Abstract
Introduction
Esophageal squamous cell carcinoma (ESCC) is a malignant tumor disease with high mortality and morbidity rates, especially for a terminal cancer. At present, the prognosis and treatment of ESCC cannot effectively control or inhibit the spread and proliferation of tumor cells. microRNAs, a class of small spliced RNAs, are essential in the regulation of tumorigenesis and tumor cell migration and proliferation. microRNAs interact with target mRNA to silence gene expression and degrade mRNA, thereby inhibiting the expression of tumor genes or impairing the expression of tumor suppressor genes.
Methods
A total of 20 human ESCC samples were collected from the Affiliated Tumor Hospital of Xinjiang Medical University. Eca109 and Kyse510 cells, which are ESCC cell lines, were subjected to FACS analysis to get side population (SP) cells and non-SP cells. Cell cycle and cell proliferation were analyzed by flow cytometry. Cell migration and invasion were detected using a transwell assay. Quantitative PCR and Western blot were performed to analyze the expression levels of ABCG2, KLF4, OCT4, and ACVR1, which are related to the stemness of stem cells. The target genes of hsa-miR-148a were predicted using TargetScan (version 7.2) and verified by a dual luciferase reporter assay. A chromatin immunoprecipitation (ChIP) assay was carried out to demonstrate direct interaction between miR-148a and ACVR1.
Results
The expression of miR-148a was significantly down-regulated in ESCC cells and significantly decreased in SP esophageal squamous cells when compared to the tumor cells. By analyzing the stem cell stemness of ESCC, overexpression of miR-148a decreased the expression of ABCG2, KLF4, SOX2, OCT4, and Nanog, indicating that miR-148a may regulate stem cell function. Target gene prediction and functional annotation of miR-148a suggested that miR-148a is involved in stem cell stemness of ESCC via ACVR1. Expression of the dual luciferase-labeled gene indicates that overexpression of miR-148a inhibits the expression of ACVR1, thereby affecting stem cell stemness.
Conclusion
miR-148a regulates the stem cell-like side populations distribution by inhibiting the expression of ACVR1 in ESCC. miR-148a may be a promising targeted therapy for ESCC.
Keywords: miR-148a, ESCC, ACVR1, cancer stem cells
Introduction
Esophageal carcinoma (EC) is a common malignant tumor of the digestive tract that seriously endangers human health. The morbidity and mortality of EC in China are the third and fourth highest for malignant tumors, respectively.1 Esophageal squamous carcinoma cell (ESCC) is one of the common forms of esophageal cancer. It mainly occurs in East Asia, especially in northern China due to long-term exposure to carcinogens in this area and because patients there are usually at a low-level nutritional status.2 Tumor development of ESCC is a complex process and is induced by a variety of factors. The abuse of alcohol and cigarettes has been reported as an important factor in promoting the development of ESCC in different countries.3–5 Genetic variation, including somatic mutations and chromosomal aberrations, can also cause tumor production in ESCC.6,7
The survival rate of patients within five years is only 5–25% when the patient is in the late stage of ESCC.8 Surgical removal of the tumor is a traditional treatment for EC. However, the anatomical location of EC makes it difficult to find early, and the invasiveness of EC is high, which leads to unsatisfactory treatment effects of surgery. Although general progress has been made in chemotherapy, molecular targeted therapy, and tumor immunotherapy in recent years, the current treatment plan has not significantly improved the prognosis of patients with EC and the 5-year survival rate is only 15–40%.9 In addition, EC easily relapses after surgery, with a local-regional recurrence rate that can be as high as 40–60%, and the retreatment effect after relapse is very poor, with a 3-year survival rate of only about 12%.10 Although diagnosis of ESCC patients and multi-directional treatment have been established, the prognosis of ESCC is still unsatisfactory, due to the low survival rate at later stages.11 Therefore, molecular mechanism and pathway research of ESCC are necessary to discover new biomarkers and targeted therapeutic drugs.
MicroRNAs (miRNAs) are a class of short-chain non-coding RNAs with 17–25 nucleic acids.12 miRNA genes are frequently located in the introns of protein-coding genes. Therefore, their expression is usually regulated by the host gene.13 The 5ʹ seed region of mature miRNAs recognizes the 3ʹ UTR region of the target mRNAs and causes translational silencing, or degradation of mRNA, by base complementation.14,15 miRNAs are also key regulators of gene transcription, and are frequently involved in the occurrence of cancers.16,17 The dysregulation of miRNA expression is very common in tumor cells, which has made miRNAs as oncogenes or tumor suppressor genes a major focus of research.18–20 Recently, some miRNAs have been found to inhibit development of ESCC by silencing tumor proliferation and expression of migration-related genes. However, there are also some miRNAs that are carcinogenic to ESCC. These miRNAs mainly inhibit the expression of important tumor suppressor genes in cancer cells, such as PTEN or SOX6.21,22 Ishibashi et al reported that miR-141-3p suppressed the expression of PHLPP2, a negative regulator of the AKT/PI3K pathway, as a target in ESCC.23
miR-148a has been reported as a tumor suppressor in multiple cancer types, including gastrointestinal, breast, ovarian, colorectal, and prostate cancer.24–27 Some important genes which involved in the process of cell proliferation, cell apoptosis, and cell migration and invasion, are targeted by miR-148a, including CCKBR, ROCK1, DNMT1, SMAD2, MMP7, BCL-2, HPIP, MET, USP4, CDC2B, ERBB3, IGF-IR, IRS1, HOTAIR, HLA-G, CAND1, MSK1, WNT1 and so on.28 Hummel et al16 reported that miR-148a was downregulated in patients with recurrent EC. They also demonstrated that in EC cell lines, upregulation of miR-148a improved sensitivity to chemotherapy. However, whether miR-148a is downregulated in primary ESCC remains unclear.
Tumor stem cells are a small group of cells with self-renewal capacity that produce malignant tumor cell progeny with extreme expansion and migration capabilities.29 Targeted therapeutic inhibition of cancer stem cells is considered a potential method to abolish the survival of tumor cells, especially in patients with ESCC, because their disease cannot be cured through surgery or chemotherapy combined with radiotherapy.30 Side population (SP) cells of ESCC have been identified as a type of tumor stem cell-like cell.31 Further research is needed to discover new biomarkers and targets for better prognosis of ESCC.
The main function of ACVR1, named activin receptor type-1, is activin binding and receptor activity. It can form a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. ACVR1 is an important activated receptor of BMP and is involved in the regulation of the BMP/Wnt signaling pathway which is very important for stem cells.32
In this study, we performed a molecular microarray of miRNA expression using the cancer genome atlas (TCGA) database and found that miR-148a is significantly associated with prognosis and disease progression in patients with ESCC. Further analysis showed that miR-148a inhibited proliferation and migration of the esophageal squamous carcinoma cell lines Eca109 and Kyse510. At the same time, functional verification was conducted on important candidate genes involved in cell cycle and differentiation. RT-PCR was used to detect the expression of miR-148a in ESCC stem cells in Eca109 and Kyse510 cell lines. The expression of ACVR1 was significantly increased in stem cells of ESCC. miR-148a may inhibits the proliferation of ESCC SP stem cells and regulates the stem cell-like side populations distribution cells by inhibiting the expression of ACVR1.
Materials and Methods
ESCC Cell Line and Case Samples
Eca109, a cell line representing Chinese ESCC, was purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences. The cell line Kyse510 representing ESCC from Japan was purchased from the Wuhan Cell Bank (CCTCC). Both cell lines were cultured in RPMI1640 medium containing 10% fetal bovine serum at 37 °C with 5% CO2. Human ESCC samples were collected and rapidly frozen in liquid nitrogen, and stored at −80 °C. All ESCC tissue samples were collected during the patient’s surgery. Prior patient consent and approval from The Affiliated Tumor Hospital of Xinjiang Medical University Ethics Committee were obtained for the use of these clinical materials for research purposes. Patient consent was written informed consent, and that this was conducted in accordance with the Declaration of Helsinki. Clinical information on the samples is summarized in Table 1. Clinical pathological tumor node-metastasis (TNM) staging was determined by the extent of tumor invasion in the esophageal wall and lymphatic and venous invasion status according to the criteria proposed by the Union for International Cancer Control (UICC) criteria.33
Table 1.
Clinical Information Summary on the ESCC Samples
| Factor | No. | No. |
|---|---|---|
| Gender | ||
| Male | 34 | 28 |
| Female | 16 | 22 |
| Age(year) | ||
| ≤60 | 38 | 33 |
| >60 | 12 | 17 |
| Clinical | ||
| I | 11 | 11 |
| II | 17 | 17 |
| III | 18 | 17 |
| IV | 4 | 5 |
| T classification | ||
| T1 | 13 | 6 |
| T2 | 15 | 12 |
| T3 | 19 | 23 |
| T4 | 3 | 9 |
| N classification | ||
| N0 | 29 | 19 |
| N1 | 21 | 31 |
| M classification | ||
| M0 | 48 | 41 |
| M1 | 2 | 9 |
| Histological differentiation | ||
| Well | 21 | 7 |
| Moderate | 20 | 19 |
| Poor | 9 | 24 |
| Vital status | ||
| Alive | 41 | 28 |
| Dead | 9 | 22 |
Cell Transfection
Cell transfection was performed for the use of Lipofectamine 2000 (Invitrogen). miRNA-148a mimic (miR-148am) and its negative control (NC) and miRNA-148a inhibitor (miR-148ai) and its negative control (iNC) plasmid (Guangzhou Ruibo, Guangzhou, China) were transferred into 80% of the EC cell lines Eca109 and Kyse510. Twelve hours after transfection, the cells were replaced with fresh cell culture medium, cell culture continued for 24 hours for RNA extraction, or 48 hours for protein extraction.
Establishment of ACVR1 Overexpression Eca109 Cells
Lentiviral expression vector pLVX-IRES-ZsGreen1 and pLL3.7 with GFP reporter and the package vectors were obtained from Dr. Jun Xu (Tongji University, Shanghai, China). The human ACVR1 cDNA was synthesized from the total RNA of the 293T cell line, which were purchased from Cell Resource Center, IBMS, CAMS/PUMC. The ACVR1 coding region with flag tag sequence was cloned into the vector to create the pLVX- ACVR1-IRES-ZsGreen1 vector. We named the ACVR1 overexpression and control cell as ACVR1 OE and control OE cells.
FACS ESCC Sorting
Eca109 and Kyse 510 cells were subjected to FACS analysis during the logarithmic growth phase. The cells were digested with 0.025% trypsin and 1 mM EDTA and rinsed in PBS. The cells were then resuspended in DMEM (with 2% FBS added) at a concentration of 1×106 cells/mL and incubated for 10 minutes at 37 °C, 5% CO2. Thereafter, the DNA-binding dye Hoechst 33,342 (Sigma) was added to the cells and they were incubated for 90 minutes under dark conditions. After incubation, the cells were washed with cold PBS and filtered through a 40 μm cell filter to obtain a single-cell suspension. After completion of FACS (FACSDiva Option; Becton Dickinson) cell analysis and sorting, Hoechst 33,342 was excited by 350 nm UV rays. 405/BP209 (Hoechst blue) and 570/BP20 (Hoechst red) fluorescence were measured using corresponding filters. Hoechst 33,342 negative cells, ie, side population (SP) cells, and Hoechst 33,342 positive cells (non-SP cells) were finally sorted into two cell subpopulations, which was used to analyze the proportion of the SP groups and verify the expression of important candidate genes.
CD90+ Expression Analysis via Flow Cytometry
Flow cytometric cell sorting was conducted using phycoerythrin (PE)-conjugated monoclonal mouse anti-human CD90 (Miltenyi Biotec GmbH, Germany). Samples were analyzed and sorted on BD FACS Canto II and FACS Aria I, respectively (BD Biosciences, USA) with data analyzed using FlowJo software (Tree Star Inc, USA).
Cell Cycle Analysis via Flow Cytometry
Cells were transfected with 10 mM miRNA-148a mimics (NC) or miRNA-148a inhibitor (iNC), and collected 48 hours after transfection. The cells were fixed with 80% cold ethanol, and the fixed cells were washed twice with PBS, then incubated in 40 μL of phosphoric acid-citrate buffer for about 30 min, washed again, and then DNA was stained with 10 μg/mL propidium iodide (PI) and 0.1% RNaseA (Sigma) for 20 min at room temperature. Measurements were performed using a FACSCalibur flow cytometer (Becton Dickinson, Co.). After excitation using a 488 nm laser, 6000 cells were obtained from each sample, and then a DNA histogram was made using CellQuest software (BD Biosciences). The fluorescence intensity of the cells measured by CellQuest was analyzed by Modfit software (Becton Dickinson), and the percentage of cells at the G1, S, and G2/M phases was determined according to the fluorescence intensity.
Cell Proliferation Analysis by Flow Cytometry
Cells were transfected with 10 mM miRNA-148a mimics (NC) or miRNA-148a inhibitor (iNC), and collected 48 hours after transfection. The cells were incubated with BrdU for 1 hour. The cultured cells were suspended in 50 μL of staining buffer, and then 100 μL of diluted DNase (300 μg/mL) was added to each tube, cells were incubated at 37 °C for 1 h, washed once, and resuspended in 50 μL of P-wash buffer to add specific cells. Immunofluorescence monoclonal antibody (BrdU monoclonal antibody 1 μL) was protected from light for 20 min at room temperature, washed once, and resuspended in 400 μL of staining buffer. The results were analyzed by flow cytometry. The flow cytometer FACSCalibur was used for detection. The excitation light was an argon ion laser 488 nm line, cells were collected by CellQuest, and the flow results were analyzed using CellQuest software (BD Biosciences).
Wound Healing and Transwell Assays
The scratch test was conducted as previously described34 and the wound size of the scratch test was observed and photographed using a microscope. Cell migration experiments were performed using a Costar chamber migration tube containing an 8 μm pore size transwell insert (Corning Incorporated, USA). The upper part of the transfer tube was covered with Matrigel (Invitrogen, USA). The migrated cells were fixed with methanol, stained with 0.1% crystal violet, and counted in five different random regions using microscopy. The 50μm scale labeled in the results pictures means 200 times magnification. Scratch experiments and cell migration experiments were performed in three effective replicates.
Western Blot
Cellular and tumor tissues proteins were extracted using RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% SDS, 0.25% Na deoxycholate, 2% Triton-X100, 1 mM PMSF, and 2μμM leupeptin). Protein concentration was determined using a Bradford protein assay. Each sample was loaded with 25 μg of protein and subjected to electrophoresis in an SDS polyacrylamide gel at 200 V for 2 hours. The separated protein was transferred to a polyvinylidene fluoride membrane. The target protein transfer efficiency was determined by Ponceau S staining. The separated protein was incubated in 5% milk for 30 minutes and incubated overnight at 4 °C. The primary antibody information is as follows: mouse anti-human ABCG2 (Santa Cruz biotechnology, sc-58,222), KLF4 (Abcam, ab75486), SOX2 (Santa Cruz Biotechnology, sc-365,823), OCT4 (Santa Cruz Biotechnology, sc-101,534), Nanog (Santa Cruz Biotechnology, sc-293,121), ACVR1 (Santa Cruz Biotechnology, sc-374,523), and β-actin (Santa Cruz Biotechnology, sc-47,778) monoclonal antibodies. Horseradish peroxidase (HRP) goat anti-mouse secondary antibody (Cell Signaling Technology, Danvers, MA, USA) was incubated for 2 hours at room temperature. X-ray film was exposed in a dark room by adding 1 mL of Supersignal West Pico Plus Chemiluminescent substrate to the membrane.
RNA Extraction and Quantitative PCR
Total RNA was extracted using TRIzol© reagent (Invitrogen). RNA quality and concentration were determined using a Nanodrop 2000 (Thermo Scientific). cDNA was synthesized according to the instructions of the High-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) and used as a template for subsequent quantitative PCR (RT-PCR). RT-PCR primers were designed by Primer3 Plus online software (https://primer3plus.com). The qRT-PCR primer sequences are shown in Table 2. The conditions of qRT-PCR were as follows: predenaturation at 95 °C for 10 minutes, denaturation at 95 °C for 15 seconds, annealing at 60 °C for 60 seconds, and a final extension at 72 °C for 40 seconds for a total of 40 cycles. GAPDH was used as an internal reference gene to calculate gene expression. The fold change was calculated by relative quantification (2−ΔΔCt). Each experiment was performed three times. For the PCR detection of miR-148a, the cell miRNA was first extracted using the Tiangen miRcute miRNA extraction and isolation kit (centrifugal column type, catalog number: DP501). Then, the RT reaction based on the stem-loop structure primers and the real-time fluorescent quantitative PCR reaction were completed using the Bulge-LoopTM miRNA qRT-PCR. The PCR program was: 95 °C for 15 min; 95 °C for 15 s, 60 °C for 60 s (40 cycles). The Bulge-Loop h-hsa-mir-148a primer set (MQPS0000384-1-100) was purchased from RiboBio Company https://www.ribobio.com/.
Table 2.
qRT-PCR Primer Sequences
| Name | Sequences (5ʹ-3ʹ) | |
|---|---|---|
| ABCG2 | Forward | TGGTGTTTCCTTGTGACACTG |
| Reverse | TGAGCCTTTGGTTAAGACCG | |
| KLF4 | Forward | GTCA GTTCA TCTGAGCG GG |
| Reverse | AGAGTTCCCATCTCAAGGCA | |
| SOX2 | Forward | GCTTAGCCTC GTCGATGAAC |
| Reverse | AACCCCAAGATGCACAACTC | |
| OCT4 | Forward | GGGAGATTGATAACTGGTGTGTT |
| Reverse | GTGTATATCCCAGGGTGATCCTC | |
| Nanog | Forward | TTTGTGGGCCTGAAGAAAACT |
| Reverse | AGGGCTGTCCTGAATAAGCAG | |
| GAPDH | Forward | ACAACTTTGGTATCGTGGAAGG |
| Reverse | GCCATCACGCCACAGTTTC | |
| ARL8B | Forward | CATCGCGTCAGGTCAATTCAG |
| Reverse | GTTGTCCTCCTATGTCCCAGA | |
| CAND1 | Forward | AGCGCCTCGTACCACATTTC |
| Reverse | TGCAGTTCCGTCATCAAATCA | |
| CDK19 | Forward | GGATTTGTTTGAGTACGAAGGGT |
| Reverse | CTACAAGCCGACATGGATATTCC | |
| ACVR1 | Forward | TGCCAAGGGGACTGGTGTAA |
| Reverse | ACTGCGAACACTACAGAGAGAA | |
| DNMT1 | Forward | AGGCGGCTCAAAGATTTGGAA |
| Reverse | GCAGAAATTCGTGCAAGAGATTC | |
| FXR1 | Forward | GAGAGAAGATTTAATGGGCCTGG |
| Reverse | GCTCAATGGCGGTAACTCCA | |
| GADD45A | Forward | GAGAGCAGAAGACCGAAAGGA |
| Reverse | CACAACACCACGTTATCGGG | |
| MYT1 | Forward | GAAGGAGCCCCGTCAAGTC |
| Reverse | AGAAGTTGCGATGATTCCCTG | |
| SIK1 | Forward | CTCCGGGTGGGTTTTTACGAC |
| Reverse | CTGCGTTTTGGTGACTCGATG | |
| SIX4 | Forward | AGCAGCTCTGGTACAAGGC |
| Reverse | CTTGAAACAATACACCGTCTCCT | |
Tumor Sphere Formation Assays
2500 Eca109 and Kyse510 sorted SP cells were seeded in 6-well plates by transfection of miRNA-148a mimics or miRNA-148a inhibitor and cultured in RPMI-1640 medium supplemented with 10% FBS. Four μg/mL heparin (Sigma), 1:50 B27 (Gibco), 20 ng/mL epidermal growth factor EGF, 20 ng/mL human basic fibroblast growth factor bFGF (Sigma-Aldrich), 100 IU/mL penicillin, and 100 μg/mL streptomycin were added to the medium, and cells were cultured for 10 days. The number of cell populations was counted using a low magnification microscope (>50 μm). A population with more than 10 cells was counted as a single cell population.
In vivo Tumorigenesis in Nude Mice
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Affiliated Tumor Hospital of Xinjiang Medical University. The experimental protocol was established according to the associated national guidelines from Ministry of Science and Technology of China. The effects of miR-148a on the in vivo tumorigenic ability was investigated by tumor xenograft experiment. A total of 1 × 106 Eca109 cells with different treatment (WT, mimic, mimic+control OE and mimic+ACVR1 OE cells) in 0.2 mL RPMI 1640 medium was subcutaneously injected into the dorsal flanks of 4–6-week-old male BALB/c nu/nu mice. The mice were maintained in a barrier facility on HEPA-filtered racks and fed with an autoclaved laboratory rodent diet. Each experimental group contained six mice. Tumor size was monitored using a caliper in the process of tumor growth and measured every 3 days. After 5 weeks, mice were killed and tumors were excised and weighed. Tumor volumes were calculated as follows: volume = (D × d2)/2, where D is the longest diameter and d is the shortest diameter.
Chromatin Immunoprecipitation (ChIP)
Cells (2×106) in a 100-mm culture dish were treated with 1% formaldehyde to cross-link proteins to DNA. The cell lysates were sonicated to shear DNA to sizes of 300–1000 bp. Equal aliquots of chromatin supernatants were incubated with 1 μg of anti-c-myc antibody (Abcam, Cambridge, MA) or an anti-IgG antibody (Millipore, Billerica, MA) overnight at 4 °C with rotation. After reverse cross-linking of protein/DNA complexes to free DNA, PCR was performed.
Bioinformatics Analysis
The following on-line software programs were used for bioinformatics analysis: The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/); TargetScan 6.2 (http://www. targetscan.org/); and miRanda (http://www.microrna.org/microrna/getGene Form.do). TCGA was used to analyze the correlation between miR-148a and ESCC progression; TargetScan 6.2 was used to analyze the biological function and predict the target genes of miR-148a.
Target Gene Prediction for miR-148a
The target genes of hsa-miR-148a were predicted using TargetScan (version 7.2),35 DIANA-microT,36 and miRDB.36 The threshold values for TargetScan, DIANA-microT, and miRDB were set to Total context++ score < −0.4 (TargetScan), miTG score >0.95 (DIANA-microT), and miRDB score > 95, respectively. Targets were functionally annotated using the UniProt database and genes involved in the development of cancer, such as inflammation, cell cycle and differentiation, and angiogenesis were screened.
Dual Luciferase Reporter Assay
A fragment in which the 3ʹ-UTR region of the ACVR1 gene interacts with miRNA-148a was cloned into the Pgl3 vector (Promega), and the antisense strand of the fragment was also cloned. The pGL3-Control vector (Promega) was used as a positive control.37,38 Stable miRNA-148a overexpression, overexpression control, miRNA-148a inhibition, and inhibition group control clones were transfected into reporter gene constructs. Firefly and Renilla luciferase signals were measured using a Dual-Luciferase assay kit (Promega).
Data Analysis
Three valid biological replicates were performed for each experiment and the experiment was repeated three times. Data are presented as mean ± standard deviation. For comparison of continuous variables between the two experimental groups, the Independent Samples t-test (equal variance not assumed) was used. For multiple group comparisons, ANOVA with post hoc Dunnett’s test was used. All statistical analyses were performed using SPSS 19.0 software.39 P < 0.05 represents statistical significance.
Results
miR-148a Was Down-Regulated in ESCC and is Significantly Associated with Prognosis
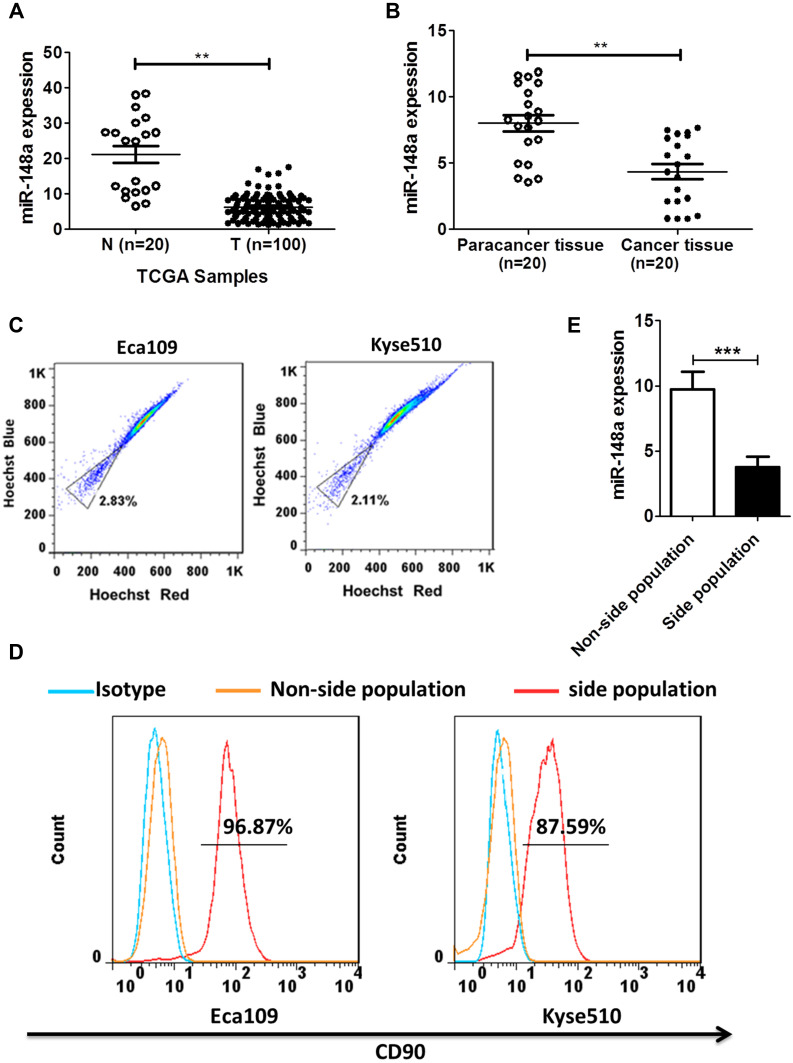
By analyzing the Cancer Genome Atlas (TCGA) microarray data set consisting of 100 primary esophageal cancer tissues and 20 normal esophageal tissues, miR-148a was significantly down-regulated in tumor tissues compared to normal tissues (Figure 1A). This indicates that the expression level of miR-148a is significantly associated with disease progression and prognosis in patients with ESCC. By verifying the clinical samples of ESCC, it was found that the expression of miR-148a in cancer tissues was significantly down-regulated when compared to that in the paracancerous adjacent tissues (p<0.01, Figure 1B). By using flow cytometric sorting, ESCC tissue cells were divided into side population (SP) group cells and non-side population (non-SP) group cells (Figure 1C). The proportion of the side groups in Eca109 and Kyse510 was similar, at 2.11% and 2.83%, respectively. CD90 is a well-known esophageal CSC marker, and the percentages of CD90+ cells in Kyse510 and Eca109 cell lines were 96.87% and 85.79%, respectively (Figure 1D). Analysis of the expression level of miR-148a in the contralateral and non-SP cells (p<0.01, Figure 1E) showed that miR-148a expression in the ESCC was significantly down-regulated.
Figure 1.
miR-148a expression was significantly down-regulated in ESCC and was closely associated with prognosis in patients with ESCC. (A) Expression profiling of miR-148a from the Cancer Genome Atlas (TCGA) datasets in primary esophageal tumors (T, n=100) and the adjacent normal tissues (ANT, n=20). (B) The expression of miR-148a in esophageal cancer tissue compared to paracancer tissue. (C) Sort side population (SP) group cells and non-side population (non-SP) using flow cytometric. (D) The percentage of CD90+ cells in the side population cells of ESCC. (E) Real-time PCR analysis of the expression of miR-148a in the side population cells of ESCC. ** indicates p < 0.01, *** indicates p < 0.001.
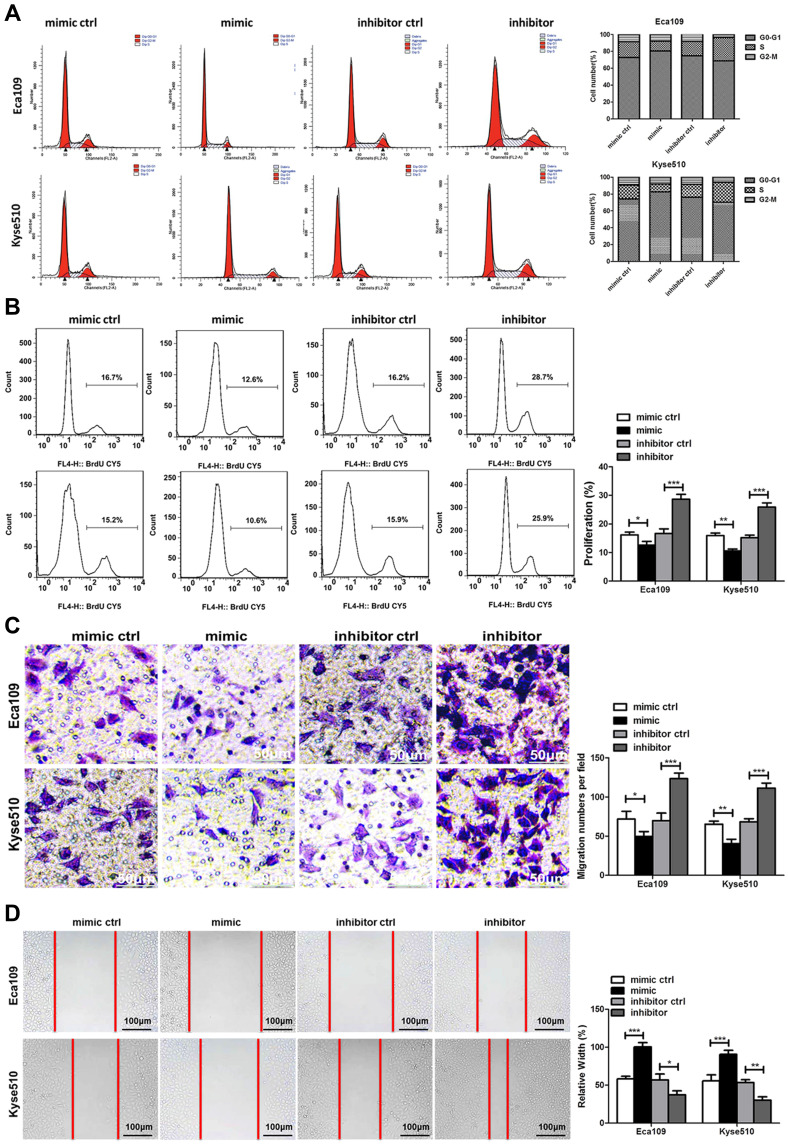
miR-148a Regulates the Cycle, Migration, and Invasion of ESCC
Flow cytometry was used to study the effect of miR-148a on the cell cycle of ESCC. As shown in Figure 2A, overexpression of miR-148a induces the restriction of ESCC in the cell cycle G1 and S phases, while inhibition of miR-148a effectively reduced the proportion of ESCC in G1 and S phases. Studies on cell proliferation found that overexpression of miR-148a significantly inhibited the proliferation of esophageal cancer cell lines Eca109 and Kyse510, while inhibition of miR-148a expression promoted EC cell proliferation compared to the control group (p<0.01, Figure 2B). To demonstrate whether miR-148a expression regulates migration and invasion of ESCC, we performed cell migration and invasion assays on ESCC cell lines Eca109 and Kyse510. As expected, miR-148a overexpression significantly reduced migration and invasion of ESCC, and inhibition of miR-148a significantly increased migration and invasion of ESCC (p<0.01, Figure 2C). The results of the scratch test further confirmed that overexpression of miR-148a significantly reduced the migration ability of ESCC (Figure 2D).
Figure 2.
miR-148a regulates the proliferation, migration, and invasion of esophageal cancer cells. (A) Cell cycle analysis using flow cytometry after transduction with the miR-148a mimic or inhibitor in Eca109 and Kyse510 cell lines. (B) Cell proliferation analysis using flow cytometry after transduction with the miR-148a mimic or inhibitor in Eca109 and Kyse510 cell lines. (C) Transwell migration assays after transduction with the miR-148a mimic or inhibitor in Eca109 and Kyse510 cell lines Scale bar, 50 μm. (D) Scratch test after transduction with the miR-148a mimic or inhibitor in Eca109 and Kyse510 cell lines. Scale bar, 100 μm. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
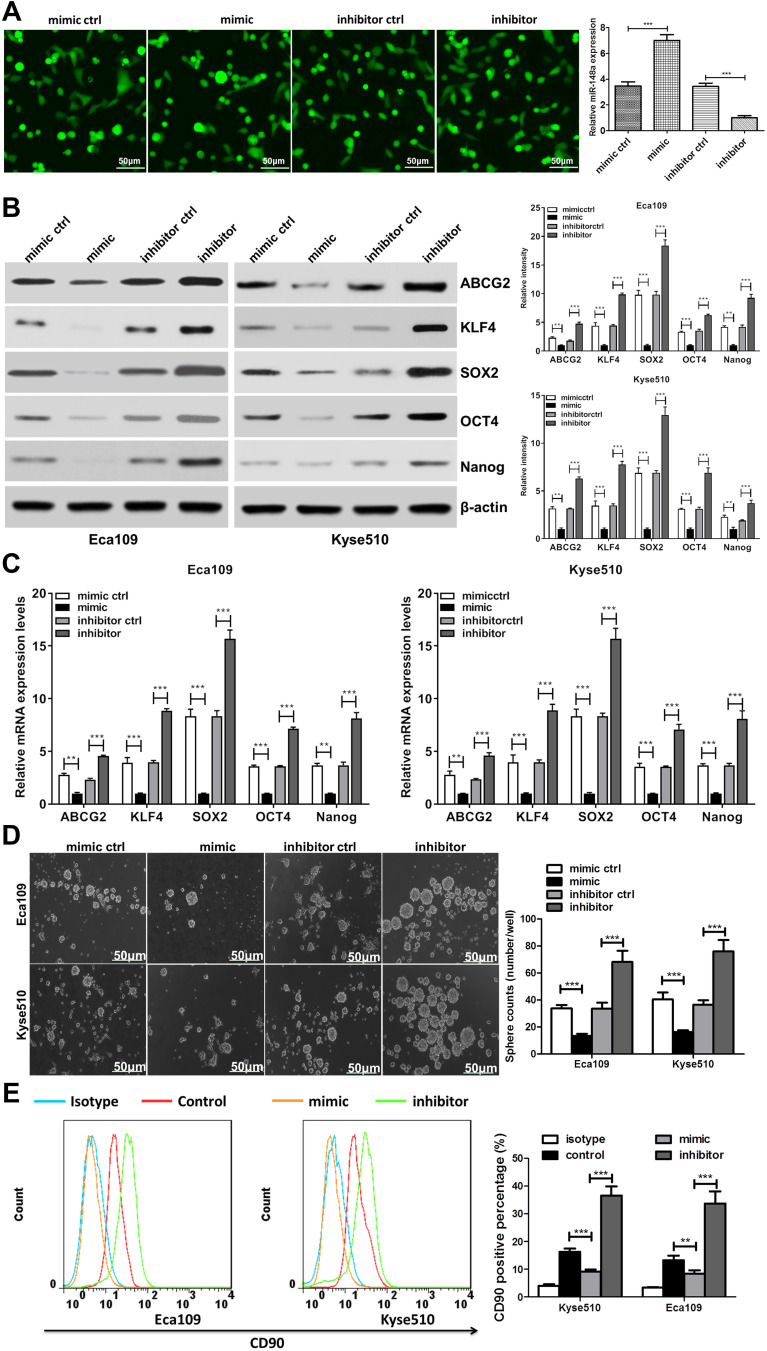
miR-148a Regulates the Stem Cell Distribution of ESCC
To investigate the role of miR-148a in the regulation of stem cells from ESCC, we analyzed the protein and mRNA expression levels of six previously published stem cell-associated genes in the ESCC lines Eca109 and Kyse510, including ABCG2, KLF4, SOX2, OCT4, and Nanog. Transfection efficiency were shown in Figure 3A. Overexpression of miR-148a significantly reduced the expression of stem cell genes, resulting in decreased activities of stem cells in ESCC. In the Eca109 cell line, the protein expression level of the stem cell genes and the mRNA level were significantly increased after the expression of miR-148a was inhibited. In the Kyse510 cell line, the expression levels of ABCG2, KLF4, SOX2, and Nanog proteins and mRNA were significantly increased (Figure 3B and C). In vitro globule assays (Figure 3D) also showed that cells overexpressing miR-148a formed fewer and smaller spheroid cells than the miR-148a mimic control group. Inhibition of miR-148a expression resulted in larger and more spheroid cells than the miR-148a inhibitor control group. Effects of miR-148a mimic and inhibitor on the distribution of CD90+ cells were determined using flow cytometric analysis. CD90+ cells were dramatically increased in miR-148a inhibitor group compared with control and mimic groups (Figure 3E). This further indicates that the expression of miR-148a has an effect of inhibiting the distribution of ESCC stem cells.
Figure 3.
miR-148a may have a regulatory effect on cancer stem cells. (A) Transfection efficiency analysis for miR-148a mimic and inhibitor. Scale bar, 50 μm (B) Western blotting analysis of the protein expression of pluripotency-associated markers in Kyse510 and Eca109 cell lines. (C) Real-time PCR analysis of the mRNA expression of pluripotency-associated markers in Kyse510 and Eca109 cell lines. (D) Representative micrographs (left) and quantification (right) of tumor spheres formation in miR-148a mimic and inhibitor cells. Scale bar, 50 μm. (E) Effects of miR-148a mimic and inhibitor cells on the distribution of CD90+ cells using flow cytometric analysis. ** indicates p < 0.01, *** indicates p < 0.001.
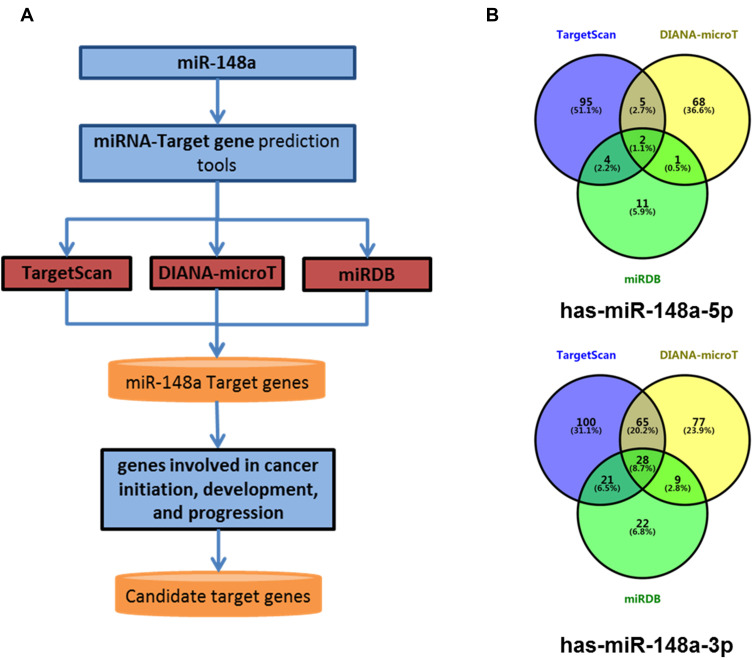
Bioinformatics Analysis for miR-148a Target Genes
To better understand the biological function of miR-148a, we used bioinformatics analysis methods to predict the target genes in the 3ʹ and 5ʹ UTR regions of miR-148a using TargetScan (version 6.2), DIANA-microT, and miRDB software (Figure 4A). The UniProt database was used to perform functional annotation of the detected target genes and showed that the genes were involved in cancer-related inflammatory responses, cell cycle and differentiation, and angiogenesis. The statistical results of the hsa-miR-148a-5p target gene prediction are summarized in Supplementary Material 1; the statistical results of the hsa-miR-148a-3p target gene prediction are summarized in Supplementary Material 2; and the results of the gene function annotation are shown in Supplementary Material 3. Target gene prediction (Figure 4B) identified 12 target genes for miR-148a 5ʹ UTR and 123 for 3ʹ UTR in the results of two or more prediction methods. There are seven genes involved in the inflammatory reaction, 10 genes involved in cell cycle and cell differentiation, and eight genes involved in angiogenesis (Table 3).
Figure 4.
Bioinformatics methods predict the target gene of miR-148a. (A) Target gene prediction of the 3ʹ and 5ʹ UTR regions of miR-148a using analysis software; (B) overlap status of hsa-miR-148a-5p target genes and hsa-miR-148a-3p target genes.
Table 3.
Genes Involved in the Inflammatory Response, Complement Activation, Cell Differentiation, and Angiogenesis
| Functional Categories | Genes |
|---|---|
| Inflammatory response | ACVR1, C5orf30, CDK19, LDLR, LIPA, RPS6KA5, and TEK |
| Cell cycle and cell differentiation | ACVR1, ARL8B, CAND1, CDK19, DNMT1, FXR1, GADD45A, MYT1, SIK1, SIX4, and SOS1 |
| Angiogenesis | ADGRB3, AGO1, DDAH1, KLF5, MEOX2, NRP1, TEK, and TGFA |
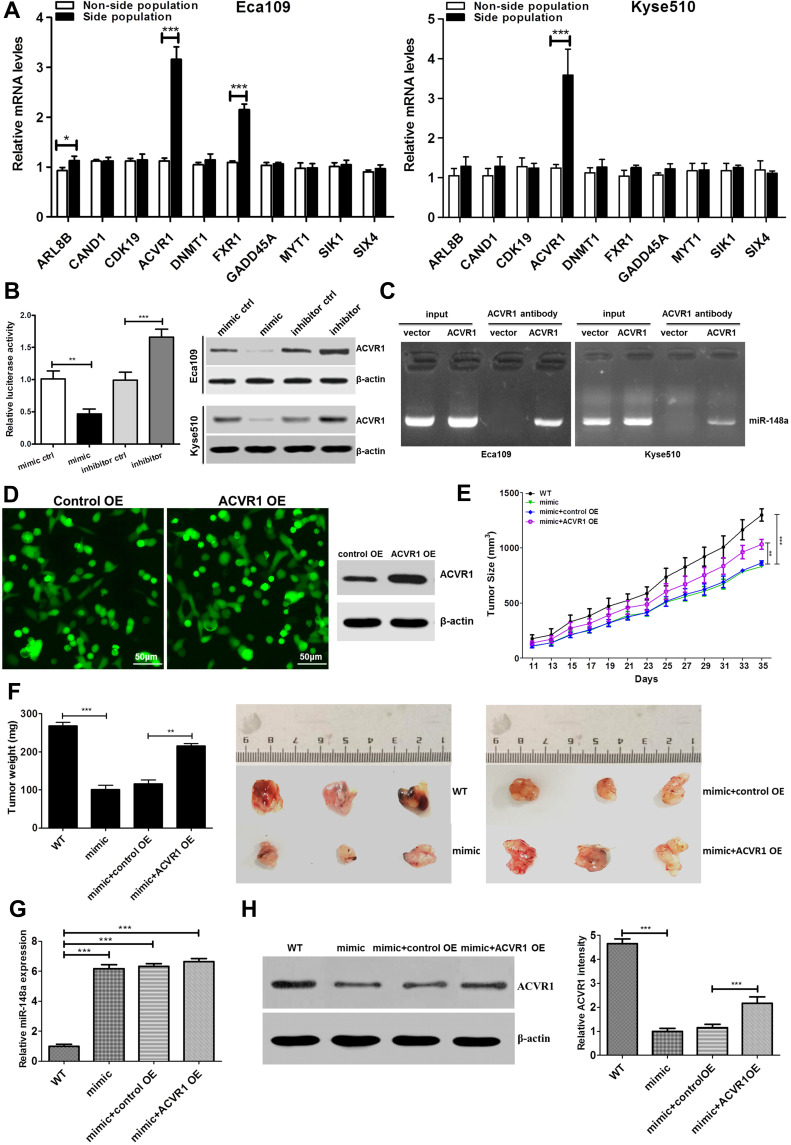
miR-148a Affects the Stem Cell Activities of ESCC by Regulating ACVR1
Functional verification was conducted of important candidate genes involved in cell cycle and differentiation. We used RT-PCR to detect the expression of target genes of hsa-miR-148a in ESCC stem cells Eca109 and Kyse510. The results showed that the expression of ACVR1 was significantly increased in stem cells of ESCC (Figure 5A). We further analyzed the expression of ACVR1 in miR-148a in ESCC stem cells using a dual luciferase reporter assay and verified the result by Western blot (p<0.01, Figure 5B). Overexpression of miR-148a significantly reduced the expression of ACVR1 in the ESCC stem cell-associated genes and inhibited the expression of ACVR1 by inhibiting the expression of miR-148a, thereby increasing the distribution of tumor stem cells. ChIP assay results showed high binding affinity of endogenous AVCR1 with miR-148a in the Kyse510 and Eca109 cell lines (Figure 5C), indicating that miR-148a directly targeted ACVR1.
Figure 5.
Upregulation of miR-148a increases tumourigenesis of ESCC cells in vivo. (A) Real-time PCR to verify target genes of hsa-miR-148 mRNA level in ESCC cell lines Eca109. (B) Luciferase activity of stem cell-associated gene ACVR1 in the indicated cells. (C) ChIP assay analysis the binding affinity of endogenous AVCR1 with miR-148a in the Kyse510 and Eca109. (D) The fluorescent image and Western blotting analysis showed that ACVR1 was overexpressed successfully. (E) Tumor volumes were measured on the indicated days. (F) Mean tumor weights and images of tumors from each group. (G) Real-time PCR analysis the level of miR-148a expression and (H) Western blotting analysis ACVR1 expression in the tumor tissue. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
ACVR1 Mitigates the miR-148a-Inhibited ESCC Tumorigenesis in vivo
The biological effect of miR-148a and AVCR1 on ESCC progression was examined using an in vivo tumor model. The Eca109/miR-148a mimic or Eca109/AVCR1 overexpression (OE) cells (Figures 3A and 5D) were subcutaneously xenografted into NOD/SCID mice. We performed the in vivo analysis following the miR-148a and AVCR1 OE transfection in the tumors bearing nude mice in WT, mimic, mimic and OE control, ACVR1 OE groups. As we expected, the level of miR-148a expression was increased significantly in mimic, mimic and OE control, ACVR1 OE group than WT group (Figure 5G). Meanwhile, ACVR1 was decreased significantly in mimic group than WT group, but comparison with mimic and OE control group, the level of ACVR1 expression was increased significantly in mimic and ACVR1 OE group (Figure 5H). As shown in Figure 5E and F, the tumors formed by Eca109/miR-148a mimic cells were smaller, in both size and weight, than the tumors formed from control cells (WT). Furthermore, the tumors formed by Eca109/miR-148a mimic and OE control cells were smaller, in both size and weight, than the tumors formed from Eca109/miR-148a mimic and AVCR1 OE cells. These results indicated that ACVR1 overexpression could mitigate the miR-148a-inhibited ESCC tumorigenesis in vivo.
Discussion
More and more reports indicate that miRNAs, as carcinogenic miRNAs or tumor suppressor miRNAs, play important roles in the development and progression of cancer.40 The first discovery of the effects of miRNAs on cancer was in a study of B-cell chronic myelogenous leukemia.41 One of the most important features of microRNAs in ESCC is the discovery that their dysregulation can cause tumor cell migration and invasion.42 However, microRNA regulation of ESCC is still largely unknown. Tumor stem cells have the ability to self-differentiate and can stably form progeny of malignant tumor cells.29 Cancer stem cells for ESCC have been isolated.30,31 By studying the regulation of microRNAs on cancer stem cells, new targeted therapeutics and diagnostic methods for disease prognosis can be identified as part of research and development on ESCC.
miR148a has variable expression and plays different biological functions in different types of cancer. Zhang et al found that miR-148a is highly expressed in colon cancer cell lines and affects tumor cell apoptosis.43 Yuan et al also found high expression of miR-148a in liver cancer malignancies and that miR-148a was significantly associated with hepatitis B.44 However, in studies of prostate malignancies, the expression level of miR-148a is significantly reduced in androgen-rejected PC3 and DU145 tumor cell lines relative to normal prostate cells and causes tumor cell tolerance and a response to androgen by inhibiting the expression of MSK1.45 In recent years, studies have found that miRNAs are essential in regulating the biological functions of normal tissue stem cells and tumor stem cell differentiation and self-renewal and regulate the stemness of normal tissue stem cells and cancer stem cells by affecting the expression of corresponding target genes.46
In the present study, we found that miR-148a was significantly lower in patients with ESCC than in normal subjects. In these patients, the expression of miR-148a was significantly down-regulated in tumor tissue compared to adjacent tissues. miR-148a may be significant for the prognosis and targeted therapy of patients with ESCC. By further studying the molecular regulation mechanism of miR-148a on ESCC, we found that overexpression of miR-148a significantly increased the amplification, migration, and invasion ability of tumor cells. Inhibition of miR-148a can effectively reduce the above-mentioned abilities of tumor cells. In addition, inhibition of miR-148a can also effectively increase apoptosis of tumor cells. This also provides strong evidence for miR-148a as a targeted therapy for ESCC.
In many tumor studies, including those on esophageal cancer, the microRNAs expression profile was found to be abnormal, regulating tumor cell proliferation, invasion, metastasis, cell cycle, differentiation and apoptosis, etc., and is closely related to tumor development, but the research for microRNA in the esophagus on squamous cell stem cell populations is very scarce. As a cell population that can continuously produce tumor cells, cancer stem cells play an important role in anticancer drugs and treatment tolerance.47,48 This was the first systematic study of the effects of miR-148a on ESCC cells and tumor stem cells to find a key method for the treatment of ESCC. We used flow cytometric sorting to screen for SP cells in ESCC, ie, ESCC stem cells.31 The target RNA and miR-148a were overexpressed and inhibited by cell transfection techniques. Overexpression of miR-148a in ESCC stem cells significantly reduced the gene expression of stem cells and affected the stem cell-like side populations distribution of stem cells. The inhibition of miR-148a effectively increased the expression of stem cell genes and its marker genes, thereby improving stem cell distribution and proliferation abilities.
By predicting the target gene of miR-148a and performing bioinformatics analysis on the target gene, we found that the miR-148a target gene participates in inflammatory reactions, cell proliferation and distribution, and angiogenesis responses related to cancer function. Among them, cell cycle and cell distribution were further studied as important functions of cancer stem cells. Functional verification analysis of these candidate genes revealed that the ACVR1 gene may be involved in the stemness of miR-148a-regulated ESCC stem cells. In a study of hepatocellular carcinoma malignant cells, miR-148a directly affected the stemness of cancer stem cells by affecting the expression of ACVR1.31 ACVR1 is an important cellular receptor of the BMP signaling pathway and belongs to the TGFb family. AVCR1 mutation was found to increase drug expression in ID1 and ID2 by causing phosphorylation of SMAD in diffuse pons gliomas, resulting in drug tolerance.49,50 Mutations in the ACVR1 gene are also found in breast cancer and progressive muscle ossification.51,52 In addition, AVCR1 was found to activate the Wnt signaling pathway, which has important effects on tumor stem cell distribution and stemness,53 so inhibition of ACVR1 can effectively reduce or inhibit the proportion of cancer stem cells. This study found that overexpression of miR-148a can effectively inhibit the expression of ACVR1, which may provide a new method for the treatment of ESCC.
Conclusions and Perspectives
In conclusion, we found that the expression of miR-148a has an important effect on the proliferation and invasion of ESCC cells and on the stem cell-like side populations distribution of ESCC stem cells. Overexpression of miR-148a inhibits the proliferation and migration of ESCC cells, and inhibition of miR-148a expression can increase the proliferation and migration of tumor cells. For overexpression of miR-148a, it is possible to reduce the differentiation and proliferation of ESCC stem cells by reducing the gene expression of ACVR1. Inhibition of the target of miR-148a increased the expression of the ACVR1 gene and increased the distribution ability of ESCC stem cells. Therefore, we may be able to diagnose the disease and determine the prognosis of patients with ESCC through the expression of miR-148a and treat ESCC by regulating the expression of miR-148a.
Funding Statement
This study was supported by the National Natural Science Foundation of China (81760499).
Data Sharing Statement
The data supporting the results in the manuscript can be obtained from the corresponding author Yisikandaer Abulimiti based on reasonable requests.
Disclosure
The authors declare that they have no competing interests in this work.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252. doi: 10.1056/NEJMra035010 [DOI] [PubMed] [Google Scholar]
- 2.Yang CS. Research on esophageal cancer in China: a review. Cancer Res. 1980;40(8 Pt 1):2633–2644. [PubMed] [Google Scholar]
- 3.Castellsague X, Munoz N, De Stefani E, et al. Independent and joint effects of tobacco smoking and alcohol drinking on the risk of esophageal cancer in men and women. Int J Cancer. 1999;82(5):657–664. doi:. [DOI] [PubMed] [Google Scholar]
- 4.Franceschi S, Talamini R, Barra S, et al. Smoking and drinking in relation to cancers of the oral cavity, pharynx, larynx, and esophagus in northern Italy. Cancer Res. 1990;50(20):6502–6507. doi: 10.1016/0165-4608(90)90155-4 [DOI] [PubMed] [Google Scholar]
- 5.Wu M, Zhao JK, Hu XS, et al. Association of smoking, alcohol drinking and dietary factors with esophageal cancer in high- and low-risk areas of Jiangsu Province, China. World J Gastroenterol. 2006;12(11):1686–1693. doi: 10.3748/wjg.v12.i11.1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin HD, Liao XY, Chen YB, et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet. 2016;98(4):709–727. doi: 10.1016/j.ajhg.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risk JM, Mills HS, Garde J, et al. The tylosis esophageal cancer (TOC) locus: more than just a familial cancer gene. Dis Esophagus. 1999;12(3):173–176. doi: 10.1046/j.1442-2050.1999.00042.x [DOI] [PubMed] [Google Scholar]
- 8.Miller JD, Jain MK, de Gara CJ, Morgan D, Urschel JD. Effect of surgical experience on results of esophagectomy for esophageal carcinoma. J Surg Oncol. 1997;65(1):20–21. doi: [DOI] [PubMed] [Google Scholar]
- 9.Chan A, Wong A. Is combined chemotherapy and radiation therapy equally effective as surgical resection in localized esophageal carcinoma? Int J Radiat Oncol Biol Phys. 1999;45(2):265–270. doi: 10.1016/s0360-3016(99)00199-6 [DOI] [PubMed] [Google Scholar]
- 10.Kong KL, Kwong DL, Chan TH, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61(1):33–42. doi: 10.1136/gutjnl-2011-300178 [DOI] [PubMed] [Google Scholar]
- 11.SEER. Cancer statistics review; 1975–2015. Available from: https://seer.cancer.gov/csr/1975_2015/. Accessed July20, 2020.
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10a):1902–1910. doi: 10.1101/gr.2722704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–5856. doi: 10.1200/jco.2009.24.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 16.Hummel R, Watson DI, Smith C, et al. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15(3):429–438. doi: 10.1007/s11605-011-1418-9 [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui R, Meng W, Sun HL, et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci U S A. 2015;112(31):E4288–97. doi: 10.1073/pnas.1502068112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HS, Lee KS, Bae HJ, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6(10):8089–8102. doi: 10.18632/oncotarget.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi: 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Zheng D, Zhang B, et al. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. J Transl Med. 2014;12(1):196. doi: 10.1186/1479-5876-12-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu T, Cao R, Li S, et al. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer. 2015;15(1):29. doi: 10.1186/s12885-015-1031-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishibashi O, Akagi I, Ogawa Y, Inui T. MiR-141-3p is upregulated in esophageal squamous cell carcinoma and targets pleckstrin homology domain leucine-rich repeat protein phosphatase-2, a negative regulator of the PI3K/AKT pathway. Biochem Biophys Res Commun. 2018;501(2):507–513. doi: 10.1016/j.bbrc.2018.05.025 [DOI] [PubMed] [Google Scholar]
- 24.Song P, Zhu H, Zhang D, et al. A genetic variant of miR-148a binding site in the SCRN1 3ʹ-UTR is associated with susceptibility and prognosis of gastric cancer. Sci Rep. 2014;4(1):7080. doi: 10.1038/srep07080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228 [DOI] [PubMed] [Google Scholar]
- 26.Xu Q, Liu LZ, Yin Y, et al. Regulatory circuit of PKM2/NF-kappaB/miR-148a/152-modulated tumor angiogenesis and cancer progression. Oncogene. 2015;34(43):5482–5493. doi: 10.1038/onc.2015.6 [DOI] [PubMed] [Google Scholar]
- 27.Tsai HL, Yang IP, Huang CW, et al. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl Res. 2013;162(4):258–268. doi: 10.1016/j.trsl.2013.07.009 [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Deng XY, Zeng XM, et al. The role of Mir-148a in cancer. J Cancer. 2016;7(10):1233–1241. doi: 10.7150/jca.14616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam F, Qiao B, Smith RA, Gopalan V, Lam AK. Cancer stem cell: fundamental experimental pathological concepts and updates. Exp Mol Pathol. 2015;98(2):184–191. doi: 10.1016/j.yexmp.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 30.Islam F, Gopalan V, Wahab R, Smith RA, Lam AK. Cancer stem cells in oesophageal squamous cell carcinoma: identification, prognostic and treatment perspectives. Crit Rev Oncol Hematol. 2015;96(1):9–19. doi: 10.1016/j.critrevonc.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 31.Huang D, Gao Q, Guo L, et al. Isolation and identification of cancer stem-like cells in esophageal carcinoma cell lines. Stem Cells Dev. 2009;18(3):465–473. doi: 10.1089/scd.2008.0033 [DOI] [PubMed] [Google Scholar]
- 32.Barruet E, Morales BM, Lwin W, et al. The ACVR1 R206H mutation found in fibrodysplasia ossificans progressiva increases human induced pluripotent stem cell-derived endothelial cell formation and collagen production through BMP-mediated SMAD1/5/8 signaling. Stem Cell Res Ther. 2016;7(7):115. doi: 10.1186/s13287-016-0372-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge CL, Wu SK, Wang WW, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6(13):10964–10977. doi: 10.18632/oncotarget.3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin L, Li Y, He T, et al. Identification of miR30b as an oncogene in renal cell carcinoma. Mol Med Rep. 2017;15(4):1837–1846. doi: 10.3892/mmr.2017.6197 [DOI] [PubMed] [Google Scholar]
- 35.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(WebServer issue):W169–73. doi: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Databaseissue):D146–52. doi: 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He H, Li S, Hong Y, et al. Kruppel-like factor 4 promotes esophageal squamous cell carcinoma differentiation by up-regulating keratin 13 expression. J Biol Chem. 2015;290(21):13567–13577. doi: 10.1074/jbc.M114.629717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng L, Xiang X, Yang F, et al. Functional evidence that the self-renewal gene NANOG regulates esophageal squamous cancer development. Biochem Biophys Res Commun. 2017;490(2):161–168. doi: 10.1016/j.bbrc.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 40.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 41.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Y, Li J, Zhu Y, et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5(22):11669–11680. doi: 10.18632/oncotarget.2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Li Y, Huang Q, et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011;18(11):1702–1710. doi: 10.1038/cdd.2011.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS One. 2012;7(4):e35331. doi: 10.1371/journal.pone.0035331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita Y, Kojima K, Ohhashi R, et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285(25):19076–19084. doi: 10.1074/jbc.M109.079525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kano M, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127(12):2804–2814. doi: 10.1002/ijc.25284 [DOI] [PubMed] [Google Scholar]
- 47.Alison MR, Lin WR, Lim SM, Nicholson LJ. Cancer stem cells: in the line of fire. Cancer Treat Rev. 2012;38(6):589–598. doi: 10.1016/j.ctrv.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 48.Li L, Liu Y, Guo Y, et al. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology. 2015;61(2):574–584. doi: 10.1002/hep.27543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor KR, Mackay A, Truffaux N, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46(5):457–461. doi: 10.1038/ng.2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buczkowicz P, Hoeman C, Rakopoulos P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–456. doi: 10.1038/ng.2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slattery ML, John EM, Torres-Mejia G, et al. Genetic variation in bone morphogenetic proteins and breast cancer risk in hispanic and non-hispanic white women: the breast cancer health disparities study. Int J Cancer. 2013;132(12):2928–2939. doi: 10.1002/ijc.27960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakahara Y, Katagiri T, Ogata N, Haga N. ACVR1 (587T>C) mutation in a variant form of fibrodysplasia ossificans progressiva: second report. Am J Med Genet A. 2014;164a(1):220–224. doi: 10.1002/ajmg.a.36219 [DOI] [PubMed] [Google Scholar]
- 53.Kamiya N, Kaartinen VM, Mishina Y. Loss-of-function of ACVR1 in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1. Biochem Biophys Res Commun. 2011;414(2):326–330. doi: 10.1016/j.bbrc.2011.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]