Flaviviruses are important emerging and reemerging human pathogens. Understanding the molecular mechanisms for viral replication and evasion of host antiviral responses is relevant to development of control strategies. Flavivirus infections produce viral noncoding RNAs, known as sfRNAs, involved in viral replication and pathogenesis. In this study, we dissected molecular determinants for Zika virus sfRNA generation in the two natural hosts, human cells and mosquitoes. We found that two RNA structures of the viral 3′ UTR operate in a cooperative manner to produce two species of sfRNAs and that the deletion of these elements has a profoundly different impact on viral replication in the two hosts. Generation of at least one sfRNA was necessary for efficient Zika virus infection of Aedes aegypti mosquitoes. Moreover, recombinant viruses with different 3′ UTR arrangements revealed an essential role of sfRNAs for productive infection in human cells. In summary, we define molecular requirements for Zika virus sfRNA accumulation and provide new ideas of how flavivirus RNA structures have evolved to succeed in different hosts.
KEYWORDS: Zika virus, flavivirus, noncoding RNAs, sfRNAs
ABSTRACT
Zika virus (ZIKV) is an emerging flavivirus, mainly transmitted by mosquitoes, which represents a global health threat. A common feature of flavivirus-infected cells is the accumulation of viral noncoding subgenomic RNAs by partial degradation of the viral genome, known as sfRNAs, involved in immune evasion and pathogenesis. Although great effort is being made to understand the mechanism by which these sfRNAs function during infection, the picture of how they work is still incomplete. In this study, we developed new genetic tools to dissect the functions of ZIKV RNA structures for viral replication and sfRNA production in mosquito and human hosts. ZIKV infections mostly accumulate two kinds of sfRNAs, sfRNA1 and sfRNA2, by stalling genome degradation upstream of duplicated stem loops (SLI and SLII) of the viral 3′ untranslated region (UTR). Although the two SLs share conserved sequences and structures, different functions have been found for ZIKV replication in human and mosquito cells. While both SLs are enhancers for viral infection in human cells, they play opposite roles in the mosquito host. The dissection of determinants for sfRNA formation indicated a strong cooperativity between SLI and SLII, supporting a high-order organization of this region of the 3′ UTR. Using recombinant ZIKV with different SLI and SLII arrangements, which produce different types of sfRNAs or lack the ability to generate these molecules, revealed that at least one sfRNA was necessary for efficient infection and transmission in Aedes aegypti mosquitoes. Importantly, we demonstrate an absolute requirement of sfRNAs for ZIKV propagation in human cells. In this regard, viruses lacking sfRNAs, constructed by deletion of the region containing SLI and SLII, were able to infect human cells but the infection was rapidly cleared by antiviral responses. Our findings are unique for ZIKV, since in previous studies, other flaviviruses with deletions of analogous regions of the genome, including dengue and West Nile viruses, accumulated distinct species of sfRNAs and were infectious in human cells. We conclude that flaviviruses share common strategies for sfRNA generation, but they have evolved mechanisms to produce different kinds of these RNAs to accomplish virus-specific functions.
IMPORTANCE Flaviviruses are important emerging and reemerging human pathogens. Understanding the molecular mechanisms for viral replication and evasion of host antiviral responses is relevant to development of control strategies. Flavivirus infections produce viral noncoding RNAs, known as sfRNAs, involved in viral replication and pathogenesis. In this study, we dissected molecular determinants for Zika virus sfRNA generation in the two natural hosts, human cells and mosquitoes. We found that two RNA structures of the viral 3′ UTR operate in a cooperative manner to produce two species of sfRNAs and that the deletion of these elements has a profoundly different impact on viral replication in the two hosts. Generation of at least one sfRNA was necessary for efficient Zika virus infection of Aedes aegypti mosquitoes. Moreover, recombinant viruses with different 3′ UTR arrangements revealed an essential role of sfRNAs for productive infection in human cells. In summary, we define molecular requirements for Zika virus sfRNA accumulation and provide new ideas of how flavivirus RNA structures have evolved to succeed in different hosts.
INTRODUCTION
The explosive epidemic of Zika virus (ZIKV) in 2016, together with the unique properties of this virus regarding transmission and disease, posed a tremendous challenge to understand its mechanisms of infection and pathogenesis (1). Due to the late appearance of symptoms in newborns from infected mothers, the dimension of the damage caused by this virus in the last few years is still unknown (2). ZIKV is an enveloped, plus-stranded RNA virus and is a member of the flavivirus genus, together with a number of important human pathogens, including dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), and Japanese encephalitis virus (JEV). The genomic RNA encodes the viral proteins but also contains a great deal of information in RNA structures present in both the translated region and the untranslated region (UTR). These RNA elements have important functions in viral translation, genome replication, immune evasion, host adaptation, and pathogenesis (for reviews, see references 3 and 4). However, understanding of the molecular mechanism by which the viral RNA elements regulate these processes is still incomplete.
The ZIKV genome is 11 kb, containing 5′ and 3′ UTRs of about 107 and 428 nucleotides, respectively. The 5′ UTR has a large Y-shaped stem-loop structure (SLA), similar to that observed in other flaviviruses, which functions as a promoter for viral polymerase binding (5). This promoter is followed by a cyclization sequence known as the 5′ UAR (upstream AUG region) that mediates long-range RNA-RNA interactions (6). Additional flavivirus cyclization sequences are located in the coding region of the capsid protein, known as DAR (downstream AUG region), CS1 (conserved sequence 1), and C1 in the case of DENV (7–11). These elements interact with complementary regions present at the viral 3′ UTR, including 3′ UAR, 3′ DAR, and 3′ CS, respectively. The 3′ UTRs of flaviviruses contain RNA structures that are different in distinct ecological or phylogenetic groups but that all share a small hairpin 3′ stem-loop (sHP-3′ SL) at the genome end (4, 12). According to the hosts that they infect, flaviviruses are divided into four groups: mosquito borne (MBFV), tick borne (TBFV), viruses that infect mammals with no known vector (NKVFV), and viruses that are insect specific (ISFV).
The 3′ UTRs of MBFV include duplications of complex RNA structures known as stem loops and dumbbells (DBs) (4). The ZIKV 3′ UTR carries conserved duplicated SLs (SLI and SLII) but bears a single DB element preceded by a unique RNA structure, absent in most MBVF, named pseudo-DB (ΨDB). SLs and DBs contain conserved sequences and structures, including stabilizing pseudoknot (PK) interactions. The compact folding of these RNA elements forms units that are known as XRN1-resistant structures (xrRNAs), due to their ability to stall exoribonuclease degradation during viral genome decay (13). This process results in the accumulation of noncoding subgenomic flavivirus RNAs (sfRNAs) (14–16). The structural details by which the SLs block XRN1 activity were elucidated by crystal structures of SLs from Murray Valley encephalitis virus (MVEV) and ZIKV (17, 18). These studies revealed a common knot-like fold of the SL that impairs XRN1 activity. DB structures also stall XRN1 when SLs are mutated or, in the case of DENV, when the virus is adapted to mosquitoes (19, 20). However, the mechanism by which DB structures block exoribonuclease-dependent degradation of the genome is still unknown.
Accumulation of sfRNAs in flavivirus infections has been shown to be implicated in viral replication, cytopathicity, and pathogenesis (14, 21–24). It has been demonstrated that sfRNAs modulate host responses against flaviviruses in humans and mosquitoes (23, 25–28). Different studies support the idea that sfRNAs act as sponges, sequestering RNA binding proteins important in antiviral responses, dysregulating cellular mRNA decay, and modulating host RNA splicing (29–32). However, the mechanisms by which these viral molecules function during flavivirus infections are still under intensive investigation (for review, see reference 33). It is important to highlight that each flavivirus produces different amounts, sizes, and numbers of sfRNAs, and the factors modulating these differences are still unclear. It is likely that different selective pressures during flavivirus evolution led to fine-tuning of the quality of sfRNAs produced by each virus depending on different antiviral responses in distinct host species, viral tropism, or other viral requirements. For instance, although DENV and ZIKV are both MBFVs and conserve two SL structures at their 3′ UTRs, DENV infection mainly accumulates a single sfRNA, while ZIKV predominantly produces two types of these molecules (18, 20). Moreover, accumulation of different amounts of sfRNAs during infection has been associated with distinct epidemiological fitness of DENV isolates (32), indicating the relevance and complexity of sfRNA functions in flavivirus infections.
In this study, we investigated molecular determinants and specific requirements for sfRNA accumulation during ZIKV infection in different hosts. To this end, we generated new infectious ZIKV clones and reporter viruses from local Argentinean isolates. Recombinant ZIKV with different 3′ UTR arrangements revealed strong cooperativity between SLI and SLII for sfRNA generation. Mutations or deletions of SLII abolished SLI exoribonuclease stalling function. In addition, ZIKV mutants that produce different types and amounts of sfRNAs revealed host-specific requirements for infection and viral propagation. The generation of at least one sfRNA was necessary for the virus to maintain the natural capacity to overcome the midgut and salivary gland barriers of Aedes aegypti mosquitoes. ZIKV lacking SLI produced large amounts of a single sfRNA, and the virus replicated in mosquito cells about 50-fold more than WT ZIKV. This virus was efficient in infecting mosquitoes, while infection in human cells was reduced. Interestingly, recombinant viruses lacking the ability to produce sfRNAs, which grow efficiently in mosquito cells, were unproductive in human cells. In summary, this work identifies molecular determinants for ZIKV sfRNA generation and demonstrates the essential role of sfRNAs for ZIKV productive infection in human cells.
RESULTS
Construction of epidemic and nonepidemic full-length ZIKV cDNA clones.
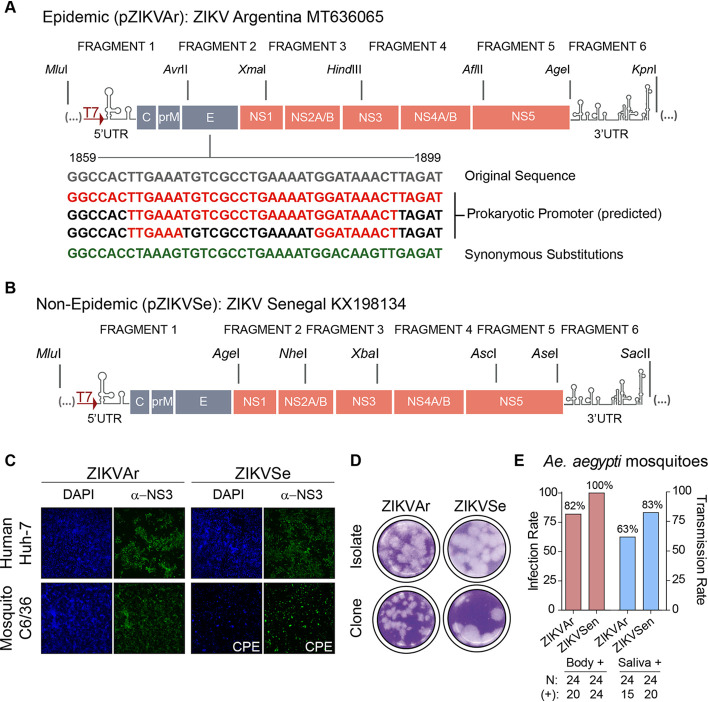
A ZIKV isolate from Argentina (epidemic virus; GenBank accession no. MT636065) was amplified in Vero cells, sequenced, and used as the template to construct a cDNA clone. The full genome was divided into 6 fragments (Fig. 1A). Each fragment (F1 to F6) was amplified by reverse transcription-PCR (RT-PCR) and individually cloned into a medium-copy-number vector, except for F2. The plasmid containing fragment F2 was consistently rescued with mutations and deletions, indicating the unstable nature of this sequence in bacteria. The mutations identified, spanning nucleotides 1859 to1899 at the envelope protein-coding sequence, disrupted the open reading frame (ORF), suggesting the presence of a cryptic promoter(s) and translation of toxic viral sequences in Escherichia coli. This problem has been previously reported for ZIKV and other flaviviruses (34, 35). Three different algorithms that predict cryptic promoters in bacteria supported the presence of such sequences in F2 (Fig. 1A). Based on this information, synonymous mutations were incorporated to disrupt the promoter. These changes were sufficient to obtain a stable F2 fragment. The complete cDNA sequence of ZIKV was then assembled in a medium-copy-number plasmid by sequential incorporation of the different fragments to obtain pZIKVAr.
FIG 1.
Design and characterization of full-length ZIKV infectious clones. (A) Representation of the strategy used for assembling a ZIKV cDNA clone from an Argentinean isolate (pZIKVAr). Restriction sites used to clone each fragment are indicated on the top. Below this schematic are shown the sequences of predicted prokaryotic promoters (red) responsible for instability in bacteria. Synonymous mutations incorporated are indicated in green. (B) Representation of the strategy used for assembling a ZIKV cDNA clone from a laboratory-adapted virus obtained from Senegal (pZIKVSe). (C) Transfections of viral RNAs generated by in vitro transcription are infectious in mosquito and human cells, as shown by immunofluorescence using antibodies against ZIKV NS3 protein. CPE, cytopathic effect. (D) The rescued ZIKVAr and ZIKVSen are infectious and form plaques in BHK cells. (E) Infection and transmission rates of the cloned viruses in A. Aegypti mosquitoes are shown. Female mosquitoes were fed with an infectious blood meal containing 1 × 106 FFU/ml of ZIKVAr or ZIKVSen.
Using a similar strategy, a second infectious clone from a laboratory-adapted ZIKV from Senegal (nonepidemic virus; GenBank accession no. KX198134) was constructed. The full-length genome was also divided into 6 fragments (Fig. 1B). Each fragment was cloned individually, without unstable sequences in E. coli. Thus, they were directly assembled into a medium-copy-number plasmid to obtain pZIKVSe. The stability of pZIKVAr and pZIKVSe plasmids was assessed by 5 successive transformations in bacteria and plasmid preparation.
To examine infectivity, T7 RNA transcripts from pZIKVAr and pZIKVSe were obtained and transfected into mosquito and human cells. RNAs were infectious, and viruses propagated in both cell types, showing profound cytopathic effect after 4 and 3 days, respectively (Fig. 1C). In addition, both viruses produced plaques that were more homogeneous with the cloned viruses than the original isolates (Fig. 1D). Titers between 107 and 108 PFU/ml were obtained from both infectious clones. ZIKVSe showed larger plaques than ZIKVAr (Fig. 1D). Viral stocks obtained from both infectious clones were used to infect A. aegypti mosquitoes. A blood meal with final titer of 1 × 106 PFU/ml was used. Both viruses showed high rate of infectivity in mosquitoes (between 80 and 100%), and both reached saliva (Fig. 1E). We conclude that the two infectious clones are suitable tools to study molecular aspects of ZIKV biology.
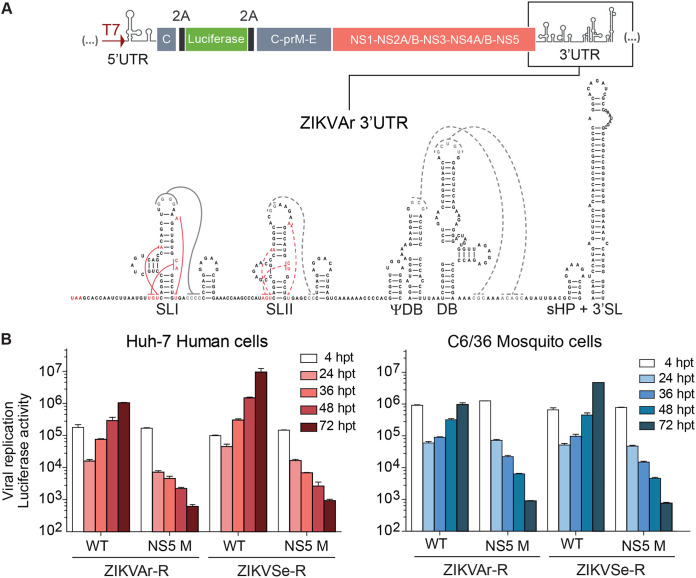
In order to dissect functions of viral RNA structures relevant for sfRNA formation, full-length ZIKV reporters carrying the luciferase gene were constructed from the infectious clones. To this end, we introduced the Renilla luciferase coding sequence flanked by foot and mouth disease virus (FMDV) 2A sites, followed by the complete viral open reading frame. In addition, viral cis-acting replication sequences present in the capsid protein coding region were included upstream of the luciferase to ensure efficient viral RNA replication (Fig. 2A). For each ZIKV reporter construct, a replication-impaired control was constructed by mutation of the GDD catalytic site of the NS5 polymerase domain (NS5 M). Transfection of transcribed RNA corresponding to epidemic (ZIKVAr-R) and nonepidemic (ZIKVSe-R) ZIKV reporters resulted in viral replication in human and mosquito cells (Fig. 2B). The reporter viruses generated infectious progeny retaining luciferase expression. Replication kinetics comparisons showed a faster replication of ZIKVSe-R than of the epidemic ZIKVAr-R in both cell types (Fig. 2B). A dynamic range of at least 3 orders of magnitude was observed when the wild type (WT) and NS5 M replication-impaired control were compared (Fig. 2B).
FIG 2.
Design and construction of ZIKV reporter systems. (A) Schematic representation of ZIKV reporter system carrying the luciferase gene flanked by FMDV 2A sequences. The predicted RNA structures in the 3′ UTR, including stem loops I and II (SLI and SLII), pseudodumbbell ΨDB, dumbbell DB, and a small hairpin 3′ stem-loop (sHP-3′ SL), are indicated. (B) Reporter ZIKVs efficiently translate and replicate in mosquito and human cells. Luciferase expression levels of ZIKVAr-R and ZIKVSen-R are shown as a function of the time. Luciferase activity was measured in transfected cells with viral RNAs corresponding to WT and replication-impaired NS5 M. The luciferase values are means ± standard deviations (representative experiment of three independent experiments). hpt, hours posttransfection.
Inverted locations of SLI and SLII in DENV and ZIKV 3′ UTRs are associated with function.
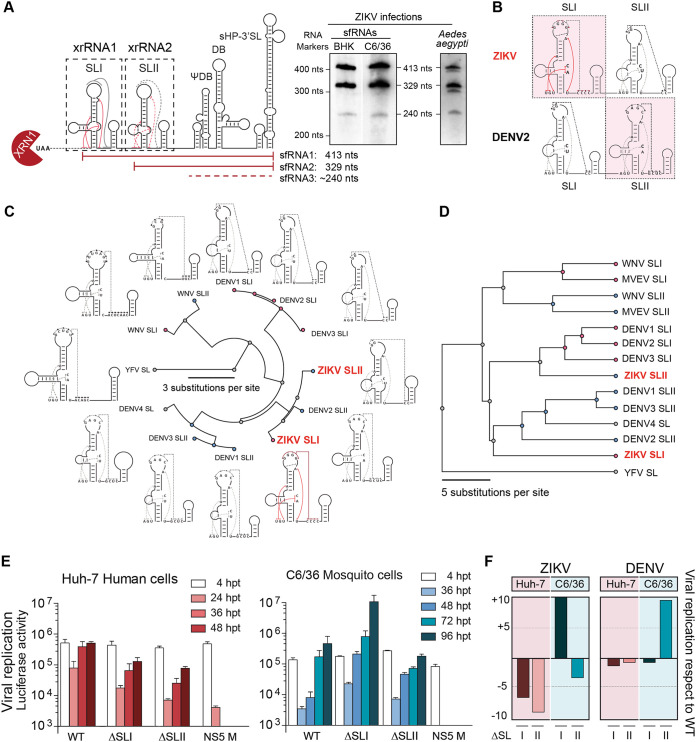
The two SL structures present at the viral 3′ UTR have been directly involved in the generation of sfRNAs for different MBFVs and in host adaptation in DENV. In the case of ZIKV infections, two main species of sfRNAs (sfRNA1 and sfRNA2) are produced in cell culture and A. aegypti mosquitoes, which are associated with xrRNA1 structure (including SLI and downstream hairpin) and xrRNA2 structure (including SLII and downstream hairpin), respectively (Fig. 3A). However, the role of the SL elements in ZIKV infection and sfRNA generation has not been systematically examined. We have previously studied functions of DENV SLI and SLII and found that they play opposite roles in infected mosquitoes, but have redundant functions in infected human cells (20, 36). Although SLI and SLII share structural elements and sequences among different MBFVs, comparison of key structural features critical for xrRNA function within these structures from DENV and ZIKV suggested that their locations may be inverted (Fig. 3B). In this regard, ZIKV SLI resembles DENV SLII, and ZIKV SLII bears sequence and structure similar to those of DENV SLI.
FIG 3.
Distinct functions of duplicated stem-loop structures present at the ZIKV 3′ UTR. (A) Schematic representation of ZIKV 3′ UTR showing the two XRN1-resistant RNAs involved in sfRNA formation (xrRNA1 and xrRNA2, including SLI and SLII, respectively). At the right are Northern blots showing the pattern of sfRNAs generated during ZIKV infection in vertebrate and invertebrate cells and A. aegypti mosquitoes as indicated. The two main sfRNAs of 413 and 329 nucleotides are shown together with a minor sfRNA of about 240 nucleotides. (B) Comparison of secondary structures of duplicated SLs from ZIKV and DENV2 based on available biochemical probing and crystal structures. (C) Fan dendrogram indicating the distance between SL structures of the indicated mosquito-borne flaviviruses (MBFVs). SLI and SLII of different MBFVs correspond to the structures included in xrRNA1 and xrRNA2, respectively, and their locations (first and second) are with respect to the 5′ end of the 3′ UTR. (D) Phylogenetic analysis based on sequence alignments of SLs from different MBFVs. (E) Distinct functions of the two SLs in human and mosquito cells, as indicated in each case. Replication of recombinant reporter ZIKV carrying deletions of xrRNA1 (ΔSLI) or xrRNA2 (ΔSLII) is shown together with the WT and a replication-impaired control NS5 M as a function of time. The luciferase values are means ± standard deviations of a representative experiment (three independent experiments were performed). (F) Roles of SLI and SLII of ZIKV and DENV in viral replication in the two hosts support inverted location of these two RNA elements.
To examine the possible inversion of the SLs in DENV and ZIKV, sequence and structural comparisons between SLs from different flaviviruses were performed (Fig. 3C and D). Both studies indicated that ZIKV SLs are closer to DENV SLs than to SLs of other flaviviruses. Secondary RNA structure similarity analyses were made for different flavivirus SLs with known structures and using the tree alignment model implemented in the RNAforester software. This method allows determination of the relative similarity between two models of secondary structures by estimating the number of changes necessary to convert one structure to another. The available crystal structures of SLs were used to guide the folding of other flavivirus SL structures (17, 18). The analysis showed that both ZIKV SLs are structurally related to DENV2 SLs, and the ZIKV SLI was clearly closer to DENV2 SLII (Fig. 3C). Moreover, by analyzing nucleotide sequence similarities by MUSCLE and using Jalview software (37), the alignment shows that ZIKV SLI is closer to DENV SLII and that ZIKV SLII is closer to DENV SLI (Fig. 3D). Based on these observations, we hypothesized that their functions may also be inverted, and this could be the reason why different types of sfRNAs are produced by the two viruses.
To evaluate the function of the ZIKV SLs, we constructed recombinant viruses in the context of the reporter system lacking each of these elements (ZIKV-RΔSLI and ZIKV-RΔSLII). In vitro-transcribed viral RNA from the WT and mutants were transfected in human and mosquito cells. Transfections in human cells indicated that the lack of either SLI or SLII reduced viral replication (Fig. 3E). The two mutant reporter viruses ZIKV-RΔSLI and ZIKV-RΔSLII consistently replicated about 10-fold less than the ZIKV-R WT. In contrast, when ZIKV-RΔSLI and ZIKV-RΔSLII RNAs were transfected into mosquito cells, the SLs showed opposite effects on viral replication (Fig. 3E). While ZIKV-RΔSLII replicated less than the ZIKV-RWT at 72 and 96 h, ZIKV-RΔSLI showed a great increase in RNA amplification, about 50-fold more than the WT (Fig. 3E). The result obtained in this experiment is the opposite of that previously reported with recombinant DENV (36). It has been previously shown that SLII of DENV was subjected to sequence variations during mosquito adaptation, and mutations in this element greatly increased viral replication. In the case of ZIKV, the deletion of SLI displayed a higher replication phenotype, suggesting functional similarities between SLI of ZIKV and SLII of DENV2 (Fig. 3F).
In conclusion, we found that ZIKV and DENV2 SLs evolved functionally equivalent structures in inverted locations of their 3′ UTRs, suggesting that SLI and SLII diverged in each virus after duplication.
Dissecting functional determinants for ZIKV sfRNA generation reveals strong cooperativity between the duplicated SL structures.
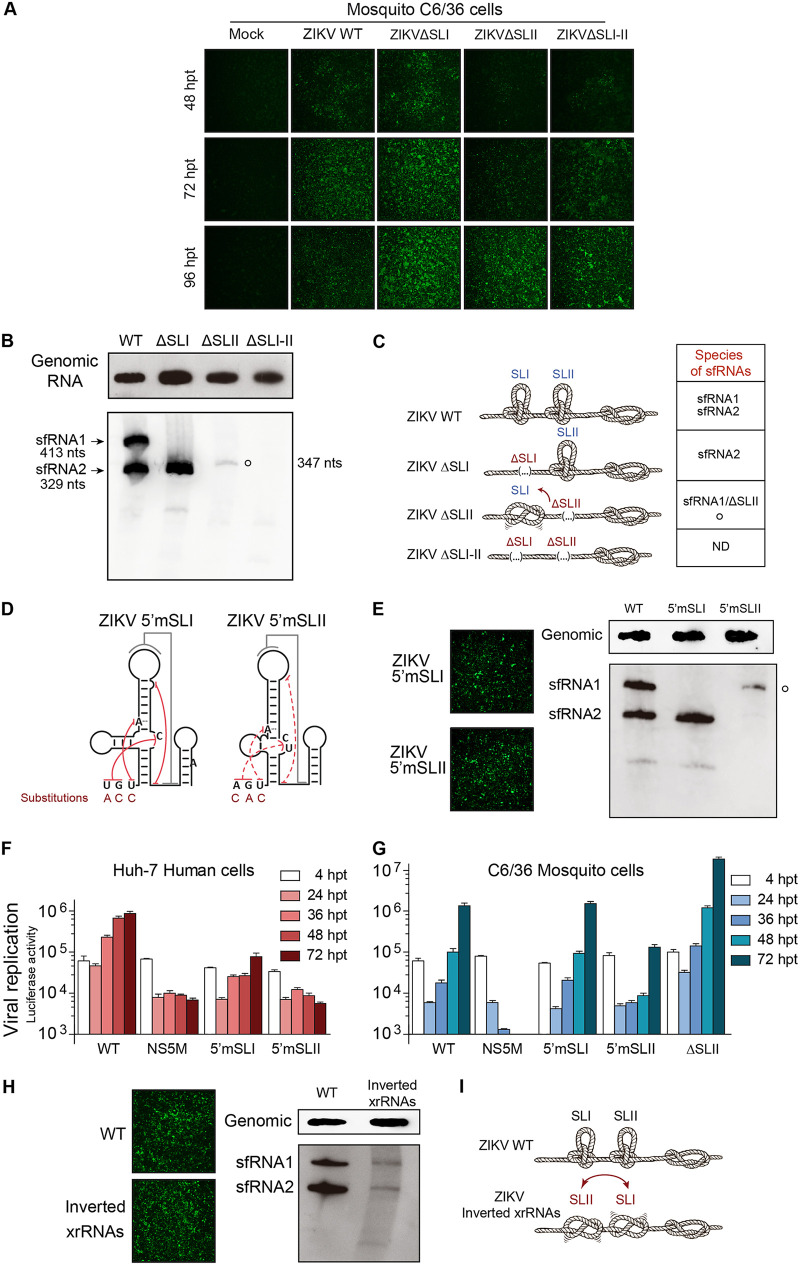
To define RNA elements required for sfRNA accumulation during ZIKV infection, we studied which types of sfRNAs are produced by viruses with different SL arrangements at the viral 3′ UTR. Recombinant viruses were designed in the context of the epidemic clone. First, we generated ZIKVΔSLI, ZIKVΔSLII, and ZIKVΔSLI-II, lacking xrRNA1, xrRNA2, and both, respectively. In vitro-transcribed RNA from these clones and the WT were transfected in C6/36 cells. Immunofluorescence (IF) analysis as a function of time showed that ZIKVΔSLI replicated faster than the WT, while ZIKVΔSLII and ZIKVΔSLI-II were slightly delayed (Fig. 4A). Total RNAs from infected cells were used to evaluate genomic RNA and to examine the quality of sfRNAs accumulated in each case. Northern blotting was performed with samples harvested at 72 h for ZIKV WT and ZIKVΔSLI and at 96 h for ZIKVΔSLII and ZIKVΔSLI-II. At the indicated times the amounts of genomic RNA, evaluated with a probe hybridizing to viral 5′ UTR sequences, were similar for WT, ΔSLII, and ΔSLI-II ZIKVs but were consistently higher for the ZIKVΔSLI mutant (Fig. 4B), in agreement with the increased RNA replication observed with the reporter virus (Fig. 3E). Extracts from ZIKV WT-infected cells showed the accumulation of two species of sfRNAs, and the recombinant ZIKVΔSLI yielded only one species (sfRNA2) of 329 nucleotides in length (Fig. 4B). sfRNA2 is the product of stalling XRN1 at the intact SLII structure of xrRNA2. However, in infections with ZIKVΔSLII, which retains an intact SLI, sfRNAs were almost undetectable. A faint band consistent with the length of the sfRNA1 (carrying a deletion of SLII) of 347 nucleotides was observed, suggesting insufficient strength of xrRNA1 alone to stop exoribonuclease degradation (Fig. 4B). Furthermore, deletion of SLI-II abolished production of sfRNAs (Fig. 4B). In this regard, it is important to mention that previous studies with other flaviviruses, including DENV and WNV, lacking the homologous SLI-SLII region, accumulated shorter sfRNA species (sfRNA3 and sfRNA4) by stalling XRN1 at DB structures (13, 14, 19, 20, 38). Thus, ZIKVΔSLI-II appears to have different properties.
FIG 4.
Dissecting the molecular determinants for ZIKV sfRNA accumulation. (A) Representative immunofluorescence showing replication and propagation of ZIKV WT and recombinants carrying deletions of each SL or both SLs. (B) SLII plays a central role in ZIKV sfRNA generation. Shown are Northern blots detecting genomic RNA (top) and sfRNAs (bottom) from infected mosquito cells with the indicated recombinant ZIKVs. The size and mobility of different sfRNA species are indicated on the left (sfRNA1 and -2). An sfRNA of 347 nucleotides in length generated by a ZIKV with SLII deletion (empty circle) is indicated on the right. (C) Illustrative representation of the SLs as knots and the species of sfRNAs produced by each virus. (D) Destabilization of SL structures alters sfRNA generation. Representation of mutants ZIKV5′mSLI and ZIKV5′mSLII, indicating the location and nucleotide substitution in each case. (E) (Left) Replication of transfected ZIKV5′mSLI and ZIKV5′mSLII RNAs in C6/36 cells shown by immunofluorescence. (Right) sfRNA pattern produced by ZIKV WT, ZIKV5′mSLI, and ZIKV5′mSLII, as indicated. The empty circle indicates the sfRNA produced by the ZIKV5′mSLII of 413 nucleotides. (F and G) Replication of ZIKVAr-R carrying the 5′mSLI and 5′mSLII mutations in human and mosquito cells, as indicated in each case, was assessed by measuring luciferase activity as a function of time. The luciferase values are means ± standard deviations from a representative experiment (three independent experiments were performed). (H) (Left) Replication of ZIKVAr WT or ZIKVAr xrRNA inverted mutant at 3 and 4 days posttransfection, shown by immunofluorescence. (Right) Northern blots showing genomic RNA (top) and sfRNAs (bottom) from infected mosquito cells with the indicated recombinant ZIKVs (WT or xrRNAs inverted mutant). (I) Illustrative representation of xrRNAs for the WT and mutant, highlighting the relevance of the location and cooperativity between the two SLs.
In summary, we found that (i) an intact SLI was not sufficient to stop the exoribonuclease activity in the absence of SLII, (ii) an intact SLII was sufficient to generate sfRNA2, and (iii) sfRNA production was absent in viruses lacking both SLs (Fig. 4C).
The strong effect of deleting SLII on the function of SLI for ZIKV sfRNA formation suggests a structural interaction between these two RNA elements. To confirm this, and to rule out unpredicted structural alterations due to the deletions introduced in the viral 3′ UTR, we designed new recombinant viruses based on crystallographic studies of flavivirus SLs (17, 18). It has been shown that nucleotides preceding the SLs base-pair with nucleotides at the three-way junction, forming an internal pseudoknot (Fig. 4D). Mutations were introduced upstream of SLI (UGU replaced by ACC) or SLII (AGU replaced by CAC), impairing in each case formation of the internal PK, ZIKV5′mSLI and ZIKV5′mSLII, respectively (Fig. 4D). These mutations were incorporated in both infectious clones and reporter ZIKV-R.
Transfection of RNA from the ZIKV WT and mutants (ZIKV5′mSLI and ZIKV5′mSLII) led to viral replication and propagation in C6/36 cells (Fig. 4E). Infection with ZIKV5′mSLI resulted in the accumulation of a single sfRNA corresponding in size to sfRNA2, indicating resistance to RNA degradation at SLII (Fig. 4E). ZIKV5′mSLII, holding an intact SLI, generated a barely detectable sfRNA (413 nucleotides long). This phenotype was similar to that observed with the deletion of the complete SLII (ZIKVΔSLII mutant). In this regard, the size difference observed in the detectable sfRNAs for ZIKVΔSLII and ZIKV5′mSLII accounts for the deletion of the SLII (compare Fig. 4B and E). These results show the requirement of a stable SLII structure for SLI function. We conclude that the two SLs display strong cooperativity for sfRNA1 production, suggesting a high-order organization of the RNA in this region of the 3′ UTR.
To evaluate the replication properties of viruses carrying the 5′mSLI and 5′mSLII substitutions, reporter ZIKV constructs were designed. RNAs corresponding to the WT, the replication-impaired control, and the two mutants (ZIKV-R 5′mSLI and 5′mSLII) were transfected into C6/36 mosquito and Huh-7 human cells, and reporter activity was evaluated as a function of time. The virus ZIKV-R 5′mSLI, which accumulated large amounts of sfRNA2, showed a delayed RNA synthesis and about 10-fold-reduced reporter activity in human cells (Fig. 4F). In contrast, the mutant ZIKV-R 5′mSLII, which was sfRNA-impaired, displayed low levels of reporter activity, supporting an important defect in replication of this virus (Fig. 4F). In mosquito cells, ZIKV-R 5′mSLI replicated similarly to the WT, while the mutant ZIKV-R 5′mSLII showed a reduction of about 10-fold in reporter activity with respect to that of the WT (Fig. 4G). These results, together with the information obtained with the recombinant and reporter viruses lacking SLI, indicate that the increased fitness of ZIKVΔSLI (also included in Fig. 4G for comparison) in mosquito cells was not due to the production of a single sfRNA (sfRNA2); the ZIKV-R 5'mSLI produced the same sfRNA2 but replicated similarly to the ZIKV-R WT (compare ZIKV-R 5′mSLI and ZIKVΔSLI).
To further assess whether location of the SLs determines the different qualities of sfRNAs produced by DENV and ZIKV, or whether the interplay between these structures requires specific locations for their function, we inverted their positions in the ZIKV 3′ UTR. To this end, the complete xrRNA2 structure (including SLII and downstream hairpin [Fig. 3A]) was placed upstream of SLI. A virus with this 3′ UTR rearrangement was infectious, and viral propagation was readily detected in mosquito cells (Fig. 4H). The genomic RNA of this recombinant virus was similar to that observed for the WT virus; however, the amounts of sfRNA1 and -2 were largely reduced (Fig. 4F). This result indicates that both SLs were inefficient to resist exoribonuclease degradation, confirming an interplay between the two structures in the WT arrangement (Fig. 4I).
In conclusion, we dissected functional elements responsible for ZIKV sfRNA formation. The data support a model in which the two SLs at the ZIKV 3′ UTR function in a cooperative manner to halt exoribonuclease activity. Importantly, we generated a recombinant ZIKV that propagates in mosquito cells without producing detectable sfRNAs.
ZIKV ability to generate sfRNAs is necessary for dual host infection.
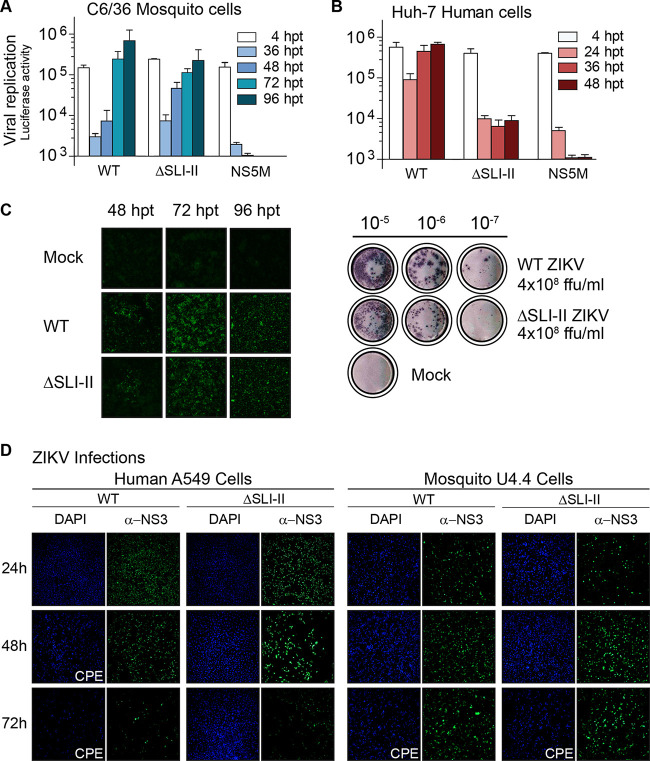
To further study the properties of the virus that lacks sfRNA accumulation, we constructed the reporter ZIKV-RΔSLI-II. Transfection of mutant, WT, or control viral RNA into insect cells showed efficient translation and RNA replication of ZIKV-RΔSLI-II (Fig. 5A). The same reporter virus showed impaired reporter activity in human cells (Fig. 5B), suggesting differential requirements of sfRNAs in the two hosts. To confirm this observation, infection and propagation properties of the ZIKVΔSLI-II in the context of the infectious clone were evaluated. Immunofluorescence as a function of time after viral RNA transfection showed efficient infection in C6/36 cells. Although ZIKVΔSLI-II showed delayed propagation with respect to that of the WT, titers above 108 infectious particles per ml, similar to those of the WT, were obtained (Fig. 5C). In contrast, no propagation of ZIKVΔSLI-II was detected in transfected Huh-7 human cells. ZIKVΔSLI-II stocks were generated in C6/36 cells, and since ZIKV does not produce plaques in these cells, focus-forming units (FFU) were evaluated to quantify the virus (Fig. 5C). To examine if the sfRNA-deficient virus was able to infect and propagate at low levels in human cells, the virus produced in mosquito cells was used to infect A549 human cells. ZIKVΔSLI-II infection was observed at 24 h by IF analysis. However, the viral antigen signal was undetectable at 72 h postinfection (hpi) (Fig. 5D). This phenotype indicates the inability of the mutant virus to sustain the infection in human cells. Together, the results show different behaviors for ZIKV-RΔSLI-II for productive propagation in mosquito and human cells.
FIG 5.
Phenotype of ZIKV lacking the ability to generate sfRNAs in human and mosquito cells (A and B) Differential requirement of sfRNA generation for ZIKV replication in the two hosts. Luciferase expression levels of ZIKVAr-R WT and ΔSLI-II are shown as a function of time in Huh-7 and C6/36 cells. The luciferase values shown are means ± standard deviations from a representative experiment (three independent experiments were performed). (C) (Left) immunofluorescence showing ZIKV antigen positivity in C6/36 cells transfected with ZIKVAr WT and ZIKVAr ΔSLI-II. (Right) The WT and mutant virus produced high-titer stocks, as measured by focus-forming units (FFU) in C6/36 cells. (D) Representative images from immunofluorescence assays showing ZIKV antigen positivity in human A549 and Dicer-competent U4.4 mosquito cells infected with ZIKVAr WT or ZIKVAr ΔSLI-II.
It has been previously reported that the mosquito C6/36 cell line is Dicer 2 deficient; thus, we hypothesized that this defect could be responsible for the permissiveness to infection of these cells. To evaluate this, we analyzed the ability of ZIKVΔSLI-II to infect the Dicer 2-competent U4.4 mosquito cells. In this regard, the ZIKVΔSLI-II was also able to infect and propagate efficiently in U4.4 cells, as determined by IF analysis (Fig. 5D), indicating that replication and propagation of ZIKVΔSLI-II are independent of the function of Dicer-2.
We conclude that a deletion at the viral 3′ UTR, which abrogates generation of sfRNAs, results in a ZIKV that infects and propagates in mosquito cells but shows profound defects in human cells.
A ZIKV that does not accumulate sfRNAs fails to control the antiviral response in human cells.
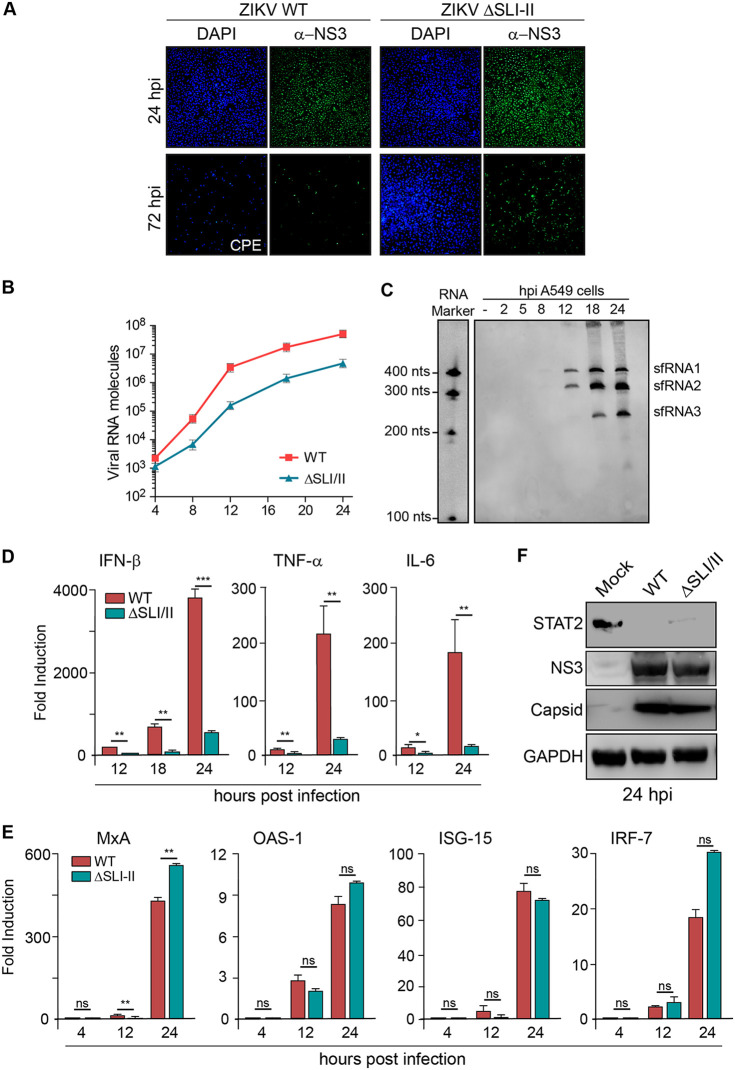
We identified a ZIKV with impaired sfRNA generation that propagated in mosquito cells but was limited in human cells. Taking into account the role of sfRNAs, we hypothesized that this virus was deficient in counteracting human antiviral responses. To study this possibility, we evaluated a time course of infection and the consequent antiviral response induced by the sfRNA-deficient ZIKV and the WT ZIKV in A549 cells. High-titer stocks of the WT and ZIKVΔSLI-II generated in mosquito cells were used to avoid secondary infections or bystander cell responses (Fig. 6A). Under these conditions, the complete monolayer was infected with both viruses at 24 hpi. However, while the WT virus showed extensive cytopathic effect and cell death at 72 hpi, cells infected with ZIKV ΔSLI-II were almost cured at this time (Fig. 6A). In this regard, positive viral antigen by IF was observed scarcely, and the monolayer was intact, as shown by 4′,6-diamidino-2-phenylindole (DAPI) staining (Fig. 6A). Quantification of intracellular accumulation of the viral genome indicated that the mutant virus infected and replicated, but the viral RNA levels were about 10-fold lower than for the WT (Fig. 6B). Kinetics of sfRNA generation for the WT virus was also determined under these conditions. This analysis indicated detectable sfRNA1 and sfRNA2 as early as 8 hpi, and at later times, sfRNA3 was also detected (Fig. 6C).
FIG 6.
Accumulation of ZIKV sfRNAs is essential for productive infection in human cells. (A) ZIKVAr ΔSLI-II infection is quickly cleared in A549 cells. Immunofluorescence shows ZIKV antigen positivity and DAPI staining in A549 cells infected at an MOI of 10 with WT or mutant ΔSLI-II virus. Cytopathic effect was observed with the WT virus at 72 hpi. (B) ZIKV RNA genome accumulation as a function of time assessed by RT-qPCR for the WT and mutant as indicated. (C) Northern blot showing the appearance of sfRNAs as a function of time from 2 to 24 h after infection of A549 cells with WT virus. RNA markers are shown on the left, and sfRNAs are indicated on the right. (D) Induction of cytokines produced in A549 cells infected with ZIKVAr WT or ZIKVAr ΔSLI-II under the same conditions as for panel A. IFN-β, TNF-α, and IL-6 mRNAs were evaluated by RT-qPCR and are expressed as fold induction with respect to the noninfected control and relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Shown are the means with SD of two replicates. Statistics were performed with an unpaired t test, with P values represented as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ns, not significant. (E) Induction of ISG in ZIKVAr WT- or ZIKVAr ΔSLI-II-infected cells. MxA, OAS-1, ISG-15, and IRF-7 mRNAs were evaluated by RT-qPCR and expressed as fold induction with respect to the control and relative to GAPDH mRNA. Shown are the means with SD of two replicates; statistics were performed as described for panel D. (F) STAT2 is efficiently degraded after 24 h of infection with WT or mutant ΔSLI-II virus in A549 cells. Western blots show human STAT2, viral structural capsid protein, nonstructural protein NS3, and GAPDH control.
Production of beta interferon (IFN-β) and proinflammatory cytokines and IFN signaling were evaluated during the first 24 h of infection. Induction of IFN-β was more than 10-fold higher for the WT than the mutant ZIKV, which correlated with higher levels of viral replication. Induction of tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) was also higher in the WT infection (Fig. 6D). Although the WT virus induced considerably higher levels of IFN, the sfRNA-deficient virus induced the same or higher levels of MxA, OAS1, ISG15, and IRF-7 mRNAs, suggesting that this mutant has a limitation in controlling IFN signaling (Fig. 6E).
Flaviviruses use diverse mechanisms to counteract the IFN signaling; some of them are well-described processes, such as NS5-mediated STAT2 degradation (39, 40). Thus, the uncontrolled induction of IFN-stimulated genes (ISGs) with the mutant virus could be associated with reduced degradation of STAT2 as a result of lower levels of NS5. To evaluate this possibility, viral proteins and STAT2 were assessed at 24 hpi. The accumulation of structural and nonstructural viral proteins was lower in the cytoplasmic extracts obtained from mutant infection than that for the WT; however, STAT2 levels were almost undetectable in both infections, indicating efficient degradation of the host protein by both viruses (Fig. 6F). These results provide evidence that the lack of sfRNAs during infection of human cells leads to an antiviral response that clears the virus. It is likely that early events after infection are triggered differentially by the sfRNA-deficient and WT viruses, since at 8 hpi, a reduced viral genome accumulation of the mutant was observed (Fig. 6B). Viral clearance could be explained by uncontrolled IFN signaling or deregulation of ISG mRNAs translation, in addition to other early intrinsic or innate responses. Further studies will be necessary to define the implication of such early events.
In summary, our findings provide conclusive evidence of the need for sfRNAs for ZIKV productive infection in human cells.
Generation of at least one sfRNA species is necessary for efficient infection and transmission of ZIKV in Aedes aegypti mosquitoes.
Natural mosquito infections with different flaviviruses produce sfRNAs that modulate antiviral responses. However, the role of these noncoding viral RNAs in mosquito infections is still unclear. To investigate the requirement of different species of sfRNAs for infection and transmission in A. aegypti, we used viral mutants deficient in generating different sfRNAs. Viral stocks of recombinant ZIKVs that produce sfRNA1 and -2, only sfRNA2, or neither of them (ZIKV WT, ZIKVΔSLI, ZIKVΔSLII, or ZIKVΔSLI-II) were generated in mosquito C6/36 cells.
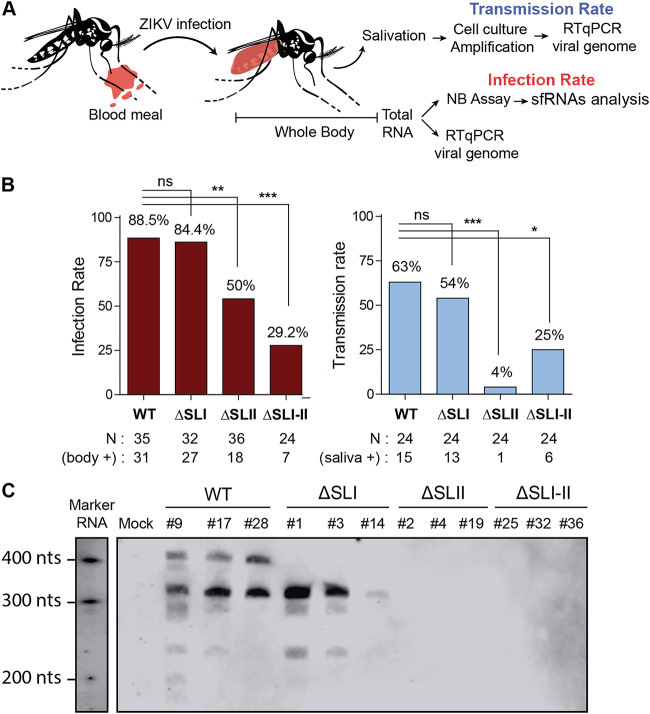
Female A. aegypti mosquitoes were fed with an infectious blood meal containing 106 FFU/ml of ZIKV WT, ZIKVΔSLI, ZIKVΔSLII, or ZIKVΔSLI-II. At 14 days postfeeding, the presence of the virus was evaluated in the body and saliva of individual mosquitoes. In addition, RNA extracts of whole mosquitoes were used to assess the presence and quality of sfRNAs by Northern blotting (Fig. 7A). The WT and the ΔSLI viruses showed high infection rates, with more than 80% of the mosquitoes ZIKV positive in three different experiments. In contrast, viruses lacking SLII or both SLs showed reduced infections rates with respect to that of WT (about 50 and 30%, respectively [Fig. 7B]). Saliva from mosquitoes was collected and used for viral amplification in cultured mosquito cells. The presence of ZIKV was assessed in the supernatant of these cultures. Using this protocol, more than 50% of the mosquitoes infected with WT or ΔSLI virus showed viral presence in saliva. Viruses lacking sfRNAs also reached saliva but with remarkable less efficiency, between 4% and 25% (Fig. 7B).
FIG 7.
At least one sfRNA is necessary for efficient ZIKV infection and transmission in A. aegypti mosquitoes. (A) Schematic overview of experimental design. (B) Female A. aegypti mosquitoes were fed with an infectious blood meal containing 1 × 106 FFU/ml of ZIKVAr WT or mutant viruses producing different qualities of sfRNAs. Mutant ZIKVAr ΔSLI, ΔSLII, or ΔSLI-II was used as indicated. Engorged females were incubated for 14 days at 28°C, and infection and transmission rates were determined by viral genome RT-qPCR of whole body or saliva. Results from a representative experiment are shown. Statistics were performed by Fisher’s exact test on cumulative data analyzing each condition in pairs. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (C) ZIKV mutants produce different kinds of sfRNAs in infected A. aegypti mosquitoes. Northern blots of RNA samples prepared from individual infected mosquitoes are shown.
Mosquitoes that were positive for ZIKV infection were analyzed to study the quality and amount of sfRNAs produced. Individual mosquitoes were used to extract total RNA and subjected to Northern blot analysis (Fig. 7C). WT ZIKV accumulated similar amounts of two species of sfRNAs and a small amount of a third species, also observed in cell culture infections, of about 250 nucleotides in length. ZIKVΔSLI produced mainly sfRNA2. In mosquitoes infected with ZIKVΔSLII or ZIKVΔSLI-II, which were positive for genomic viral RNA, no sfRNAs were detected, confirming a central role for SLII in sfRNA formation.
We conclude that production of sfRNA1 is not required for ZIKV to complete a successful transmission cycle. Importantly, production of at least one sfRNA is necessary for efficient infection and transmission of ZIKV in A. aegypti mosquitoes.
DISCUSSION
In this study, we identified molecular requirements for ZIKV sfRNA generation, providing new information about the relevance of different species of sfRNAs for infecting human cells and A. aegypti mosquitoes. A complex interplay between different RNA structures present at the viral 3′ UTR for production of sfRNAs was found. Importantly, we generated new recombinant ZIKVs incapable of producing sfRNAs that were able to infect and propagate in mosquito cells. In contrast, the sfRNA-impaired virus was rapidly cleared in human cells, demonstrating the essential role of sfRNAs in counteracting antiviral responses in these cells. Interestingly, A. aegypti mosquito infections using a battery of ZIKV mutants producing different kinds and amounts of sfRNAs revealed that at least one species of sfRNA is sufficient for infection and transmission, and viruses that did not produce sfRNAs infected mosquitoes inefficiently. Finally, distinct spatial arrangement of ZIKV and DENV RNA structures at the viral 3′ UTR were defined, supporting specialization of RNA elements for viral infection, with possible implications in virulence and viral transmission.
Important human flaviviruses, including DENV, YFV, ZIKV, and WNV, produce different kinds of sfRNAs, with different lengths and amounts (15, 18–20). A plethora of activities have been proposed for these molecules, and likely they play multiple roles during infection. It has been reported that they suppress type I IFN response in vertebrates, inhibit RNA interference (RNAi) and Toll pathways in insects, impair cellular RNA turnover, and regulate cellular splicing (for review, see references 33 and 41). Different studies have searched for the molecular targets of sfRNA in viral RNA sensing and IFN response pathways. In this regard, binding of DENV sfRNAs to G3BP1, G3BP1, and CAPRIN1 was proposed to modulate translation of ISGs (25). Also, sfRNA binding to TRIM25 was reported to reduce viral sensing by RIG-I (32). More recently, it has been reported that sfRNA binding to FMRP and DDX6 (ME31B in mosquitoes) could restrict antiviral activities of these proteins (29, 31, 42, 43). Because flaviviruses infect a wide range of hosts (humans, birds, bats, and insects, among others), sfRNAs should counteract different types of antiviral responses or facilitate viral processes in different environments. Thus, the distinct kinds of sfRNAs accumulated in flavivirus infections likely fulfill virus-specific needs. Although an important body of work about molecular determinants and functions of sfRNAs from different MBFVs has been generated, it is necessary to understand the mechanisms, functions, and evolution of sfRNAs in each virus.
Using recombinant ZIKV, we dissected requirements for sfRNA formation during infection. WT ZIKV mainly accumulates sfRNA1 and sfRNA2. We found that sfRNA1 formation requires the integrity not only of SLI but also of SLII. Point mutations that destabilize the SLII structure or deletion of this element impaired accumulation of both sfRNA1 and sfRNA2, suggesting strong cooperativity between the two SLs. In this regard, a recent study analyzed the conformation of complete sfRNAs from ZIKV by small-angle X-ray scattering (SAXS) and computational modeling (44). This report indicated that the region of the 3′ UTR containing SLI and SLII (xrRNA1-2) forms compact structures with reduced flexibility in solution, providing a structural basis for the functional cooperativity observed in this study with recombinant ZIKVs. Importantly, previous functional studies using DENV and WNV have shown coupling between the two SLs for sfRNA formation (20, 38). Using DENV, we have previously shown that deletion of SLII reduces SLI activity. In this case, xrRNA1 was still active and a distinctive pattern of sfRNAs was observed in infected cells (20, 45). In addition, infections with recombinant WNV with SLIV deletion (analogous to SLII in DENV and ZIKV) reduced function of the upstream SL and resulted in the accumulation of smaller sfRNAs (14, 19). It is important to mention that in the case of ZIKV, the lack of SLII almost abolished sfRNA accumulation, a property that was found to be unique for this virus (Fig. 4B). Together, these studies highlight the conservation of high-order RNA structures involved in sfRNA formation in different MBFVs. Of note, vaccine candidates for DENV and ZIKV with deletions within the DB structures have been shown to produce lower levels of sfRNAs, suggesting additional interactions between different regions of the 3′ UTR, in this case between SLs and DB elements (29, 46). RNA-RNA contacts between different domains of the 3′ UTR or interaction with proteins could facilitate the coupling observed. Further structural and functional studies will be necessary to define the nature of this cooperativity.
Our studies indicate that the two SL structures in the ZIKV genome diverged to acquire distinct functions. Deletion or mutations of SLI or SLII have different impacts on ZIKV replication (Fig. 3). Interestingly, opposite functions were found for the two SL structures for ZIKV infections in mosquito cells (Fig. 3E). Similar opposed functions were previously observed with DENV, but the SLs displayed an inverted location in the 3′ UTR (Fig. 3) (36). While SLII of ZIKV and SLI of DENV acted as enhancers of infection, SLI of ZIKV and SLII of DENV displayed strong inhibitory activity on viral RNA replication. In this regard, we have previously reported that adaptation of DENV to mosquito cells or competition experiments in infected Aedes mosquitoes led to positive selection of mutations and deletions specifically within SLII. Increased viral fitness was observed with cloned DENV variants adapted to mosquito with mutations in SLII (20). We have previously suggested that a change in sfRNA patterns (due to SLII mutations) was the cause of increased DENV replication in mosquitoes. Our data using ZIKV suggest that changes in the sequences of SLI rather than alteration in sfRNAs could be the cause of the increased viral fitness. In this regard, a ZIKV mutation in SLI (ZIKV5′mSLI) that alters sfRNA pattern, similar to that observed with the SLI deletion mutant, does not increase viral replication (Fig. 4E and G), suggesting that deletion of specific inhibitory sequences and not the accumulation of sfRNA2 may explain the phenotype observed.
It is intriguing why ZIKV and DENV have evolved functions of the SL structures in inverted locations. We can speculate that a ZIKV with a similar SL arrangement as that observed in the DENV 3′ UTR will be less fit in nature. In this case, the ZIKV SLII would be under selective pressure in mosquitoes, incorporating mutations that would impair production of both sfRNAs (Fig. 4B) and reduce the viral ability to infect and be transmitted by mosquitoes (Fig. 7). In contrast, in the ZIKV arrangement, if variations in SLI are associated with mosquito adaptation, this virus will still accumulate large amounts of sfRNA2 (Fig. 4B and E) for efficient infection and transmission (Fig. 7). In the case of DENV, the natural 3′ UTR arrangement is viable because mutations in SLII still leads to accumulation of sfRNAs, maintaining high fitness in mosquitoes (20). Thus, SL specialization in the two viruses could be under selective pressure to produce the necessary amounts and kinds of sfRNAs for each virus. Our studies support distinct functions of SLI and SLII in the two viruses, while maintaining, in both cases, SLII as a central regulator for sfRNA generation.
ZIKVs lacking the ability to generate sfRNAs replicated and propagated efficiently in mosquito cells, but infections were unproductive in human cells (Fig. 5A and B). This indicates that by manipulating the viral 3′ UTR it is possible to restrict replication to a single host. Interestingly, infection of human cells with a ZIKV lacking sfRNA production showed an unexpected phenotype. When a high multiplicity of infection (MOI) was used, infection was observed by IF analysis; however, this was followed by a quick elimination of the virus (Fig. 6A). It is likely that the cellular antiviral response controlled the propagation of the sfRNA-deficient virus. The levels of IFN and proinflammatory cytokines induced after infection correlated well with viral replication levels, which were about 10-fold higher for the WT than the mutant virus (Fig. 6B and E). However, the strong induction of ISGs observed with the sfRNA-deficient virus suggested a lack of control of IFN signaling (Fig. 6E). It is unclear whether this is a main cause of virus elimination. Additional intrinsic antiviral responses, controlled by sfRNAs in the WT, can be also relevant in limiting infection with the mutant virus. Importantly, we demonstrated for the first time an absolute requirement of sfRNAs for ZIKV propagation in human cells. This is a unique property of ZIKV, because previous reports showed that sfRNA-deficient flaviviruses, including WNV, YFV, and DENV, were able to sustain productive infections in human cells (15, 20, 23).
In A. aegypti mosquitoes, we found that at least one sfRNA was necessary for high infection and transmission rates. Viruses with deletions of SLII alone or in combination with SLI were severely impaired. Northern blot analysis of individual mosquitoes infected with these viruses indicated that in samples that were quantitative PCR (qPCR) positive for viral genome, no sfRNAs were detected. In contrast, mosquitoes infected with WT or ΔSLI virus accumulated sfRNA1 and sfRNA2 or only sfRNA2, respectively. Interestingly, a recent report has demonstrated a role for ZIKV sfRNAs in binding host antiviral proteins in mosquitoes (42). In that study, a ZIKV producing sfRNA2 but lacking sfRNA1 was employed. Using this virus, a significant role was assigned to sfRNA1 for infection and transmission. We found that a ZIKV producing only sfRNA2 (lacking sfRNA1) showed efficient infection and transmission rates in A. aegypti. However, although the mutations used in this study and in the previous study produced only sfRNA2, the recombinant viruses used carried different mutations in their 3′ UTRs. It is possible that the defect observed previously is due to the mutation introduced rather than the lack of sfRNA1. Also, because deletion of SLI displays a replication advantage in cell culture, we cannot rule out that sequences within SLI could be detrimental in adult mosquitoes, which may be masked by additional roles of SLI in sfRNA accumulation. Nevertheless, further studies will be necessary to dissect multiple functions of these RNA structures in mosquito infections. We conclude that ZIKV impaired in sfRNA formation displays strong defects in overcoming mosquito infection barriers, which is essential for viral transmission.
In this work, we delivered new genetic tools to dissect ZIKV molecular biology and defined functions of sfRNAs in viral infections in the two hosts. We believe that dissecting the roles of viral RNA determinants of relevant flaviviruses will provide opportunities for rational antiviral strategies.
MATERIALS AND METHODS
Ethics statement.
Human blood used to feed mosquitoes was obtained from healthy volunteer donors. Healthy donor recruitment was organized by local investigator assessment using medical history, laboratory results, and clinical examinations. Biological samples were supplied through participation of healthy volunteers at the ICAReB biobanking platform (BB-0033-00062/ICAReB platform/Institut Pasteur, Paris/BBMRI AO203/[BIORESOURCE]) of the Institut Pasteur to the CoSImmGen and Diagmicoll protocols which have been approved by the French Ethical Committee (CPP) Ile-de-France I. The Diagmicoll protocols were declared to the French Research Ministry under reference no. DC 2008–68 COL 1.
Construction of ZIKV infectious clones.
A clinical isolate of 2016 from Argentina, ARCB116141 (GenBank accession no. MK637519), obtained in the Maiztegui Institute (INEVH, Pergamino, Buenos Aires, Argentina), and a lab-adapted virus from Senegal, A1C1/V2 (GenBank accession no. KX198134), were used for full-genome sequencing and cDNA cloning. Viral isolates were grown in A549 human cells, and RNA was purified from supernatants with TRIzol (Thermo Fisher Scientific) and reverse transcribed using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega). cDNA fragments were amplified by PCR and amplicons were sequenced using oligonucleotides spanning the complete ZIKV genome (see Table S1 in the supplemental material). The 5′ and 3′ UTRs were sequenced by ligation of the 5′ and 3′ ends of the genome. For this end, viral RNA decapping using RNA 5′ pyrophosphohydrolase (New England BioLabs [NEB]) was followed by ligation with T4 RNA ligase (NEB). The RNA was reverse transcribed and cDNA was amplified by PCR using the oligonucleotides AVG2137 (CCATGATCTGTATATAACACTTATTCATCCCCA) and AVG2008 (CTGAGTCAAAAAACCCCACGCG). For full-length ZIKV cDNA assembly, six PCR amplicons were cloned into pBR322 as depicted in Fig. 1A. Cryptic E. coli promoters were predicted with the publicly available software of the Berkeley Drosophila Genome Project, Softberry Inc. Software, and the Genome2D web server. Putative promoters were disrupted by introducing synonymous changes. In addition, in order to facilitate assembly and reverse genetic studies, specific sites for restriction enzymes were inserted or removed by synonymous substitutions. A modified T7 promoter (5′-TAATACGACTCACTATAAG-3′) was inserted upstream of the viral sequence and a unique restriction site was inserted at the 3′ end for linearization and runoff transcription.
Construction of ZIKV reporter virus.
In order to design a ZIKV reporter virus, we used a strategy similar to the one previously reported for DENV (45). In brief, we inserted the Renilla luciferase gene flanked by the autoproteolytic FMDV 2A peptide. At the N terminus of the polyprotein, the cis-acting elements for genome replication present in the capsid coding sequence were duplicated, followed by the luciferase gene and the complete viral ORF. The cDNA clones from the two reporter systems were fully sequenced.
RNA transcription and transfection and viral infection.
Plasmids containing the full-length sequences of the epidemic and nonepidemic isolates, pZIKVAr and pZIKVSe, were linearized with KpnI and SacII, respectively, and used for in vitro transcription using T7 RNA polymerase (Ambion) in the presence of m7GpppA cap analog (NEB). RNA quantification was assessed using a Qubit4 (Invitrogen) and RNA integrity was confirmed on 1% agarose gels. RNA transcripts were transfected into C6/36HT and A549 cells using Lipofectamine 2000 and Opti-MEM media (Invitrogen). Viruses were harvested as a function of time. To generate viral stocks, C6/36HT cells were used.
Cell culture.
ZIKV infections were performed using the following cell lines. C6/36HT cells (ATCC CRL-1660), an Aedes albopictus cell line adapted to grow at 33°C, and U4.4 cells (A. albopictus) were cultured in Leibovitz’s L-15 medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml of penicillin, 100 μg/ml of streptomycin, 0.3% tryptose phosphate broth, 0.02% glutamine, 1% minimal essential medium (MEM) nonessential amino acid solution, and 0.25 μg/ml of amphotericin B (Fungizone). A549 cells (human epithelial lung cell line) were cultured in Dulbecco’s modified Eagle’s medium with nutrient mixture F-12 (DMEM–F-12) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Huh-7 cells (human hepatocyte cell line) were cultured in Dulbecco’s modified Eagle’s high-glucose medium (4500 mg/liter) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin.
Recombinant ZIKVs.
Mutations were introduced in the full-length cDNA of ZIKV ARCB116141 pZIKVAr, replacing the AgeI-KpnI fragment with the respective fragment derived from overlapping PCRs containing the desired mutations using the oligonucleotides described in Table S1. The deletions were designed as follows: ZIKVΔSLI lacked nucleotides 10399 to 10466, ZIKVΔSLII nucleotides 10482 to 10547, and ZIKVΔSLI-II nucleotides 10399 to 10547. For recombinant reporter viruses, the mutations were introduced in the context of the pZIKVAr-R using the same restriction enzymes and strategy as described for pZIKVAr.
Immunofluorescence assays.
C6/36HT or human A549 cells infected with WT or recombinant ZIKV were grown in 24-well plates containing 1-cm2 coverslips. At various times posttransfection, coverslips were removed, and cells were fixed with methanol for 15 min at –20°C. For detection of viral antigens, a polyclonal anti-NS3 was used. Alexa Fluor 488-conjugated rabbit anti-mouse immunoglobulin G (Molecular Probes) was employed to detect the primary antibody under the same conditions.
Viral titration.
Viral titers were determined by plaque assays in BHK-21 cells or by colorimetric-based focus forming units (FFU) in C6/36HT cells. For the latter, cells were prepared in 24-well plates, infected with serial 10-fold dilutions, and covered with a semisolid L-15 medium with 0.8% carboxymethyl cellulose (Sigma-Aldrich). After incubation at 33°C for 8 to 9 days, medium was removed and cells were fixed with 4% saccharose paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature and blocked in 250 μl/well of B buffer (2.5% bovine serum, Internegocios; 0.1% Tween 20-PBS) at room temperature for 1 h. The blocking buffer was then replaced with 250 μl/well of rabbit polyclonal anti-NS3 antibody (1:500 in blocking buffer) for 1 h at room temperature. After being washed three times with 0.1% Tween 20-PBS, the cells were subjected to alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma; 1:500 in blocking buffer) for 1 h. For chromogenic development, we used 200 μl/well of 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT) color development substrate (Promega) according to the manufacturer’s instructions. Chromogenic development was monitored at room temperature, and the reaction was stopped by rinsing twice with 200 ml/well of PBS.
RNA quantification.
For qRT-PCR of viral RNA, total RNA samples were used for reverse transcription as previously described (47). Reactions were performed in duplicates in 96-well plates using 2 μl of the RT reaction mixture as the template, 5 μl of FastStart SYBR green Master 2× mix (Roche), a 300 nM concentration of each primer, and RNase-free water to 10 μl. The primers AVG1117 (5′-ACAAGTCGAACAACCTGGTCCAT-3′) and AVG1118 (5′-GCCGCACCATTGGTCTTCTC-3′) were targeted to amplify nucleotides 9937 to 10113 within the NS5 coding sequence. For quantification of IFN-β, TNF, IL-6, and ISGs, qRT-PCR was used; the oligonucleotides employed are detailed in Table S1.
Viral RNA structure comparison and sequence alignments.
RNA structure comparison was performed using RNAforester, which estimates RNA structural similarities based on the tree alignment model (48), and sequence alignments via the MUSCLE algorithm and UPGMA (unweighted pair group method using average linkages) tree construction method.
Northern blot assay.
For ZIKV sfRNA detection, total RNA was obtained using TRIzol from infected cells and separated on 1% agarose, 2% formaldehyde, or 5% polyacrylamide 7 M urea gels. RNA was then transferred onto a nylon membrane (Hybond-N; GE Healthcare) using a semidry blotting apparatus and UV cross-linked. For detection of genomic RNA (gRNA), samples were loaded into a slot blot apparatus using nylon membranes (Hybond-N; GE Healthcare). Then, membranes were blocked overnight (ON) at 68°C in DIG Easy Hyb granule (Sigma-Aldrich) hybridization solution. Uniformly digoxigenin-labeled RNA probes were obtained by in vitro transcription using the DIG Northern starter kit (Sigma-Aldrich). Two RNA probes were designed (i) for detecting genomic RNA that hybridize the 5′ UTR and (ii) for detecting both genomic RNA and sfRNAs that hybridize the 3′ UTR. The 5′ probe was complementary to nucleotides 1 to 483 of the viral genome, and the 3′ probe was complementary to the complete 3′ UTR (nucleotides 10373 to 10807). Northern blots were visualized with the ImageQuant LAS 4000 (GE Healthcare Life Sciences). For detection of sfRNAs in A. aegypti mosquitoes, RNAs from individual mosquitoes were obtained by TRIzol extraction and processed as the samples obtained from cell culture.
Mosquitoes.
Experiments were performed with A. aegypti female mosquitoes belonging to the 17th generation from wild mosquitoes collected in Kamphaeng Phet Province (Thailand). The insectary conditions for mosquito maintenance were 28°C, 70% relative humidity, and a 12-h light and 12-h dark cycle. Adults were maintained with permanent access to 10% sucrose solution.
Experimental ZIKV infections in vivo.
Wild-type and recombinant ZIKVs that produce different qualities of sfRNAs were obtained in mosquito C6/35 HT cells. For experimental mosquito infections, 6- to 8-day-old female mosquitoes were deprived of sucrose for 24 h and transferred to a biosafety level 3 insectary. Washed rabbit erythrocytes resuspended in phosphate-buffered saline were mixed 2:1 with prediluted viral stock and supplemented with 10 mM ATP (Sigma-Aldrich). The viral stock was prediluted in Leibovitz’s L-15 medium with 10% FBS, 0.1% penicillin-streptomycin, and 1% sodium bicarbonate (Gibco) to reach an infectious titer ranging from 5 × 106 to 1 × 107 focus-forming units per ml of blood using a focus-forming assay in C6/36 cells as described above. A control blood meal was prepared identically except that a supernatant of mock-inoculated cells replaced the viral suspension. Mosquitoes fed on the infectious or control blood meal for 30 min through a membrane feeding system (Hemotek Ltd.) set at 37°C with a piece of desalted pig intestine as the membrane. Following the blood meal, fully engorged females were selected and incubated at 28°C and 70% relative humidity under a 12-h light and 12-h dark cycle with permanent access to 10% sucrose. After 14 days, mosquitos were used to evaluate infection in the body and to evaluate virus in saliva. After removing legs and wings, mosquitoes were salivated in a tip containing 20 μl of FBS for 30 min. Mosquito saliva was amplified in C6/36 cells for 5 days. Total RNA from C6/36 cells and supernatants was extracted with the Quick-RNA Viral 96 kit (Zymo Research). Total RNA from body, legs, and wings was extracted using TRIzol reagent (Ambion) for viral RNA detection.
Data availability.
The sequence of the ZIKV isolate from Argentina (epidemic virus) is available in GenBank under accession number MT636065.
Supplementary Material
ACKNOWLEDGMENTS
A.V.G., M.M.G.L.L., and L.D.B. are members of the Argentinean Council of Investigation (CONICET). H.M.P. and G.S.C.N. were granted CONICET fellowships. This work was supported by NIH (NIAID) grants R01.AI095175 and PICT-2017-1717, NIH grant PICT-2015-2555 to A.V.G., and the French Government’s Investissement d’Avenir program Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant ANR-10-LABX-62-IBEID) to M.C.S.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Weaver SC, Charlier C, Vasilakis N, Lecuit M. 2018. Zika, chikungunya, and other emerging vector-borne viral diseases. Annu Rev Med 69:395–408. doi: 10.1146/annurev-med-050715-105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbeito-Andres J, Pezzuto P, Higa LM, Dias AA, Vasconcelos JM, Santos TMP, Ferreira J, Ferreira RO, Dutra FF, Rossi AD, Barbosa RV, Amorim CKN, De Souza MPC, Chimelli L, Aguiar RS, Gonzalez PN, Lara FA, Castro MC, Molnar Z, Lopes RT, Bozza MT, Vianez J, Barbeito CG, Cuervo P, Bellio M, Tanuri A, Garcez PP. 2020. Congenital Zika syndrome is associated with maternal protein malnutrition. Sci Adv 6:eaaw6284. doi: 10.1126/sciadv.aaw6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goertz GP, Abbo SR, Fros JJ, Pijlman GP. 2018. Functional RNA during Zika virus infection. Virus Res 254:41–53. doi: 10.1016/j.virusres.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Villordo SM, Carballeda JM, Filomatori CV, Gamarnik AV. 2016. RNA structure duplications and flavivirus host adaptation. Trends Microbiol 24:270–283. doi: 10.1016/j.tim.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You S, Falgout B, Markoff L, Padmanabhan R. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J Biol Chem 276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- 9.Friebe P, Harris E. 2010. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J Virol 84:6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu ZY, Kwok CK, Qin CF, MacRae IJ, Goodfellow I, Marioni JC, Kudla G, Miska EA. 2018. COMRADES determines in vivo RNA structures and interactions. Nat Methods 15:785–788. doi: 10.1038/s41592-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV. 2015. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol 89:3430–3437. doi: 10.1128/JVI.02677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gritsun TS, Gould EA. 2007. Origin and evolution of 3′UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv Virus Res 69:203–248. [DOI] [PubMed] [Google Scholar]
- 13.Kieft JS, Rabe JL, Chapman EG. 2015. New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: conservation, folding, and host adaptation. RNA Biol 12:1169–2017. doi: 10.1080/15476286.2015.1094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. 2010. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol 84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urosevic N, van Maanen M, Mansfield JP, Mackenzie JS, Shellam GR. 1997. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol 78:23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- 17.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. 2014. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi PY, Nix JC, Beckham JD, Kieft JS. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354:1148–1152. doi: 10.1126/science.aah3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filomatori CV, Carballeda JM, Villordo SM, Aguirre S, Pallares HM, Maestre AM, Sanchez-Vargas I, Blair CD, Fabri C, Morales MA, Fernandez-Sesma A, Gamarnik AV. 2017. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog 13:e1006265. doi: 10.1371/journal.ppat.1006265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Liu H, Zou J, Zhang B, Yuan Z. 2014. Dengue virus subgenomic RNA induces apoptosis through the Bcl-2-mediated PI3k/Akt signaling pathway. Virology 448:15–25. doi: 10.1016/j.virol.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Donald CL, Brennan B, Cumberworth SL, Rezelj VV, Clark JJ, Cordeiro MT, Freitas de Oliveira Franca R, Pena LJ, Wilkie GS, Da Silva Filipe A, Davis C, Hughes J, Varjak M, Selinger M, Zuvanov L, Owsianka AM, Patel AH, McLauchlan J, Lindenbach BD, Fall G, Sall AA, Biek R, Rehwinkel J, Schnettler E, Kohl A. 2016. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl Trop Dis 10:e0005048. doi: 10.1371/journal.pntd.0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. 2012. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol 86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q-Y, Li X-F, Niu X, Li N, Wang H-J, Deng C-L, Ye H-Q, Huang X-Y, Chen Q, Xu Y-P, Dong H-L, Li X-D, Zhao H, Shi P-Y, Yuan Z-M, Gong P, Fang X, Qin C-F, Zhang B. 2020. Short direct repeats in the 3′ untranslated region are involved in subgenomic flaviviral RNA production. J Virol 94:e01175-19. doi: 10.1128/JVI.01175-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bidet K, Dadlani D, Garcia-Blanco MA. 2014. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. 2012. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J Virol 86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goertz GP, Fros JJ, Miesen P, Vogels CBF, van der Bent ML, Geertsema C, Koenraadt CJM, van Rij RP, van Oers MM, Pijlman GP. 2016. Noncoding subgenomic flavivirus rna is processed by the mosquito RNA interference machinery and determines West Nile virus transmission by Culex pipiens mosquitoes. J Virol 90:10145–10159. doi: 10.1128/JVI.00930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pompon J, Manuel M, Ng GK, Wong B, Shan C, Manokaran G, Soto-Acosta R, Bradrick SS, Ooi EE, Misse D, Shi PY, Garcia-Blanco MA. 2017. Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog 13:e1006535. doi: 10.1371/journal.ppat.1006535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soto-Acosta R, Xie X, Shan C, Baker CK, Shi PY, Rossi SL, Garcia-Blanco MA, Bradrick S. 2018. Fragile X mental retardation protein is a Zika virus restriction factor that is antagonized by subgenomic flaviviral RNA. Elife 7:e39023. doi: 10.7554/eLife.39023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. 2012. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalski D, Ontiveros JG, Russo J, Charley PA, Anderson JR, Heck AM, Geiss BJ, Wilusz J. 2019. Zika virus noncoding sfRNAs sequester multiple host-derived RNA-binding proteins and modulate mRNA decay and splicing during infection. J Biol Chem 294:16282–16296. doi: 10.1074/jbc.RA119.009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slonchak A, Khromykh AA. 2018. Subgenomic flaviviral RNAs: what do we know after the first decade of research. Antiviral Res 159:13–25. doi: 10.1016/j.antiviral.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Pu SY, Wu RH, Yang CC, Jao TM, Tsai MH, Wang JC, Lin HM, Chao YS, Yueh A. 2011. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J Virol 85:2927–2941. doi: 10.1128/JVI.01986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Münster M, Płaszczyca A, Cortese M, Neufeldt C, Goellner S, Long G, Bartenschlager R. 2018. A reverse genetics system for Zika virus based on a simple molecular cloning strategy. Viruses 10:368. doi: 10.3390/v10070368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villordo SM, Filomatori CV, Sanchez-Vargas I, Blair CD, Gamarnik AV. 2015. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog 11:e1004604. doi: 10.1371/journal.ppat.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife 3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep 17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke BD, Roby JA, Slonchak A, Khromykh AA. 2015. Functional non-coding RNAs derived from the flavivirus 3′ untranslated region. Virus Res 206:53–61. doi: 10.1016/j.virusres.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 42.Goertz GP, van Bree JWM, Hiralal A, Fernhout BM, Steffens C, Boeren S, Visser TM, Vogels CBF, Abbo SR, Fros JJ, Koenraadt CJM, van Oers MM, Pijlman GP. 2019. Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc Natl Acad Sci U S A 116:19136–19144. doi: 10.1073/pnas.1905617116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol 8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X. 2019. Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution. EMBO Rep 20:e47016. doi: 10.15252/embr.201847016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Borba L, Villordo SM, Marsico FL, Carballeda JM, Filomatori CV, Gebhard LG, Pallares HM, Lequime S, Lambrechts L, Sanchez Vargas I, Blair CD, Gamarnik AV. 2019. RNA structure duplication in the dengue virus 3' UTR: redundancy or host specificity? mBio 10:e02506-18. doi: 10.1128/mBio.02506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustos-Arriaga J, Gromowski GD, Tsetsarkin KA, Firestone C-Y, Castro-Jiménez T, Pletnev AG, Cedillo-Barrón L, Whitehead SS. 2018. Decreased accumulation of subgenomic RNA in human cells infected with vaccine candidate DEN4Delta30 increases viral susceptibility to type I interferon. Vaccine 36:3460–3467. doi: 10.1016/j.vaccine.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 47.Byk LA, Iglesias NG, De Maio FA, Gebhard LG, Rossi M, Gamarnik AV. 2016. Dengue virus genome uncoating requires ubiquitination. mBio 7:e00804-16. doi: 10.1128/mBio.00804-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hochsmann M, Voss B, Giegerich R. 2004. Pure multiple RNA secondary structure alignments: a progressive profile approach. IEEE/ACM Trans Comput Biol Bioinform 1:53–62. doi: 10.1109/TCBB.2004.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence of the ZIKV isolate from Argentina (epidemic virus) is available in GenBank under accession number MT636065.