Abstract
The emerging and re-emerging viral infections are constant threats to human health and wellbeing. Several strategies have been explored to develop vaccines against these viral diseases. The main effort in the journey of development of vaccines is to neutralize the fusion protein using antibodies. However, significant efforts have been made in discovering peptides and small molecules that inhibit the fusion between virus and host cell, thereby inhibiting the entry of viruses. This class of inhibitors is called entry inhibitors, and they are extremely efficient in reducing viral infection as the entry of the virus is considered as the first step of infection. Nevertheless, these inhibitors are highly selective for a particular virus as antibody-based vaccines. The recent COVID-19 pandemic lets us ponder to shift our attention towards broad-spectrum antiviral agents from the so-called ‘one bug-one drug’ approach. This review discusses peptide and small molecule-based entry inhibitors against class I, II, and III viruses and sheds light on broad-spectrum antiviral agents.
Graphic Abstract
Keywords: Entry inhibitor, Viral infection, Membrane fusion
Viral Infections
In the current scenario, emerging and re-emerging viral infections are the major pandemic threats to humankind. The rate of replication of virion and the rate of transmission play crucial roles for any viral infections. Most of the viruses are zoonotic in nature, which transmit from animals to humans. HIV, filoviruses (such as Ebola and Marburg), henipaviruses (such as Hendra and Nipah), coronaviruses (such as severe acute respiratory syndrome (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-CoV), COVID-19), and influenza are some popular examples of zoonoses. In addition, some virus families like flaviviridae (such as Dengue virus (DENV), bunyaviridae (Hanta virus), and arenaviridae (Junin and Lassa virus) are animal or arthropod-borne, and also cause deadliest viral diseases (Vigant et al. 2015). A summary of some viral epidemic is shown in Table 1.
Table 1.
Different viral diseases and their impact on humankind
| Viral disease | People infected | People died | Year(s) | References |
|---|---|---|---|---|
| COVID-19 | 20 million | > 5,00,000 | 2019–Present | Johns Hopkins University database |
| MERS | 2494 | 858 | 2012–2019 | WHO website |
| Dengue | 100 million | 38,000 | 2016 | Bloom and Cadarette (2019) |
|
Influenza (Swine flu) |
700 million to 1.4 billion | 575,000 | 2009–2010 | Saunders-Hastings and Krewski (2016) |
| SARS | 8098 | 774 | 2002–2003 | Olsen et al. (2003) |
| HIV/AIDS | 75 million | 32 million | 1960–2008 | WHO website |
| Small Pox | 130,000 | 26,000 | 1974 | Bloom and Cadarette (2019) |
|
Influenza (Hong Kong flu) |
> 500 million | 500,000–2 million | 1968–1970 | Saunders-Hastings and Krewski (2016) |
|
Influenza (Asian flu) |
> 500 million | 1–2 million | 1957–1958 | Saunders-Hastings and Krewski (2016) |
|
Influenza (Spanish flu) |
500 million | 50 million | 1918–1919 | Taubenberger and Morens (2006) |
Initiatives have been taken to develop antiviral agents to intervene viral infection, and most of the strategies are based on the ‘one bug-one drug’ approach. However, this strategy is not adequate for responding to an ever-increasing number of emerging and re-emerging disease-causing viruses. In addition, there are many mammalian viruses present in the wildlife reservoir which remain to be discovered (Vigant et al. 2015). Again, the tendency of drug resistance is a leading parameter to cause failure of the above approach.
This review discusses the efficient ways to block the entry of viruses, thereby providing protection against viral infection. Though the morphology, genome structure, and life cycle of each virus is different, most of the viruses follow a common pathway for their entry into the human cells. Therefore, targeting those conserved features could be a successful strategy to defy the fusion between virus and host cell membranes.
Viral Entry into Host Cells
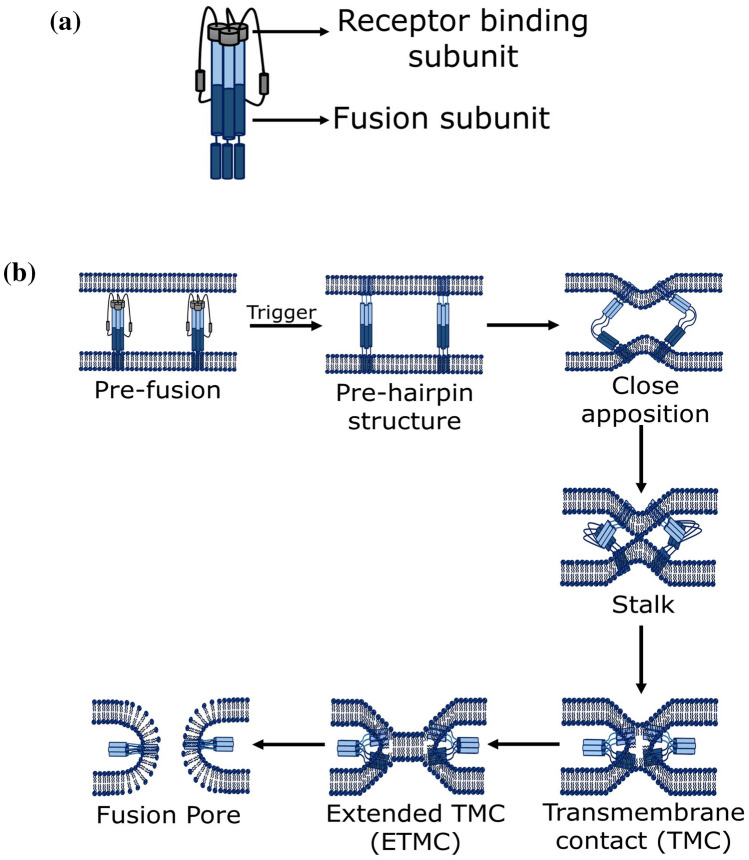
A majority of viral pathogens that cause emerging and re-emerging infectious diseases are membrane-enveloped viruses. Enveloped viruses are characterized by a lipid bilayer, known as envelope, covering the virus particle i.e. virion. The virion consists of an outer protein coat called capsid which surrounds the inner core that comprises nucleic acids (either RNA or DNA). One or more glycoproteins are generally decorated on the viral envelope. A general structure of an enveloped virus is shown in Fig. 1.
Fig. 1.
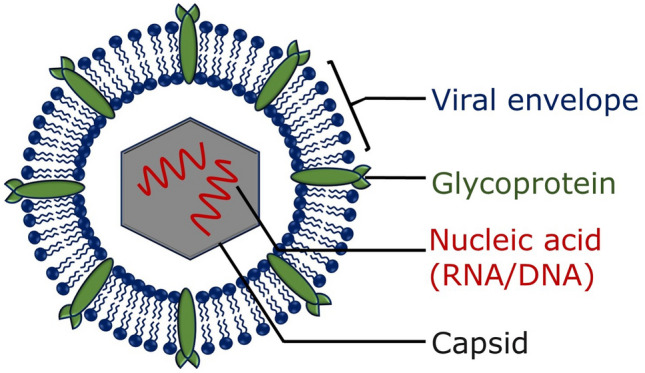
Schematic representation of an enveloped virus
Viruses are obligate intracellular parasites, which carry out their replication within the host cell, and cannot multiply outside a living cell. Once the virus enters into a host cell, it competes with the immune response of the target and disrupts a variety of cellular proteins and chemical pathways to survive and replicate (Mazzon and Marsh 2019). Therefore, delivery of the viral genome inside the host cell is the earliest stage for any viral infection. At this stage, enveloped viruses first bind to specific surface receptors of the target cell membrane non-covalently, followed by fusion between virus and host cell. This fusion step may occur at the cell surface or after internalization of the virus particle by endocytosis. In either case, fusion is catalysed by one type of viral glycoproteins (in case of influenza viruses, retroviruses) or combinations of multiple viral glycoproteins (in case of herpesviruses, paramyxoviruses). Viral glycoproteins (fusogens), when activated by non-covalent interactions with surface receptors and/or by acidic endosomal pH, result in large-scale conformational change which drives the fusion process (Mas and Melero 2013; Melikyan 2008; White et al. 2008). In essence, virus-cell fusion is, therefore, the step at which the virus particle loses its individuality. HIV and herpes simplex virus fuse at the cell membrane, whereas influenza, polio, hepatitis C, and foot-and-mouth disease viruses enter into the cell via endocytosis followed by endosomal fusion.
The structures of pre-fusion and post-fusion conformations of many viral fusogens have been studied using x-ray crystallography and cryo-electron microscopy. It has been believed that the post-fusion conformation is more stable compared to the pre-fusion conformation (Mas and Melero 2013; White et al. 2008). Moreover, there is a high kinetic barrier between the pre- and post-fusion conformation. Upon protonation (at low endosomal pH) or binding to the receptor and/or co-receptor leads to conformational change of the fusogen, and the energy liberated during this transition successfully drives the fusion process (Harrison 2015; Mas and Melero 2013). The free energy profile for membrane fusion process considering the lipid-stalk model is shown in Fig. 2.
Fig. 2.
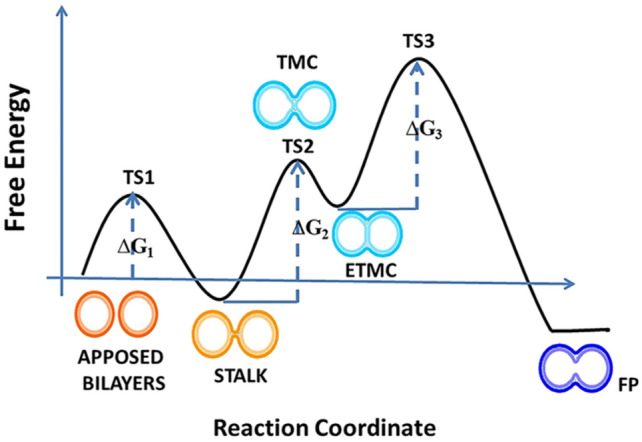
Schematic representation of the fusion reaction profile considering the lipid-stalk model of membrane fusion. The figure has been
adapted from reference Meher et al. (2019b) with permission
The sequence of viral glycoprotein (fusion protein) differs from virus to virus; however, it has been observed that all viral fusion proteins share some common structural characteristics. Usually, upon activation, all viral fusion proteins undergo conformational rearrangement to assume pre-hairpin structure. The lifespan of the pre-hairpin structure varies from seconds to minutes depending upon the virus (Floyd et al. 2008; Munoz-Barroso et al. 1998). Interestingly, the conformational rearrangement of the fusion protein exposes the hydrophobic segment, known as fusion peptide, to the membrane surface (Mas and Melero 2013; Pattnaik et al. 2018). The exposed hydrophobic fusion peptide is ultimately bound to the target membrane, whereas transmembrane domain (TMD) region of the fusion protein remains rooted in viral membranes. Thus, bridging of viral and target membrane occurs by two different portions of fusion protein, followed by the collapsing of the pre-hairpin bridge that results into close apposition of the two lipid bilayers. The close apposition of two lipid bilayers causes merging of the outer leaflets of each bilayer to form the semi-stable stalk intermediate (Pattnaik et al. 2018; Yang and Huang 2003). The stalk follows the formation of a transmembrane contact (TMC), and extended transmembrane contact (ETMC). Finally the ETMC opens to form a fusion pore leaving a highly stable post-fusion hairpin conformation inserted into the target membrane (Lee and Lentz 1997; Pattnaik et al. 2018; Siegel 1999). Figure 3 shows the schematic representation of a fusion process.
Fig. 3.
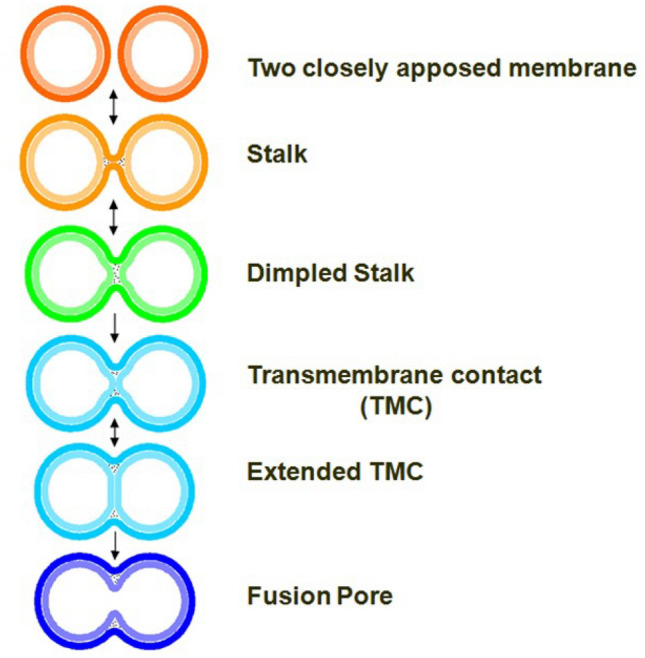
Schematic representation of stepwise fusion process. The figure has been
adapted from reference Pattnaik et al. (2018) with permission
There may be one or more viral fusion protein interacting with the target membrane to form the pre-hairpin intermediate as the bending energy depends on the composition of the membrane (Pattnaik et al. 2018; Tristram-Nagle and Nagle 2007). The requirement of number of trimeric proteins to induce fusion differs from virus to virus (Harrison 2015; Kielian and Rey 2006).
Viral Fusion Proteins
Generally, viral fusion proteins are classified into three different classes, class I, II, and III. Despite little diversity in structural features, they all form a trimeric hairpin which was confirmed by their high-resolution structures (Earp et al. 2005; Harrison 2005; Kielian and Rey 2006; Lamb and Jardetzky 2007; Weissenhorn et al. 2007; White et al. 2008). Structurally all the fusion proteins contain three C-terminal regions that wrap the central N-terminal trimeric core (White et al. 2008).
Class I Fusion Protein
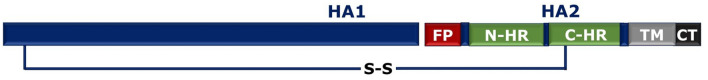
Class I fusion proteins have been shown to maintain homology within a particular family; however, there is no sequence homology across families of class I fusion proteins. However, all class I fusion proteins are trimers in pre-fusion as well as post-fusion conformations. They are being characterized by a single chain precursor which becomes fusion active by proteolytic cleavage. The cleavage results into two fragments, N-terminal fragment which is generally the receptor-binding domain (e.g. the HA1 fragment of influenza or the gp120 fragment of HIV-1) and the C-terminal fusion facilitating fragment (e.g. HA2 fragment of influenza or the gp41 fragment of HIV-1). The central N-terminal is trimeric α-helical coiled coil adorned by three C-terminal helices easing formation of six-helix bundle (6HB) (White et al. 2008). Formation of this 6HB provides the energy required to overcome coulombic repulsion between two apposing membranes (Mas and Melero 2013). The general pathway of membrane fusion by class 1 fusion protein is shown in Fig. 4. Generally, the fusion peptide locates at the N-terminal of the class 1 fusion protein, However, in influenza HA and paramyxovirus F protein its location differs (Pattnaik et al. 2018; White et al. 2008). A schematic representation of class I fusion protein from N to C-terminal is shown in Fig. 5. The group of enveloped viruses carrying class I fusion proteins includes retroviridae family (Human immunodeficiency virus HIV), orthomyxoviradae virus family (Influenza A, B, C, and D), coronaviridae family (SARS), filoviridae family (Ebola), and paramyxoviridae family (Sendai virus) (Rey and Lok 2018). Different class I fusion protein, location of fusion peptide, and their fusion subunits are shown in Table 2 (White et al. 2008).
Fig. 4.
Schematic representation of a the fusion protein, b membrane fusion steps followed by class 1 fusion protein
Fig. 5.
Schematic representation of a class I fusion protein. Here FP represents fusion peptide, NHR and CHR represent N- and C-heptad regions, respectively, and TMD represents transmembrane domain
Table 2.
Different class I fusion protein
| Class I virus family | Viruses | Fusion protein | Fusion subunit | Fusion peptide location | Fusion pH |
|---|---|---|---|---|---|
| Retroviridae | Human immunodeficiency virus (HIV) | gp160 | gp41 | N-terminal (mostly) | Neutral/low |
| Orthomyxoviridae | Influenza virus | HA | HA2 | N-terminal | Low |
| Coronaviradae | Severe acute respiratory syndrome (SARS) virus | S | S2 | Internal | Neutral/low |
| Filoviridae | Ebola virus | GP | GP2 | Internal | Low |
| Paramyxoviridae | Sendai virus | F | F1 | N-terminal | Neutral |
| Arenaviridae | Lassa virus | GP, SSP | GP2 | N-terminal | Low |
Entry of HIV
The human immunodeficiency virus (HIV) expresses an envelope glycoprotein gp160 which mediates the entry of HIV. The schematic representation of HIV envelope protein is shown in Fig. 6. Initially, gp160 undergoes receptor-mediated proteolytic cleavage into two subunits viz., gp120 (the surface protein) and gp41 (the transmembrane protein) (Meher et al. 2019b; Skehel and Wiley 2000; White et al. 2008). The heterodimer, gp120 and gp41 organize on the viral membrane as a trimer of three gp41 subunits and three gp120 subunit through non-covalent interaction (Berkhout et al. 2012). First, the surface subunit gp120 binds to the CD4 receptor and undergoes a substantial conformational change leading to higher affinity towards the chemokine co-receptor CCR5 or CXCR4. This induces the activation of a spring-loaded conformational change of gp41. This conformational change leads to the pre-hairpin conformation where the protein gp41 holds both the viral membrane and host cell membrane together through transmembrane domain (TMD) and hydrophobic fusion peptide, respectively (Fig. 4b). In addition to the TMD and fusion peptide, gp41 contains two heptad regions NHR and CHR and their interaction proceeds to the formation of six-helix bundle (6HB) which brings the apposed membrane into closer contact (Fig. 4b) (Chan and Kim 1998). The change in free energy during the formation of 6HB facilitates the formation of fusion pore and hence HIV entry is possible (Berkhout et al. 2012).
Fig. 6.
Schematic representation of HIV envelope protein. Here, FP represents fusion peptide, NHR and CHR represent N- and C-heptad regions, respectively, and TM represents transmembrane domain
Entry of Influenza Virus
The major glycoprotein on the influenza envelope virus is Hemagglutinin (HA) and it is synthesized as a precursor polypeptide, HA0. The schematic representation of influenza virus hemagglutinin is shown in Fig. 7. HA0 is activated upon proteolytic cleavage into two subunits, HA1 (the receptor-binding subunit) and HA2 (the fusion subunit), linked by a disulphide bond (Meher and Chakraborty 2017; Prabhu et al. 2009; Stevens et al. 2006). Sialic acid present on the glycolipids or glycoproteins of the host cell acts as the receptor for HA1 binding, followed by the internalization of the virus through endocytosis (Bullough et al. 1994; Kadam et al. 2017). The acidic endosomal environment (pH ~ 5.3) promotes protonation of HA1 subunit that triggers the electrostatic repulsion between HA1 and HA2 resulting in the separation of HA1 from HA2. This detachment of HA1 from HA2 promotes drastic conformational change of HA2 subunit followed by relocation of fusion peptide over 100 Å from its original location (Bullough et al. 1994; Carr and Kim 1993). This major conformational change exposes the fusion peptide (20–25 amino acids in the N-terminal of the HA2 protein), which binds to the target membrane (Fig. 4b) (Gething et al. 1986; Meher and Chakraborty 2017). The further conformational change of HA2 brings the transmembrane domain close to the fusion peptide resulting hairpin structure (Fig. 4b) following the further step of viral entry.
Fig. 7.
Schematic representation of influenza virus hemagglutinin. Here FP represents fusion peptide, NHR and CHR represent N- and C-heptad regions, respectively, and TM represents transmembrane domain
Paramyxovirus Entry
Paramyxoviruses cause critical human diseases, ranging from lower respiratory tract diseases in infants caused by human parainfluenza virus type 1 (HPIV1), type 2 (HPIV2), and type 3 (HPIV3) to highly lethal central nervous system diseases caused by the emerging paramyxoviruses such as Hendra virus and Nipah virus (Collins et al. 1996; Porotto et al. 2009; Williams et al. 2004).
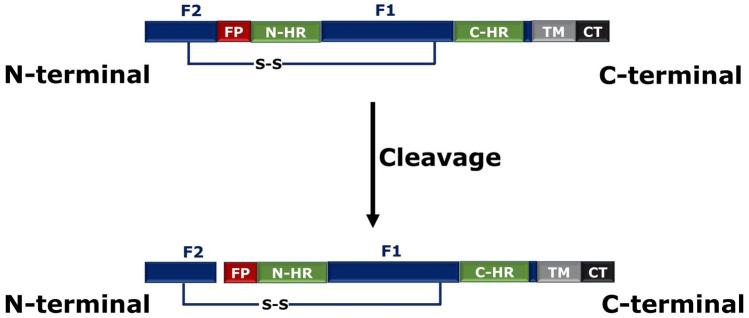
The fusion protein for Paramyxovirus entry is known as ‘F’ protein. The function of F protein is quite different from other class 1 fusion proteins. There are some other receptor-binding proteins such as HN in HPIV3 and simian virus 5. The fusion protein F is produced as an uncleaved form, F0, after cleavage F2 (N-terminal) and F1 (C-terminal) are produced which are linked by a disulphide bond previously. During this process, the fusion peptide (N-terminal segment) that was located in the internal region of F1 domain gets exposed (Dutch et al. 2000; Mas and Melero 2013). F1 is equivalent to HA2 chain of influenza virus. In addition to fusion peptide, F1 has two heptad regions (NHR and CHR) in its ectodomain. The location of NHR (or HRA) is adjacent to the fusion peptide, whereas CHR (or HRB) is proximal to the transmembrane region (C-terminus of F1). Upon binding to the receptor, F protein is activated followed by a series of conformational changes leading to separation of CHR coil and refolding of NHR coil to form an elongated trimeric coiled coil (Mas and Melero 2013). The fusion peptide, now at the N-terminal of the F1 segment, is inserted into the target membrane to form the pre-hairpin intermediate (Fig. 4b). This is followed by elongation of fusion peptide and some conformational changes. During this process, the CHR gets inverted and form six-helix bundle with NHR and promotes fusion between viral and host cell (Dutch 2010). The schematic representation of F protein and its cleavage-induced activation is shown in Fig. 8.
Fig. 8.
Schematic representation of F protein cleavage. Here FP represents fusion peptide, NHR and CHR represent N- and C-heptad regions, S–S represents the disulphide bond and TM represents transmembrane domain
SARS-CoV Entry
The coronavirus spike protein (S) plays a crucial role for severe acute respiratory syndrome coronavirus (SARS-CoV) entry into the target cell. The S protein undergoes proteolytic cleavage to produce S1 and S2 subunits associated by non-covalent bond. However, this proteolytic cleavage is not an absolute requirement for inducing membrane fusion (Masters 2006). The interaction between S1 spike protein and the cell surface receptor protein, angiotensin-converting enzyme 2 (ACE2) is the primary step for the membrane fusion (Masters 2006; Taguchi and Shimazaki 2000). The receptor binding induces conformational changes in S2 subunit leading to exposure of N-terminal fusion peptide from the interior of S2 subunit, and positions it to interact with the host cell membrane (Cavanagh and Davis 1986; Tripet et al. 2004). The S2 protein contains fusion peptide (FP) at N-terminal region, internal fusion peptide (IFP), and pre-transmembrane peptide (PTM). In addition to these, it comprises highly conserved two heptad regions, NHR (or HR1) and CHR (or HR2), and transmembrane domain at the C-terminus (Bosch et al. 2004). Atomic resolution structure of HR1 and HR2 complex showed the formation of a 6HB, which drives the fusion process (Deng et al. 2006; Hakansson-McReynolds et al. 2006; Meher et al. 2019a; Supekar et al. 2004; Xu et al. 2004). The schematic representation of S protein and its S1/S2 cleavage site is shown in Fig. 9.
Fig. 9.
Schematic representation of S protein. Here FP represents fusion peptide, NHR and CHR represent N- and C-heptad regions, SP and TM represent signal peptide and transmembrane domain, respectively
Class II Viral Fusion Proteins
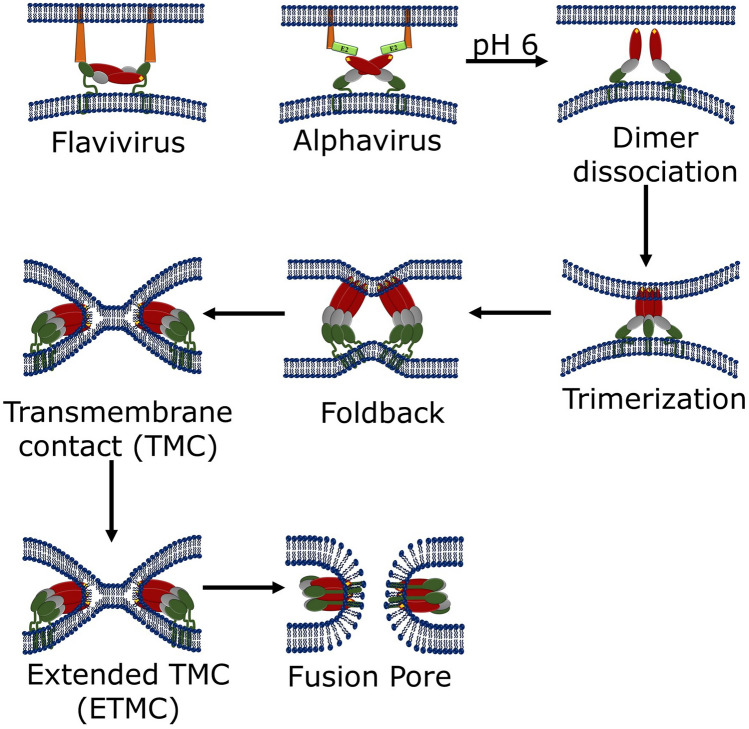
Generally, class II fusion proteins do not maintain sequence homology, but their secondary and tertiary structures are conserved. Interestingly, the structures of the class II fusion proteins are different from class I fusion proteins (Kielian 2006; Lescar et al. 2001). Basically, the class II fusion proteins are extended finger like molecules containing three globular domains having mostly β-sheets structures (Fig. 10). The dimeric pre-fusion structures of class II fusion proteins form homotrimers in the post-fusion condition (Kielian 2006). The domain I of class II viral fusion protein is a β-barrel, which contains the N-terminus. The extension of two adjacent β-strands of domain I results in long finger-like structure called domain II. At the tip of domain II, a highly conserved hydrophobic fusion loop (represented as yellow diamond in Fig. 11) is present (Kielian 2006; Mas and Melero 2013). In between domain I and domain II, there exists a hinge region, which is flexible and allows different angles of flexibility between two domains (Kielian 2006; Mukhopadhyay et al. 2005). The domain III participates in the cell surface receptor recognition. Domain III is terminated in a segment called “stem” and this stem connects domain III with transmembrane anchor (Harrison 2015). Different class II fusion proteins and location of fusion peptide and their fusogenic subunits are shown in Table 3 (Harrison 2015; White et al. 2008).
Fig. 10.
Schematic representation of stepwise fusion process by class II fusion protein
Fig. 11.
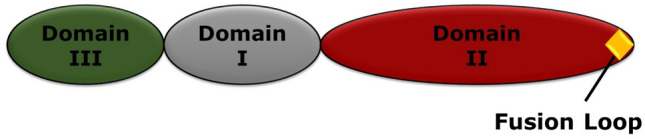
Schematic representation of class II fusion protein. Here, grey, red, and green colours represent domains I, II, and III respectively. The fusion loop is shown by yellow diamond (Color figure online)
Table 3.
Different class II fusion proteins
| Class II virus family | Virus | Fusion protein | Fusion subunit | Fusion peptide location | Fusion pH |
|---|---|---|---|---|---|
| Flaviviridae | Dengue virus | E | E | Internal | Low |
| Alphaviridae | Semliki Forest Virus | E1 | E1 | Internal | Low |
| Bunyaviridae | Hantavirus |
GN/GC (GN/GC) |
GC | Internal | Low |
The class II viral fusion protein (such as E of flaviviruses and E1 for alphaviruses) forms 1:1 dimer with another chaperone protein (prM and pE2 for flaviviruses and alphaviruses, respectively). The initial step of the fusion process is the cleavage of the dimer by the signal proteases and furin in the trans-Golgi network (TGN). The monomeric fusion protein undergoes pH-induced conformational change in endosome. This results into exposure of fusion loop towards the target membrane for bridging the target and viral membranes (Guirakhoo et al. 1991; Harrison 2015; Lobigs and Garoff 1990). The structure of post-fusion homotrimer reveals that the fusion loops insert only marginally into the target membrane (Modis 2013). Few examples of class II viral fusion have been described in the subsequent sections.
Flavivirus Entry
The fusion protein for flavivirus entry is known as E protein, and is synthesized and folds co-translationally with another regulatory chaperone protein, prM (Kielian 2006). The fusion protein E binds the receptor leading to clathrin-mediated endocytosis. The acidic endosomal environment (pH 6) triggers the conformational change facilitating exposure of the fusion loop at the tip of domain II (Mancini et al. 2000; Modis 2013; Zhang et al. 2003). The domains I and II bend 30° with respect to each other resulting swing away of fusion loop from viral surface towards the target membrane. This domain movement releases the constraints executed by the close-packing of E on the viral membrane and hence allowing free diffusion of fusion protein E. Flaviviral E inserts as monomer to the target membrane and this membrane insertion promotes lateral reshuffling of E monomers to form trimer (Modis 2013). This is comparable with the pre-hairpin intermediate of class I fusion protein-mediated fusion process where the two different regions of each E protein are inserted into the two membranes to be fused. There is a flexible hinge region between domain I and domain II and at this stage there is rotation of 33° of Domain III with respect to Domain I, bringing the C-terminal stem region and fusion loop peptide (inserted into the target membrane) together. This refolding brings the two membranes together and thus drives fusion (White et al. 2008).
Alphavirus Entry
The fusion protein for alphavirus entry is known as E1 protein. It is being produced and folds co-translationally with another regulatory chaperone protein, p62 (Kielian 2006). The chaperone binds the receptor followed by internalization of the virus to the endosome. Upon acidic endosomal environment, the dissociation of the protector protein E2 occurs resulting in the release of constrains and exposure of the fusion loops. At this stage there is 15-degree hinge movement of domain II relative to domain I resulting swing towards nearest three-fold symmetry axis in virus causing trimerization with adjacent E1. Alphaviral E1 inserts as trimer to the target membrane (Modis 2013). The E1 protein undergoes similar conformational rearrangement as flavivirus E protein, leading to fusion of viral and endosomal membranes (Mas and Melero 2013).
Class III Fusion Proteins
Class III fusion proteins have structural features similar to both class I and class II fusion proteins. In pre-fusion state they are trimeric, and mostly contain α-helical coiled-coil structure like class I fusion proteins. In addition, their fusion domains are similar to class II fusion proteins, the fusion loops are found at the tip of extended β-strands. Unlike class I fusion proteins, class III fusion proteins do not require proteolytic processing of either a protein precursor (as in class I) or a regulatory protein (as in class II) to become fusion active, rather they are expressed from individual m-RNA (Mas and Melero 2013; White et al. 2008). Class III fusion proteins are identified in vesicular stomatitis virus (VSV) glycoprotein G, herpes simplex virus glycoprotein B (gB), and baculoviruses. The pre-fusion and post-fusion structures of VSV-G have been studied but only the post-fusion structure of other two viruses are reported so far (Rey and Lok 2018). The schematic representation of VSV-G protein is shown in Fig. 12.
Fig. 12.
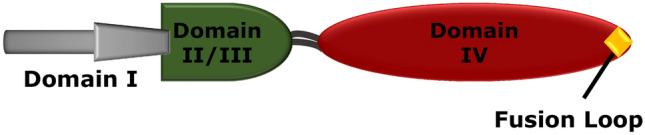
Schematic representation of VSV glycoprotein. The fusion loop is shown by a diamond on Domain IV
Vesicular Stomatitis Virus Entry
The structures of G protein of VSV in high-pH (pre-fusion) and low-pH (post-fusion) conformations indicate the structural and functional similarity with influenza virus hemagglutinin and/or flavivirus E (Harrison 2008). Upon acidification, a number of conformational changes occur in the fusion protein leading to the exposure of the fusion peptide to the viral membrane. This is followed by the bending of the C-terminus to the N-terminus, forming the six-helix bundle (6HB), and thereby bringing both membranes to the close apposition. The low pH-induced conformational changes are reversible in case of VSV-G. There is thermodynamic equilibrium between pre-fusion (high pH) and post-fusion (low pH) states, and this equilibrium shifts towards the post-fusion state at low pH (White et al. 2008). There is rotation of domain I and domain II with respect to domain IV and a little reshuffling of domain III occurs during transition from pre-fusion to post-fusion conformation. This transition is very similar to HA2 refolding as in case of influenza. The rotation of domain I and domain II, and folding of C-terminal can be correlated to the central coiled-coil extension in case of influenza (Harrison 2015).
Class I Viral Entry Inhibitors
Peptide-Based Inhibitors
Generally, the peptide-based inhibitors prevent the formation of six-helix bundle (6HB), which is a trademark for the class I viral fusion. Therefore, most of the peptide inhibitors have been designed by mimicking the sequence of NHR and CHR, so that the inhibitor molecule interacts with the complementary helix, and blocks the 6HB formation. The discovery of Enfuvirtide (T20) to inhibit entry of HIV gathers the momentum for the development of peptide-based fusion inhibitors. As a result, several peptide-based fusion inhibitors have been developed in last couple of decades with improved stability as well as better pharmacokinetics to block viral entry.
Inhibiting HIV Entry
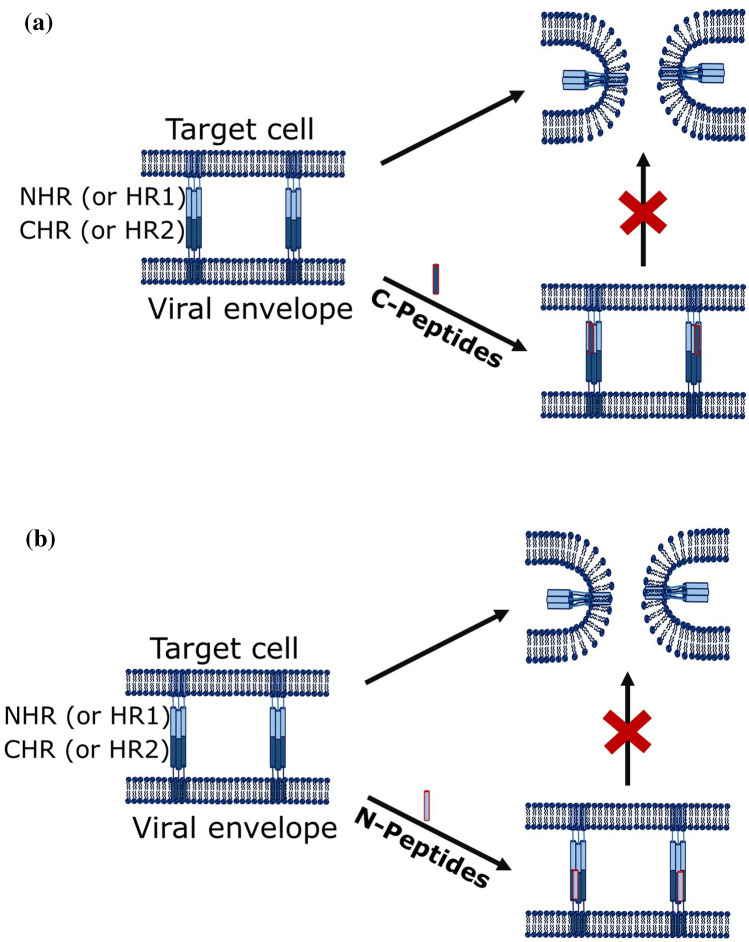
Peptide-based fusion inhibitors for HIV are classified into two categories viz., C-peptides and N-peptides. C-peptides mimics the C-terminal heptad region (or HR2) sequence to target the NHR (or HR1) of gp41, whereas N-peptides are developed from N-terminal heptad region to target the CHR region of gp41 (Fig. 13) to block the formation of 6HB. Both the C- and N-peptides have potential to inhibit HIV entry, but poor solubility of N-peptides (as they are hydrophobic in nature) makes them less potent as inhibitors (Baldwin et al. 2003; Lu et al. 1995; Root et al. 2001). T20, T1249, Cp32M, Sifuvirtide, and T2635 are some examples of C-peptides and IQN17, IQN23, and NCCG-gp41 are some examples of N-peptides.
Fig. 13.
Schematic representation of mechanism of inhibition by a C-peptides and b N-peptides
T20 (also known as Enfuvirtide or Fuzen) is the first peptide-based fusion inhibitor developed for inhibiting HIV entry. It is a 36-amino acid residue peptide derived from α-helical carboxy terminal heptad domain (CHR) of gp41. It was designed to target the complimentary NHR, thereby, preventing 6HB formation and stops the progress of the subsequent steps of fusion (Wild et al. 1994a, b). The poor bioavailability and lower pharmacokinetic efficacy (plasma half-life of about 4 h) lead to requirement of higher doses of T20 (Berkhout et al. 2012; Kilby et al. 1998, 2002; Lalezari et al. 2003; Stocker et al. 2006; Zhang et al. 2002). T1249 is another peptide, designed based on CHR sequence of gp41 that inhibits HIV entry. T1249 is a 39-amino acid long peptide, which is designed by extending the N-terminal of T20. Interestingly, T1249 has better efficacy than T20 and is sensitive against T20-resistant HIV-1 variants (Berkhout et al. 2012; Lalezari et al. 2005).
The low serum half-life of T20 and T1249 necessitates further development of peptide-based inhibitors with better serum half-life. The next-generation inhibitors such as Cp32M, Sifuvirtide, and T2635 are having more helical structure than that of T20 and T1249, and showed better binding with the NHR. In addition, these peptides demonstrated better stability as well as higher serum half-life. Moreover, these peptides are active against virus variants that are resistant to T20 and T1249 (Eggink et al. 2009, 2011; Wang et al. 2009c; Yao et al. 2012).
Work from several groups has shown that CHR of gp41 also acts as a target for HIV entry inhibition. N-peptides (based on NHR) are weak inhibitors (with IC50 values in micromolar range) as compared to C-peptides, which show IC50 values in nanomolar range. This could be attributed to the fact that N-peptides are hydrophobic, aggregation prone, and poorly soluble in aqueous medium. Efforts have been made to solve this problem, and to develop inhibitors with better IC50 values. The first effort was to develop a chimeric peptide with higher solubility and higher affinity towards CHR. This has been achieved by linking NHR to a GCN4-based trimerization motif. Peptides such as IQN17 (trimeric coiled-coil GCN4-pIQI fused with N-terminus of a 17-residue NHR (N17)), IQN23 (trimeric coiled-coil GCN4-pIQI fused with N-terminus of a 23-residue NHR (N23)) were developed to bind CHR with higher affinity (Baldwin et al. 2003; Berkhout et al. 2012; Bianchi et al. 2005; Eckert and Kim 2001; Eckert et al. 1999). Another chimeric N-peptide NCCG-gp41 was derived from the NHR of gp41 that interacts with the CHR. The trimer of the chimeric peptide forms a stable complex with the CHR via engineered intermolecular disulphide bond, thereby demonstrates IC50 values in nano molar range (Baldwin et al. 2003; Berkhout et al. 2012; Eckert and Kim 2001; Louis et al. 2001). A preformed 5-helix peptide containing three NHR helices and two CHR are joined by short peptide linkers to form a 5-HB. This 5-HB interacts with free CHR of gp41 to form the 6HB in the pre-fusion state with nanomolar IC50 value, thereby restricting gp41 to form the post-fusion conformation (Baldwin et al. 2003; Berkhout et al. 2012; Root et al. 2001). There are several mutated NHR-based peptides that demonstrate anti-HIV activity. N36Mut(e,g) is one such example that shows nanomolar IC50 value. However, this peptide does not affect the 6HB formation directly, rather disrupts the trimeric NHR coiled-coil structure of the pre-hairpin intermediate (Berkhout et al. 2012; Bewley et al. 2002).
Conjugating cholesterol to the inhibitory peptide enhances the membrane partitioning of the peptide to the model membrane as well as in human blood cell, thereby suggesting better inhibitory effect of the cholesterol-conjugated peptide than the parent peptide (Hollmann et al. 2013; Ingallinella et al. 2009). In addition, the cholesterol-conjugated peptides show improved pharmacokinetics (Pessi 2015). Artificial peptides conjugated with cholesterol and pocket-specific small molecules demonstrate improved antiviral activity, even against the strains that are resistant to enfuvirtide (Wang et al. 2014).
Several fusion inhibitors have been designed to inhibit the interaction between gp120 and the host cell receptor, CD4 as the interaction between CD4 and gp120 is crucial for the docking of the virus on the cell surface. Peptide 12p1 (RINNIPWSEAMM) inhibits the interaction between CD4 and gp120 from three different HIV strains (Ferrer and Harrison 1999). This indicates that 12p1 binds a conserved site on gp120, and demonstrates broad-spectrum viral entry against multiple strains. The conserved regions of gp120 have also been utilized to develop peptide and protein-based inhibitors to block the interaction between gp120 and CD4 to avoid the viral entry, which has been extensively reviewed by Pu et al. (2019).
Inhibiting Influenza Entry
Unlike HIV, there are several challenges in developing peptide-based inhibitors for influenza virus. Influenza virus fuses in the presence of acidic endosomal pH. Therefore, the delivery of the inhibitory peptide to the endosome makes the problem more difficult. In addition, stability of the peptide in lower endosomal pH is an added problem (Yang et al. 2013).
Tagging cholesterol with the peptide enhances the partitioning of the peptide to the endosome and reveals anti-influenza properties. Peptides derived from the influenza hemagglutinin (HA) are generally incompetent to inhibit viral entry. However, it has been found that peptides, with a GSGSG linker, and the cholesterol group at C-terminal are potent inhibitors for influenza entry. These peptides trap HA in a transient intermediate state and prevent the subsequent steps to proceed for completion of fusion (Lee et al. 2011). Again, cholesterol-conjugated peptides such as S-KKWK and its derivatives (cholesterol is conjugated to the N-terminus of the short peptide KKWK) exhibited potent and broad anti-influenza A virus (IAV) activity in vitro and in vivo. S-KKWK binds to a conserved hydrophobic pocket on HA2 subunit, thereby preventing the six-helix bundle formation, and required conformational rearrangement of the HA2 subunit, which is an essential step for membrane fusion (Lin et al. 2017).
Generally, peptide inhibitors bind to the highly conserved stem epitope to block the low pH-induced conformational rearrangements required for membrane fusion. A library of cyclic peptides bind to this stem epitope, and inhibit influenza entry by stabilizing the pre-fusion conformation as shown in Fig. 14 (Kadam et al. 2017).
Fig. 14.
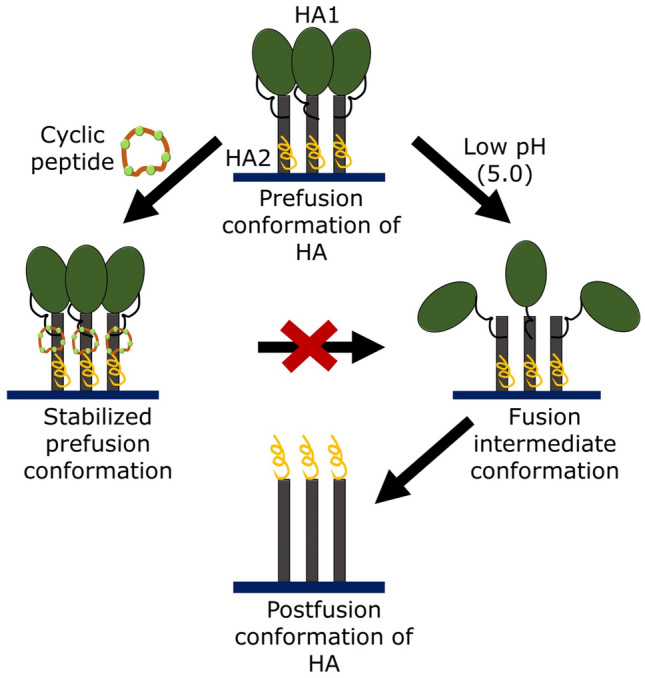
Schematic representation of mechanism of HA conformational change inhibition by cyclic peptides
Inhibiting Paramyxovirus Entry
Like HIV and influenza, 6HB is the common feature of the entry of paramyxoviruses. This common mechanism of viral entry is suggestive of a common target to inhibit viral membrane fusion. Indeed, many potent inhibitors are designed and developed utilizing the heptad region (HR) of many paramyxoviruses’ fusion proteins. Peptides derived from conserved heptad repeat (HR) regions of paramyxovirus fusion (F) proteins inhibit viral fusion by disrupting the formation of 6HB (Fig. 13), which is essential for driving the fusion reaction towards completion (Porotto et al. 2009).
Peptides derived from CHR regions of many paramyxoviruses, including Sendai virus, measles virus, Newcastle disease virus (NDV), respiratory syncytial virus (RSV), simian virus 5 (SV5), Hendra virus (HeV), and Nipah virus (NiV), can inhibit the entry of the homologous viruses (Joshi et al. 1998; Lambert et al. 1996; Porotto et al. 2006, 2010; Rapaport et al. 1995; Wild and Buckland 1997; Yao and Compans 1996; Young et al. 1997, 1999). Peptides such as WT, G12A, Ran, SV-163, and SV-473 were derived from the fusion peptide, and the two HR domains of Sendai virus F protein. It has been reported that SV-473 derived from the CHR inhibits fusion between virus with human red blood cells, though the mechanism is yet to be elucidated (Rapaport et al. 1995). Peptides synthesized from CHR from the F proteins of human parainfluenza virus type 2 (PI2) and type 3 (PI3) showed potential to inhibit virus-induced cell fusion, and control the spread of viral infection. In addition, the longer peptides (34 amino acids for PI2F or 35 amino acids for PI3F), which are extended from CHR to the transmembrane domain showed broad inhibition of cell fusion (Yao and Compans 1996).
Human metapneumovirus (hMPV) was discovered in 2001 (van den Hoogen et al. 2001). Synthetic peptides from NHR and CHR were induced to form a thermostable (melting temperature, 90 °C) helical structure, and showed to form hexameric coiled-coil structure. The peptide synthesized from NHR region demonstrates significant inhibitory effect against hMPV entry with around 50 nM IC50 value (Miller et al. 2007). CHR region-derived peptides of the F protein of HPIV3 also display inhibitory effects against HPIV and HeV/NiV entry (Porotto et al. 2006, 2010). Interestingly, the conjugation of cholesterol to the CHR-derived peptide localizes at the site of fusion and deactivates the fusion protein, thereby, enhancing the inhibitory efficiency of the peptide. The cholesterol-tagged peptides create a protective antiviral shield by targeting the fusion protein directly at the site of fusion, therefore elevate the inhibitory potential against paramyxoviruses (Porotto et al. 2010).
Inhibiting SARS-CoV Entry
The inhibitory properties of synthetic peptides derived from NHR and CHR were also tested for SARS-CoV. The CHR peptide was shown to be a potent inhibitor of mouse hepatitis virus (MHV) entry into the cell, confirmed from biological assays. CHR of MHV binds to the NHR region of the spike protein, and inhibits the subsequent interaction with native CHR, consequently, blocks the entry of SARS-CoV (Bosch et al. 2003). Interestingly, the NHR-derived peptides do not show any inhibitory effect. However, CHR peptides of SARS-CoV have reduced efficacy compared to the CHR peptides of the murine coronavirus mouse hepatitis virus (Bosch et al. 2004). Another CHR-based peptide CP-1 having low alpha-helicity also inhibits virus infection in the micromolar range. It has been demonstrated that the equimolar mixture of CP-1 and NP-1 (NHR-based peptide) forms a 6HB, which indicates that the CP-1 is capable to form 6HB with the native NHR, thereby, impeding the formation of post-fusion structure of the spike protein resulting in fusion inhibition (Liu et al. 2004).
A CHR-derived peptide EK1 targets the NHR of human coronavirus (HCoV) S protein, which proved to be effective in inhibiting infection of five different HCoVs, including SARS-CoV and MERS-CoV, and three SARS-related CoVs (SARS-CoVs) (Xia et al. 2019). In addition, the same group have reported that EK1 peptide is effective against SARS-CoV-2 S protein-mediated membrane fusion (Xia et al. 2020b). Again, cholesterol conjugation of EK1 (EK1C4) increases the inhibitory efficacy (Xia et al. 2020a). A higher helical stability of NHR as well as efficient interaction with CHR site to form a stable 6HB structure, which leads to enhanced fusion capacity, was found in the fusion protein of SARS-CoV-2. Modified HR2 sequence-derived peptide IPB01 with a cholesterol group, known as IPB02 has high ability in inhibiting SARS-CoV-2 as well as SARS-CoV (Zhu et al. 2020).
Some synthetic peptides outside the spike protein heptad repeat regions show antiviral activities. Zheng et al. have hypothesized that agents that interfered with, or bound competitively with some specific protein domains would disrupt the function of S protein resulting inhibition of SARS-CoV infections. They synthesized four derivative peptides of S protein fragments that showed antiviral activities in a cell line. In addition, two of these peptides reduced SARS-CoV infectivity over 10,000-fold through pre-incubation. Again, the infections were completely inhibited in the presence of three peptides (Zheng et al. 2005).
The binding of SARS-CoV spike (S) protein to the receptor i.e. cellular angiotensin-converting enzyme 2 (ACE2) is the first step in SARS-CoV infection. Therefore, peptides which block spike protein and ACE2 interaction may inhibit SARS-CoV entry. Small peptides derived from S protein such as SP-4 (residues 192–203), SP-8 (residues 483–494), and SP-10 (residues 668–679) significantly blocked the interaction between S protein and ACE2 with IC50 values of 4.30, 6.99, and 1.88 nM, respectively. Using alanine scanning mutagenesis, Han et al. suggested that charged amino acids between residues 22 and 57 were crucial for inhibitory efficiency, and two peptides (residues 22–44 and 22–57) derived from ACE2 showed antiviral activity with IC50 values of 50 and 6 μM, respectively. In addition, a peptide containing two segments of ACE2 i.e. residues 22–44 and 351–357 linked together by glycine revealed a potent antiviral activity with IC50 of about 0.1 µM (Han et al. 2006). This has further shown that the serine protease TMPRSS2 plays a pivotal role in priming the S protein, which is important for the entry of SARS-CoV (Glowacka et al. 2011; Matsuyama et al. 2010; Shulla et al. 2011). Therefore, it has been postulated that the inhibition of TMPRSS2 could lead to inhibit the entry of SARS-CoV (Kawase et al. 2012). Camostat mesylate, an inhibitor for TMPRSS2 partially blocks the SARS-CoV-2 entry, however, application of camostat mesylate with E-64d (an inhibitor for CatB/L) completely inhibits the viral entry (Hoffmann et al. 2020). This result indicates that the CatB/L, cysteine protease, also plays an important role in viral entry, and this protein could also be considered as a potential target for development of SARS-CoV-2 entry inhibitors.
Small-Molecule Inhibitors
Even though peptide-based fusion inhibitors have taken the centre stage, however, they have several limitations. Therefore, significant efforts have been made towards developing small-molecule fusion inhibitors.
Inhibiting HIV Entry
The surface protein (gp120), transmembrane protein (gp41), receptor (CD4), and chemokine co-receptor (CCR5 or CXCR4) are actively involved in HIV entry, therefore, these proteins are targets for developing HIV inhibitors. The interaction between the CD4 receptor and surface protein gp120 is the preliminary step of HIV entry. Therefore, quite a few small molecules target gp120 and inhibit HIV entry by blocking the interaction between gp120 and CD4. Small molecules such as BMS-378806 (Chen et al. 2005), BMS-488043 (Ho et al. 2006), NBD-556, NBD-557 (Zhao et al. 2005), and NBD-09027 (Curreli et al. 2012) are coming in this category.
Small-molecule inhibitors such as 5M030, 5M038, and 5M041 (Frey et al. 2006), ADS-J1 (Wang et al. 2009a), a number of N-substituted pyrrole derivatives (NB-2, NB-64, A12, GLS-22, and GLS-23) (Jiang et al. 2004; Liu et al. 2008; Wang et al. 2010), furan-based molecules (NB-293, NB-206) (Katritzky et al. 2009), and indole-based compounds (Balogh et al. 2009; Zhou et al. 2011, 2014) target the deep hydrophobic pocket on the NHR trimer core, and thereby inhibit the formation of 6HB and block membrane fusion (He et al. 2007, 2008; Liu et al. 2007). The binding site of CD4 is exposed at the native state of gp120, while the CHR-binding sites in gp41 are exposed only after the completion of binding of gp120 to the receptor CD4. However, this exposure is transient and lasts for few seconds to minutes. But, the CHR-binding sites of gp41, particularly the hydrophobic pocket region, are more conserved as compared to CD4 binding site in gp120. Consequently, small-molecule inhibitors targeting conserved gp41 pocket region are expected to have broader antiviral spectrum than those targeting gp120 (Lu et al. 2016).
Chemokine receptors are important cofactor for HIV entry and act as an important target for antiviral small molecule. Significant efforts have been given to develop molecules that block the interaction between gp120 and chemokine receptor, and control the expression of chemokine receptor in the target cells to inhibit HIV entry. Two positively charged peptides, ALX40-4C and T22, and a bicyclam compound called AMD3100 bind directly to CXCR4, thereby, blocking its interaction with gp120 and inhibit entry of HIV (Chan and Kim 1998). Maraviroc is a US FDA and European Medicines Agency (EMA)-approved novel small molecule that blocks gp120 interaction with CCR5 with potent anti-HIV activity (Xu et al. 2014). Ibalizumab (Trogarzo), a monoclonal antibody binds to the CD4 receptor and blocks its interaction with gp120, and inhibits HIV entry which was also approved by FDA (Beccari et al. 2019).
Inhibiting Influenza Entry
Two molecules, MBX2329 and MBX2546, with amino alkyl phenol ether and sulfonamide scaffolds, respectively, inhibit entry of influenza virus with IC50 value of 0.3–5.9 μM. They also inhibit a wide spectrum of influenza A viruses, which includes the 2009 pandemic influenza virus A/H1N1/2009, highly pathogenic avian influenza (HPAI) virus A/H5N1, and oseltamivir-resistant A/H1N1 strains. Both MBX2329 and MBX2546 bind to stem region of the HA trimer and inhibit HA-mediated fusion (Basu et al. 2014).
Generally, HA inhibitors are of two types, the first type blocks the interaction of HA1 with the neuraminic acid (Neu) receptors present on the surface of the target host cells, and the other disturbs the HA2-mediated fusion process by inhibiting the low pH-induced conformational change of HA2. Compounds such as BMY-27709 (Luo et al. 1996, 1997; Wu et al. 2017) and stachyflin (Wu et al. 2017; Yagi et al. 1999; Yoshimoto et al. 1999, 2000) come under the first category.
The indole alkaloids such as Neoechinulin B bind to the influenza envelope HA and disrupt its interaction with the sialic acid receptor, and the attachment of viruses to the target host cells (Chen et al. 2015). Thiazolides such as Nitazoxanide blocks the maturation of viral HA and hence active against influenza viruses (Ashton et al. 2010; Belardo et al. 2015; Haffizulla et al. 2014). Three different saponins with 3-O-β-chacotriosyl residue showed significant inhibitory activity for H5N1 entry into cells with IC50 value of 7.22–9.25 mM (Song et al. 2009).
Inhibiting Paramyxovirus Entry
Measles virus (MV) uses both CD46 and CD150 as cellular receptors (de Vries et al. 2010). Plemper et al. suggested that a microdomain in the Measles virus (MV) fusion protein (F protein) is structurally conserved in the paramyxovirus family (Plemper et al. 2003). Therefore, this conserved microdomain is an important target for the development of antiviral molecules. The first compound of this kind was 5-amino-2-benzylbenzoxazole (B17/OX-1), which is having an IC50 value of 50 μM against MV. OX-1 also prevents the early step of lipid mixing, or hemifusion formation (Plemper et al. 2004). An acyclic variant of OX-1 (4-nitro-2-phenylaceylaminobenzamide, AS-48) demonstrated approximately 10 to 15-fold increase in antiviral activity (Plemper et al. 2005).
Inhibiting SARS-CoV Entry
Simmons et al. proposed a three-step process for SARS-CoV infection. Receptor binding is the first step followed by conformational change of spike S protein, and cathepsin L proteolysis within the endosome. They targeted the last step for inhibition of SARS-CoV replication by small molecule. Compound MDL28170 inhibited the infection with IC50 value of 2.5 nM (Simmons et al. 2005). Using the molecule fingerprint searching method and molecular dynamics simulation Wang et al. suggested that a compound (MOL376) derived from a Chinese medicine herb with the therapeutic efficacy on the human body suitably binds to the binding pocket of cathepsin L and hence it might become a capable inhibitor for SARS-CoV infection (Wang et al. 2007).
Based on the concept of chemical genetics, Kao et al. identified 104 compounds with anti-SARS-CoV activity. Out of these, 2 compounds interact with the SARS-CoV main protease (Mpro), 7 interact with helicase (Hel), and 18 target spike (S) protein-ACE2-mediated viral entry. Three selected compounds that showed better efficacies are MP576, HE602, and VE607, which target the SARS-CoV main protease, helicase, and ACE2-mediated viral entry, respectively, with IC50 value in micromolar range (Kao et al. 2004).
Class II Viral Entry Inhibition
The control of class II viral infections by vaccination is still not efficient. For example, CYD-TDV (chimeric yellow fever virus-tetravalent dengue vaccine) developed by Sanofi Pasteur was first dengue vaccine, and got the licence in December 2015 for use in individuals between 9 and 45 years of age, living in endemic areas. However, its average efficiency was 50%, 39%, 75%, and 77% for DENV-1, DENV-2, DENV-3, and DENV-4, respectively (Capeding et al. 2014; Chew et al. 2017; Villar et al. 2015).
Several strategies have been taken to develop inhibitors for class II viruses. One such approach is similar to the antiviral compound against HIV. The peptides derived from stem region of the fusion core may bind to the trimeric core of the post-fusion conformations and disrupt the final folding of the fusion protein which is essential for membrane fusion (Kielian 2006; Modis 2013). Although the exact inhibition mechanism is unknown, it is believed that DN59 (a peptide corresponding to stem domain of DENV) and WN83 (designed from overlapping regions of E protein domain I/II junction) may interfere with intramolecular interaction between stem and other portions of class II viral fusion protein. It has been reported that DN59 is active against both dengue and West Nile virus, but WN83 is only active against West Nile virus (Hrobowski et al. 2005).
The other strategy to inhibit class II viral infection is to target the hinge region of the fusion protein. Hinge region is a flexible region which undergoes conformational change during virus maturation and conversion from neutral to acidic pH to induce trimer formation. In flavivirus, the pH-dependent fusion is also affected by mutation in the hinge region, while in alphaviruses the cholesterol dependency was reduced by mutation in the hinge region. A hydrophobic pocket was identified in the dengue virus E, and its crystal structure at neutral pH suggests that a detergent molecule n-octyl-β-glucoside is present in this pocket. Blocking this pocket to reduce the hinge flexibility may inhibit class II fusion protein-induced membrane fusion (Kielian 2006; Modis 2013). High-throughput docking with this hydrophobic pocket led to the identification of compound 6 (Wang et al. 2009b) and compound 1662G07 (Schmidt et al. 2012) to block dengue virus entry. It was reported that compound 6 had an average 50% effective concentration of 119 nM against dengue virus serotype 2 in human cell line (Wang et al. 2009b). Poh et al. identified a small molecule, NITD448, which binds to the hydrophobic pocket and demonstrates antiviral efficiency with IC50 of 6.5 μM (Poh et al. 2009).
Class III Viral Entry Inhibition
IFITM3 and Tetherin are located on the plasma membrane. Tetherin restricts infections by retroviruses and filoviruses whereas IFITM3 inhibits influenza virus and flavivirus infection. In addition, Weidner et al. reported that both IFITM3 and Tetherin inhibit VSV infections. IFITM3 blocks the infection after endocytosis during or before primary transcription, while Tetherin inhibits the release of virion particles from the infected cells. Again, the N-terminal 21 amino acids residue and C-terminal transmembrane region of IFITM3 are essential for its antiviral function (Weidner et al. 2010). The lysosomal inhibitor chloroquine also inhibits VSV infection to 50%, when added after 1.5 h of infection (Miller and Lenard 1980).
Broad-Spectrum Antiviral Agents: Need of the Hour
Viral infections are major pandemic threats in all time to the human civilization. Emerging infectious diseases pose a constant menace to global health and economy. Vaccination is the only way to get away from the cruelty of these emerging and re-emerging viruses, however, the vaccines are highly specific to a particular virus even to a particular strain. The traditional strategy of ‘one bug-one drug’ in antiviral drug development is inadequate for responding to an increasing diversity of viruses that cause deadly diseases in humans (Vigant et al. 2015). Till date more than 20 million people are suffering from coronavirus disease 2019 (COVID-19) that claimed more than half a million lives, and stalled the livelihood of uncountable. Therefore, the development of broad-spectrum antiviral agent is the need of the hour to combat continuously emerging viral diseases. As viruses use their own fusion machinery, it differs from virus to virus, which makes it challenging to develop broad-spectrum antiviral agent by targeting the fusion machine of the virus. However, the fusion process follows some common pathway in terms of evolution of membrane structure and conformation (Jahn et al. 2003). This prompted us to develop certain class of peptides or small molecules that inhibit the structural changes in the membrane that is essential for fusion process. In search of peptide-based broad-spectrum antiviral agent by targeting the membrane, we recently developed coronin 1-derived tryptophan–aspartic acid (WD)-containing peptides, which show significant inhibition of polyethylene glycol (PEG)-mediated fusion of model membranes (Pattnaik and Chakraborty 2018, 2019).
Pathogenic mycobacteria survive within macrophages for years by evading lysosomal degradation (Jayachandran et al. 2007; Pieters 2008). In addition to different biochemical processes, the role of phagosomal coat protein coronin 1 cannot be undermined. Coronin 1 is recruited to the phagosomal membranes only when live mycobacteria are present in the phagosome (Gruenberg and Stenmark 2004; Pieters et al. 2013). This clearly suggests the putative function of coronin 1 in inhibiting fusion between phagosome and lysosome. Interestingly, in coronin 1, the positions of tryptophan and aspartic acid are conserved across the species (Eckert et al. 2011). Therefore, the inhibitory effect of tryptophan–aspartic acid (WD)-containing peptides derived from coronin 1 were investigated. It has been found that two peptides, TG-23 and GL-22, substantially reduce the PEG-mediated fusion of small unilamellar vesicles by inhibiting water penetration into the membrane, which is important for membrane fusion (Pattnaik and Chakraborty 2018). Though these results are preliminary, but displays promise for the development of broad-spectrum antiviral agents that can target and modify the membrane physical properties to avoid fusion between virus and the host cell.
Concluding Remark and Future Perspectives
It is not very difficult to identify emerging viral diseases and the genetic fingerprint of viral fusion proteins, however, the discovery of vaccine or drugs against these viruses is extremely time consuming. Moreover, some viral infections such as the ongoing COVID-19 pandemic spread at an extraordinary rate, which overtakes the development of suitable therapies. Thus, the traditional approach of ‘one bug-one drug’ is not sufficient for combating emerging viral diseases. Therefore, in our opinion, more proactive research on broad-spectrum antiviral agents might be helpful to deal with future pandemic situations.
Acknowledgements
This work was supported by research grant from the Science and Technology Department, Government of Odisha. H.C. thanks the University Grants Commission for UGC-Assistant Professor position. G.P.P acknowledges Science and Technology Department, Government of Odisha for Research Fellowship. We thank members of Chakraborty laboratory for their comments and discussions.
Compliance with Ethical Standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ashton LV, Callan RL, Rao S, Landolt GA. In vitro susceptibility of canine influenza A (H3N8) virus to nitazoxanide and tizoxanide. Vet Med Int. 2010;2010:891010. doi: 10.4061/2010/891010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin CE, Sanders RW, Berkhout B. Inhibiting HIV-1 entry with fusion inhibitors. Curr Med Chem. 2003;10:1633–1642. doi: 10.2174/0929867033457124. [DOI] [PubMed] [Google Scholar]
- Balogh E, Wu D, Zhou G, Gochin M. NMR second site screening for structure determination of ligands bound in the hydrophobic pocket of HIV-1 gp41. J Am Chem Soc. 2009;131:2821–2823. doi: 10.1021/ja8094558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Antanasijevic A, Wang M, Li B, Mills DM, Ames JA, Nash PJ, Williams JD, Peet NP, Moir DT, Prichard MN, Keith KA, Barnard DL, Caffrey M, Rong L, Bowlin TL. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza a virus fusion. J Virol. 2014;88:1447–1460. doi: 10.1128/JVI.01225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari MV, Mogle BT, Sidman EF, Mastro KA, Asiago-Reddy E, Kufel WD. Ibalizumab, a novel monoclonal antibody for the management of multidrug-resistant HIV-1 infection. Antimicrob Agents Chemother. 2019;63:e00110–e00119. doi: 10.1128/AAC.00110-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardo G, Cenciarelli O, La Frazia S, Rossignol JF, Santoro MG. Synergistic effect of nitazoxanide with neuraminidase inhibitors against influenza A viruses in vitro. Antimicrob Agents Chemother. 2015;59:1061–1069. doi: 10.1128/AAC.03947-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, Eggink D, Sanders RW. Is there a future for antiviral fusion inhibitors? Curr Opin Virol. 2012;2:50–59. doi: 10.1016/j.coviro.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Louis JM, Ghirlando R, Clore GM. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J Biol Chem. 2002;277:14238–14245. doi: 10.1074/jbc.M201453200. [DOI] [PubMed] [Google Scholar]
- Bianchi E, Finotto M, Ingallinella P, Hrin R, Carella AV, Hou XS, Schleif WA, Miller MD, Geleziunas R, Pessi A. Covalent stabilization of coiled coils of the HIV gp41 N region yields extremely potent and broad inhibitors of viral infection. Proc Natl Acad Sci USA. 2005;102:12903–10908. doi: 10.1073/pnas.0502449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom DE, Cadarette D. Infectious disease threats in the twenty-first century: strengthening the global response. Front Immunol. 2019;10:549. doi: 10.3389/fimmu.2019.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch BJ, Martina BE, Van Der Zee R, Lepault J, Haijema BJ, Versluis C, Heck AJ, De Groot R, Osterhaus AD, Rottier PJ. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci USA. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon IK, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384:1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Cavanagh D, Davis PJ. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J Gen Virol. 1986;67:1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Chen X, Si L, Liu D, Proksch P, Zhang L, Zhou D, Lin W. Neoechinulin B and its analogues as potential entry inhibitors of influenza viruses, targeting viral hemagglutinin. Eur J Med Chem. 2015;93:182–195. doi: 10.1016/j.ejmech.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Chew MF, Poh KS, Poh CL. Peptides as therapeutic agents for dengue virus. Int J Med Sci. 2017;14:1342–1359. doi: 10.7150/ijms.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P, Chanock R, McIntosh K (1996) Parainfluenza viruses. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE (eds) Fields virology. Lippincott-Raven Publishers, Philadelphia, pp 1205–1241
- Curreli F, Choudhury S, Pyatkin I, Zagorodnikov VP, Bulay AK, Altieri A, Kwon YD, Kwong PD, Debnath AK. Design, synthesis, and antiviral activity of entry inhibitors that target the CD4-binding site of HIV-1. J Med Chem. 2012;55:4764–4775. doi: 10.1021/jm3002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RD, Lemon K, Ludlow M, McQuaid S, Yuksel S, van Amerongen G, Rennick LJ, Rima BK, Osterhaus AD, de Swart RL, Duprex WP. In vivo tropism of attenuated and pathogenic measles virus expressing green fluorescent protein in macaques. J Virol. 2010;84:4714–4724. doi: 10.1128/JVI.02633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Liu J, Zheng Q, Yong W, Lu M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure. 2006;14:889–899. doi: 10.1016/j.str.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE. Entry and fusion of emerging paramyxoviruses. PLoS Pathog. 2010;6:e1000881. doi: 10.1371/journal.ppat.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch RE, Jardetzky TS, Lamb RA. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci Rep. 2000;20:597–612. doi: 10.1023/a:1010467106305. [DOI] [PubMed] [Google Scholar]
- Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, Hammesfahr B, Kollmar M. A holistic phylogeny of the coronin gene family reveals an ancient origin of the tandem-coronin, defines a new subfamily, and predicts protein function. BMC Evol Biol. 2011;11:268. doi: 10.1186/1471-2148-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci USA. 2001;98:11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- Eggink D, Bontjer I, Langedijk JP, Berkhout B, Sanders RW. Resistance of human immunodeficiency virus type 1 to a third-generation fusion inhibitor requires multiple mutations in gp41 and is accompanied by a dramatic loss of gp41 function. J Virol. 2011;85:10785–10797. doi: 10.1128/JVI.05331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Langedijk JP, Bonvin AM, Deng Y, Lu M, Berkhout B, Sanders RW. Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J Biol Chem. 2009;284:26941–26950. doi: 10.1074/jbc.M109.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Harrison SC. Peptide ligands to human immunodeficiency virus type 1 gp120 identified from phage display libraries. J Virol. 1999;73:5795–5802. doi: 10.1128/jvi.73.7.5795-5802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci USA. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci USA. 2006;103:13938–13943. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ, Doms RW, York D, White J. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72:1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- Haffizulla J, Hartman A, Hoppers M, Resnick H, Samudrala S, Ginocchio C, Bardin M, Rossignol JF. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect Dis. 2014;14:609–618. doi: 10.1016/S1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson-McReynolds S, Jiang S, Rong L, Caffrey M. Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state. J Biol Chem. 2006;281:11965–11971. doi: 10.1074/jbc.M601174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Cheng J, Li J, Qi Z, Lu H, Dong M, Jiang S, Dai Q. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: implications for designing novel anti-HIV fusion inhibitors. J Virol. 2008;82:6349–6358. doi: 10.1128/JVI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Liu S, Jing W, Lu H, Cai D, Chin DJ, Debnath AK, Kirchhoff F, Jiang S. Conserved residue Lys574 in the cavity of HIV-1 Gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J Biol Chem. 2007;282:25631–25639. doi: 10.1074/jbc.M703781200. [DOI] [PubMed] [Google Scholar]
- Ho HT, Fan L, Nowicka-Sans B, McAuliffe B, Li CB, Yamanaka G, Zhou N, Fang H, Dicker I, Dalterio R, Gong YF, Wang T, Yin Z, Ueda Y, Matiskella J, Kadow J, Clapham P, Robinson J, Colonno R, Lin PF. Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J Virol. 2006;80:4017–4025. doi: 10.1128/JVI.80.8.4017-4025.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann A, Matos PM, Augusto MT, Castanho MA, Santos NC. Conjugation of cholesterol to HIV-1 fusion inhibitor C34 increases peptide-membrane interactions potentiating its action. PLoS One. 2013;8:e60302. doi: 10.1371/journal.pone.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingallinella P, Bianchi E, Ladwa NA, Wang YJ, Hrin R, Veneziano M, Bonelli F, Ketas TJ, Moore JP, Miller MD, Pessi A. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc Natl Acad Sci USA. 2009;106:5801–5806. doi: 10.1073/pnas.0901007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Jayachandran R, Sundaramurthy V, Combaluzier B, Mueller P, Korf H, Huygen K, Miyazaki T, Albrecht I, Massner J, Pieters J. Survival of mycobacteria in macrophages is mediated by coronin 1-dependent activation of calcineurin. Cell. 2007;130:37–50. doi: 10.1016/j.cell.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Jiang S, Lu H, Liu S, Zhao Q, He Y, Debnath AK. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob Agents Chemother. 2004;48:4349–4359. doi: 10.1128/AAC.48.11.4349-4359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SB, Dutch RE, Lamb RA. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- Kadam RU, Juraszek J, Brandenburg B, Buyck C, Schepens WBG, Kesteleyn B, Stoops B, Vreeken RJ, Vermond J, Goutier W, Tang C, Vogels R, Friesen RHE, Goudsmit J, van Dongen MJP, Wilson IA. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao RY, Tsui WH, Lee TS, Tanner JA, Watt RM, Huang JD, Hu L, Chen G, Chen Z, Zhang L, He T, Chan KH, Tse H, To AP, Ng LW, Wong BC, Tsoi HW, Yang D, Ho DD, Yuen KY. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritzky AR, Tala SR, Lu H, Vakulenko AV, Chen QY, Sivapackiam J, Pandya K, Jiang S, Debnath AK. Design, synthesis, and structure-activity relationship of a novel series of 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl)furans as HIV-1 entry inhibitors. J Med Chem. 2009;52:7631–7639. doi: 10.1021/jm900450n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M. Class II virus membrane fusion proteins. Virology. 2006;344:38–47. doi: 10.1016/j.virol.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- Kilby JM, Lalezari JP, Eron JJ, Carlson M, Cohen C, Arduino RC, Goodgame JC, Gallant JE, Volberding P, Murphy RL, Valentine F, Saag MS, Nelson EL, Sista PR, Dusek A. The safety, plasma pharmacokinetics, and antiviral activity of subcutaneous enfuvirtide (T-20), a peptide inhibitor of gp41-mediated virus fusion, in HIV-infected adults. AIDS Res Hum Retrovir. 2002;18:685–693. doi: 10.1089/088922202760072294. [DOI] [PubMed] [Google Scholar]
- Lalezari JP, Bellos NC, Sathasivam K, Richmond GJ, Cohen CJ, Myers RA, Jr, Henry DH, Raskino C, Melby T, Murchison H, Zhang Y, Spence R, Greenberg ML, Demasi RA, Miralles GD. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J Infect Dis. 2005;191:1155–1163. doi: 10.1086/427993. [DOI] [PubMed] [Google Scholar]
- Lalezari JP, Eron JJ, Carlson M, Cohen C, DeJesus E, Arduino RC, Gallant JE, Volberding P, Murphy RL, Valentine F, Nelson EL, Sista PR, Dusek A, Kilby JM. A phase II clinical study of the long-term safety and antiviral activity of enfuvirtide-based antiretroviral therapy. AIDS. 2003;17:691–698. doi: 10.1097/00002030-200303280-00007. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR., Jr Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lentz BR. Evolution of lipidic structures during model membrane fusion and the relation of this process to cell membrane fusion. Biochemistry. 1997;36:6251–6259. doi: 10.1021/bi970404c. [DOI] [PubMed] [Google Scholar]
- Lee KK, Pessi A, Gui L, Santoprete A, Talekar A, Moscona A, Porotto M. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J Biol Chem. 2011;286:42141–42149. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- Lin D, Luo Y, Yang G, Li F, Xie X, Chen D, He L, Wang J, Ye C, Lu S, Lv L, Liu S, He J. Potent influenza A virus entry inhibitors targeting a conserved region of hemagglutinin. Biochem Pharmacol. 2017;144:35–51. doi: 10.1016/j.bcp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- Liu K, Lu H, Hou L, Qi Z, Teixeira C, Barbault F, Fan BT, Liu S, Jiang S, Xie L. Design, synthesis, and biological evaluation of N-carboxyphenylpyrrole derivatives as potent HIV fusion inhibitors targeting gp41. J Med Chem. 2008;51:7843–7854. doi: 10.1021/jm800869t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Jing W, Cheung B, Lu H, Sun J, Yan X, Niu J, Farmar J, Wu S, Jiang S. HIV gp41 C-terminal heptad repeat contains multifunctional domains. Relation to mechanisms of action of anti-HIV peptides. J Biol Chem. 2007;282:9612–9620. doi: 10.1074/jbc.M609148200. [DOI] [PubMed] [Google Scholar]
- Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante CR, Xiong H, Farmar J, Debnath AK, Tien P, Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis JM, Bewley CA, Clore GM. Design and properties of N(CCG)-gp41, a chimeric gp41 molecule with nanomolar HIV fusion inhibitory activity. J Biol Chem. 2001;276:29485–29489. doi: 10.1074/jbc.C100317200. [DOI] [PubMed] [Google Scholar]
- Lu L, Yu F, Cai L, Debnath AK, Jiang S. Development of small-molecule HIV entry inhibitors specifically targeting gp120 or gp41. Curr Top Med Chem. 2016;16:1074–1090. doi: 10.2174/1568026615666150901114527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Luo G, Colonno R, Krystal M. Characterization of a hemagglutinin-specific inhibitor of influenza A virus. Virology. 1996;226:66–76. doi: 10.1006/viro.1996.0628. [DOI] [PubMed] [Google Scholar]
- Luo G, Torri A, Harte WE, Danetz S, Cianci C, Tiley L, Day S, Mullaney D, Yu KL, Ouellet C, Dextraze P, Meanwell N, Colonno R, Krystal M. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J Virol. 1997;71:4062–4070. doi: 10.1128/jvi.71.5.4062-4070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Mas V, Melero JA. Entry of enveloped viruses into host cells: membrane fusion. Subcell Biochem. 2013;68:467–487. doi: 10.1007/978-94-007-6552-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Marsh M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Research. 2019;8:1628. doi: 10.12688/f1000research.19694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meher G, Bhattacharjya S, Chakraborty H. Membrane cholesterol modulates oligomeric status and peptide-membrane interaction of severe acute respiratory syndrome coronavirus fusion peptide. J Phys Chem B. 2019;123:10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- Meher G, Chakraborty H. Organization and dynamics of Trp14 of hemagglutinin fusion peptide in membrane mimetic environment. Chem Phys Lipids. 2017;205:48–54. doi: 10.1016/j.chemphyslip.2017.04.010. [DOI] [PubMed] [Google Scholar]
- Meher G, Sinha S, Pattnaik GP, Ghosh Dastidar S, Chakraborty H. Cholesterol modulates membrane properties and the interaction of gp41 fusion peptide to promote membrane fusion. J Phys Chem B. 2019;123:7113–7122. doi: 10.1021/acs.jpcb.9b04577. [DOI] [PubMed] [Google Scholar]
- Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DK, Lenard J. Inhibition of vesicular stomatitis virus infection by spike glycoprotein. Evidence for an intracellular, G protein-requiring step. J Cell Biol. 1980;84:430–437. doi: 10.1083/jcb.84.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Tollefson S, Crowe JE, Jr, Williams JV, Wright DW. Examination of a fusogenic hexameric core from human metapneumovirus and identification of a potent synthetic peptide inhibitor from the heptad repeat 1 region. J Virol. 2007;81:141–149. doi: 10.1128/JVI.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y. Class II fusion proteins. Adv Exp Med Biol. 2013;790:150–166. doi: 10.1007/978-1-4614-7651-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SJ, Chang HL, Cheung TY, Tang AF, Fisk TL, Ooi SP, Kuo HW, Jiang DD, Chen KT, Lando J, Hsu KH, Chen TJ, Dowell SF. Transmission of the severe acute respiratory syndrome on aircraft. N Engl J Med. 2003;349:2416–2422. doi: 10.1056/NEJMoa031349. [DOI] [PubMed] [Google Scholar]
- Pattnaik GP, Chakraborty H. Coronin 1 derived tryptophan-aspartic acid containing peptides inhibit membrane fusion. Chem Phys Lipids. 2018;217:35–42. doi: 10.1016/j.chemphyslip.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Pattnaik GP, Chakraborty H. Cholesterol alters the inhibitory efficiency of peptide-based membrane fusion inhibitor. Biochim Biophys Acta Biomembr. 2019;1861:183056. doi: 10.1016/j.bbamem.2019.183056. [DOI] [PubMed] [Google Scholar]
- Pattnaik GP, Meher G, Chakraborty H. Exploring the mechanism of viral peptide-induced membrane fusion. Adv Exp Med Biol. 2018;1112:69–78. doi: 10.1007/978-981-13-3065-0_6. [DOI] [PubMed] [Google Scholar]
- Pessi A. Cholesterol-conjugated peptide antivirals: a path to a rapid response to emerging viral diseases. J Pept Sci. 2015;21:379–386. doi: 10.1002/psc.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. Coronin 1 in innate immunity. Subcell Biochem. 2008;48:116–123. doi: 10.1007/978-0-387-09595-0_11. [DOI] [PubMed] [Google Scholar]
- Pieters J, Muller P, Jayachandran R. On guard: coronin proteins in innate and adaptive immunity. Nat Rev Immunol. 2013;13:510–518. doi: 10.1038/nri3465. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Doyle J, Sun A, Prussia A, Cheng LT, Rota PA, Liotta DC, Snyder JP, Compans RW. Design of a small-molecule entry inhibitor with activity against primary measles virus strains. Antimicrob Agents Chemother. 2005;49:3755–3761. doi: 10.1128/AAC.49.9.3755-3761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Erlandson KJ, Lakdawala AS, Sun A, Prussia A, Boonsombat J, Aki-Sener E, Yalcin I, Yildiz I, Temiz-Arpaci O, Tekiner B, Liotta DC, Snyder JP, Compans RW. A target site for template-based design of measles virus entry inhibitors. Proc Natl Acad Sci USA. 2004;101:5628–5633. doi: 10.1073/pnas.0308520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Lakdawala AS, Gernert KM, Snyder JP, Compans RW. Structural features of paramyxovirus F protein required for fusion initiation. Biochemistry. 2003;42:6645–6655. doi: 10.1021/bi034385k. [DOI] [PubMed] [Google Scholar]
- Poh MK, Yip A, Zhang S, Priestle JP, Ma NL, Smit JM, Wilschut J, Shi PY, Wenk MR, Schul W. A small molecule fusion inhibitor of dengue virus. Antivir Res. 2009;84:260–266. doi: 10.1016/j.antiviral.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Porotto M, Doctor L, Carta P, Fornabaio M, Greengard O, Kellogg GE, Moscona A. Inhibition of hendra virus fusion. J Virol. 2006;80:9837–9849. doi: 10.1128/JVI.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porotto M, Yokoyama CC, Orefice G, Kim HS, Aljofan M, Mungall BA, Moscona A. Kinetic dependence of paramyxovirus entry inhibition. J Virol. 2009;83:6947–6951. doi: 10.1128/JVI.00416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porotto M, Yokoyama CC, Palermo LM, Mungall B, Aljofan M, Cortese R, Pessi A, Moscona A. Viral entry inhibitors targeted to the membrane site of action. J Virol. 2010;84:6760–6768. doi: 10.1128/JVI.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu N, Prabakaran M, Ho HT, Velumani S, Qiang J, Goutama M, Kwang J. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J Virol. 2009;83:2553–2562. doi: 10.1128/JVI.02165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Wang Q, Xu W, Lu L, Jiang S. Development of protein- and peptide-based HIV entry inhibitors targeting gp120 or gp41. Viruses. 2019;11:705. doi: 10.3390/v11080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Lok SM. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell. 2018;172:1319–1334. doi: 10.1016/j.cell.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- Saunders-Hastings PR, Krewski D. Reviewing the history of pandemic influenza: understanding patterns of emergence and transmission. Pathogens. 2016;5:66. doi: 10.3390/pathogens5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Lee K, Yang PL, Harrison SC. Small-molecule inhibitors of dengue-virus entry. PLoS Pathog. 2012;8:e1002627. doi: 10.1371/journal.ppat.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel DP. The modified stalk mechanism of lamellar/inverted phase transitions and its implications for membrane fusion. Biophys J. 1999;76:291–313. doi: 10.1016/S0006-3495(99)77197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Song G, Yang S, Zhang W, Cao Y, Wang P, Ding N, Zhang Z, Guo Y, Li Y. Discovery of the first series of small molecule H5N1 entry inhibitors. J Med Chem. 2009;52:7368–7371. doi: 10.1021/jm900275m. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Stocker H, Kloft C, Plock N, Breske A, Kruse G, Herzmann C, Schulbin H, Kreckel P, Weber C, Goebel F, Roeling J, Staszewski S, Plettenberg A, Moecklinghoff C, Arasteh K, Kurowski M. Pharmacokinetics of enfuvirtide in patients treated in typical routine clinical settings. Antimicrob Agents Chemother. 2006;50:667–673. doi: 10.1128/AAC.50.2.667-673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar VM, Bruckmann C, Ingallinella P, Bianchi E, Pessi A, Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc Natl Acad Sci USA. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F, Shimazaki YK. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J Gen Virol. 2000;81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B, Howard MW, Jobling M, Holmes RK, Holmes KV, Hodges RS. Structural characterization of the SARS-coronavirus spike S fusion protein core. J Biol Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristram-Nagle S, Nagle JF. HIV-1 fusion peptide decreases bending energy and promotes curved fusion intermediates. Biophys J. 2007;93:2048–2055. doi: 10.1529/biophysj.107.109181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigant F, Santos NC, Lee B. Broad-spectrum antivirals against viral fusion. Nat Rev Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramirez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortes Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med. 2015;372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- Wang C, Shi W, Cai L, Lu L, Yu F, Wang Q, Jiang X, Xu X, Wang K, Xu L, Jiang S, Liu K. Artificial peptides conjugated with cholesterol and pocket-specific small molecules potently inhibit infection by laboratory-adapted and primary HIV-1 isolates and enfuvirtide-resistant HIV-1 strains. J Antimicrob Chemother. 2014;69:1537–1545. doi: 10.1093/jac/dku010. [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Z, Guo A, Mao Q, Lu H, An X, Xia C, Li X, Debnath AK, Wu S, Liu S, Jiang S. ADS-J1 inhibits human immunodeficiency virus type 1 entry by interacting with the gp41 pocket region and blocking fusion-active gp41 core formation. Antimicrob Agents Chemother. 2009;53:4987–4998. doi: 10.1128/AAC.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QY, Patel SJ, Vangrevelinghe E, Xu HY, Rao R, Jaber D, Schul W, Gu F, Heudi O, Ma NL, Poh MK, Phong WY, Keller TH, Jacoby E, Vasudevan SG. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53:1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RR, Yang LM, Wang YH, Pang W, Tam SC, Tien P, Zheng YT. Sifuvirtide, a potent HIV fusion inhibitor peptide. Biochem Biophys Res Commun. 2009;382:540–544. doi: 10.1016/j.bbrc.2009.03.057. [DOI] [PubMed] [Google Scholar]
- Wang SQ, Du QS, Zhao K, Li AX, Wei DQ, Chou KC. Virtual screening for finding natural inhibitor against cathepsin-L for SARS therapy. Amino Acids. 2007;33:129–135. doi: 10.1007/s00726-006-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lu H, Zhu Q, Jiang S, Liao Y. Structure-based design, synthesis and biological evaluation of new N-carboxyphenylpyrrole derivatives as HIV fusion inhibitors targeting gp41. Bioorg Med Chem Lett. 2010;20:189–192. doi: 10.1016/j.bmcl.2009.10.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner JM, Jiang D, Pan XB, Chang J, Block TM, Guo JT. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Hinz A, Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C, Dubay JW, Greenwell T, Baird T, Jr, Oas TG, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild TF, Buckland R. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J Gen Virol. 1997;78:107–111. doi: 10.1099/0022-1317-78-1-107. [DOI] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE., Jr Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sun Q, Zhang C, Yang S, Li L, Jia Z. Progress of small molecular inhibitors in the development of anti-influenza virus agents. Theranostics. 2017;7:826–845. doi: 10.7150/thno.17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, Wang Q, Du L, Tan W, Wilson IA, Jiang S, Yang B, Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S, Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GG, Guo J, Wu Y. Chemokine receptor CCR5 antagonist maraviroc: medicinal chemistry and clinical applications. Curr Top Med Chem. 2014;14:1504–1514. doi: 10.2174/1568026614666140827143745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lou Z, Liu Y, Pang H, Tien P, Gao GF, Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi S, Ono J, Yoshimoto J, Sugita K, Hattori N, Fujioka T, Fujiwara T, Sugimoto H, Hirano K, Hashimoto N. Development of anti-influenza virus drugs I: improvement of oral absorption and in vivo anti-influenza activity of Stachyflin and its derivatives. Pharm Res. 1999;16:1041–1046. doi: 10.1023/a:1018983715982. [DOI] [PubMed] [Google Scholar]
- Yang J, Li M, Shen X, Liu S. Influenza A virus entry inhibitors targeting the hemagglutinin. Viruses. 2013;5:352–373. doi: 10.3390/v5010352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang HW. A rhombohedral phase of lipid containing a membrane fusion intermediate structure. Biophys J. 2003;84:1808–1817. doi: 10.1016/S0006-3495(03)74988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Compans RW. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- Yao X, Chong H, Zhang C, Qiu Z, Qin B, Han R, Waltersperger S, Wang M, He Y, Cui S. Structural basis of potent and broad HIV-1 fusion inhibitor CP32M. J Biol Chem. 2012;287:26618–26629. doi: 10.1074/jbc.M112.381079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto J, Kakui M, Iwasaki H, Fujiwara T, Sugimoto H, Hattori N. Identification of a novel HA conformational change inhibitor of human influenza virus. Arch Virol. 1999;144:865–878. doi: 10.1007/s007050050552. [DOI] [PubMed] [Google Scholar]
- Yoshimoto J, Kakui M, Iwasaki H, Sugimoto H, Fujiwara T, Hattori N. Identification of amino acids of influenza virus HA responsible for resistance to a fusion inhibitor, Stachyflin. Microbiol Immunol. 2000;44:677–685. doi: 10.1111/j.1348-0421.2000.tb02549.x. [DOI] [PubMed] [Google Scholar]
- Young JK, Hicks RP, Wright GE, Morrison TG. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology. 1997;238:291–304. doi: 10.1006/viro.1997.8834. [DOI] [PubMed] [Google Scholar]
- Young JK, Li D, Abramowitz MC, Morrison TG. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J Virol. 1999;73:5945–5956. doi: 10.1128/jvi.73.7.5945-5956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Nieforth K, Lang JM, Rouzier-Panis R, Reynes J, Dorr A, Kolis S, Stiles MR, Kinchelow T, Patel IH. Pharmacokinetics of plasma enfuvirtide after subcutaneous administration to patients with human immunodeficiency virus: inverse Gaussian density absorption and 2-compartment disposition. Clin Pharmacol Ther. 2002;72:10–19. doi: 10.1067/mcp.2002.125945. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Zheng BJ, Guan Y, Hez ML, Sun H, Du L, Zheng Y, Wong KL, Chen H, Chen Y, Lu L, Tanner JA, Watt RM, Niccolai N, Bernini A, Spiga O, Woo PC, Kung HF, Yuen KY, Huang JD. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir Ther. 2005;10:393–403. [PubMed] [Google Scholar]
- Zhou G, Sofiyev V, Kaur H, Snyder BA, Mankowski MK, Hogan PA, Ptak RG, Gochin M. Structure–activity relationship studies of indole-based compounds as small molecule HIV-1 fusion inhibitors targeting glycoprotein 41. J Med Chem. 2014;57:5270–5281. doi: 10.1021/jm500344y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Wu D, Snyder B, Ptak RG, Kaur H, Gochin M. Development of indole compounds as small molecule fusion inhibitors targeting HIV-1 glycoprotein-41. J Med Chem. 2011;54:7220–7231. doi: 10.1021/jm200791z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yu D, Yan H, Chong H, He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J Virol. 2020;94:e00635-20. doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]