Significance
In 2016/2017, highly pathogenic avian influenza (HPAI) virus of the subtype H5 spilled over into wild birds and caused the largest known HPAI epidemic in Europe, affecting poultry and wild birds. During its spread, the virus frequently exchanged genetic material (reassortment) with cocirculating low-pathogenic avian influenza viruses. To determine where and when these reassortments occurred, we analyzed Eurasian avian influenza viruses and identified a large set of H5 HPAI reassortants. We found that new genetic material likely came from wild birds across their migratory range and from domestic ducks not only in China, but also in central Europe. This knowledge is important to understand how the virus could adapt to wild birds and become established in wild bird populations.
Keywords: highly pathogenic avian influenza, emerging infectious diseases, phylogenetic analysis, poultry, wild birds
Abstract
Highly pathogenic avian influenza (HPAI) viruses of the H5 A/goose/Guangdong/1/96 lineage can cause severe disease in poultry and wild birds, and occasionally in humans. In recent years, H5 HPAI viruses of this lineage infecting poultry in Asia have spilled over into wild birds and spread via bird migration to countries in Europe, Africa, and North America. In 2016/2017, this spillover resulted in the largest HPAI epidemic on record in Europe and was associated with an unusually high frequency of reassortments between H5 HPAI viruses and cocirculating low-pathogenic avian influenza viruses. Here, we show that the seven main H5 reassortant viruses had various combinations of gene segments 1, 2, 3, 5, and 6. Using detailed time-resolved phylogenetic analysis, most of these gene segments likely originated from wild birds and at dates and locations that corresponded to their hosts’ migratory cycles. However, some gene segments in two reassortant viruses likely originated from domestic anseriforms, either in spring 2016 in east China or in autumn 2016 in central Europe. Our results demonstrate that, in addition to domestic anseriforms in Asia, both migratory wild birds and domestic anseriforms in Europe are relevant sources of gene segments for recent reassortant H5 HPAI viruses. The ease with which these H5 HPAI viruses reassort, in combination with repeated spillovers of H5 HPAI viruses into wild birds, increases the risk of emergence of a reassortant virus that persists in wild bird populations yet remains highly pathogenic for poultry.
Infection with highly pathogenic avian influenza (HPAI) virus of the H5 A/goose/Guangdong/1/96 (Gs/Gd) lineage can cause severe disease in birds and in mammals, including people (1, 2). Since the beginning of the century, H5 HPAI viruses have repeatedly spilled over from poultry to free-living wild birds, especially in Asia. This spillover in regions with a high poultry density and intensive interaction between wild bird and poultry populations has altered the epidemiology of H5 HPAI viruses in several ways. First, migration of HPAI virus-infected wild birds acts as a new route of long-distance spread of H5 HPAI viruses into countries with bird populations that were free of these viruses (3). Second, direct or indirect contact with infected wild birds is a new route of HPAI virus incursion into poultry farms (4–7). Third, H5 HPAI virus infection is a new source of considerable mortality in wild birds themselves, and it may substantially affect population dynamics of wild birds and threaten highly protected species like the white-tailed eagle (Haliaeetus albicilla) and the peregrine falcon (Falco peregrinus) (8, 9). Because we have a poor understanding of wild birds as a new niche for H5 HPAI viruses, it is difficult to design efficient surveillance programs, as well as effective prevention and control measures.
Since 1996, the Gs/Gd lineage of H5 HPAI viruses has evolved rapidly and is now highly diverse. Following the emergence of Gs/Gd, the lineage has evolved into numerous genetically distinct clades (10, 11). Several of these clades have spread via wild birds from Asia to Europe since 2004: clade 1 in 2004, clades 2.2 and 2.2.1 from 2005 to 2007, clade 2.3.2 from 2008 to 2010, and clade 2.3.4.4 from 2014 to 2019 (1, 3, 12, 13). Gs/Gd lineage H5 HPAI viruses are now endemic in areas of Asia and continue to evolve, so new epidemics are likely to occur.
The routes by which H5 HPAI viruses are carried over long distances, as well as the particular migratory species involved, are still only roughly known. From wintering grounds of migratory birds in Southeast Asia, H5 HPAI viruses are carried north to breeding grounds on the northern parts of the Eurasian and North American continents and then west to wintering grounds in Europe, east to wintering grounds in North America, or back south to wintering grounds in Asia (3). The species involved are thought to be long-distance migrants of the family Anatidae (ducks, geese, and swans). Several species in this family (e.g., Eurasian wigeon [Mareca penelope], Eurasian teal [Anas crecca], and northern pintail [Anas acuta]) have migratory routes that correspond to the observed pattern of virus spread (3), have been found infected with H5 HPAI virus at different locations along their respective migratory routes (3), and can be infected with and excrete H5 HPAI virus—at least under laboratory conditions—without showing detectable clinical signs (1, 14–17).
In 2016 and 2017, H5 HPAI viruses (belonging to cluster B, Gochang like) spread again widely across Eurasia, causing the largest and most widespread HPAI epidemic ever recorded in Europe: between October 2016 and August 2017, 1,207 individual HPAI virus outbreaks in poultry holdings were reported in 24 European Union countries, and 1,590 wild bird mortality events were recorded in 29 countries of the European Union and in Switzerland (18). In contrast, relatively few wild birds and poultry farms were affected in 2014/2015: between November 2014 and February 2015, H5N8 HPAI was detected in only 11 poultry farms (turkey, duck, chicken) and other holdings and in a small number of wild waterfowl across five European countries, as well as in two mute swans (Cygnus olor) in Sweden (3, 19), indicating a marked difference between epidemics.
During the 2016 to 2017 epidemic, five reassortant viruses were detected in Germany alone, to which the H5 HPAI virus contributed at least the clade 2.3.4.4 hemagglutinin (HA) gene segment and often, also that clade’s corresponding matrix (M) and nonstructural (NS) gene segments, while cocirculating low-pathogenic avian influenza (LPAI) viruses contributed gene segments coding for the other viral proteins (20). The genetic diversity generated through reassortment plays an important role in the evolution of influenza viruses (21) and may provide the opportunity for the adaptation of H5 HPAI virus to wild bird populations. Despite this, we know little about the genesis and spread of these reassortants. We therefore performed extensive phylogeographic and epidemiological analyses of the 2016 to 2017 H5 HPAI virus epidemic, including analysis of the temporal and geographical spread of the reassortants and their individual gene segments, and of the avian species involved. Our main goals were to estimate where and when these reassortments occurred and to identify which avian influenza viruses from which host species provided gene segments for reassortment. We used genetic sequences of avian influenza viruses obtained from poultry and wild birds worldwide and shared through public databases and epidemiological data obtained from the World Organization for Animal Health. We focused on HPAI viruses of H5 clade 2.3.4.4 collected between May 2016 and July 2017. Data were discussed within the Global Consortium of H5N8 and Related Influenza Viruses (3). The aim of this consortium is to foster data exchange and global analysis of H5Nx avian influenza epidemics (3).
Results
Classification of Whole-Genome Sequences of HPAI Virus H5Nx into Reassortants per Gene Segment.
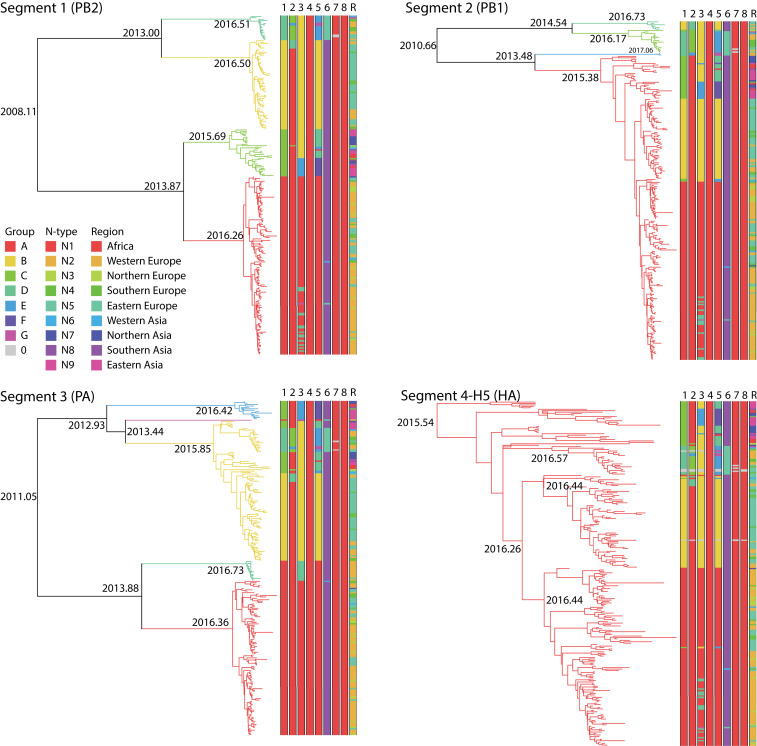
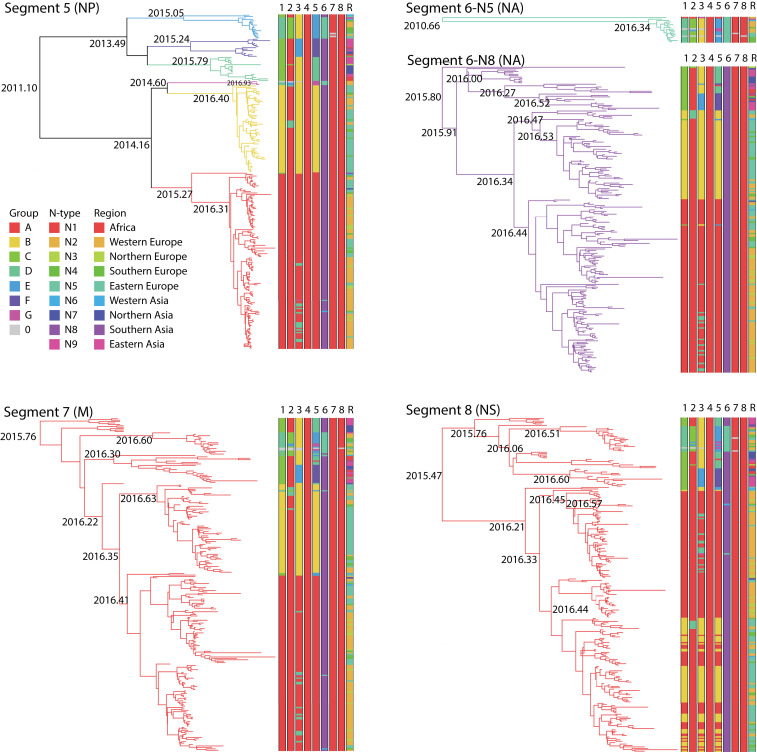
Sequence data for individual gene segments 1 (polymerase basic 2, PB2), 2 (polymerase basic 1, PB1), 3 (polymerase acidic, PA), 5 (nucleoprotein, NP), 7 (M), and 8 (NS) were classified into groups based on phylogenetic similarity, and sequence data for gene segment 6 (neuraminidase, NA) were classified by subtype (Materials and Methods). Using BEAST (22), time-scaled trees were inferred for each segment or subtype for segment 6 (Figs. 1 and 2). The discontinuous character of the phylogenetic groups was consistent with the importation of novel sequences (i.e., reassortment or genetic shift), suggesting that these groups corresponded to reassortants (Table 1). These results suggested that segment 4 (the HA gene) had four main combinations (A to D) with segment 1, three (A, C, D) with segment 2, four (A, B, D, E) with segment 3, five (A, B, D, E, F) with segment 5, two (N8 and N5) with segment 6, and one (A) with segments 7 and 8. The phylogenetic analysis of the eight gene segments indicated that, in some cases, multiple gene segments transferred in the same reassortment event. An example is the simultaneous reassortment of gene segments 1, 3, and 5 (Figs. 1 and 2 and Table 1).
Fig. 1.
Bayesian time-resolved phylogenetic trees for gene segments 1 to 4 of H5NX HPAI viruses sampled between May 2016 and July 2017 and for which whole genomes were sequenced. Distinct clades (groups) in the trees are given different colored branches. The number of distinct groups differs per gene segment: four for segment 1, three for segment 2, five for segment 3, and one for segment 4. Furthermore, all eight segments of each virus are represented as eight parallel bars at the tips of each tree, with the colors in each segment's bar corresponding to the branches of the individual gene sequences to indicate reassortment. The final bar represents the geographic locations of the sequences in the tree.
Fig. 2.
Bayesian time-resolved phylogenetic trees for gene segments 5 to 8 of H5NX HPAI viruses sampled between May 2016 and July 2017 and for which whole genomes were sequenced. For segment 6, there are separate phylogenetic trees for subtypes N8 and N5. Distinct clades (groups) in the trees are given different colored branches. The number of distinct groups differs per gene segment: five for segment 5, two for segment 6, and one for segments 7 and 8. Furthermore, all eight segments of each virus are represented as eight parallel bars at the tips of each tree, with the colors in each segment’s bar corresponding to the branches of the individual gene sequences to indicate reassortment. The final bar represents the geographic locations of the sequences in the tree.
Table 1.
The names of the main reassortants and the phylogenetic groups within each gene segment
| Reassortant | Date of first detection (full-genome sequence) | Estimated date of origin and 95% highest posterior density CIs | No. of sequences | Phylogenetic group per gene segment | |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||||
| No. | Name | Group | PB2 | PB1 | PA | HA | NP | NA | M1 | NS1 | |||
| 1 | CABAD8AA | I | 5/1/16 | 3 February 2016 | 16 | C | A | B | A | D | 8 | A | A |
| 11/15–4/16 | |||||||||||||
| 2 | CAEAF8AA | I | 9/10/16 | 9 August 2016 | 13 | C | A | E | A | F | 8 | A | A |
| 5/16–9/16 | |||||||||||||
| 3 | AAAAA8AA | II | 8/27/16 | 13 July 2016 | 109 | A | A | A | A | A | 8 | A | A |
| 5/16–8/16 | |||||||||||||
| 4 | AADAA8AA | II | 11/11/16 | 3 October 2016 | 14 | A | A | D | A | A | 8 | A | A |
| 8/16–10/16 | |||||||||||||
| 5 | DCBAE5AA | III | 10/1/16 | 26 July 2016 | 15 | D | C | B | A | E | 5 | A | A |
| 5/16–9/16 | |||||||||||||
| 6 | BABAB8AA | IV | 10/19/16 | 7 September 2016 | 56 | B | A | B | A | B | 8 | A | A |
| 7/16–10/16 | |||||||||||||
| 7 | BDBAB8AA | IV | 12/2/16 | 4 November 2016 | 6 | B | D | B | A | B | 8 | A | A |
| 9/16–11/16 | |||||||||||||
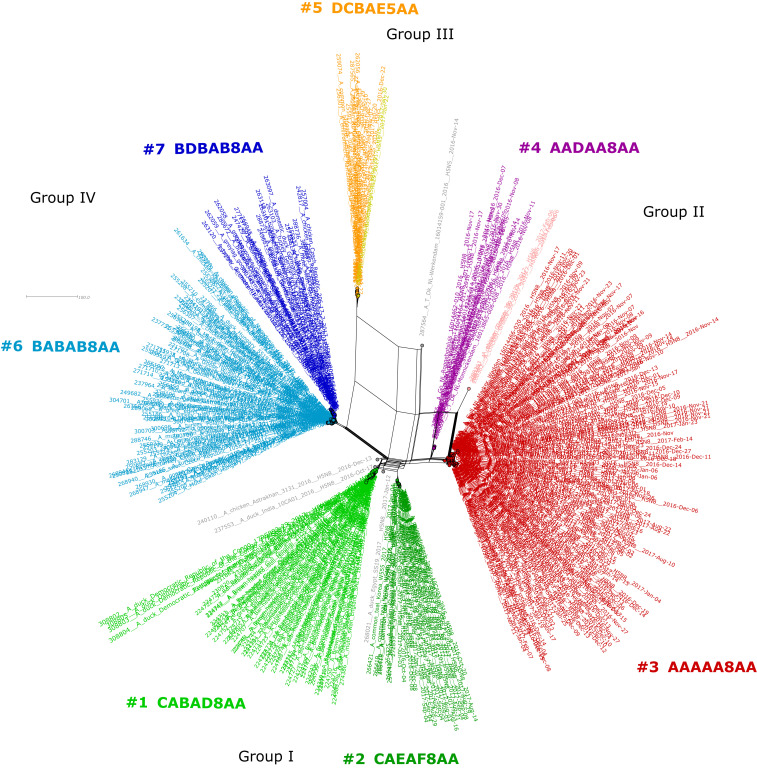
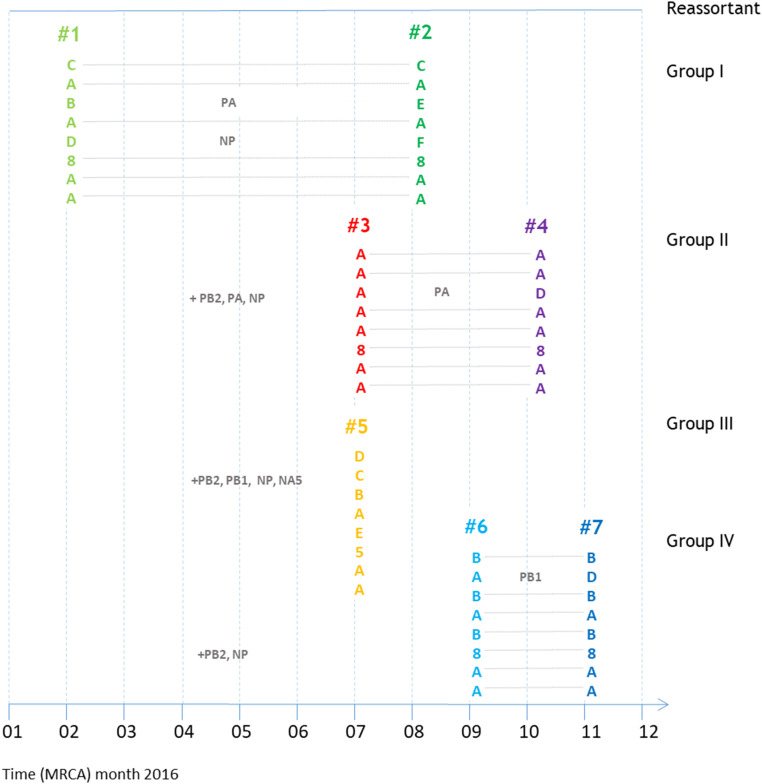
By Influenza Reassortment analysis via Supernetworks (IRIS), a total of 446 full-genome sequences of clade 2.3.4.4 H5 HPAI viruses could be divided into 11 distinct reassortants, 7 of which were common. Supernetworks are calculated from all eight segment-sorted maximum-likelihood trees and visualize the phylogenetic relationships of each HPAI virus where taxa are represented by nodes and their relationships as edges (23). This corresponded very closely to the reassortants distinguished by BEAST analysis. The reassortants formed four groups: group I, first detected in May 2016; group II, first detected in August 2016 and including the reassortant that caused the main epidemic in wild birds and poultry in Europe; group III, first detected in December 2016 and including a change of the NA serotype from eight to five; and group IV, also first detected in December 2016 (Figs. 3 and 4 and Table 1).
Fig. 3.
Supernetwork of 2016/2017 H5 HPAI viruses, based on full-genome sequences, generated by using maximum-likelihood (ML) phylogenetic trees of sorted sequences according to segments. The eight ML trees of the segments were used to calculate a supernetwork. Each reassortant is indicated by a different color and assembled into four groups. The seven most common reassortants are numbered 1 to 7. Less common reassortants (total: four) are indicated in gray. Each circle represents full genome of HPAI virus, and the edges represent their phylogenetic relationship. Details as names, collection dates, and coordinates are given in SI Appendix, Table S8.
Fig. 4.
Overview of the main reassortments described in this manuscript. Group designations and colors correspond to those in Fig. 3. The positions of the reassortants correspond to the estimated dates of origin in Table 1. Dotted lines join the gene segments that are related.
Spatiotemporal Distribution of the Main Reassortants.
The clade H5N8 HPAI virus was widespread in Eurasia in 2016 and extended into North Africa (Fig. 5). During 2016 and 2017, the virus spread widely from east Asia (SI Appendix, Table S1) westward to Europe, eastward to north Asia, and southward to south Asia and Africa between the second quarter (3-mo division) of 2016 and the second quarter of 2017. In the second quarter of 2016, the virus was detected sporadically in north-central China (Qinghai Lake) and the border between Russia and Mongolia (Uvs-Nuur Lake) (SI Appendix, Fig. S1). The westward spread to Europe started with infrequent detections in south-central Russia (Chany Lake and Kurgan in western Siberia) in the third quarter of 2016 (SI Appendix, Fig. S2) and west Russia (Tatarstan) in the fourth quarter of 2016 (SI Appendix, Fig. S3). Subsequently, the virus was detected at great frequency in all parts of Europe (north, south, east, west, central) (SI Appendix, Table S1) in the fourth quarter of 2016 and the first quarter of 2017 (SI Appendix, Fig. S4), followed by infrequent detections in the second quarter of 2017 (SI Appendix, Fig. S5). The virus also was detected in North Africa (Egypt) and south Asia (India) in the fourth quarter of 2016, in central Africa (Congo) in the second quarter of 2017, in north Asia (Kamchatka, Russia) in the fourth quarter of 2016, and in east Asia (South Korea) from the fourth quarter of 2016 to the second quarter of 2017. The species affected were primarily wild birds in the second and third quarters of 2016, both wild birds and poultry in the fourth quarter of 2016 and first quarter of 2017, and poultry in the second quarter of 2017 (SI Appendix, Figs. S1–S5). There was no apparent spatial association between the distribution of sampled viruses and the density of the poultry population in Eurasia (SI Appendix, Fig. S6). During the period 2016/2017, H5N8 HPAI viruses were not detected in other parts of the world, including North and South America.
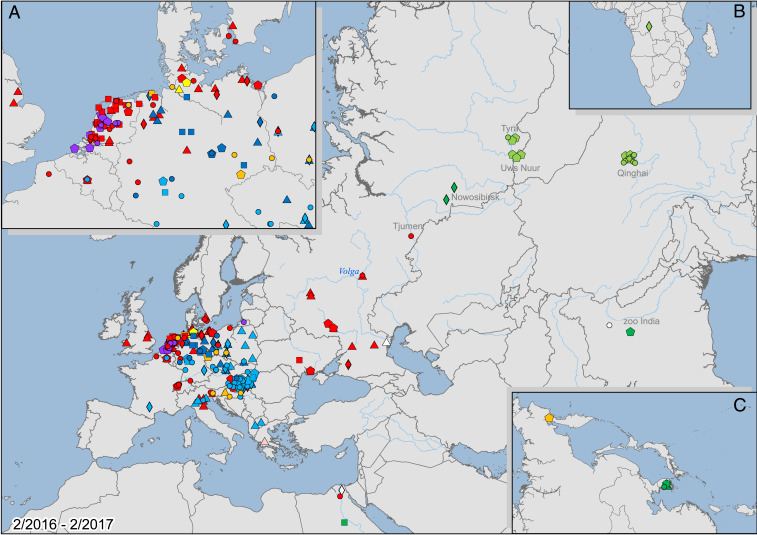
Fig. 5.
Geographical distribution of detected H5NX HPAI viruses with full-genome sequences between the second quarter of 2016 and the second quarter of 2017. Viruses were detected from the east coast of Asia to the northwest coast of Europe, southern to northern Africa, and in south Asia. A–C show magnified maps of (A) the northwest coast of Europe, (B) southern Africa, and (C) the east coast of Asia. Lambert conformal conic projection. Light green: reassortant 1; dark green: reassortant 2; red: reassortant 3; purple: reassortant 4; orange: reassortant 5; light blue: reassortant 6; dark blue: reassortant 7; circle: Wild-ans; square: Wild-ans-long; pentagon: Wild-other; rhomboid: Dom-ans; triangle: Dom-gal. Map image credit: Copyright © 1995–2020 Esri. All rights reserved. Published in the United States of America.
Inferred Hosts, Dates, and Locations of Origin of the Seven Main Reassortants.
Reassortant 1 (CABAD8AA; n = 16).
This is the most ancestral reassortant in our analyses. The origin dates are estimated to be November 2015 for the most recent common grand ancestor (MRCGA) and February 2016 for the most recent common ancestor (MRCA) (SI Appendix, Fig. S7). The gene segments are inferred to originate from west, south, and east China for the MRCGA and north China for the MRCA. The inferred hosts of nearly all gene segments of both the MRCGA and MRCA are either long-range anseriform migrants or wild anseriforms. The exception is gene segment 6 (NA), for which the inferred hosts are domestic anseriforms. These dates and locations suggest that the MRCA of CABAD8AA originated at the end of the wintering period 2015/2016 or beginning of spring migration 2016, with gene segment 6 contributed by domestic anseriforms from east China and remaining gene segments contributed by wild anseriforms (including long-range migrants) from wintering locations in west, south, and east China. It cannot be ruled out that some gene segments (e.g., 3 and 7) were carried farther south during autumn migration 2015 to unsampled wintering locations in south Asia and then back to north China during spring migration.
The first actual CABAD8AA detections were from May 2016 on Qinghai Lake in China and Uvs-Nuur Lake at the Mongolian–Russian border (24, 25), just north of the median inferred location of origin of the MRCA. This suggests that this reassortant was carried farther north during spring migration 2016, presumably by wild birds on the way to breeding locations in north Russia. The first detected virus (A/Brown-headed_Gull/Qinghai/ZTO1-LU/2016) had an HPAI virus H5 clade 2.3.4.4b-derived backbone of segments 4, 6, and 8, combined with segments 1, 2, 3, 5, and 7 from different Asian low pathogenic avian influenza (LPAI) viruses (24, 25). After May 2016, this reassortant was not detected until May 2017, when it reemerged in domestic ducks in the Democratic Republic of Congo (26).
Reassortant 2 (CAEAF8AA; n = 13).
The new gene segments in this reassortant, compared with CABAD8AA, are segments 3 (PA) and 5 (NP). The inferred hosts of these new gene segments could be wild anseriforms, long-range anseriform migrants, other wild birds, or domestic anseriforms (SI Appendix, Fig. S8). The origin dates are estimated to be May 2016 for the MRCGA and August 2016 for the MRCA. The gene segments are inferred to originate from almost the full breadth of Eurasia (west Poland to east China) for the MRCGA and from a narrower breadth of Eurasia (east Azerbaijan to west China, but most locations in the region centered on the Kazakhstan–China border) for the MRCA. These dates and locations suggest that the MRCA of CAEAF8AA originated during premigratory aggregation or autumn migration in the Kazakhstan–China border region.
The first actual CAEAF8AA detection (A/gadwall/Chany/97/2016) was from September 2016 in Russia. It was later found in wild birds at widely dispersed locations: from Italy to Korea and from Russia to Egypt and India. Except for Russia, these sites might be wintering locations of wild migratory birds.
Reassortant 3 (AAAAA8AA; n = 109).
This is the main virus detected in the 2016 to 2017 epidemic in Europe. The new gene segments in this reassortant, compared with CAEAF8AA, are segments 1 (PB2), 3 (PA), and 5 (NP). The inferred hosts of these new gene segments are long-range anseriform migrants (segment 1), other wild birds (segment 3), or either of these two groups (segment 5) (SI Appendix, Fig. S9). The origin dates are estimated to be June 2016 for the MRCGA and July 2016 for the MRCA. The gene segments are inferred to originate from Belarus to India for the MRCGA and from a much more restricted area (Belarus to west Kazakhstan) for the MRCA. These dates and locations suggest that the MRCA of AAAAA8AA originated during premigratory aggregation or early autumn migration 2016 somewhere between Belarus and Kazakhstan.
The first actual AAAAA8AA detection (A/gadwall/Kurgan/2442/2016) was on 27 August 2016 in central Russia at the longitude of the Ural Mountains. The second detection occurred on 2 October 2016 in Tatarstan, Russia, several hundred kilometers westward. From November 2016 onward, this virus was detected in wild birds in coastal regions of the Baltic and North Seas, as well as in wild birds on Lake Constance, Lake Biel, and Lake Neuchâtel, farther south in Europe. It also was found in wild birds in Ukraine, south Russia (Krasnodar), Hungary, and Italy. This suggests that the virus was carried west during autumn migration of wild birds in 2016, on the way to wintering locations in central, west, and south Europe.
Throughout Europe, this virus caused both massive die-offs in wild birds and outbreaks in poultry farms, lasting until at least summer 2017 (8, 20, 27–33).
Reassortant 4 (AADAA8AA; n = 14).
The new gene segment in this reassortant, compared with AAAAA8AA, is segment 3 (PA). The inferred hosts of this gene segment are other wild birds (SI Appendix, Fig. S10). The origin dates are estimated to be August 2016 for the MRCGA and October 2016 for the MRCA. The gene segments of the MRCA are inferred to originate from Germany to west Russia. These dates and locations suggest that the MRCA of AADAA8AA originated during late autumn migration 2016.
The actual detections of AADAA8AA were first in the Netherlands in November 2016 and then in Kaliningrad Oblast, Russia, in February 2017. This suggests that, following its generation in autumn 2016, this reassortant was restricted to a limited number of wintering locations in central and west Europe.
Reassortant 5 (DCBAE5AA; n = 15).
The new gene segments in this reassortant, compared with CABAD8AA, are segments 1 (PB2), 5 (NP), and 6 (NA). The inferred hosts of these gene segments are wild anseriforms (segments 1 and 5), with ancestry of segment 6 remaining unclear, although segment 6 clustered most closely with those of LPAI viruses of the subtypes H9N5 or H7N5 found in 2015 in shorebirds in Asia (A/common redshank/Singapore/F83-1/2015, KU144675; A/black-tailed godwit/Bangladesh/24734/2015, KY635758) (SI Appendix, Fig. S11). The origin dates are estimated to be June 2016 for the MRCGA and July 2016 for the MRCA. The gene segments are inferred to originate from west-central Russia to northwest China to Thailand for the MRCGA and from south-central Russia for the MRCA. These dates and locations suggest that the MRCA of DCBAE5AA originated during the breeding season or premigratory aggregation 2016.
The actual detections of DCBAE5AA were in Georgia, the Czech Republic, Italy, and Germany from December 2016 to February 2017, suggesting that the reassortant was carried by wild birds during autumn migration 2016 to wintering locations in Europe. This virus caused outbreaks in poultry farms in Croatia and Germany. The reassortant also was detected in eastern Eurasia—on the Kamchatka Peninsula, Russia—in October 2016, which it likely reached via a different migration route.
Reassortant 6 (BABAB8AA; n = 56).
This is the second most common virus detected during the 2016 to 2017 epidemic in Europe, after AAAAA8AA. The new gene segments in this reassortant, compared with CABAD8AA, are segments 1 (PB2) and 5 (NP). The inferred host of segment 1 is unclear (either domestic anseriforms or other wild birds), and the inferred host of segment 5 is domestic anseriforms (SI Appendix, Fig. S12). The origin dates are estimated to be July 2016 for the MRCGA and September 2016 for the MRCA. The gene segments are inferred to originate from Ukraine to west Russia for the MRCGA and from several hundred kilometers due west—Hungary to Ukraine—for the MRCA. These dates and locations suggest that the MRCA of BABAB8AA originated during autumn migration 2016.
The first actual detections of BABAB8AA were in Croatia, Hungary, and France from October 2016. This virus was later detected farther north, in Germany and Poland in winter 2016 to 2017 (20, 33). This suggests that the reassortant spread in a restricted part of Europe during late autumn migration and the wintering period and potentially involved spillback of gene segments from domestic anseriforms to wild birds, possibly in Hungary.
Reassortant 7 (BDBAB8AA; n = 6).
The new gene segment in this reassortant, compared with BABAB8AA, is segment 2 (PB1). The inferred hosts of this gene segment are wild anseriforms or long-range anseriform migrants (SI Appendix, Fig. S13). The origin dates are November 2016 for both the MRCGA and the MRCA. However, gene segment 2 has an earlier origin date, May 2016, suggesting that it reached west Europe in a two-step process: transport to breeding location in Siberia during spring migration 2016 and transport to wintering location in west Europe during autumn migration 2016. The gene segments of the MRCGA are inferred to originate from a restricted region—Germany to Ukraine—except for gene segment 2, from west China; those of the MRCA are inferred to originate from an even more restricted region, Poland to Ukraine. These dates and locations suggest that the MRCA of BDBAB8AA originated during late autumn migration 2016 or wintering period 2016 to 2017.
The actual detections of BDBAB8AA were first in Poland and Germany, from December 2016 onward. This suggests that the reassortant originated locally in Europe due to a reassortment of its predecessor, BABAB8AA, with a gene segment from a wild anseriform, likely a long-range migrant from Siberia. This virus caused outbreaks in poultry farms in Poland, Germany, and Russia.
Summary of the Genesis and Spread of Seven Main Reassortants.
In summary (Table 2 and SI Appendix, Fig. S13 and Table S2), most of the new gene segments for the reassortant viruses originated from wild birds (wild anseriforms, including long-range anseriforms, or other wild birds), and the dates and locations of origin correspond with areas used by wild birds during different phases of their migratory cycles: wintering period to spring migration in north China, Mongolia, and Russia (Siberia) for reassortant 1; breeding period to autumn migration in Belarus/Kazakhstan/China/Russia for reassortants 2, 3, and 5; autumn migration in Hungary/Germany/Ukraine/Russia for reassortants 4 and 6; and autumn migration to wintering period in Poland/Ukraine for reassortant 7. The only new gene segments not thought to originate from wild birds were gene segment 6 of reassortant 1 and gene segment 5 of reassortant 6; these gene segments were inferred to originate from domestic anseriforms.
Table 2.
Overview of spatiotemporal spread of reassortants of H5 HPAI viruses in the 2016/2017 epidemic from time of origin of MRCA to 1 July 2017
| No. | Reassortant | Time of origin of MRCA | Country of origin of MRCA | Host origin of new gene segments | Subsequently detected spread of virus | |||||
| Month in 2016 | Phase of life cycle* | Europe† | Northern Africa | Southern Africa‡ | South Asia | East Asia | ||||
| 1 | CABAD8AA | February | M, W | China | Not relevant | X | X | |||
| 3 | AAAAA8AA | July | M, B | Belarus to Kazakhstan | Wild bird | X | ||||
| 5 | DCBAE5AA | July | M, B | Russia | Wild bird | X | X | |||
| 2 | CAEAF8AA | August | M | Azerbaijan to China | Not determined | X | X | X | X | |
| 6 | BABAB8AA | September | M, W | Hungary to Ukraine | Wild bird and poultry | X | ||||
| 4 | AADAA8AA | October | M, W | Germany to Russia§ | Wild bird | X | ||||
| 7 | BDBAB8AA | November | M, W | Poland to Ukraine | Wild bird | X | ||||
B, breeding; M, migration; W, wintering; X, virus detected.
Phase of life cycle of long-distance migratory birds breeding in Siberia.
Ural Mountains taken as geographical border between Europe and Asia.
South border of Sahara taken as geographical border between northern and southern Africa.
Kaliningrad.
Migratory Patterns of Wild Birds Found Positive for H5N8 HPAI Virus in the European Union.
Based on reports to the Office International des Epizooties (OIE), H5N8 HPAI viruses were detected in a total of 56 species found dead in the European Union between 1 October 2016 and 5 July 2017 (18). Of these 56 species, 14 species had migratory populations that wintered in the European Union and bred at longitudes at least 60°E, which is at the longitude of the Ural Mountains (Table 3). Of these 14 species, 13 were water birds belonging to the family Anatidae (including ducks, geese, and swans), while 1 species belonged to the family Turdidae (thrushes).
Table 3.
Wild bird species in which H5N8 HPAI virus was detected in dead birds found in the European Union between 1 October 2016 and 5 July 2017 and reported to the OIE and in which at least some populations migrate long distance
| Species migrating long distance* | No. of H5N8-positive dead birds | Estimated population size | Ref. | |
| Common name | Scientific name | |||
| Mute swan† | C. olor | 1,217 | 495,000 | 34, p. 46 |
| Tufted duck | Aythya fuligula | 190 | 1,800,000 | 34, p. 187 |
| Whooper swan | Cygnus cygnus | 149 | 57,000 | 34, p. 51 |
| Eurasian wigeon† | M. penelope | 89 | 1,810,000 | 34, p. 117 |
| Mallard† | Anas platyrhynchos | 55 | 3,250,000 | 34, p. 132 |
| Greater white-fronted goose | Anser albifrons | 15 | 1,365,000 | 34, p. 66 |
| Common pochard | Aythya ferina | 10 | 1,700,000 | 34, p. 177 |
| Common teal† | A. crecca | 7 | 2,960,000 | 34, p. 123 |
| Common shelduck | Tadorna tadorna | 2 | 155,000 | 34, p. 101 |
| Lesser white-fronted goose | Anser erythropus | 2 | 63,000 | 34, p. 70 |
| Red-crested pochard | Netta rufina | 2 | 250,000 | 34, p. 171 |
| Bewick’s swan | Cygnus bewickii | 2 | 17,000 | 34, p. 55 |
| Common goldeneye | Bucephala clangula | 1 | 420,000 | 34, p. 217 |
| Common fieldfare | Turdus pilaris | 1 | 42,850,000 | 35 |
Here defined as wintering in the European Union and breeding at least beyond longitude 60°E, at the level of the Ural Mountains.
Those species that were also detected positive for H5N8 in 2014/2015 in Europe.
Discussion
The overall impact of the 2016/2017 epidemic of H5N8 HPAI virus (belonging to cluster B, Gochang like) differed markedly from the 2014/2015 epidemic of H5N8 HPAI virus (belonging to cluster A, Buan/Donglim like), both in geographical focus and the breadth of the host species affected. The 2016/2017 epidemic caused the largest recorded HPAI epidemic in poultry in Europe (18, 36) but did not spread to North America or Japan. In contrast, the 2014/2015 epidemic resulted in only limited poultry mortality in Europe and Japan (3) but caused major losses in the United States and to a lesser extent, in Canada, with over 48 million heads of poultry lost or destroyed (37). During the 2016/2017 epidemic, there was high mortality in free-living wild birds in Europe. In the Netherlands alone, ∼13,600 wild birds of 71 species were found dead during the epidemic (8). Considering an estimated detection rate of only 10 to 25%, the actual mortality probably was much higher and represented substantial percentages of wintering populations of several species in the Netherlands: 5% of tufted ducks and Eurasian wigeons, 2 to 10% of greater black-backed gulls, and 11 to 39% of peregrine falcons (8). Also during the 2016/2017 epidemic, H5N8 HPAI virus caused unusually high mortality in white-tailed eagles, with 17 laboratory-confirmed fatal infections in Germany between November 2016 and April 2017 (9), and in a well-monitored population of mute swans (C. olor) in the United Kingdom, causing an age-adjusted mortality of 143 per 1,000 birds (38). In most cases, the effect of this mortality of wild birds at the population level was not clear. However, the mortality rate of tufted ducks in parts of the Netherlands was so high (estimated at 25% of the local population) that population dynamics might have been affected substantially (8). In contrast, high wild bird mortality in Europe was not recorded in 2014/2015 (3). In the United States in 2014/2015, the majority of wild ducks did not show clear clinical signs of disease, although there was high mortality of wild raptor species (eagles, hawks, falcons, and owls) and wild geese (39). Experimental infections confirmed that the 2016/2017 H5N8 HPAI virus was substantially more virulent in domestic ducks than that from 2014/2015 (40).
We speculate that the high virulence of the H5N8 HPAI virus in 2016/2017 may be a side effect of selection for high excretion in wild water birds. Higher virus excretion not only results in more efficient transmission among hosts but also in increased severity of clinical signs. According to the intermediate virulence (or trade-off) hypothesis, there is selective advantage for higher excretion up to the point that higher transmission is counteracted by increased clinical signs and even death (41). It is at this point of intermediate virulence that the virus has the greatest evolutionary fitness. This fitness level depends in part on the level of population immunity, which will depend on the proportion of naïve birds and the presence of other circulating strains (38). It remains to be seen whether virulence of the H5N8 HPAI virus will change as it further evolves within the international metapopulation of wild birds and poultry. It also remains to be seen how the zoonotic potential of the H5N8 HPAI virus will change; based on experimental ferret infections with three of the viruses discussed here, the adaption of H5N8 HPAI virus to avian hosts was associated with a reduced zoonotic potential (40, 42).
The range of wild bird species involved in the 2016/2017 epidemic in Europe differed from that in 2014/2015, although nonuniform sampling over time and space cannot be ruled out as a potential bias. In 2014/2015, only four wild bird species were found positive for H5N8 in Europe. In contrast, 56 species were found positive in Europe in 2016/2017 (18). Of these 56 species, 14 had migratory populations that wintered in the European Union and bred at longitudes at least 60°E (Table 3). These 14 species included the same species as found in 2014/2015 (Eurasian wigeon, common teal, mallard, and mute swan) but also, 10 others. Thus, in 2016/2017 there was evidence for many more wild bird species that could have transported H5N8 viruses from breeding areas in Siberia to wintering areas in Europe. Which of these species transport virus remains to be determined and requires more detailed investigation of the candidate hosts, both in the context of avian influenza epidemiology and long-range movement ecology.
Reassortment frequency was much higher in the 2016/2017 epidemic than in the 2014/2015 epidemic and than in the European epidemic of H5N1 HPAI virus in 2006/2007. In 2016/2017, 11 reassortants were detected (Fig. 3), of which 7 were common (Fig. 4 and Table 1). In 2014/2015, three reassortants were detected in North America (10) and none elsewhere (3). For the reassortment of gene segments from two influenza viruses to occur, both viruses need to infect the same host cell at the same time. Possible reasons for the high frequency of reassortment in 2016/2017 may be related to the extent and timing of the epidemic, the host range infected by H5N8 HPAI virus, target tissues infected by H5N8 virus, and the ease with which the H5N8 HPAI virus reassorts with other influenza viruses (e.g., due to special features of some gene segments). The extent of the epidemic in wild birds was much greater in Europe in 2016/2017 than in 2014/2015 and may explain the higher frequency of reassortants in 2016/2017. Similarly, the fact that the 2014/2015 epidemic was greater in the United States than in Europe may explain the occurrence of reassortants in the United States (10, 43). Based on estimated MRCAs, the epidemic in Europe in 2016/2017 (MRCA of most common reassortant 3: July) started 1 mo earlier than in 2014/2015 (MRCA for HA and NA: August), so it may have coincided better with the peak of LPAI virus infection in wild water birds in autumn (44), thereby increasing the chance of reassortment between H5N8 HPAI virus and LPAI virus. The likelihood of the two peaks coinciding depends on how long it takes a virus to reach peak prevalence after its appearance in a wild waterfowl population. Although the host range of wild water birds infected with H5N8 HPAI virus in Europe in 2016/2017 was larger than in 2014/2015, dabbling duck species, particularly common teal and mallard, were found infected with H5N8 HPAI virus in both epidemics (Table 3). These latter species are considered to have the highest prevalence of LPAI virus infection (45) and therefore, considered to be important “mixing vessels” between HPAI virus and LPAI virus. Based on experimental infections of Pekin ducks, the H5N8 HPAI virus of 2016/2017 had greater tropism for the small intestine than that of 2014/2015 (40) and therefore, may have had a greater chance for reassortment with LPAI viruses, which mainly target the intestine in wild water birds (46). Even before 2016/2017, there was clear evidence that—unlike other clades of H5 HPAI virus, which are mostly found as H5N1—the H5 HPAI segment of the clade 2.3.4.4 viruses reassorted frequently, acquiring NA segments including N5, N2, N8, and N6 (3, 13). This indicates that the gene constellation of clade 2.3.4.4 H5 HPAI viruses is promiscuous, allowing efficient reassortment with gene segments from cocirculating LPAI viruses (3, 20). The range of wild bird species in which H5N8 HPAI virus was detected, the important role of wild birds as donors of new gene segments for reassortant viruses, and the inferred timing and location of reassortment events provide valuable support for continued surveillance of avian influenza in wild bird populations and provide information to improve the efficiency of such surveillance.
The origin of gene segments of the reassortant viruses was not only wild birds but also poultry. Specifically, gene segment 6 of reassortant 1 and gene segments 1, 2, 5, and 6 of reassortant 6 were inferred to originate from domestic anseriforms (Table 2). Transfer of LPAI virus gene segments between domestic anseriforms and wild birds has been observed before in Asia (47). Based on phylogenetic analysis, most of the reassortants of HPAI virus H5 clade 2.3.4.4 observed between 2006 and 2012 were generated in domestic anseriforms and particularly, domestic anseriforms in eastern Asia (China) (3). However, this spillback from domestic birds to wild birds has also been observed in Europe and implies that poultry populations in Europe form an additional source of viral genetic material for reassortment of circulating H5 HPAI clade 2.3.4.4 viruses.
An intriguing aspect of the 2016/2017 reassortants is the lack of reassortment involving gene segments 7 (M) and 8 (NS) in the time span of this study. Among the seven most common reassortants, segments 1, 2, 3, and 5 had three to five reassortments, segment 6 had only two reassortments, and segments 7 and 8 had no reassortments at all (Table 1). We did not consider segment 4 (HA) because the selection of genome sequences for analysis was based on segment 4 belonging to H5 HPAI virus clade 2.3.4.4, so any reassortments of this gene segment would automatically have been discounted. In the three reassortants detected during the 2014/2015 epidemic in North America (39), reassortment was observed for gene segments 2, 3, and 6 but not for gene segments 1, 5, and 7. Therefore, the lack of reassortment of gene segment 7 (M1) is common to both the 2014/2015 and the 2016/2017 reassortants. It appears that some gene segments (1 to 3, 5, 6) of the H5 HPAI 2.3.4.4 virus clade can be switched without noticeable loss in fitness, while switching was not observed for other segments (7, perhaps 8). It remains to be determined whether 1) selective epistasis occurs between segments 4 and 7, such that particular combinations are especially fit, or 2) there is some mechanistic reason why reassortment events do not occur between segments 4 and 7; for example, the viral RNA packaging signals in the segments might play a role (48, 49). Further factors have to be identified by future studies using, for example, reverse genetics and in vivo experiments with the different reassortants. Caveats in a global phylogeographic analysis such as this one are that the dataset depends on the availability of sequence information and that sampling of poultry and wild birds may be biased and differ per country. However, the large amount of data collected and analyzed with different techniques likely helped to reduce any study-related bias.
In conclusion, the 2016/2017 H5NX HPAI virus has caused the largest known HPAI epidemic in Europe, with high mortality in both poultry and wild birds, involvement of at least 14 long-distance migratory bird species, and the generation of an unusually high number of reassortant viruses. Further studies are required 1) to understand the relationship between level of virulence of the 2016/2017 H5NX HPAI virus and its fitness in wild bird populations (e.g., by comparative experimental infections); 2) to determine the overlap in breeding areas in Siberia among long-distance migratory birds wintering in Africa, Europe, Asia, and North America (e.g., through satellite telemetry); and 3) to determine the viral, host-related, and environmental circumstances that favor the generation of reassortants between H5NX HPAI and LPAI viruses (e.g., by experimental exchange of gene segments among genetically engineered viruses). This knowledge is critical to evaluate the risk of evolution of an H5NX HPAI virus that is able to be maintained in wild water bird populations in absence of its presence in poultry. Should H5NX HPAI become established in wild water bird populations, eradication will no longer be possible, and this disease will continue to pose a global risk for animal and human health in the foreseeable future.
Materials and Methods
Sequence Data.
Sequence data obtained from field isolates collected from domestic and wild birds were contributed by the partners from 25 member countries in the Global Consortium for H5N8 Avian Influenza Viruses. The data were shared via the EpiFlu database from the Global Initiative on Sharing All Influenza Data (GISAID) and the International Nucleotide Sequence Database Collaboration (INSDC). Additional viral sequences from both GISAID and INSDC were also used, including other clade 2.3.4.4 sequences and non-H5 sequences. Data used included viral sequence data, host species, and date and location of sampling. Where specific location or information on province or city level was not uploaded, it was extracted by matching country and outbreak date with entries from official sources. The data used for analysis consisted of 1) 240 isolates (232 complete genomes, 8 partial genomes) from 25 different countries during the period May 2016 to July 2017, including viruses of the subtypes H5N8 (218), H5N5 (21), and H5N6 (1), from domestic (93) and wild birds (147) (Figs. 1 and 2 and SI Appendix, Table S3) and 2) an expanded dataset of 251 to 279 sequences per internal segment, 284 sequences for segment 4 (H5), 236 sequences for segment 6 (N8), and 20 sequences for segment 6 (N5) (SI Appendix, Table S4). The data providers are summarized and acknowledged (SI Appendix, Table S5). The additional sequences were chosen from Basic Local Alignment Search Tool (BLAST) analyses to be genetically close to the different groups (labeled groups A to G) (Figs. 1 and 2) but from earlier time points and not restricted to any particular subtype. Specifically, BLAST analyses were performed on an early representative of each group in each segment, retrieving up to 500 sequences from GISAID. Neighbor-joining trees were created for all of the unique retrieved sequences and original dataset, and sequences within the larger clades encompassing the clades of the original data and also including sequences from earlier time points were selected. The earliest sequences in the extended datasets were from 2014 for segments 1, 5, 6-N8, 7, and 8; autumn 2011 for segments 2 and 3; autumn 2009 for segment 4; and spring 2015 for segment 6-N5.
Phylogenetic Analysis Using BEAST.
Time-scaled trees.
Bayesian time-resolved phylogenetic trees were estimated using BEAST 1.10.4 (22) using the SRD06 nucleotide substitution model with a four-category gamma distribution model of site-specific rate variation and separate site partitions for codon positions 1 + 2 vs. position 3 with HKY (Hasegawa–Kishino–Yano) substitution models on each, an uncorrelated relaxed clock with a log-normal distribution, and two coalescent tree priors [constant population size and the flexible sky grid tree prior (50)].
For dataset 1, at least two independent Markov Chain Monte Carlo (MCMC) chains were run for each internal segment, for segment 4-H5, and for segment 6-N8. Each chain consisted of 200,000,000 steps and was sampled every 20,000 steps, and the first 10% of samples were discarded as burn-in. Segment N5 was run with only 10,000,000 steps, with sampling at every 1,000 steps, because of its much smaller number of sequences. The MCMC settings were chosen to achieve a post burn-in effective sample size of at least 200, which is the accepted standard in BEAST analyses. A post burn-in sample of 1,000 posterior trees was obtained for each segment (or N subtype) in the dataset.
Time-scaled trees with group labels.
Segments 1, 2, 3, and 5 were divided into well-supported monophyletic distinct groups with labels A to G, with A having the greatest number of sequences; segment 6 (the NA gene) was divided into NA subtypes and was thus N8 or N5. Genetic diversity in segments 4, 7, and 8 was limited, and a distinct clade structure similar to segments 1, 2, 3, and 5 was not present, so these were placed in only one group (labeled A). The average genetic pairwise genetic distances within groups were 0.003 to 0.006 for segments 1, 2, 3, and 5; 0.006 to 0.008 for segments 4, 6 (N8), 7, and 8; and 0.014 for segment 6 (N5). The latter was due to a single more distant sequence (Fig. 2). The average pairwise genetic distances between groups were larger than 0.025 in all cases.
Therefore, the annotation for each reassortant consisted of a letter for each of the gene segments 1 to 5, 7, and 8 and a numeral for gene segment 6 (e.g., CABAD8AA). These groups and subtypes were mapped as discrete traits onto the posterior sample of time-scaled trees of all segments, and the number of label changes was estimated in BEAST [using a Markov Jumps model (51) with an asymmetric transition rate matrices] to indicate the relative frequency of reassortment of the internal segments.
Time-scaled trees with spatial coordinates.
The expanded dataset 2 was split into groups on a per-segment basis, generating several smaller datasets (SI Appendix, Table S6). Time-scaled trees of these groups were inferred with BEAST using the SRD06 model, a relaxed log-normal clock model, and constant population size models. Groups that were adjacent to each other in a neighbor-joining tree of the expanded dataset and that shared a common outgroup in the expanded dataset were combined for the purposes of inferring BEAST trees (e.g., segment 1 groups A and C, segment 3 groups A and B). In this way, closely related sequences from the autumn and winter 2016/2017 epidemics along with other previously isolated and closely related sequences not from those epidemics were analyzed together with the result that analyses of diverse groups and trees containing long branches were avoided. For each dataset, the post burn-in independent BEAST MCMC chains were combined and then down sampled to create posterior tree sets containing 1,000 trees. As before, MCMC settings were chosen to achieve a post burn-in effective sample size of at least 200.
Phylogeographic continuous trait spatial diffusion models were inferred for each of the datasets using the 1,000 posterior trees as an empirical tree distribution and MCMC chain lengths of 1,100,000 steps, sampling every 1,000 steps. Different diffusion models were tested using the N8 data, and the homogeneous Brownian motion model with latitude and longitude of the sampling locations was selected for use with all. In cases where exactly the same latitude and longitude for isolates were reported, a small random jitter of mean 0.001° was applied. A video illustrating the temporal and spatial spread across Eurasia of seven main H5 reassortant viruses is shown in Movie S1.
Origin of reassortments estimation.
Summary maximum clade credibility (MCC) trees for the post burn-in posterior time-scaled trees with spatial location reconstructions were created using TreeAnnotator. The MCC trees contained estimates of the time and location (and the 95% highest posterior density CIs of these values) at each internal (ancestral) node. Estimates of the origin of a reassortant genotype were made for each of its constituent segments by finding the MRCA nodes for all of the sequences of each reassortant code (e.g., AAAAA8AA) in each of the corresponding MCC trees and reporting the time and location of these identified internal tree nodes. The time and location of the immediate ancestors (MRCGAs) of these identified MRCAs are also reported in order to indicate the overall direction.
Host species origin of reassortments estimation.
In addition to the time-scaled trees with spatial coordinates, the inferred host type of the MRCA and the MRCGA nodes were also estimated by using a five-state structured coalescent approximation model [MASCOT (52)] for host species: domestic anseriform birds (Dom-ans), domestic galliform birds (Dom-gal), long-range migratory wild anseriform birds (Wild-ans-long), other wild anseriform birds (Wild-ans), and other wild birds (Wild-other) (SI Appendix, Table S7). The MRCGA and MRCA host categories were extracted in order to indicate whether a gene segment was established in a particular host category or whether it was switching between host categories. Those species that have both long-distance and short-distance/resident populations—mallard, tufted duck, mute swan, and common goldeneye were placed in the “Wild-ans” category to be conservative. Additionally, the one peacock sample was classified with the “Wild-other” birds. Results from these models are reported as probabilities for each host-type state at the node of interest. These structured coalescent approximate models are less susceptible to sampling bias than the simpler discrete trait models and are considered to be appropriate for analyzing cross-species transmission in avian influenza (52).
Phylogenetic Analysis Using IRIS.
For IRIS, full-genome sequences were retrieved from the Influenza Research database (https://www.fludb.org/) and the EpiFlu database (https://www.gisaid.org/) (SI Appendix, Table S8). We acknowledge the laboratories for providing sequence information via EpiFlu (SI Appendix, Table S5). Full-genome sequences of influenza A H5 clade 2.3.4.4 viruses were manually curated and annotated using Geneious v11.1.5 (Biomatters). The sequences were sorted into segments, and segment-based multiple alignment using fast Fourier transform (53) was performed. Subsequently, maximum-likelihood phylogenetic analyses were performed using Randomized Axelerated Maximum Likelihood (54), including 1,000 bootstrap replicates. SplitsTree4 (55) was used to generate a supernetwork (23) from the resulting trees. Supernetworks are calculated as consensus split networks obtained as a combination of all eight generated segment-sorted maximum-likelihood trees. They represent and visualize the phylogenetic relationships of each influenza virus where taxa are represented by nodes and their relationship as edges. Data were filtered to omit sequences with a collection date after 1 July 2017.
Maps.
Maps were created with ArcGIS Desktop 10.5.1 (ESRI) using the locations recorded in the GISAID database and background layers from ArcGIS Online (https://www.arcgis.com/index.html) and the Gridded Livestock of the World (GLW 3) (56) to show date and location of sampling of the virus sequences used for the above phylogenetic analysis by IRIS (SI Appendix, Table S8). Birds sampled at the same location were manually shifted slightly to avoid completely overlapping shapes in the map display.
Annual Migration Patterns of Wild Birds Involved in H5NX HPAI Epidemics.
Our phylogenetic analyses indicate that migrating wild birds carried HPAI virus H5NX to Europe in 2016/2017. Therefore, we first determined which wild bird species had been reported positive for H5NX virus in the European Union between 1 October 2016 and 5 July 2017, based on information extracted from the World Organization for Animal Health (57) and summarized by the European Food Safety Authority (18). Subsequently, we selected those species that had populations that migrated long distances (i.e., wintering in the European Union and breeding beyond at least longitude 60°E). We also reported estimates for the size of these populations (Table 3).
Supplementary Material
Acknowledgments
We thank Nicole Reimer and Patrick Wysocki for technical assistance in the preparation of the maps and Franz Conraths for critical review of the manuscript. This work was supported by European Union Horizon 2020 COllaborative Management Platform for detection and Analyses of (Re-) emerging and foodborne outbreaks in Europe (COMPARE) Grant 643476 and Dynamics of avian influenza in a changing world (DELTA-FLU) Grant 727922, by the Biotechnology and Biosciences Research Council (BBSRC) Institute Strategic Programme Grant: Control of Infectious Diseases BBS/E/D/20002173, and by the Scottish Government Rural and Environment Science and Analytical Services Division as part of the Centre of Expertise on Animal Disease Outbreaks (EPIC). S.J.L. is also supported by a University of Edinburgh Chancellor’s Fellowship. The map in Fig. 5 was created using ArcGIS® software by Esri. ArcGIS® and ArcMapTM are the intellectual property of Esri and are used herein under license. Copyright © Esri. All rights reserved. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Government.
Footnotes
The authors declare no competing interest.
A complete list of the Global Consortium for H5N8 and Related Influenza Viruses can be found in SI Appendix.
This article is a PNAS Direct Submission.
Data deposition: Sequences are shared via the EpiFlu database from the Global Initiative on Sharing All Influenza Data and the International Nucleotide Sequence Database Collaboration.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001813117/-/DCSupplemental.
Contributor Information
Collaborators: Samantha J. Lycett, Anne Pohlmann, Christoph Staubach, Valentina Caliendo, Steven van Borm, Andrew Breed, Francois-Xavier Briand, Ian Brown, Ádám Dán, Thomas DeLiberto, Sophie von Dobschuetz, Ron Fouchier, Marius Gilbert, Sarah Hill, Charlotte Kristiane Hjulsager, Hon Ip, Marion Koopmans, Lars Erik Larsen, Dong-Hun Lee, Mahmoud Mohamed Naguib, Isabella Monne, Oliver Pybus, Andrew Ramey, Vladimir Savic, Kirill Sharshov, Alexander Shestopalov, Chang-Seon Song, Mieke Steensels, David Swayne, Edyta Świętoń, XiuFeng Wan, Siamak Zohari, Mark Woolhouse, Martin Beer, and Thijs Kuiken
References
- 1.van den Brand J. M. A. et al., Wild ducks excrete highly pathogenic avian influenza virus H5N8 (2014-2015) without clinical or pathological evidence of disease. Emerg. Microbes Infect. 7, 67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton T. C., The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses 10, E461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lycett S. J. et al.; Global Consortium for H5N8 and Related Influenza Viruses , Role for migratory wild birds in the global spread of avian influenza H5N8. Science 354, 213–217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Globig A. et al., Epidemiological and ornithological aspects of outbreaks of highly pathogenic avian influenza virus H5N1 of Asian lineage in wild birds in Germany, 2006 and 2007. Transbound. Emerg. Dis. 56, 57–72 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Bouwstra R. J. et al., Phylogenetic analysis of highly pathogenic avian influenza A(H5N8) virus outbreak strains provides evidence for four separate introductions and one between-poultry farm transmission in The Netherlands, November 2014. Euro Surveill. 20, 21174 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Hanna A. et al., Genetic characterization of highly pathogenic avian influenza (H5N8) virus from domestic ducks, England, November 2014. Emerg. Infect. Dis. 21, 879–882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conraths F. J. et al., Highly pathogenic avian influenza H5N8 in Germany: Outbreak investigations. Transbound. Emerg. Dis. 63, 10–13 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Kleyheeg E. et al., Deaths among wild birds during highly pathogenic avian influenza A(H5N8) virus outbreak, The Netherlands. Emerg. Infect. Dis. 23, 2050–2054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krone O. et al., White-tailed sea eagle (Haliaeetus albicilla) die-off due to infection with highly pathogenic avian influenza virus, subtype H5N8, in Germany. Viruses 10, E478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D. H. et al., Highly pathogenic avian influenza viruses and generation of novel reassortants, United States, 2014-2015. Emerg. Infect. Dis. 22, 1283–1285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization , Evolution of the influenza A(H5) haemagglutinin: WHO/OIE/FAO H5 Working Group reports a new clade designated 2.3.4.4. https://www.who.int/influenza/gisrs_laboratory/h5_nomenclature_clade2344/en/. Accessed 31 July 2020.
- 12.Smith G. J. D., Donis R. O.; World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5 Evolution Working Group , Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. Influenza Other Respir. Viruses 9, 271–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lycett S. J., Duchatel F., Digard P., A brief history of bird flu. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown J. D., Stallknecht D. E., Beck J. R., Suarez D. L., Swayne D. E., Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 12, 1663–1670 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keawcharoen J. et al., Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14, 600–607 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemeth N. M. et al., Experimental infection of bar-headed geese (Anser indicus) and ruddy shelducks (Tadorna ferruginea) with a clade 2.3.2 H5N1 highly pathogenic avian influenza virus. Vet. Pathol. 50, 961–970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon J. H. et al., Experimental infection of H5N1 and H5N8 highly pathogenic avian influenza viruses in Northern Pintail (Anas acuta). Transbound. Emerg. Dis. 65, 1367–1371 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Brown I. et al., Scientific report on the avian influenza overview October 2016–August 2017. EFSA J. 15, e05018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harder T. et al., Influenza A(H5N8) virus similar to strain in Korea causing highly pathogenic avian influenza in Germany. Emerg. Infect. Dis. 21, 860–863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pohlmann A. et al., Swarm incursions of reassortants of highly pathogenic avian influenza virus strains H5N8 and H5N5, clade 2.3.4.4b, Germany, winter 2016/17. Sci. Rep. 8, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steel J., Lowen A. C., Influenza A virus reassortment. Curr. Top. Microbiol. Immunol. 385, 377–401 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Suchard M. A. et al., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Lee D. H. et al., Novel reassortant clade 2.3.4.4 avian influenza A(H5N8) virus in wild aquatic birds, Russia, 2016. Emerg. Infect. Dis. 23, 359–360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M. et al., Highly pathogenic avian influenza A(H5N8) virus in wild migratory birds, Qinghai Lake, China. Emerg. Infect. Dis. 23, 637–641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Twabela A. T., et al. , Highly pathogenic avian influenza A(H5N8) virus, Democratic Republic of the Congo, 2017. Emerg. Infect. Dis. 24, 1371–1374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poen M. J. et al., Lack of virological and serological evidence for continued circulation of highly pathogenic avian influenza H5N8 virus in wild birds in The Netherlands, 14 November 2014 to 31 January 2016. Euro Surveill. 21, 30349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beerens N. et al., Multiple reassorted viruses as cause of highly pathogenic avian influenza A(H5N8) virus epidemic, The Netherlands, 2016. Emerg. Infect. Dis. 23, 1974–1981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fusaro A. et al., Genetic diversity of highly pathogenic avian influenza A(H5N8/H5N5) viruses in Italy, 2016-17. Emerg. Infect. Dis. 23, 1543–1547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohlmann A. et al., Outbreaks among wild birds and domestic poultry caused by reassorted influenza A(H5N8) clade 2.3.4.4 viruses, Germany, 2016. Emerg. Infect. Dis. 23, 633–636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selim A. A. et al., Highly pathogenic avian influenza virus (H5N8) clade 2.3.4.4 infection in migratory birds, Egypt. Emerg. Infect. Dis. 23, 1048–1051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulatti P. et al., Integration of genetic and epidemiological data to infer H5N8 HPAI virus transmission dynamics during the 2016-2017 epidemic in Italy. Sci. Rep. 8, 18037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Świętoń E., Śmietanka K., Phylogenetic and molecular analysis of highly pathogenic avian influenza H5N8 and H5N5 viruses detected in Poland in 2016-2017. Transbound. Emerg. Dis. 65, 1664–1670 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Scott D. A., Rose P. M., Atlas of Anatidae Populations in Africa and Western Eurasia, (Wetlands International, 1996). [Google Scholar]
- 35.Cramp S., Handbook of the Birds of Europe, the Middle East and North Africa: The Birds of the Western Palearctic Volume V: Tyrant Flycatchers to Thrushes, (Oxford University Press, New York, NY, 1988). [Google Scholar]
- 36.Alarcon P. et al., Comparison of 2016-17 and previous epizootics of highly pathogenic avian influenza H5 Guangdong lineage in Europe. Emerg. Infect. Dis. 24, 2270–2283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.USDA , Epidemiologic and other analyses of HPAI-affected poultry flocks: September 9, 2015 report. https://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/downloads/Epidemiologic-Analysis-Sept-2015.pdf. Accessed 31 July 2020.
- 38.Hill S. C. et al., Comparative micro-epidemiology of pathogenic avian influenza virus outbreaks in a wild bird population. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ip H. S., et al. , High rates of detection of clade 2.3.4.4 highly pathogenic avian influenza H5 viruses in wild birds in the Pacific Northwest during the winter of 2014-15. Avian Dis. 60 (suppl.), 354–358 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Grund C. et al., A novel European H5N8 influenza A virus has increased virulence in ducks but low zoonotic potential. Emerg. Microbes Infect. 7, 132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alizon S., Hurford A., Mideo N., Van Baalen M., Virulence evolution and the trade-off hypothesis: History, current state of affairs and the future. J. Evol. Biol. 22, 245–259 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Pohlmann A. et al., Genetic characterization and zoonotic potential of highly pathogenic avian influenza virus A(H5N6/H5N5), Germany, 2017-2018. Emerg. Infect. Dis. 25, 1973–1976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee D. H. et al., Transmission dynamics of highly pathogenic avian influenza virus A(H5Nx) clade 2.3.4.4, North America, 2014-2015. Emerg. Infect. Dis. 24, 1840–1848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk J. G. et al., Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. J. Anim. Ecol. 83, 266–275 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsen B., et al. , Global patterns of influenza A virus in wild birds. Science 312, 384–388 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Kuiken T., Is low pathogenic avian influenza virus virulent for wild waterbirds? Proc. Biol. Sci. 280, 20130990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J., Xie D., Nie Z., Xu B., Drummond A. J., Inferring host roles in bayesian phylodynamics of global avian influenza A virus H9N2. Virology 538, 86–96 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Gerber M., Isel C., Moules V., Marquet R., Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 22, 446–455 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Essere B. et al., Critical role of segment-specific packaging signals in genetic reassortment of influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 110, E3840–E3848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gill M. S. et al., Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 30, 713–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minin V. N., Suchard M. A., Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 56, 391–412 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Müller N. F., Rasmussen D., Stadler T., MASCOT: Parameter and state inference under the marginal structured coalescent approximation. Bioinformatics 34, 3843–3848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kloepper T. H., Huson D. H., Drawing explicit phylogenetic networks and their integration into SplitsTree. BMC Evol. Biol. 8, 22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert M. et al., Global distribution data for cattle, buffaloes, horses, sheep, goats, pigs, chickens and ducks in 2010. Sci. Data 5, 180227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.OIE , Data from “Update on avian influenza in animals (types H5 and H7).” Avian Influenza Portal. https://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/. Accessed 31 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.