Abstract
The state of latency occurs when a microbe’s persistence in a host produces host damage without perturbing homeostasis sufficiently to cause clinical symptoms or disease. The mechanisms contributing to latency are diverse and depend on the nature of both the microbe and the host. Latency has advantages for both host and microbe. The host avoids progressive damage caused by interaction with the microbe that may translate into disease, and the microbe secures a stable niche in which to survive. Latency is clinically important because some latent microbes can be transmitted to other hosts, and it is associated with a risk for recrudescent microbial growth and development of disease. In addition, it can predispose the host to other diseases, such as malignancies. Hence, latency is a temporally unstable state with an eventual outcome that mainly depends on host immunity. Latency is an integral part of the pathogenic strategies of microbes that require human (and/or mammalian) hosts, including herpesviruses, retroviruses, Mycobacterium tuberculosis, and Toxoplasma gondii. However, latency is also an outcome of infection with environmental organisms such as Cryptococcus neoformans, which require no host in their replicative cycles. For most microbes that achieve latency, there is a need for a better understanding and more investigation of host and microbial mechanisms that result in this state.
This article discusses the complex state of latency in microbial pathogenesis using the damage-response framework (DRF) as a conceptual guide. The DRF puts forth an integrated theory of microbial pathogenesis and infectious diseases that incorporates the role of the host, as well as the microbe, to define the outcome of host-microbe relationships. The DRF was first proposed in 1999 as a solution to the late-20th-century problem of explaining the increase in human disease with microbes that were thought to be nonpathogenic or very rare causes of disease in those with intact immunity (1–3). These microbes often resided on the skin and/or mucosal surfaces of normal people, e.g., staphylococci, enterococci, enteric Gram-negatives, and Candida spp., and included those that were previously characterized as “opportunists” because they were nearly always observed as causes of disease in patients with immune impairment, e.g., Pneumocystis, Cryptococcus, Histoplasma, and Toxoplasma spp. During this time, there was also a recrudescence of adult disease caused by viruses with the capacity for latency, e.g., Epstein-Barr virus, cytomegalovirus, and other herpesviruses. There were three main factors driving the emergence of these microbes: (a) breach of barriers as reflected by the increased use of plastic catheters and advances in surgery; (b) exogenous immunosuppression as a consequence of advances in the treatment/management of malignancy, inflammatory diseases, and organ transplantation with cytotoxic and/or immunosuppressive agents; and (c) endogenous immunosuppression brought on by the AIDS pandemic. Collectively, these factors led to an unprecedented number of individuals with acquired immune impairments ranging from loss of barrier immunity to deficits in humoral and/or cellular immunity.
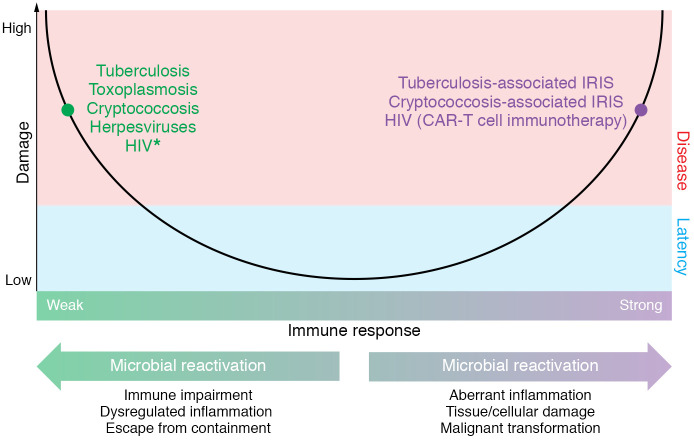
The DRF provided a tractable and integrated theory that clinicians and researchers could use to explain, predict, and propose new approaches for the therapy and study of the pathogenesis of common, emerging, and reemerging microbes (3). The basic tenet of the DRF is that the relevant outcome of host-microbe interaction is host damage, which is a function of the host response and can be mediated by host or microbial factors or both. As illustrated in Figure 1, the relationship between host damage and the host immune response is depicted by a simple, upright parabola, whereby damage is plotted on the y axis and the host response, ranging from weak to strong, is plotted on the x axis. The threshold for disease, a clinical state, is represented by a horizontal line across the y axis. This schema makes it possible to explain several important principles, including that (a) microbial infection may result in damage without causing clinical disease and (b) host damage and disease occur in the setting of either an insufficient or an exuberant immune response to a microbe. While the latter concept was novel when the DRF was first proposed, today it is common to attribute clinical disease to host inflammation, even in the setting of microbial clearance. In fact, we attribute this change in emphasis to the success of the DRF in reframing questions of microbial pathogenesis.
Figure 1. Microbial latency in the context of the DRF.
The DRF views latency as an outcome of host-microbe interaction. In this state, the host is asymptomatic because the degree of damage resulting from host-microbe interaction is not sufficient to perturb homeostasis. All forms of latency involve some damage to the host at the cellular and/or tissue level. We posit that reactivation can occur at either horn of the parabola. Reactivation of latent microbes such as M. tuberculosis, C. neoformans, T. gondii, and HSV is often associated with conditions that impair the immune response. Reactivation is manifested clinically by recurrence of disease in the setting of immune suppression or weak immunity (the left side of the parabola). However, strong immune responses that result in dysregulation of inflammation may also affect the host-microbe relationship and lead to reactivation, e.g., in tuberculosis and cryptococcosis. This is manifested clinically by a recurrence of disease. For example, reactivation of tuberculosis with caseous necrosis that does not eradicate mycobacteria stems from a strong immune response, and tuberculosis-associated and cryptococcosis-associated immune reconstitution inflammatory syndromes (IRIS) result in immune damage in the setting of an augmented immune response to microbial antigens. Although some of these antigens are likely to be material released from dead microbes, ability of both tuberculosis and cryptococcosis to recur after treatment means that viable cells remain in tissue. HIV (left side of the parabola) is marked with an asterisk because this organism continues to damage the immune system during its latent quiescent state and thus differs from the others as creating its own conditions for the progression to AIDS. HIV (right side of the parabola) is a strategic target for reactivation to reduce the reservoir of latently infected CD4+ T cells, e.g., with chimeric antigen receptor (CAR) T cells (55).
A major contribution of the DRF to clinical practice and research was its ability to account for different outcomes of infection with a single microbe without invoking confusing and inconsistent terminology, e.g., pathogen, nonpathogen, opportunist, primary pathogen, colonizer, commensal (2, 4). To describe the state of host-microbe interaction, the DRF puts forth five states of microbial existence in a host: (a) infection, (b) colonization, (c) commensalism, (d) disease, and (e) latency (also referred to as chronicity and/or persistence) (2). According to the DRF, there are no pathogens, commensals, etc.; there are only microbes and hosts whose interaction results in one of these states. The DRF proposes that these states are continuous and differ from one another only as a function of damage in the host and time. The states of colonization and commensalism can transition to the state of disease in the setting of immune impairment, e.g., loss of barrier immunity, medications that impair immunity, and primary or acquired immune compromise. Similarly, the state of latency can transition to that of disease, although neither the clinical nor the microbial triggers for loss of latency are fully understood. Notably, the DRF depicts latency as a chronic and/or persistent state that differs from the states of colonization and disease as a function of host damage and time. Colonization and latency are states in which the host damage that occurs does not perturb homeostasis to a degree that results in clinical disease. As a result, each state lies below the line on the DRF parabola that delineates clinical disease. However, latency and colonization differ as a function of time. Colonizing microbes either are eliminated or transition to the state of disease, after which some may enter a state of latency. In contrast, latent microbes remain in a host over time after the host has experienced an episode of disease.
The concept of latency is useful for clinicians and basic scientists alike. For clinicians, understanding the state of a microbe in a host can inform an understanding of disease pathogenesis and prognosis, as well as the use of antimicrobial agents, making this an evidence-based decision, as well as decisions on implementation of infection control measures (5). For researchers, accounting for the state of the microbe in a host makes it possible to devise more precise questions to unravel pathogenesis and virulence.
What is latency?
Clinically, at the bench, and in experimental models, the state of latency is one of quiescence, whereby there are no clinical symptoms in the host and there is no evidence of microbial growth, despite the continued presence of the microbe in the host. For microbes, the state of latency may be due to a genetic change that blocks replication entirely, a slow rate of replication that may not be detectable with available tools, or sequestration that escapes detection. None of these strategies produce a degree of damage that translates into clinical disease. Generally, the state of latency is invoked for microbes formerly associated with known acquisition and/or clinical illness. As such, there is no clinical evidence of the microbe, but it remains present in the host and the risk of recrudescent disease looms. Recrudescent disease is often characterized as “reactivation.” Traditional examples of latent microbes include Mycobacterium tuberculosis and herpesviruses. The state of latency contrasts with microbial states that feature either acquisition without clinical symptoms or disease, e.g., colonization or commensalism, or acquisition followed by continued microbial growth resulting in symptoms, disease, elimination, or death of the host.
Classical, functional views of latency, whereby there is no clinical disease and no evidence of microbial growth, represent the tip of a complex iceberg. M. tuberculosis is a microbe that is universally accepted as capable of latency. On the other hand, based on its clinical course, human immunodeficiency virus (HIV) was originally believed to enter a state of latency after an initial illness that was followed by a long asymptomatic state. However, advances in understanding the biology of the virus and its interaction with the host revealed that instead of being quiescent, the virus continues to replicate during the asymptomatic period between infection and AIDS. This period of containment by the host immune system resulted in a clinically latent state that lasted until the immune system had sustained a degree of damage that could not prevent the emergence of clinical disease, often from an intercurrent infection. Another way in which HIV latency is characterized is as the state in which reservoirs of virus may remain after administration of agents to induce a cure (6).
Latency in the context of the DRF
The state of latency is referred to as a state of persistence to convey that certain latent microbes continue to replicate in, and/or are recovered as “infectious” particles from, their hosts (7–9). Thus, clinical latency can occur in the setting of either persistence, a state characterized by ongoing microbial (usually viral) replication, albeit at a reduced rate; or latency, a state characterized by microbial presence without ongoing replication. While persistence best describes the state of HIV and other microbes in hosts that exhibit clinical latency despite ongoing viral replication, microbial persistence and latency each result in an outcome of host-microbe interaction that does not feature clinical signs or symptoms. From the viewpoint of the DRF, persistence and latency are comparable states, with the caveat that persistent and latent microbes have different biological strategies for survival and for immune evasion and may evoke different host immune responses. As above, latency is not associated with a degree of damage that perturbs homeostasis or translates into clinical disease. While this concept may appear simplistic, it provides the flexibility that is necessary to incorporate and explain emerging evidence that microbes not previously thought to exhibit latency can exhibit periods of clinical quiescence. One example of this phenomenon is Ebola virus, which may recrudesce after recovery from disease (10); another is the growing recognition that adenoviruses may persist and reactivate in immunosuppressed hosts (9).
One variable that affects the state of latency is time. For certain infectious diseases, the term “latency period” is used to describe the time between microbial acquisition and clinically evident disease. Historically, recognition of this period made it possible to identify that the yellow fever virus was transmitted to humans by mosquitoes following an extrinsic latency period (11). This usage of the term “latency period” is often used synonymously with “incubation period.” Although “latency period” does not represent classical latency, it is important to account for this period, because it delineates a time during which a microbe can be transmitted from person to person while the transmitting individual is asymptomatic. This period has been an intense focus of the investigation of transmission of the 2019 novel Coronavirus (12, 13). Challenges to our understanding of this period have increased since the 2010s, when it was discovered that Ebola virus disease could recrudesce (14, 15) and that Zika virus could persist in semen months after initial acquisition (16, 17). A common thread that links Ebola and Zika to prolonged existence in a host is the presence of host reservoirs that contain and sequester these microbes. While these reservoirs remain incompletely understood, from a clinical standpoint, the presence of these viruses in an asymptomatic host blurs the line between microbial persistence, latency, and clinical disease. From the standpoint of microbial pathogenesis, the state of a “quiescent” microbe in a host is crucially important for epidemiology, diagnosis, and prevention strategies, as well as approaches to therapy and vaccine design.
Why is latency important?
Latency provides a mechanism for a microbe to establish an ongoing or persistent niche in a host. For the microbe and the host, latency offers a relatively safe reservoir. It allows the microbe to travel with the host with the possibility of reaching new populations. The presence of a latent microbe signals the potential for reactivation and future disease. This may or may not occur. In humans, death of the host with a latent microbe generally signals the death of the microbe. However, in feral hosts, death can enable the microbe to find new hosts if a carcass is eaten or returns to the soil when the host dies and decomposes, provided the microbe remains viable. In this regard, it has been suggested that Coccidioides immitis reached South America after being carried by migrating humans (18), possibly in a latent state. For the individual carrying a latent microbe, there is the potential for damage that translates into another disease. For example, lung inflammation from latent M. tuberculosis can progress to scar formation and subsequent carcinoma. In experimental Cryptococcus neoformans infection in rats, control of pneumonia is associated with persistence of fungal cells in granulomas that alters the pulmonary immune response to promote airway hyperresponsiveness (19). Cryptococcal antigens may also trigger asthma in children (20). These examples show that establishment of the latent state can alter immune and inflammatory responses in the organs involved and predispose the host to very different diseases.
Why do certain microbes elicit latency?
Not all microbes exhibit the capacity for latency. In this respect, microbes with prolonged incubation periods between infection and disease (clinical latency) differ from microbes with classically defined persistence or latency. The former microbes are clinically quiescent during the period of clinical latency because the microbial burden and/or immune response does not elicit sufficient damage to affect homeostasis and translate into clinical disease. In contrast, microbes that exhibit “classical” latency or persistence infect the host and enter this state without necessarily causing clinical symptoms or progressing to disease. The microbial mechanisms and immune evasion strategies that lead to the state of latency are complex and varied. Some microbes capable of establishing the classical state of latency, such as M. tuberculosis and C. neoformans, enter a state characterized by a host response that features persistence of the microbe within tissue granulomas. The initial state of infection with these microbes may not be associated with a state of disease. Other microbes, such as herpesviruses, assume a state whereby their genomes are maintained in host cells and viral progeny are not produced, but possess the potential for reactivated viral replication (21). Although recent evidence suggests that certain arboviruses, e.g., Zika virus, may remain sequestered in body fluids for a prolonged period (22–24), more data are needed to delineate this state. Nonetheless, clinical latency of arboviruses (17, 25) as well as Ebola virus (26, 27) is important to recognize because of the potential for viral transmission, particularly in endemic areas. This underscores the epidemiological and public health importance of distinguishing clinical from microbial latency and characterizing molecular mechanisms that govern and maintain the latter for therapy and vaccine development (28, 29).
From a microbiological perspective, the capacity for latency benefits the microbe by enabling it to establish a niche from which it is not likely to be eliminated. However, if the state of latency transitions to a state of renewed growth that translates into disease, there is the potential for microbial elimination via the host immune response, antimicrobial therapy, or death of the host. From a host perspective, when a microbe enters a state of latency, this is usually a better outcome than progression to disease, with the caveat that latency is marked by the potential for microbial recrudescence and development of disease. There is also the risk of viral malignant transformation (30–33). Thus, while the state of latency is a low-cost, possibly zero-sum solution for host and microbe alike, this cost-benefit calculation assumes that the state of the microbe and the state of the host remain constant. The host state may be altered by intercurrent disease, the development of impaired immunity, senescence, or the myriad factors that perturb host-microbe interaction (34). For the microbe, there is always the possibility that immunity will clear the microbe, or that other factors will impair the latent/persistent state.
Why certain microbes exhibit latency while others do not is largely unknown. When one surveys the qualities of microbes capable of exhibiting the state of latency, one is struck by their diversity and the different pathogenic strategies they may employ. Perhaps the ability to establish a niche drives the eventual outcome of host-microbe interaction to latency. In considering this possibility, it is important to distinguish microbes that require a human niche to survive from those that do not. Herpesviruses are examples of the former. The latter include microbes that can live in the environment or nonhuman hosts. The need for latency and the mechanisms that lead to its occurrence differ for each type of microbe. For microbes that require a human host, the need for latency is paramount, because failure to establish latency risks loss of life. For microbes that can live outside of a human host, there may be no need for latency. Rather, it may be part of a virulence mechanism that provides a niche for survival. Consistent with these relative needs, microbes that require human hosts to survive employ complex molecular mechanisms to ensure their ability to persist or remain (microbiologically) latent without producing viable progeny. On the other hand, microbes with a broader host range may employ different mechanisms, similar to those they might employ in the environment, which ensure their ability to live with limited means, such as an ability to enter and live within host cells. This applies to some fungal microbes that do not require animal hosts to survive, such as C. neoformans and Histoplasma capsulatum (35–38).
Although the mechanism(s) of latency for different microbes vary, there are three general microbial persistence strategies. First, there is persistence in granulomas; this is exhibited by M. tuberculosis and C. neoformans (39–42). For these microbes, one a bacterium and one a fungus, microbial cells survive despite eliciting a robust tissue inflammatory response, because the microbe employs an immune evasion strategy that protects it from elimination by the host. This strategy involves entering macrophages and/or interfering with microbicidal immune responses. Second, there is persistence that co-opts host cells for replication; this is exhibited by herpesviruses and retroviruses (43–47). For these viruses, persistence is part of the viral life cycle in the host. Persistence of Toxoplasma gondii in animal cells also reflects this strategy (48, 49). For such microbes, there is complete dependence on the host for survival, and latency is part of the microbe’s replicative and survival strategies. Third, there is microbial persistence in protected sites whereby immunity does not eradicate microbes; this is exemplified by Zika virus and Ebola virus persistence, although to date, there is no evidence that these microbes exhibit classical latency (see above).
Examples of latency in phylogenetically distant microbes
The series on Latency in Infectious Disease in the JCI features Reviews on five microbes capable of establishing latency: human immunodeficiency virus (HIV) (50); herpes simplex virus (51); Mycobacterium tuberculosis; Toxoplasma gondii (52); and Cryptococcus neoformans (53). In considering the topic of latency in microbial pathogenesis, one can identify similarities among these very different types of microbes, with the caveat that each host-microbe relationship that results in microbial latency is unique with respect to microbial biology, host immunity, and how latency is established and maintained. These five microbes illustrate these distinctions, as each can persist in a host it has entered without causing clinical symptoms, yet the outcome and details of their pathogenic processes are very different. Although aspects of latency with each of these microbes are briefly mentioned above, this section considers their similarities and differences through the lens of the DRF.
Viruses
HIV and herpes simplex (HSV) are viruses that establish lifelong relationships with their hosts, during which the host can experience extended periods without clinical disease. However, these viruses differ in their lifestyles, host locations in which they reside, and mechanisms of virulence and host damage. Both viruses are acquired by infection of cutaneous or mucocutaneous surfaces, including by sexual transmission, although HIV can be acquired by inoculation into the blood (e.g., by injection, and/or historically by transfusion). HIV initially causes a systemic process characterized by viral replication in immune cells that may be marked by a clinical syndrome with fever, adenopathy, rash, and other manifestations. This initial response is followed by a quiescent clinical period that can last for years, and this period has been referred to as latency. If untreated, HIV continues to replicate in host cells at a rate that depends on host immune mechanisms until it results in progressive damage to the immune system, severe immunosuppression characterized by CD4+ T cell depletion, and a cascade of immunological deficits that also affect antibody and innate immunity. Hence, this long asymptomatic period is not a stable state. Instead, it inexorably progresses to disease and is thus different from the latency states described for other microbes, for which infection can be stable for the life of the host. The immune impairment stemming from the ability of HIV to persist and continue to replicate leads to recrudescence or reactivation of classically latent microbes, such as C. neoformans and T. gondii, which the host normally controls with immunological mechanisms that are destroyed by HIV-mediated damage. HSV is like HIV in that it may be acquired by sexual transmission and may reactivate in the setting of immune suppression. However, unlike HIV, which resides in CD4+ T (and potentially other immune) cells, latent HSV resides in neurons, commonly affecting the trigeminal (HSV-1) and sacral (HSV-2) ganglia. During latency, HSV persists in these cells by downregulating host immune molecules to avoid triggering a virus-clearing immune response. Hence, in latency, HSV-mediated damage is limited to infected cells, whereas reactivation leads to viral replication and axonal transport to mucocutaneous sites, where a lytic cellular infection is manifested as blisters, erosions, and painful lesions. Both HIV and HSV are transmissible during latent and disease states.
Bacterium
M. tuberculosis is a bacterium that is acquired by inhalation of aerosolized organisms emitted by a host with disease (tuberculosis). Pulmonary infection, which follows inhalation, is then followed by either clearance, latency, or disease. If the initial host immune response fails to clear the mycobacterial inoculum, it is contained in the lungs by a granulomatous tissue response that can be asymptomatic and lead to a latent state in which the microbe persists in granulomas. The size of the granuloma varies in different individuals and can range from microscopic to visible by chest radiography. The establishment and maintenance of M. tuberculosis latency results in cellular, tissue, and organ damage, with the extent of damage varying depending on the individual host and the nature of the host-microbe interaction, including such variables as inoculum, host genetics, host nutrition, and location of infection in the lung. The persistence of viable organisms in granulomatous lesions presents a future danger to the host from reactivation of mycobacteria growth and progression to disease. Potential mechanisms of reactivation include interference with host defense by drugs that affect the immune response, such as steroids and immunomodulatory biologics; coinfection with agents that affect immunity, such as HIV; and aging, as there is a lifetime risk of M. tuberculosis reactivation, which increases with time. The persistence of inflammation in the lung can also result in malignant transformation and the development of scar carcinomas. M. tuberculosis is not transmissible during latency. Transmission requires a level of bacillary burden that occurs in disease, which is marked by mycobacterial proliferation in lung tissue, tissue destruction, and the formation of infectious aerosols during expectoration and coughing.
Parasite
T. gondii is an apicomplexan parasite that can infect numerous hosts but requires cats to complete its replicative cycle. Human infection is acquired by ingestion of T. gondii–infected animal tissue, which results in gastrointestinal infection followed by widespread dissemination to organs and skeletal muscle. The process of infection and dissemination is often asymptomatic, with the latent state being established as the parasite infects individual cells in a complex cellular biological process that involves the formation of a specific parasitic vacuole in which T. gondii persists until the tissues are ingested by another host. If host immunity is impaired, the parasite can reactivate and replicate in tissue, creating a necrotic lesion. Before the current era of antiretroviral therapy, patients with HIV infection and advanced immunosuppression (AIDS) often developed cerebral toxoplasmosis, which appeared as ring-enhancing lesions in CT scans. Hence, the state of latency of T. gondii is associated with cellular damage to parasite-infected cells. In addition, there is some evidence that high tissue parasitic loads can impair organ function, as suggested by the association of infection with some psychiatric disorders.
Fungus
C. neoformans is a soil fungus that is the causative agent of cryptococcosis, a life-threatening disease that usually presents as meningoencephalitis. Unlike HIV, HSV, M. tuberculosis, and T. gondii, C. neoformans has no need for an animal host in any part of its replicative cycle. As such, its ability to cause human disease appears to represent an instance of accidental virulence (54), whereby traits honed for environmental survival allow it to survive in humans. Despite their large phylogenetic distance from one another, the pathogenesis of C. neoformans has remarkable similarities to that of M. tuberculosis. Both begin with an initial pulmonary infection that either is cleared from the lung or becomes latent in a granuloma (for C. neoformans, a cryptococcoma; for M. tuberculosis, a tuberculoma). Cryptococcomas are sites of granulomatous inflammation that are associated with damage to lung tissue. Like M. tuberculosis, C. neoformans is a facultative intracellular microbe that replicates in macrophages, in which it produces cellular damage. Fungal cells can remain viable in lung tissue for years and reactivate to cause progressive replication and disease years after initial infection. Most individuals with latent C. neoformans infection are entirely asymptomatic. However, as with M. tuberculosis, herpesviruses, and T. gondii, the latent state of C. neoformans can progress to disease when there is a perturbation of host immunity that impairs cellular, humoral, and/or innate immune mechanisms used for microbial containment.
In summary, for each of the microbes discussed, the state of latency is associated with persistent host immune system (cellular and/or tissue) damage that does not affect homeostasis in a way that results in clinical symptoms or disease. Of the five microbial examples, HIV differs from the others in that the quiescent asymptomatic period that is characterized as a period of latency is accompanied by progressive viral replication and inexorable damage to the immune system that inevitably leads to disease and death if untreated. In contrast, the most common outcome of latent HSV, M. tuberculosis, T. gondii, and C. neoformans in immunologically normal individuals is a continued asymptomatic period of clinical latency that endures unless this state of host-microbe interaction is disrupted by factors that impair host immunity or other homeostatic mechanisms. Persons with immune impairment due to HIV or the use of cytotoxic or lymphocytic drugs and/or biologics for malignancy, inflammatory (rheumatological) disease, or solid organ transplantation are at risk for reactivation of M. tuberculosis, T. gondii, and C. neoformans. The common denominator in each of these host-microbe relationships is that the asymptomatic period associated with latency is unstable and fraught with potential for future danger. On the other hand, latency may last for the life of the host. Historically the latent state has received relatively little attention compared with the state of disease. This probably reflects the challenges involved in studying the state of latency, which is usually clinically quiescent, stable, and difficult to study without perturbing. Currently there is a paucity of animal models to investigate latency, and in vitro systems are unlikely to fully recapitulate the in vivo state. Future studies should develop such models with the goal of further defining the host and microbial attributes that contribute to persistence, stability with the potential for instability, and, in some cases, instability that characterize latent microbes. This may reveal new strategies to control the progression from latency to disease and develop vaccines for microbes with the potential for latency. However, accomplishing this will probably require the development of new tools and techniques to study the state of latency without perturbing it. Given the importance of latency in microbial pathogenesis, a renewed focus on this state should be a research priority, for it is likely to provide new actionable insights into the host-microbe relationship that could be used to benefit human health.
Version 1. 08/17/2020
Electronic publication
Version 2. 09/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(9):4525–4531.https://doi.org/10.1172/JCI136221.
Contributor Information
Liise-anne Pirofski, Email: l.pirofski@einsteinmed.org.
Arturo Casadevall, Email: acasadevall@jhu.edu.
References
- 1.Casadevall A, Pirofski LA. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun. 1999;67(8):3703–3713. doi: 10.1128/IAI.67.8.3703-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall A, Pirofski LA. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect Immun. 2000;68(12):6511–6518. doi: 10.1128/IAI.68.12.6511-6518.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A, Pirofski LA. The damage-response framework of microbial pathogenesis. Nat Rev Microbiol. 2003;1(1):17–24. doi: 10.1038/nrmicro732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pirofski LA, Casadevall A. The meaning of microbial exposure, infection, colonisation, and disease in clinical practice. Lancet Infect Dis. 2002;2(10):628–635. doi: 10.1016/S1473-3099(02)00398-5. [DOI] [PubMed] [Google Scholar]
- 5.Godbout EJ, Madaline T, Casadevall A, Bearman G, Pirofski LA. The damage response framework and infection prevention: from concept to bedside. Infect Control Hosp Epidemiol. 2020;41(3):337–341. doi: 10.1017/ice.2019.354. [DOI] [PubMed] [Google Scholar]
- 6.Rose R, et al. Eradication of HIV from tissue reservoirs: challenges for the cure. AIDS Res Hum Retroviruses. 2018;34(1):3–8. doi: 10.1089/aid.2017.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurz S, Steffens HP, Mayer A, Harris JR, Reddehase MJ. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71(4):2980–2987. doi: 10.1128/JVI.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goic B, Saleh MC. Living with the enemy: viral persistent infections from a friendly viewpoint. Curr Opin Microbiol. 2012;15(4):531–537. doi: 10.1016/j.mib.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Radke JR, Cook JL. Human adenovirus infections: update and consideration of mechanisms of viral persistence. Curr Opin Infect Dis. 2018;31(3):251–256. doi: 10.1097/QCO.0000000000000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy H. British nurse makes “full recovery” from reactivated Ebola virus. BMJ. 2015;351:h6133. doi: 10.1136/bmj.h6133. [DOI] [PubMed] [Google Scholar]
- 11.Staples JE, Monath TP. Yellow fever: 100 years of discovery. JAMA. 2008;300(8):960–962. doi: 10.1001/jama.300.8.960. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiura H, et al. The extent of transmission of novel coronavirus in Wuhan, China, 2020. J Clin Med. 2020;9(2):E330. doi: 10.3390/jcm9020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JR, et al. Retinal pigment epithelial cells are a potential reservoir for Ebola virus in the human eye. Transl Vis Sci Technol. 2017;6(4):12. doi: 10.1167/tvst.6.4.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Griensven J, et al. Evaluation of convalescent plasma for Ebola virus disease in Guinea. N Engl J Med. 2016;374(1):33–42. doi: 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkinson B, et al. Presence and persistence of Zika virus RNA in semen, United Kingdom, 2016. Emerging Infect Dis. 2017;23(4):611–615. doi: 10.3201/eid2304.161692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Counotte MJ, et al. Sexual transmission of Zika virus and other flaviviruses: a living systematic review. PLoS Med. 2018;15(7):e1002611. doi: 10.1371/journal.pmed.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher MC, et al. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc Natl Acad Sci U S A. 2001;98(8):4558–4562. doi: 10.1073/pnas.071406098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman DL, Davis J, Bommarito F, Shao X, Casadevall A. Enhanced allergic inflammation and airway responsiveness in rats with chronic Cryptococcus neoformans infection: potential role for fungal pulmonary infection in the pathogenesis of asthma. J Infect Dis. 2006;193(8):1178–1186. doi: 10.1086/501363. [DOI] [PubMed] [Google Scholar]
- 20.Goldman DL, Li X, Tsirilakis K, Andrade C, Casadevall A, Vicencio AG. Increased chitinase expression and fungal-specific antibodies in the bronchoalveolar lavage fluid of asthmatic children. Clin Exp Allergy. 2012;42(4):523–530. doi: 10.1111/j.1365-2222.2011.03886.x. [DOI] [PubMed] [Google Scholar]
- 21.Speck SH, Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8(1):100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira DBL, et al. Persistence and intra-host genetic evolution of Zika virus infection in symptomatic adults: a special view in the male reproductive system. Viruses. 2018;10(11):E615. doi: 10.3390/v10110615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biava M, et al. Persistence of ZIKV-RNA in the cellular fraction of semen is accompanied by a surrogate-marker of viral replication. Diagnostic implications for sexual transmission. New Microbiol. 2018;41(1):30–33. [PubMed] [Google Scholar]
- 24.Musso D, et al. Detection of Zika virus RNA in semen of asymptomatic blood donors. Clin Microbiol Infect. 2017;23(12):1001.e1–1001.e3. doi: 10.1016/j.cmi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Bujalance S, et al. Persistence and infectivity of Zika virus in semen after returning from endemic areas: report of 5 cases. J Clin Virol. 2017;96:110–115. doi: 10.1016/j.jcv.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Whitmer SLM, et al. Active Ebola virus replication and heterogeneous evolutionary rates in EVD survivors. Cell Rep. 2018;22(5):1159–1168. doi: 10.1016/j.celrep.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velásquez GE, Aibana O, Ling EJ, Diakite I, Mooring EQ, Murray MB. Time from infection to disease and infectiousness for Ebola virus disease, a systematic review. Clin Infect Dis. 2015;61(7):1135–1140. doi: 10.1093/cid/civ531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, et al. Cost-effectiveness of increasing access to contraception during the Zika virus outbreak, Puerto Rico, 2016. Emerging Infect Dis. 2017;23(1):74–82. doi: 10.3201/eid2301.161322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinchoff J, et al. Evidence-based process for prioritizing positive behaviors for promotion: Zika prevention in Latin America and the Caribbean and applicability to future health emergency responses. Glob Health Sci Pract. 2019;7(3):404–417. doi: 10.9745/GHSP-D-19-00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarzmann F, Jäger M, Prang N, Wolf H. The control of lytic replication of Epstein-Barr virus in B lymphocytes (Review) Int J Mol Med. 1998;1(1):137–142. doi: 10.3892/ijmm.1.1.137. [DOI] [PubMed] [Google Scholar]
- 31.Tsao SW, Tsang CM, To KF, Lo KW. The role of Epstein-Barr virus in epithelial malignancies. J Pathol. 2015;235(2):323–333. doi: 10.1002/path.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samimi M, Touzé A. Merkel cell carcinoma: the first human cancer shown to be associated with a polyomavirus. Presse Med. 2014;43(12 pt 2):e405–e411. doi: 10.1016/j.lpm.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins N, Irusen EM, Koegelenberg CF. Pulmonary scar carcinoma in South Africa. S Afr Med J. 2017;107(4):320–322. doi: 10.7196/SAMJ.2017.v107i4.12158. [DOI] [PubMed] [Google Scholar]
- 34.Casadevall A, Pirofski LA. What is a host? Attributes of individual susceptibility. Infect Immun. 2018;86(2):e00636-17. doi: 10.1128/IAI.00636-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaragoza O. Basic principles of the virulence of Cryptococcus. Virulence. 2019;10(1):490–501. doi: 10.1080/21505594.2019.1614383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kronstad J, et al. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryotic Cell. 2012;11(2):109–118. doi: 10.1128/EC.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woods JP. Histoplasma capsulatum molecular genetics, pathogenesis, and responsiveness to its environment. Fungal Genet Biol. 2002;35(2):81–97. doi: 10.1006/fgbi.2001.1311. [DOI] [PubMed] [Google Scholar]
- 38.Nemecek JC, Wüthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312(5773):583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 39.Pieters J. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe. 2008;3(6):399–407. doi: 10.1016/j.chom.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Goldman DL, Lee SC, Mednick AJ, Montella L, Casadevall A. Persistent Cryptococcus neoformans pulmonary infection in the rat is associated with intracellular parasitism, decreased inducible nitric oxide synthase expression, and altered antibody responsiveness to cryptococcal polysaccharide. Infect Immun. 2000;68(2):832–838. doi: 10.1128/IAI.68.2.832-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salgame P. Host innate and Th1 responses and the bacterial factors that control Mycobacterium tuberculosis infection. Curr Opin Immunol. 2005;17(4):374–380. doi: 10.1016/j.coi.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Brunet K, Alanio A, Lortholary O, Rammaert B. Reactivation of dormant/latent fungal infection. J Infect. 2018;77(6):463–468. doi: 10.1016/j.jinf.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Guan X, Zhang M, Fu M, Luo S, Hu Q. Herpes simplex virus type 2 immediate early protein ICP27 inhibits IFN-β production in mucosal epithelial cells by antagonizing IRF3 activation. Front Immunol. 2019;10:290. doi: 10.3389/fimmu.2019.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grey F. Role of microRNAs in herpesvirus latency and persistence. J Gen Virol. 2015;96(pt 4):739–751. doi: 10.1099/vir.0.070862-0. [DOI] [PubMed] [Google Scholar]
- 45.Buehler J, et al. Opposing regulation of the EGF receptor: a molecular switch controlling cytomegalovirus latency and replication. PLoS Pathog. 2016;12(5):e1005655. doi: 10.1371/journal.ppat.1005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mbonye U, Karn J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr HIV Res. 2011;9(8):554–567. doi: 10.2174/157016211798998736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, et al. Two cellular microRNAs, miR-196b and miR-1290, contribute to HIV-1 latency. Virology. 2015;486:228–238. doi: 10.1016/j.virol.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 48.Holmes MJ, Augusto LDS, Zhang M, Wek RC, Sullivan WJ. Translational control in the latency of apicomplexan parasites. Trends Parasitol. 2017;33(12):947–960. doi: 10.1016/j.pt.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menard KL, Haskins BE, Denkers EY. Impact of Toxoplasma gondii infection on host non-coding RNA responses. Front Cell Infect Microbiol. 2019;9:132. doi: 10.3389/fcimb.2019.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dufour C, Gantner P, Fromentin R, Chomont N. The multifaceted nature of HIV latency. J Clin Invest. 2020;130(7):3381–3390. doi: 10.1172/JCI136227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen JI. Herpesvirus latency. J Clin Invest. 2020;130(7):3361–3369. doi: 10.1172/JCI136225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao XY, Ewald SE. The molecular biology and immune control of chronic Toxoplasma gondii infection. J Clin Invest. 2020;130(7):3370–3380. doi: 10.1172/JCI136226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alanio A. Dormancy in Cryptococcus neoformans: 60 years of accumulating evidence. J Clin Invest. 2020;130(7):3353–3360. doi: 10.1172/JCI136223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, Martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryotic Cell. 2007;6(12):2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herzig E, et al. Attacking latent HIV with convertible CAR-T cells, a highly adaptable killing platform. Cell. 2019;179(4):880–894.e10. doi: 10.1016/j.cell.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]