Abstract
Although whole-genome sequencing has provided novel insights into Neisseria meningitidis, many open reading frames have only been annotated as hypothetical proteins with unknown biological functions. Our previous genetic analyses revealed that the hypothetical protein, NMB1345, plays a crucial role in meningococcal infection in human brain microvascular endothelial cells; however, NMB1345 has no homology to any identified protein in databases and its physiological function could not be elucidated using pre-existing methods. Among the many biological technologies to examine transient protein-protein interaction in vivo, one of the developed methods is genetic code expansion with non-canonical amino acids (ncAAs) utilizing a pyrrolysyl-tRNA synthetase/tRNAPyl pair from Methanosarcina species: However, this method has never been applied to assign function-unknown proteins in pathogenic bacteria. In the present study, we developed a new method to genetically incorporate ncAAs-encoded photocrosslinking probes into N. meningitidis by utilizing a pyrrolysyl-tRNA synthetase/tRNAPyl pair and elucidated the biological function(s) of the NMB1345 protein. The results revealed that the NMB1345 protein directly interacts with PilE, a major component of meningococcal pili, and further physicochemical and genetic analyses showed that the interaction between the NMB1345 protein and PilE was important for both functional pilus formation and meningococcal infectious ability in N. meningitidis. The present study using this new methodology for N. meningitidis provides novel insights into meningococcal pathogenesis by assigning the function of a hypothetical protein.
Introduction
Neisseria meningitidis is a fastidious Gram-negative microorganism that generally exists in the non-invasive so-called “carrier state” at a rate of 0.4–25% in human populations [1, 2]. However, N. meningitidis exhibits the abilities to cross the epithelial layer of the upper respiratory tract, infiltrate the bloodstream, evade the defenses of the human immune system, adhere to the endothelial layers of peripheral and brain vessels, cross the brain-blood barrier, and replicate in cerebrospinal fluid [3]. Although epidemiological analyses previously suggested that a human genetic polymorphism [4] and climate conditions [5] are important for predicting the outcomes of infection, the reasons why invasive meningococcal diseases (IMD) occur in some individuals, but not in others remain unclear [6].
Classical analyses revealed the factors exposed on meningococcal surfaces, such as the polysaccharide capsule, pili, Opa, and Opc, (reviewed in [7–9]), and genome mining with whole-genome sequencing (WGS) identified the following meningococcal pathogenic factors: NhhA, NadA [10], App [11], NalP [12], MspA [13], TspA [14], adhesion complex protein (ACP) [15], and fHbp [16]. Among the factors described above, meningococcal pili, which are mainly composed of the major pilin, PilE, and three minor pilins, PilV, PilX, and ComP, play the most important roles in the meningococcal infection processes involved in interactions with human epithelial and endothelial cells (reviewed in [17, 18]). To elucidate the molecular mechanisms underlying meningococcal pathogenesis in more detail, functional genomics by linking genotypes to phenotypes have allowed investigation of the relationships between gene transcript abundance or deficiencies and the capacity to function under various physiological conditions to be investigated, by genome-wide signature-tagged mutagenesis (STM) with an infant rat infection model [19], with human cultured cells [20], and in different media [21], microarrays with infection in human cultured cells [22], comparative genomics [23, 24], transcriptomics [25–28], and proteomics [29–31]. However, the mechanisms by which N. meningitidis causes septicemia and meningitis in humans cannot be completely explained by these characterized factors [32].
Although progressive WGS recently identified many genes in the meningococcal genome, more than half of those in the annotated open reading frame have remained as function-unknown hypothetical proteins. One of the difficulties associated with the characterization of meningococcal hypothetical proteins may be the limited number of analytical methods applicable to N. meningitidis, including tools to directly manipulate meningococcal components [33, 34]. Under these conditions, we also conducted STM to identify the factors responsible in the sequence type (ST)-2032 N. meningitidis strain, known for its high infectious ability in human brain microvascular endothelial cells (HBMEC) [35]. Among the STM mutants with highly defective infectious abilities in HBMEC, we identified PilV [36] of the meningococcal pilus and GltT-GltM glutamate transporter [37, 38] as meningococcal pathogenic factors for infection in host cells. However, several STM mutants were identified as disruptants of hypothetical genes, and we noted that, one disruptant of a hypothetical gene annotated as NMB1345 in the N. meningitidis MC58 genome [39], exhibited largely defective internalization ability into HBMEC. However, the deduced amino acid sequence of NMB1345 lacked homology with any function-known proteins from other species. We previously attempted to examine protein-protein interactions using already existing methods, such as the two-hybrid system or a pull-down assay, to find some clues about the function of the NMB1345 protein but were unsuccessful. Therefore, more powerful strategies are needed to elucidate the function of the NMB1345 protein in N. meningitidis.
One of the appropriate methods for our study is genetic code expansion with non-canonical amino acids (ncAAs) utilizing a pyrrolysyl-tRNA synthetase/tRNAPyl pair from Methanosarcina mazei (MmPylRS/T) [40]. The genetic code expansion allows ncAAs containing fluorophores, photocaged groups, photocrosslinkers, metal ion-chelating groups, and other groups to be site-specifically incorporated into target proteins in vivo in a broad range of species from Escherichia coli to eukaryotic systems, such as Caenorhabditis elegans, fruit flies, and mice (reviewed in [41]), thereby enabling in vivo examinations of biological functions. Protein photocrosslinking via the incorporation of photoreactive ncAAs has emerged as a superior strategy for identifying physiological interaction partner(s) and their functions [42, 43], and in combination with mass spectrometry (MS), these approaches have resolved biological issues that are difficult or impossible to address using the majority of currently available methods [42]. However, the question of whether the orthogonality of the tRNAPyl-PylRS pair is specific or compatible with the translational system in N. meningitidis has not been addressed.
In the present study, we applied genetic code expansion to N. meningitidis and developed a method for the genetic incorporation of photoreactive ncAAs into N. meningitidis in order to identify the physiological function of the NMB1345 protein. After establishing this method, we performed site-specific incorporation with one of the most widely used photoreactive amino acids, p-benzoyl-L-phenylalanine (pBPa) [44], into the NMB1345 protein in N. meningitidis. By using crosslinking in N. meningitidis followed by purification and MS analyses of endogenous proteins crosslinked to the NMB1345 protein, we found that the NMB1345 protein specifically interacted with PilE, a major component of meningococcal pili, and thus designated NMB1345 as PilE associated molecule A (PamA) in the present study. We further identified the amino acid residue of PilE crosslinked to PamA K273 and further found that the site-directed PilE mutation reduced both the interaction with PamA and the ability to internalize into HBMEC. Considering these results, we speculated that PamA plays an important role in the formation of meningococcal pili. This is the first study to successfully incorporate ncAAs into N. meningitidis in order to determine the physiological function of a protein with an unknown function and structure in pathogenic bacteria.
Materials and methods
Bacterial growth conditions
N. meningitidis strains HT1125 [35] and its derivatives, H44/76 [45] as well as derivatives harboring IncQ plasmids (Table 1) were routinely grown on GC agar plates at 37 oC in a 5% CO2 atmosphere [46]. E. coli strains were grown in Luria-Bertani (LB) broth liquid cultures (Becton- Dickinson) or on LB plate (LB liquid medium containing 1.5% agar) or in MagicMedia (Invitrogen) at the indicated temperatures. When required, antibiotics were added at the following concentrations: kanamycin at 150 μg/ml, chloramphenicol at 5 μg/ml, erythromycin at 4 μg/ml, and spectinomycin at 75 μg/ml for N. meningitidis; kanamycin at 50 μg/ml, chloramphenicol at 25 μg/ml, and ampicillin at 50 μg/ml for E. coli. The N. meningitidis and E. coli strains used in the present study are listed in Table 2.
Table 1. IncQ plasmids introduced into N. meningitidis strains which results were shown in this study.
| Plasmid | Relative properties | Antibiotic selection marker | References |
|---|---|---|---|
| pHT128 | Derivative of pGSS33, an IncQ broad-host -range vector | Tet, Cml | [33] |
| pHT1212 | Derivative of pHT128 carrying a lacIq-Ptac-MmPylRS[Y306A/Y384F] and two Plpp-tRNApyl genes | Tet, Cml | This study |
| pHT1262 | Derivative of pHT128 carrying a lacIq-MmPylRS([A302T/N346T/C348T/W417C]) and two Plpp-tRNApyl genes | Tet, Cml | This study |
| pHT936 | Derivative of pHT128 carrying lacIq-Ptac-gst+ genes | Cml | This study |
| pHT1355 | Derivative of pHT1212 carrying a Ptac-gst E51amb gene | Cml | This study |
| pHT1356 | Derivative of pHT1212 carrying a Ptac-gst F52amb gene | Cml | This study |
| pHT1357 | Derivative of pHT1262 carrying a Ptac-gst E51amb gene | Cml | This study |
| pHT1358 | Derivative of pHT1262 carrying a Ptac-gst F52amb gene | Cml | This study |
| pHT1263 | Derivative of pHT1262 carrying a pamA+-His6 gene | Cml | This study |
| pHT1270 | Derivative of pHT1262 carrying a pamA K208amb-His6 gene | Cml | This study |
| pHT1274 | Derivative of pHT1262 carrying a pamA K278amb-His6 gene | Cml | This study |
| pHT1276 | Derivative of pHT1262 carrying a pamA K309amb-His6 gene | Cml | This study |
| pHT1279 | Derivative of pHT1262 carrying a pamA K341amb-His6 gene | Cml | This study |
| pHT1282 | Derivative of pHT1262 carrying a pamA K382amb-His6 gene | Cml | This study |
| pHT1284 | Derivative of pHT1262 carrying a pamA K395amb-His6 gene | Cml | This study |
| pHT1388 | Derivative of pHT1262 carrying a pamA K278amb-StrepTag2-His6 gene | Cml | This study |
Tet and Cml stand for tetracycline and chloramphenicol resistance marker, respectively.
Table 2. Strains used in this study.
| N. meningitidis strains | |||
| HT1125 | |||
| Strain | Genotype | Parent strain | Reference |
| HT1125 | ΔsiaB-ΔsiaD::kan (ST-2032) | [35] | |
| HT1572 | ΔsiaB-ΔsiaD::kan pamA::Tn-spc | HT1125 | This study |
| HT1736 | ΔsiaB-ΔsiaD::kan pamA::Tn-spc ggt::pamA+-cat | HT1572 | This study |
| HT1822 | ΔsiaB-ΔsiaD::kan ΔpamA::spc | HT1125 | This study |
| HT2224 | ΔsiaB-ΔsiaD::kan ΔpamA::spc ggt::pamA+-cat | HT1822 | This study |
| HT2215 | ΔsiaB-ΔsiaD::kan pamA’-lacZ-ermC | HT1822 | This study |
| HT2217 | ΔsiaB-ΔsiaD::kan pamA’-phoA-ermC | HT1822 | This study |
| HT1744 | ΔsiaB-ΔsiaD::kan pilE+-cat (translational fusion) | HT1125 | This study |
| HT2167 | ΔsiaB-ΔsiaD::kan pilE I12A-cat (translational fusion) | HT1125 | This study |
| HT2218 | ΔsiaB-ΔsiaD::kan pilF+-FLAG-ermC | HT1125 | This study |
| HT2219 | ΔsiaB-ΔsiaD::kan pilF+-FLAG -ermCΔpamA::spc | HT1822 | This study |
| HT2136 | ΔsiaB-ΔsiaD::kan pilP+-FLAG -ermC | HT1125 | This study |
| HT2137 | ΔsiaB-ΔsiaD::kan pilP+-FALG-ermCΔpamA::spc | HT1822 | This study |
| HT2132 | ΔsiaB-ΔsiaD::kan pilM+-HA -ermC | HT1125 | This study |
| HT2133 | ΔsiaB-ΔsiaD::kan pilM+-HA -ermC ΔpamA::spc | HT1822 | This study |
| HT2211 | ΔsiaB-ΔsiaD::kan pilX+-FLAG-ermC | HT1125 | This study |
| HT2212 | ΔsiaB-ΔsiaD::kan pilX+-FLAG -ermC ΔpamA::spc | HT1822 | This study |
| HT1688 | ΔsiaB-ΔsiaD::kan pilE::ermC | HT1125 | [36] |
| HT1822 | ΔsiaB-ΔsiaD::kan ΔpilV::ermC | HT1125 | [36] |
| HT2230 | ΔsiaB-ΔsiaD::kan pilE+-cat pilX+-FLAG-ermC | HT1744 | This study |
| HT2231 | ΔsiaB-ΔsiaD::kan pilE I12A-cat pilX+-FLAG-ermC | HT2167 | This study |
| H44/76 | |||
| Strain | Genotype | Parent strain | Reference |
| H44/76 | wild type strain (ST-32, Serogroup B) | [33] | |
| HT1001 | H44/76 gyrA (NalR) | H44/76 | This study |
| HT1940 | ΔpamA::spc | H44/76 | This study |
| HT2095 | ΔpamA::spc pilE+-FLAG-ermC | HT1940 | This stud |
| HT2014 | ΔpamA::spc pilE::ermC | HT1940 | This study |
| HT2162 | ΔpamA::spc pilE+-ermC (translational fusion) | HT1940 | This study |
| HT2166 | ΔpamA::spc pilE I12A-ermC (translational fusion) | HT1940 | This study |
| HT1015 | pilE::ermC | H44/76 | [35] |
| Escherichia coli strains | |||
| Strain | Genotype | References | |
| JM109 | endA1 gyrA96 hsdR17(rk-mk+) mcrB+ recA1 relA1 supE44 thi-1Δ(lac-proAB) F'[traD36 proAB lacIqZΔM15] | Nippon gene | |
| BL21 | F–dcm ompT hsdSB(rB–mB–) gal | NEB | |
| BL21(DE3) | F–dcm ompT hsdSB(rB–mB–) gal λ(DE3) | NEB | |
Production of anti-PamA protein rabbit antiserum
All PCR experiments were performed with PrimeSTAR Max GXL DNA polymerase (Takara Bio, Japan). The approximately 3.5-kb PCR fragment containing the pamA gene amplified from N. meningitidis HT1125 genomic DNA with the primer set (NMB1345-21 and NMB1345-2) (S1 Table) was cloned into the pTWV228 vector (Takara Bio, Japan), resulting in pHT922 (S2 Table), which is the ancestral plasmid in the present study. A ΔN-pamA gene, in which the N-terminal hydrophobic region consisting of the first 23 amino acids was removed (S1 Fig), was amplified by PCR with the primer set (NMB1345(pET303)-1(NsiI) and NMB1345(pET303)-2(BH)) (S1 Table) and cloned into the same site of the pET303/CT-His expression plasmid (Invitrogen) to construct pHT934 (S3 Table). Regarding the production of the ΔN-PamA protein for rabbit immunization, a 1 L culture of E. coli strain BL21 (NEB) harboring the plasmid was grown in MagicMedia at 37°C overnight, and the cells were collected. The subsequent purification of the recombinant protein and generation of polyclonal rabbit antibodies to the putative hydrophilic domain of the PamA protein were performed as described previously [46].
Construction of meningococcal mutants
In order to construct the N. meningitidis pamA deletion mutant, the 6.3-kb PCR fragment was amplified with the primers (NMB1345-3 and NMB1345-4) (S1 Table) from pHT922, in which a 1.5-kb DNA fragment containing the coding region of the pamA gene was completely removed, and ligated with a spectinomycin-resistance gene (spc) after phosphorylation to construct pHT930. A 3-kb DNA fragment, in which the pamA structural gene was replaced with the spc gene, was amplified with the primers (NMB1345-21 and NMB1345-2) (S1 Table) from pHT930 and transformed into N. meningitidis strain HT1125, and spectinomycin-resistant (SpcR) clones were selected, resulting in the ΔpamA mutant HT1822 (Table 2).
The ΔpamA mutant complemented with the pamA+ gene at the ggt allele was constructed as follows: A chloramphenicol-resistance gene (cat) was inserted into the SmaI site downstream of the pamA gene of pHT922 to generate pHT923 (S2 Table). A 4.6-kb PCR fragment containing the pamA+-cat gene was amplified from pHT923, in which the meningococcal ggt structural gene was cloned in pTWV228 with primers (M13-RV-ggt-5′and M13-47-ggt-3′) (S1 Table) and cloned into the BstXI site of pHT195 (at the middle of the ggt coding region) by In-Fusion cloning (Clontech), resulting in the plasmid pHT924. A 6.3-kb DNA fragment containing the ggt::pamA+-cat genes was amplified from pHT924 with primers (ggt-3 and ggt-4) (S1 Table), transformed into N. meningitidis ggt::pamA+-cat mutants, and chloramphenicol-resistant (Cmlr) clones were selected, resulting in the pamA deletion mutant HT2224, which were ectopically complemented with the pamA+ genes expressed from their own promoter at the ggt allele (Table 1).
Since we have demonstrated that the pilE-cat translational fusion could work as well as the wild-type PilE+ N. meningitidis strain [36], an N. meningitidis I12A pilE mutant, in which Ile at position 12 was substituted with Ala, was constructed as described previously. In brief, pHT872 (pilE+-cat translational fusion on pGEM-3z) (S2 Table) [36] was site-directed mutagenized with primers (pilE-I12A-1 and pilE I12A-2) (S1 Table) to construct pHT1536 (S2 Table). After confirming its sequence, a 1.5-kb PCR fragment containing the pilE I12A-cat gene was amplified from pHT1536 by a primer set (T7 and SP6) (S1 Table) and transformed into HT1125. Cmlr clones were selected and the mutation was confirmed by direct sequencing as for the pilE I12A-cat N. meningitidis mutant HT2167 (Table 1).
To construct the pilE+-ermC or pilE I12A-ermC translational fusion in N. meningitidis strain H44/76 harboring the IncQ plasmid, a 0.9-kb DNA fragment from HT1125 chromosomal DNA containing the structural gene of the wild-type pilE gene and its upstream region was amplified with both primers (pGEM-3z-(SmaI)-2(15mer)-pilE-51 and pilE-52) (S1 Table). A 0.2-kb DNA fragment of the pilE downstream region was also amplified with a primer set (pilE-53 and pilE-18) (S1 Table), and a 0.75-kb DNA fragment of the ermC gene containing the SD sequence was amplified from pHT24 with a primer set (ermC-21 and ermC-22) (S1 Table). The three PCR fragments were cloned into the SmaI site of pGEM-3z to construct pHT1504 (S2 Table). The N. meningitidis I12A pilE mutant was constructed as described above using the primer set (pilE-I12A-1 and pilE I12A-2) (S1 Table) with pHT1504 as the template. After the sequence was confirmed, a 1.5-kb PCR fragment containing the pilE I12A-ermC genes was amplified from pHT1536 (S2 Table) with universal primers (T7 and SP6) and transformed into HT1125. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilE I12A-ermC N. meningitidis mutant HT2166 (Table 1). A 1.5-kb PCR fragment containing the pilE+-ermC genes amplified from pHT1504 (S2 Table) or the pilE I12A-ermC genes amplified from pHT1536 (S2 Table) by universal primers (T7 and SP6) was transformed into HT1940. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilE+-ermC HT2162 or pilE I12A-ermC HT2166 N. meningitidis mutants in the ΔpamA::spc genetic background (Table 1).
The addition of a FLAG tag to the pilE gene at the 3′-terminus on its chromosomal locus was achieved as follows: a 1.6-kb PCR fragment containing the pilE allele in H44/76 was amplified with a primer set (pilE-11and pilE-12) (S1 Table), and after phosphorylation, it was cloned into pMW119 to construct pHT1419 (S2 Table). A 5.8-kb PCR fragment was amplified with a primer set (pilE-17 and FLAG’(15mer)-pilE-18) (S1 Table) from pHT24, and a 1-kb PCR fragment containing the ermC gene was also amplified with a primer set (pilE-17’(15mer)-M13-47 and FLAG-RV-M) (S1 Table) from pHT24. The two PCR fragments were ligated by In-Fusion Cloning to construct pHT1420, in which the FLAG tag was fused to the pilE structural gene followed by an ermC selection marker (S2 Table). A 2.6-kb PCR fragment containing the pilE+-FLAG-ermC genes was amplified from pHT1420 with a primer set (pilE-11 and pilE-12) and transformed into HT1940. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilE+-FLAG-ermC N. meningitidis mutant HT2095 (Table 1).
The pilE insertional mutant HT2014 was constructed by transforming HT1940 with 500 ng of HT1015 (H44/76 pilE::ermC) [35] chromosomal DNA, followed by the selection of the Ermr strain HT2014 (Table 1).
The addition of a FLAG tag to the pilF gene at the 3′-terminus on its chromosomal locus was performed as follows: a 1.8-kb PCR fragment containing the pilF allele in the H44/76 N. meningitidis strain was amplified with a primer set (pMW(SmaI)-up(15mer)-pilF-1 and pMW(SmaI)-down(15mer)-pilF-2) (S1 Table) and cloned into the SmaI site of pMW119 by In-Fusion cloning to construct pHT1449 (S1 Table). A 6-kb PCR fragment amplified with a primer set (pilF-3 and FLAG’(15mer)-pilF-4) (S1 Table) from pHT1499 and a 1-kb PCR fragment containing the ermC gene amplified with a primer set (pilF-3’(15mer)-M13-47 and FLAG-RV-M) (S1 Table) from pHT24 were ligated by In-Fusion Cloning to construct pHT1457, in which the FLAG tag was fused to the pilF structural gene followed by an ermC selection marker (S1 Table). A 2.8-kb PCR fragment containing the pilF+-FLAG-ermC genes was amplified from pHT1457 with a primer set (pMW(SmaI)-up(15mer)-pilF-1 and pMW(SmaI)-down(15mer)-pilF-2) (S1 Table), and then transformed into HT1125 and HT1822. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilF+-FLAG-ermC N. meningitidis mutants HT2218 and HT2219, respectively (Table 1).
A 3.9-kb DNA fragment containing partial pilM-pilN+-pilO+-pilP+ genes in H44/76 N. meningitidis was amplified with a primer set (pMW(SmaI)-up(15mer)-pilMNOP-3 and pMW(SmaI)-down(15mer)-pilMNOP-4) (S1 Table) and cloned into the SmaI site of pMW119 by In-Fusion cloning to construct pHT1450 (S2 Table).
The addition of a FLAG tag to the pilP gene at the 3’ terminus on its chromosomal locus was achieved as follows: a 7.2-kb DNA fragment amplified with a primer set (pilP-3 and FLAG’(15mer)-pilP-2) (S1 Table) from pHT1450 and a 1-kb PCR fragment containing the ermC gene amplified with a primer set (pilP-3’(15mer)-M13-47 and FLAG-RV-M) (S1 Table) from pHT24 were ligated by In-Fusion cloning to construct pHT1454 (S2 Table), in which the FLAG tag was fused to the pilP structural gene at the 3’ terminus followed by an ermC selection marker. A 4.9-kb PCR fragment containing the pilP+-FLAG-ermC genes was amplified from pHT1454 with a primer set (pMW(SmaI)-up(15mer)-pilMNOP-3 and pMW(SmaI)-down(15mer)-pilMNOP-4)(S1 Table) and transformed into HT1125 and HT1822. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilP+-FLAG-ermC N. meningitidis mutants HT2136 and HT2137, respectively (Table 1).
The addition of the HA tag to the pilM gene at the 3’ terminus on its chromosomal locus was achieved; however, the ORFs of the pilM (1.1 kb), pilN (0.6 kb), and pilO (0.6 kb) genes were very close or overlapped, and thus an ermC selection marker was inserted downstream of the pilO gene. A 7.1-kb PCR fragment containing the partial pilM gene and pMW119 from pHT1450 was amplified with a primer set (pilO-3 and HA’-pilM-2) (S1 Table). A 1.2-kb PCR fragment was amplified with a primer set (HA’(15mer)-pilN-1 and pilO-2) from pHT1450 and a 1-kb ermC gene fragment was also amplified with a primer set (pilO-2’(15mer)-M13-RV and pilO-3(15mer)-M13-47) (S1 Table) from pHT24. The three PCR fragments were ligated by In-Fusion Cloning to construct pHT1453, in which the HA tag was fused to the pilM structural gene with an ermC selection marker (S2 Table). A 2.6-kb PCR fragment containing the pilM+-HA pilN+pilO+-ermC-pilP genes was amplified from pHT1453 with a primer set (pMW(SmaI)-up(15mer)-pilMNOP-3 and pMW(SmaI)-down(15mer)-pilMNOP-4) (S1 Table) and transformed into HT1125 and HT1822. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilM+-HA-ermC N. meningitidis mutants HT2132 and HT2133, respectively.
The addition of a FLAG tag to the pilX gene at the 3’ terminus on its chromosomal locus was achieved as follows: a 0.5-kb pilX DNA region containing the 400-bp upstream and 160-bp downstream regions of the pilX ochre codon in H44/76 was amplified with a primer set (pMW(SmaI)-up(15mer)-pilX-3 and pMW(SmaI)-down(15mer)-pilX-6) (S1 Table), and then cloned into the SmaI site of pMW119 by In-Fusion cloning to construct pHT1594. A 4.7-kb PCR fragment amplified with a primer set (M13-47′(15mer)-pilX-5 and FLAG′(15mer)-pilX-8) (S1 Table) from pHT1594, and a 1-kb PCR fragment containing the ermC gene amplified with a primer set (FLAG-RV-M and M13-47) (S1 Table) from pHT24 were ligated by In-Fusion Cloning to construct pHT1597, in which the FLAG tag was fused to the pilX structural gene followed by an ermC selection marker (S2 Table). A 1.5-kb PCR fragment containing partial pilX-FLAG-ermC genes was amplified from pHT1597 with a primer set (pMW(SmaI)-up(15mer)-pilX-3 and pMW(SmaI)-down(15mer)-pilX-6) (S1 Table) and transformed into HT1125 and HT1822. Ermr clones were selected and the mutation was confirmed by direct sequencing as for the pilX+-FLAG-ermC N. meningitidis mutants HT2211 and HT2212, respectively (Table 1).
pamA-lacZ translational fusion N. meningitidis mutants were constructed as follows: a 3-kb PCR fragment containing the lacZ gene was amplified from E. coli strain W3110 [47] chromosomal DNA with the primers (lacZ-21 and M13-RV’(15mer)-lacZ-22) (S1 Table) and a 1-kb ermC gene fragment was amplified with universal primers (M13-RV and M13-47) (S1 Table) from pHT24 [33]. The two PCR fragments were cloned into a 7.2-kb DNA fragment (designated Fragment C) amplified from pHT922 containing the upstream and downstream regions of the pamA gene with the first 408-bp region including the N-terminal membrane spanning region (designated pamA′) with a primer set (M13-47′(15mer)-nmb1345-61 and lacZ′-21(15mer)-nmb1345-62) (S1 Table) by In-Fusion cloning to construct pH1605 (S2 Table and Fig 2B).
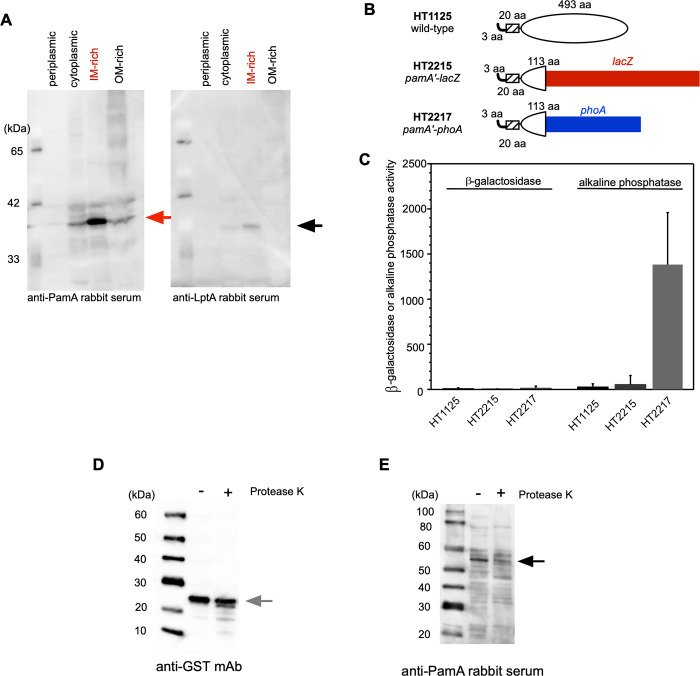
Fig 2. Characterization of the biochemical properties of the PamA protein in N. meningitidis.
(A) Western blot analyses of subcellular fractions of the PamA protein in N. meningitidis strain HT1125. A whole-cell lysate periplasmic fraction (P) isolated by CHCl3 shock, cytoplasmic and periplasmic fraction (CP), IM-enriched fraction, and outer membrane (OM)-enriched fraction were prepared by the sonication method from the N. meningitidis strain, as described previously [45]. Western blotting with anti-PamA (left) and anti-LptA (right) [55] rabbit antisera. LptA is a control for the proper isolation of the IM-rich fraction. Black and gray arrows show PamA and LptA, respectively. (B) Membrane topology of the PamA protein using the pamA′-lacZ and pamA′-phoA fused N. meningitidis strain. The hatched region consists of the 20 amino acid residues corresponding to the putative membrane-spanning region (S1 Fig). The first 408-bp region of the pamA gene was fused to the lacZ or phoA gene, and the gene was incorporated into the pamA locus on the meningococcal chromosome. (C) β-Galactosidase and alkaline phosphatase activities. (D,E) Western blot of spheroplasted N. meningitidis strain H44/76 harboring pHT936 (an IncQ plasmid expressing the gst+ gene, Table 1) followed by a treatment with Proteinase K. Aliquots of the sample were analyzed by Western blotting with anti-GST mAb (D) and anti-PamA rabbit serum (E). Black and gray arrows show PamA and GST, respectively.
pamA′-phoA translational fusion N. meningitidis mutants were constructed as follows: a 1.5-kb PCR fragment containing the phoA gene was amplified from E. coli strain W3110 chromosomal DNA with the primers (phoA-21 and M13-RV’(15mer)-phoA-2) (S1 Table), and a 1-kb ermC gene fragment was amplified with universal primers (M13-RV and M13-47) from pHT24 [33]. The two PCR fragments were cloned into Fragment C by In-Fusion Cloning to construct pH1603 (S2 Table). PCR fragments amplified with nmb1345-5 and nmb1345-21 (S1 Table) from pHT1605 containing the pamA′-lacZ-ermC-downstream region of the pamA gene or from pHT1603 containing the pamA′-phoA-ermC-downstream region of the pamA gene were transformed to HT1125, and erythromycin-resistant clones were selected, as for the pamA′-lacZ::ermC or pamA′-phoA::ermC N. meningitidis mutants on its chromosome HT2215 and HT2217, respectively (Table 1 and Fig 2B).
Assessment of host cell-associated and internalized bacteria
The infection of HBMEC with N. meningitidis was performed as described previously [46]. Results are expressed as means ± the standard deviation (SD), and bacterial numbers were statistically compared using the two-tailed Student’s t-test.
Western blotting
SDS-PAGE and Western blotting were performed as described previously [48].
Observations of meningococci and ezrin accumulation by immunofluorescence staining
HBMEC monolayer preparations, N. meningitidis infections of these layers, and the preparation of immunostaining samples were performed as described previously [46]. Infected HBMEC and attached meningococci were observed using an ECLIPSE E600 microscope (Nikon) with a 100× oil immersion objective.
Construction of IncQ plasmids for N. meningitidis
The mutated M. mazei pylT (tRNAPyl) and mutated pylRS genes for mAzZLys (PylRS[Y306A/Y384F]) [49] and pBPa (PylRS A302T/N346T/C348T/W417C]) [50] (MmpBpaRS) were derived from pCDF-Pyl-Fx1 [49]. The four point mutations were generated by three-step mutations with the following three primer sets (A302-T and A302-7-R) (S1 Table), (N346T-C348T-F and N346-C348-R) (S1 Table), and (W417C-F and W417C-R) (S1 Table) using the Mutagenesis Basal Kit (Takara Bio). The plasmid constructed from pCDF-Pyl-Fx1 as described above was named pCDF-Pyl-Fx3 in this study (S3 Table). A 2-kb PCR fragment containing one copy of the pylRS gene (Y306A/Y384F) and two copies of the pylT genes expressed from the E. coli lpp promoter [49] (pylRS (Y306A/Y384F)/T) from pCDF-Pyl-Fx1 or a 2-kb PCR fragment containing one copy of the MmpBPaRS genes and two copies of the pylT genes expressed from the E. coli lpp promoter [49] (MmpBPaRS/T) from pCDF-Pyl-Fx3 were amplified by PCR with a primer set (ptac-12-pylRS-1 and M13-47-reverse-lppT-2) (S1 Table), and then cloned into pTTQ18, an expression vector with a strong tac promoter (Ptac) controlled by the LacI protein [51] using In-Fusion cloning to construct pHT1208 and pHT1261, respectively (S2 Table). The PCR fragment containing the lacIq-Ptac-pylRS[Y306A/Y384F]/T or lacIq-Ptac-MmpBPaRS/T genes was amplified by PCR with primers (M13-47 reverse and ptac-12) (S1 Table), and after phosphorylation, the fragment was cloned into the blunted SacI and PstI sites of pHT128, an IncQ plasmid [33], resulting in pHT1212 (lacIq-Ptac-pylRS[Y306A/Y384F]/T) and pHT1262 (lacIq-Ptac-MmpBPaRS/T), respectively (Table 1 and S4 Table).
PCR fragments containing the gst gene derived from pGEX-6P1 (GE Healthcare) with an amber codon at position Glu 51 (gst E51) or Phe 52 (gst F52) were constructed as follows: the gst E51 or gst F52 mutation was introduced by site-directed mutagenesis with two sets of primers (gst E51amb-1 and gst E51amb-2 for gst E51) (gst F52amb-1 and gst F52amb-2 for gst F52) (S1 Table) using the PrimeSTAR Mutagenesis Basal Kit. After the sequence was confirmed, a 0.5-kb PCR fragment containing Ptac-gstE51amb or Ptac-gstF52amb was amplified with a primer set (tetA-1-gst-up and gst-down-tetA-2) (S1 Table) and cloned into the SalI site of pHT1212 or pHT1216 to construct pHT1355, pHT1356, pHT1357, and pHT1358, respectively (Table 1).
To construct a series of amber mutations in the pamA gene, the His6 tag was initially introduced into the 3’ terminus of the native pamA gene as follows: a 7.2-kb PCR fragment was amplified from pHT922 with a primer set (NMB1345-1 and NMB1345-2-(His)6) (S1 Table), phosphorylated, and then self-ligated to construct pHT1213 (Table 1).
Amber mutation series (pamA K-amb) plasmids (see S1 Fig) were constructed by site-directed mutagenesis of pHT1213 (S2 Table) with primer sets (S1 Table). The pamA K-amb gene on pTWV228 was confirmed by sequencing and the plasmid is listed in S2 Table. A 2.2-kb PCR fragment containing the pamA K-amb gene was amplified with a primer set (tetA-NMB1345-up and tetA-NMB1345-down) (S1 Table) from the pTWV228-based plasmids listed in S2 Table, and cloned into the SalI site of pHT1216 or pHT1262 by In-Fusion cloning to construct IncQ plasmids containing the lacIq-Ptac-pylRS[Y306A/Y384F]/T and pamA K-amb genes (pHT1212) or lacIq-Ptac-MmpBPaRS/T and pamA K-amb genes (pHT1262) listed in Table 1.
The addition of StrepTag2-His6 synthetic DNA (S3 Fig) to the pamA K278amb gene was achieved as follows: a 6.5-kb PCR fragment amplified from pHT1242 with a primer set (spacer-1(15mer)-nmb1345-12 and His6′(15mer)-nmb1345-11) (S1 Table) was ligated to StrepTag2-His6 synthetic DNA by In-Fusion cloning to construct pHT1293 (S2 Table). A 2.4-kb PCR fragment containing the pamAK278amb-StrepTag2-His6 gene was amplified with a primer set (tetA-NMB1345-up and tetA-NMB1345-down) (S1 Table) from pHT1293 and cloned into the SalI site of pHT1262 by In-Fusion cloning, to construct pHT1388 (Table 1).
The introduction of IncQ plasmids into N. meningitidis strains was performed as described previously [33].
UV crosslinking of whole N. meningitidis cells expressing the PamA Lys-amber (pamA K-amb) mutants by pyrrolysine-based amber suppression with pBPa
The N. meningitidis H44/76 ΔpamA::spc mutant (HT1940) (Table 2) harboring IncQ plasmids containing lacIq -Ptac-MmpBPaRS/T genes and pamA K-amber genes (Table 1) was cultivated on GC agar plates containing 1 mM IPTG and 1 mM pBPa at 37°C in 5% CO2 for 18 hours. To identify the optimal pamA K-amb mutants for pyrrolyl-based amber suppression followed by photocrosslinking, N. meningitidis transformants grown one-fourth of a GC plate were suspended in 50 μl PBS, transferred into the wells of a 96-well plate/strain, and irradiated with 365 nm UV using a FL-365-SD UV lamp stand (Opticode, Japan) at a distance of 2 cm on ice for 2 hours. After harvesting, UV-irradiated bacteria were resuspended in 400 μl of urea buffer (50 mM Tris-HCl pH7.5, 8 M urea, 1% Triton X-100, and 100 mM NaCl) and then transferred into a 1.5-mL tube. Bacteria were disrupted by sonication, and the soluble fraction was obtained by centrifugation. PamA K-amb-His6 proteins with expression suppressed by pBPa were purified with 200 μL TALON resin (Clontech). The purified fraction (approx. 250 μl) was precipitated by TCA/acetone at -80 oC overnight, and the precipitate was finally dried and suspended in 25 μl 1 × SDS buffer. Aliquots were analyzed by SDS-PAGE.
Identification of endogenous proteins crosslinked with PamA K278(pBPa) in N. meningitidis
To identify endogenous protein(s) crosslinked to PamA K278(pBPa) in N. meningitidis, an synthetic DNA fragment containing twin Strep-Tag (StrepTag2) [52] was further introduced into pHT1274, resulting in pHT1388 (S3 Fig and Table 2). N. meningitidis HT1940 harboring pHT1388 was grown on approximately 200 GC agar plates containing 1 mM IPTG and 1 mM pBPa at 37°C in 5% CO2 for 18 hours. The bacteria from 3 GC agar plates were suspended in 0.5 mL PBS, transferred into the wells of a 12-well plate (more than 50 wells in total), and irradiated with 365 nm UV on ice for 2 hours. The irradiated bacteria from 2 wells were transferred into a 15-mL tube and harvested by centrifugation. The collected bacteria were suspended in 5 mL IP Lysis buffer (25 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol) and disrupted by sonication followed by centrifugation at 12,000 rpm at 4°C for 30 min. The supernatant (soluble fraction) was collected into one tube. Crosslinked proteins were purified by two column chromatography steps using Ni-Sepharose High Performance (GE Healthcare) and Strep-Tactin Sepharose (Iba, Germany). The purified fraction was concentrated approximately 70-fold using a VIVASPIN TURBO 15 (MW 50k) Centrifuge Filter unit (Sartorius), to a final volume of approximately 100 μL. An aliquot of the sample was analyzed by 4–12% NuPAGE (Invitrogen), and immunoblotting with an anti-His6 monoclonal antibody (mAb) (Wako, Japan). All proteins were also visualized using the CBB staining kit (APRO SCIENCE, Japan) and bands that were also detected by immunoblotting were excised. These bands were subjected to trypsin digestion and peptide extraction. Extracted peptides were dried and resuspended in H2O and 0.1% (w/w) trifluoroacetic acid for MS. The LC-MS/MS analysis was performed with an Advance nanoLC (Michrom Biosources) coupled to an LTS Mass Spectrometer (Thermo Fisher Scientific). Proteins were identified by running MASCOT (http://www.matrixscience.com) against the NCBI database (NCBInr 20160202). Candidates with molecular masses that were consistent with the difference between the crosslinked complexes and the PamA protein were selected. The accuracy of each interaction between PamA K278(pBPa) and the candidate was further examined by constructing N. meningitidis HT1940 harboring pHT1388 mutants, in which the FLAG tag was added to the candidate gene on its chromosome, as in the pilX-FLAG N. meningitidis strain (Table 1) described above.
Identification of the counterpart of the amino acid residue of PilE crosslinked with PamA K278(pBPa)
A DNA fragment containing ΔN-pamA K278amb was amplified from pHT1242 with a primer set (NMB1345(pGEX)-1(BH) and NMB1345(pGEX)-2(XhoI)) (S1 Table) and cloned into the same sites of pMAL-c2 (NEB) to construct pHT1473 (S3 Table). A 0.5-kb PCR fragment containing the Gln6-ΔN-PilE gene, in which the first 9 amino acids (PilE leader peptides in N. meningitidis) were replaced with 6 glutamine residues (Gln6) (see the Results section), was amplified with a primer set ((Gln)6-pilE-45 and pCDF-1b(PacI)(15mer)-pilE-44) (S1 Table) from H44/76 N. meningitidis chromosomal DNA, and designated as fragment B. To clone the PCR fragment into pCDF-1b-kan (S3 Table) [53], a 3.6-kb DNA fragment was amplified from pCDF-1b-kan with a primer set (pCDF-1b (downstream of PacI) and (Gln)6(15mer)-pCDF-1b-2) (S1 Table) and ligated to PCR fragment B by In-Fusion Cloning to construct pHT1447 (S3 Table), in which the ΔN-(Gln)6-PilE protein was expressed from the T7 promoter (PT7). A 0.6-kb PCR fragment containing the PT7- ΔN-(Gln)6-pilE gene was amplified from pHT1447 with a primer set (pSTV28(SmaI-5′)-pCDF-1b-1 and pSTV28(SmaI-3′)-pCDF-1b-2) (S1 Table) and cloned into the SmaI site of pSTV28 (Takara Bio) by In-Fusion Cloning to construct pHT1474 (S3 Table). E. coli BL21(DE3) cells harboring pCDF-Pyl-Fx3, pHT1473, and pHT1474 were cultivated in 100 mL MagicMedia containing 1 mM pBPa at 30°C overnight. The harvested bacteria were suspended in 1.25 mL PBS and divided into 5 aliquots (0.25 mL/tube), transferred into the wells of a 24-well plate/aliquot, and irradiated with 365 nm UV light on ice for 2 hours. UV-irradiated bacteria were collected again in one tube and disrupted by suspension in 1 mL B-PER (Thermo) containing protease inhibitor cocktail (Nacalai Tesque, Japan), followed by centrifugation at 12,000 rpm at 4°C for 30 min. The MBP-ΔN-PamA K278(pBPa) protein crosslinked to Gln6-ΔN-PilE [as well as uncrosslinked MBP-ΔN-PamA K278(pBPa)] was purified with amylose resin (NEB). The purified fraction was concentrated approximately 70-fold using a VIVASPIN TURBO 15 (MW 50k) Centrifuge Filter unit (Sartorius) to a final volume of approximately 250 μL. An aliquot of the sample was analyzed by 4–12% NuPAGE (Invitrogen), and the bands corresponding to the complex containing MBP-ΔN-PamA K278(pBPa) crosslinked to Gln6-ΔN-PilE were excised. In addition, an uncrosslinked band corresponding to MBP-ΔN-PamA K278(pBPa) was isolated for a comparative analysis. The band corresponding to the Gln6-ΔN-PilE protein was also excised. The amino acid residue of Gln6-ΔN-PilE crosslinked to ΔN-PamA K278(pBPa) was identified by comparing peptides from the three bands as described previously [54].
Transmission Electron Microscopy (TEM)
Negatively stained samples were prepared for TEM as described previously [55]. Electron microscopy was performed with a Hitachi H-7600 Transmission Electron Microscope.
Measurement of the ratio of pilus components to PilE in purified pili
Meningococcal pili were prepared as described previously [36]. The relative amounts of PilF-FLAG, PilP-FLAG, PilM-FLAG, PilV, and PilX-FLAG to PilE in purified pili were examined by Western blotting with anti-FLAG mAb (Wako, Japan), anti-PilV [36], or anti-PilE rabbit serum [33] with appropriate dilutions using a quantification program in Fusion Capt 17 in Fusion Solo 7S (M&S Instruments). Results are expressed as means ± SD. The adhered and internalized bacterial numbers and the relative amounts of pilus components to PilE in purified pili were compared using the two-tailed Student’s t-test, and P values of <0.05 were considered significant.
Results
Genetic evidence that a hypothetical protein annotated as NMB1345 is important for N. meningitidis internalization into HBMEC
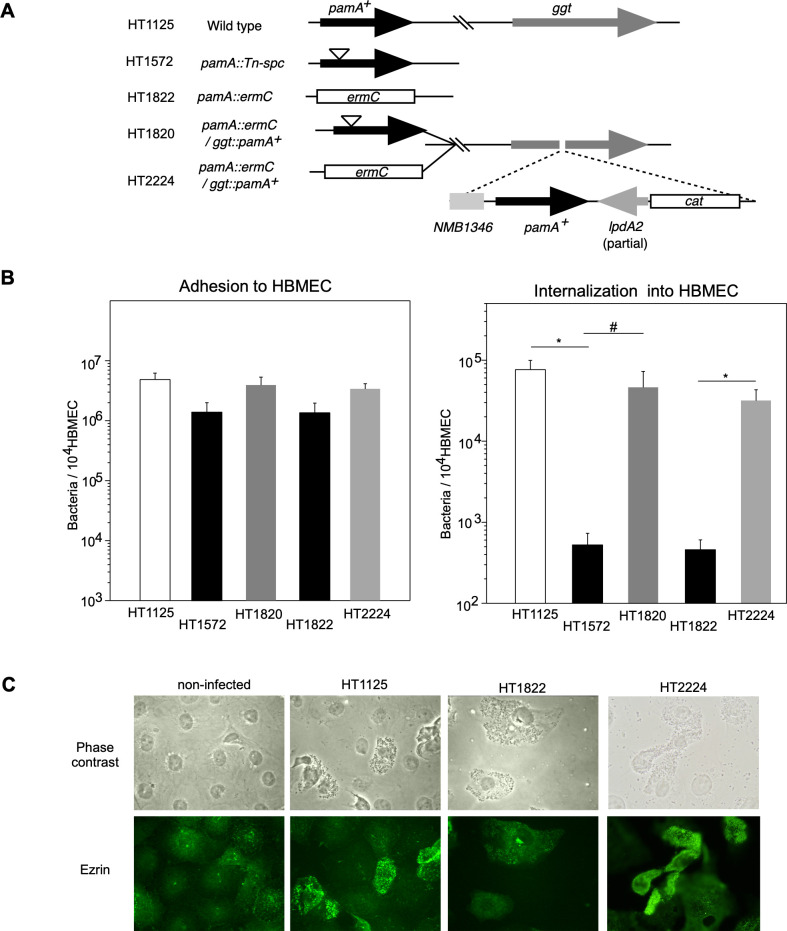
During the course of our research on the infectious abilities of the ST-2032 N. meningitidis strain mutagenized by STM to HBMEC, we found that a transposon mutant, in which the gene annotated as NMB1345 (renamed as PamA in this study) in the N. meningitidis MC58 genome [39] was disrupted and resulted in defective infectious ability in HBMEC (Fig 1A). To confirm the relationship between the mutation and infectious defect in the mutant, we constructed a null mutant (HT1822 ΔpamA::spc) and an ectopically complemented strain with the wild-type pamA+ gene at the ggt locus (HT2224) and examined their infectious activities in HBMEC (Fig 1B). While the insertional and null mutants adhered to HBMEC less efficiently than the wild-type strain HT1125, the number of internalized bacteria largely decreased to approximately 1/100 of HT1125 and the defect in internalized bacterial numbers was recovered in HT2224. These results indicate that the mutation in the pamA gene affects meningococcal infection in HBMEC, particularly internalization.
Fig 1. Characterization of ΔpamA N. meningitidis mutant.
(A) Schematic representation of the wild-type, insertion, and deletion mutants in the pamA gene and ectopic complementation of the pamA+ gene at the ggt locus in N. meningitidis strains. (B) Effect of ΔpamA mutation on N. meningitidis infection of HBMEC. The adherence (left) and internalization (right) of pamA N. meningitidis mutants to HBMEC, and the effects of complementation of the pamA+ gene in the pamA deletion mutant on bacterial infection. Each value is the mean ± standard error of the mean (CFU per 104 HBMEC) from at least four experiments. Open, filled, light gray, and dark gray bars indicate the bacterial number of N. meningitidis wild-type pamA+ (HT1125), pamA::Tn-spc (HT1572), and ΔpamA::spc (HT1822), and pamA- mutants in which the pamA+ gene was ectopically complemented (HT1736 and HT2224), respectively (see S1 Table). *P<0.01, #P<0.05, significantly different from the pamA+ strain or pamA- mutants complemented with the pamA+ gene. (C) Immunofluorescence microscopy showing the accumulation of ezrin beneath N. meningitidis-infected HBMEC. The HBMEC monolayer was infected with wild-type pamA+ (middle-left), ΔpamA (middle-right), and ΔpamA/pamA+ (right) N. meningitidis strains. A non-infected HBMEC monolayer is also shown in the left panels. Bacteria and HBMEC were observed by phase-contrast microscopy (upper panels). Ezrin was immunostained with an anti-ezrin mAb and Alexa Fluor 488-conjugated rabbit anti-mouse IgG (green channel, lower panels).
We also investigated host cell cytoskeleton rearrangements caused by meningococcal infection by monitoring the localization of ezrin, because the accumulation of ezrin beneath meningococci on HBMEC is required for bacterial internalization into host cells [36, 37] (Fig 1C). While ezrin was widely distributed throughout non-infected cells, it was condensed at the site of bacterial attachment in HBMEC infected with wild-type N. meningitidis strain HT1125. In contrast, ezrin condensation was not observed in cells infected with the ΔpamA N. meningitidis mutant HT1822, but was detected in HT2224 (ΔpamA/pamA+). These results are consistent with previous findings, and strongly suggest that PamA participates in the meningococcal-elicited bacteria-induced reorganization of the host cell cytoskeleton upon N. meningitidis infection.
Characterization of the biochemical properties of PamA in N. meningitidis
The deduced amino acid sequence of the PamA protein consists of 516 residues (S1 Fig) with a predicted molecular mass of 57 kDa. The hydrophobicity analysis indicated that the PamA protein had a one membrane-spanning region at its N terminus (S1 Fig), suggesting that the PamA protein is localized at the membrane in N. meningitidis. To clarify the localization in N. meningitidis, cellular proteins were biochemically fractionated using a differential detergent solubilization method [45] and analyzed by Western blotting (Fig 2A). LptA, a meningococcal inner membrane (IM) protein [55], was largely detected in the 4% Triton X-100 soluble (IM-enriched) fraction, supporting the proper fractionation in this experiment. Under the same conditions, the PamA protein was mainly enriched in the IM-rich fraction. This result suggests that PamA is located at the IM in N. meningitidis.
We also investigated the membrane topology of the PamA molecule in N. meningitidis by constructing N. meningitidis mutants, in which the first N-terminal 136 amino acid residues of PamA including the membrane-spanning region, termed pamA′, were fused to the lacZ or phoA gene [56] on the chromosome (Fig 2B)(see Materials and Methods). While the wild-type HT1125 and pamA′-lacZ N. meningitidis HT2215 strains (Table 2) did not exhibit β-galactosidase activity, the pamA′-phoA N. meningitidis strain HT2217 (Table 2) showed alkaline phosphatase activity (Fig 2C). Since the alkaline phosphatase activity of PhoA was detected only in the periplasmic space [57], these genetic results suggest that the hydrophobic region of the PamA protein faces the periplasmic side. We also biochemically examined the membrane topology, using a Proteinase K digestion experiment with the spheroplasted N. meningitidis strain expressing glutathione S-transferase (GST) from an IncQ plasmid (Fig 2D and 2E). Under the condition where the cellular protein GST was mostly protected from Proteinase K digestion (Fig 2D), PamA was largely digested (Fig 2E). Collectively, these results suggest that PamA is localized at the IM and faces the periplasmic space in N. meningitidis.
Incorporation of ncAAs into proteins in living N. meningitidis
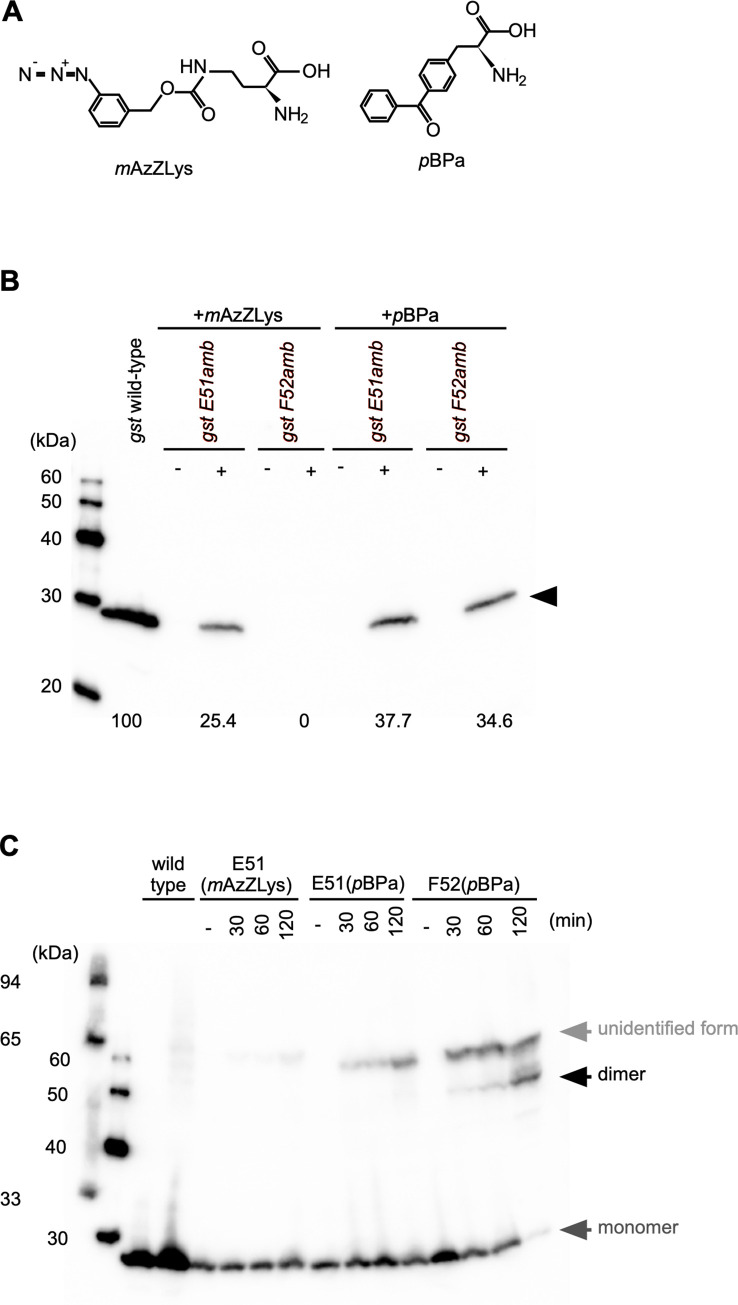
To attempt the application of genetic code expansion to N. meningitidis for the incorporation of ncAAs with a photocrosslinker into the PamA protein, we used two mutated MmPylRS/T genes to genetically encode ncAAs in N. meningitidis. A previous study reported that a modified MmPylRS gene for E. coli, MmPylRS[Y306A/Y384F] [49] is applicable to Nε-(m-azidobenzyloxycarbonyl)-L-lysine (mAzZLys) (Fig 3A, left), which is a photocrosslinker excited by ultraviolet (UV) irradiation [58], and MmPylRS([A302T/N346T/C348T/W417C]) [50] (MmpBPaRS) is optimized for p-benzoyl-L-phenylalanine (pBPa) (Fig 3A, right), which is also one of the widely used photocrosslinkers [44].
Fig 3. Incorporation of ncAAs into N. meningitidis proteins monitored by Western blotting.
(A) Chemical structures of ncAAs, mAzZLys (Left) or pBPa (right) used in this study. (B) Estimation of the efficiency of pyrrolysine-based amber suppression with mAzZLys or pBPa for GST E51amb or GST F52amb in N. meningitidis. Bacterial extracts equivalent to an OD600 of 0.1 were analyzed by Western blotting with an anti-GST mAb. The horizontal black arrow shows the full-length GST protein. Numbers indicate relative suppression efficiency when wild-type GST expression is defined as 100%. (C) Detection of irradiation- and time-dependent GST dimerization of GST F52 (pBPa) in N. meningitidis. Bacterial extracts equivalent to an OD600 of 0.05, irradiated with UV light for the indicated times, were analyzed by Western blotting with an anti-GST mAb. Dark gray and black arrows show the monomer and dimer forms of the GST protein, respectively. The light gray arrow shows an unspecified form of GST generated by UV irradiation.
We examined the incorporation of mAzZLys and pBPa in N. meningitidis by biochemically monitoring the suppression of the amber codon at position Glu51 or Phe52 of GST (GST E51amb or GST F52amb) by Western blotting (Fig 3B). The expression of both the gst E51amb and gst F52amb mutants was suppressed by pBPa, while the gst F52amb mutant was not suppressed by mAzZLys. The suppression efficiency could be considered as approximately 25% of the wild-type GST protein with pBPa in the gst E51amb mutant, and 37% and 35% with mAzZLys and pBPa in the gst F52amb mutant, respectively. Since the incorporation efficiency of ncAAs is approximately 35–50% in E. coli [59], this result suggested that genetic code expansion by the two modified MmPylRS/T genes was efficiently conducted in N. meningitidis (Fig 3B). These results obtained by biochemical and genetic pyrrolysine-based amber suppression suggest that mAzZLys and pBPa are incorporated into the GST protein in N. meningitidis. The proper incorporation of pBPa into a protein in N. meningitidis, as determined by an MS analysis, was examined with PamA K278 (pBPa)(see a later section, S5 Fig) since the amounts of GST proteins in N. meningitidis were too low to purify.
We further examined the UV crosslinking by using whole bacterial cells (Fig 3C). Longer UV irradiation increased the amount of the dimer form of GST, and the majority of GST F52(pBPa) was fixed as the dimer form for 2 hours because GST is a dimeric enzyme [44, 60, 61]. On the other hand, the dimerization of the GST E51amb(mAzZLys) and GST E51(pBPa) proteins was not observed by photocrosslinking, while GST E51(mTmdZLys) dimerized in E. coli [62], indicating that the processes involved in UV crosslinking with ncAAs do not always function well in N. meningitidis. Collectively, these results demonstrated that physiological transient protein-protein interactions can be detected by crosslinking via the incorporation of photoreactive ncAAs in N. meningitidis and showed that the incorporation of ncAAs by pyrrolysine-based amber suppression could be performed in N. meningitidis.
Incorporation of a genetically encoded photocrosslinking amino acid into the PamA protein to identify the accompanying protein in N. meningitidis
Since it was not possible to predict the three-dimensional structure of PamA by Phyre2 [63], due to the lack of homology to any other protein identified to date, we were unable to speculate which amino acid residues are exposed on the outside of the PamA molecule in N. meningitidis. Therefore, the identities of suitable amino acid residues for the incorporation of photoreactive ncAAs into the PamA protein remained unclear. Thus, in the present study, we focused on the lysine (Lys or K) residue for amber (UAG) substitution because lysine is primarily located on protein surfaces [64, 65]. Forty Lys residues are present in the PamA protein (S1 Fig), and we constructed three pamA amber mutants (designated as pamA K-amb) at K148 (pamA K148amb), K273 (pamA K273amb), and K388 (pamA K388amb) to estimate the proper position(s) in the PamA protein for ncAAs incorporation. N. meningitidis strain H44/76 ΔpamA (HT1940) expressing the MmPylRS[Y306A/Y384F]/T genes and one of the three pamA K-amb genes on an IncQ plasmid was cultivated in the presence of mAzZLys, and whole bacterial extracts were analyzed by Western blotting (S2A Fig). The full length and a sufficient amount of PamA K148amb by incorporation of mAzZLys [(PamA K148(mAzZLys)], PamA K273(mAzZLys), and PamA K388(mAzZLys) were only observed in the presence of mAzZLys, suggesting that mAzZLys is incorporated into the PamA protein by pyrrolysine-based amber suppression in N. meningitidis. We also examined UV-irradiated whole bacterial cells by Western blotting, faint bands appeared to correspond to proteins crosslinked to PamA (mAzZLys), were detected in the PamA K273(mAzZLys) and PamA K388(mAzZLys) samples (S2B Fig). The same result was also obtained for N. meningitidis strain HT1940 expressing the MmpBPaRS/T genes and one of the three pamA K-amb genes on an IncQ plasmid, cultivated in the presence of pBPa (S3 Fig). These results indicate that the Lys residues located from position Lys148 to the carboxy-terminal end (S1 Fig) were localized on the surface of the PamA molecule in N. meningitidis. Moreover, the amber suppression appeared to be more efficiently for PamA K273(pBPa) and PamA K388(pBPa) with pBPa than with mAzZLys (Fig 4 and S3 Fig). Thus, the photocrosslinking experiments by pyrrolysine-based amber suppression described hereafter were conducted with the pBPa and MmpBPaRS/T genes.
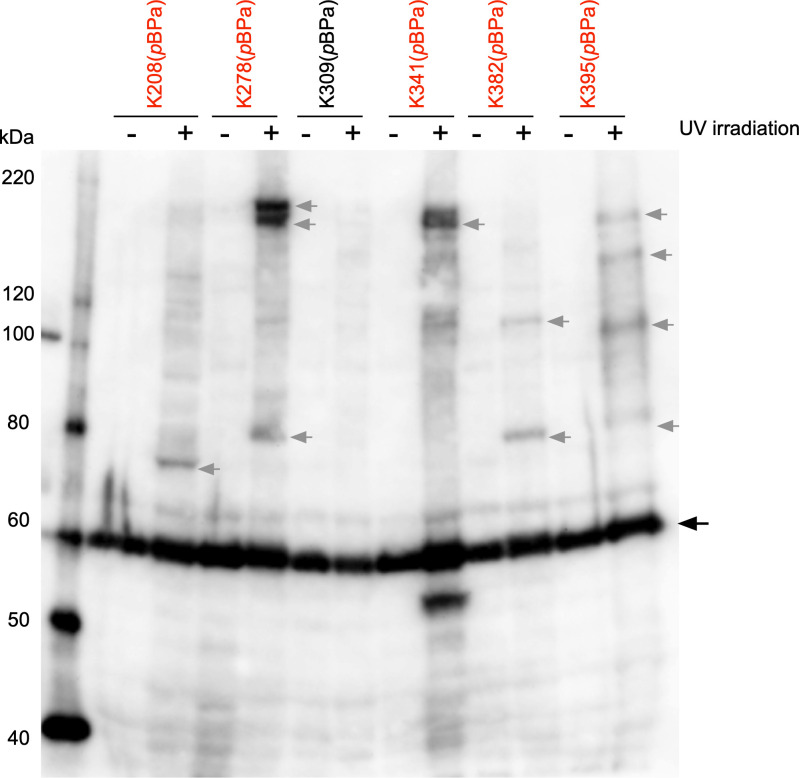
Fig 4. PamA K208(pBPa), PamA K278(pBPa), PamA K314(pBPa), PamA K382(pBPa) and PamA K395(pBPa) proteins crosslinked to unidentified endogenous proteins in N. meningitidis.
UV crosslinking of Pam K208amb, K278amb, K309amb, K341amb, K395amb, and K382amb mutants expressed by pyrrolysine-based amber suppression with pBPa in ΔpamA N. meningitidis. Bacteria that grew on one-fourth of a GC agar plate in the presence of 1 mM pBPa were crosslinked by UV light irradiation for 2 hours and then the PamA K(pBPa)-His6 protein was purified. An aliquot was analyzed by anti-His6 mAb. The black arrow shows the full-length PamA K-amb protein expressed by pyrrolysine-based amber suppression with pBPa, and the gray arrow indicates protein complexes crosslinked between PamA K(pBPa) and unidentified endogenous proteins in N. meningitidis.
To confirm the proper position(s) for the incorporation of pBPa into the PamA protein, we constructed 26 pamA K-amber mutants, in which each Lys residue located from position Lys148 to the carboxy-terminal end in the PamA protein was replaced with the amber codon (S1 Fig, Table 1 and S4 Table). UV-irradiated N. meningitidis strain HT1940 cells expressing the MmpBPaRS/T and pamA K-amber-His6 genes were analyzed by Western blotting (S3 Fig). Some PamA(pBPa) mutants seemed to be crosslinked to some endogenous protein in N. meningitidis (shown in red in S3 Fig), and the more detailed analysis revealed that the following six pamA K-amber mutants with expression suppressed by pBPa [K208(pBPa), K278(pBPa), K309(pBPa), K341(pBPa), K382(pBPa), and K395(pBPa)] appeared to be crosslinked to some endogenous proteins in N. meningitidis (shown in red in Fig 4) while K309(pBPa) was eliminated. In the present study, we focused only on the proteins crosslinked to PamA K278(pBPa), because the three bands with the highest intensities were found in the PamA K278(pBPa) sample.
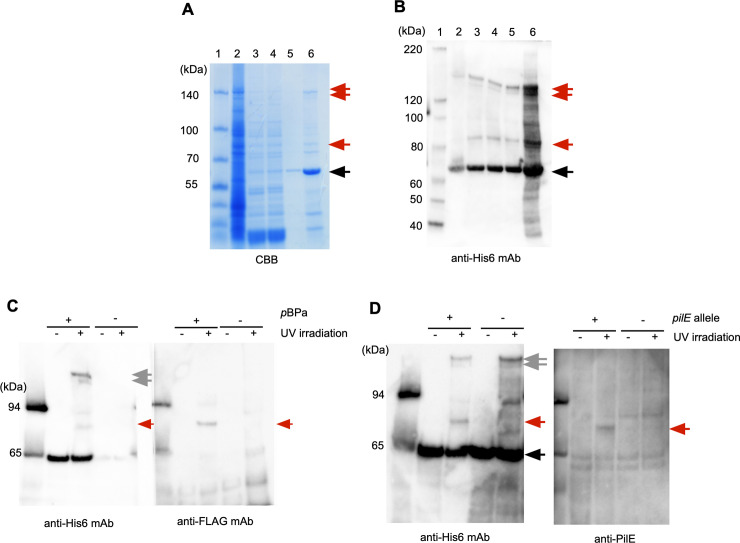
To characterize the 150-, 140-, and 85-kDa protein complexes that contained an endogenous protein crosslinked to PamA K278(pBPa), we added a Twin-Strep tag (Strep2) [66] and His6 tag at the carboxy-terminal end (S4 Fig) and purified the three corresponding protein complexes from N. meningitidis strain HT1940 harboring pHT1388 grown on approximately 200 plates as described in the Materials and Methods. During the purification process, we also obtained the purified PamA K278 (pBPa) protein expressed in N. meningitidis. The incorporation of pBPa at K278amb of PamA-Strep2 -His6, was confirmed by MS analyses (S5 Fig). This result demonstrates that an ncAA was selectively incorporated into a protein in N. meningitidis.
The three corresponding protein complexes were subjected to a gel-based proteomic analysis using MS (Fig 5A and 5B). A compilation of the results obtained using MASCOT (MATRIX SCIENCE, Japan) suggested several candidates, and we selected those with higher scores and numbers of matched peptides for more reliable results in the present study (Table 3). Since the molecular mass of PamA-Strep2-His6 is approximately 62 kDa due to the addition of Strep2-His6 at the carboxy-terminal end (S4 Fig), the bands corresponding to 150- and 140-kDa were presumed to contain counterparts with a molecular mass of approximately 100 kDa. PamA is listed as the top score of the MASTCOT analysis (Table 3) for the 150 and 140 kDa bands, providing support for the presence of this protein in the two bands. On the other hand, we did not identify any candidate with a deduced molecular mass from 80 to 90 kDa by MASCOT (Table 3). To examine the possibility that the observed molecular mass on SDS-PAGE was different from the calculated molecular mass in the database, we constructed N. meningitidis mutants that expressed the FLAG-tagged proteins of 5 candidates with estimated molecular masses of 75–160 kDa for 150 kDa, and those of 9 candidates with estimated molecular masses of 48–74 kDa (shown in bold in Table 3). Crosslinked samples from the N. meningitidis mutants expressing the FLAG-tagged protein, MmpBPaRS/T and PamA K278(pBPa)-Strep2-His6, were examined by Western blotting; however, none of these candidates were crosslinked to PamA K278(pBPa) in N. meningitidis. Considering these results, the two proteins presented by the 150- and 140-kDa bands with the highest intensities on Western blotting apparently correspond to the homo-trimer of the PamA protein (see Discussion and S12 Fig).
Fig 5. Purification and identification of an endogenous protein crosslinked to PamA K278(pBPa)-Strep2-His6.
Proteins in each purification step were analyzed by 4–12% SDS-PAGE and staining with the CBB staining kit (A) and Western blotting with anti-His6 mAb (B). Lane 1, molecular mass standards; lane 2, N. meningitidis crude soluble extract; lane 3, after the Ni-Sepharose column; lane 4, after dialysis with IP Lysis buffer; lane 5, after the Strep-Tactin Sepharose column; lane 6, after concentration with VIVASPIN TURBO 15. Black and red arrows indicate K278(pBPa)-Strep2-His6 and the unidentified endogenous proteins crosslinked to PamA K278(pBPa)-Strep2-His6, respectively. (C) The PilE-FLAG protein was crosslinked to PamA K278(pBPa) in N. meningitidis. The black arrow shows the full-length PamA K278amb protein expressed by pyrrolysine-based amber suppression with pBPa, and the gray arrow indicates the homotrimer (See text) in N. meningitidis. The red arrow shows the complex of PilE-FLAG crosslinked to PamA K278(pBPa)-Strep2-His6 in N. meningitidis. +/- indicates the presence or absence of pBPa, and irradiation or no irradiation with UV light, respectively. (D) The crosslinked band was reacted with anti-PilE rabbit serum. Black and gray arrows show PamA K278(pBPa)-Strep2-His6 and the putative homotrimer complex of PamA K278(pBPa), respectively, in N. meningitidis. The red arrow shows the complex of PilE crosslinked with PamA K278(pBPa)-Strep2-His6 in N. meningitidis. +/- indicates the presence or absence of the pilE gene, and irradiation or no irradiation with UV light, respectively.
Table 3. Summary of the protein candidates crosslinked to PamA in N. meningitidis identified by mass spectrometry.
| 1. Protein identified from the photocrosslinking products for a molecular weight around 150kDa | |||||||
| Accession number | Protein name | N. meningitidis strain | Estimated Molecular weight | Number of matched peptides | MASCOT score | Sequence coverage | |
| gi|325136153 | hypothetical protein NMBM0579_0886 | NMBM0579_0886 | 57kDa | 102 | 1843 | 25% | |
| gi|325144519 | hypothetical protein | NMBM01240013_0931 | 57kDa | 99 | 1758 | 24% | |
| gi|325140435 | hypothetical protein | CU385 | 57kDa | 93 | 1718 | 24% | |
| gi|488174415 | hypothetical protein | 57kDa | 93 | 1920 | 22% | ||
| gi|485353984 | DNA-directed RNA polymerase, β subunit | 73696 | 150kDa | 24 | 1436 | 22% | |
| gi|488170470 | hypothetical protein | 57kDa | 71 | 1394 | 20% | ||
| gi|6977941 | App protein | 160kDa | 7 | 425 | 7% | ||
| gi|915797040 | adhesin | 160kDa | 6 | 390 | 6% | ||
| gi|83270238 | AusI | 158kDa | 4 | 288 | 4% | ||
| gi|389605917 | NAD(P) transhydrogenase subunit alpha | α522 | 54kDa | 2 | 162 | 2% | |
| gi|488141712 | carbon starvation protein A | 75kDa | 3 | 146 | 3% | ||
| gi|896381024 | hypothetical protein | 57kDa | 8 | 119 | 2% | ||
| gi|488158877 | hypothetical protein | 147kDa | 2 | 109 | 2% | ||
| gi|254672742 | alanine or glycine: cation symporter, AGCS family | α275 | 45kDa | 2 | 109 | 1% | |
| 2. Protein identified from the photocrosslinking products for a molecular weight around 140kDa. | |||||||
| Accession number | Protein name | N. meningitidis strain | Estimated Molecular weight | Number of matched peptides | MASCOT score | Sequence coverage | |
| gi|325136153 | hypothetical protein NMBM0579_0886 | NMBM0579_0886 | 57kDa | 80 | 1852 | 25% | |
| gi|120866758 | hypothetical protein NMC1281 | FAM18 | 56kDa | 69 | 1863 | 23% | |
| gi|501178984 | DNA-directed RNA polymerase β subunit | 154kDa | 28 | 1808 | 27% | ||
| gi|325140435 | hypothetical protein NMBCU385_0826 | CU385 | 57kDa | 72 | 1803 | 22% | |
| gi|325144519 | hypothetical protein | NMBM01240013_0931 | 57kDa | 78 | 1782 | 24% | |
| gi|488171514 | hypothetical protein | 57kDa | 70 | 1656 | 23% | ||
| gi|488170470 | hypothetical protein | 57kDa | 54 | 1267 | 18% | ||
| gi|488186495 | hypothetical protein | 147kDa | 12 | 800 | 12% | ||
| gi|488186095 | immunoglobulin A1 protease | 121kDa | 9 | 653 | 9% | ||
| gi|488151215 | peptidylprolyl isomerase | 56kDa | 8 | 500 | 8% | ||
| gi|316985319 | filamentous hemagglutinin family N-terminal domain protein | H44/76 | 261kDa | 8 | 496 | 8% | |
| gi|488154218 | heme biosynthesis operon protein HemX | 48kDa | 7 | 490 | 7% | ||
| gi|488151215 | peptidylprolyl isomerase | 56kDa | 8 | 465 | 8% | ||
| gi|488146114 | D-lactate dehydrogenase | 64kDa | 5 | 363 | 5% | ||
| gi|389605276 | UPF0141 inner membrane protein yhjW | 62kDa | 6 | 351 | 5% | ||
| gi|488170091 | peptidase | 200kDa | 3 | 202 | 3% | ||
| gi|120866107 | putative protein-export membrane protein | FAM18 | 66kDa | 3 | 176 | 3% | |
| gi|732853 | IgA1 protease | 54kDa | 3 | 168 | 3% | ||
| gi|488141712 | carbon starvation protein A | 74kDa | 3 | 165 | 3% | ||
| gi|316985875 | 4Fe-4S binding domain protein | 146kDa | 3 | 150 | 3% | ||
| gi|389605917 | NAD(P) transhydrogenase subunit α | 54kDa | 2 | 140 | 2% | ||
| gi|488141355 | arginine decarboxylase | 71kDa | 2 | 129 | 2% | ||
| gi|349520 | pilus structural subunit, partial | 17kDa | 2 | 128 | 2% | ||
| gi|488143525 | protein translocase component YidC | 60kDa | 2 | 125 | 2% | ||
| gi|254670993 | 1-deoxy-D-xylulose 5-phosphate synthase | 69kDa | 2 | 114 | 2% | ||
| gi|304337210 | methionine-R-sulfoxide reductase | 59kDa | 2 | 112 | 2% | ||
| gi|4838369 | NatD | 52kDa | 2 | 109 | 2% | ||
| 3. Protein identified from the photocrosslinking products for a molecular weight around 85kDa. | |||||||
| Accession number | Protein name | N. meningitidis strain | Estimated Molecular weight | Number of matched peptides | MASCOT score | Sequence coverage | |
| gi|2460281 | outer membrane protein Omp85 | 88kDa | 63 | 2799 | 41% | ||
| gi|488170971 | membrane protein | 88kDa | 58 | 2599 | 38% | ||
| gi|325136153 | hypothetical protein NMBM0579_0886 | 57kDa | 63 | 1774 | 24% | ||
| gi|325144519 | hypothetical protein NMBM01240013_0931 | 57kDa | 60 | 1692 | 23% | ||
| gi|488171514 | hypothetical protein | 57kDa | 56 | 1619 | 22% | ||
| gi|488174415 | hypothetical protein | 57kDa | 56 | 1577 | 22% | ||
| gi|325141117 | LPS-assembly protein LptD | CU385 | 87kDa | 21 | 1440 | 19% | |
| gi|488147360 | elongation factor G | 77kDa | 27 | 1398 | 20% | ||
| gi|325141117 | putative organic solvent tolerance protein | CU385 | 87kDa | 20 | 1149 | 18% | |
| gi|409107079 | type IV pilus biogenesis and competence protein PilQ | 80kDa | 17 | 973 | 15% | ||
| gi|209363328 | hemoglobin receptor, partial | 87kDa | 16 | 909 | 14% | ||
| gi|488154195 | ATP-dependent Clp protease ATP-binding subunit ClpA | 85kDa | 21 | 905 | 16% | ||
| gi|30017077 | TonB-dependent siderophore receptor FetA | 73kDa | 14 | 793 | 13% | ||
| gi|120866913 | RNA polymerase sigma factor | 75kDa | 11 | 616 | 10% | ||
| gi|325142240 | 2-oxoglutarate dehydrogenase E1 component | 102kDa | 10 | 496 | 8% | ||
| gi|488150134 | GNAT family N-acetyltransferase | 89kDa | 7 | 421 | 7% | ||
| gi|496712676 | fimbrial protein | 18kDa | 9 | 307 | 4% | ||
| gi|645213765 | peptidase | 88kDa | 5 | 295 | 5% | ||
| gi|402319479 | hypothetical protein NMEN93004_1215 | 18kDa | 6 | 294 | 5% | ||
| gi|488144915 | phosphoenolpyruvate synthase | 87kDa | 5 | 293 | 5% | ||
| gi|254673965 | putative efflux system transmembrane protein | 113kDa | 5 | 249 | 4% | ||
| gi|496706569 | transferrin-binding protein 2 | 74kDa | 5 | 239 | 4% | ||
| gi|896272027 | pilus assembly protein PilQ | 75kDa | 5 | 222 | 4% | ||
| gi|45245 | periplasmic iron-binding protein | 34kDa | 4 | 204 | 4% | ||
| gi|121052755 | GTP pyrophosphokinase | 86kDa | 3 | 199 | 3% | ||
Proteins shown in italics corresponds to the PamA protein itself.
Proteins shown in bold indicates the candidates examined for crosslinking to PamA K278(pBPa) in N. meningitidis by constructing the FLAG-tagged candidate mutants, but the crosslinking could not be detected.
The protein shown underlined is PilE crosslinked to PamA K278(pBPa) in N. meningitidis, which was confirmed in this study.
We then characterized the 85-kDa band, which could contain an approximately 20 kDa endogenous protein crosslinked to PamA K278(pBPa). LC-MS/MS followed by MASCOT analyses revealed two possible candidates, a fimbrial protein (17 kDa) and the hypothetical protein NMEN93004_1215 (18 kDa) (Table 3). The BLAST homology search indicated that while we could not find any protein corresponding to NMEN93004_1215 in the databases, the fimbrial protein (accession number gi|496712676) was 95% identical to the protein NMBH4476_0018 annotated as “type IV pilus assembly protein PilA” in N. meningitidis strain H44/76 with the highest E-value (Table 3). Further computational analyses revealed that the “PilA protein” annotated in GenBank was 96% identical to PilE, a major component of meningococcal pili (reviewed in [17]). To confirm that the 85-kDa complex was composed of PilE and K278(pBPa)-Strep2-His6, we constructed the pilE-FLAG N. meningitidis strain HT2095 and examined the crosslinked samples by Western blotting with an anti-FLAG mAb (Fig 5C). The analysis revealed that PilE-FLAG was present in the 85-kDa band, with dependence on pBPa and UV irradiation (Fig 5C). We further examined crosslinked samples from the wild-type (HT1940 harboring pHT1388) and pilE-insertional mutant (HT2014 harboring pHT1388) by Western blotting with anti-PilE rabbit serum [33] (Fig 5D). The 85-kDa band was also observed with anti-PilE rabbit serum in the pilE+ genetic background only with pBPa and UV irradiation. Considering these results, we conclude that the 85-kDa band contains the PilE protein, indicating that PamA interacts with PilE in N. meningitidis.
Mapping the amino acid residue in PilE that crosslinks to the PamA protein
To further clarify the interaction between PamA and PilE, we attempted to identify the amino acid residue of PilE that crosslinks with PamA K278(pBPa). Since mapping was difficult and tedious [42], particularly for the endogenous proteins, the examination was conducted with recombinant proteins in E. coli.
We examined the interaction of two truncated recombinants, ΔN-PamA K278amb fused to maltose-binding protein (MBP) (MBP-ΔN-PamA K278amb) and ΔN-PilE with six glutamines (Gln6) in E. coli (S6 Fig). The crosslinked complex between MBP-ΔN-PamA K278(pBPa) and Gln6-ΔN-PilE was purified from E. coli and investigated using a gel-based proteomic analysis with LC-MS/MS (S7–S9 Figs) [42]. The peptides containing pBPa (IEVGK*[pBPa]LAFSTK), corresponding to aa 639 to 650 of MBP-ΔN-PamA K278(pBPa) (corresponding to aa 274 to 284 of native PamA), from crosslinked and uncrosslinked samples were analyzed by MS/MS. As shown in S9 Fig, the sequence was read from the annotated b and y ions of a crosslinked peptide until isoleucine (Ile) at position 19. This result indicated that the region around Ile19 in Gln6-ΔN -PilE is the site crosslinked to K278(pBPa) in MBP-ΔN-PamA.
Genetic elucidation of the interaction between PamA and PilE in N. meningitidis and its important for meningococcal internalization into HBMEC
Ile at position 19 in Gln6-ΔN-PilE corresponded to position 12 in native PilE in N. meningitidis (S10 Fig) because prepilin is processed by the prepilin peptidase PilD during translocation at the IM in N. meningitidis [36, 67]. Multiple alignments showed that the N-terminal region of PilE around Ile12 is highly conserved among neisserial species, including N. gonorrhoeae and N. lactamica, implicating the importance of the region in PilE (S10A Fig) [68].
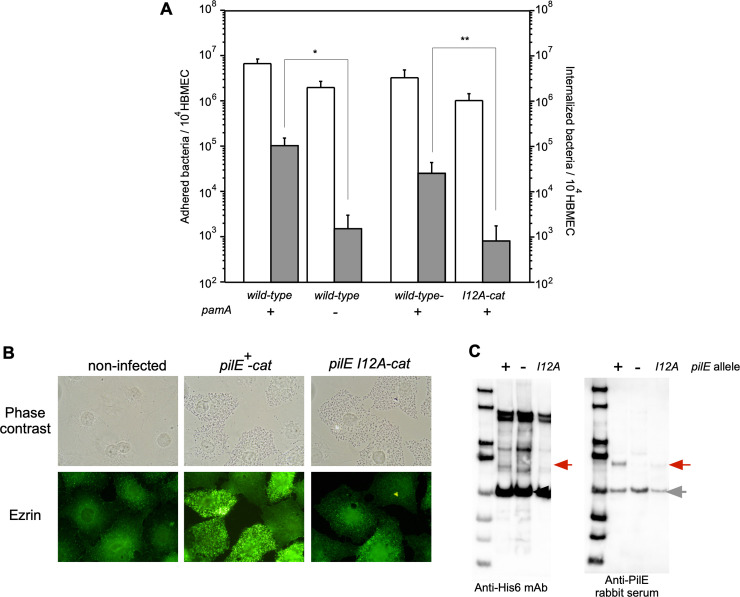
To further examine the functional interaction between PamA and the region containing the PilE I12 residue in N. meningitidis, the meningococcal pilE I12A-cat mutant HT2167 was constructed (Table 1), and we monitored its infectious ability and the accompanying host cell’s cytoskeleton rearrangement by the localization of ezrin. The pilE I12A N. meningitidis mutant was less efficiently internalized into HBMEC, while the adhesion remained unchanged (Fig 6A), and ezrin did not accumulate beneath the pilE I12A-cat mutant (Fig 6B). Western blotting with UV-irradiated samples purified from HT2167 expressing the MmpBPaRS/T and pamA K278amb genes also confirmed that the interaction between PilE and PamA K278(pBPa) is markedly reduced by the pilE I12A mutation (Fig 6C), providing genetic evidence for the interaction between PamA and PilE in N. meningitidis. Furthermore, since the phenotypes appeared to be mostly identical to that of the ΔpamA N. meningitidis mutant (Figs 1 and 6A), these results indicated that PamA and PilE interact functionally with each other in N. meningitidis and that this interaction is important for meningococcal infection in HBMEC.
Fig 6. The pilE I12A N. meningitidis mutant also showed reduced internalization into HBMEC, the induction of cytoskeleton rearrangements, and interactions with PamA in N. meningitidis.
(A) A I12A mutation in the pilE gene affects N. meningitidis internalization into HBMEC. Adherence (white) and internalization (gray) of N. meningitidis strains in HBMEC. Each value is the mean ± standard error of the mean (CFU per 104 HBMEC) from at least four experiments. The bars indicate the bacterial numbers of N. meningitidis wild-type pamA+ (HT1125, left), ΔpamA::spc (HT1822, middle-left), pilE+-cat (HT1744 middle-right), and pilE I12A-cat (HT2167, right), respectively (see S1 Table). *P<0.01, **P<0.02, significantly different from the pamA+ strain or pilE+-cat strain. (B) Immunofluorescence microscopy showing the accumulation of ezrin beneath N. meningitidis strains. The HBMEC monolayer was infected with wild-type pilE+-cat (middle) and pilE I12A-cat (right) N. meningitidis strains. A non-infected HBMEC monolayer is also shown in the left panels. Bacteria and HBMEC were observed by phase-contrast microscopy (upper panels). Ezrin was immunostained with anti-ezrin mAb and Alexa Fluor 488-conjugated rabbit anti-mouse IgG (green channel, lower panels). (C) PilE I12A was crosslinked to PamA K278(pBPa) less efficiently than wild-type PilE in N. meningitidis. Black and red arrows show the crosslinked complex between PamA K278(pBPa) and PilE, and the gray arrow indicates an unidentified band that crossreacted with anti-PilE rabbit serum. +/- indicates the presence or absence of the pilE gene, respectively.
Characterization of pili in the ΔpamA N. meningitidis mutant
The results described above suggested that PamA affects the functions of meningococcal pili, resulting in a negative impact on meningococcal infection in HBMEC. To further clarify the effects of PamA on pili, we investigated the function of pili in N. meningitidis. Electron microscopy with negative staining showed that the ΔpamA N. meningitidis mutant was similarly piliated to the wild-type strain HT1125 (S11A Fig, left and middle-left). Meningococcal pili are known to play a role not only in the infection of host cells [17, 69], but also in meningococcal natural competence, switching motility, and aggregation [70, 71]. An examination of pilus function in natural competence and aggregation revealed that the pili of the ΔpamA N. meningitidis mutant retain similar functions to those of the wild-type pilE N. meningitidis strain (S11B and S11C Fig). These results suggest that PamA does not participate in pilus formation itself or in functions related to natural competence and aggregation. It is important to note that the pilE I12A-cat N. meningitidis mutant is also as functional as the wild-type pilE-cat N. meningitidis strain while its piliation number its apparently smaller possibly due to translational fusion with the cat gene (S11 Fig and Fig 6).
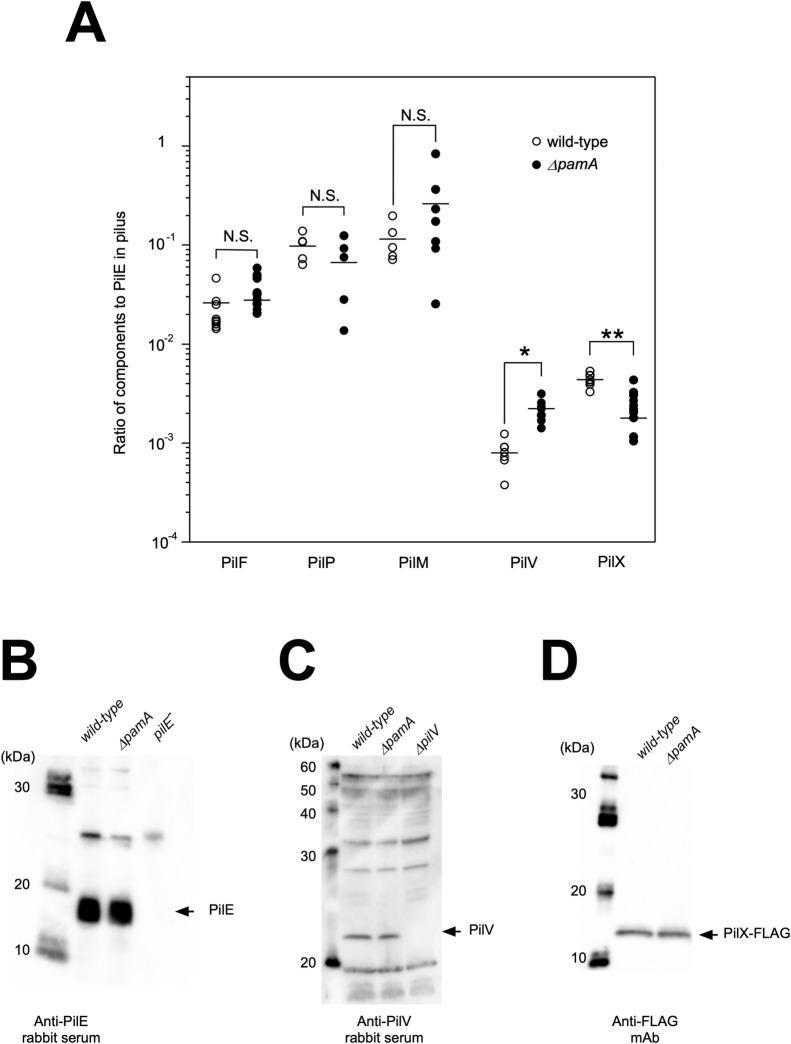
We examined the effects of PamA on meningococcal pili in more detail. Although the main component of the meningococcal pilus is PilE, its formation requires more than 20 proteins [72–74]. To investigate the effects of the pamA mutation on pili, we initially assessed the pilus protein levels in HT1125 and HT1822 using LC-MS/MS with Tandem Mass Tag (TMT) labeling [38] (S5 Table), and found that the amounts of some pilus components appeared to differ between HT1125 and HT1822. However, accurate comparisons of the protein amounts, particularly that for PilE, by TMT are methodologically difficult because N. meningitidis has 8 non-expressed pilS genes [75], according to the deduced amino acids recorded as Protein in the database. Therefore, we directly monitored the contents of pilus components, particularly for three minor pilins: PilX for aggregation in and adherence to human cultured cells [71, 76, 77], PilV to trigger plasma membrane reorganization [36, 71, 78], and ComP for natural competence [79]. PilF, PilM, and PilP, located on the meningococcal inner membrane for pilus formation [8, 67, 69, 80, 81], were also monitored. The relative amounts of the components to the PilE subunit in purified pili were measured by Western blotting followed by quantification (Fig 7). The relative ratios of PilF, PilP, and PilM to PilE did not significantly differ between the ΔpamA and wild-type N. meningitidis strains. On the other hand, in ΔpamA pili the relative amount of PilV increased, whereas that of PilX decreased (Fig 7A). It is important to note that the amount of ComP was too low in the whole cell extract or purified pili to detect by WB, even when the comP-FLAG N. meningitidis strain or anti-ComP rabbit antiserum was used (data not shown). We also confirmed that the expressions of PilE, PilV, and PilX-FLAG were not affected by the ΔpamA mutation (Fig 7B). Considering these results, the pamA mutation may cause unusual proportions of PilV and PilX to PilE in meningococcal pili, probably by improper sorting of PilE during pili formation, which might affect meningococcal infection in human cultured cells.
Fig 7. Disruption of the interaction between PamA and PilE leads to abnormal proportions of PilV and PilX relative to PilE in meningococcal pili.
(A) Ratio of pilus components to PilE in purified pili. The relative amounts of PilF-FLAG (HT2218 and HT2219), PilP-FLAG (HT2136 and HT2137), PilM-HA (HT2132 and HT2133), PilV (HT1125 and HT1822), and PilX-FLAG (HT2211 and HT2212) to PilE in purified pili were measured by Western blotting with appropriately diluted samples followed by quantification of the band intensity using the 30-kDa band in the molecular marker as an internal golden standard. The values obtained were divided by the value of PilE in the same sample. The significance of differences was examined by the t-test. (B, C, D) The pamA mutation does not affect the expression of the PilE (B), PilV (C), or PilX-FLAG (D) protein in N. meningitidis. Bacterial extracts equivalent to OD600 value of 0.0025 (for PilE) or 0.05 (for PilV and PilX-FLAG) were analyzed by Western blotting with anti-PilE and anti-PilV rabbit sera, and an anti-FLAG mAb, respectively. Black arrows indicate PilE, PilV, and PilX-FLAG, respectively.
Discussion
While progressive genome sequencing has recently provided novel insights into N. meningitidis, more than 50% of the annotated ORFs remain as “hypothetical proteins”, and their biological functions are currently unclear. This may be due to an incomplete understanding of meningococcal pathogenesis, which is largely attributable to the lack of analytical methods for N. meningitidis [34]. While the results of our genetic analyses indicated that the hypothetical protein NMB1345 (renamed PamA in the present study) in N. meningitidis plays an important role in meningococcal internalization into human cultured cells, the novel methodologies currently available for meningococci did not provide any insights into the function of PamA in N. meningitidis. Therefore, the incorporation of photoreactive ncAAs by pyrrolysine-based amber suppression was applied to a meningococcal protein with an unknown function, which implicated its physiological function in N. meningitidis.
The incorporation of ncAAs by pyrrolysine-based amber suppression has been used for site-specific incorporation into target proteins in vivo in a broad range of species from E. coli to eukaryotic systems (reviewed in [41]). However, regarding pathogenic bacteria, only a few studies have successfully incorporated ncAAs into EPEC [82], Shigella flexinelli [82], Salmonella typhimurium [83], M. tuberculosis [84], and B. cereus [85]. Moreover, these bacteria were only subjected to examinations of the incorporation of ncAAs into a well-characterized protein (e.g. GST or GFP), with a low molecular mass (approx. 10 kDa) and a known function [86], or a peptide [85], produced from expression vectors. Moreover, X-ray structure information for target proteins will also be advantageous for photoreactive ncAAs incorporation because the proper positions to incorporate ncAAs may be more easily predicted. Even when the X-ray structure of a target protein is unknown, a lower molecular mass is more advantageous because it is easier to scan the optimal site for ncAA incorporation in the target protein. Therefore, genetic photocrosslinker incorporation has not yet been applied to analyze a protein with unknown function or three-dimensional structure is unknown. Furthermore, some disadvantages in the application of photocrosslinking ncAAs incorporation combined with MS to Neisseria species are the very few expression vectors for neisseriae [33, 34] [87] and the limited culture methods in liquid medium due to autolysis [88]. To the best of our knowledge, this is the first study to demonstrate the incorporation of ncAAs into N. meningitidis. Moreover, after observing that ncAA-incorporated GST proteins are crosslinked as the dimer form in N. meningitidis (Fig 3E), we incorporated pBPa into the PamA protein in N. meningitidis (S5 Fig) and found that this meningococcal hypothetical protein with no physiological and structural information is closely associated with PilE in N. meningitidis.
Our strategy of incorporating a genetic photocrosslinker coupled with MS in N. meningitidis revealed that the meningococcal hypothetical protein PamA is associated with PilE in N. meningitidis. Further physicochemical analyses with PamA and PilE recombinants in E. coli identified the crosslinked regions between PamA K278(pBPa) PilE (S7–S9 Figs), and genetic analyses using the pilE I12A N. meningitidis mutant suggested that the interaction between PamA and PilE in N. meningitidis is functionally important for meningococcal pilus formation and infection to HBMEC (Fig 6). While further studies are needed, the PamA interaction with the N-terminal region of PilE around the Ile12 residue (S10 Fig) may be important to maintain the function of meningococcal pili by PamA in N. meningitidis.
The function of meningococcal PamA has not been clearly discerned in this study. The present results showed that PamA is not required for pilus formation itself (S11A Fig) but participates to keep optimal proportions of PilV and PilX to PilE in meningococcal pili (Fig 7). This result does not seem to contradict previous findings showing that the proper ratio of PilV and PilX to PilE in pili leads to pilus conformational changes that trigger plasma membrane reorganization [36, 71, 78, 76, 77]. However, we could not eliminate the possibility that more essential dysregulation(s) other than the imbalance of PilV and PilX to PilE in meningococcal pili by the pamA mutant were not identified in this study. We focused on three bands crosslinked with PamA K278(pBPa) with the highest intensity among the five PamA K-amb (pBPa) mutants, while the interaction between PamA and PilE found was identified from a band with the lowest intensity (Fig 4). On the other hand, the two protein complexes with the highest intensities (approximately 150 and 140 kDa) crosslinked to PamA K278(pBPa) were not identified in any protein other than PamA itself by peptide mass fingerprinting (Table 3). Although further analyses are needed, the two bands appeared to correspond to the homotrimer of PamA because intramolecular crosslinking was also detected in the MBP-ΔN-PamA K278(pBPa) recombinant protein in E. coli (S12 Fig). The endogenous proteins crosslinked to the PamA K208(pBPa), PamA K341(pBPa), PamA K382(pBPa), and PamA K395(pBPa) mutants have yet to be identified (Fig 4) and their characterization will provide more information about the novel function of PamA in N. meningitidis.
Surprisingly, unidentified genes involved in pilus function are still being discovered, although more than 20 proteins involved in pilus synthesis and function have already been identified by genetic [72–74] and WGS [32, 39] analyses. Therefore, our strategy using genetic screening by STM combined with genetically-encoded photocrosslinking amino acid incorporation and an MS analysis, could be a powerful method to identify new pathogenic factors in N. meningitidis. BLAST searches revealed pamA gene homologues in only three neisserial species: N. meningitidis, N. gonorrhoeae, and N. lactamica (S13 Fig). N. meningitidis and N. gonorrhoeae are closely related bacterial pathogens that share many common genes with a high degree of sequence identity (typically greater than 95%), and DNA relatedness studies clustered commensal N. lactamica with these two pathogenic Neisseria spp. [89]. Since the sequences of the pamA genes appeared to be more divergent among the three species than within N. meningitidis (S13 Fig), the pamA gene may have been acquired at a later stage of evolution by the three neisserial species, and the restricted possession of the pamA gene in the three neisserial species may reflect the physiological function of PamA.
While further studies are needed to clarify the functions of PamA in Neisseria, it is important to note that the application of new methodology to N. meningitidis provided novel insights into the physiological function of a meningococcal hypothetical protein with no obvious information such as its predicted biological function or three-dimensional structure. Our established system for ncAA incorporation by pyrrolysine-based amber suppression into N. meningitidis will facilitate a broad range of N. meningitidis studies, particularly the mapping of protein-protein interaction networks or characterization of the functions of hypothetical proteins in vivo. Nevertheless, as shown in the present study, in vivo research involving the combination of STM and photoreactive ncAAs incorporation by pyrrolysine-based amber suppression with an MS analysis will reveal meningococcal virulence factors that are difficult to identify using most currently available methods. The ncAA incorporation system described in the present study may become one of the strongest driving advancements for studies on meningococcal biology, pathogenesis, and vaccine development.
Supporting information
(GenBank Accession No LC511747). Lysine (K) residues are shown in red and the K residues replaced with amber codons in this study are indicated in bold red. Positions of K residues are shown as blue numbers. The grey bar indicates the putative hydrophobic region identified by SOSUI [90], which region were deleted to construct the PamA recombinants in E. coli in this study.
(PDF)
(A) Detection of PamA K148amb, K273 and K388amb expressed by pyrrolysine-based amber suppression with mAzZLys in N. meningitidis. Bacteria grown on one-fourth of a GC agar plate in the presence or absence of 0.3 mM mAzZLys and extracts equivalent to OD600 of 0.1 were analyzed by Western blotting with anti-PamA rabbit serum. +/- indicates the presence or absence of mAzZLys. The black arrow shows the full-length PamA protein. (B) Detection of complexes containing PamA K(mAzZLys) crosslinked to an endogenous protein in N. meningitidis. Bacteria grown on one-fourth of a GC agar plate in the presence of 0.3 mM mAzZLys were crosslinked by UV light irradiation, and mixed with SDS buffer. Aliquots were analyzed by Western blotting with an anti-His6 mAb. +/- indicates presence or absence of mAzZLys (upper), treatment or nontreatment with UV irradiation (lower). Black and grey arrows show the full-length PamA protein and putative complexes corresponding to PamA K(mAzZLys) crosslinked to an endogenous protein, respectively.
(PDF)
UV crosslinking of 27 PamA K-amb mutants expressed by pyrrolysine-based amber suppression with pBPa in ΔpamA N. meningitidis. The black arrow shows the full-length PamA K-amb protein expressed by pyrrolysine-based amber suppression with pBPa. PamA K-amb mutants shown in red were subjected to a more detailed analysis in Fig 4.
(PDF)
(A) Amino acid sequence of the C-terminal region of PamA K278-Strep2-His6. (B) Nucleotide sequence of the C-terminal region of PamA K278-Strep2-His6. The same colors in A and B corresponds to the same region in amino acid (A) and nucleotide (B) sequences.
(PDF)
(A) Amino acid sequence of PamA with Strep2-His6 tag at C-terminus. The tryptic peptide containing a ncAA, pBPa is highlighted in red and the pBPa at position 278 is represented as X. (B) The incorporation of pBPa at position 278 was confirmed by MALDI-TOF MS analysis of the tryptic peptide IEVGXLAFSTK (X represents pBPa). The observed (obsd) molecular masses agreed well with the calculated (calcd) masses. (C) MALDI-TOF MS/MS analysis of the tryptic peptide of shown in B. Tandem mass spectrum of the peptide IEVGXLAFSTK (X = pBPa). The sequence can be read from the annotated b (red) or y (blue) ion series.
(PDF)
Recombinant proteins crosslinked in E. coli were purified with amylose resin and concentrated to 500 μl as described in the Materials and Methods. Aliquots (approximately 1 μl) were fractionated by SDS-PAGE and analyzed by staining with a CBB staining kit (A), and Western blotting with anti-MBP (B) and anti-His6 (C) mAbs. Prestained molecular mass standards are shown by grey numbers with asterisks, since the apparent molecular masses were different from the actual masses due to abnormal mobility in the gel. +/- indicates treatment or non-treatment with UV irradiation, respectively. Black, blue and red arrows indicate PamA ΔN-K278amb (no pyrrolysine-based amber suppression with pBPa), PamA ΔN-K278(pBPa)-Strep2-His6 (full-length) and the complex crosslinked between PamA K278(pBPa)-Strep2-His6 and ΔN-Gln6-PilE, respectively. (D) Schematic figure of the crosslinking between PamA ΔN-K278(pBPa)-Strep2-His6 and Gln6-ΔN-PilE recombinant proteins in E. coli.
(PDF)
(A) Liquid chromatogram of uncrosslinked MBP-ΔN-PamA K278(pBPa) and the Gln6-ΔN-PilE protein (upper), and the complex crosslinked between MBP-ΔN-PamA K278(pBPa) and Gln6-ΔN-PilE (lower). The fraction eluted at 12.38 min from the uncrosslinked sample contained the peptide IEVGK(pBPa)LAFSTK corresponding to position 639 to 650 of MBP-ΔN-PamA K278(pBPa) (positions 274 to 284 for native PamA), determined by an MS analysis. (B) Enlarged liquid chromatogram after 14 min of elution. The fraction eluted at 17.32 min from the crosslinked sample contained the peptide MQQQQQQFTLIELMIVIAIVGILAAVALPAYQDYTARAQVSEAILLAEGQK, corresponding to the first 51 amino acids of Gln6-ΔN-PilE at the N terminus, which was crosslinked to the peptide IEVGK(pBPa)LAFSTK from the MBP-ΔN-PamA K278(pBPa) protein. (C) MS spectrum of the LC fraction eluted at 17.32 min that contained the cross-linked peptide between MQQQQQQFTLIELMIVIAIVGILAAVALPAYQDYTARAQVSEAILLAEGQK from Gln6-ΔN-PilE and IEVGK(pBPa)LAFSTK from MBP-ΔN-PamA K278(pBPa)(obsd:1380.9367 [M+H]5+).
(PDF)
The sequence was read from the annotated b and y ion series; b3, y4, y5, y6, y7, y8, y9, and y10 ions were observed.
(PDF)
The sequence was read from the annotated b and y ion series; b3, y4, y5, y6, y7, y8, y9, and y10 ions of the peptide IEVGK(pBPa)LAFSTK from MBP-ΔN-PamA K278(pBPa) (shown in red), and b3, b6, b9, b10, b11, b12, b13, b14, b15, b16, b17, b18, y4, y5, y6, y7, y8, y9, y10, y11, y12, y13, y14, y15, y16, y17, y18, y19, y20, y21, y22, y23, y24, y25, y26, y27, y28, y29, y30, y31, and y32 ions of the peptide MQQQQQQFTLIELMIVIAIVGILAAVALPAYQDYTARAQVSEAILLAEGQK from Gln6-ΔN-PilE (shown in blue) were observed.
(PDF)
(A) Amino acid sequence alignment of PilE proteins from N. meningitidis (Nm), N. gonorrhoeae (Ng) and N. lactamica (Nl). The N-terminal half of the α-helix (α1-N) responsible for pilin assembly is shown by red (residues 1–14; α1:1–14) and cyan (residues 15–28; α1:15–28) bars, respectively. The αβ-loop, protruding from the globular domain to form a ridge on the subunit surface is indicated as a yellow bar. The D-region containing the hypervariable loop that protrudes as a second ridge on the globular domain is indicated as a light green bar. Red numbers indicate the positions of amino acids at which the functions of α1-N are divided. Ile at position 12 is indicated by a red vertical arrow. N. meningitidis HT1125 PilE (DDBJ accession no. AB698857), N. meningitidis H44/76 PilE (GenBank accession no. CP002420), N. gonorrhoeae MS11 PilE (EMBL accession no. CAI08338) and N. lactamica NLA_1780 type IV pilus assembly protein PilA (GenBank accession no. FN995097). (B) X-ray crystal structure of gonococcal PilE [91]. α1:1–14, α1:15–28, αβ-loop and D-region are shown in red, cyan, yellow and light green, respectively (the colors in B correspond to those in A). Ile at position 12 is indicated as a space-filling molecule.
(PDF)
(A) Electron micrographs showing piliation of N. meningitidis strains HT1125 (wild-type), HT1822 (ΔpamA), HT1744 (pilE+-cat), and HT2167 (pilE I12A-cat). Upper and lower panels show magnifications of 20,000 and 50,000, respectively. Scale bars shown in black (upper) and white (lower) represent 200 nm. (B) Quantification of the competence for DNA transformation in N. meningitidis wild-type, and pilE-, ΔpamA, pilE+-cat and pilE I12A-cat mutants. Equivalent numbers of recipient cells were transformed using 0.5 μg of chromosomal DNA purified from the NalR (nalidixic acid resistant) strain HT1001, and NalR transformants were counted. Results are expressed as numbers of NalR transformants per 1 μg DNA and ± standard deviation from at least 8 independent experiments. (C) Aggregation as assessed by phase-contrast microscopy. Aggregates of N. meningitidis strains HT1125 (wild-type), HT1156 (pilE-), HT1822 (ΔpamA), HT1744 (pilE+-cat) and HT2167 (pilE I12A-cat) were observed after 4 hours of incubation in RPMI containing 10% fetal bovine serum.
(PDF)
MS/MS spectrum of the peptide IEVGK(pBPa)LAFSTK (at positions 274 to 284 for native PamA) crosslinked to LNELVNLVTDLQIGAFINPNGSIAPS (at positions 247 to 273 for native PamA) in MBP-ΔN-PamA K278(pBPa). The crosslinked site was analyzed by SIM-XL (http://patternlabforproteomics.org/sim-xl). The sequence can be read from the annotated b and y ion series: b3, y3, y4, y5, y6 ions of the peptide IEVGK(pBPa)LAFSTK from MBP-ΔN-PamA K278(pBPa) (shown in red), and b3, b4, b5, b6, b7, b8, b10, b13, b19, b21, y4, y5, y6, y7, y8, y9, y10, y11, y12, y13, y14, y15 ions of a peptide LNELVNLVTDLQIGAFINPNGSIAPS from MBP-ΔN-PamA K278(pBPa) (blue) were observed, respectively. We could not confirm whether pBPa at position 278 was crosslinked to Leu at position 257 or Gln at position 258 (shown in yellow overlay).
(PDF)
The pamA genes (the genes encoding “conserved hypothetical protein”) from three neisserial species were analyzed by Molecular Evolutionary Genetics Analysis (MEGA) X [92]. The evolutionary history was inferred using the Neighbor-Joining method [93]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [94] and are in the units of the number of base substitutions per site. The robustness of the NJ method was tested by bootstrapping with 500 replicates of data, and the percentages are shown at the nodes. This analysis involved 19 nucleotide sequences of genes encoding N. meningitidis HT1125 PamA (DDBJ accession no. LC511747), N. meningitidis H44/76 conserved hypothetical protein (GenBank accession no. CP002420), N. meningitidis MC58 hypothetical protein (GenBank accession no. AE002098), N. meningitidis M04-240196 conserved hypothetical protein (GenBank accession no. CP002423), N. meningitidis alpha14 conserved hypothetical protein (GenBank accession no. AM889136), N. meningitidis NZ-05/33 conserved hypothetical protein (GenBank accession no. CP002424), N. meningitidis M01-240149 conserved hypothetical protein (GenBank accession no. CP002421), N. meningitidis alpha710 hypothetical protein (GenBank accession no. CP001561), N. meningitidis 053442 conserved hypothetical protein (GenBank accession no. CP000381), N. meningitidis 8013 conserved hypothetical protein (GenBank accession no. FM999788), N. meningitidis M01-240355 conserved hypothetical protein (GenBank accession no. CP002422), N. meningitidis 510612 hypothetical protein (GenBank accession no. CP007524), N. meningitidis WUE2694 conserved hypothetical protein (GenBank accession no. FR774048), N. meningitidis G2136 conserved hypothetical protein (GenBank accession no. CP002419), N. meningitidis Z2491 hypothetical protein (GenBank accession no. AL157959), N. meningitidis FAM18 hypothetical protein (GenBank accession no. AM421808), N. gonorrhoeae FA1090 hypothetical protein (RefSeq accession no. NC_002946), N. gonorrhoeae NCCP11945 conserved hypothetical protein (GenBank accession no. CP001051) and N. lactamica putative secreted protein (GenBank accession no. FN995097).
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Pili isolated from N. meningitidis strains HT1125 (wild-type) and HT1822 (ΔpamA) were subjected to trypsin digestion. Labelling of the peptides and analysis by LC-MS/MS were performed as described previously [38]. Proteins were identified by running MASCOT against the NCBI database (NCBInr 20160202). Ratio of the protein levels were expressed as the value of relative amount in HT1822 divided into those in HT1125. The results from four independent experiments were shown in this table.
(DOCX)
(PDF)
Acknowledgments
We thank Dr. Masatomo Morita for advice about phylogenetic analyses.
Data Availability
All relevant data are within the paper and its Supporting Information files. The nucleotide sequence of PamA from N. meningitidis strain HT1125 has been deposited in the DNA Data Bank of Japan (DDBJ) (accession code: LC511747).
Funding Statement
This work was supported by JSPS KAKENHI Grant Numbers 15K08485 and 19K07550 (to H.T.), AMED under Grant Number 18fk0108071j0101 (to H.T.), by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from AMED under Grant Number JP16am0101022 (to S.Y.), and under Grant Number JP17am0101081 (to S.Y.).
References
- 1.Takahashi H, Haga M, Sunagawa T, Saitoh T, Kitahara T, Matsumoto S, et al. Meningococcal carriage rates in healthy individuals in Japan determined using Loop-Mediated Isothermal Amplification and oral throat wash specimens. J Infect Chemother. 2016;22(7):501–4. 10.1016/j.jiac.2015.12.016 [DOI] [PubMed] [Google Scholar]
- 2.Chang Q, Tzeng YL, Stephens DS. Meningococcal disease: changes in epidemiology and prevention. Clinical epidemiology. 2012;4:237–45. 10.2147/CLEP.S28410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coureuil M, Jamet A, Bille E, Lecuyer H, Bourdoulous S, Nassif X. Molecular interactions between Neisseria meningitidis and its human host. Cell Microbiol. 2019;21(11):e13063 10.1111/cmi.13063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouwer MC, Read RC, van de Beek D. Host genetics and outcome in meningococcal disease: a systematic review and meta-analysis. The Lancet Infectious diseases. 2010;10(4):262–74. 10.1016/S1473-3099(10)70045-1 [DOI] [PubMed] [Google Scholar]
- 5.Codjoe SN, Nabie VA. Climate change and cerebrospinal meningitis in the Ghanaian meningitis belt. International Journal of Environmental Research and Public Health. 2014;11(7):6923–39. 10.3390/ijerph110706923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peak IR, Srikhanta Y, Dieckelmann M, Moxon ER, Jennings MP. Identification and characterisation of a novel conserved outer membrane protein from Neisseria meningitidis. FEMS Immunol Med Microbiol. 2000;28(4):329–34. 10.1111/j.1574-695X.2000.tb01494.x [DOI] [PubMed] [Google Scholar]
- 7.Virji M. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat Rev Microbiol. 2009;7(4):274–86. 10.1038/nrmicro2097 [DOI] [PubMed] [Google Scholar]
- 8.Coureuil M, Lecuyer H, Bourdoulous S, Nassif X. A journey into the brain: insight into how bacterial pathogens cross blood-brain barriers. Nat Rev Microbiol. 2017;15(3):149–59. 10.1038/nrmicro.2016.178 [DOI] [PubMed] [Google Scholar]
- 9.Schubert-Unkmeir A. Molecular mechanisms involved in the interaction of Neisseria meningitidis with cells of the human blood-cerebrospinal fluid barrier. Pathog Dis. 2017;75(2). 10.1093/femspd/ftx023 [DOI] [PubMed] [Google Scholar]
- 10.Capecchi B, Adu-Bobie J, Di Marcello F, Ciucchi L, Masignani V, Taddei A, et al. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol Microbiol. 2005;55(3):687–98. 10.1111/j.1365-2958.2004.04423.x [DOI] [PubMed] [Google Scholar]
- 11.Serruto D, Adu-Bobie J, Scarselli M, Veggi D, Pizza M, Rappuoli R, et al. Neisseria meningitidis App, a new adhesin with autocatalytic serine protease activity. Mol Microbiol. 2003;48(2):323–34. 10.1046/j.1365-2958.2003.03420.x [DOI] [PubMed] [Google Scholar]
- 12.van Ulsen P, van Alphen L, ten Hove J, Fransen F, van der Ley P, Tommassen J. A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol. 2003;50(3):1017–30. 10.1046/j.1365-2958.2003.03773.x [DOI] [PubMed] [Google Scholar]
- 13.Turner DP, Marietou AG, Johnston L, Ho KK, Rogers AJ, Wooldridge KG, et al. Characterization of MspA, an immunogenic autotransporter protein that mediates adhesion to epithelial and endothelial cells in Neisseria meningitidis. Infect Immun. 2006;74(5):2957–64. 10.1128/IAI.74.5.2957-2964.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldfield NJ, Bland SJ, Taraktsoglou M, Dos Ramos FJ, Robinson K, Wooldridge KG, et al. T-cell stimulating protein A (TspA) of Neisseria meningitidis is required for optimal adhesion to human cells. Cell Microbiol. 2007;9(2):463–78. 10.1111/j.1462-5822.2006.00803.x [DOI] [PubMed] [Google Scholar]
- 15.Hung MC, Heckels JE, Christodoulides M. The adhesin complex protein (ACP) of Neisseria meningitidis is a new adhesin with vaccine potential. MBio. 2013;4(2). 10.1128/mBio.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72(4):2088–2100. 10.1128/iai.72.4.2088-2100.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coureuil M, Jamet A, Bille E, Lecuyer H, Bourdoulous S, Nassif X. Molecular interactions between Neisseria meningitidis and its human host. Cell Microbiol. 2019:e13063 10.1111/cmi.13063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dos Santos Souza I, Maissa N, Ziveri J, Morand PC, Coureuil M, Nassif X, et al. Meningococcal disease: A paradigm of type-IV pilus dependent pathogenesis. Cell Microbiol. 2020;22(4):e13185 10.1111/cmi.13185 [DOI] [PubMed] [Google Scholar]
- 19.Sun YH, Bakshi S, Chalmers R, Tang CM. Functional genomics of Neisseria meningitidis pathogenesis. Nat Med. 2000;6(11):1269–73. 10.1038/81380 [DOI] [PubMed] [Google Scholar]
- 20.Exley RM, Sim R, Goodwin L, Winterbotham MS, M.C., Read RC, Tang CM. Identification of meningococcal genes necessary for colonization of human upper airway tissue. Infect Immun. 2009;77(1):45–51. 10.1128/IAI.00968-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendum TA, Newcombe J, Mannan AA, Kierzek AM, McFadden J. Interrogation of global mutagenesis data with a genome scale model of Neisseria meningitidis to assess gene fitness in vitro and in sera. Genome Biol. 2011;12(12):R127 10.1186/gb-2011-12-12-r127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietrich G, Kurz S, Hubner C, Aepinus C, Theiss S, Guckenberger M, et al. Transcriptome analysis of Neisseria meningitidis during infection. J Bacteriol. 2003;185(1):155–64. 10.1128/jb.185.1.155-164.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hotopp JCD, Grifantini R, Kumar N, Tzeng YL, Fouts D, Frigimelica E, et al. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152(Pt 12):3733–49. 10.1099/mic.0.29261-0 [DOI] [PubMed] [Google Scholar]
- 24.Joseph B, Schneiker-Bekel S, Blom J, Claus H, Linke B, Schwarz RF, et al. Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J Bacteriol. 2010;192(20):5363–77. 10.1128/JB.00883-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph B, Frosch M, Schoen C, Schubert-Unkmeir A. Transcriptome analyses in the interaction of Neisseria meningitidis with mammalian host cells. Methods Mol Biol. 2012;799:267–93. 10.1007/978-1-61779-346-2_17 [DOI] [PubMed] [Google Scholar]
- 26.Hey A, Li MS, Hudson MJ, Langford PR, Kroll JS. Transcriptional profiling of Neisseria meningitidis interacting with human epithelial cells in a long-term in vitro colonization model. Infect Immun. 2013;81(11):4149–59. 10.1128/IAI.00397-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Echenique-Rivera H, Muzzi A, Del Tordello E, Seib K, Francois P, Rappuoli R, et al. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 2011;7(5):e1002027 10.1371/journal.ppat.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedman AK, Li MS, Langford PRF, Kroll JS. Transcriptional profiling of serogroup B Neisseria meningitidis growing in human blood: an approach to vaccine antigen discovery. PLoS One. 2012;7(6):e39718 10.1371/journal.pone.0039718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernardini G, Braconi D, Santucci A. The analysis of Neisseria meningitidis proteomes: Reference maps and their maps and their applications. Proteomics. 2007;7(16):2933–46. 10.1002/pmic.200700094 [DOI] [PubMed] [Google Scholar]
- 30.van Alen T, Claus H, Zahedi RP, Groh J, Blazyca H, Lappann M, et al. Comparative proteomic analysis of biofilm and planktonic cells of Neisseria meningitidis. Proteomics. 2010;10(24):4512–21. 10.1002/pmic.201000267 [DOI] [PubMed] [Google Scholar]
- 31.Kanova E, Jimenez-Munguia I, Majerova P, Tkacova Z, Bhide K, Mertinkova P, et al. Deciphering the Interactome of Neisseria meningitidis With Human Brain Microvascular Endothelial Cells. Front Microbiol. 2018;9:2294 10.3389/fmicb.2018.02294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siena E, Bodini M, Medini D. Interplay Between Virulence and Variability Factors as a Potential Driver of Invasive Meningococcal Disease. Comput Struct Biotechnol J. 2018;16:61–9. 10.1016/j.csbj.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi H, Watanabe H. A broad-host-range vector of incompatibility group Q can work as a plasmid vector in Neisseria meningitidis: a new genetical tool. Microbiology. 2002;148(Pt 1):229–36. 10.1099/00221287-148-1-229 [DOI] [PubMed] [Google Scholar]
- 34.Soriani M. Unraveling Neisseria meningitidis pathogenesis: from functional genomics to experimental models. F1000Res. 2017;6:1228 10.12688/f1000research.11279.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi H, Kim KS, Watanabe H. Differential in vitro infectious abilities of two common Japan-specific sequence-type (ST) clones of disease-associated ST-2032 and carrier-associated ST-2046 Neisseria meningitidis strains in human endothelial and epithelial cell lines. FEMS Immunol Med Microbiol. 2008;52(1):36–46. 10.1111/j.1574-695X.2007.00342.x [DOI] [PubMed] [Google Scholar]
- 36.Takahashi H, Yanagisawa T, Kim KS, Yokoyama S, Ohnishi M. Meningococcal PilV potentiates Neisseria meningitidis type IV pilus-mediated internalization into human endothelial and epithelial cells. Infect Immun. 2012;80(12):4154–66. 10.1128/IAI.00423-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi H, Kim KS, Watanabe H. Meningococcal internalization into human endothelial and epithelial cells is triggered by the influx of extracellular L-glutamate via GltT L-glutamate ABC transporter in Neisseria meningitidis. Infect Immun. 2011;79(1):380–92. 10.1128/IAI.00497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi H, Yanagisawa T, Kim KS, Yokoyama S, Ohnishi M. Multiple Functions of Glutamate Uptake via Meningococcal GltT-GltM L-Glutamate ABC Transporter in Neisseria meningitidis Internalization into Human Brain Microvascular Endothelial Cells. Infect Immun. 2015;83(9):3555–67. 10.1128/IAI.00654-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287(5459):1809–15. 10.1126/science.287.5459.1809 [DOI] [PubMed] [Google Scholar]
- 40.Willis JCW, Chin JW. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat Chem. 2018;10(8):831–7. 10.1038/s41557-018-0052-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L. Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts. Acc Chem Res. 2017;50(11):2767–75. 10.1021/acs.accounts.7b00376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holding AN. XL-MS: Protein cross-linking coupled with mass spectrometry. Methods. 2015;89:54–63. 10.1016/j.ymeth.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 43.Dumas A, Lercher L, Spicer CD, Davis BG. Designing logical codon reassignment—Expanding the chemistry in biology. Chem Sci. 2015;6(1):50–69. 10.1039/c4sc01534g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A. 2002;99(17):11020–4. 10.1073/pnas.172226299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi H, Watanabe H. Post-translational processing of Neisseria meningitidis g-glutamyl aminopeptidase and its association with inner membrane facing to the cytoplasmic space. FEMS Microbiol Lett. 2004;234(1):27–35. 10.1016/j.femsle.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 46.Takahashi H, Watanabe H, Kim KS, Yokoyama S, Yanagisawa T. The Meningococcal Cysteine Transport System Plays a Crucial Role in Neisseria meningitidis survival in human brain microvascular endothelial cells. mBio. 2018;9(6):e02332–18. 10.1128/mBio.02332-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi H, Inada T, Postma P, Aiba H. CRP down-regulates adenylate cyclase activity by reducing the level of phosphorylated IIA(Glc), the glucose-specific phosphotransferase protein, in Escherichia coli. Mol Gen Genet. 1998;259(3):317–26. 10.1007/s004380050818 [DOI] [PubMed] [Google Scholar]
- 48.Takahashi H, Hirose K, Watanabe H. Necessity of meningococcal gamma-glutamyl aminopeptidase for the Neisseria meningitidis growth in rat cerebrospinal fluid (CSF) and CSF mimicking medium. J Bacteriol. 2004;186(1):244–7. 10.1128/jb.186.1.244-247.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode. Chem Biol. 2008;15(11):1187–97. 10.1016/j.chembiol.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 50.Lacey VK, Kouie GV, Noel JP, Wang L. Expanding the library and substrate diversity of the pyrrolysyl-tRNA synthetase. Chembiochem. 2013;14(16):2100–5. 10.1002/cbic.201300400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark MJ. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51(2–3):255–67. 10.1016/0378-1119(87)90314-3 [DOI] [PubMed] [Google Scholar]
- 52.Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or. Capturing of proteins. Nat Protoc. 2007;2(6):1528–35. 10.1038/nprot.2007.209 [DOI] [PubMed] [Google Scholar]
- 53.Yanagisawa T, Takahashi M, Mukai T, Sato S, Wakamori M, Shirouzu M, et al. Multiple site-specific installations of Nepsilon-monomethyl-L-lysine into histone proteins by cell-based and cell-free protein synthesis. Chembiochem. 2014;15(12):1830–8. 10.1002/cbic.201402291 [DOI] [PubMed] [Google Scholar]
- 54.Masuda A, Dohmae N. Examination of an absolute quantity of less than a hundred nanograms of proteins by amino acid analysis. Anal Bioanal Chem. 2013;405(25):8073–81. 10.1007/s00216-013-7056-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi H, Carlson RW, Muszynski A, Choudhury B, Kim KS, Stephens DS, et al. Modification of lipooligosaccharide with phosphoethanolamine by LptA in Neisseria meningitidis enhances meningococcal adhesion to human endothelial and epithelial cells. Infect Immun. 2008;76(12):5777–89. 10.1128/IAI.00676-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsey ME, Hackett KT, Kotha C, Dillard JP. New complementation constructs for inducible and constitutive gene expression in Neisseria gonorrhoeae and Neisseria meningitidis. Appl Environ Microbiol. 2012;78(9):3068–78. 10.1128/AEM.07871-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyle-Vavra S, Seifert HS. Shuttle mutagenesis: a mini-transposon for producing PhoA fusions with exported proteins in Neisseria gonorrhoeae. Gene. 1995;155(1):101–6. 10.1016/0378-1119(94)00890-5 [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi A, Matsuda T, Ohtake K, Yanagisawa T, Yokoyama S, Fujiwara Y, et al. Incorporation of a Doubly Functionalized Synthetic Amino Acid into Proteins for Creating Chemical and Light-Induced Conjugates. Bioconjug Chem. 2016;27(1):198–206. 10.1021/acs.bioconjchem.5b00602 [DOI] [PubMed] [Google Scholar]
- 59.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395(2):361–74. 10.1016/j.jmb.2009.10.030 [DOI] [PubMed] [Google Scholar]
- 60.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124(31):9026–7. 10.1021/ja027007w [DOI] [PubMed] [Google Scholar]
- 61.Fabrini R, De Luca A, Stella L, Mei G, Orioni B, Ciccone S, et al. Monomer-dimer equilibrium in glutathione transferases: a critical re-examination. Biochemistry. 2009;48(43):10473–82. 10.1021/bi901238t [DOI] [PubMed] [Google Scholar]
- 62.Yanagisawa T, Kuratani M, Seki E, Hino N, Sakamoto K, Yokoyama S. Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase. Cell Chem Biol. 2019;26(7):936–49. 10.1016/j.chembiol.2019.03.008 [DOI] [PubMed] [Google Scholar]
- 63.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, Tan M, Xie Z, Dai LF, Y. C, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7(1):58–63. 10.1038/nchembio.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sokalingam S, Raghunathan G, Soundrarajan N, Lee SG. A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS One. 2012;7(7):e40410 10.1371/journal.pone.0040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korndorfer IP, Skerra A. Improved affinity of engineered streptavidin for the Strep-tag II peptide is due to a fixed open conformation of the lid-like loop at the binding site. Protein Sci. 2002;11(4):883–93. 10.1110/ps.4150102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61(6):1510–22. 10.1111/j.1365-2958.2006.05341.x [DOI] [PubMed] [Google Scholar]
- 68.Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, Craig L. Structure of the Neisseria meningitidis Type IV pilus. Nat Commun. 2016;7:13015 10.1038/ncomms13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carbonnelle E, Hill DJ, Morand P, Griffiths NJ, Bourdoulous S, Murillo I, et al. Meningococcal interactions with the host. Vaccine. 2009;27 Suppl 2:B78–89. 10.1016/j.vaccine.2009.04.069 [DOI] [PubMed] [Google Scholar]
- 70.Brown DR, Helaine S, Carbonnelle E, Pelicic V. Systematic functional analysis reveals that a set of seven genes is involved in Fine-Tuning of the Multiple Functions Mediated by Type IV Pili in Neisseria meningitidis. Infect Immun. 2010;78(7):3053–63. 10.1128/IAI.00099-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imhaus AF, Dumenil G. The number of Neisseria meningitidis type IV pili determines host cell. EMBO J. 2014;33(16):1767–83. 10.15252/embj.201488031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carbonnelle E, Hélaine S, Prouvensier L, Nassif X, Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step. Mol Microbiol. 2005;55(1):54–64. 10.1111/j.1365-2958.2004.04364.x [DOI] [PubMed] [Google Scholar]
- 73.Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68(4):827–37. 10.1111/j.1365-2958.2008.06197.x [DOI] [PubMed] [Google Scholar]
- 74.Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol. 2017;15(6):365–79. 10.1038/nrmicro.2017.40 [DOI] [PubMed] [Google Scholar]
- 75.Wormann ME, Horien CL, Bennett JS, Jolley KA, Maiden MC, Tang CM, et al. Sequence, distribution and chromosomal context of class I and class II pilin genes of Neisseria meningitidis identified in whole genome sequences. BMC Genomics. 2014;15:253 10.1186/1471-2164-15-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol. 2005;55(1):65–77. 10.1111/j.1365-2958.2004.04372.x [DOI] [PubMed] [Google Scholar]
- 77.Brissac T, Mikaty G, Dumenil G, Coureuil M, Nassif X. The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect Immun. 2012;80(9):3297–306. 10.1128/IAI.00369-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mikaty G, Soyer M, Mairey E, Henry N, Dyer D, Forest KT, et al. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 2009;5(2):e1000314 10.1371/journal.ppat.1000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolfgang M, van Putten JP, Hayes SF, Koomey M. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol. 1999;31(5):1345–57. 10.1046/j.1365-2958.1999.01269.x [DOI] [PubMed] [Google Scholar]
- 80.Hill DJ, Griffiths NJ, Borodina E, Virji M. Cellular and molecular biology of Neisseria meningitidis colonization and invasive disease. Clin Sci (Lond). 2010;118(9):547–64. 10.1042/CS20090513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V. Large-scale study of the interactions between proteins involved in type IV pilus in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol. 2012;84(5):857–73. 10.1111/j.1365-2958.2012.08062.x [DOI] [PubMed] [Google Scholar]
- 82.Lin S, Zhang Z, Xu H, Li L, Chen S, Li J, et al. Site-specific incorporation of photo-cross-linker and bioorthogonal amino acids into enteric bacterial pathogens. J Am Chem Soc. 2011;133(50):20581–7. 10.1021/ja209008w [DOI] [PubMed] [Google Scholar]
- 83.Gan Q, Lehman BP, Bobik TA, Fan C. Expanding the genetic code of Salmonella with non-canonical amino acids. Sci Rep. 2016;6:39920 10.1038/srep39920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang F, R S., Guo J, Shen W, Shen W, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PLoS One. 2010;5(2):e9354 10.1371/journal.pone.0009354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo X, Zambaldo C, Liu T, Zhang Y, Xuan W, Wang C, et al. Recombinant thiopeptides containing noncanonical amino acids. Proc Natl Acad Sci U S A. 2016;113(13):3615–20. 10.1073/pnas.1602733113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang M, Lin S, Song X, Liu J, Fu Y, Ge X, et al. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7(10):671–7. 10.1038/nchembio.644 [DOI] [PubMed] [Google Scholar]
- 87.O' Dwyer CA, Langford PR, Kroll JS. A novel neisserial shuttle plasmid: a useful new tool for meningococcal research. FEMS Microbiol Lett. 2005;251(1):143–7. 10.1016/j.femsle.2005.07.036 [DOI] [PubMed] [Google Scholar]
- 88.Spence JM, Wright L, Clark VL. Laboratory maintenance of Neisseria gonorrhoeae. Curr Protoc Microbiol. 2008;Chapter 4:Unit 4A.1 10.1002/9780471729259.mc04a01s8 [DOI] [PubMed] [Google Scholar]
- 89.Guibourdenche M, Popoff MY, Riou JY. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis. Ann Inst Pasteur Microbiol. 1986;137B(2):177–85. 10.1016/s0769-2609(86)80106-5 [DOI] [PubMed] [Google Scholar]
- 90.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane. Bioinformatics. 1998;14(4):378–9. 10.1093/bioinformatics/14.4.378 [DOI] [PubMed] [Google Scholar]
- 91.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, et al. Type IV pilus structure by cryo-electron microscopy and crystallography. Mol Cell. 2006;23(5):651–62. 10.1016/j.molcel.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 92.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–9. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 94.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining. Proc Natl Acad Sci U S A. 2004;101(30):11030–5. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]