Abstract
Summary
iBioProVis is an interactive tool for visual analysis of the compound bioactivity space in the context of target proteins, drugs and drug candidate compounds. iBioProVis tool takes target protein identifiers and, optionally, compound SMILES as input, and uses the state-of-the-art non-linear dimensionality reduction method t-Distributed Stochastic Neighbor Embedding (t-SNE) to plot the distribution of compounds embedded in a 2D map, based on the similarity of structural properties of compounds and in the context of compounds’ cognate targets. Similar compounds, which are embedded to proximate points on the 2D map, may bind the same or similar target proteins. Thus, iBioProVis can be used to easily observe the structural distribution of one or two target proteins’ known ligands on the 2D compound space, and to infer new binders to the same protein, or to infer new potential target(s) for a compound of interest, based on this distribution. Principal component analysis (PCA) projection of the input compounds is also provided, Hence the user can interactively observe the same compound or a group of selected compounds which is projected by both PCA and embedded by t-SNE. iBioProVis also provides detailed information about drugs and drug candidate compounds through cross-references to widely used and well-known databases, in the form of linked table views. Two use-case studies were demonstrated, one being on angiotensin-converting enzyme 2 (ACE2) protein which is Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike protein receptor. ACE2 binding compounds and seven antiviral drugs were closely embedded in which two of them have been under clinical trial for Coronavirus disease 19 (COVID-19).
Availability and implementation
iBioProVis and its carefully filtered dataset are available at https://ibpv.kansil.org/ for public use.
Contact
vatalay@metu.edu.tr
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The ChEMBL database (version 25) has 1 879 206 distinct small molecule compounds with 12 482 target proteins and 15 506 670 reported bioactivities (Mendez et al., 2019). Even if only the data in ChEMBL are considered, there are more than 11 billion possible compound-target protein pairs to be tested in vitro experimentally. Unfortunately, public databases or datasets have limited coverage as only partial information is available regarding the compound–target interaction space, mainly due to high costs and labor requirements associated with large-scale screening experiments. Therefore, prior knowledge about the eventual target proteins or cellular signaling events, in which a small molecule is involved, becomes crucial for novel drug-target discovery (Rifaioglu et al., 2019). Furthermore, the representation of drugs and their targets in databases lack the comparative holistic view of the molecular action on multiple targets and structural similarity of the compounds.
A small number of studies have recently become available to visualize the chemical space and the compound bioactivity space (Awale and Raymond, 2016; Gaspar et al., 2015; Gütlein et al., 2012; Janssen et al., 2019; Karlov et al., 2019). Janssen et al. (2019) and Karlov et al. (2019) made visualization tools available, only for pre-computed datasets. The tool developed by Awale and Raymond (2016) performs visualization by principal component analysis (PCA) which is a linear and global method and may thus miss non-linear and local relations among the input drug molecules. Another study, Gaspar et al. (2015) only presents their results and no tool is made available. The tool built by Gütlein et al. (2012) does not involve target proteins; however, the user can apply clustering on the compounds to observe their groupings.
We describe a tool called iBioProVis, which uses a map-based method to embed a given set of compounds, particularly active compounds in the context of their cognate target proteins, as points onto a real-coordinate 2D space, based on the structural descriptors of the compounds. iBioProVis allows the interactive visualization of these embeddings. The sources of the set of compounds are not restricted; the compounds may be coming from a list of user-defined compounds indicated as canonical SMILES strings (e.g. the source of this list can be the output of a machine learning method, which predicts interacting compounds to the target of interest), drugs from DrugBank or target proteins’ active compounds that are extracted from a reliable compound-target bioactivity measurement dataset, which is a carefully processed and filtered subset of the ChEMBL (v25) database. The output is the 2D embedding of the input set of compounds. By looking at the distribution of compounds as points in this embedding, the user can infer that the compounds that are close to each other may possess similar protein target characteristics. We use the extended connectivity fingerprint (Rogers and Hahn, 2010) with bond diameter four (ECFP4) as the compound descriptor and PCA and t-Stochastic Neighbor Embedding (t-SNE) to generate the 2D embeddings. We also provide a reliable compound-target bioactivity measurement dataset, which is a carefully processed and filtered subset of ChEMBL (v25) database, to be used with iBioProVis.
iBioProVis is an interactive web-based visualization tool and it has advantages when compared with existing studies and tools. First of all, the computation is performed in real-time and visualization can be done for a variety of input set of compounds; i.e. iBioProVis is not restricted in terms of datasets. iBioProVis is a web-based tool and it does not require any installation. Since the local neighborhood is essential for drug discovery and drug repurposing, iBioProVis employs t-SNE. Additionally, a PCA projection of the compounds is provided as well, for comparison. Furthermore, visual analysis of the compound bioactivity space is made possible in several contexts such as target proteins, drugs and drug candidate compounds. The user has the option to select the compounds from a reliable filtered compound-target bioactivity measurement dataset, which is a carefully processed and filtered subset of ChEMBL (v25) database.
2 Materials and methods
iBioProVis has its own bioactivity dataset, processed and filtered from the ChEMBL (v25) database, which originally contains a total of 15 506 670 data points (i.e. bioactivity measurements; Mendez et al., 2019). After the application of several filtering and pre-processing steps (which are outlined in the Supplementary Material and at https://ibpv.kansil.org/dataset) to generate the iBioProVis compound-target protein dataset, the number of bioactivity measurements was reduced to 890 886 which contains 3803 unique target proteins and 581 442 unique compounds. The whole dataset is available for download at https://ibpv.kansil.org/dataset. If the user desires, iBioProVis embedding operations are applied to this filtered dataset. Upon a user submission of target protein identifier(s), iBioProVis first extracts ECFP4 for the compounds of the given target protein(s), to be used as compound feature vectors. The tool then generates a distance matrix for the given compounds, based on the Tanimoto coefficient. The distance matrix becomes the input to the t-SNE algorithm which produces the 2D embeddings of the compound feature vectors (van der Maaten and Hinton, 2008). Finally, these 2D embeddings are plotted as a scatter plot and the point that corresponds to each compound is color-labeled based on the target protein that the compound is reported to bind to. It is also possible to give the representations of drugs or compounds of interest in canonical SMILES notations during the input phase, to obtain their 2D embeddings along with the binders of the given target proteins. Once the embedding process is completed and displayed, the user is able to select a set of compounds on the constructed plot and observe their ChEMBL identifiers and the target proteins that they actively bind to. Several cross-references to widely used and well-known biological databases are also provided so that the user can easily relate the entities and navigate to those databases by clickable links. The cross-referenced databases are UniProt, IntAct, PubChem, DrugBank and Clinical Trials. These steps taken to generate the t-SNE embeddings are given in Algorithm 1 in the Supplementary Material and its expected complexity is O(nlogn) where n is the total number of compounds. PCA projection of the input compounds is provided as well, and the worst-case time complexity is O(n2) when PCA is used. The Bokeh library is employed to generate interactive and user-friendly visualizations (Bokeh Development Team, 2019).
3 Web interface and case studies: β-adrenergic receptors and angiotensin-converting enzyme 2
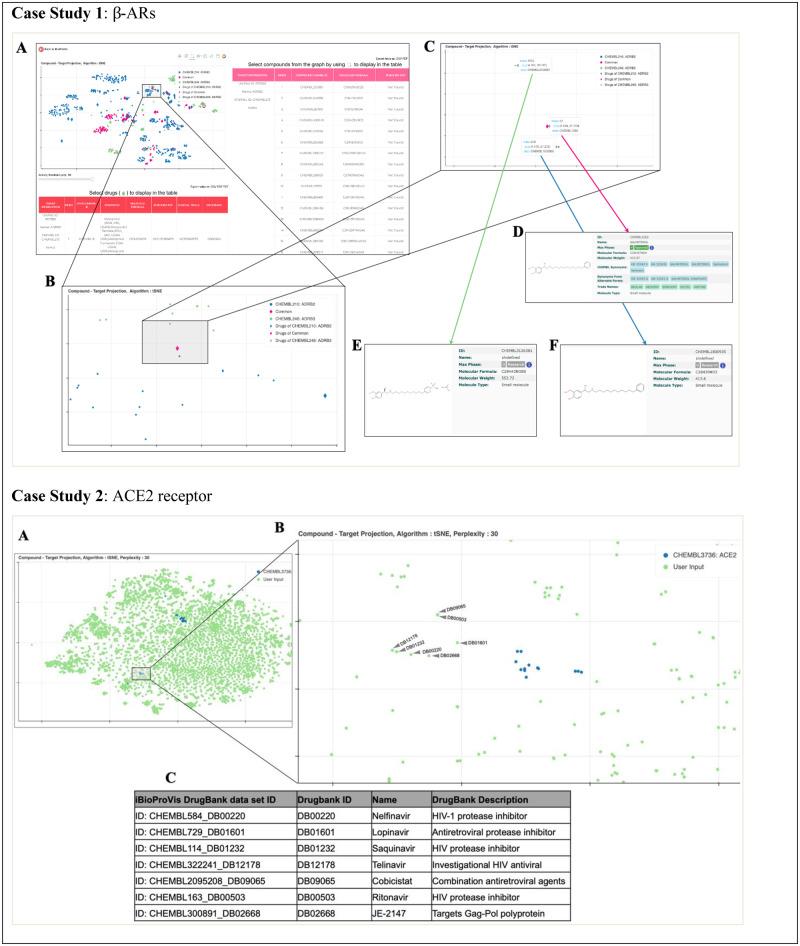
A sample web interface embedding is demonstrated in Figure 1. The active compounds are colored either in blue or in green. Additional user-input compounds are shown in red and drugs (approved and experimental drugs found in the DrugBank database) are represented by diamond shapes. When a user selects a set of compounds, the information about these compounds and their target proteins is shown in two different tables (side table: compounds, bottom table: drugs), where the compounds (rows) are grouped by their respective target proteins. An additional group is created for the user-input compounds since their target information is not presented. This information is shown under the ‘Target Information’ column. iBioProVis provides UniProt protein accessions, gene names and ChEMBL identifiers for the target proteins. In addition to these, compound ChEMBL ID, molecular formulas and PubChem cross-references are given under this table, for the selected compounds. The second (bottom) table is reserved to present only the approved or experimental drugs in the user’s selection. Here, iBioProVis provides drug names, and clinical trial cross-references in addition to the aforementioned information. At the top right side of the plot, there are buttons for easy navigation on the plot such as pan, box zoom, box select, wheel zoom, tap, reset and save. There is a bioactivity value filter at the bottom of the plot, which can be used interactively to remove the compounds that do not satisfy the selected bioactivity threshold [against the corresponding target protein(s)].
Fig. 1.
Comparative interactive embedding output of iBioProVis Case Study 1: β-ARs with β-adrenergic receptors (β-ARs). (A) ADRB2 (blue nodes) and ADRB3 (green nodes) with data tables of drugs (diamonds) and ChEMBL compounds (round circles). Common small molecules acting on β-ARs are represented with magenta-colored nodes. (B and C) Interactive zoom in to visual clusters and the nodes (D–F) Compounds in close vicinity share similar molecular structures. Case Study 2: ACE2 receptor binding 58 compounds (blue nodes) and 6516 small molecule drugs (green nodes) from DrugBank were embedded together (A). Compounds targeting ACE2 are clustered in two distinct groups. Seven antiviral drugs were closely embedded with one of the ACE2 cluster (B and C). The web link to the iBioProVis projection for Case Study 2 is given in the Supplementary Material. (Color version of this figure is available at Bioinformatics online.)
The first use-case example of iBioProVis is demonstrated on β-adrenergic receptors (β-ARs) ADRB2 (Beta-2 adrenergic receptor) and ADRB3 (Beta-3 adrenergic receptor) (Fig. 1). Recent studies have shown that isoform-specific activation of β-ARs is associated with distinct cellular events in various tissues. Hence, targeting β-ARs selectively is important for their molecular pathology-specific actions. Small molecules targeting ADRB2 and ADRB3 with affinities < 10 µM were embedded with iBioProVis’ interactive web interface (Fig. 1A and B). Molecules with similar structures are located in close vicinity, although they are reported to act on different isoforms of β-ARs (blue versus green nodes in Fig. 1C). Molecules shared by both proteins are represented by magenta-colored nodes. As seen in Figure 1, Salmeterol (CHEMBL1263) targets both isoforms of β-ARs, whereas compound CHEMBL1800935 targets the ADRB2 protein and CHEMBL3126381 targets the ADRB3 protein. All three compounds possess very similar structure. This specific example clearly demonstrates that although CHEMBL1800935 has been reported to act on ARDB2, iBioProVis embedding indicates that this compound may also act on ARDB3. Hence, by examining the embedding by iBioProVis, one can hypothesize ARDB3 as a new target of CHEMBL1800935. The same argument is valid for compound CHEMBL3126381 which acts on ARDB2 but may also act on ARDB3.
We selected the angiotensin-converting enzyme 2 (ACE2) receptor, which has been reported as the SARS-CoV-2 spike protein receptor for viral entry, as the second use-case demonstration (Hoffmann et al., 2020). ACE2 receptor (CHEMBL3736) is associated with 58 compounds which satisfy the iBioProVis bioactivity dataset criteria (Supplementary Material). The 58 compounds were embedded together with 6516 small molecule drugs from the DrugBank database (Fig. 1, Case Study 2; Wishart et al., 2018). There are two significantly separated clusters of ACE2 receptor binding compounds. Although compounds of the cluster at the upper right side are defined as thiol-based ACE2 inhibitors, the cluster of compounds in Figure 1, Case Study 2, panel B are from another study which designed ACE2 peptidase activity inhibitors (Deatonn et al., 2008; Mores et al., 2008). These experimental inhibitors were closely embedded with seven antiviral-protease inhibitors among which Lopivanir and Ritonavir are currently under clinical trials for use against SARS-CoV-2 (Cao et al., 2020; Harrison, 2020).
4 Conclusion
iBioProVis is an unprecedented tool that can be utilized for virtual screening and for chemical genomics. It can be used for several purposes, including the investigation and analysis of how active compounds of different target proteins are distributed on a 2D space, as well as the prediction of bioactivity profiles for new or uncharacterized compounds, based on the features of compounds with known bioactivity information. Furthermore, it may provide insight to drug repurposing studies by identifying the compounds that are embedded close to an approved drug, especially when those compounds are known binders of a different target protein.
Funding
This work was supported by Scientific and Technological Council of Turkey (TÜBİTAK) Grant number: EEEAG-116E930.
Conflict of Interest: none declared.
Supplementary Material
References
- Awale M., Raymond J.L. (2016) Web-based 3D-visualization of the DrugBank chemical space. J. Cheminformatics, 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokeh Development Team. (2019) Bokeh: Python library for interactive visualization. http://www.bokeh.pydata.org.
- Cao M.D. et al. (2020) A trial of Lopinavir-Ritonavir in adults hospitalized with severe Coivd-19. N. Engl. J. Med., 382, doi:10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton D.N. et al. (2008) Thiol-based angiotensin-converting enzyme 2 inhibitors: P1 modifications for the exploration of the S1 subsite. Bioorg. Med. Chem. Lett., 18, 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar H.A. et al. (2015) Chemical data visualization and analysis with incremental generative topographic mapping: big data challenge. J. Chem. Inf. Model, 55, 84–94. [DOI] [PubMed] [Google Scholar]
- Gütlein M. et al. (2012) CheS-Mapper—chemical space mapping and visualization in 3D. J. Cheminformatics, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C. (2020) Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol., 1787–1799, doi:10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- Hoffmann M. et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A.P. et al. (2019) Drug discovery maps, a machine learning model that visualizes and predicts kinome-inhibitor interaction landscapes. J. Chem. Inf. Model, 59, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlov D.S. et al. (2019) Chemical space exploration guided by deep neural networks. RSC Advances, 9, 5151–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez D. et al. (2019) ChEMBL—towards direct deposition of bioassay data. Nucleic Acids Res., 47, D930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mores A. et al. (2008) Development of potent and selective phosphinic peptide inhibitors of angiotensin-converting enzyme. J. Med. Chem., 51, 2216–2226. [DOI] [PubMed] [Google Scholar]
- Rifaioglu A.S. et al. (2019) Recent applications of deep learning and machine intelligence on in-silico drug discovery. Brief. Bioinform., 20, 1878–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D., Hahn M. (2010) Extended-connectivity fingerprints. J. Chem. Inf. Model, 50, 742–754. [DOI] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G.E. (2008) Visualizing data using t-SNE. J. Mach. Learn. Res., 9, 2579–2605. [Google Scholar]
- Wishart D.S. et al. (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res., 46, 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.