Highlights
-
•
The Angiotensin system has been implicated in the pathogenesis of COVID-19.
-
•
Functional ACE/ACE2 polymorphisms might contribute to the outcome of COVID-19.
-
•
Severe COVID-19 was associated with hypertension, male gender, and ACE-DD genotype.
-
•
The ACE2 polymorphism was not associated with the disease outcome.
-
•
ACE2 showed no coding variants that could explain an increased risk of COVID-19.
Abbreviations: Covid-19, coronavirus disease 19; ACE, angiotensin converting enzyme; RAAS, renin-angiotensin aldosterone system; Ang, angiotensin; RFLP, restriction fragment length polymorphism; ICU, Intensive Care Unit
Keywords: COVID-19, Angiotensin converting enzyme, Gene polymorphism, Genetic association
Abstract
The Angiotensin system is implicated in the pathogenesis of COVID-19. First, ACE2 is the cellular receptor for SARS-CoV-2, and expression of the ACE2 gene could regulate the individuaĺs susceptibility to infection. In addition, the balance between ACE1 and ACE2 activity has been implicated in the pathogenesis of respiratory diseases and could play a role in the severity of COVID-19. Functional ACE1/ACE2 gene polymorphisms have been associated with the risk of cardiovascular and pulmonary diseases, and could thus also contribute to the outcome of COVID-19.
We studied 204 COVID-19 patients (137 non-severe and 67 severe-ICU cases) and 536 age-matched controls. The ACE1 insertion/deletion and ACE2 rs2285666 polymorphism were determined. Variables frequencies were compared between the groups by logistic regression. We also sequenced the ACE2 coding nucleotides in a group of patients.
Severe COVID-19 was associated with hypertension male gender (p < 0.001), hypertension (p = 0.006), hypercholesterolaemia (p = 0.046), and the ACE1-DD genotype (p = 0.049). In the multiple logistic regression hypertension (p = 0.02, OR = 2.26, 95%CI = 1.12–4.63) and male gender (p = 0.002; OR = 3.15, 95%CI = 1.56–6.66) remained as independent significant predictors of severity. The ACE2 polymorphism was not associated with the disease outcome. The ACE2 sequencing showed no coding sequence variants that could explain an increased risk of developing COVID-19.
In conclusion, an adverse outcome of COVID-19 was associated with male gender, hypertension, hypercholesterolemia and the ACE1 genotype. Our work suggested that the ACE1-I/D might influence COVID-19 severity, but the effect was dependent on the hypertensive status. This result requires further validation in other large cohorts.
1. Introduction
The SARS-CoV-2 responsible for the COVID-19 pandemic is a new coronavirus of the subgenus Sarbecovirus (Gorbalenya et al., 2020). Like SARS-CoV, the SARS-CoV-2 is an Angiotensin I converting enzyme 2 (ACE2)-tropic virus, and the “spike” (S) protein of the viral envelope would thus bind to the nasopharyngeal mucosa and alveolar pneumocytes that express ACE2 at their surface (Yan et al., 2020, Shang et al., 2020, Hoffmann et al., 2020). The clinical spectrum of this disease, termed COVID-19, ranges from mild to very severe cases (Fu et al., 2020, Rivieccio et al., 2020). It has been hypothesized that viral infection drives an exacerbated inflammatory response, leading to severe lung injury that may require ICU admission, mechanical ventilation and increases the risk of multi-organ failure and death (Jose and Manuel, 2020).
The Renin-Angiotensin-Aldosterone system (RAAS) seems to play an important role in the pathogenesis of COVID-19 (Ingraham et al., 2020). The angiotensin-converting enzyme (ACE or ACE1) catalyzes the synthesis of Angiotensin-II (Ang-II) from Ang-I, and ACE2 hydrolyzes Ang-II into Ang-1–7. Ang-II binds to the AT1-receptor driving vasoconstriction, fibrosis, inflammation, thrombosis, among other responses; while Ang-1–7 binds to the AT2-receptor with increased vasodilation and reduced fibrosis, inflammation, and thrombosis. The ACE and ACE2 are thus seen as opposite players in the balance that determines the risk of developing hypertension and cardiovascular disease. In the lung, ACE2 drives a protective response by reducing oedema, permeability and pulmonary damage (Imai et al., 2005, Bao et al., 2020, Annoni et al., 2019, Kuba et al., 2005). Of note, hypertension and cardiovascular disease are frequent comorbidities in COVID-19, and are strongly associated with the risk of hospitalization and death in individuals exposed to SARS-CoV-2 (Zhou et al., 2020, Guo et al., 2020, Shi et al., 2020, Bonow et al., 2019).
Both, acquired and inherited factors associated with differences in the expression and function of the RAAS components could explain the risk of developing COVID-19 and adverse events. For instance, ACE2 expression in the lungs markedly decreases with age and is greater in men than in women (Xie et al., 2006). This could explain the higher risk for adverse outcomes in elderly and male. In general, conditions related with a reduced ACE2 expression would increase the risk for hypertension, cardiac hypertrophy, and heart failure (Patel et al., 2012, Luo et al., 2019). In opposition, a high activity of ACE would increase the risk of lung and cardiovascular disease by increased activity of the Ang-II/AT1R axis. Common variants in the two ACE genes have been associated with the risk of hypertension, heart disease, renal failure, and pulmonary disease. In fact, the ACE insertion/deletion (I/D) is one of the best characterised human polymorphisms. Individuals with a D/D genotype showed the highest blood ACE levels, and this increased expression would explain the higher risk for cardiovascular and respiratory disease among individuals who are deletion-homozygous. This polymorphism has been related with the outcome in acute respiratory distress syndrome (ARDS) by some authors, and also with the progression of pneumonia in SARS (Marshall et al., 2002, Matsuda et al., 2012, Villar et al., 2008, Itoyama et al., 2004).
The ACE2 gene is on chromosome X and several single nucleotide polymorphisms (SNPs) have been investigated as risk factors for hypertension and heart failure, including a G to A change at nucleotide + 4 of intron 3 (SNP rs2285666) (van der Merwe et al., 2008, Zhang et al., 2018, Lu et al., 2012, Chiu et al., 2004). The fact that ACE2 is on chromosome X has been seen as a disadvantage for male carriers of alleles linked to a lower ACE2 expression, and could explain the higher prevalence of severe COVID-19 among males. SARS-CoV down-regulates myocardial ACE2 expression and this could explain the myocardial inflammation and damage and adverse cardiac outcomes in patients with SARS (Oudit et al, 2009).
Our current knowledge supports a role for the ACE/ACE2 imbalance in the pathogenesis of COVID-19. In this context, variants at these genes associated with differences in gene expression and protein function might explain the individuaĺs predisposition to manifest the disease symptoms and the risk for hospitalization and adverse events. Moreover, some authors have hypothesised that regional differences in allele frequencies could explain the different rate of the incidence and mortality (Yamamoto et al., 2020 Jul, Devaux et al., 2020, Delanghe et al., 2020, Cao et al., 2020, Hatami et al., 2020).
Our purpose was to determine whether two common functional ACE and ACE2 variants were associated with susceptibility and outcome in COVID-19.
2. Patients and methods
2.1. Study cohorts
We collected the anthropometric and clinical data of 204 patients who required hospitalization due to COVID-19 (mean age 64.77 years, range 24–95). All the study participants were Caucasian from the region of Asturias (Northern Spain, total population 1 million), and positive for SARS-Cov-2 (PCR test from nasal swabs or tracheobronchial aspirates). Severe cases (n = 67) were defined as those in need of critical care support, including high-flow oxygen, positive-pressure ventilation (either invasive or non-invasive) or vasoactive drugs. We also studied 536 healthy population controls matched with the patients for age (n = 536; mean age 70.01 years, range 50–81). The presence of comorbidities (hypertension, diabetes, hypercholesterolaemia) was obtained from the participants medical records. The study was approved by the Ethics Committee of Principado de Asturias (Oviedo, Spain). All the patients or their representatives gave their consent to participate.
2.2. Genotyping
The I/D polymorphism (rs4646994) in intron 16 of the ACE gene was genotyped by polymerase chain reaction (PCR) followed by agarose gel electrophoresis to visualise the two alleles, as reported (Alvarez et al., 1998a, Alvarez et al., 1998b, Coto et al., 2010). For the ACE2 rs2285666 A/G SNP the PCR fragments were digested with the restriction enzyme AluI and electrophoresis on agarose gels (Suppl. figures).
2.3. ACE2 sequencing
The ACE2 coding exons of 60 male patients (30 severe and 30 non-severe) were amplified with primers designated from exon flanking introns (Suppl. Table 1). These fragments were sequenced with Sanger BigDye chemistry in a capillary ABI3130xl equipment, and the sequences for each patient compared with the ACE2 reference sequence (www.ensembl.org).
2.4. Statistical analysis
All the patients and controls data were collected in an excel file and following the requirements of the Ethical Committee. The statistical analysis was performed with the R-project free software (www.r-project.org). The logistic regression (linear generalized model, LGM) was used to compare mean values and frequencies between the groups.
3. Results
Compared to age-matched controls, patients with COVID-19 did not differ in the frequency of diabetes, hypertension, and the ACE-DD genotype (Table 1 ). We found a non-significantly lower frequency of hypercholesterolemia in the patients. We compared these variables in severe COVID-19 (patients who required mechanical ventilation and/or ICU supportive care) and mild-disease patients. Male sex, hypertension, hypercholesterolemia, and the ACE-DD genotype frequencies were significantly higher in the severe group (Table 1). The multiple logistic regression-LGM showed that hypertension (p = 0.02; OR = 2.26, 95%CI = 1.12–4.63) and male gender (p = 0.002; OR = 3.15, 95%CI = 1.56–6.66) remained as independent significant predictors of severity.
Table 1.
Main clinical values and genotype frequencies in total patients and controls.
| Severe Disease N = 67 |
MildDisease N = 137 |
p | All COVID N = 204 |
Controls N = 536 |
p | |
|---|---|---|---|---|---|---|
| Mean age | 65.76 | 64.56 | 0.53 | 64.96 | 69.88 | AV |
| Male | 53 (79%) | 72 (53%) | <0.001 | 125 (61%) | 248 (46%) | AV |
| Hypertensives | 41 (61%) | 56 (41%) | 0.006 | 97 (48%) | 228 (43%) | 0.22 |
| Diabetes | 15 (22%) | 21 (15%) | 0.21 | 36 (18%) | 123 (23%) | 0.13 |
| Hyperchols | 29 (43%) | 40 (29%) | 0.046 | 69 (34%) | 220 (41%) | 0.07 |
| ACE I/D | ||||||
| DD | 31 (46%) | 44 (32%) | 0.049* | 75 (37%) | 195 (36%) | 0.92* |
| ID | 31 (46%) | 76 (56%) | 107 (52%) | 256 (48%) | ||
| II | 5 (8%) | 17 (12%) | 22 (11%) | 85 (16%) | ||
| ACE2# | ||||||
| A-carriers | 52 (78%) | 94 (69%) | 0.18 | 146 (72%) | 374 (75%) | 0.33 |
| G | 15 (22%) | 43 (31%) | 58 (28%) | 124 (25%) |
AV: adjust variable, #498 controls genotyped, A-carriers = male A and female AA + AG.
* DD vs ID + II.
We then compared the values in men and women. In men, hypertension, hypercholesterolemia and the ACE-DD were significantly increased in severe patients (Table 2 ). The ACE2 rs2285666 alleles did not differ between the two patients groups and were non-significantly higher to the control frequencies. The same analysis performed among female patients revealed no differences between the severe and mild cases, although we observed a trend toward a higher risk for severity among hypertension and hypercholesterolaemia, and a non-significantly higher frequency of the A-allele in the two patients groups compared to controls (Suppl. Table 2). The results in this female cohort would be limited by the reduced size of the severe cases (n = 14).
Table 2.
Values in the male patients and controls.
| Male | ||||||
|---|---|---|---|---|---|---|
| Severe N = 53 |
Mild N = 72 |
p | Covid N = 125 |
Control N = 248 |
p | |
| Mean age | 67.19 | 63.89 | 0.13 | 64.77 | 70.01 | AV |
| Hypertensives | 34 (64%) | 26 (36%) | 0.002 | 60 (48%) | 97 (39%) | 0.10 |
| Diabetics | 12 (23%) | 12 (17%) | 0.42 | 24 (19%) | 47 (19%) | 1 |
| Hiperlchols | 25 (47%) | 18 (25%) | 0.01 | 43 (34%) | 82 (33%) | 0.79 |
| ACE2# | ||||||
| G | 43 (81%) | 58 (81%) | 0.92 | 101 (81%) | 200 (85%) | 0.25 |
| A | 10 (19%) | 14 (19%) | 24 (19%) | 34 (15%) | ||
| ACE I/D | ||||||
| DD | 28 (53%) | 25 (35%) | 0.043* | 53 (42%) | 85 (34%) | 0.13 |
| ID | 23 (43%) | 43 (60%) | 66 (53%) | 123 (50%) | ||
| II | 2 (4%) | 4 (5%) | 6 (5%) | 40 (16%) | ||
AV: adjust variable, #234 male controls genotyped.
* DD vs ID + II.
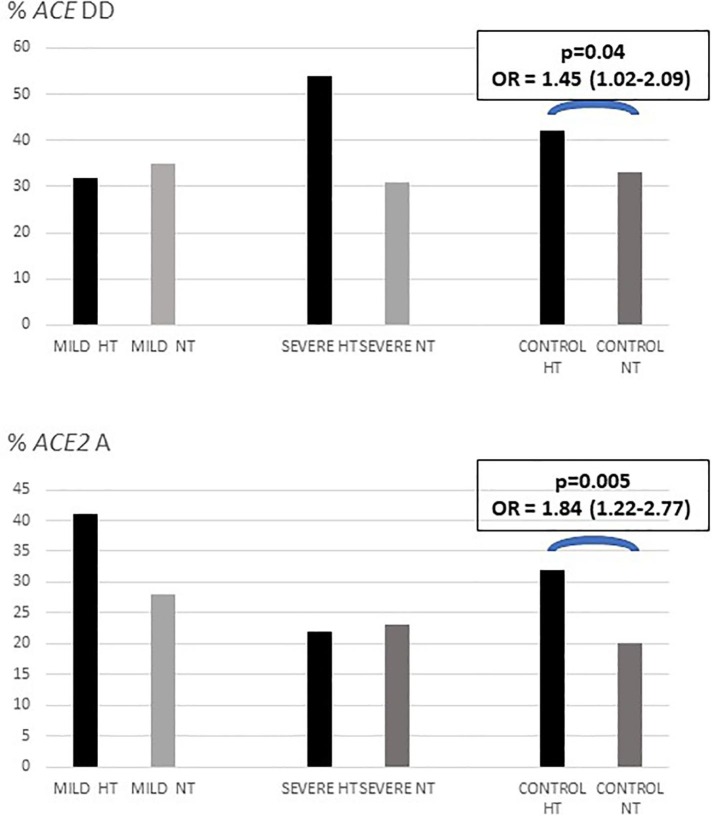
To better understand the relationship between the two ACE polymorphisms and hypertension, we compared the genotype frequencies between hypertensives and normotensives in male and female patients and controls (Fig. 1 ; Suppl. Table 3). In males, the ACE-DD genotype had a higher frequency among hypertensives in the three groups, without statistically significant differences. The ACE2-A allele was significantly increased in the hypertensive controls (21% vs. 10%; p = 0.02), with no significant differences between severe and non-severe COVID-19 cases. In the female cohort, the DD frequencies did not differ between hypertensive and normotensive in the three groups. The rs2285666 A allele was significantly more frequent among the hypertensive controls (p = 0.04). These results suggested that the ACE-DD genotype and ACE2-A carriers could be at a higher risk for hypertension in our population, and genotype and allele differences between the severe and non-severe COVID-19 cases could be attribute to their association with hypertension.
Fig. 1.
Frequencies of ACE-DD and ACE2 rs2285666 A-carriers according to hypertension in COVID-19 patients and population controls. HT = hypertensives, NT = normotensives.
We sequenced the ACE2 coding and immediate intronic flanking regions in a group of patients, and the only variant identified was rs2285666. This suggested a very low genetic variability in the SARS-CoV-2 receptor, and a significant absence of common variants that could increase the risk for infection in our population.
4. Discussion
Since the outbreak of the COVID-19 pandemic several authors have speculated about the role of the ACE and ACE2 gene polymorphisms in disease susceptibility and severity. Because ACE2 is the SARS-CoV-2 receptor, a functional variant that increased gene expression could be associated with a higher number of membrane-bound viral binding sites, increasing the vulnerability of carriers to infection. These risk variants might be particularly adverse in males, who carry only one copy of X-linked ACE2 gene. The ACE2 rs2285666 SNP was in the intronic-consensus splicing nucleotides and could thus affect the processing of ACE2 total RNA to mRNA and, eventually, the amount of the protein. At least one study has reported higher levels of circulating ACE2 levels in men than in women (Sama et al., 2020). One study has investigated the effect of rs2285666 on serum ACE2 levels, and found significantly higher levels in A-carriers compared to G-homozygotes (Wu et al., 2017). A different intronic SNP in strong linkage disequilibrium with rs2285666 (rs879922) has been associated with ACE2 gene expression (Zhang et al., 2018). We thus hypothesised that these functional ACE2 variants could modify the disease outcome. In our study, the A-allele frequency was non-significantly higher in patients vs. controls but was associated with hypertension in both, male and female controls. Lower ACE2 levels should be harmful for patients with lung disease, and the frequency observed in COVID-19 patients could thus be the balance between the negative association with viral infection (lower expression in the airway epithelia) and a positive association with respiratory and cardiovascular disease (lower expression in lung and other organs).
The ACE2 A allele has frequencies of 0.15 and 0.19 in our elderly controls. According to the human gene variation databases, this frequency is lower that the reported among unselected Caucasians (0.20–0.25) including the Spanish population (0.24). This lower frequency could be characteristic of our population, but might also reflect a reduction of the lifespan for rs2285666 A carriers.
Functional variants in receptors for other viruses confer resistance to infection. One the best characterised is CCR5, the cellular receptor for HIV. A common variant that determines the absence of the receptor (CCR5-Δ32) confers a complete resistance to HIV-infection among homozygotes, while reduces the disease progression in heterozygotes. Approximately 1% of the Caucasians are CCR5-Δ32 homozygotes and they have a non-significant reduction in lifespan (Alvarez et al., 1998a, Alvarez et al., 1998b). On the contrary, ACE2 pathogenic variants are very rare at a population scale, and the complete absence of the receptor would be incompatible with life in humans. Moreover, according to the human genome variation databases, there are no common missense changes in the coding ACE2 sequence. At a minor allele frequency > 0.01% only four missense changes have been reported (all with global frequencies < 1%), and rs2285666 was the only variant that could affect splicing (Suppl. Table 4). We sequenced the exon and intron-flanking sequence in 60 patients and rs2285666 was the only identified variant. Therefore, it is unlikely that the ACE2 coding variants have a significant effect on susceptibility to SARS-CoV-2 infection. Of course, this does not exclude that variants in other gene regions are related with gene expression and the amount of protein.
ACE2 expression is regulated by Ang-II through the AT1R-pathway (Koka et al., 2008, Xiao et al., 2013). ACE could thus modulate the ACE2 expression and activity through the regulation of Ang-II levels. In this scenario the ACE2 gene might be down-regulated in ACE1-DD homozygotes, who have increased ACE activity. There are two mechanisms by which functional ACE variants could modulate the risk of developing and the clinical outcome of SARS-CoV-2 infection. The reduction of ACE2 expression could protect against viral infection but would also reduce the beneficial effect of ACE2 in the lung and other organs. At the same time, ACE would enhance the deleterious Ang-II/AT1R response. The ACE-DD has been associated with and increased risk of respiratory distress system by some authors, but not confirmed by others (Matsuda et al., 2012, Villar et al., 2008, Itoyama et al., 2004). This polymorphism was investigated in 44 Vietnamese SARS cases and 103 healthy controls who had been exposed to SARS-CoV and 50 controls without contact with SARS-patients (non-exposed) (Itoyama et al., 2004). There were no significant differences for DD-frequency among the groups, suggesting that this polymorphism has no effect on the risk for SARS-CoV infection. However, the frequency of the D allele was significantly higher in hypoxemic than in the non-hypoxemic patients, and could thus contribute to the progression of pneumonia in SARS. In our study we did not find differences for DD-frequencies between COVID-19 and controls, thus confirming the lack of association with the risk of developing COVID-19 symptoms. However, we did not study individuals exposed who remained asymptomatic and we could thus not exclude an effect in the resistance to viral infection. We found a significant higher risk for a severe form of COVID-19 in males. This association was not found among females, although the number of severe-disease women was very low (n = 14). We confirmed that this genotype was associated with the risk for hypertension in our male controls, with a trend for association with hypertension in the patients. Thus, we concluded that the deleterious effect of the ACE polymorphism on COVID-19 outcome was likely due to its association with hypertension.
Our study has several limitations, mainly the reduced sample size of the patients and of female severe cases in particular. This limits the statistical interpretation of the significant and non-significant associations. Also, we did not study subjects exposed to the virus who did not show disease symptoms. These individuals would be resistant to SARS-CoV-2 infection and are crucial for the identification of gene variants associated with disease susceptibility.
In conclusion, our study suggested that the ACE-ID polymorphism was associated with the risk of developing severe COVID-19 depending on the hypertension status. The ACE2 rs2285666 variant was associated with hypertension in our elderly population, without significant difference between mild and severe COVID-19 patients.
Contributorship. All the authors contributed to this work by recruiting the patients and performing the genetic and statistical analysis. JG, GMA and EC wrote the ms. All the authors approved the submission of this ms.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by a grant from the Spanish Plan Nacional de I+D+I Ministerio de Economía y Competitividad and the European FEDER, grant ISCIII-Red de Investigación Renal-REDINREN RD16/9/5 (EC).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2020.145102.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alvarez V., López-Larrea C., Coto E. Mutational analysis of the CCR5 and CXCR4 genes (HIV-1 co-receptors) in resistance to HIV-1 infection and AIDS development among intravenous drug users. Hum. Genet. 1998;102:483–486. doi: 10.1007/s004390050726. [DOI] [PubMed] [Google Scholar]
- Alvarez R., Reguero J.R., Batalla A. Angiotensin-converting enzyme and angiotensin II receptor 1 polymorphisms: association with early coronary disease. Cardiovasc. Res. 1998;40:375–379. doi: 10.1016/s0008-6363(98)00179-5. [DOI] [PubMed] [Google Scholar]
- Annoni F., Orbegozo D., Rahmania L. Angiotensin-converting enzymes in acute respiratory distress syndrome. Intensive Care Med. 2019;45:1159–1160. doi: 10.1007/s00134-019-05600-6. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Bonow R.O., Fonarow G.C., O’Gara P.T., Yancy C.W. Association of coronavirus disease 2019 (COVID-19) with myocardial injury and mortality. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1105. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6 doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R.W.K., Tang N.L.S., Hui D.S.C. ACE2 gene polymorphisms do not affect outcome of severe acute respiratory syndrome. Clin. Chem. 2004;50:1683–1686. doi: 10.1373/clinchem.2004.035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coto E., Palacín M., Martín M. Functional polymorphisms in genes of the Angiotensin and Serotonin systems and risk of hypertrophic cardiomyopathy: AT1R as a potential modifier. J. Transl. Med. 2010;8 doi: 10.1186/1479-5876-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanghe J.R., Speeckaert M.M., De Buyzere M.L. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. 2020;58:1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Wang B., Yuan T., Chen X. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: a systematic review and meta-analysis. J. Infect. 2020;80:656–665. doi: 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Fan Y., Chen M. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID- 19) JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1017. epub ahead of press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatami N., Ahi S., Sadeghinikoo A. Worlwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: an ecological meta-regression. Endocrine. 2020 doi: 10.1007/s12020-020-02381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(271–280) doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham NE, Barakat AG, Reilkoff R, et al. Understanding the renin-angiotensin-aldosterone-SARS-CoV-axis: a comprehensive review. EurResp J. 2000912. [DOI] [PMC free article] [PubMed]
- ItoyamaS K.N., Quy T. ACE1 polymorphism and progression of SARS. Biochem Biophys Res Commun. 2004;323:1124–1129. doi: 10.1016/j.bbrc.2004.08.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose RJ, Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020; S2213-2600: 30216-2. [DOI] [PMC free article] [PubMed]
- Koka V., Huang X.R., Chung A.C.K. Angiotensin II up-regulates angiotensin I-converting enzyme (ACE), but down-regulates ACE2 via the AT1-ERK/p38 MAP kinase pathway. Am. J. Pathol. 2008;172:1174–1183. doi: 10.2353/ajpath.2008.070762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Yang Y., Wang Y. ACE2 gene polymorphism and essential hypertension: an updated meta-analysis involving 11,051 subjects. Mol. Biol. Rep. 2012;39:6581–6589. doi: 10.1007/s11033-012-1487-1. [DOI] [PubMed] [Google Scholar]
- Luo Y., Liu C., Guan T. Association of ACE2 genetic polymorphisms with hypertension-related target organ damages in south Xinjiang. Hypertens. Res. 2019;42:681–689. doi: 10.1038/s41440-018-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R.P., Webb S., Bellingan G.J. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2002;166:646–650. doi: 10.1164/rccm.2108086. [DOI] [PubMed] [Google Scholar]
- Matsuda A., Kishi T., Jacob A. Association between insertion/deletion polymorphism in angiotensin-converting enzyme gene and acute lung injury/acute respiratory distress syndrome: a meta-analysis. BMC Med. Genet. 2012;13(76) doi: 10.1186/1471-2350-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit G.Y., Kassiri Z., Jiang C. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V.B., Bodiga S., Fan D., Das S.K., Wang Z., Wang W., Basu R., Zhong J., Kassiri Z., Oudit G.Y. Cardioprotective effects mediated by angiotensin II type 1 receptor blockade and enhancing angiotensin 1–7 in experimental heart failure in angiotensin-converting enzyme 2-null mice. Hypertension. 2012;59:1195–1203. doi: 10.1161/HYPERTENSIONAHA.112.191650. [DOI] [PubMed] [Google Scholar]
- Rivieccio B.A., Luconi E., Boracchi P. Heterogeneity of COVID-19 outbreak in Italy. Acta Biomed. 2020;91:31–34. doi: 10.23750/abm.v91i2.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama I.E., Ravera A., Santema B.T. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur. Heart J. 2020;41:1810–1817. doi: 10.1093/eurheartj/ehaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Qin M., Shen B. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.0950. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe L., Cloete R., Revera M. Genetic variation in angiotensin-converting enzyme 2 gene is associated with extent of left ventricular hypertrophy in hypertrophic cardiomyopathy. Hum. Genet. 2008;124:57–61. doi: 10.1007/s00439-008-0524-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar J., Flores C., Pérez-Méndez L. Angiotensin-converting enzyme insertion/deletion polymorphism is not associated with susceptibility and outcome in sepsis and acute respiratory distress syndrome. Intensive Care Med. 2008;34:488–495. doi: 10.1007/s00134-007-0937-z. [DOI] [PubMed] [Google Scholar]
- Wu Y.H., Li J.Y., Wang C., Zhang L.M., Qiao H. The ACE2 G8790A polymorphism: involvement in type 2 diabetes mellitus combined with cerebral stroke. J. Clin. Lab. Anal. 2017;31 doi: 10.1002/jcla.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Haack K.K.V., Zucker I.H. Angiotensin II regulates ACE and ACE2 in neurons through p38 mitogen-activated protein kinase and extracellular signal-regulated kinase 1/2 signaling. Am. J. Physiol. Cell Physiol. 2013;304:C1073–C1079. doi: 10.1152/ajpcell.00364.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Chen J., Wang X., Zhang F., Liu Y. Age- and gender-related difference of ACE2 expression in rat lung. Life Sci. 2006;78:2166–2171. doi: 10.1016/j.lfs.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Ariumi Y., Nishida N., Yamamoto R., Bauer G., Gojobori T., Shimotohno K., Mizokami M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;3 doi: 10.1016/j.gene.2020.144944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cong M., Wang N. Association of angiotensin-converting enzyme 2 gene polymorphism and enzymatic activity with essential hypertension in different gender: a case–control study. Medicine (Baltimore) 2018;97 doi: 10.1097/MD.0000000000012917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.