Motif analysis on evolutionarily conserved regions in laminin enabled the discovery of novel ligands for hydrogel development.
Abstract
Biologically active ligands (e.g., RGDS from fibronectin) play critical roles in the development of chemically defined biomaterials. However, recent decades have shown only limited progress in discovering novel extracellular matrix–protein–derived ligands for translational applications. Through motif analysis of evolutionarily conserved RGD-containing regions in laminin (LM) and peptide-functionalized hydrogel microarray screening, we identified a peptide (a1) that showed superior supports for endothelial cell (EC) functions. Mechanistic studies attributed the results to the capacity of a1 engaging both LM- and Fn-binding integrins. RNA sequencing of ECs in a1-functionalized hydrogels showed ~60% similarities with Matrigel in “vasculature development” gene ontology terms. Vasculogenesis assays revealed the capacity of a1-formulated hydrogels to improve EC network formation. Injectable alginates functionalized with a1 and MMPQK (a vascular endothelial growth factor–mimetic peptide with a matrix metalloproteinase–degradable linker) increased blood perfusion and functional recovery over decellularized extracellular matrix and (RGDS + MMPQK)–functionalized hydrogels in an ischemic hindlimb model, illustrating the power of this approach.
INTRODUCTION
While notable progress has been made to derive natural biomaterials [e.g., decellularized extracellular matrix (dECM)] to treat ischemic diseases and repair wounds during the past decade, their undefined nature limits mechanistic insights into their therapeutic effectiveness and the potential for further improvement (1, 2). On the other hand, chemically defined biomaterials provide a synthetic strategy to develop biomaterials with targeted function (3–5). Engrafting the essential active components (ligands) of ECM proteins [e.g., RGDS from fibronectin (Fn-RGD)] onto chemically defined biomaterials has been a widely used strategy to engage endothelial cell (EC) integrins (e.g., αiibβ3 and αvβ3) to promote EC vascular network formation (the essential step for therapeutic vascularization) (6–8). However, limited capacities of RGDS to engage EC integrins have greatly restricted the development of chemically defined biomaterials with comparable vascularization capacities of natural biomaterials (3, 4). This clearly states an unmet need to identify novel potent ligands for therapeutic vascularization and biomaterials research as a whole.
Significant efforts have been devoted to screening for novel cell-adhesive peptide ligands. However, the low throughput, along with a lack of strategies to identify a suitable pool of candidates, has limited discovery and applications of the new ligands in therapeutic vascularization and biomaterials research. Laminin (LM) is known to play a critical role in supporting EC functions for vascularization (9, 10). LM-enriched natural biomaterials (i.e., Matrigel) also show excellent properties for EC vascular network formation, leading to its use as a gold standard substrate for EC functional assays (11). Although extensive screening efforts in LM has led to a variety of cell-adhesive peptides (12–14), these peptides have rarely been used in biomaterials research and therapeutic vascularization, suggesting their limited improvement over the widely used RGDS. Here, by leveraging motif analysis of evolutionarily conserved RGD-containing regions in LM with functional screening, we discovered an LM-derived peptide (α1) that showed strong support for EC fundamental functions over commonly used RGDS. The mechanistic studies showed that α1 can target both Fn- and LM-targeting integrins and generate a similar profile of “vasculature development” gene ontology (GO) term with Matrigel. Vasculogenesis assay revealed the capacity of α1-modified substrates to significantly improve EC “clustering” for vasculogenic network formation, when compared to RGDS and two sequentially similar peptides (i.e., A99 and PA26) identified by previous screening studies. Injectable alginate functionalized with α1 and MMPQK [a vascular endothelial growth factor (VEGF)–mimetic peptide with a matrix metalloproteinase (MMP)–degradable linkage] showed enhanced properties to restore blood flow in a mouse hindlimb ischemia model when compared to dECM as well as the alginates functionalized with RGDS + MMPQK.
RESULTS
Identification of α1 through bioinformatic analysis on evolutionarily conserved sequences in LM
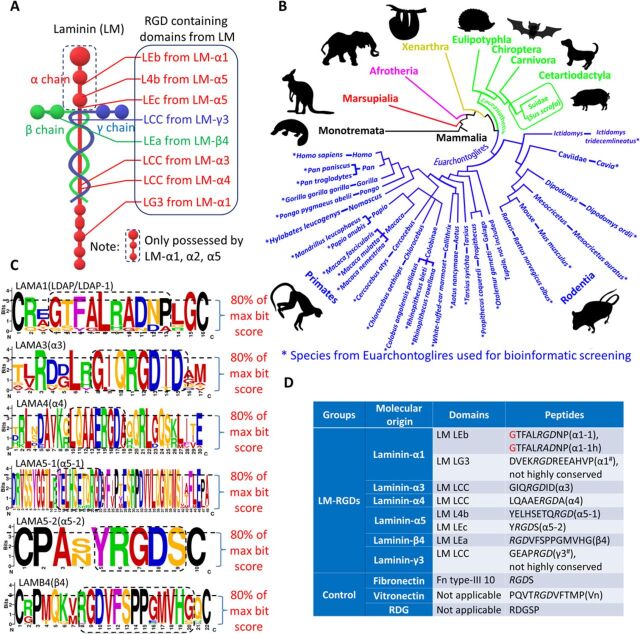
RGD motifs were expressed in eight regions among total 12 different LM-α, β, and γ subunits, including laminin coiled-coil (LCC) domains (regions for triplet formation) (15), epidermal growth factor–like (LEa/LEb/LEc) domains (potentially involved in mitogenic function and regulating the development of certain cell lineages) (16), IV (L4a/L4b) domain (unknown functions), and globular (LG1-5) domains (Fig. 1A). To identify the evolutionarily conserved LM-RGD sequences, we first aligned RGD-containing regions of LM subunits from Euarchontoglires species (the superorder of Primates and Glires, specific species, and their taxonomy are shown in Fig. 1B; proteins are shown in table S1) (17). The evolutionary conservation probability was calculated and visualized for each position in the sequences using bitmap-based motif analysis (Fig. 1C and fig. S1, A to D) (18). Using 80% value of max bit value as the filtering criteria, we determined the highly conserved regions around “RGD” site followed by selecting the amino acid with highest occurring frequency for each of conserved positions (Fig. 1C). With this method, we identified six highly conserved RGD-containing sequences from LM. Uniquely, the motif from LM-α1 contains two derivations at the RGD position (RGD version: GTFALRGDNP and RAD version: GTFALRADNP). As RAD is a known cell-adhesive variant of RGD (19, 20), both of them were included for functional screening. Combined with two Fn-RGDs, Vn-RGD, and two nonconserved LM-RGDs (fig. S1, E and F), a library of 12 RGD-containing peptide sequences was constructed (Fig. 1D).
Fig. 1. Evolutionary conservation and motif analysis enable identification of LM-derived RGD peptides for microarray construction.
(A) LM structural diagram with location of all RGD-containing domains. (B) Schematic diagram showing the species of Euarchontoglires used for bioinformatic alignment screening (*) in context of their phylogenetic relationship. Different colors indicate each subfamily from Mammalia. (C) Bitmap plots of motif analysis of the highly conserved sequences among species in Euarchontoglires. Dashed-line boxes from bitmap plots determine the highly conserved positions around/at RGD (80% of max bit score set as the filter criteria). The most repeated amino acid (signified by size of letter) from each conserved peptide position was used as the representative amino acid for synthesis. (D) The list of identified consensus LM-RGD sequences from motif analysis and their correspondent origin domains (Fn-RGD and Vn-RGD included as the positive controls) for LM-RGD microarray screening. Glycine in red as the linker for the peptides from LM-α1 was optimized by following experiments.
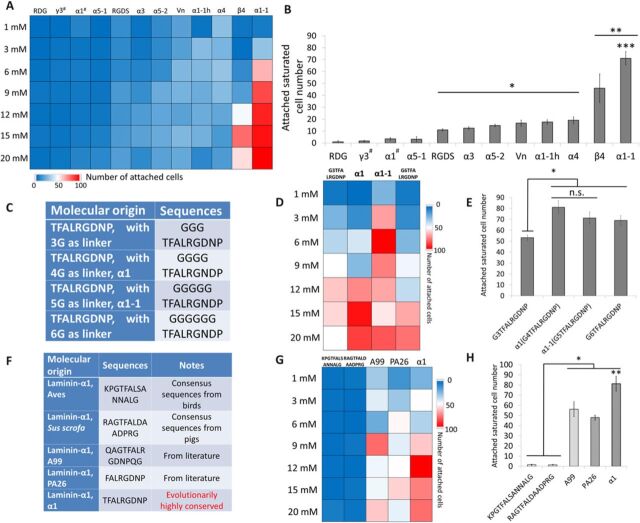
To evaluate the functions of these new peptides, we used a newly established peptide-functionalized hydrogel microarray to screen human umbilical vein EC (HUVEC) adhesion (Fig. 2A and fig. S2A) (21). Sigmoidal relationships between the numbers of attached HUVECs versus peptide concentrations were established (fig. S2B), and the saturated attached cell number of each peptide was used to examine binding activities of the LM-RGD peptides (Fig. 2B). While the two nonconserved LM-RGDs showed minimal HUVEC-adhesive properties, six of the seven conserved peptides showed significant HUVEC attachment. Among them, four conserved LM-RGD peptides showed similar adhesive properties with RGDS. There are also two novel evolutionarily conserved LM-RGD peptides (β4 and α1-1) showing significantly higher cell attachment than RGDS. Among all six peptides, α1-1 (G5TFALRGDNP) showed the highest activities for HUVEC attachment.
Fig. 2. Hydrogel microarrays facilitate discovery of LM-derived peptide α1 for improved EC attachment.
(A and B) Heat map of HUVEC attachment and quantification of saturated attachment (from sigmoidal plots of peptide concentration versus attached cell number) on the LM-RGD–derived hydrogel microarray; n ≥ 10. * indicates significantly more HUVEC attachment than on RDG, α1#-, γ3#-, and α5-1–modified hydrogels; P < 0.05. ** indicates significantly more HUVEC attachment than remaining groups; P < 0.05. *** indicates significantly more HUVEC attachment than on β4-modified hydrogels; P < 0.05. (C) Library of glycine-linker optimization for the peptides from LM-α1. (D and E) Heat map of HUVEC attachment and quantification of saturated attachment on the hydrogel microarray for glycine-linker optimization; n ≥ 10. * indicates significant differences between groups. n.s. indicates no significant difference. (F) Library of reported α1-similar sequences or α1-correspondent sequences from the species out of Euarchontoglires. (G and H) Heat map of HUVEC attachment and quantification of the saturated attachment on the α1-based hydrogel microarray; n ≥ 10. * indicates significantly more HUVEC attachment than on hydrogels modified with the correspondent motifs from Aves and Sus scrofa; P < 0.05. ** indicates significantly more HUVEC attachment on α1-modified poly(ethylene glycol) (PEG) hydrogels than on PA26- (22) and A99-modified (22) hydrogels; P < 0.05. All plots represent means ± SD.
The linker between the ligands and materials plays a critical role for cell surface receptors to engage ligands. The linker used here is the previously optimized 4-glycine linker (21). However, the identified evolutionarily conserved sequence, α1-1 (G5TFALRGDNP), starts with additional glycine. To elaborate the linker for the most concise sequence with fully exhibited activities, a library of the identified sequence with a different length of glycine linker (3-glycine, 4-glycine, 5-glycine, and 6-glycine) has been designed for EC adhesion screening (Fig. 2C). Only 3-glycine–linker sequence has shown insufficient capacities to fully exhibit the activities of our identified sequence, while 4-glycine–linker, 5-glycine–linker and 6-glycine–linker sequences all showed similar activities compared with α1-1 (the identified sequence) (Fig. 2, D and E). In summary, no significant difference was found between the 4-glycine–linker and 5-glycine–linker sequences, indicating that G4TFALRGDNP (α1) is the most concise sequence. Therefore, G4TFALRGDNP (α1) was used for later experiments.
To further evaluate the use of evolutionary conservation as a peptide selection rationale for functional screening, we compared the HUVEC attachment on α1 to the corresponding sequences found in Aves (biological class of birds) and Sus scrofa (porcine, a common translational model organism) (Fig. 2, F to H). Those sequences from Aves and S. scrofa presented minimal or no effects to HUVECs, consistent with the facts that these species are taxonomically further than rodents/primates to human (shown in Fig. 1B) (17). Notably, the α1 (TFALRGDNP) showed significantly higher cell-adhesive capacities than the sequentially similar LM-derived RGD peptides from previous reports, A99 (QAGTFALRGDNPQG) (22) and PA26 (FALRGDNP) (22) (Fig. 2, F to H). All these results showed the highest activity of α1 among all the peptides for EC adhesion and supported the rationale of evolutionary conservation motif analysis for potential ligand selections.
α1-modified hydrogels promote EC fundamental functions in vitro
We validated the microarray-based findings through the two-dimensional (2D) HUVEC culture assays on hydrogel substrates inside 96-well plates (fig. S3, A and D). Poly(ethylene glycol) (PEG) hydrogel and alginate hydrogel were used here to represent the hydrogels formed through two distinct cross-linking mechanisms [ultraviolet (UV)–initiated and Ca2+-coordinated cross-linking, respectively]. Alginate here was functionalized with peptides through established copper-assisted azide-alkyne cycloaddition (CuAAC) “click” chemistry, which is highly efficient and cytocompatible (fig. S4, A to F). Consistent with the microarray results, α1-functionalized PEG and alginate hydrogels showed significantly higher cell attachment than RGDS (fig. S3, B and E). HUVECs on α1-modified PEG substrates also had a significantly larger spreading area (fig. S3C). Since the soluble form of the peptides can inhibit cell attachment on immobilized form via competitive binding to integrins (22), the inhibitory effects of soluble α1 and RGDS peptides on HUVEC attachment to α1-functionalized alginate substrates were also examined. The soluble α1 showed a significant stronger capacity to inhibit HUVEC attachment compared to RGDS (fig. S5A). Furthermore, the enhanced EC function was further supported by the faster HUVEC migration on the α1-functionalized alginate substrates (fig. S5, B to E). In addition, the mechanical properties of the peptide-functionalized hydrogels were examined and showed no significant difference among each hydrogel group, suggesting that the difference in EC attachment and function comes mainly from the ligands, instead of mechanical properties (fig. S5F).
Mechanistic investigations of α1-functionalized hydrogels
To explore the fundamental mechanisms underlying the improved capacities of α1-functionalized substrates for EC functions, we performed blocking experiments of integrins reported to be involved in LM- and Fn-mediated adhesion: α1, α2, α3, and α6 (LM-binding); αv and β3 (Fn-binding); and β1 (both LM- and Fn-binding) (22). EDTA and nonantibody blocking was used as a positive and negative control, respectively. The blocking experiments also included RGDS-functionalized substrates and β4 (the second-best peptide from screening)–functionalized substrates. Blocking Fn-binding integrins (αv and β3) led to significantly reduced EC attachment on all RGDS and α1- and β4-functionalized substrates (Fig. 3A). While blocking LM-binding integrins, α3 and α6 did not affect HUVEC attachment on RGDS-modified substrates; it did affect HUVEC attachment on LM-RGD (α1 and β4)–modified substrates. When compared to β4-modified substrates, α1-modified substrates showed much wider spectrum of the sensitivities to almost all the LM-binding integrins (α1, α2, α3, α6, and β1) used in the experiment, which could potentially explain the activity difference between these two peptides for EC adhesion. This result indicated an important role of LM-binding integrin for HUVEC attachment onto LM-RGD–modified substrates and demonstrated that α1 can interact with both Fn- and LM-binding integrins (Fig. 3A).
Fig. 3. α1-modified hydrogel binds to both Fn- and LM-binding integrins of ECs.
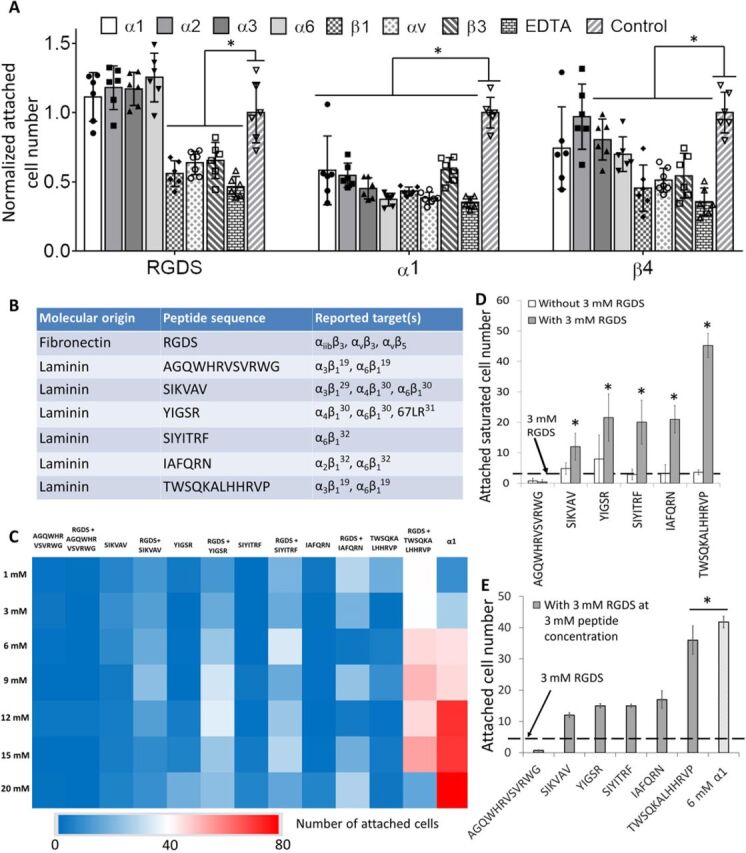
(A) Quantifications of normalized attached HUVEC (with/without integrin blocking) number on α1-/RGDS-/β4-modified alginate hydrogels; n = 6. * indicates significant difference compared to no integrin blocking; P < 0.05. (B) The library of selected non-RGD, LM-binding-integrin targeted peptides for hydrogel microarray screening. (C) Heat map of HUVEC attachment on the hydrogel microarray of the non-RGD, LM-binding-integrin targeted peptides with/without additional 3 mM RGDS. α1-modified PEG hydrogel set as the positive control. (D) Quantification of the saturated attached HUVEC number based on the sigmoidal plots of peptide concentration versus attached cell number from the microarray of LM-binding-integrin targeted peptides with/without additional 3 mM RGDS; n ≥ 10. * indicates significant difference compared to “without 3 mM RGDS”; P < 0.05. (E) Quantification of the average attached HUVEC number at 3 mM LM-binding-integrin targeted, non-RGD peptides with additional 3 mM RGDS; n ≥ 10. α1-modified PEG hydrogel set (6 mM) acts as the positive control as the total peptide concentration is fixed at 6 mM. * indicates significant difference compared to rest of groups; P < 0.05. All plots represent means ± SD/SEM.
We then hypothesized that the ability of α1 to engage both Fn- and LM-binding integrins synergistically improves EC adhesion and leads to its superior property to support EC functions. To test this hypothesis, we conducted a combinatorial microarray screening to evaluate synergistic effects between Fn-binding integrin and LM-binding integrins by using RGDS and LM-integrin–targeting non-RGD peptides (14, 23–26) (Fig. 3B). While the LM-integrin–targeting non-RGD peptides alone showed very low attachment of HUVECs, the combination of the non-RGD peptides with a low concentration of RGDS (3 mM) led to significantly increased attachment of HUVECs (Fig. 3, C and D), showing the synergy between Fn- and LM-integrin–targeting peptides to improve EC adhesion. This was further supported by the high expression of both Fn- and LM-binding integrins in ECs (27), along with the well-known important roles of Fn- and LM-binding integrins (e.g., αvβ3 and α6, respectively) in vascularization (28, 29). Notably, fixing the total peptide(s) concentration at 6 mM (i.e., 3 mM non-RGD + 3 mM RGDS), α1 still showed the superior activities for HUVEC adhesion than most combination groups, supporting its suitability to support HUVEC functions (Fig. 3E).
RNA sequencing analyses of ECs in α1-functionalized hydrogels
To gain a holistic view and comparison of genetic changes of ECs cultured in α1- and RGDS-functionalized alginate hydrogels, RNA sequencing (RNA-seq) was performed. HUVECs cultured in α1 hydrogel showed 2500 differentially expressed (DE) genes (adjusted P < 0.05 and fold change > 1.5 or < −1.5) compared to those in RGDS hydrogels (fig. S6A and table S2).
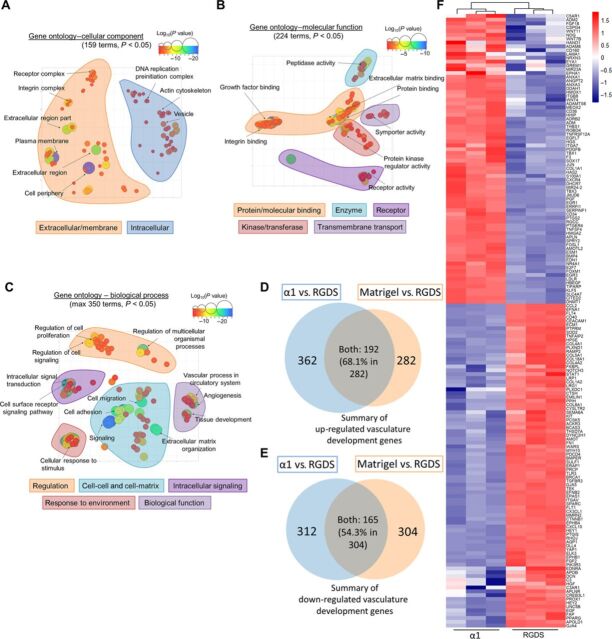
DE genes were analyzed using GO terms that provide a context of cellular component (CC), molecular function (MF), and biological process (BP). Then, all significant terms were used and visualized in semantic similarity–based scatterplots using REduce and VIsualize Gene Ontology (REViGO), where selected GO terms and GO term groupings were highlighted to indicate overall similarities and spatial organization. GO-CC REViGO analysis with all 159 significant terms (P < 0.05) revealed that the α1 genetic profile showed significantly higher activity than RGDS relating mainly to the extracellular space and cell membrane, e.g., receptor complex (P = 2.2 × 10−5), integrin complex (P = 1.3 × 10−2), and extracellular region (P = 6.40 × 10−14) (Fig. 4A). GO-MF terms with all 224 significant terms (P < 0.05) showed significantly higher activity relating to protein binding, enzyme, and receptor activity, e.g., integrin binding (P = 2.3 × 10−4), peptidase regulator (P = 7.8 × 10−3), and receptor binding (P = 2.8 × 10−10), in α1 compared to RGDS (Fig. 4B). GO-CC and GO-MF classification supported the improved properties of α1 over RGDS for ECs reflected by their differences in integrin binding and increased cell-matrix interactions. For GO-BP, the analysis resulted in a total of 1913 significant terms (P < 0.05). REViGO visualization with the top 350 terms showed significant grouping relating to cell-matrix and cell-cell interactions, such as “cell migration” (P = 5.2 × 10−15) and “cell adhesion” (P = 3.3 × 10−9), revealing enhanced vascular biological functions, such as vasculature development (P = 3.7 × 10−14) (Fig. 4C). Notably, these results from GO-BP analyses are consistent with the outcomes of in vitro assays discussed above (see figs. S2, S3, and S5).
Fig. 4. RNA-seq analyses to characterize vasculogenic material-cell interaction in α1-modified hydrogel.
(A to C) Visualization of all (max 350 terms) significant (P < 0.05) GO terms of DE genes (fold change > 1.5 or < −1.5, adjusted P < 0.05) from HUVECs in α1- versus RGDS-functionalized hydrogels. GO terms of CC, MF, and BP are organized in semantic similarity–based scatterplots using REViGO that combines redundant terms into a single, representative term based on a simple clustering algorithm relying on semantic similarity measures. (D and E) The Venn diagram of up-regulated (D) and down-regulated (E) genes from transcriptomic assay of α1/RGDS and Matrigel/RGDS in the vasculature development GO-BP term. (F) Heat map of DE genes assembled from the GO-BP vasculature development term (GO:0001944) gene list showing the gene expression profile of α1-modified hydrogels over RGDS-modified hydrogels.
A detailed examination of all the DE genes in the vasculature development GO-BP term (GO:0001944) showed a unique, specific α1-based transcriptomic signature (Fig. 4F). In the signature, several groups of gene expression were increased in α1 over RGDS, including genes related to ECM remolding (e.g., SPARC, COL1A2, and COL4A1), integrin and its downstream proteins (e.g., ITGA1, ITGAV, and PIK3R3), and growth factors/receptors (e.g., EGF, FGF2, and FLT1). Notably, in comparison to previous work using 3D culture of HUVECs in Matrigel versus RGDS-modified PEG hydrogel, transcriptomic comparison revealed a ~60% overlap of DE genes from α1-modified hydrogel and Matrigel when they were compared to RGDS-modified hydrogels, supporting a similar provascular potential of α1-modified hydrogels (LM-derived) and Matrigel (containing ~60% LM) (Fig. 4, D and E, and fig. S7). Other vascularization highly related GO-BP terms, such as “angiogenesis” (GO:0001525), “vasculogenesis” (GO:0001570), and “positive regulation of vasculature development” (GO:1904018) were also compared, showing ~60, ~60, and ~65% similarities with Matrigel (fig. S6, B to G).
α1-formulated hydrogels promote EC vascular network formation
While in vitro analysis above showed enhanced EC attachment, spreading, and migration in α1-modified hydrogels, we next developed α1-formulated hydrogel to test its vasculogenic properties. Considering the importance of cosignaling between integrin ligands and growth factors to improve vascularization (1), we compared vascular morphogenesis in 3D hydrogels modified with MMPQK [a VEGF-mimetic peptide (QK) with an MMP-degradable linker] (30) only, α1 only, and α1 + MMPQK (fig. S8, A to D). The formulation of α1 + MMPQK promoted the highest HUVEC vascular network formation among all the groups, which is evidenced by the quantification of total HUVEC network length, number of branches, and number of branch points. Therefore, the formulation of α1 + MMPQK was used for the later experiments to promote vascularization.
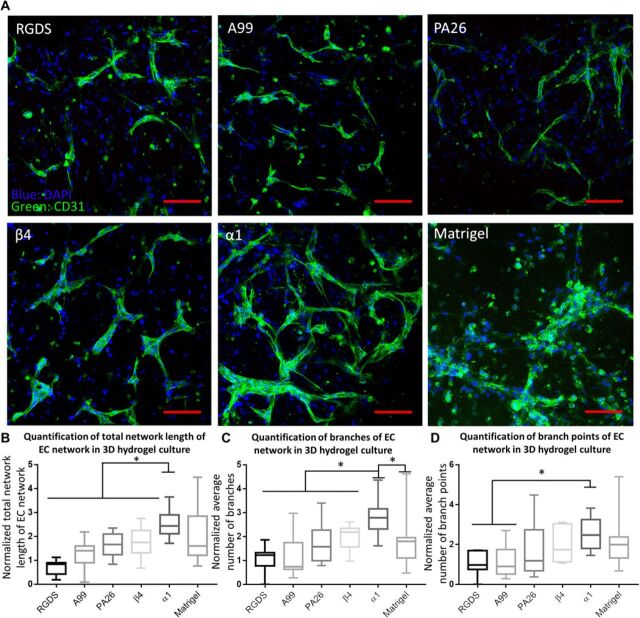
To compare the results from our screening strategy with the previous ones, we also prepared A99 + MMPQK and PA26 + MMPQK and included them as controls (table S5). As shown in Fig. 5, A to D, HUVECs exhibited elongated morphology in all six different hydrogels after 4 days’ culture. Among them, HUVECs showed the least number of branches, branch points, and shortest total network length in RGDS-formulated hydrogels. A99-, PA26-, and β4-formulated hydrogel supported HUVEC network with a moderate number of branches, branch points, and fair total network length. In contrast, α1-formulated hydrogel exhibited the highest support for HUVECs network formation, demonstrated by the highest number of branches, branch points, and maximum total network length in hydrogel. Notably, when compared to the HUVEC network formed in Matrigel (31), α1-formulated hydrogel still showed more branches and branch points as well as the similar total network length. There were lots of sprouting HUVEC “clusters” found in α1-formulated hydrogel and Matrigel. When zooming into these sprouting HUVEC clusters, largely elongated HUVECs were found to connect with other distant HUVECs to form EC networks (fig. S8, E to H). All these phenomena are the essential indication for active vasculogenesis, consistent with the previous report (32). In summary, α1-formulated hydrogel showed superior properties to promote EC vasculogenic network formation, indicating the substantial vasculogenic potential for the translational application.
Fig. 5. The vasculogenesis assay on RGDS-, A99-, PA26-, β4-, and α1-modified hydrogels revealed the capacity of α1-modified hydrogel to promote vasculogenic network formation.
(A) Representative images of HUVEC vascular network in RGDS-, A99-, PA26-, β4-, and α1-modified hydrogels and Matrigel (scale bar, 100 μm). Green, CD31; blue, 4′,6-diamidino-2-phenylindole (DAPI). Sprouting HUVEC clusters were found with elongated HUVECs in β4- and α1-modified hydrogels and Matrigel. (B to D) Quantification of total network length, branches, and branch points of EC network in 3D hydrogel culture (RGDS-, A99-, PA26-, β4-, and α1-modified hydrogels and Matrigel); n ≥ 11 for each group. * indicates significant difference; P < 0.05. All plots represent means ± SD.
α1-formulated hydrogel improved functional recovery of ischemic injury
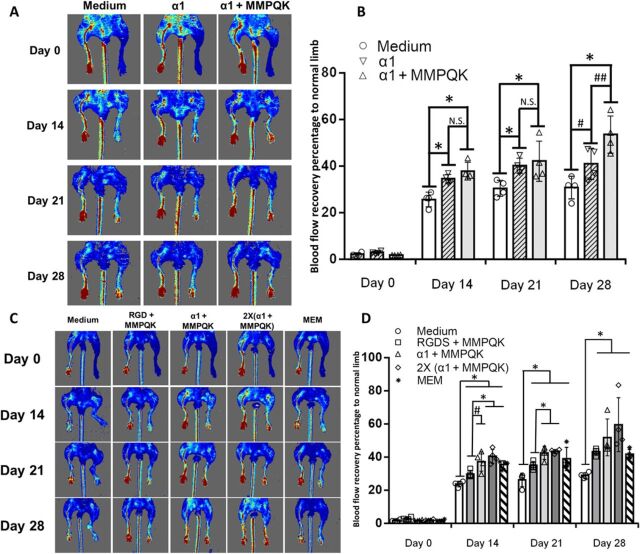
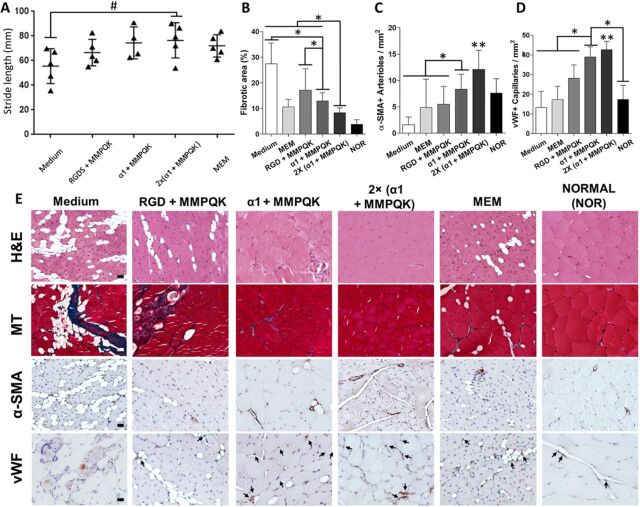
A murine hindlimb ischemia model was used to evaluate the therapeutic vasculogenic potential of α1-formulated injectable hydrogels (33). A preliminary experiment was performed between a media injection group and a blank alginate (nonpeptide conjugated)–injected group, showing no difference (fig. S9A). While injection of α1 and α1 + MMPQK hydrogels after ischemic injury showed a substantial increase in blood restoration compared to media injection, the α1 + MMPQK group showed higher recovery than the α1-only group by day 28. In addition, the formulation of α1 + MMPQK showed consistently significant recovery for the blood flow compared to medium-treated groups at days 14, 21, and 28 (Fig. 6, A and B). Therefore, the formulation of α1 + MMPQK was used for the remaining experiments. A further investigation of the therapeutic effects of α1-formulated hydrogels was performed with a range of hydrogel formulations examined in the same model (the second batch in fig. S9B). RGDS + MMPQK–modified alginates were used to represent the widely used RGDS-based hydrogels for vascularization (6, 7, 34). While Matrigel was not included because of its mouse tumor origin, dECM from mouse skeletal muscle (MEM) was included to represent natural source-derived materials (35). Double-dosed α1 + MMPQK [i.e., 2×(α1 + MMPQK)] was also incorporated to examine the dose effect of peptides on hydrogels. All the results are shown in Fig. 6, C and D. While the medium-injected group (medium) plateaued at ~30% blood flow recovery through days 21 and 28, MEM and RGDS + MMPQK both showed modest improvement of ~38% blood flow recovery at day 21 and ~43% recovery at day 28. α1 + MMPQK treatment displayed significantly improved blood flow recovery (~43%) at day 21 with further increase (~52%) by day 28. In addition, increasing the peptide dose with 2×(α1 + MMPQK) exhibited blood flow recovery ~43% at day 21 and even higher blood flow recovery (~60%) by day 28. Furthermore, α1-formulated hydrogel and MEM treatments showed improved functional recovery of ischemic limb as measured by gait analysis (Fig. 7A). Histological analysis of the harvested limb muscle tissues showed a significant decrease in muscle degeneration and fibrotic area (Fig. 7, B and E, and fig. S9C) and significant increase in artery and capillary blood vessel development (Fig. 7, C to E, and fig. S9, D and E) in α1-formulated hydrogels than medium, RGDS + MMPQK, and MEM. Overall, these data support the therapeutic vascularization effects of α1-formulated hydrogels to improve functional and structural recovery after ischemic injury.
Fig. 6. Enhanced recovery of blood perfusion after ischemic hindlimb injury using α1-formulated hydrogel.
(A) Representative images of laser Doppler perfusion imaging on the ischemic hindlimb after injecting the different formulations (medium, α1 only, and α1 + MMPQK) of alginate hydrogels for serial analysis (left leg, normal; right leg, ischemic). (B) Quantification of blood flow recovery percentage of ischemic limb to normal limb (n = 4 to 5). * indicates significance difference; P < 0.05. N.S. indicates no significant difference between the groups; P > 0.05. # indicates improved blood flow of the α1 hydrogel–treated group compared to medium-treated group at day 28; P = 0.053. ## indicates the improved blood flow of α1 + MMPQK hydrogel–treated group compared to α1-treated group at day 28; P = 0.054. (C) Representative images of laser Doppler perfusion imaging on the ischemic hindlimb after injecting the formulations of the second batch (groups shown in fig. S9B) alginate hydrogels for serial analysis (left leg, normal; right leg, ischemic). (D) Quantification of blood flow recovery percentage of ischemic limb to normal limb (n = 4 to 5). * indicates significance difference; P < 0.05. # indicates the improved blood flow of α1 + MMPQK hydrogel–treated ischemic legs compared to RGDS + MMPQK hydrogel ischemic legs at day 14; P = 0.052.
Fig. 7. Improved functional and morphological recovery from ischemic hindlimb injury using α1-formulated hydrogel.
(A) Gait analysis of the stride length on ischemic hindlimb (n = 4 to 5). # indicates improved stride length of 2×(α1 + MMPQK) hydrogel–treated ischemic legs compared to medium groups (significant functional improvement) at day 14; P = 0.053. (B to D) Quantification of fibrotic areas (%) from Masson’s trichrome (MT) analysis (B), anti–smooth muscle actin–positive (α-SMA+) arteries (C), and vWF-positive (vWF+) capillaries (D) from treated ischemic muscle tissue sections and controls at 4 weeks after surgery, n ≥ 20 for the quantification. * indicates significance difference; P < 0.05. ** indicates significantly more α-SMA+ arteries and vWF+ capillaries of 2×(α1 + MMPQK)–treated ischemic legs compared to remaining groups at day 28; P < 0.05. The significant analyses of all these histological assays are shown in fig. S9 (C to E). (E) Representative images of hematoxylin and eosin (H&E), MT, and immunohistochemical staining (α-SMA and vWF) of treated ischemic muscle tissue sections and controls at 4 weeks after surgery (scale bars, 100 μm). All plots represent means ± SD/SEM.
DISCUSSION
Using biologically derived ligands to represent the long and complex proteins in ECM is a widely used biomimetic approach in general biomedical research (3–5). The strategy to select peptide segments from large ECM proteins as biological ligands is critical because literature has shown the amino acids surrounding receptor-binding core (e.g., RGD) significantly alter the activities and/or targeted receptors of the ligands (36). In addition, the use of mathematic permutation to identify suitable ligands from LM can lead to millions of leads and make it impractical. This highlighted that the current lack of rational ligand candidates’ selection strategy, coupled with the low throughput of screening cell-materials interactions, has limited the identification of novel potent ligands for biomedical applications (12, 13, 37). With the progress of bioinformatics and proteomics, we leveraged motif analysis to screen a vascularization-related ECM protein, LM, from different species of mammals to identify the evolutionarily conserved RGD-containing motifs for therapeutic vascularization. The RGD-containing motifs were selected because of its EC-adhesive capacity and its synergistic potential with VEGF receptor activation (1, 38). By focusing on the species from Euarchontoglires, we identified seven conserved peptides, six of which showed significant HUVEC-adhesive capacities (successful rate, ~86%), comparing to the nonconserved LM-RGDs with minimal HUVEC-adhesive properties. Four of the six peptides showed similar HUVEC-adhesive properties with RGDS, while two (β4 and α1) of them exhibited fold improvement compared to RGDS. These results highlighted biological significance of evolutionarily conserved motifs and supported the effectiveness of the approach using evolutionarily conserved motifs for peptide ligand development. This is further supported by the similar cell attachment profile of human adipose-derived stem cells (hADSCs), a cell type widely used in regenerative medicine applications, on the LM-RGD peptide–functionalized hydrogel microarray (fig. S10).
As the further validation of the screening strategy, we compared the highest EC-adhesive peptide, α1, with the corresponding sequences from Aves and S. scrofa as well as previously reported, sequentially similar sequences (A99 and PA26). Low HUVEC attachment on Aves- and S. scrofa sequences–modified surfaces indicated the necessity of ligand selection from taxonomic similar species (i.e., Euarchontoglires) for maximal human cell function. This is notable regards to the use of commonly available porcine dECM for potential translational human applications (39). The significantly improved performance of α1-modified hydrogel compared to A99- and PA26-modified hydrogels confirmed through microarray screen, as well as 3D vasculogenic assay, indicated that evolutionarily conserved α1 (TFALRGDNP) is the essential active part of the RGD-containing loop from LEb of LM α1 and is a functionally distinct peptide. The addition (A99: QAGTFALRGDNPQG) or elimination (PA26: FALRGDNP) of amino acid(s) on the highly conserved α1 sequence reduced its capacities to support HUVECs for vascularization. Collectively, these results demonstrated the power of combining evolutionarily conserved motif analysis with functional screening to rapidly identify essential core sequences from these LM-RGD–containing domains.
The results of integrin blocking experiments and combinatorial LM- and Fn-integrin–targeted microarray provided fundamental insights toward α1 to engage both LM- and Fn-targeted integrins to improve EC vasculogenic potential. This is further supported by the similar vasculature development GO terms of α1-functionalized hydrogel to Matrigel, an LM-enriched and Fn-containing matrix, and gold standard material for in vitro EC functional assays. Combining this with the superior ability of α1-formulated hydrogel to enhance functional recovery of mouse ischemic hindlimb over the MEM, our results demonstrated the remarkable potential of the unique α1 for therapeutic vascularization. In addition to the ischemic hindlimb disease model, which is a model relevant to peripheral vascular disease (40), we expect that the α1 peptide will have a wide range of applications on treating wounds and ischemic diseases (e.g., cerebral ischemia and myocardial infarction). As the rheological requirements of injectable hydrogels are similar to those of extrudable hydrogels (e.g., shear thinning) for bioprinting, the injectable α1 + MMPQK hydrogels can be further engineered and applied as a vasculogenic bioink to fabricate vascularized tissue engineering constructs. In addition, since previous studies mostly focused on the angiogenic effects of the biomimetic peptides from ECM (41), our work would also potentially bring a new perspective for the field to reconsider the different effects of the identified ECM-derived peptides for vessel regeneration through vasculogenesis, which is also an important wound healing and regeneration mechanism.
SUMMARY AND OUTLOOK
To identify novel ECM-derived ligands for biomaterials development, here, we performed a virtual screening of 199 different LM subunits from around 28 taxonomic human-similar species from Euarchontoglires to identify evolutionarily conserved RGD-containing ECM motifs and tested their EC adhesion capabilities using our peptide-functionalized hydrogel microarray platform. Among the six active LM-RGD peptides, α1 displayed superior properties to support HUVEC functions for vascularization over commonly used RGDS. The mechanistic investigations attributed this to the ability of α1 for targeting both Fn- and LM-targeting integrins. RNA-seq showed that α1-modified hydrogel exhibited high potential in transcriptomic profile of vasculature development and 60% similarities to Matrigel. The vasculogenesis assay revealed the capacity of α1-formulated hydrogel to promote vasculogenic “cluster” extensions and connections for EC vascular network formation. Injectable alginate functionalized with α1 + MMPQK more effectively improved blood flow restoration in a mouse hindlimb ischemia model over dECM as well as the alginates functionalized with RGDS + MMPQK. These results demonstrated the translational potential of α1 in treating ischemic diseases and repairing wounds. It also illustrated the power of our platform to rapidly identify novel peptide ligands with great potency for therapeutic vasculogenesis. We expect that the combined use of evolutionary conserved motif analysis, peptide functionalized hydrogel microarrays, and transcriptomic analysis offers a fast, effective, and comprehensive manner to explore, identify, and study novel chemically defined biomaterials for therapeutic applications.
MATERIALS AND METHODS
Materials and instrument
All chemicals used for this study were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Microarray spotting pins (946MP9B) were purchased from Arrayit Corporation (Sunnyvale, CA). A custom-designed microarrayer was assembled and produced by BioDot (Irvine, CA). The liquid chromatography–mass spectrometry (LC-MS) system used is Thermo Fisher LCQ Fleettm Ion Trap Mass Spectrometer. Proton nuclear magnetic resonance (1H-NMR) was recorded on BRUKER AV500-III spectrometers. Primary and secondary antibodies were purchased from Abcam (Cambridge, UK). The primers for reverse transcription polymerase chain reaction were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). Sodium alginate (LF20/40) was purchased from FMC BioPolymer (Philadelphia, PA).
Cell culture
HUVECs (Lonza, Basel, Switzerland) were cultured in EGM-2 BulletKit Medium (Lonza, Basel, Switzerland). The medium supplements contained 2% bovine serum albumin (BSA), hFBF-B, VEGF, R3–insulin-like growth factor-1, ascorbic acid, heparin, fetal bovine serum, hEGF, and GA-1000. hADSCs were cultured in ADSC Growth Medium (Lonza, Basel, Switzerland). For the rest of the assays (unless otherwise stated), Dulbecco’s modified Eagle’s medium (DMEM) (11054020, Thermo Fisher Scientific Inc., USA) with 10% fetal bovine serum (26140079; Thermo Fisher Scientific Inc., USA) was used to exclude the influence from growth factors. Growth medium was changed every other day, and cells were passaged every 6 days. All experiments were conducted using both passage 4 (P4) HUVECs and hADSCs.
Bioinformatics-assisted ECM protein screening for highly conserved sequences and motif analysis
Bioinformatics-assisted ECM protein screening for highly conserved sequences was performed using the following database: UniProt database, supported by European Bioinformatics Institute (EMBL-EBI), the SIB Swiss Institute of Bioinformatics, and the Protein Information Resource. The specific sequence for each ECM protein/ECM protein subunit was collected from mammalian species (e.g., human, mouse, rat, and chimpanzee) and further categorized into two groups based on two major mammalian superorders: the proteins from Euarchonotoglires superoder (e.g., human and mouse) and the proteins from Laurasiatheria superorder (e.g., pig). The sequences in these two groups were aligned through the tool of Clustal Omega from EMBL-EBI (42). The algorithm was described by Sӧding (43). The conserved sequences were selected using two different algorithms based on their residing domains (fig. S1A). Specifically, the sequences from EGF-like domains were chopped out from the first cysteine to the neighboring cysteine, which form a ring structure with the previous cysteine; while the sequences from non-EGF–like domains were chopped out from the first highly conserved amino acids to the last highly conserved amino acids, for demonstration, two (or more) unconserved amino acids are included for motif analysis (fig. S1B). The algorithm for judgment of amino acid conservation is described by Sӧding (43). In particular, the selected conserved sequences from the species of Euarchonotoglires superoder were further categorized into two categories, primates and rodents for the calculation of sequence conservation. The sequence conservation at a particular position in the alignment, Rsequence, was calculated using the equation in fig. S1 (C and D) with small sample corrections (n ≤ 50) (44), and the alignment results were visualized through WebLogo (45). The analysis results were generated through WebLogo, and 80% value of max bit value was applied as the filtering criteria to generate the highly conserved sequences. The most repeated amino acid (signified by size of letter) from each conserved peptide position was used as the representative amino acid for synthesis. The sequences were then included into the library for screening. The highly conserved sequences from other species (i.e., birds) were also created with same algorithm for screening.
Synthesis and characterization of N-terminal unmodified peptides and methacrylated peptides
Peptides used in this work were synthesized by solid-phase peptide synthesis (SPPS). The SPPS was conducted using the standard procedure described in Novabiochem peptide synthesis manual. N-terminal unmodified peptides were cleaved from resin right after the deprotection once reaching the designed sequences. The preparation of methyacrylated peptides was demonstrated through our previous publication (21). Briefly, 2-isocyanatoethyl methacrylate [3 equivalent (eq) dissolved in N,N′-dimethylformamide (DMF)] was used to react with the terminal amine group of the peptide chain (1 eq) before they were cleaved from the resin. All the peptides were purified by a flash column and characterized by LC-MS.
Single peptide–based microarray fabrication and screening
Methacrylated peptides were dissolved in DMF at predesignated ratios (1, 3, 6, 9, 12, 15, and 20 mM) and mixed with poly(ethylene glycol) diacrylate (PEGDA) [containing 1% dimyristoylphosphatidic acid (DMPA) as initiator] [DMF solution of methacrylated peptide: PEGDA, 1:1 (v/v)] and then transferred into a 384-well plate for microarray fabrication. The microarrays were printed in a humid Ar atmosphere on epoxy monolayer-coated glass slides (Xenopore XENOSLIDE E, Hawthorne, NJ), which were first dip-coated in 4% (v/v) poly(hydroxyethyl methacrylate) [poly(HEMA)] using a customized microarrayer (Biodot). Spots were polymerized via 10-s exposure to long-wave UV (365 nm), dried at <50 mtorr for at least 7 days. Before use, the chips were sterilized by UV for 30 min for each side and then washed with phosphate-buffered saline (PBS) twice for 15 min to remove residual monomer or solvent. HUVECs (Lonza, Basel, Switzerland) and hADSCs (Lonza, Basel, Switzerland) were seeded onto each array at the density of 13,000 cells/cm2 and cultured for 12 hours, respectively. They were then fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI) (R37605, Thermo Fisher Scientific Inc.) for cell number counting and phalloidin (1:50 dilution; A12379, Thermo Fisher Scientific Inc.) for F-actin for the lead peptide identification.
Combinatorial microarray fabrication and screening
Methacrylated peptides and combinations were dissolved in DMF at predesignated ratios and mixed with PEGDA (containing 1% DMPA as initiator) [DMF solution of methacrylated peptide: PEGDA, 1:1 (v/v)] and then transferred into a 384-well plate for microarray fabrication. The rest of the fabrication processes were the same as single peptide–based microarray fabrication described above.
Preparation of peptide-functionalized PEG hydrogel substrates in 96-well plates
Methacrylated peptides (MA-G4RGDS and MA-G4TFALRGDNP) were dissolved in DMF to prepare 15 mM peptide solutions and mixed with PEGDA (containing 1% DMPA as initiator) [DMF solution of methacrylated peptide: PEGDA, 1:1 (v/v)]. The mixed solution was transferred into 96-well plates (50 μl in each well). The 96-well plates were exposed under long-wave UV (365 nm) in a humid Ar atmosphere for 5 min to prepare peptide-modified PEG hydrogel substrates and dried at <50 mtorr for at least 7 days. Before use, the hydrogel substrates were swelled with PBS buffer. HUVECs were seeded at the density of 25,000 cells/cm2 onto the hydrogel substrates and cultured for 12 hours. They were then fixed and stained with DAPI for cell number counting and phalloidin for F-actin to estimate cell spreading.
Alginate synthesis, oxidation, and peptide conjugation on alginate
Sodium alginate was prepared using the method established by Bouhadir et al. (46). The final oxidation of the alginate was 5%. The peptides were conjugated onto the oxidized alginate by the CuAAC click chemistry (47). The peptide-functionalized alginates were purified through repeated precipitations in ethanol (three times) (46). The completion of the conjugation reactions was verified by comparing the LC-MS of a copper-peptide-ligands complex before the reaction and the LC-MS of the liquid residual after the alginate precipitation. The conjugation efficacy was examined through 1H-NMR of peptide-conjugated alginate by comparing the characteristic peak of the proton on alginate backbone and the proton on the triazole. 0.125 mmol/g (peptide/alginate powder) was used as the peptide functionalization ratio for the rest of experiments unless otherwise stated. Peptide was also functionalized through 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) chemistry as the control using the established method in the reference (48).
Preparation of alginate substrates in 96-well plates
The Ca2+-containing gelatin substrates for alginate cross-linking were prepared in 96-well plates according to the literature (49). Fifty microliters of peptide-functionalized (RGDS and α1, respectively) 1% (w/v) alginate solution was transferred into the Ca2+-containing gelatin-coated 96-well plates and gelled for 40 min. Then, the well plates were transferred into an incubator for 10 min to melt the Ca2+-containing gelatin substrates. After Ca2+-containing substrates were removed, HUVECs were seeded onto the hydrogel substrates at the density of 25,000 cells/cm2 and cultured for 12 hours. Then, they were fixed and stained with DAPI for cell number counting and phalloidin for F-actin.
EC viability and proliferation on peptide-functionalized alginate hydrogels
The Ca2+-containing gelatin substrates for alginate cross-linking were prepared in 96-well plates according to the literature (49). Fifty microliters of α1-functionalized (through EDC or click chemistry, respectively) 1% (w/v) alginate solution was transferred into the Ca2+-containing gelatin-coated 96-well plates and gelled for 40 min. Then, the ECs (25,000 cells/cm2) were seeded on the surface and left for 1 hour. The well plates were transferred into the incubator for 10 min to melt the Ca2+-containing gelatin substrates. After Ca2+-containing substrates were removed, EGM-2 BulletKit Medium were added and the ECs were cultured for 4 days. The cell numbers were counted at days 0, 2, and 4, and the viability was examined through the LIVE/DEAD Cell Imaging Kit (R37601, Thermo Fisher Scientific Inc.), according to the manufacturer’s instructions.
Antibody blocking experiments on alginate substrates
The antibody blocking experiments were conducted according to the protocol from a previously published study (22). Specifically, the integrin antibodies and their combinations [no antibody, anti-integrin α1 (FB12, MAB1973, EMD Millipore, USA), α2 (P1E6, MAB1950Z, EMD Millipore, USA), α3 (P1B5, CP11L, EMD Millipore, USA), α6 (GoH3, 14-0495-82, Thermo Fisher Scientific Inc.), αv (P3G8, MAB1953, EMD Millipore, USA), β1 (6S6, MAB2253, EMD Millipore, USA), β3 (25E11, MAB1957, EMD Millipore, USA), EDTA, and anti-integrin α6 + anti-integrin αv] were aliquoted into predesigned ratios (final ratios in the mixture: 1:20 for anti-integrin α6, 1:33 for the rest of the anti-integrin antibodies, 5 mM EDTA) with PBS. The HUVECs were incubated with these antibodies for 15 min, sedimented by low-speed centrifugation, suspended in 100 μl of serum-free DMEM and 0.02% BSA, and then seeded onto the 1% (w/v) peptide-functionalized alginates at the density of 25,000 cells/cm2. The wells were transferred into the incubator for 2 hours. After that, unattached cells were rinsed away, and the attached cells were then fixed and stained with DAPI for cell number counting and phalloidin for F-actin.
Soluble-peptide blocking experiments on alginate substrates
The soluble-peptide blocking experiments were conducted according to the protocol from a previously published study (22). Specifically, soluble peptides (GGGGRGDS, GGGGTFALRGDNP, synthesized through SPPS) were aliquoted at the concentration of 0.5 mg/ml with PBS. The HUVECs were incubated in their respective peptide solution for 20 min, sedimented by low-speed centrifugation, suspended in 100 μl of serum-free DMEM and 0.02% BSA, and then seeded onto the 1% (w/v) α1-functionalized alginates at the density of 25,000 cells/cm2. No peptide-blocked group was used as the control. The wells were incubated for 2 hours. After that, unattached cells were rinsed away, and the attached cells were then fixed and stained with DAPI for cell number counting and phalloidin for F-actin.
EC migration assay
Oxidized (5%), peptide-functionalized alginate was mixed with 1% oxidized blank alginate in the ratio 1:1 (w/w) to prepare 1% (w/v) alginate aqueous solution. The solution was placed on agarose substrate containing 100 mM Ca2+ to form alginate gel. HUVECs (500,000 cells/cm2) were seeded onto the surfaces of the cross-linked alginates. The HUVECs on the alginates were incubated for 12 hours. The media were then removed, and the alginates were rinsed with PBS three times and then supplemented with the fresh media. A 10-ul pipette tip was used to create a noncell-attached area by scratching the surfaces of alginates. The cell migration was monitored at the scratched areas on different time points and plotted to calculate the cell migration speed. HUVECs were cultured with EGM supplemented with mitomycin C (5 μg/ml).
Vasculogenesis assay
The Ca2+-containing gelatin substrates for alginate cross-linking was prepared as the previous literature mentioned (49). In the first-batch experiments, alginate modified with the peptide combinations (MMPQK only, α1 only, and α1 + MMPQK) was dissolved into HUVEC media to prepare 2% (w/v) peptide-functionalized alginate solution. No peptide functionalized alginate hydrogel was used as the control. For the second batch, alginate modified with the peptide combinations (RGDS + MMPQK, A99 + MMPQK, PA26 + MMPQK, β4 + MMPQK, and α1 + MMPQK) was dissolved into HUVEC media to prepare 2% (w/v) peptide-functionalized alginate solution. For both batches, the solution was mixed with HUVECs (2 million cells/ml) and hADSCs (1 million cells/ml). One hundred microliters of mixture of the alginate and HUVECs was transferred into the Ca2+-containing gelatin-coated 96-well plates and gelled for 40 min. Then, the well plates were transferred into incubator for 10 min to melt the Ca2+-containing gelatin substrates. After Ca2+-containing substrates were removed, media was added into each well and cultured for 4 days. Media were changed every day. The cells were then fixed and stained with DAPI and anti-CD31 (1:200 dilution; 550389, BD Biosciences). The fluorescent pictures were taken by a Leica TCS SP5 confocal microscope system. The quantification of number of branches, number of branch points, number of lumens, total lumen covered areas, and total network length of EC network are through ImageJ with the plug “Angiogenesis Analyzer.” The 3D reconstruction was accomplished by Imaris (Oxford Instruments, UK.).
The test of mechanical properties of the hydrogel
Rheological measurements were performed using an AR-G2 rheometer (TA Instruments). For characterization of peptide-functionalized hydrogels, an 8-mm standard steel parallel plate geometry with a measurement gap of 1 mm was used. A dynamic oscillatory frequency sweep from 0.5 to 100 rad/s at 1% strain was used to compare storage and loss modulus between hydrogel groups. A solvent trap was used to keep gels hydrated during testing. For characterization of peptide-functionalized alginate hydrogel, a 25-mm standard steel parallel plate geometry with a measurement gap of 1 mm was used. A shear stress ramp was swept from 0.01 to 100 Pa, sampling every 3 s for a 4-min duration. A shear rate ramp was swept from 0.01 to 100 (1/s), sampling every 3 s for a 4-min duration. A dynamic oscillatory frequency sweep from 0.01 to 10 Hz at 1% strain was used to measure storage modulus, loss modulus, and complex viscosity. A 2-min equilibration step was performed for all measurements, and the temperature was 25°C throughout the measurement period for all procedures.
Hindlimb ischemia surgery and biomaterials injection
The hindlimb ischemia surgery was introduced as previous literature demonstrated (50). All experiments using this model were reviewed and approved by the Institutional Animal Care and Use Committee of Yonsei University (approval number: IACUC-A-201612-518-01). Briefly, mice (C57BL/6, 11-week-old male; OrientBio, Seongnam, Korea) were anesthetized with ketamine (100 mg/kg; Yuhan, Seoul, Korea) and xylazine (20 mg/kg; Bayer Korea, Ansan, Korea). After that, the femoral artery and its branches were ligated with 6-0 prolene sutures (Ethicon, Somerville, NJ, USA). The arteries were then excised from the femoral artery’s proximal origin to the distal point of bifurcation of the saphenous and popliteal arteries (33). Mice were randomly distributed into multiple groups (as shown in fig. S9B, n = 4 to 5 for each group) and injected with 50 ul of a mixture (1:1 ratio) of the 4% (w/w) alginate gel and calcium gluconate solution (112 mg/ml) (or media as control). dECM hydrogel from mouse skeletal muscle (MEM) was prepared as previously reported (51) and also served as a control group. The mice were examined by serial scanning of the hindlimb surface blood flow with the laser Doppler perfusion imager (Moor Instruments, Devon, UK) on days 0, 14, 21, and 28 after treatments. Digital color-coded images were analyzed to quantify blood flow from the knee joint to the toe region, and perfusion rate was calculated by the ratio of the ischemic limb to the normal limb. The functional recovery of ischemic limb was examined by a gait analysis as previously reported (52). The limbs of each mouse were painted with colors using nontoxic inks and encouraged to walk on a paper. The interval between ischemic injured hindlimb footprints was measured on day 14 after hydrogel treatments. The ischemic muscles were harvested 28 days after the biomaterials injection, fixed with 4% paraformaldehyde (Sigma-Aldrich) overnight, and embedded in paraffin and sectioned (4 μm). The sectioned samples were stained using hematoxylin and eosin to quantify the muscle degeneration as well as tissue inflammation (53) and Masson’s trichrome to examine fibrotic area in the ischemic regions (53). Immunohistochemistry was performed on the capillaries and arterioles using anti–von Willebrand factor (vWF) (1:200 dilution, AB7356, Millipore) and anti–smooth muscle α-actin (SMA) (1:200 dilution; MAB1522, Millipore) following standard histological procedures. The density of the capillaries and arterioles was quantified by counting vWF-positive microvessels and SMA-positive microvessels, respectively. The sections were counterstained with hematoxylin (Sigma-Aldrich) and imaged with an inverted microscope (IX71, Olympus, Tokyo, Japan).
RNA sequencing
Total RNA was isolated from cell-containing (HUVEC seeding density, 2 million cells/ml) peptide-modified alginate [at alginate concentration 1% (w/v)] hydrogels after 24 hours of culture according to the kit and protocol of an Omega Bio-tek E.Z.N.A. including the use of the Homogenizer Columns during the homogenization step (Omega Bio-tek, Norcross, GA). For each group (n = 3), 50-μl hydrogels were made using 2 million cells/ml. To prepare RNA-seq libraries, the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA) was used; 100 to 200 ng of total input RNA was used in accordance with the manufacturer’s protocol. High-throughput sequencing (HTS) was performed using an Illumina HiSeq 2000 with each mRNA library sequenced to a minimum depth of ~50 million reads. A single-end 50-cycle sequencing strategy was used. Data were subjected to Illumina quality control procedures (>80% of the data yielded a Phred score of 30). RNA-seq data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE126717.
Secondary analyses were carried out on an OnRamp Bioinformatics Genomics Research Platform as previously described (OnRamp Bioinformatics, San Diego, CA, USA) (54). OnRamp’s Advanced Genomics Analysis Engine uses an automated RNA-seq workflow to process data, including (i) FastQC to perform data validation and quality control; (ii) CutAdapt (55) to trim and filter adapter sequences, primers, poly-A tails, and other unwanted sequences; (iii) TopHat2 (56) to align mRNA sequencing reads to hg19 genome using the ultrahigh-throughput splice-aware short-read aligner Bowtie2 (57); (iv) HTSeq (58) to establish counts that represent the number of reads for each transcript; and (v) DESeq2 (59) to perform DE analysis, which enabled the inference of differential signals with robust statistical power. Transcript count data from DESeq2 analysis of the samples were sorted according to their adjusted P value (or q value), which is the smallest false discovery rate (FDR) at which a transcript is called significant. FDR is the expected fraction of false-positive tests among significant tests and was calculated using the Benjamini-Hochberg multiple testing adjustment procedure. Advaita Bio’s iPathwayGuide was used to perform further characterization, including a summary of DE genes (α1 compared to RGDS groups), GO, and pathway analysis for genes with at least 1.5-fold change in a positive or negative direction and a q value of <0.05. The DE gene list of our study can be found in table S2.
GO terms with P < 0.05 from the Advaita Bio’s iPathwayGuide output (table S3) were then visualized in semantic similarity–based scatterplots using REViGO that combines redundant terms into a single, representative term based on a simple clustering algorithm relying on semantic similarity measures (60). For each GO category, all significant GO terms or the maximum number (i.e., 350) were used, and P value was notated by size and color of circle. Selected GO terms were labeled to indicate overall spatial organization, and spatially similar terms were subjectively grouped into larger categories (i.e., colored grouping). Vasculature development genes were defined according to the GO BP vasculature development term (GO:0001944). Heat maps were made using the pheatmap package in R using regularized-logarithm transformed counts (rlog function within DESeq2 after correcting for sequencing depth). When comparing the genes between our study and the publically available Matrigel study (GSE93511), RSEM (RNA-Seq by Expectation-Maximization)–expected counts of 3D hydrogel HUVEC samples, PEG-RGDS (GEO sample IDs: 7_In_PEG_HUVEC, 17_In_PEG_HUVEC) and Matrigel (GEO sample IDs: 19_In_MG_HUVEC, 9_In_MG_HUVEC), from GSE93511 were rounded to whole numbers, filtered for shared genes, corrected for sequencing depth, and rlog-transformed before all up-regulated/down-regulated vasculature development genes (i.e., part of GO BP vasculature development term, GO:0001944) were included in a supplementary heat map and ordered by fold change from our experiment. The Venn diagram comparison of up-regulated/down-regulated vasculature development genes included all vasculature development genes from each dataset (significant and nonsignificant) generated. The lists of vasculature development genes from each study can be found in the table S4.
Statistical analysis
The results were shown in the means ± SD/SEM and analyzed using Sigmaplot, GraphPad, and Excel statistical software. All the analysis separately used unpaired Student’s t tests. A confidence interval of 95% was used.
Supplementary Material
Acknowledgments
We would like to acknowledge W. da Silveira for insight into RNA-seq/GSEA analysis. Funding: This work was supported in part by Brain Korea 21(BK21) PLUS program. Eun Je Jeon is fellowship awardee by BK21 PLUS program. The work was supported by the National Institutes of Health (R01 HL133308 and 8P20 GM103444), the NIH Cardiovascular Training Grant (T32 HL007260), and the grants (2017R1A2B3005994 and 2018M3A9H1021382) from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT), Republic of Korea and the Institute for Basic Science (IBS-R026-D1). This study used the services of the Morphology, Imaging, and Instrumentation Core, which is supported by NIH-NIGMS P30 GM103342 to the South Carolina COBRE for Developmentally Based Cardiovascular Diseases and was supported, in part, by the Genomics Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313). This work was also supported in part by the National Science Foundation EPSCoR program under NSF award number EPS-0903795 and number OIA-1655740. This work was supported in part by Brain Korea 21(BK21) PLUS program. E.J.J. is fellowship awardee by BK21 PLUS program. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect those of the National Science Foundation. Author contributions: J.J., S-W.C., and Y.M. conceived the study with assistance from J.C.C., M.J.Y., and S.G.; J.J., E.J., and D.J.R. designed the experiments with M.L., S-W.C., and Y.M.; J.J. supervised all of the experiments, led the data analyses and manuscript preparation with S-W.C. and Y.M. Bioinformatics-assisted ECM protein screening and motif analysis were performed by J.J.; J.J. and R.W.B. synthesized all of the peptides and materials used in study. J.J., X.L., and J.C.C. performed the LC-MS and 1H-NMR for the characterization of the peptides and materials. J.J. performed the peptide-functionalized microarray screening, in vitro cell adhesion, migration, antibody blocking and soluble-peptide blocking experiments. J. J. and R.W.B. performed the test of mechanical properties of the hydrogel and the vasculogenesis assay. J.J. performed in vitro immunofluorescence staining, confocal imaging and image analysis with assistance from M.L., R.C., and D.J.R.; J.J. and D.J.R. performed all of the RNA-seq analysis of a1/RGDS and public datasets (Matrigel/RGDS), including meta-analysis and GSEA. J.J., E.J., M.L., S.L., Y.J., S-W.C., and Y.M. designed the animal experiments. E.J., S.L., and Y.J. performed the animal experiments, gait analysis and immunohistochemistry staining. J.J. and E.J. made the data analysis and data visualization for the results of the animal experiments, gait analysis and immunohistochemistry staining. J.C., M.J.Y., S.G., S-W.C., and Y.M. supervised the efforts, including the manuscript preparation. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Correspondence and requests for materials should be addressed to seungwoocho@yonsei.ac.kr and mei@clemson.edu.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/28/eaaz5894/DC1
REFERENCES AND NOTES
- 1.Briquez P. S., Clegg L. E., Martino M. M., Gabhann F. M., Hubbell J. A., Design principles for therapeutic angiogenic materials. Nat. Rev. Mater. 1, 15006 (2016). [Google Scholar]
- 2.Carmeliet P., Jain R. K., Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutolf M. P., Hubbell J. A., Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 23, 47–55 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Rosales A. M., Anseth K. S., The design of reversible hydrogels to capture extracellular matrix dynamics. Nat. Rev. Mater. 1, 15012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khetan S., Guvendiren M., Legant W. R., Cohen D. M., Chen C. S., Burdick J. A., Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 12, 458–465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen E. H., Zanotelli M. R., Schwartz M. P., Murphy W. L., Differential effects of cell adhesion, modulus and VEGFR-2 inhibition on capillary network formation in synthetic hydrogel arrays. Biomaterials 35, 2149–2161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali S., Saik J. E., Gould D. J., Dickinson M. E., West J. L., Immobilization of cell-adhesive laminin peptides in degradable PEGDA hydrogels influences endothelial cell tubulogenesis. BioRes. Open Access 2, 241–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Acuña R., Quirós M., Huang S., Siuda D., Spence J. R., Nusrat A., García A. J., PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat. Protoc. 13, 2102–2119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iorio V., Troughton L. D., Hamill K. J., Laminins: Roles and utility in wound repair. Adv. Wound Care 4, 250–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis G. E., Senger D. R., Endothelial extracellular matrix: Biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ. Res. 97, 1093–1107 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Hughes C. S., Postovit L. M., Lajoie G. A., Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886–1890 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Nomizu M., Yokoyama F., Suzuki N., Okazaki I., Nishi N., Ponce M. L., Kleinman H. K., Yamamoto Y., Nakagawa S., Mayumi T., Identification of homologous biologically active sites on the N-terminal domain of laminin alpha chains. Biochemistry 40, 15310–15317 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Nomizu M., Kuratomi Y., Song S.-Y., Ponce M. L., Hoffman M. P., Powell S. K., Miyoshi K., Otaka A., Kleinman H. K., Yamada Y., Identification of cell binding sequences in mouse laminin γ1 chain by systematic peptide screening. J. Biol. Chem. 272, 32198–32205 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Katagiri F., Ishikawa M., Yamada Y., Hozumi K., Kikkawa Y., Nomizu M., Screening of integrin-binding peptides from the laminin α4 and α5 chain G domain peptide library. Arch. Biochem. Biophys. 521, 32–42 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Sasaki M., Kleinman H. K., Huber H., Deutzmann R., Yamada Y., Laminin, a multidomain protein. The A chain has a unique globular domain and homology with the basement membrane proteoglycan and the laminin B chains. J. Biol. Chem. 263, 16536–16544 (1988). [PubMed] [Google Scholar]
- 16.Engel J., EGF-like domains in extracellular matrix proteins: Localized signals for growth and differentiation? FEBS Lett. 251, 1–7 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Ranwez V., Delsuc F., Ranwez S., Belkhir K., Tilak M.-K., Douzery E. J. P., OrthoMaM: A database of orthologous genomic markers for placental mammal phylogenetics. BMC Evol. Biol. 7, 241 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider T. D., Stephens R. M., Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierschbacher M. D., Ruoslahti E., Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. U.S.A. 81, 5985–5988 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genové E., Shen C., Zhang S., Semino C. E., The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function. Biomaterials 26, 3341–3351 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Jia J., Coyle R. C., Richards D. J., Berry C. L., Barrs R. W., Biggs J., Chou C. J., Trusk T. C., Mei Y., Development of peptide-functionalized synthetic hydrogel microarrays for stem cell and tissue engineering applications. Acta Biomater. 45, 110–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tashiro K.-i., Sephel G. C., Greatorex D., Sasaki M., Shirashi N., Martin G. R., Kleinman H. K., Yamada Y., The RGD containing site of the mouse laminin A chain is active for cell attachment, spreading, migration and neurite outgrowth. J. Cell. Physiol. 146, 451–459 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Freitas V. M., Vilas-Boas V. F., Pimenta D. C., Loureiro V., Juliano M. A., Carvalho M. R., Pinheiro J. J. V., Camargo A. C. M., Moriscot A. S., Hoffman M. P., SIKVAV, a laminin α1-derived peptide, interacts with integrins and increases protease activity of a human salivary gland adenoid cystic carcinoma cell line through the ERK 1/2 signaling pathway. Am. J. Pathol. 171, 124–138 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frith J. E., Mills R. J., Hudson J. E., Cooper-White J. J., Tailored integrin–extracellular matrix interactions to direct human mesenchymal stem cell differentiation. Stem Cells Dev. 21, 2442–2456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graf J., Ogle R. C., Robey F. A., Sasaki M., Martin G. R., Yamada Y., Kleinman H. K., A pentapeptide from the laminin B1 chain mediates cell adhesion and binds to 67000 laminin receptor. Biochemistry 26, 6896–6900 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Nomizu M., Kim W. H., Yamamura K., Utani A., Song S. Y., Otaka A., Roller P. P., Kleinman H. K., Yamada Y., Identification of cell binding sites in the laminin α1 chain carboxyl-terminal globular domain by systematic screening of synthetic peptides. J. Biol. Chem. 270, 20583–20590 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Conforti G., Dominguez-Jimenez C., Zanetti A., Gimbrone M. A. Jr., Cremona O., Marchisio P. C., Dejana E., Human endothelial cells express integrin receptors on the luminal aspect of their membrane. Blood 80, 437–446 (1992). [PubMed] [Google Scholar]
- 28.Bouvard C., Gafsou B., Dizier B., Galy-Fauroux I., Lokajczyk A., Boisson-Vidal C., Fischer A.-M., Helley D., α6-integrin subunit plays a major role in the proangiogenic properties of endothelial progenitor cells. Arterioscler. Thromb. Vasc. Biol. 30, 1569–1575 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Kerr J. S., Mousa S. A., Slee A. M., αvβ3 integrin in angiogenesis and restenosis. Drug News Perspect. 14, 143 (2001). [PubMed] [Google Scholar]
- 30.Patterson J., Hubbell J. A., Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials 31, 7836–7845 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Schwartz M. P., Hou Z., Bai Y., Ardalani H., Swanson S., Steill J., Ruotti V., Elwell A., Nguyen B. K., Bolin J., Stewart R., Thomson J. A., Murphy W. L., A genome-wide analysis of human pluripotent stem cell-derived endothelial cells in 2D or 3D culture. Stem Cell Rep. 8, 907–918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blinder Y. J., Freiman A., Raindel N., Mooney D. J., Levenberg S., Vasculogenic dynamics in 3D engineered tissue constructs. Sci. Rep. 5, 17840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H.-J., Jin Y., Shin J., Yang K., Lee C., Yang H. S., Cho S.-W., Catechol-functionalized hyaluronic acid hydrogels enhance angiogenesis and osteogenesis of human adipose-derived stem cells in critical tissue defects. Biomacromolecules 17, 1939–1948 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Bidarra S. J., Barrias C. C., Fonseca K. B., Barbosa M. A., Soares R. A., Granja P. L., Injectable in situ crosslinkable RGD-modified alginate matrix for endothelial cells delivery. Biomaterials 32, 7897–7904 (2011). [DOI] [PubMed] [Google Scholar]
- 35.DeQuach J. A., Lin J. E., Cam C., Hu D., Salvatore M. A., Sheikh F., Christman K. L., Injectable skeletal muscle matrix hydrogel promotes neovascularization and muscle cell infiltration in a hindlimb ischemia model. Eur. Cell. Mater. 23, 400–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Healy J. M., Murayama O., Maeda T., Yoshino K., Sekiguchi K., Kikuchi M., Peptide ligands for integrin. alpha.v.beta.3 selected from random phage display libraries. Biochemistry 34, 3948–3955 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Nomizu M., Kuratomi Y., Malinda K. M., Song S.-Y., Miyoshi K., Otaka A., Powell S. K., Hoffman M. P., Kleinman H. K., Yamada Y., Cell binding sequences in mouse laminin α1 chain. J. Biol. Chem. 273, 32491–32499 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Martino M. M., Tortelli F., Mochizuki M., Traub S., Ben-David D., Kuhn G. A., Müller R., Livne E., Eming S. A., Hubbell J. A., Engineering the growth factor microenvironment with fibronectin domains to promote wound and bone tissue healing. Sci. Transl. Med. 3, 100ra89 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Ott H. C., Matthiesen T. S., Goh S.-K., Black L. D., Kren S. M., Netoff T. I., Taylor D. A., Perfusion-decellularized matrix: Using nature's platform to engineer a bioartificial heart. Nat. Med. 14, 213–221 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Carmeliet P., Angiogenesis in life, disease and medicine. Nature 438, 932–936 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Bouïs D., Kusumanto Y., Meijer C., Mulder N. H., Hospers G. A. P., A review on pro-and anti-angiogenic factors as targets of clinical intervention. Pharmacol. Res. 53, 89–103 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Söding J., Protein homology detection by HMM–HMM comparison. Bioinformatics 21, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A., Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188, 415–431 (1986). [DOI] [PubMed] [Google Scholar]
- 45.Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouhadir K. H., Lee K. Y., Alsberg E., Damm K. L., Anderson K. W., Mooney D. J., Degradation of partially oxidized alginate and its potential application for tissue engineering. Biotechnol. Prog. 17, 945–950 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Hein J. E., Fokin V. V., Copper-catalyzed azide–alkyne cycloaddition (CuAAC) and beyond: New reactivity of copper (I) acetylides. Chem. Soc. Rev. 39, 1302–1315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia J., Richards D. J., Pollard S., Tan Y., Rodriguez J., Visconti R. P., Trusk T. C., Yost M. J., Yao H., Markwald R. R., Mei Y., Engineering alginate as bioink for bioprinting. Acta Biomater. 10, 4323–4331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pataky K., Braschler T., Negro A., Renaud P., Lutolf M. P., Brugger J., Microdrop printing of hydrogel bioinks into 3D tissue-like geometries. Adv. Mater. 24, 391–396 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Hellingman A. A., Bastiaansen A. J. N. M., de Vries M. R., Seghers L., Lijkwan M. A., Löwik C. W., Hamming J. F., Quax P. H. A., Variations in surgical procedures for hind limb ischaemia mouse models result in differences in collateral formation. Eur. J. Vasc. Endovasc. Surg. 40, 796–803 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Jin Y., Lee J. S., Kim J., Min S., Wi S., Yu J. H., Chang G.-E., Cho A.-N., Choi Y., Ahn D.-H., Cho S.-R., Cheong E., Kim Y.-G., Kim H.-P., Kim Y., Kim D. S., Kim H. W., Quan Z., Kang H.-C., Cho S.-W., Three-dimensional brain-like microenvironments facilitate the direct reprogramming of fibroblasts into therapeutic neurons. Nat. Biomed. Eng. 2, 522 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Jayabal S., Ljungberg L., Erwes T., Cormier A., Quilez S., Jaouhari S. E., Watt A. J., Rapid onset of motor deficits in a mouse model of spinocerebellar ataxia type 6 precedes late cerebellar degeneration. eNeuro 2, ENEURO.0094-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang F., Cho S.-W., Son S. M., Bogatyrev S. R., Singh D., Green J. J., Mei Y., Park S., Bhang S. H., Kim B.-S., Langer R., Anderson D. G., Genetic engineering of human stem cells for enhanced angiogenesis using biodegradable polymeric nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 107, 3317–3322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis-Turak J., Courtney S. M., Hazard E. S., Glen W. B. Jr., da Silveira W. A., Wesselman T., Harbin L. P., Wolf B. J., Chung D., Hardiman G., Genomics pipelines and data integration: Challenges and opportunities in the research setting. Expert Rev. Mol. Diagn. 17, 225–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 56.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/28/eaaz5894/DC1