Abstract
Nowadays, exposure to infectious diseases caused by pathogenic viruses has become one of the major human concerns in health fields. In the meantime, hepatitis viruses are associated with health problems, especially in liver tissue. So far, several types of these viruses have been known including: HAV, HBV, HCV, HDV, HEV, and HGV. Nevertheless, it seems that hepatitis C is the major viral infection among all of the hepatitis infections. The cirrhosis and hepatocellular carcinoma are known as the most important pathological complications of this virus, from which seven genotypes have been identified. However, among these genotypes, the incidence rate of genotypes 1 and 3 is more than others. In this review, we have investigated the relationship between all HCV genotypes and therapeutic responses against them. Regarding heterogeneity between hepatitis C genotypes, it is not possible to access an effective vaccine against this virus, and treatment is the only applicable strategy. Response to treatment is different among genotypes, and it has resulted that each genotype has a specific therapeutic regimen of itself. Therefore, it seems that determination of hepatitis C genotype, as a key tool, is essential in controlling therapeutic regimen, improving local control programs and eventually producing an effective vaccine.
Keywords: Hepatitis C virus, Antiviral agent, Hepatitis virus, Interferon
Introduction
Hepatitis C, is a non-cytopathic enveloped, with a positive-sense single-stranded RNA virus, which belongs to genus Hepacivirus, and Flaviviridae family. This virus is classified into seven different genotypes and causes both phases of liver infections, acute and chronic (cirrhosis and hepatocellular carcinoma) [46]. The whole genome of hepatitis C has about 9.4 kbp length, which is initially encoded to a large polyprotein, which is cleaved to ten proteins by viral protease, and contains three structural (core, E1, and E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). It is notable that, NS3/4A protease and NS5B RNA polymerase, significantly affect the pathogenesis of HCV [44]. In addition, the 5’ terminal region of genomic RNA contains highly conserved sequences, which are used for molecular diagnosis of this virus. Current studies have shown that genomic RNA has different lengths among different hepatitis C virus genotypes; for example, genome size in genotype 1 is 9.4 kbp, while it is 9.099 kbp and 9.063 kbp in genotypes 2 and 3 respectively. What is clear is that the differences in genomic RNA length can affect some properties of the virus such as pathogenicity, clinical manifestations, response to treatment and also inability to produce anti-HCV vaccines [44, 46]. According to recent reports of the World Health Organization (WHO), approximately 150–170 million people are infected with acute hepatitis C forms, and 75–85% of them develop chronic forms of the disease. Unfortunately, 1–5% of the patients die due to the disease progression to hepatocellular carcinoma (HCC). Annually, 3–4 million new cases of hepatitis C are reported worldwide; thereby, based on WHO programs, this disease must be eradicated by 2030 [31, 44]. According to literature reviews, the treatment outcomes of HCV infection are exclusively dependent on HCV genotypes [31, 46]. Therefore, the aim of this study is to provide a comprehensive discussion about the determinative role of HCV genotypes on efficient treatment of HCV infection.
HCV and response to treatment
Hepatitis C virus is transmitted through different routes, such as blood transfusion, injections, sexual contact, surgery, and tattoo. The HCV Real-time PCR and anti-HCV antibody tests are accounted for as the most important diagnostic tests, which routinely used in laboratories. However, these tests only determine HCV infection, without differentiating the current and previous infections; and their results are not reliable for therapeutic monitoring [44]. In general, for evaluating response to treatment, several beneficial items are being used; such as (1) sustained virological response (SVR), which indicates viral load after the treatment period (almost 24 weeks), (2) decrease in liver enzymes especially ALT and (iii) histological investigation of liver tissue biopsy [24]. Interferon and guanosine analog (Ribavirin) are being used for hepatitis C treatment in the last two decades. Polyethylene glycol interferon (Peg-IFN) was introduced in the last decade, which was a better compound with more half-life compared to conventional IFN. Nonetheless, some studies have shown that a standard regimen including IFN (subcutaneous injection three times a week) and Ribavirin (800–1200 mg/day) for 24 weeks, is not effective for all HCV patients and response to treatment is different among them [24, 44].
SVR ration of different HCV genotypes
Many studies have shown that different hepatitis C genotypes have different responses to treatment. IFN and Ribavirin complex regimen is significantly effective among infected patients with genotype 1 (SVR more than 90%). However, based on Zuberi et al. genotypes 2 and 3 have a poor response to treatment of SVR 27.8% and 58.8% respectively [24, 39]. In other studies, it was shown that genotypes 1 and 4 responded well to the combination therapy with IFN plus Ribavirin, while genotypes 2 and 3 had variable responses. Furthermore, genotypes 5 and 6 have an appropriate response to Peg-IFN and Ribavirin for 48 weeks (Table 1) [7, 24]. Regard to different outcomes of treatment between different genotypes of hepatitis C, recent efforts have focused on direct-acting antivirals (DAAs) compounds such as Telaprevir, Boceprevir, Grazoprevir (NS3/4A protease inhibitors), Elbasvir (NS5A inhibitor) and Sofosbuvir (NS5B Polymerase inhibitor). Three compounds Telaprevir and Boceprevir, and Peg-IFN were prescribed for treatment of genotype 1, and SVR results were 60–70%. Sofosbuvir along with Peg-IFN and Ribavirin had admissible results in the treatment of genotypes 1 and 4. In addition, the treatment of three genotypes 1, 4 and 6 with Grazoprevir, Elbasvir, and Ribavirin had acceptable results. Clearly, like conventional treatments, response to treatment with DAAs depends on hepatitis C genotype. For example, none of the anti-protease NS3/4A has been effective in the treatment of genotype 3; on the other hand, Sofosbuvir has effectively treated genotype 2, and SVR is 97% [40]. Recent studies have shown that Telaprevir in the treatment of genotype 4, and a combination of Sofosbuvir, Peg-IFN, and Ribavirin in the treatment of genotype 3, have been associated with satisfactory results. But in general, in genotypes 2 and 3, the response to treatment is poor in comparison with genotypes 1 and 4; thus, the treatment period is longer (48 weeks). Moreover, genotypes 5, 6 and 7 entirely are not been studied, because they merely are there in specific endemic regions [32]. Nowadays, we face with some resistant hepatitis C genotypes to DAAs, such as NS3 protease, NS5B polymerase, and NS5A polymerase inhibitors, which the most resistant is of them is genotype 1a [35].
Table 1.
Summary of studies describing the treatment of different genotypes of hepatitis C virus
Geographical distribution of genotypes
Generally, genotypes 1 and 3 are the most common genotypes worldwide. Relatively, about 46% and 30% of hepatitis C cases are related to genotypes 1 and 3, respectively. However, some genotypes are dominant in particular geographic areas. For example, genotype 4 in Egypt, the Middle East and Central Africa; and genotypes 5 and 6 in South Africa and Asia are the predominant genotypes [36]. As in Iran, genotypes 1a, 3a, and 1b are the most common [31, 44]. However, there has been a recent increase in the prevalence of genotype 3a, but a decrease in genotypes 1a and 1b [31]. According to geographic studies in Iran, the most common genotypes include genotype 1a in the north and south, genotype 3a in the north and the center, genotype 1b in the south and the west, as well as genotype 2 in the west. These geographic patterns are different from the patterns obtained in the Middle East; however, they are similar to the patterns in North America [27]. According to the review of literature, HCV-3 infected patients incompletely response to treatment with ration treatment of HCV 1a infection is about 48 weeks [16, 34]. Based on the present interferon-free guideline, infected cases with HCV-3 which develop to cirrhosis, to get better results, must be treated by Daclatasvir plus Sofosbuvir with or without RBV for a period of 24 weeks. Although the therapeutic protocols are various in HCV-1 infections, the best strategy for containment of HCV-1 outbreak is consisting of the beginning of Ribavirin for 24 weeks, which must be followed by anti-viral susceptibility tests [22, 23]. Nowadays, there are various clinical studies recommendations for the performance of the best therapeutic regimens for HCV 1/3 infected cases. Zeuzem et al. have shown that the use of Glecaprevir plus Pibrentasvir regimen for 8–12 weeks has efficient therapeutic results for HCV genotypes 1 or 3 infections, but it may that studies are heterogeneous and to be continued throughout the worldwide [47].
In Iran, genotype 1 is isolated from patients engaged with hemophilia, thalassemia, hemodialysis, and from transplant recipients. Genotype 1b is isolated from alcohol consumers, blood recipients and patients admitted to surgical wards. Genotypes 1a, 3a and 4 are isolated from injecting drug users (IDUs) and hemodialysis patients, respectively [44]. It is worth mentioning that in Iran, either of genotypes own a specific regimen of themselves, but in our country, due to expensive production costs and limited access to DAA drugs, the conventional protocol for treatment all patients contain prescription of IFN along with Ribavirin, but of note, the results of this method of treatment are failed in some cases [40, 44].
DAAs and IDSA recommendations
Presently, it is demonstrated that DAAs therapy is more acceptable than conventional ones (interferon-based regimens) because of their high efficacy, patient tolerance, and clearance rate and a short course of treatment [14, 42]. DAAs compounds were first approved by the FDA in 2011. Now, there are 11 different antiviral agents in DAAs category, which are FDA-approved for efficiently treating HCV infections [9, 18]. In recent years, an updated recommendation has been issued by the IDSA (Infectious Diseases Society of America) containing standard guidelines to manage HCV infection [17]. Based on IDSA recommendations, there are several important factors such as patient’s underlying conditions, cirrhosis status, co-infection with other infectious diseases in particular HIV, HCV genotype and drug-susceptibility testing of HCV strains which should be considered for efficient treatment of HCV infections (Table 2) [9, 20]. Overall, managing HCV infection is challenging and depends on viral characterizations (genotype, type of resistant, infection with multiple genotypes) or epigenetic evidence of HCV infected cases. For example, various genome-wide association studies (GWAS) have revealed that single nucleotide polymorphisms (SNPs) of IL28B gene associated with spontaneous clearance (SC) and sustained virological response (SVR) after PEG-IFN-a/RBV therapy [17, 38, 43, 45]. There are 3 main SNPs of IL28B gene including rs809917, rs12979860 and rs12980275 which are located in regulatory, transcription initiation or promoter region of interleukin 28B (IL28B) gene on chromosome 19 and encodes IFN-λ3 which can effect on expression of IL28B cytokine and decrease antiviral defense and spontaneous clearance of HCV infections [19, 28]. The result of one study indicated that in Asian population, the patients with CC polymorphism of rs12079860 SNP show spontaneous clearance and SVR rate 2-fold increase in comparison with the patients with TT genotype [33]. Also it has been shown that rs12979860 C/C genotype is associated with SVR in Caucasian, Asian, European, Hispanic and African-American populations and the genotype of rs8099917 has been reported to be the strongest predictor marker in genotype 2 and 3 [8, 25]. Moreover, It has been demonstrated that the polymorphism of the rs8099917 SNP like TG/GG allele in HCV patients in Asian population had high SVR rate and spontaneous clearance, but rs8099917 GG/GG genotype indicated no SRV rate. [6, 12, 33].Therefore, it seems necessary to understand the extensive aspects of HCV infection for efficient management and treatment of HCV infected cases.
Table 2.
Highlighted recommendations by IDSA for treatment HCV genotypes
| HCV genotype(s) | Therapeutic drug(s) | Duration of treatment (weeks) | Treated subjects | With or without cirrhosis |
|---|---|---|---|---|
| HCV genotype 1 | Elbasvir/Grazoprevir | 12 | Naive | without cirrhosis |
| Glecaprevir/Pibrentasvir | 8–12 | HIV negative Sofosbuvir failure | Without cirrhosis | |
| Ledipasvir/sofosbuvir | 12–24 | DAAs failure (particularly NS5A inhibitor) | Without cirrhosis | |
| Sofosbuvir/Velpatasvir | 12–24 | DAAs failure | Without cirrhosis | |
| Sofosbuvir/velpatasvir/voxilaprevir | 12 | NS5A failure | Without cirrhosis | |
| HCV genotype 2 | Daclatasvir plus Sofosbuvir | 12 | Post-liver transplant | Without cirrhosis |
| Glecaprevir/Pibrentasvir | 8–12 | Transplantation | Without cirrhosis | |
| Sofosbuvir/velpatasvir | 12–24 | DAAs failure, transplantation | Without cirrhosis | |
| Sofosbuvir/Velpatasvir/Voxilaprevir | 12 | NS5A failure | With cirrhosis | |
| HCV genotype 3 | Daclatasvir plus Sofosbuvir | 12 | Naive | Without cirrhosis |
| Glecaprevir/Pibrentasvir | 8–12 | Naïve | With cirrhosis | |
| Sofosbuvir plus Elbasvir/Grazoprevir | 12 | Naive | Compensated cirrhosis | |
| Sofosbuvir/velpatasvir | 12 | Naïve | Without cirrhosis | |
| Sofosbuvir/velpatasvir/voxilaprevir | 12 | DAAs failure | Without cirrhosis | |
| HCV genotype 4 | Daclatasvir plus sofosbuvir | 12 | HIV/HCV coinfection | Decompensated cirrhosis |
| Elbasvir/Grazoprevir | 12 | Naive | Without cirrhosis | |
| Glecaprevir/Pibrentasvir | 8–12 | Naive, peg-IFN plus ribavirin | With cirrhosis | |
| Ledipasvir/Sofosbuvir | 12–24 | Naïve, Sofosbuvir failure | Without cirrhosis | |
| Sofosbuvir/Velpatasvir | 12–24 | NS3 protease inhibitor failure | Without cirrhosis | |
| HCV genotypes 5 and 6 | Glecaprevir/Pibrentasvir | 8–12 | Naive, peg-IFN plus ribavirin | With cirrhosis |
| Ledipasvir/Sofosbuvir | 12–24 | Naive, transplantation, Sofosbuvir failure | Without cirrhosis | |
| Sofosbuvir/Velpatasvir | 12–24 | Naive, DAAs failure | Without cirrhosis | |
| Sofosbuvir/Velpatasvir/Voxilaprevir | 12 | NS5A failure | Without cirrhosis | |
| Regardless genotype | Daclatasvir plus sofosbuvir | 12 | HCV/HIV coinfection | With cirrhosis |
| Mixed genotypes | Glecaprevir/pibrentasvir or sofosbuvir/velpatasvir |
The new generation anti-HCV drugs
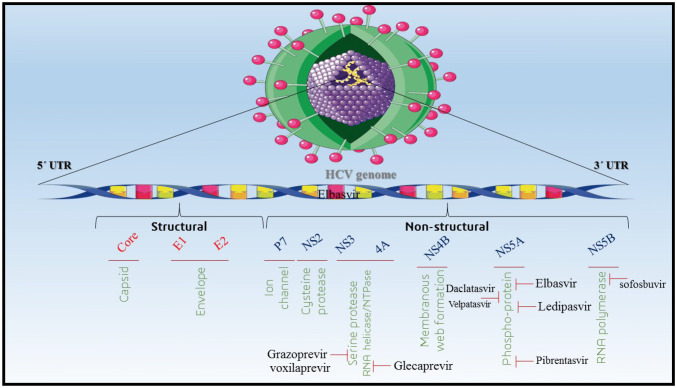
Zepatier (Elbasvir 50 mg/ Grazoprevir 100 mg) is FDA approved in May 2015 which is containing Elbasvir (an inhibitor of hepatitis C virus’s NS5A protein) and Grazoprevir (an NS3/4A inhibitor) recommended for HCV genotypes including 1a, 1b as well as 4 without cirrhosis [1]. According to the literatures, it has been revealed that initiation of treatment with this drug has SVR in 94–97% of cases against genotype 1 and 97–100% of cases against genotype 4. Moreover, the total of SVR for hepatitis without cirrhotic was 97% while it was 95.7% for cirrhotic [2]. Mavyret (Glecaprevir, an HCV NS3/4A protease inhibitor/Pibrentasvir, an HCV NS5A inhibitor) is a FDA approved drug from 2017 which is recommended against wide range of HCV genotypes 2–6 without cirrhosis or with mild cirrhosis [5]. In clinical trials, Mavyret was associated with SVR 92–100% (treatment course 12 months) for HCV genotype 2 [10]. Daclatasvir plus Sofosbuvir (NS5A/B RNA Polymerase inhibitors) was appropriate for HCV genotypes 2 and 3 patients with SVR 94–100% in different human trials [37]. In a cohort study in Egypt, it has been shown that Daclatasvir plus Sofosbuvir dad achieved to SVR 96% for HCV genotype 4 infection after 12 months of treatment [3]. Harvoni (Ledipasvir 90 mg/Sofosbuvir 400 mg) is as a RNA polymerase inhibitor which is the first FDA approved hepatitis therapeutic regimen free of interferon and Ribavirin [26]. As noted it can improve the SVR rate from 94 to 99% against genotypes 1, 4, 5 and 6 [21]. In the Fig. 1, some of effective anti HCV drugs and their effects on different compartments of HCV virus has been noted.
Fig. 1.
Antiviral drugs against hepatitis C virus. Different types of anti-viral drugs have various effects on the HCV replication, including: Grazoprevir and Voxilaprevir inhibit serine protease; Glecaprevir inhibits RNA helicase\NTPase; Daclatasvir, Velpatasvir, Elbasvir, Ledipasvir and Pibrentasvir inhibit phosphor-protein; Sofosbuvir inhibits RNA polymerase
Concluding Remarks
Nowadays, approximately 2–3% of the world population is infected with the hepatitis C virus, in which 5–10% of them develop severe liver disorders, such as cirrhosis and hepatocellular carcinoma. Regarding heterogeneity between hepatitis C genotypes, it is not possible to access an effective vaccine against this virus, and treatment is the only applicable strategy. Response to treatment is different among different genotypes, and it results that each genotype has a specific therapeutic regimen of itself. Therefore, given the above mentioned, it seems that determination of hepatitis C genotype, as a key tool, is essential in controlling therapeutic regimen, improving local control programs and eventually producing an effective vaccine. Determinative factors in SVR rate in HCV genotypes are including: SNP polymorphism in HCV [29], Polymorphism in human genome particularly SNP in immune system [11, 41], Viral load, Body response to medicine, and HCV genotype genome polymorphism; for example genotype 1 and 3 infection are associated with more aggressive liver disease, with increased risk for cirrhosis and fibrosis, as well as greater risk for hepatocellular carcinoma.
Funding
None to be declared.
Compliance with ethical standards
Conflict of interest
None to be declared.
Ethical considerations
The Ethics Committee of Mashhad University of Medical Sciences approved the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed M. Era of direct acting anti-viral agents for the treatment of hepatitis C. World J Hepatol. 2018;10(10):670. doi: 10.4254/wjh.v10.i10.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed H, et al. Meta-analysis of grazoprevir plus elbasvir for treatment of hepatitis C virus genotype 1 infection. Ann Hepatol. 2018;17(1):18–32. doi: 10.5604/01.3001.0010.7532. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed OA, et al. Sofosbuvir plus daclatasvir in treatment of chronic hepatitis C genotype 4 infection in a cohort of Egyptian patients: an experiment the size of Egyptian village. Int J Hepatol. 2018;2018:9616234. doi: 10.1155/2018/9616234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Naamani K, Al Sinani S, Deschênes M. Epidemiology and treatment of hepatitis C genotypes 5 and 6. Can J Gastroenterol Hepatol. 2013;27(1):e8–e12. doi: 10.1155/2013/624986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asselah T, et al. Efficacy of glecaprevir/pibrentasvir for 8 or 12 weeks in patients with hepatitis C virus genotype 2, 4, 5, or 6 infection without cirrhosis. Clin Gastroenterol Hepatol. 2018;16(3):417–426. doi: 10.1016/j.cgh.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 6.Cariani E, et al. Translating pharmacogenetics into clinical practice: interleukin (IL) 28B and inosine triphosphatase (ITPA) polymophisms in hepatitis C virus (HCV) infection. Clin Chem Lab Med. 2011;49(8):1247–1256. doi: 10.1515/CCLM.2011.618. [DOI] [PubMed] [Google Scholar]
- 7.Cartwright EJ, Miller L. Novel drugs in the management of difficult-to-treat hepatitis C genotypes. Hepat Med. 2013;5:53. doi: 10.2147/HMER.S48545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T-Y, et al. Impact of serum levels and gene polymorphism of cytokines on chronic hepatitis C infection. Transl Res. 2007;150(2):116–121. doi: 10.1016/j.trsl.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Chung RT, et al. Hepatitis C guidance 2018 update: AASLD-IDSA recommendations for testing, managing, and treating hepatitis C virus infection. Clin Infect Dis. 2018;67(10):1477–1492. doi: 10.1093/cid/ciy585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen DE. Clinical development of Viekira Pak to Mavyret. In: Sofia M, editor. HCV: the journey from discovery to a cure. Topics in medicinal chemistry. Cham: Springer; 2019. [Google Scholar]
- 11.De Re V, et al. Clinical significance of polymorphisms in immune response genes in hepatitis C-related hepatocellular carcinoma. Front Microbiol. 2019;10:475. doi: 10.3389/fmicb.2019.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Marco V, et al. Role of IL-28B and inosine triphosphatase polymorphisms in efficacy and safety of Peg-Interferon and ribavirin in chronic hepatitis C compensated cirrhosis with and without oesophageal varices. J Viral Hepatitis. 2013;20(2):113–121. doi: 10.1111/j.1365-2893.2012.01637.x. [DOI] [PubMed] [Google Scholar]
- 13.El Kassas M, et al. Elbasvir and grazoprevir for chronic hepatitis C genotypes 1 and 4. Expert Rev Clin Pharmacol. 2016;9(11):1413–1421. doi: 10.1080/17512433.2016.1233813. [DOI] [PubMed] [Google Scholar]
- 14.Fabrizi F, Dixit V, Messa P. Antiviral therapy of symptomatic HCV-associated mixed cryoglobulinemia: meta-analysis of clinical studies. J Med Virol. 2013;85(6):1019–1027. doi: 10.1002/jmv.23562. [DOI] [PubMed] [Google Scholar]
- 15.Fung J, et al. Chronic hepatitis C virus genotype 6 infection: response to pegylated interferon and ribavirin. J Infect Dis. 2008;198(6):808–812. doi: 10.1086/591252. [DOI] [PubMed] [Google Scholar]
- 16.Gane E, et al. High efficacy of ABT-493 and ABT-530 treatment in patients with HCV genotype 1 or 3 infection and compensated cirrhosis. Gastroenterology. 2016;151(4):651–659.e1. doi: 10.1053/j.gastro.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 18.Geddawy A, et al. Direct acting anti-hepatitis C virus drugs: clinical pharmacology and future direction. J Transl Int Med. 2017;5(1):8–17. doi: 10.1515/jtim-2017-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgel P, et al. Virus–host interactions in hepatitis C virus infection: implications for molecular pathogenesis and antiviral strategies. Trends Mol Med. 2010;16(6):277–286. doi: 10.1016/j.molmed.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Grebely J, et al. Recommendations for the management of hepatitis C virus infection among people who inject drugs. Int J Drug Policy. 2015;26(10):1028–1038. doi: 10.1016/j.drugpo.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gritsenko D, Hughes G. Ledipasvir/Sofosbuvir (harvoni): improving options for hepatitis C virus infection. Pharm Ther. 2015;40(4):256. [PMC free article] [PubMed] [Google Scholar]
- 22.Halota W, et al. Recommendations for the treatment of hepatitis C in 2017. Clin Exp Hepatol. 2017;3(2):47. doi: 10.5114/ceh.2017.67782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hézode C, et al. Daclatasvir plus sofosbuvir with or without ribavirin in genotype 3 patients from a large French multicenter compassionate use program. Hepatology. 2015;62:314A–314A. [Google Scholar]
- 24.Jamall IS, et al. Is pegylated interferon superior to interferon, with ribavarin, in chronic hepatitis C genotypes 2/3? World J Gastroenterol. 2008;14(43):6627. doi: 10.3748/wjg.14.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiménez-Sousa MA, et al. Meta-analysis: implications of interleukin-28B polymorphisms in spontaneous and treatment-related clearance for patients with hepatitis C. BMC Med. 2013;11(1):6. doi: 10.1186/1741-7015-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keating GM. Ledipasvir/Sofosbuvir: a review of its use in chronic hepatitis C. Drugs. 2015;75(6):675–685. doi: 10.1007/s40265-015-0381-2. [DOI] [PubMed] [Google Scholar]
- 27.Khodabandehloo M, Roshani D. Prevalence of hepatitis C virus genotypes in Iranian patients: a systematic review and meta-analysis. Hepatitis monthly. 2014;14(12):e22915. doi: 10.5812/hepatmon.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SU, et al. Association between IL28B polymorphisms and spontaneous clearance of hepatitis B virus infection. PLoS ONE. 2013;8(7):e69166. doi: 10.1371/journal.pone.0069166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapa D, et al. Hepatitis C virus genetic variability, human immune response, and genome polymorphisms: which is the interplay? Cells. 2019;8(4):305. doi: 10.3390/cells8040305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawitz E, et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368(20):1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 31.Mahmud S, Akbarzadeh V, Abu-Raddad LJ. The epidemiology of hepatitis C virus in Iran: systematic review and meta-analyses. Sci Rep. 2018;8(1):150. doi: 10.1038/s41598-017-18296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta V, et al. Impact of direct acting antiviral therapy for treatment of hepatitis C genotypes 1, 3 and 4: a real life experience from India. J Clin Exp Hepatol. 2018;8(1):7–14. doi: 10.1016/j.jceh.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagliaccetti NE, Robek MD. Interferon-λ in HCV infection and therapy. Viruses. 2010;2(8):1589–1602. doi: 10.3390/v2081589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearlman BL. Extended-therapy duration for chronic hepatitis C, genotype 1: the long and the short of it. World J Gastroenterol. 2008;14(23):3621. doi: 10.3748/wjg.14.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poveda E, et al. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res. 2014;108:181–191. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 36.Prakash S, et al. Distribution of hepatitis C genotypes in Uttar Pradesh, India; rare genotype 4 detected. J Med Virol. 2018;90(12):1875–1881. doi: 10.1002/jmv.25277. [DOI] [PubMed] [Google Scholar]
- 37.Rashid S, Shehzad A, Rafique S. Frequency of side effects of sofosbuvir and daclatsavir in patients of chronic Hepatitis C. J Rawalpindi Med Coll. 2019;23(3):138–142. [Google Scholar]
- 38.Rauch A, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):1338–1345.e7. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 39.Robinson CW. A Comparison of the nutritional status and physical activity levels of persons with and without the hepatitis C virus. Howard University; 2015.
- 40.Sakr AA, Hanifi JM, Lin MV. Successful treatment of mixed hepatitis C genotypes in a cirrhotic patient with an all-oral, interferon-free regimen. ACG Case Rep J. 2017;4:e16. doi: 10.14309/crj.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer EA, Chung RT. The impact of human gene polymorphisms on HCV infection and disease outcome. In: Seminars in liver disease. ©Thieme Medical Publishers; 2011. [DOI] [PubMed]
- 42.Sise ME, et al. Treatment of hepatitis C virus–associated mixed cryoglobulinemia with direct-acting antiviral agents. Hepatology. 2016;63(2):408–417. doi: 10.1002/hep.28297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suppiah V, et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat Genet. 2009;41(10):1100. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 44.Taherkhani R, Farshadpour F. Epidemiology of hepatitis C virus in Iran. World J Gastroenterol. 2015;21(38):10790. doi: 10.3748/wjg.v21.i38.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka Y, et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 46.Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13(2):223–235. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeuzem S, et al. Glecaprevir–pibrentasvir for 8 or 12 weeks in HCV genotype 1 or 3 infection. N Engl J Med. 2018;378(4):354–369. doi: 10.1056/NEJMoa1702417. [DOI] [PubMed] [Google Scholar]