). Images were acquired with a Keyence microscope.
ChIP assays.
ChIP assays were performed as per the manufacturer’s protocol (ab500, Abcam) with some modifications (10). Antibodies against Brd4 (A301-985A100) were from Bethyl Laboratories, and H4K16Ac (catalog 61529) and p300 (catalog 61903) were from Active Motif. ChIP-DNA was amplified by real-time PCR with primers listed in Table 1, using SYBR Green PCR Master Mix (Life Technologies). Results are normalized to input DNA.
Bleomycin induced lung fibrosis in aged mice.
Eighteen-month-old healthy C57BL mice were used. A single dose of normal saline or bleomycin sulfate at 1.5 U/kg body weight was oropharyngeal instilled. OTX015 was administrated by oral gavage twice daily at 25 mg/kg (35) starting at week 3 (day 21) after bleomycin injury till week 6 (day 42). All mice were sacrificed at the end of week 6 after bleomycin injury. The mouse lungs were prepared for whole lung tissues lysates, hydroxyproline analysis, histological studies for lung tissues with H&E stains, and for primary lung fibroblasts culture. The primary lung fibroblasts were collected at passage 1 for whole cell lysate for Western blots. Some of the mice, before sacrificed were subjected to lung micro-CT scan. Lysates from whole lung tissues and from primary murine lung fibroblasts cultures collected for Western blots were used as described above.
Mice lung micro-CT Imaging.
The noncontrast micro-CT images of mice were acquired in supine position using in vivo small animal micro-CT (MILabs). During acquisition, the animals were under isoflurane anesthesia and prospectively gated for a single inspiratory phase of respiration. All images were acquired at ultrafocused magnification with the following parameters: tube voltage (55 kV), tube current (0.19 mA), scan angle (360o), and with 20-ms exposure. The respiration gating is set for a rising trigger with a 25-ms delay, 100-ms duration, 100-ms inhibition, 1080 mV threshold, and 80 mV hysteresis. The trigger is set for when the signal first crosses over the hysteresis and intersects with the specified threshold. All images were reconstructed using MILabs Reconstruction Software at 40 μ-voxel resolution at a single respiratory phase at 0.2 phase and 0.1 window. A region-growing algorithm was applied for segmentation of lung parenchyma from the micro-CT images. In region-growing segmentation, a seed point was selected within the lungs and lungs are grown from the seed point based on the density thresholds of (–1024 HU, –100 HU). The lung mask was then applied to identify regions of aerated lung (areas < –550 HU) (36). Additionally, 15th percentile of lung density (60) was estimated to statistically evaluate the changes in lung density across saline, bleomycin, and bleomycin with OTX015-treated groups. The closer the 15th percentile value of the lung mask to the density of air (–1024 HU), the higher the percentage of aerated lung.
Hydroxyproline analysis.
Mouse lung tissues were collected and hydroxyproline content was analyzed by the QuickZyme Total Collagen Assay from QuickZyme BioSciences. Briefly, the lung tissues were dried in an oven at 70°C for 48 hours and then hydrolyzed in 6 N HCL at 95°C for 20 hours. Then hydroxyproline content were measured following the manufacturer’s protocol using collagen as standards.
Statistics.
Data are presented as the mean ± SD. All data were statistically analyzed using GraphPad Prism 5.0. One-way ANOVA was used to compare multiple groups; comparisons between 2 groups were performed with the Student’s paired 2-tailed t test. A P value of less than 0.05 was considered to be statistically significant.
Study approval.
Deidentified human tissues were obtained from the University of Alabama at Birmingham (UAB) Airway Tissue Procurement Program for primary culture of lung fibroblasts, which was approved by the UAB Institutional Review Board. All animal studies were performed in accordance with protocols approved by the UAB Institutional Animal Care and with Committee.
Author contributions
YYS conceived and coordinated the study, designed the experiments, analyzed the data, and wrote the manuscript; XL, ZX, and QJZ performed the experiments and analyzed the data; DS, SB, and SMR established the protocol and analyzed the mouse CT imaging data; and VJT conceived and coordinated the study, analyzed the data, and edited the manuscript.
Supplementary Material
Acknowledgments
This work was funded by NIH grant R01 AG050567 (to YYS); NIH grants P30 DK072482 and R35 HL135816 (to SR); and NIH grants R01 AG046210 and P01 HL114470 and VA Merit Award I01BX003056 (to VJT). We thank the UAB Health Services Endowment grant for partial funding of the micro-CT used in this study.
Version 1. 06/16/2020
In-Press Preview
Version 2. 07/23/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(14):e137127.https://doi.org/10.1172/jci.insight.137127.
Contributor Information
Yan Y. Sanders, Email: yansanders@uabmc.edu.
Q. Jennifer Zhou, Email: jenzh@uab.edu.
Zheyi Xiang, Email: xzy566@uab.edu.
Denise Stanford, Email: denisestanford@uabmc.edu.
Sandeep Bodduluri, Email: sbodduluri@uabmc.edu.
Steven M. Rowe, Email: SRowe@peds.uab.edu.
Victor J. Thannickal, Email: vjthan@uab.edu.
References
- 1.Thannickal VJ. Mechanistic links between aging and lung fibrosis. Biogerontology. 2013;14(6):609–615. doi: 10.1007/s10522-013-9451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King TE, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 3.Richeldi L, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 4.Thannickal VJ, et al. Myofibroblast differentiation by transforming growth factor-beta1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. J Biol Chem. 2003;278(14):12384–12389. doi: 10.1074/jbc.M208544200. [DOI] [PubMed] [Google Scholar]
- 5.Hecker L, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med. 2009;15(9):1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horowitz JC, Thannickal VJ. Epithelial-mesenchymal interactions in pulmonary fibrosis. Semin Respir Crit Care Med. 2006;27(6):600–612. doi: 10.1055/s-2006-957332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyu X, Hu M, Peng J, Zhang X, Sanders YY. HDAC inhibitors as antifibrotic drugs in cardiac and pulmonary fibrosis. Ther Adv Chronic Dis. 2019;10:2040622319862697. doi: 10.1177/2040622319862697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez IE, Eickelberg O. The impact of TGF-β on lung fibrosis: from targeting to biomarkers. Proc Am Thorac Soc. 2012;9(3):111–116. doi: 10.1513/pats.201203-023AW. [DOI] [PubMed] [Google Scholar]
- 9.Sanders YY, et al. SMAD-independent down-regulation of caveolin-1 by TGF-β: effects on proliferation and survival of myofibroblasts. PLoS One. 2015;10(2):e0116995. doi: 10.1371/journal.pone.0116995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders YY, Liu H, Liu G, Thannickal VJ. Epigenetic mechanisms regulate NADPH oxidase-4 expression in cellular senescence. Free Radic Biol Med. 2015;79:197–205. doi: 10.1016/j.freeradbiomed.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Zhao QD, et al. NADPH oxidase 4 induces cardiac fibrosis and hypertrophy through activating Akt/mTOR and NFκB signaling pathways. Circulation. 2015;131(7):643–655. doi: 10.1161/CIRCULATIONAHA.114.011079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang S, Kisseleva T, Brenner DA. The Role of NADPH Oxidases (NOXs) in Liver Fibrosis and the Activation of Myofibroblasts. Front Physiol. 2016;7:17. doi: 10.3389/fphys.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong BY, et al. Oxidative stress caused by activation of NADPH oxidase 4 promotes contrast-induced acute kidney injury. PLoS One. 2018;13(1):e0191034. doi: 10.1371/journal.pone.0191034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piera-Velazquez S, Makul A, Jimenez SA. Increased expression of NAPDH oxidase 4 in systemic sclerosis dermal fibroblasts: regulation by transforming growth factor beta. Arthritis Rheumatol. 2015;67(10):2749–2758. doi: 10.1002/art.39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Augsburger F, et al. Pharmacological characterization of the seven human NOX isoforms and their inhibitors. Redox Biol. 2019;26:101272. doi: 10.1016/j.redox.2019.101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alqahtani A, et al. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA. 2019;5(3):FSO372. doi: 10.4155/fsoa-2018-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders YY, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39(5):610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol. 2011;45(1):16–23. doi: 10.1165/rcmb.2010-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders YY, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186(6):525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Epigenomics of idiopathic pulmonary fibrosis. Epigenomics. 2012;4(2):195–203. doi: 10.2217/epi.12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dakhlallah D, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187(4):397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115(6):2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez R, Meslamani J, Zhou MM. The bromodomain: from epigenome reader to druggable target. Biochim Biophys Acta. 2014;1839(8):676–685. doi: 10.1016/j.bbagrm.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12(5):659–665. [PMC free article] [PubMed] [Google Scholar]
- 26.Ding N, et al. BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci USA. 2015;112(51):15713–15718. doi: 10.1073/pnas.1522163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X, et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am J Pathol. 2013;183(2):470–479. doi: 10.1016/j.ajpath.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 28.Zhou B, et al. Brd4 inhibition attenuates unilateral ureteral obstruction-induced fibrosis by blocking TGF-β-mediated Nox4 expression. Redox Biol. 2017;11:390–402. doi: 10.1016/j.redox.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar K, et al. BET inhibitors block pancreatic stellate cell collagen I production and attenuate fibrosis in vivo. JCI Insight. 2017;2(3):e88032. doi: 10.1172/jci.insight.88032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anand P, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirguet O, et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013;56(19):7501–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 32.Amorim S, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3(4):e196–e204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 33.Boudreau HE, Ma WF, Korzeniowska A, Park JJ, Bhagwat MA, Leto TL. Histone modifications affect differential regulation of TGFβ- induced NADPH oxidase 4 (NOX4) by wild-type and mutant p53. Oncotarget. 2017;8(27):44379–44397. doi: 10.18632/oncotarget.17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hecker L, et al. Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci Transl Med. 2014;6(231):231ra47. doi: 10.1126/scitranslmed.3008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berenguer-Daizé C, et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int J Cancer. 2016;139(9):2047–2055. doi: 10.1002/ijc.30256. [DOI] [PubMed] [Google Scholar]
- 36.De Langhe E, et al. Quantification of lung fibrosis and emphysema in mice using automated micro-computed tomography. PLoS One. 2012;7(8):e43123. doi: 10.1371/journal.pone.0043123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parr DG, Stoel BC, Stolk J, Stockley RA. Validation of computed tomographic lung densitometry for monitoring emphysema in alpha1-antitrypsin deficiency. Thorax. 2006;61(6):485–490. doi: 10.1136/thx.2005.054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17(1):164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders YY, Hagood JS, Liu H, Zhang W, Ambalavanan N, Thannickal VJ. Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. Eur Respir J. 2014;43(5):1448–1458. doi: 10.1183/09031936.00095113. [DOI] [PubMed] [Google Scholar]
- 41.Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann Oncol. 2017;28(8):1776–1787. doi: 10.1093/annonc/mdx157. [DOI] [PubMed] [Google Scholar]
- 42.Stock CJW, et al. Bromodomain and Extraterminal (BET) Protein Inhibition Restores Redox Balance and Inhibits Myofibroblast Activation. Biomed Res Int. 2019;2019:1484736. doi: 10.1155/2019/1484736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernard K, et al. NADPH Oxidase 4 (Nox4) Suppresses Mitochondrial Biogenesis and Bioenergetics in Lung Fibroblasts via a Nuclear Factor Erythroid-derived 2-like 2 (Nrf2)-dependent Pathway. J Biol Chem. 2017;292(7):3029–3038. doi: 10.1074/jbc.M116.752261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ijaz T, et al. Coordinate activities of BRD4 and CDK9 in the transcriptional elongation complex are required for TGFβ-induced Nox4 expression and myofibroblast transdifferentiation. Cell Death Dis. 2017;8(2):e2606. doi: 10.1038/cddis.2016.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JE, et al. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat Commun. 2017;8(1):2217. doi: 10.1038/s41467-017-02403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T, Kamikawa YF, Donohoe ME. Brd4’s Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep. 2018;25(7):1756–1771. doi: 10.1016/j.celrep.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Floyd SR, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498(7453):246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stratton MS, Haldar SM, McKinsey TA. BRD4 inhibition for the treatment of pathological organ fibrosis. F1000Res. 2017;6:F1000 Faculty Rev-1015. doi: 10.12688/f1000research.11339.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang X, et al. BET bromodomain proteins mediate downstream signaling events following growth factor stimulation in human lung fibroblasts and are involved in bleomycin-induced pulmonary fibrosis. Mol Pharmacol. 2013;83(1):283–293. doi: 10.1124/mol.112.081661. [DOI] [PubMed] [Google Scholar]
- 51.Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40(3):362–382. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Habiel DM, Hogaboam CM. Heterogeneity of Fibroblasts and Myofibroblasts in Pulmonary Fibrosis. Curr Pathobiol Rep. 2017;5(2):101–110. doi: 10.1007/s40139-017-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piquereau J, et al. The BET Bromodomain Inhibitor I-BET-151 Induces Structural and Functional Alterations of the Heart Mitochondria in Healthy Male Mice and Rats. Int J Mol Sci. 2019;20(7):E1527. doi: 10.3390/ijms20071527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525(7570):543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borbely G, Haldosen LA, Dahlman-Wright K, Zhao C. Induction of USP17 by combining BET and HDAC inhibitors in breast cancer cells. Oncotarget. 2015;6(32):33623–33635. doi: 10.18632/oncotarget.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raghu G, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thannickal VJ, Fanburg BL. Activation of an H2O2-generating NADH oxidase in human lung fibroblasts by transforming growth factor beta 1. J Biol Chem. 1995;270(51):30334–30338. doi: 10.1074/jbc.270.51.30334. [DOI] [PubMed] [Google Scholar]
- 58.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(-) lung fibroblasts. Am J Respir Cell Mol Biol. 2007;36(2):226–235. doi: 10.1165/rcmb.2006-0178OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Liu H, Hock T, Thannickal VJ, Sanders YY. Histone deacetylase inhibition downregulates collagen 3A1 in fibrotic lung fibroblasts. Int J Mol Sci. 2013;14(10):19605–19617. doi: 10.3390/ijms141019605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tani H, et al. Autoregulation by the right coronary artery in dogs with open chests; comparison with the left coronary artery. Pflugers Arch. 1990;416(4):442–447. doi: 10.1007/BF00370752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.