Abstract
Although several drugs have been designed in the last few years to target specific key pathways and functions in colorectal cancer (CRC), the backbone of CRC treatment is still made up of compounds which rely on DNA damage to accomplish their role. DNA damage response (DDR) and checkpoint pathways are intertwined signaling networks that arrest cell cycle, recognize and repair genetic mistakes which arise during DNA replication and transcription, as well as through the exposure to chemical and physical agents that interact with nucleic acids. The good but highly variable activity of DNA damaging agents in the treatment of CRC suggests that intrinsic alterations in DDR pathways and cell cycle checkpoints may contribute differentially to the way cancer cells react to DNA damage. In the present review, our aim is to depict the recent advances in understanding the molecular basis of the activity of DNA damaging agents used for the treatment of CRC. We focus on the known and potential drug targets that are part of these complex and intertwined pathways. We describe the potential role of the checkpoints in CRC, and how their pharmacological manipulation could lead to chemopotentiation or synergism with currently used drugs. Novel therapeutic agents playing a role in DDR and checkpoint inhibition are assessed. We discuss the possible rationale for combining PARP inhibition with DNA damaging agents, and we address the link between DDR and EGFR pathways in CRC.
Keywords: Colorectal cancer, Chk2, cell cycle, DNA damage response, irinotecan, oxaliplatin
INTRODUCTION: COLORECTAL CANCER, DNA DAMAGE RESPONSE PATHWAYS, AND THE CELL CYCLE CHECKPOINTS
Replication of DNA by living organisms requires order and control to ensure correct transmission of all the information coded by nucleic acids from one generation to the next. Indeed, since the first self-reproducing nucleic acids appeared on the Earth, in the likely shape of RNA molecules [1, 2], billions of years of evolution have contributed to the development of one of the most precise and complex machineries of the cell: the DNA damage response (DDR) pathways.
Broadly speaking, DDR pathways are intertwined signaling networks that recognize and repair mistakes that unavoidably arise during DNA replication and transcription, as well as through the exposure of living beings to natural and artificial chemical and physical agents that interact with nucleic acids, damaging them and endangering their fundamental function, i.e. propagating correct genetic information across generations of organisms (prevention of germinal mutations), and across generations of cells inside the same multi-cellular organism [3]. Within the DDR pathways, the eukaryotic cell possesses checkpoint control mechanisms that can arrest the various phases of cell replication and allow for the repair of DNA damage (either endogenously generated by replication itself or through exogenous agents) [4–7]. These processes determine whether the cells repair their genetic material, thus completing the synthesis of sister chromatids and mitosis, or undergo programmed cell death (apoptosis).
Bert Vogelstein defined cancer as “a genetic disease of the somatic cell” [8]. Such a synthetic definition is particularly appropriate to define the genesis of colorectal cancer (CRC). Through either chromosomal instability (CIN, possibly mediated by APC (adenomatous polyposis coli) loss of function or by other mechanisms [9]) or abnormal accumulation of mutations (such as in mismatch repair (MMR) deficiency [10, 11]), colon mucosa cells bearing DNA alterations may survive and proliferate, eventually accumulating enough aberrancies in key genes to allow them to duplicate without control, escape normal cell-cell and cellmatrix inhibitions, acquire an aberrant behavior, and generate tumors.
CRC carcinogenesis is possibly one of the best studied and characterized pathological phenomena in humans [12]. This is due in part to the relative easiness of detection and removal of early, pre-cancerous lesions through instrumental procedures, which has allowed the identification of sequential phases of CRC development. Also, familial clusters of CRC have provided insights into the function of specific genes involved in CRC development, the loss of which results in CRC with extremely high frequency compared to subjects that do not bear such mutations (see Table 1). The germline-mutated genes which charachterize these syndromes are often part of DDR pathways, or directly influence chromosomal stability: emblematic cases are the association of APC/Wnt pathway function loss with CIN [13, 14], and that of MMR deficiency with microsatellite instability and the so called “mutator phenotype” (i.e., a cancer characterized by an increased mutation rate compared to healthy tissues) [15, 16]. Although several drugs have been designed in the last few years to target specific key pathways and functions in CRC (see Bagnasco L. et al., and Ballestrero A. et al., in the present issue of CCDT), the backbone of CRC treatment is still made up of compounds which rely on DNA damage to accomplish their role. Such agents (irinotecan, oxaliplatin, and 5-fluorouracil) yield highly variable, but often important results in patients affected by CRC. This suggests that intrinsic alterations of CRC subtypes may contribute differentially to the way cancer cells react to DNA damage. Indeed, both familial and sporadic CRC is often characterized by aberrancies in DDR pathways (resulting in increased chromosomal or microsatellite instability) and these may explain, in part, such phenotypes of sensitivity/resistance.
Table 1.
Germline mutations in colorectal familial syndromes.
| Syndrome | Involved Gene(s) | Affected Process | Actively Affecting DNA Integrity ? | References |
|---|---|---|---|---|
| Familial adenomatous polyposis (FAP) | APC | APC/β-catenin/Wnt pathway | Yes, chromosomal instability, aneuploidy | [13, 14, 20] |
| MYH-associated polyposis (MAP) | MYH | Base excision repair | Yes, chromosomal instability, aneuploidy | [13, 21–23] |
| Human non-polyposis colon cancer (HNPCC) | MSH2, MLH1, PMS1, PMS2, MSH6, TGFBR2, MLH3 | Mismatch repair | Yes, microsatellite instability, increased mutation rate | [15, 24, 25] |
| Oligodontia-colorectal cancer syndrome | AXIN2 | APC/β-catenin/Wnt pathway | Not described | [14, 20, 26] |
| Familial Finnish colorectal cancer predisposition | CHEK2 | Cell cycle checkpoint | Yes, DNA integrity maintenance | [27, 28] |
| Peutz - Jeghers syndrome | STK11 | Wnt pathway, cell cycle | Not described | [29–33] |
Here, we review the best characterized mechanisms that, in CRC, confer susceptibility/resistance to two DNA damaging chemotherapeutics that are commonly used for the treatment of CRC, irinotecan and oxaliplatin. We describe the potential role of checkpoint pathways, the manipulation of which may lead to chemopotentiation or synergism with currently used drugs, and we discuss the possible rationale for combining PARP inhibition with DNA damaging agents in CRC. The intertwining of DDR pathways with EGFR pathway activation is also addressed. We will not deal with the issue of sensitivity/resistance of CRC to 5-fluorouracil (5-FU), because of the incompletely ascertained relative role of DDR and checkpoint pathways in the response to this agent, compared to its more widely studied mechanism of thymidylate synthase blocking and RNA synthesis interference. We refer the reader to in-depth reviews dealing with such topic (see [17–19]).
PHENOTYPES OF SUSCEPTIBILITY/RESISTANCE TO DNA DAMAGING AGENTS IN CRC: IRINOTECAN AND OXALIPLATIN
Irinotecan
Irinotecan (Camptostar®, Pfizer Inc.) is a semisynthetic analogue of camptothecin (CPT) [34], an anticancer compound isolated from the barks of Camptotheca Acuminata. CPT was discovered in 1966 during systematic in vitro screenings with plant-derived compounds [35]. Camptothecins are topoisomerase I (Top1) inhibitors, acting as interface traps between the enzyme Top1 and DNA, and stabilizing such ternary macromolecular complex. As a consequence, DNA double-strand breaks (DSBs) are generated, and massive DNA damage ensues [34]. Following two large phase III clinical trials, irinotecan was approved as first line treatment of CRC, in combination with 5-FU and leucovorin [36, 37]; it is also active in several other types of solid tumors [38, 39]. Irinotecan is scarcely active per se, but needs to be activated into the highly potent metabolite SN-38 through enzymatic hydrolysis by carboxylesterase 1 (CES1) [40], CES2 [41], and butyrylcholinesterase (BCHE) [42]. These enzymes are widely expressed in tissues. On the other hand, inactivation of SN-38 occurs through glucuronidation into the inactive compound SN-38G by liver UDP glucuronosyltransferases 1A1, 1A7 and 1A9 (UGT1A1, UGT1A7, and UGT1A9) [43–47], and elimination via biliary fluids of irinotecan and its metabolites is due to efflux pumps like ABCB1 and ABCB2 (irinotecan, SN-38, and SN-38G), as well as ABCG2 (SN-38) [48–51]. Needless to say, several studies have assessed the association of variants and polymorphisms of the aforementioned genes with the high variability in activity and toxicity of irinotecan in clinical practice. As a result, the UGT1A1*28 genotype is now accepted as a predictor of irinotecan toxicity, and the FDA recommends its testing before starting irinotecan-containing regimens [52]. Likewise, an ABCG2 single nucleotide polymorphism (SNP) has been associated with increased myelosuppression upon irinotecan-based regimen administration [53]. Clearly, prospective studies assessing multiple variables that may confer predisposition to increased toxicity or to diminished activity of irinotecan are warranted to establish a panel of recognized predictors of toxicity that this compound may cause in patients. Apart from variability in the genes that mediate the pharmacokinetic response to irinotecan, other cancer-cell specific factors must play a role in determining the activity of Top1 inhibitors. This can be evinced by the high variability of CPT toxicity in large in vitro screenings, like those performed in the NCI-60 panel of cancer cell lines [54]. Although this in vitro variation is due, in part, to the overexpression of efflux pumps by cancer cells, the noncamptothecin-like Top1 inhibitors (e.g., the indenoisoquinolines [55]), which are not multi-drug resistance pump substrates, exhibit variability in their in vitro activity. By themselves, compounds like irinotecan do not create irreversible complexes with Top1 and DNA. Only when advancing replication forks collide with such macromolecular complexes, there is generation of DSBs, with activation of DDR, cell cycle block, and cell death. TOP1 mRNA and protein expression levels have been demonstrated to correlate directly, in vitro, with the activity of Top1 inhibitors [56]. Dosing Top1 levels in cancer may therefore predict irinotecan response. Finally, several enzymes contribute to Top1–DNA complex removal and DNA integrity preservation. Tyrosyl-DNA phosphodiesterase (TDP1) hydrolyzes the phosphodiester bond at a DNA 3’-end linked to a tyrosyl moiety and has been implicated in the repair of Top1-DNA covalent complexes. TDP1 possesses the ability to hydrolyze and remove other 3’ DNA lesions, like 3’ abasic sites and 3’ phosphodycolate, suggesting its activity as a DNA repair enzyme [57]. Thus, inhibiting TDP1 may synergize with Top1 inhibition or, alternatively, TDP1 overexpression may be a resistance factor to Top1 inhibition in cancer cells [58]. Likewise, siRNA-silencing of ERCC1 (excision repair cross complementation group 1), an endonuclease involved in nucleotide excision repair (NER), was recently shown by our group to potentiate the effects of CPT in concomitance with the blockade of PARP by Veliparib (Abbott, Inc.) [59], a PARP1/2 inhibitor currently undergoing clinical trials (http://clinicaltrials.gov). Clearly, DDR pathways play an important role in determining cancer sensitivity to Top1 inhibitors. On the one hand, knowing the presence of alterations in DDR components, via either somatic mutations, or expression modifications may be useful in predicting the response of CRC to irinotecan. On the other hand, such gene products may be susceptible to selective inhibition with the rationale of potentiating the effect of Top1 inhibitors.
Oxaliplatin
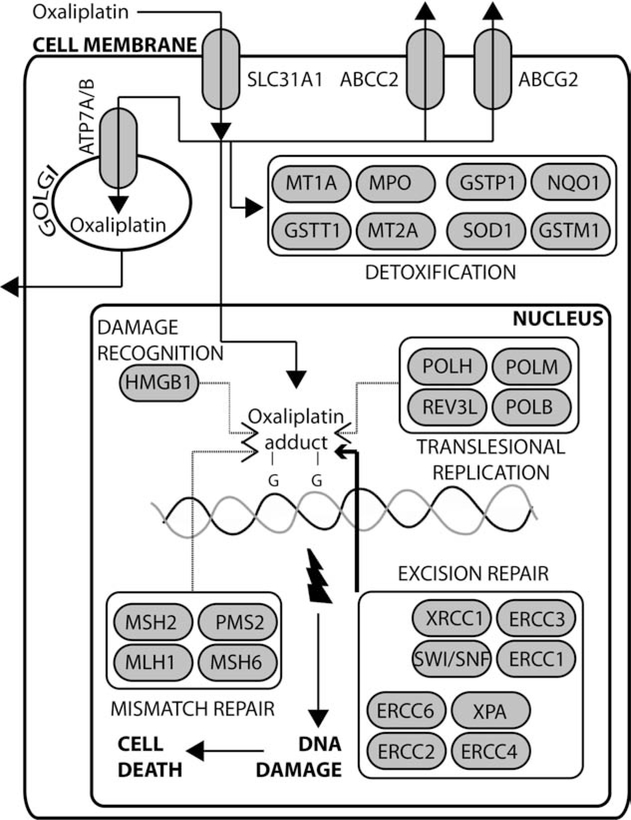
Oxaliplatin was discovered in 1976 at Nagoya City University, in the effort of finding water-soluble platinum-based compounds that could have good antitumor activity, yet lacking cross-resistance with cisplatin [60]. Following the observation of high activity against in vitro models of CRC [61], and of the synergistic effect of combining oxaliplatin with 5-FU in vitro and in vivo [62], great efforts were placed in the clinical development of this compound in patients affected by CRC [63]. The combination of oxaliplatin with 5-FU and leucovorin was then confirmed as a valuable treatment option for CRC in phase III studies conducted in patients with advanced disease [64, 65] and in the adjuvant setting of lymph-node positive, non-metastatic CRC [66, 67]. Moreover, second-line infusional 5-FU, leucovorin, and oxaliplatin (FOLFOX4) was recently shown to be non-inferior to irinotecan in metastatic CRC, with a better time-to-progression and response rate and fewer grade III side effects than irinotecan alone [68]. Oxaliplatin is now approved for stage III and metastatic CRC, in combination with 5-FU and leucovorin; the addition of the antiangiogenetic monoclonal antibody bevacizumab to this drug combination has led to even better results in terms of overall survival in patients with metastatic CRC (see Bagnasco L., et al., in the present issue of CCDT). Although exhibiting different spectra of activity, for only partially understood reasons [63, 69, 70], platinum coordination complexes are thought to act mainly by covalently binding to nucleophilic sites on cellular DNA (especially with the N7 position of guanine – GG intrastrand crosslinks), via inter- and intrastrand crosslinks, and DNA-protein crosslinks [71]. Therefore, these compounds prevent cell division and growth [71], and disrupt vital processes like DNA replication and transcription [72], finally leading to cell death by apoptosis [69, 73]. Like irinotecan, pharmacokinetics of oxaliplatin influence its activity (see Fig. 1). Platinum compounds enter the cell through an active copper transporter, SLC31A1 [74], and are actively expelled by cytoplasm member transporters (ABCC2 and ABCG2 [75–78]) and through trans-Golgi network copper pumps (ATP7A and ATP7B [79, 80]). While increased expression of such efflux pumps has been repeatedly associated with resistance to platinum-base compounds, SLC31A1-mediated influx of oxaliplatin is probably among the determinants of the ototoxicity observed in oxaliplatin treated patients [81]. Also, several genes involved in detoxification processes (e.g., MPO, SOD1, GSTM1, NQO1, GSTP1, and MT), are able to lower the concentration of platinum-based compounds inside the cell, and specific alleles of these genes may contribute to differences in sensitivity and toxicity observed among treated patients [71].
Fig. (1).
Oxaliplatin pathways: thin dotted arrows indicate pathways that are not deemed relevant in oxaliplatin DDR compared with cisplatin (this diagram has been reproduced with modifications from http://www.pharmgkb.org. Permission has been given by PharmGKB and Stanford University; copyright PharmGKB).
A unique feature of oxaliplatin, compared to cisplatin, is its ability to form bulkier adducts to DNA, and such lesions differ from those induced by cisplatin concerning patterns of distortion of the DNA helix, and of hydrogen bonding to neighboring DNA segments [82]. Such oxaliplatin properties may explain in part its different spectrum of activity compared to other compounds of the same class. Indeed, oxaliplatin-GG adducts possess distinct minor conformations compared to cisplatin-GG ones [83]. As a consequence, high-mobility group proteins like HMGB1, which recognize platinum-DNA adducts and stimulate DDR response [84], show smaller binding affinity to oxaliplatin-GG adducts and are probably less important in determining the response of cancer cells to its toxic effects [83]. Once platinum adducts are established, the MMR and the NER pathway are involved in the subsequent management of the generated lesions, while translesional replication polymerases can proceed through DNA lesions and continue DNA replication (see Fig. 1). Intrguingly, while non-functional NER components (in particular polymorphisms of ERCC1, XPS, and XRCC1 genes), which repair bulky DNA lesions, are associated with increased sensitivity of CRC to platinum-based compounds [85–87], overexpression of members of NER pathways such as ERCC1 is associated with poor response to platinum-based chemoradiotherapy in non-smallcell lung cancer [88]. The link between ERCC1 expression and poor response to oxaliplatin-based chemotherapy has also been reported in CRC [89]. In contrast with cisplatin, oxaliplatin-mediated activity does not seem to depend on a functional MMR pathway to induce cell apoptotic response [63, 69, 90]: this aspect of oxaliplatin-mediated toxicity may explain its activity in CRC, where the MMR pathway members are thought to be deficient in up to 15% of sporadic CRC [91, 92]. Finally, the greater efficiency of bypass of oxaliplatin-GG adducts by translesional replication polymerases like POLH may in part explain the better mutagenicity profile of oxaliplatin versus cisplatin [70], but increases in replicative bypass, which may play a role in cisplatin resistance [93], have not been shown to induce such phenotype toward oxaliplatin [69]. In conclusion, both pharmacokinetics (i.e., host and tumor-specific properties) and pharmacodynamics (in our case, tumor-specific properties) can contribute to sensitivity/resistance to oxaliplatin in CRC patients. Anyway, DDR pathways, in particular the NER complex, are likely a key element in determining such phenotypes in CRC.
CELL CYCLE CHECKPOINTS AND CRC
Cellular molecular surveillance systems, defined checkpoints, allow the detection of damaged DNA and temporarily arrest cell cycle progression to enable DNA repair and prevent further damage. This phenomenon provides the rationale to inhibit checkpoints in order to limit the time available for repair and consequently increase the sensitivity of cells to DNA damaging agents [94].
Overview
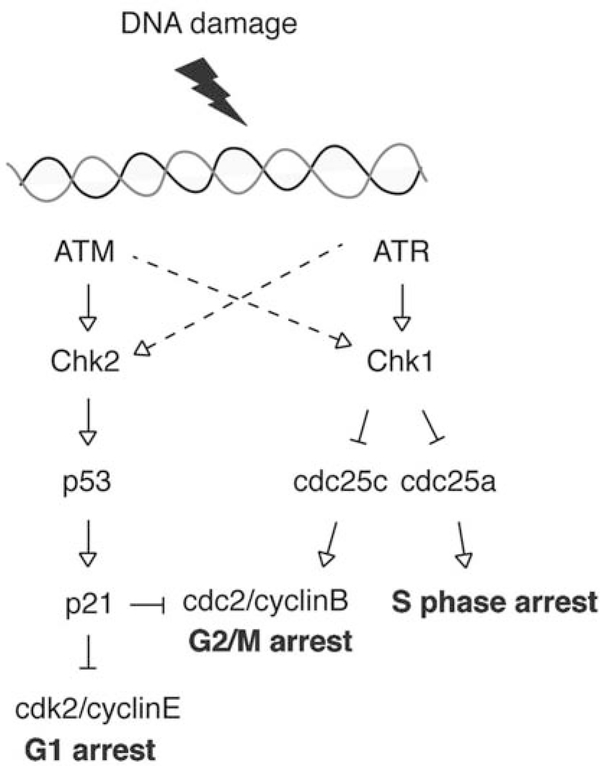
The cycle of DNA replication, chromosome separation and division is regulated by mechanisms that maintain the order of these steps and ensure that each step is orderly carried out. Key to these processes are the cell cycle checkpoints, where cells may arrest their replication to ensure that DDR mechanisms have time to operate prior to continuing through the cycle into mitosis [95–97]. The G1/S checkpoint is regulated by checkpoint kinase 2 (Chk2) and p53 and the intra-S and G2/M checkpoint is controlled by checkpoint kinase 1 (Chk1) (see Fig. 2). The ATM (ataxia telangiectasia mutated)-Chk2 pathway is mainly activated by DSBs produced by ionizing radiations or DNA damaging agents, but can also be activated by replication-mediated DSBs [98]. The ATR (ATR and Rad3 homolog)-Chk1 pathway is mainly activated in the case of replication-mediated DSBs [98]. Checkpoint kinases play a key role in maintaining the integrity of the genome [99]. In cancer cells, defects in the checkpoint proteins and checkpoint control mechanisms are frequent [100] and responsible for tumorigenesis. Loss of the checkpoints may be a cause of genomic instability in cancer cells [101]. Tumors exhibit three kinds of chromosomal alterations: rearrangements in the structure of individual chromosomes, whole chromosome aneuploidy, and the generation of polyploid cells [101].
Fig. (2).
Cell cycle checkpoints.
Several aberrations in the cell cycle checkpoints have been shown to have prognostic significance in CRC [102]. First, we will list the various alterations in the cell cycle checkpoints encountered in CRCs. Then, we will give an overview of the potential utility to target the remaining functional checkpoints for treatment purposes.
Alterations in the Cell Cycle Checkpoints
Mitotic Checkpoints
Mitotic checkpoint deficiencies can contribute to CIN and tumor formation in mammals [103]. Mice with deficiencies in the mitotic checkpoint proteins MAD2, BubR1 and Bub3 present CIN, and some of them have susceptibility to cancer [104–107]. BubR1+/− mice present an increase of incidence of colon adenocarcinomas when exposed to carcinogens [105]. In most CRCs, a CIN leading to an abnormal chromosome number, aneuploidy, is consistently associated with the loss of function of a mitotic checkpoint [108, 109]. One mutation of BUB1B (encoding BubR1) in an aneuploid CRC was a somatic deletion, resulting in the removal of the kinase domain. Such alteration was similar to the truncating mutations found in mosaic variegated aneuploidy (MVA) disease, a rare recessive disease characterized by mutations in BUB1B [110]. Lengauer and Wang observed that the spindle checkpoints genes BUB1 and BUB1B are mutated in some CIN tumors of individuals with CRC [111]. Moreover, other studies in CRC demonstrated the presence of mutations in established or putative mitotic checkpoints genes such as BUB1, ZW10, ZWILCH and KNTC1 (a.k.a. Rod) [112–114]. Even though these mutations occur rarely in CRCs, nonmutational reduction in the expression of such genes has been reported in a small percentage of colorectal tumors, and increase in expression has been reported as well in several CRCs [113, 115–117]. In CRC biopsies obtained from 16 patients, Menssen et al., observed that elevated expression of c-MYC correlated with increased MAD2 levels [118]. c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1, and this may have potential implications in the origin of CIN [118]. Taxanes, which disrupt microtubule function and inhibit mitosis [119], have failed to demonstrate significant clinical benefit in phase II trials in CRC [120]. The high incidence of CIN in this disease, coupled with alterations in spindle checkpoint regulators in vivo, may explain the disappointing results associated with such category of anti-cancer drugs [120].
CHEK1
The presence of a nucleotide stretch of nine consecutive nucleotides in the coding region of CHEK1 led to the hypothesis that tumors with defects in MMR and hence with microsatellite instability could accumulate mutations in the CHEK1-coding locus. Indeed in CRC, in which microsatellite instability is a frequent event, insertion/deletion of one nucleotide in CHEK1 coding region could be found [100, 121, 122], resulting in Chk1 protein truncation and inactivation. The resulting truncated Chk1 proteins are predicted to be defective due to the lack of the C-terminal end of the catalytic domain and the complete loss of the SQrich regulatory domain [123]. Bertoni et al., showed that 1 of 10 CRCs with high frequency of microsatellite instability carry the (A)9 CHEK1 mutation resulting in a truncated protein [122, 124]. Furlan et al., reported this same incidence of CHEK1 mutations in an independent cohort [121].
CHEK2
The checkpoint kinase 2 gene (CHEK2), identified as a breast cancer susceptibility gene, could also be a candidate gene for CRC susceptibility. Liu et al., screened for mutations of all 14 exons of CHEK2 in 56 CRC cell lines [125]. CHEK2 mutation in CRC was a low frequency event. Ten cell lines out of the 56 analyzed had sequence variations within the exons, and only DLD-1/HCT-15 showed a heterozygous missense mutation, R145W [125]. Based on sequence analysis, DLD-1/HCT-15 was found to carry one mutant allele (R145W) and one wild-type allele of CHEK2 [126]. The R145W mutation, which was in the forkhead homology-associated (FHA) domain keeps some basal kinase activity, but cannot be phosphorylated by ATM and cannot be activated following DNA damage. Because the FHA domain is involved in protein-protein interactions, this mutation could affect the association of Chk2 with other proteins [127]. Using next-generation sequencing and targeted resequencing across the panel of 60 established cancer cell lines from the NCI Anticancer Screen (NCI60), we confirmed the two point mutations, R145W (see above) and A247D (which disrupts a residue in the catalytic motif), in the HCT-15 cell line [128]. Recently, the incidence of the CHEK2 1100delC mutation in familial and non-familial CRC patients was studied. At the molecular level, the 1100delC mutation eliminates the kinase domain and activity of Chk2 [127]. To evaluate the importance of 1100delC in CRC, Kilpivaara et al., studied the frequency of the 1100delC allele in 662 CRC patients, including 149 familial and 513 non-familial cases [129]. The frequency was 2.6% for the whole patient cohort, 1.3% for familial and 2.9% for non-familial, which is not significantly higher than the expected frequency in the population (1.9%). Moreover, they observed a low frequency of the loss of the wt allele (17.6%) in the germline mutation carrier tumors, and the loss of the mutant allele in one tumor. Their study suggested no significant association between germline CHEK2 1100delC and familial or sporadic CRC, but a low penetrance effect of CHEK2 in CRC pathogenesis could not be ruled out. Moreover, another study by Meijers-Heijboer et al., reported that the frequency of the 1100delC mutation was somewhat higher in MMR gene mutation carriers and in patients without MMR gene mutations compared with the controls. In addition, they reported that CHEK2 1100delC mutation was absent in 95 familial adenomatous polyposis families [130]. de Jong et al., studied the mutation in 629 unselected CRCs, 230 controls and 105 selected CRCs diagnosed before age 50. In this study, the unselected CRC patients showed no increased frequency of the CHEK2 1100delC genotype compared to controls [131]. However, after stratifying unselected CRC patients according to defined genetic risk, a significantly increased frequency was observed [131]. There was no association of the 1100delC genotype with tumor localization, gender or age at diagnosis [131]. van Puijenbroek et al., studied 564 familial colorectal tumors. They found that only a small percentage of patients presenting loss of CHEK2 expression have the CHEK2 1100delC mutation [132]. The authors also concluded that Chk2 protein abrogation is not caused by the CHEK2 germline variants R117G, R137Q, R145W, I157T and R180H in familial CRC [132]. Schutte et al., reported a relatively high frequency of the 1100delC variant in families predisposed to combined breast and colon cancer [133]. In 2011, a meta-analysis of CHEK2 1100delC variant, found an association of CHEK2 1100delC variant with unselected CRC and familial CRC [134]. In conclusion, CHEK2 mutations probably contribute, albeit with low penetrance, to CRC susceptibility. Moreover, it is noteworthy that not only Chk2 but other proteins implicated in G1/S are affected in CRC: p21 waf1/cip1 protein, which is a p53-inducible inhibitor of the cyclin-dependent kinases, and cyclins D and E which are key regulators of G1 progression. Abnormal p21 mRNA and protein levels have been reported in CRC [135–137]. Overexpression of cyclin D1 may be a marker of aggressive biological behaviour in CRC, and has been correlated with advanced stage [135]. Tort et al., studied the alteration of several components along the p16-cyclin D1pRb-E2F-cyclin E pathway in human colorectal adenomas, concluding that cyclin D1 and cyclin E showed distinct clinical variables and biological behavior, with potential implications for individualized management of tumors with elevated cyclin D1 versus cyclin E [138]. Furthermore, p53 aberrations are among the commonest genetic changes in the development of CRC [139].
Therefore, several alterations of the checkpoints can be present in CRC, and may have an impact on its prognosis and possibly on the design of targeted treatments.
Targeting the Checkpoints
The common occurrence of checkpoint defects in cancer cells provides the rationale for targeting checkpoint pathways for therapeutic intervention [123]. When cancer cells harbor some checkpoint defects, they exhibit a greater dependence on the remaining checkpoint processes. Unlike normal cells with a full arsenal of checkpoints responses, tumors with defects in some checkpoint(s) could be deprived of their remaining checkpoint pathways(s), and consequently driven into death due to accumulation of excessive DNA damage [123]; a potential strategy could be targeting p53-deficient cancers that lack the G1 checkpoint and inhibiting the checkpoints kinases known to participate in the G2 checkpoint, such as ATM/ATR or Chk1 [123]. Consequently, checkpoint inhibition is an opportunity to selectively increase the effect of DNA damaging agents, when the checkpoint kinase is the kinase that remains intact.
Different chemical categories of checkpoint inhibitors have been identified: aminopyrazoles, indazoles, tricyclic compounds, ureas, carbamates, diazepinones, pyrimidines, benzimidazole quinolones, macrocyclic compounds, etc. [140–142]. Checkpoint inhibitors modulate the cellular response to DNA damage, leading to checkpoint abrogation, inhibition of DNA repair and changes in the regulation of cell death.
UCN-01 (7-hydroxy-staurosporine) targets Chk1, but also potently inhibits a number of other kinases including Chk2 and a number of the cyclin-dependent kinases. It is currently believed that the potentiation of DNA-damaging agents is primarily driven by the inhibition of Chk1 and, hence, abrogation of cell cycle arrest [143, 144]. In CRC cells, the complete growth arrest induced by WMC26 (bisimidazoacridonesin, a potential bisintercalating anticancer drug with CRC selectivity in vitro [145]) sensitizes to apoptotic death induced by UCN-01 [146]. This suggests that combining WMC26 and Chk1 inhibition may be an attractive treatment method for CRC, that exploits the highly tumor-selective activity of WMC26 [146]. Furuta et al., showed that UCN-01 inactivates p21, allowing the repair of replication-mediated DSBs induced by Top1 [147]. This study provided the rationale for using camptothecins in cells with functional defects in the p21 pathway, and for combining camptothecins and cell cycle checkpoint inhibitors, such as UCN-01, in cancer chemotherapy [147]. The results by Petersen et al., suggest that p53-dependent G1 arrest in both first and second cell cycles may protect human cancer cells from cell death after treatment with ionizing radiation (IR) and Chk1 inhibitors. Hence the idea to combine Chk1 inhibitors with IR to selectively target p53-deficient cancer cells becomes strongly attractive [148]. Xiao et al., showed that Chk1 downregulation sensitizes tumor cells to the toxicity of paclitaxel in cell proliferation assays; Chk1 inhibition facilitates paclitaxel-induced M-phase entry by activation of Cdc2 kinase and accumulation of cyclin B1, the required cofactor for Cdc2 kinase activity [149]. Therefore, in addition to its role as an enforcer of S and G2checkpoints in response to genotoxic stress, Chk1 also plays a protective role in mitotic checkpoint to lessen mitotic catastrophe, and thereby limits cell death. Moreover, Xiao et al., demonstrated that 5-FU activates Chk1 and induces an early S-phase arrest, through Chk1-mediated CDC25A proteolysis leading to inhibition of CDK2 [150]. Chk1 elimination stabilizes CDC25A protein product and results in the abrogation of the S checkpoint and resumption of DNA synthesis, which in turn leads to excessive accumulation of DSBs [150]. As a result, downregulation of Chk1 potentiates 5-FU efficacy through the induction of premature chromosomal condensation followed by apoptosis [150]. As a consequence, Chk1 inhibition not only potentiates the toxicity of conventional DNA-damaging agents such as IR and topoisomerase inhibitors, but also enhances the toxicity of anti-microtubule agents [149] and antimetabolites in cancer cell lines [150]. All these studies suggest that inhibiting the G2 checkpoint is a “key element” to sensitize p53-deficient CRC cells to DNA damaging agents, anti-microtubule agents or antimetabolites.
Concerning Chk2, Castedo et al., showed that inhibition of Chk2 facilitates the induction of mitotic catastrophe in HCT116 cells, thus sensitizing proliferating cells to chemotherapy-induced apoptosis [151]. Chk2 deficiency is associated with defective S-phase checkpoint, prolonged G2 arrest, and hypersensitivity to CPT [152]. Therefore there is a potential value for testing Chk2 status in colorectal tumors treated with Top1 inhibitor clinically. Theoretically, the combination of Chk2 inhibitors with oxaliplatin may further sensitize CRC cells to oxaliplatin treatment. Paradoxically, these inhibitors produced an antagonistic effect on the response to oxaliplatin, which was reversed by the reintroduction of Chk2 [153]. These observations may have important implications for the design of trials foreseeing the use of oxaliplatin in CRC therapy combined with Chk2 inhibitors [153]. At comparable concentrations in human colon cancer cells, cisplatin slowed down the replication phase and activated the G2/M checkpoint, whereas oxaliplatin activated the G1/S checkpoint and completely blocked the G2/M transition [154]. Chk2 inhibitors have been developed as sensitizers for chemotherapeutic agents. However, Chen et al., reported that Chk2 activation alone led to potent inhibition of cancer cell proliferation; consequently, indirect activation of checkpoint kinases may be a novel approach for cancer therapy in selected genomic contexts, such as p53 wild-type CRCs [155]. Recently, we observed that in TRAIL (tumor necrosis factor-related apoptosis-inducing ligand)-treated colon carcinoma cells, Chk2 is activated and amplified the TRAIL-induced apoptosis by a feedback loop, activating the upstream caspase-8 [156]. From a therapeutic standpoint, our findings suggest that the level of Chk2 and activated P-Chk2-T68 in tumor tissues could have a prognostic value for predicting the efficiency of a TRAIL-based therapy. Moreover, our results provide a rationale for combining TRAIL with DNA-damaging agents. Indeed, DNA damage might sensitize tumor cells to TRAIL by preactivating Chk2 and the positive feedback loop that amplifies TRAIL-induced apoptosis. Such a mechanism may partly account for the known synergism between TRAIL and DNA damaging agents [157, 158]. These studies show the multifaceted nature of Chk2: indeed, Chk2 inhibitors could sensitize to chemotherapeutic agents, but in selected genomic context the indirect activation of Chk2 could itself be an approach for cancer therapy (for an in-depth analysis of the variability and the potential genomic determinants of Chk2 status in vitro, see also [128]).
Recently, some Chk inhibitors (for both Chk1 and Chk2) have shown their efficiency in enhancing DNA damaging agents in colon carcinoma cells [159, 160].
Inactivation of p53 increases the cytotoxicity of CPT in HCT116, which demonstrates a critical role of p53 as a G1 checkpoint regulator after CPT-induced DNA damage and suggests a rationale for the selectivity of CPT toward p53-deficient tumors [161]. Filder et al., showed that PG490, triptolide, sensitizes tumor cells to Top1 inhibitors by blocking p53-mediated induction of p21 [162]. They also demonstrated that PG490–88, a derivative of triptolide, causes tumor regression of CRC xenografts when administered alone, and synergizes with irinotecan (a camptothecin analog) to cause tumor regression [163]. In 2003, PG490–88 underwent a phase I clinical trial in patients affected by solid tumors [163]. PG490–88 caused tumor regression in some cases in combination with DNA-damaging agents, suggesting its role as an antineoplastic agent and chemosensitizer for the treatment of patients with solid tumors [163].
During the mitotic spindle checkpoint arrest, Sayed et al., identified two novel roles for casein kinase 2 (CK2): a role in maintaining increased cyclinB/cdc2 kinase activity, as a component of G2 arrest; and a role in p53-mediated apoptosis [164]. Kaestner et al., showed that treatment with MLN8054 (a selective Aurora Kinase A inhibitor) in human CRC cell lines induces defects in mitotic spindle assembly, which causes a transient spindle checkpoint-dependent mitotic arrest [165]. The cell cycle arrest is not maintained due to the ability of MLN8054 to override the spindle checkpoint. Subsequently, MLN8054-treated cells exit from mitosis and activate a p53-dependent postmitotic G1 checkpoint, which subsequently induces p21 and Bax, leading to G1 arrest followed by the induction of apoptosis. ZM447439, an Aurora Kinase B inhibitor, also interferes with normal chromosome alignment during mitosis and overrides the mitotic spindle checkpoint but allows a subsequent endoreduplication, although the compound potently activates the p53-dependent postmitotic G1 checkpoint [165].
In summary, the discovery and development of checkpoint inhibitors is an area of intense interest and active research. The preclinical data provide confidence that these inhibitors will be useful in clinical practice, mostly in combination with DNA-damaging therapies, for the improvement of CRC treatment.
PARP INHIBITORS IN CRC THERAPY: PRECLINICAL FINDINGS AND CLINICAL PERSPECTIVES
Poly(ADP-ribose) polymerases (PARP) are a family of 17 structurally related mammalian enzymes (reviewed in [166–169]). PARP1 is the most abundant member of the PARP family. It is normally associated with chromatin, and PARP1 activation is associated with transcription activation, in some cases in association with DNA cleavage by topoisomerase II beta (TOP2B) [170, 171]. PARP1 is strongly activated by DNA damage, including SSBs and DSBs. PARP catalyzes poly(ADP-ribose) (PAR) polymer formation transferring by nicotinamide adenine dinucleotide (NAD+). PARP polymerizes PAR to substrate proteins (histone, topoisomerase enzymes among others) including itself. The automodified PARP binds to DNA and recruits XRCC1, DNA ligase III (LIG3), DNA polymerase β (POLB) to the DNA damage sites and is thereby involved in base excision repair (BER) [172–174]. PARP, along with XRCC1, has also been implicated in the alternative non-homologous end joining (NHEJ) pathway of DSB repair [175, 176]. Finally, PARP inhibits the conversion of SSBs to DSBs [59]. Thus, PARPs have an important role in the maintenance of genomic integrity, making them promising targets for pharmacological strategy [177, 178]. PARP inhibitors are now used in an increasing number of clinical trials [179]. Because BRCA1 and BRCA2 are defective in many cancers and necessary for DSB repair, inhibition of PARP1 is selectively effective in BRCA1/BRCA2-deficient cancer cells [180, 181]. Thus, PARP inhibitors provide a novel way to treat BRCA-deficient cells in combination with chemotherapy and radiation, while sparing normal cells.
PARP is closely related with CRC. In the early stage of colorectal carcinogenesis, PARP1 mRNA overexpression was detected in 64 (70.3%) of the 91 tumors analyzed (65 adenomas and 26 submucosal (pT1) cancers). PARP1 overexpression is significantly correlated with tumor size and histopathology [182]. In leucocytes of patients with familial adenomatous polyposis (FAP), who are genetically predisposed to colon cancer, stimulation of PARP activity upon radiation was absent comparing with that of healthy volunteers [183].
Single nucleotide polymorphisms (SNPs) in the PARP1 gene could also modify the CRC risk. In an ancillary analysis of the Singapore Chinese Health Study, which 1,176 healthy control and 310 cancer patients (180 colon and 130 rectum cancers), a positive association between the PARP1 codon 940 Lys/Arg and Arg/Arg genotypes and CRC risk was observed [184]. Polymorphisms in Val742 (Val742Ala) modifies the association of high red-meat content diets with risk of CRC [185]. These findings suggest that PARP1 might play an important role in tumorigenesis of CRC.
PARP inhibitors potentiate the cytotoxicity of several DNA damaging agents including ionizing radiation, alkylating agents and Top1 inhibitors (reviewed in [167, 178]). We recently published the results of the combination of veliparib with Top1 inhibitors in human cancer cell models [59]. Veliparib (ABT-888) synergized with CPT and induced DSBs. Moreover, siRNA knockdown of XPF or ERCC1 enhanced the antiproliferative effects of veliparib in camptothecin-treated cells, suggesting the involvement of XPF-ERCC1 in Top1-induced DNA damage as an alternative pathway from PARP and TDP1, and providing the rationale for combining inhibition of PARP1 and Top1 in cancer with XPF-ERCC1 deficiency.
The tricyclic benzimidazole AG14361, a potent PARP inhibitor enhanced the radiosensitivity of CRC xenografts [186]. AG14361 (5 mg/kg) was intra-peritoneally administered daily for 5 days to CD-1 nude mice bearing LoVo or SW620 (CRC cell lines) xenografts, followed by 2 Gy irradiation. Under such treatment schedule, tumor growth was delayed by 37 days compared with a delay of 19 days (with local tumor irradiation alone) (P = 0.008). Veliparib potentiated radiation (2 Gy/d x 10) in a HCT116 CRC xenografted model, with a median survival time of 36 days compared with 23 days (P < 0.036, log-rank test) from radiation alone [187].
Beyond radiosensitization, PARP inhibitors also synergize with chemotherapy, such as temozolomide (TMZ) and irinotecan in colorectal tumor models. ABT-888 facilitates the conversion of TMZ-induced SSBs to DSBs and potentiates the cytotoxic effect of TMZ in the HCT116 cell line [188]. Oral administration of GPI-15427 enhances the antitumor efficacy of TMZ and irinotecan combination in HT-29 and LoVo xenografts [189]. Also GPI 15427 showed a protective effect on irinotecan-induced severe intestinal injury of jejunum and delayed diarrhea [189]. GPI-15427 did not exacerbate myelotoxicity induced by full dose TMZ.
Based on these preclinical data, four PARP inhibitors, olaparib (AZD2281, Astra-Zeneca, Inc.), veliparib (ABT888, Abbott, Inc.), iniparib (SSAR240550-BSI-201, SanofiAventis, Inc.) and CEP-9722 (Cephalon Inc.) have entered the clinical phase of experimentation in CRC tratment. According to the information from http://clinicaltrials.gov, several clinical trials are undergoing (last accessed January 20th, 2012). In a phase II trial, olaparib is used alone to treat previously-treated patients with stage IV, measurable CRC, stratified by microsatellite instability status, whereas a phase I trial assessing the safety/efficacy of olaparib/irinotecan combination in locally advanced or metastatic CRC has recently been completed. A phase I trial of veliparib in combination with irinotecan, 5-FU, Folinic Acid (FOLFIRI) to treat patients with advanced solid tumors including CRC was started in February 2010, and another phase I study, which evaluates the association of veliparib with CapeOX (capecitabine and oxaliplatin) in advanced solid tumors is ongoing. Also, a phase II study of veliparib in combination with TMZ in patients with heavily pretreated, metastatic CRC has been ongoing in Georgetown University Medical Center since October 2009 and will be completed in December 2012. Moreover, the phase I study of evaluating the safety and pharmacokinetics of iniparib and the study of CEP-9722 in combination with gemcitabine and cisplatin have also been initiated in patients with advanced solid tumors. Results from these studies will help to clarify the real potential of PARP inhibition as a strategy to sensitize CRC to current DNA damaging agents on clinic.
INTERACTIONS BETWEEN THE EGFR AND DDR PATHWAYS
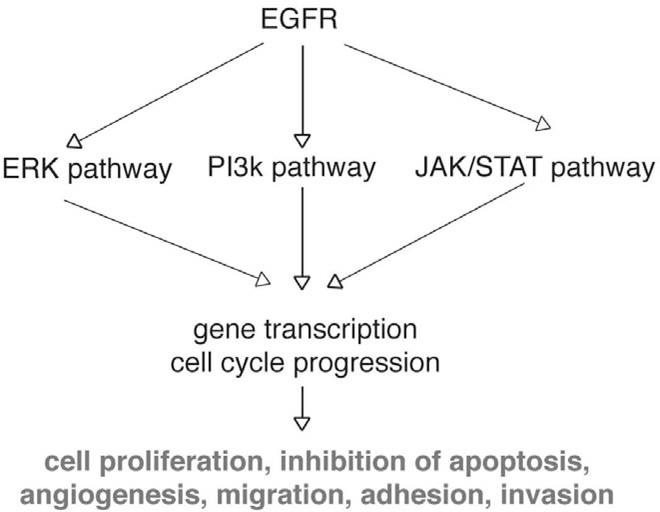
The epidermal growth factor receptor (EGFR) has emerged as a relevant molecular target for cancer. The activation of EGFR results in activation of a series of downstream signaling pathways that ultimately regulate key cellular processes such as proliferation, survival, adhesion, migration and differentiation [see the review by Ballestrero A. et al., in the present issue of CCDT and [190] (Fig. 3)]. EGFR is frequently dysregulated in CRC, and the overexpression of the receptor confers a poor prognosis [190–193]. Thus, EGFR has become an attractive target in the treatment of CRC. Buisine et al., investigated the polymorphic simple sequence repeat in EGFR intron 1 (CASSR I) in CRC [194]. They detected mutations in CA-SSR I in 86 % of MSI-H (microsatellite instability with high frequency) CRCs and in 0 % of MSS (microsatellite stable) CRCs [194]. This indicates that EGFR gene could be a novel putative specific target of MMR-deficient CRCs [194]. Of interest, no association could be observed between EGFR expression and CA-SSR I in tumor or normal tissues [194]. Thus, CA-SSR I sequence does not contribute to the regulation of EGFR transcription in colon, and should not be considered as a promising predictive marker for response to EGFR inhibitors in patients with CRC [194]. Several strategies to inhibit the EGFR and its downstream signaling pathways are available in preclinical and clinical studies. They include monoclonal antibodies against the extracellular domain of the receptor and small-molecule inhibitors of its tyrosine kinase activity (see Ballestrero A. et al., in the present issue of CCDT, [190], and ClinicalTrials.gov website). Erlotinib, a selective EGFR tyrosine kinase inhibitor, reduces thymidylate synthase (TS) activity in EGFR-expressing cells, probably due to cell cycle arrest in the G1 phase [195]. Combining erlotinib with pyrimidine trifluorothymidine (TFT) shuts down even more TS, possibly due to cell cycle interference [195]; consequently combining EGFR blockade with fluoropyrimidines has a molecular rationale in CRC. Hiro et al., showed that 5-FU-induced activation of EGFR followed by radiation in chemo/radioresistant SW480 cells results in up-regulation of ERCC1 [196]. On the contrary, 5-FU-induced degradation of EGFR followed by radiation in radiosensitive CRC cell lines resulted in down-regulation of ERCC1 [196]. These data suggest that there is a complementary interaction between EGFR and ERCC1, and that 5-FU-induced EGFR activation confers protection against radiation through activation of NER pathway [196]. Among the proteins activated by EGFR is the transcription factor STAT3. Vigneron et al., observed that the EGFR-STAT3 oncogenic pathway up-regulates the EME1 endonuclease, reducing DNA damage after Top1 inhibition [197]. These results suggest the rationale for combining anti-EGFR therapy with Top1 inhibitors to sensitize drug-resistant colorectal tumors [197]. Gefitinib (Iressa®, Astra-Zeneca, Inc. and Teva, Inc.), a selective EGFR kinase inhibitor enhances the antitumor effect of oxaliplatin in CRC cell lines [198]. Gefitinib not only enhances cellular Pt-DNA adducts but also markedly inhibits their removal, which prolongs oxaliplatin pro-apoptotic effects [198]. Recently, the combination of a dimeric Affibody molecule (ZEGFR:1907)2 targeting EGFR with radiation has shown a decrease in the survival of HCT116 cells [199].
Fig. (3).
EGFR pathways.
EGFR inhibition seems a logical therapeutic strategy to enhance CRC treatment with DNA damaging agents, and clinical and translational studies need to be pursued to completely understand the possible relationship between EGFR inhibition and DDR pathway modulation.
CONCLUDING REMARKS
In the present review, we have tried to depict the relationship between members of two essential cellular pathways, DDR and checkpoint, with CRC. We have explored several known and potential determinants that could be of importance in better targeting this disease, and we have illustrated the rationale lying at the foundation of two currently used DNA damaging agents, irinotecan and oxaliplatin. Clearly, our picture is only a blurred image of the magnificient fresco that makes up such complicated and wondrously intertwined pathways as DDR and checkpoints are. These last years have seen tremendous improvements in our understanding of DDR and checkpoints, as well as great and positive changes in CRC treatment, but a much greater effort needs to be undertaken to unveil the determinants of CRC resistance to chemoterapeutical agents, to optimize treatment strategies, and to identify new targets in the management of this disease.
ACKNOWLEDGEMENTS
This work was in part supported by an AIRC “My First AIRC Grant”, ID No. 10570.
ABBREVIATIONS
- APC
adenomatous polyposis coli
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3 homolog
- BER
base excision repair
- CIN
chromosomal instability
- CRC
colorectal camcer
- Chk1
checkpoint kinase 1
- Chk2
checkpoint kinase 2
- DDR
DNA damage response
- DSB
DNA double-strand break
- EGFR
epidermal growth factor receptor
- ERCC1
excision repair cross complementation group 1
- FAP
familial adenomatous polyposis
- FHA
forkhead homology-associated
- 5-FU
5-fluorouracil
- MMR
mismatch repair
- MSI-H
microsatellite instability with high frequency
- MSS
microsatellite stable
- MVA
mosaic variegated aneuploidy
- NAD
nicotinamide adenine dinucleotide
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- PAR
poly(ADP-ribose)
- PARP
poly(ADP-ribose) polymerase
- SNP
single nucleotide polymorphism
- SSB
single-strand break
- TFT
trifluorothymidine
- TMZ
temozolomide
- Top1
topoisomerse
- TRAIL
tumor necrosis factor-related inducing ligand apoptosis-
- TS
thymidylate synthase
Footnotes
CONFLICT OF INTEREST
None.
REFERENCES
- [1].Joyce GF The antiquity of RNA-based evolution. Nature 2002, 418 (6894), 214–221. [DOI] [PubMed] [Google Scholar]
- [2].Shechner DM; Grant RA; Bagby SC; Koldobskaya Y; Piccirilli JA; Bartel DP Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 2009, 326 (5957), 12711275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jackson SP; Bartek J The DNA-damage response in human biology and disease. Nature 2009, 461 (7267), 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kastan MB; Bartek J Cell-cycle checkpoints and cancer. Nature 2004, 432 (7015), 316–323. [DOI] [PubMed] [Google Scholar]
- [5].Stracker TH; Usui T; Petrini JH Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst). 2009, 8 (9), 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Conti C; Seiler JA; Pommier Y The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle 2007, 6 (22), 27602767. [DOI] [PubMed] [Google Scholar]
- [7].Pommier Y; Sordet O; Rao VA; Zhang H; Kohn KW Targeting chk2 kinase: molecular interaction maps and therapeutic rationale. Curr. Pharm. Des. 2005, 11 (22), 2855–2872. [DOI] [PubMed] [Google Scholar]
- [8].Vogelstein B; Kinzler KW Cancer genes and the pathways they control. Nat. Med. 2004, 10 (8), 789–799. [DOI] [PubMed] [Google Scholar]
- [9].Michor F; Iwasa Y; Vogelstein B; Lengauer C; Nowak MA, Can chromosomal instability initiate tumorigenesis? Semin. Cancer Biol. 2005, 15 (1), 43–49. [DOI] [PubMed] [Google Scholar]
- [10].Li GM Mechanisms and functions of DNA mismatch repair. Cell Res. 2008, 18 (1), 85–98. [DOI] [PubMed] [Google Scholar]
- [11].Jiricny J The multifaceted mismatch-repair system. Nat. Rev. Mol.Cell. Biol. 2006, 7 (5), 335–346. [DOI] [PubMed] [Google Scholar]
- [12].Cummings OW Pathology of the adenoma-carcinoma sequence: from aberrant crypt focus to invasive carcinoma. Semin. Gastrointest. Dis 2000, 11 (4), 229–237. [PubMed] [Google Scholar]
- [13].Cardoso J; Molenaar L; de Menezes RX; van Leerdam M; Rosenberg C; Moslein G; Sampson J; Morreau H; Boer JM; Fodde R Chromosomal instability in MYH- and APC-mutant adenomatous polyps. Cancer Res. 2006, 66 (5), 2514–2519. [DOI] [PubMed] [Google Scholar]
- [14].Hadjihannas MV; Behrens J CIN By WNT: growth pathways, mitotic control and chromosomal instability in cancer. Cell Cycle 2006, 5 (18), 2077–2081. [DOI] [PubMed] [Google Scholar]
- [15].Popat S; Hubner R; Houlston RS Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin.Oncol. 2005, 23 (3), 609–618. [DOI] [PubMed] [Google Scholar]
- [16].Loeb LA A mutator phenotype in cancer. Cancer Res. 2001, 61 (8), 3230–3239. [PubMed] [Google Scholar]
- [17].Longley DB; Harkin DP; Johnston PG 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3 (5), 330–338. [DOI] [PubMed] [Google Scholar]
- [18].Sampath D; Rao VA; Plunkett W Mechanisms of apoptosis induction by nucleoside analogs. Oncogene 2003, 22 (56), 90639074. [DOI] [PubMed] [Google Scholar]
- [19].Wyatt MD; Wilson DM 3rd, Participation of DNA repair in the response to 5-fluorouracil. Cell Mol. Life Sci. 2009, 66 (5), 788799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Segditsas S; Tomlinson I Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006, 25 (57), 75317537. [DOI] [PubMed] [Google Scholar]
- [21].Parker AR; Sieber OM; Shi C; Hua L; Takao M; Tomlinson IP; Eshleman JR Cells with pathogenic biallelic mutations in the human MUTYH gene are defective in DNA damage binding and repair. Carcinogenesis 2005, 26 (11), 20102018. [DOI] [PubMed] [Google Scholar]
- [22].Al-Tassan N; Chmiel NH; Maynard J; Fleming N; Livingston AL; Williams GT; Hodges AK; Davies DR; David SS; Sampson JR; Cheadle JP Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat. Genet. 2002, 30 (2), 227–232. [DOI] [PubMed] [Google Scholar]
- [23].Sieber OM; Lipton L; Crabtree M; Heinimann K; Fidalgo P; Phillips RK; Bisgaard ML; Orntoft TF; Aaltonen LA; Hodgson SV; Thomas HJ; Tomlinson IP Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 2003, 348 (9), 791–799. [DOI] [PubMed] [Google Scholar]
- [24].Venkatachalam R; Ligtenberg MJ; Hoogerbrugge N; de Bruijn DR; Kuiper RP; Geurts van Kessel A The epigenetics of (hereditary) colorectal cancer. Cancer Genet. Cytogenet. 2010, 203 (1), 1–6. [DOI] [PubMed] [Google Scholar]
- [25].Jasperson KW; Tuohy TM; Neklason DW; Burt RW Hereditary and familial colon cancer. Gastroenterology 2010, 138 (6), 2044–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lammi L; Arte S; Somer M; Jarvinen H; Lahermo P; Thesleff I; Pirinen S; Nieminen P Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004, 74 (5), 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kilpivaara O; Alhopuro P; Vahteristo P; Aaltonen LA; Nevanlinna H CHEK2 I157T associates with familial and sporadic colorectal cancer. J. Med. Genet. 2006, 43 (7), e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Reinhardt HC; Yaffe MB Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 2009, 21 (2), 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jeghers H; Mc KV; Katz KH Generalized intestinal polyposis and melanin spots of the oral mucosa, lips and digits; a syndrome of diagnostic significance. N. Engl. J. Med. 1949, 241 (26), 1031–1036. [DOI] [PubMed] [Google Scholar]
- [30].Farmer RG; Hawk WA; Turnbull RB Jr. The spectrum of the Peutz-Jeghers syndrome. report of 3 cases. Am. J. Dig. Dis.1963, 8, 953–961. [DOI] [PubMed] [Google Scholar]
- [31].Hemminki A; Tomlinson I; Markie D; Jarvinen H; Sistonen P; Bjorkqvist AM; Knuutila S; Salovaara R; Bodmer W; Shibata D; de la Chapelle A; Aaltonen LA Localization of a susceptibility locus for Peutz-Jeghers syndrome to 19p using comparative genomic hybridization and targeted linkage analysis. Nat. Genet. 1997, 15 (1), 87–90. [DOI] [PubMed] [Google Scholar]
- [32].Jenne DE; Reimann H; Nezu J; Friedel W; Loff S; Jeschke R; Muller O; Back W; Zimmer M Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat. Genet. 1998, 18 (1), 38–43. [DOI] [PubMed] [Google Scholar]
- [33].Hemminki A; Markie D; Tomlinson I; Avizienyte E; Roth S; Loukola A; Bignell G; Warren W; Aminoff M; Hoglund P; Jarvinen H; Kristo P; Pelin K; Ridanpaa M; Salovaara R; Toro T; Bodmer W; Olschwang S; Olsen AS; Stratton MR; de la Chapelle A; Aaltonen LA A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 1998, 391 (6663), 184–187. [DOI] [PubMed] [Google Scholar]
- [34].Pommier Y Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 2006, 6 (10), 789–802. [DOI] [PubMed] [Google Scholar]
- [35].Wall ME; Wani MC; Taylor H Isolation and chemical characterization of antitumor agents from plants. Cancer Treat. Rep. 1976, 60 (8), 1011–1030. [PubMed] [Google Scholar]
- [36].Saltz LB; Cox JV; Blanke C; Rosen LS; Fehrenbacher L; Moore MJ; Maroun JA; Ackland SP; Locker PK; Pirotta N; Elfring GL; Miller LL Irinotecan plus fluorouracil and leucovorin for metastatic colorectal Cancer Irinotecan Study Group. N. Engl. J. Med. 2000, 343 (13), 905–914. [DOI] [PubMed] [Google Scholar]
- [37].Douillard JY; Cunningham D; Roth AD; Navarro M; James RD; Karasek P; Jandik P; Iveson T; Carmichael J; Alakl M; Gruia G; Awad L; Rougier P Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet 2000, 355 (9209), 1041–1047. [DOI] [PubMed] [Google Scholar]
- [38].Xu Y; Villalona-Calero MA Irinotecan: mechanisms of tumor resistance and novel strategies for modulating its activity. Ann. Oncol. 2002, 13 (12), 1841–1851. [DOI] [PubMed] [Google Scholar]
- [39].Song Y; Shao Z; Dexheimer TS; Scher ES; Pommier Y; Cushman M Structure-based design, synthesis, and biological studies of new anticancer norindenoisoquinoline topoisomerase I inhibitors. J. Med. Chem. 2010, 53 (5), 1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wu MH; Yan B; Humerickhouse R; Dolan ME Irinotecan activation by human carboxylesterases in colorectal adenocarcinoma cells. Clin. Cancer Res. 2002, 8 (8), 2696–2700. [PubMed] [Google Scholar]
- [41].Charasson V; Bellott R; Meynard D; Longy M; Gorry P; Robert J Pharmacogenetics of human carboxylesterase 2, an enzyme involved in the activation of irinotecan into SN-38. Clin. Pharmacol. Ther. 2004, 76 (6), 528–535. [DOI] [PubMed] [Google Scholar]
- [42].Morton CL; Wadkins RM; Danks MK; Potter PM The anticancer prodrug CPT-11 is a potent inhibitor of acetylcholinesterase but is rapidly catalyzed to SN-38 by butyrylcholinesterase. Cancer Res. 1999, 59 (7), 1458–1463. [PubMed] [Google Scholar]
- [43].Boige V; Mendiboure J; Pignon JP; Loriot MA; Castaing M; Barrois M; Malka D; Tregouet DA; Bouche O; Le Corre D; Miran I; Mulot C; Ducreux M; Beaune P; Laurent-Puig P Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000–05. J. Clin. Oncol 2010, 28 (15), 25562564. [DOI] [PubMed] [Google Scholar]
- [44].McLeod HL; Sargent DJ; Marsh S; Green EM; King CR; Fuchs CS; Ramanathan RK; Williamson SK; Findlay BP; Thibodeau SN; Grothey A; Morton RF; Goldberg RM Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J. Clin. Oncol 2010, 28 (20), 3227–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Freyer G; Duret A; Milano G; Chatelut E; Rebischung C; Delord JP; Merrouche Y; Lledo G; Etienne MC; Falandry C Pharmacogenetic tailoring of irinotecan-based first-line chemotherapy in metastatic colorectal cancer: results of a pilot study. Anticancer Res. 2011, 31 (1), 359–366. [PubMed] [Google Scholar]
- [46].Jinno H; Saeki M; Saito Y; Tanaka-Kagawa T; Hanioka N; Sai K; Kaniwa N; Ando M; Shirao K; Minami H; Ohtsu A; Yoshida T; Saijo N; Ozawa S; Sawada J Functional characterization of human UDP-glucuronosyltransferase 1A9 variant, D256N, found in Japanese cancer patients. J. Pharmacol. Exp. Ther. 2003, 306 (2), 688–693. [DOI] [PubMed] [Google Scholar]
- [47].Monaghan G; Ryan M; Seddon R; Hume R; Burchell B Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet 1996, 347 (9001), 578581. [DOI] [PubMed] [Google Scholar]
- [48].Chu XY; Suzuki H; Ueda K; Kato Y; Akiyama S; Sugiyama Y Active efflux of CPT-11 and its metabolites in human KB-derived cell lines. J. Pharmacol. Exp. Ther. 1999, 288 (2), 735–741. [PubMed] [Google Scholar]
- [49].Mathijssen RH; Marsh S; Karlsson MO; Xie R; Baker SD; Verweij J; Sparreboom A; McLeod HL Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin. Cancer Res. 2003, 9 (9), 3246–3253. [PubMed] [Google Scholar]
- [50].Pillot GA; Read WL; Hennenfent KL; Marsh S; Gao F; Viswanathan A; Cummings K; McLeod HL; Govindan R A phase II study of irinotecan and carboplatin in advanced non-small cell lung cancer with pharmacogenomic analysis: final report. J. Thorac Oncol. 2006, 1 (9), 972–978. [PubMed] [Google Scholar]
- [51].Ulrich CM; Robien K; McLeod HL Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat. Rev. Cancer 2003, 3 (12), 912–920. [DOI] [PubMed] [Google Scholar]
- [52].O’Dwyer PJ; Catalano RB Uridine diphosphate glucuronosyltransferase (UGT) 1A1 and irinotecan: practical pharmacogenomics arrives in cancer therapy. J. Clin. Oncol. 2006, 24 (28), 4534–4538. [DOI] [PubMed] [Google Scholar]
- [53].Cha PC; Mushiroda T; Zembutsu H; Harada H; Shinoda N; Kawamoto S; Shimoyama R; Nishidate T; Furuhata T; Sasaki K; Hirata K; Nakamura Y Single nucleotide polymorphism in ABCG2 is associated with irinotecan-induced severe myelosuppression. J. Hum. Genet. 2009, 54 (10), 572–580. [DOI] [PubMed] [Google Scholar]
- [54].Shoemaker RH The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6 (10), 813–823. [DOI] [PubMed] [Google Scholar]
- [55].Kiselev E; Dexheimer TS; Pommier Y; Cushman M Design, synthesis, and evaluation of dibenzo[c,h][1,6]naphthyridines as topoisomerase I inhibitors and potential anticancer agents. J. Med. Chem. 2010, 53 (24), 8716–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pfister TD; Reinhold WC; Agama K; Gupta S; Khin SA; Kinders RJ; Parchment RE; Tomaszewski JE; Doroshow JH; Pommier Y Topoisomerase I levels in the NCI-60 cancer cell line panel determined by validated ELISA and microarray analysis and correlation with indenoisoquinoline sensitivity. Mol. Cancer Ther. 2009, 8 (7), 1878–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Das BB; Dexheimer TS; Maddali K; Pommier Y Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (46), 19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dexheimer TS; Antony S; Marchand C; Pommier Y Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 2008, 8 (4), 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang YW; Regairaz M; Seiler JA; Agama KK; Doroshow JH; Pommier Y Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011, 39 (9), 3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kidani Y; Noji M; Tashiro T Antitumor activity of platinum(II) complexes of 1,2-diamino-cyclohexane isomers. Gann 1980, 71 (5), 637–643. [PubMed] [Google Scholar]
- [61].Rixe O; Ortuzar W; Alvarez M; Parker R; Reed E; Paull K; Fojo T Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug-resistant cell lines and in the cell lines of the National Cancer Institute’s Anticancer Drug Screen panel. Biochem. Pharmacol. 1996, 52 (12), 1855–1865. [DOI] [PubMed] [Google Scholar]
- [62].Mathe G; Kidani Y; Segiguchi M; Eriguchi M; Fredj G; Peytavin G; Misset JL; Brienza S; de Vassals F; Chenu E Oxalato-platinum or 1-OHP, a third-generation platinum complex: an experimental and clinical appraisal and preliminary comparison with cis-platinum and carboplatinum. Biomed. Pharmacother. 1989, 43 (4), 237–250. [DOI] [PubMed] [Google Scholar]
- [63].Di Francesco AM; Ruggiero A; Riccardi R, Cellular and molecular aspects of drugs of the future: oxaliplatin. Cell Mol. Life Sci. 2002, 59 (11), 1914–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].de Gramont A; Figer A; Seymour M; Homerin M; Hmissi A; Cassidy J; Boni C; Cortes-Funes H; Cervantes A; Freyer G; Papamichael D; Le Bail N; Louvet C; Hendler D; de Braud F; Wilson C; Morvan F; Bonetti A Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal Cancer J. Clin. Oncol. 2000, 18 (16), 2938–2947. [DOI] [PubMed] [Google Scholar]
- [65].Rothenberg ML; Oza AM; Bigelow RH; Berlin JD; Marshall JL; Ramanathan RK; Hart LL; Gupta S; Garay CA; Burger BG; Le Bail N; Haller DG Superiority of oxaliplatin and fluorouracil-leucovorin compared with either therapy alone in patients with progressive colorectal cancer after irinotecan and fluorouracil-leucovorin: interim results of a phase III trial. J. Clin. Oncol. 2003, 21 (11), 2059–2069. [DOI] [PubMed] [Google Scholar]
- [66].Andre T; Boni C; Mounedji-Boudiaf L; Navarro M;Tabernero J; Hickish T; Topham C; Zaninelli M; Clingan P; Bridgewater J; Tabah-Fisch I, de Gramont, A , Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon. Cancer N. Engl. J. Med 2004, 350 (23), 2343–2351. [DOI] [PubMed] [Google Scholar]
- [67].Andre T; Boni C; Navarro M; Tabernero J; Hickish T; Topham C; Bonetti A; Clingan P; Bridgewater J; Rivera F; de Gramont A Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J. Clin. Oncol. 2009, 27 (19), 3109–3116. [DOI] [PubMed] [Google Scholar]
- [68].Kim GP; Sargent DJ; Mahoney MR; Rowland KM Jr.; Philip PA; Mitchell E; Mathews AP; Fitch TR; Goldberg RM; Alberts SR; Pitot HC Phase III noninferiority trial comparing irinotecan with oxaliplatin, fluorouracil, and leucovorin in patients with advanced colorectal carcinoma previously treated with fluorouracil: N9841. J. Clin. Oncol. 2009, 27 (17), 2848–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Raymond E; Faivre S; Woynarowski JM; Chaney SG, Oxaliplatin: mechanism of action and antineoplastic activity. Semin. Oncol. 1998, 25 (2 Suppl 5), 4–12. [PubMed] [Google Scholar]
- [70].Chaney SG; Campbell SL; Bassett E; Wu Y Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit. Rev. Oncol. Hematol. 2005, 53 (1), 3–11. [DOI] [PubMed] [Google Scholar]
- [71].Marsh S; McLeod H; Dolan E; Shukla SJ; Rabik CA; Gong L; Hernandez-Boussard T; Lou XJ; Klein TE; Altman RB Platinum pathway. Pharmacogenet. Genomics 2009, 19 (7), 563–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Graham J; Mushin M; Kirkpatrick P Oxaliplatin. Nat. Rev. Drug Discov. 2004, 3 (1), 11–2. [DOI] [PubMed] [Google Scholar]
- [73].Jamieson ER; Lippard SJ Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999, 99 (9), 2467–2498. [DOI] [PubMed] [Google Scholar]
- [74].Kruh GD Lustrous insights into cisplatin accumulation: copper transporters. Clin. Cancer Res. 2003, 9 (16 Pt 1), 5807–5809. [PubMed] [Google Scholar]
- [75].Yoh K; Ishii G; Yokose T; Minegishi Y; Tsuta K; Goto K; Nishiwaki Y; Kodama T; Suga M; Ochiai A Breast cancer resistance protein impacts clinical outcome in platinum-based chemotherapy for advanced non-small cell lung. Cancer Clin. Cancer Res. 2004, 10 (5), 1691–1697. [DOI] [PubMed] [Google Scholar]
- [76].Li XQ; Li J; Shi SB; Chen P; Yu LC; Bao QL Expression of MRP1, BCRP, LRP and ERCC1 as prognostic factors in non-small cell lung cancer patients receiving postoperative cisplatin-based chemotherapy. Int. J. Biol. Markers 2009, 24 (4), 230–237. [DOI] [PubMed] [Google Scholar]
- [77].Glaysher S; Yiannakis D; Gabriel FG; Johnson P; Polak ME; Knight LA; Goldthorpe Z; Peregrin K; Gyi M; Modi P; Rahamim J; Smith ME; Amer K; Addis B; Poole M; Narayanan A; Gulliford TJ; Andreotti PE; Cree IA Resistance gene expression determines the in vitro chemosensitivity of non-small cell lung cancer (NSCLC). BMC Cancer 2009, 9, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Moyer AM; Sun Z; Batzler AJ; Li L; Schaid DJ; Yang P; Weinshilboum RM Glutathione pathway genetic polymorphisms and lung cancer survival after platinum-based chemotherapy. Cancer Epidemiol. Biomarkers Prev. 2010, 19 (3), 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Fukushima-Uesaka H; Saito Y; Maekawa K; Kurose K; Sugiyama E; Katori N; Kaniwa N; Hasegawa R; Hamaguchi T; Eguchi-Nakajima T; Kato K; Yamada Y; Shimada Y; Yoshida T; Yamamoto N; Nokihara H; Kunitoh H; Ohe Y; Tamura T; Ura T; Saito M; Muro K; Doi T; Fuse N; Yoshino T; Ohtsu A; Saijo N; Matsumura Y; Okuda H; Sawada J Genetic polymorphisms of copper- and platinum drugefflux transporters ATP7A and ATP7B in Japanese cancer patients. Drug Metab. Pharmacokinet. 2009, 24 (6), 565–574. [DOI] [PubMed] [Google Scholar]
- [80].Samimi G; Safaei R; Katano K; Holzer AK; Rochdi M; Tomioka M; Goodman M; Howell SB Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin. Cancer Res. 2004, 10 (14), 4661–4669. [DOI] [PubMed] [Google Scholar]
- [81].More SS; Akil O; Ianculescu AG; Geier EG; Lustig LR; Giacomini KM Role of the copper transporter, CTR1, in platinum-induced ototoxicity. J. Neurosci. 2010, 30 (28), 95009509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sharma S; Gong P; Temple B; Bhattacharyya D; Dokholyan NV; Chaney SG Molecular dynamic simulations of cisplatin- and oxaliplatin-d(GG) intrastand cross-links reveal differences in their conformational dynamics. J. Mol. Biol. 2007, 373 (5), 11231140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ramachandran S; Temple BR; Chaney SG; Dokholyan NV Structural basis for the sequence-dependent effects of platinum-DNA adducts. Nucleic Acids Res. 2009, 37 (8), 2434–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Freimuth RR; Xiao M; Marsh S; Minton M; Addleman N; Van Booven DJ; McLeod HL; Kwok PY Polymorphism discovery in 51 chemotherapy pathway genes. Hum. Mol. Genet. 2005, 14 (23), 3595–3603. [DOI] [PubMed] [Google Scholar]
- [85].Pare L; Marcuello E; Altes A; del Rio E; Sedano L; Salazar J; Cortes A; Barnadas A; Baiget M Pharmacogenetic prediction of clinical outcome in advanced colorectal cancer patients receiving oxaliplatin/5-fluorouracil as first-line chemotherapy. Br. J. Cancer 2008, 99 (7), 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Liang J; Jiang T; Yao RY; Liu ZM; Lv HY; Qi WW The combination of ERCC1 and XRCC1 gene polymorphisms better predicts clinical outcome to oxaliplatin-based chemotherapy in metastatic colorectal cancer. bCancer chemotherapy and pharmacology 2010, 66 (3), 493–500. [DOI] [PubMed] [Google Scholar]
- [87].Ruzzo A; Graziano F; Loupakis F; Rulli E; Canestrari E; Santini D; Catalano V; Ficarelli R; Maltese P; Bisonni R; Masi G; Schiavon G; Giordani P; Giustini L; Falcone A; Tonini G; Silva R; Mattioli R; Floriani I; Magnani M Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J. Clin. Oncol 2007, 25 (10), 1247–1254. [DOI] [PubMed] [Google Scholar]
- [88].Hwang IG; Ahn MJ; Park BB; Ahn YC; Han J; Lee S; Kim J; Shim YM; Ahn JS; Park K ERCC1 expression as a prognostic marker in N2(+) nonsmall-cell lung cancer patients treated with platinum-based neoadjuvant concurrent chemoradiotherapy. Cancer 2008, 113 (6), 1379–1386. [DOI] [PubMed] [Google Scholar]
- [89].Kim SH; Kwon HC; Oh SY; Lee DM; Lee S; Lee JH; Roh MS; Kim DC; Park KJ; Choi HJ; Kim HJ Prognostic value of ERCC1, thymidylate synthase, and glutathione S-transferase pi for 5-FU/oxaliplatin chemotherapy in advanced colorectal cancer. Am. J. Clin. Oncol. 2009, 32 (1), 38–43. [DOI] [PubMed] [Google Scholar]
- [90].aisman A; Varchenko M; Umar A; Kunkel TA; Risinger JI; Barrett JC; Hamilton TC; Chaney SG The role of hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin resistance: correlation with replicative bypass of platinum-DNA adducts. Cancer Res. 1998, 58 (16), 3579–3585. [PubMed] [Google Scholar]
- [91].Kane MF; Loda M; Gaida GM; Lipman J; Mishra R; Goldman H; Jessup JM; Kolodner R Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997, 57 (5), 808–811. [PubMed] [Google Scholar]
- [92].Iacopetta B; Grieu F; Amanuel B Microsatellite instability in colorectal cancer. Asia Pac. J. Clin. Oncol. 2010, 6 (4), 260–269. [DOI] [PubMed] [Google Scholar]
- [93].Albertella MR; Green CM; Lehmann AR; O’Connor MJ A role for polymerase eta in the cellular tolerance to cisplatininduced damage. Cancer Res. 2005, 65 (21), 9799–9806. [DOI] [PubMed] [Google Scholar]
- [94].Indovina P; Giordano A Targeting the checkpoint kinase WEE1: selective sensitization of cancer cells to DNA-damaging drugs. Cancer Biol. Ther. 2010, 9 (7), 523–525. [DOI] [PubMed] [Google Scholar]
- [95].Hartwell LH; Weinert TA Checkpoints: controls that ensure the order of cell cycle events. Science 1989, 246 (4930), 629–634. [DOI] [PubMed] [Google Scholar]
- [96].Pommier Y; Kohn KW [Cell cycle and checkpoints in oncology: new therapeutic targets]. Med Sci (Paris). 2003, 19 (2), 173–186. [DOI] [PubMed] [Google Scholar]
- [97].Hartwell LH; Kastan MB Cell cycle control and cancer. Science 1994, 266 (5192), 1821–1828. [DOI] [PubMed] [Google Scholar]
- [98].Pommier Y; Sordet O; Rao VA; Zhang H; Kohn KW Targeting chk2 kinase: molecular interaction maps and therapeutic rationale. Curr. Pharm. Des. 2005, 11 (22), 2855–2872. [DOI] [PubMed] [Google Scholar]
- [99].Reinhardt HC; Yaffe MB Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 2009, 21 (2), 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Damia G; Broggini M Cell cycle checkpoint proteins and cellular response to treatment by anticancer agents. Cell Cycle 2004, 3 (1), 46–50. [PubMed] [Google Scholar]
- [101].Hartwell L; Weinert T; Kadyk L; Garvik B Cell cycle checkpoints, genomic integrity, and cancer. Cold Spring Harb. Symp. Quant. Biol. 1994, 59, 259–263. [DOI] [PubMed] [Google Scholar]
- [102].McKay JA; Douglas JJ; Ross VG; Curran S; Loane JF; Ahmed FY; Cassidy J; McLeod HL; Murray GI Analysis of key cell-cycle checkpoint proteins in colorectal tumours. J. Pathol. 2002, 196 (4), 386–393. [DOI] [PubMed] [Google Scholar]
- [103].Kops GJ; Weaver BA; Cleveland DW On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 2005, 5 (10), 773–785. [DOI] [PubMed] [Google Scholar]
- [104].Michel LS; Liberal V; Chatterjee A; Kirchwegger R; Pasche B; Gerald W; Dobles M; Sorger PK; Murty VV; Benezra R MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 2001, 409 (6818), 355–359. [DOI] [PubMed] [Google Scholar]
- [105].Dai W; Wang Q; Liu T; Swamy M; Fang Y; Xie S; Mahmood R; Yang YM; Xu M; Rao CV Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004, 64 (2), 440–445. [DOI] [PubMed] [Google Scholar]
- [106].Baker DJ; Jeganathan KB; Cameron JD; Thompson M; Juneja S; Kopecka A; Kumar R; Jenkins RB; de Groen PC; Roche P; van Deursen JM BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004, 36 (7), 744–749. [DOI] [PubMed] [Google Scholar]
- [107].Babu JR; Jeganathan KB; Baker DJ; Wu X; Kang-Decker N, van Deursen JM , Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 2003, 160 (3), 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Dalton WB; Yang VW Mitotic origins of chromosomal instability in colorectal cancer. Curr. Colorectal Cancer Rep. 2007, 3 (2), 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Cahill DP; Lengauer C; Yu J; Riggins GJ; Willson JK; Markowitz SD; Kinzler KW; Vogelstein B Mutations of mitotic checkpoint genes in human cancers. Nature 1998, 392 (6673), 300–303. [DOI] [PubMed] [Google Scholar]
- [110].Hanks S; Coleman K; Reid S; Plaja A; Firth H; Fitzpatrick D; Kidd A; Mehes K; Nash R; Robin N; Shannon N; Tolmie J; Swansbury J; Irrthum A; Douglas J; Rahman N Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004, 36 (11), 11591161. [DOI] [PubMed] [Google Scholar]
- [111].Lengauer C; Wang Z From spindle checkpoint to cancer. Nat. Genet. 2004, 36 (11), 1144–1145. [DOI] [PubMed] [Google Scholar]
- [112].Wang Z; Cummins JM; Shen D; Cahill DP; Jallepalli PV; Wang TL; Parsons DW; Traverso G; Awad M; Silliman N; Ptak J; Szabo S; Willson JK; Markowitz SD; Goldberg ML; Karess R; Kinzler KW; Vogelstein B; Velculescu VE; Lengauer C Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004, 64 (9), 2998–3001. [DOI] [PubMed] [Google Scholar]
- [113].Shichiri M; Yoshinaga K; Hisatomi H; Sugihara K; Hirata Y Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival. Cancer Res. 2002, 62 (1), 13–17. [PubMed] [Google Scholar]
- [114].Imai Y; Shiratori Y; Kato N; Inoue T; Omata M Mutational inactivation of mitotic checkpoint genes, hsMAD2 and hBUB1, is rare in sporadic digestive tract cancers. Jpn. J. Cancer Res. 1999, 90 (8), 837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Rimkus C; Friederichs J; Rosenberg R; Holzmann B; Siewert JR; Janssen KP Expression of the mitotic checkpoint gene MAD2L2 has prognostic significance in colon cancer. Int. J. Cancer 2007, 120 (1), 207–211. [DOI] [PubMed] [Google Scholar]
- [116].Li GQ; Zhang HF Mad2 and p27 expression profiles in colorectal cancer and its clinical significance. World J.Gastroenterol. 2004, 10 (21), 3218–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Li GQ; Li H; Zhang HF Mad2 and p53 expression profiles in colorectal cancer and its clinical significance. World J. Gastroenterol. 2003, 9 (9), 1972–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Menssen A; Epanchintsev A; Lodygin D; Rezaei N; Jung P; Verdoodt B; Diebold J; Hermeking H c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle 2007, 6 (3), 339352. [DOI] [PubMed] [Google Scholar]
- [119].Rodriguez-Antona C Pharmacogenomics of paclitaxel. Pharmacogenomics. 2010, 11 (5), 621–3. [DOI] [PubMed] [Google Scholar]
- [120].Swanton C; Tomlinson I; Downward J Chromosomal instability, colorectal cancer and taxane resistance. Cell Cycle 2006, 5 (8), 818–823. [DOI] [PubMed] [Google Scholar]
- [121].Furlan D; Casati B; Cerutti R; Facco C; Terracciano L; Capella C; Chiaravalli AM Genetic progression in sporadic endometrial and gastrointestinal cancers with high microsatellite instability. J. Pathol. 2002, 197 (5), 603–609. [DOI] [PubMed] [Google Scholar]
- [122].Bertoni F; Codegoni AM; Furlan D; Tibiletti MG; Capella C; Broggini M CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer 1999, 26 (2), 176–180. [PubMed] [Google Scholar]
- [123].Bartek J; Lukas J Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3 (5), 421–429. [DOI] [PubMed] [Google Scholar]
- [124].Lewis KA; Bakkum-Gamez J; Loewen R; French AJ; Thibodeau SN; Cliby WA Mutations in the ataxia telangiectasia and rad3-related-checkpoint kinase 1 DNA damage response axis in colon cancers. Genes Chromosomes Cancer 2007, 46 (12), 1061–1068. [DOI] [PubMed] [Google Scholar]
- [125].Liu WD; Zhong BY; Zhang YD; Choi GS Mutation analysis of the checkpoint kinase 2 gene in colorectal cancer cell lines. Chin. Med. J. (Engl). 2007, 120 (23), 2119–2123. [PubMed] [Google Scholar]
- [126].Bell DW; Varley JM; Szydlo TE; Kang DH; Wahrer DC; Shannon KE; Lubratovich M; Verselis SJ; Isselbacher KJ; Fraumeni JF; Birch JM; Li FP; Garber JE; Haber DA Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999, 286 (5449), 2528–2531. [DOI] [PubMed] [Google Scholar]
- [127].Wu X; Webster SR; Chen J Characterization of tumorassociated Chk2 mutations. J. Biol. Chem. 2001, 276 (4), 29712974. [DOI] [PubMed] [Google Scholar]
- [128].Zoppoli G; Solier S; Reinhold WC; Liu H; Connelly JW Jr.; Monks A; Shoemaker RH; Abaan OD; Davis SR; Meltzer PS; Doroshow JH; Pommier Y CHEK2 genomic and proteomic analyses reveal genetic inactivation or endogenous activation across the 60 cell lines of the US National Cancer Institute. Oncogene 2012, 31 (4), 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kilpivaara O; Laiho P; Aaltonen LA; Nevanlinna H, CHEK2 1100delC and colorectal cancer. J. Med. Genet. 2003, 40 (10), e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Meijers-Heijboer H; Wijnen J; Vasen H; Wasielewski M; Wagner A; Hollestelle A; Elstrodt F; van den Bos R; de Snoo A; Fat GT; Brekelmans C; Jagmohan S; Franken P; Verkuijlen P; van den Ouweland A; Chapman P; Tops C; Moslein G; Burn J; Lynch H; Klijn J; Fodde R; Schutte M The CHEK2 1100delC mutation identifies families with a hereditary breast and colorectal cancer phenotype. Am. J. Hum.Genet. 2003, 72 (5), 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].de Jong MM; Nolte IM; Te Meerman GJ; van der Graaf WT; Mulder MJ; van der Steege G; Bruinenberg M; Schaapveld M; Niessen RC; Berends MJ; Sijmons RH; Hofstra RM; de Vries EG; Kleibeuker JH Colorectal cancer and the CHEK2 1100delC mutation. Genes Chromosomes Cancer 2005, 43 (4), 377–382. [DOI] [PubMed] [Google Scholar]
- [132].van Puijenbroek M; van Asperen CJ; van Mil A; Devilee P; van Wezel T; Morreau H Homozygosity for a CHEK2*1100delC mutation identified in familial colorectal cancer does not lead to a severe clinical phenotype. J. Pathol. 2005, 206 (2), 198–204. [DOI] [PubMed] [Google Scholar]
- [133].Schutte M; Seal S; Barfoot R; Meijers-Heijboer H; Wasielewski M; Evans DG; Eccles D; Meijers C; Lohman F; Klijn J; van den Ouweland A; Futreal PA; Nathanson KL; Weber BL; Easton DF; Stratton MR; Rahman N Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am. J. Hum. Genet. 2003, 72 (4), 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Xiang HP; Geng XP; Ge WW; Li H Meta-analysis of CHEK2 1100delC variant and colorectal cancer susceptibility. Eur. J. Cancer 2011, 47 (17), 2546–2551. [DOI] [PubMed] [Google Scholar]
- [135].Ioachim E Expression patterns of cyclins D1, E and cyclin-dependent kinase inhibitors p21waf1/cip1, p27kip1 in colorectal carcinoma: correlation with other cell cycle regulators (pRb, p53 and Ki-67 and PCNA) and clinicopathological features. Int. J. Clin. Pract 2008, 62 (11), 1736–1743. [DOI] [PubMed] [Google Scholar]
- [136].Zirbes TK; Baldus SE; Moenig SP; Nolden S; Kunze D; Shafizadeh ST; Schneider PM; Thiele J; Hoelscher AH; Dienes HP Prognostic impact of p21/waf1/cip1 in colorectal cancer. Int. J. Cancer 2000, 89 (1), 14–18. [DOI] [PubMed] [Google Scholar]
- [137].Yasui W; Akama Y; Yokozaki H; Semba S; Kudo Y; Shimamoto F; Tahara E Expression of p21WAF1/CIP1 in colorectal adenomas and adenocarcinomas and its correlation with p53 protein expression. Pathol. Int. 1997, 47 (7), 470–477. [DOI] [PubMed] [Google Scholar]
- [138].Tort F; Bartkova J; Sehested M; Orntoft T; Lukas J; Bartek J Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006, 66 (21), 10258–10263. [DOI] [PubMed] [Google Scholar]
- [139].Rodrigues NR; Rowan A; Smith ME; Kerr IB; Bodmer WF; Gannon JV; Lane DP p53 mutations in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A. 1990, 87 (19), 7555–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Prudhomme M Novel checkpoint 1 inhibitors. Recent Pat Anticancer Drug Discov. 2006, 1 (1), 55–68. [DOI] [PubMed] [Google Scholar]
- [141].Janetka JW; Ashwell S; Zabludoff S; Lyne P Inhibitors of checkpoint kinases: from discovery to the clinic. Curr. Opin. Drug Discov. Devel. 2007, 10 (4), 473–486. [PubMed] [Google Scholar]
- [142].Collins I; Garrett MD Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr. Opin. Pharmacol. 2005, 5 (4), 366–373. [DOI] [PubMed] [Google Scholar]
- [143].Wang Q; Fan S; Eastman A; Worland PJ; Sausville EA; O’Connor PM UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J. Natl. Cancer Inst. 1996, 88 (14), 956–965. [DOI] [PubMed] [Google Scholar]
- [144].Ashwell S; Zabludoff S DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin. Cancer Res. 2008, 14 (13), 4032–4037. [DOI] [PubMed] [Google Scholar]
- [145].Hernandez L; Cholody WM; Hudson EA; Resau JH; Pauly G; Michejda CJ Mechanism of action of bisimidazoacridones, new drugs with potent, selective activity against colon cancer. Cancer Res. 1995, 55 (11), 2338–2345. [PubMed] [Google Scholar]
- [146].Cholody WM; Kosakowska-Cholody T; Michejda CJ Bisimidazoacridones induce a potent cytostatic effect in colon tumor cells that sensitizes them to killing by UCN-01. Cancer Chemother. Pharmacol. 2001, 47 (3), 241–249. [DOI] [PubMed] [Google Scholar]
- [147].Furuta T; Hayward RL; Meng LH; Takemura H; Aune GJ; Bonner WM; Aladjem MI; Kohn KW; Pommier Y p21CDKN1A allows the repair of replication-mediated DNA double-strand breaks induced by topoisomerase I and is inactivated by the checkpoint kinase inhibitor 7-hydroxystaurosporine. Oncogene 2006, 25 (20), 2839–2849. [DOI] [PubMed] [Google Scholar]
- [148].Petersen L; Hasvold G; Lukas J; Bartek J; Syljuasen RG p53-dependent G(1) arrest in 1st or 2nd cell cycle may protect human cancer cells from cell death after treatment with ionizing radiation and Chk1 inhibitors. Cell Prolif. 2010, 43 (4), 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Xiao Z; Xue J; Semizarov D; Sowin TJ; Rosenberg SH; Zhang H Novel indication for cancer therapy: Chk1 inhibition sensitizes tumor cells to antimitotics. Int. J. Cancer 2005, 115 (4), 528–538. [DOI] [PubMed] [Google Scholar]
- [150].Xiao Z; Xue J; Sowin TJ; Rosenberg SH; Zhang H A novel mechanism of checkpoint abrogation conferred by Chk1 downregulation. Oncogene 2005, 24 (8), 1403–1411. [DOI] [PubMed] [Google Scholar]
- [151].Castedo M; Perfettini JL; Roumier T; Yakushijin K; Horne D; Medema R; Kroemer G The cell cycle checkpoint kinase Chk2 is a negative regulator of mitotic catastrophe. Oncogene 2004, 23 (25), 4353–4361. [DOI] [PubMed] [Google Scholar]
- [152].Jobson AG; Lountos GT; Lorenzi PL; Llamas J; Connelly J; Cerna D; Tropea JE; Onda A; Zoppoli G; Kondapaka S; Zhang G; Caplen NJ; Cardellina JH 2nd; Yoo SS; Monks A; Self C; Waugh DS; Shoemaker RH; Pommier Y Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide]. J. Pharmacol. Exp. Ther. 2009, 331 (3), 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Pires IM; Ward TH; Dive C Oxaliplatin responses in colorectal cancer cells are modulated by CHK2 kinase inhibitors. Br. J. Pharmacol. 2010, 159 (6), 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Voland C; Bord A; Peleraux A; Penarier G; Carriere D; Galiegue S; Cvitkovic E; Jbilo O; Casellas P Repression of cell cycle-related proteins by oxaliplatin but not cisplatin in human colon cancer cells. Mol. Cancer Ther. 2006, 5 (9), 2149–2157. [DOI] [PubMed] [Google Scholar]
- [155].Chen CR; Wang W; Rogoff HA; Li X; Mang W; Li CJ Dual induction of apoptosis and senescence in cancer cells by Chk2 activation: checkpoint activation as a strategy against cancer. Cancer Res. 2005, 65 (14), 6017–6021. [DOI] [PubMed] [Google Scholar]
- [156].Solier S; Sordet O; Kohn KW; Pommier Y Death receptorinduced activation of the Chk2- and histone H2AX-associated DNA damage response pathways. Mol. Cell Biol. 2009, 29 (1), 6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Wendt J; von Haefen C; Hemmati P; Belka C; Dorken B; Daniel PT TRAIL sensitizes for ionizing irradiation-induced apoptosis through an entirely Bax-dependent mitochondrial cell death pathway. Oncogene 2005, 24 (25), 4052–4064. [DOI] [PubMed] [Google Scholar]
- [158].Ray S; Shyam S; Fraizer GC; Almasan A S-phase checkpoints regulate Apo2 ligand/TRAIL and CPT-11-induced apoptosis of prostate cancer cells. Mol. Cancer Ther. 2007, 6 (4), 1368–1378. [DOI] [PubMed] [Google Scholar]
- [159].Aris SM; Pommier Y Potentiation of the novel Topoisomerase I inhibitor indenoisoquinoline LMP-400 by the cell checkpoint and Chk1-Chk2 inhibitor, AZD7762. Cancer Res. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Riesterer O; Matsumoto F; Wang L; Pickett J; Molkentine D; Giri U; Milas L; Raju U A novel Chk inhibitor, XL-844, increases human cancer cell radiosensitivity through promotion of mitotic catastrophe. Invest. New Drugs 2011, 29 (3), 514–522. [DOI] [PubMed] [Google Scholar]
- [161].Gupta M; Fan S; Zhan Q; Kohn KW; O’Connor PM; Pommier Y Inactivation of p53 increases the cytotoxicity of camptothecin in human colon HCT116 and breast MCF-7 cancer cells. Clin. Cancer Res. 1997, 3 (9), 1653–1660. [PubMed] [Google Scholar]
- [162].Chang WT; Kang JJ; Lee KY; Wei K; Anderson E; Gotmare S; Ross JA; Rosen GD Triptolide and chemotherapy cooperate in tumor cell apoptosis. A role for the p53 pathway. J. Biol. Chem. 2001, 276 (3), 2221–2227. [DOI] [PubMed] [Google Scholar]
- [163].Fidler JM; Li K; Chung C; Wei K; Ross JA; Gao M; Rosen GD PG490–88, a derivative of triptolide, causes tumor regression and sensitizes tumors to chemotherapy. Mol. Cancer Ther. 2003, 2 (9), 855–862. [PubMed] [Google Scholar]
- [164].Sayed M; Pelech S; Wong C; Marotta A; Salh B Protein kinase CK2 is involved in G2 arrest and apoptosis following spindle damage in epithelial cells. Oncogene 2001, 20 (48), 69947005. [DOI] [PubMed] [Google Scholar]
- [165].Kaestner P; Stolz A; Bastians H Determinants for the efficiency of anticancer drugs targeting either Aurora-A or Aurora-B kinases in human colon carcinoma cells. Mol. Cancer Ther. 2009, 8 (7), 2046–2056. [DOI] [PubMed] [Google Scholar]
- [166].Redon CE; Nakamura AJ; Zhang YW; Ji JJ; Bonner WM; Kinders RJ; Parchment RE; Doroshow JH; Pommier Y Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin. Cancer Res. 2010, 16 (18), 4532–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Rouleau M; Patel A; Hendzel MJ; Kaufmann SH; Poirier GG PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 2010, 10 (4), 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kim MY; Zhang T; Kraus WL Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005, 19 (17), 1951–1967. [DOI] [PubMed] [Google Scholar]
- [169].Schreiber V; Dantzer F; Ame JC; de Murcia G Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006, 7 (7), 517–528. [DOI] [PubMed] [Google Scholar]
- [170].Ju BG; Lunyak VV; Perissi V; Garcia-Bassets I; Rose DW; Glass CK; Rosenfeld MG A topoisomerase IIbetamediated dsDNA break required for regulated transcription. Science 2006, 312 (5781), 1798–1802. [DOI] [PubMed] [Google Scholar]
- [171].Lis JT; Kraus WL Promoter cleavage: a topoIIbeta and PARP1 collaboration. Cell 2006, 125 (7), 1225–7. [DOI] [PubMed] [Google Scholar]
- [172].Caldecott KW XRCC1 and DNA strand break repair. DNA Repair (Amst). 2003, 2 (9), 955–969. [DOI] [PubMed] [Google Scholar]
- [173].Masson M; Niedergang C; Schreiber V; Muller S; Menissierde Murcia J; de Murcia G XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998, 18 (6), 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].El-Khamisy SF; Masutani M; Suzuki H; Caldecott KW A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003, 31 (19), 5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Audebert M; Salles B; Calsou P Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004, 279 (53), 55117–55126. [DOI] [PubMed] [Google Scholar]
- [176].Iliakis G Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother. Oncol. 2009, 92 (3), 310–315. [DOI] [PubMed] [Google Scholar]
- [177].Rouleau M; Patel A; Hendzel MJ; Kaufmann SH; Poirier GG PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 2010, 10 (4), 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Drew Y; Plummer R PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist. Updat. 2009, 12 (6), 153–156. [DOI] [PubMed] [Google Scholar]
- [179].Annunziata CM; O’Shaughnessy J Poly (adp-ribose) polymerase as a novel therapeutic target in cancer. Clin. Cancer Res. 2010, 16 (18), 4517–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Bryant HE; Schultz N; Thomas HD; Parker KM; Flower D; Lopez E; Kyle S; Meuth M; Curtin NJ; Helleday T Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434 (7035), 913–917. [DOI] [PubMed] [Google Scholar]
- [181].Farmer H; McCabe N; Lord CJ; Tutt AN; Johnson DA; Richardson TB; Santarosa M; Dillon KJ; Hickson I; Knights C; Martin NM; Jackson SP; Smith GC; Ashworth A Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434 (7035), 917–921. [DOI] [PubMed] [Google Scholar]
- [182].Nosho K; Yamamoto H; Mikami M; Taniguchi H; Takahashi T; Adachi Y; Imamura A; Imai K; Shinomura Y Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur. J. Cancer 2006, 42 (14), 2374–2381. [DOI] [PubMed] [Google Scholar]
- [183].Cristovao L; Lechner MC; Fidalgo P; Leitao CN; Mira FC; Rueff J Absence of stimulation of poly(ADP-ribose) polymerase activity in patients predisposed to colon cancer. Br. J.Cancer 1998, 77 (10), 1628–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Stern MC; Conti DV; Siegmund KD; Corral R; Yuan JM; Koh WP; Yu MC DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol. Biomarkers Prev. 2007, 16 (11), 2363–2372. [DOI] [PubMed] [Google Scholar]
- [185].Brevik A; Joshi AD; Corral R; Onland-Moret NC; Siegmund KD; Le Marchand L; Baron JA; Martinez ME; Haile RW; Ahnen D; Sandler RS; Lance P; Stern MC Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol. Biomarkers Prev. 2010, 19 (12), 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Calabrese CR; Almassy R; Barton S; Batey MA; Calvert AH; Canan-Koch S; Durkacz BW; Hostomsky Z; Kumpf RA; Kyle S; Li J; Maegley K; Newell DR; Notarianni E; Stratford IJ; Skalitzky D; Thomas HD; Wang LZ; Webber SE; Williams KJ; Curtin NJ Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl. Cancer Inst. 2004, 96(1),56–67. [DOI] [PubMed] [Google Scholar]
- [187].Donawho CK; Luo Y; Penning TD; Bauch JL; Bouska JJ; Bontcheva-Diaz VD; Cox BF; DeWeese TL; Dillehay LE; Ferguson DC; Ghoreishi-Haack NS; Grimm DR; Guan R; Han EK; Holley-Shanks RR; Hristov B; Idler KB; Jarvis K; Johnson EF; Kleinberg LR; Klinghofer V; Lasko LM; Liu X; Marsh KC; McGonigal TP; Meulbroek A; Olson AM; Palma JP; Rodriguez LE; Shi Y; Stavropoulos JA; Tsurutani AC; Zhu GD; Rosenberg SH; Giranda VL; Frost DJ ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 2007, 13 (9), 27282737. [DOI] [PubMed] [Google Scholar]
- [188].Liu X; Shi Y; Guan R; Donawho C; Luo Y; Palma J; Zhu GD; Johnson EF; Rodriguez LE; Ghoreishi-Haack N; Jarvis K; Hradil VP; Colon-Lopez M; Cox BF; Klinghofer V; Penning T; Rosenberg SH; Frost D; Giranda VL Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of singlestranded DNA damages to double-stranded DNA breaks. Mol. Cancer Res. 2008, 6 (10), 1621–1629. [DOI] [PubMed] [Google Scholar]
- [189].Tentori L; Leonetti C; Scarsella M; Muzi A; Mazzon E; Vergati M; Forini O; Lapidus R; Xu W; Dorio AS; Zhang J; Cuzzocrea S; Graziani G Inhibition of poly(ADP-ribose) polymerase prevents irinotecan-induced intestinal damage and enhances irinotecan/temozolomide efficacy against colon carcinoma. FASEB J. 2006, 20 (10), 1709–1711. [DOI] [PubMed] [Google Scholar]
- [190].Amador ML; Hidalgo M Epidermal growth factor receptor as a therapeutic target for the treatment of colorectal cancer. Clin. Colorectal Cancer 2004, 4 (1), 51–62. [DOI] [PubMed] [Google Scholar]
- [191].Hemming AW; Davis NL; Kluftinger A; Robinson B; Quenville NF; Liseman B; LeRiche J Prognostic markers of colorectal cancer: an evaluation of DNA content, epidermal growth factor receptor, and Ki-67. J. Surg. Oncol 1992, 51 (3), 147–152. [DOI] [PubMed] [Google Scholar]
- [192].Mayer A; Takimoto M; Fritz E; Schellander G; Kofler K; Ludwig H The prognostic significance of proliferating cell nuclear antigen, epidermal growth factor receptor, and mdr gene expression in colorectal cancer. Cancer 1993, 71 (8), 2454–2460. [DOI] [PubMed] [Google Scholar]
- [193].Bronte G; Terrasi M; Rizzo S; Sivestris N; Ficorella C; Cajozzo M; Di Gaudio F; Gulotta G; Siragusa S; Gebbia N; Russo A EGFR genomic alterations in cancer: prognostic and predictive values. Front. Biosci. (Elite Ed). 2011, 3, 879–887. [DOI] [PubMed] [Google Scholar]
- [194].Buisine MP; Wacrenier A; Mariette C; Leteurtre E; Escande F; Aissi S; Ketele A; Leclercq A; Porchet N; Lesuffleur T Frequent mutations of the CA simple sequence repeat in intron 1 of EGFR in mismatch repair-deficient colorectal cancers. World J. Gastroenterol. 2008, 14 (7), 1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Bijnsdorp IV; Kruyt FA; Fukushima M; Smid K; Gokoel S; Peters GJ Molecular mechanism underlying the synergistic interaction between trifluorothymidine and the epidermal growth factor receptor inhibitor erlotinib in human colorectal cancer cell lines. Cancer Sci. 2010, 101 (2), 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Hiro J; Inoue Y; Toiyama Y; Miki C; Kusunoki M Mechanism of resistance to chemoradiation in p53 mutant human colon cancer. Int. J. Oncol. 2008, 32 (6), 1305–1310. [DOI] [PubMed] [Google Scholar]
- [197].Vigneron A; Gamelin E; Coqueret O The EGFR-STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res. 2008, 68 (3), 815–825. [DOI] [PubMed] [Google Scholar]
- [198].Xu JM; Azzariti A; Severino M; Lu B; Colucci G; Paradiso A Characterization of sequence-dependent synergy between ZD1839 (“Iressa”) and oxaliplatin. Biochem. Pharmacol. 2003, 66 (4), 551–563. [DOI] [PubMed] [Google Scholar]
- [199].Haggblad Sahlberg S; Spiegelberg D; Lennartsson J; Nygren P; Glimelius B; Stenerlow B The effect of a dimeric Affibody molecule (ZEGFR:1907)2 targeting EGFR. Int. J. Oncol. 2012, 40 (, 176–184. [DOI] [PubMed] [Google Scholar]