Abstract
Retinal pigment epithelium (RPE) differentiated from human induced pluripotent stem cells, called induced retinal pigment epithelium (iRPE), is being explored as a cell-based therapy for the treatment of retinal degenerative diseases, especially age-related macular degeneration. The success of RPE implantation is linked to the use of biomimetic scaffolds that simulate Bruch's membrane and promote RPE maturation and integration as a functional tissue. Due to difficulties associated with animal protein-derived scaffolds, including sterility and pro-inflammatory responses, current practices favor the use of synthetic polymers, such as polycaprolactone (PCL), for generating nanofibrous scaffolds. In this study, we tested the hypothesis that plant protein-derived fibrous scaffolds can provide favorable conditions permissive for the maturation of RPE tissue sheets in vitro. Our natural, soy protein-derived nanofibrous scaffolds exhibited a J-shaped stress–strain curve that more closely resembled the mechanical properties of native tissues than PCL with significantly higher hydrophilicity of the natural scaffolds, favoring in vivo implantation. We then demonstrate that iRPE sheets growing on these soy protein scaffolds are equivalent to iRPE monolayers cultured on synthetic PCL nanofibrous scaffolds. Immunohistochemistry demonstrated RPE-like morphology and functionality with appropriate localization of RPE markers RPE65, PMEL17, Ezrin, and ZO1 and with anticipated histotypic polarization of vascular endothelial growth factor and pigment epithelium-derived growth factor as indicated by enzyme-linked immunosorbent assay. Scanning electron microscopy revealed dense microvilli on the cell surface and homogeneous tight junctional contacts between the cells. Finally, comparative transcriptome analysis in conjunction with principal component analysis demonstrated that iRPE on nanofibrous scaffolds, either natural or synthetic, matured more consistently than on nonfibrous substrates. Taken together, our studies suggest that the maturation of cultured iRPE sheets for subsequent clinical applications might benefit from the use of nanofibrous scaffolds generated from natural proteins.
Impact statement
Induced retinal pigment epithelium (iRPE) from patient-derived induced pluripotent stem cells (iPSCs) may yield powerful treatments of retinal diseases, including age-related macular degeneration. Recent studies, including early human clinical trials, demonstrate the importance of selecting appropriate biomaterial scaffolds to support tissue-engineered iRPE sheets during implantation. Electrospun scaffolds show particular promise due to their similarity to the structure of the native Bruch's membrane. In this study, we describe the use of electroprocessed nanofibrous soy protein scaffolds to generate polarized sheets of human iPSC-derived iRPE sheets. Our evaluation, including RNA-seq transcriptomics, indicates that these scaffolds are viable alternatives to scaffolds electrospun from synthetic polymers.
Keywords: iRPE, nanofibrous scaffold, electrospinning, soy protein, human iPSCs, RNA-seq
Introduction
The retinal pigment epithelium (RPE) is a monolayer of cells that cover the back of the retina and support the light-sensing photoreceptors. In age-related macular degeneration (AMD), a leading cause of blindness in developed countries, RPE senescence, dysfunction, and cell loss are a key aspect of the retinal pathophysiology.1 As such, cell therapies based on the replacement of lost RPE by cell transplantation offer hopes for treatment and are the subject of intense current investigations.2,3 Recent advances in the generation of diverse somatic cell types, including RPE, from pluripotent stem cells (PSCs) have created unprecedented opportunities to study cell therapies for degenerative diseases, such as those affecting the retina.4 Indeed, in one of the first clinical trials of stem cell-derived therapy, patients with AMD were injected subretinally with RPE cells generated from stem cells in vitro.5,6
Several protocols have been described for the derivation of RPE from induced pluripotent stem cells (iPSCs) through modulation of developmentally-relevant signaling pathways.7–12 RPE derived from iPSCs (called induced retinal pigment epithelium [iRPE]) could be delivered to the recipient eye in one of two approaches1: injection of dissociated cells5,13 or2 implantation of an intact monolayer of RPE as a sheet.14–18 Dissociated cell suspension makes the surgical procedure simpler, but loss of cell–cell contact negatively impacts functional engraftment. RPE sheets maintain appropriate cell polarity and organization that favor graft survival, integration, and functionality but require more sophisticated surgical methods.
Preclinical and early clinical studies to date suggest that both approaches are safe.5,13,14,17,18 Progress in handling and surgical placement of RPE sheets combined with encouraging early data from first human studies suggest that RPE sheet transplantation may be a preferred strategy for future clinical application.19
A current challenge for this approach is the selection and fabrication of a scaffold material that serves as a carrier for the RPE sheet. In addition to being biodegradable and biocompatible, the ideal scaffold biomaterial should manifest biophysical properties that are conducive to RPE maturation in vitro and functional engraftment in vivo following transplantation. In its native environment, the RPE monolayer is basally attached to Bruch's membrane, a unique pentalaminar structure composed of two collagenous zones and a central elastic lamina. The mechanical and biophysical properties of Bruch's membrane have been extensively studied and should guide investigations into potential scaffold materials for RPE culture in vitro.
Electrospun nanofibrous scaffolds are structurally similar to native Bruch's membranes making them a promising candidate for RPE implantation.20 The diameters of electrospun nanofibers, in hundreds of nanometers, are similar to those of the collagen fibers in Bruch's membrane and exhibit comparable mechanical and biophysical properties, including stiffness and nonlinear elasticity. With an array of natural and synthetic biomaterials available for their manufacture, each with its own advantages and disadvantages, electrospun scaffolds offer a range of biochemical, structural, and mechanical characteristics.
Although promising, naturally derived scaffolds, including those produced from animal collagen, bacterial cellulose, and amniotic tissue, have potential problems with purity, sterility, reproducibility, and scalability for clinical applications. Synthetic scaffolds, frequently from polycaprolactone (PCL), poly(L-lactic acid) (PLLA), and poly(lactic-co-glycolic acid) (PLGA), avoid many of the issues associated with natural scaffolds.20 However, the mechanical properties of synthetic scaffolds, especially stiffness and elasticity, widely differ from those of natural scaffolds and the native Bruch's membrane. Furthermore, synthetic scaffolds, while usually biologically inert, may lack biocompatibility and biochemical cues, such as the release of bioactive peptides or the presence of protein motifs common to the extracellular matrix, characteristic of naturally derived scaffolds.21
Plant protein-based scaffolds are increasingly being investigated for their use in tissue engineering and regenerative medicine applications. Natural in origin, plant protein scaffolds exhibit many key biochemical and biomimetic characteristics absent in synthetic scaffolds, while avoiding the limitations of many animal and bacteria-derived natural scaffolds.22
In particular, nanofibrous scaffolds produced from natural soy protein isolate are highly reproducible, cost-effective, implantable, and biocompatible.23 These scaffolds exhibit mechanical and biophysical properties similar to those of native epithelial tissues and promote anti-inflammatory responses in vivo.22–24 Soy scaffolds have been used for seeding human dermal fibroblasts for skin regeneration25–27 and mesenchymal stem cells,28 showing favorable structural and cell biological characteristics. In addition, degradation products of these scaffolds are unlikely to trigger strong immune reactions.25,27
In this study, we explored the suitability of a novel electroprocessed soy scaffold to promote iRPE monolayer sheet maturation. In this study, we report that the soy scaffold performs comparably to PCL substrates commonly used for growing iRPE sheets. Furthermore, we demonstrate the utility of RNA-seq as an approach for a comprehensive assessment of transcriptomic profiles of iRPE sheets and show reduced batch-to-batch variation when the cells are cultured on nanofibrous scaffolds of either type versus commonly used nonfibrous substrates.
Methods
Soy scaffolds were prepared by blow electrospinning, as this method possesses the ability to produce uniform fibers from aqueous solutions. Therefore, 10% (w/v) water-soluble soy (CLARISOY™, ADM, Chicago, IL, >90% protein purity per manufacturer) and 1.25% (w/v) poly(ethylene oxide) (Mw 900,000; Sigma-Aldrich) were dissolved in deionized water overnight. The solution was placed in a 3 mL syringe (BD) and extruded at 1 mL/h through a blunted 25 gauge needle (McMaster-Carr, Princeton, NJ) in line with a modified polypropylene, barbed T-connector (Cole-Parmer) held under 10 psi of pressurized air, 20 cm from a grounded, circular copper target (2 cm diameter) with a 5 kV electric field. Scaffolds were spun for 3 min before removal. Diagrams describing the difference between traditional electrospinning and blow electrospinning are shown in Supplementary Figure S1.
After manufacturing, the blow electrospun soy scaffolds were crosslinked. The crosslinking solution was prepared by dissolving 100 mM EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) and 100 mM NHS (N-hydroxysuccinimide) (Thermo Fisher Scientific) in 95% ethanol (Decon Labs, Inc.). Freshly prepared soy scaffolds were placed in individual wells of a 12-well plate (Corning), and 500 μL of crosslinking solution was added to each. The plate was placed on an orbital shaker for 4 h. After 4 h, the solution was aspirated, and the scaffolds washed thrice with 1 × DPBS (Dulbecco's phosphate-buffered saline) (Corning) for 5 min each under continuous shaking. Crosslinked scaffolds were stored in DPBS at 4°C for up to 1 week.
To prepare control scaffolds, 7.5% (w/v) PCL (Mn 80,000) (Sigma-Aldrich) was dissolved in 1,1,1,3,3,3-hexafluoro-2-isopropanol (HFIP). The solution was placed in a 3 mL Norm-Ject syringe (BD) and extruded through a 20 gauge, blunt-tip needle (McMaster-Carr) at a rate of 1 mL/h using a KD Scientific syringe pump under 15 kV, 10 cm from a grounded, circular copper target (2 cm diameter). Errant fibers were removed as needed. Scaffolds were spun for 5 min before removal. PCL scaffolds were evaluated identically to soy.
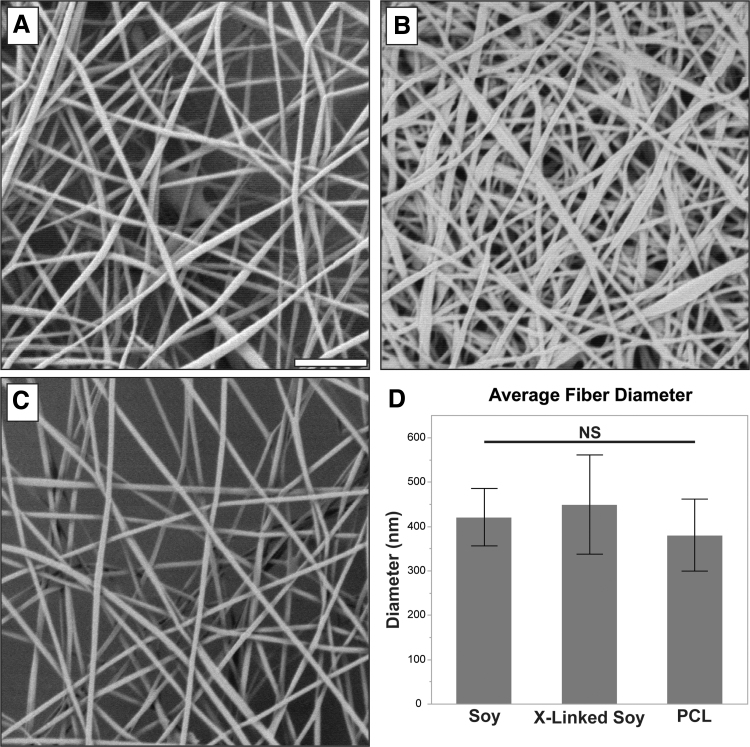
Scaffold samples taken from different batches of each material type were imaged in a 9320B 8500B field emission scanning electron microscope (SEM) (Keysight Technologies, Santa Rosa, CA) at 10,000 × magnification. Ten fibers from 10 images per batch were measured using ImageJ. The frequency of measured fiber diameters and the average of each batch were calculated and averaged across three batches per scaffold type to obtain overall fiber thickness. The soy scaffold was evaluated before and after crosslinking to assess swelling of the fibers upon crosslinking. A total of five samples of each material prepared on different days were evaluated. SEM examination confirmed that all scaffold fabrication procedures generated uniform, nanofibrous sheets of similar diameters (∼400 nm).
Figure 1A and C shows representative SEM images of the soy and PCL scaffolds, respectively, before crosslinking. Both the PCL and soy showed relatively consistent fiber diameters with no apparent splattering, spraying, or voids in the scaffold. No significant difference was detected in the fiber diameters (Fig. 1D). The soy scaffold postcrosslinking is shown in Figure 1B. Although some swelling is apparent, it did not produce a significant increase in fiber diameters or change fiber morphology.
FIG. 1.
Characteristics of soy and PCL scaffolds. (A) A representative SEM of a soy scaffold. (B) A representative SEM of a soy scaffold postcrosslinking. (C) A representative SEM of a PCL scaffold. (D) Observed diameter (mean ± SD) of a PCL scaffold, soy scaffold, and soy scaffold after crosslinking (X-linked). Scale bar = 5 μm. PCL, polycaprolactone; SD, standard deviation; SEM, scanning electron microscope.
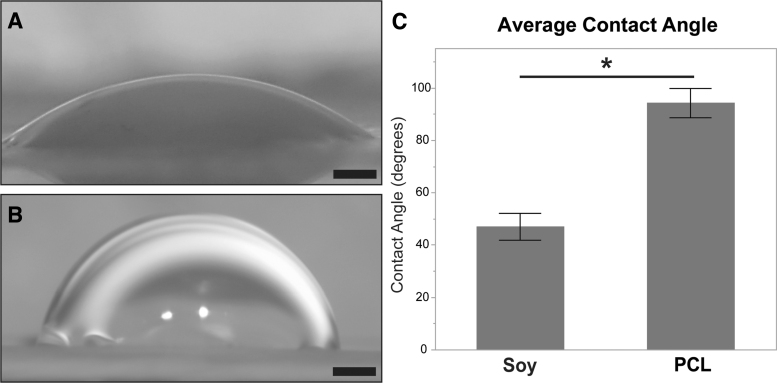
Contact angles were measured on dry scaffolds (after crosslinking in the case of soy). Scaffolds were affixed to microscope slides, and three 3 μL drops of deionized water were placed directly onto the surface across the length of the scaffold. The drops were allowed to settle for 60 s before imaging with a stereomicroscope (Leica Camera). The contact angles were evaluated using the Contact Angle plugin for ImageJ. Three samples from three batches per scaffold type were evaluated. A total of 10 measurements were taken from different samples. Contact angle measurements showed that, in contrast to the observed similarity of fiber diameters, the water contact angles for each material type exhibited significant differences. Figure 2A and B shows the appearance of water droplets on the crosslinked soy scaffolds and on the PCL, respectively. As shown in Figure 2C, soy scaffolds exhibited significantly smaller contact angles than PCL scaffolds (∼50° and 90°, respectively) indicating that the soy scaffolds were more hydrophilic (wettable) and thus more favorable to cell attachment.29
FIG. 2.
Surface hydrophobicity. (A) A 3 μL water droplet on the soy scaffold. (B) A 3 μL water droplet on the PCL scaffold. (C) Water droplet contact angle (mean ± SD) observed for the PCL and soy (after crosslinking) scaffolds. Scale bar = 500 μm. Asterisk indicates significance for p < 0.05.
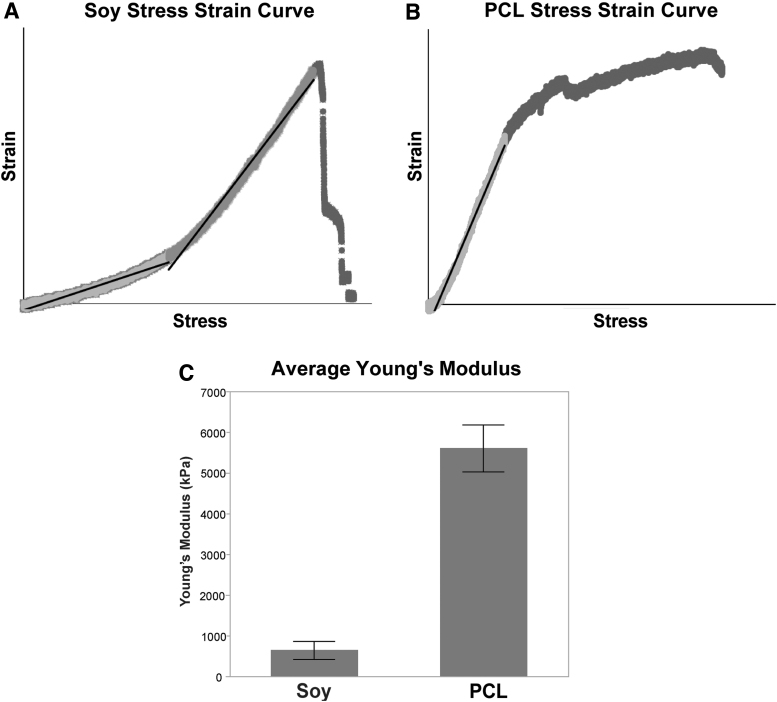
To evaluate their mechanical properties, scaffolds were cut into rectangles 3 cm long by 1 cm wide. Each sample had its thickness evaluated as above. The rectangles were placed into a tensile tester (Instron, Norwood, MA) and affixed in tight-fitting grips. The samples were drawn in the tensile tester at a rate of 1 mm per minute until rupture. The Young's Moduli were determined from the slope of linear segments of the stress–strain curves. Three replicates each of scaffolds prepared on separate days were evaluated. Young's moduli testing demonstrated substantial differences between the mechanical properties of PCL and soy scaffolds (Fig. 3). The soy scaffold exhibited a J-shaped behavior typical for natural matrices with Young's modulus, a measure of stiffness, increasing as increasing load was applied. The Young's modulus of the soy scaffolds was ∼644 kPa in a relaxed state (Fig. 3C). The linear stress–strain curve of the PCL scaffold, typical for synthetic materials, indicated an average Young's modulus of ∼5.6 MPa (Fig. 3C). A summary of the mechanical properties of the two nanofibrous scaffold types is shown in Table 1. Additional information regarding scaffold appearance and thickness is presented in Supplementary Figure S2.
FIG. 3.
Young's moduli of scaffolds. (A) The stress–strain curve of the crosslinked soy scaffold. (B) The stress–strain curve of the PCL scaffold. (C) Comparison of Young's modulus (mean ± SD) of the two scaffold types (kPa).
Table 1.
A Summary of the Mechanical Properties of the Soy and Polycaprolactone Nanofibrous Scaffolds
| Material type | Average fiber diameter (nm) | Average contact angle (degrees) | Average young's modulus (kPa) | Nonlinear stress–stain curve? |
|---|---|---|---|---|
| Crosslinked soy | 449.7 | 47.0 | 644.8 | Yes |
| PCL | 380.75 | 94.2 | 5609.1 | No |
All values provided for soy are postcrosslinking.
PCL, polycaprolactone.
A novel disinfection and mounting approach was used in preparation for cell culture. Scaffolds were disinfected immediately before cell seeding by soaking in 70% ethanol for 1 h followed by three consecutive rinses with PBS. After disinfection, scaffolds were immediately mounted to Transwell™ membranes for culture. Scaffolds were affixed against the Transwell membranes using 12 mm Snapwell™ 0.4 μm Inserts (Corning). The Snapwell inserts were detached from their holders and the scaffolds placed directly against the membranes. The inserts were then reattached to the holders, which mechanically held the scaffolds to the membrane without the use of chemical adhesives (Supplementary Fig. S3). The choice was made to not remove the Transwell component of the inserts, in contrast to the method described by McHugh et al.30 This controlled for possible error due to higher nutrient availability in the electrospun groups should the electrospun scaffolds have higher porosity than the Transwell.
Experiment
Experimental design
The aim of the experiment was to test the usefulness of electroblown soy scaffolds for the culture of iPSC-derived iRPE compared to existing substrates. We evaluated the phenotypic outcomes of iRPE matured on soy versus that of commonly used electrospun PCL scaffolds (as a nanofibrous control) and Transwell membranes (as a control for high permeability). PCL scaffolds were selected as a nanofibrous control because of their common use, fiber diameters, and swelling characteristics similar to those of soy scaffolds and their nonacidic degradation products. For RNA-seq transcriptomics, iRPE was also cultured on laminin-coated chamber slides. This latter control was included to provide an additional baseline for the behavior of iRPE under traditional culture methods and to provide a more comparative analysis of the significance of iRPE grown on nanoporous versus nanofibrous surfaces. The experiments were carried out according to the methodology detailed above. In brief, iRPE was differentiated and matured from iPSC, seeded onto the various substrates, and cultured for 8 weeks after confluence. The ensuing iRPE sheets were either fixed and visualized by SEM and confocal immunohistochemistry or processed for biochemical/RNA-seq transcriptomics analyses.
iPSC lines and maintenance
iPSC 377E is a human iPSC line reprogrammed from skin biopsies using integration-free Sendai virus carrying the Yamanaka factors. This line's genome integrity and pluripotency have been previously established.31 iPSCs were maintained in feeder-free conditions in Essential 8 medium (Thermo Fisher Scientific) and on human embryonic stem cell-qualified Matrigel™ (Corning) coated six-well plates. Cells were passaged every 3–4 days using an ethylenediaminetetraacetic acid (EDTA)-based protocol.32
RPE differentiation and maturation
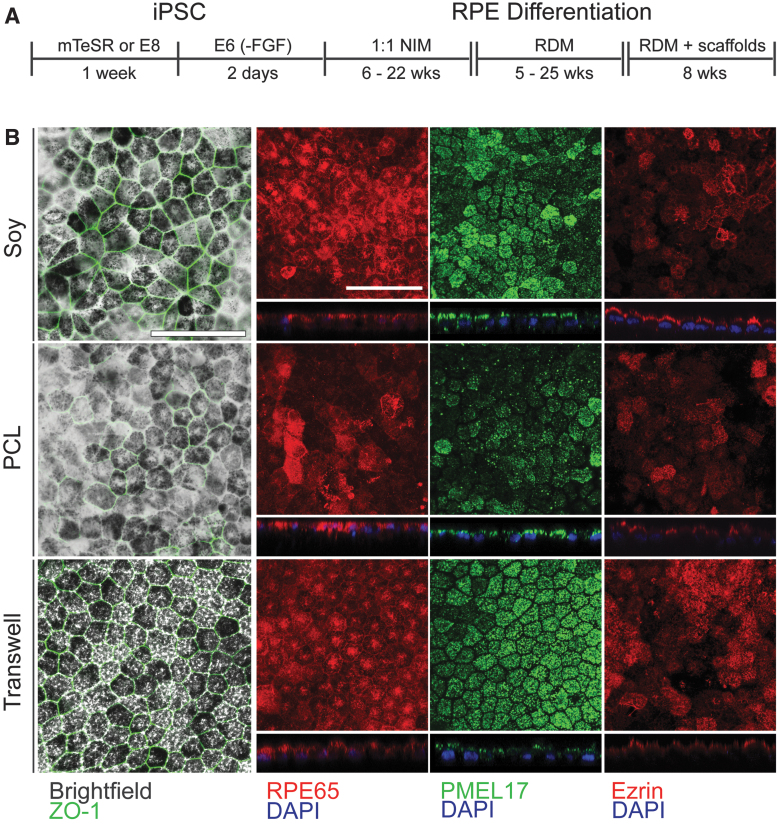
The protocols to differentiate iPSCs into RPEs were modified from published work14,33 as follows. Figure 4A describes a simplified timeline of the differentiation and maturation with major reagents. Once iPSC cultures reached 100% confluence, cells were placed in Essential 6 medium (Thermo Fisher Scientific). After 48 h, the cells were placed into Neural Induction Media (NIM: DMEM/F12 (1:1), 1% N2 Supplement, 1 × nonessential amino acids, 1% antibiotic–antimycotic (Thermo Fisher Scientific), and 20 μg/mL heparin (MilliporeSigma)). The medium was changed every 2 days for up to 22 weeks. Cell patches with apparent RPE morphology and dark pigmentation were manually picked with tungsten needles, dissociated into single cells with 0.25% Trypsin/EDTA (Thermo Fisher Scientific) exposure for 5 min, and seeded on laminin (5 μg/mL, rhLN521, BioLamina)-coated dishes at 200,000 cells per 1.12 cm2 in Retinal Differentiation Media (RDM: DMEM:F12 3:1, 2% B27 with vitamin A, 2 mM GlutaMAX, 1% Antibiotic–antimycotic (Thermo Fisher Scientific), and 10% fetal bovine serum (FBS) (Atlanta Biologics)) with 10 μM ROCK Inhibitor (RI), Y-27632 (Tocris), for the first 24 h. FBS was removed once cell–cell junctions reappeared as assessed by visualization of epithelial-like hexagonal morphology and tight cellular packing under light microscopy (up to 1–1.5 weeks after seeding).
FIG. 4.
Differentiation of RPE on scaffolds. (A) Medium types used over time for the differentiation of iPSCs into RPE. (B) Fluorescence and brightfield microscopy of fully differentiated RPE cells on the three material types. Phalloidin staining and immunostaining using antibodies against PMEL17, RPE65, and ZO-1 are shown as indicated. For all figures, scale bar = 50 μm. iPSC, induced pluripotent stem cell; RPE, retinal pigment epithelium.
After 5–25 weeks RPEs were trypsinized and seeded onto laminin-coated scaffolds at a density of 178,000 cells per cm2 in RDM with RI and 10% FBS. RI was removed after 24 h. FBS was removed once cell–cell junctions reappeared at ∼7–10 days. RPEs were cultured on scaffolds for 8 weeks and collected for processing. RPE differentiation studies were performed in triplicate from three separate cell stocks. Cells were simultaneously plated onto all conditions described below. All maturation and culture time points were kept identical for each batch, independent of which material type the cells were seeded on.
Immunohistochemistry
For immunostaining, cells were fixed in 4% paraformaldehyde for 20 min followed by blocking (2.5%TX-100, 1%BSA, 5%DKY serum, 1 × PBS) at room temperature for 1 h. Primary antibodies were incubated overnight at 4°C (0.5%TX-100, 1%BSA, 1 × PBS). Secondary antibodies (488 or 568 Alexa Fluor Invitrogen Ms or Rb IgG 1:1000) were incubated for 1 h at room temperature and mounted with Fluoromount-G (SouthernBiotech 0100–01). Primary antibodies used were as follows: PMEL17 (1:250) (Novus Biological NBP2–44520), RPE65 (1:250) (Custom Dr. Redmond NIH NEI), ZO-1 (1:50) (Invitrogen 40–2200), Ezrin (1:250) (Thermo Scientific MS-661-P1ABX), DAPI (1:1000) (Invitrogen D1306), and F-actin (1:250) (405 Phalloidin).
Enzyme-linked immunosorbent assay
Apical and basolateral supernatants of the cell cultures on the Transwell plates covered with the various substrates were analyzed for the polarized production of vascular endothelial growth factor (VEGF) and pigment epithelium-derived growth factor (PEDF) by enzyme-linked immunosorbent assay (ELISA). Human VEGF (Cat# KHG0111; Thermo Fisher) and human PEDF ELISAs (Cat.# RD191114200R; BioVendor) were performed according to the manufacturer's protocols. A total of three replicates were performed for this experiment.
RNA isolation
RNA isolation using the RNeasy Mini Kit (Qiagen) was performed according to the manufacturer's protocol. RNA integrity numbers were obtained using an Agilent Technologies, Inc., Bioanalyzer following the manufacturer's protocol. RNA was isolated from three replicates for each condition cultured on different days.
RNA-seq analysis
Libraries for RNA-seq were constructed from 100 ng of total RNA using the TruSeq Stranded mRNA Kit (Illumina, San Diego, CA). Paired-end sequencing of 125 bases was performed on a HiSeq 2500 (Illumina), and only reads passing chastity filtering were kept for further analysis. Gene-level quantitation was performed as previously reported.34 Briefly, reads were aligned and quantitated to Ensembl v94 annotation using Kallisto v0.44.035 and Tximport v1.8.0.36 Analysis was performed in the R v3.6.1 environment.
Genes identified for heatmap generation for RPE signature genes were obtained from Soneson et al.37 Genes for specific biological processes (vitamin A and melanin metabolic processes and phagocytosis) were obtained from gene ontology (GO) (geneontology.org). Principal component analysis (PCA) was performed using the prcomp function in R on the gene-level log2 CPM (exon counts per million reads) values. The top 500 gene loadings contributing to PC1 and PC2 (Principal Components 1 and 2) of the PCA were used for gene enrichment analysis using gProfileR v0.6.7.38 All raw and processed data can be accessed through the NCBI GEO database with the accession number GSE153645.
Statistical analysis
The comparison of mechanical properties was evaluated using an unpaired Student's t-test in most instances. For comparisons, including crosslinked soy, a one-way analysis of variance (ANOVA) was performed. Total VEGF amounts and average PEDF concentrations were plotted using Prism7 software with a one-way ANOVA after confirming that the data were normally distributed, using the Shapiro–Wilk method. In all cases, results were considered statistically significant for p-values <0.05.
Experimental results
RPE maturation and characterization
The differentiation/maturation protocol, the different media used, and the timeline of their application are shown in Figure 4A. Culture of iRPE cells according to this protocol yielded consistent maturation on all three substrates (soy scaffold, PCL scaffold, Transwell). Figure 4B shows a comparison of RPE maturation on the three material types across an array of phenotypic RPE markers using both brightfield (pigmentation) and fluorescence microscopy (ZO-1, RPE65, PMEL17, Ezrin).
Qualitatively, the appearance of the brightfield/ZO-1 overlay showed similarities between soy and Transwell, compared to PCL, particularly in the sharpness of the distribution of the ZO-1 tight junction protein between adjacent cells. Similarly, the pigmentation for the soy and Transwell conditions appeared darker than the PCL. This soy-Transwell similarity was also observed in terms of the patterning and distribution of the RPE markers RPE65 (centers and perimeters fluorescing for the soy and Transwell, compared to a more random distribution for the PCL) and PMEL17 (more even distribution of fluorescence).
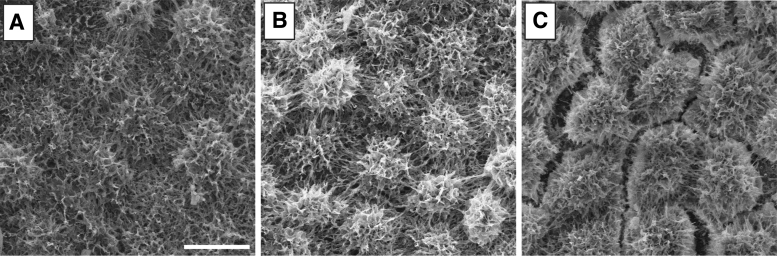
SEM images of differentiated RPE cells cultured on soy (Fig. 5A) and PCL scaffolds (Fig. 5B), as well as Transwell surfaces alone (Fig. 5C), show general similarities across all three substrates. Cells on all three material types produced confluent monolayers exhibiting highly ciliated surfaces, characteristic of mature RPE. The appearance of voids between cells of the Transwell may be due to cracking during the desiccation process on a relatively noncompliant surface.
FIG. 5.
A representative SEM of RPE on scaffolds. (A) Differentiated RPE cells on a crosslinked soy scaffold. (B) Differentiated RPE cells on a PCL scaffold. (C) Differentiated RPE cells on a Transwell™ surface. Scale bar = 10 μm.
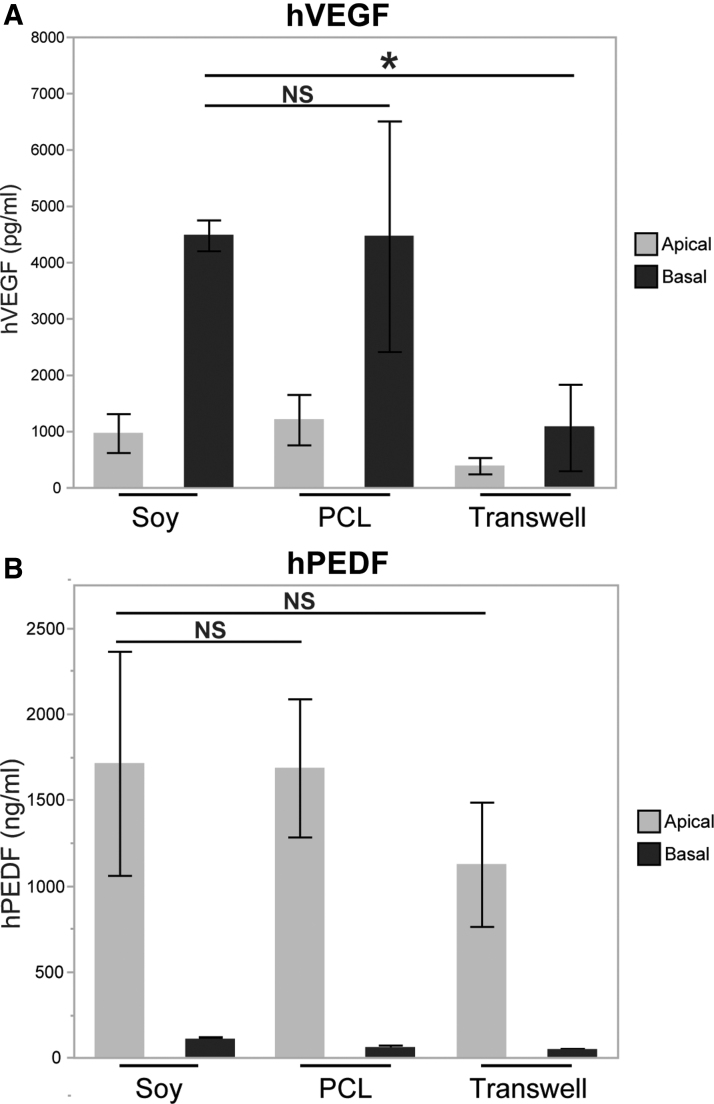
Functional cell polarization was assessed on the three materials by measuring apical and basal secretion of PEDF and VEGF using ELISA. In all three conditions, polarized secretion was observed on the basal side for VEGF (Fig. 6A) and on the apical side for PEDF (Fig. 6B), in line with the physiological roles of each protein. Although the PCL and soy scaffold values tended to be higher in both conditions for the basal secretion of VEGF and the apical secretion of PEDF only the difference between Transwell and soy basal VEGF was statistically significant. This was likely due to the high signal and low variability observed in the soy basal VEGF.
FIG. 6.
ELISA results. (A) ELISA values for hVEGF on the apical and basal sides of RPE differentiated on three material types. (B) ELISA values for hPEDF on the apical and basal sides of RPE differentiated on three material types. Values reported as mean ± SD. Asterisk indicates significance for p < 0.05. ELISA, enzyme-linked immunosorbent assay; hPEDF, human pigment epithelium-derived growth factor; hVEGF, human vascular endothelial growth factor.
Transcriptome analysis
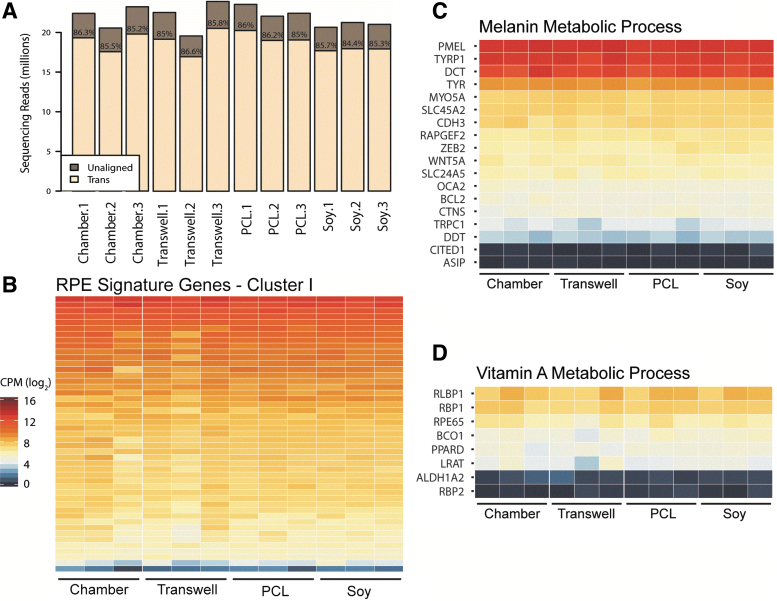
Figure 7 depicts the expression levels of mRNA transcripts frequently evaluated when investigating RPE differentiation and function. The data in Figure 7 broadly indicate similarities between all trials and batches tested. Figure 7A shows the sequencing depth found per RNA sample and the total percentage of each sample aligned to known gene annotations, typical quality control metrics for RNA sequencing. A range of 17–20 million aligned reads per sample was observed with a range of reads aligned to known genes between 84.4% and 86.6%, indicating good quality transcriptome data obtained for all samples in all conditions. Shown in Figure 7B is a heatmap of the expression of RPE signature genes (see Supplementary Table S1) from three independent batches of the four growth conditions, with Figure 7C and D showing the heatmaps of, respectively, the melanin metabolic GO biological process and vitamin A metabolic process, key pathways that are critical for native RPE function. Expression levels of each key gene appeared approximately equivalent across all samples. Similar expression of key marker genes demonstrates maintenance of RPE fate and expression of mature markers in all substrate conditions.
FIG. 7.
Similarity in transcriptomes (determined by RNA-seq). (A) Sequencing depth per sample. Bar height is the sequencing depth per sample, whereas “Trans” indicated the percentage of reads aligning to the known gene annotations used in calculating gene expression. (B) Gene expression of RPE signature cluster I genes from Strunnikova et al.38 Gene expression values are presented as log2 CPM. List of genes is shown in Supplementary Table S1. (C, D) Gene expression values of genes of two GO pathways important to functional RPE, melanin metabolic process (C) (GO:0006582) and vitamin A metabolic process (D) (GO:0006776). Scale is similar to that featured in panel B. CPM, exon counts per million; GO, gene ontology.
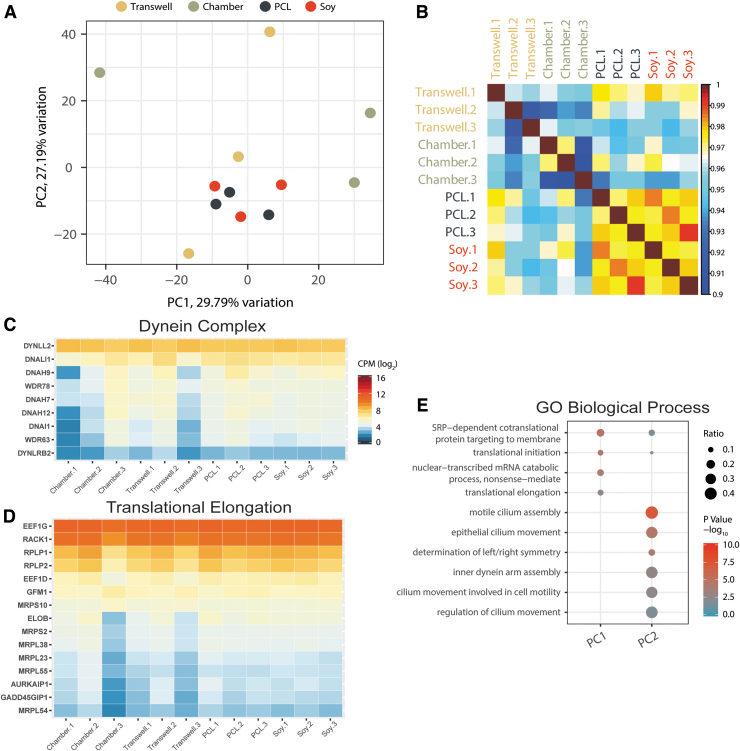
We did, however, observe broader differences in expression levels and batch-to-batch variation between the four conditions using RNA-seq (Fig. 8). Figure 8A shows a PCA using log2 CPM values from 12,674 genes from each sample. The PCA suggests smaller variance when the cells were grown on the nanofibrous scaffolds (PCL and soy), but substantially larger variance for the other two nonfibrous substrates (chamber and Transwell). This is complemented by a Pearson correlation of the log2 CPM values for each sample shown in Figure 8B. Figure 8C and D shows heatmaps of two examples of GO biological processes (dynein complex and translational elongation, respectively) that were identified to have the largest degrees of variability as per the PCA.
FIG. 8.
Variability in transcriptomes. (A) PCA using the log2 CPM values from 12,674 expressed genes. (B) Pearson correlation of each sample using the expressed gene log2 CPM values. “Transwell.1” is shown on the exterior to indicate similarities to the nanofibrous conditions. (C, D) Expression values of genes of two GO example pathways showing the highest batch variation, dynein complex (C) (GO:0030286) and translational elongation (D) (GO:0006414). Scale for both is equal to that shown in (C). (E) GO biological process enrichment for the top 500 genes that contribute to the factor loadings for each of the principal components 1 and 2. Replicates “1,” “2,” and “3” shown in (B), (C), and (D) indicate the first, second, and third batch replicates that were differentiated concurrently for each substrate type. PCA, principal component analysis.
Notably, the largest difference was observed between the “Chamber” and “Transwell” conditions across different batches. In contrast, variance in global transcriptional profiles in any of the “PCL” or “Soy” samples across different batches was much lower than for “Chamber” or “Transwell.” Figure 8E describes the top GO biological processes contributing to the variation observed in the principal components. The highest observed difference in expression was detected in genes associated with cilium assembly and RNA translation.
Discussion
Mechanical properties of the tissue environment constitute an important aspect of the cells' milieu, arising from distinct physical properties of cells and extracellular matrix components.39 Incorporation of bioengineered materials with appropriate mechanical and/or biochemical cues into cell therapy approaches promises to create permissive conditions for successful engraftment by maintaining key aspects of the cell niche. In the context of RPE transplantation, various substrates have been used to maintain the integrity of the cell sheets for implantation and recapitulate properties of the native Bruch's membrane. These include synthetic polymers,11,14,18 dissolvable collagen gels,8 and human amniotic membranes.17
Ideal materials for RPE transplantation should have structural and physical properties similar to those of Bruch's membrane, that is, provide enough mechanical support during the transplantation procedure, be biodegradable to allow the subsequent recreation of RPE-Bruch's membrane interface, and be sufficiently scalable for cost-effectiveness. While each of the materials used so far may present some advantageous properties, optimal substrates are likely yet to be identified.
Substantial disagreement persists in the assessment of the mechanical properties of Bruch's membrane in situ. Descriptions of the Young's modulus range from as low as 1 kPa40 to nearly 20 MPa,41 with some authors suggesting that it may not be measurable at all.42 Despite this variation, at least two authors have suggested that the true modulus across multiple species is likely in the single-digit MPa, with some indication that the stiffness increases from sub-MPa ranges during development.43–45 Using similar testing methodologies as in the current study, Wang et al. demonstrated that the average Young's modulus of the porcine Bruch's membrane/choroid complex is ∼1.60 MPa increasing to 2.44 MPa when lightly stressed.44 This is similar to the 0.644 to 2.5 MPa increase observed for the soy scaffolds in this study (Fig. 3A, C).
Furthermore, White et al. evaluated the response of RPE-like ARPE-19 cells on photopolymerized hydrogel scaffolds made of Poly(ethylene glycol) diacrylate with stiffnesses ranging from 0.06 to 1.2 MPa.46 They demonstrated that growing cells on scaffolds with higher Young's moduli resulted in significantly increased expression of IL-6 and IL-8 pro-inflammatory cytokines that are specifically associated with decreased implant efficacy. Taken together, these data suggest that soy scaffolds, with a lower Young's modulus value resembling that of native Bruch's membrane, may be more favorable for iRPE culture in vitro than other synthetic materials currently in use.
The importance of matching the biomechanical properties of implanted scaffolds and engineered tissues to their target environments has long been recognized for successful integration and long-term functionality.47–49 Of particular interest is the role that the stiffness of their surrounding matrix plays for differentiation and maturation of stem cells.50 While the Young's modulus of the soy scaffold more closely matches that of Bruch's membrane than PCL, this single parameter is not a comprehensive measure of the mechanical characteristics of the substrate types.
Changes in mechanic properties of scaffolds and fibers while being under mechanical loading is a critical feature of biological tissues. It is intrinsically linked to both external forces placed on the tissue and more relevant to tissues that experience limited loading by internal forces initiated by cells during migration, remodeling, or by strain originating from the cytoskeleton.51,52
While the PCL scaffolds exhibit a relatively constant Young's modulus with increasing load, the soy scaffolds show a nonlinear modulus that increases as more force is applied. This produces a “J-shaped” stress–strain curve that is a fundamental characteristic of biological materials.53 These differences suggest that the mechanical properties of the soy scaffolds may promote improved integration and clinical outcomes if translated to human implantation.
This difference in physical characteristics between the scaffolds also translates to differences in handling. Unsupported by larger structures, cell-laden PCL and soy scaffolds handle remarkably differently. The PCL exhibits physical characteristics similar to traditional plastic, resilient to deformation but requiring significant force to straighten or move. By contrast, soy scaffolds behave more like delicate connective fascia that become more rigid when stretched. In our hands, soy scaffold sheets were less brittle and were easier to handle, unfold, or reposition without risking damage to the epithelial sheet. Although future experiments will be required, we see indications that this difference may have contributed to the larger gaps between cells in the SEM images in Figure 5B and C (PCL and Transwell) versus Figure 5A (soy). This difference may translate to simpler implantation procedures if naturally derived materials such as soy find use in clinical applications.
Interestingly, substrates with lower hydrophobicity have been tied to decreased laminin adsorption, which in turn led to reduced expression of signature RPE markers.54 Decreased RPE marker expression was not observed with soy compared to PCL scaffolds despite the significantly lower hydrophobicity of soy scaffolds. We speculate that this may be due to the presence of lunasin, a polypeptide commonly found in soy isolate, that possesses the RGD-like motif common to both collagens and laminins.55,56
Lunasin peptides in soy scaffolds thus could engage cell surface integrins and compensate for reduced laminin adsorption and improve cell adherence. Because lunasin is present throughout the soy and covalently bound to the scaffold through crosslinking, this optimized binding is unlikely to decrease during degradation over long-term implantation and may provide further argument for its application as a Bruch's membrane analog for iRPE therapies.
Taken together with the aforementioned advantages of plant versus animal protein-based materials,22 soy scaffolds impart unique properties relevant to clinical translation that are superior to any other material type described in the literature. Notably, although the soy protein isolates are predominately composed of the major constitutive proteins glycinin and β-conglycinin, soy protein isolates contain a multitude of bioactive proteins, peptide fragments, and domains with unique properties.57 Future studies will be aimed at identifying the specific activities of these components and their applications in tissue engineering.
As in general with cells differentiated from hESCs or iPSCs, cross-comparison of the results between reports and laboratories differentiating and maturing iRPE remains challenging.58 For this reason, a panel of assays, including structural (electron microscopy), molecular (set of marker gene expression determined by quantitative polymerase chain reaction), and physiological (adenosine triphosphate-induced calcium signaling, electrical responses, and transepithelial fluid transport) characteristics, has been proposed to set standards for iRPE maturity, particularly with a view toward their future clinical applications.58,59
While key markers and functional assays are widely used to characterize and to control the quality of RPEs intended for cell therapy, whole transcriptome analysis has been used less widely for the assessment of the extent of iRPE differentiation.60,61 Key markers of RPE maturity, such as pigmentation and junction formation, are critical quality indicators. However, these alone do not provide comprehensive information about cell states. Likely many non-RPE specific genes may be important in the context of successful transplantation, such as changes in metabolic activity affecting long-term RPE-photoreceptor interactions.
Application of RNA-seq may facilitate the detection of other potentially detrimental variations due to batch variability, differences in maturation time points, or differences in the success of iPSC induction.62–64 Previous analyses of gene expression in RPE focused on the identification of distinct gene signatures in development and differentiation38,60,65 and have not assessed iRPEs grown on biomaterial scaffolds. By comparative transcriptome analysis of iRPE grown on scaffolds, we were able to evaluate not only the expression of key marker genes but also to identify batch-to-batch variability in gene expression profiles of iRPE cultured on the various substrates.
Performing the final maturation of iRPE monolayers on nanofibrous scaffolds appears to reduce overall variance in transcriptional profiles, suggesting that iRPE sheets grown on nanofibrous scaffolds may provide a more consistent cell product. This may be the result of clustering of focal adhesions more like in native tissues, enabling improved cytoskeletal arrangements. Taken together, we propose that whole transcriptome analysis represents a valuable quality control measure for cell products in translational research studies.
Intriguingly, no large-scale differences in gene expression profiles among iRPE grown on different substrates were found in RPE signature genes. Transcripts typical of RPE appear to be robustly expressed in all samples, consistent with efficient differentiation into the desired cell type. However, analysis of the highest variability genes revealed enrichment for processes related to ciliogenesis. A recent report identified signaling from primary cilia as important for RPE maturation.66 As cells exit the cell cycle, emerging primary cilia restrain the activity of canonical Wnt signaling, which in turn could promote RPE maturation.
Our results suggest that culturing iRPE on nanofibrous biomaterial scaffolds, whether synthetic or natural, allows a more uniform expression of ciliogenesis related genes, which may result in more consistent maturation when translated to clinical applications. Our data indicate that broader phenotypic changes may be present in our cultured iRPE than is revealed in the transcriptome. Future directions of this study will incorporate high-throughput proteomic profiling to present a more complete picture of the phenotypic differences between soy-based and synthetic scaffolds.
Conclusion
Our study describes a novel application of soy-based nanofibrous scaffold for the maturation of RPE differentiated from human iPSCs and applies RNA-sequencing to evaluate the quality of differentiation on the various substrates. We demonstrate that the soy-based scaffold can match or exceed the outcomes of commonly used synthetic standards while exhibiting superior mechanical and biochemical properties that may enhance integration and compatibility postimplantation. In addition, we find lower batch-to-batch variation in transcriptional profiles with culture on synthetic and nonsynthetic nanofibrous scaffolds. Our studies suggest that iRPE maturation on naturally derived soy scaffolds, which are mechanically softer, more compliant, and with favorable biochemical features, is likely to lead to improved clinical outcomes. These data warrant future in vivo experimentation with soy protein scaffolds in iRPE transplantation to evaluate the role of bioactive components of soy proteins in the improved outcomes.
Supplementary Material
Acknowledgments
The authors thank Dr. Michael Redmond, NIH NEI, for providing the generous gift of his custom RPE65 antibody and Mr. Tyler Pfister, NIH NEI (with Dr. Brian Brooks lab), for help with SEM processing. The authors are grateful to Dr. Kapil Bharti, NIH NEI, for helpful discussions and Dr. Joel Sheffield, Dept. Biology, CST, Temple University, for using their SEM equipment. The authors acknowledge Dr. Robert Fariss, NIH NEI, and Dr. Jennifer Kielczewski, NIH NEI, for assistance with confocal imaging. The authors also thank Mr. Tom Pohida, NIH CIT, and Dr. Nicole Morgan, NIH NIBI, for help developing the electrospinning methodology and Mr. Chris Panebianco, Ms. Vasudha Surampudi, and Dr. Dawn Henke, all NIH NEI, for helping with scaffold fabrication.
Disclosure Statement
No competing financial interests exist.
Funding Information
These studies were supported, in part, by grants from the Intramural Research Program of the National Eye Institute (ZIAEY000450 and ZIAEY000474), the Nation Eye Institute (ZIAEY000490), and the National Aeronautics and Space Administration (Grant # 80NSSC18K1480) and utilized the high-performance computational capabilities of the Biowulf Linux cluster at NIH (http://biowulf.nih.gov). PIL is the Laura H. Carnell Professor of Bioengineering at Temple University.
Supplementary Material
References
- 1. Ambati J., and Fowler B.J.. Mechanisms of age-related macular degeneration. Neuron 75, 26, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. da Cruz L., Chen F.K., Ahmado A., Greenwood J., and Coffey P.. RPE transplantation and its role in retinal disease. Prog Retin Eye Res 26, 598, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Ben M'Barek K., Habeler W., Regent F., and Monville C.. Developing cell-based therapies for RPE-associated degenerative eye diseases. In: Bharti K., ed. Pluripotent Stem Cells in Eye Disease Therapy. Cham: Springer International Publishing, 2019, p. 55. [DOI] [PubMed] [Google Scholar]
- 4. Stern J.H., Tian Y., Funderburgh J., et al. Regenerating eye tissues to preserve and restore vision. Cell Stem Cell 22, 834, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwartz S.D., Hubschman J.-P., Heilwell G., et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 379, 713, 2012 [DOI] [PubMed] [Google Scholar]
- 6. Schwartz S.D., Tan G., Hosseini H., and Nagiel A.. Subretinal transplantation of embryonic stem cell-derived retinal pigment epithelium for the treatment of macular degeneration: an assessment at 4 years. Invest Ophthalmol Vis Sci 57, ORSFc1, 2016 [DOI] [PubMed] [Google Scholar]
- 7. Lane A., Philip L.R., Ruban L., et al. Engineering efficient retinal pigment epithelium differentiation from human pluripotent stem cells. Stem Cells Transl Med 3, 1295, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Idelson M., Alper R., Obolensky A., et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell 5, 396, 2009 [DOI] [PubMed] [Google Scholar]
- 9. Buchholz D.E., Pennington B.O., Croze R.H., Hinman C.R., Coffey P.J., and Clegg D.O.. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl Med 2, 384, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maruotti J., Sripathi S.R., Bharti K., et al. Small-molecule-directed, efficient generation of retinal pigment epithelium from human pluripotent stem cells. Proc Natl Acad Sci U S A 112, 10950, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pennington B.O., Clegg D.O., Melkoumian Z.K., and Hikita S.T.. Defined culture of human embryonic stem cells and xeno-free derivation of retinal pigmented epithelial cells on a novel, synthetic substrate. Stem Cells Transl Med 4, 165, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Croze R.H., Buchholz D.E., Radeke M.J., et al. ROCK inhibition extends passage of pluripotent stem cell-derived retinal pigmented epithelium. Stem Cells Transl Med 3, 1066, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carr A.J., Vugler A.A., Hikita S.T., et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One 4, e8152, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. da Cruz L., Fynes K., Georgiadis O., et al. Phase 1 clinical study of an embryonic stem cell-derived retinal pigment epithelium patch in age-related macular degeneration. Nat Biotechnol 36, 328, 2018 [DOI] [PubMed] [Google Scholar]
- 15. Kamao H., Mandai M., Okamoto S., et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep 2, 205, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma R., Khristov V., Rising A., et al. Clinical-grade stem cell–derived retinal pigment epithelium patch rescues retinal degeneration in rodents and pigs. Sci Transl Med 11, eaat5580, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ben M'Barek K., Habeler W., Plancheron A., et al. Human ESC–derived retinal epithelial cell sheets potentiate rescue of photoreceptor cell loss in rats with retinal degeneration. Sci Transl Med 9, eaai7471, 2017 [DOI] [PubMed] [Google Scholar]
- 18. Kashani A.H., Lebkowski J.S., Rahhal F.M., et al. A bioengineered retinal pigment epithelial monolayer for advanced, dry age-related macular degeneration. Sci Transl Med 10, eaao4097, 2018 [DOI] [PubMed] [Google Scholar]
- 19. Falkner-Radler C.I., Krebs I., Glittenberg C., et al. Human retinal pigment epithelium (RPE) transplantation: outcome after autologous RPE-choroid sheet and RPE cell-suspension in a randomised clinical study. Br J Ophthalmol 95, 370, 2011 [DOI] [PubMed] [Google Scholar]
- 20. Hotaling N.A., Khristov V., Wan Q., et al. Nanofiber scaffold-based tissue-engineered retinal pigment epithelium to treat degenerative eye diseases. J Ocul Pharmacol Ther 32, 272, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White C.E., and Olabisi R.M.. Scaffolds for retinal pigment epithelial cell transplantation in age-related macular degeneration. J Tissue Eng 8, 2041731417720841, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Har-el Y.E., Gerstenhaber J.A., Brodsky R., Huneke R.B., and Lelkes P.I.. Electrospun soy protein scaffolds as wound dressings: enhanced reepithelialization in a porcine model of wound healing. Wound Med 5, 9, 2014 [Google Scholar]
- 23. Timnak A., Gerstenhaber J.A., Dong K., Har-El Y.E., and Lelkes P.I.. Gradient porous fibrous scaffolds: a novel approach to improving cell penetration in electrospun scaffolds. Biomed Mater 13, 065010, 2018 [DOI] [PubMed] [Google Scholar]
- 24. Ramji K., and Shah R.N.. Electrospun soy protein nanofiber scaffolds for tissue regeneration. J Biomater Appl 29, 411, 2014 [DOI] [PubMed] [Google Scholar]
- 25. Ahn S., Chantre C.O., Gannon A.R., et al. Soy protein/cellulose nanofiber scaffolds mimicking skin extracellular matrix for enhanced wound healing. Adv Healthc Mater 7, e1701175, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin L., Perets A., Har-el Y.E., et al. Alimentary ‘green’ proteins as electrospun scaffolds for skin regenerative engineering. J Tissue Eng Regen Med 7, 994, 2013 [DOI] [PubMed] [Google Scholar]
- 27. Barkay-Olami H., and Zilberman M.. Novel porous soy protein-based blend structures for biomedical applications: microstructure, mechanical, and physical properties. J Biomed Mater Res B Appl Biomater 104, 1109, 2016 [DOI] [PubMed] [Google Scholar]
- 28. Chien K.B., and Shah R.N.. Novel soy protein scaffolds for tissue regeneration: material characterization and interaction with human mesenchymal stem cells. Acta Biomater 8, 694, 2012 [DOI] [PubMed] [Google Scholar]
- 29. Webb K., Hlady V., and Tresco P.A.. Relative importance of surface wettability and charged functional groups on NIH 3T3 fibroblast attachment, spreading, and cytoskeletal organization. J Biomed Mater Res 41, 422, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McHugh K.J., Tao S.L., and Saint-Geniez M.. Porous poly(ɛ-caprolactone) scaffolds for retinal pigment epithelium transplantation. Invest Ophthalmol Vis Sci 55, 1754, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaya K.D., Chen H.Y., Brooks M.J., et al. Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal. Mol Vis 25, 663, 2019 [PMC free article] [PubMed] [Google Scholar]
- 32. Beers J., Linask K.L., Chen J.A., et al. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci Rep 5, 11319, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fanelli G., Gonzalez-Cordero A., Gardner P.J., et al. Human stem cell-derived retinal epithelial cells activate complement via collectin 11 in response to stress. Sci Rep 7, 14625, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swaroop M., Brooks M.J., Gieser L., Swaroop A., and Zheng W.. Patient iPSC-derived neural stem cells exhibit phenotypes in concordance with the clinical severity of mucopolysaccharidosis I. Hum Mol Genet 27, 3612, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brooks M.J., Chen H.Y., Kelley R.A., et al. Improved retinal organoid differentiation by modulating signaling pathways revealed by comparative transcriptome analyses with development in vivo. Stem Cell Rep 13, 891, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bray N.L., Pimentel H., Melsted P., and Pachter L.. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525, 2016 [DOI] [PubMed] [Google Scholar]
- 37. Soneson C., Love M.I., and Robinson M.D.. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strunnikova N.V., Maminishkis A., Barb J.J., et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet 19, 2468, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wells R.G. The role of matrix stiffness in regulating cell behavior. Hepatology 47, 1394, 2008 [DOI] [PubMed] [Google Scholar]
- 40. Fisher R.F. The influence of age on some ocular basement membranes. Eye 1, 184, 1987 [DOI] [PubMed] [Google Scholar]
- 41. Chan W.H., Hussain A.A., and Marshall J.. Youngs modulus of Bruchs membrane: implications for AMD. Invest Ophthalmol Vis Sci 48, 2187, 2007 [Google Scholar]
- 42. Friedman E. Update of the vascular model of AMD. Br J Ophthalmol 88, 161, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Candiello J., Balasubramani M., Schreiber E.M., et al. Biomechanical properties of native basement membranes. FEBS J 274, 2897, 2007 [DOI] [PubMed] [Google Scholar]
- 44. Wang X., Teoh C.K.G., Chan A.S.Y., Thangarajoo S., Jonas J.B., and Girard M.J.A.. Biomechanical properties of Bruch's membrane-choroid complex and their influence on optic nerve head biomechanics. Invest Ophthalmol Vis Sci 59, 2808, 2018 [DOI] [PubMed] [Google Scholar]
- 45. McKee C.T., Last J.A., Russell P., and Murphy C.J.. Indentation versus tensile measurements of young's modulus for soft biological tissues. Tissue Eng B Rev 17, 155, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. White C., DiStefano T., and Olabisi R.. The influence of substrate modulus on retinal pigment epithelial cells. J Biomed Mater Res A 105, 1260, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Discher D.E., Janmey P., an dWang Y.-l.. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139, 2005 [DOI] [PubMed] [Google Scholar]
- 48. Baiguera S., Urbani L., and Del Gaudio C.. Tissue engineered scaffolds for an effective healing and regeneration: reviewing orthotopic studies. Biomed Res Int 2014, 398069, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Badylak S.F., Freytes D.O., and Gilbert T.W.. Extracellular matrix as a biological scaffold material: structure and function. Acta Biomater 5, 1, 2009 [DOI] [PubMed] [Google Scholar]
- 50. Discher D.E., Mooney D.J., and Zandstra P.W.. Growth factors, matrices, and forces combine and control stem cells. Science (New York, NY) 324, 1673, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han Y.L., Ronceray P., Xu G., et al. Cell contraction induces long-ranged stress stiffening in the extracellular matrix. Proc Natl Acad Sci U S A 115, 4075, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guilak F., Butler D.L., Goldstein S.A., and Baaijens F.P.T.. Biomechanics and mechanobiology in functional tissue engineering. J Biomech 47, 1933, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu B., Rollo B., Stamp L.A., et al. Non-linear elasticity of core/shell spun PGS/PLLA fibres and their effect on cell proliferation. Biomaterials 34, 6306, 2013 [DOI] [PubMed] [Google Scholar]
- 54. Surrao D.C., Greferath U., Chau Y.Q., et al. Design, development and characterization of synthetic Bruch's membranes. Acta Biomater 64, 357, 2017 [DOI] [PubMed] [Google Scholar]
- 55. Cam A., and de Mejia E.G.. RGD-peptide lunasin inhibits Akt-mediated NF-κB activation in human macrophages through interaction with the αVβ3 integrin. Mol Nutr Food Res 56, 1569, 2012 [DOI] [PubMed] [Google Scholar]
- 56. Shidal C., Inaba J.-I., Yaddanapudi K., and Davis K.R.. The soy-derived peptide Lunasin inhibits invasive potential of melanoma initiating cells. Oncotarget 8, 25525, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chatterjee C., Gleddie S., and Xiao C.-W.. Soybean bioactive peptides and their functional properties. Nutrients 10, 1211, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miyagishima K.J., Wan Q., Miller S.S., and Bharti K.. A basis for comparison: sensitive authentication of stem cell derived RPE using physiological responses of intact RPE monolayers. Stem Cell Transl Investig 4, e1497, 2017 [PMC free article] [PubMed] [Google Scholar]
- 59. Miyagishima K.J., Wan Q., Corneo B., et al. In pursuit of authenticity: induced pluripotent stem cell-derived retinal pigment epithelium for clinical applications. Stem Cells Transl Med 5, 1562, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liao J.L., Yu J., Huang K., et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum Mol Genet 19, 4229, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brandl C., Zimmermann S.J., Milenkovic V.M., et al. In-depth characterisation of Retinal Pigment Epithelium (RPE) cells derived from human induced pluripotent stem cells (hiPSC). Neuromolecular Med 16, 551, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Volpato V., Smith J., Sandor C., et al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: A multi-site Omics study. Stem Cell Rep 11, 897, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morrison G., Liu C., Wing C., Delaney S.M., Zhang W., and Dolan M.E.. Evaluation of inter-batch differences in stem-cell derived neurons. Stem Cell Res 16, 140, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun C., Zhang J., Zheng D., Wang J., Yang H., and Zhang X.. Transcriptome variations among human embryonic stem cell lines are associated with their differentiation propensity. PLoS One 13, e0192625, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Z., Zhang Y., Xiao H., Liang X., Sun D., and Peng S.. A gene expression profile of the developing human retinal pigment epithelium. Mol Vis 18, 2961, 2012 [PMC free article] [PubMed] [Google Scholar]
- 66. May-Simera H.L., Wan Q., Jha B.S., et al. Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep 22, 189, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.