In this review, Rambout and Maquat discuss known roles of the nuclear cap-binding complex (CBC) during the transcription of genes that encode proteins, stitching together past studies from diverse groups to describe the continuum of CBC-mediated checks and balances in eukaryotic cells.
Keywords: CBP80, CBP20, CBC, transcription preinitiation complex, transcription initiation, promoter-proximal pausing, transcription elongation, RNA 3′ end formation, transcription termination, pre-mRNA splicing, alternative splicing
Abstract
The largely nuclear cap-binding complex (CBC) binds to the 5′ caps of RNA polymerase II (RNAPII)-synthesized transcripts and serves as a dynamic interaction platform for a myriad of RNA processing factors that regulate gene expression. While influence of the CBC can extend into the cytoplasm, here we review the roles of the CBC in the nucleus, with a focus on protein-coding genes. We discuss differences between CBC function in yeast and mammals, covering the steps of transcription initiation, release of RNAPII from pausing, transcription elongation, cotranscriptional pre-mRNA splicing, transcription termination, and consequences of spurious transcription. We describe parameters known to control the binding of generic or gene-specific cofactors that regulate CBC activities depending on the process(es) targeted, illustrating how the CBC is an ever-changing choreographer of gene expression.
The nuclear cap-binding complex (CBC), which is a heterodimer conserved from Saccharomyces cerevisiae to Homo sapiens, is composed of two cap-binding proteins (CBPs). CBP20 directly binds the m7G cap at the 5′ end of RNA polymerase II (RNAPII)-synthesized transcripts, while CBP80 stabilizes the binding of CBP20 to the cap and serves as an interaction platform for numerous factors that control virtually every step of gene expression (Gonatopoulos-Pournatzis and Cowling 2014; Müller-McNicoll and Neugebauer 2014). RNAPII-synthesized transcripts bound by the CBC include precursor and processed mRNAs, long noncoding RNAs (lncRNAs), promoter upstream transcripts (PROMPTs), enhancer RNAs (eRNAs), immature small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) of intergenic origin, and primary-microRNAs (pri-miRNAs). Whereas our lab discovered and contributed to elucidating the role of the CBC in the cytoplasm during the pioneer round of translation and nonsense-mediated mRNA decay (NMD) (for review, see Maquat et al. 2010; Ryu and Kim 2017; Kurosaki et al. 2019), our current research focuses on the role of the CBC during gene transcription by RNAPII (Cho et al. 2018). Here, we review known roles of the CBC in the nucleus during the transcription of genes that encode proteins, stitching together past studies from diverse groups to describe the continuum of CBC-mediated checks and balances in eukaryotic cells.
Chromatin-associated steps in the synthesis and processing of protein-coding transcripts
The transcription of eukaryotic protein-coding genes is a stepwise process that can be divided into fundamental stages, all of which are regulated by the CBC: preinitiation complex assembly, transcription initiation, promoter-proximal pausing, processive transcription elongation, and transcription termination coupled to pre-mRNA 3′ end processing. The process of pre-mRNA splicing, being largely cotranscriptional, is also presented in this review.
Preinitiation complex assembly
The first step of gene transcription is assembly at the core promoter of a preinitiation complex (PIC) composed of RNAPII and general transcription factors (GTFs). PIC assembly is positively and negatively controlled by a plethora of transcription factors and cofactors as well as chromatin remodelers that regulate promoter accessibility (Eychenne et al. 2017; Nogales et al. 2017).
Transcription initiation
Transcription cofactors subsequently convey activating cues leading to phosphorylation of the RNAPII carboxy-terminal domain (CTD) at serine 5 (S5) and S7 by the cyclin-dependent kinase 7 (CDK7) subunit of the GTF TFIIH (Haberle and Stark 2018). CTD phosphorylation enables RNAPII to escape the promoter and begin pre-mRNA synthesis; i.e., to initiate transcription. Despite being essential to the process of gene transcription in both lower and higher eukaryotes (Schier and Taatjes 2020), PIC assembly and transcription initiation are preferentially targeted for regulating gene expression in lower eukaryotes, such as the budding yeast S. cerevisiae (Hahn and Young 2011), while promoter-proximal pausing has emerged as the early step that largely regulates gene expression in metazoans (Adelman and Lis 2012).
Promoter-proximal pausing and pre-mRNA capping
In higher eukaryotes, RNAPII often pauses 25–50 bp downstream from the transcription start site (TSS), awaiting additional signals to engage in processive transcription (Core and Adelman 2019). As the nascent transcript emerges from the exit channel of RNAPII, RNAPII stalling is induced by recruitment of (1) the two-subunit DSIF (dichloro-1-β-D-ribofuranosylbenzimidazole [DRB] sensitivity-inducing factor) complex composed of SPT4 and SPT5, and (2) the four-subunit NELF (negative elongation factor) complex composed of NELF-A, NELF-B, either the NELF-C or NELF-D isoform of the NELFCD gene, and NELF-E. Particularly relevant to this review, promoter-proximal pausing and accessibility of the 5′ end of pre-mRNA as it emerges from the RNAPII exit channel coincide to allow pre-mRNA 5′ end capping (Rasmussen and Lis 1993; Martinez-Rucobo et al. 2015) and CBC recruitment to the 5′ cap (Visa et al. 1996; Listerman et al. 2006; Glover-Cutter et al. 2008). Genome-wide recruitment of the CBC to 5′ caps is facilitated by NELF (Aoi et al. 2020), which directly binds the CBC via the NELF-E subunit (Narita et al. 2007).
Even though promoter-proximal pausing is less prevalent in lower eukaryotes, in S. cerevisiae, promoter-proximal accumulation of RNAPII can be observed following genetic inhibition of the first catalytic step of pre-mRNA capping (Lahudkar et al. 2014). Nonetheless, such pausing does not coincide with CBC recruitment, which occurs more than ∼110 bp downstream from the TSS (Lidschreiber et al. 2013).
Transcription elongation
Recruitment of the positive transcription elongation factor b (P-TEFb) to promoters, so that its catalytic subunit CDK9 phosphorylates the DSIF and NELF pausing complexes and S2 of the RNAPII CTD, releases RNAPII from promoter-proximal pausing (Core and Adelman 2019). RNAPII then engages in processive transcription elongation, which is further established by the kinase activity of CDK12 (Chirackal Manavalan et al. 2019).
Transcription elongation can be interrupted by additional pausing events. As one important example, RNAPII can pause at stable first nucleosomes (i.e., +1 nucleosomes) in mammals (Barski et al. 2007; Chiu et al. 2018; Aoi et al. 2020), in fruit fly (Mavrich et al. 2008; Weber et al. 2014), and in the fission yeast Schizosaccharomyces pombe (Booth et al. 2016). Like promoter-proximal pausing, release from +1 nucleosome pausing relies on P-TEFb (Booth et al. 2018; Chiu et al. 2018). However, pausing at +1 nucleosomes is more transient (t1/2 < 15 sec) than pausing at promoter-proximal regions (t1/2 = 2–30 min) (Core and Adelman 2019). At least in mammals, +1 nucleosomes feedback to promoter-proximal pausing (Jimeno-González et al. 2015) as well as pre-mRNA splicing and early transcription termination (Chiu et al. 2018). On one hand, stable +1 nucleosomes enhance promoter-proximal pausing by augmenting NELF recruitment (Jimeno-González et al. 2015). On the other hand, RNAPII pausing at +1 nucleosomes located downstream from CpG islands has been proposed to cause premature transcription termination in cases where the U1 small nuclear ribonucleoprotein (snRNP) fails to recognize the 5′ splice site of the first intron (Chiu et al. 2018).
Pre-mRNA splicing
Consistent with extensive physical interactions between the transcription machinery and splicing factors, more than 75% of splicing events occur cotranscriptionally (Neugebauer 2019). Posttranscriptional events are more likely to characterize constitutive splicing of 3′-terminal introns and alternative splicing (Neugebauer 2019). Whereas cotranscriptional splicing kinetics may vary greatly between species (Neugebauer 2019), the core splicing machinery is globally well-conserved (Will and Lührmann 2011). First, 5′ splice site recognition by U1 snRNP, SF1 (BBP in yeast) binding to the branchpoint sequence, and U2AF (Mud2p in yeast) binding to the polypyrimidine tract and 3′ splice site result in formation of the early (E) complex (commitment complex in yeast). Second, U2 snRNP replaces SF1/BBP to form the A complex. Third, recruitment of the U4/U6•U5 tri-snRNP forms the B complex, which is subsequently extensively remodeled to form the B* complex that catalyzes the first splicing reaction. The C complex subsequently finalizes splicing, releasing the lariat intron.
Transcription termination and pre-mRNA 3′ end processing
Transcription termination, which occurs downstream from genes, is triggered by cleavage and polyadenylation specificity factor (CPSF) together with cleavage stimulation factor (CstF) recognition of a bipartite polyadenylation signal (PAS) in nascent pre-mRNA (Neve et al. 2017). Subsequent assembly of the complete 3′ end cleavage and polyadenylation (CPA) complex, which includes cleavage factors I and II (CFI and CFII), results in pre-mRNA cleavage and addition of a nontemplated poly(A) tail by poly(A) polymerase (PAP). RNAPII pausing during transcription termination, as well as the binding of proteins, can influence the selection of cryptic PASs that result in 3′ untranslated regions (3′UTRs) of different sizes and functions in a process called alternative polyadenylation (Bentley 2014).
While this cleavage and polyadenylation pathway typifies the majority of transcripts produced by protein-coding genes, replication-dependent histone (RDH) mRNAs are not polyadenylated. Instead, these short intronless transcripts, which are expressed during S phase of the cell cycle, typically end in a 3′ stem–loop structure (Marzluff et al. 2008). The most recent model characterizing 3′ end processing of RDH pre-mRNAs involves the recruitment of CPSF and the RNA endonucleolytic Integrator complex (Takahashi et al. 2020).
Transcription termination per se is the release of RNAPII from the gene following pre-mRNA 3′ end cleavage. Briefly, two not mutually exclusive models for termination prevail (Porrua and Libri 2015; Eaton et al. 2020). In the “allosteric model,” RNAPII undergoes conformational changes that result in its dissociation from the DNA template. In the “torpedo model,” 5′ to-3′ degradation of the 3′ cleavage product of the nascent transcript results in disassembly of the transcription elongation complex.
The CBC promotes transcription initiation and elongation
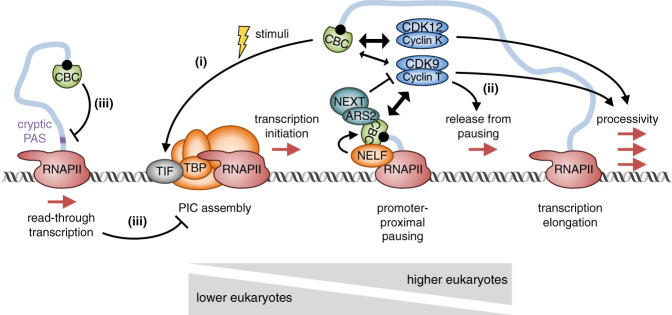
In S. cerevisiae, the CBC (yeast CBC [yCBC]) that is bound to the 5′ cap of elongating transcripts facilitates PIC assembly for subsequent rounds of transcription initiation, particularly at promoters induced by environmental changes, for example, promoters induced by glucose (Lahudkar et al. 2011) or osmotic shock (Fig. 1, step i; Li et al. 2016). CBC-mediated PIC formation and, subsequently, transcription initiation rely on the direct interaction of Cbp20 protein (Cbp20p) and/or Cbp80p with gene-specific transcription initiation factors (TIFs), such as Mot1p (Lahudkar et al. 2011), Hot1p–Hog1p (Li et al. 2016), or Rap1p (Li et al. 2016). These TIFs are recruited to the core promoters of target genes and are thought to promote transcription initiation through the subsequent recruitment of TATA-box-binding protein (TBP), a component of the GTF TFIID. Thus, yCBC-mediated transcription initiation may be limited to genes whose promoter includes sequences recognized by transcription factors or cofactors that also bind Cbp20p and/or Cbp80p. Likewise, in the model plant Arabidopsis thaliana, the CBC (plant CBC [pCBC]) transactivates the flowering gene FLC through the coactivator FRIGIDA (FRI) (Bezerra et al. 2004; Geraldo et al. 2009), which promotes FLC transcription by recruiting the GTF TFIID subunit TAF14 as well as chromatin modifiers (Choi et al. 2011). A role for pre-mRNA capping in transcription reinitiation in budding yeast is supported by the finding that capping enzymes Cet1p–Ceg1p and Abd1p promote PIC reassembly following promoter escape in an in vitro-reconstituted transcription initiation system (Fujiwara et al. 2019).
Figure 1.
Model for the regulation of transcription initiation and elongation by the CBC. (Step i) In budding yeast and plants, upon stimulation (yellow lightning), the CBC bound to the 5′ cap (black ball) of an elongating transcript (blue line) promotes the recruitment of transcription initiation factors (TIFs) and TATA-box-binding protein (TBP)-like factors to the promoter, thereby facilitating preinitiation complex (PIC) assembly and transcription reinitiation. (Step ii) The CBC recruits the elongation factors CDK9 and CDK12 to release RNAPII from promoter-proximal and/or +1 nucleosome pausing (in mammals) and/or increase RNAPII processivity (in yeast and mammals). (Step iii) In budding yeast, the CBC promotes read-through transcription by inhibiting the selection of cryptic polyadenylation signals (PASs), thereby inhibiting the transcription of downstream genes. Wedges at the bottom of the figure represent the preferred mode of transcriptional regulation in lower versus higher eukaryotes.
In Drosophila melanogaster, knockdown (KD) of Cbp80 decreases total RNAPII occupancy at the promoter of inducible genes while increasing occupancy of initiating RNAPII(pS5) relative to elongating RNAPII(pS2) (Kachaev et al. 2019). These results indicate that fruit fly Cbp80 not only promotes transcription (re)initiation but also facilitates release from promoter-proximal pausing. Such coupling is proposed to be mediated by the interaction of Cbp80 with the poly(A)-binding protein-interacting protein 2 (Paip2) and the TBP-related factor Trf2 (Kachaev et al. 2019). Interestingly, Cbp80-mediated expression of the fruit fly DHR3 gene following ecdysone treatment is associated with recruitment of Paip2 to only one of many alternative DHR3-gene promoters that is specifically induced by ecdysone (Kachaev et al. 2019), raising the possibility that the CBC also participates in selecting alternative promoters through its interaction with Paip2. Here again, this mechanism may depend on promoter sequences that are targeted by the binding of yet-to-be identified transcription factors to the CBC and/or to Paip2.
In humans, NELF-E directly binds CBP80 in vitro in a cap-dependent manner (Schulze and Cusack 2017), each NELF subunit coimmunoprecipitates with CBP80 in cell lysates (Gebhardt et al. 2015)—with at least the NELF-E–CBP80 coimmunoprecipitation (co-IP) being partially resistant to RNase A treatment (Narita et al. 2007)—and NELF-C is required for the recruitment of CBP80 to cellular promoters (Aoi et al. 2020). Loss of CBP80 at promoter regions upon NELF-C depletion is concomitant with (1) genome-wide shifting of RNAPII(pS2) occupancy from promoter-proximal pause sites to downstream pause sites that seem to be associated with +1 nucleosomes, and (2) decreased RNAPII(pS2) occupancy on gene bodies (Aoi et al. 2020). Results from precision nuclear run-on sequencing (PRO-seq) that mapped genome-wide occupancy of elongating RNAPII may indicate that RNAPII pausing at +1 nucleosomes is stabilized upon loss of the CBC (Aoi et al. 2020). Results from PRO-cap that measured the abundance of 5′ capped nascent transcripts together with results from ChIP-Seq that mapped occupancy on DNA for the decapping enzyme DCP2 and the 5′-to-3′ exonuclease XRN2 suggest that RNAPII pausing at +1 nucleosomes upon loss of the CBC is followed by pre-mRNA decapping by DCP2 and, subsequently, pre-mRNA decay by XRN2 (Aoi et al. 2020). The idea that the CBC protects pre-mRNAs from 5′ end surveillance and cotranscriptional decay is not new (Gonatopoulos-Pournatzis and Cowling 2014).
Human cell studies demonstrated that CBP80 and CBP20 coimmunoprecipitate with the CDK9 and cyclin T1 subunits of P-TEFb in the presence of RNase A, and also recruit CDK9 to promoter-proximal as well as distal regions of target genes (Lenasi et al. 2011). Impairment of P-TEFb recruitment following CBP20-KD or CBP80-KD is associated with lower levels of elongating RNAPII(pS2) on promoter-proximal and/or coding regions of a subset of target genes concomitantly with higher levels of short initiating transcripts (Lenasi et al. 2011). Thus, the CBC appears to control release from promoter-proximal pausing in a gene-specific manner, possibly through interactions with sequence-specific transcription factors or cofactors. Notably, similar observations were made using a CBC-responsive intronless reporter (Lenasi et al. 2011), arguing against the possibility that the decrease in transcription elongation following CBC-KD is an indirect consequence of defects in pre-mRNA splicing (see below), even though P-TEFb has also been implicated in pre-mRNA splicing (Barboric et al. 2009).
Together, these results indicate that, in humans, NELF and P-TEFb control release from promoter-proximal pausing and processive elongation in a CBC-dependent manner (Fig. 1, step ii). Thus, processive elongation would occur only if the nascent pre-mRNA is properly capped and the cap is subsequently bound by the CBC, consistent with the idea that promoter-proximal pausing not only functions as a regulator of gene expression, but also as a pre-mRNA surveillance mechanism (Rambout et al. 2020). Positive effects of the CBC on transcription elongation may be inhibited by the key CBC cofactors arsenite resistance protein 2 (ARS2) and the nuclear exosome targeting (NEXT) complex (see below), whose individual KD globally increases RNAPII occupancy in gene bodies relative to promoter-proximal pause sites (Iasillo et al. 2017). This function of ARS2 and NEXT is likely independent of their role in the cotranscriptional 3′-to-5′ decay of aberrant transcripts (see below), since KD of the nuclear exosome subunit RRP40 did not alter RNAPII occupancy (Iasillo et al. 2017).
Despite the lack of evidence for promoter-proximal pausing and the absence of NELF orthologs in S. cerevisiae (Lidschreiber et al. 2013), the yCBC similarly recruits Bur1p and Bur2p, orthologs of mammalian P-TEFb subunits CDK9 and cyclin T (Bartkowiak et al. 2010), to promoter-proximal regions and, depending on the gene analyzed, also to the central region and 3′ end of genes (Hossain et al. 2013; Lidschreiber et al. 2013). Additionally, the yCBC genetically interacts with and recruits Ctk1p and Ctk2p, but only to gene bodies (Hossain et al. 2013; Lidschreiber et al. 2013). Ckt1p and Ckt2p are orthologs of mammalian CDK12 and cyclin K, respectively (Bartkowiak et al. 2010). yCBC-mediated recruitment of the Bur and Ctk complexes correlates with sustained levels of elongating RNAPII(pS2) and phospho-CTD-dependent histone H3K36 trimethylation (H3K36me3), which is an epigenetic mark of active transcription (Hossain et al. 2013; Lidschreiber et al. 2013). Notably, effects of Cbp80p on RNAPII processivity are not universally evident (Wong et al. 2007), suggesting confinement to specific genes and/or contexts. Gene-specific Cbp80p-dependent control of RNAPII elongation rates may rely on intragenic features, possibly in conjunction with DNA-binding transcription factors and/or RNA-binding proteins, rather than or in addition to promoter features (Rambout et al. 2018).
The CBC promotes constitutive splicing of first introns at different steps of spliceosome assembly, depending on the intron
Historically, the first reported molecular function of the CBC pertained to its contribution to the splicing of an in vitro-transcribed and m7G capped single intron-containing pre-mRNA in nuclear extracts of human cells (Izaurralde et al. 1994). These findings were later extended to budding yeast (Colot et al. 1996; Lewis et al. 1996a) and plants (Kuhn et al. 2007) using similar in vitro experimental designs as well as in vivo approaches. Generally speaking, the CBC preferentially promotes cotranscriptional splicing of first introns (Görnemann et al. 2005; Bittencourt et al. 2008) characterized by low splicing efficiencies (Colot et al. 1996; Laubinger et al. 2008) independently, at least to some extent, of the length of the intron or its position relative to the 5′ cap (Lewis et al. 1996b; Qiu et al. 2007; Hallais et al. 2013). Several molecular mechanisms underlying CBC-dependent control of pre-mRNA splicing have been proposed, the relative prevalence of which is under debate. However, they appear to depend on the particular gene rather than the organism under study. We review below proposed mechanisms.
Initial studies indicated that the CBC promotes first intron removal by facilitating recognition of the 5′ splice site by U1 snRNP and, subsequently, formation of the E complex in the case of humans (Lewis et al. 1996b) or the corresponding commitment complex in the case of budding yeast (Fig. 2, step i; Colot et al. 1996; Lewis et al. 1996a; Fortes et al. 1999b; Görnemann et al. 2005). In yeast, yCBC-dependent recruitment of U1 snRNP possibly relies on physical and genetic interactions between the yCBC and the U1 snRNP protein Luc7p (Fortes et al. 1999a). Intriguingly, despite conserved protein–protein interactions between yeast and plants, while pCBC preferentially targets first introns, plant LUC7 preferentially targets last introns (de Francisco Amorim et al. 2018). Nonetheless, first introns that are partially retained in pCBC mutants are completely spliced out when LUC7 is mutated (de Francisco Amorim et al. 2018), indicating that LUC7 counteracts pCBC function at first introns whose splicing is inefficient (de Francisco Amorim et al. 2018). In humans, the accessory CBC protein NCBP3 (see below), which does not exist in plants or yeast, directly interacts with LUC7L2 (Das and Yu 2012), one of three human orthologs of yeast Luc7p (Hosono 2019). This suggests that some type of complicated interplay between the CBC and LUC7 proteins also exists in mammals.
Figure 2.
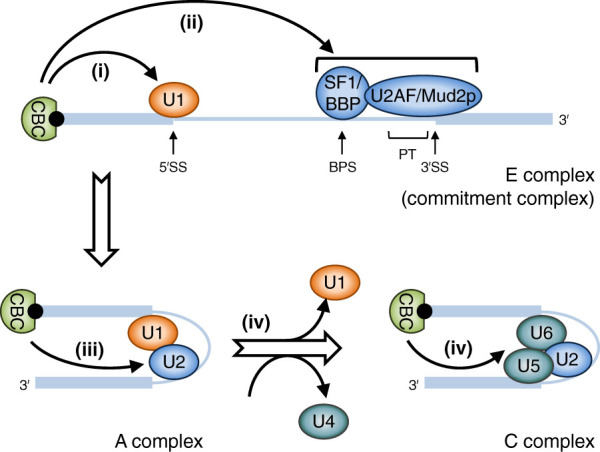
Model for the CBC-dependent removal of first introns. (Step i) The CBC facilitates the recruitment of U1 snRNP (U1) to the 5′ splice site (5′SS) of the first intron. (Step ii) The CBC promotes recruitment of the BBP–Mud2p (yeast)/SF1–U2AF (mammals) complex to the 3′ end of the first intron, which is comprised of the branchpoint sequence (BPS), the polypyrimidine tract (PT), and the 3′ splice site (3′SS). (Step iii) The CBC promotes recruitment of U2 snRNP (U2) to the 3′ end of the first intron. (Step iv) The CBC catalyzes the replacement of U1 snRNP by U6 snRNP (U6) at the 3′ splice site. Thick horizontal blue lines represent the first and second exons, and the intervening thin blue line represents the first intron.
While the CBC-dependent U1 snRNP recruitment model is supported by numerous independent studies, in vivo assays indicate that, at least in yeast, it may be true for only a limited number of genes (Görnemann et al. 2005). At least four additional mechanisms for how the CBC promotes gene expression through pre-mRNA splicing have been proposed. One mechanism may prevail over others depending on the particular gene; for example, 5′ splice site characteristics (Bragulat et al. 2010) or, as discussed above, intron length or intron distance from the 5′ cap, as well as promoter features.
In one, the yCBC may complete formation of the commitment complex through its physical and genetic interaction with the branchpoint recognition complex BBP–Mud2p (orthologous to mammalian SF1–U2AF65) (Fig. 2, step ii; Fortes et al. 1999b; Qiu et al. 2012). In a second mechanism, the yCBC appears to facilitate U2 snRNP recruitment, presumably to the branchpoint so as to promote formation of the A complex (Fig 2, step iii; Bragulat et al. 2010). Possibly supporting the idea that this second mechanism is conserved in mammals, the splicing function of the human CBC can be functionally replaced by the expression of serine-arginine-rich (SR) proteins, members of a family of splicing factors that bridge 5′ and 3′ splice sites (Lewis et al. 1996b). In a third mechanism, both yeast and human CBCs support splicing by promoting cotranscriptional replacement of U1 snRNP by U6 snRNP, which occurs concomitantly with recruitment of the U4/U6•U5 tri-snRNP during B complex formation that is subsequently remodeled in the C complex (Fig 2, step iv; O'Mullane and Eperon 1998; Görnemann et al. 2005). This scenario may constitute the most prevalent role of the CBC in pre-mRNA splicing in yeast and mammals. In yet another mechanism, a study using human cells proposed that the CBC recruits U1 snRNP to 5′ splice sites via the U4/U6•U5 tri-snRNP, whose many subunits robustly coimmunoprecipitate with CBP80 in the presence of RNase A (Pabis et al. 2013).
In A. thaliana, the pCBC teams up with SERRATE (SE), the ortholog of mammalian ARS2 (see below), and the plant-specific cap-associated protein FRI to support intron 1 splicing (Laubinger et al. 2008; Geraldo et al. 2009). Other regulators of CBC-dependent splicing include the tumor suppressor and NMD inhibitor MARVELD1 in humans (Hu et al. 2013; Wang et al. 2013; Shi et al. 2014) and ubiquitinated histone H2B in budding yeast (Hérissant et al. 2014).
As expected, the ability of the CBC to influence splicing can be altered by posttranslational modifications that regulate its affinity for the 5′ cap. For example, addition of GTP to CBP20 appears to recruit the GTP-binding protein Cdc42 (Wilson et al. 1999, 2000). Cdc42 in turn activates Cdc42-activated S6 kinase RPS6KB1, which phosphorylates CBP80 so as to increase CBC binding to the 5′ cap of a reporter pre-mRNA, thereby increasing its splicing (Wilson et al. 1999, 2000).
Splicing, in particular of first introns, stimulates transcription elongation (Fong and Zhou 2001; Chathoth et al. 2014), U1 snRNP-dependent transcription reinitiation (Damgaard et al. 2008), H3K4 trimethylation (i.e., acquisition of H3K4me3, which is a chromatin mark of active transcription) (Bieberstein et al. 2012), and selection of weak alternative promoters within the first intron of genes being transcribed (Fiszbein et al. 2019). Among genes characterized by CBC-regulated pre-mRNA splicing, some but not all are also regulated transcriptionally by the CBC in uncharacterized mechanisms (Geraldo et al. 2009; Lenasi et al. 2011; Pabis et al. 2013). Future studies that characterize genes based on the extent to which their transcriptional activation and pre-mRNA splicing efficiencies are mechanistically coupled, and whether coupling is mediated by transcriptional effects on splicing, splicing effects on transcription, or both, are sure to lend important information on how gene expression is constitutively and, possibly, conditionally controlled by the CBC.
The CBC regulates alternative splicing
In addition to facilitating constitutive splicing of first introns, the CBC regulates hundreds of alternative splicing events in human cells (Lenasi et al. 2011) and plants (Raczynska et al. 2010, 2014; Kanno et al. 2020). In humans, one proposed mechanism involves CBC-dependent recruitment of P-TEFb, phosphorylation of RNAPII S2, followed by the recruitment of the SR splicing factor 2/alternative splicing factor (SF2/ASF, now renamed SRSF1) on a splicing enhancer so as to promote exon inclusion (Lenasi et al. 2011). Whether CBC-p-TEFb-dependent control of RNAPII elongation rates contributes to alternative pre-mRNA splicing has not been directly tested. Nonetheless, gene-specific recruitment of P-TEFb by the CBC (Lenasi et al. 2011) could explain how the CBC regulates only a subset of alternative splicing events. Heterogeneous nuclear ribonucleoprotein F (hnRNPF) is another possible regulator of CBC-dependent control of alternative splicing (Gamberi et al. 1997). The possible involvement of sequence-specific RNA-binding proteins such as SRSF1 (Anczuków et al. 2015) or structure-specific RNA-binding protein such as hnRNPF (Huang et al. 2017) suggests that RNA features influence the CBC-dependent control of alternative pre-mRNA splicing.
RNA-seq analyses of 5-wk-old A. thaliana leaves knocked out for CBP80 or CBP20 (Raczynska et al. 2010) or expressing SE harboring a point mutation (Raczynska et al. 2014) indicate that the three proteins cooperate to regulate an overlapping set of alternative splicing events, primarily selecting alternative 5′ splice sites of first introns. In contrast, 2-wk-old A. thaliana seedlings expressing a CBP80 harboring a point mutation predominantly manifest intron retention (Kanno et al. 2020). The findings that the CBP80 knock-out plant and the CBP80 point mutant plant exhibit different defects on alternative splicing could be explained by different domains of CBP80 affecting different CBP80 functions and/or differences in the age and type of tissues analyzed.
Clearly, the role of the CBC in alternative splicing is largely understudied. In particular, it remains to be shown whether CBC function in alternative splicing is mechanistically connected to its well-characterized role in constitutive splicing, is influenced by transcription elongation, or involves interactions with dedicated alternative splicing factors. While indirect, the role of the CBC and/or its cofactor PHAX in the synthesis (Yeh et al. 2017) and nuclear export of snRNAs (Izaurralde et al. 1995; Ohno et al. 2000; Segref et al. 2001) also contributes to alternative pre-mRNA splicing (Garcia et al. 2016; Yeh et al. 2017).
The CBC promotes selection of proximal 3′ end cleavage sites of structured transcripts from intronless genes
Immunodepletion of CBP80 from human HeLa-cell nuclear extracts reduces the stability of the CPSF–CstF complex on in vitro-transcribed and capped intronless pre-mRNA (Flaherty et al. 1997). As a consequence, PAS cleavage of pre-mRNA is reduced by 75% in vitro, but the efficiency with which a precleaved pre-mRNA undergoes polyadenylation remains unaffected (Flaherty et al. 1997). A role for the CBC per se in pre-mRNA 3′ end cleavage was demonstrated by 3′ end cleavage being partially restored following the addition of recombinant CBP80 and CBP20 (Flaherty et al. 1997). Despite the cotranscriptional nature of RNA 3′ end processing, these results indicate that CBP80 requires neither ongoing transcription nor sequential cotranscriptional recruitment of cleavage factors to facilitate the cleavage step of 3′ end processing. Since the reporter used was intronless, these results also rule out that the CBC functions in 3′ end processing depend on its functions in pre-mRNA splicing.
CBP80-promoted PAS cleavage relies on the physical association of pre-mRNA 5′ and 3′ ends (Flaherty et al. 1997). Physical and functional interactions of CBP80 at the 5′ cap with CPSF–CstF at the 3′ end of transcripts are not direct and require additional cofactors (Flaherty et al. 1997). Characterized cofactors (Fig. 3) include NELF and ARS2, whose direct binding to the CBC are mutually exclusive (Schulze and Cusack 2017). Another cofactor is CFI, which coimmunoprecipitates with ARS2 in a partially RNase A-sensitive manner (Machitani et al. 2020) and is predicted to bind directly to CBP20 of the CBC via its CFIm25 small subunit (Yang et al. 2011).
Figure 3.
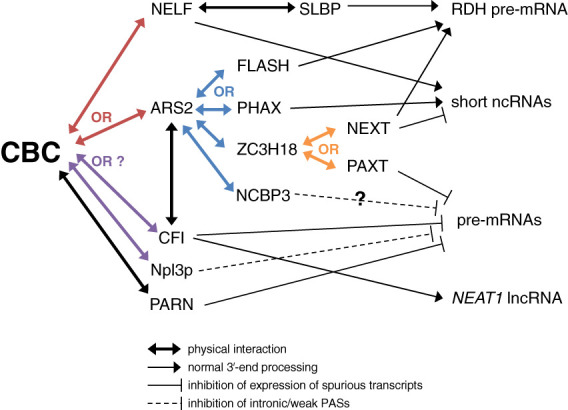
The CBC serves as a hub for many cofactors. Illustration of how the CBC teams up with a number of cofactors that often bind in mutually exclusive mechanisms to differentially regulate gene expression. Interactions and their effects on four types of transcripts are exemplified. The color of each “OR” specifies mutually exclusive interactions designated by lines with divergent arrowheads of the same color.
CBC–NELF
Data deriving from in vitro assays as well as chromatin immunoprecipitation (ChIP) and colocalization experiments using human HeLa cells have provided insight into CBC–NELF-dependent 3′ end processing of RDH pre-mRNAs (Narita et al. 2007). Results indicate that NELF-E connects 5′ cap-bound CBC and 3′ end-associated histone stem–loop-binding protein (SLBP) so as to process the 3′ end of RDH pre-mRNAs in specialized nuclear bodies that reside close to the associated RDH genes (Narita et al. 2007) and overlap with Cajal bodies (Takahashi et al. 2020). Depletion of CBP80, NELF-E, or SLBP results in use of a distal PAS and the accumulation of long and abnormal polyadenylated mRNA species (Narita et al. 2007). A model was proposed involving NELF recruitment to RDH gene promoters followed by the recruitment of the CBC to the 5′ caps and then SLBP to the 3′ stem–loop of RDH pre-mRNAs (Narita et al. 2007). This model implies that NELF does not dissociate from RNAPII following release from promoter-proximal pausing, which notably is not prevalent on RDH genes (Anamika et al. 2012).
CBC–ARS2
In both humans and mice, KD of the CBC cofactor ARS2 also results in the accumulation of polyadenylated RDH mRNAs, which it binds directly (Gruber et al. 2012; Andersen et al. 2013; Hallais et al. 2013; O'Sullivan et al. 2015). The mechanism by which the CBC–ARS2 complex (also referred to as CBCA) controls RDH mRNA processing involves the recruitment of either of at least two different processing factors to the proper 3′ cleavage site: (1) Fas-associated death domain (FADD)-like interleukin 1β (IL-1β)-converting enzyme (FLICE) associated with a huge protein (FLASH) (Hallais et al. 2013; Yang et al. 2013; O'Sullivan et al. 2015), or (2) the zinc finger protein ZC3H18, which directly binds ARS2 and also recruits NEXT (Andersen et al. 2013). Whereas the CBC–ARS2–FLASH complex is likely to be specific for the processing of RDH transcripts, the CBC–ARS2–NEXT complex (also referred to as CBCN) does likewise plus also promotes the maturation of human telomerase RNA (Tseng et al. 2015) and inhibits pervasive transcription (see below). Regardless of its cofactor, CBC–ARS2 stimulates 3′ end RDH pre-mRNA cleavage when the transcript 5′ cap and 3′ end are in close proximity, typically <0.5–1 kb apart (Hallais et al. 2013). This distance requirement may reflect gradual disruption or remodeling of the CBC–ARS2 complex during transcription elongation (Müller-McNicoll and Neugebauer 2014). In Drosophila, Ars2 does not bind FLASH and is dispensable for 3′ end processing of RDH pre-mRNAs (Sabath et al. 2013).
CBC–NELF- and/or CBC–ARS2-dependent processing of RDH pre-mRNAs is possibly reminiscent of the CBC-mediated 3′ end processing of pre-snRNAs (Müller-McNicoll and Neugebauer 2014; Takahashi et al. 2020). Like RDH transcripts, snRNAs are short, intronless, and highly structured transcripts that are not controlled by promoter-proximal pausing and not polyadenylated unless a downstream PAS is used (Guiro and Murphy 2017). 3′ end processing of RDH pre-mRNAs and pre-snRNAs appears to depend on the recruitment of CPSF and integrator via interactor of little elongation complex (LEC) ELL subunit 1 (ICE1) (Takahashi et al. 2020). Since ICE1 directly binds CBP80 and coimmunoprecipitates with the above-mentioned CBC cofactors (i.e., NELF, SLBP, ARS2, FLASH, and ZC3H18) in the presence of benzonase, a handoff model was proposed in which promoter-recruited LEC is escorted to 3′ end processing sites through the CBC (Takahashi et al. 2020).
Nonetheless, CBC–NELF- and CBC–ARS2-dependent processing of RDH pre-mRNAs differ in other ways. In humans, where CBC–NELF function is not associated with the slowing of RNAPII by NELF at the stem–loop-associated cleavage site (Narita et al. 2007), CBC–ARS2 function is reduced by a potent inhibitor of P-TEFb, the ncRNA 7SK (Gruber et al. 2012). This suggests that RNAPII elongation rates may play a role in CBC–ARS2- but not CBC–NELF-mediated 3′ end RNA processing. Adding to regulatory complexity, promoters may differ in their ability to assemble the mutually exclusive CBC–ARS2 and CBC–NELF complexes given that ARS2, but not NELF, is required for proper RDH pre-mRNA cleavage when a CMV promoter is used to drive gene expression (Hallais et al. 2013).
Through its association with CFI, ARS2 promotes the selection of a proximal PAS in the intronless lncRNA NEAT1, which otherwise ends with a triple-helix structure (Machitani et al. 2020). The 3′ end of both NEAT1 lncRNA isoforms is located several kilobases downstream from their common promoter (Machitani et al. 2020); that is, a distance at which CBC–ARS2 fails to regulate RDH 3′ end processing (Hallais et al. 2013). Therefore, ARS2-dependent mechanisms controlling the processing of short intronless RNAs such as RDH pre-mRNAs, and long intronless RNAs such as NEAT1 lncRNAs, are undoubtedly very different.
The CBC inhibits selection of cryptic or first intron PASs
In budding yeast, the yCBC controls alternative transcription termination by inhibiting the selection of weak polyadenylation sites (Figs. 3,4). As examples, genetic deletion of the Cbp80p-encoding gene CBC1 results in selection of the proximal-most of several cryptic PASs of cyc1-512 RNA (Das et al. 2000) and RNA14 RNA (Wong et al. 2007) as well as use of the weak PAS of mutated gal10-Δ56 RNA (Wong et al. 2007). In the cases of RNA14 and gal10-Δ56 RNAs, CBC binding to Npl3p, one of the most abundant hnRNPs in yeast (Shen et al. 2000), inhibits recruitment of the CFIA complex (the yeast ortholog of mammalian CFII) to the weak PAS (Wong et al. 2007). The RNA-binding protein Npl3p inhibits the selection of the weak PAS not by controlling RNAPII occupancy (Wong et al. 2007) but by competing with the polyadenylation/termination factors for PAS binding (Fig. 4A; Bucheli and Buratowski 2005). Since deletion of Cbp80p and Npl3p did not affect recruitment of the CFIA complex to the strong termination site of those genes tested, “antiterminator” functions of the CBC appear to apply only to weak PASs (Wong et al. 2007). Notably, Cbp80p-dependent inhibition of the weak PAS of mutated gal10-Δ56 RNA results in read-through transcription into the promoter of the downstream gene, therefore indirectly preventing PIC formation at that downstream promoter (Fig. 1, step iii; Wong et al. 2007).
Figure 4.
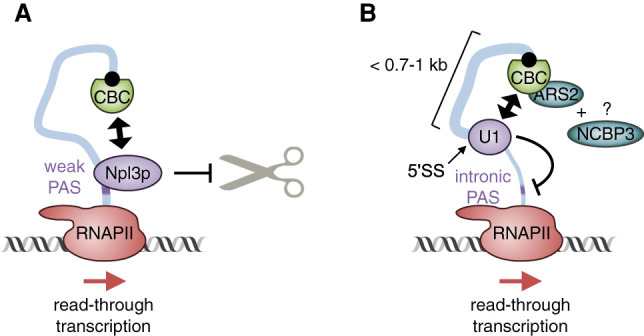
Models for the inhibition of weak polyadenylation sites by the CBC. (A) In budding yeast, the yCBC recruits Npl3p to weak PASs, thereby inhibiting CFIA binding and premature 3′ end processing. (B) In humans, CBC–ARS2 inhibits the selection of intronic PASs by facilitating the recruitment of U1 snRNP to the 5′ splice site of the first intron. We predict that this mechanism depends on the CBC–ARS2 cofactor NCBP3.
In human cells, the CBC inhibits transcription termination associated with intronic PASs in a mechanism involving competition between 3′ end processing and splicing (Fig. 4B). For example, tethering CBP20 or CBP80 to the first exon of adeno-associated virus type 5 RNA inhibits transcription termination within the first intron, presumably by inhibiting use of a cryptic PAS and promoting recognition and use of the 5′ splice site (Qiu et al. 2007). Along the same lines, mutation of the 5′ splice site of the first intron of NR3C1 pre-mRNA activates a cryptic intronic PAS in a CBP80- and ARS2-promoted manner (Hallais et al. 2013). Reminiscent of CBC–ARS2-dependent processing of RDH pre-mRNAs (see above), CBC–ARS2-dependent competition of 3′ end processing and splicing depends on the distance between the CBC-bound 5′ cap and the 5′ splice site, with a longer distance (>0.7–1 kb) inhibiting coupling (Qiu et al. 2007; Hallais et al. 2013). The CBC also represses the use of cryptic PASs in first introns in A. thaliana (Kuhn et al. 2007), but the underlying molecular mechanisms have not been investigated.
Reports converge toward a model in which CBC–ARS2 forms a complex with different factors depending on the targeted RNA species: CBC–ARS2–FLASH is at the 5′ cap of RDH transcripts (see above), CBC–ARS2–PHAX (also referred to as CBCAP) is at the cap of pre-snRNAs or other short RNA species, and CBC–ARS2–NCBP3 is at the 5′ cap of mRNAs and, to a lesser extent, lncRNAs (Gebhardt et al. 2015; Giacometti et al. 2017). Considering that neither FLASH nor PHAX inhibits the selection of intronic PASs of protein-coding genes (Hallais et al. 2013), we consider it likely that NCBP3 functions with CBC and ARS2 to do so (Fig. 3). Supporting this idea, like the CBC and ARS2, NCBP3 associates with U1 snRNP, which functions in a process called “telescripting“ (So et al. 2019). NCBP3 also associates with the transcription and export (TREX) complex (Gebhardt et al. 2015), which manifests numerous roles in gene expression that include pre-mRNA splicing and 3′ end processing (Heath et al. 2016).
RNA surveillance: the CBC suppresses the synthesis or promotes the degradation of spurious transcripts
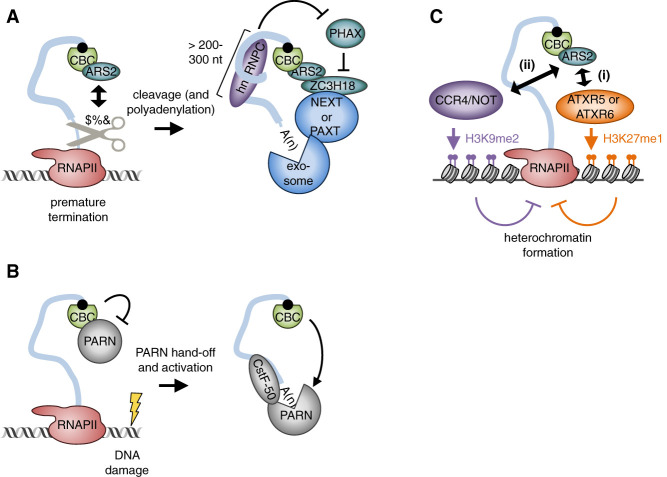
Several reports describe how CBC–ARS2 suppresses pervasive transcription of both short noncoding transcription units encoding mRNA-like PASs (e.g., 3′ extended snRNAs and PROMPTs) and protein-coding genes by acting on their pre-mRNAs at the first intron 5′ splice site in humans (Andersen et al. 2013; Iasillo et al. 2017) and possibly in yeast (Figs. 3, 5A; Colot et al. 1996). In one proposed mechanism, CBC–ARS2 recruits the 3′ end CFI cleavage factor CLP1 to promote cleavage of unwanted RNAs (Hallais et al. 2013). In another probably complementary mechanism, CBC–ARS2 recruits ZC3H18 that, following early transcription termination, mobilizes either the NEXT complex, in the case of normally short transcripts (<1 kb) (Lubas et al. 2011; Andersen et al. 2013; Giacometti et al. 2017), or the poly(A) tail exosome targeting (PAXT) complex, in the case of normally longer (up to 10–100 kb) and polyadenylated transcripts (Meola et al. 2016). Targeted RNAs are subsequently degraded by the nuclear exosome. Just as ARS2 competes with NELF for CBC binding (Fig. 3, red double arrows), ZC3H18 competes with the export factor PHAX for ARS2 binding (Fig. 3, blue double arrows), thereby determining the fate of the targeted RNA; i.e., nuclear decay or increased expression, the latter presumably via enhanced RNA 3′ end processing (Giacometti et al. 2017). This type of competition is not likely to be prevalent on pre-mRNAs >200–300 nt, which is the length of unstructured RNA wrapped around a heterotetramer of hnRNPC1/C2, because hnRNPC1/C2 inhibits PHAX binding to the CBC (McCloskey et al. 2012). The CBC–ARS2–ZC3H18 complex seems to be the functional ortholog of the yeast Nrd1–Nab3–Sen1 (NNS) complex, which also promotes early transcription termination in a “distance to cap”-dependent manner (Porrua and Libri 2015).
Figure 5.
Models for the inhibition of spurious transcription by the CBC. (A) CBC–ARS2 recruits the CFI constituent CLP1 to promote the cleavage of spurious transcripts. CBC–ARS2 then recruits the exosome via ZC3H18 and NEXT or PAXT so as to degrade the cleaved transcript. CBC–ARS2 binding to ZC3H18, and thus recruitment of the exosome, is inhibited by PHAX, whose binding to CBC–ARS2 is itself inhibited by hnRNPC1/C2, a molecular ruler that wraps strings of 200–300 nt of unstructured RNA. The CBC often prevents spurious transcription by acting on cap-proximal PASs typically found in first introns of pre-mRNAs. (B) The CBC hands-off PARN to CstF-50 at sites of DNA damage, thereby releasing from inhibition its deadenylase activity to initiate RNA decay. (C, step i) In plants, CBC–ARS2 (that is, pCBC-SE) recruits the H3K27 monomethyl (me1) transferases ATXR5 and ATXR6 to promote heterochromatin formation and inhibit spurious transcription of transposable elements. (Step ii) In fission yeast, yCBC–ARS2 promotes H3K9 dimethylation (me2) through its binding to CCR4–NOT, thereby promoting heterochromatin formation and inhibiting spurious transcription of retrotransposons and coding genes containing cryptic introns.
In mammalian cells, the CBC likewise prevents accumulation of early terminated mRNAs by depositing the poly(A)-specific ribonuclease (PARN) at sites of DNA damage following UV exposure (Figs. 3, 5B; Cevher et al. 2010). In a proposed model, 5′ cap-bound CBP80 hands-off PARN to 3′ end-associated CstF-50 following RNAPII stalling at sites of DNA damage (Cevher et al. 2010). While CBP80 binding to PARN inhibits PARN deadenylase activity (Balatsos et al. 2006), PARN becomes active when bound to CstF-50 so as to prevent polyadenylation and, eventually, initiate RNA decay in the nucleus (Cevher et al. 2010). While not the focus of this review, release of CBP80-mediated inhibition of PARN following DNA damage is also critical for the degradation of snoRNAs within nucleoli (Duan et al. 2019).
In plants and fission yeast, ARS2 shuts down spurious transcription by promoting heterochromatin formation (Fig. 5C). SE, the plant ortholog of ARS2, prevents transcription of transposable elements by recruiting the histone methyltransferases ATXR5 and ATXR6, thereby promoting H3K27 monomethylation (H3K27me1) and chromatin compaction (Ma et al. 2018). In S. pombe, the ARS2 ortholog Pir2 cooperates with the multifunctional CCR4–NOT complex to induce H3K9 dimethylation (H3K9me2)-mediated heterochromatin formation at retrotransposon loci and developmentally regulated genes containing cryptic introns (Sugiyama et al. 2016).
Alternatives to the cap-bound CBP20–CBP80 CBC
An important caveat to the interpretation of results we report and analyze in this review is that a significant number of publications limited their experimental design to the study of CBP80 or CBP20, assuming they do not exist independently of the CBC heterodimer at the 5′ cap of transcripts. For example, the finding that CBP80 and CBP20 are found at a 1:1 molar ratio in the B and C splicing complexes supports the idea that the CBC is the functional unit of CBP80- and CBP20-dependent control of pre-mRNA splicing (Schmidt et al. 2014). However, several lines of evidence indicate that this assumption may not universally hold.
First, <30% of CBP20 iCLIP reads mapped to the first 100 nt of bound RNAs in HeLa Kyoto cells (Giacometti et al. 2017). In this experiment, CBP20 derived from a bacterial artificial chromosome (BAC) transgene; i.e., the entire CBP20 gene including introns and regulatory sequences. This raises the possibility that the non-cap-associated iCLIP reads typify the binding properties of CBP20S, an alternative splicing isoform of CBP20 lacking most of its RNA recognition motif (Pabis et al. 2010). Unlike full-length CBP20, CBP20S does not bind CBP80 or the m7G-cap, but nonetheless binds mRNA and localizes to active transcription sites (Pabis et al. 2010). Another possibility is that non-cap-associated iCLIP reads are the result of mRNP loops that bridge the mRNA 5′ cap and body.
Second, NCBP3 was initially described as a stress-associated alternative cap-binding protein that teams up with CBP80 at the 5′ cap of mRNAs and lncRNAs so as to form a NCBP3–CBP80 CBC as an alternative to the canonical CBP20–CBP80 CBC (Gebhardt et al. 2015). While these conclusions were challenged by additional in vitro work describing NCBP3 as a CBP20–CBP80 CBC accessory protein (Schulze et al. 2018), independent PAR-CLIP experiments showed that NCBP3 binds not only cap-proximal 5′UTRs but also coding sequences (CDS), 3′UTRs, and introns (Baltz et al. 2012). Whether NCBP3 is a 5′ cap-binding protein that forms an alternative NCBP3–CBP80 CBC, a CBP20–CBP80 CBC accessory protein, an RNA-binding protein, or all three, depending on the transcript or its metabolic step, is an open question.
Third, mass-spectrometry analysis of protease-treated oligo(dT)-pulldowns identified a short peptide consisting of amino acids 312–323 of human CBP80 as an RNA-binding motif (RBM) (Castello et al. 2016). We mapped this RBM to the first α helix of the central repeat (310–422) of the three evolutionary conserved MIF4G (middle portion of [the cytoplasmic cap-binding protein] eukaryotic translation initiation factor 4G [eIF4G]) domains of CBP80 (Mazza et al. 2001). MIF4G domains are found in numerous RNA metabolic factors and, while RNA-binding properties have been reported, these domains have been characterized largely as protein–protein interaction platforms (Marintchev et al. 2009; Clerici et al. 2014; Raisch et al. 2018). Intriguingly, the central RBM-containing MIF4G domain of CBP80 is structurally most similar to the central MIF4G domain of eIF4G (Marintchev and Wagner 2005), which binds eIF4A next to its RNA-binding surface (Marintchev et al. 2009). Thus, it possible that crosslinks of the CBP80 RBM to RNA do not reflect direct RNA binding, but instead close proximity to another RNA-binding domain. Considering the RBM of CBP80 is buried within the cap-bound CBP20–CBP80 CBC structure (Calero et al. 2002; Cho et al. 2018), extensive remodeling would be required to make it accessible so as to be able to bind directly to or be found in close proximity of RNA. Whether this would involve CBP20S, NCBP3, or another undefined RNA-binding protein remains unknown.
Conclusion and perspectives
As illustrated throughout this review, the CBC serves as a binding platform for a plethora of factors controlling essentially every step of gene expression, including chromatin accessibility, transcript synthesis and processing, and nuclear RNA decay. Promoter features, the presence and positions of introns, distance from the transcription start site to the 5′-most splicing or 3′ end cleavage site, and RNAPII processivity constitute some of the identified determinants defining factors with which the CBC cooperates to control the fate of the RNA to whose 5′ cap it is bound. Under certain circumstances, even the fate of transcripts synthesized from nearby transcription start sites is regulated.
Comparing data obtained from studies of different species suggests that the CBC and some of its cofactors have undergone convergence in how they have evolved to regulate gene transcription (Fig. 1). In lower eukaryotes, such as S. cerevisiae, which lack prototypical promoter-proximal pausing, the yCBC typically promotes gene transcription by facilitating PIC formation: It recruits TIFs and TBP-like factors to promoters. The yCBC also enhances transcription elongation throughout gene bodies via recruitment of the Bur complex to promoter-proximal regions and the Bur or Ctk complex to gene bodies. In higher eukaryotes such as H. sapiens, in which promoter-proximal pausing has emerged as an important nexus of gene control, the CBC promotes gene transcription via P-TEFb so as to release RNAPII from promoter-proximal pausing and possibly ensure RNAPII processivity throughout gene bodies.
The CBC preferentially promotes the removal of first introns, possibly characterized by specific sequences, and regulates numerous alternative splicing events, in particular those involving first introns. Even though roles for the CBC in pre-mRNA splicing emerged almost three decades ago, it is unclear why and to what extent different mechanisms prevail in different contexts. These mechanisms include enhanced 5′ splice site recognition by U1 snRNP, enhanced recruitment of the SF1/BBP–Mud2p/U2AF complex to 3′ intronic sequences, replacement of SF1/BBP by U2 snRNP, and replacement of U1 snRNP by U6 snRNP (Fig. 2).
Function of the CBC in transcript 3′ end processing is tightly regulated by mutually exclusive and combinatorial interactions whose network extends to alternative splicing, alternative polyadenylation, and RNA surveillance, the latter of which includes the control of pervasive transcription (Figs. 3–5). Globally, it appears that the CBC promotes RNA cleavage at proximal sites of intronless and structured transcripts, such as RDH pre-mRNAs, pre-snRNAs, and NEAT1 lncRNAs. The CBC also prevents early pre-mRNA 3′ end processing and polyadenylation—and thus early transcription termination—at cryptic sites in first introns, unless splicing is impaired, in which case the CBC ensures nuclear decay of the aberrant transcript.
ARS2 obviously plays a central role in CBC-dependent 3′ end processing of nascent pre-mRNAs, and possibly pre-mRNA splicing. Whereas it is known that ARS2 tends to partner with different cofactors depending on the RNA species it targets, how this sorting takes place remains largely uncharacterized. Localization of the processing site within RNA seems to be a critical factor, impacting CBC-dependent processing in numerous ways. The location of the processing site could reflect one of a number of determinants, including physical distance from the TSS, length of the elongating transcript, time since transcription was initiated, or sites at which competition with other RNA processing events takes place. As an example, elongating transcript length controls identity of the CBC cofactors through recruitment of the hnRNPC1/C2 molecular ruler (McCloskey et al. 2012). As another example, CBC-mediated 3′ end processing competes with pre-mRNA splicing, in which case RNAPII pausing, and thus the time elapsed since transcription initiation, and U1 telescripting likely play important roles. Others have proposed that promoter features control the identity of CBC partners (Hallais et al. 2013), echoing the current model by which transcription factor (and cofactor) binding to promoters as well as gene bodies can control the fate of nascent transcripts by regulating RNAPII elongation rates and/or by binding pre-mRNA to either recruit or block one or more processing factors (Rambout et al. 2018). Along these lines, we found that the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α) associates with the CBC at the 5′ cap of nascent transcripts to induce gene transcription (Cho et al. 2018) and prevent cytoplasmic accumulation of intron 1-containing transcripts (Rambout et al. 2020). We predict that PGC-1α represents one of many mammalian transcriptional cofactors that control CBC activity in a gene-dependent manner.
Despite the three decades that have passed since the “canonical” CBP20–CBP80 cap-binding complex was first discovered (Ohno et al. 1990), many fascinating questions remain unanswered and are ripe for future discoveries of the regulatory interplay between genes and their transcripts. Important issues that merit investigation in the future include expanding on evidence that the CBC operates on only a subset of genes: What defines these genes and the mechanisms by which CBC function can vary among these genes remain largely unknown. Another topic worthy of pursuit derives from emerging evidence that CBP80 may function independently of CBP20 and/or the 5′ cap.
Acknowledgments
We apologize to those colleagues whose work we could not cite due to space limitations. This work was supported by American Heart Association (AHA) Postdoctoral Fellowship 18POST33960339 to X.R. and by National Institutes of Health (NIH) grant R01 GM059514 to L.E.M.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.339986.120.
References
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anamika K, Gyenis A, Poidevin L, Poch O, Tora L. 2012. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS One 7: e38769 10.1371/journal.pone.0038769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, et al. 2015. SRSF1-regulated alternative splicing in breast cancer. Mol Cell 60: 105–117. 10.1016/j.molcel.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20: 1367–1376. 10.1038/nsmb.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Smith ER, Shah AP, Rendleman EJ, Marshall SA, Woodfin AR, Chen FX, Shiekhattar R, Shilatifard A. 2020. NELF regulates a promoter-proximal step distinct from RNA Pol II pause-release. Mol Cell 78: 261–274.e5. 10.1016/j.molcel.2020.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balatsos NA, Nilsson P, Mazza C, Cusack S, Virtanen A. 2006. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC). J Biol Chem 281: 4517–4522. 10.1074/jbc.M508590200 [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 2009. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci 106: 7798–7803. 10.1073/pnas.0903188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316. 10.1101/gad.1968210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM. 2004. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J 40: 112–119. 10.1111/j.1365-313X.2004.02194.x [DOI] [PubMed] [Google Scholar]
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. 2012. First exon length controls active chromatin signatures and transcription. Cell Rep 2: 62–68. 10.1016/j.celrep.2012.05.019 [DOI] [PubMed] [Google Scholar]
- Bittencourt D, Dutertre M, Sanchez G, Barbier J, Gratadou L, Auboeuf D. 2008. Cotranscriptional splicing potentiates the mRNA production from a subset of estradiol-stimulated genes. Mol Cell Biol 28: 5811–5824. 10.1128/MCB.02231-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth GT, Wang IX, Cheung VG, Lis JT. 2016. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res 26: 799–811. 10.1101/gr.204578.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth GT, Parua PK, Sansó M, Fisher RP, Lis JT. 2018. Cdk9 regulates a promoter-proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nat Commun 9: 543 10.1038/s41467-018-03006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragulat M, Meyer M, Macias S, Camats M, Labrador M, Vilardell J. 2010. RPL30 regulation of splicing reveals distinct roles for Cbp80 in U1 and U2 snRNP cotranscriptional recruitment. RNA 16: 2033–2041. 10.1261/rna.2366310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli ME, Buratowski S. 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. Embo J 24: 2150–2160. 10.1038/sj.emboj.7600687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. 2002. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol 9: 912–917. 10.1038/nsb874 [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. 2016. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 63: 696–710. 10.1016/j.molcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE. 2010. Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. Embo J 29: 1674–1687. 10.1038/emboj.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chathoth KT, Barrass JD, Webb S, Beggs JD. 2014. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell 53: 779–790. 10.1016/j.molcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirackal Manavalan AP, Pilarova K, Kluge M, Bartholomeeusen K, Rajecky M, Oppelt J, Khirsariya P, Paruch K, Krejci L, Friedel CC, et al. 2019. CDK12 controls G1/S progression by regulating RNAPII processivity at core DNA replication genes. EMBO Rep 20: e47592 10.15252/embr.201847592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, Sharp PA. 2018. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol Cell 69: 648–663.e7. 10.1016/j.molcel.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Rambout X, Gleghorn ML, Nguyen PQT, Phipps CR, Miyoshi K, Myers JR, Kataoka N, Fasan R, Maquat LE. 2018. Transcriptional coactivator PGC-1α contains a novel CBP80-binding motif that orchestrates efficient target gene expression. Genes Dev 32: 555–567. 10.1101/gad.309773.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I. 2011. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303. 10.1105/tpc.110.075911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Deniaud A, Boehm V, Gehring NH, Schaffitzel C, Cusack S. 2014. Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res 42: 2673–2686. 10.1093/nar/gkt1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708. 10.1101/gad.10.13.1699 [DOI] [PubMed] [Google Scholar]
- Core L, Adelman K. 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33: 960–982. 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278. 10.1016/j.molcel.2007.11.035 [DOI] [PubMed] [Google Scholar]
- Das J, Yu H. 2012. HINT: high-quality protein interactomes and their applications in understanding human disease. BMC Syst Biol 6: 92 10.1186/1752-0509-6-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Guo Z, Russo P, Chartrand P, Sherman F. 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol Cell Biol 20: 2827–2838. 10.1128/MCB.20.8.2827-2838.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Francisco Amorim M, Willing EM, Szabo EX, Francisco-Mangilet AG, Droste-Borel I, Maček B, Schneeberger K, Laubinger S. 2018. The U1 snRNP subunit LUC7 modulates plant development and stress responses via regulation of alternative splicing. Plant Cell 30: 2838–2854. 10.1105/tpc.18.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan TL, He GJ, Hu LD, Yan YB. 2019. The intrinsically disordered C-terminal domain triggers nucleolar localization and function switch of PARN in response to DNA damage. Cells 8: 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JD, Francis L, Davidson L, West S. 2020. A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev 34: 132–145. 10.1101/gad.332833.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eychenne T, Werner M, Soutourina J. 2017. Toward understanding of the mechanisms of Mediator function in vivo: focus on the preinitiation complex assembly. Transcription 8: 328–342. 10.1080/21541264.2017.1329000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszbein A, Krick KS, Begg BE, Burge CB. 2019. Exon-mediated activation of transcription starts. Cell 179: 1551–1565.e17. 10.1016/j.cell.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci 94: 11893–11898. 10.1073/pnas.94.22.11893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933. 10.1038/414929a [DOI] [PubMed] [Google Scholar]
- Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj IW. 1999a. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev 13: 2425–2438. 10.1101/gad.13.18.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. 1999b. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol Cell Biol 19: 6543–6553. 10.1128/MCB.19.10.6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara R, Damodaren N, Wilusz JE, Murakami K. 2019. The capping enzyme facilitates promoter escape and assembly of a follow-on preinitiation complex for reinitiation. Proc Natl Acad Sci 116: 22573–22582. 10.1073/pnas.1905449116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberi C, Izaurralde E, Beisel C, Mattaj IW. 1997. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol Cell Biol 17: 2587–2597. 10.1128/MCB.17.5.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EL, Wen Y, Praveen K, Matera AG. 2016. Transcriptomic comparison of Drosophila snRNP biogenesis mutants reveals mutant-specific changes in pre-mRNA processing: implications for spinal muscular atrophy. RNA 22: 1215–1227. 10.1261/rna.057208.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Habjan M, Benda C, Meiler A, Haas DA, Hein MY, Mann A, Mann M, Habermann B, Pichlmair A. 2015. mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nat Commun 6: 8192 10.1038/ncomms9192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo N, Bäurle I, Kidou S, Hu X, Dean C. 2009. FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150: 1611–1618. 10.1104/pp.109.137448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti S, Benbahouche NEH, Domanski M, Robert MC, Meola N, Lubas M, Bukenborg J, Andersen JS, Schulze WM, Verheggen C, et al. 2017. Mutually exclusive CBC-containing complexes contribute to RNA Fate. Cell Rep 18: 2635–2650. 10.1016/j.celrep.2017.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78. 10.1038/nsmb1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Cowling VH. 2014. Cap-binding complex (CBC). Biochem J 457: 231–242. 10.1042/BJ20131214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63. 10.1016/j.molcel.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. 2012. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell 45: 87–98. 10.1016/j.molcel.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiro J, Murphy S. 2017. Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol 7: 170073 10.1098/rsob.170073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Stark A. 2018. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol 19: 621–637. 10.1038/s41580-018-0028-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189: 705–736. 10.1534/genetics.111.127019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche Nel H, Gostan T, Vandermoere F, Robert MC, Cusack S, et al. 2013. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 20: 1358–1366. 10.1038/nsmb.2720 [DOI] [PubMed] [Google Scholar]
- Heath CG, Viphakone N, Wilson SA. 2016. The role of TREX in gene expression and disease. Biochem J 473: 2911–2935. 10.1042/BCJ20160010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissant L, Moehle EA, Bertaccini D, Van Dorsselaer A, Schaeffer-Reiss C, Guthrie C, Dargemont C. 2014. H2B ubiquitylation modulates spliceosome assembly and function in budding yeast. Biol Cell 106: 126–138. 10.1111/boc.201400003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono N. 2019. Genetic abnormalities and pathophysiology of MDS. Int J Clin Oncol 24: 885–892. 10.1007/s10147-019-01462-6 [DOI] [PubMed] [Google Scholar]
- Hossain MA, Chung C, Pradhan SK, Johnson TL. 2013. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol Cell Biol 33: 785–799. 10.1128/MCB.00947-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Li Y, Li P. 2013. MARVELD1 inhibits nonsense-mediated RNA decay by repressing serine phosphorylation of UPF1. PLoS One 8: e68291 10.1371/journal.pone.0068291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Harvey SE, Hu X, Cheng C. 2017. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev 31: 2296–2309. 10.1101/gad.305862.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iasillo C, Schmid M, Yahia Y, Maqbool MA, Descostes N, Karadoulama E, Bertrand E, Andrau JC, Jensen TH. 2017. ARS2 is a general suppressor of pervasive transcription. Nucleic Acids Res 45: 10229–10241. 10.1093/nar/gkx647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668. 10.1016/0092-8674(94)90530-4 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712. 10.1038/376709a0 [DOI] [PubMed] [Google Scholar]
- Jimeno-González S, Ceballos-Chávez M, Reyes JC. 2015. A positioned +1 nucleosome enhances promoter-proximal pausing. Nucleic Acids Res 43: 3068–3078. 10.1093/nar/gkv149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachaev ZM, Lebedeva LA, Shaposhnikov AV, Moresco JJ, Yates JR 3rd, Schedl P, Shidlovskii YV. 2019. Paip2 cooperates with Cbp80 at an active promoter and participates in RNA polymerase II phosphorylation in Drosophila. FEBS Lett 593: 1102–1112. 10.1002/1873-3468.13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Venhuizen P, Wu MT, Chiou P, Chang CL, Kalyna M, Matzke AJM, Matzke M. 2020. A collection of pre-mRNA splicing mutants in Arabidopsis thaliana. G3 10: 1983–1996. 10.1534/g3.119.400998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Breton G, Schroeder JI. 2007. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J 50: 1049–1062. 10.1111/j.1365-313X.2007.03110.x [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, Maquat LE. 2019. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20: 406–420. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR. 2011. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res 39: 2188–2209. 10.1093/nar/gkq1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahudkar S, Durairaj G, Uprety B, Bhaumik SR. 2014. A novel role for Cet1p mRNA 5′-triphosphatase in promoter proximal accumulation of RNA polymerase II in Saccharomyces cerevisiase. Genetics 196: 161–176. 10.1534/genetics.113.158535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. 2008. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci 105: 8795–8800. 10.1073/pnas.0802493105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, Peterlin BM, Barboric M. 2011. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb). J Biol Chem 286: 22758–22768. 10.1074/jbc.M111.235077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Görlich D, Mattaj IW. 1996a. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res 24: 3332–3336. 10.1093/nar/24.17.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. 1996b. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev 10: 1683–1698. 10.1101/gad.10.13.1683 [DOI] [PubMed] [Google Scholar]
- Li T, De Clercq N, Medina DA, Garre E, Sunnerhagen P, Pérez-Ortín JE, Alepuz P. 2016. The mRNA cap-binding protein Cbc1 is required for high and timely expression of genes by promoting the accumulation of gene-specific activators at promoters. Biochim Biophys Acta 1859: 405–419. 10.1016/j.bbagrm.2016.01.002 [DOI] [PubMed] [Google Scholar]
- Lidschreiber M, Leike K, Cramer P. 2013. Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol Cell Biol 33: 3805–3816. 10.1128/MCB.00361-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822. 10.1038/nsmb1135 [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. 10.1016/j.molcel.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Ma Z, Castillo-González C, Wang Z, Sun D, Hu X, Shen X, Potok ME, Zhang X. 2018. Arabidopsis Serrate coordinates histone methyltransferases ATXR5/6 and RNA processing factor RDR6 to regulate transposon expression. Dev Cell 45: 769–784.e6. 10.1016/j.devcel.2018.05.023 [DOI] [PubMed] [Google Scholar]
- Machitani M, Taniguchi I, Ohno M. 2020. ARS2 regulates nuclear paraspeckle formation through 3′-end processing and stability of NEAT1 long non-coding RNA. Mol Cell Biol 40: e00269–00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Hwang J, Sato H, Tang Y. 2010. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb Symp Quant Biol 75: 127–134. 10.1101/sqb.2010.75.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. 2005. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry 44: 12265–12272. 10.1021/bi051271v [DOI] [PubMed] [Google Scholar]
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136: 447–460. 10.1016/j.cell.2009.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJ, Hemann M, Herzog F, Stark H, Cramer P. 2015. Molecular basis of transcription-coupled pre-mRNA capping. Mol Cell 58: 1079–1089. 10.1016/j.molcel.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008. Nucleosome organization in the Drosophila genome. Nature 453: 358–362. 10.1038/nature06929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S. 2001. Crystal structure of the human nuclear cap binding complex. Mol Cell 8: 383–396. 10.1016/S1097-2765(01)00299-4 [DOI] [PubMed] [Google Scholar]
- McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. 2012. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science 335: 1643–1646. 10.1126/science.1218469 [DOI] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, et al. 2016. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell 64: 520–533. 10.1016/j.molcel.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Müller-McNicoll M, Neugebauer KM. 2014. Good cap/bad cap: how the cap-binding complex determines RNA fate. Nat Struct Mol Biol 21: 9–12. 10.1038/nsmb.2751 [DOI] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26: 349–365. 10.1016/j.molcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Neugebauer KM. 2019. Nascent RNA and the coordination of splicing with transcription. Cold Spring Harb Perspect Biol 11: a032227 10.1101/cshperspect.a032227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve J, Patel R, Wang Z, Louey A, Furger AM. 2017. Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol 14: 865–890. 10.1080/15476286.2017.1306171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Louder RK, He Y. 2017. Structural insights into the eukaryotic transcription initiation machinery. Annu Rev Biophys 46: 59–83. 10.1146/annurev-biophys-070816-033751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Kataoka N, Shimura Y. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res 18: 6989–6995. 10.1093/nar/18.23.6989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198. 10.1016/S0092-8674(00)80829-6 [DOI] [PubMed] [Google Scholar]
- O'Mullane L, Eperon IC. 1998. The pre-mRNA 5′ cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5′ splice sites. Mol Cell Biol 18: 7510–7520. 10.1128/MCB.18.12.7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan C, Christie J, Pienaar M, Gambling J, Nickerson PE, Alford SC, Chow RL, Howard PL. 2015. Mutagenesis of ARS2 domains to assess possible roles in cell cycle progression and microRNA and replication-dependent histone mRNA biogenesis. Mol Cell Biol 35: 3753–3767. 10.1128/MCB.00272-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M, Neufeld N, Shav-Tal Y, Neugebauer KM. 2010. Binding properties and dynamic localization of an alternative isoform of the cap-binding complex subunit CBP20. Nucleus 1: 412–421. 10.4161/nucl.1.5.12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M, Neufeld N, Steiner MC, Bojic T, Shav-Tal Y, Neugebauer KM. 2013. The nuclear cap-binding complex interacts with the U4/U6.U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA 19: 1054–1063. 10.1261/rna.037069.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D. 2015. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 16: 190–202. 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Pintel D. 2007. Distance-dependent processing of adeno-associated virus type 5 RNA is controlled by 5′ exon definition. J Virol 81: 7974–7984. 10.1128/JVI.00714-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu ZR, Chico L, Chang J, Shuman S, Schwer B. 2012. Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA. RNA 18: 1996–2011. 10.1261/rna.033746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, Szweykowska-Kulinska Z, Brown JW, Jarmolowski A. 2010. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res 38: 265–278. 10.1093/nar/gkp869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska KD, Stepien A, Kierzkowski D, Kalak M, Bajczyk M, McNicol J, Simpson CG, Szweykowska-Kulinska Z, Brown JW, Jarmolowski A. 2014. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res 42: 1224–1244. 10.1093/nar/gkt894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisch T, Sandmeir F, Weichenrieder O, Valkov E, Izaurralde E. 2018. Structural and biochemical analysis of a NOT1 MIF4G-like domain of the CCR4–NOT complex. J Struct Biol 204: 388–395. 10.1016/j.jsb.2018.10.009 [DOI] [PubMed] [Google Scholar]
- Rambout X, Dequiedt F, Maquat LE. 2018. Beyond transcription: roles of transcription factors in pre-mRNA splicing. Chem Rev 118: 4339–4364. 10.1021/acs.chemrev.7b00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambout X, Cho H, Maquat LE. 2020. Transcriptional coactivator PGC-1α binding to newly synthesized RNA via CBP80: a nexus for co- and posttranscriptional gene regulation. Cold Spring Harb Symp Quant Biol 84: 040212 10.1101/sqb.2019.84.040212 [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci 90: 7923–7927. 10.1073/pnas.90.17.7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu I, Kim YK. 2017. Translation initiation mediated by nuclear cap-binding protein complex. BMB Rep 50: 186–193. 10.5483/BMBRep.2017.50.4.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath I, Skrajna A, Yang XC, Dadlez M, Marzluff WF, Dominski Z. 2013. 3′-end processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA 19: 1726–1744. 10.1261/rna.040360.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AC, Taatjes DJ. 2020. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34: 465–488. 10.1101/gad.335679.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Gronborg M, Deckert J, Bessonov S, Conrad T, Lührmann R, Urlaub H. 2014. Mass spectrometry-based relative quantification of proteins in precatalytic and catalytically active spliceosomes by metabolic labeling (SILAC), chemical labeling (iTRAQ), and label-free spectral count. RNA 20: 406–420. 10.1261/rna.041244.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WM, Cusack S. 2017. Structural basis for mutually exclusive co-transcriptional nuclear cap-binding complexes with either NELF-E or ARS2. Nat Commun 8: 1302 10.1038/s41467-017-01402-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WM, Stein F, Rettel M, Nanao M, Cusack S. 2018. Structural analysis of human ARS2 as a platform for co-transcriptional RNA sorting. Nat Commun 9: 1701 10.1038/s41467-018-04142-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Mattaj IW, Ohno M. 2001. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7: 351–360. 10.1017/S1355838201002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EC, Stage-Zimmermann T, Chui P, Silver PA. 2000. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J Biol Chem 275: 23718–23724. 10.1074/jbc.M002312200 [DOI] [PubMed] [Google Scholar]
- Shi M, Wang S, Yao Y, Li Y, Zhang H, Han F, Nie H, Su J, Wang Z, Yue L, et al. 2014. Biological and clinical significance of epigenetic silencing of MARVELD1 gene in lung cancer. Sci Rep 4: 7545 10.1038/srep07545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Di C, Cai Z, Venters CC, Guo J, Oh JM, Arai C, Dreyfuss G. 2019. A complex of U1 snRNP with cleavage and polyadenylation factors controls telescripting, regulating mRNA transcription in human cells. Mol Cell 76: 590–599.e4. 10.1016/j.molcel.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Thillainadesan G, Chalamcharla VR, Meng Z, Balachandran V, Dhakshnamoorthy J, Zhou M, Grewal SIS. 2016. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol Cell 61: 747–759. 10.1016/j.molcel.2016.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Ranjan A, Chen S, Suzuki H, Shibata M, Hirose T, Hirose H, Sasaki K, Abe R, Chen K, et al. 2020. The role of Mediator and Little Elongation Complex in transcription termination. Nat Commun 11: 1063 10.1038/s41467-020-14849-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Wang HF, Burns AM, Schroeder MR, Gaspari M, Baumann P. 2015. Human telomerase RNA processing and quality control. Cell Rep 13: 2232–2243. 10.1016/j.celrep.2015.10.075 [DOI] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol 133: 5–14. 10.1083/jcb.133.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hu J, Yao Y, Shi M, Yue L, Han F, Zhang H, He J, Liu S, Li Y. 2013. MARVELD1 regulates integrin β1-mediated cell adhesion and actin organization via inhibiting its pre-mRNA processing. Int J Biochem Cell Biol 45: 2679–2687. 10.1016/j.biocel.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. 2014. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell 53: 819–830. 10.1016/j.molcel.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KF, Fortes P, Singh US, Ohno M, Mattaj IW, Cerione RA. 1999. The nuclear cap-binding complex is a novel target of growth factor receptor-coupled signal transduction. J Biol Chem 274: 4166–4173. 10.1074/jbc.274.7.4166 [DOI] [PubMed] [Google Scholar]
- Wilson KF, Wu WJ, Cerione RA. 2000. Cdc42 stimulates RNA splicing via the S6 kinase and a novel S6 kinase target, the nuclear cap-binding complex. J Biol Chem 275: 37307–37310. 10.1074/jbc.C000482200 [DOI] [PubMed] [Google Scholar]
- Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG. 2007. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol Cell Biol 27: 6520–6531. 10.1128/MCB.00733-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Gilmartin GM, Doublié S. 2011. The structure of human cleavage factor Im hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol 8: 748–753. 10.4161/rna.8.5.16040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. 2013. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol Cell Biol 33: 28–37. 10.1128/MCB.00653-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CS, Chang SL, Chen JH, Wang HK, Chou YC, Wang CH, Huang SH, Larson A, Pleiss JA, Chang WH, et al. 2017. The conserved AU dinucleotide at the 5′ end of nascent U1 snRNA is optimized for the interaction with nuclear cap-binding-complex. Nucleic Acids Res 45: 9679–9693. 10.1093/nar/gkx608 [DOI] [PMC free article] [PubMed] [Google Scholar]