Abstract
Hepatitis B, a viral infection that affects the liver, is thought to affect over 257 million people worldwide, and long-term infection can lead to life-threatening issues such as cirrhosis or liver cancer. Chronic hepatitis B develops by the interaction between hepatitis B virus (HBV) and host immune response. However, questions of how HBV-infected cells thwart immune system defenses remain unanswered. Extracellular vesicles (EVs) are used for cellular communication, carrying cargoes such as RNAs, proteins, and lipids and delivering them intracellularly after being endocytosed by target cells. HBV-infected liver cells secrete several types of EVs into body fluids such as complete and incomplete virions, and exosomes. We previously demonstrated that monocytes that incorporated EVs moved to immunoregulatory phenotypes via up-regulation of PD-L1, an immunocheckpoint molecule, and down-regulation of CD69, a leukocyte activation molecule. In this study, we transfected mice with HBV using hydrodynamic injection and studied the effects of EVs secreted by HBV-infected liver cells. EVs secreted from cells with HBV replication strongly suppressed the immune response, inhibiting the eradication of HBV-replicating cells in the mice transfected with HBV. EVs were systemically incorporated in multiple organs, including liver, bone marrow (BM), and intestine. Intriguingly, the BM cells that incorporated EVs acquired intestinal tropism and the dendritic cell populations in the intestine increased. These findings suggest that the EVs secreted by HBV-infected liver cells exert immunosuppressive functions, and that an association between the liver, bone marrow, and intestinal tract exists through EVs secreted from HBV-infected cells.
Keywords: HBV, extracellular vesicles, hydrodynamic HBV transfection mouse model, PD-L1, dendritic cells, hepatitis B virus (HBV, Hep B), extracellular vesicles, immunosuppression, dendritic cell, viral immunology
Hepatitis B virus (HBV) infection is a major global health burden. Approximately 2 billion people are currently infected with HBV, and among them, more than 257 million are chronic HBV carriers (1, 2). Chronic hepatitis B is a risk for cirrhosis and hepatocellular carcinoma, and more than 700,000 people die from HBV-related diseases each year (3).
HBV is not considered cytopathic. The control of viral infection and the degree of liver damage depend on the complex interaction between viral replication and the host's immune response (1). Multiple mechanisms of host immune responses, including robust CD8+ T cell and neutralizing antibody responses, are extremely important for viral clearance (4, 5). However, these host immune responses are usually dysregulated in persistent HBV infection. The dysregulation mechanism is still not completely understood. One of the possible mechanisms by which HBV-infected hepatocytes interact and communicate with the host immune system and other uninfected cells is through extracellular vesicles (EVs) (6, 7).
EVs are secreted from various cell types to the extracellular milieu. EVs are either released directly from the plasma membrane or during fusion between multivesicular bodies (MVBs) and the plasma membrane (8, 9). EVs that are released from MVBs are termed exosomes. These nanoscale membrane-enclosed vesicles carry a variety of bio-macromolecules, such as proteins, mRNA, microRNA, and other noncoding RNAs (10, 11). Similar to EVs, viruses are released from the plasma membrane, through several pathways, and even through the MVB route (12). Therefore, it is suggested that some viruses can utilize the exosomal pathway for cell-to-cell spread to avoid the immune system surveillance (12). In recent years, it has been reported that the contents of EVs are modulated by viral infection (13), and EVs secreted from viral infected cells have the potential to control various immune responses. For example, EVs released from hepatitis C virus-infected cells promoted the proliferation of T follicular regulatory cells and regulated germinal center responses by limiting T follicular helper cells and B cells (14). Epstein Bar virus (EBV) encodes its own small RNAs packed into the EVs, which are then used to manipulate the immune system to establish a tumor microenvironment (15).
Recently, the role of EVs in the liver microenvironment in response to HBV infection have been reported. Kouwaki et al. (16) found that HBV infection of hepatocytes increased immunoregulatory microRNA levels in EVs to regulate innate immune responses to the virus. Yang et al. (17) demonstrated that exosomes circulating in the sera of chronic hepatitis B (CHB) patients contained both HBV nucleic acids and proteins. These exosomes ingested by NK cells played a role in NK-cell dysfunction thus in HBV transmission (17). Our previous work revealed that HBV-replicating cells secreting EVs targeted peripheral blood monocytes and significantly up-regulated programmed death 1 ligand-1 (PD-L1) protein expression, a critical immune regulatory molecule, and down-regulated CD69, a leukocyte activation molecule (18). However, these roles of EVs secreted from HBV-infected cells were studied only with in vitro experiments. Hence, the biological significance of EVs secreted from HBV-infected hepatocytes remains elusive, which was the aim of this study.
In this study, the role of EVs secreted from HBV-infected cells in the regulation of the viral infection was investigated in an in vivo system. We evidenced that EVs were immunosuppressive to inhibit the eradication of HBV-replicating cells in the hydrodynamic HBV transfection mouse model. Furthermore, EVs were systemically incorporated in multiple organs, including the liver, bone marrow (BM), and intestine. BM cells that incorporated EVs migrated to the intestine.
Result
Characterization of EVs secreted from HBV-replicating cells and -nonreplicating cells
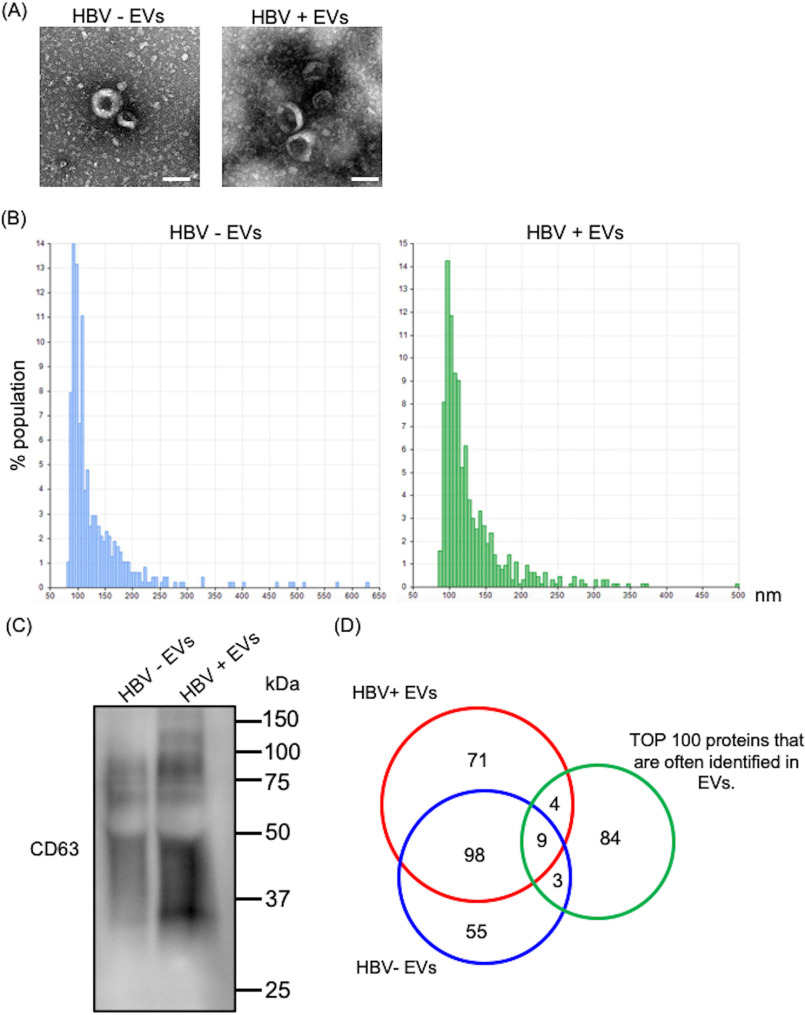
EVs were isolated from the culture supernatants of HBV-replicating (HepAD38) or HBV-nonreplicating (HepG2) cells. These EVs were characterized using transmission EM, nanoparticle tracking analysis, and LC–MS proteomic analysis. These EVs showed the typical exosome-like round morphology by transmission EM (Fig. 1A). The qNano analysis of these EVs indicated that the vesicles were ∼100 nm in diameter (Fig. 1B). The presence of CD63, which is common EV/exosome marker, was analyzed by Western blotting (Fig. 1C). 169 protein groups and 153 protein groups were identified in HBV-replicating and HBV-nonreplicating cells, respectively, using LC–MS proteomic analysis (Fig. 1D). The identified proteins information are listed in Tables S1 and S2. Of 169 protein groups detected in HBV-replicating cells EVs, 13 were present in the top 100 proteins that are often identified in EVs (from Vesiclepedia of EVs proteome). Moreover, of 153 protein groups detected in HBV-nonreplicating cells EVs, 12 were present in the top 100 proteins that are often identified in EVs (Fig. 1D).
Figure 1.
Characterization of EVs secreted from HepAD38 or HepG2 cells. A, electron microscope image of EVs isolated from HBV-replicating and -nonreplicating cells culture medium by ultracentrifugation. White bar indicates 100 μm. B, qNano analysis of EVs isolated from HBV-replicating and -nonreplicating cells culture medium. C, the presence of CD63 was analyzed by Western blotting (representative of three experiments). D, after LC–MS proteomic analysis of the EVs, 169 protein groups and 153 protein groups were identified in HBV-replicating and HBV-nonreplicating cells, respectively; 98 protein groups were common between EVs from HBV-replicating cells and EVs from HBV-nonreplicating cells.
EVs secreted from HBV-replicating cells had in vivo immunosuppressive effects
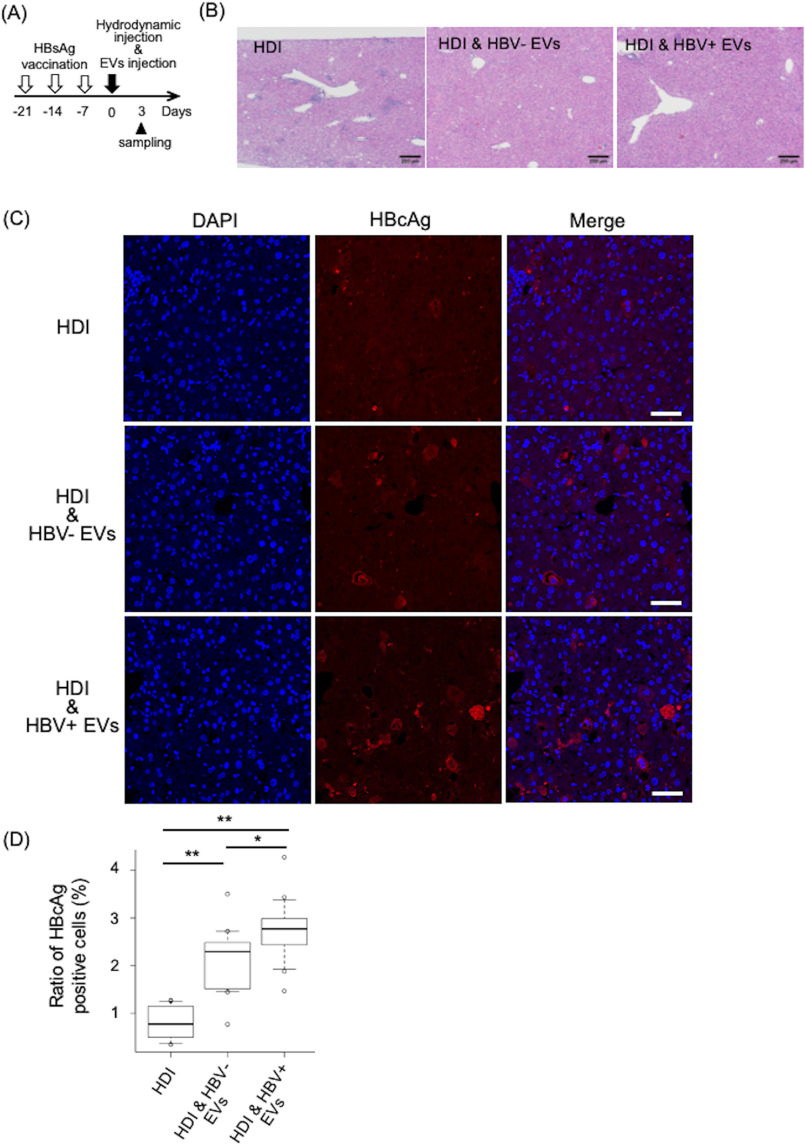
The in vivo function of EVs secreted from HBV-replicating cells was investigated with an in vivo mouse model of HBV transfection by using hydrodynamic injection (HDI). HDI is a technique that is used deliver nucleic acids into live mice. Delivery through this method results in the in vivo transfection of foreign DNA primarily in the liver. The HBV HDI mouse model is useful tool for studying the host immune response, the pathophysiology of HBV-related disease, and several antiviral drugs (19–21). Inflammation was induced by immunizing mice with a licensed aluminum-based recombinant HBV surface antigen (HBsAg) vaccine before introduction of HBV replicon plasmid, as HDI alone causes almost no liver inflammation (22). Timeline for the experiment of the HBV HDI mouse model is shown in Fig. 2A. 3 days after HDI, the histological examination revealed remarkably less infiltration of mononuclear cells in the liver of mice treated with EVs derived from HepAD38 cells regardless of HBV induction, as a difference to that observed in mice without the treatment (Fig. 2B). Moreover, the number of hepatocytes positive for the HBV core antigens (HBcAg) slightly increased in the liver of mice treated with the EVs derived from the HBV-induced HepAD38 than that observed without HBV induction (Fig. 2, C and D). Moreover, HBcAg-positive hepatocytes were barely detected in liver sections from mice without treatment (Fig. 2, C and D). The results suggested that HBV-replicating cells escaped from immune cells in the liver of the mice treated with EVs, especially after HBV induction, indicating that EVs derived from HepAD38 cells had immunosuppressive functions and that more activity was obtained by HBV replication.
Figure 2.
EVs secreted from HepAD38 cells had immunosuppressive effects. A, timeline for HBsAg immunization followed by HDI. B, HE staining and C, immunofluorescence of mouse liver sections obtained 3 days after injection of HDI alone, HDI, and HBV-EVs or HDI and HBV+ EVs. Sections were stained to detect HBcAg (red) and nuclei using DAPI (blue). D, HBcAg-positive cells were counted in 15 fields from 3 mice. Ratio to total cells were calculated. Statistical analysis was performed using one-way ANOVA and subsequent Tukey's HSD method. *, p < 0.05, **, p < 0.01. Black and white bars indicate 200 and 50 μm, respectively.
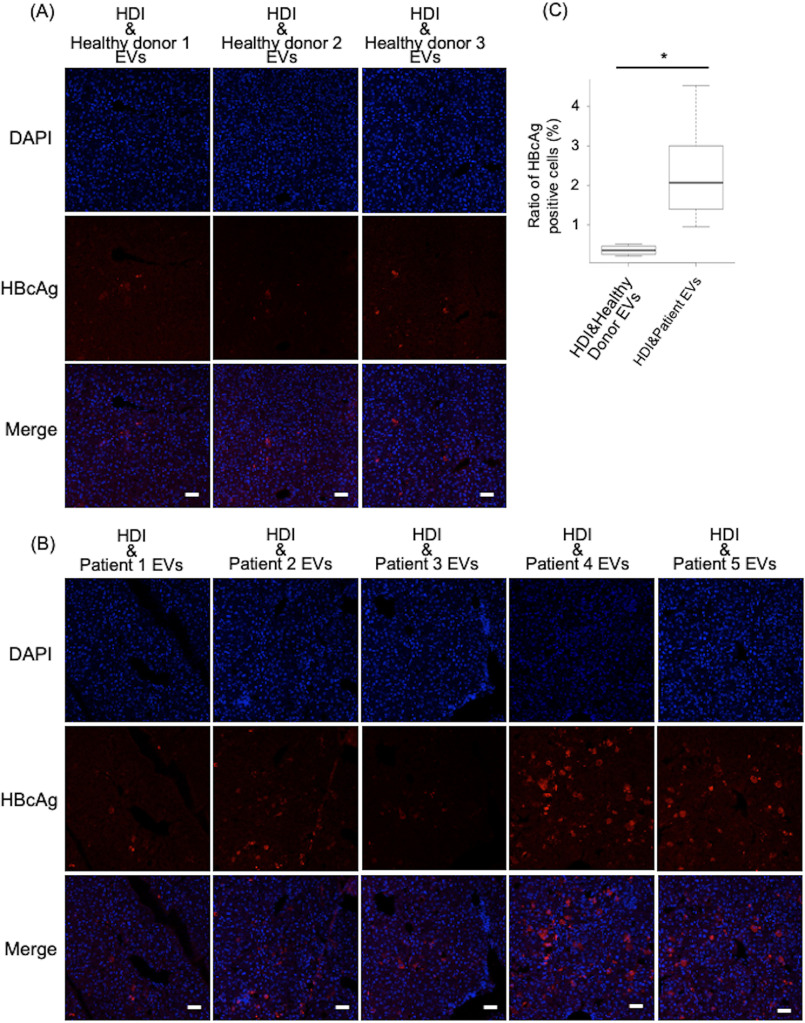
As EVs derived from tumor cells suppressed the immune responses (23–25), possibly EVs derived from the cells after HBV induction were masked by the tumor characteristics. Then to confirm that HBV infection itself caused EVs to exert immunosuppressive activity, the EVs collected from the sera of healthy donors and HBV patients was tested. These EVs were characterized using nanoparticle tracking analysis (Fig. S1). Clinical data of the patients are listed in the Table 1. Few if any HBcAg-positive hepatocytes were detected in the liver of the mice treated with EVs from healthy donors (Fig. 3A), whereas significant HBcAg-positive hepatocytes were observed in those of HBV patients (Fig. 3, B and C). Furthermore, HBcAg was not detected in the liver of mice treated with EVs derived from HBV patients alone without HDI (Fig. S2). These results suggested that HBV infection potentiated the EV immunosuppressive ability. Though the difference of the ability of the EVs secreted from HBV-replicating cells and nonreplicating cells was small, the data in Fig. 3 provides more evidence that the tiny change might be biologically relevant.
Table 1.
Clinical data of the patients
| Patient number | Sex | Age | HBV genotype | NRTIs treatmenta | HBV DNA (log copies/ml) | HBsAg IU/ml |
|---|---|---|---|---|---|---|
| 1 | M | 27 | NAb | No | NA | 45.7 |
| 2 | M | 21 | NA | No | NA | NA |
| 3 | M | 43 | NA | No | NA | NA |
| 4 | M | 26 | C | No | 9.32 | >500 |
| 5 | M | 34 | C | No | 8.94 | NA |
aNRTI, nucleotide reverse transcriptase inhibitors.
bNA, not abilable.
Figure 3.
EVs from HBV patients had immunosuppressive effects. A and B, immunofluorescence of mouse liver sections obtained 3 days after injection of HDI and healthy donors EVs injection or HDI and HBV patients EVs injection. Sections were stained to detect HBcAg (red) and nuclei using DAPI (blue). Two mice were used per each healthy donors EVs injection or HBV patients EVs injection group, and 5 fields were counted in each mouse. *, p < 0.05, Student's t test. White bars indicate 50 μm.
Still patient samples showed variable HBcAg positivity. Few HBcAg-positive hepatocytes were detected in the liver of the mice treated with EVs from one HBV patient (patient 1) with massive HBcAg-positive hepatocytes in patients 4 and 5. It may depend on the disease phases. The function of the EVs may be different between patients in the immune tolerant phase and patients in the immune active phase.
PD-L1 expression on dendritic cells in the liver-draining lymph node increased after EVs derived from an HBV patient were injected
Liver-draining lymph nodes (LNs) induced an anti-HBV–specific immune response responsible for HBV clearance in HBV plasmid-injected mice (26). Thus, we investigated liver-draining LNs in mice 3 days after HDI and EVs injection.
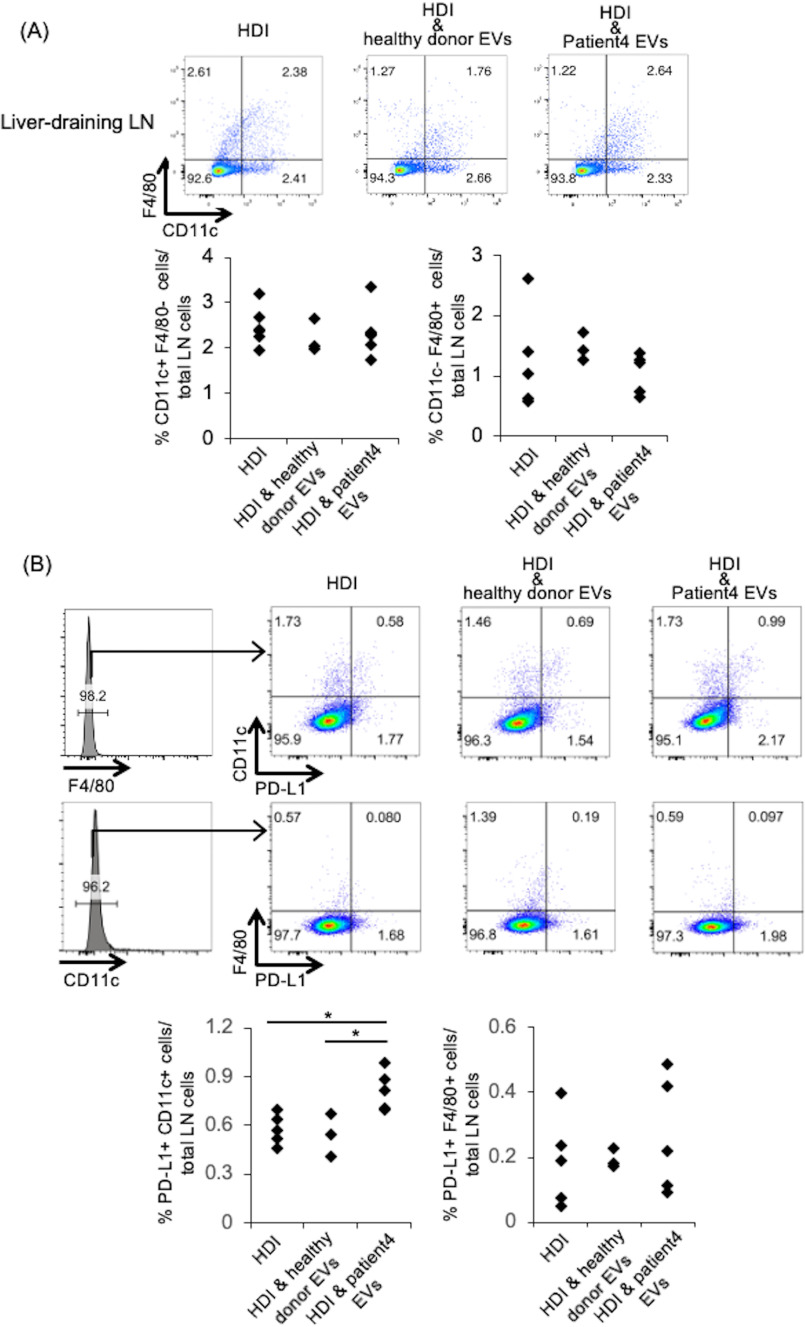
The percentages of CD3+ CD4+, CD3+ CD8+ T and B220+ B cells in liver-draining LNs in mice did not change after injection of EVs derived from an HBV patient (Figs. S3 and S4). Furthermore, the percentages of CD11c+ dendritic cells (DCs) and F4/80+ macrophages in liver-draining LNs in mice were no different after injection of EVs derived from an HBV patient (Fig. 4A). These results indicated that EVs derived from an HBV patient did not affect the ratio of immune cells in liver-draining LNs.
Figure 4.
The ratio of PD-L1+ CD11c+ DCs increased in liver-draining LN after injection of EVs from a HBV patient. Cells from liver-draining LN were isolated 3 days after HDI and analyzed. A, the frequencies of F4/80+ macrophages and CD11c+ DCs in mice treated with HDI alone, HDI and healthy donors EVs and HDI and HBV patients EVs were detected by flow cytometry (top). B, the ratio of PD-L1+ CD11c+ cells and PD-L1+ F4/80+ cells to total LN cells was investigated for all the above conditions (bottom). Statistical analysis was performed using one-way ANOVA and subsequent Tukey's HSD method. *, p < 0.05.
Previously, we reported that HBV-replicating cells secreting EVs targeted antigen presentation cells, which expressed almost no PD-L1 and significantly induced PD-L1 protein expression (18). Furthermore, in vitro studies have shown that in chronic HBV infection, blockade of the PD-1–PD-L1 axis can increase both the production of HBV antibodies (27) and the numbers and functionality of HBV-specific T cells (28, 29). Next, we investigated PD-L1 expression in liver-draining LN. The expression of PD-L1 in the liver-draining LN showed no difference in B cells (Fig. S5). CD3+ PD-L1+ T cells increased in the mice injected with HBV patient-derived EVs, compared with those with healthy donor-derived EVs (Fig. S5), Furthermore, the ratio of CD11c+ PD-L1+ DCs increased significantly in mice injected with HBV patient-derived EVs (Fig. 4B), compared with that in mice injected with healthy donor-derived EVs. The up-regulation of PD-L1 in CD11c+ DCs may be one of the mechanisms inducing suppression of CD4+ or CD8+ T cells.
EVs derived from HBV-replicating cells were systemically engulfed in mice
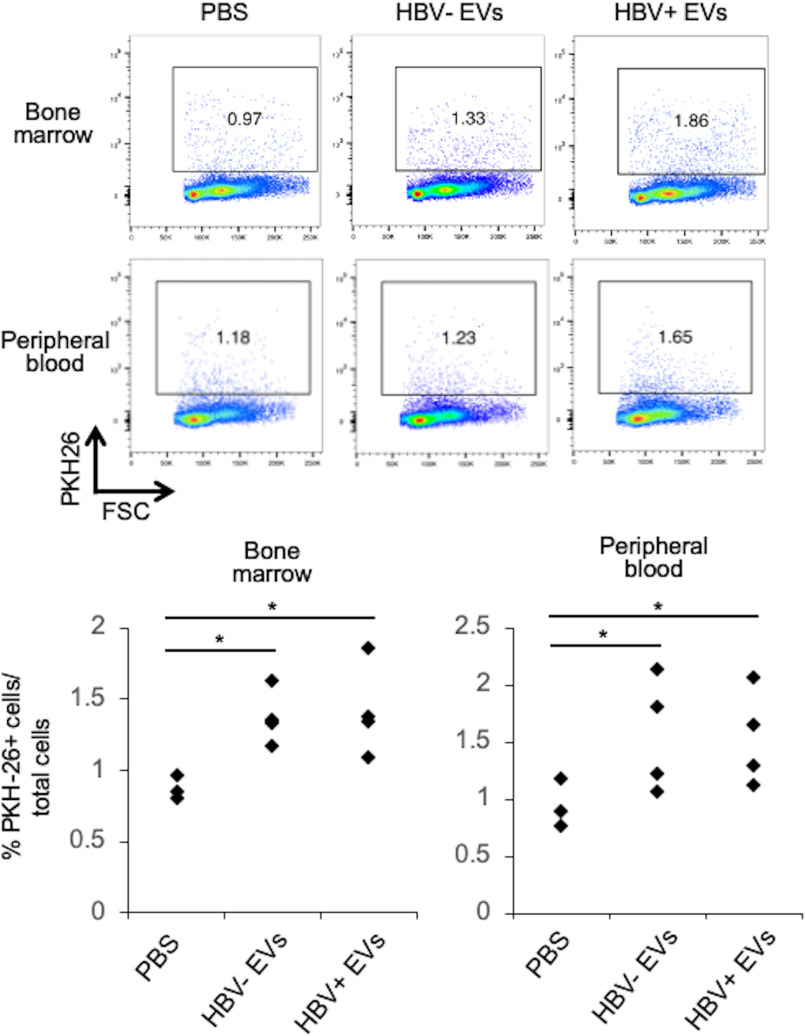
Previously, we evidenced that a significant amount of the EVs secreted by EBV-infected tumor cells were engulfed by local macrophages in the tumor microenvironment and bone marrow monocytes (15). Therefore, the systemic biodistribution of the EVs derived from HBV-replicating cells was examined. PKH-26–labeled EVs secreted from HepAD38 cells were injected in the tail vein of mice, and after 24 h, cryosections were prepared for each organ and observed with a confocal laser microscopy. According to Fig. S6, EVs mainly accumulated in liver, liver-draining LNs, and spleen. Also, cells that ingested EVs were detected in plasma and in particular, in the bone marrow (Fig. 5). These results suggested that EVs secreted from HBV-replicating cells were locally and systemically engulfed.
Figure 5.
EVs accumulated in bone marrow. Cells from BM and peripheral blood were isolated 24 h after injection of PKH-26–labeled EVs secreted from HepAD38 cells. The frequencies of PKH-26–positive cells were detected by flow cytometry.
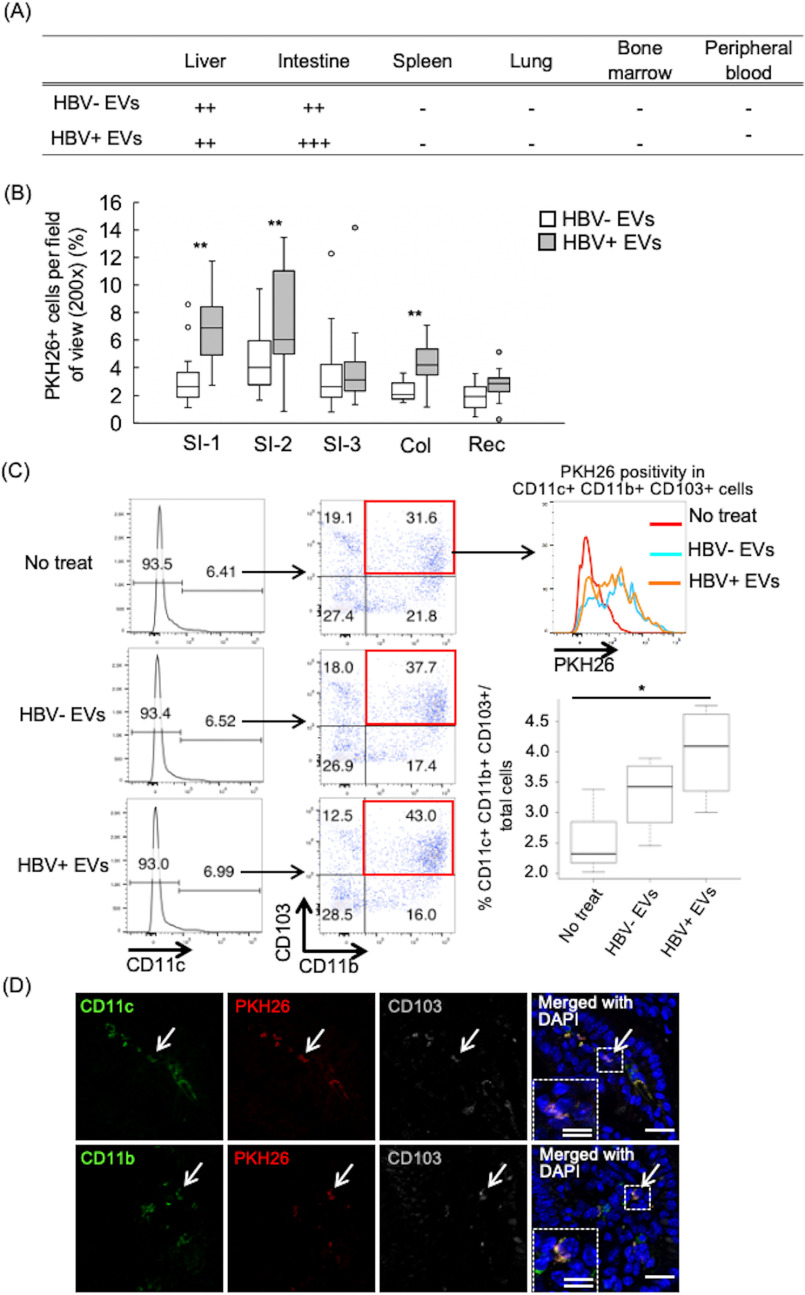
Bone marrow cells that incorporated EVs secreted from HBV-replicating cells acquired intestinal tropism
Later, we investigated the significance of EVs uptake in bone marrow. BM cells were collected from three mice. PKH-26-labeled EVs secreted from HBV-replicating cells or -nonreplicating cells were added into the BM cells culture. After 24 h, the entire BM cells including PKH-26–positive and –negative cells were transplanted into three recipient mice. After 24 h, cryosections were prepared for each organ and observed with a confocal laser microscopy. Transplanted BM cells with PKH-26–labeled EVs (BM-EVs) were detected in the liver and in particular, in the intestine (Fig. 6A), especially in the small intestine near the stomach and colon (Fig. 6B). Next, we investigated whether population of the bone marrow cells change by taking up EVs from HBV-replicating cells. The proportion of CD11b+ CD11c+ CD103+ cells to entire BM cells, immunoregulatory DC populations in the intestine, was increased after HBV-replicating cells derived EVs added into BM cells culture, and the majority of those cells were PKH-26 positive (Fig. 6C). Furthermore, CD11c+ CD103+ or CD11b+ CD103+ BM-EVs cells, which indicated the regulatory DC-like cells, were detected in the small intestine 24 h after transplanting BM-EVs (Fig. 6D). These results suggested existence of the association between the liver, bone marrow, and intestinal tract through EVs secreted from HBV-replicating cells and that bone marrow cells incorporating EVs secreted from HBV-replicating cells may be involved in intestinal immunity.
Figure 6.
Bone marrow cells that incorporated EVs secreted from HBV-replicating cells acquired intestinal tropism and the frequencies of CD11c+ CD11b+ CD103+ DCs increased after HBV replicating cells-derived EVs uptake. A, the entire BM cells treated by PKH-26–labeled EVs secreted from the HepAD38 cell with HBV replication were transplanted into recipient mice. PKH-26–positive cells were detected in each tissue 24 h after transplant. B, PKH-26–positive cells were counted in the small intestine, colon, and rectum. The small intestine was observed in three places from the near side from the stomach. SI-1, SI-2, and SI-3 indicate the positions of 5, 10, and 15 cm from the stomach, respectively. Three mice were treated per group, and 10 fields were counted in each mouse. **, p < 0.01, Student's t test. C, a total of 10 μg of PKH-26–labeled EVs secreted from HepAD38 cell with HBV replication were added into bone marrow cells culture. After 24 h, the CD11c+ CD11b+ CD103+ were analyzed by flow cytometry and the frequency of those cells in the whole bone marrow cells were calculated and plotted in a graphic. Statistical analysis was performed using one-way ANOVA and subsequent Tukey's HSD method, *, p < 0.05. D, immunofluorescence of mouse small intestine sections obtained 24 h after injection of BM cells incorporating PKH-26–labeled EVs secreted from HepAD38 cells. Sections were stained to show CD11b, CD11c, CD103, nuclei using DAPI. PKH26+ CD11c+ CD103+ cells and PKH26+ CD11b+ CD103+ cells are indicated by white arrows. Dotted areas are shown at higher magnifications and are indicated by white arrows with a broken line. White single and double bars indicate 20 and 10 μm, respectively.
Discussion
In this study, we showed that the EVs secreted from HBV-replicating hepatocytes had immunosuppressive functions that inhibit the eradication of HBV-replicating hepatocytes. Furthermore, we identified a novel axis of liver–BM–gut mediated by the EVs from HBV-replicating cells, which possibly exerts critical functions in chronic hepatitis B.
One important consideration in this type of studies is the method of EVs preparation, because it significantly affects its heterogenous composition. The EVs obtained in this study were collected by ultracentrifugation, which is still a gold standard method. The fraction of the EVs included incomplete and complete forms of the HBV virus, except for exosomes (18). As HBV does not infect rodents, the presence of HBcAg observed in the liver of mice treated the EVs derived from HBV-replicating hepatocytes was not due to extra HBV infection. The incomplete form mainly consisted of two types. The first type was the classical HBsAg spheres and filaments of 20 nm in diameter (HBsAg particles). These subviral particles were composed of only the viral surface proteins and were found in the blood of infected individuals at up to 100,000-fold in excess over the complete virions (at 1014/ml) (30). The second class is empty genome-free virions, which contain the surface proteins enclosing the viral capsid but no genome, and are typically found at 100-fold higher levels over the complete virions in the blood of infected individuals (31, 32). In CHB patients, free HBsAg particles bind to anti-HBs antibodies, working as “decoy” for HBV-infected hepatocyte to escape from immune attack. In contrast, as the hydrodynamic HBV transfection model induces immune reaction by vaccination of HBsAg, the HBsAg on the subviral particles should induce an immunoreactive reaction. Previously we reported that subviral particles strongly induced PD-L1 in monocytes (18). Accordingly, the function of the subviral particles might contribute to the immunosuppressive functions of the EVs.
The results on EVs in HBV infection obtained using only the in vitro experiments are still controversial. Kowaki et al. (16) reported that exosomes induce NKG2D ligand expression in macrophages by stimulating MyD88, TICAM-1, and MAVS-dependent pathways, whereas the HBV-infected hepatocytes increase the levels of immunomodulatory microRNAs in EVs and exosomes, which migrate to macrophages and suppress IL-12p35 mRNA expression to counteract the innate immune response of the host. Yang et al. (17) demonstrated that exosomes circulating in the sera of CHB patients ingested by NK cells were shown to play a role in NK-cell dysfunction, and thus also in HBV transmission. According to our results, and for the first time we showed that EVs from HBV-replicating cells were immunosuppressive to persistence HBV infection with in vivo experiments.
HBV-specific T cells are exhausted in chronic HBV infection. It has been reported that not only PD-1 but also various immune checkpoint molecules such as CTLA-4 (33), TIM-3 (34), and LAG-3 (35) are involved in HBV-specific T cells exhaustion. In our experiments using the HDI mouse model, EVs secreted from HBV-replicating cells induced PD-L1 up-regulation on DCs in liver-draining LN, but the difference was small. This suggests that the immunosuppressive effect of EVs secreted from HBV-replicating cells may be related not to the PD-1–PD-L1 axis but to other immune checkpoint molecules or another mechanism. Hence, further study is needed in the future.
EVs incorporated by BM cells specifically migrated to proximal intestine and colon. Recently it was reported that stromal and dendritic cell gene signatures and polarization of T cells against the same luminal antigen differed between gut LNs, with the proximal small intestine-draining gut LNs preferentially producing tolerogenic responses and the distal gut LNs stimulating pro-inflammatory T cell responses (36). The dendritic cells that engulfed EVs in the proximal intestine might enhance immune regulatory functions of the gut, whereas those in the colons might affect the gut flora, which has been reported to be critical for onset of CHB (37). Therefore, immunoregulatory function of the EVs through the liver–BM–gut axis might affect the gut microbiota or intestinal immunity.
In summary, the EVs derived from HBV-infected cells exerted immunosuppressive functions and the association between liver, bone marrow, and intestinal tract through EVs secreted from HBV-infected cells existed. However, further investigation is needed for the elucidation of its biological significance.
Experimental procedures
Ethics statement
This study was performed in strict accordance with the ethical guidelines of the Declaration of Helsinki and use of human material was approved by the local ethics committees of Tokai University. Sera samples were prepared from healthy donor and HBV patients after written informed consent. Studies on mice were conducted in accordance with both the Guidelines for Animal Experimentation of the Japanese Association for Laboratory Animal Science and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Cells
HepAD38 cells that were derived from the HepG2 cell line and support tetracycline-inducible HBV replication were kindly provided by Dr. Watashi (38). HepAD38 cells were used as HBV-replicating cells, and HepG2 cells were used as HBV-nonreplicating cells in this study. HepAD38 cells were cultured in DMEM/F-12 (Life Technologies) and HepG2 were cultured in DMEM high glucose (Life Technologies) supplemented with 10% doxycycline-free fetal bovine serum (FBS), 100 units/ml of penicillin, and 100 µg/ml of streptomycin (Life Technologies) at 37 °C humidified air that contained 5% CO2. Furthermore, maintenance of HBV replication in HepAD38 cells was maintained without doxycycline.
We used healthy human blood samples. PBMCs were isolated from heparinized whole blood of healthy individuals using lymphoprep (Axis-Shield, Oslo, Norway). Isolated PBMCs were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% (v/v) FBS, 100 units/ml of penicillin, and 100 mg/ml of streptomycin in 24-well–plates.
BM cells were collected from C57BL/6j mice (6 week) purchased from CLEA Japan (Tokyo, Japan). The animals were kept in the Tokai University Laboratory Animal Center in specific pathogen-free conditions. All mice were used according to guidelines for experimental animal use specified by the Tokai University. Collected BM cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) FBS, 100 units/ml of penicillin, and 100 mg/ml of streptomycin in 6- or 24-well–plates.
EVs isolation
Sera samples were prepared from healthy donor and HBV patients using standard operating procedures (39). EVs were isolated from cell culture supernatant and healthy donor and HBV patients' serum samples. To remove bovine EVs from the FBS, FBS was ultracentrifuged at 110,000 × g for 18 h at 4 °C using a Type 70.1 Ti (Beckman Coulter, Brea, CA). The supernatant was filtered through a 0.22-μm filter (Merck Millipore, KGaA, Darmstadt, Germany). HepG2 cells and HepAD38 cells with HBV replication were cultured in EV-free DMEM high glucose medium or DMEM/F-12 medium for 3 days. Culture supernatants and serum were centrifuged at 1500 × g for 15 min at room temperature. To thoroughly remove cellular debris, the supernatants were filtered through a 0.22-μm filter. For EV preparation, the filtered supernatants were ultracentrifuged at 110,000 × g for 70 min at 4 °C. The pellets were washed with 8 ml of PBS after ultracentrifugation and resuspended in PBS. Collected EVs were labeled with PKH26 dye (Sigma-Aldrich). Then, the amount of protein in EVs were measured by using BCA protein assay kit (TAKARA, Shiga, Japan). Electron microscope image of EVs were obtained by transmission EM JEM-1400 (JEOL, Tokyo, Japan). The measurement of size and distribution was based on tunable resistive pulse sensing and carried out using a qNano Gold system (Izon Science Ltd, Christchurch, New Zealand), which combined the tunable nanopores with proprietary data capture and analysis software (Izon Control Suite version 3.3.2.2001; Izon Science).
Western blotting analysis
EV pellets were lysed in RIPA assay buffer (Wako) for 30 min on ice. Protein samples were electrophoresed on a 12% SDS–polyacrylamide gel and blotted onto polyvinylidene difluoride membranes (Bio-Rad Laboratories). The blots were blocked with PVDF Blocking Reagent (TOYOBO, Osaka, Japan) for 1 h at room temperature. The blocked membranes were incubated with the appropriate primary antibodies diluted in Can Get Signal solution 1 (TOYOBO) for 1 h at room temperature, followed by incubation with horseradish peroxidase (HRP)-conjugated corresponding secondary antibodies diluted in Can Get Signal solution 2 (TOYOBO) for 1 h at room temperature. Subsequently, the blots were incubated with Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) for a few seconds, and chemiluminescence was detected using the ChemiDoc Touch system (Bio-Rad). The following antibodies were used: mouse anti-CD63 (Santa Cruz Biotechnology, Santa Cruz, CA) and sheep anti-mouse IgG HRP (Sigma).
Animal experiments
C57BL/6j mice (6 weeks old) were used. For HDI, mice were transferred to a biosafety level 3 laboratory in Soka University after gaining permission from the Soka University Committee. Recombinant HBsAg vaccine (MSD, Tokyo, Japan) was administered three times to mice by intraperitoneal injections of 0.5 mg of HBsAg. HDI was performed according to the procedure of Liu et al. (40). Briefly, 15 μg of plasmid DNA HBV-pPB (41) was injected into the tail vein within 5-7 s in a volume of saline equivalent to 10% of the body weight. 40 μg of EVs were injected into the tail vein 2 h after HDI. Liver tissue samples were collected 3 days after HDI and fixed in 4% paraformaldehyde for histologic analyses. Liver-drainning LNs were collected and used for FACS analysis.
For examination of the systemic biodistribution of EVs, 40 μg of PKH26-labeled EVs were injected in the tail vein. After 24 h, liver, liver-draining LNs, spleen, and intestine were collected and fixed in 4% paraformaldehyde for histologic analyses. PBMCs and BM cells were collected and used for FACS analysis.
For investigating the significance of EVs uptake in bone marrow, BM cells were collected from C57BL/6j mice (6 weeks old). PKH-26–labeled EVs secreted from HepAD38 cells with HBV replication were added into the BM cells culture. After 24 h, whole BM cells were transplanted into recipient mice. PKH-26–positive cells were detected in each tissue 24 h after transplant.
Protein identification by LC–MS
EVs samples dissolved with EzApply (ATTO, Tokyo, Japan) were heat denatured and then subjected to SDS-PAGE. The gel was stained with EzStain Silver (ATTO). Gel lanes were excised and cut into 20 slices per lane. Every gel slice was decolorized using a solution of 30 mm potassium ferricyanide and 100 mm sodium thiosulfate, washed with 30% acetonitrile, and then subjected to in-gel trypsin (cutting at C-terminal of lysine and arginine) digestion. Extracted tryptic peptides were then purified using C18 StageTip according to a previous report (42). The purified peptide mixture was separated by a prominence nano-liquid chromatography system (Shimadzu, Kyoto, Japan). The sample was first loaded onto the trap column L-column micro ODS (CERI's, Tokyo, Japan) at a flow rate of 0.04 ml/min with mobile phase C (water, 0.1% formic acid) for 6 min, then the trapped samples were separated in a C18 picofrit column (100-μm internal diameter, 5-μm bead size, Nikkyo Technos Co., Tokyo, Japan). The HPLC gradient was 2–40% mobile phase B (acetonitrile/water, 95/5, 0.1% formic acid) in mobile phase A (acetonitrile/water, 2/98, 0.1% formic acid) at a flow rate of 300 nl/min for 60 min. Data-dependent acquisitions were carried out on a LC–MS-IT-TOF (Shimadzu) equipped with a nano electrospray ionization (nESI) source in positive ion mode. The scan range was 500–1500 m/z. Up to 3 precursor ions were selected for collision-induced dissociation fragmentation. Peak lists were generated using LCMSsolusion version 3 (Shimadzu). The threshold score for accepting individual spectra was set at 25. The resulting data were submitted onto the Mascot search engine (http://www.matrixscience.com; Mascot Server ver2.4.01) where a search was performed on the Swiss-Prot database (20,366 entries; 1-2020) to determine candidate peptides. Up to 1 missed cleavage was permitted. Mass tolerance for the precursor ion and its fragment ions was set to 0.05 Da. Meanwhile, fixed modification was set to the carbamidomethyl of cysteine; and variable modification was set to oxidation of methionine.
For statistical evaluation of the date obtained, false discovery rate (FDR) was used. The FDR was calculated by searching candidates in reversed protein database. Protein ID with FDR estimate of < 9% were accepted as valid IDs.
Histologic analysis
For histologic analyses, fixed tissues were embedded in paraffin or optimum cutting temperature compound (Sakura Finetechnical, Tokyo, Japan) as previously described (43). Paraffin-embedded tissues were sectioned and stained with hematoxylin-eosin. For immunostaining, frozen sections were stained using the anti-HBcAg (Thermo Fisher Scientific, Waltham, MA), FITC anti-mouse CD11b (Biolegend, San Diego, CA), FITC anti-mouse CD11c (Biolegend), and anti-mouse biotin-conjugated CD103 (Invitrogen) antibodies. Nuclei were visualized with DAPI (H-1200; Vector Laboratories, Burlingame, CA). As a secondary antibody, anti-rabbit IgG H&L Alexa Fluor 594 (abcam) and APC streptavidin (Biolegend) was used. Stained sections were mounted with gold antifade reagent (Invitrogen) and examined using the Zeiss LSM700 scanning laser confocal microscope.
Flow cytometry
Fluorescently labeled antibodies and reagents were purchased from Biolegend. Cells were stained with the indicated monoclonal antibodies for surface antigens according to a standard protocol. The antibodies used were FITC anti-mouse PD-1, FITC anti-mouse CD11c, PE anti-mouse CD8α, PE anti-mouse PD-L1, APC anti-mouse CD4, APC anti-mouse B220, APC anti-mouse F4/80, and APC-Cy7 anti-mouse CD3. Flow cytometric analysis was performed on a FACS Verse flow cytometer (BD Bioscience), and the data were analyzed using FlowJo software.
Statistical analysis
Date were analyzed by R. Differences between means were evaluated using one-way ANOVA and subsequent Tukey's HSD method for multiple comparisons or Student's t test for the analysis of the significance between two groups. Values of p < 0.05 were considered significant. Data are presented as the mean ± S.D.
Date availability
Mass data files (.mzXML) were deposited and made publicly available with jPOSTrepo, which is an international standard date repository for proteomes (44) for HepAD38 EVs and Immunosuppression via extracellular vesicles in hepatitis B 12 HepG2 EVs and assigned the jPOST ID JPST000781 and ProteomeXchange PXD018292.
Supplementary Material
Acknowledgments
We thank the Support Center for Medical Research and Education, Tokai University, for technical assistance.
This article contains supporting information.
Author contributions—M. K. and A. K. conceptualization; M. K., M. O., K. K., H. D. K., M. M., and T. K. resources; M. K. and M. I. data curation; M. K. formal analysis; M. K. and A. K. funding acquisition; M. K. and Y. Y. validation; M. K., Y. Y., and M. I. investigation; M. K. and Y. Y. visualization; M. K., K. K., M. M., and A. K. methodology; M. K. and A. K. writing-original draft; M. K. and A. K. project administration; M. K., Y. Y., M. O., K. K., M. I., H. D. K., M. M., T. K., and A. K. writing-review and editing.
Funding and additional information—This study was supported by 2019 Tokai University School of Medicine Research Aid (to M. K.), JSPS KAKENHI Grant JP18K15131 (to M. K.), and Research Program on Hepatitis from Japan Agency for Medical Research and Development Grant 19fk0210054s0201 (to A. K.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- HBV
- hepatitis B virus
- EVs
- extracellular vesicles
- BM
- bone marrow
- MVBs
- multivesicular bodies
- EBV
- Epstein Bar virus
- CHB
- chronic hepatitis B
- PD-L1
- programmed death ligand-1
- HDI
- hydrodynamic injection
- HBsAg
- hepatitis B surface antigen
- HBcAg
- hepatitis B core antigen
- LNs
- lymph nodes
- DCs
- dendritic cells
- HRP
- horseradish peroxidase
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- PBMC
- peripheral blood mononuclear cell
- FDR
- false discovery rate
- DAPI
- 4′,6-diamidino-2-phenylindole
- APC
- allophycocyanin.
References
- 1. Liaw Y. F., and Chu C. M. (2009) Hepatitis B virus infection. Lancet 373, 582–592 10.1016/S0140-6736(09)60207-5 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization. Global Hepatitis Report (2017) WHO, Geneva [Google Scholar]
- 3. Tuaillon E., Lozano C., Kuster N., Poinso A., Kania D., Nagot N., Mondain A. M., Pageaux G. P., Ducos J., Van de Perre P., and Reynes J. (2012) Long-term hepatitis B virus surface antigen decay in HIV-1/hepatitis B virus-coinfected adults initiating a tenofovir-containing regimen. J. Clin. Microbiol. 50, 3096–3098 10.1128/JCM.00971-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dandri M., and Locarnini S. (2012) New insight in the pathobiology of hepatitis B virus infection. Gut 61, i6–i17 10.1136/gutjnl-2012-302056 [DOI] [PubMed] [Google Scholar]
- 5. Guidotti L. G., Isogawa M., and Chisari F. V. (2015) Host-virus interactions in hepatitis B virus infection. Curr. Opin. Immunol. 36, 61–66 10.1016/j.coi.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson M. R., Kashanchi F., and Jacobson S. (2016) Exosomes in viral disease. Neurotherapeutics 13, 535–546 10.1007/s13311-016-0450-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raab-Traub N., and Dittmer D. P. (2017) Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 15, 559–572 10.1038/nrmicro.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raposo G., and Stoorvogel W. (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kowal J., Tkach M., and Thery C. (2014) Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125 10.1016/j.ceb.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 10. Jia S., Zocco D., Samuels M. L., Chou M. F., Chammas R., Skog J., Zarovni N., Momen-Heravi F., and Kuo W. P. (2014) Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Rev. Mol. Diagn. 14, 307–321 10.1586/14737159.2014.893828 [DOI] [PubMed] [Google Scholar]
- 11. Imani Fooladi A. A., and Mahmoodzadeh Hosseini H. (2014) Biological functions of exosomes in the liver in health and disease. Hepat. Mon. 14, e13514 10.5812/hepatmon.13514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meckes D. G., Jr. (2015) Exosomal communication goes viral. J. Virol. 89, 5200–5203 10.1128/JVI.02470-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jia X., Chen J., Megger D. A., Zhang X., Kozlowski M., Zhang L., Fang Z., Li J., Chu Q., Wu M., Li Y., Sitek B., and Yuan Z. (2017) Label-free proteomeic analysis of exosomes derived from inducible hepatitis B virus-replicating HepAD38 cell line. Mol. Cell. Proteomics 16, S144–S160 10.1074/mcp.M116.063503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cobb D. A., Kim O. K., Golden-Mason L., Rosen H. R., and Hahn Y. S. (2018) Hepatocyte-derived exosomes promote T follicular regulatory cell expansion during hepatitis C virus infection. Hepatology 67, 71–85 10.1002/hep.29409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higuchi H., Yamakawa N., Imadome K. I., Yahata T., Kotaki R., Ogata J., Kakizaki M., Fujita K., Lu J., Yokoyama K., Okuyama K., Sato A., Takamatsu M., Kurosaki N., Alba S. M., et al. (2018) Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood 131, 2552–2567 10.1182/blood-2017-07-794529 [DOI] [PubMed] [Google Scholar]
- 16. Kouwaki T., Fukushima Y., Daito T., Sanada T., Yamamoto N., Mifsud E. J., Leong C. R., Tsukiyama-Kohara K., Kohara M., Matsumoto M., Seya T., and Oshiumi H. (2016) Extracellular vesicles including exosomes regulate innate immune responses to hepatitis B virus infection. Front. Immunol. 7, 335 10.3389/fimmu.2016.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y., Han Q., Hou Z., Zhang C., Tian Z., and Zhang J. (2017) Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell. Mol. Immunol. 14, 465–475 10.1038/cmi.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kakizaki M., Yamamoto Y., Yabuta S., Kurosaki N., Kagawa T., and Kotani A. (2018) The immune function of extracellular vesicles in hepatitis B virus-infected hepatocytes. PLoS ONE 13, e0205886 10.1371/journal.pone.0205886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ketzinel-Gilad M., Zauberman A., Nussbaum O., Shoshany Y., Ben-Moshe O., Pappo O., Felig Y., Ilan E., Wald H., Dagan S., and Galun E. (2006) The use of the hydrodynamic HBV animal model to study HBV biology and anti-viral therapy. Hepatol. Res. 34, 228–237 10.1016/j.hepres.2006.01.008 [DOI] [PubMed] [Google Scholar]
- 20. Yang D., Liu L., Zhu D., Peng H., Su L., Fu Y. X., and Zhang L. (2014) A mouse model for HBV immunotolerance and immunotherapy. Cell. Mol. Immunol. 11, 71–78 10.1038/cmi.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tzeng H. T., Tsai H. F., Chyuan I. T., Liao H. J., Chen C. J., Chen P. J., and Hsu P. N. (2014) Tumor necrosis factor-alpha induced by hepatitis B virus core mediating the immune response for hepatitis B viral clearance in mice model. PLoS ONE 9, e34420 10.1371/journal.pone.0103008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsumoto T., Takahashi K., Inuzuka T., Kim S. K., Kurosaki T., Kawakami S., Chiba T., Seno H., and Marusawa H. (2017) Activation of TNF-α-AID axis and co-inhibitory signals in coordinateon with Th1-type immunity in a mouse model recapitulating hepatitis B. Antiviral. Res. 139, 138–145 10.1016/j.antiviral.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 23. Clayton A., Mitchell J. P., Court J., Mason M. D., and Tabi Z. (2007) Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 67, 7458–7466 10.1158/0008-5472.CAN-06-3456 [DOI] [PubMed] [Google Scholar]
- 24. Wieckowski E. U., Visus C., Szajnik M., Szczepanski M. J., Storkus W. J., and Whiteside T. L. (2009) Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor- reactive activated CD8+ T lymphocytes. J. Immunol. 183, 3720–3730 10.4049/jimmunol.0900970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gutiérrez-Vázquez C., Villarroya-Beltri C., Mittelbrunn M., and Sánchez-Madrid F. (2013) Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 251, 125–142 10.1111/imr.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng M., Yu J., and Tian Z. (2013) Characterization of the liver-draining lymph nodes in mice and their role in mounting regional immunity to HBV. Cell. Mol. Immunol. 10, 143–150 10.1038/cmi.2012.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salimzadeh L., Le Bert N., Dutertre C. A., Gill U. S., Newell E. W., Frey C., Hung M., Novikov N., Fletcher S., Kennedy P. T., and Bertoletti A. (2018) PD-1 blockade partially recovers dysfunctional virus-specific B cells in chronic hepatitis B infection. J. Clin. Invest. 128, 4573–4587 10.1172/JCI121957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boni C., Fisicaro P., Valdatta C., Amadei B., Di Vincenzo P., Giuberti T., Laccabue D., Zerbini A., Cavalli A., Missale G., Bertoletti A., and Ferrari C. (2007) Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 81, 4215–4225 10.1128/JVI.02844-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fisicaro P., Valdatta C., Massari M., Loggi E., Biasini E., Sacchelli L., Cavallo M. C., Silini E. M., Andreone P., Missale G., and Ferrari C. (2010) Anti-viral intrahepatic T-cell responses can be restored by blocking programmed death-1 pathway in chronic hepatitis B. Gastroenterology 138, 682–693, e1–e4 10.1053/j.gastro.2009.09.052 [DOI] [PubMed] [Google Scholar]
- 30. Blumberg B. S. (1977) Australia antigen and the biology of hepatitis B. Science 197, 17–25 10.1126/science.325649 [DOI] [PubMed] [Google Scholar]
- 31. Ning X., Nguyen D., Mentzer L., Adams C., Lee H., Ashley R., Hafenstein S., and Hu J. (2011) Secretion of genome-free hepatitis B virus: single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 7, e1002255 10.1371/journal.ppat.1002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luckenbaugh L., Kitrinos K. M., Delaney W. E., and Hu J. (2015) Genome-free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J. Viral Hepat. 22, 561–570 10.1111/jvh.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao H., Zhang R., and Zhang W. (2018) CTLA-4 interferes with HBV-specific T cell immune response. Int. J. Mol. Med. 42, 703–712 10.3892/ijmm.2018.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mohammadizad H., Shahbazi M., Hasanjani Roushan M. R., Soltanzadeh-Yamchi M., and Mohammadnia-Afrouzi M. (2019) TIM-3 as a marker of exhaustion in CD8+ T cells of active hepatitis B patients. Microb. Pathog. 128, 323–328 10.1016/j.micpath.2019.01.026 [DOI] [PubMed] [Google Scholar]
- 35. Ye B., Li X., Dong Y., Wang Y., Tian L., Lin S., Liu X., Kong H., and Chen Y. (2017) Increasing LAG-3 expression suppresses T-cell function in chronic hepatitis B: a balance between immunity strength and liver injury extent. Medicine (Baltimore) 96, e5275 10.1097/MD.0000000000005275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Esterhazy D., Canesso M. C. C., Mesin L., Muller P. A., de Castro T. B. R., Lockhart A., ElJalby M., Faria A. M. C., and Mucida D. (2019) Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 569, 126–130 10.1038/s41586-019-1125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang Y., and Cai Y. (2017) Gut microbiota and hepatitis-B-virus-induced chronic liver disease: implications for faecal microbiota transplantation therapy. J. Hosp. Infect. 96, 342–348 10.1016/j.jhin.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 38. Ogura N., Watashi K., Noguchi T., and Wakita T. (2014) Formation of covalently closed circular DNA in Hep38.7-Tet cells, a tetracycline inducible hepatitis B virus expression cell line. Biochem. Biophys. Res. Commun. 452, 315–321 10.1016/j.bbrc.2014.08.029 [DOI] [PubMed] [Google Scholar]
- 39. Tuck M. K., Chan D. W., Chia D., Godwin A. K., Grizzle W. E., Krueger K. E., Rom W., Sanda M., Sorbara L., Stass S., Wang W., and Brenner D. E. (2009) Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J. Proteome Res. 8, 113–117 10.1021/pr800545q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu F., Song Y., and Liu D. (1999) Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 10.1038/sj.gt.3300947 [DOI] [PubMed] [Google Scholar]
- 41. Kim H. Y., Park G. S., Kim E. G., Kang S. H., Shin H. J., Park S., and Kim K. H. (2004) Oligomer synthesis by priming deficient polymerase in hepatitis B virus core particle. Virology 322, 22–30 10.1016/j.virol.2004.01.009 [DOI] [PubMed] [Google Scholar]
- 42. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 43. Kakizaki M., Kashiwazaki H., and Watanabe R. (2014) Mutant murine hepatitis virus-induced apoptosis in the hippocampus. Jpn. J. Infect. Dis. 67, 9–16 10.7883/yoken.67.9 [DOI] [PubMed] [Google Scholar]
- 44. Okuda S., Watanabe Y., Moriya Y., Kawano S., Yamamoto T., Matsumoto M., Takami T., Kobayashi D., Araki N., Yoshizawa A. C., Tabata T., Sugiyama N., Goto S., and Ishihama Y. (2017) jPOSTrepo: an international standard data repository for proteomes. Nucleic Acid Res. 45, D1107–D1111 10.1093/nar/gkw1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.