Significance
Upon binding of effectors, allosteric molecules change their structures and responses to the downstream molecules, which can be viewed as the molecular if−then device. Simulating binding of two pioneer transcription factors (TFs), Sox2 and Oct4, to a nucleosome, which is the fundamental unit of genome folding, we found that a nucleosome acts as a new type of allosteric molecule. Free nucleosomes exhibited rotation-coupled sliding of their DNA among metastable positions. The Sox2 binding on them selected a specific rotational phase of its motif, inducing global sliding of nucleosomal DNA. Consequently, the repositioned DNA affected the accessibility of another TF, Oct4, or the second molecule of Sox2 at a distant region within the nucleosome, which thus is a long-distance allosteric effect.
Keywords: allostery, Sox2, Oct4, pioneer factor, coarse-grained molecular dynamics
Abstract
While recent experiments revealed that some pioneer transcription factors (TFs) can bind to their target DNA sequences inside a nucleosome, the binding dynamics of their target recognitions are poorly understood. Here we used the latest coarse-grained models and molecular dynamics simulations to study the nucleosome-binding procedure of the two pioneer TFs, Sox2 and Oct4. In the simulations for a strongly positioning nucleosome, Sox2 selected its target DNA sequence only when the target was exposed. Otherwise, Sox2 entropically bound to the dyad region nonspecifically. In contrast, Oct4 plastically bound on the nucleosome mainly in two ways. First, the two POU domains of Oct4 separately bound to the two parallel gyres of the nucleosomal DNA, supporting the previous experimental results of the partial motif recognition. Second, the POUS domain of Oct4 favored binding on the acidic patch of histones. Then, simulating the TFs binding to a genomic nucleosome, the LIN28B nucleosome, we found that the recognition of a pseudo motif by Sox2 induced the local DNA bending and shifted the population of the rotational position of the nucleosomal DNA. The redistributed DNA phase, in turn, changed the accessibility of a distant TF binding site, which consequently affected the binding probability of a second Sox2 or Oct4. These results revealed a nucleosomal DNA-mediated allosteric mechanism, through which one TF binding event can change the global conformation, and effectively regulate the binding of another TF at distant sites. Our simulations provide insights into the binding mechanism of single and multiple TFs on the nucleosome.
Transcription factors (TFs) read the sequence information coded in genomic DNA to regulate gene expression. However, the targets of eukaryotic TFs are often masked by nucleosomes, which not only act as fundamental units of genome packaging but also contribute to gene regulation (1). In the core structure of a nucleosome, two copies of four types of histone proteins (H2A, H2B, H3, and H4) form an octameric complex and are wrapped by ∼147 DNA base pairs (bp) for ∼1.7 turns (called nucleosomal DNA) (2). The nucleosomal DNA is inaccessible to most TFs, with some exceptions, including the pioneer TFs (3). The pioneer TFs can recognize their target sequences even in nucleosomes, which has been implicated to be related to their biological functions in cell differentiation and reprogramming and thus is now under intensive study (4). For example, high-throughput protein microarray experiments have shown that the pioneer TFs are different from normal TFs in the preference of the secondary structure they use to recognize the nucleosome (5). Comparison of chromatin immunoprecipitation sequence and MNase results have revealed distinctive binding patterns and sequence preferences for the pioneer TFs to identify their nucleosomal targets (6). On the other side, a recently developed high-throughput technique, nucleosome consecutive affinity purification–systematic evolution of ligands by exponential enrichment (NCAP-SELEX), has unveiled some standard features of the general TF−nucleosome binding, such as the preference for nucleosomal DNA ends or the dyad, the parallel binding to two gyres, and the periodic binding pattern (7). Despite the fruitful experimental results, a structure-based quantitative understanding of the TF−nucleosome interacting mechanism is still missing. Moreover, how TF binding interplays with the dynamics of the nucleosomal DNA and histones remains unclear.
Combinatorial binding of multiple TFs on DNA targets adds a higher layer of gene regulation (8). TFs achieve the cooperativity in many ways, such as direct interaction through DNA-binding domains (9), or allostery through DNA conformational changes (10, 11). When the nucleosome is involved, alternative mechanisms emerge that facilitate nonspecific long-distance cooperative binding of TFs (12). Nucleosomes undergo spontaneous dynamics such as partial unwrapping (13), gaping (14), and rotation-coupled sliding of the nucleosomal DNA (15–18). These dynamics can be manipulated by TFs to regulate their binding mutually (19, 20). For example, binding of one TF to the entry/exit changes the accessibility of inner nucleosomal DNA and then affects the association rate of another TF (21). In extreme cases, TFs can displace a nucleosome and expose a range of ∼147 bp of DNA to other TFs (22). This type of DNA unwrapping mediated cooperativity has been illustrated and analyzed by both experimental and theoretical studies (19, 21, 22). In contrast, the possible relationship between nucleosomal DNA sliding and the TF target search process is rarely considered.
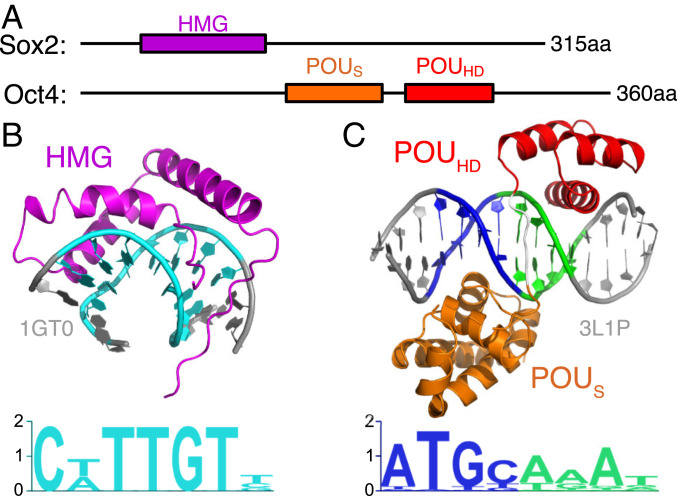
Here we used two pioneer factors, Sox2 and Oct4, as examples to study how TFs search for their targets in the nucleosome. Sox2 and Oct4 are two of the four key TFs that cause the conversion of somatic cells into the induced pluripotent stem cells (23). Sox2 recognizes the DNA sequence “CTTTGTT” with its high-mobility group (HMG) domain and causes a sharp bending of DNA by intercalating into the DNA minor groove (Fig. 1 A and B) (24). The DNA-binding domain in Oct4 comprises two POU (named after the homology regions found in the TFs Pit-1, Oct-1, Oct-2, and Unc-86) (25) domains, POU-specific (POUS) and POU-homeo (POUHD). Each of the two POU domains, via interactions in the DNA major groove, recognizes four DNA base pairs, adding up to the 8-bp motif “ATGCAAAT” (Fig. 1 A and C) (26). Interestingly, the crystal structures of the complex of Sox2, Oct4, or Sox2/Oct4 with a linear (naked) DNA do not fit well with the canonical nucleosome structure: The DNA curvature upon Sox2 binding is much higher than nucleosomal DNA, and the DNA-encircling binding pattern of Oct4 would clash with the histones (6). Experiments have shown that both TFs sacrificed some specificity and bound to partial target motifs for the binding to nucleosomal DNA (6). Furthermore, nucleosome-binding assays have also revealed the dependence of Sox2 binding probability on the rotational positioning of the target motif in the nucleosomal DNA (7, 27). During the review process of the current work, two groups reported cryoelectron microscopy (cryo-EM) structures of Sox2−nucleosome (28) and Sox2−Oct4−nucleosome (29) complexes, respectively. Interestingly, these structures suggested different pioneer TF−nucleosome binding mechanisms, which was dependent on DNA sequence and the positioning of TFs’ target motifs. However, these structures revealed a common feature of Sox2 induced sharp bending of nucleosomal DNA (28, 29).
Fig. 1.
The DNA-binding domains of Sox2 and Oct4. (A) The full-length Sox2 protein (315 amino acids [aa]) uses the HMG domain (79 aa, purple) to bind to DNA. The Oct4 protein (360 aa) has two DNA-binding domains, POUS (85 aa, orange) and POUHD (51 aa, red) connected by a linker of 15 aa. (B) The DNA-binding complex structure (Upper) and sequence motif (Lower) for the Sox2 HMG domain. The complex structure is based on PDB entry 1GT0. DNA sequence piece “CTTTGTT” is colored in cyan. (C) The complex structure (Upper) of Oct4 binding to its consensus sequence (Lower). The consensus sequences of the POUS and POUHD domains are colored in blue and green, respectively. The structure is based on PDB entry 3L1P.
Despite the above-mentioned experimental results, the dynamic target searching mechanism is unknown. Here we used molecular dynamics (MD) simulations to investigate the binding of Sox2 and Oct4 on nucleosomes. A recent computational work used all-atom MD simulations to study the nucleosomal DNA features and their relationship with Oct4 binding (30). However, due to the computational cost, the sampling of conformations was limited and highly dependent on the initial configurations in the simulations (30). Here we employed the coarse-grained (CG) models that have gained success in achieving the balance between accuracy and efficiency (31). Notably, we recently developed a method, which incorporates both the position weight matrix and the complex structural information (PWMcos), to model the sequence-specific interactions between proteins and DNA (32). Combined with the latest versions of the CG models for protein [the AICG2+ (33)] and DNA [the 3SPN.2C (34)], we validated the CG MD method for many protein−DNA complexes, investigating the diffusion of TFs on naked DNAs (35), as well as the sliding and partial unwrapping of the nucleosomal DNA (16, 17).
In this work, we utilized the latest CG models to perform MD simulations on Sox2 and Oct4 binding to both naked DNAs and nucleosomes. We first confirmed that, in our simulations, Sox2 and Oct4 successfully found their consensus sequences in naked DNAs. We then inserted the TF target sequences into different locations of a strongly positioning [“601” (36)] nucleosome and showed that Sox2 accessed its target motif only at certain rotational phasing positions, whereas Oct4 had its two POU domains adopting a two-gyre binding pattern on the nucleosomal DNA. We also found conformations where the POUS domain contacted the acidic patch on the histones. Finally, by putting multiple Sox2/Oct4 on a genomic nucleosome, the LIN28B nucleosome, we revealed an allostery mechanism in which the first bound Sox2 resulted in local bending and global sliding of the nucleosomal DNA and consequently changed the exposure/burying of distant binding sites for other TFs to bind.
Results
Sox2 and Oct4 Bind to Naked DNAs with Different Mechanisms.
In the present work, we employed a CG model to study the DNA binding dynamics of Sox2 and Oct4. The CG model has been carefully calibrated with respect to many experimental data on protein dynamics (33), DNA mechanics (34), TF-binding on naked DNA (32, 35), and nucleosome dynamics (17). In our models, each amino acid in a protein is represented by one particle located at its position (33), and each nucleotide is simplified as three particles, each representing phosphate, sugar, and base (34). For protein−DNA interactions, we considered both direct and indirect readouts. The former included DNA sequence-dependent interactions between amino acids and nucleobases (32). The latter included sequence-nonspecific electrostatic and excluded volume interactions. With these models, we simulated the binding and diffusion of Sox2 and Oct4 on different DNA sequences.
Before putting the TFs onto nucleosomes, we first studied the binding of Sox2 and Oct4 to naked double-stranded DNAs (dsDNAs). The DNA sequences used for Sox2 and Oct4 binding are both taken from the Hoxb1 regulatory element and are the same as those used in previous NMR studies (37, 38) (Table 1).
Table 1.
Sequences of the dsDNAs used in the simulations for Sox2/Oct4 binding to naked DNAs
| System | DNA sequence |
| Sox2-naked DNA binding | CAGTGTCTTTGTCATGCTAATGCTAGGTG |
| Oct4-naked DNA binding | CATTTGTCATGCTAATGCTTGGTG |
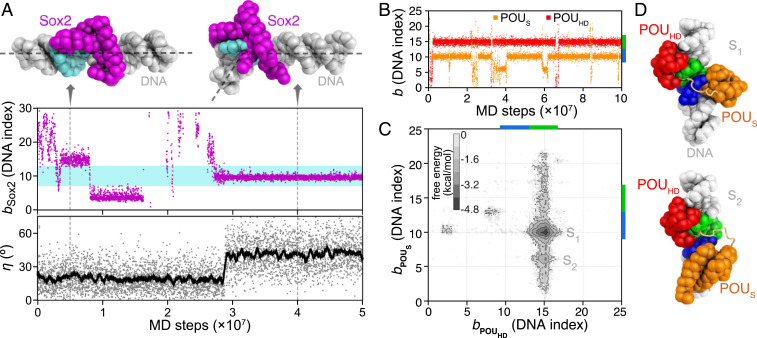
Fig. 2A shows a representative trajectory for Sox2. After the initial binding to a nonspecific site, Sox2 diffused along DNA (Fig. 2 A, Upper). During the diffusion, Sox2 paused at pseudo motifs (the binding position and 15) with relatively long dwell times. Eventually, Sox2 was bound to its consensus sequence with the highest stability (after MD steps). By monitoring the bending angle of DNA (Fig. 2 A, Lower), we found that, only when Sox2 was bound to the consensus sequence, it induced a sharp bending of DNA up to , whereas, while Sox2 was in the nontarget regions, DNA was bent to . These results are consistent with previous experimental observations of Sox2 binding-induced DNA bend (24, 39). Besides, the relatively straight DNA conformation during Sox2 sliding in our simulations can be essential for the efficiency of target search (35, 40, 41). Overall, these results show the ability of our model to describe the diffusion and the recognition of sequence-specific DNA-binding protein on the rugged landscape of the real genomic sequence.
Fig. 2.
MD simulations of Sox2 and Oct4 binding to naked DNAs (see Table 1 for DNA sequences). (A) A representative trajectory of the Sox2 binding and induced bending of DNA. The DNA binding position bSox2 (see Methods for definition) of Sox2 (purple dots in Middle) and the bending angle η of DNA (see SI Appendix, Supplementary Methods for definition; gray dots in Lower) are plotted as functions of MD steps. The cyan region in Middle (DNA indices 7 to 13) represents the consensus sequence for Sox2 binding. The black line in Lower is the moving average of the DNA bending angle data. (Upper) Snapshots of Sox2-DNA sequence-nonspecific (Left, MD step ) and specific binding (Right, MD step ). The dashed lines on the structures show the direction of DNA double helix axes. (B) Representative time series of the Oct4 POUS (orange) and POUHD (red) binding positions on DNA. The consensus sequences for POUS and POUHD are marked by blue and green bars on the right vertical axis. (C) The 2D free energy surface of binding positions of the POUS and POUHD domains. (D) Representative structures of the two free energy minima (S1 and S2). The color scheme of the structures is the same as Fig. 1.
For Oct4, we show, in Fig. 2B, the binding positions of the two POU domains as functions of simulation time from a representative simulation trajectory. After their rapid recognition of the consensus sequences, both POUS and POUHD can transiently leave their motif positions. Clearly, the simulated binding affinity of the POUHD is higher than that of the POUS, which is consistent with the previous experimental results (42). These results suggest a unique binding mechanism for Oct4 in which one domain (POUHD) with higher DNA binding affinity acts as an anchor, effectively increasing the local concentration of the weaker domain (POUS). Based on 20 independent MD trajectories, we plotted the two-dimensional (2D) free energy surface projected onto the coordinates that describe the POUS and POUHD binding positions ( and ; Fig. 2C). POUHD had an overwhelmingly high binding probability at its consensus target. In contrast, POUS had a second highly populated binding position (S2, with DNA sequence “ATGA” in the complementary strand) in addition to its consensus sequence (S1, “ATGC”). We show the representative structures of these two states in Fig. 2D. The plastic spatial arrangement of the two POU domains on DNA suggests possibly adaptable binding patterns of Oct4 in different cellular environments.
Sox2 Binding Position on the 601 Nucleosome Is Dependent on the Rotational Phasing.
We next considered the binding of TFs onto nucleosomes. Although our final target was to get an integrated picture of TF binding collaborating with the dynamics of the nucleosome, we would like to first concentrate on the target search process of TFs, without considering drastic DNA sliding in the nucleosome. Therefore, we started from the binding of Sox2 on the 601 nucleosome, whose DNA sequence is designed to achieve the high affinity and specificity of nucleosome positioning (36). Consistently, with our CG models, we did not observe the evident sliding of the 601 nucleosome larger than 2 bp during the MD simulations (17).
One of the most critical features of the nucleosome as a TF blocker is the rotational phasing of the nucleosomal DNA (43–45). The pattern of DNA wrapping around the histones introduces a 10-bp periodicity of DNA being buried or exposed in nucleosomes. As revealed by previous experiments, the intrinsic phasing of DNA, and the rotational position of the TF target motif, play essential roles in determining the TF binding sites (7, 27, 43). Inspired by these experiments, we designed our target nucleosomal DNA sequence by inserting the Sox2 target motifs into the strong positioning 601 nucleosomes; we inserted the 7-bp Sox2 motif (“CTTTGTT”) at 10 different positions of the 601 sequence, the DNA index of the first site being from 95 to 104 (hereafter referred to as BS95 to BS104; SI Appendix, Fig. S1). These 10 sequences covered all of the possible rotational positions inside a nucleosome.
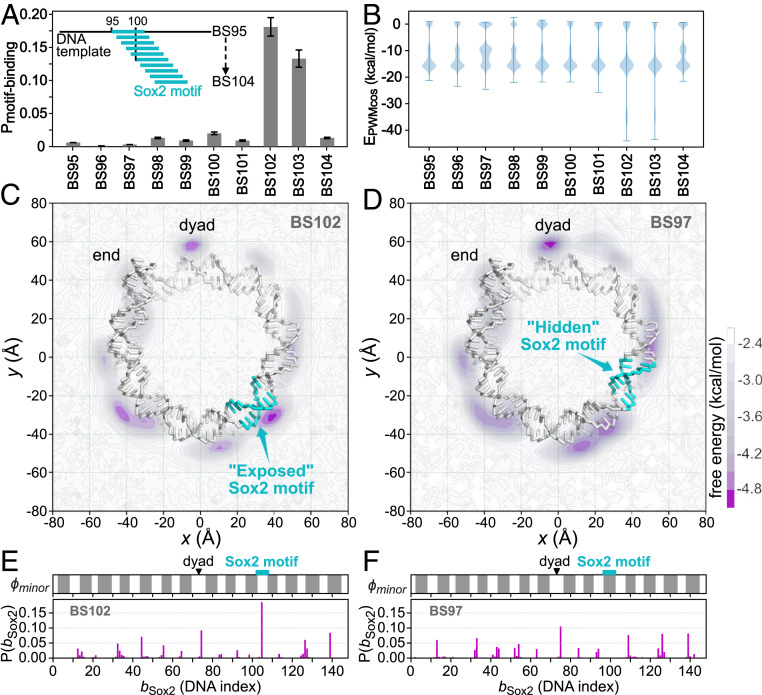
We performed MD simulations of Sox2 binding to all of these modified 601 nucleosomes. For each, we ran 20 independent trajectories for MD steps. From these simulations, we analyzed the probability for Sox2 to find the target motif and the energy contribution of the sequence-specific interactions (Fig. 3 A and B). We found that Sox2 recognized its consensus motif only in BS102 and BS103, where the motifs were put at some specific rotational positions (Fig. 3A). In the other cases, the sequence-specific−type interaction energy between Sox2 and the nucleosomal DNA (>−20 kcal/mol) was much weaker than in the consensus motif binding case (∼−35 kcal/mol, BS101 and BS102) (Fig. 3B).
Fig. 3.
Sox2 binding to the modified 601 nucleosomes, in which the Sox2 consensus sequence “CTTTGTT” was inserted into the 601 sequence at different positions, with in the name BS meaning the insertion position. (A) Probability of Sox2 recognizing its consensus motif on the 10 modified 601 nucleosomes. Error bars are based on bootstrapping (SI Appendix, Supplementary Methods). (B) Distribution of the sequence-specific interaction energy between Sox2 and the nucleosomal DNA . (C) The 2D free energy surface of Sox2 center-of-mass (COM) on the BS102 nucleosome. DNA is shown as sticks, and histone proteins are hidden for clarity. The Sox2 motif is colored cyan. The purple intensity represents free energy value as depicted in the color bar. (D) The same as C but for Sox2 binding on the BS97 nucleosome. (E) One-dimensional distribution of the mean Sox2 binding positions on the BS102 nucleosomal DNA (, Lower) and the accessibility of the nucleosomal DNA minor groove (, Upper). The exposed regions that are accessible to Sox2 are shown in white , whereas the histone covered regions are shown in gray (see SI Appendix, Supplementary Methods for the detailed definition). The dyad and the Sox2 motif insertion positions are marked by a black triangle and a cyan rectangle, respectively. (F) The same as E but for the BS97.
In Fig. 3 C−F, we show more detailed analysis of Sox2 binding to two of the above-described nucleosomes, BS102 and BS97. The former has the highest Sox2 binding probability to the consensus motif (Fig. 3A), while the latter has the motif positioned 5 bp shifted from the former. Fig. 3 C and D shows the 2D distribution of the center-of-mass (COM) of Sox2 around the BS102 and BS97 nucleosomes, respectively.
In the BS102, the minor groove of the Sox2 target motif was facing outward from the nucleosome and was adequately exposed for TF binding (Fig. 3C). Consequently, we found that Sox2 was able to recognize its consensus motif target and had the highest population around this region (Fig. 3C and Movie S1). To quantitatively measure the dependence of Sox2 binding on the rotational position of the target motif, we introduced a binary score to describe the phase of the nucleosomal DNA minor groove, with representing the exposed region and for the buried region (SI Appendix, Supplementary Methods and Fig. S2). In Fig. 3E, we plotted the exposure score as well as the binding probabilities of Sox2 on the BS102: The Sox2 binding positions fit well into the exposed DNA positions. In particular, the consensus motif in the BS102 is in the exposed region of nucleosomal DNA, and Sox2 was bound to this site with the highest probability (Fig. 3E).
Conversely, in the BS97, the Sox2 motif was facing inward and covered by the histones (Fig. 3D). Sox2 did not find its target motif in the BS97. Instead, we found that Sox2 bound to a wide range of exposed DNA minor grooves in a sequence-nonspecific manner (Fig. 3F), consistent with the energy analysis (Fig. 3B). Interestingly, we found that Sox2 had a preference for the dyad in the BS97 (Fig. 3D and Movie S2). Our degree-of-exposure analysis showed that there was a wider exposed region at dyad compared with other parts of the nucleosome (Fig. 3D). By looking at the structure, we found that, at the regions where two gyres of DNA wrap around the histones, the accessible surface for Sox2 binding is limited by both histones and the other gyre of DNA (SI Appendix, Fig. S3A), whereas, near dyad, where only one gyre of DNA is present, the Sox2 accessible surface is larger (SI Appendix, Fig. S3B). Considering that, on BS97, Sox2 was bound nonspecifically to the nucleosomal DNA, entropy played the crucial role in the distribution of Sox2 binding position. Therefore, we concluded that the dyad was preferred by Sox2 because of more possible rotational positions to bind to. Our results provide explanations for the previous observations of the preference of the nucleosome dyad by the Sox family proteins (7).
These results together quantitatively show that the Sox2 binding position on the nucleosome is highly dependent on the rotational position of its target motif. The periodic binding and the preference for the dyad are consistent with previously proposed general features for TF recognition of nucleosomal targets (7).
We also investigated the conformational changes of the nucleosomes upon Sox2 binding. Similar to the binding of Sox2 to a naked DNA (Fig. 2A), we found that the recognition of the consensus motif by Sox2 was coupled with sharper bending of nucleosomal DNA and local disruption of DNA−histone contacts (SI Appendix, Fig. S4). Interestingly, the Sox2 binding-induced local distortion of DNA curvature was also observed in the recently resolved cryo-EM structures of Sox2−nucleosome complexes (28, 29). Our simulation results are quantitatively consistent with these experiments and provide direct support to Sox2’s recognition of its target on the nucleosomal DNA (SI Appendix, Fig. S4). On the other side, due to the use of a very strong positioning sequence here and the relatively deep position of Sox2 motif at SHL3, we did not observe large-scale conformational changes, such as DNA unwrapping or disassembly of the nucleosome. Consistently, a cryo-EM structure of Sox2−Oct4−601 nucleosome complex showed that Sox2 binding at SHL5 also caused local DNA bending only (29).
Oct4 Binds to the 601 Nucleosome with Flexibly Different Patterns.
Similar to Sox2, we conducted simulations of Oct4 binding to the 601 nucleosomes with the Oct4 target motif inserted at 10 consecutive rotational positions, BS95 to BS104 (for each nucleosome, 20 trajectories of MD steps). However, in contrast to Sox2, Oct4 did not exhibit clear rotation phase-dependent binding to the target motif. This is probably because the 8-bp long consensus sequence of Oct4 cannot be sufficiently exposed in any of the rotation phase. Instead, we found a much more diverse binding of Oct4 to the nucleosome using the two POU domains in plastic manners, which was not sensitive to the rotation phase of the target motif. Since each POU domain recognizes only a 4-bp motif, there can be several consensus or pseudoconsensus motifs, by chance, in a nucleosomal DNA, which results in diverse binding. Of numerous binding patterns, we found the following two modes outstanding.
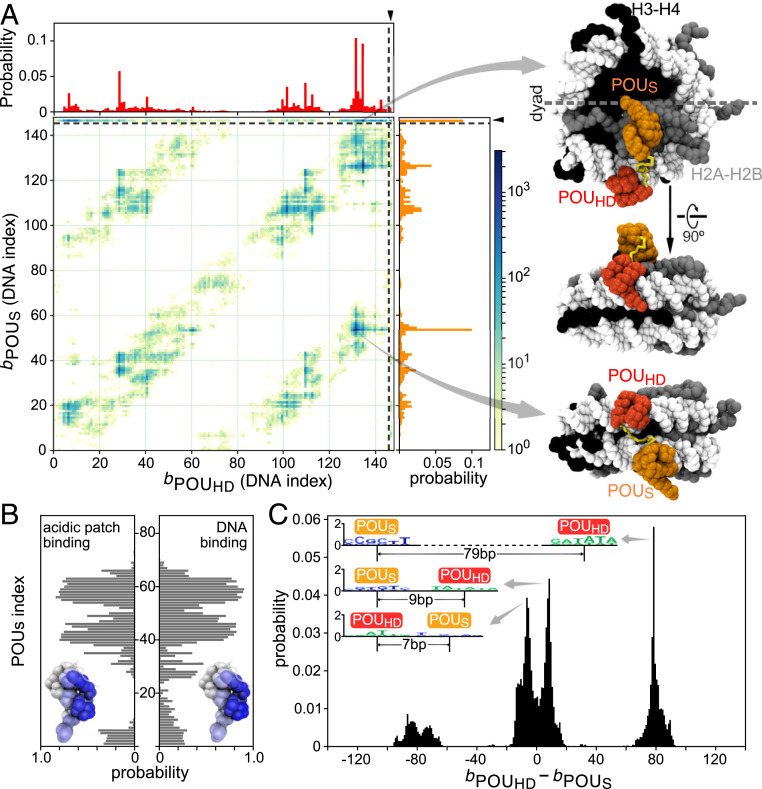
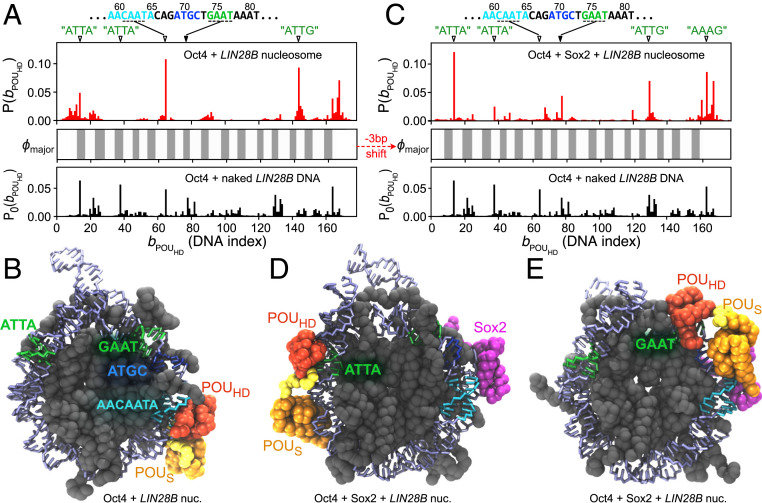
In the first mode found, interestingly, the POUS domain was bound to the “acidic patch” on the H2A−H2B histone dimers (46), whereas POUHD recognized its pseudoconsensus DNA sequences (a representative structure depicted at the top of Fig. 4 A, Right; also see Movie S3). Fig. 4A shows probability distributions of Oct4 binding to the BS104 nucleosome (the 8-bp target motif was put at the DNA indices 104 to 111 of the 601 nucleosome) in one and two dimensions, where the mean binding positions of two POU domains are shown as the DNA indices 1 to 147, in addition to the acidic patch binding as the extraindex 148. The probability distribution of the POUS binding position (the vertical 1D plot in Fig. 4A) shows a high peak at index 148, indicating its binding to acidic patch. The recognition of the acidic patch by the POUS is surprising but reasonable, considering there are electrostatic attractions between the positively charged surface on the POUS domain and the negatively charged acidic patch (46). To gain a better understanding of the binding interface, we analyzed the contact probability between the amino acids in the POUS domain and the histone. We found that POUS used the same surface to bind nonspecifically to the nucleosomal DNA and to recognize the acidic patch (Fig. 4B). Notably, we did not observe stable binding of the POUHD to the acidic patch. Interestingly, the POUHD domain has more positive net charges than the POUS domain . Therefore, we propose that the positive charge distribution and the geometric shape of the DNA-binding surface in the POUS domain are uniquely suitable and specific for the recognition of the acidic patch. We suggest that this property of Oct4 might contribute to its biological functions, such as the regulation of chromatin structure through altering nucleosome packing or chaperone binding, by blocking the accessibility of the acidic patch (46, 47).
Fig. 4.
Oct4 binding to the 601 nucleosomes with different patterns. (A) (Left) The 2D distribution of the POUHD and POUS binding positions on the nucleosomal DNA and the histone for the BS104 nucleosome. One-dimensional distributions for each domain are plotted at the top (POUHD) and on the right (POUS), respectively, of the main graph. On the axis of protein binding positions, 1 to 147 represents the DNA base pair index, whereas 148 indicates the binding of POU domains on the acidic patch of histone H2A−H2B (separated by the dashed lines). (Right) Representative snapshots of the most populated binding configurations. The top two structures show the binding pattern with POUS domain binding to the acidic patch and the POUHD domain binding on DNA. The bottom structure shows a binding pattern in which the two domains bind to two gyres of the nucleosomal DNA. (B) Probability of each residue in the POUS domain used to bind (Left) the acidic patch and (Right) the nucleosomal DNA. Results averaged over the 10 modified 601 nucleosomes, BS95 to BS104. (C) Probability distribution of the distance between the binding positions of two POU domains . Illustrative sequence logos are shown in Inset for the highest peaks ( and 79 bp). Results averaged over the 10 nucleosomes, BS95 to BS104.
In the second outstanding binding mode, each of the two POU domains was bound to the different gyres of DNA in the nucleosome. We depicted a snapshot (the bottom structure in Fig. 4 A, Right) in which the POUHD domain was bound on the “ATTG” (DNA indices 130 to 134), while the POUS domain was bound to the “GCAC” (DNA indices 50 to 53) that is only one base different from the consensus motif (Movie S4). In this two-gyre binding pattern, the binding sites of the two POU domains had a gap of ∼80 bp, which can be seen as the off-diagonal high-density belts in the 2D plot of Fig. 4A. Oct4 used these patterns to bind to a wide range of different locations on any of the simulated nucleosomes, BS95 to BS104. As we have shown in Fig. 2, the linker between the two POU domains is flexible, and thus the two domains recognized their target motifs plastically within the restrained distance. Consequently, Oct4 should be able to simultaneously bind to two distant target sites in the context of the nucleosome. We further analyzed the distance between the binding sites of the two POU domains and plotted the distance distribution in Fig. 4C. As can be seen in both the local and the nonlocal binding patterns, the two POU domains can have different orders. On the 601 nucleosome, the most probable binding pattern is that the POUHD domain binds to a site ∼80 bp downstream of the POUS binding site (Fig. 4 C, Inset), although this order should be DNA sequence-dependent and thus is not universal. We propose that this type of noncanonical motif with a long spacer can be a possible subject for the high-throughput experiments that determine TF binding motifs in the real genome. Moreover, our results also provided direct structural evidence for the previously found “gyre-spanning” TF−nucleosome binding (7) and extended this mechanism by showing that the binding pattern of a multidomain TF can be more flexible than simply recognizing the parallel DNA gyres (Fig. 4C).
We also note that, in contrast to Sox2, both POU domains of Oct4 disfavored the binding to the dyad region of the 601 nucleosome (Fig. 4A).
Spontaneous Sliding of the LIN28B Nucleosome.
In the previous two sections, we showed that the rotational position of nucleosomal DNA modulates the binding pattern of TFs, especially in the case of Sox2. However, in those simulations, we used the 601 sequence, which almost prohibited the sliding motion of the nucleosomal DNA. We next move to a nucleosome formed in a regulatory region of the human lin28b gene, which is important for reprogramming and pluripotency. The nucleosome designated as the LIN28B nucleosome hereafter includes both Sox2 and Oct4 motifs (6). In the simulation, we included the 177-bp sequence taking into consideration possible nucleosome sliding, in which the Sox2 and Oct4 target sequences are found at 59 to 65 (“AACAATA”) and 69 to 77 (“ATGCTGAAT”, which contains a 1-bp gap between two halves and a 1-bp difference in the second half, relative to the canonical motif “ATGCAAAT”), respectively (see SI Appendix, Supplementary Methods for the whole DNA sequence used in the simulation). Before simulating Sox2/Oct4 binding to the LIN28B nucleosome, we performed simulations of Sox2/Oct4 binding on the naked DNA of the same sequence. Our results showed that Sox2 was able to find its consensus sequence among many pseudo motifs, whereas Oct4 favored the several DNA motifs similar to the POUHD consensus motif, for example, “ATTA” at 15 to 18 and 39 to 42, “GAAT” at 74 to 77, “ATTG” at 131 to 134, and “AAAG” at 164 to 167 (SI Appendix, Fig. S5). These results are consistent with previous DNase experiment observations that Sox2 and Oct4 used both specific and nonspecific binding on the LIN28B DNA (6). Particularly, a recent all-atom MD simulation work confirmed that Oct4 stably bound to the “ATTA” at 39 to 42 and to the “GAAT” at 74 to 77 (30). Interestingly, we noticed that the DNA sequence at 13 to 22 (“GTATTAACAT”) is identical to that at 37 to 46, which suggests similar affinities to the “ATTA” sites at 15 to 18 and 39 to 42 for the POUHD domain. Indeed, our simulation results showed similar binding probabilities of the POUHD domain to these two sites (SI Appendix, Fig. S5B).
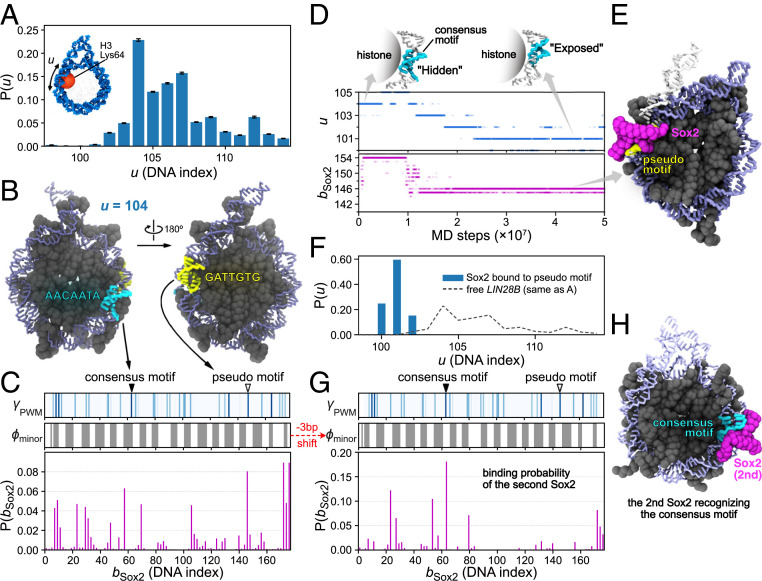
As shown in the cases of Sox2/Oct4 binding to the 601 nucleosome, the rotational positioning of the target motifs in the nucleosomal DNA affected the binding probability. However, different from the strongly positioning 601 sequence, we have to consider possible rotation-coupled sliding of the LIN28B DNA in the nucleosome. Therefore, we performed MD simulations to study the spontaneous DNA sliding in the LIN28B nucleosome. To monitor the DNA sliding behavior, we employed a coordinate , which is defined as the index of the closest phosphate to H3 Lys64 (practically, we chose one of the two Lys64 residues, as shown in Fig. 5 A, Inset). We computed the distribution of the DNA sliding position from MD simulations, which shows a most highly populated state at the sliding position (Fig. 5A and SI Appendix, Fig. S6). This result is consistent with a structure-based prediction (48) (SI Appendix, Fig. S6D). In the following simulations for Sox2/Oct4−nucleosome binding, we used the state of the position as the initial structure for the LIN28B nucleosome.
Fig. 5.
Sox2 binds to both pseudo and consensus motifs in the LIN28B nucleosome through DNA allostery. (A) Distribution of the DNA positioning u that describes the sliding of DNA in LIN28B nucleosome; is defined by the index of the phosphate that is the closest to Lys64 in the histone H3 (the red ball in Inset). Error bars are based on bootstrapping (SI Appendix, Supplementary Methods). (B) Structure of the LIN28B nucleosome at the most populated sliding position . The consensus motif (“AACAATA” at DNA index 59 to 65, cyan) has its minor groove facing “inward,” whereas a pseudo motif (“GATTGTG” at 144 to 150, yellow) has its minor groove exposed. (C) Binding probability of Sox2 on the LIN28B nucleosome starting from the state (Lower). Binding energy score (represented by blue intensity) and the exposure score (, gray for 0 and white for 1) (Upper and Middle). The positions of the consensus and the pseudo motifs in B are indicated by the solid and the blank triangles. (D) A representative time series of the Sox2 binding position (, Lower) and the sliding of nucleosomal DNA (, Middle) in the LIN28B nucleosome. Two DNA structures are shown to illustrate the rotational position of the consensus motif at and , respectively (Upper). (E) A representative structure of Sox2 binding on the pseudo motif. We show a fraction of end DNA from the superimposed free nucleosome (white) to compare with the sharply bent DNA caused by Sox2 binding. (F) Distribution of the sliding coordinate in the simulations that have one Sox2 binding on the pseudo motif as the initial structure. (G) DNA contact probability of the second Sox2, analyzed from the simulations in which one Sox2 was bound to the pseudo motif in the initial structure. (H) A representative structure of the two Sox2−LIN28B nucleosome bindings, with the second Sox2 binding on the consensus motif.
Sox2 Preferred a Pseudo Motif over the Consensus Sequence on LIN28B Nucleosome.
We then turned to simulate the binding of Sox2 onto the LIN28B nucleosome, starting from the highest population position. As clarified above, the rotational position of the Sox2 target motif determines the binding pattern and position of Sox2. We thus first take a look at the Sox2 motif position on the LIN28B nucleosome (Fig. 5B). In the position, the consensus motif of Sox2 (“AACAATA”) at DNA indices 59 to 65 was mostly buried. In contrast, we found that a distant pseudo motif (“GATTGTG”) at DNA indices 144 to 150 near the exit point was in a more exposed state. Quantitatively, we aligned the Sox2 binding energy score with the exposure extent of the LIN28B sequence for the state (Fig. 5C). The results suggested a higher probability for Sox2 to bind to the pseudo motif than the consensus sequence. To test this possibility, we put Sox2 around the LIN28B nucleosome and tracked the binding and target searching process of Sox2 in MD simulations.
We analyzed the distribution of Sox2’s binding position (Fig. 5C). Within the nucleosome, Sox2 had the highest probability to bind to the pseudo motif located at DNA index 146 (Fig. 5 B and C). On the other hand, we observed no binding event of Sox2 to the occluded consensus motif. These results confirmed that the intrinsic rotational positioning of the LIN28B sequence might not favor Sox2's recognition of its consensus motif, but instead afford a possibility to bind a second site.
Besides, Sox2 had a high contact probability to the linker DNA (Fig. 5C). This result is reasonable because the linker DNA was outside of the nucleosome core part. The strong electrostatic interaction then dominated the attraction of Sox2 to the exposed regions of DNA. Note that, at the state , the 177-bp nucleosomal DNA had one end of linker DNA longer than the other (Fig. 5B). Correspondingly, one end had a higher binding probability of Sox2 than the other one (Fig. 5C).
Sox2 Pseudo Motif Binding-Induced Nucleosomal DNA Sliding.
In addition to the analysis of Sox2 binding positions, we also looked into the dynamics of Sox2 as well as the nucleosome. Fig. 5D shows a representative time series of Sox2 searching on DNA and the sliding of the nucleosomal DNA (the sliding position ) (also see Movie S5). After the initial binding to the nucleosome, Sox2 bound to a position near the entry/exit . After MD steps, Sox2 moved to the pseudo motif at . We found that the binding of Sox2 at the pseudo motif induced the sharp bending of DNA, whose curvature was larger than the regular bending of nucleosomal DNA (Fig. 5E). We observed this type of stable binding of Sox2 at the pseudo motif and the consequent sharp bending of DNA in 15 out of 50 independent simulations (see SI Appendix, Fig. S7 for the other trajectories).
Interestingly, we noticed that the binding event of Sox2 to the pseudo motif highly correlated with the subsequent sliding of nucleosomal DNA. In the example trajectory shown in Fig. 5D, the DNA slid from the position toward after the binding of Sox2 to . We also observed the same behavior of nucleosomal DNA sliding in all of the 15 trajectories where the Sox2 pseudo motif binding occurred (SI Appendix, Fig. S7). After DNA sliding to the position, the Sox2−nucleosome complex was stable, and we did not observe any backward sliding. These results show that the Sox2 binding-induced local conformational change of DNA biased the DNA sliding in a unidirectional way, from initially to a new position at , where the Sox2−DNA−histone complex achieved a stable conformation.
It is then straightforward to consider the change in the phasing of the consensus motif as a consequence of the DNA sliding. Interestingly, we found that, as DNA slid from to , the originally “hidden” consensus motif (“AACAATA” at 59 to 65) changed to a more “exposed” state (see the structures in Fig. 5 D, Upper). These results suggested that the Sox2 pseudo motif binding (at ) might increase the accessibility of the distant consensus motif (DNA indices 59 to 65) for a second Sox2 to recognize.
To verify our hypothesis, we performed MD simulations, in which we added one additional Sox2 molecule to the LIN28B nucleosome where one Sox2 already bound at the pseudo motif. In Fig. 5 F–H, we show the results of the second Sox2 interacting with the LIN28B nucleosome. As expected, we found that, during these simulations, the nucleosomal DNA was almost “locked” in a state of b ≈ 101 (Fig. 5F), in which the consensus motif was adequately exposed and ready for binding. As a consequence, the second Sox2 recognized its consensus motif at , indicated by the contact probability of the second Sox2 (Fig. 5 G and H and Movie S6). These results revealed an allosteric regulation of TF binding through the rotational position changes of the nucleosomal DNA (see more in Discussion).
Oct4 Binding to the LIN28B Nucleosome.
As discussed in the previous section, the Sox2 binding-induced rotational position changes of the nucleosomal DNA could affect the target searching processes of other proteins. We then tried to find out whether Oct4 had the same effect or not.
We first performed MD simulations of a single Oct4 binding to the LIN28B nucleosome. As the initial structure of these simulations, we used the nucleosome at the position and put Oct4 at different positions around the nucleosome. Based on the analysis of 50 independent -step MD trajectories, we plotted the probability distribution of the binding position of the POUHD domain in Fig. 6 A, Upper. As a control, in Fig. 6A, we also plotted the distribution of the POUHD domain binding to the naked LIN28B DNA (Fig. 6 A, Lower, , also shown in SI Appendix, Fig. S5B), as well as the exposure score of the major groove of the nucleosomal DNA (Fig. 6 A, Middle, , defined in SI Appendix, Supplementary Methods) at the state. We found that the highly populated “ATTA” sites (at 15 to 18 and 39 to 42) and the canonical “GAAT” (at 74 to 77) in Oct4-naked DNA binding were no longer favored binding positions in the LIN28B nucleosome. Instead, the “CAAT” sequence piece (61 to 64) inside the Sox2 target motif became the most probable binding site. Note that, in the nucleosome, the major groove of the Sox2 motif, especially the “CAAT” piece, was facing outward (Fig. 5B) and was well exposed for the POUHD to bind. In Fig. 6B, we showed a representative structure of Oct4 binding to the LIN28B nucleosome, in which the POUHD domain was stably binding to the “CAAT” and the POUS domain was nonspecifically contacting the major groove of nearby DNA sequences (also see Movie S7). This binding pattern of the two POU domains is consistent with what we observed in the Oct4-naked DNA binding (Fig. 2 B and C). In addition to the “CAAT” binding site, a second-highest probable binding site emerged as “ATTG” at DNA indices 145 to 148. Notably, we found that the difference between Oct4 binding to the naked and the nucleosomal LIN28B DNA could be well explained by the rotational positioning and the exposure extent of the target binding sites in DNA ( in Fig. 6 A, Middle). For example, the high-affinity “ATTA” sites were mostly buried in the LIN28B nucleosomal DNA and became low-affinity sites in the POUHD−nucleosome binding, whereas the weaker binding sites in the naked LIN28B DNA, such as the “CAAT” and the “ATTG” pieces, were favored by the POUHD in the nucleosome because of their higher exposure extent (Fig. 6A).
Fig. 6.
The Oct4 binding position on the LIN28B nucleosome with and without Sox2 prebound to its pseudo motif. (A) Probability distribution of the binding position of the POUHD domain when Oct4 was bound to the free LIN28B nucleosome (Upper, red bars). Middle shows the exposure score of DNA major groove (; defined in SI Appendix, Supplementary Methods). As a control, we show the binding probability of the POUHD domain to the naked LIN28B DNA (Lower). Correspondingly, on the top of Upper, we show the sequences and positions of possible high-affinity sites for the POUHD binding, which are “ATTA” (15 to 18), “ATTA” (39 to 42), “CAAT” (61 to 64), “GAAT” (74 to 77), “ATTG” (131 to 134), “ATTG” (145—148), and “AAAG” (164 to 167). (B) A representative structure of Oct4 binding to the free LIN28B nucleosome with the highest probability. The POUHD domain recognizes the sequence of “CAAT” (61 to 64) in the major groove of the Sox2 consensus motif. (C) The same as A except that Upper shows the distribution of the POUHD binding positions on the Sox2-bound LIN28B nucleosome, in which the nucleosomal DNA was “fixed” at u = 101 by Sox2-binding. (D) A representative structure of Oct4 binding to the Sox2-bound LIN28B nucleosome, with the POUHD recognizing the “ATTA” piece (15 to 18). (E) A representative structure of POUHD binding to the canonical consensus sequence, “GAAT” (74 to 77), in the Sox2-bound LIN28B nucleosome. The color scheme of the structures in C–E is the same as in A and B.
As discussed above, the sliding-coupled rotational positioning of the nucleosomal DNA affected the binding probability of Oct4. Therefore, it is important to find out the rotation and sliding features of the LIN28B nucleosomal DNA upon Oct4 binding. However, in contrast to the significant changes induced by Sox2, we did not observe compelling sliding of the LIN28B nucleosomal DNA upon Oct4 binding. The probability distribution of the positioning in the Oct4−nucleosome complex showed a single peak at (SI Appendix, Fig. S8). These results suggested that, in the competition with the histones, Oct4 binding was not strong enough to change the rotational positioning of the nucleosomal DNA.
We next tried to find out the possible allosteric regulatory effect of Sox2 pseudo motif binding on the Oct4 binding position. We added one Oct4 molecule to the Sox2−nucleosome complex, in which the Sox2 had been bound to the pseudo motif and the sliding was therefore constrained at (Fig. 5E). We performed 50 independent MD simulations, in which the Oct4 was initially randomly located around the Sox2−nucleosome complex. The resulting contact probability distribution of the POUHD domain on the nucleosomal DNA (Fig. 6 C, Upper) exhibited a clearly different pattern from the results of Oct4 binding to the LIN28B nucleosome without Sox2 (Fig. 6 A, Upper; see SI Appendix, Fig. S9 for the 2D contact probability of both POU domains). We found that the 3-bp sliding of the nucleosomal DNA from to prominently changed the exposure/burying of the POUHD binding sites (Fig. 6 A and C, Middle). Consequently, the POUHD was able to find the newly exposed DNA sequences such as the “ATTA” at 15 to 18 (Fig. 6D and Movie S8) or the “GAAT” at 74 to 77 (Fig. 6E). These results showed that the prebinding of Sox2 to the pseudo motif regulated the rotational positioning of the nucleosomal DNA, which, in turn, adjusted the binding position of Oct4.
Discussion
Sox2 and Oct4 Bind on Nucleosomes as Pioneer Factors.
Pioneer TFs are different from normal factors by the ability to recognize their target DNA sequences in the closed chromatin structure (3, 5, 49). Previous experiments have tried to link the pioneer factors' nucleosome-binding capability with their biological function to regulate the pluripotency of cells (50–52). Although many nucleosome-binding features of Sox2 and Oct4 have been supported by our MD simulation results, such as the dependence on the rotational positioning (7) (Fig. 3) and the partial motif recognition (6) (Fig. 4), there is missing information about how these pioneer factors open the compact chromatin structure. Our results suggest that Sox2 binding can regulate the accessibility of other TFs’ targets in the nucleosomal DNA. Sox2, together with the subsequently bound TFs, may interplay with the intrinsic free energy landscape of nucleosomes (53) and cause possible large-scale nucleosome conformational changes. Interestingly, the recently solved cryo-EM structures of nucleosome bound by Sox2/Sox11 and Sox2+Oct4 revealed sequence-dependent different pioneer TF−nucleosome binding mechanisms (28, 29). On a modified 601 nucleosome, Sox2 and Oct4 together recognized their consensus motif at SHL-6 and bent the entry/exit region of DNA away from histone (29), whereas, when Sox2 bound to its motif at SHL+5, the DNA conformational change was localized (29), consistent with our simulation results (SI Appendix, Fig. S4). In contrast, the structure of Sox2/Sox11 binding to SHL+2 of another nucleosome showed that Sox2/Sox11 facilitated the detachment of terminal DNA in the second gyre (28). These results showed that Sox2 binding could affect nucleosome structure in different ways, which is apparently dependent on the DNA sequence.
Besides, recent studies of the intrinsically disordered activation domains of TFs suggested transcription regulation and genome organization mechanisms based on the liquid−liquid phase separation (54, 55). These findings suggest that, in addition to the pioneer factors’ nucleosome-binding features shown in the current work, study of the disordered activation domains may be necessary to understand biological functions of the pioneer factors more thoroughly.
Nucleosome Acts as an Allosteric Scaffold for TF Binding.
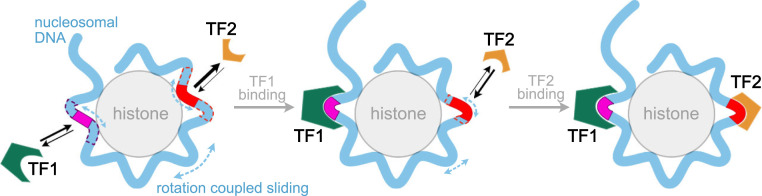
Nucleosome structures are flexible enough to be regulated by other factors. The spontaneous nucleosome dynamics such as partial unwrapping and rotation-coupled sliding of the nucleosomal DNA has been well documented by both experiments and MD simulations (13, 15–17). The partial unwrapping of DNA opens up more space for TF to bind (13), whereas TF binding, in turn, changes the accessibility of deeper nucleosomal DNA for additional TF binding (21). These regulations can be considered as allosteric regulation (12). Here our simulation results provided clues for another possible allosteric mechanism of collaborative TF−nucleosome binding. We show that one TF (Sox2) binding on its target can change the rotational positioning of the nucleosomal DNA and thus regulate the exposure extent of distant binding sites for another TF (Sox2 or Oct4) to recognize (Fig. 7). Compared with the unwrapping-related TF−nucleosome binding cooperativity, the sliding-mediated mechanism is energetically more favorable, considering that there is less breakup of hydrogen bonds between histone and DNA (12). Besides, the sliding-mediated allostery may affect more distant TF binding (bp, the length of nucleosomal DNA) than the unwrapping-related one (tens of base pairs) (12). Interestingly, our finding of Sox2’s allosteric regulation on Oct4 binding provided an insightful explanation for previous findings that Sox2 reduced the search time and increased the residence time of Oct4 (56). We suggest possible high-resolution experiments, such as fluorescence resonance energy transfer (57), to test the Sox2 controlled nucleosomal DNA sliding and the allosteric regulation of TF binding.
Fig. 7.
Allosteric regulation of TF binding through nucleosomal DNA. (Left) TF binding probability to their targets in nucleosomal DNA is dependent on both DNA sequence and the rotational position of the target site. The TF1’s target is more exposed than the TF2’s target. Nucleosomal DNA undergoes spontaneous rotation-coupled sliding. (Middle) Binding of TF1 restricts the sliding motion of the nucleosomal DNA and changes the exposure extent of a distant binding site for TF2, which, in turn, results in a higher binding probability of TF2 (Right).
Local Induced Fit and Global Conformational Selection.
As discussed above, Sox2 binding led to much sharper bending of DNA than observed in normal nucleosome (Fig. 5E and SI Appendix, Fig. S4). Therefore, on a local scale, we considered this as an induced fit mechanism for Sox2−nucleosome binding. On a larger landscape, the spontaneous sliding of the LIN28B nucleosomal DNA covered a wide range of the rotational positioning (Fig. 5A), including u ≈ 101 and u ≈ 101, at which the Sox2’s consensus motif was highly exposed. Indeed, Sox2 had a probability to find its consensus motif when nucleosome slid to these states (see SI Appendix, Fig. S10 for results of the simulations starting from ). However, these states were less populated, and the spontaneous sliding of nucleosomal DNA is relatively slow (16, 17), suggesting that free LIN28B nucleosome was not optimized for Sox2 to recognize its consensus motif. In comparison, Sox2 binding at the pseudo motif drastically narrowed the rotational positioning to u = 100 to ∼102 (Fig. 5F) and increased the exposure probability of Sox2’s consensus motif. Consequently, Sox2’s binding probability to its target was up-regulated. These results show that the binding of Sox2 to the pseudo motif induced the local conformational changes and selected the global positioning of nucleosomal DNA. This type of TF-regulated nucleosome allostery also emphasizes the importance of pseudo motifs in TF’s target search process.
Validation of Our CG Models.
Apparently, the CG models we used here sacrificed atomistic details of interactions to gain sampling efficiency. Although our simulation results are consistent with experimental results (6, 7, 28, 29), a high-resolution picture of the physical interactions such as hydrogen bonds and salt bridges was not provided. To get such information, reconstruction of all-atom structures from CG models and all-atom simulations might be helpful. As an example, we rebuilt the all-atom structures for the POUS domain and the H2A−H2B dimer and performed simulations to check the stability of the structure of the POUS recognizing the acidic patch (SI Appendix, Fig. S11). We found that the POUS−H2A−H2B ternary complex was stable during 50-ns all-atom simulations. We also located several key residues in the POUS domain that contributed to the binding with H2A−H2B (SI Appendix, Fig. S11).
Conclusion
We used CG MD simulations to investigate the binding mechanisms of pioneer TFs Sox2 and Oct4 on the nucleosomes. We first studied the Sox2 binding on a strongly positioning sequence 601 and found that the binding position of Sox2 was highly dependent on the rotational positioning of the target motif. Sox2 could only recognize the exposed motifs, whereas, when the consensus sequence was buried, Sox2 preferentially bound to the dyad among all of the nonspecific binding sites. As for Oct4, we found a bridging binding pattern in which the two POU domains parallelly bound to the two gyres of nucleosomal DNA. Our simulations also showed a possibility of the POUS domain binding on the acidic patch on histone. By studying the binding of Sox2 and Oct4 on the LIN28B nucleosome, we proposed a nucleosomal DNA-mediated allostery mechanism which combined the induced fit and conformational selection scenarios: Sox2 binding can induce the local distortion of DNA, which, consequently, selects the global rotational positioning of the nucleosomal DNA. The redistributed sliding phase then allosterically regulates the binding of Oct4 and the second Sox2.
Methods
Reference Structures.
In the present work, we studied the binding of Sox2 and Oct4 to both naked DNAs and nucleosomes. We used Protein Data Bank (PDB) entries 1GT0 and 3L1P as the reference structures of the DNA-binding domains of Sox2 and Oct4, respectively. We used the 3DNA package (58) to build the reference structures of B-form dsDNAs. As the template structure of nucleosome, we used PDB entry 1KX5. To construct nucleosome structures with different DNA sequences, we fixed the histone structure and changed DNA from that in 1KX5 to the target sequences, followed by an energy minimization.
Protein and DNA Models.
In our CG MD simulations, we used the AICG2+ model (33) for protein, by which every amino acid is represented by one particle located at the atom. The energy function of the AICG2+ is expressed as , where involves all bonded interactions, is the -type structure-based term, and is the excluded volume interaction (33). For protein−protein interfaces where there are no -type interactions, namely, between Sox2 and histone and between Oct4 and histone, we applied electrostatic interactions modeled by the Debye−Hückel theory. More details of the protein CG modeling can be found in SI Appendix, Supplementary Methods.
For CG DNA, we employed the 3SPN.2C model (34), with every nucleotide depicted by three particles, each representing phosphate (P), sugar (S), and base (B). Notably, the potentials and parameters in this model have been well tuned to capture the sequence-dependent geometric, mechanical, and thermodynamic properties of dsDNA (34).
Protein−DNA Interactions.
The dominant part of protein-DNA interactions is the electrostatics, for which we used the Debye−Hückel model. The charge of phosphate was set to when we were calculating DNA interaction with protein, whereas, for internal DNA interactions, the phosphate charge was set to [default value in the 3SPN.2C model (34)]. For accuracy, we employed the restrained electrostatic potential from atomic charges to CG charges (RESPAC) method (59) to calculate the partial charge distribution on proteins. As for the excluded volume effect, we used residue-type-dependent radii for both protein and DNA particles (35). Besides, we used the newly developed PWMcos model for Sox2 and Oct4 to enable their specific recognition of DNA sequences (32). Accordingly, we applied a sequence-nonspecific variation of the PWMcos model to represent the hydrogen bonds in histone−DNA interactions (17).
MD Simulations.
All of the MD simulations were conducted by Langevin dynamics at temperature T = 300 K, with the friction coefficient and a step size of ∼1 ps.
We first calibrated parameters in the PWMcos model for Sox2 and Oct4 by matching their simulated dissociation constant with DNA to the experimental results (37, 38). These simulations were carried out at the ionic concentration of 150 mM, which was also used in experiments. We monitored the distance between the COM of protein and DNA, based on which we then calculated from the probabilities of the bound and unbound states (35) (see more details about the calibration in SI Appendix, Supplementary Methods). The PWM data for Sox2 and Oct4 was downloaded from the JASPAR database (60).
After parameter calibration, we used our model to simulate binding and sliding of Sox2 and Oct4 on naked dsDNAs. The binding position of Sox2 or a POU domain on DNA is represented by a coordinate , where is the index of DNA nucleotide from which at least one particle (P, S, or B) is within 10 Å of a , and is the total number of the DNA nucleotides in contact with the corresponding protein.
We then used the strong positioning 601 sequence (36) as a template and inserted target motifs of Sox2 (7 bp) into different positions of the 601 nucleosomal DNA (147 bp). We performed simulations of Sox2 binding to nucleosomes with these designed DNA sequences. A full list of the simulated nucleosomal DNA sequences can be found in SI Appendix, Supplementary Methods and Fig. S1. In these simulations, we added a constraint between the COM of Sox2 and the COM of histone octamer so that their distance was limited to be smaller than 350 Å. All of these simulations were conducted at ionic concentration of 200 mM. For every sequence, we performed 20 independent simulations (in total, independent runs), each for MD steps. In each simulation, we placed Sox2 at a different position around the nucleosome as the initial structure. We also applied the same methods to simulate the Oct4 binding on the modified 601 nucleosomes.
We then studied the binding of Sox2 and Oct4 on the LIN28B nucleosome (see SI Appendix, Supplementary Methods for the DNA sequence), which contains the target motifs for both TFs. We first performed extensive MD simulations (64 independent runs, each for steps; see SI Appendix, Supplementary Methods for more details) to get the equilibrated sampling of the rotational positioning of the nucleosomal DNA (Fig. 5A and SI Appendix, Fig. S6). Then we put Sox2 and Oct4 around the LIN28B nucleosome, starting from the most populated rotational position of , to study the binding and target searching process of the TFs ( steps runs for each TF). In the simulations of two TFs (Sox2 + Sox2 or Sox2 + Oct4) binding to the LIN28B nucleosome, we used the Sox2−nucleosome complex (Fig. 5E), where one Sox2 was bound to a pseudo motif, as the initial structure and added the other TF (Sox2 or Oct4) around the nucleosome ( steps runs for either Sox2 or Oct4). All of the above simulations for TF binding on the LIN28B nucleosome were conducted at temperature 300 K and ionic strength of 200 mM. Besides, we also performed test simulations at higher temperature (310 K) and salt concentration (410 mM) (SI Appendix, Fig. S12).
All of the simulations were performed with the CafeMol MD package (61).
Supplementary Material
Acknowledgments
We thank Giovanni Brandani for many insightful discussions and for CG modeling of nucleosomes. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 16KT0054 (to S.T.) and 16H01303 (to S.T.), the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) as “Priority Issue on Post-K computer” (S.T.), and the RIKEN Pioneering Project “Dynamical Structural Biology” (S.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005500117/-/DCSupplemental.
Data Availability.
The Software CafeMol can be downloaded from www.cafemol.org/. Data included in the main text and SI Appendix are sufficient to reproduce the work. All of the large-volume simulation trajectories are stored on local servers and can be obtained upon reasonable request.
References
- 1.Onufriev A. V., Schiessel H., The nucleosome: From structure to function through physics. Curr. Opin. Struct. Biol. 56, 119–130 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Magnani L., Eeckhoute J., Lupien M., Pioneer factors: Directing transcriptional regulators within the chromatin environment. Trends Genet. 27, 465–474 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Iwafuchi-Doi M., Zaret K. S., Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez Garcia M. et al., Structural features of transcription factors associating with nucleosome binding. Mol. Cell 75, 921–932.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soufi A. et al., Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu F. et al., The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chronis C. et al., Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgunova E., Taipale J., Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 47, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Jolma A. et al., DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kim S. et al., Probing allostery through DNA. Science 339, 816–819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takada S., Brandani G. B., Tan C., Nucleosomes as allosteric scaffolds for genetic regulation. Curr. Opin. Struct. Biol. 62, 93–101 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Li G., Levitus M., Bustamante C., Widom J., Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Ngo T. T. M., Ha T., Nucleosomes undergo slow spontaneous gaping. Nucleic Acids Res. 43, 3964–3971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaus A., Owen-Hughes T., Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol. Cell. Biol. 23, 7767–7779 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niina T., Brandani G. B., Tan C., Takada S., Sequence-dependent nucleosome sliding in rotation-coupled and uncoupled modes revealed by molecular simulations. PLoS Comput. Biol. 13, e1005880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandani G. B., Niina T., Tan C., Takada S., DNA sliding in nucleosomes via twist defect propagation revealed by molecular simulations. Nucleic Acids Res. 46, 2788–2801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaytan A. K. et al., Coupling between histone conformations and DNA geometry in nucleosomes on a microsecond timescale: Atomistic insights into nucleosome functions. J. Mol. Biol. 428, 221–237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tims H. S., Gurunathan K., Levitus M., Widom J., Dynamics of nucleosome invasion by DNA binding proteins. J. Mol. Biol. 411, 430–448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polach K. J., Widom J., Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J. Mol. Biol. 254, 130–149 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Gibson M. D., Gatchalian J., Slater A., Kutateladze T. G., Poirier M. G., PHF1 Tudor and N-terminal domains synergistically target partially unwrapped nucleosomes to increase DNA accessibility. Nucleic Acids Res. 45, 3767–3776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirny L. A., Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 22534–22539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okita K., Ichisaka T., Yamanaka S., Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hou L., Srivastava Y., Jauch R., Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 63, 2–12 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Ryan A. K., Rosenfeld M. G., POU domain family values: Flexibility, partnerships, and developmental codes. Genes Dev. 11, 1207–1225 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Jerabek S., Merino F., Schöler H. R., Cojocaru V., OCT4: Dynamic DNA binding pioneers stem cell pluripotency. Biochim. Biophys. Acta 1839, 138–154 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Kraus W. L., Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 65, 589–603.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodonova S. O., Zhu F., Dienemann C., Taipale J., Cramer P., Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 580, 669–672 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Michael A. K. et al., Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 368, 1460–1465 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Huertas J., MacCarthy C. M., Schöler H. R., Cojocaru V., Nucleosomal DNA dynamics mediate Oct4 pioneer factor binding. Biophys. J. 118, 2280–2296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takada S. et al., Modeling structural dynamics of biomolecular complexes by coarse-grained molecular simulations. Acc. Chem. Res. 48, 3026–3035 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Tan C., Takada S., Dynamic and structural modeling of the specificity in protein-DNA interactions guided by binding assay and structure data. J. Chem. Theory Comput. 14, 3877–3889 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Li W., Wang W., Takada S., Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc. Natl. Acad. Sci. U.S.A. 111, 10550–10555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman G. S., Hinckley D. M., Lequieu J. P., Whitmer J. K., de Pablo J. J., Coarse-grained modeling of DNA curvature. J. Chem. Phys. 141, 165103 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Tan C., Terakawa T., Takada S., Dynamic coupling among protein binding, sliding, and DNA bending revealed by molecular dynamics. J. Am. Chem. Soc. 138, 8512–8522 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Takayama Y., Clore G. M., Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc. Natl. Acad. Sci. U.S.A. 108, E169–E176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takayama Y., Clore G. M., Interplay between minor and major groove-binding transcription factors Sox2 and Oct1 in translocation on DNA studied by paramagnetic and diamagnetic NMR. J. Biol. Chem. 287, 14349–14363 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaffidi P., Bianchi M. E., Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem. 276, 47296–47302 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Kamagata K., Mano E., Ouchi K., Kanbayashi S., Johnson R. C., High free-energy barrier of 1D diffusion along DNA by architectural DNA-binding proteins. J. Mol. Biol. 430, 655–667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacherjee A., Levy Y., Search by proteins for their DNA target site: 1. The effect of DNA conformation on protein sliding. Nucleic Acids Res. 42, 12404–12414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klemm J. D., Pabo C. O., Oct-1 POU domain-DNA interactions: Cooperative binding of isolated subdomains and effects of covalent linkage. Genes Dev. 10, 27–36 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Cui F., Zhurkin V. B., Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nucleic Acids Res. 42, 836–847 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahu G. et al., p53 binding to nucleosomal DNA depends on the rotational positioning of DNA response element. J. Biol. Chem. 285, 1321–1332 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto S. et al., DNA damage detection in nucleosomes involves DNA register shifting. Nature 571, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalashnikova A. A., Porter-Goff M. E., Muthurajan U. M., Luger K., Hansen J. C., The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 10, 20121022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Q., Yang R., Korolev N., Liu C. F., Nordenskiöld L., Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail-H2A acidic patch interaction. J. Mol. Biol. 429, 2075–2092 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Alharbi B. A., Alshammari T. H., Felton N. L., Zhurkin V. B., Cui F., nuMap: A web platform for accurate prediction of nucleosome positioning. Genomics Proteomics Bioinformatics 12, 249–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaret K. S., Lerner J., Iwafuchi-Doi M., Chromatin scanning by dynamic binding of pioneer factors. Mol. Cell 62, 665–667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobersch S., Rubio K., Barreto G., Pioneer factors and architectural proteins mediating embryonic expression signatures in cancer. Trends Mol. Med. 25, 287–302 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Mayran A., Drouin J., Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meers M. P., Janssens D. H., Henikoff S., Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol. Cell 75, 562–575.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B., Zheng W., Papoian G. A., Wolynes P. G., Exploring the free energy landscape of nucleosomes. J. Am. Chem. Soc. 138, 8126–8133 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Boija A. et al., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabari B. R. et al., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J. et al., Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deindl S. et al., ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 152, 442–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu X.-J., Olson W. K., 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terakawa T., Takada S., RESPAC: Method to determine partial charges in coarse-grained protein model and its application to DNA-binding proteins. J. Chem. Theory Comput. 10, 711–721 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Mathelier A. et al., JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42, D142–D147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenzaki H. et al., CafeMol: A coarse-grained biomolecular simulator for simulating proteins at work. J. Chem. Theory Comput. 7, 1979–1989 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Software CafeMol can be downloaded from www.cafemol.org/. Data included in the main text and SI Appendix are sufficient to reproduce the work. All of the large-volume simulation trajectories are stored on local servers and can be obtained upon reasonable request.