SUMMARY
Paneth cells are the primary source of C-type lysozyme, a β−1,4-N-acetylmuramoylhydrolase that enzymatically processes bacterial cell walls. Paneth cells are normally present in human cecum and ascending colon, but are rarely found in descending colon and rectum; Paneth cell metaplasia in this region and aberrant lysozyme production are hallmarks of IBD pathology. Here we examined the impact of aberrant lysozyme production in colonic inflammation. Targeted disruption of Paneth cell lysozyme (Lyz1) protected mice from experimental colitis. Lyz1-deficiency diminished intestinal immune responses to bacterial molecular patterns and resulted in the expansion of lysozyme-sensitive mucolytic bacteria, including Ruminococcus gnavus, a Crohn’s disease-associated pathobiont. Ectopic lysozyme production in colonic epithelium suppressed lysozyme-sensitive bacteria and exacerbated colitis. Transfer of R. gnavus into Lyz1−/− hosts elicited a type 2 immune response, causing epithelial reprograming and enhanced anti-colitogenic capacity. In contrast, in lysozyme-intact hosts, processed R. gnavus drove pro-inflammatory responses. Thus, Paneth cell lysozyme balances intestinal anti- and pro-inflammatory responses, with implications for IBD.
Keywords: Lysozyme, Paneth cell, Type 2 immunity, Colitis, Lyz1, Ruminococcus gnavus
eTOC Blurb
Paneth cell metaplasia to the colon and rectum and aberrant lysosome production are hallmarks of inflammatory bowel disease in humans. Using mouse models were Lyz1 is deleted or ectopically expressed, Yu, Balasubramanian et al show that Paneth cell lysozyme regulates the abundance of mucolytic commensal bacteria and thereby the intestinal inflammatory response.
Graphical Abstract
INTRODUCTION
Intestinal Paneth cells and certain myeloid cells produce the C-type lysozyme, a β−1,4-N-acetylmuramoylhydrolase that enzymatically processes bacterial cell walls. The products of lysozyme processing, such as muramyl dipeptide (MDP) can be important agonists of pattern recognition receptors (PRR), notably NOD-like receptors (NLR) (Balasubramanian and Gao, 2017). Biochemical studies of lysozyme identified a helix-loop-helix bactericidal domain mediating its membrane-permeabilizing action (Canfield and Liu, 1965; Ibrahim et al., 2001b). It is distinct from lysozyme’s enzymatic domain (Ibrahim, 1998), illustrating a unique dual functionality of lysozyme that distinguishes it from other antimicrobial peptides.
Paneth cells secrete lysozyme into the intestinal lumen (Bel et al., 2017), constituting the primary source of luminal lysozyme that directly encounters commensal bacteria. Macrophages and neutrophils are major sources of lysozyme within the intestinal lamina propria (LP). In humans, macrophage- and Paneth cell-derived lysozyme is encoded by a single LYZ gene on chromosome 12q15. This gene is located in the vicinity of an Ulcerative Colitis (UC) risk locus harboring IFN-γ, IL26, and IL22 (Jostins et al., 2012; Silverberg et al., 2009). Hereditary LYZ mutation causes familial amyloidosis (Pepys et al., 1993) and patients carrying mutant LYZ exhibit gastritis and inflammatory bowel disease (IBD) symptoms such as abdominal pain, malabsorption, diarrhea, and weight loss (Girnius et al., 2012; Jean et al., 2014).
Whereas Paneth cells are absent in rodent colonic epithelium, these cells are normally present in human cecum and ascending (i.e., right) colon. However, they are rarely found in human descending colon and rectum and Paneth cell metaplasia in this region is a hallmark of IBD pathology (Singh et al., 2020; Tanaka et al., 2001). Clinical studies demonstrated a correlation between aberrant lysozyme production and IBD. Increased fecal lysozyme was reported in UC patients over 70 years ago (Meyer et al., 1947, 1948). Subsequent studies suggested that fecal and blood lysozyme levels are excellent indicators for IBD activity (Di Ruscio et al., 2017; Klass and Neale, 1978). Colonic epithelia of UC patients exhibit elevated expression of lysozyme messenger (m)RNA and protein, primarily in metaplastic Paneth-like cells (Fahlgren et al., 2003), which has been speculated to represent the host’s attempt to control the increased bacterial adherence to the intestinal epithelial cell (IEC) surface. Paneth cells in Crohn’s Disease (CD) patients with polymorphisms in ATG16L1 (T300A) or NOD2 alleles exhibit aberrant lysozyme packaging, with lysozyme-containing secretory granules abnormally dispersed within the cytoplasm or degraded (Cadwell et al., 2008; VanDussen et al., 2014). This aberrant lysozyme granular morphology in Paneth cells is predictive of the timing of CD recurrence after surgery (Liu et al., 2017; VanDussen et al., 2014). Another major CD susceptibility gene product, Leucine-Rich Repeat Kinase 2 (LRRK2), selectively packages lysozyme but not REG3 or defensins into the dense core secretory granules in Paneth cells (Zhang et al., 2015). Loss of LRRK2 results in lysozyme mis-trafficking and degradation, a deficiency speculated to be responsible for the increased Listeria monocytogenes invasion in Lrrk2−/− mice (Zhang et al., 2015).
Although abnormal Paneth cell morphology and lysozyme production are observed in IBD pathology, whether aberrant lysozyme production per se impacts disease susceptibility has not been directly examined. Here we used multiple mouse models to address this question, and found that lysozyme produced by Paneth cells defined certain populations of gut commensal bacteria. Ectopic lysozyme production exacerbated experimental colitis, whereas lysozyme deficiency altered the mucosal immune profile via an altered bacterial landscape.
RESULTS
MDP and NLR signaling is diminished in the intestine of Lyz1−/− mice
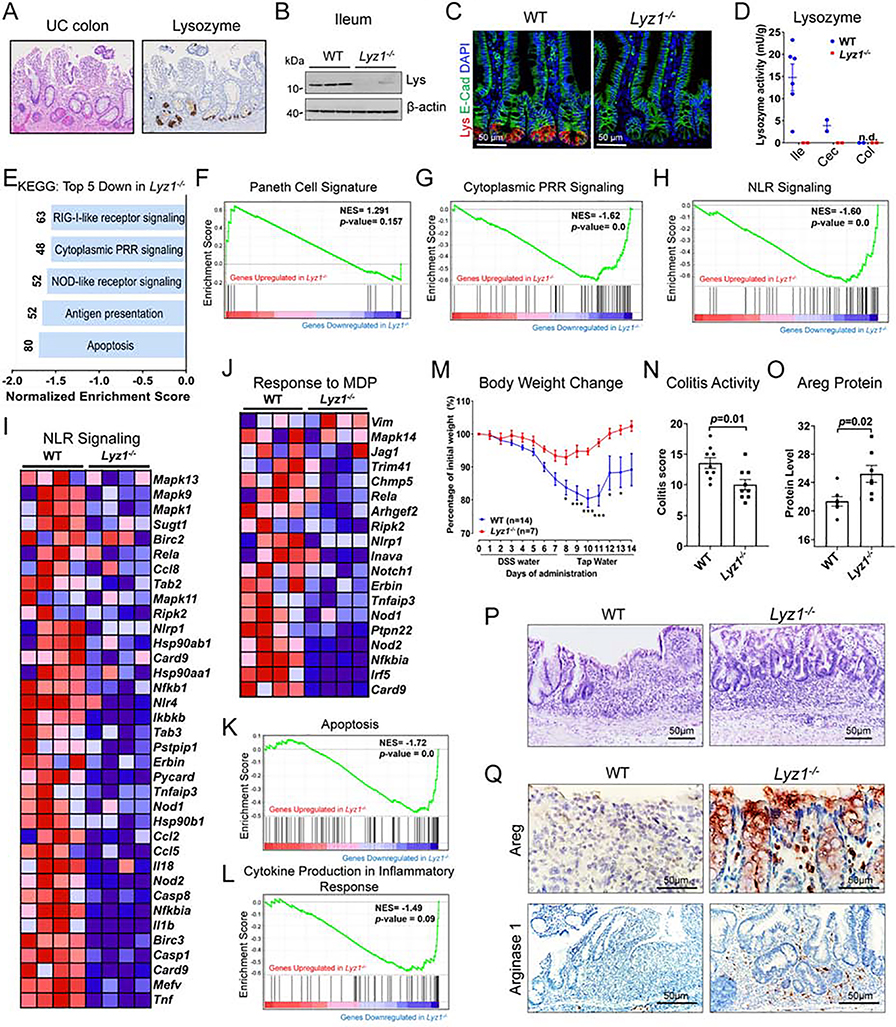
The physiological function of intestinal luminal lysozyme is unclear. While normal human left colons do not have lysozyme-expressing Paneth cells, metaplastic Paneth cells in IBD patients’ left colons expressed lysozyme (Fig. 1A, Fig. S1A, p=0.00012). Mouse Lyz1 and Lyz2 encode for lysozyme in Paneth cells and in leukocytes, respectively (Markart et al., 2004a). We targeted Lyz1 (Yu et al., 2018), and developed Lyz1−/− mice to study the function of Paneth cell lysozyme. Loss of intestinal lysozyme in Lyz1−/− mice was confirmed at the level of mRNA (Fig. S1B), protein (Fig. 1B–C, Fig. S1C), and enzymatic activity (Fig. 1D). Lyz1−/− mice did not show changes in body weight or activity compared to their wild type (WT) littermates. All comparisons in this study were conducted among littermates.
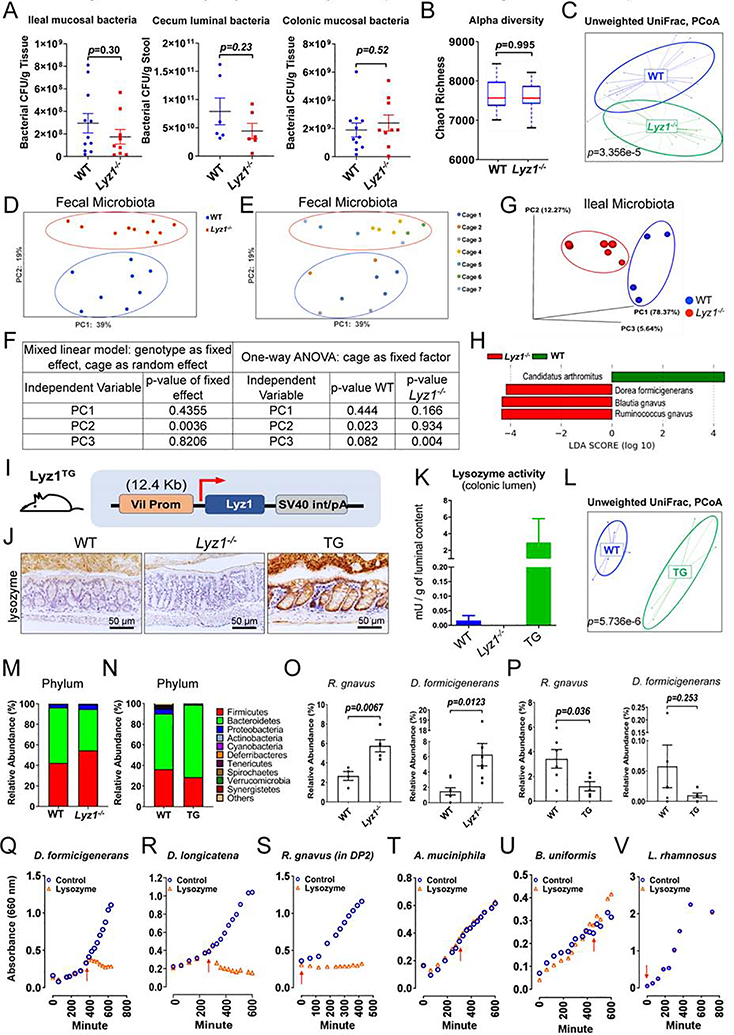
Figure 1. Lyz1 deficiency diminishes NLR signaling and reduces inflammation during experimental colitis.
(A) Representative IBD patient left colon with metaplastic Paneth cells positive for lysozyme. (B-C) Loss of lysozyme protein expression in Lyz1−/− ileum by Western blotting (B; N=3) and by immunostaining (C; lysozyme in red; N>20). (D) Enzymatic activity of lysozyme in WT and Lyz1−/− ileum, cecum and proximal colon luminal contents was measured by a fluorometric assay (N=2–6 per genotype). (E) Top 5 pathways identified by KEGG analysis of differential gene expression by bulk RNAseq of WT and Lyz1−/− ileum (N=4 for each genotype for E-L). (F) No change of Paneth cell signature in Lyz1−/− ileum in GSEA analysis. (G-H) Significant reduction of cytoplasmic PRR and NLR signaling in Lyz1−/− ileum in GSEA analysis (p=0.0). (I-J) Differenatial expression of genes involved in NLR signaling, in particular to MDP, in Lyz1−/− ileum. (K-L) Suppressed apoptosis and inflammatory cytokine production in Lyz1−/− ileum in GSEA analysis. (M) Body weight change in WT and Lyz1−/− mice during 3% DSS treatment and recovery. (N) Histological colitis activity scores in WT and Lyz1−/− mice after DSS treatment. (O) Areg protein quantitation from immune-stained sections of DSS-treated WT and Lyz1−/− mouse colons. (P) Representative H&E staining of the distal colons of DSS-treated WT and Lyz1−/− mice. (Q) Representative immunostaining of Areg and Arginase 1 in the colon of DSS-treated WT and Lyz1−/− mice. All bar graphs display mean ± SEM from at least two independent experiments. See also Figure S1.
Bulk RNA sequencing analysis of the ileum of adult WT and Lyz1−/− mice revealed 404 transcripts whose expression was increased and 447 transcripts whose expression was decreased in Lyz1−/− mice (p<0.05, Fig. S1D). Lyz1 was abolished as expected (Fig. S1E). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed the top 5 decreased modules in Lyz1−/− intestines to represent apoptosis, antigen presentation, NLR signaling, cytoplasmic PRR signaling, and RIG1-like receptor signaling (p=0.0, Fig. 1E). Gene Set Enrichment Analysis (GSEA) suggested that Paneth cell transcriptome was preserved in the Lyz1−/− ileum (p>0.05, Fig. 1F), a finding validated by the preserved expression of Paneth cell-specific markers such as Mmp7 (Fig. S1F). Signature gene sets for cytoplasmic PRR signaling (Fig. 1G), especially the NLR signaling (Fig. 1H, I), were reduced in Lyz1−/− intestines. We detected a reduced cellular response to MDP (Fig. 1J), likely reflecting a lack of lysozyme-processing of peptidoglycan. GSEA and gene ontology analysis further identified suppression of cytokine production, suppression of inflammatory response, and suppression of ROS signaling in Lyz1−/− intestines (Fig. 1K–L, Fig. S1G–H). Thus, Lyz1 deficiency diminishes the intestinal mucosal response to bacterial molecular patterns.
Lyz1 deficiency ameliorated dextran sulfate sodium (DSS)-induced intestinal inflammation
Paneth cell-derived lysozyme is secreted into the intestinal lumen (Bel et al., 2017) and excreted with feces (Meyer et al., 1947, 1948). In line with this, lysozyme immunoreactivity was detected in WT adult mouse colonic lumen, and it was lost in Lyz1−/− mice (Fig. S1C). Given the correlation between lysozyme and intestinal inflammation, we then tested the impact of lysozyme deficiency on experimental colitis. DSS in drinking water induces colitis in mice (Okayasu et al., 1990). DSS administration in WT mice robustly elevated Lyz1 and Lyz2 expression in inflamed colons whereas the same treatment elevated Lyz2 in Lyz1−/− mouse colons (Fig. S1I, J). Compared to WT littermates, colitis in Lyz1−/− mice was ameliorated, with rapid body weight recovery (Fig. 1M) and lower pathology scores (Fig. 1M, N, P). This observed protection in the Lyz1−/− mice was accompanied by elevated Areg and Arg-1 (Fig. 1O, Q, and Fig. S1K), two biomarkers associated with mucosal repair and downstream of the type 2 immune response (Pesce et al., 2009; Zaiss et al., 2006). Thus, Lyz1 deficiency ameliorated mucosal inflammation during DSS colitis.
Lyz1-deficient intestines had increased population of goblet and tuft cells
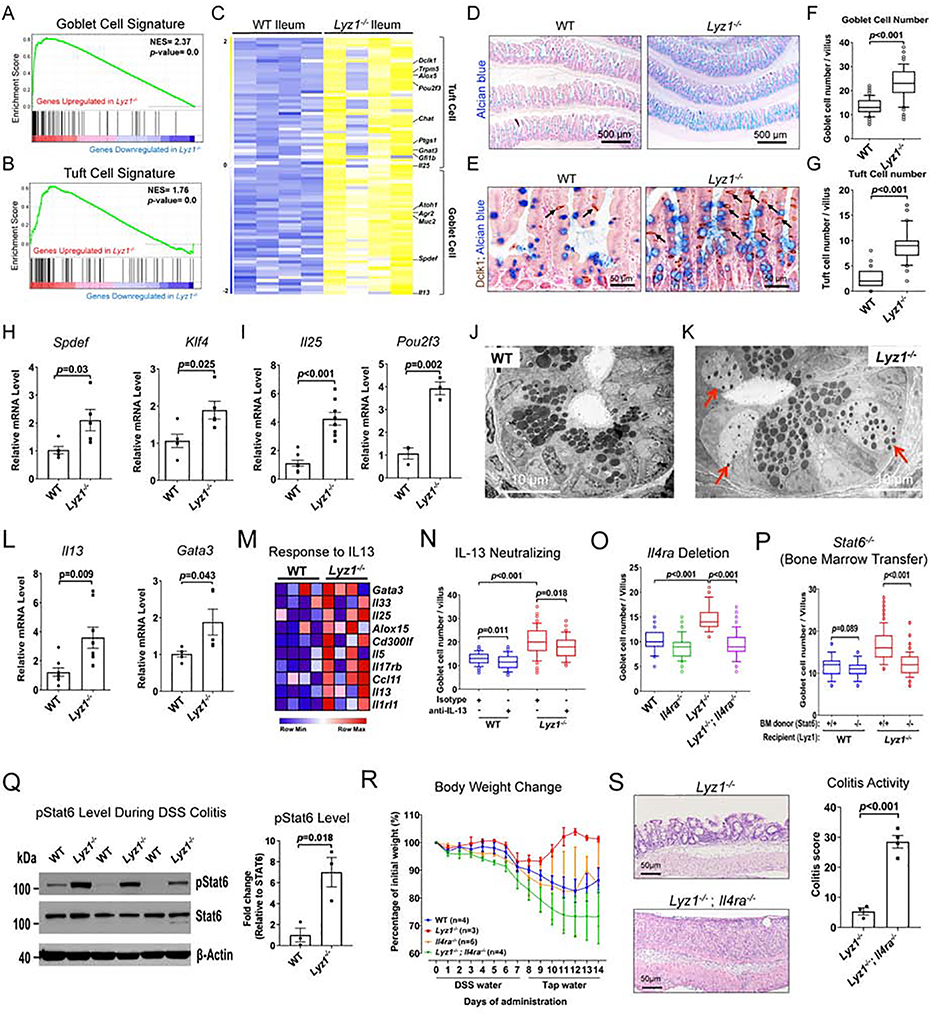
We sought to investigate the above observed protective mechanism in the Lyz1−/− mice. RNAseq showed increased goblet and tuft cell transcripts and a reduced enterocyte transcriptome (Fig. 2A–C and Fig. S2A). More tuft and goblet cells in Lyz1−/− ileum were validated by staining (Fig. 2D–G, Fig. S2B) and increased transcripts of Klf4 and Spdef for goblet cells (Fig. 2H)(Gregorieff et al., 2009; Katz et al., 2002); and Il25 and Pou2f3 for tuft cells (Fig. 2I)(Gerbe et al., 2016; von Moltke et al., 2016). A pronounced goblet cell hyperplasia in the Lyz1−/− ileum was accompanied by abnormally smaller Paneth cell electron-dense granules surrounded by expanded electron-lucent halos formed by packaging of Muc2 mucin (compare Lyz1−/− in Fig. 2K to WT in 2J and Fig. S2C, D) (Stahl et al., 2018).
Figure 2. Elevated type 2 immune response in Lyz1−/− intestinal mucosa mediates anti-colitogenic protection.
(A-B) Elevated goblet and tuft cell signatures in Lyz1−/− ileum in GSEA analysis (p=0.0). (C) Differential expression of goblet and tuft cell specific genes along with IL-25 and IL-13 in WT and Lyz1−/− ileum (N=4 for each genotype). (D-E) Representative Alcian blue (goblet) and DCLK1 (tuft) staining in WT and Lyz1−/− ileum. (F-G) Quantification of the number of goblet and tuft cells per villus in WT and Lyz1−/− littermates (N=5 mice per genotype). (H-I) Real-time qPCR for goblet and tuft cell specific genes (N=3~6 per genotype). (J-K) Transmission electron microscopy of the WT and Lyz1−/− ileal crypts demonstrating cells with granules characteristic of both goblet and Paneth cells. The electron dense granules were surrounded by expanded halos (red arrows) in abnormal Paneth cells in the Lyz1−/− (representative of N=3 for each genotype). (L) qPCR analysis of IL-13 and Gata3 mRNA in WT and Lyz1−/− ileum (N=3–6 for each genotype). (M) Differential expression of IL-13 responsive genes in WT and Lyz1−/− ileum (N=4 per genotype). (N) Goblet cell numbers (counted from 50 villi per field of vision per mouse) in WT or Lyz1−/− mice treated with neutralizing anti-IL-13 antibody, anti- CD90.2, or isotype control (N=2 for each condition per genotype). (O) Goblet cell numbers (counted from 50 villi per field of vision per mouse) in WT, Lyz1−/−, Il4ra−/−, or Lyz1−/−; Il4ra−/− mice (N=3 for each genotype). (P) Goblet cell numbers (counted from 50 villi per field of vision per mouse) in WT or Lyz1−/− bone marrow chimeras with hematopietic cells from Stat6+/+ or Stat6−/− donors (total N=4 mice for each condition; 2 independent experiments). (Q) Western blotting analysis of pStat6 from the colons of DSS-treated WT and Lyz1−/− mice (N=3 for each genotype). (R) Body weight change in WT, Lyz1−/−, Il4ra−/−, or Lyz1−/−; Il4ra−/− mice treated by 3% DSS and during recovery. (S) Representative H&E staining and colitis histological activity score in DSS-treated Lyz1−/− and Lyz1−/−; Il4ra−/− mice. All bar graphs display mean ± SEM from at least two independent experiments. See also Figure S2.
Tuft cell-derived IL-25 instructs goblet cell differentiation via promoting type 2 cytokine-producing cells in the intestine (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016). RNAseq and qPCR showed elevated Il25, Il13, Gata3, and Il4 (Fig. 2C, I, L, and Fig. S2E), along with an IL-13 signaling gene set (Fig. 2M). However, qPCR did not detect significant changes in cytokines or transcription factors for type 1, type 3, or T-regulatory cells (Fig. S2F–H). Thus, a skewed type 2 immune profile existed in Lyz1−/− intestines consistent with an activated goblet and tuft cell program.
The anti-colitogenic protection in the Lyz1−/− mice requires IL-13-IL-4Ra-Stat6 axis
IL-13 secreted from ILC2s promotes goblet and tuft cell differentiation (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016). Lyz1−/− mice treated with neutralizing anti-IL-13 antibody (Proust et al., 2003) had reduced goblet and tuft cells when compared to isotype control-treated Lyz1−/− littermates (Fig. 2N, and Fig. S2I). IL-13 signals through IL-4Ra receptor (Doran et al., 2017; Rael and Lockey, 2011). The epithelial phenotype of Lyz1-/; Il4ra−/−- mice was reversed to that of WT littermates (Fig. 2O, and Fig. S2J). IL-13 drives type 2 immune response via Stat6 (Doran et al., 2017; Rael and Lockey, 2011). We transplanted bone marrow from Stat6−/− mice into total body irradiated Lyz1−/− recipients (carrying WT Stat6 in IECs). Chimeras with Stat6−/− bone marrow showed a reduction in goblet cells compared to Lyz1−/− recipients reconstituted by WT bone marrow (Fig. 2P and Fig. S2K). Thus, the IL-13-IL-4Ra-Stat6 axis was responsible for the epithelial repopulation in Lyz1−/− intestines in homeostasis.
Areg and Arg1 are critical components of type 2 immune signaling (Pesce et al., 2009; Zaiss et al., 2006); and both were elevated in the colon of DSS-treated Lyz1−/− mice (Fig. 1Q). We examined pStat6 in DSS-treated colons and found a 7-fold increase of this type 2 immune response indicator (Kaplan et al., 1996) in the inflamed colons of Lyz1−/− mice compared to WT littermates (p=0.018, Fig. 2Q). Areg promotes barrier function and tissue repair, protecting against DSS colitis (Chen et al., 2018). We reasoned that the observed type 2 immune activation might mediate the anti-colitogenic effects of Lyz1-deficiency, similar to helminth therapy of autoimmune disease (Smallwood et al., 2017), and promote mucosal healing (Gause et al., 2013). With an underlying premise that Il4ra is indispensable for type 2 immune response (Noben-Trauth et al., 1997), we challenged Lyz1−/− ;Il4ra−/− mice with DSS. Compared to Lyz1−/− littermates, DSS-treated Lyz1−/−;Il4ra−/− mice exhibited an exacerbated colitis with a complete loss of body weight recovery (Fig. 2R) with worsened colitis score (Fig. 2S). Thus, an enhanced type 2 immune response mediated protection during DSS colitis in Lyz1−/− mice.
scRNAseq revealed immune-activated ILC2 in Lyz1−/− ileal LP
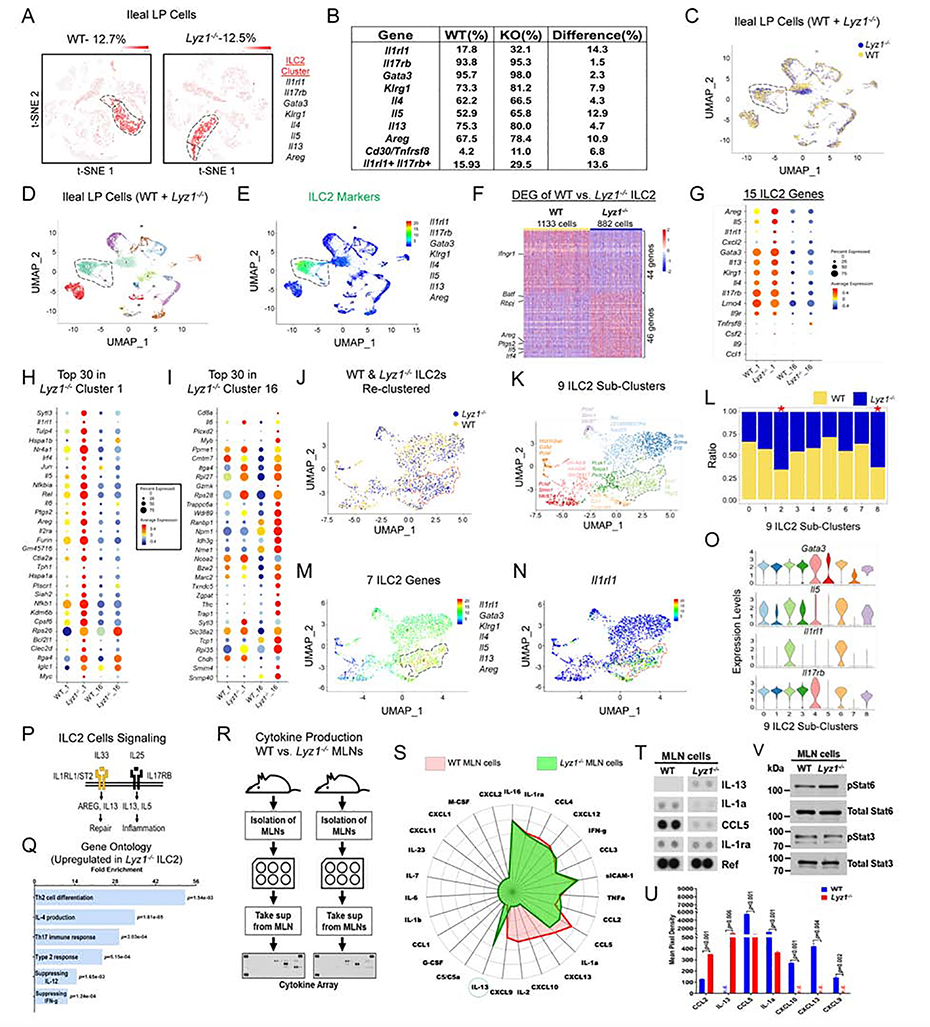
To define Lyz1−/− intestinal immune cell composition, we performed a single cell RNA sequencing (scRNAseq) analysis using ileal LP of separately housed Lyz1−/− and WT littermates. Unsupervised separate clustering of WT and Lyz1−/− samples identified 23 and 19 clusters, respectively (Fig. S3A). WT ILC2 clusters contained 1,017 cells out of 8,011 total LP cells, while Lyz1−/− ILC2 clusters had 844 cells out of 6,733 total LP cells (Fig. 3A). The ILC2 percentage was unchanged (12.7% vs. 12.5%). However, compared to WT LP, Lyz1−/− LP displayed a 14.3%, 12.9%, 10.9% and 4.7% increase in Il1rl1+Il33r+, Il5+, Areg+, and Il13+ cells, respectively (Fig. 3B).
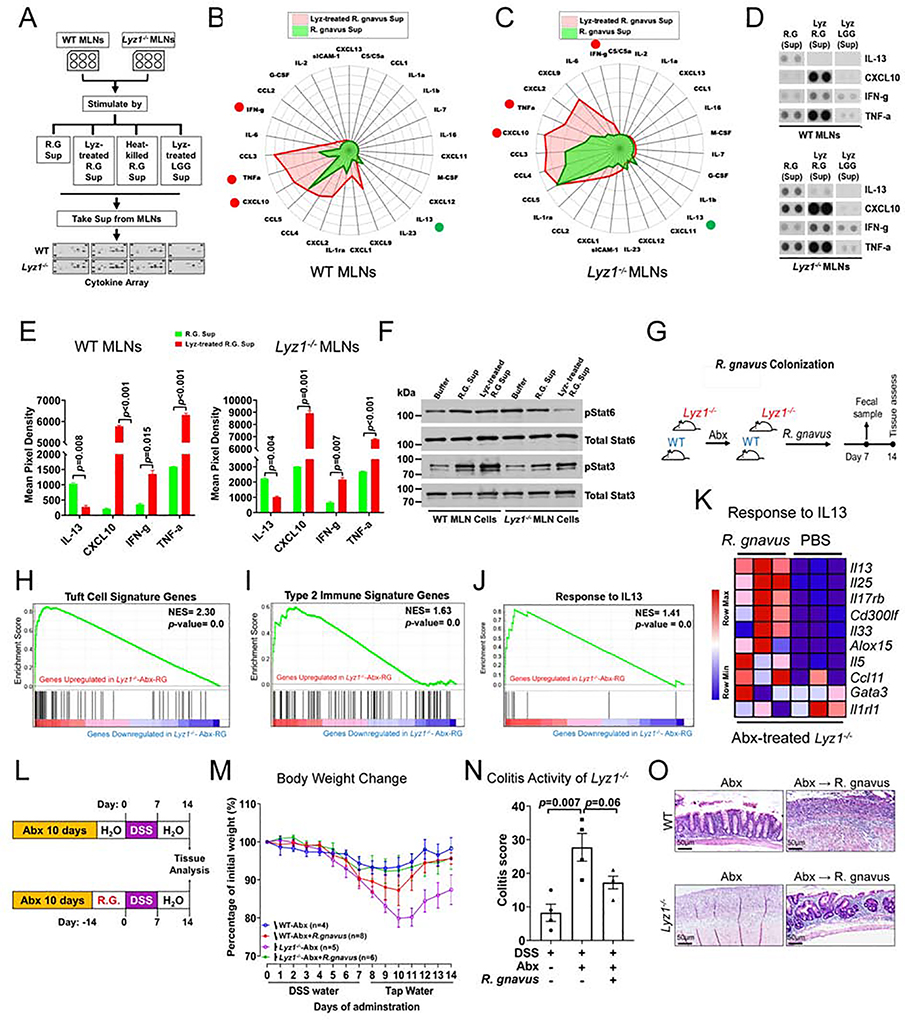
Figure 3. scRNAseq reveals immune-activated ILC2 in Lyz1−/− ileal lamina propria.
(A) Unsupervised separate clustering (t-SNE plot) of LP cells identified ILC2 populations in WT mice (total of 1,017 cells, 12.7% of all LP cells) and in Lyz1−/− mice (844 cells, 12.5% of all LP cells). (B) Table summarizing the percentage of ILC2 cells that expressed indicated genes in WT and Lyz1−/− LP. (C-D) Uniform manifold approximation and projection (UMAP) of combined clustering of WT and Lyz1−/− LP cells (WT, 8767 cells; Lyz1−/−, 7492 cells) resulted in 23 distinct clusters. (E) ILC2s (clusters 1 &16) identified based on the indicated set of signature genes. (F) 90 differentially expressed genes between WT and Lyz1−/− ILC2 population in cluster 1. (G-I) Representative differentially expressed genes in WT and Lyz1−/− ILC2s. Dot size reflects the percentage of cells in the cluster that express the gene; color indicates the average expression of the gene. (G) Differential expression of 15 ILC2 signature genes by WT and Lyz1−/− ILC2s. (H) Top 30 genes increased in Lyz1−/− cluster 1. (I) Top 30 genes increased in Lyz1−/− cluster 16. (J-O) All WT and Lyz1−/− ILC2 cells (a total of 2281 cells) were further partitioned into 9 sub-clusters colored by genotype (J) or by cluster (K), with top 3 differentially expressed genes indicated next to each sub-cluster. (L) Bar graph with the relative distribution of WT and Lyz1−/− ILC2 cells in each sub-cluster. The Lyz1−/− ILC2-dominated sub-clusters 2 & 8 were denoted by an asterisk. (M) UMAP projection with 7 indicated signature genes elevated in sub-cluster 2 of Lyz1−/− ILC2. (N) ILC2 UMAP projection with highlighetd relative expression of Il1rl1 mRNA. (O) Violin plots of indicated gene expression across 9 ILC2 sub-clusters. (P) Schematic illustration of ILC2 signaling through IL1RL1 and IL17RB receptors in tissue repair and inflammation. (Q) Gene ontology (GO) categories (sorted by P value) with top pathways overrepresented in Lyz1−/− ILC2 compared to WT ILC2 in cluster 1. (R-S) Experimental design and radar plot with differential cytokine/chemokine secretion by WT (red) and Lyz1−/− (green) MLN cells at steady state. IL-13 highlighted by a circle. (T) Representative dot blots showing elevated IL-13 production by Lyz1−/− MLN cells. (U) Quantification of cytokine/chemokine production from two independent experiments. The bar graph displays mean ± SEM. (V) Western blots for pStat6 and pStat3 using MLN cell lysates from the experiment shown in S-U. See also Figure S3.
Combined clustering of all WT and Lyz1−/− LP cells revealed 23 clusters (Fig. 3C, D). Clusters 1 and 16 were identified as ILC2s based on signature genes (Fig. 3E, Fig. S3B). Distinct transcriptomic profiles of WT and Lyz1−/− ILC2s were evident through heatmap analysis (Fig. 3F, Fig. S3C). Known ILC2 signatures genes (Wallrapp et al., 2017), Areg, Il5, and Il1rl1 were highly elevated in Lyz1−/− ILC2s (Fig. 3F–I), along with others with documented roles in promoting type 2 immune response, e.g., Ptgs2 and Irf4 (Kalinski, 2012; Williams et al., 2013).
WT and Lyz1−/− ILC2s could be further partitioned into 9 sub-clusters (Fig. 3J, 3K), with sub-clusters 2 and 8 dominated by Lyz1−/− ILC2s (Fig. 3L). The expression of 7 ILC2 signature genes (Il1rl1, Gata3, Klrg1, Il4, Il5, Il13, and Areg) was elevated in sub-cluster 2 (Figure. 3M, O, and Fig. S3D), with Il1rl1 being highly expressed (Figure. 3N, O). ILC2 cells signal through Il1rl1-Il33r, and drive Areg production for tissue repair (Herbert et al., 2019; Kabata et al., 2018) (Fig. 3P). Expansion of this subset in Lyz1−/− suggested a distinct heterogeneity between WT and Lyz1−/− ILC2s. Gene ontology analysis of Lyz1−/− versus WT ILC2 revealed significant induction of type 2 immune responses in Lyz1−/− mice (Fig. 3Q, Fig. S3E). In contrast to Lyz1−/− ILC2s, induction of Th1 cytokine, activation of IFN-γ and reactive oxygen species were revealed in WT ILC2s (Fig. 3Q, Fig. S3F).
Using a dot blot array, we examined cytokines secreted from single cell suspensions of mesenteric lymph nodes (MLNs) isolated from separately housed WT and Lyz1−/− littermates in homeostasis (Fig. 3R, Fig. S3G). After 24 hours of in vitro culture, Lyz1−/− MLNs secreted about 700-fold higher amount of IL-13 compared to WT MLNs (Fig. 3S–U). The secretion of CCL2 (MCP-1) known to control Th2 polarization (Gu et al., 2000) was also increased in Lyz1−/− MLNs (Fig. 3S, U). WT MLNs produced more IL-1a and CCL5 than Lyz1−/− MLNs (Fig. 3S–U). CXCL-9, CXCL-10, CXCL-13, and IL-2 were below detection limit in Lyz1−/− MLNs (Fig. 3S, U and Fig. S3H), consistent with a reduced inflammatory response (Fig. 1L). Lyz1−/− MLN cell lysates had elevated pStat6 compared to WT MLN (Fig. 3V and Fig. S3I). Thus, Lyz1−/− intestinal mucosa had an activated type 2 immune profile.
Altered mucosal immunity in Lyz1−/− mice is microbiota-dependent
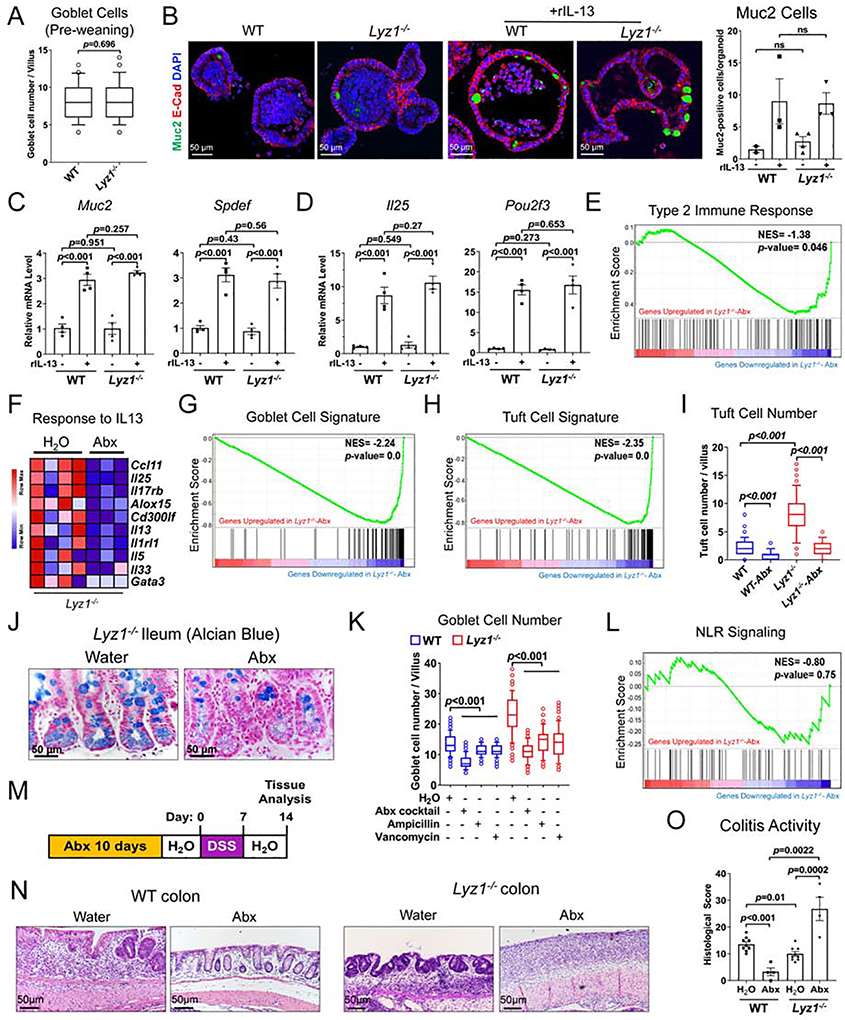
We sought to determine the signal upstream of the observed mucosal changes. We noticed that pre-weaning Lyz1−/− pups showed no difference of goblet cell composition from their WT littermate (Fig. 4A), suggesting that the mucosal changes occurred in adulthood. Enteroids cultured from WT and Lyz1−/− adult mice in regular ENR medium had equivalent goblet and tuft cell numbers or specific transcripts (Fig. 4B–D). When recombinant IL-13 (Huaux et al., 2003) was added, similar goblet and tuft cell differentiation was observed in WT and Lyz1−/− enteroids (Fig. 4B–D, Suppl. 4A–B). Thus, the observed goblet and tuft cell phenotype in Lyz1−/− mice was not epithelial cell-intrinsic and required non-epithelial signals.
Figure 4. Altered mucosal immunity in Lyz1−/− mice is microbiota-dependent.
(A) Goblet cell numbers (counted from 50 villi per field of vision per mouse) in WT or Lyz1−/− mice based on Alcian blue staining of ileal sections of 14 day-old WT and Lyz1−/− mice (N=4 for each genotype from 2 independent experiments). (B) The effect of genotype and IL-13 treatment on goblet cell maturation (Muc2+ cells in green) in ileal entroids from WT and Lyz1−/− mice. (C-D) qPCR analysis of mRNA expression of goblet- and tuft cell-specific genes in ileal WT and Lyz1−/− enteroids. (E) GSEA analysis of genes related to type 2 immune response in the ileum of untreated or Abx-treated Lyz1−/− mice (N=3–4, bulk RNAseq). (F) Differential expression of IL-13-responsive genes in the ileum of untreated or Abx-treated Lyz1−/− mice. (G-H) GSEA analysis of goblet and tuft cell gene signatures in the ileum of untreated or Abx-treated Lyz1−/− mice. (I) The effects of genotyope and Abx treatment on DCLK1+ tuft cell numbers (counted from 50 villi per field of vision per mouse; N=4–5 for each condition). (J) Representative alcian blue staining of the ileum of untreated or Abx-treated Lyz1−/− mice. (K) Goblet cell numbers (counted from 50 villi per field of vision per mouse) in WT or Lyz1−/− mice treated with regular water, or water with ampicillin, vancomycin, or an Abx cocktail (N=2–4 in each group). (L) Unaltered NLR signaling gene signature in untreated or Abx-treated Lyz1−/− mice by GSEA analysis. (M) Schematic of the experimental design for panels N-O. (N-O) Representative H&E images of distal colon and colitis activitivity scores in WT and Lyz1−/− mice treated as in panel M (N=3–7 in each group). All bar graphs display mean ± SEM from at least two independent experiments. See also Figure S4.
We then treated separately housed WT and Lyz1−/− adult littermates with a cocktail of antibiotics (Abx) in drinking water (Rakoff-Nahoum et al., 2004) to test the role of Lyz1−/− gut microbiota. Bulk RNAseq showed that Abx-treated Lyz1−/− ileum had a reduced type 2 immune signature (Fig. 4E). IL-13 responsive gene set was markedly reduced Abx-treated mouse ileum (Fig. 4F). Abx treatment also reduced goblet (Fig. 4G) and tuft cell signatures in the Lyz1−/− ileum (Fig. 4H). These results were validated by immunostaining and qPCR (Fig. 4I–K, Fig. S4C). Importantly, Abx treatment did not affect NLR signaling in Lyz1−/− mice, consistent with a lack of lysozyme-mediated cell-wall processing in Lyz1−/− mice (Fig. 4L). When Abx-treated WT and Lyz1−/− mice were challenged by DSS (Fig. 4M), a more exacerbated colitis was found in Abx-treated Lyz1−/− mice than in those with regular water (Fig. 4N–O). Thus, gut microbiota of Lyz1−/− mice was required for the type 2 immune profile and the anti-colitogenic effect.
Lyz1 deficiency changed the gut microbiota landscape
Despite lysozyme’s reported bactericidal function, Lyz1−/− mice showed no change in luminal bacterial load in cecum or mucosa-associated bacteria in ileum and colon (Fig. 5A). All analyses were performed on littermate WT and Lyz1−/− mice from Lyz1+/− breeding pairs, and no commercial WT mice were used in our experiments. 16S rRNA amplicon profiling of fecal and ileal content showed unchanged α-diversity between WT and Lyz1−/− microbiota (Fig. 5B, Fig. S5A). However, unweighed UniFrac analysis revealed statistically significant difference in fecal (p=3.356E-5, Fig. 5C) and ileal (p=0.014, Fig. S5B) bacterial composition. To determine the cage versus genotype effects, WT and Lyz1−/− mice from the same breeding parents were co-housed for 3 weeks, separated into different cages according to genotype (total of 8 WTs in 3 cages; 10 Lyz1−/−s in 4 cages), and tested after 1-month of separate housing to assess bacterial composition. Principal coordinate analyses (PCoA) of fecal bacterial compositions revealed a significant separation driven by genotype when cage was considered as a random effect in a mixed linear model (Fig. 5D, F) (McCafferty et al., 2013). One-way ANOVA showed that cage effects only contributed to differences among mice of the same genotypes (Fig. 5E, F). PCoA showed a separation of ileal luminal bacteria of separately housed WT and Lyz1−/− mice (Fig. 5G). Linear Discriminant Analysis (LDA) for ileal luminal bacteria revealed an expansion of Ruminococcus gnavus, Blautia gnavus, and Dorea formicigenerans and a reduction of Candidatus Arthromitus (segmented filamentous bacteria) in Lyz1−/− mice (Fig. 5H). LDA of fecal bacteria also showed relative expansion of D. formicigenerans and reduction of Candidatus Arthromitus (Fig. S5C), while LDA of ileal mucosa-associated bacteria identified a reduced abundance of Candidatus Arthromitus (Fig. S5D). Thus, Lyz1 deficiency altered gut bacterial landscape.
Figure 5. Paneth cell lysozyme deficiency or overproduction alters gut microbiota composition.
(A) Bacterial loads were determined in ileal and colonic mucosa and cecal lumen in WT and Lyz1−/− mice (N=6–10 in each group). (B-H) 16S amplicon profiling of fecal or ileal luminal microbiota from WT and Lyz1−/− mice. (B) Chao1 index (alpha diversity) (fecal microbiota, n=10– 12). (C) Unweighted UniFrac analysis of fecal microbiota from WT and Lyz1−/− mice (n=14–16). (D-E) PCoA analysis of fecal microbiota with consideration for genotype and cage effect (total of 8 WTs in 3 cages; 10 Lyz1−/−s in 4 cages). (F) Mixed linear model analysis using genotype as fixed and cage as random effect. One-way ANOVA showed that cage effects contributed to differences among mice of the same genotype. (G) PCoA analysis of ileal luminal microbiota from separately housed adult Lyz1−/− and WT littermates. (H) LDA of ileal luminal bacteria with species contracting (Candidatus Arthromitus) or expanding (R. gnavus, B. gnavus, and D. formicigenerans) in Lyz1−/−mice. (I) Schematic diagram of the Villin-Lyz1TG transgenic (TG) construct. (J) Lysozyme immunohistochemistry for WT, Lyz1−/−, and TG mouse colons (representative of N>3 for each genotype). (K) Lysozyme enzymatic activity in the colonic lumen of WT, Lyz1−/−, and TG mice (N=3 for each genotype). (L) Unweighted UniFrac analysis (16S amplicon profiling) of WT and TG mouse fecal microbiota. (M) Relative phyla abundance in the feces of separately housed WT and Lyz1−/− mice (N=4–8). (N) Relative phyla abundance of WT and TG mice (N=5–6). (O) Relative abundance of R. gnavus and D. formicigenerans in WT and Lyz1−/− fecal microbiota (N=4–10 per genotype). (P) Relative abundance of R. gnavus and D. formicigenerans in WT and TG fecal microbiota (N=5–6). (Q-V) Growth sensitivity of selected bacteria to lysozyme based on OD 660nm readings. Red arrowhead indicated the addition of 200 μg/ml lysozyme (data from 2–4 independent experiments). All bar graphs display mean ± SEM from at least two independent experiments. See also Figure S5.
Ectopic lysozyme production in colon reduced lysozyme-sensitive bacteria
To obtain detailed insights into how epithelial lysozyme might shape bacterial composition, we developed a second mouse model: Villin-Lyz1TG transgenic mice (TG). We used a 12.4 Kb Villin-1 promoter to drive Lyz1 expression (Fig. 5I), in order to model ectopic lysozyme production seen in human UC (Fig. 1A). Two independent TG founders were validated (Fig. S5E) and were separately maintained on pure C57BL/6 background. TG colony with their WT littermates were maintained separately from the Lyz1−/− colony, therefore WT mice were not shared between studies. TG mice showed an epithelial cell specific lysozyme production in colon (Fig. 5J, Fig. S5F–H), and in small intestinal villi (Fig. S5I). Strong lysozyme immunoreactivity in TG mouse colonic lumen echoed a robust lysozyme enzymatic activity (Fig. 5K), providing us the rationale to examine the impact of ectopic lysozyme on colonic microbial landscape. When challenged by DSS, TG mice exhibited an exacerbated colitis compared to WT littermates (Supp. Fig. 5K–M).
Fecal bacterial 16S rRNA amplicon profiling did not detect a change in α-diversity (Fig. S5J). Unweighed UniFrac analysis showed a strong separation between WT and TG microbiota (p=5.736E-6, Fig. 5K). Parallel examination suggested that Lyz1−/− mouse fecal microbiota had an expansion in Firmicutes and reduction in Bacteroidetes compared to their WT counterparts (Fig. 5M), while these phyla in TG mice altered in an opposite direction compared to their WT controls (Fig. 5N).
Thirty-six species showed a relative increase in Lyz1−/− feces (Table. S1), and 25 of them belonged to Firmicutes phylum represented by Dorea, Ruminococcus, Robinsoniella, Acetatifactor, and Tyzzerella genera (Fig. S5N). Species were decreased in TG, with 19 belonging to Firmicutes (Fig. S5O). Cross-comparison revealed 13 species expanded in Lyz1−/− but decreased in TG (Table. S2). Ranking by relative abundances gave rise to top 9 species with >1,000 amplicons detected in at least one genotype (Table. S2, Table. S3). Among them, R. gnavus, D. formicigenerans, M schaedleri, A. muciniphila were also expanded in Lyz1−/− ileal luminal microbiota (Fig. S5P, Table. S3, Table. S4). Similar comparison revealed 7 species increased in TG but decreased in Lyz1−/− mice, with the top one being Lactobacillus murinus (Table. S5). Candidatus Arthromitus showed an expansion in TG but a reduction in Lyz1−/− mice (Fig. 5H, Table. S5, Fig. S5C–D). LDA independently identified D formicigenerans and L. murinus as top expanded species in Lyz1−/− and TG fecal microbiota, respectively (Fig. S5C and 5Q).
Among the top expanded species in Lyz1−/−, R. gnavus, D. formicigenerans, M. schaedleri, and A. muciniphila were mucolytic bacteria (Png et al., 2010; Tailford et al., 2015b) (Robertson et al., 2005) (Table. S3). Both D. formicigenerans and R. gnavus contain genes for sialic acid metabolism and mucin degradation (Tailford et al., 2015a). Expansions of these species were observed in 3 independent 16S rRNA sequencing experiments (Fig. 5O, P).
The reverse alterations of certain species under lysozyme deficiency and overproduction suggested their sensitivities to luminal lysozyme. We thus tested the growth sensitivity to lysozyme for a selective panel of culturable bacteria. In anaerobic condition, D. formicigenerans was highly sensitive to lysozyme at the exponential growth phase upon exposure to 20 μg/mL of hen egg lysozyme (Fig. 5Q, Fig. S5R). A separate Dorea genus member, D. longicatena, showed similar lysozyme sensitivity (Fig. 5R). R. gnavus growth in L-YHBHI.4 medium was unchanged when exposed to lysozyme during exponential or lag phases (Fig. S5S). However, when cultured in DP2 defined media (see Methods), the growth of R. gnavus was prevented by lysozyme added at the time of or 30 min after inoculation of culture (Fig. 5S). Addition of lysozyme 60 min after inoculation only partially prevented growth of R. gnavus. If lysozyme was added 4 hr after inoculation of the culture, the bacteria were largely resistant (Fig. S5T), indicating potential density-dependent resistance similar to collective antibiotics resistance reported previously (Brook, 1989; Tan et al., 2012). When the same time course was conducted for R. gnavus cultured in L-YHBHI.4, the lysozyme remained ineffective in suppressing its growth, indicating an environment-dependent lysozyme sensitivity. A. muciniphila was insensitive to lysozyme during any growth phase (Fig. 5T). Likewise, B. uniformis, despite expansion in Lyz1−/− (Fig. S5C), and L. rhamnosus were both resistant to lysozyme (Fig. 5U, V). These distinct growth responses of commensal bacteria to lysozyme indicated drastically different lysozyme-mediated bacterial killing (Ellison and Giehl, 1991; Ibrahim et al., 2001a; Laible and Germaine, 1985).
Microbiota from Lyz1−/− mice does not offer anti-colitogenic protection when transferred into “lysozyme-sufficient” recipients
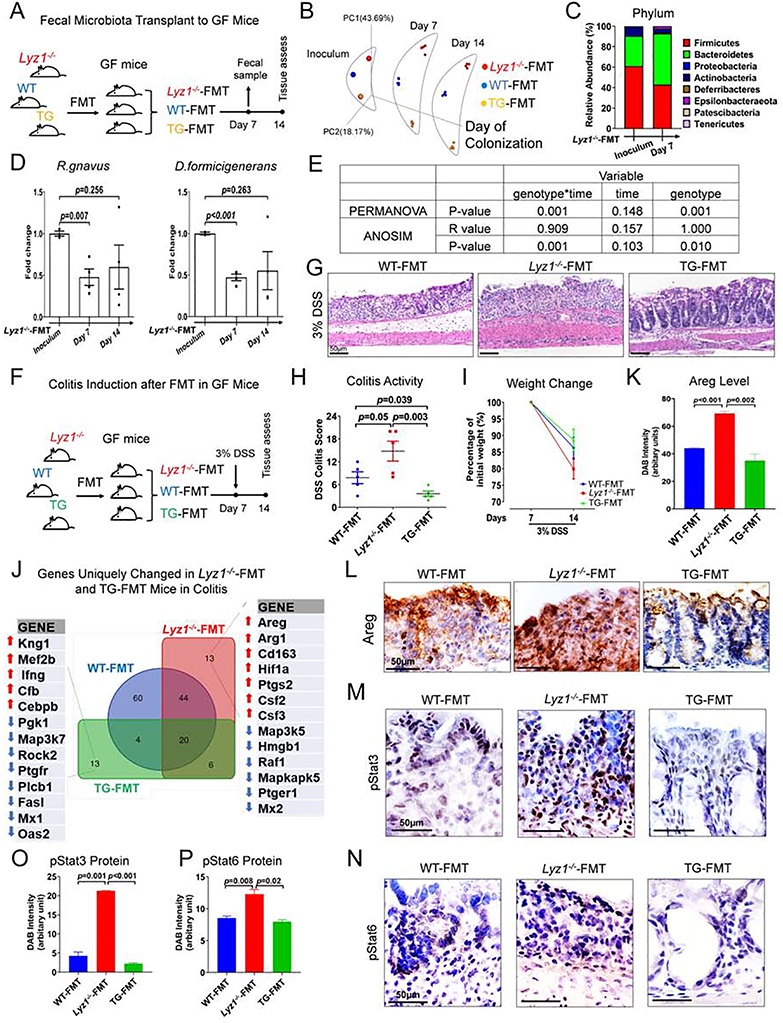
To test if the anti-inflammatory activity of Lyz1−/− microbiota could be transferred to recipients with intact Lyz1, we performed fecal microbiota transfer (FMT) using germ-free (GF) C57BL/6 recipients. Bacterial suspensions pooled from the ceca of WT (N=3), Lyz1−/− (N=3), or TG (N=3) mice were gavaged to WT GF mice (N=5 for each genotype of FMT, Fig. 6A). 16S rRNA amplicon profiling validated colonization after a week and successful maintenance of distinct WT, Lyz1−/−, or TG microbiota in individual GF recipients (Fig. 6B). Following FMT of Lyz1−/− microbiota, abundance of Firmicutes was reduced, while Bacteroidetes expanded after a week (Fig. 6C). qPCR showed reduced abundance of R. gnavus and D. formicigenerans in Lyz1−/−-FMT recipients 1-week after transfer (Fig. 6D), likely due to the intact host lysozyme activity. Despite this partial reduction, PERMANOVA and ANOSIM analysis (Ericsson et al., 2018), using genotype or duration of colonization as variables, showed that the genotype of FMT donors played the dominant role in defining the observed separation rather than the duration of colonization (Fig. 6E).
Figure 6. Lyz1-deficient microbiota transplanted to Lyz1-intact host, promotes inflammation in experimental colitis.
(A) Experimental design schematics. (B) PCoA showed the maintenance of diverse communities in GF mice with FMT from the 3 genotypes. (C) Averaged relative abundance of phyla in the Lyz1−/−-FMT inoculum and in the colonized WT GF mice 7 days later. (D) Relative abundance of D. formicigenerans and R. gnavus in fecal microbiota of Lyz1−/−-FMT mice, 7 and day 14 post-FMT as compared to inoculum. (E) In PERMANOVA and ANOSIM analysis, the source/genotype of the inoculum, but not the duration of colonization, determined the microbial differences. (F) Experimental design of DSS colitis in ex-GF mice after FMT with different microbiota (N=5 ex-GF mice-FMT donor genotype). (G-H) Representative H&E images of distal colons and colitis activity scores in DSS-treated ex-GF mice. (I) Body weight changes of ex-GF mice before and after DSS treatment. (J) NanoString analysis of inflammation-related gene expression in the proximal colons of ex-GF mice before and after DSS treatment. (K-L) Areg immunohistochemistry in the colons of DSS-treated ex-GF mice. (M-P) Representative pStat3 and pStat6 immunohistochemistry and semi-quantitative analysis in the colons of DSS-treated ex-GF mice (n=5 in each group). Data are represented as mean ± SEM in D, H, K, O and P from at least two independent experiments. See also Figure S6.
To test whether FMT of Lyz1−/− or TG microbiota might modulate the response to DSS, GF mice transplanted with WT, Lyz1−/−, or TG microbiota were treated with regular water or 3% DSS (Fig. 6F). Unexpectedly, Lyz1−/−-FMT mice exhibited an exacerbated colitis (Fig. 6G, H), loss of 20% of body weight on average (N=5, Fig. 6I), and a 20% mortality at the end of the study. All WT-FMT and TG-FMT mice survived the treatment. TG-FMT mice exhibited a degree of protection judged by histology (Fig. 6G, H) and body weight change (p>0.05, Fig. 6I).
NanoString transcriptome analysis using nCounter Inflammation Panel (Mouse v2) of DSS-treated mouse colonic mucosa showed that inflammatory genes were generally elevated in all DSS-treated mice (Fig. S6A–C). Among 13 genes affected exclusively in Lyz1−/−-FMT mice were elevated inflammatory transcripts: Hif1a, Ptgs2, Csf2, Csf3, and anti-inflammatory Areg, Arg-1, and Cd163 (Fig. 6J). Immunostaining verified Areg elevations in Lyz1−/−-FMT colons (Fig. 6K, L). pStat6 and pStat3 proteins were also elevated in Lyz1−/−-FMT, compared to WT-FMT and TG-FMT colons (Fig. 6M–P and Fig. S6D, E). Thus, transferring Lyz1−/− microbiota to lysozyme-sufficient recipients exacerbated colitis likely through enhanced bacterial processing by host lysozyme. This increased inflammatory response likely masked Areg’s tissue repair activity.
TG-FMT mouse colonic mucosa had reduced inflammatory response compared to WT-FMT or Lyz1−/−-FMT mice, exhibited by reduced MAPK, NF-κB, T-cell receptor, and natural killer cell cytotoxicity pathways (Fig. S6F). Oas2, Mx1, and cell death-related genes Fasl and Rock2, were among the most reduced transcripts in TG=FMT mice during DSS colitis (Fig. 6J). Thus, transfer of Lyz1−/− microbiota to lysozyme-sufficient hosts did not transfer protection.
Lysozyme-processed and non-processed R. gnavus induced different immune responses
We postulated that lysozyme-sensitive bacteria might elicit varying immune responses dependent upon lysozyme processing. We chose R. gnavus to test this hypothesis due to its expansion in IBD patients (Hall et al., 2017; Joossens et al., 2011; Sartor and Mazmanian, 2012; Willing et al., 2010). Lyz1−/− mice carrying high abundance of R. gnavus also showed more goblet cells (Fig. S7A). We tested the impact of live R. gnavus and lysozyme-processed R. gnavus on intestinal MLN cells. Supernatants were collected from live R. gnavus or lysozyme-treated R. gnavus, applied to WT or Lyz1−/− MLN cells, and examined for cytokine production using dot blots (Fig. 7A and Fig. S7B). Supernatants of non-processed R. gnavus elicited a 5-fold induction of IL-13, among other cytokines from MLN cells from WT (Fig. 7B, D, E) and Lyz1−/− mice (Fig. 7C, D, E). In contrast, lysozyme-processed R. gnavus failed to induce IL-13, and instead induced a range of inflammatory cytokines: TNFa, CXCL1, CXCL2, CXCL9, CXCL10, CCL3, IL-6, and IFNγ (Fig. 7B, D, E). Different from lysozyme-processed R. gnavus, heat-killed R. gnavus induced a confined panel of cytokines from WT and Lyz1−/− MLNs (Fig. S7C, D). Lysozyme-treated L. rhamnosus (a lysozyme-resistant control, Fig. 5V) was the least effective (Fig. S7E, F), suggesting that the observed cytokine induction was specific to lysozyme-processed R. gnavus, and not due to lysozyme alone (Fig. 7D). Live R. gnavus supernatants also promoted pStat6, whereas lysozyme-processed R. gnavus induced pStat3 (Fig. 7F, Fig. S7G, H) in WT and Lyz1−/− MLNs. Thus, live and lysozyme-processed R. gnavus elicited different MLN cytokine profile.
Figure 7. Distinct inflammatory cytokine induction by R. gnavus in lysozyme’s presence or absence.
(A) Experimental design for data in panels B-G. (B-C) Radar plots of cytokine/chemokine concentrations in the media of from WT and Lyz1−/− MLN cells stimulated by supernatants from lysozyme-treated R. gnavus (red) versus untreated live R. gnavus culture (green). (D-E) Representative dot blots and summary quantification from two independent experiments (each in 2 replicates). (F) Western blots of pStat6 and pStat3 in lysates of WT and Lyz1−/− MLN cells treated analogous to B-E. (G) Experimental design for data in panels H-K. (H-J) Bulk RNAseq of ileal mucosa 14-days after R. gnavus gavage (n=3). Increased tuft cell signature, type 2 immune response, and IL-13 response in R. gnavus colonized mice in GSEA analysis. (K) Differential expression of IL-13 responsive genes in R. gnavus-colonized Lyz1−/− compared to PBS-gavaged Lyz1−/− mice. (L) Experimental design for data in panels M-O showing that DSS colitis was induced in R. gnavus colonized WT and Lyz1−/− littermates following Abx treatment. (M) Body weight changes during DSS exposure and recovery in Abx-precleared WT and Lyz1−/− mice with or without R. gnavus association (n number indicated; 2 independent experiments). (N) Colitis activity scores in the colon of DSS-treated Lyz1−/− mice with or withour R. gnavus association. (O) Representative H. & E. images of DSS colons of Abx-treated and R. gnavus colonized WT and Lyz1−/− mice. All bar graphs display mean ± SEM. See also Figure S7.
To test if R. gnavus elicits distinct immune response in vivo in Lyz1-sufficient versus - deficient hosts, WT and Lyz1−/− littermates were first treated by Abx, followed by colonization with R. gnavus (Fig. 7G), or L. rhamnosus (as a lysozyme-resistant control). Successful colonization was confirmed by PCR (Fig. S7I, J). Bulk RNAseq showed that R. gnavus colonization of Abx-treated Lyz1−/−- mice restored the type 2 responses (Fig. 7H–K), which had been diminished by Abx treatment (Fig. 4E, F, H). qPCR validated the increased IL-13 and IL-25 after R. gnavus colonization of Abx-treated Lyz1−/− mice (Fig. S7K, L). Immunostaining confirmed that R. gnavus, but not L. rhamnosus, induced expansion of goblet and tuft cells (Fig. S7M–O). Of note, although R. gnavus increased goblet and tuft cells in WT mice (Fig. S7M–O), it elicited a more potent type 2 response in Lyz1−/− than in WT mice (Fig. S7P).
To assess the modulatory effects of processed and uprocessed R. gnavus on experimental colitis, WT and Lyz1−/−- Abx-treated mice were first colonized with R. gnavus and then treated with DSS (Fig. 7L). Abx-treated Lyz1−/− mice developed severe colitis (Fig. 4N–O), however, R. gnavus colonization ameliorated it (Fig. 7M–O). R. gnavus colonization of Abx-treated WT mice exacerbated colitis (Fig. 7M–O). We concluded that host Lyz1 status balanced the pro- or anti-inflammatory effects of lysozyme-sensitive bacteria.
DISCUSSION
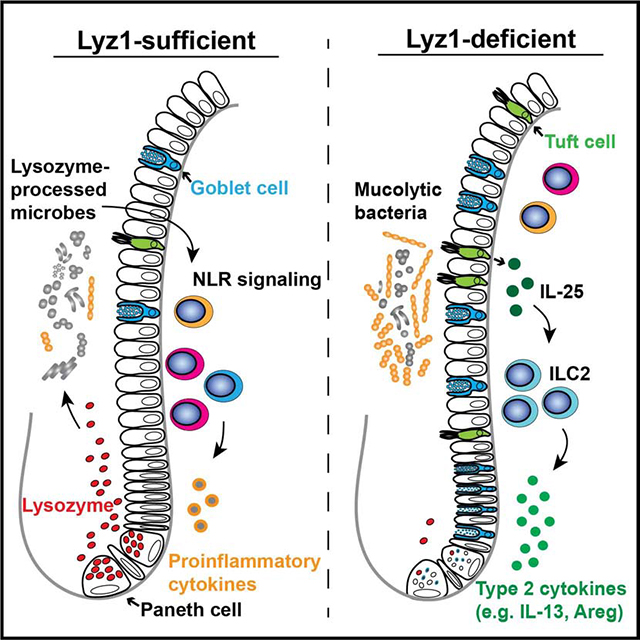
Abnormalities in Paneth cell lysozyme granules and secretory defects were reported in CD patients (Liu et al., 2017; VanDussen et al., 2014) and in mice carrying CD susceptibility alleles (Bel et al., 2017). In UC, metaplastic Paneth cells in left colon are a feature of this disease (Singh et al., 2020; Tanaka et al., 2001). We found that these metaplastic Paneth cells expressed lysozyme. We used Lyz1-deficient and -overexpressing mice to model these aberrant lysozyme productions, and revealed that luminal lysozyme abundance controls the composition of ileal and colonic microbiota, regulating mucosal inflammatory responses at steady-state and during experimental colitis.
Prior in vitro work suggested that lysozyme displayed bactericidal activity towards Gram-positive and Gram-negative species (Ibrahim, 1998) (Ellison and Giehl, 1991; Ibrahim et al., 2001a; Laible and Germaine, 1985). We found that bacterial load and alpha diversity in multiple anatomical regions were not affected by lysozyme deficiency or overproduction, which was counterintuitive of the notion that lysozyme is non-specific antimicrobial peptide (Ragland and Criss, 2017). Our data suggest that distinct bacterial populations possess differential sensitivity to lysozyme. For example, Lactobacilli were resistant to lysozyme, while Dorea were very sensitive. Among lysozyme-sensitive species, the growth of D. formicigenerans versus R. gnavus also exhibited variable responses to lysozyme. Thus, a fine-tuned microbial or host-microbe factors may exist to determine the distinct lysozyme-sensitive profiles of individual commensal species.
A diminished intestinal lysozyme secretion in certain IBD patients may allow a selective expansion of species such as R. gnavus (Hall et al., 2017; Joossens et al., 2011; Sartor and Mazmanian, 2012; Willing et al., 2010). Conversely, ectopic production of lysozyme by metaplastic Paneth cells in inflamed UC colons may suppress lysozyme-sensitive species but nevertheless drive inflammation through bacterial cell-wall processing. Such interplay between host lysozyme and microbiota may also exist in Lyz2−/− mice that lack lysozyme M in macrophages and neutrophils. Lyz2−/− mice displayed higher susceptibility to pulmonary infection by M. luteus or S. pneumonia (Ganz et al., 2003; Markart et al., 2004b; Shimada et al., 2008). These studies suggested that the exacerbated inflammation in Lyz2−/− mice were due to the failure of lysozyme M to inactivate peptidoglycan. However, Lyz1−/− ileal mucosa had a diminished mucosal sensing and response to bacterial peptidoglycan at steady state, accompanied by reduced basal inflammatory response. Thus, the different sites where these lysozymes are produced may contribute to the observed differences. Monocytic lysozyme M may mediate direct bacterial killing to prevent infection spreading, while luminal lysozyme P, with an intact barrier, may primarily process bacterial cell-wall to alert the immune system.
Lyz1-deficiency led to changes in both epithelial and immune cell compartments. The reshaped gut microbiota in Lyz1−/− mice unexpectedly promoted a type 2 response responsible for goblet and tuft cell expansion. One of the lysozyme-sensitive species, R. gnavus, induced a similar type 2 response when colonizing the Lyz1−/− mice, supporting the homeostasis whereby certain commensal bacteria modulate the inflammatory tone as reported in the lungs (Chua et al., 2018). Simultaneous expansion of two phylogenically related R. gnavus and D. formicigenerans in Lyz1−/− mice may link to their expression of genes for sialic acid metabolism and mucin degradation (Tailford et al., 2015a). Degradation of mucin coupled with the induction of mucin-producing goblet cells by mucolytic bacterial species (Png et al., 2010; Tailford et al., 2015b) may promote syntrophy by fueling the assimilation of mucin monosaccharides by other bacteria (Sartor and Mazmanian, 2012; Willing et al., 2010). R. gnavus also promoted goblet and tuft cell program in WT mice, yet it did not exert protection. We speculate that when barrier function is compromised, lysozyme-processed R. gnavus may escalate the inflammation that overrides the protective type 2 response. These data collectively support a model whereby the expansion of certain IBD-related species such as R. gnavus may promote tissue healing when luminal lysozyme is reduced (Laurent et al., 2017; Pulendran and Artis, 2012).
The exact molecular identities of the bacterial products that skewed the type 2 immune response in Lyz1−/− mice are unknown. Live, non-processed R. gnavus, or similar bacteria, may generate products eliciting cytokines such as IL-13. Notably, tuft cells sense gut microbial metabolites through taste receptors and GPR91 to promote an ILC2 immune circuit (Howitt et al., 2016; Lei et al., 2018; Nadjsombati et al., 2018; Schneider et al., 2018). Bacterially derived succinate engages GPR91 to initiate type 2 immune response (Lei et al., 2018; Nadjsombati et al., 2018; Schneider et al., 2018). R. gnavus may not be a strong succinate producer based on pilot study. Helminths are capable of inducing tuft cell hyperplasia and type 2 response in Gpr91-deficient mice (Nadjsombati et al., 2018), indicating multiple gut biome-initiated pathways promoting differential mucosal immune response.
Based on our analysis, it is reasonable to propose that processing of sensitive bacterial cell walls by lysozyme drives inflammatory response in mucosal immune cells. Elevated lysozyme production during active colitis is expected to enhance bacterial processing and killing, which prevents bacterial infiltration but exacerbating type 1 and 3 inflammation.
A study found that co-culture of intestinal organoids with Th1 cells increased Paneth cell specific genes, while IL-13 treatment increased tuft cell differentiation (Biton et al., 2018) leading to a notion that distinct Th cytokines guide MHCII+Lgr5+ stem cells to specific epithelial cell types. Paneth cells possibly monitor and direct the intestinal type 1 immunity via lysozyme while goblet and tuft cells coordinate with type 2 immunity. In so doing, the Th1-Paneth cell axis is balanced by goblet-tuft-Th2 circuit to maintain gut homeostasis.
LIMITATIONS OF STUDY
Currently, no murine model precisely models the lysozyme-producing metaplastic Paneth cells in distal colon of IBD patients. The Villin-Lyz1 transgenic mice developed in this study were not colon-specific and, as such, lysozyme was overexpressed in epithelia throughout the entire intestinal tract. As the abundance of luminal lysozyme in these mice was expected to be greater than produced solely by metaplastic Paneth cells in human IBD, the experimental colitis exhibited in Villin-Lyz1 mice should be interpreted with caution. The colitis study used in current study was limited to the well-established DSS model. Additional research will elucidate the role of lysozyme in various inflammatory models, in particular during pathogen infection. The insights into the impact of other lysozyme-sensitive bacterial species remain limited. Future work will define species-specific impact, under lysozyme-deficient and sufficient conditions, on mucosal inflammatory response. The mechanism of the anti-inflammatory activities of Villin-Lyz1 mouse gut microbiota, when transferred to gnotobiotic mice, remained unclear. Future work will identify the specific species mediating such protection.
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to, and will be fulfilled by, the Lead Contact, Nan Gao at Rutgers University-Newark. email: ngao@newark.rutgers.edu.
Materials availability
Mouse lines generated in this study are available upon request.
Data and code availability
The scRNA-seq data generated in this study are deposited in Gene Expression Omnibus(GEO) with accession number GSE151152. The bulk RNA-seq data generated in this study are deposited in GEO with accession number GSE151151.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Lyz1−/− mice were derived in a previously published study (Yu et al., 2018) where the Lyz1 gene was disrupted by a knock-in gene cassette “H2BmCherry-IRES-CreER”. The mouse allele was backcrossed to C57BL/6 for 8 generations. To generate Villin-Lyz1TG mice, mouse Lyz1 coding sequence was amplified using ileal tissue cDNA and inserted at XhoI and AgeI sites of a 12.4kbVillin-ΔATG vector (Addgene, Plasmid #19358), in which a 12.4kb-length Villin promoter directs the Lyz1 expression in intestinal epithelial cells (Madison et al., 2002). The construct was digested with PmeI to remove pUC18 vector backbone. The linearized DNA fragment containing Villin promoter/ Lyz1CDS/SV40 polyA signal sequences was separated by 0.8% agrose gel, purified and subjected to DNA dialysis in microinjection buffer (8 mM Tris-HCl, 0.15 mM EDTA). After dialysis, the DNA fragment was diluted to 2ng/μl with microinjection buffer and used for pronuclear microinjection on C57BL/6 genetic background. 6 founders were identified using genotyping PCR. Two founder lines were confirmed and maintained separately on C57BL/6 background. The expression of Lyz1TG was tested on F1 pups by real-time PCR, western blot and immunofluorescence. Stat6−/− (Stock No. 005977) and Il4ra−/− (Stock No. 003514) mice were purchased from Jackson Laboratory.
All animal experiments were approved and overseen by the Institutional Animal Care and Use Committee of Rutgers University and University of Arizona. Animal experiments were conducted in accordance with NIH guidelines and US federal laws. All mice were housed in individually ventilated cages under specific pathogen free conditions at Rutgers University Newark animal facility or University of Arizona gnotobiotic facility accredited by the Association of Assessment and Accreditation of Laboratory Animal Care International. All mice were maintained on a 12-hour light/dark cycle and fed by food and water ad libitum. All experiments were performed on littermates, and data were typically reported on the basis of multiple experiments of independent litters. When WT and Lyz1−/− mice were separately housed for an experiment, it was reported as separately housed littermates in the text and figure legend. WT mice throughout the study were generated from the same colonies where Lyz1−/− or TG mice were produced. No commercial WT mice were used as controls in any experiment.
Human Samples
The human tissue samples were de-identified and procured under approved Institutional Review Board (Princeton Medical Center: BN2239; BN2294).
Bacterial strains
Akkermansia muciniphila (DSMZ, DSM-22959), Bacteroides uniformis (DSMZ, DSM-6597), and Dorea formicigenerans (DSMZ, DSM-3992) were grown in appropriated L-YHBHI.4 (Liquid Yeast extract Hemin Brain Heart Infusion.4) growth medium. Dorea longicatena (DSMZ, DSM-13814) was grown in 104 PYG modified medium. Ruminococcus gnavus (in-house INRA-Micalis collection (Dabard et al., 2001) was grown in either L-YHBHI.4 or DP2 growth medium (Table. S6). For bacteria colonization, R. gnavus (ATCC, Catalog No. 29149) was cultured in AnaeroGRO Chopped Meat Glucose Broth (Hardy Diagnostics, Catalog No. AG19H). All anaerobic cultures were conducted at 37°C using the Hungate culture method (Hungate, 1950) at Institut Micalis, INRA, France or Dr. Haggblom’s lab at Rutgers-New Brunswick. Lactobacillus rhamnosus GG (LGG, ATCC, Catalog No. 53103) was aerobically cultured in commercial Lactobacilli MRS Broth (BD Biosciences, Catalog No. 288130) in our own lab. For each bacterial specie, 24h growth media were inoculated with a 1:9 dilution to a duplicate set of young culture of similar composition.
METHOD DETAILS
DSS Experimental Colitis
Adult littermates (N=3–14/group) were administered with 3% dextran sulfate sodium (DSS, Colitis grade, 36–50KDa, MP Biologics, SKU 0216011080) in tap water for 7 days and recovered with tap water for another 7 days. Colon tissues were harvested for further pathological analysis. Colonic damage was scored blindly by a GI pathologist as described previously (Chassaing et al., 2014).
Briefly, H&E sections were blindly scored by a certificated pathologist. Scores (0–4) were assigned based on the severity of epithelial injury and leukocyte infiltration into the mucosa, submucosa and muscularis. These three scores were multiplied by an extended factor to assess the extent of the change: 1 for focal, 2 for patchy, and 3 for diffuse, and summed to achieve the final score out of the maximal score of 36. For Figure 6H, colitis score of the dead Lyz1−/−-FMT mouse was assumed to be equal to the worst colitis score among the Lyz1−/−-FMT mice.
In vivo IL-13 Neutralization
For IL-13 neutralization, 8–12-week-old mice (N=2 for each genotype in each treatment group) were administrated i.p. with indicated antibody or isotype control at a dose of 250ug/mouse on Day 0 and Day 3. Intestinal tissues were collected on Day 5 for further analysis. IL-13 neutralizing antibody (Catalog No. MAB413) and Rat IgG2a Isotype control (Catalog No. MAB006) were purchased from R&D systems.
Bone Marrow Transfer
To study the role of lamina propria immune cells in Lyz1−/− phenotypes, we derived chimera mice using bone marrow transfer. Briefly, bone marrow from femur and tibias were harvested from gender and age-matched wild type (N=4) and Stat6−/− mice (N=4) to generate single cell suspensions (5×106 cells/100μl). Recipient mice (WT, N=6; Lyz1−/−, N=6) received two rounds of whole-body irradiation (6 Gy), 3 hr apart. Then, bone marrow single cells (100 μl) were injected into the irradiated recipients through ophthalmic venous plexus. After 2 mo reconstitution, recipient mice were euthanized, and intestinal tissues were collected for further analysis.
Lysozyme Resistance Assay
To determine whether certain bacterial species were sensitive to lysozyme, 200 μg/L sterile chicken egg white lysozyme solution (Sigma, Catalog No. L6876) was added to each young culture media duplicates at the time of the inoculation of the bacteria into the young culture or during the exceptional phase. The growth of each bacteria was measured every 30min or 1h by absorbance (A660) for 8–10h.
Intestinal Lysozyme Activity Assay
To measure the activity of endogenous lysozyme, we used fluorometric based lysozyme activity assay kit (Abcam, Catalog No. ab211113) according to the manufacturer’s instructions. Briefly, luminal contents were centrifuged at 12,000×g for 5 min at 4°C and supernatants were subjected to lysozyme activity assay. 10μl of each supernatant was mixed with 4μl lysozyme substrate in 60μl assay buffer and incubated for 1hr at 37°C. After incubation, 50μl of lysozyme stop buffer was added into each well and subjected to fluorescence measurement (Ex/Em=360/445nm). Serial dilutions of 10μM 4-Methylumbelliferone (4-MU) were used for standard curve preparation. Variation in fluorescence in each sample was applied to the standard curve to get the amount of 4-MU generated during the reaction. The lysozyme activity in the supernatants is calculated as the amount of 4-MU generated in the reaction divided by the reaction time and sample volume.
Antibiotics Treatment and Bacteria Colonization
Age- and gender-matched mice (N=16 for each genotype) were fed with autoclaved tap water containing antibiotics cocktail for one week and recovered on autoclaved tap water without antibiotics for 3 days. Fecal pellets were collected for DNA extraction and real-time PCR using bacterial universal primer set was conducted to ensure bacteria depletion after antibiotics treatment. Then, R. gnavus or LGG suspension (1×108/mouse) in 1.5% NaHCO3 (pH8.0) was gavaged into each mouse (N=10 for R. gnavus colonization in each genotype, N=4 for LGG colonization in each genotype). After 1 wk, fecal pellets were collected for R. gnavus quantification using real-time PCR with R. gnavus specific primer set.
Bacterial DNA Extraction, Sequencing and Analysis
Mouse feces, ileal luminal, or cecum luminal contents were freshly collected and frozen at - 80°C. Genomic DNA was isolated using Qiagen QIAamp DNA Stool Mini Kit (Catalog No. 51604) or Invitrogen PureLink Microbiome DNA purification kit (Catalog No. A29789) according to the manufacturer’s instructions. Purified DNA samples were sent to PrimeBio Research Institute LLC for barcoded 16s rDNA library construction and next-generation sequencing using Invitrogen Ion Torrent Sequencing kit. Briefly, 3 ng of the purified bacteria DNA was used for 16s rRNA PCR amplification. The amplicons were generated with the Ion 16s Metagenomics Kit (Thermofisher, Catalog No. A26216), which amplifies seven hypervariable regions (V2, V3, V4, V6, V7, V8, and V9) of bacterial 16S rRNA. After the 16s rRNA PCR amplification, the Ion Plus Fragment Library Kit (Thermofisher, Catalog No. 4471252) was used for library construction. The constructed libraries were templated with Ion PGM™ Template Hi-Q Kit (Thermofisher, Catalog No. A27739), and then sequenced with Ion PGM™ Hi-Q™ Sequencing Kit (Thermofisher, Catalog No. A25592). The sequencing reads were processed by Ion Torrent Suites and Ion Reporter software Metagenomics 16S w1.1 workflow, which enables the identification, at the genus or species level, of microbes present in complex polybacterial samples via the curated MicroSEQ™ ID 16S rRNA reference database and the curated Greengenes database. In brief, FASTQ quality files were split by samples according to the barcode and BAM files were generated by Ion Torrent Suites. The BAM files were uploaded into Ion Reporter, and reads were filtered by primer and length. Unique reads were kept, and abundance was calculated over a set of thresholds. Taxonomical classification was performed using multistage BLAST search of reads against two databases mentioned above, and OTU tables were created.
Metastats (http://metastats.cbcb.umd.edu/detection.html) was used to detect differentially abundant features between microbial communities of different groups, that is, features that are enriched or depleted in one population versus another. Metastats employs the false discovery rate to improve specificity in high-complexity environments, and separately handles sparsely sampled features using Fisher’s exact test. QIIME (qiime.org) was used for microbial ecological Chao1 richness (alpha diversity) and PCoA (beta diversity) assay. Linear Discriminant Analysis (LDA) was performed using LDA Effect size (LEfSe) analysis tool (https://huttenhower.sph.harvard.edu/galaxy/). Briefly, the non-parametric factorial Kruskal-Wallis sum-rank test was used to detect features with significant differential abundance with respect to the species level. Biological significance was subsequently evaluated using a set of pairwise tests among subclasses by unpaired Wilcoxon rank-sum test. Then, LEfSe was applied to estimate the effect size of each differentially abundant feature to perform dimension reduction.
To examine cage effects, WT and Lyz1−/− mice from the same breeding parents were co-housed for 3 weeks, separated into different cages according to genotype (e.g. WT cages and Lyz1−/− cages), and tested after 1-month of separate housing to assess bacterial composition. Based on a mixed linear model and one-way ANOVA (McCafferty et al., 2013), cage and genotype effects were calculated.
Fecal Microbiota Transplantation and Gnotobiotic Mice
9–10-week old female germ-free C57BL/6J mice maintained in flexible isolators at the University of Arizona gnotobiotic facility were used according to an approved IACUC protocol 07–126 (to P.R.K.). Study was limited to female mice to reduce aggression upon cage transfer and experimental grouping. Fecal microbiota transplantation (FMT) was performed by administering fecal slurry prepared from pooled stools from either WT (N=9), Lyz1−/− (N=9) or Villin-Lyz1TG (N=10) mice by oral gavage with 100mL of suspension and by painting the abdominal pelt with the same amount (Day 0). Upon colonization, mice were randomly assigned to control or DSS group (N=5) and transferred to Tecniplast iso-positive ventilated cages (one FMT group/treatment/cage). Mice were allowed to acclimate to the new environment for 7 days (Day 7), after which control mice were continued on autoclaved water, while DSS group received 3% DSS in drinking water for another 7 days (till day 14). Body weight was monitored daily. Mice were euthanized on day 14 and colons were harvested for RNA isolation or fixed in buffered formalin for histological analysis.
Fecal samples from all ex-GF mice were collected at day 0 (the colonization day, serve as a negative control), and then at day 7, and day 14. Fecal pellets were stored at −80°C until use. Genomic DNA was extracted and purified with PowerFecal Pro DNA kit (Qiagen, Cat No. 51804) according to the manual provided by the manufacturer. The samples were homogenized using provided lysis buffer and the tubes pre-filled with the 96 well plate shaker (Mo-Bio, Cat No. 11996) with the 2 ml adapters (Mo-Bio, Cat No 11990) two times 10 minutes at speed 30Hz each at 4°C. The hypervariable V4 region of the 16S rRNA gene was amplified by PCR from each sample using barcoded primers (515F and 806R). The forward primer is common for all samples, whereas the reverse primers, consisting of Golay barcodes, were unique for each sample. Both reverse and forward primes are extended with the sequencing primer pads, linkers, and Illumina adapters (Caporaso et al., 2012). The PCR was performed on LightCycler 96 (Roche) with MyFiTM Mix (Bioline Meridian, Cat No. BIO-25050) in the final volume 40μL. Amplicons were quantified using Quant-It PicoGreen dsDNA Assay kit (ThermoFisher Scientific, Cat No. P7589), according to the manufacturer’s protocol. Equal amount of amplified DNA (240ng) from each sample were pooled and cleaned using UltraClean PCR Clean-Up Kit (MoBio, Cat No. 12500). Pooled amplicons were diluted, denatured with NaOH at final concentration 0.1N, and 6.75 pmols of the pooled library was sequenced at our laboratory on MiSeq platform (Illumina) using custom primers (Caporaso et al., 2012). Due to the limited sequence diversity among 16S rRNA amplicons, 5% of the PhiX Sequencing Control V3 (Illumina, Cat No. FC-110–3001) made from phiX174, was added to the run. The pooled 16S rRNA library was subjected to the paired-end sequencing using 2 × 150bp MiSeq Reagent Kit V2 (Illumina, Cat No. MS-102–2002).
De-multiplexing was done using idemp script (https://github.com/yhwu/idemp). Filtering, dereplication, sample inference, chimera identification, and merging of paired-end reads was done with a reference-free Divisive Amplicon Denoising Algorithm 2 (Dada2) R package (Callahan et al., 2017). The ASVs taxonomy was assigned using RDP classifier against SILVA database (Quast et al., 2013) release 132 (https://www.arb-silva.de/documentation/release-132/). The vegan package (https://CRAN.R-project.org/package=vegan) was used as a tool for diversity analysis, ordination methods, for the analysis of dissimilarities, and statistical analysis (Oksanen et al., 2019). The obtained results were visualized with ggplot2 package (Hadley).
Tissue Collection, Fixation and Histochemistry
Mouse intestinal tissues were collected and fixed in 4% paraformaldehyde or 10% neutral formalin overnight. Then tissues were transferred into 70% ethanol and subjected to paraffin embedding at the Histology Core Facility at Rutgers University Hospital Cancer Center. 5μm sections were sliced from paraffin blocks, rehydrated and subjected to H & E staining. To highlight mucin-producing goblet cells, rehydrated sections were subjected to Alcian blue staining. Alcian blue solution (pH2.5) was applied onto slides and incubated for 30 min at room temperature. Then, slides were washed in running water for 2 min and counterstained with Vector Nuclear Fast Red (Vector Labs, Catalog No. H-3403) for 2 min. Slides were rinsed in running water for 5 min and then subjected to dehydration and mounting with Cytoseal-60 (ThermoFisher Scientific, Catalog No. 8310–4). Images were collected by Nikon TE2000D with NIS Elements D version 4.4 and analyzed by ImageJ software.
Transmission Electron Microscopy
The procedure for transmission electron microscopy was described previously (Gao and Kaestner, 2010; Yu et al., 2014). Briefly, 1–2mm ileal tissues were harvested from age-and gender-matched mice (N=3 for each genotype) and immediately immersed in 0.1M sodium cacodylate containing 2% glutaraldehyde (EM grade) and 2% paraformaldehyde for overnight fixation at cold room. Next day, samples were washed twice with 0.1M sodium cacodylate, then post fixed with 1% buffered OsO4 and stained en bloc with 0.1% uranyl acetate. The samples were subjected to dehydration using increasing concentrations of ethanol and then in propylene oxide. After balanced in EMBed-812/propylene oxide (1:1) and 100% EMBed-812, the samples were subjected to embedding using EMBed-812 kit (Electron Microscopy Sciences, Catalog No.14120).
Immunofluorescence and Immunohistochemistry
The procedures for immunofluorescence and immunohistochemistry have been previously described (Yu et al., 2018). Briefly, 5μm paraffin embedded sections were subjected to rehydration and antigen retrieval. Slides were doused into the sub-boiling antigen retrieval buffer (1μM citric acid, pH 6.0 or 1μM EDTA, pH8.0) for 20 min and then immediately transferred into running water. To block endogenous peroxidase activity, slides were incubated in 3% H2O2 solution diluted in methanol for 10 min. Then, slides were blocked with PBS buffer containing 0.1% Triton X-100, 2%BSA and 2% normal donkey serum for 2hr at room temperature, and then probed with indicated antibodies at 4°C overnight. Next day, slides were washed with PBS and incubated with either fluorescent dye-conjugated secondary antibodies (immunofluorescence) or Polymer HPR-conjugated secondary antibodies (immunohistochemistry).
For Immunofluorescence, slides were incubated with indicated secondary antibodies for 1 hr at room temperature and then followed by DAPI or Topro3 nuclear counterstaining. Then slides were washed with PBS, air-dried and mounted with Prolong Gold Antifade Mountant (Invitrogen, Catalog No. P36930). Images were collected using Zeiss LSM510 with 40x oil lens and analyzed by ImageJ software.
For Immunohistochemistry, ImmPRESS HRP anti-rabbit or anti-mouse IgG polymer (Vector Labs, Catalog No. MP-7401 and MP-7402 respectively) were used. After 1 hr incubation, slides were washed with PBS and subjected to DAB development. Hematoxylin QS (Vector Labs, Catalog No. H-3404) was used for nuclear counterstaining. Then, slides were dehydrated and mounted with Cytoseal-60.
Mesenteric Lymph Nodes Isolation and Cytokine Detection
Mesenteric lymph nodes (MLNs) adjacent to ileum and colon were collected and meshed against 70μm cell strainer. Single cell suspensions were centrifuged, counted and seeded into 6-well plate at 3×106/well density. The MLN cells were cultured in RPMI1640 containing 10%FBS, 2 mM l-glutamine, 1xpenicillin/streptomycin solution, 20 mM HEPES, MEM nonessential amino acids, 1 mM sodium pyruvate and β-mercaptoethanol. The indicated supernatants and lysozyme lysis buffer were added into each well (1:50 v/v). After 24 hr coincubation, cytokines in culture media were detected using commercially available cytokine array kit (R &D systems, Catalog No. ARY006) according to the manufacturer’s instructions. All the experiments were performed under the same conditions to reduce the variabilities. Quantification of each cytokine from two duplicate dots were conducted using ImageJ. The MLN cells were harvested in 2x SDS buffer containing 1x Proteinase Inhibitor cocktail (Roche, Catalog No. 5892791001) and 1x Phosphatase Inhibitor cocktail (Roche, Catalog No. 4906845001), denatured under 95°C for 15 min and subjected to western blot analysis.
Intestinal Crypts Isolation and In Vitro Culture
The procedure for Intestinal crypts isolation and in vitro culture was conducted as previously described (Yu et al., 2014; Yu et al., 2018). Briefly, mouse ileal fragments were harvested, washed in PBS and incubated in PBS containing 5mM EDTA for 40 min. After vigorous vortexing, crypt suspensions were passed through 70μm cell strainer and centrifuged at 200×g for 5 min at 4°C. Then, intestinal crypts were counted, and resuspended in Matrigel (Corning, Catalog No. 354230). After Matrigel polymerization, IntestiCult organoid growth media (StemCell Technologies, Catalog No. 06005) was added to each well After 3-day culture, recombinant mouse IL-13 (R&D systems, Catalog No. 413-ML) was added or not and incubated for 2 more days. Then, intestinal organoids were subjected to fixation by 4% paraformaldehyde followed by immunofluorescence.
RNA Extraction, Reverse transcription and Real-time PCR
Tissue RNA isolation was conducted using RNeasy Mini Kit (Qiagen, Catalog No. 74104) according to the manufacturer’s instructions. Then 1μg of RNA were subjected to reverse transcription using Maxima First Strand cDNA Synthesis Kit for RT-qPCR (ThermoFisher, Catalog No. K1641). Real-time PCR was performed as previously described with indicated primer sets listed in Table. S7.
Bulk RNA-Seq Analysis
Mouse Ileum fragments (3–4 mice/genotype/treatment) were collected and subjected to RNA extraction. RNA samples were submitted for sequencing using BGISEQ500 (BGI-US). Sequenced files were aligned to mm9 genome index using Kallisto (v0.45.0) (Bray et al., 2016) with default settings and 1000 bootstraps. Reads were normalized in DESeq2 (Build 3.9) using default settings in R. RNA-seq data are deposited in GEO (accession # under being processed). For Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) performed on goblet cell gene signatures (Haber et al., 2017), Tuft signature genes (Nadjsombati et al., 2018), Paneth cell signature genes, and ILC2/Th2 cell signature genes (Robinette et al., 2015), pre-ranked files of differentially expressed genes were calculated by the rank metric = -log(p-value)* SIGN(logFC) (Jia et al., 2015). Heatmap of select goblet cell and Tuft cell genes was generated to visualize fpkms of individual wildtype and Lyz1−/− replicates. For GSEA performed on cytoplasmic pattern recognition receptor (PRR) signaling, NOD-like receptor (NLR) signaling, apoptosis, cytokine production in inflammatory response, and response to IL-13, corresponding molecular signature databases were used to determine significance of differential expression between conditions. One thousand permutations were performed for each gene list tested, normalized enrichment scores (NES) and nominal P values are reported for genes signatures. Heatmaps generated by GSEA are on a relative min max scale. Nominal p-value < 0.05 was considered to indicate significant enrichment.
Single cell dissociation and sorting
The distal ileum of adult WT and Lyz1−/− mice (separately housed) were dissected and briefly rinsed in ice-cold PBS (Fisher Scientific, SH30256LS). The tissue was opened longitudinally, rinsed in ice-cold PBS, and further sliced to 1–2 mm pieces. The pieces were rinsed in 30mL of ice-cold PBS by inverting the falcon tube 10–15 times. The pieces were then transferred into fresh 30ml of ice-cold PBS. This step was repeated until the solution remained clear after inverting. Each piece was then transferred to 30 mL crypt isolation buffer, containing 5 mM EDTA (Invitrogen, Catalog No.AM9260G), 2% BSA (Sigma, Catalog No.A3294), and HBSS Ca/Mg-free (Sigma, Catalog No.H9394), and allowed to shake at 37°C for 15 minutes. The tubes were subsequently vigorously shaken to release the epithelial layer from the pieces after which the solution was discarded. This was repeated in another 30ml of crypt isolation buffer. The pieces were then incubated in DMEM/F12 medium (ThermoFisher, 12634–010) for 10 minutes on a petri dish, at room temperature. The solution was discarded, and the pieces were thoroughly minced. 10ml of Hank’s balanced salt solution with calcium and magnesium (Sigma, Catalog No.55037C) containing 100 U/mL Collagenase II (ThermoFisher, Catalog No.17101015), 500 U/mL DNase I (Sigma, Catalog No.DN25), was added to the minced tissue and allowed to shake at 37°C for 30 minutes. When tissues completely digested, the solution was passed through a 70μM cell strainer into a 50mL falcon tube. The tubes were centrifuged at 200g for 10 minutes. The pellets were resuspended in 1mL of FACS solution (2% BSA in PBS). The single-cell suspensions were stained with DAPI and immediately subjected to sorting by BD Biosciences Aria II Flow Cytometer (BD FACSAria II). Single viable lamina propria lymphocytes and mesenchymal cells were gated by forward scatter, side scatter and by negative staining for DAPI.
Droplet-based Single Cell RNA-Seq
Cell number and viability were determined by a Propidium Iodide based fluorescence assay using Moxi GO II System, Orflo Prod#MXG102 (ORFLO Technologies, LLC). Droplet-based single-cell partitioning and single-cell RNA-Seq libraries were generated using the Chromium Single-Cell 3′ Reagent v3 Kit (10X Genomics, Pleasanton, CA) on the 10X Chromium Controller as per the manufacturer’s protocol. Live cells in single cell suspension with 95% viability were mixed with Gel beads, RT reagents and Partitioning Oil into a Single-Cell 3′ Chip and loaded onto 10X Chromium Controller for GEM generation. Briefly, the protocol includes RT, cleanup, cDNA amplification, fragmentation, end repair & A-tail prep, adapter ligation and incorporation of sample indices into finished libraries, which are compatible with Illumina next-generation sequencing platforms. Sample quantification and quality control were determined using Qubit Fluorometer (Invitrogen, Life Technologies) and TapeStation (Agilent Technologies, Santa Clara CA) respectively. cDNA libraries were sequenced on Illumina NovaSeq 6000 sequencer (Illumina, San Diego, CA) with a configuration of 28/8/0/91-bp for cell barcode, sample barcode and mRNA reads respectively as recommendation by 10X Genomics. The Chromium Single Cell Software is used to analyze and visualize single cell 3’ RNA-seq data produced by the 10X Chromium Platform. The 10X chromium software package includes Cell Ranger Pipelines and Loupe Cell Browser. Cell Ranger pipelines use raw 10X single cell sequencing data from an Illumina sequencer and perform demultiplexing, unique barcode processing, and single cell 3’ gene counting. The WT and Lyz1−/− count matrix files generated by Cell Ranger were imported to Partek flow software for secondary data analysis. The single cell analysis for this paper was generated using Partek® Flow®, version 9.0, build 9.0.20.0202. Copyright ©; 2020 Partek Inc., St. Louis, MO, USA. For each cell, quality control metrics such as number of read counts, number of detected genes, and percentage of mitochondrial reads were calculated and used to filter out low quality cells. The data was then log2+1 normalized based on counts per million and the genes that were not expressed in at least 99% of the cells were excluded from the data set. For figures 3A–C and 3E–J, the WT and Lyz1−/− datasets were analyzed separately. For figures 3K– L, and Suppl. Fig 3A–D, the data was first scaled to remove batch effect, then the merged dataset was used for further analyses. Principal components (PCs) were calculated for the samples and the scree plot was used to determine the number of PCs to be used. Based on the PC value, cells were divided into clusters in an unsupervised fashion. The ANOVA test was used to identify differentially expressed genes (DEGs) for each cluster by comparing genes in all the clusters, filtering genes that were increased by 1.5-fold, and then sorting them by ascending p-value. The clusters were then visualized using t-Distributed Stochastic Neighbor Embedding (t-SNE). Based on previous literature, a list of the genes indicated in figure 3B was curated and used to identify ILC2 populations to study the differences in type 2 response between the WT and Lyz1−/− mice. The ILC2 population in WT and Lyz1−/− was lasso selected and classified. DEGs were identified for the sub clusters in ILC2 population for each genotype and the signature DEG genes (FDR set up ≤1e-8, fold change>2) were visualized by generating a heat map. Then, trajectory analysis using Monocle 2 was performed on these ILC2 cells. DEGs were identified for each of the states by running gene specific analysis in Partek.
nCounter Analysis of Gene Expression
Colonic mucosal RNA and DNA was isolated using AllPrep DNA/RNA Mini Kit (Qiagen, Catalog No. 80204) using the protocol previously described (Harrison et al., 2018). The protocol was slightly modified to include a bead-beating step. Expression of inflammation-related genes was assessed using nCounter Inflammation Panel (Mouse v2) codecs, nCounter Master Kit (NanoString Technologies, Seattle, WA), and the NanoString nCounter® Analysis System at the University of Arizona Genetics Core facility. Data were analyzed using nSolver Analysis software v4.0 with the nCounter Advanced Analysis module v. 2.0.115 (NanoString Technologies).
Quantification and Statistical Analyses
All graphic data and statistical analyses were conducted using Graphpad Prism (https://www.graphpad.com) and Microsoft Excel Office 365. The data were reported as mean ± SEM in bar graphs. Statistical comparisons between two groups were undertaken using unpaired two-tailed Student’s t test. p-value was indicated in corresponding graphs. The immunofluorescent data were reported from 3 to 5 sections of each animal; 2 to 16 mice were used in each experiment unless stated differently. The significant level of enrichment score in the GSEA was computed by an empirical phenotype-based permutation test conducted to reserve the complex correlation structure of gene expression patterns. The phenotype labels were permuted and then the enrichment score of the gene set were recalculated for the permuted data, thus resulting in a null distribution for the enrichment score. The nominal p value of the resulting enrichment score was empirically computed relative to this null distribution as previously described (Subramanian et al., 2005).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-IL-13 | R &D Systems | Cat#MAB413; RRID:AB_2124171 |
| Rat IgG2a isotype control | R&D Systems | Cat#MAB006; RRID:AB_357349 |
| Rabbit anti-Lysozyme | BioGenex | Cat#AR024-10R |
| Rabbit anti-Mucin 2 (Muc2) | Santa Cruz Biotech | Cat#sc-15334; RRID:AB_2146667 |
| Rabbit anti-DCLK1 | Abcam | Cat#ab37994; RRID:AB_873538 |
| Goat anti-MMP7 | Santa Cruz Biotech | Cat# sc-8832; RRID:AB_649337 |
| Mouse anti-E-Cadherin (E-Cad) | BD Biosciences | Cat#610404; RRID:AB_397786 |
| Mouse anti-Amphiregulin (Areg) | Santa Cruz Biotech | Cat# sc-74501; RRID:AB_1118939 |
| Rabbit anti-Arginase 1 | Cell Signaling | Cat#93668; RRID:AB_2800207 |
| Rabbit anti-phospho-Stat3 (pStat3) | Cell Signaling | Cat#9145; RRID:AB_2491009 |
| Rabbit anti-phospho-Stat6 (pStat6) | Thermo Fisher Scientific | Cat#700247; RRID:AB_2532305 |
| Donkey anti-mouse IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A21202; RRID:AB_141607 |
| Donkey anti-rabbit IgG, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10042; RRID:AB_2534017 |
| ImmPRESS HRP anti-rabbit IgG polymer | Vector Labs | Cat# MP-7401; RRID:AB_2336529 |
| ImmPRESS HRP anti-mouse IgG polymer | Vector Labs | Cat#MP-7402; RRID:AB_2336528 |
| Bacterial and Virus Strains | ||
| Akkermansia muciniphila | DSMZ | DSM-22959 |
| Bacteroides uniformis | DSMZ | DSM-6597 |
| Dorea formicigenerans | DSMZ | DSM-3992 |
| Ruminococcus gnavus | DSMZ, ATCC | FRE1, ATCC29149 |
| Lactobacillus rhamnosus GG | ATCC | ATCC53103 |
| Dorea longicatena | DSMZ | DSM-13814 |
| Biological Samples | ||
| UC colon biopsy samples | Provided by Dr. Lanjing Zhang, Princeton University Hospital | N/A |
| CD colon and ileum biopsy samples | Provided by Dr. Lanjing Zhang, Princeton University Hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant IL-13 | R&D Systems | Cat#413-ML |
| Chicken egg white lysozyme | Sigma | Cat#L6876 |
| Dextran sulfate sodium (DSS, Colitis grade) | MP Biologics | SKU 0216011080 |
| Sodium cacodylate buffer, 0.2M, ph7.4 | Electron Microscopy Sciences | Cat# 11650 |
| 8% Glutaraldehyde (EM grade) | Electron Microscopy Sciences | Cat#16019 |
| 16% Paraformaldehyde (EM grade) | Electron Microscopy Sciences | Cat#15700 |
| 4% Osmium Tetroxide (OsO4) | Electron Microscopy Sciences | Cat#19150 |
| Proteinase Inhibitor cocktail | Roche | Cat#5892791001 |
| Phosphatase Inhibitor cocktail | Roche | Cat#4906845001 |
| Matrigel (growth factor reduced) | Corning | Cat#354230 |
| IntestiCult organoid culture medium | StemCell Technologies | Cat#06005 |
| Vector Nuclear Fast Red | Vector Labs | Cat# H-3403 |
| EMBed-812 kit | Electron Microscopy Sciences | Cat#14120 |
| Prolong Gold Antifade Mountant | Thermo Fisher Scientific | Cat# P36930 |
| Hematoxylin QS | Vector Labs | Cat# H-3404 |
| Cytoseal 60 | Thermo Fisher Scientific | Cat#8310-4 |
| 4’,6-Diamidino-2-Phenylindole (DAPI) | Thermo Fisher Scientific | Cat#D1306 |
| Critical Commercial Assays | ||
| Lysozyme activity assay kit | Abcam | Cat#ab211113 |
| QIAamp DNA Stool Mini Kit | Qiagen | Cat#51604 |
| PureLink Microbiome DNA purification kit | Thermo Fisher Scientific | Cat# A29789 |
| Ion 16s Metagenomics Kit | Thermo Fisher Scientific | Cat#A26216 |
| Cytokine array kit | R&D Systems | Cat# ARY006 |
| Maxima First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | Cat# K1641 |
| AllPrep DNA/RNA Mini Kit | Qiagen | Cat#80204 |
| Deposited Data | ||
| Single-cell RNA-seq | This paper | GEO: GSE151152 |
| Bulk-cell RNA-seq | This paper | GEO: GSE151151 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Lyz1H2BmCherry-IRES-CreERT2 | Yu et al., 2018 | N/A |
| Mouse: Villin-Lyz1TG transgenic | This study | N/A |
| Mouse: B6.129S2(C)-Stat6tm1Gru/J (Stat6KO) | Kaplan et al., 1996 | Jax #005977 |
| Mouse: BALB/c-Il4ratm1Sz/J (Il4RαKO) | Noben-Trauth et al., 1997 | Jax #003514 |
| Oligonucleotides | ||
| Primers for Dorea formicigenerans, see Table S7 | Kurakawa et al., 2015 | N/A |
| Primers for Ruminococcus gnavus, see Table S7 | Png et al.,2010 | N/A |
| Primers for Bacterial 16s rRNA, see Table S7 | Randall et al., 2010 | N/A |
| Primers for Tritrichomonas muris, see Table S7 | Howitt et al., 2016 | N/A |
| Primers for Mouse Lyz1, see Table S7 | This paper | N/A |
| Primers for Mouse Mucin 2 (Muc2), see Table S7 | This paper | N/A |
| Primers for Mouse Klf4, see Table S7 | This paper | N/A |
| Primers for Mouse Spdef, see Table S7 | This paper | N/A |
| Primers for Mouse Il4, see Table S7 | This paper | N/A |
| Primers for Mouse Il13, see Table S7 | This paper | N/A |
| Primers for Mouse Il17a, see Table S7 | This paper | N/A |
| Primers for Mouse Il17f, see Table S7 | This paper | N/A |
| Primers for Mouse Il25, see Table S7 | This paper | N/A |
| Primers for Mouse Il33, see Table S7 | This paper | N/A |
| Primers for Mouse Tslp, see Table S7 | This paper | N/A |
| Primers for Mouse Tnfa, see Table S7 | This paper | N/A |
| Primers for Mouse Pou2f3, see Table S7 | This paper | N/A |
| Primers for Mouse Gata3, see Table S7 | This paper | N/A |
| Primers for Mouse Tbx21 (Tbx), see Table S7 | This paper | N/A |
| Primers for Mouse Rorc (Rorγt), see Table S7 | This paper | N/A |
| Primers for Mouse Foxp3, see Table S7 | This paper | N/A |
| Primers for Mouse Tgfb1, see Table S7 | This paper | N/A |
| Primers for Mouse Amphiregulin (Areg), see Table S7 | This paper | N/A |
| Primers for Mouse Hprt, see Table S7 | This paper | N/A |
| Software and Algorithms | ||
| Metastats | White et al., 2009 | http://metastats.cbcb.umd.edu/detection.html |
| QIIME (v.1.9.1) | Caporaso et al., 2010 | https://qiime.org |
| LDA Effect size (LEfSe) analysis tool | Afgan et al., 2018; Segeta et al., 2011 | https://huttenhower.sph.harvard.edu/galaxy/ |
| Kallisto | Bray et al., 2016 | https://pachterlab.github.io/kallisto/ |
| nSolver Analysis software v4.0 with the nCounter Advanced Analysis module (v.2.0.115) | NanoString Technologies | https://www.nanostring.com/products/analysis-software/nsolver |
| GSEA | Subramanian et al., 2005; Mootha et al., 2003 | http://software.broadinstitute.org/gsea/index.jsp |
| GO Enrichment Analysis | Ashburner et al., 2000; Mi et al., 2019 | http://geneontology.org/ |
| Origin Pro 2019 | OriginLab | https://www.originlab.com/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Graphpad Prism (v.7.0) | Graphpad | https://www.graphpad.com/ |
| Cell Ranger Single Cell Software Suite (v3.0.1) | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Partek Flow Software (v.9.0.20.0202) | Partek | In house license |
| Seurat (R toolkit) | Butler et al., 2018 | https://satijalab.org/seurat v.3.1.0 |
Highlights.
Abnormal production of Paneth cell lysozyme is a feature of human IBD
Intestinal luminal lysozyme determines the abundance of mucolytic commensal bacteria
Processing lysozyme-sensitive bacterial cell wall drives colitis in Lyz1+/+ hosts
Lyz1−/− mice have reduced NLR signaling but an anti-colitogenic type 2 immune response
ACKNOWLEDGEMENTS
The authors are indebted to Ivaylo I. Ivanov (Columbia University) and Mi-Na Kweon (University of Ulsan College of Medicine) for helpful discussion. We thank Peter Romanienko and Ghassan Yehia (Rutgers Cancer Institute of New Jersey Genome Editing Shared Resource P30CA072720-5922) for deriving Lyz1−/− and Villin-Lyz1 mice, Luke Fritzk (Digital Imaging and Histology Core) for histology, Sukhwinder Singh and Tammy Mui- Galenkamp (Flow Cytometry and Immunology Core) for FACS analysis, Seema Husain (The Genomics Center) for scRNA sequencing, and the excellent technical support from PrimBio Research Institute (Exton, PA). This work was supported by NIH grants (DK102934, DK119198, AT010243, CA178599), NSF/BIO/IDBR grants (1353890, 1952823), ACS Scholar Award (RSG-15-060-01-TBE), Rutgers IMRT award to N.G.; Rutgers Chancellor’s SEED grant (to E.M.B.); NIH grant R01CA190558 (to M.P.V.); R01AI134040 (to G.S.Y.); 5R01DK109711, PANDA Endowment in Autoimmune Diseases (to P.R.K.); NSF grants (IOS-1456673, IOS-1754783 to R.P.F.); NIH F31 DK121428 (to S. B.); NJCCR fellowship DCHS19PPC038 (to J. F.); and a CCFA Career Development Award 406794 (to S.Y.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46, W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian I, and Gao N (2017). From sensing to shaping microbiota: insights into the role of NOD2 in intestinal homeostasis and progression of Crohn’s disease. Am J Physiol Gastrointest Liver Physiol 313, G7–G13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, and Hooper LV (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brook I (1989). Inoculum effect. Rev Infect Dis 11, 361–368. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. (2008). A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, and Holmes SP (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11, 2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield RE, and Liu AK (1965). THE DISULFIDE BONDS OF EGG WHITE LYSOZYME (MURAMIDASE). J Biol Chem 240, 1997–2002. [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Aitken JD, Malleshappa M, and Vijay-Kumar M (2014). Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol 104, Unit 15 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Yang W, Huang X, Cao AT, Bilotta AJ, Xiao Y, Sun M, Chen L, Ma C, Liu X, et al. (2018). Neutrophils Promote Amphiregulin Production in Intestinal Epithelial Cells through TGF-beta and Contribute to Intestinal Homeostasis. Journal of immunology 201, 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HH, Chou HC, Tung YL, Chiang BL, Liao CC, Liu HH, and Ni YH (2018). Intestinal Dysbiosis Featuring Abundance of Ruminococcus gnavus Associates With Allergic Diseases in Infants. Gastroenterology 154, 154–167. [DOI] [PubMed] [Google Scholar]
- Dabard J, Bridonneau C, Phillipe C, Anglade P, Molle D, Nardi M, Ladire M, Girardin H, Marcille F, Gomez A, et al. (2001). Ruminococcin A, a new lantibiotic produced by a Ruminococcus gnavus strain isolated from human feces. Appl Environ Microbiol 67, 4111–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio M, Vernia F, Ciccone A, Frieri G, and Latella G (2017). Surrogate Fecal Biomarkers in Inflammatory Bowel Disease: Rivals or Complementary Tools of Fecal Calprotectin? Inflammatory bowel diseases 24, 78–92. [DOI] [PubMed] [Google Scholar]
- Doran E, Cai F, Holweg CTJ, Wong K, Brumm J, and Arron JR (2017). Interleukin-13 in Asthma and Other Eosinophilic Disorders. Front Med (Lausanne) 4, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison RT 3rd, and Giehl TJ (1991). Killing of gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest 88, 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Gagliardi J, Bouhan D, Spollen WG, Givan SA, and Franklin CL (2018). The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci Rep 8, 4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren A, Hammarstrom S, Danielsson A, and Hammarstrom ML (2003). Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin Exp Immunol 131, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T, Gabayan V, Liao HI, Liu L, Oren A, Graf T, and Cole AM (2003). Increased inflammation in lysozyme M-deficient mice in response to Micrococcus luteus and its peptidoglycan. Blood 101, 2388–2392. [DOI] [PubMed] [Google Scholar]
- Gao N, and Kaestner KH (2010). Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev 24, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause WC, Wynn TA, and Allen JE (2013). Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat Rev Immunol 13, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, et al. (2016). Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girnius S, Skinner M, Spencer B, Prokaeva T, Bartholomew C, O’Hara C, Seldin DC, and Connors LH (2012). A new lysozyme tyr54asn mutation causing amyloidosis in a family of Swedish ancestry with gastrointestinal symptoms. Amyloid 19, 182–185. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Stange DE, Kujala P, Begthel H, van den Born M, Korving J, Peters PJ, and Clevers H (2009). The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology 137, 1333–1345 e1331–1333. [DOI] [PubMed] [Google Scholar]
- Gu L, Tseng S, Horner RM, Tam C, Loda M, and Rollins BJ (2000). Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature 404, 407–411. [DOI] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley W Ggplot2 : elegrant graphics for data analysis, Second edition. edn.
- Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. (2017). A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 9, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CA, Laubitz D, Ohland CL, Midura-Kiela MT, Patil K, Besselsen DG, Jamwal DR, Jobin C, Ghishan FK, and Kiela PR (2018). Microbial dysbiosis associated with impaired intestinal Na(+)/H(+) exchange accelerates and exacerbates colitis in ex-germ free mice. Mucosal Immunol 11, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert DR, Douglas B, and Zullo K (2019). Group 2 Innate Lymphoid Cells (ILC2): Type 2 Immunity and Helminth Immunity. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, et al. (2016). Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaux F, Liu T, McGarry B, Ullenbruch M, and Phan SH (2003). Dual roles of IL-4 in lung injury and fibrosis. J Immunol 170, 2083–2092. [DOI] [PubMed] [Google Scholar]
- Hungate RE (1950). The anaerobic mesophilic cellulolytic bacteria. Bacteriological reviews 14, 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim HR (1998). On the novel catalytically-independent antimicrobial function of hen egg-white lysozyme: a conformation-dependent activity. Nahrung 42, 187–193. [DOI] [PubMed] [Google Scholar]
- Ibrahim HR, Matsuzaki T, and Aoki T (2001a). Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS letters 506, 27–32. [DOI] [PubMed] [Google Scholar]
- Ibrahim HR, Thomas U, and Pellegrini A (2001b). A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J Biol Chem 276, 43767–43774. [DOI] [PubMed] [Google Scholar]
- Jia Z, Zhang X, Guan N, Bo X, Barnes MR, and Luo Z (2015). Gene Ranking of RNASeq Data via Discriminant Non-Negative Matrix Factorization. PLoS One 10, e0137782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean E, Ebbo M, Valleix S, Benarous L, Heyries L, Grados A, Bernit E, Grateau G, Papo T, Granel B, et al. (2014). A new family with hereditary lysozyme amyloidosis with gastritis and inflammatory bowel disease as prevailing symptoms. BMC Gastroenterol 14, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, and Vermeire S (2011). Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabata H, Moro K, and Koyasu S (2018). The group 2 innate lymphoid cell (ILC2) regulatory network and its underlying mechanisms. Immunological reviews 286, 37–52. [DOI] [PubMed] [Google Scholar]
- Kalinski P (2012). Regulation of immune responses by prostaglandin E2. J Immunol 188, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH, Schindler U, Smiley ST, and Grusby MJ (1996). Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 4, 313–319. [DOI] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, and Kaestner KH (2002). The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129, 2619–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass HJ, and Neale G (1978). Serum and faecal lysozyme in inflammatory bowel disease. Gut 19, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ogata K, Matsuda K, Tsuji H, Kubota H, Takada T, Kado Y, Asahara T, Takahashi T, and Nomoto K (2015). Diversity of Intestinal Clostridium coccoides Group in the Japanese Population, as Demonstrated by Reverse Transcription-Quantitative PCR. PLoS One 10, e0126226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible NJ, and Germaine GR (1985). Bactericidal activity of human lysozyme, muramidase-inactive lysozyme, and cationic polypeptides against Streptococcus sanguis and Streptococcus faecalis: inhibition by chitin oligosaccharides. Infect Immun 48, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent P, Jolivel V, Manicki P, Chiu L, Contin-Bordes C, Truchetet ME, and Pradeu T (2017). Immune-Mediated Repair: A Matter of Plasticity. Front Immunol 8, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W, Ren W, Ohmoto M, Urban JF Jr., Matsumoto I, Margolskee RF, and Jiang P (2018). Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A 115, 5552–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TC, Naito T, Liu Z, VanDussen KL, Haritunians T, Li D, Endo K, Kawai Y, Nagasaki M, Kinouchi Y, et al. (2017). LRRK2 but not ATG16L1 is associated with Paneth cell defect in Japanese Crohn’s disease patients. JCI Insight 2, e91917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL (2002). Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 277, 33275–33283. [DOI] [PubMed] [Google Scholar]
- Markart P, Faust N, Graf T, Na CL, Weaver TE, and Akinbi HT (2004a). Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem J 380, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markart P, Korfhagen TR, Weaver TE, and Akinbi HT (2004b). Mouse lysozyme M is important in pulmonary host defense against Klebsiella pneumoniae infection. Am J Respir Crit Care Med 169, 454–458. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, and Fodor AA (2013). Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J 7, 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Gellhorn A, and et al. (1947). Lysozyme in chronic ulcerative colitis. Proc Soc Exp Biol Med 65, 221. [DOI] [PubMed] [Google Scholar]
- Meyer K, Gellhorn A, and et al. (1948). Lysozyme activity in ulcerative alimentary disease; lysozyme activity in chronic ulcerative colitis. Am J Med 5, 496–502. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, et al. (2018). Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49, 33–41 e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth N, Shultz LD, Brombacher F, Urban JF Jr., Gu H, and Paul WE (1997). An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci U S A 94, 10838–10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, and Nakaya R (1990). A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 98, 694–702. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, et al. (2019). The vegan package. Community ecology package, 25–4. [Google Scholar]
- Pepys MB, Hawkins PN, Booth DR, Vigushin DM, Tennent GA, Soutar AK, Totty N, Nguyen O, Blake CC, Terry CJ, et al. (1993). Human lysozyme gene mutations cause hereditary systemic amyloidosis. Nature 362, 553–557. [DOI] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, and Wynn TA (2009). Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog 5, e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, and Florin TH (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105, 2420–2428. [DOI] [PubMed] [Google Scholar]
- Proust B, Nahori MA, Ruffie C, Lefort J, and Vargaftig BB (2003). Persistence of bronchopulmonary hyper-reactivity and eosinophilic lung inflammation after anti-IL-5 or -IL-13 treatment in allergic BALB/c and IL-4Ralpha knockout mice. Clin Exp Allergy 33, 119–131. [DOI] [PubMed] [Google Scholar]
- Pulendran B, and Artis D (2012). New paradigms in type 2 immunity. Science 337, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glockner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rael EL, and Lockey RF (2011). Interleukin-13 signaling and its role in asthma. World Allergy Organ J 4, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland SA, and Criss AK (2017). From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog 13, e1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, and Medzhitov R (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. [DOI] [PubMed] [Google Scholar]
- Randall L, Lemma F, Rodgers J, Vidal A, and Clifton-Hadley F (2010). Development and evaluation of internal amplification controls for use in a real-time duplex PCR assay for detection of Campylobacter coli and Campylobacter jejuni. J Med Microbiol 59, 172–178. [DOI] [PubMed] [Google Scholar]
- Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, and Lee A (2005). Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 55, 1199–1204. [DOI] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, and Immunological Genome C (2015). Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 16, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB, and Mazmanian SK (2012). Intestinal Microbes in Inflammatory Bowel Diseases. Am J Gastroenterol Suppl 1, 15–21. [Google Scholar]
- Schneider C, O’Leary CE, von Moltke J, Liang HE, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, and Locksley RM (2018). A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 174, 271–284 e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada J, Moon SK, Lee HY, Takeshita T, Pan H, Woo JI, Gellibolian R, Yamanaka N, and Lim DJ (2008). Lysozyme M deficiency leads to an increased susceptibility to Streptococcus pneumoniae-induced otitis media. BMC Infect Dis 8, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, et al. (2009). Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet 41, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Balasubramanian I, Zhang L, and Gao N (2020). Metaplastic Paneth Cells in Extra-Intestinal Mucosal Niche Indicate a Link to Microbiome and Inflammation. Front Physiol 11, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood TB, Giacomin PR, Loukas A, Mulvenna JP, Clark RJ, and Miles JJ (2017). Helminth Immunomodulation in Autoimmune Disease. Front Immunol 8, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Tremblay S, Montero M, Vogl W, Xia L, Jacobson K, Menendez A, and Vallance BA (2018). The Muc2 mucin coats murine Paneth cell granules and facilitates their content release and dispersion. Am J Physiol Gastrointest Liver Physiol 315, G195–G205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh D, and Juge N (2015a). Mucin glycan foraging in the human gut microbiome. Front Genet 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le Gall G, de Vos WM, Taylor GL, and Juge N (2015b). Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nature communications 6, 7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Smith RP, Srimani JK, Riccione KA, Prasada S, Kuehn M, and You L (2012). The inoculum effect and band-pass bacterial response to periodic antibiotic treatment. Mol Syst Biol 8, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Saito H, Kusumi T, Fukuda S, Shimoyama T, Sasaki Y, Suto K, Munakata A, and Kudo H (2001). Spatial distribution and histogenesis of colorectal Paneth cell metaplasia in idiopathic inflammatory bowel disease. J Gastroenterol Hepatol 16, 1353–1359. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Liu TC, Li D, Towfic F, Modiano N, Winter R, Haritunians T, Taylor KD, Dhall D, Targan SR, et al. (2014). Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 146, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, and Locksley RM (2016). Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour RE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, et al. (2017). The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Nagarajan N, and Pop M (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5, e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Tjota MY, Clay BS, Vander Lugt B, Bandukwala HS, Hrusch CL, Decker DC, Blaine KM, Fixsen BR, Singh H, et al. (2013). Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun 4, 2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, and Engstrand L (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854 e1841. [DOI] [PubMed] [Google Scholar]
- Yu S, Nie Y, Knowles B, Sakamori R, Stypulkowski E, Patel C, Das S, Douard V, Ferraris RP, Bonder EM, et al. (2014). TLR sorting by Rab11 endosomes maintains intestinal epithelial-microbial homeostasis. The EMBO journal 33, 1882–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Tong K, Zhao Y, Balasubramanian I, Yap GS, Ferraris RP, Bonder EM, Verzi MP, and Gao N (2018). Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell 23, 46–59 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss DM, Yang L, Shah PR, Kobie JJ, Urban JF, and Mosmann TR (2006). Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science 314, 1746. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Pan Y, Yan R, Zeng B, Wang H, Zhang X, Li W, Wei H, and Liu Z (2015). Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat Immunol 16, 918–926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data generated in this study are deposited in Gene Expression Omnibus(GEO) with accession number GSE151152. The bulk RNA-seq data generated in this study are deposited in GEO with accession number GSE151151.