Abstract
We present a microsphere-based flow cytometry assay that quantifies the ability of plasma to inhibit the binding of spike protein to angiotensin-converting enzyme 2. Plasma from 22 patients who had recovered from mild coronavirus disease 2019 (COVID-19) and expressed anti–spike protein trimer immunoglobulin G inhibited angiotensin-converting enzyme 2–spike protein binding to a greater degree than controls. The degree of inhibition was correlated with anti–spike protein immunoglobulin G levels, neutralizing titers in a pseudotyped lentiviral assay, and the presence of fever during illness. This inhibition assay may be broadly useful to quantify the functional antibody response of patients recovered from COVID-19 or vaccine recipients in a cell-free assay system.
Keywords: serology, COVID-19, SARS-COV-2, ACE2, Spike protein, binding inhibition, neutralization
Using a rapid, microsphere-based assay, the authors quantified the ability of coronavirus disease 2019 convalescent plasma to inhibit binding of severe acute respiratory syndrome coronavirus 2 spike protein to its receptor, angiotensin-converting enzyme, demonstrating inhibition in >90% of recovered patients.
In late 2019 a novel severe acute respiratory syndrome (SARS)–like coronavirus, SARS coronavirus 2 (SARS-CoV-2), emerged in Wuhan, China [1]. The disease it causes, coronavirus disease 2019 (COVID-19), was declared a pandemic by the World Health Organization in March 2020 [2]. The cryo-electron microscopic structure of the spike glycoprotein, a viral surface protein that mediates cell entry of coronaviruses, was revealed by 2 group of investigators [3, 4] as a trimeric structure with ≤1 of 3 receptor-binding domains (RBDs) in the “up” state, capable of binding to its target. Similar to the 2003 SARS-CoV-1 virus, angiotensin-converting enzyme 2 (ACE2) serves as the receptor necessary and sufficient for infection of the target cell [3–7].
Nearly all patients who recover from SARS-CoV-2 infection produce immunoglobulin (Ig) M and IgG antibodies against the spike protein [7–9], and a large number of serological tests have been produced and marketed. However, it is unclear how effective the detected antibodies are at neutralizing viral activity. For example, a study measuring in vitro inhibition of ACE2-RBD binding with an enzyme-linked immunosorbent assay (ELISA) showed that only 3 of 26 recovered patients (11.5%) had strongly inhibited binding [7], while a larger study using pseudotyped lentivirus showed significant neutralization of infectivity in 165 of 175 recovered patients (94.3%) [9]. Indeed, human monoclonal antibodies that neutralize SARS-CoV-2 infectivity have been showed to bind epitopes both within [7] and outside the RBD [10], so it is unclear whether RBD-based measurements capture the full repertoire of inhibition of viral infectivity. Pseudoviruses that use the native spike protein to infect cells appear to identify antibodies that neutralize viral entry [9], but these assays are technically demanding, require specialized biosafety facilities, and may be difficult to scale up for population-level testing.
The objective of the current study was to design an overnight, cell-free assay to quantify a plasma sample’s ability to inhibit the binding of ACE2 to a recombinant COVID-19 spike protein. The assay is based on immunoprecipitation detected by flow cytometry (IP-FCM) technology, a highly sensitive and reagent-efficient method for detecting protein-protein interactions using minimal amounts of biomaterial [11, 12]. We found that though only a minority of persons who have recovered from symptoms of COVID-19 produce antibodies that inhibit the RBD binding to ACE2 in vitro, almost all (22 of 24) produce antibodies that potently inhibit the prefusion trimer binding to ACE2. Our results provide a new, relatively rapid, and high-throughput method to quantify circulating levels of functional anti–SARS-CoV-2 antibodies, and they suggest that the entire spike protein as opposed to the RBD should be used when characterizing SARS-CoV-2 immunity.
METHODS
Human Samples
Negative controls consisted of banked samples collected from healthy adults before January 2020. Controls ranged in age from 19 to 66 years (median, 37 years) and 18 of 30 were female (60%). Plasma samples from adults recovered from COVID-19 came from the Seattle Children’s Research Institute SARS2 Recovered Cohort. All patients reported testing polymerase chain reaction (PCR) positive for SARS-CoV-2. The day of sample collection ranged from 14 to 73 days after symptom onset, with a median of 36 days. All SARS-CoV-2 infections were symptomatic, ranging from very mild to moderate illness; no patients required hospitalization or supplemental oxygen. Of 24 patients, 12 experienced fever and 16 experienced cough. All patients provided informed consent, and parent studies were approved by the Fred Hutchison Cancer Research Center or Seattle Children’s Hospital institutional review board.
Protein Purification
The RBD construct (AA 319–541; UniProt P0DTC2) was cloned into the pCDNA3.4 protein expression vector with an 8xHis tag and tandem AviTag. The SARS-CoV-2 trimer [3] and ACE2 constructs were generously provided by Jason McLellan (University of Texas at Austin) and Barney Graham (National Institutes of Health) [3]. Proteins were expressed in 293F cells in antibiotic-free, serum-free media, as described elsewhere [13]. Briefly, DNA was transfected using PEI Max and grown for 5 days at 37°C and 5% carbon dioxide. The recombinant proteins were purified by NiNTA affinity chromatography and polished by size exclusion chromatography for size on a Superdex 200 16/600.
Magnetic Bead Coupling
MagPlex microspheres (Luminex MC100XX-01, where XX encodes the bead region; all regions are compatible) were coupled as described elsewhere [14]. Briefly, 250 μL of magnetic beads were magnetically separated using a magnetic tube rack (New England Biolabs; catalog no. S1506S) for 60 seconds and washed 3 times with 250 μL of 2-(N-morpholino)ethanesulfonic acid (MES) buffer (50 mmol/L MES, pH 6.0, with 1 mmol/L ethylenediaminetetraacetic acid, in double-distilled water; stored at 4°C and used at room temperature [RT]).
Magnetic beads were resuspended in 200 μL of MES buffer and 25 μL of freshly prepared 50 mg/mL sulfo-N-hydroxysulfosuccinimide (NHS) (ThermoScientific; no 24510) in MES buffer was added to the beads and briefly vortexed. Then 25 μL of 50 mg/mL freshly made 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (Pierce) in MES buffer was added, and the tube was quickly vortexed. The tube was covered to protect from light and shaken at 1000 rpm for 20 minutes at RT on a pulsing vortexer.
Beads were then washed twice in phosphate-buffered saline (PBS), and incubated with 25 ug of ACE2, trimer, or RBD protein in 250 μL of PBS, on a 1000-rpm vortexer for 2 hours at RT protected from light. Beads were then washed in PBS and quenched in 750 μL of blocking storage solution (1% bovine serum albumin [BSA] in PBS and 0.01% sodium azide) for 30 minutes at RT, on a 1000-rpm vortexer protected from light. Finally, beads were stored in 100 μL of blocking storage solution at 4°C until use.
IgG Detection with IP-FCM
IP-FCM was performed as described elsewhere [11, 12]. Plasma samples from both SARS-CoV-2–positive and SARS-CoV-2–negative adults was heated to 56°C for 1 hour and then spun at 13 000g for 10 minutes at 4°C. The supernatant was diluted (1:1000 unless otherwise indicated) in cold flow cytometry immunoprecipitation (Fly-P) Buffer (50 mmol/L Tris [pH 7.4], 100 mmol/L sodium chloride, 1% BSA, and 0.01% sodium azide) and distributed into wells of a 96-well plate at a volume of 50 μL per well, in duplicate. To each well, approximately 2.5 × 103 magnetic beads were added. All plasma samples were also run in parallel using BSA-coupled magnetic beads to determine the baseline nonspecific signal generated from each sample.
The wells of the 96-well plate were then capped, and the plate left to rotate at RT for 2 hours. A magnetic plate washer was used to wash the beads 3 times (Bio-Plex Pro Wash Station; BioRad) before incubation with 50 μL of 1:200 anti–human IgG antibodies conjugated to phycoerythrin (PE) (Jackson ImmunoResearch; 709-116-149, lot 145536) in Fly-P Buffer protected from light for 30 minutes at RT. The plate was washed as described above, and samples were resuspended in 50 μL of cold Fly-P Buffer. Beads were then read on an Acea Novocyte flow cytometer with the following gating strategy: gating of beads using forward scatter height versus side scatter height, eliminating doublets using forward scatter height versus forward scatter area, and detecting PE fluorescence using fluorescence channel 2 (488-nm excitation; 572/28-nm detection). Background-subtracted median fluorescence intensity (MFI) was calculated for each individual by subtracting the BSA-coupled bead MFI from the RBD- or trimer-coupled bead MFI.
ACE2–Spike Protein Inhibition Assay
RBD or trimer protein was biotinylated by adding 1 μL of freshly-dissolved sulfo-NHS-Biotin (ThermoScientific; 21217) for 30 minutes on ice. The reaction was quenched with 100 mmol/L Tris-hydrochloride and excess biotin was removed by 3 washes in a 10K molecular weight cutoff Amicon Ultra spin filter (Millipore). Biotinylated RBD or trimer protein was added to 2.5 × 103 magnetic beads in a total volume of 50 μL, in duplicate. To measure inhibition of binding, soluble unlabeled ACE2 or diluted, heat-inactivated plasma samples were added to each well, maintaining a final reaction volume of 50 μL. Plasma was diluted 1:50 in FlyP buffer unless otherwise indicated.
Each well of the plate was capped and mixed end-over-end at 4°C overnight. The next day the plate was washed as described above and incubated with 50 μL of 1:200 streptavidin-conjugated PE (BioLegend; 405204) in Fly-P Buffer protected from light for 30 minutes at RT. The plate was washed again, and samples were resuspended in 50 μL of cold Fly-P Buffer for detection on the flow cytometer, with gating as described above. Data were expressed either as MFI or as the percentage of inhibition, calculated as 100 × (1 − [MFI of sample]/[MFI of wells without inhibitor added]).
50% Inhibitory Concentration Determination using Pseudotyped Lentivirus
Full-length SARS-CoV-2 S protein was used to create pseudo-typed lentivirus as described elsewhere [15], with slight modifications. Plasmids encoding S and human immunodeficiency virus type 1 structural genes were cotransfected into 293T cells using PEI Max and grown at 32°C for 3 days before harvesting supernatants. Next, 293T cells stably expressing ACE2 were plated in 96-well plates at a density of 104 per well 24 hours before viral challenge, and polybrene was added at 2 ug/mL for 30 minutes before viral challenge.
Heat-inactivated plasma samples from COVID-19–positive patients or pre–COVID-19 controls were serially diluted from 1:50–1:109 350 and coincubated with virus for 60 minutes before plating on cells. The plates were incubated at 37°C for 65 hours, the media removed, and 100 μl of Steady Glo (Promega) added. Relative light unit (RLU) values were read on a luminometer, and the percentage of neutralization was calculated as (RLUvirus − RLUvirus+plasma)/(RLUvirus) × 100. We selected a subset of total samples to span the range of inhibition for both COVID-19–positive and control samples, and all samples analyzed are reported. The 50% inhibitory concentration (IC50) values were extrapolated from a nonlinear regression model and represent the reciprocal plasma dilution at which 50% viral inhibition was recorded.
IgG Depletion Assay
Patient plasma was diluted 1:50 in Fly-P Buffer and incubated with 2 μL of RBD- or trimer-coupled carboxy-modified latex beads (CML; see Supplementary Material) overnight at 4°C. The next day, the beads were pelleted at 13 000 rpm for 1 minute, removed, and discarded. A 2-μL aliquot of plasma was reserved, and 2 μL of new, RBD-or trimer-coupled CML beads were added. This process was repeated for a total of 3 overnight depletions. On the fourth day, 25 μL of the RBD- or trimer-depleted plasma was run on the ACE2-spike binding assay, as described above.
Statistical Analyses
Technical replicates were averaged and data were imported with Prism 8.0 software (GraphPad). Comparisons between 2 groups were made using 2-tailed t tests, and among 3 groups with 1-way analysis of variance (ANOVA) followed by post hoc tests comparing all columns, corrected for multiple comparisons by the Tukey method. Correlations and half-maximal effective concentrations were determined using simple linear regressions or 4-parameter logistic regressions with default settings, and r2 values are reported.
RESULTS
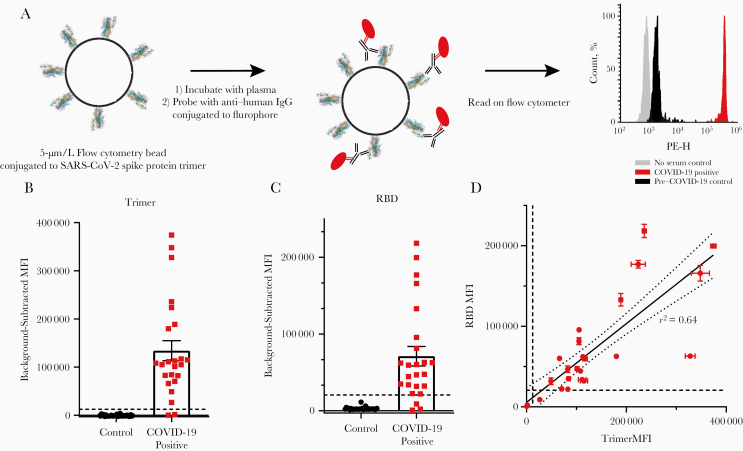
Luminex MagPlex microspheres coupled to either the recombinant RBD fragment [16] or a spike protein trimer construct [3] were incubated with plasma specimens, and then with anti–human IgG antibodies conjugated to PE (Figure 1A). SARS-CoV-2 antibodies were stringently defined as “positive” at a median fluorescence intensity (MFI) greater than the highest control value plus 5 standard deviations (dashed lines in Figure 1B and 1C). Using this definition, 22 of 24 COVID-19–positive samples (91.6%) were classified as “seropositive” for trimer reactivity, and 2 of 24 (87.5%) as seropositive for RBD. There was a moderate correlation between RBD and trimer immunoreactivity (Figure 1D; r2 = 0.64). During assay development, we performed experiments using CML microspheres on a subset of the total plasma samples. We present those data and CML-specific methods in Supplementary Figure 1 to compare two different microsphere types, and to note the good correlation between trimer (r2 = 0.85) and RBD (r2 = 0.75) values for the 12 samples that were measured using both microsphere types (Supplementary Figure 1F and 1G). While we chose to use MagPlex beads owing to higher average fluorescence values, lower nonspecific binding, and the availability of magnetic plate washing, CML beads are also a viable, less expensive option.
Figure 1.
Detection of anti–severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immunoglobulin G (IgG) using receptor-binding domain (RBD) and trimer constructs. A, Graphic representation of the methods. Phycoerythrin height (PE-H) indicates fluorescence intensity. B, C, IgG levels in plasma samples from 24 coronavirus disease 2019 (COVID-19)–positive and 30 pre–COVID-19 controls were measured using trimer-conjugated (B) or RBD-conjugated (C) microspheres. Dashed lines represent the cutoff for positive sample designation, calculated as maximum control value plus 5 standard deviations, at a median fluorescence intensity (MFI) of 12 432 for trimer and 27 119 for RBD. D, The MFIs of IgG measured on the trimer and RBD assays were significantly correlated. Dashed lines represent cutoffs for positive designation; the solid line represent a linear regression, with 95% confidence intervals represented by dotted lines (r2 = 0.64); the slope is significantly nonzero (F[1,46] = 82.4; P < .001).
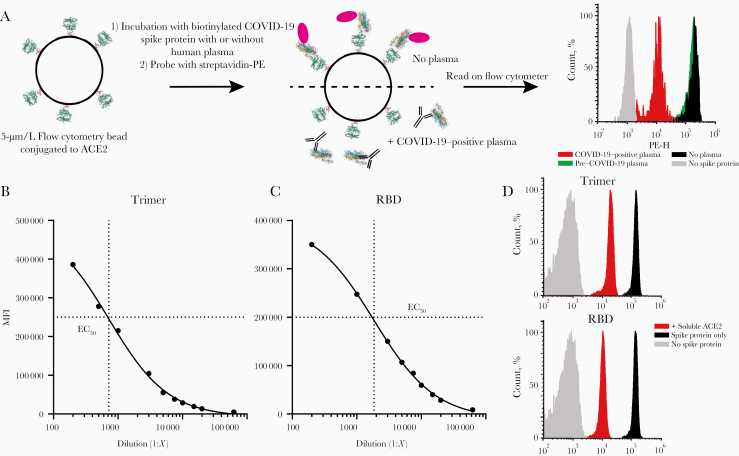
To quantify the ability of convalescent plasma to inhibit the spike protein binding to ACE2, we coupled human ACE2 protein to MagPlex microspheres incubated with biotinylated RBD or trimer, followed by incubation with streptavidin-PE (Figure 2A). Both trimer and RBD demonstrated strong binding to ACE2 (Figure 2B and 2C), and the half-maximal effective concentration was used to determine the dilution of spike proteins added to the binding inhibition assay. As a specificity control, unlabeled human ACE2 inhibited binding of both RBD and trimer by >90% (Figure 2D). After optimizing the plasma dilution (Supplementary Figure 2C and 2D), we evaluated the same plasma samples for their ability to inhibit the ACE2-spike interaction. Analyzed as a group, the MFI of control plasma samples did not differ significantly from that of no-plasma controls in the trimer or RBD assays (left graphs in Figure 3A and 3B, respectively), while COVID-19–positive samples significantly inhibited binding (trimer ANOVA F[2,57] = 52.4, P < .001, COVID-19–positive vs no plasma and pre–COVID-19 P < .001 by Tukey post hoc test; RBD ANOVA F[2,57] = 5.538, P = .006, COVID-19–positive vs pre–COVID-19 P = .004 by Tukey post hoc test). Similarly, when data were expressed as the percentage of inhibition, COVID-19–positive samples produced greater inhibition than pre–COVID-19 samples for both trimer and RBD (P < .001 and P = .002, respectively, by 2-tailed t test; Figure 3A and 3B, right graphs).
Figure 2.
Development of the angiotensin-converting enzyme 2 (ACE2)–spike protein binding inhibition assay. A, Graphic representation of the methods. Phycoerythrin (PE) height (PE-H) indicates fluorescence intensity. B, C, Half-maximal effective concentration (EC50) of the ACE2-trimer (B) or ACE2–receptor-binding domain (RBD) (C) binding reaction was determined by serial dilution of the trimer or RBD fragment. D, Soluble, unlabeled ACE2 was added to the trimer or RBD binding assay (red histograms), which resulted in a >90% reduction in the median fluorescence intensity (MFI) compared with binding assays without soluble ACE2 (black histograms). RBD or trimer was omitted to establish background MFI (gray histograms).
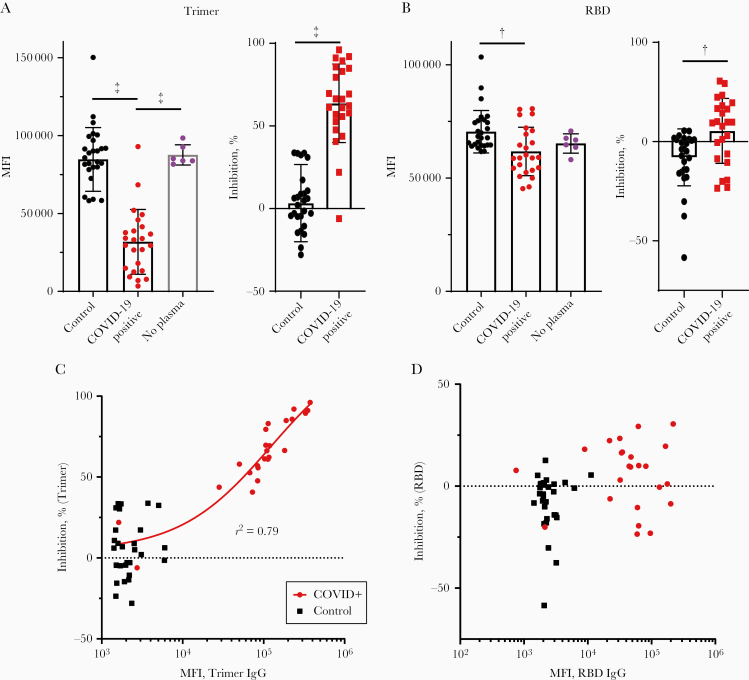
Figure 3.
Inhibition of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein and angiotensin-converting enzyme 2 (ACE2) binding by recovered patient plasma. A, Compared with a no-plasma control (purple) or pre–coronavirus disease 2019 (COVID-19) control plasma (black), COVID-19–positive plasma significantly inhibited trimer-ACE2 binding. The same data are expressed as median fluorescence intensity (MFI) (left) or the percentages inhibition against the no-plasma control mean (right). ‡P < .001. B, Compared with pre–COVID-2019 plasma (black) COVID-19–positive plasma significantly inhibited receptor-binding domain (RBD)–ACE2 binding (but not compared with a no-plasma control (purple)). The same data are expressed as MFI (left) or percentage of inhibition versus the no-plasma control mean (right). †P < .005. C, Individual values for each sample obtained from the anti-trimer immunoglobulin G (IgG) assay (MFI values) plotted against percentage inhibition of the trimer-ACE2 binding inhibition assay. A regression line for all COVID-19–positive samples is shown. D, Similar to C, with MFI from the anti-RBD IgG assay MFI plotted against percentage inhibition on the RBD-ACE2 binding inhibition assay for all samples.
However, when data were examined by individual, important differences emerge between the trimer- and RBD-based assays. For the trimer inhibition assay, all 22 plasma samples that tested positive for anti-trimer IgG (as defined above) showed greater inhibition than any of the negative control samples (Figure 3A). Conversely, in the RBD assay, only 9 of 24 samples showed greater inhibition than the highest negative control, although 21 of 24 were seropositive (Figure 3B). Moreover, in the trimer assay there was a strong correlation between the amount of anti-trimer IgG and the percentage inhibition among COVID-19–positive (r2 = 0.79) but not negative control (r2 = 0.04) samples (Figure 3C). There was no such correlation in the RBD assay (COVID-19–positive samples, r2 = 0.047; negative controls, r2 = 0.034) (Figure 3D). There was also no correlation between the percentage inhibition of plasma samples in the RBD and trimer assays (COVID-19–positive samples, r2 = 0.036; pre–COVID-19 samples, r2 = 0.011). Experiments performed using CML microspheres during assay development using a subset of the total plasma showed similar results, although with a smaller dynamic range (Supplementary Figure 2), and there was good correlation between trimer values for the 12 samples that were measured using both microsphere types (r2 = 0.85; Supplementary Figure 2K).
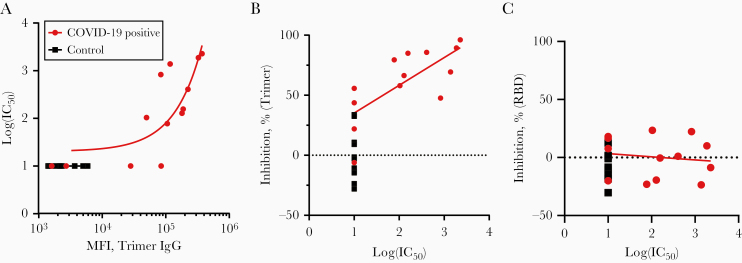
The trimer has several modifications, including a fibritin trimerization domain and elimination of the Furin cleavage site [3, 17], which could potentially induce artifacts. We selected a subset of COVID-19–positive samples that spanned the range of the trimer inhibition assay and performed a neutralizing titer assay using a pseudotyped lentivirus to determine an IC50 for each sample. Nine of 13 COVID-19–positive samples (9 of 11 IgG-positive samples) showed IC50 values greater than the minimum detectable value (1:50), which was correlated with anti-trimer IgG levels (Figure 4A; r2 = 0.60). Among all COVID-19–positive samples, there was a moderate correlation between IC50 value and percentage of inhibition in the trimer inhibition assay (Figure 4B; r2 = 0.52); there was no correlation for the RBD inhibition assay (Figure 4C; r2 = 0.02). Samples with trimer inhibition <60% showed a range of IC50 values (<1:50 to 1:830), while all samples with inhibition >60% showed IC50 plasma dilutions of at least 1:77, to a maximum of 1:2277. It is unclear whether the differences between the 2 data sets represent limitations of the trimer inhibition assay, the lentiviral pseudovirus assay, or both. However, these data demonstrate that, at least for strongly inhibiting samples, there is moderate agreement between the cell-based and microsphere-based assays.
Figure 4.
Comparison of the angiotensin-converting enzyme 2 (ACE2)–trimer binding inhibition assay with a pseudovirus neutralization assay. A, The log(50% inhibitory concentration [IC50]) value obtained from a lentiviral pseudovirus assay for 13 coronavirus disease 2019 (COVID-19)–positive and 12 control samples is plotted against anti-trimer immunoglobulin G (IgG) data (median fluorescence intensity [MFI]). B, C, A regression line for all COVID-19–positive samples is shown (r2 = 0.60). Percentage inhibition on the trimer-ACE2 binding inhibition assay (B) or receptor-binding domain (RBD)–ACE2 binding inhibition assay (C) is plotted against log(IC50) values from the lentiviral pseudovirus assay for 13 COVID-19–positive and 12 control samples. A regression line for all COVID-19–positive samples is shown (trimer, r2 = 0.52; RBD, r2 = 0.02).
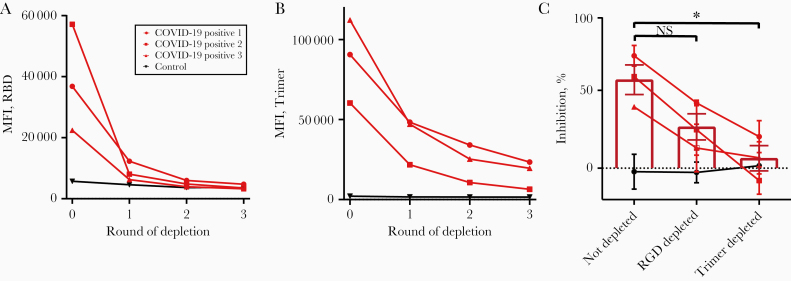
To determine whether inhibition of binding was caused by antibodies binding within or outside the RBD, we next depleted the plasma of 1 negative control and 3 COVID-19–positive samples by incubating with large numbers of trimer- or RBD-conjugated CML beads for 3 consecutive overnight incubations until the IgG levels detected in the plasma plateaued (Figure 5A and 5B), and we performed the trimer-ACE2 inhibition assay on the depleted plasma. Inhibition was partially (although nonsignificantly) reduced by RBD depletion, and it was reduced to a greater degree by trimer depletion (Figure 5C) (ANOVA F[2,6] = 8.51, P = .02, not depleted vs RBD depleted P = .11, not depleted vs trimer depleted P = .02, RBD depleted vs trimer depleted P = .30 by Tukey post hoc test). RBD depletion was more efficient than trimer depletion in terms of reduction in detectable anti-SARS-CoV-2 IgG (RBD average depleted MFI, 3845 vs 3629 for control; trimer average depleted MFI, 16 481 vs 1545 for control) (Figure 5A and 5B), but trimer depletion was more effective at removing the neutralizing capacity of the plasma. These data suggest that spike protein trimer neutralizing antibodies bind both within and outside the RBD.
Figure 5.
Depletion of receptor-binding domain (RBD)–binding antibodies only partially lowers angiotensin-converting enzyme 2 (ACE2)–trimer inhibition. A, B, Plasma samples from 3 coronavirus disease 2019 (COVID-19)–positive and 1 control individual were depleted of RBD-binding (A) or trimer-binding I (B) immunoglobulins by incubation with 3 rounds of RBD- or trimer-conjugated carboxy-modified latex (CML) beads. After each round, a sample was removed and assayed for remaining anti-RBD (A) or anti-trimer immunoglobulin G (IgG) (B). C, COVID-19–positive, not-depleted plasma inhibited trimer-ACE2 binding. Anti-RBD-depleted plasma was less efficient at inhibiting trimer-ACE2 binding, although the difference was not significant by analysis of variance (ANOVA). Anti-trimer–depleted plasma showed a significantly reduced ability to inhibit ACE2-trimer binding, with the mean overlapping with control levels (ANOVA F[2,6] = 8.51, P = .02, not depleted vs RBD depleted P = .11, not significant [NS]; not depleted vs trimer depleted). *P = .02; RBD depleted vs trimer depleted P = .30 by Tukey post hoc test.
COVID-19–positive patients were between 28 and 63 years of age (median, 42 years of age) and were 71% female (17 of 24 patients). Age was associated with a trend toward increased anti-trimer IgG and inhibition (per-year MFI difference, 3816 [P = .09]; inhibition difference, 1% [P = .07]), although this may have been driven by the 2 patients negative for anti-SARS2 IgG, who were both relatively young (aged 28 and 41 years). Sex, days since symptom onset, and cough were not associated with anti-trimer IgG or inhibition. In contrast, fever was strongly associated with both anti-trimer IgG and inhibition in both unadjusted (MFI difference, 103 982 [P = .007]; inhibition difference, 20% [P = .04]) and adjusted models that accounted for patient age, sex, and days from symptom onset (adjusted MFI difference, 103 530 [P = .01]; adjusted inhibition difference, 20% [P = .03]). Both anti-SARS2 IgG–negative patients had very mild disease without fever, and follow-up testing using the commercial Cellex platform as well as laboratory-developed ELISAs using spike protein and nucleoprotein (n = 3 laboratories) failed to detect anti-SARS2 antibodies.
Discussion
The acquisition of antiviral immunity to SARS-CoV-2 will be a critical prerequisite for a return to normalcy from the current pandemic. As of this writing, it is still unclear whether infection or vaccination will lead to robust, long-lasting immunity [18]. While some early data suggested ineffective immune responses, such as low levels of ACE2-RBD inhibition by convalescent serum in an ELISA-style binding assay [7] or the detection of viral RNA in recovered patients [19], more recent data suggest effective antiviral antibody activity in the majority of patients. For example, serum from 94% of recovered patients effectively inhibited pseudotyped lentiviral infection of susceptible cells [9], similar to our rates; 4 rhesus macaques experimentally infected with SARS-CoV-2 were resistant to reinfection [20]; and a recent report suggests that the viral RNA detected in recovered patients is not associated with active infection or the ability to transmit COVID-19 [21]. Our data, demonstrating in vitro inhibition of ACE2-trimer binding in 92% of recovered patients, adds to this growing literature collectively suggesting that infection with SARS-CoV-2 results in robust immunity, at least in the short term.
Several limitations to our study should be noted. We focus solely on binding of ACE2 to the spike protein and ignore other potential antigens or other immune mechanisms such as inhibition of protease cleavage that might prevent viral entry into the cell. While the correlation between microsphere-based ACE2-trimer binding inhibition and cell-based pseudovirus neutralization was moderate, 2 samples that showed <60% binding inhibition showed undetectable neutralization. The relationships between microsphere inhibition, pseudotyped lentivirus IC50, and live SARS-CoV-2 virus IC50s require further study. Moreover, it is unclear what level of inhibition (in psuedovirus or bead-based assays) is correlated with functional resistance to reinfection. Follow-up studies tracking inhibition over time while simultaneously monitoring patients for reinfection will be necessary, given the ethical impossibility of experimental human inoculation. Finally, our SARS-CoV-2 PCR-positive, antibody-negative samples leave many questions. For example, did they clear the virus through mechanisms beyond our detection, such as antibodies targeted to alternative viral proteins or non–B-cell–dependent mechanisms? Or were their PCR results false-positives? Most importantly, are they susceptible to future infections? Follow-up with these or similar individuals will be important.
In conclusion, the bead-based ACE2-trimer binding inhibition assay presented here could be broadly useful in the settings of routine clinical evaluation of functional immunity in recovered patients, selecting the most potent postconvalescent plasma for use as therapy, and evaluating the functionality of anti-SARS-CoV-2 antibodies produced in response to experimental vaccines currently under development. In light of our results, we propose that future studies should favor trimer-style constructs for serological assays.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Jason McLellan and Barney Graham for their generous gift of the severe acute respiratory syndrome coronavirus 2 recombinant trimer and human angiotensin-converting enzyme 2 expression constructs; Lee Nelson of the Fred Hutchinson Cancer Research Center for providing negative control samples; Jesse Bloom of the Fred Hutchinson Cancer Research Center for providing the lentiviral pseudovirus plasmids; Juliane Gust for technical assistance; the scientists and support staff at Seattle Children’s Research Institute who enabled coronavirus disease 2019 (COVID-19) research to continue during the pandemic, especially Sean Sandin and Marnie Chinn; and the recovered COVID-19–positive plasma donors.
Author contributions. S. E. P. S. conceived the study. E. P. G., W. E. H., D. N. S., and S. E. P. S. planned experiments. E. P. G. performed all microsphere-based experiments. N. D., O. T., and V.V. designed, expressed, and purified recombinant proteins and performed QC and antigenic properties, and conducted neutralization assays. W. E. H., Y. J., and L. F. conducted the Seattle Children’s SARS2 Recovered Cohort sample collection and provided samples and deidentified patient data. E. P. G., W. E. H., D. N. S., and S. E. P. S. analyzed the data. E. P. G., W. E. H., L. F., and S. E. P. S. wrote the manuscript, and all authors read and approved the manuscript.
Financial support. This work was supported by the University of Washington Institute for Translational Heath Sciences (COVID-19 research award via grant UL1 TR002319), the Research Integration Hub at Seattle Children’s Research Institute (COVID-19 award to S. E. P. S.), the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (grant K08 AI135072 and grant R01 AI140951 to D. N. S.), and the Burroughs Wellcome Fund (CAMS 1017213 grant to W. E. H.).
Potential conflicts of interest. A provisional patent application has been filed by Seattle Children’s Research Institute on the assays reported in this article. The authors report no other potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed 2020; 91:157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020; 181:281–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 2020; 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020; 5:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X, Li R, Pan Z, et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol 2020; 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 1–4. [DOI] [PubMed] [Google Scholar]
- 9. Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv [Preprint]. 20 April 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.03.30.20047365v2. [Google Scholar]
- 10. Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 2020; 11:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith SE, Bida AT, Davis TR, et al. IP-FCM measures physiologic protein-protein interactions modulated by signal transduction and small-molecule drug inhibition. PLoS One 2012; 7:e45722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis TR, Schrum AG. IP-FCM: immunoprecipitation detected by flow cytometry. J Vis Exp 2010; e2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fisher BS, Dambrauskas N, Trakhimets O, et al. Oral immunization with HIV-1 envelope SOSIP trimers elicits systemic immune responses and cross-reactive anti-V1V2 antibodies in non-human primates. PLoS One 2020; 15:e0233577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown EA, Neier SC, Neuhauser C, Schrum AG, Smith SEP. Quantification of protein interaction network dynamics using multiplexed co-immunoprecipitation. J Vis Exp 2019; e60029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crawford KHD, Eguia R, Dingens AS, et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 2020; 12 : 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pallesen J, Wang N, Corbett KS, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 2017; 114:E7348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirkcaldy RD, King BA, Brooks JT. COVID-19 and postinfection immunity: limited evidence, many remaining questions. JAMA 2020; 323:2245–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xing Y, Mo P, Xiao Y, Zhao O, Zhang Y, Wang F. Post-discharge surveillance and positive virus detection in two medical staff recovered from coronavirus disease 2019 (COVID-19), China, January to February 2020. Eurosurveillance 2020; 25:2000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bao L, Deng W, Gao H, et al. Lack of reinfection in rhesus macaques infected with SARS-CoV-2. bioRxiv [Preprint]. 1 May 2020. Available from https://www.biorxiv.org/content/10.1101/2020.03.13.990226v2.article-info. [Google Scholar]
- 21. Roy S. COVID-19 reinfection: myth or truth? Sn Compr Clin Med 2020; 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.