Abstract
BACKGROUND
Mechanistic endophenotypes can inform process models of psychopathology and aid interpretation of genetic risk factors. Smaller total brain and subcortical volumes are associated with ADHD and provide clues to its development. This study evaluates whether common genetic risk for ADHD is associated with total brain volume and hypothesized subcortical structures in children.
METHODS
Children 7–15 years old were recruited for a case-control study (N=312, N=199 ADHD). Children were assessed with a multi-informant, best-estimate diagnostic procedure and motion-corrected MRI measured brain volumes. Polygenic scores were computed based on discovery data from the Psychiatric Genomics Consortium (N=19,099 ADHD, N=34,194 controls) and the ENIGMA+CHARGE consortium (N=26,577).
RESULTS
ADHD was associated with smaller total brain volume, and altered volumes of caudate, cerebellum, putamen, and thalamus after adjustment for total brain volume; however, effects were larger and statistically reliable only in boys. Total brain volume was associated with an ADHD polygenic score (β=−0.147 (−0.27, −0.03)), and mediated a small proportion of the effect of polygenic risk on ADHD diagnosis, (average ACME=0.0087, p=0.012). This finding was stronger in boys (average ACME=0.019, p=0.008). In addition, we confirm genetic variation associated with whole brain volume, via an intracranial volume polygenic score.
CONCLUSION
Common genetic risk for ADHD is not expressed primarily as developmental alterations in subcortical brain volumes, but appears to alter brain development in other ways, as evidenced by total brain volume differences. This is among the first demonstrations of this effect using molecular genetic data. Potential sex differences in these effects warrant further examination.
INTRODUCTION
Attention Deficit Hyperactivity Disorder (ADHD) is associated with alterations in brain development, including smaller total brain and subcortical structure volumes (Hoogman et al. 2017; Shaw et al. 2018). However, variability in findings is substantial (Valera et al. 2007; Frodl & Skokauskas 2012). Included in this uncertainty are the effects of gender due to a preponderance of male samples (Frodl & Skokauskas 2012), of medication due to often limited information on medication history (Frodl & Skokauskas 2012), and potential variation with development (Shaw et al. 2018). Nonetheless, smaller total brain volume appears to be a feature of ADHD, especially at younger ages (Castellanos et al. 2002; Greven et al. 2015). Crucially for etiological models, it remains unclear how structural changes in ADHD relate to the disorder’s genetic liability. Moving that question forward is the central aim of the current study.
The genetic underpinnings of ADHD (and brain development) are complex, involving both common and rare genetic variants, as well as likely epigenetic effects (Faraone & Larsson 2019). While both total brain and subcortical volumes appear to have substantial heritability (Blokland et al. 2012), whether the genetic factors influencing ADHD susceptibility also play a role in brain development related to ADHD is largely unknown.
Here we investigate the effects of common DNA variants using a polygenic risk score (PGS), an approach that has proven fruitful in studies of ADHD (Riglin et al. 2016) and other disorders (Whalley et al. 2015; Wang et al. 2017). Recent large discovery datasets have enabled effective confirmation tests of pathophysiological models using neuro-biological features of ADHD, such as executive functioning (Nigg et al. 2018).
Total brain volume is a potentially important indicator of neurodevelopmental processes and provides the foundation for comparative brain development in childhood pathologies. Subcortical structures are potentially important for their specific functional associations, as well as clues to developmental timing. Differential volumetric loss may reflect regional differences in gene expression (Hess et al. 2017). Five meta-analyses (Valera et al. 2007; Ellison-Wright et al. 2008; Nakao et al. 2011; Frodl & Skokauskas 2012; Norman et al. 2016) and one major mega-analysis and meta-analysis with over 1000 cases and controls (Hoogman et al. 2017) have consolidated what is known about total brain and subcortical volumes in ADHD. Norman et al. reported that ADHD was associated with smaller volume in the putamen/globus pallidus and the right caudate nucleus (Norman et al. 2016). The ENIGMA consortium mega-analysis found that ADHD children under age 15 years had reliably smaller total brain (d=0.14), putamen, caudate, amygdala, and nucleus accumbens volumes—again with small effect sizes (d=0.13–0.19) (Hoogman et al. 2017). They reported that sex differences and medication effects were not reliable. Although the ENIGMA study did not examine cerebellum, a large NIH study reported smaller cerebellar volume in ADHD (Wyciszkiewicz et al. 2017), therefore we also examined cerebellum.
Yet for all of these findings, their relation to polygenic risk for ADHD is almost unstudied. One recent study examined ADHD polygenic risk and subcortical brain volumes in children with and without traumatic brain injury, but did not examine the correlation of polygenic risk with subcortical brain volume itself (Stojanovski et al. 2019). Another recent study examined the relationship between polygenic scores for several psychiatric disorders and brain volumes in a population cohort, and found caudate volume associated with ADHD polygenic risk (Alemany et al. 2019). However, ours is the first report to examine the subcortical structures proposed in Hoogman et al. (2017) for ADHD, and total brain volume, in relation to ADHD polygenic risk in a large, well-characterized case-control study.
The small effect sizes seen in the ENIGMA meta-analysis may be due to heterogeneity in the disorder (Fair et al. 2012; Costa Dias et al. 2015) or heterogeneity in study designs (e.g. age, scanners, etc). The studies pooled in the meta-analyses and mega-analysis mostly have relatively small sample sizes. There remains a need for more study of girls, and it is notable that none of the meta-analyses considered motion artifact as a potential source of variation (Savalia et al. 2017), which we account for in the current study. However, the major gap we note, and our primary focus, is the need to clarify how common genetic risk for ADHD relates to putative ADHD brain correlates.
The dearth of studies that can consider sex-specific effects is important. Sex differences may be crucial for understanding mediators of genetic influences on ADHD. ADHD is more common in males for unclear reasons (Martel 2013) and distinct features of development in girls are important, yet often overlooked (Hinshaw 2018). Brain development differs normatively in boys and girls due in part to hormonal influences (Martel 2013; Király et al. 2016), suggesting that developmental vulnerabilities that differ by sex may be related to ADHD (Wang et al. 2018). Yet, common genetic liability appears to be similar in boys and girls with ADHD as measured by a polygenic score, (Martin et al. 2018). Taken together, these findings suggest that genetic risk for ADHD may operate via different mechanisms in boys and girls—exemplifying etiological heterogeneity in ADHD. Accordingly, we planned to evaluate sex-by-diagnosis interactions carefully.
Our primary aim was to discover whether total brain volume or any major subcortical volume is related to ADHD genetic risk, while secondarily considering sex effects and using updated motion correction methodology.
METHODS
Participants
Participants were 501 children with reliable MRI data, of whom 312 were unrelated and of northern European ancestry. All reported results are from analyses conducted on this genetically homogeneous subset of 312 children. The children’s average age was 10.2 years. ADHD was deliberately oversampled to ensure adequate clinical variation to detect genetic signal and to enable us to examine ADHD heterogeneity. To preserve the representativeness of the sample, we did not oversample for sex or other demographics. Thus, we expected groups to differ on sex ratio and possibly on socioeconomic standing.
Recruitment and Diagnostic Assignment
Human subjects and ethics approvals were obtained from the local University Institutional Review Board. A parent/legal guardian provided written informed consent, and children provided written assent. After screening, a clinical evaluation was conducted using standardized, well-normed rating scales from parent and teacher, parent semi-structured clinical interview, child intellectual testing, and clinical observation. Best estimate research diagnoses and final eligibility were established by a team of two experienced clinicians (a child psychiatrist and a child psychologist) who independently arrived at the diagnosis. See the Supplemental Materials for further details and exclusion criteria. The flow chart for participation eligibility is depicted in Supplemental Figure S1.
MRI acquisition and processing
Details of the MRI acquisition are in the Supplemental Materials. All data were processed following slightly modified pipelines developed by the Human Connectome Project (Mills et al. 2017; Miranda-Dominguez et al. 2017). These pipelines require the use of FSL (Jenkinson et al. 2012) and FreeSurfer (Fischl 2012). Gain field distortion corrected T1-weighted volumes were first aligned to the Montreal Neurological Institute (MNI) AC-PC axis and then non-linearly normalized to the MNI atlas. Later, the T1-weighted volume was re-registered to the MNI template (Fonov et al. 2011) using boundary-based registration (Greve & Fischl 2009) and segmented using the recon-all procedure in FreeSurfer.
The images went through a manual QC protocol developed by the Developmental Cognition and Neuroimaging Lab at Oregon Health & Science University. Post-FreeSurfer models were visually inspected, using the BrainSprite Viewer (https://github.com/surchs/brainsprite), to determine if the processing pipeline would accurately extract volumetric measurements. MRIs were included only if: 1) the T1 was properly aligned with the MNI registration atlas, 2) delineation between white and grey matter was achieved with minimal error, 3) blurriness and artifacts from movement did not distort segmentation, and 4) there was no significant warping of the T1 image.
We addressed the issue of motion during MRI acquisition in two ways. First, subjects whose scans did not pass the manual QC procedure were removed. Second, an estimate of the amount of motion during the scan (average framewise displacement) was included as a covariate in the regression models.
Motion estimation
Blood oxygen level-dependent (BOLD) data acquired after the structural scans was utilized as a best estimate of motion, based on a recently published procedure (Savalia et al. 2017). Because participants tend to move at similar rates throughout a run (Dosenbach et al. 2017), motion during the BOLD scans can be utilized to estimate motion during structural scans. Details of the motion estimation procedure are in the Supplemental Materials.
Polygenic score computation
The ADHD polygenic score was based on results from a genome-wide association study (GWAS) meta-analysis of subjects of European-ancestry (19,099 ADHD cases, 34,194 controls) conducted by the Psychiatric Genomics Consortium (Demontis et al. 2019), and has been described previously (Nigg et al. 2018). Polygenic scores for brain volume measures were based on a GWAS of 30,717 subjects of European-ancestry conducted by the ENIGMA Consortium (Hibar et al. 2015). An intracranial volume polygenic score was also created from the combined ENIGMA+CHARGE consortium data (Adams et al. 2016).
The polygenic scores represent the cumulative effect of all trait-associated (GWAS p<0.5) single nucleotide polymorphisms (SNPs) across the genome. The score for each individual is a weighted-average of all their trait-associated alleles, where the weight for each allele is the effect size (β or log of odds ratio) of the SNP association. Details of how the polygenic scores were constructed, as well as genotype processing and QC, are available in the Supplemental Materials.
Data analysis
Regression models with total brain volume as the outcome were adjusted for age, sex, and average framewise displacement (FD). Regression models with subcortical volumes (nucleus accumbens, amygdala, caudate, cerebellum, hippocampus, pallidum, putamen, thalamus) as the outcome were also adjusted for total brain volume (TBV; equations 1–3 below). Volume measures were the average of left and right volumes, following the ENIGMA study (Hoogman et al. 2017), and were standardized for all analyses. Cohen’s d measures, calculated using equation 10 in (Nakagawa & Cuthill 2007), are reported for the effect size of categorical variables (diagnosis and medication use). Standardized regression coefficients are reported for the effect sizes of the polygenic scores.
We included sex interaction terms for all variables of interest (diagnosis, PGS, medication) because of the evidence for differences in brain development between the sexes. We performed secondary sex-stratified analyses, regardless of whether a sex interaction was significant, to aid the interpretation of sex effects and for completeness of reporting. However, differences in sex-specific effects should be interpreted with caution when the sex-interaction term was not significant in the primary analysis. Given the limitations of sample size and the small effects being studied, we report effect estimates and 95% confidence intervals for all nominally significant effects. Unadjusted and FDR-adjusted p-values, corrected for the 9 volumes tested, are reported in Tables 2 and 3. Results for the following regression models are reported (note: models 1–3 were also done with TBV as the outcome):
volume ~ age + sex + FD + TBV + diagnosis + sex ∗ diagnosis
volume ~ age + sex + FD + TBV + medication + sex ∗ medication
volume ~ age + sex + FD + TBV + PGSADHD + sex ∗ PGSADHD
TBV ~ age + sex + FD + PGSICV + sex ∗ diagnosis
TBV ~ age + sex + FD + PGSICV + sex ∗ PGSADHD
A statistical mediation analysis was performed to test the plausibility of the hypothesis that genetic risk for ADHD is mediated through differences in brain volume. The analysis was done with the mediation package in R (Tingley et al. 2014), with 500 bootstrapped simulations used to estimate model parameters.
RESULTS
Overview and sample description
Table 1 provides the clinical and demographic description of the sample as well as the raw MRI data for all available single scans (N=501) and the European ancestry subgroup (N=312) utilized in the reported analyses. Diagnostic groups did not differ significantly in age or income (Wilcoxon p-values >0.1). However, as expected, the ADHD group had a higher proportion of males (p<0.05).
Table 1:
Raw data sample descriptions (mean, SD)
| Full MRI Sample | European Ancestry Subsample | |||
|---|---|---|---|---|
| Control | ADHD | Control | ADHD | |
| N | 208 | 293 | 113 | 199 |
| N(%) boys | 100 (48%) | 206(70%) | 60(53%) | 140(70%) |
| Age at scan | 10.31(1.6) | 10.26(1.6) | 10.23(1.4) | 10.12(1.5) |
| Family Income | 4.8(1.6) | 4.4(1.9) | 4.95(1.7) | 4.59(1.8) |
| Parent Conners (T) | ||||
| Inattention | 46.5(7.8) | 75.0(11.9) | 46.3(7.2) | 75.3(12.1) |
| Hyperactivity | 48.1(9.3) | 71.3(15.2) | 48.2(10.0) | 71.4(15.0) |
| Exec Function | 47.2(7.3) | 71.1(12.1) | 46.9(7.2) | 71.8(12.5) |
| Aggression | 48.8(8.7) | 54.7(12.5) | 49.2(8.9) | 54.1(12.2) |
| Teacher Conners (T) | ||||
| Inattention (T) | 46.9(7.7) | 67.1(11.4) | 47.3(7.5) | 66.6(11.2) |
| Hyperactivity (T) | 47.5(7.2) | 67.9(16.1) | 47.5(7.3) | 66.8(16.1) |
| Learning/ex (T) | 44.7(4.9) | 58.2(9.9) | 44.8(5.0) | 58.0(9.8) |
| Aggression (T) | 47.4(5.6) | 57.6(15.4) | 47.6(6.9) | 56.1(14.8) |
| MRI scores | ||||
| Mean framewise displacement | .180(.18) | .381(.53) | .167(.14) | .409(.61) |
| Total brain volume | 1211159(116433) | 1205166(107971) | 1229251(120601) | 1213321(105598) |
| Cerebellar volume | 108787(10834) | 108991(9631) | 110689(10816) | 108895(9411) |
| Thalamus | 15596(1487) | 15631(1464) | 15744(1479) | 15712(1484) |
| Caudate | 8804(1036) | 8842(1094) | 8795(1046) | 8882(1081) |
| Putamen | 12850(1369) | 12646(1262) | 12919(1497) | 12607(1221) |
| Globus pallidus | 3731(375) | 3676(359) | 3741(394) | 3684(362) |
| Hippocampus | 8117(783) | 8027(789) | 8214(768) | 8089(806) |
| Amygdala | 3262(438) | 3285(425) | 3333(423) | 3301(428) |
| Nucleus Accumb. | 1214(174) | 1191(161) | 1221(161) | 1208(159) |
| Polygenic Risk score | NA | NA | .473(.16) | .512(.143) |
|
|
||||
Conners Scores are T-scores based on national norms; ADHD medications are listed in Supplemental Table S1. PGS=polygenic score as explained in the text. All volume measures are given in mm3. Family income measures are based on the following scale: 1=less than $25,000, 2=$25,000–$35,000, 3=$35,000–$50,000, 4=$50,000–$75,000, 5=$75,000–$100,000, 6=$100,000–$130,000, 7=more than $150,000.
Primary Analyses
Brain Volumes Associated with ADHD
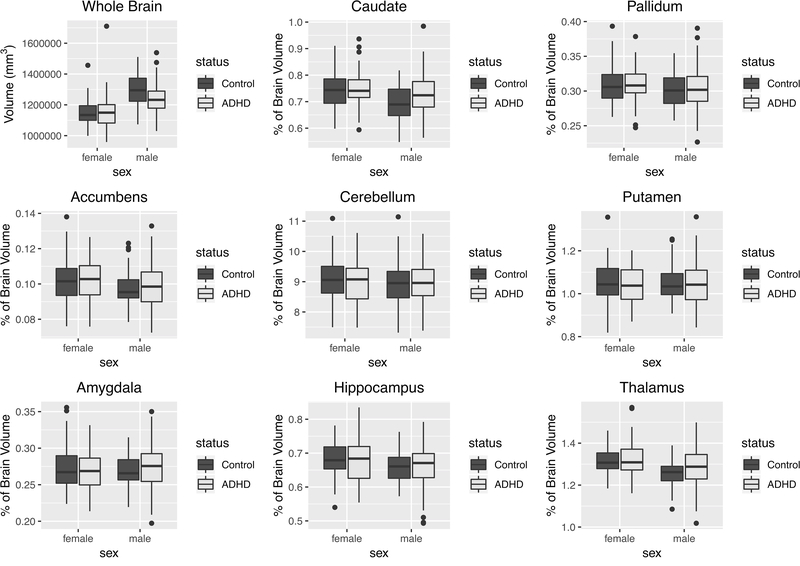
Distributions of whole brain volume and the percentage of total brain volume for each subcortical region are shown in Figure 1. The distributions of the raw volumes for all structures are shown in Supplemental Figure S2. Whole brain volume was smaller in ADHD cases compared to controls, but only among males (sex-by-diagnosis interaction p=0.0068, corrected p=.062); within males d=−0.594 (−0.68, −0.23)). Table 2 shows the results of analyses testing the association between ADHD and brain volume measures in the entire cohort, as well as the results of sex-stratified analyses.
Figure 1.
Whole brain and subcortical volumes (as percentage of total brain volume) stratified by sex and ADHD status. For each subcortical region, the averages of left and right volumes are reported.
Table 2.
Brain volumes associated with ADHD status.
| Region | ADHD d‡ | P (FDR)‡ | ADHD d | P (FDR) | Sex * ADHD P (FDR) | ADHD d, males | P (FDR), males | ADHD d, females | P (FDR), females |
|---|---|---|---|---|---|---|---|---|---|
| Whole Brain | −0.320 | 0.00747 (0.0388) | −0.455 | 1.56e-4 (0.00140) | 0.00679 (0.0611) | −0.595 | 1.80e-4 (0.00162) | 0.0556 | 0.773 (0.947) |
| Accumbens | 0.0115 | 0.923 (0.984) | 0.00571 | 0.962 (0.962) | 0.967 (0.967) | −0.00255 | 0.987 (0.987) | 0.0130 | 0.947 (0.947) |
| Amygdala | −0.00243 | 0.984 (0.984) | 0.0492 | 0.680 (0.765) | 0.503 (0.730) | 0.0475 | 0.761 (0.857) | −0.0617 | 0.751 (0.947) |
| Caudate | 0.314 | 0.00862 (0.0388) | 0.309 | 0.00993 (0.0447) | 0.380 (0.730) | 0.350 | 0.0261 (0.0783) | 0.159 | 0.415 (0.947) |
| Cerebellum | −0.227 | 0.0574 (0.103) | −0.262 | 0.0285 (0.0770) | 0.252 (0.730) | −0.327 | 0.0376 (0.0847) | 0.0748 | 0.700 (0.947) |
| Hippocampus | −0.0826 | 0.488 (0.627) | −0.105 | 0.378 (0.486) | 0.585 (0.730) | −0.144 | 0.358 (0.461) | −0.0493 | 0.800 (0.947) |
| Pallidum | −0.106 | 0.372 (0.558) | −0.117 | 0.329 (0.486) | 0.649 (0.730) | −0.164 | 0.296 (0.444) | 0.0258 | 0.894 (0.947) |
| Putamen | −0.252 | 0.0352 (0.103) | −0.253 | 0.0342 (0.0770) | 0.434 (0.730) | −0.377 | 0.0169 (0.0759) | −0.0356 | 0.854 (0.947) |
| Thalamus | 0.235 | 0.0488 (0.103) | 0.236 | 0.0482 (0.0868) | 0.469 (0.730) | 0.256 | 0.102 (0.184) | 0.213 | 0.273 (0.947) |
Effect and p-value from model without the sex interaction term (main effect).
Effects in bold are statistically significant after FDR correction (α = 0.05) for the 9 volumes tested.
Cerebellum and putamen volumes were smaller in ADHD cases compared to controls, after adjusting for total brain volume (cerebellum d=−0.262 (−0.49, −0.04); putamen d=−0.253 (−0.48, −0.03)).
Caudate and thalamus volumes were larger in ADHD cases (caudate d=0.309 (0.09, 0.53); thalamus d=0.236 (0.01, 0.46)).
Although a small literature has examined the association of ADHD medication use with structural brain volumes (Nakao et al. 2011; Frodl & Skokauskas 2012), the effect of medication use has not been investigated extensively in genetic-imaging studies of ADHD. Of our final sample of 312 children, 29% (46% of ADHD cases) had a history of ADHD medication use, consistent with previous community sampling rates (Visser et al. 2013); the majority either mixed amphetamine salts or methylphenidate preparations (Supplemental Table S1). Total brain and thalamus volumes were smaller among ADHD patients with a history of medication use compared to medication-naïve ADHD patients (Supplemental Table S2). For total brain volume, this medication effect was similar in males and females. However, for thalamus volume the medication effect was significant only among boys (sex-by-medication use interaction p=0.034). For both total brain and thalamus volumes, there was a slight trend towards smaller volume with longer duration of medication use, but these effects were not statistically reliable (Supplemental Figure S3).
Patients with a history of medication use are slightly older than medication-naïve patients (mean age=10.4 vs. 9.2 years), but age was included in the regression model and does not explain the medication effects. There was no difference in income between the groups.
Because the decision to treat ADHD patients is correlated with symptom severity, we examined whether the observed association might be due to higher symptom scores in the medicated subjects. (Note: both parent and teacher reported ADHD Rating Scale total symptom measures were significantly associated with whole brain volume (p=2e-4, β=−0.247 (−0.38, −0.12), and p=0.0021, β=−0.196 (−0.32, −0.07), respectively). Within ADHD cases, the effect size of medication history on brain volume remains essentially unchanged after accounting for parent-reported total symptom scores on the ADHD Rating scale in the model, although significance of the total brain volume association is reduced (d=−0.255 (−0.54, 0.03) for total brain volume, and d=−0.339 (−0.62, −0.06) for thalamus volume). The sex-by-medication use interaction remained significant for thalamus volume (p=0.034).
As a second check on the possible effect of symptom severity, we accounted for lifetime comorbid psychiatric disorders in the model. The association between medication use and brain volumes remained significant (d=−0.321 (−0.60, −0.04) for total brain volume, and d=−0.368 (−0.65, −0.09) for thalamus volume), and again the sex-by-medication use interaction remained significant for thalamus volume (p=0.036).
Finally, the association between total brain volume and ADHD status remains significant (males d=−0.439 (−0.78, −0.10), sex-by-diagnosis interaction p=0.012) when analyzing medication-naïve subjects only, indicating that the case-control differences are not due to medication use.
ADHD Polygenic Score Analysis
We have previously shown a PGS for ADHD to be significantly associated with ADHD diagnosis in a sample that largely overlaps the cohort studied here (N=514, Nagelkerke R2=0.045, p=1.1e-5) (Nigg et al. 2018). We tested whether this ADHD PGS was associated with brain volume measures, and whether associations with this PGS were different between the sexes (Table 3). We observed an association of smaller whole brain volumes with higher polygenic risk for ADHD (β=−0.147 (−0.27, −0.03)). This association remains significant after accounting for ADHD diagnosis (β=−0.127 (−0.25, −0.01)), suggesting the association is not simply explained by ADHD cases having higher polygenic scores, and is consistent with a liability threshold model of the disorder.
Table 3.
Brain volumes associated with PGC ADHD polygenic score.
| Region | PGS β‡ | P (FDR)‡ | PGS β | P (FDR) | Sex * PGS P (FDR) | PGS β, males | P (FDR), males | PGS β, females | P (FDR), females |
|---|---|---|---|---|---|---|---|---|---|
| Whole Brain | −0.107 | 0.0380 (0.342) | −0.147 | 0.0187 (0.168) | 0.254 (0.512) | −0.150 | 0.0207 (0.186) | −0.0231 | 0.787 (0.886) |
| Accumbens | 0.0676 | 0.184 (0.433) | 0.0305 | 0.622 (0.800) | 0.295 (0.512) | 0.0285 | 0.658 (0.741) | 0.137 | 0.106 (0.238) |
| Amygdala | 0.0682 | 0.127 (0.433) | 0.0515 | 0.344 (0.753) | 0.592 (0.592) | 0.0505 | 0.380 (0.731) | 0.0992 | 0.171 (0.309) |
| Caudate | 0.0108 | 0.811 (0.811) | 0.0368 | 0.502 (0.753) | 0.406 (0.512) | 0.0261 | 0.655 (0.741) | −0.0379 | 0.581 (0.748) |
| Cerebellum | −0.0283 | 0.515 (0.663) | 0.0181 | 0.733 (0.818) | 0.126 (0.423) | 0.0278 | 0.622 (0.741) | −0.119 | 0.071 (0.213) |
| Hippocampus | 0.0501 | 0.258 (0.464) | 0.0731 | 0.176 (0.753) | 0.455 (0.512) | 0.0744 | 0.179 (0.652) | 0.00241 | 0.975 (0.975) |
| Pallidum | 0.0598 | 0.192 (0.433) | 0.0128 | 0.818 (0.818) | 0.141 (0.423) | 0.0123 | 0.837 (0.837) | 0.149 | 0.0377 (0.170) |
| Putamen | 0.0335 | 0.466 (0.663) | −0.0630 | 0.254 (0.753) | 0.00241 (0.0217) | −0.0729 | 0.217 (0.652) | 0.224 | 0.00148 (0.0134) |
| Thalamus | −0.00907 | 0.798 (0.811) | −0.0322 | 0.456 (0.753) | 0.350 (0.512) | −0.0392 | 0.406 (0.731) | 0.0355 | 0.485 (0.727) |
Effect and p-value from model without the sex interaction term (main effect).
Whole brain volume statistically mediates a small but reliable proportion of the effect of the ADHD PGS on ADHD diagnosis (average ACME=0.0087, p=0.012; proportion of effect mediated=0.157), when accounting for sex and age. The effect is most robust, however, within boys (average ACME=0.019, p=0.008; proportion of effect mediated=0.42).
None of the subcortical volumes were associated with the ADHD PGS after adjusting for total brain volume (all p-values >0.1), in our full sample. However, we did observe a significant sex-by-PGS interaction for putamen volume (p=0.00241). Among females only, we observed a significant association of increased putamen volume with increased genetic risk for ADHD (β=0.224 (0.09, 0.36)). Given that putamen volume was not associated with ADHD diagnosis in females, this result should be followed-up with a larger sample size.
Brain Volume Polygenic Score Analysis
Total brain volume and intracranial volume are highly heritable traits (Blokland et al. 2012). Therefore, it is possible that genetic factors unrelated to ADHD contribute to the observed associations between total brain volume and both ADHD diagnosis and the ADHD PGS. To test this hypothesis, we constructed a polygenic score for intracranial volume (ICV PGS) using results from the ENIGMA+CHARGE GWAS of brain volume in primarily healthy subjects (Adams et al. 2016).
As expected, total brain volume was significantly associated with the ICV PGS (p=2.65e-6; Supplemental Table S5). However, the ADHD PGS and the ICV PGS were not significantly correlated (p=0.958), suggesting that common genetic variants that influence brain volume differences in the general population are at least partially distinct from those that influence ADHD-related volume differences. Furthermore, after adjusting for the ICV PGS, both the association between total brain volume and ADHD diagnosis (males d=−0.613 (−0.90, −0.33); sex-by-diagnosis interaction p=0.00266) and the association between total brain volume and ADHD PGS (β=−0.148 (−0.27, −0.03)) remain significant.
Polygenic scores for putamen (p=0.00524) and thalamus (p=0.0018) were also significantly associated with their corresponding volumes in our cohort. Results of the polygenic analyses for all subcortical volumes are reported in the Supplemental Materials.
DISCUSSION
Our results provide some of the first evidence related to common genetic risk and subcortical and total brain volume in ADHD. Overall, findings provide little support for the hypothesis that genetic effects drive subcortical volume effects in ADHD. On the other hand, more support was observed for genetic risk playing a role in overall brain development (represented by total brain volume) in ADHD pathophysiology. In the current sample, however, this effect was essentially confined to boys.
The genetic factors that contribute to ADHD susceptibility, and how those factors contribute to the pathophysiology of ADHD, are only beginning to be investigated using molecular genetic measures. Examining the relationship between genetic factors and disease endophenotypes, such as brain imaging measures, may help to uncover the mechanisms of genetic risk. We found that the association between ADHD polygenic risk and ADHD diagnosis may be due, in small part, to the effects of genetic variants on whole brain size. That said, the fact that the polygenic risk for ADHD explains only a small part of the variation in total brain volume, and was not associated with subcortical structure volumes, implies the need for further examination of the role of environmental exposures in ADHD associated brain findings. Of course, it is also likely that other types of genetic variation, apart from common SNPs, and epigenetic variation influence disease endophenotypes and susceptibility.
Furthermore, the small mediation effect we observed shows that our data is consistent with genetic risk being mediated through brain development processes, but does not prove a causal relationship between brain volume and ADHD. It is possible that genetic risk factors act through other mechanisms, and that smaller brain volume is a result of the disorder. Nonetheless, results are suggestive and consistent with an etiological model linking genetic risk with global neurodevelopment in ADHD.
We also observed a significant association between brain volume and an intracranial volume PGS derived from a GWAS of primarily healthy subjects. While this polygenic score was reassuringly associated with total brain volume in our cohort, it was not associated with ADHD. Although interpreting a null p-value is hazardous, this finding suggests that common genetic variation that influences normal variation in brain volume is largely distinct from the genetics that influence volume differences related to the disorder. A recent analysis with large sample sizes did find a small but significant genetic correlation between ADHD and ICV, using the LD-score regression technique (Klein et al. 2019). It is possible that we were simply underpowered to detect these shared polygenic effects in our cohort. Nevertheless, genetic factors associated with ICV did not explain the association between brain volume and the ADHD PGS in our dataset.
Sex-specific effects have not been much investigated in genetic-imaging studies of ADHD, and when studied the results have been inconsistent. Some studies have shown fewer ADHD-associated volume differences among females (Qiu et al. 2009), while others have shown no gender differences (Castellanos et al. 2002; Hoogman et al. 2017). The recent large ENIGMA consortium meta-analysis reported no differential sex effects for any of the brain volumes studied (Hoogman et al. 2017), although it should be noted that the two male-only samples included in that meta-analysis showed the qualitatively largest ADHD-related differences in intracranial volume.
We observed a reliable sex-by-diagnosis interaction for total brain volume, with a much larger effect size in boys than in girls. While sex-by-diagnosis interactions for subcortical regions were not significant after controlling for total brain volume, it was potentially interesting that in stratified analyses, effects were qualitatively larger in boys, suggesting larger samples might detect such effects.
Overall, our results with a relatively large, homogenous group of girls and boys, suggest that volumetric differences between ADHD cases and controls, as well as those associated with polygenic risk for ADHD, occur more clearly among males.
These sex-specific effects may provide a clue to the differential incidence, trajectories and clinical presentations seen for male and female ADHD patients (Martel 2013), considering what is known about differences in brain development between the sexes (Király et al. 2016). Common genetic risk for ADHD may interact with other biological systems differentially in boys and girls, leading to ADHD by partially different pathophysiologies.
We observed a significant association between history of ADHD medication use and smaller whole brain volume. This appears to be conceptually explainable by the fact that medication use is associated with worse ADHD severity. However, correcting for ADHD symptom count did not eliminate the effect, suggesting that medication history may be a marker for some unaccounted-for aspect of disorder severity in our cohort. This finding contrasts with previous studies that have shown either no association of subcortical brain volumes with history of medication treatment (Hoogman et al. 2017) or have shown that higher percentages of medicated subjects is associated with fewer differences between ADHD cases and controls (Nakao et al. 2011; Frodl & Skokauskas 2012). It should be noted, however, that these previous studies included patients with a much larger age range than ours. Castellanos et al. found that both medicated and unmedicated ADHD patients had reduced total cerebral volume compared to controls. Medicated patients showed a smaller difference in volume than unmedicated patients, but the difference between the two patient groups was not significant (Castellanos et al. 2002). Importantly, however, the medication effect seen in our cohort did not account for our genetic findings on brain volume.
Consistent with previous findings (Norman et al. 2016; Hoogman et al. 2017; Wyciszkiewicz et al. 2017), cerebellum and putamen volumes were smaller in ADHD patients, although the difference in cerebellum volume for ADHD patients seen here is smaller than that observed previously (Wyciszkiewicz et al. 2017). The effect size for putamen seen here (d=−0.252 for the full sample) was within the range seen among children in the recent ENIGMA study (95% CI (−0.28, −0.09)) (Hoogman et al. 2017).
In contrast with previous studies (Hoogman et al. 2017; Wang et al. 2018), however, we observed larger caudate and thalamus volumes in ADHD compared to controls. While this is puzzling, prior studies have shown heterogeneity in effect directions for ADHD and subcortical structures across populations with different etiologies (Stojanovski et al. 2019), and it may be that our sample of cases differed in unknown ways from others. Nonetheless, these results should be interpreted with caution, and follow-up studies will be needed to clarify the direction and size of these effects or moderators of this effect.
The current study was larger than almost all prior single-sample, single-site studies of ADHD subcortical brain volumes. Thus, it was well-powered (80%) to detect moderate differences between ADHD cases and controls (Cohen’s d > 0.38). Although a previous large meta-analysis found effects larger than this (e.g. d = 0.485 for total cerebral volume) (Valera et al. 2007), other studies have suggested smaller effect sizes are likely (Hoogman et al. 2017). In addition to its respectable sample size, several advantages of the current dataset add to the importance of the results and distinguish it from multi-site meta-analyses. First, the cohort analyzed (N=312) is well-characterized and genetically homogeneous (Nigg et al. 2018). Second, all imaging data was acquired and processed with consistent methods, and a validated technique to correct for motion during MRI acquisition was used. We believe our results provide an important additional resource for consideration in relation to meta-datasets, particularly in light of the uncertainty known to exist when combining genetically heterogeneous datasets (Ni et al. 2018).
Even so, several limitations to this study should be noted. First, our sample includes nearly twice as many males as females. This imbalance complicates our interpretation of sex effects, given that statistical power was reduced in our female-only analyses. However, this imbalance cannot account for the significant sex-by-diagnosis interaction for total brain volume or for the qualitatively larger effect sizes in boys. For subcortical structures, the large differences in effect estimates between sexes call for further studies. Second, given the non-randomized design, and the relatively small number of subjects with medication history, our study is not optimal for testing medication effects. Nonetheless, thorough testing of possible confounding with demographic variables, symptom severity, and comorbidities lends confidence to our findings. As already stated, causality cannot be assumed for any of the correlations noted.
Heterogeneity among patient samples is an ongoing challenge in ADHD research, particularly when studying the small effect sizes seen in imaging and genetic studies. The current study provides important insights into ways of reducing the impact of sample heterogeneity, as well as breaking new ground regarding genetic risk and ADHD brain development. First, we have shown that common genetic variants, associated both with ADHD and brain volume in the general population, are associated with total brain volume in an ADHD patient cohort. Accounting for genetic factors may help explain differences between studies and provide more accurate estimates of non-genetic effects. Second, our analyses include proper control of motion during MRI acquisition. Correction for motion bias has typically been lacking in ADHD imaging studies and may contribute to inconsistencies across studies.
Overall, our findings provide key insights into mechanistic endophenotypes of ADHD. Importantly, we show an association between common genetic risk for ADHD and total brain volume, as well as significant differences between boys and girls when examining ADHD-associated brain volume differences. We believe these findings will help move the field towards a better understanding of brain development and genetic risk related to ADHD.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elizabeth Nousen and Jessica Tipsord for data processing and project management support.
FINANCIAL SUPPORT
This work was supported by funding from the National Institutes of Mental Health (J.N., R01MH099064 and R01MH086654), (D.F. and J.N. R01MH115357), (D.F. R01MH096773).
CONFLICT OF INTEREST
Dr. Nikolas has received research grants for non-pharmacological research from Shire (USA-000594). Dr. Fair is non-shareholding Vice President and Chief Scientific Officer (CSO) of Nous Imaging Inc, and co-inventor of Framewise Integrated Real Time Motion Monitoring. Dr. Nigg receives royalties from Guilford Press for two books, What Causes ADHD (2006) and Getting Ahead of ADHD (2017). In the past year, Dr. Faraone received income, potential income, travel expenses continuing education support and/or research support from Lundbeck, Rhodes, Arbor, KenPharm, Ironshore, Shire, Akili Interactive Labs, CogCubed, Alcobra, VAYA, Sunovion, Genomind and Neurolifesciences. With his institution, Dr. Faraone has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. In previous years, Dr. Faraone received support from: Shire, Neurovance, Alcobra, Otsuka, McNeil, Janssen, Novartis, Pfizer and Eli Lilly. Dr. Faraone receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health, Oxford University Press: Schizophrenia: The Facts and Elsevier: ADHD: Non-Pharmacologic Interventions. Dr. Faraone is principal investigator of www.adhdinadults.com. Dr. Faraone is supported by the K.G. Jebsen Centre for Research on Neuropsychiatric Disorders, University of Bergen, Bergen, Norway, the European Union’s Seventh Framework Programme for research; technological development and demonstration under grant agreement no 602805, the European Union’s Horizon 2020 research and innovation programme under grant agreement No 667302 and National Institute of Mental Health grants 5R01MH101519 and U01 MH109536-01. All other authors report no competing interests.
Footnotes
ETHICAL STANDARDS
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
REFERENCES
- Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Rentería ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivières S, Beecham AH, Jahanshad N, Wittfeld K, Van der Lee SJ, Abramovic L, Alhusaini S, Amin N, Andersson M, Arfanakis K, Aribisala BS, Armstrong NJ, Athanasiu L, Axelsson T, Beiser A, Bernard M, Bis JC, Blanken LM, Blanton SH, Bohlken MM, Boks MP, Bralten J, Brickman AM, Carmichael O, Chakravarty MM, Chauhan G, Chen Q, Ching CR, Cuellar-Partida G, Den Braber A, Doan NT, Ehrlich S, Filippi I, Ge T, Giddaluru S, Goldman AL, Gottesman RF, Greven CU, Grimm O, Griswold ME, Guadalupe T, Hass J, Haukvik UK, Hilal S, Hofer E, Hoehn D, Holmes AJ, Hoogman M, Janowitz D, Jia T, Kasperaviciute D, Kim S, Klein M, Kraemer B, Lee PH, Liao J, Liewald DC, Lopez LM, Luciano M, Macare C, Marquand A, Matarin M, Mather KA, Mattheisen M, Mazoyer B, McKay DR, McWhirter R, Milaneschi Y, Mirza-Schreiber N, Muetzel RL, Maniega SM, Nho K, Nugent AC, Olde Loohuis LM, Oosterlaan J, Papmeyer M, Pappa I, Pirpamer L, Pudas S, Pütz B, Rajan KB, Ramasamy A, Richards JS, Risacher SL, Roiz-Santiañez R, Rommelse N, Rose EJ, Royle NA, Rundek T, et al. (2016). Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nature neuroscience 19, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany S, Jansen PR, Muetzel RL, Marques N, El Marroun H, Jaddoe VW, Polderman TJ, Tiemeier H, Posthuma D, White T (2019). Common polygenic variations for psychiatric disorders and cognition in relation to brain morphology in the general pediatric population. Journal of the American Academy of Child & Adolescent Psychiatry, 58(6), 600–607. [DOI] [PubMed] [Google Scholar]
- Blokland GAM, de Zubicaray GI, McMahon KL, Wright MJ (2012). Genetic and Environmental Influences on Neuroimaging Phenotypes: A Meta-Analytical Perspective on Twin Imaging Studies. Twin research and human genetics : the official journal of the International Society for Twin Studies 15, 351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos FX, Lee PP, Sharp W, Jeffries NO, Greenstein DK, Clasen LS, Blumenthal JD, James RS, Ebens CL, Walter JM, Zijdenbos A, Evans AC, Giedd JN, Rapoport JL (2002). Developmental trajectories of brain volume abnormalities in children and adolescents with attention-deficit/hyperactivity disorder. JAMA 288, 1740–1748. [DOI] [PubMed] [Google Scholar]
- Costa Dias TG, Iyer SP, Carpenter SD, Cary RP, Wilson VB, Mitchell SH, Nigg JT, Fair DA (2015). Characterizing heterogeneity in children with and without ADHD based on reward system connectivity. Developmental Cognitive Neuroscience 11, 155–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Belliveau R, Bybjerg-Grauholm J, Bækved-Hansen M, Cerrato F, Chambert K, Churchhouse C, Dumont A, Eriksson N, Gandal M, Goldstein J, Grove J, Hansen CS, Hauberg M, Hollegaard M, Howrigan DP, Huang H, Maller J, Martin AR, Moran J, Pallesen J, Palmer DS, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Ripke S, Robinson EB, Satterstrom FK, Stevens C, Turley P, Won H, Con -ADHD Working Group of the Psychiatric Genomics, Lifecourse & -Early, Epidemiology (EAGLE) G, Team −23andMe Research, Andreassen OA, Burton C, Boomsma D, Cormand B, Dalsgaard S, Frsanke B, Gelernter J, Geschwind D, Hakonarson H, Hsaavik J, Kranzler H, Kuntsi J, Langley K, Lesch K-P, Middeldorp C, Reif A, Rohde LA, Roussos P, Schachar R, Sklar P, Sonuga-Barke E, Sullivan PF, Thapar A, Tung J, Waldman I, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Daly MJ, Faraone SV, Børglum AD, Neale BM (2019). Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. Nature Genetics 51, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NUF, Koller JM, Earl EA, Miranda-Dominguez O, Klein RL, Van AN, Snyder AZ, Nagel BJ, Nigg JT, Nguyen AL, Wesevich V, Greene DJ, Fair DA (2017). Real-time motion analytics during brain MRI improve data quality and reduce costs. NeuroImage 161, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison-Wright I, Ellison-Wright Z, Bullmore E (2008). Structural brain change in Attention Deficit Hyperactivity Disorder identified by meta-analysis. BMC psychiatry 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Bathula D, Nikolas MA, Nigg JT (2012). Distinct neuropsychological subgroups in typically developing youth inform heterogeneity in children with ADHD. Proceedings of the National Academy of Sciences of the United States of America 109, 6769–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Larsson H (2019). Genetics of attention deficit hyperactivity disorder. Molecular Psychiatry 24, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B (2012). FreeSurfer. NeuroImage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL, Brain Development Cooperative Group (2011). Unbiased average age-appropriate atlases for pediatric studies. NeuroImage 54, 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Skokauskas N (2012). Meta-analysis of structural MRI studies in children and adults with attention deficit hyperactivity disorder indicates treatment effects. Acta Psychiatrica Scandinavica 125, 114–126. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B (2009). Accurate and robust brain image alignment using boundary-based registration. NeuroImage 48, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greven CU, Bralten J, Mennes M, O’Dwyer L, van Hulzen KJE, Rommelse N, Schweren LJS, Hoekstra PJ, Hartman CA, Heslenfeld D, Oosterlaan J, Faraone SV, Franke B, Zwiers MP, Arias-Vasquez A, Buitelaar JK (2015). Developmentally stable whole-brain volume reductions and developmentally sensitive caudate and putamen volume alterations in those with attention-deficit/hyperactivity disorder and their unaffected siblings. JAMA psychiatry 72, 490–499. [DOI] [PubMed] [Google Scholar]
- Hess JL, Akutagava-Martins GC, Patak JD, Glatt SJ, Faraone SV (2017). Why is there selective subcortical vulnerability in ADHD? Clues from postmortem brain gene expression data. Molecular Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, Aribisala BS, Armstrong NJ, Bernard M, Bohlken MM, Boks MP, Bralten J, Brown AA, Chakravarty MM, Chen Q, Ching CRK, Cuellar-Partida G, den Braber A, Giddaluru S, Goldman AL, Grimm O, Guadalupe T, Hass J, Woldehawariat G, Holmes AJ, Hoogman M, Janowitz D, Jia T, Kim S, Klein M, Kraemer B, Lee PH, Olde Loohuis LM, Luciano M, Macare C, Mather KA, Mattheisen M, Milaneschi Y, Nho K, Papmeyer M, Ramasamy A, Risacher SL, Roiz-Santiañez R, Rose EJ, Salami A, Sämann PG, Schmaal L, Schork AJ, Shin J, Strike LT, Teumer A, van Donkelaar MMJ, van Eijk KR, Walters RK, Westlye LT, Whelan CD, Winkler AM, Zwiers MP, Alhusaini S, Athanasiu L, Ehrlich S, Hakobjan MMH, Hartberg CB, Haukvik UK, Heister AJGAM, Hoehn D, Kasperaviciute D, Liewald DCM, Lopez LM, Makkinje RRR, Matarin M, Naber MAM, McKay DR, Needham M, Nugent AC, Pütz B, Royle NA, Shen L, Sprooten E, Trabzuni D, van der Marel SSL, van Hulzen KJE, Walton E, Wolf C, Almasy L, Ames D, Arepalli S, Assareh AA, Bastin ME, Brodaty H, Bulayeva KB, Carless MA, Cichon S, Corvin A, et al. (2015). Common genetic variants influence human subcortical brain structures. Nature 520, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw SP (2018). Attention Deficit Hyperactivity Disorder (ADHD): Controversy, Developmental Mechanisms, and Multiple Levels of Analysis. Annual Review of Clinical Psychology 14, 291–316. [DOI] [PubMed] [Google Scholar]
- Hoogman M, Bralten J, Hibar DP, Mennes M, Zwiers MP, Schweren LSJ, van Hulzen KJE, Medland SE, Shumskaya E, Jahanshad N, Zeeuw P de, Szekely E, Sudre G, Wolfers T, Onnink AMH, Dammers JT, Mostert JC, Vives-Gilabert Y, Kohls G, Oberwelland E, Seitz J, Schulte-Rüther M, Ambrosino S, Doyle AE, Høvik MF, Dramsdahl M, Tamm L, van Erp TGM, Dale A, Schork A, Conzelmann A, Zierhut K, Baur R, McCarthy H, Yoncheva YN, Cubillo A, Chantiluke K, Mehta MA, Paloyelis Y, Hohmann S, Baumeister S, Bramati I, Mattos P, Tovar-Moll F, Douglas P, Banaschewski T, Brandeis D, Kuntsi J, Asherson P, Rubia K, Kelly C, Martino AD, Milham MP, Castellanos FX, Frodl T, Zentis M, Lesch K-P, Reif A, Pauli P, Jernigan TL, Haavik J, Plessen KJ, Lundervold AJ, Hugdahl K, Seidman LJ, Biederman J, Rommelse N, Heslenfeld DJ, Hartman CA, Hoekstra PJ, Oosterlaan J, Polier G von, Konrad K, Vilarroya O, Ramos-Quiroga JA, Soliva JC, Durston S, Buitelaar JK, Faraone SV, Shaw P, Thompson PM, Franke B (2017). Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. The Lancet. Psychiatry 4, 310–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM (2012). FSL. NeuroImage 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Király A, Szabó N, Tóth E, Csete G, Faragó P, Kocsis K, Must A, Vécsei L, Kincses ZT (2016). Male brain ages faster: the age and gender dependence of subcortical volumes. Brain Imaging and Behavior 10, 901–910. [DOI] [PubMed] [Google Scholar]
- Klein M, Walters RK, Demontis D, Stein JL, Hibar DP, Adams HH, Bralten J, Roth Mota N, Schachar R, Sonuga-Barke E, Mattheisen M, Neale BM, Thompson PM, Medland SE, Børglum AD, Faraone SV, Arias-Vasquez A, Franke B (2019). Genetic Markers of ADHD-Related Variations in Intracranial Volume. American Journal of Psychiatry 176, 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel MM (2013). Sexual selection and sex differences in the prevalence of childhood externalizing and adolescent internalizing disorders. Psychological Bulletin 139, 1221–1259. [DOI] [PubMed] [Google Scholar]
- Martin J, Walters RK, Demontis D, Mattheisen M, Lee SH, Robinson E, Brikell I, Ghirardi L, Larsson H, Lichtenstein P, Eriksson N, 23andMe Research Team, Psychiatric Genomics Consortium: ADHD Subgroup, iPSYCH–Broad ADHD Workgroup, Werge T, Mortensen PB, Pedersen MG, Mors O, Nordentoft M, Hougaard DM, Bybjerg-Grauholm J, Wray NR, Franke B, Fasraone SV, O’Donovan MC, Thapar A, Børglum AD, Neale BM (2018). A Genetic Investigation of Sex Bias in the Prevalence of Attention-Deficit/Hyperactivity Disorder. Biological Psychiatry 83, 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills BD, Miranda-Dominguez O, Mills KL, Earl E, Cordova M, Painter J, Karalunas SL, Nigg JT, Fair DA (2017). ADHD and attentional control: Impaired segregation of task positive and task negative brain networks. Network Neuroscience 2, 200–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA (2017). Heritability of the human connectome: A connectotyping study. Network Neuroscience 2, 175–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Cuthill IC (2007). Effect size, confidence interval and statistical significance: a practical guide for biologists. Biological Reviews of the Cambridge Philosophical Society 82, 591–605. [DOI] [PubMed] [Google Scholar]
- Nakao T, Radua J, Rubia K, Mataix-Cols D (2011). Gray matter volume abnormalities in ADHD: voxel-based meta-analysis exploring the effects of age and stimulant medication. The American Journal of Psychiatry 168, 1154–1163. [DOI] [PubMed] [Google Scholar]
- Ni G, Moser G, Wray NR, Lee SH (2018). Estimation of Genetic Correlation via Linkage Disequilibrium Score Regression and Genomic Restricted Maximum Likelihood. American Journal of Human Genetics 102, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT, Gustafsson HC, Karalunas SL, Ryabinin P, McWeeney SK, Faraone SV, Mooney MA, Fair DA, Wilmot B (2018). Working Memory and Vigilance as Multivariate Endophenotypes Related to Common Genetic Risk for Attention-Deficit/Hyperactivity Disorder. Journal of the American Academy of Child and Adolescent Psychiatry 57, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman LJ, Carlisi C, Lukito S, Hart H, Mataix-Cols D, Radua J, Rubia K (2016). Structural and Functional Brain Abnormalities in Attention-Deficit/Hyperactivity Disorder and Obsessive-Compulsive Disorder: A Comparative Meta-analysis. JAMA psychiatry 73, 815–825. [DOI] [PubMed] [Google Scholar]
- Qiu A, Crocetti D, Adler M, Mahone EM, Denckla MB, Miller MI, Mostofsky SH (2009). Basal Ganglia Volume and Shape in Children With Attention Deficit Hyperactivity Disorder. American Journal of Psychiatry 166, 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglin L, Collishaw S, Thapar AK, Dalsgaard S, Langley K, Smith GD, Stergiakouli E, Maughan B, O’Donovan MC, Thapar A (2016). Association of Genetic Risk Variants With Attention-Deficit/Hyperactivity Disorder Trajectories in the General Population. JAMA psychiatry 73, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savalia NK, Agres PF, Chan MY, Feczko EJ, Kennedy KM, Wig GS (2017). Motion-related artifacts in structural brain images revealed with independent estimates of in-scanner head motion. Human Brain Mapping 38, 472–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Ishii-Takahashi A, Park MT, Devenyi GA, Zibman C, Kasparek S, Sudre G, Mangalmurti A, Hoogman M, Tiemeier H, von Polier G, Shook D, Muetzel R, Chakravarty MM, Konrad K, Durston S, White T (2018). A multicohort, longitudinal study of cerebellar development in attention deficit hyperactivity disorder. Journal of Child Psychology and Psychiatry, and Allied Disciplines [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski S, Felsky D, Viviano JD, Shahab S, Bangali R, Burton CL, Devenyi GA, O’Donnell LJ, Szatmari P, Chakravarty MM, Ameis S (2019). Polygenic Risk and Neural Substrates of Attention-Deficit/Hyperactivity Disorder Symptoms in Youths With a History of Mild Traumatic Brain Injury. Biological Psychiatry, 85(5), 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley D, Yamamoto T, Hirose K, Keele L, Imai K (2014). mediation: R Package for Causal Mediation Analysis. Journal of Statistical Software, 59(5), 1–38.26917999 [Google Scholar]
- Valera EM, Faraone SV, Murray KE, Seidman LJ (2007). Meta-analysis of structural imaging findings in attention-deficit/hyperactivity disorder. Biological Psychiatry 61, 1361–1369. [DOI] [PubMed] [Google Scholar]
- Visser SN, Blumberg SJ, Danielson ML, Bitsko RH, Kogan MD (2013). State-Based and Demographic Variation in Parent-Reported Medication Rates for Attention-Deficit/Hyperactivity Disorder, 2007–2008. Preventing Chronic Disease 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhang X, Li A, Zhu M, Liu S, Qin W, Li J, Yu C, Jiang T, Liu B (2017). Polygenic risk for five psychiatric disorders and cross-disorder and disorder-specific neural connectivity in two independent populations. NeuroImage. Clinical 14, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Q, Li S, Li G, Zuo C, Liao S, Long Y, Li S, Joshi RM (2018). Gender differences in anomalous subcortical morphology for children with ADHD. Neuroscience Letters 665, 176–181. [DOI] [PubMed] [Google Scholar]
- Whalley HC, Hall L, Romaniuk L, Macdonald A, Lawrie SM, Sussmann JE, McIntosh AM (2015). Impact of cross-disorder polygenic risk on frontal brain activation with specific effect of schizophrenia risk. Schizophrenia Research 161, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyciszkiewicz A, Pawlak MA, Krawiec K (2017). Cerebellar Volume in Children With Attention-Deficit Hyperactivity Disorder (ADHD). Journal of Child Neurology 32, 215–221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.