Abstract
Rabies virus infection is an endemic disease which remains central to public health issues. The presence of epigenetics associated with the over-expression of B7-H1 in mice brain infected with rabies virus was investigated for the first time. A significant increase (p < 0.05) in mRNA level of B7-H1 as the disease progressed was observed. The percentage of methylated region was significantly (p < 0.05) higher in infected tissues relative to uninfected. DNA methyltransferase (DNMT) and histone acetylase (HAT) activities were also significantly (p < 0.05) higher in most infected brain tissues. HAT had a relatively higher proportion than DNMT when compared to the normal. Paradoxically, it can be inferred that the rabies virus uses epigenetic mechanisms as a means of manipulating host genes, as there was an increase in global DNMT and HAT activities with concomitant increase in B7-H1 promoter methylation and expression.
Keywords: Rabies virus infection, B7-H1, Promoter region, DNA methylation, Histone acetylation
Rabies virus (RABV) causes about 55,000 human deaths globally each year and the vast majority of deaths (84%) occur in rural areas [25]. It is an endemic disease which remains an important public health issue in Nigeria and other African countries [18]. The rabies virus is a bullet shaped negative single stranded Ribonucleic Acid (RNA) virus and a member of the genus Lyssavirus in the Rabdoviridae family. It is neurotropic and causes fatal encephalitis in mammals [22]. The virus consists of 12 kb strand RNA genome which encodes for five different proteins [2]. The virus is usually transmitted by the saliva of an infected animal after bites or scratches [18]. It then enters the central nervous system (CNS) through retrograde transport and undergoes rapid viral replication from where it reaches the salivary glands and excreted in saliva, at this stage it can be transmitted to a new host [4]. Once the disease is established in the brain, it leads to nearly 100% mortality rate in humans if timely post-exposure prophylaxis is not administered [22]. The rabies virus (RABV) has developed an immuno-subversive strategy by up-regulating the expression of molecules such as B7-H1 [13]. B7 homolog 1 (B7-H1) also known as programmed death ligand-1(PD-L1) or CD274 is a B7 family member that inhibits T-cell activation and cell-mediated toxic function of T cells. It is a surface glycoprotein known to be expressed on macrophages, immune privileged sites such as maternal-foetal barrier and on a majority of tumor cells [13]. It is up-regulated on rabies infected cells, thereby initiating a negative feedback mechanism that ultimately leads to T-cell exhaustion in infiltrating T-cells. The B7-H1 molecule binds to its receptor, programmed death 1 (PD-1) on T cells, limiting the activity of the T-cell receptor (TCR) [1]. Epigenetic modifications such as gene promoter methylation has been shown to be associated with B7-H1 expression in several diseases [7] except that in RABV infection, it has not been reported. Epigenetics is used to describe those mechanisms that are able to modify the expression levels of selected genes without necessarily altering their DNA sequence based on the variability of writers, erasers and readers.[8]. Epigenetic regulation is crucial during early development and differentiation and its dysregulation has been implicated in cancers and a number of viral infections [19]. These epigenetic mechanisms involve an interaction between DNA methylation, histone modification and mechanisms mediated by non-coding RNA molecules which to regulate transcription of genes [23]. Viruses manipulate host defense mechanism using epigenetic mechanisms in order to avoid viral proteins recognition by the immune system during latent infection [19]. The mechanism by which the rabies virus up regulates B7-H1 to favor its neuroinvasiveness has not been fully understood and there is presently no cure or means of managing the disease once it has reached the brain. Previous studies have indicated that T cells entering the NS underwent apoptosis in the course of RABV infection by the expression of B7-H1 [4, 13, 24]. Also mice lacking B7-H1 have a significant survival which clearly shows that B7-H1 is critically involved in the strategies used by RABV to escape the host immune response [12]. Interestingly, the mechanism by which RABV explore epigenetic strategy in relation to B7-H1 has not been established. Consequently, in this study we investigated the epigenetic alterations associated with B7-H1 in Challenge virus standard (CVS) strain and Nigerian street rabies virus (SRV) infected mice. Interestingly, our findings paradoxically indicate that increase in expression of B7-H1 during rabies infection is associated with simultaneous methylation of the DNA and histone acetylation.
The laboratory RABV strains CVS and SRV (SV470) were obtained from National Veterinary Research Institute (NVRI), Vom, Jos, Plateau State, Nigeria. A total of thirty-four weeks old mice were used. The experiment was carried out in three groups which includes; Group 1; CVS infected mice, Group 2; SRV infected mice and Group 3; Normal control. Mice in each experimental group were inoculated intramuscularly in the hind limb with 0.03 ml of 10% suspension of rabid mice brain. Disease progression was evaluated by monitoring clinical signs and mortality. Mice were chosen at random from each group on days 5, 7, 9, 11 and 13 post infections and humanly sacrificed by mild anesthesia. The harvested mice brain samples were subjected to the direct fluorescent antibody test (DFAT) as previously described [16], to test for the presence of Lyssavirus antigens. Also, total RNA was extracted from 25 mg of mice brain tissue using Isolate II RNA mini kit (Bioline Reagent Ltd., UK) according to manufacturer’s protocol. RNA quality was measured using a Nanodrop Spectrophotometer (ND 1000, USA). Furthermore, exactly 500 ng of isolated RNA was converted to cDNA using SensiFAST cDNA Synthesis kit (Bioline Reagent Ltd., UK) according to manufacturer’s protocol. For quantification of mRNA, 2 × SensiFAST SYBR® No-ROX Mix RT-PCR (Bioline Reagent Ltd., UK) was used according to manufacturer’s instructions. Primers for 18S ribosomal RNA used as reference gene and B7-H1 are listed in Table 1. The real-time PCR data were analyzed using the 2−∆∆CT relative quantification method. DNA was equally extracted from 25 mg of mice brain tissues using Isolate II Genomic DNA kit (Bioline Reagent Ltd., UK) and quality checked using Nanodrop Spectrophotometer (ND 1000, USA). Bisulfite modification was carried out on the isolated DNA sample using EZ DNA methylation kit according to manufacturer’s protocol. As a control for efficiency of bisulfite conversion, 2 µl of converted DNA was used to carry out a conventional PCR against primer for non-converted DNA and checked using agarose gel electrophoresis. However, no amplicon was observed indicating a successful conversion. DNA was eluted in 30 µl of the provided elution buffer. Bisulfite converted DNA was used for MSqPCR using the 2 × SensiFAST SYBR® No-ROX Mix RT-PCR according to manufacturer’s instructions. Primers specific for fully methylated and fully unmethylated B7-H1 promoter sequences (mB7H1, uB7H1), positive controls (100% values) and standard curves unmethylated DNA (uDNA) and CG Genome Universal Methylated DNA (mDNA) (S7821; Millipore) were used. For reference gene, a primer pair corresponding to a specific β-actin sequence was chosen. Within this β-actin sequence no CpG sites are present. Thus, the cytosines are always unmethylated, irrespective of methylation signals in other gene regions. Primers used for mB7H1, uB7H1, and β-actin are listed in Table 1. Methylation was calculated as percentage methylated region (PMR) as follows:
Table 1.
List of Primers used for mRNA level and methylation analyses
| Genes | Forward primer | Reverse primer | Reference |
|---|---|---|---|
| B7-H1 | 5′-GGATGCTTCTCAATGTGACCAGCAG-3′ | 5′-GGATGTGTTGCAGGCAGTTCTGGG-3′ | [12] |
| mB7-H1 | 5′-TTTTTATCGAGAGCGTTTTTTTCGT-3′ | 5′-AAAAAATAAATCTTCCCTTACCGTC-3′ | Methprimer software |
| uB7-H1 | 5′-TTTTTTATTGAGAGTGTTTTTTTTGT-3′ | 5′-AAAAAATAAATCTTCCCTTACCATC-3′ | Methprimer software |
| β-actin | 5′-GGGTTGGTTTGTATATTGATTTGAG-3′ | 5′-TAAAAAACCTATAACCCTCCCACTAA-3′ | Methprimer software |
| 18S rRNA | 5′-GGGAGCCTGAGAAACGGC-3′ | 5′-GGGTCGGGAGTGGGTAATTT-3′ | [12] |
Percentage methylated region of B7-H1 (PMR) = (Mean mB7H1 − Mean β-actin)/(Mean mDNA − Mean β-actin) × 100.
Percentage unmethylated region of B7-H1 (PUR) = (Mean uB7H1 − Mean β-actin)/(Mean uDNA − Mean β-actin) × 100 [10].
Mice brain tissues were used for colorimetric measurement of HAT and DNMT activities using the Mouse Hat1 (histone acetyltransferase type B catalytic subunit) ELISA kit and Mouse DNA methyltransferase (DNAM) ELISA Kit according to manufacturer’s instructions. To end with, each experiment was carried and repeated at least three times and where appropriate, values were presented as mean ± standard deviation. Student t-test and one way ANOVA as the case may be were used to determine statistical difference. The level of significance was assessed by Tukey’s honestly significant difference (HSD) at p < 0.05.
Rabies virus (RABV) antigen (N protein) was qualitatively detected in CVS and SRV inoculated mice from day 5 to 13 post infection (Table 2). This was supported by some critical clinical manifestation with non-specifically presence of ruffled fur, paralyzed hind limb and total paralysis. As seen from Fig. 1, significant (p < 0.05) increase in B7-H1 mRNA level was observed which was directly proportional to the increase in days of post infection in both CVS and SRV-induced mice. The Percentage Methylated Region (PMR) in all the infected samples were higher in comparison to the normal in both CVS SRV-induced mice (70.6%) as indicated in Fig. 2, and the increase was statistically significant (p < 0.05) and the opposite was observed in the percentage unmethylated region (PUR) (Fig. 2). This is evident at day 11 with highest percentage of 85.1 in CVS and 88.5 for SRV at day 13 post-infections for CVS. Overall, these data showed that the rate of DNA methylation in the B7-H1 locus increased in the rabies infected brain tissue compared to the non-infected. The DNMT activity (Fig. 3) of the rabies infected tissues CVS group were mostly higher (p < 0.05) than the control group except day 9 which had a lower activity while the SRV had a higher DNMT activity in the control group compared to the infected tissues in day 5 and 9 only. Similarly, the HAT activity (Fig. 4) in the infected samples in the CVS group was significantly (p < 0.05) higher in most samples in comparison to the control even though day 5 was comparatively lower. While HAT activity in the SRV group was higher than the control except for day 13, the increase though was not statistically high as that of CVS. Overall in both DNMT and HAT, there was a variation in their activities. The DNMT activity in all the infected samples had a proportion of approximately 1 except for samples from SRV day 13 while the proportion of HAT activity was very high in the CVS group compared to the SRV group (Table 3). Overall the proportion of HAT activity is higher than DNMT activity in both groups.
Table 2.
Correlation of clinical manifestation in CVS and SRV infected mice with antigen detection
| Days post infection | Clinical manifestation | Antigen detection results | Age of mice (days) | ||
|---|---|---|---|---|---|
| Ruffled fur | Paralyzed hind limb | Total paralysis | DFAT | ||
| CVS | |||||
| 5 | − | − | − | + | 33 |
| 7 | + | − | − | + | 35 |
| 9 | + | + | − | + | 37 |
| 11 | + | + | + | + | 39 |
| 13 | + | + | + | + | 41 |
| SRV | |||||
| 5 | − | − | − | + | 33 |
| 7 | − | − | − | + | 35 |
| 9 | + | − | − | + | 37 |
| 11 | + | + | − | + | 39 |
| 13 | + | + | + | + | 41 |
Fig. 1.
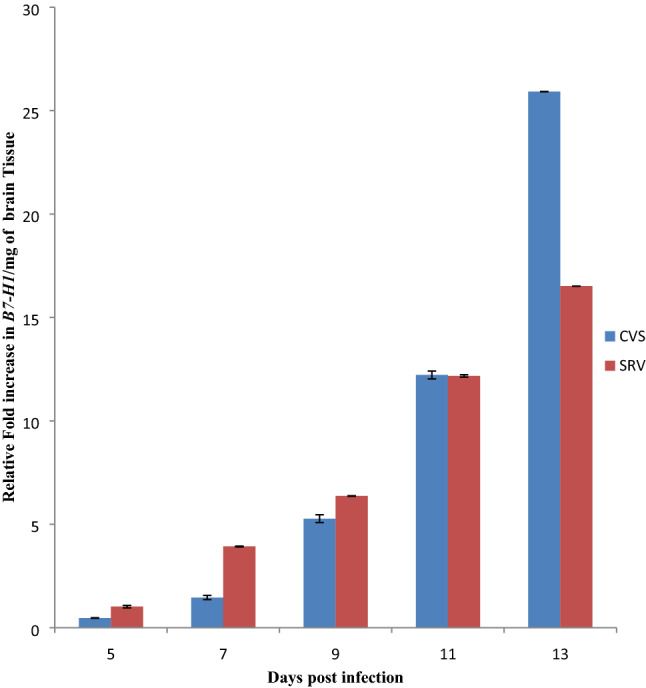
Expression profile of B7-H1 gene in the brain tissue of RABV infected mice by quantitative PCR. The mean with different superscript are statistically significant (p < 0.05). CVS challenge virus standard strain, SRV Nigerian street rabies virus, RABV rabies virus
Fig. 2.
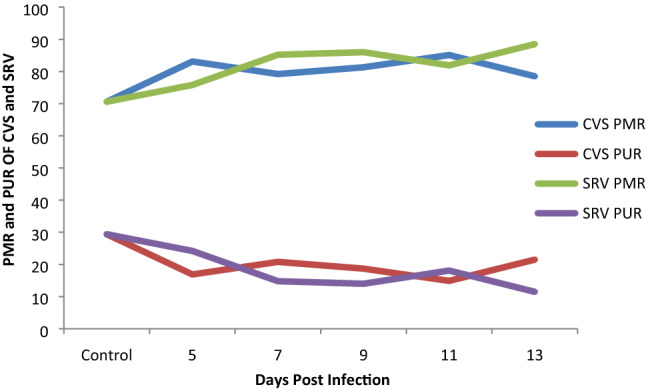
Percentage methylated and unmethylated region of B7-H1 promoter in CVS and SRV infected mice brain tissues. PMR percentage methylated region, PUR percentage unmethylated region
Fig. 3.
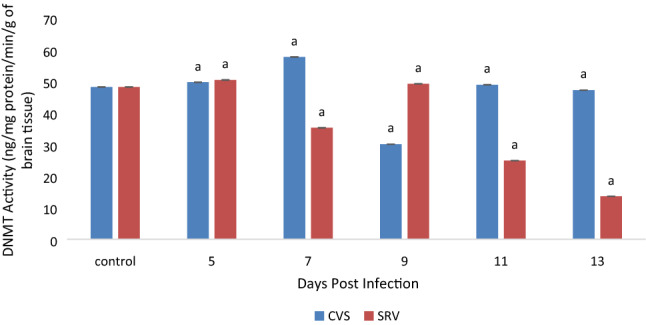
Level of DNA methyltransferase (DNMT) activity in CVS and SRV infected mice brain tissue. Mean values with superscript (a) are statistically significant as compared with the control (p < 0.05). CVS challenge virus standard strain, SRV Nigerian street rabies virus
Fig. 4.

The level of histone acetyltransferase (HAT) activity in CVS and SRV infected mice brain tissue. Mean values with superscript (a) are statistically significant as compared with the control (p < 0.05). CVS challenge virus standard strain, SRV Nigerian street rabies virus
Table 3.
Proportion of DNMT and HAT activity in CVS and SRV infected mice as compared with the control
| Days post infection | DNMT (sample activity/control activity) | HAT (sample activity/control activity) | ||
|---|---|---|---|---|
| CVS | SRV | CVS | SRV | |
| 5 | 1.00 | 1.04 | 0.48 | 1.04 |
| 7 | 1.19 | 0.73 | 7.28 | 9.12 |
| 9 | 0.62 | 1.02 | 9.12 | 1.52 |
| 11 | 1.01 | 0.52 | 8.00 | 1.72 |
| 13 | 0.97 | 0.28 | 15.12 | 0.76 |
CVS challenge virus standard strain, SRV Nigerian street rabies virus
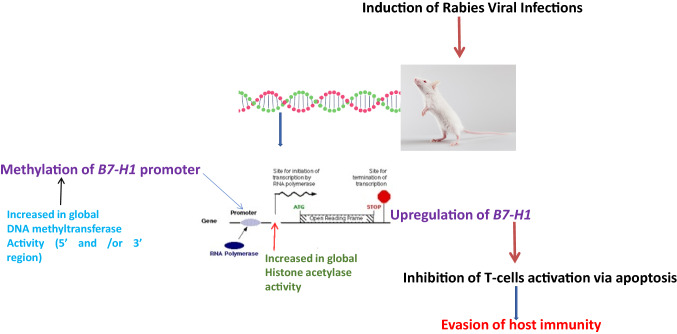
Rabies is a neglected tropical disease present on all continents, except Antarctica with over 3.7 million disability-adjusted life years (DALYs) lost every year [18]. The progression of rabies virus infection is determined by a number of complex virus host interactions [20]. Several studies have shown that RABV up-regulates the expression of immune-subversive molecules such as B7-H1 [12]. This was confirmed in our study as there was a statistically progressive increase in B7-H1 expression from day 5 to day 13 post infection in both CVS and SRV infected mice brain tissues as the disease progressed, indicating that there was over-expression of B7-H1 during rabies infection in comparison to the non-infected tissues. The mechanism by which B7-H1 is up-regulated in the course of RABV infection is not fully understood and several studies have indicated different ways by which B7-H1 expression is regulated via signaling pathways like STAT 3 [6], interferon activation [5], CSN5 activity [14] and epigenetic factors such as DNA methylation [7], HDAC activity [5] and ubiquitination of B7-H1 [14]. Many studies have also shown that viruses such as Epstein Barr virus (EBV), Human Papiloma virus (HPV) interfere with host epigenetic machinery [19], but there is currently no study on the epigenetic mechanisms the RABV uses particularly in respect to B7-H1 up-regulation. These epigenetic modifications are usually complex and intertwined and investigating them has led to novel findings and discovery of new mechanisms of action [9]. From the MSqPCR results, there was an increase in methylation of the B7-H1 promoter in both strains as we observed a higher PMR (with the highest being 88.55%) in all the samples obtained from the infected tissues compared to the non-infected tissue (70.6%). This was unexpected as most studies on DNA methylation indicated that methylation of the promoter region is inversely correlated with gene expression. However, data and results from recent studies [3, 9, 15] have shown that DNA methylation might have a paradoxical role in expression and that hypermethylation may lead to activation of transcription. This seemed to be the case in our study as DNA methylation was directly related with gene expression. A number of mechanisms have been observed to be implicated in situations of direct correlation between DNA methylation and gene expression which could give reasons for this phenomenon. A study suggested that methylated CpG islands in the promoter region and also in intragenic 3′ regions in the gene may be flanked by regions which are binding sites for transcription factors conferring cis and trans regulatory mechanisms that may counteract the effect of DNA methylation leading to activation of expression [9,3]. Another study indicated the use of an alternate promoter where, methylation blocks access of the transcription machinery to the CpG island allowing the use of the basic core promoter of the gene, while in situations where the CpG island is unmethylated, transcription starts from the CpG island in preference to the basic core promoter [9]. Interestingly, hydroxylation of already methylated CpG islands by Ten-eleven translocation family (TET) of protein have been reported to reactivate gene expression [17], which may likely be the case as currently observed from our findings. To further confirm the results from the methylation analysis, we measured the level of activity of global DNA methyltransferase and noticed that the activity of the enzyme mostly increased in the infected samples in both strains as observed in the PMR values obtained. This confirms that during RABV infection there is methylation of DNA, which is in agreement with our results obtained from the methylation analysis. The DNMT activity might not be on the promoter region of B7-H1 but probably at the 3′ region and also on other genes and may lead to silencing or expression of these genes. It also agrees with studies which have indicated that in diseases, particularly viral infections there is increased DNA methylation which is associated with regulation of genes involved in control of the cell cycle, apoptosis and tumour suppression [23]. Other factors like histone modifications could also play a significant role [21]. The level of HAT activity was also measured. Acetylation occurs at specific lysines in the N-terminal histone tails and is associated with transcriptional activity, by reducing the net positive charge of the histones and weakens interactions with DNA allowing transcription factor interact with the gene [26]. We observed an increase in HAT activity, signifying that acetylation of histones is actively going on during infection with RABV. This might be associated with differentially expressed genes during RABV infection involved in regulation of cell metabolism, protein synthesis, synaptic activity, and cell growth and differentiation as indicated in a study [20], though in our study, the increase in CVS was higher than in SRV which might be associated with it been a highly pathogenic strain [14]. To ascertain which of the enzymes played a more prominent role during rabies virus infection, the proportion of their activity was determined, and it indicated that the proportion of DNMT activity in all the samples was almost on the same level as they all gave a value of approximately 1 except in SRV day 13. This shows that, even though there was DNMT activity during the infection, the increase was not progressive and almost remained constant, indicating that the methylation of DNA most likely happened at the onset of infection. While the proportion of HAT activity was high, indicating that a large number of histones are acetylated during RABV infection, leading to an increase in the number of genes being expressed and probably the expression of these genes during the infection are associated with histone acetylation [11]. Paradoxically, our findings depicted an increase in B7-H1 promoter methylation at the onset of RABV infection and also increase in global DNMT and HAT activity ironically leading to an increase in expression of the gene during rabies virus infection favoring the transition from heterochromatin to euchromatin, suggesting a likely dual role of DNA methylation (Fig. 5). The results gotten from this study indicate that the RABV virus uses epigenetic mechanisms as a tool for manipulating its host genes for its survival. We can then infer that paradoxically, the increase in expression of B7-H1 during rabies infection is associated with methylation of the DNA and histone acetylation. Further studies need to be carried out to investigate the mechanism by which methylation of B7-H1 promoter region leads to gene expression by carrying out detailed molecular studies such as methylated DNA Immuno-precipitation (MeDIP), chromatin immuno-precipitation (chIP), and analysis of B7-H1 gene data throughout the region downstream and upstream of the Translation start site.
Fig. 5.
Proposed mechanism of epigenetic modifications associated with B7-H1 in mice-induced with rabies virus infection
Acknowledgements
We wish to thank and appreciate Africa Center of Excellence for Neglected Tropical Diseases and Forensic Biotechnology, (ACENTDFB) Ahmadu Bello University (ABU), Zaria for funding this important project. We also wish to acknowledge Dr. David Dantong of the Department of Microbiology, Faculty of Veterinary Medicine, University of Abuja, Nigeria for the help he has rendered in getting us CVS and SRV strains of the rabies virus used in this study.
Author contributions
Conception: AM, CEO and GSNK; Design: AM, CEO, MMA and GSNK; Execution: CEO, AM, JCA; Interpretation: AM, CEO, GSNK, JKPK; Writing and Proof reading: CEO, AM, GSNK, MMA, AM, JCA, JKPK.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest..
Human and animal rights
Animal housing and experimental protocols were performed according to the guidelines of the Nigerian council on Animal Care with the approval of the Ahmadu Bello University animal care committee.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Afreen S, Dermime S. The immunoinhibitory B7-H1 molecule as a potential target in cancer: killing many birds with one stone. Hematol Oncol Stem Cell Ther. 2014;7(1):1–17. doi: 10.1016/j.hemonc.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Albertini AAV, Ruigrok RWH, Blondel D. Rabies virus transcription and replication. Adv Virus Res. 2011;79:1–22. doi: 10.1016/B978-0-12-387040-7.00001-9. [DOI] [PubMed] [Google Scholar]
- 3.Anastasiadi D, Codina AE, Piferrer F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet Chromatin. 2018;11:1–17. doi: 10.1186/s13072-018-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopy D, et al. Ambivalent role of the innate immune response in rabies virus pathogenesis. J Virol. 2011;85(13):6657–6668. doi: 10.1128/JVI.00302-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng R, et al. HDAC is indispensable for IFN-γ-induced B7-H1 expression in gastric cancer. Clin Epigenet. 2018;10(153):1–14. doi: 10.1186/s13148-018-0589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L, et al. PARP1 suppresses the transcription of PD-L1 by poly (ADP-ribosyl)ating STAT3. Cancer Immunol Res. 2019;7(1):136–149. doi: 10.1158/2326-6066.CIR-18-0071. [DOI] [PubMed] [Google Scholar]
- 7.Goltz D, et al. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2017;6(1):1–4. doi: 10.1080/2162402X.2016.1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez-Díaz E, Jordà M, Peinado MA, Rivero A. Epigenetics of host-pathogen interactions: the road ahead and the road behind. Public Libr Sci Pathog. 2012;8(11):1–10. doi: 10.1371/journal.ppat.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halpern KB, Vana T, Walker MD. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J Biol Chem. 2014;289(34):23882–23892. doi: 10.1074/jbc.M114.573469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hattermann K, Mehdorn HM, Mentlein R, Schultka S, Held-Feindt J. A methylation-specific and SYBR-green-based quantitative polymerase chain reaction technique for O6-methylguanine DNA methyltransferase promoter methylation analysis. Anal Biochem. 2008;377(1):62–71. doi: 10.1016/j.ab.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Kurdistani SK, Tavazoie S, Grunstein M, Hall B, Angeles L. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Lafon M, et al. Detrimental contribution of the immuno-inhibitor B7-H1 to rabies virus encephalitis. J Immunol. 2008;180:7506–7515. doi: 10.4049/jimmunol.180.11.7506. [DOI] [PubMed] [Google Scholar]
- 13.Lafon M. Evasive strategies in rabies virus infection. Adv Virus Res. 2011;79:33–53. doi: 10.1016/B978-0-12-387040-7.00003-2. [DOI] [PubMed] [Google Scholar]
- 14.Lu C, Liu K. Epigenetic regulation of PD-L1 expression and pancreatic cancer response to checkpoint immunotherapy. Transl Cancer Res. 2017;6(Suppl 3):652–654. doi: 10.21037/tcr.2017.05.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masatani T, et al. Importance of rabies virus nucleoprotein in viral evasion of interferon response in the brain. Microbiol Immunol. 2013;57(7):511–517. doi: 10.1111/1348-0421.12058. [DOI] [PubMed] [Google Scholar]
- 16.Meslin FX, Kaplan MM, Koprowski H. Laboratory techniques in rabies. 4. Geneva, Switzerland: World Health Organization; 1996. [Google Scholar]
- 17.Miranda-Gonçalves V, Lameirinhas A, Henrique R, Jerónimo C. Metabolism and epigenetic interplay in cancer: regulation and putative therapeutic targets. Front Genet. 2018;9:427. doi: 10.3389/fgene.2018.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otolorin RG, et al. A review on human deaths associated with rabies in Nigeria. J Vaccines Vaccin. 2015;06(01):1–6. [Google Scholar]
- 19.Poreba E, Broniarczyk JK, Gozdzicka-Jozefiak A. Epigenetic mechanisms in virus-induced tumorigenesis. Clin Epigenet. 2011;2(2):233–247. doi: 10.1007/s13148-011-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prosniak M, Hooper DC, Dietzschold B, Koprowski H. Effect of rabies virus infection on gene expression in mouse brain. Proc. Nat. Acad. Sci. 2001;98(5):2758–2763. doi: 10.1073/pnas.051630298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy MA, Natarajan R. Recent developments in epigenetics of acute and chronic kidney diseases. Int Soc Nephrol. 2015;88(2):250–261. doi: 10.1038/ki.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srithayakumar V, Sribalachandran H, Rosatte R, Nadin-Davis SA, Kyle CJ. Innate immune responses in raccoons after raccoon rabies virus infection. J Gen Virol. 2014;95(Pt 1):16–25. doi: 10.1099/vir.0.053942-0. [DOI] [PubMed] [Google Scholar]
- 23.Steele N, Finn P, Brown R, Plumb JA. Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br J Cancer. 2009;100:758–763. doi: 10.1038/sj.bjc.6604932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubol S, Kasisith J, Pitidhammabhorn D, Tepsumethanol V. Screening of pro-apoptotic genes upregulated in an experimental street rabies virus-infected neonatal mouse brain. Microbiol Immunol. 2005;49(5):423–431. doi: 10.1111/j.1348-0421.2005.tb03746.x. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. World Malaria Report 2013. 2013.
- 26.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272(45):28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]