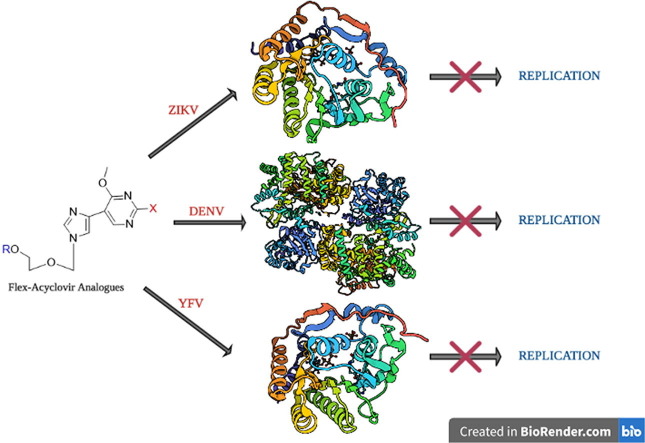
Graphical abstract
Keywords: Flavivirus, Dengue, Zika, Yellow Fever, Methyltransferase, Nucleoside, Fleximers, Acyclovir
Abstract
Flaviviruses, such as Dengue (DENV) and Zika (ZIKV) viruses, represent a severe health burden. There are currently no FDA-approved treatments, and vaccines against most flaviviruses are still lacking. We have developed several flexible analogues (“fleximers”) of the FDA-approved nucleoside Acyclovir that exhibit activity against various RNA viruses, demonstrating their broad-spectrum potential. The current study reports activity against DENV and Yellow Fever Virus (YFV), particularly for compound 1. Studies to elucidate the mechanism of action suggest the flex-analogue triphosphates, especially 1-TP, inhibit DENV and ZIKV methyltransferases, and a secondary, albeit weak, effect on the DENV RNA-dependent RNA polymerase was observed at high concentrations. The results of these studies are reported herein.
1. Introduction
Within the Flaviviradae family, the genus Flavivirus contains over 70 viruses with a growing number of unclassified members.a Many Flavivirus members are known to cause severe disease, such as Dengue hemorrhagic fever, sometimes associated to human mortality.1, 2, 3, 4 Members of this genus, including Dengue virus (DENV), West Nile Virus (WNV), Zika Virus (ZIKV), Yellow Fever Virus (YFV), and tick-borne encephalitis virus (TBEV), represent a tremendous health burden.1, 2, 3, 4, 5 Of these lethal flaviviruses, DENV poses the most severe threat with over 50 million documented cases, and between 12,500 and 25,000 deaths reported annually.3, 6, 7 Recently it was discovered that ZIKV infections in pregnant women have led to numerous infant abnormalities including microcephaly and severe brain malformations, as well as the development of Guillain-Barré syndrome in adults.2, 5, 7, 8, 9 Furthermore, more recent studies have demonstrated that ZIKV infections are sexually transmittable, with a detectable viral load in semen up to 26 weeks post symptomatic onset.10, 11, 12, 13, 14, 15, 16 Unfortunately, due to increased globalization, it is inevitable that new, undiscovered flaviviruses will continue to spread, endangering populations worldwide. As a result, new and viable therapeutic options need to be developed in order to better combat these emerging infections.
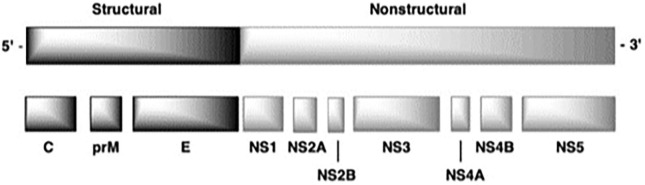
Flaviviruses are single stranded, positive-sense RNA viruses, with capped genomes of approximately 11 Kb in length.3, 17, 18 The genome contains an untranslated 5′-region followed by a single open reading frame and an untranslated 3′-end region.3, 17, 19 The open reading frame encodes for three structural proteins (capsid, pre-membrane, and envelope), as well as seven non-structural (NS) proteins forming the replication transcription complex. Among these NS proteins, the NS3 multifunctional protein has been shown to harbor serine protease, 5′-RNA triphosphatase (RTPase), nucleoside triphosphatase (NTPase), and helicase activities. The NS5 protein is another multifunctional protein essential for virus replication, which is responsible for the RNA-dependent-RNA-polymerase (RdRp) and methyl transferase (MTase) activities needed for RNA capping (Fig. 1 ).1, 2, 3, 17, 18, 19, 20
Fig. 1.
General structure of the flavivirus genome including 5′ and 3′ untranslated regions and the polyprotein processing of both the structural and nonstructural protein regions.17, 18
Of the seven flavivirus NS proteins, one of the most important targets for drug design is the NS5 protein, which is the most conserved protein of the flaviviruses and plays an essential role in viral replication and capping. The C-terminal domain of the NS5 protein contains the RdRp domain and the N-terminal domain is responsible for the S-adenosyl-l-methionine (SAM) dependent N7 and 2′-O-MTase activity for the viral RNA.6, 21, 22, 23 The aforementioned MTase activities modify the cap structure of the flaviviral RNA through N7-methylation of the 5′-guanine of the cap structure and 2′-O-methylation of the first transcribed adenosine nucleotide (N7MEGpppA2′OMe-RNA) (Fig. 2 ).6, 21, 22, 23, 24
Fig. 2.
The canonical RNA capping pathway of eukaryotic cells requires four main enzyme activities: (i) RTPase (NS3 in flaviviruses) that hydrolyzes the 5′-triphosphate end of the nascent RNA transcript into a 5′-diphosphate;25 (ii) RNA guanylyltransferase (putatively NS5) which then transfers the GMP moiety of GTP to the 5′-nucleotide diphosphate end;26 (iii) the RNA (guanine-N7)-MTase) that methylates the N7 position on the 5′-guanine; and (iv) the RNA (nucleoside-2′-O)-MTase which methylates the 2′-OH (a conserved adenosine in flaviviruses) of the subsequent nucleotide, resulting in cap-1 structure for the viral RNA.6, 21, 22, 24, 27, 28, 29 Both methylation reactions are catalyzed by a single MTase domain and SAM is used as a methyl donor, generating S-adenosyl-l-homocysteine (SAH) as a by-product.28, 29
All flaviviral MTases share a conserved Rossmann-fold structure consisting of a SAH/SAM binding site, a cap/GTP binding site, and an RNA-binding pocket.28, 29, 30 Studies have shown that the presence of the methylated 5′ cap is essential for the protection and stability of the viral RNA throughout the viral replication cycle; thus, disruption of the MTase activity would interfere with viral replication.6, 21, 22 Indeed, it has been demonstrated that the N7-methylation of flaviviral RNA cap structure is essential for viral mRNA translation into protein, whereas the 2′-O methylation is a “marker of self” limiting the detection of viral RNA by the host innate immune sensors of the RIG-like family such as RIG-1 and MDA5.28, 29, 31 As such, the essential roles played by viral MTases during the viral life cycle demonstrate the great potential of these enzymes as viable targets for drug design.
While the N-terminal domain of the NS5 protein is responsible for cap-MTase activities, the C-terminal domain of the protein is responsible for the RdRp activity.4, 32, 33 Unlike most polymerases flaviviral RdRp utilizes a de novo initiation mechanism, wherein a 5′-triphosphate AG RNA dinucleotide is first synthesized by the polymerase, even in the absence of RNA template. This AG dinucleotide is next used by the polymerase as a primer for RNA polymerization.4, 32, 33, 34 Proper function of the RdRp is critical for flaviviral replication, thus, impeding the ability of RdRp to synthesize viral RNA is also an attractive target for drug design. Furthermore, a therapeutic that could disrupt both the MTase activity as well as the RdRp activity of the NS5 protein could prove to be a highly effective broad-spectrum inhibitor for the treatment of numerous flaviviruses.
Unfortunately, there are currently no FDA approved therapeutics for treating flavivirus infections.8, 19 Similarly, vaccine development for flaviviruses has been challenging, especially for DENV due to the necessity to provide a vaccine that would be effective against all four serotypes.8, 35, 36 Furthermore, if a serotype of DENV is not fully protected against, a patient is more likely to develop severe Dengue hemorrhagic fever or Dengue shock syndrome.35, 36 As such, broad spectrum therapeutics are needed in order to better combat these viral infections.
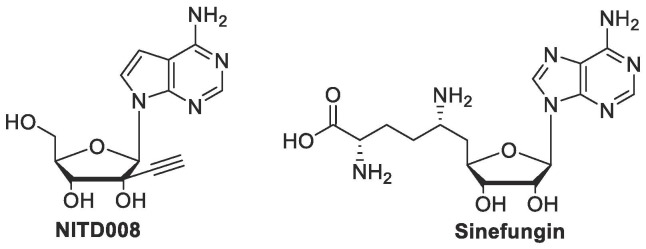
Recent studies have focused on either developing novel therapeutics or repurposing previously approved drugs in order to expedite the development process.7, 8, 37, 38, 39, 40 Of these therapeutics, nucleoside analogues initially garnered much attention due to their ability to disrupt the function of important viral replication enzymes.38, 41 One example of a potent nucleoside analogue is NITD008 (Fig. 3 ), an adenosine mimic that has demonstrated the ability to inhibit the RdRp domain of all four serotypes of DENV with an average EC50 value of 0.64 µM.5, 38, 42 While these initial studies were promising, various studies found that NITD008 is not a viable option for prophylaxis against DENV, as preclinical studies have demonstrated cytotoxicity associated with NITD008 treatment.5, 38
Fig. 3.
Early examples of antiflaviviral nucleoside inhibitors.
Another example is Sinefungin (Fig. 3), a natural SAM/SAH mimic that has demonstrated potent antiviral activity against numerous viral MTases, including those of flaviviruses with an IC50 value of 0.03 µM against N7 methylations and 0.041 µM against 2′-O-methylations in DENV.42, 43 Unfortunately, Sinefungin has not been pursued further as a flavivirus therapeutic due to its low selectivity for viral MTases versus human MTases.37, 43 While these analogues ultimately proved ineffective as potential therapeutics, they demonstrated the potential scope for utilizing nucleoside analogues in anti-flavivirus therapeutics.
Over the past two decades, the Seley-Radtke lab has focused on developing flexible purine base nucleoside analogues termed “fleximers”.44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57 These compounds feature a purine ring that is “split” into the imidazole and pyrimidine moieties, with a single carbon-carbon bond between the C4 of the imidazole and the C5 of the pyrimidine (proximal fleximers), or the C5 of the imidazole and the C6 of the pyrimidine (distal fleximers) (Fig. 4 ).44, 45, 46, 47
Fig. 4.
Structure of proximal and distal guanosine fleximers compared to natural guanosine.43, 44, 45, 46
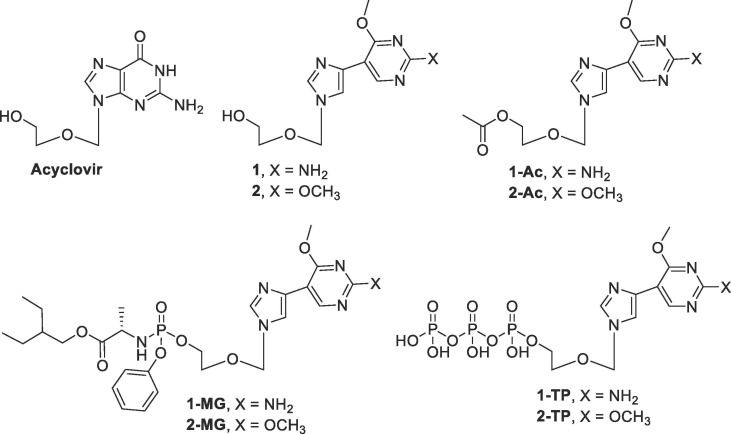
These nucleoside analogues retain the hydrogen bonding and stacking elements necessary for nucleoside recognizing enzymes, while allowing for alternative interactions in the enzyme binding site.44, 45, 46, 47, 49, 50, 51 This inherent flexibility allows for free rotation around the carbon-carbon bond between the imidazole and pyrimidine rings, thereby increasing the rotational degrees of freedom and allowing the fleximer to interact with other binding site moieties that were previously unattainable by the parent purine nucleoside.46, 47, 49, 50, 51 Due to these interesting characteristics, the Seley-Radtke lab has recently applied the fleximer approach to FDA-approved nucleoside inhibitors in order to create more potent analogues for antiviral therapeutics. Acyclovir (ACV), for example, is an FDA-approved acyclic nucleoside analogue mainly used to treat herpes simplex virus and varicella zoster virus infections.58, 59, 60
Previously, fleximer analogues were synthesized utilizing the sugar moiety found in ACV, where broad spectrum screening of the Flex-ACV analogues revealed compound 1 to be active (10.1 μM) against HCoV-NL63, an endogenous strain of human coronavirus (CoV) that displays similar symptoms to the common cold.53 Further analysis of compound 1 and its acetylated prodrug 1-Ac (Fig. 5 ) demonstrated low micromolar in vitro antiviral activity against both Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) - two deadly human coronaviruses for which there is currently no cure. Compound 1-Ac exhibited activity against MERS at 3.4 μM (in Vero) and 11.9 μM against SARS, while 1 inhibited HCoV-NL63 at 8.8 µM. These findings were ground-breaking since these compounds were the first nucleoside analogues to exhibit low micromolar levels of anti-coronavirus activity.53
Fig. 5.
Structure of the target fleximer analogues compared to the parent analogue Acyclovir.
These promising results prompted further investigation of these analogues against other viruses such as filoviruses, particularly given the dual anti-CoV and anti-Ebola activity recently noted by the nucleoside analogue Remdesivir.61, 62 In vitro antiviral testing revealed that compound 1, 1-Ac, and the phosphoramidate prodrug 1-MG were all active against Ebola (EBOV) virus, with compound 1 exhibiting the greatest activity (EC50 = 2.2 ± 0.3 µM).54 These results were quite interesting as they suggest the potential for dual activity for compound 1 and 1-Ac against both CoVs53 and EBOV.54 Further studies also revealed promising anti-EBOV activity for compounds 2 and 2-Ac, with the acetylated analogue 2-Ac demonstrating an EC50 value of 8.2 ± 1.8 µM (unpublished data).
Due to the remerging prevalence of DENV and ZIKV throughout the world, the ability of the Flex-ACV compounds to inhibit those viruses was pursued. Congruently, the compounds were also analyzed further in an effort to elucidate their mechanism of action as well as to explore the design of more potent compounds. Herein, the synthesis, antiviral activity against both DENV and ZIKV, and biological studies designed to uncover their potential mechanism of action for several analogues are described.
2. Results
2.1. Chemistry
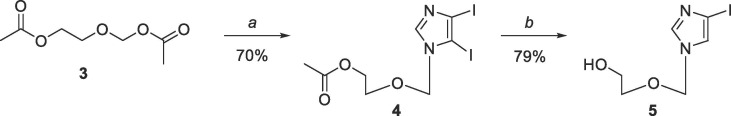
The compounds for this study were chosen based on the previous results for compounds 1, 1-Ac, and 1-MG against MERS, SARS,53 and EBOV,54 as well as unpublished results for compound 2 and 2-Ac against EBOV. The previously reported organometallic coupling procedures used by our group53, 54 to couple the two heterocyclic moieties involved tedious and multiple purification processes to remove the tin from the Stille coupling methodology, which led to very poor yields. As a result, attention turned to the Suzuki coupling methodology, which resulted in much cleaner reactions, facile purifications, as well as greatly improved yields. Starting with Scheme 1 , coupling the imidazole to the commercially available 2-[(acetyloxy)methoxy]ethyl acetate (3) using BSA and TMS-triflate gave 4, which, following selective deiodination, gave iodoimidazole 5.54
Scheme 1.
Reagents and conditions: (a) 4,5-diiodoimidazole, BSA, TMSOTf, ACN, rt for 4 h then 80 °C for 18 h; (b) 30% EtOH, 5 eq Na2SO3, 120 °C, overnight.
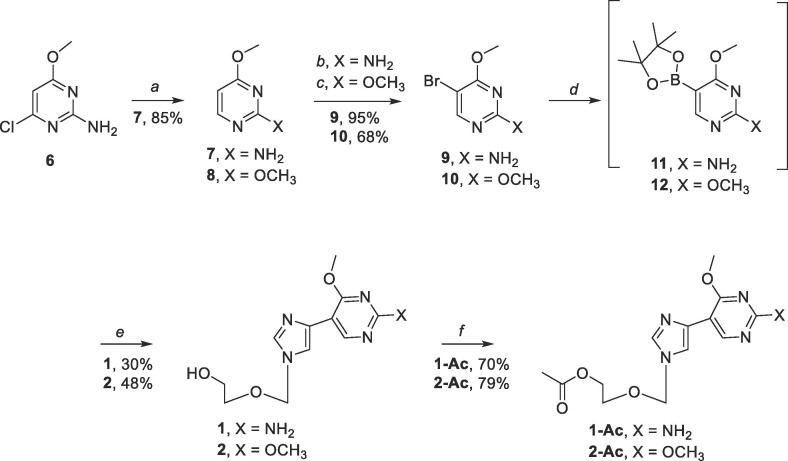
Compound 9 was synthesized starting with commercially available 2-amino-4-chloro-6-methoxypyrimidine for series 1 (Scheme 2 ).63 Similarly, compound 10 was synthesized starting with commercially available 2,4-dimethoxypyrimidine for series 2.64 Subsequent Suzuki-Miyaura cross-coupling of 9 and 5 gave 1 (30% over two steps), and coupling of 10 with 5 provided 2 (48% over two steps) (Scheme 2), each by way of 11 or 12 as the in-situ intermediate for the modified Suzuki-Miyaura couplings.63 Compounds 1 and 2 were then used to synthesize the acetate protected prodrugs 1-Ac and 2-Ac respectively (Scheme 2).65
Scheme 2.
Reagents and conditions: (a) DIPEA, 10% Pd/C, H2, rt, 4 h; (b) NBS, CHCl3, rt, dark, 5 h; (c) Br2, NaHCO3, 50% MeOH, rt, 3 h; (d) pin2B2, KOAc, Pd(PPh3)4, DME, 90 °C, overnight; (e) 5, Pd(PPh3)4, NaHCO3, 90 °C, 4 h; (f) 1 or 2, Ac2O, DMAP, DMF, rt, 3 h.
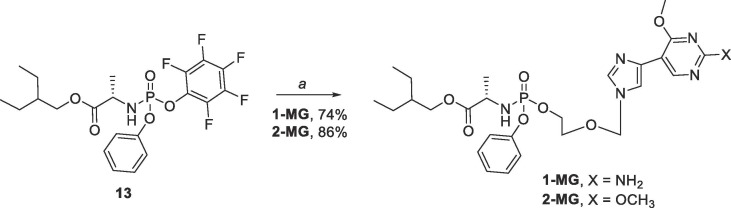
Synthesis of the phosphoramidate prodrugs 1-MG and 2-MG began with commercially available l-alanine and utilized procedures previously described by our lab54 as well as those found in the literature66 to yield the 2- ethylbutyl ((perfluorophenoxy)(phenoxy)phosphoryl)-l-alaninate intermediate 13 (Scheme 3 ). Reaction of this intermediate with either 1 or 2 and tert-butyl magnesium chloride afforded the phosphoramidate prodrugs 1-MG and 2-MG as diastereomeric mixtures in moderate yields (74% and 86% respectively).
Scheme 3.
Reagents and conditions: (a) 1 or 2, tBuMgCl, THF, rt, overnight.
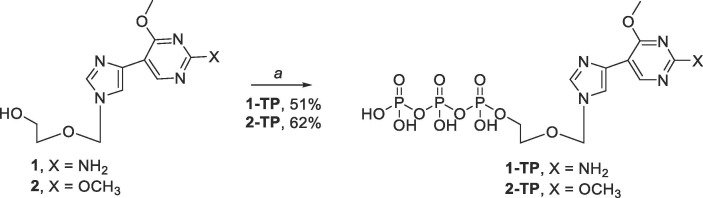
Finally, synthesis of the triphosphate analogues 1-TP and 2-TP were accomplished using a modified procedure by Hollenstein et. al. which utilized 2-chloro-1,3,2-benzo-dioxaphosphorin-4-one (SalPCl) and tributylammonium pyrophosphate.67 The methodology developed by Hollenstein et. al. noted important differences that ultimately greatly increased overall yields. For instance, prior to the reaction it is important that the fleximer nucleoside be coevaporated with anhydrous pyridine then dried in vacuo overnight, instead of storing the fleximer nucleoside in dried pyridine, 1,4-dioxane, and molecular sieves overnight. Furthermore, proper handling of SalPCl is important. SalPCl is a commercially available reagent that is typically a glassy green solid, however, once exposed to moisture, develops a powdery white coating on the outside of the crystals that should be scraped off prior to addition or the reaction goes poorly. Finally, in order to maximize the yield, HPLC purification should be done immediately to make the entire purification process more facile.
Synthesis of the triphosphate analogues 1-TP and 2-TP began with the addition of SalPCl to the fleximer scaffold to give a phosphite intermediate (Scheme 4 ).67 Then, addition of tributylammonium pyrophosphate and tributylamine induced cyclization of the phosphate moieties. After stirring at room temperature for 45 min, I2 and water were added to the reaction mixture in order to promote the oxidation of the α phosphorous from a P(III) to a P(V) center.67 Finally, the excess iodide was quenched with 10% sodium thiosulfate and the crude reaction was purified via HPLC to give either 1-TP or 2-TP. Following purification, triphosphates 1-TP and 2-TP were obtained as the triethylamine salts. As the triethylamine salts were not suitable for the enzymatic assays, these compounds were converted to their sodium salt forms using a Dowex 50Wx2 Na.+ ion exchange column. This produced both triphosphates in good yields (50% for 1-TP and 62% for 2-TP). We have repeated this approach numerous times now and the yields have stayed consistent.
Scheme 4.
Reagents and conditions: (a) i. 1 or 2, SalPCl, Pyr, 1,4-dioxane, rt, 45 min; ii. tributyl ammonium pyrophosphate, tributylamine, DMF, rt, 45 min; iii. I2, H2O, Pyr, rt, 30 min.
2.2. Antiviral activity
The potent antiviral activity demonstrated by compounds 1, 1-Ac, and 1-MG against a wide array of viruses including SARS-CoV,53 MERS-CoV,53 as well as filoviruses such as Ebola54 prompted further investigation with these analogues against additional viruses. These analogues, as well as the dimethoxy analogues 2 and 2-Ac, were then screened against various flaviviruses including DENV, ZIKV, and YFV. The analogues were analyzed utilizing a visual cytopathic effect assay on Vero76 cells infected with the live-virus isolates of DENV (New Guinea C), ZIKV (MR766), and YFV (17D).
The results showed that several flex-analogues demonstrated moderate to potent antiviral activity against all the flaviviruses tested, with compound 1 demonstrating the greatest antiviral activity against DENV (EC50 = 0.057 µM) (Table 1 ). Compound 1 also demonstrated potent antiviral activity against YFV (EC50 = 0.37 µM) with a selective index (SI) of 4.6. Although this is not ideal, preliminary minimum tolerated dose (MTD) studies have revealed no toxicity up to 250 mgs/kg, and we are currently pursuing those studies further to also explore the ProTide analogues.
Table 1.
Antiviral activity of flex-analogues against various flaviviruses including Dengue (DENV), Zika (ZIKV), and Yellow Fever Virus (YFV) in Vero76 cells.
| Compound | DENV |
ZIKV |
YFV |
|||
|---|---|---|---|---|---|---|
| aEC50 | bCC50 | EC50 | CC50 | EC50 | CC50 | |
| 1 | 0.057 | 1.20 | >1.80 | >1.80 | 0.37 | 1.7 |
| 1-Ac | 6.1 | 65 | >100 | >100 | >57 | 57 |
| 1-MG | >57 | 57 | >31 | 31 | >55 | 55 |
| 2 | >50 | 50 | >76 | 76 | >58 | 58 |
| 2-Ac | 19 | 53 | 79 | >100 | >58 | 58 |
| 2-MG | >53 | 53 | >56 | 56 | >53 | 53 |
Values are reported in µM.
EC50: effective concentration showing 50% inhibition of virus-induced CPE.
CC50: cytotoxic concentration showing 50% inhibition of cell survival.
A significant decrease in toxicity was observed with the acetate protected analogue 1-Ac against DENV compared to the parent analogue 1 (CC50 = 65 µM and CC50 = 1.2 µM respectively), however, a decrease in activity was also observed as 1-Ac demonstrated an EC50 of 6.1 µM. While not as potent as compound 1, compound 2-Ac demonstrated moderate activity against DENV with an EC50 of 19 µM, and little associated cytotoxicity. None of the analogues tested demonstrated any antiviral activity against ZIKV, and only analogue 1 demonstrated activity against YFV.
These results suggest that compound 1 could potentially act as a broad spectrum antiviral therapeutic across a wide range of viral families including coronaviruses, filoviruses, and now flaviviruses.
2.3. Inhibition NS5 activities: RdRp and MTase activity
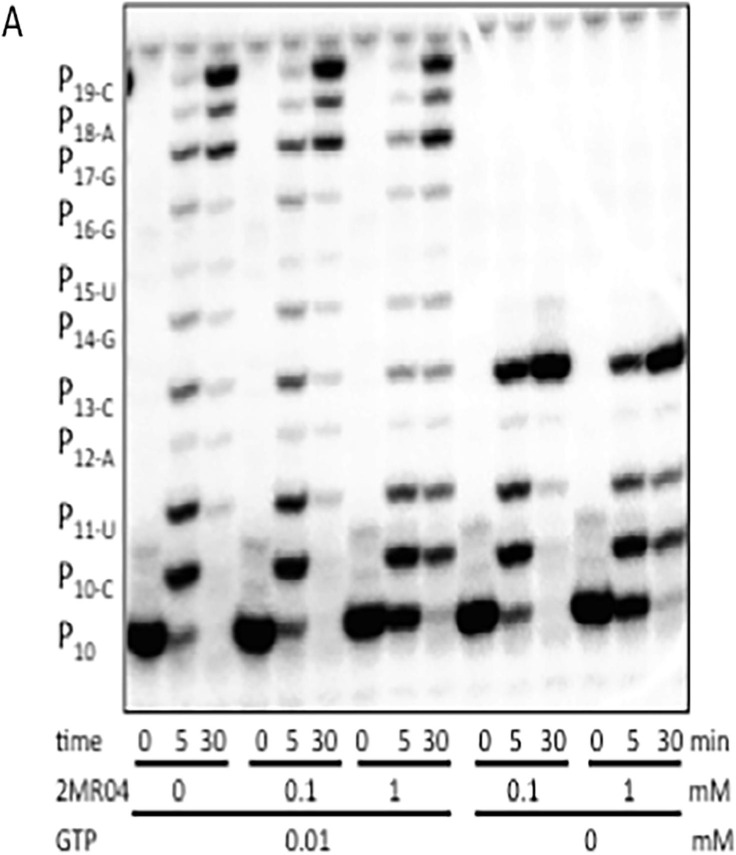
As nucleotide analogues may act as competitive inhibitors, chain terminators or mutagenic nucleotides incorporated into RNA, 1-TP and 2-TP were tested for their ability to inhibit or be incorporated into RNA using DENV RdRp. As shown in Fig. 6 for 1-TP (2MR04), very weak inhibition was observed at a high concentration of 1 mM in the presence of all four NTPs and also in the absence of GTP. In absence of GTP 1-TP is not significantly incorporated in the growing chain (see right part of Fig. 6) thus does not act as a chain terminator. Similarly, we did not observe incorporation of 1-TP or 2-TP instead of A, C or U (not shown). As no inhibition or incorporation was observed at concentrations below 200 μM, it is unlikely that the RdRp activity is significantly targeted by these compounds.
Fig. 6.
Effect of increasing concentrations of 1-TP (2MR04) on DENV polymerase activity. An elongation complex was formed of DENV NS5 and a primer/template combination corresponding to the 5’ end of DENV2 genome and the 3’ end of the antigenome. Primer elongation reactions were done in presence of all NTPs or in absence of GTP to test the incorporation of 1-TP as a GTP analogue. Reaction mixtures were analyzed by denaturating PAGE and substrate (P10) and product bands were visualized by autoradiography.
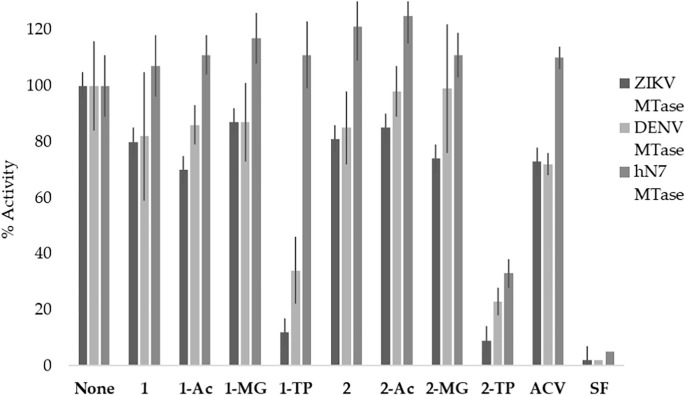
In that regard, as mentioned previously, the MTase activity for flavivirus NS5 is an interesting and important target for the development of antiviral therapeutics. In the flaviviruses, the MTases and RdRp are in the same protein complex. However, unlike viral RdRps, which demonstrate a high mutation rate68, 69, the viral MTase structure is highly conserved across most flavivirus species,21, 70 making viral MTases an attractive target for drug design. As such, compounds 1, 1-Ac, 1-MG, and 1-TP were analyzed for activity against DENV, ZIKV, and human N7 MTases utilizing a radioactive filter-binding assay (Fig. 7 ). The inhibition of the 2′-O-MTase activity of DENV and ZIKV, and that of the human N7 (RNMT) MTases was first analyzed against 50 µM of compound. Briefly, the MTases were incubated with synthetic RNA substrates (GpppAC5), radioactive 3H-SAM, and a Flex-analogue at 30 °C for 30 min.27 The reaction products were then filtered on DEAE membranes and the radioactivity transferred on the RNA was quantified. Sinefungin was utilized as an inhibitory control due to its known inhibition of both viral and human MTases.37, 42, 43
Fig. 7.
Percent inhibition of ZIKV and DENV MTase by series 1 and 2 (50 µM). None of the compounds inhibited human N7 MTase, suggesting that these analogues selectively inhibit the viral MTases.
While compound 1 did not exhibit a significant inhibitory effect on the different MTases activities, the triphosphate form 1- TP inhibited both DENV MTase and ZIKV MTase at 34% and 12% respectively (Fig. 7). The triphosphate analogue 2-TP demonstrated the greatest inhibitory activity against ZIKV MTase at 9%. Furthermore, none of the analogues tested inhibited human N7 MTase activity, which also suggests these analogues selectively inhibit viral MTases.
Analogues 1-TP and 2-TP were then further analyzed in order to determine IC50 values against the MTases (Table 2 ). This data was congruent with the previous MTase data where compounds 1-TP and 2-TP demonstrated a greater inhibitory effect against ZIKV MTase compared to DENV MTase. The triphosphate 2-TP was most potent against ZIKV MTase (0.15 µM) whereas the triphosphate 1-TP (IC50 = 1.7 µM) was still active against ZIKV MTase but to a lesser degree than 2-TP. This data suggests that the antiviral activity seen with compound 1 is due to inhibition of the MTase activity rather than inhibition of the viral polymerases, since nucleosides must first be converted by kinases to the corresponding triphosphates in order to be active against and/or recognized.
Table 2.
Inhibition of MTase activity of compounds 1-TP and 2-TP against DENV NS5-MTase, ZIKV NS5-MTase, and human N7 MTase.
| Compound | ZIKV MTase IC50 | DENV MTase IC50 | hN7 MTase IC50 |
|---|---|---|---|
| 1-TP | 1.7 µM | 8.4 µM | 49 µM |
| 2-TP | 0.15 µM | 1.1 µM | 13 µM |
2.4. Computational molecular modeling studies
In order to gain further insights on the mechanism of action of the fleximers, their predicted binding to DENV (PDB ID 4V0R), ZIKV (PDB ID 5GOZ), YFV (PBD ID 3EVD), and human N7 (PDB ID 5E9W) MTase crystal structures were evaluated using a series of docking simulations. In particular, the cap/GTP binding site of these enzymes was explored since the triphosphates 1-TP and 2-TP were the most active against both ZIKV and DENV MTases. Furthermore, as this binding site is highly conserved among flaviviruses,70 it was hypothesized that if the fleximers efficiently bind in this site, they could potentially serve as broad spectrum inhibitors.
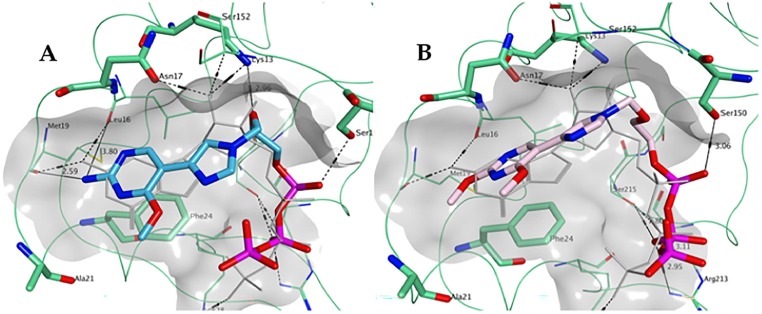
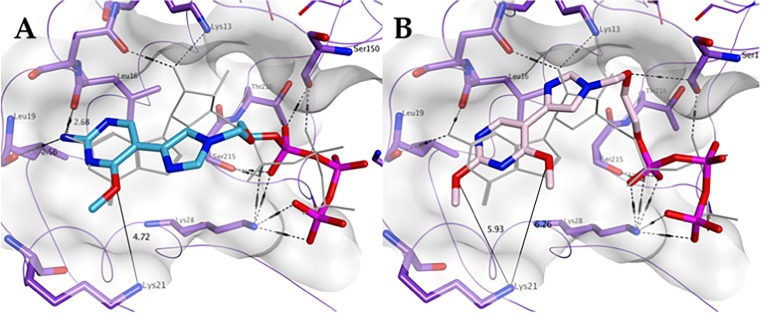
As shown in Fig. 8 A, 1-TP is predicted to maintain most of the key hydrogen bonding and stacking interactions shown by GTP in the ZIKV MTase structure demonstrating a very similar spatial occupation of the pocket overall. Notably, the hydrogen bonding between the free amine group of the fleximer with Met19 and Leu16 as well as between the oxygen in the sugar moiety of the fleximer and Lys13 of the enzyme binding site appears to be similar to the corresponding groups in GTP.
Fig. 8.
Predicted binding of A) 1-TP (carbon atoms in light blue) and B) 2-TP (carbon atoms in pink) to the GTP pocket of ZIKV NS5 MTase (PDB ID 5GOZ). Co-crystallized GTP is shown in light grey.
The triphosphate analogue 2-TP was also analyzed (Fig. 8B), in order to assess the potential effect of the replacement of the free amine at the 2-position with a methoxy group on binding to the ZIKV MTase. According to the docking results obtained, this modification is associated with the potential loss of hydrogen bonding with the backbone of Met19 and Leu16. However, the flex-nucleobase was still oriented such a way that it interacted with Phe24. In the case of both 1-TP and 2-TP, the triphosphate moiety was placed in the same region observed for GTP, and overall both compounds were predicted to occupy the pocket in a similar fashion to GTP.
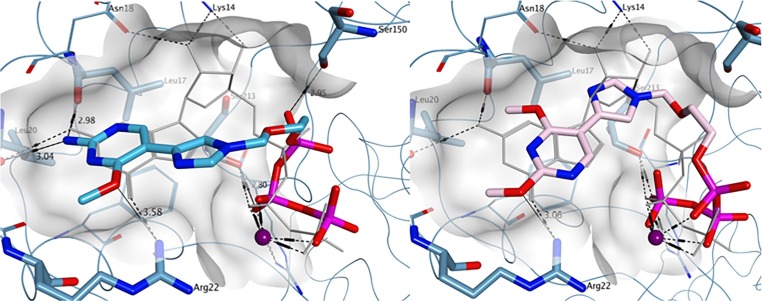
While the ZIKV MTase GTP binding site shows the presence of an alanine at position 21, the residue in the corresponding position is replaced by an arginine (Arg22) within DENV-3 and DENV-4, and by a lysine (Lys22) in DENV-1 and DENV-2 (Fig. 9 A, DENV-3, PDB ID 4V0R). The arginine (or lysine) lateral chain allows for an additional hydrogen bond with the methoxy group of compound 1-TP, which could potentially explain its increased antiviral activity against DENV compared to ZIKV. When compared to the binding interactions found with 1-TP, 2-TP also displayed similar potential hydrogen bonding interaction between the 4-methoxy group and Arg22. As seen for ZIKV MTase, the replacement of the amine group in 1-TP with the methoxy group in 2-TP led to a loss of hydrogen bond formation with Leu17 and Leu20 (Fig. 9B). Moreover, the overall binding of both 1-TP and 2-TP was consistent with the conformation observed for co-crystallized GTP. In summary, the molecular modeling results obtained for both ZIKV and DENV MTases are in accordance with the experimental data found in the enzymatic MTase assay. The lack of significant antiviral activity displayed by 2-TP could also be explained by poor phosphorylation of the parent fleximer analogue to its triphosphate form, likely due to the role of the nucleobase amine group in substrate recognition by the phosphorylating enzymes.
Fig. 9.
Predicted binding of A) 1-TP (carbon atoms in light blue) and B) 2-TP (carbon atoms in pink) to the GTP pocket of DENV NS5 MTase (PDB ID 4V0R). Co-crystallized GTP is shown in light grey.
Similar to the DENV-1 and DENV-2 MTase binding sites, the YFV GTP MTase binding site (PDB ID 3EVD) showed the presence of a lysine residue, Lys21, in proximity to the nucleobase subsite of co-crystallized GTP. The results of the simulations revealed that the triphosphate 1-TP is still predicted to maintain key hydrogen bonding interactions with Leu19 and Leu16. However, unlike the DENV-3 binding site, the Lys21 lateral chain does not appear to be at an optimum distance to interact with the 4-methoxy group (Fig. 10 A). This supports the decrease in activity seen with compound 1 against DENV and YFV (0.057 µM compared to 0.37 µM). By comparison, 2-TP is unable to form any substantial hydrogen bonding interactions with Leu19, Leu16, or Lys21 (Fig. 10B).
Fig. 10.
Predicted binding of A) 1-TP (carbon atoms in light blue) and B) 2-TP (carbon atoms in pink) to the GTP pocket of YFV NS5 MTase (PDB ID 3EVD). Co-crystallized GTP is shown in light grey.
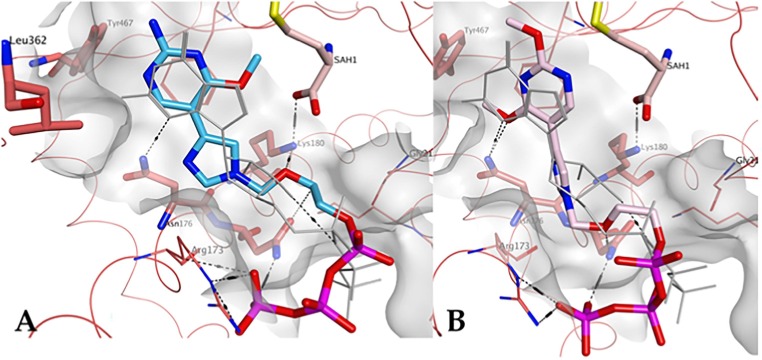
Finally, the potential interactions between the fleximer triphosphate analogues and the human mRNA cap guanine-N7 GTP binding site were analyzed (PDB ID 5E9W; GTP coordinates as defined in the E. cuniculi Ecm1 crystal structure 1RI1). The GTP binding pocket of human N7 MTase is significantly different from the one found in flaviviruses: it more closely resembles the SAM/SAH binding site and possesses a different amino acid residue composition. In line with the experimental data obtained, 1-TP was not predicted to have strong binding interactions in this site, even though the fleximer can adopt a similar general orientation in comparison with the natural ligand GTP (Fig. 11 A). The flex-nucleobase occupied a larger region of space than that defined by the GTP guanine moiety, and the residues surrounding this region of the pocket do not participate in H-bond interactions. Moreover, there are no other notable interactions with the flex-nucleobase of the scaffold, thus supporting the experimental data observed for the reduced inhibition of this enzyme. In contrast, docking results in the GTP binding pocket of human N7 MTase would suggest a better interaction of 2-TP to this enzyme in comparison with 1-TP, as the presence of the two methoxy groups appear to allow formation of a hydrogen bond with Asn176 (Fig. 11B).
Fig. 11.
Predicted binding of A) 1-TP (carbon atoms in ight blue) and B) 2-TP (carbon atoms in pink) to the GTP pocket of human mRNA cap guanine-N7 MTase GTP binding site (PDB ID 5E9W). GTP is shown in light grey.
3. Conclusions
The design and synthesis of new and more effective antiviral drugs is of critical importance to the biomedical field in order to treat viruses such as flaviviruses. While ongoing studies have identified various therapeutics as potential treatments for diseases caused by flaviviruses, there are currently no FDA approved vaccines (except for YFV, however this vaccine has been associated with serious adverse effects.71) or treatment, and as such, it is critical that an effective treatment option is developed. The flex-analogues reported in this study have demonstrated moderate activity against various flaviviruses, with analogue 1 being most active against DENV and YFV. While the mechanism of action has yet to be fully elucidated, these preliminary studies have shown that compound 1-TP inhibits the DENV and ZIKV MTases with IC50 values of 8.4 µM and 1.7 µM respectively, potentially by binding in the GTP binding site of this enzyme. These results are promising due to the highly conserved nature of flavivirus MTases. Further research is currently underway in order to fully elucidate their mechanism(s) of action as well as to screen these analogues against other flaviviruses such as West Nile Virus and Tick-Borne Encephalitis, in order to see if these analogues could serve as broad-spectrum treatments against additional flaviviruses.
4. Experimental
4.1. Chemical synthesis
4.1.1. General Information
All reactions were performed using oven-dried glassware under a nitrogen atmosphere with magnetic stirring. Reagents were purchased from Sigma-Aldrich, Alfa Aesar, and Combiblocks. Solvents were either purchased as anhydrous or were dried using the MBRAUN solvent purification system (MB-SPS). Reactions were monitored by thin layer chromatography (TLC) using EMD silica gel 60 F254 coated glass-backed TLC plates and visualized with a UV lamp and/or KMnO4 stain. Column chromatography was performed on a Combiflash®Rf automated chromatography using REdiSep Rf silica. High Pressure Liquid Chromatography (HPLC) purification was performed on an LC-8A Shimadzu preparation liquid chromatograph using a Zorbax SB C18 column 9.4x250 mm, 5 µm. Samples were lyophilized utilizing a Labconco Freezone 1 L lyophilizer. 1H NMR spectra was recorded on a JOEL Eclipse ECX (400 MHz) spectrometer. Chemical shifts are reported in parts per million (δ ppm) from tetramethylsilane with the solvent resonance as an internal standard. Data for the 1H NMR are reported as follows; br = broad, s = singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublets, m = multiplet. 13C NMR were recorded on a JOEL Eclipse ECX (126 MHz) or a Bruker Advance III HD (101 MHz) spectrometer with complete proton decoupling. Data for 13C NMR are reported in terms of chemical shifts (δ ppm) with the solvent resonance as the internal standard. 31P NMR was recorded on a JOEL Eclipse ECX (162 MHz) spectrometer with complete proton decoupling using 85% phosphoric acid as an external standard. Data for 31P NMR are reported in terms of chemical shift (δ ppm) (multiplicity, integration), with multiplicities reported the same as for 1H NMR. Mass spectrometry was performed on a Bruker AmaZon X quadrupole ion trap mass spectrometer using electrospray ionization (ESI). The purity of all compounds tested in biological assays were verified by elemental analyses performed by Atlantic Microlabs.
4.1.2. Synthesis of 2-((4,5-diiodo-1H-imidazol-1-yl)methoxy)ethyl acetate (4)
4,5-diiodoimidazole (10.0 g, 31.25 mmol) was suspended in anh. acetonitrile (200 mL) under nitrogen and stirred at room temperature for 10 mins. Then bis(trimethylsilyl)acetamide (46.0 mL, 187.68 mmol) and 3 (6.0 mL, 37.46 mmol) were added and the solution was allowed to stir at room temperature under direct nitrogen bubble for 4 h. Trimethylsilyl trifluoromethanesulfonate (8.2 mL, 47.22 mmol) was then added slowly dropwise over 5 min. The reaction was then heated to 80 °C under direct nitrogen bubble for 18 h. After cooling to room temperature, the reaction was cooled further in a salt/ice bath to 0 °C and quenched with NaHCO3 (100 mL). After stirring for 10 min, the reaction was extracted 5x with CH2Cl2 (200 mL). The combined organic layers were dried with MgSO4, gravity filtered, and the solvent was removed in vacuo to afford an amber oil with yellow solid. The crude mixture was then purified by flash column chromatography on silica gel (40–80% EtOAc/Hexanes) to give the pure product as a white solid with yellow oil (9.5 g, 70%); Rf = 0.39 (1:1 EtOAc/Hexanes); 1H NMR (400 MHz, CDCl3) δ 7.61 (s, 1H), 5.18 (s, 1H), 3.93–3.95 (t, J = 4.6 Hz, 2H), 3.45–3.47 (t, J = 4.6 Hz, 2H), 1.82 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 170.67, 142.06, 97.32, 82.71, 77.93, 66.69, 62.76, 21.07; MS (ESI + ) m/z calcd for C8H10I2N2O3 [M + H+]: 436.99, found: 436.9.
4.1.3. Synthesis of 2-((4-iodo-1H-imidazole-1-yl)methoxy)ethan-1-ol (5)
To a solution of compound 4 (5.6 g, 12.87 mmol) in 30% EtOH and H2O (125 mL total volume) was added Na2SO3 (8.17 g, 64.82 mmol). The solution was then refluxed at 120 °C for 18 h. After cooling to room temperature, the reaction was concentrated in vacuo to remove the EtOH and was extracted 4x with CH2Cl2. The combined organic layers were dried with MgSO4 and gravity filtered. The solvent was removed in vacuo to afford a white solid. The crude product was then purified by flash column chromatography on silica gel (0–20% CH3OH in CH2Cl2) to give the pure product as a white solid (2.73 g, 79%); Rf = 0.79 (1:3 CH3OH/CH2Cl2); 1H NMR (400 MHz, DMSO‑d 6) δ 7.74 (s, 1H), 7.46 (s, 1H), 5.31 (s, 2H), 4.65–4.68 (t, J = 10.5 Hz, 1H), 3.42–3.47 (m, 2H), 3.35–3.37 (m, 2H); 13C NMR (126 MHz, DMSO‑d 6) δ 140.5, 125.5, 83.4, 76.2, 70.7, 60.3; MS (ESI + ) m/z calcd for C6H9IN2O2 [M + H+]: 269.05, found 269.0.
4.1.4. Synthesis of 4-methoxypyrimidine-2-amine (7)
Commercially available 2-amino-4-chloro-6-methoxy-pyrimidine (8.0 g, 50.13 mmol) was suspended in 3:2 EtOAc: CH3OH (175 mL total volume). After addition of diisopropylethylamine (18 mL, 100.25 mmol) and 10% Pd/C (1.6 g, 20% w/w), the reaction was mixed on a hydrogenator for 4 h. Upon completion, the reaction was filtered over a celite pad, washed with EtOAc (100 mL), and the solvent was removed in vacuo to give a pale yellow/white solid. The crude product was then purified by flash column chromatography on silica gel (40–80% EtOAC in Hexanes) to give the pure product as a white solid (5.3 g, 85%); Rf = 0.34 (1:1 EtOAc/Hexanes); 1H NMR (400 MHz, DMSO‑d 6) δ 7.90–7.91 (d, J = 5.5 Hz, 1H), 6.52 (s, 2H)5.94–5.95 (d, J = 5.5 Hz, 1H), 3.74 (s, 3H); 13C NMR (126 MHz, DMSO‑d 6) δ 170.16, 164.08, 158.99, 96.58, 53.25; MS (ESI + ) m/z calcd for C5H7N3O [M + H+]: 126.13, found 126.2.
4.1.5. Synthesis of 5-bromo-4-methoxypyrimidin-2-amine (9)
To a solution of compound 7 (1.5 g, 11.99 mmol) in chloroform, N-bromosuccinimide (2.14 g, 11.99 mmol) was added. After stirring in the dark for 5 h, the solution was added to CH2Cl2 (200 mL) and 1 M NaOH (100 mL). Upon mixing the layers were separated and the organic layer was washed with brine (100 mL). The organic layer was then dried with MgSO4 and gravity filtered. The solvent was removed in vacuo to afford a yellow/white solid. The crude product was then purified by flash column chromatography on silica gel (0–5% CH3OH in CH2Cl2) to give the pure product as a fluffy white solid (2.4 g, 95%); Rf = 0.59 (1:1 EtOAc/Hexanes); 1H NMR (400 MHz, DMSO‑d 6) δ 8.05 (s, 1H), 6.78 (s, 2H), 3.83 (s, 3H); 13C NMR (126 MHz, DMSO‑d 6) δ 165.17, 162.91, 159.48, 91.02, 54.44; MS (ESI + ) m/z calcd for C5H6BrN3O [M+]: 204.03, found: 204.0.
4.1.6. Synthesis of 5-bromo-2,4-dimethoxypyrimidine (10)
NaHCO3 (2 g, 23.81 mmol) was slurried with compound 8 (1.74 mL, 14.27 mmol) in 50% CH3OH in water (45 mL total volume). Br2 (1.32 mL, 25.69 mmol) was added dropwise over 1 h with stirring. After 30 min of Br2 addition, additional NaHCO3 (3.5 g, 41.66 mmol) was added and the reaction mixture was stirred at room temperature for 2 h. The resulting white precipitate was vacuum filtered and washed with CH3OH (20 mL) and dried under vacuum to give the product as a shiny white solid (2.12 g, 68%); Rf = 0.97 (1:1 EtOAc/Hexanes); 1H NMR (400 MHz, CDCl3) δ 8.28 (s, 1H), 4.03 (s, 3H), 3.96 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 166.86, 164.36, 159.23, 98.26, 55.33, 55.00; MS (ESI + ) m/z calcd for C6H7BrN2O2 [M+]: 219.04, found: 219.0.
4.1.7. Synthesis of 2-((4-(2-amino-4-methoxypyrimidin-5-yl)-1H-imidazol-1-yl) methoxy)ethan-1-ol (1)
To a dry Schlenk flask, compound 9 (0.50 g, 2.45 mmol), bis(pinacolato)diboron (0.75 g, 2.94 mmol), and potassium acetate (0.72 g, 7.35 mmol), were added and suspended in dimethoxy ethane (50 mL). Then the solution was subjected to freeze–pump-thaw cycles under N2 (3x). Tetrakis(triphenylphosphine)-palladium(0) (0.30 g, 0.25 mmol) was added to the reaction flask and subjected to one more freeze-pump-thaw cycle. The reaction was then heated to 90 °C overnight with stirring. The reaction was then cooled to room temperature and 5 (0.65 g, 2.45 mmol), tetrakis(triphenylphosphine)palladium(0) (0.14 g, 0.12 mmol), and aqueous NaHCO3 (20 mL) were added. The flask was evacuated and pumped with N2 and heated to 90 °C for 4 h. After stirring, the reaction was cooled to room temperature and the solvent was removed in vacuo. The residue was suspended in CH2Cl2/CH3OH and NaHCO3 was gravity filtered away from the solution. Solvent was removed in vacuo to give the crude product as an orange oil. The crude product was purified via flash column chromatography (0–10% CH3OH in CH2Cl2) to afford a yellow oil. The residue was then washed with minimal aqueous sodium thiosulfate and extracted with EtOAc 3-4x. The organic layer was dried with MgSO4, gravity filtered, and the solvent was removed in vacuo to give a pink solid. The product was further purified via flash column chromatography on silica gel (5–10% CH3OH in CH2Cl2) to afford the product as a light pink solid (0.19 g, 30%); Rf = 0.26 (1:9 CH3OH/CH2Cl2); 1H NMR (400 MHz, DMSO‑d 6) δ 8.59 (s, 1H), 7.77 (s, 1H), 7.37 (s, 1H), 6.52 (s, 2H), 5.34 (s, 2H), 4.64–4.66 (t, J = 11.0 Hz, 1H), 3.91 (s, 3H), 3.41–3.44 (t, J = 10.0 Hz, 2H), 3.36–3.38 (t, J = 9.6 Hz, 2H); 13C NMR (126 MHz, DMSO‑d 6) δ 165.78, 162.24, 155.48, 138.08, 134.72, 117.18, 104.65, 76.21, 70.39, 60.44, 53.67; MS (ESI + ) m/z calcd for C11H15N5O3 [M + H+]: 266.27, found 266.2; Elemental analysis: Anal. Calcd for C11H15N5O3: C, 49.81; H, 5.70; N, 26.40. Found: C, 49.74; H, 5.76; N, 26.12.
4.1.8. Synthesis of 2-((4-(2,4-dimethoxypyrimidine-5-yl)-1H-imidazol-1-yl) methoxy)ethan-1-ol (2)
To a dry Schlenk flask, compound 10 (0.50 g, 2.28 mmol), bis(pinacolato)diboron (0.69 g, 2.74 mmol), and potassium acetate (0.67 g, 6.84 mmol) were added and suspended in dimethoxy ethane (50 mL). Then the solution was subjected to freeze–pump-thaw cycles under N2 (3x). Tetrakis(triphenylphosphine)-palladium(0) (0.26 g, 0.22 mmol) was added to the reaction flask and subjected to one more freeze-pump-thaw cycle. The reaction was heated to 90 °C overnight with stirring. The reaction was then cooled to room temperature and 5 (0.61 g, 2.28 mmol), tetrakis(triphenylphosphine)palladium(0) (0.13 g, 0.11 mmol), and aqueous NaHCO3 (20 mL) were added. The flask was evacuated and pumped with N2 and heated to 90 °C for 4 h. After stirring, the reaction was cooled to room temperature and the solvent was removed in vacuo. The residue was suspended in CH2Cl2/CH3OH and NaHCO3 was gravity filtered away from the solution. Solvent was removed in vacuo to give the crude product as a yellow solid with a yellow oil. The crude product was purified via flash chromatography on silica gel (0–5% CH3OH in CH2Cl2) to afford the product as a white solid (0.31 g, 48%); Rf = 0.38 (1:9 CH3OH/CH2Cl2); 1H NMR (400 MHz, CD3OD) δ 8.77 (s, 1H), 7.87 (s, 1H), 7.63 (s, 1H), 5.44 (s, 2H), 4.10 (s, 3H), 3.98 (s, 3H), 3.62–3.64 (t, J = 9.1 Hz, 2H), 3.51–3.64 (t, J = 9.6 Hz, 2H); 13C NMR (126 MHz, CD3OD) δ 167.13, 163.70, 154.60, 137.89, 133.13, 118.77, 108.89, 76.54, 70.07, 60.57, 54.08, 53.48; MS (ESI + ) m/z calcd for C12H16N4O4 [M + H+]: 281.28, found: 281.2; Elemental analysis: Anal. calcd for C12H16N4O4: C, 51.42; H, 5.75; N, 19.99. Found: C, 50.99; H, 5.68; N, 19.72.
4.1.9. Synthesis of 2-((4-(2-amino-4-methoxypyrimidin-5-yl)-1H-imidazol-1-yl)methoxy)ethyl acetate (1-Ac)
Compound 1 (0.050 g, 0.19 mmol) was suspended in anh. DMF (2 mL) and treated with acetic anhydride (0.053 mL, 0.56 mmol) and dimethylaminopyridine (0.0023 g, 0.019 mmol). The reaction was stirred at room temperature for 3 h. After stirring, the solvent was removed in vacuo to give the crude product as a clear oil. The crude product was purified via flash column chromatography on silica gel (0–5% CH3OH in CH2Cl2) to afford the product as a light pink solid (0.040 g, 70%); Rf = 0.28 (1:9 CH3OH/CH2Cl2); 1H NMR (400 MHz, CD3OD) δ 8.54 (s, 1H), 7.48 (s, 1H), 5.40 (s, 2H), 4.13–4.15 (t, J = 9.6 Hz, 2H), 4.00 (s, 3H), 3.64–3.66 (t, J = 9.6 Hz, 2H), 1.96 (s, 3H); 13C NMR (126 MHz, CD3OD) δ 171.27, 166.39, 161.86, 153.87, 137.44, 134.30, 117.40, 104.76, 76.25, 66.53, 62.92, 52.78, 19.33; MS (ESI + ) m/z calcd for C13H17N5O4 [M + H+]: 308.31, found: 308.2; Elemental analysis: Anal. calcd for C13H17N5O4 + 0.3% CH3OH: C, 50.18; H, 5.78; N, 22.08. Found: C, 50.41; H, 5.79; N, 22.10.
4.1.10. Synthesis of 2-((4–2,4-dimethoxypyrimidin-5-yl)-1H-imidazol-1-yl)methoxy) ethyl acetate (2-Ac)
Compound 2 (0.050 g, 0.18 mmol) was suspended in anh. DMF (2 mL) and treated with acetic anhydride (0.051 mL, 0.53 mmol) and dimethylaminopyridine (0.0022 g, 0.018 mmol). The reaction was stirred at room temperature for 3 h. After stirring, the solvent was removed in vacuo to give the crude product as an orange oil. The crude product was purified via flash chromatography on silica gel (0–5% CH3OH in CH2Cl2) and placed on a vacuum pump overnight to afford the product as a peach colored solid (0.076 g, 86%); Rf = 0.41 (1:9 CH3OH/CH2Cl2); 1H NMR (400 MHz, CD3OD) δ 8.77 (s, 1H), 7.86 (s, 1H), 7.60 (s, 1H), 5.42 (s, 2H), 4.13–4.16 (t, J = 9.2 Hz, 2H), 4.09 (s, 3H), 3.97 (s, 3H), 3.66–3.68 (t, J = 9.6 Hz, 2H), 1.96 (s, 3H); 13C NMR (126 MHz, CD3OD) δ 171.23, 167.09, 163.70, 154.62, 137.89, 133.19, 118.70, 108.81, 76.32, 66.62, 62.92, 54.11, 53.50, 19.35; MS (ESI + ) m/z calcd for C14H18N4O5 [M+H+]: 323.32, found: 323.2; Elemental analysis: Anal. calcd for C14H18N4O5 + 0.4% H2O: C, 51.18; H, 5.79; N, 16.93. Found: C, 51.03; H, 5.75; N, 17.00.
4.1.11. Synthesis of 2-ethylbutyl((2-((4-(2-amino-4-methoxypyrimidin-5-yl)-1H-imidazol-1-yl)methoxy)-ethoxy)-(phenoxy)phosphoryl)alaninate (1-MG)
To a solution of 1 (0.04 g, 0.15 mmol) in anh·THF (3 mL) under a nitrogen atmosphere was added tertbutylmagnesium chloride (1 M in THF, 0.24 mL, 0.24 mmol) dropwise, and the reaction was stirred at room temperature for 30 min. In a separate flask, 13 (0.08 g, 0.17 mmol) was suspended in anh·THF (4 mL) and added to the reaction vessel. The reaction was allowed to stir at room temperature under a nitrogen atmosphere overnight. After stirring overnight, the reaction was quenched with CH3OH (2 mL) and stirred for another 5 min before removal of solvent in vacuo. The crude mixture was then purified by flash column chromatography on silica gel (0–10% CH3OH in CH2Cl2) to give the pure product as a pink oil (0.06 g, 74% mix of diastereomers); Rf = 0.49 (1:9 CH3OH/CH2Cl2); 1H NMR (400 MHz, CD3OD) δ 8.64 (s, 1H), 7.79–7.80 (d, J = 5.4 Hz, 1H), 7.49 (s, 1H), 7.26–7.30 (m, 2H), 7.10–7.16 (m, 3H), 5.38–5.41 (app d, 2H), 4.13–4.22 (m, 2H), 3.86–4.04 (m, 6H), 3.65–3.70 (m, 2H), 1.40–1.48 (m, 1H), 1.27–1.35 (m, 7H), 0.82–0.86 (m, 6H); 13C NMR (126 MHz, CD3OD) δ 173.85, 166.46, 161.66, 153.50, 150.83, 137.48, 134.14, 129.38, 124.72, 120.17, 120.10, 117.4, 104.86, 76.31, 67.44, 67.38, 66.70, 65.69, 52.88, 50.51, 50.22, 40.41, 22.92, 19.31, 19.25, 19.21, 19.14, 10.02; 31P NMR (162 MHz, CD3OD) δ 4.39, 4.17; MS (ESI + ) m/z calcd for C26H37N6O7P [M + H+]: 577.25, found: 577.3; Elemental analysis: Anal. calcd for C26H37N6O7P + 1.3% CH3OH: C, 53.04; H, 6.88; N, 13.59. Found: C, 53.31; H, 6.66; N, 13.35.
4.1.12. Synthesis of 2-ethylbutyl ((2-((4-(2,4-dimethoxypyrimidin-5-yl)-1H-imidazol-1-yl)methoxy)ethoxy)-(phenoxy)-phosphoryl)alaninate (2-MG)
To a solution of 2 (0.042 g, 0.15 mmol) in anh·THF (3 mL) under a nitrogen atmosphere was added tertbutylmagnesium chloride (1 M in THF, 0.30 mL, 0.30 mmol) dropwise, and the reaction was stirred at room temperature for 30 min. In a separate flask, 13 was suspended in anh·THF (4 mL) and added to the reaction vessel. The reaction was allowed to stir at room temperature under a nitrogen atmosphere overnight. After stirring overnight, the reaction was quenched with CH3OH (2 mL) and stirred for another 5 min before removal of solvent in vacuo. The crude mixture was then purified twice by flash column chromatography on silica gel (0–10% CH3OH in CH2Cl2) to give the product as a yellow oil (0.075 g, 86% mix of diastereomers); Rf = 0.48 (1:9 CH3OH/CH2Cl2); 1H NMR (400 MHz, CDCl3) δ 8.96 (s, 1H), 7.58–7.60 (d, J = 5.9 Hz, 1H), 7.44–7.45 (d, J = 2.7 Hz, 1H), 7.23–7.27 (m, 2H), 7.07–7.16 (m, 3H), 5.26–5.28 (app. d, 2H), 4.13–4.21 (m, 2H), 3.92–4.05 (m, 8H), 3.72–3.82 (m, 1H), 3.58–3.63 (m, 2H), 1.41–1.59 (m, 1H), 1.24–1.34 (m, 8H), 0.79–0.84 (m, 6H); 13C NMR (126 MHz, CDCl3) δ 173.65, 166.94, 163.72, 155.64, 150.67, 137.30, 134.72, 129.70, 125.03, 120.20, 118.06, 109.32, 67.53, 65.66, 54.9, 54.16, 50.28, 40.25, 23.21, 21.16, 11.03; MS (ESI + ) m/z calcd for C27H38N5O8P [M + H+]: 592.60, found: 592.3; Elemental Analysis: Anal. calcd for C27H38N5O8P + 1% CH3OH: C, 53.93; H, 6.79; N, 11.23. Found: C, 53.89; H, 6.55; N, 11.02.
4.1.13. General procedure for triphosphate synthesis (1-TP and 2-TP)66
Nucleoside 1 or 2 (0.3 g, 0.1 mmol) was coevaporated with anh. Pyridine and placed in a vacuum overnight. Simultaneously, tributylammonium pyrophosphate (0.13 mmol) was dried in a separate flask under vacuum overnight. The dry nucleoside was dissolved in anh. Pyridine (0.2 mL) and anh. 1,4-dioxane (0.4 mL) under a N2 filled balloon. Then 2-chloro-1,3,2-benzodioxaphosphorin-4-one (0.12 mmol) was added and the reaction was stirred at room temperature for 45 min (Ensure that the white powder impurity is scraped off of the 2-chloro-1,3,2-benzodioxaphosphorin-4-one crystal, which should either be a glassy green or white solid, before use). Then anh. DMF (0.17 mL) and tributylamine (0.23 mmol) were added to the tributylammonium pyrophosphate flask and sonicated to mix. The solution was added to the reaction mixture and stirred at room temperature for 45 min. In a separate vial, iodine (0.041 g, 0.16 mmol) was added to pyridine (0.98 mL) and H2O (20 µL) and sonicated to mix. The contents were then added to the reaction flask and stirred at room temperature for 30 min. The reaction was quenched with 10% Na2S2O3 (3 mL). The solvent was removed in vacuo while keeping the temperature of the water bath below 30 °C. Then H2O (4 mL) was added and the reaction sat for 30 min at room temperature. Solvent was removed under vacuum to afford crude triphosphate as a yellow oil. Products were purified via reverse phase HPLC (0–100% 0.1 M TEAB with 30% MeCN in 0.1 M TEAB, flow rate = 3.5 mL/min). Products were then purified further via ion-exchange chromatography utilizing Dowex 50WX2 Na+ charged resin. Column was washed with ultra-pure H2O, 1 M HCl, ultra-pure H2O, 1 M NaOH, and finally ultra-pure H2O before the sample was run through the column and washed with more ultra-pure H2O. All samples were then freeze dried by lyophilization for testing.
4.1.13.1. Characterization of 2-((4-(2-amino-4-methoxypyrimidin-5-yl)-1H-imidazol-1-yl)methoxy)ethyl tetrahydrogen triphosphate (1-TP)
Product is a white solid after lyophilization (0.029 g, 50.9%); 1H NMR (400 MHz, D2O) δ 8.3 (s, 1H), 7.9, (s, 1H), 7.5 (s, 1H), 5.4 (s, 2H), 4.7 (s, 2H), 4.0 (m, 2H), 3.8 (s, 3H), 3.7 (m, 2H); 31P NMR (162 MHz, D2O) δ −9.5 (d, 1P), −10.5 (m, 1P), –22.6 (t, 1P); MS (ESI + ) m/z calcd for C11H18N5O12P3 [M−H]: 504.02, found: 503.98.
4.1.13.2. Characterization of 2-((4-(2,4-dimethoxypyrimidin-5-yl)-1H-imidazol-1-yl)methoxy)ethyl tetrahydrogen triphosphate (2-TP)
Product is a white solid after lyophilization (0.035 g, 62%); 1H NMR (400 MHz, D2O) δ 8.4, (s, 1H), 7.9 (s, 1H), 7.6 (s, 1H), 5.4 (s, 2H), 4.0 (m, 2H), 3.9 (s, 3H), 3.8 (s, 3H), 3.6 (m, 2H); 13C NMR (126 MHz, D2O) δ 167.3, 163.4, 153.9, 138.4, 133.6, 119.4, 109.4, 76.3, 68.2, 64.9, 55.0, 54.6; 31P NMR (162 MHz, D2O) δ −6.2 (d, 1P), −10.4 (m, 1P), –22.1 (t, 1P); MS (ESI + ) m/z calcd for C12H19N4O13P3 [M−H]: 519.22, found: 519.01.
4.2. Enzyme production and purification
The coding sequence corresponding to the ZIKV and DENV-3 MTase domain were cloned in fusion with hist6-Tag in pQE30 expression vector as previously described.72, 73 and the corresponding protein were produced Escherichia coli T7 Express Iq (New England BioLabs). Cells were grown in Terrific Brother at 97 °C until the optical density of 0.6 at 600 nm (OD600) and the Mtase expression was induced by adding 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) before overnight expression at 17 °C. The bacteria were pellets and lysed in (50 mM Tris-HCl [pH 8], 300 mM NaCl, 5% glycerol, 0.1% Triton, 10 µg/ml DNase I, 2 tablets of EDTA-free antiprotease cocktail [Roche], 0.25 mg/ml lysozyme). After sonication the clarified proteins were purified by immobilized metal affinity chromatography (IMAC, GE Healthcare), and after, several washes of elution were performed in 50 mM Tris-HCl, 300 mM NaCl, and 250 mM imidazole (pH 8.0). The eluted proteins were then separated by gel filtration on a 16/60 Superdex 200 (GE Healthcare) equilibrated in a mixture of 10 mM HEPES, 500 mM NaCl, 5% glycerol, and 1 mM dithiothreitol (DTT [pH 7.5]).
For the NS5 polymerase encoding both MTase and RdRp activities, the gene coding for a N-terminal His6-tagged DENV NS5 (serotype 2, strain New Guinea C) was cloned in a pQE30 expression plasmid as described previously in Selisko et. al.74 Expression and purification was as described by Potisopon et. al.75 The synthetic ZIKV NS5 gene cloned in a pQE30 expression plasmid was obtained from Genescript. The sequence used was from strain H/PF/2013 from French Polynesia, Genbank acc# KJ776791. The ZIKV NS5, carrying His6-tag at its N-terminus, was produced as described above for DENV NS5 with the exception that IMAC TALON beads were washed with 1.5 M NaCl. SEC was performed using a Superdex S75 HR 16/20 column (GE Healthcare) with SEC buffer 50 mM HEPES pH 7.5, 750 mM NaCl, 10% glycerol, 10 mM DTT. After the second purification step, proteins were concentrated up to around 8 mg/ml (78 µM NS5) and stored at −20 °C after adding glycerol to a final concentration of 40%. Protein purity was higher than 95% as judged by SDS-PAGE. Protein stock concentrations were determined by absorbance measurements at 280 nm using a Nanodrop 2000 (Thermo Scientific).
4.3. DENV and ZIKV 2′-O-MTase assay
DENV-3 and ZIKV 2′-O-MTase activity was followed by incubating the MTase (0.5 µM) with small capped RNA substrate GpppAC5 in the presence of [3H]AdoMet.76 The MTase activity assay was performed in 20 µL samples containing 40 mM Tris-HCl pH 7.5, 5 mM DTT, 10 µM AdoMet (0.2–2 µCi [3H]AdoMet), 0.5 µM of MTase, 1 µM GpppAC5, and the inhibitors. The reaction was incubated at 30 °C for 30 min and stopped by 20-fold filution in an ice-cold 100 µM AdoHcy solution. Samples were then transferred onto a DEAE membrane (DEAE Filtermat; Wallac) by a Filtermat Harvester (Packard Instruments) washed with 0.01 M ammonium formate (pH 8.0), water, and ethanol, and the radioactivity transferred onto RNA was measured using a Wallac 1450 MicroBeta Trilux Liquid Scintillation Counter.72
The inhibitor concentration at 50% activity (IC50) was determined by performing DENV-3 and ZIKV MTase assays, in the presence of a serial dilution of the inhibitor. All data points were measure in triplicate. The IC50 values were determined using Prism software and adjusted to a logistic dose-response function: % activity = 100/(1 + [I]/IC50)b, where b corresponds to the slope factor and [I] to inhibitor concentration.77
4.4. Molecular modeling
All molecular modeling studies were performed on a Viglen Genie Intel®Core™ i7-3770 vPro CPU@ 3.40 GHz × 8 running Ubuntu 14.04. The MTase structures were downloaded from the PDB data bank (http://www.rcsb.org/; PDB codes 5GOZ,78 4V0R,79 3EVD,80 and 5E9W.81). GTP coordinates for the human N7 MTase crystal structure 5E9W were obtained from the E. cuniculi cap-MTase Ecm1 crystal structure 1RI1, which is in complex with GTP.70 This was done after structural superimposition of 5E9W with 1RI1, which reveals a highly conserved architecture and amino acid residue composition for the GTP binding pockets of the two proteins. Hydrogen atoms were added to the proteins, using the Protonate 3D routine of the Molecular Operating Environment (MOE).82 Ligand structures were built with MOE and minimized using the MMFF94x force field until a RMSD gradient of 0.05 kcal mol−1 Å−1 was reached. The docking simulations were performed using PLANTS applying the following parameters: search algorithm: aco_ants 20, aco_evap 0.15, aco_sigma 2.0; binding site: 5GOZ: bindingsite_center [5.952 45.608–22.492], bindingsite_radius 9; 4V0R: bindingsite_center [-28.826–19.856–30.783], bindingsite_radius 9; 3EVD: bindingsite center [27.897 24.789–44.440], bindingsite radius 9; 5E9W: bindingsite center [15.299 37.929 95.164], binding radius 9; cluster algorithm: cluster_rmsd 2.0, cluster_structures 30; scoring function: chemplp.83 Docking results were visually inspected using MOE.
Author Contributions
Synthesis, J.E.T, C.D.W, and A.F.; enzyme assays, C.V., B.M., W.A., B.S., B.C., E.D., and B.C.; computational modeling, M.B. and A.B.; writing- original draft preparation, J.E.T. and K.L.S.R.; writing- review and editing, K.L.S.R., B.C., E.D., M.C., M.B., and A.B.; K.L.S.R planned, designed, and organized the study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by National Institutes of Health: NIGMS T32 GM066706 (KSR) and NIAID R21AI135252-01 (KSR, JET, CDW).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2020.115713.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Pierson T.C., Diamond M.S. Nat Microbiol. 2020;5:796–812. doi: 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldescu V., Behnam M.A.M., Vasilakis N., Klein C.D. Nat Rev Drug Discov. 2017 doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay S., Kuhn R.J., Rossmann M.G. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 4.Lu G., Gong P. Virus Res. 2017;234:34–43. doi: 10.1016/j.virusres.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 5.García L.L., Padilla L., Castaño J.C. Virol J. 2017;14:95. doi: 10.1186/s12985-017-0761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollati M., Alvarez K., Assenberg R. Antiviral Res. 2010;87:125–148. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatelain G., Debing Y., De Burghgraeve T. Eur J Med Chem. 2013;65:249–255. doi: 10.1016/j.ejmech.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 8.Barrows N.J., Campos R.K., Powell S.T. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honein M.A., Dawson A.L., Petersen E.E. JAMA. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- 10.Joguet G., Mansuy J.M., Matusali G. Lancet Infect Dis. 2017 [Google Scholar]
- 11.D'Ortenzio E., Matheron S., Yazdanpanah Y. N Engl J Med. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy M. BMJ. 2016;352 [Google Scholar]
- 13.Hills S.L., Russell K., Hennessey M. MMWR Morb Mortal Wkly Rep. 2016;65:215–216. doi: 10.15585/mmwr.mm6508e2. [DOI] [PubMed] [Google Scholar]
- 14.Musso D., Roche C., Robin E., Nhan T., Teissier A., Cao-Lormeau V.M. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansuy J.M., Dutertre M., Mengelle C. Lancet Infect Dis. 2016;16:405. doi: 10.1016/S1473-3099(16)00138-9. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson B., Hearn P., Afrough B. Detection of Zika Virus in Semen. Emerg Infect Dis. 2016;22:940. doi: 10.3201/eid2205.160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bidet K., Garcia-Blanco M.A. Biochem. J. 2014;462:215–230. doi: 10.1042/BJ20140456. [DOI] [PubMed] [Google Scholar]
- 18.Selisko B., Wang C., Harris E., Canard B. Curr Opin Virol. 2014;9:74–83. doi: 10.1016/j.coviro.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sampath A., Padmanabhan R. Antiviral Res. 2009;81:6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck C., Jimenez-Clavero M.A., Leblond A. Int J Environ Res Public Health. 2013;10:6049–6083. doi: 10.3390/ijerph10116049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milani M., Mastrangelo E., Bollati M. Antiviral Res. 2009;83:28–34. doi: 10.1016/j.antiviral.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bollati M., Milani M., Mastrangelo E. J Mol Biol. 2009;385:140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Liu, L.; Dong, H.; Chen, H.; Zhang, J.; Ling, H.; Li, Z.; Shi, P. Y.; Li, H. Front Biol (Beijing). 2010, 5, 286-303. [DOI] [PMC free article] [PubMed]
- 24.Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benarroch D., Selisko B., Locatelli G.A., Maga G., Romette J.L., Canard B. Virology. 2004;328:208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Issur M., Geiss B.J., Bougie I. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egloff M.P., Decroly E., Malet H. J Mol Biol. 2007;372:723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Dong H., Zhang B., Shi P.Y. Antiviral Res. 2008;80:1–10. doi: 10.1016/j.antiviral.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H., Chang D.C., Hua M.H. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin W.H., Kihara D. Methods Mol Biol. 1958;2019:1–13. doi: 10.1007/978-1-4939-9161-7_1. [DOI] [PubMed] [Google Scholar]
- 31.Brecher M., Chen H., Liu B. PLoS ONE. 2015;10 [Google Scholar]
- 32.Choi K.H., Rossmann M.G. Curr Opin Struct Biol. 2009;19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Tarantino D., Cannalire R., Mastrangelo E. Antiviral Res. 2016;134:226–235. doi: 10.1016/j.antiviral.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Kao C.C., Singh P., Ecker D.J. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 35.Low J.G., Ooi E.E., Vasudevan S.G. J Infect Dis. 2017;215:S96–S102. doi: 10.1093/infdis/jiw423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzelnick L.C., Coloma J., Harris E. Lancet Infect Dis. 2017;17:e88–e100. doi: 10.1016/S1473-3099(16)30473-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim S.P., Sonntag L.S., Noble C. J Biol Chem. 2011;286:6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Z., Chen Y.L., Schul W. Proc Natl Acad Sci U S A. 2009;106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Zou P., Wu F., Lu L., Jiang S. Front Med. 2017;11:449–461. doi: 10.1007/s11684-017-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haviernik J., Štefánik M., Fojtíková M. Viruses. 2018;10:184. doi: 10.3390/v10040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H., Liu L., Jones S.A. Antiviral Res. 2013;97:232–239. doi: 10.1016/j.antiviral.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim S.P., Noble C.G., Shi P.Y. Antiviral Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Chung K.Y., Dong H., Chao A.T., Shi P.Y., Lescar J., Lim S.P. Virology. 2010;402:52–60. doi: 10.1016/j.virol.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Seley K.L., Zhang L., Hagos A. Fleximers. Org Lett. 2001;3:3209–3210. doi: 10.1021/ol0165443. [DOI] [PubMed] [Google Scholar]
- 45.Seley K.L., Zhang L., Hagos A., Quirk S. J Org Chem. 2002;67:3365–3373. doi: 10.1021/jo0255476. [DOI] [PubMed] [Google Scholar]
- 46.Seley K.L., Salim S., Zhang L., O'Daniel P.I. J Org Chem. 2005;70:1612–1619. doi: 10.1021/jo048218h. [DOI] [PubMed] [Google Scholar]
- 47.Seley K.L., Salim S., Zhang L. Org Lett. 2005;7:63–66. doi: 10.1021/ol047895v. [DOI] [PubMed] [Google Scholar]
- 48.Seley K.L., Quirk S., Salim S., Zhang L., Hagos A. Bioorg Med Chem Lett. 2003;13:1985–1988. doi: 10.1016/s0960-894x(03)00331-7. [DOI] [PubMed] [Google Scholar]
- 49.Polak M., Seley K.L., Plavec J. J Am Chem Soc. 2004;126:8159–8166. doi: 10.1021/ja0498078. [DOI] [PubMed] [Google Scholar]
- 50.Quirk S., Seley K.L. Biochem. 2005;44:13172–13178. doi: 10.1021/bi051288d. [DOI] [PubMed] [Google Scholar]
- 51.Quirk S., Seley K.L. Biochem. 2005;44:10854–10863. doi: 10.1021/bi0503605. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z., Jochmans D., Ku T., Paeshuyse J., Neyts J., Seley-Radtke K.L. ACS Infect Dis. 2015;1:357–366. doi: 10.1021/acsinfecdis.5b00029. [DOI] [PubMed] [Google Scholar]
- 53.Peters H.L., Jochmans D., de Wilde A.H. Bioorg Med Chem Lett. 2015;25:2923–2926. doi: 10.1016/j.bmcl.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yates M.K., Raje M.R., Chatterjee P. Bioorg Med Chem Lett. 2017;27:2800–2802. doi: 10.1016/j.bmcl.2017.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wauchope O.R., Velasquez M., Seley-Radtke K. Synthesis (Stuttg) 2012;44:3496–3504. doi: 10.1055/s-0032-1316791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmermann S.C., Sadler J.M., O'Daniel P.I., Kim N.T., Seley-Radtke K.L. Nucleosides Nucleotides Nucleic Acids. 2013;32:137–154. doi: 10.1080/15257770.2013.771187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann S.C., O'Neill E., Ebiloma G.U., Wallace L.J., De Koning H.P., Seley-Radtke K.L. Molecules. 2014;19:21200–21214. doi: 10.3390/molecules191221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elion G.B. J Med Virol Suppl. 1993;1:2–6. doi: 10.1002/jmv.1890410503. [DOI] [PubMed] [Google Scholar]
- 59.Elion G.B., Furman P.A., Fyfe J.A., de Miranda P., Beauchamp L., Schaeffer H.J. Proc Natl Acad Sci U S A. 1977;74:5716–5720. doi: 10.1073/pnas.74.12.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reardon J.E., Spector T. J Biol Chem. 1989;264:7405–7411. [PubMed] [Google Scholar]
- 61.Warren T.K., Jordan R., Lo M.K. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siegel D., Hui H.C., Doerffler E. J Med Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 63.Burger M.T., Pecchi S., Wagman A. ACS Med Chem Lett. 2011;2:774–779. doi: 10.1021/ml200156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nadège B., Srinivas Reddy D., Paul K. Org Lett. 2008;10:1715–1718. [Google Scholar]
- 65.Balzarini, J.; Camarasa, M.J.; Vel ´azquez, S.; PRODRUGS CLEAVABLE BY CD26. WO2004/98644, 2004.
- 66.Ross B.S., Reddy P.G., Zhang H.R., Rachakonda S., Sofia M.J. J Org Chem. 2011;76:8311–8319. doi: 10.1021/jo201492m. [DOI] [PubMed] [Google Scholar]
- 67.Hollenstein, M.; Smith, C. C.; Räz, M. J Vis Exp. 2014. [DOI] [PMC free article] [PubMed]
- 68.Combe M., Sanjuán R. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elena S.F., Sanjuán R. J Virol. 2005;79:11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D. Mol Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 71.de Menezes Martins, R.; Fernandes Leal Mda, L.; Homma, A. Hum Vaccin Immunother. 2015, 11, 2183-7. [DOI] [PMC free article] [PubMed]
- 72.Coutard B., Barral K., Lichiere J. J Virol. 2017:91. doi: 10.1128/JVI.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barral K., Sallamand C., Petzold C. Antiviral Res. 2013;99:292–300. doi: 10.1016/j.antiviral.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Selisko B., Dutartre H., Guillemot J.C. Virology. 2006;351:145–158. doi: 10.1016/j.virol.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 75.Potisopon S., Ferron F., Fattorini V., Selisko B., Canard B. Antiviral Res. 2017;140:25–36. doi: 10.1016/j.antiviral.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 76.Peyrane F., Selisko B., Decroly E. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkl1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeLean A., Munson P.J., Rodbard D. Am J Physiol. 1978;235:E97–E102. doi: 10.1152/ajpendo.1978.235.2.E97. [DOI] [PubMed] [Google Scholar]
- 78.Zhang C., Feng T., Cheng J. Biochem Biophys Res Commun. 2017;492:624–630. doi: 10.1016/j.bbrc.2016.11.098. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y., Soh T.S., Zheng J. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geiss B.J., Thompson A.A., Andrews A.J. J Mol Biol. 2009;385:1643–1654. doi: 10.1016/j.jmb.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varshney D., Petit A.P., Bueren-Calabuig J.A. Nucleic Acids Res. 2016;44:10423–10436. doi: 10.1093/nar/gkw637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.ULC, C. C. G. Molecular Operating Environment (MOE), 2013.08, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2018.
- 83.Korb, O.; Stützle, T.; Exner, T. E. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. In Ant Colony Optimization and Swarm Intelligence: 5th International Workshop, ANTS 2006, Brussels, Belgium, September 4-7, 2006. Proceedings, Dorigo, M.; Gambardella, L. M.; Birattari, M.; Martinoli, A.; Poli, R.; Stützle, T., Eds. Springer Berlin Heidelberg: Berlin, Heidelberg, 2006; pp 247-258.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.