In this review, Gencel-Augusto and Lozano summarize the data on p53 mutants with a functional tetramerization domain that form mixed tetramers and in some cases have dominant-negative effects (DNE) that inactivate wild-type p53. They conclude that the DNE is mostly observed after DNA damage but fails in other contexts.
Keywords: tetramerization, transcription factor, tumor suppressor
Abstract
The p53 tumor suppressor functions as a tetrameric transcription factor to regulate hundreds of genes—many in a tissue-specific manner. Missense mutations in cancers in the p53 DNA-binding and tetramerization domains cement the importance of these domains in tumor suppression. p53 mutants with a functional tetramerization domain form mixed tetramers, which in some cases have dominant-negative effects (DNE) that inactivate wild-type p53. DNA damage appears necessary but not sufficient for DNE, indicating that upstream signals impact DNE. Posttranslational modifications and protein–protein interactions alter p53 tetramerization affecting transcription, stability, and localization. These regulatory components limit the dominant-negative effects of mutant p53 on wild-type p53 activity. A deeper understanding of the molecular basis for DNE may drive development of drugs that release WT p53 and allow tumor suppression.
The p53 tumor suppressor is the most often mutated gene in cancer. Additionally, other mechanisms, such as high levels of MDM2 and MDM4 also inhibit p53 in a mutually exclusive relationship with p53 alterations, suggesting that this pathway is likely dampened in all cancers (Wasylishen and Lozano 2016). Since p53 is a potent transcription factor that functions as a tetramer to regulate (both up and down) hundreds of genes, it can form mixed tetramers with mutant p53 sometimes obliterating wild-type (WT) p53 activity. In addition, posttranslational modifications (PTMs) and protein–protein interactions influence p53 activity. In some cases, mutant p53 is able to inhibit WT p53 function as a dominant negative, and in other cases it is clear that mutant p53 cannot inactivate WT p53. This review summarizes the data and concludes that the dominant-negative effect (DNE) is mostly observed after DNA damage but fails in other contexts.
p53 as a tetrameric transcription factor
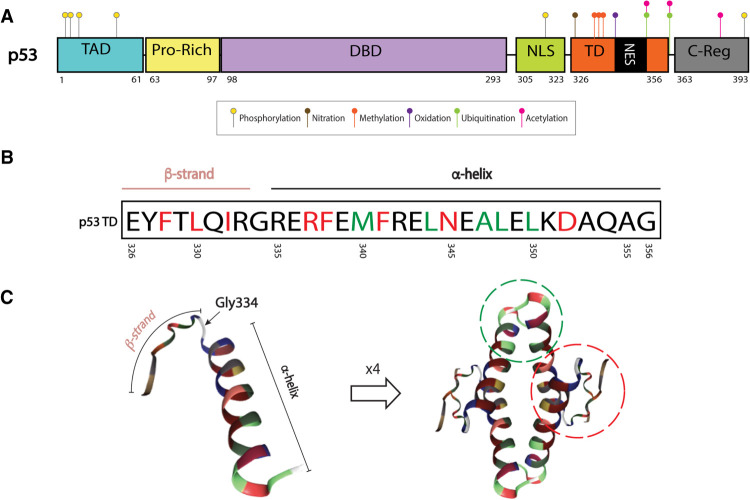
The first indication that p53 forms high molecular weight complexes came from identification of an ∼200-kDa entity that interacts with SV40 large-T antigen (McCormick et al. 1981). Later, chemical crosslinking, gel filtration chromatography, and zonal velocity gradient centrifugation assays confirmed the tetrameric nature of p53 (Friedman et al. 1993). The C-terminal end of p53 (residues 311-367), which includes the tetramerization domain (TD) (Fig. 1A), forms high molecular weight complexes (Pavletich et al. 1993). To date, the structure of the p53 TD (amino acids 326-356) has been resolved by X-ray crystallography and NMR approaches (Clore et al. 1994; Jeffrey et al. 1995; Mittl et al. 1998; Mora et al. 2008).
Figure 1.
p53 tetramerization domain (TD) structure. (A) Map of p53 protein. Posttranslational modifications (PTMs) known to be important for tetramerization dynamics and TD-regulated processes: serine 315 and 392 phosphorylation (yellow), tyrosine 327 nitration (brown), arginine 333, 335, and 337 methylation (orange), methionine 340 oxidation (purple), lysine 351 and 357 ubiquitination (green), and lysine 351 and 357 acetylation (pink). PTMs that require tetramerization: serine 6, 9, 15, and 46 phosphorylation (yellow) and lysine 382 acetylation (pink). Ubiquitination in C-terminal lysines (dependent on tetramerization) not shown. (B) The p53 TD is composed of a β strand, a tight turn (Gly334), and an α-helix. Residues involved in dimer and tetramer formation are depicted in red and green, respectively (Chène 2001) (C) Monomer or single peptide (left) and tetramer (right) forms of the p53 TD. Monomer–monomer interactions of β sheets and α helices are depicted by a red dotted line. Dimer–dimer interactions of α helices are depicted by green dotted line (residues outside of these circles can also be involved in stabilizing each complex). Structures extracted from PDB ID 1AIE (Mittl et al. 1998; Berman et al. 2003).
The secondary structure of a single peptide of the TD is comprised of a β strand, a tight turn, and an α-helix. Monomers dimerize through their antiparallel β sheets and a double-helical bundle. Dimers tetramerize through their α helices, forming a four-helical bundle (Mateu and Fersht 1998). Hydrogen bonds and salt bridges are critical for dimer formation, while tetramer formation is mainly dictated by hydrophobic interactions (Fig. 1 B,C; Chène 2001).
After DNA damage or stress (such as inflammation, oncogene activation, nutrient deprivation, hypoxia (Harris and Levine 2005), p53 tetramers are observed in the nucleus suggesting that this feature is essential for its transcriptional activity (Gaglia et al. 2013). The DNA-binding domain (DBD) of p53 recognizes two sequential decameric motifs (or two half-sites) of DNA, each comprised of 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ (Pu and Py stand for purine and pyrimidine, respectively), separated by up to 13 bp (el-Deiry et al. 1992). Each dimer binds one decameric sequence, but tetramerization is essential for stabilization of the p53–DNA complex. In fact, tetrameric p53 increases binding affinity to its DNA sequence by a 1000-fold relative to the monomer (Weinberg et al. 2004). Due to the nature of the p53 response element, the formation of p53 dimers during protein translation (Nicholls et al. 2002) and the dynamics of the p53–DNA interaction, p53 is generally known as “a dimer of dimers”(Clore et al. 1994; Lee et al. 1994; Jeffrey et al. 1995). More recently, D'Abramo et al. (2016) built an atomistic model for the tetrameric form of p53 (residues 92-360) with a consensus 18-bp DNA sequence. By molecular dynamics, they determined that continuous long-range communications exist between the TD and DBD; the TD modulates conformation of the DBD. This evidence supports an induced-fit mechanism of p53-DNA binding for which the TD-containing C-terminal portion of the protein is important.
Monomers, dimers, and tetramers of p53 exist in cells. Gaglia et al. (2013) show that in a resting state, p53 is present mainly as dimers (∼59%), followed by monomers (∼28%), and lastly, tetramers (∼13%) through fluorescence correlation spectroscopy (FCS). This finding is consistent with a previous report by Rajagopalan et al. (2011) using in vitro FCS that suggests the main oligomeric state of p53 in resting conditions is a dimer. The in vitro association of dimers into tetramers is a slow process, which suggests that it requires additional players and/or modifications (Natan et al. 2009). Consistent with the concept that tetramers are essential for p53 function as a transcription factor, about 93% of p53 abundance in cells are tetramers after DNA damage (Gaglia et al. 2013). Tetramerization is a process that can be influenced by the increase of p53 protein concentration in the cell (Johnson et al. 1995; Mateu and Fersht 1998; Kamada et al. 2016) (mainly through inhibition of the E3 ligase Mdm2). Additionally, other factors seem to contribute to p53 tetramerization as Gaglia and Lahav (2014) report functional tetramers are formed prior to the increase in p53 protein levels, upon DNA damage.
Regulation of the p53 oligomeric state
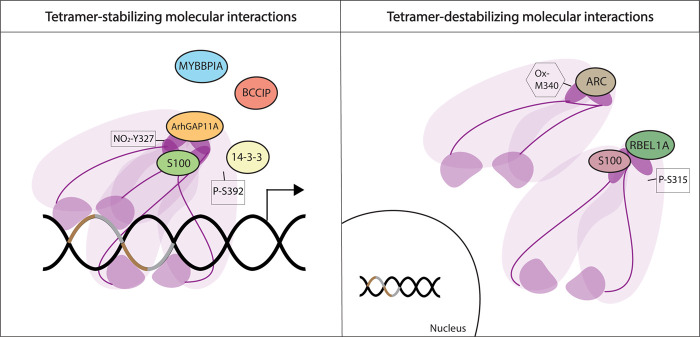
Several studies show that specific posttranslational modifications (PTMs) and interacting proteins regulate p53 tetramerization. Some interactions are able to stabilize tetrameric forms of p53, while others have the ability to impede tetramer formation (Fig. 2).
Figure 2.
Regulation of p53 oligomeric state. Protein interactions and posttranslational modifications (PTMs) regulate p53 oligomerization by either stabilizing p53 tetramers or inhibiting their formation. Interacting proteins are depicted as circles. Only ArhGAP11A and some S100 members have been shown to directly bind the p53 TD and stabilize it. ARC, RBEL1A, and other S100 members bind directly to the p53 TD and impede tetramer formation (exemplified here as steric blockage of oligomerization of two dimers). Some PTMs also regulate tetramer formation, positively or negatively (nitration [NO2], oxidation [Ox], and phosphorylation [P]). Purple denotes p53, TD is dark purple color (see Fig. 5, below). Brown and gray DNA pieces denote a p53 response element.
Proteins that promote p53 tetrameric status
For example, MYBBPIA, a protein involved in rRNA transcriptional regulation and processing, regulates p53 oligomerization. In the presence of ectopically expressed MYBBPIA, p53 is mainly found in high molecular weight fractions. This high molecular weight fraction decreases when a mutant MYBBPIA incapable of binding p53 was expressed, suggesting a function for this protein in stabilizing tetramers, although the investigators did not confirm a direct role (Ono et al. 2014). Similarly, BCCIP, a protein that interacts with p21 and BRCA2, modulates p53 tetramer formation. BCCIP knockdown cells fail to transactivate the p53 target genes CDKN1A (p21) and MDM2 in response to DNA damage, and p53 does not efficiently bind to the promoters of these genes. On the other hand, BCCIP overexpression led to increased tetramer formation as compared with normal cells. However, a direct interaction between BCCIP and p53 could not be confirmed suggesting that it is either a very unstable interaction or its effect on p53 tetramer formation is indirect (Meng et al. 2007). A third protein, ArhGAP11A is capable of binding the TD of p53 through its RhoGAP domain. A tetrameric form of p53 is necessary for this interaction, since p53 mutants incapable of forming tetramers do not bind ArhGAP11A. ArhGAP11A enhances p53 function possibly through stabilizing tetramer conformation, augmenting DNA-binding ability (Xu et al. 2013). Furthermore, members of the 14-3-3 family enhance the binding of p53 to sequence-specific DNA by stabilizing tetramers (Rajagopalan et al. 2008; Schumacher et al. 2010). Finally, members of a family of calcium-binding proteins known as S100 are able to bind the TD of p53 enhancing tetramer formation and activation of p53 function (van Dieck et al. 2009a,b). Thus, numerous proteins either directly or indirectly enhance p53 tetramer formation.
Proteins that inhibit p53 tetrameric status
RBEL1A, a Ras-like GTPase that is overexpressed in cancers, interacts with the TD of p53 and suppresses oligomer formation of an ectopically expressed C-terminal fragment of p53 that includes the TD. Silencing of RBEL1A results in p53 oligomerization and increased target gene expression (Lui et al. 2015). Another protein, ARC, which is elevated in breast cancers, binds directly to the TD of p53. Binding of ARC exposes the Nuclear Export Signal (NES) contained in the p53 TD, allowing for p53 nuclear exclusion (Foo et al. 2007). Importantly, Gaglia and Lahav (2014) report that disruption of the p53-ARC interaction led to abnormally high levels of p53 in the nucleus and increased target gene expression. This finding suggests that the ARC-p53 TD interaction may be a protective mechanism in normal cells, sequestering p53 until it is needed as in response to DNA damage (Gaglia and Lahav 2014). While some members of S100 family of calcium-binding proteins can promote p53 oligomerization, others can inhibit it. This process is dependent on protein concentration as well as S100 member type (van Dieck et al. 2009a). Notably, certain PTMs in p53 can alter S100 protein affinity (van Dieck et al. 2009b). Thus, the cellular milieu in which p53 is present will determine the oligomeric state of p53 and its location.
PTMs influence p53 tetramerization
Besides protein interactions as important influencers of p53 oligomerization, certain PTMs also regulate tetramer formation (Figs. 1, 2). For example, phosphorylation of p53S392, although not in the TD itself, regulates tetramerization. A phosphorylated peptide promotes tetramerization by almost 10 times compared with an unphosphorylated peptide. Phosphorylation of p53S315 reverses this effect (Sakaguchi et al. 1997). Importantly, p53S392 is differentially phosphorylated by diverse stress signals, indicating that the signals upstream of phosphorylation can dictate tetramerization (Kapoor and Lozano 1998; Lu et al. 1998; Cox and Meek 2010). A nitric oxide-dependent p53 response occurs on p53Y327 by nitration and stabilizes the p53 tetramer, thus increasing transcriptional activity (Yakovlev et al. 2010). In contrast, Nomura et al. (2009) reported that oxidation of p53M340 in the TD destabilizes tetramers by forming a loose structure that makes it more sensitive to enzyme degradation. Laptenko et al. (2015) report no effect in tetramer formation for substitution of lysines in the C-terminal regulatory domain (p53K370, K372, K373, K381, K382, and K386) by arginine or glutamine suggesting that their modification does not regulate tetramerization. Moreover, numerous other modifications of the C terminus occur, but these have not been studied with regard to tetramerization (Meek and Anderson 2009).
In summary, tetramer formation is a multifactorial process that serves as another layer of regulation of p53 function. Many proteins bind the p53 TD and regulate tetramerization, a process required for p53 transcription function. Numerous PTMs also occur in the TD that regulate p53 tetramerization, and thus transcriptional activity; many others have not been studied. The timing and mechanisms for p53 tetramerization require more in-depth study.
p53 tetramerization domain (TD) mutants
Most p53 alterations target the DBD (∼80% in germline, >90% in somatic p53 alterations) (Bouaoun et al. 2016). However, as a transcription factor whose functional unit is a tetramer (Friedman et al. 1993; McLure and Lee 1998), TD mutations are another mechanism in which cancers can abrogate WT p53 function. Mutations in this region account for ∼20% of germline mutations in Li-Fraumeni syndrome (LFS), a cancer-predisposition disorder characterized by the presence of a germline p53 mutation (Malkin et al. 1990; Petitjean et al. 2007; Bouaoun et al. 2016), and ∼2% of somatic p53 mutations (Bouaoun et al. 2016). Considering the length of each domain (DBD ∼200 amino acids, TD 30 amino acids), the mutation frequency in both is similar in the germline (∼0.4%/residue in DBD, ∼0.66%/residue in TD), suggesting that both domains are essential for p53 function, further implicating a critical role for the TD in tumor suppression (Kamada et al. 2011).
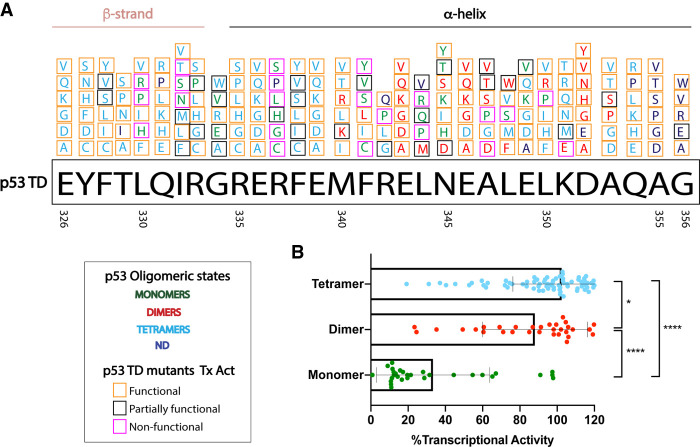
TD mutations can impair the ability of p53 to form tetramers, resulting in monomeric or dimeric forms of the protein. Most of these alterations are missense mutations (∼87%). A majority of TD residues are found mutated in cancer. Functionally speaking, many of these substitutions are predicted to disrupt the ability of p53 to properly tetramerize, depending on the location of the residue and the type of amino acid substitution (Fig. 3A; Kato et al. 2003; Kawaguchi et al. 2005; Bouaoun et al. 2016). To date, several in vitro approaches have been carried out with the intention of characterizing the effects of TD mutations on p53 function (Mateu and Fersht 1998; Chène and Bechter 1999a; Kato et al. 2003; Kawaguchi et al. 2005; Imagawa et al. 2009; Kamada et al. 2011; Giacomelli et al. 2018). By taking advantage of yeast-based assays using a reporter system where expression is driven by the p53 response elements of eight target genes (CDKN1A [p21], MDM2, BCL2L4 [Bax], GADD45, SFN [14-3-3 σ], TP53AIP1, PMAIP1 [Noxa], and RRM2B [p53R2]), Kato et al. (2003) and Kawaguchi et al. (2005) determined that impairment of tetramerization is associated with loss of transcriptional activity (Fig. 3B), although there are some exceptions to this. For example, p53I332T is still able to form tetramers but is transcriptionally dead. Perhaps this mutant affects the conformation of the DNA-binding domain, interaction with the transcriptional machinery, or p53 localization. In contrast, p53N345Y/K/H is unable to form tetramers, yet it is transcriptionally functional in yeast. Some of the mutations were predicted to be transcriptionally hyperactive (p53R335H, p53F338Y, and p53A353V) and to retain a tetrameric structure. The p53 R335H mutation has been found in cancer and reported as functional in different studies (Imagawa et al. 2009; Giacomelli et al. 2018). How cancers are able to tolerate a hyperactive form of p53 remains to be explored but it may interact differently with the repertoire of proteins that affect TD function or may affect nuclear localization. Imagawa et al. (2009) described another reporter system to assay for transcriptional activity of 43 TD mutants (40 found in cancer), this time in a mammalian system (H1299 and A549 cell lines). They used fluorescence intensity distribution analysis to analyze the monomeric fraction for each of the mutants. Generally, p53 TD mutants lacked transactivation when monomer fraction was >90% (L330H, R337C/P, F341C, R342P, and L344P/R). A previously described dimer mutant also shows relative lack of activity (A347T). Last, Kamada et al. (2011) designed peptides for 49 cancer-relevant TD mutations and analyzed their stability and oligomeric structure. They found that eight of 49 peptides displayed nontetrameric structures (monomers: L330P, L330R, R337P, R342P, and L344P; dimers: F341C, L344R, and A347T) with substitutions in hydrophobic residues being more detrimental to tetrameric structure than solvent-exposed ones. In a different study, seven of these residues (with the exception of A347T) are predicted to be nonfunctional as measured by resistance to nutlin or sensitivity to etoposide treatment of A549 cells with exogenous mutant p53 expression (Giacomelli et al. 2018).
Figure 3.
p53 TD missense mutants and their outcomes. (A) p53 TD residues 326–356 are illustrated along with their substitutions to several other amino acids. The oligomeric state outcome for each substitution is font color-coded for each residue. (Light-blue) Tetramers; (red) dimers; (green) monomers; (dark-blue) no data for oligomeric status. Additionally, the transcriptional activity (Tx Act) of these substitutions is highlighted in color-coded squares. (Pink) Nonfunctional; (black) partially functional; (orange) fully functional. Tx Act was based on transactivation of reporter driven by eight p53 response elements in yeast system (Kato et al. 2003; Kawaguchi et al. 2005); data are available at IARC TP53 database and genomics data (Bouaoun et al. 2016). (B) Each TD mutant analyzed (categorized by oligomeric state) is represented by a dot that depicts an average transactivation value of eight p53 response elements. Nontetrameric p53 mutants associated with decreased Tx Act. One-way ANOVA, followed up by Sidak's multiple comparisons test for monomer–dimer, dimer–tetramer, and monomer–tetramer pairs. (*) P < 0.05; (****) P < 0.0001.
Even though these studies have provided valuable insights to TD mutant functions, they have caveats such as superphysiological protein levels (overexpression experiments), nonphysiological temperature (yeast-based assays), and system-specific differences (yeast vs. mammalian experimental conditions), which can impact PTMs and the presence of proteins that influence p53 activity. The only in vivo model that currently exists to understand the physiological relevance of p53 oligomerization impairment corresponds to p53R337H, the most common alteration in the TD in LFS. This founder mutation is highly widespread in southern and southeastern populations in Brazil and shows partial penetrance with onset of adrenal carcinomas in children (Ribeiro et al. 2001; Palmero et al. 2008). In a knock-in mouse model, the presence of the p53R337H allele did not affect overall survival or de novo tumorigenesis but did predispose animals to Diethyl nitrosamine-induced liver tumorigenesis, in a mutant allele dose-dependent manner. p53R337H preferentially forms monomers (∼ 60%), is physiologically stable and transcriptionally impaired (Park et al. 2018).
Importantly, the oligomeric status of p53 TD mutants can impact clinical outcome in LFS patients. While monomeric p53 TD mutants have a 100% cancer penetrance and yield a median survival time of ∼33 yr, multimeric p53 TD mutants (dimers or tetramers) exhibit a cancer penetrance of ∼80% and 51-yr median survival. Notably, monomeric mutants exhibit worse outcomes than hot spot p53 DBD mutants (penetrance: ∼90%; median survival: ∼47 yr) (Fischer et al. 2018), highlighting the importance of the p53 TD in tumor suppression and the need to understand the mechanism of action of TD mutants in physiologically-relevant settings. The abnormally high cellular levels of cancer-prevalent p53 TD mutants (Rollenhagen and Chène 1998; Katz et al. 2018; Park et al. 2018) may confer additional activities that can contribute to disease severity and possibly explain disease outcomes.
The TD plays a role in many aspects of p53 regulation; alterations in the p53 TD may not simply abolish tetramer formation or alter sequence-specific DNA binding. p53 TD PTMs and cellular localization can contribute to changes in interdomain and intermolecular interactions, modifying p53 behavior and therefore contributing to tumor development despite potential transcriptional ability. More studies on how different oligomerization states affect p53 interactions with other proteins, PTMs and DNA binding need to be carried out, as they could have important therapeutic applications. As there is precedent for small molecules to restore WT p53 conformation of p53 DBD mutants (Bykov et al. 2002), the design of small molecules that aim at recovering tetrameric structures of the protein could serve as therapeutic approaches for p53 TD mutant cancers (Gordo et al. 2008). Additionally, molecules that bind to specific oligomeric states could also be used as therapeutic strategy in cancers that retain WT p53. Molecules that bind preferentially to WT p53 tetramers offer the potential of stabilizing this active state and promoting subsequent PTMs and interactions necessary for proper p53 function. For example, Gabizon et al. (2012) discovered a PKCα-derived peptide that interacts with the C-terminal portion of p53 in its tetrameric form. This interaction increases tetramer stability in vitro (Gabizon et al. 2012; Gabizon and Friedler 2014). In contrast, molecules that bind preferentially to nontetrameric forms of p53 could serve to trap mutant p53 (rendering it unable to form mixed tetramers with WT p53) and block its dominant-negative effect over WT p53.
Noncanonical p53 TD functions
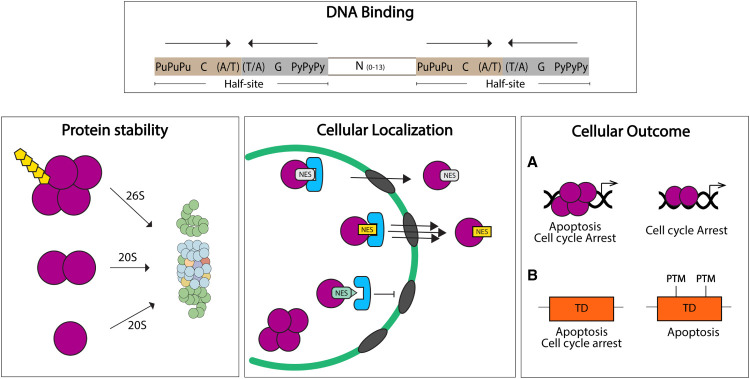
DNA binding is just one of many processes regulated by the TD, as it can also influence p53 protein stability, localization, and cellular outcomes (Fig. 4). The TD does so by two mechanisms: One is by creating a proper quaternary structure for PTMs and/or protein interactions to occur. It is known that some PTMs required for p53 activation only occur upon tetramer formation. For example, S15 phosphorylation (which disrupts interactions with the E3 ligase Mdm2) is dependent on tetramerization of murine p53 (Warnock et al. 2008). Likewise, S6, S9, and S46 phosphorylation, as well as K382 acetylation (Fig. 1), are markedly reduced in tetramerization-impaired mutants of p53 (Warnock et al. 2008). The preferred existence of PTMs only on tetrameric p53 also suggests that certain protein interactions may only occur upon tetramerization of p53. Therefore, oligomeric status is an important regulator of p53 activity. The second mechanism by which the TD influences noncanonical activities is by direct modification of its residues. Some PTMs in TD residues or TD-interacting proteins can also influence processes that ultimately affect p53 activity (Jansson et al. 2008; Kruse and Gu 2009; Muscolini et al. 2009, 2011; Beckerman et al. 2016).
Figure 4.
The TD regulates several p53 functions. DNA Binding. The p53 response element is composed by two half-sites where two independent dimers bind and interact as a tetramer, increasing p53-DNA affinity and transactivation. Protein stability: Tetramerization is required for ubiquitin-dependent degradation of p53 (mediated by 26S proteasome). Dimers and monomers are mainly degraded by ubiquitin-independent mechanisms (20S proteasome). Cellular localization: p53 TD nuclear export signal (NES) controls p53 localization. Monomers expose the NES and retain p53 in the cytoplasm. Tetramerization allows for NES masking and nuclear localization. Mutations in the NES could regulate nuclear export (yellow and light green) by modifying affinity to the export machinery. Cellular outcome: (A) Model for oligomeric state-dependent outcomes: tetramers have full p53 activities, with the potential of initiating both apoptotic and cell cycle arrest programs. Nontetrameric p53 could have partial p53 target gene activation, favoring a certain cellular outcome (e.g., cell cycle arrest). (B) Model for TD PTM-dependent outcomes: Specific PTMs in TD residues can modulate the p53 cellular response between apoptosis and cell cycle arrest.
Cellular localization
Another layer of regulation of p53 function consists of ensuring appropriate subcellular localization (Shaulsky et al. 1991). p53 has a hydrophobic, leucine-rich sequence, which makes up a nuclear export signal (NES) (residues 340–351) contained in the TD (Fig. 1). The NES controls p53 cellular localization depending on its oligomeric state: in nontetrameric forms of p53 (monomers and possibly dimers), the NES is exposed and p53 exists in the cytoplasm. Consistently, this NES is buried upon tetramerization, facilitating nuclear localization of p53 and consequent transcriptional activation (Fig. 4; Stommel et al. 1999).
Some PTMs in TD residues also promote or inhibit p53 nuclear export (mechanism 2 above). Ubiquitination at residues K351 and K357, catalyzed by MSL2, an E3 ubiquitin ligase, are implicated in promoting cytoplasmic localization of p53 (Kruse and Gu 2009). A cancer-related single amino acid substitution, p53K351N, was found to abrogate p53K351 ubiquitination and p53 nuclear export (Muscolini et al. 2009, 2011). Additionally, substitution of TD arginine residues 333, 335, and 337 with lysines (p53 KKK) altered p53 cellular localization. While ∼60% of WT p53 transfected SAOS-2 cells have nuclear p53 staining, only 26% of p53 KKK transfected cells exhibit nuclear p53. PTMs in these residues, such as arginine methylation, probably affect the oligomeric status and thus contribute to increased cytoplasmic localization and decreased transcriptional activity (Jansson et al. 2008).
Given the importance of p53 nuclear localization for its function as a transcription factor, alterations that impact nuclear localization will have severe defects. p53 TD peptides are able to block hyperactive export of p53 in neuroblastoma cells and lead to increased p53 nuclear localization (Stommel et al. 1999). This presents a therapeutic possibility that needs to be explored, especially in light of the fact that some tumors have cytoplasmic WT p53 (Moll et al. 1992, 1995). Similarly, blocking the nuclear export machinery to elevate nuclear p53 levels could further enhance its activity but may have untold effects on other proteins (Kanai et al. 2007).
p53 protein stability
p53 is mainly regulated at the protein level through protein–protein interactions and PTMs. For example, the main negative regulator of p53, MDM2, ubiquitinates p53 and targets it for proteasomal degradation (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997). Importantly, MDM2-mediated p53 ubiquitination is dependent on tetramerization (Fig. 4). Ubiquitin is observed on p53 WT as compared with C-terminal deletion mutants (p53 ΔC-term). Ubiquitin levels can be rescued after fusing p53 ΔC-term to a dimerization domain of another protein, restoring oligomerization capacity (Maki 1999). Because the main ubiquitylation sites of p53 are located in the C terminus, the best tools to demonstrate tetramerization dependence of MDM2-mediated p53 degradation are oligomerization-defective mutants with a functional C-terminal regulatory domain. Lomax et al. (1998) showed that p53L344P (monomer) has reduced ability to bind MDM2. Furthermore, p53 TD mutants L330R/E/P (monomers) and E343A, E344A, A347T, and L348A (dimers) are underubiquitylated by MDM2 and preferably degraded by the ubiquitin independent 20S proteasome (Hjerpe et al. 2010; Lang et al. 2014; Katz et al. 2018). Surprisingly, Katz et al. (2018) report that p53 dimers are more efficiently bound by MDM2 than monomers and degraded in a ubiquitin-independent manner. Similarly, Hjerpe et al. (2010) show that some oligomerization-defective p53 proteins exhibit different levels of MDM2-mediated proteasome degradation in a ubiquitin-independent manner. Although this challenges our current understanding of p53 degradation mechanism by MDM2, Kulikov et al. (2010) report that MDM2 directly binds to the proteasome, enhancing p53–proteasome association and leading to efficient degradation. This potentially explains why p53 is still degraded in the presence of MDM2 even without polyubiquitin chains. If preferential binding of MDM2 to p53 dimers is confirmed, it offers a therapeutic approach to inhibit the oncogenic potential of p53 DBD mutants (cancer hot spot mutants) by inducing dimerization and thus their degradation (Katz et al. 2018). Finally, whether MDM2 plays an active role in modulating the p53 oligomerization state remains an open question.
Cell fate control
p53 initiates several cellular programs such as apoptosis, cell cycle arrest, and senescence (Martinez et al. 1991; Yonish-Rouach et al. 1991; Brown et al. 1997; Riley et al. 2008). How p53 makes this decision depends on many factors that are not fully understood (Vousden and Prives 2009). Increasing evidence suggests that PTMs in some TD residues may act as molecular switches between p53-dependent cellular programs of apoptosis and cell cycle arrest, thereby regulating cellular outcomes (Fig. 4). For example, p53 KKK, a p53 mutant deficient in methylation at residues R333, R335, and R337, abrogates the ability of SAOS-2 cells to undergo cell cycle arrest but not apoptosis. This mutant cannot transactivate CDKN1A (p21) or GADD45 and is deficient at binding their promoters, but remains functional for activation of the apoptosis inducer APAF1 (Jansson et al. 2008). More recently, Beckerman et al. (2016) describes a role for p53 acetylation (p53K351Q/R and p53K357Q/R) in fine tuning target gene activation between growth arrest and apoptosis programs. Mimicking acetylation of these residues (by their substitution to glutamine) results in apoptosis but impairs cell cycle arrest. These data imply that DNA-binding affinity is important as CDKN1A (p21) has one of the strongest p53 response elements.
Additionally, p53 oligomeric status may also impact selective transcriptional program activation. Fischer et al. (2016) tested different p53 TD variants and demonstrated that a p53 mutant that forms only dimers is not able to initiate an apoptotic program, failing to induce TP53AIP1 expression, while it retains cell cycle arrest functions mediated by CDKN1A (p21) expression (Fischer et al. 2016). This is supported by a comprehensive study of amino acid substitutions of the p53 TD and their effects on oligomeric status and p53 binding element-dependent transcription. While some of the mutations (L330H/R, I332N/S, R337S/G/C/P, F341Y/S/C/V, R342P, L344P/R, A347P, L348S, and L350P) yielded a transcriptionally inactive p53 protein, others had differential target gene activation. For example, dimeric mutants A347D and K351E could transactivate p53R2 but not other genes tested (Kato et al. 2003; Kawaguchi et al. 2005).
Important for cell fate decisions as well as for p53 transcriptional activity and stability, acetylation of C-terminal lysines of p53 is dependent on tetramer formation. Itahana et al. (2009) show that the acetyltransferase p300 efficiently acetylates tetrameric p53 but cannot do so for tetramer-defective mutants of p53. Importantly, nonacetylated forms of p53 were defective in CDKN1A (p21) transactivation (Itahana et al. 2009). Similarly, the kinase Chk1, which phosphorylates p53 at the N terminus and promotes its dissociation from MDM2, requires tetramerization of p53 as deletion in the p53 TD abrogates Chk1-dependent phosphorylation of these residues (Shieh et al. 2000).
Thus, activation of p53 is heavily influenced by PTMs and protein interactions. The evidence presented above places the TD as an essential domain for regulating p53 tumor suppressive activities, in addition to reports that show the DBD fine-tunes cell fate decisions between cell cycle and apoptotic responses (Sykes et al. 2006; Tang et al. 2006). Cancer-related TD mutants need to be studied in physiologically relevant model systems to understand their full potential in regulating p53 function.
The mutant p53 dominant-negative effect (DNE) is an elusive phenotype
A dominant-negative effect (DNE) is defined as the interaction of two proteins in which one eliminates the functionality of the other and, in the context of p53, is based on the ability of WT and DNA-binding (core) domain mutants to form tetramers. Therefore, the TD is the basis of the DNE of mutant over WT p53. However, the DNE is an elusive phenotype as increasing reports show contradictory evidence that mutant p53 cannot undermine WT p53 function.
Early evidence that mutant p53 has a DNE comes from findings that mutant and WT p53 interact and this interaction diminishes WT p53 activity (Eliyahu et al. 1988; Halevy et al. 1989; Milner and Medcalf 1991). Consistent with the TD being a key factor for a DNE, p53 DBD mutants with additional alterations in the TD are no longer able to diminish WT p53 function. Assays with C-terminal truncation of p53 DBD mutants yields no complexes between WT and mutant proteins (Milner and Medcalf 1990). p53 DBD mutants (p53C135S and p53V143A) with additional TD alterations that favor monomeric or dimeric forms of p53 lost their transformation ability in the presence of WT p53 (Slingerland et al. 1993). Likewise, p53R175H with a mutation in the TD (p53R175H, L330A) lost its effect over WT, as measured by DNA binding. Furthermore, coexpression of WT and monomeric p53 TD mutants (R342P or L344P) does not affect p53 target gene transactivation (Chène et al. 1997; Chène and Bechter 1999b). More recently, the addition of the L344P mutation (monomer) into p53R273H abrogates the ability of mutant p53 to inhibit WT p53-dependent transactivation of BCL2L4 (Bax), MDM2, and PMAIP1 (Noxa) in SAOS-2 cells (Xu et al. 2011). These studies support the crucial role of a functional TD in p53 mutant proteins to exert a DNE over WT p53.
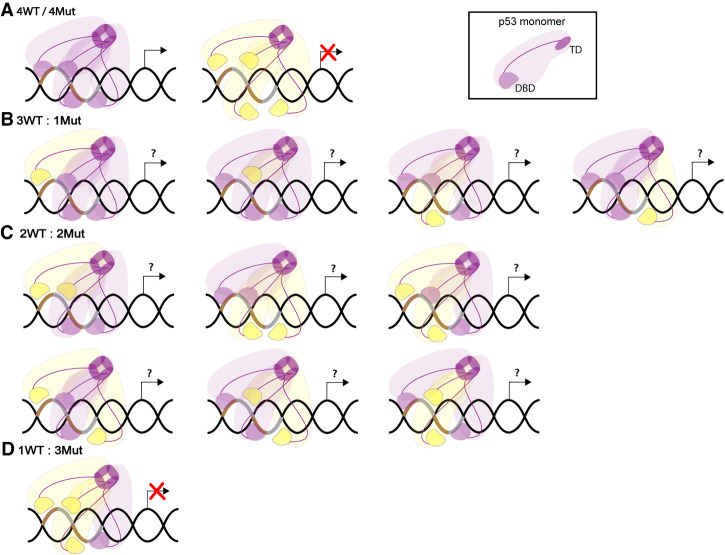
By definition, the DNE assumes that mutant and WT p53 alleles are expressed identically. Tetramers of p53 can be mixed when mutant and WT proteins are present (Fig. 5). As p53 tetramerization potentiates DNA-binding efficiency and consequently transcription, four functional DNA-binding domains are required for maximal p53 activity (Weinberg et al. 2004). Thus, the number of mutant and WT p53 molecules composing each tetramer can influence the degree of inactivation of the complex (Chan et al. 2004). In vitro, mixed tetramers of WT and the p53R273H mutant are observed (2WT:2Mut) and they are able to bind a p53 response element, although with less affinity than a WT homotetramer (Natan et al. 2009). The position of mutant p53 within a mixed tetramer could also impact transcriptional outcome depending on the nature of the response element; however, this has not been experimentally addressed. Furthermore, PTMs of mutant p53 will influence tetramerization and protein stability, also affecting tetramer composition and transcriptional output. The binding efficiency between WT and mutant p53 proteins could also dictate whether they differentially form tetramers. In addition, the binding efficiency of WT or mutant p53 molecules into homotetramers using FRET, showed that WT p53 oligomerized better than p53 mutants (V143A, R175H, R248Q, R249S, and R273H), a phenomena termed “dominant-positive effect” that could overshadow the DNE (Walerych et al. 2018). Certainly, p53 mutants with a nonfunctional TD (achieved either by direct mutations in the TD, or by the indirect effect on protein structure of a mutation in another p53 domain) will not form tetramers and will not display DNE (Milner and Medcalf 1990; Slingerland et al. 1993; Chène et al. 1997; Chène and Bechter 1999b; Xu et al. 2011).
Figure 5.
Mutant and WT p53 form mixed tetramers. (A) Homotetramers of p53 WT (purple) or p53 DBD mutant (yellow) proteins are formed when these have a functional TD. p53 WT tetramers allow efficient binding to p53 response element (brown and gray DNA segments) and are transcriptionally functional, while mutant tetramers are not. (B) Mixed tetramers composed of three p53 WT and one p53 DBD mutant proteins. (C) Mixed tetramers composed by two p53 WT and two p53 DBD mutant proteins. (B,C) Locations of mutant proteins can vary and potentially have different impacts on transcription. (D) Mixed tetramers composed of one p53 WT and three p53 DBD mutant proteins are transcriptionally inactive (Chan et al. 2004).
Many assays take advantage of overexpression systems to examine a role for mutant p53 in inhibiting WT p53 activity (Lavigueur et al. 1989; Farmer et al. 1992; Kern et al. 1992; Harvey et al. 1995; Hegi et al. 2000; Klein et al. 2000; Chan et al. 2004; Xu et al. 2011; Aubrey et al. 2018; Walerych et al. 2018). However, these approaches are often criticized, since superphysiological levels of mutant p53 overestimate the DNE, and thus these data are not discussed here. Here we discuss studies using vectors that allow equal transcription of mutant and WT p53 at comparable amounts, and cell lines with heterozygous p53 mutations. In addition, in vivo studies in mice and humans with heterozygous p53 mutations are discussed. If the effects of mutant p53 on WT p53 function are within 10% of a p53-null readout, we considered them DNE. Key studies are summarized in Tables 1 and 2.
Table 1.
When DNE is observed
Table 2.
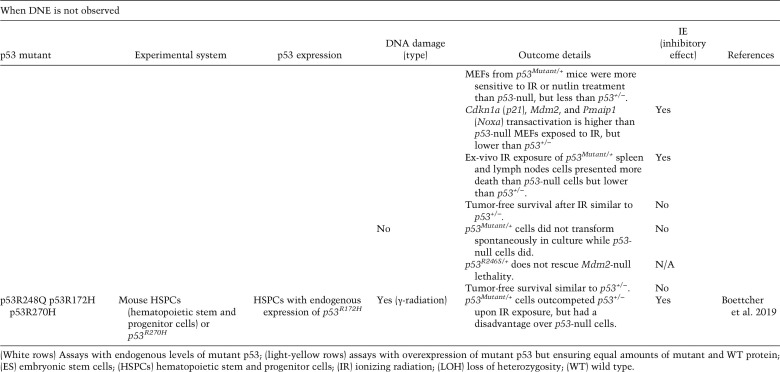
When DNE is not observed
When DNE is observed
Studies of mutant p53 expressed from its endogenous locus in p53 heterozygous mice have provided insights on the DNE (Table 1). A knock-in mouse model heterozygous for the p53R172H mutation (p53R175H in humans) contains one mutant and one WT p53 allele similar to LFS patients whom inherit a mutant TP53 allele (Lang et al. 2004; Malkin 2011). After ionizing radiation (IR), p53R172H/+ embryos show an apoptotic response in the embryonic hypothalamus that is similar to p53-null embryos providing clear evidence for inactivation of WT p53; that is, a DNE (Lang et al. 2004). In a different study, murine ES cells with p53R270H and p53P275S heterozygous mutations show increased resistance to DNA-damage, similar to p53-null cells that can be explained by delayed transactivation of p53 target genes such as Bcl2l4 (Bax) and Ccng1 (cyclin G1) (de Vries et al. 2002). Lee and Sabapathy (2008) examined mouse ES cells heterozygous for the p53 R246S mutation and report a DNE in various assays. p53R246S/+ ES cells were resistant to cell death and were unable to transactivate Pmaip1 (Noxa), Cdkn1a (p21), and Mdm2 similar to p53-null ES cells after doxorubicin treatment. The inability of differentiated p53R246S/+ ES cells to arrest during the cell cycle after DNA damage was also similar to p53-null cells. In addition, ES cells introduced into the livers of scid mice formed tumors similar to p53-null ES cells. Last, γ radiation of p53R246S/+ mice showed apoptosis in the thymus similar to p53-null mice and led to blood counts that were similar upon recovery (Lee et al. 2012). One other study of UV treatment of p53R270H/+ mice led to tumor-free survival similar to p53-null mice (14 mice per group) (Wijnhoven et al. 2007). These in vivo experiments using various p53 mutant heterozygous mice show a DNE upon treatment with various kinds of DNA damage. It should be noted that DNA damage allows PTMs on both mutant and WT p53 proteins (Harris and Levine 2005; Suh et al. 2011) possibly altering tetramer formation, protein stability, and localization.
Boettcher et al. (2019) recently reported a role for mutant p53 DNE in myeloid malignancies. By taking advantage of CRISPR-engineered AML isogenic cell lines (p53+/+, p53+/−, p53R248Q/+, p53R248Q/−, and p53−/−) expressing mutant and WT p53 from their endogenous loci, the investigators demonstrate that after DNA damage (daunorubicin), mutant p53R248Q in p53R248Q/+ cells inhibits the ability of WT p53 to activate CDKN1A transcription similar to p53-null cells. In these experiments, p53R248Q/+ cells were comparable with p53-null cells with regard to cell cycle arrest and apoptosis outcomes after daunorubicin treatment. These data clearly show that in human cell lines, mutant p53 inhibits WT p53 activity in response to DNA damage, a clear example of DNE.
When DNE is not observed
The most incontrovertible evidence that DNE does not occur in vivo includes studies with Mdm2-null mice. Mice lacking Mdm2 are embryo lethal and this phenotype is completely rescued by concomitant deletion of p53 but not by p53+/− or, more importantly, p53R172H/+. In fact, p53R172H/+ and p53R246S/+ mutants that show clear DNE under certain contexts, are embryonic lethal upon Mdm2 deletion (Lang et al. 2004; Terzian et al. 2008; Lee et al. 2012), suggesting that the DNE does not occur during embryonic development in the absence of Mdm2. Using p53 hypo-morphic alleles, expression of ∼16% of WT p53 (p53neo/neo) in an Mdm2-null background remains embryonic lethal but ∼7% WT p53 (p53neo/−) delays lethality of mice until weaning age (Wang et al. 2011). Therefore, any p53 mutant allele that partially inhibits WT p53 function to ∼16% or more of WT p53 activity would fail to rescue lethality. This minimal threshold suggests that mutant p53R172H does not inhibit WT p53 in a dominant-negative (DN) fashion in embryogenesis.
Moreover, in tumors from p53R172H/+ mice, the DNE is not apparent as analyzed by tumor free survival and loss of heterozygosity (LOH). p53R172H/+ and p53R270H/+ mice have survival curves that are intermediate between WT and p53-null (Lang et al. 2004; Olive et al. 2004). Since a DNE should inactivate the WT p53 protein, the survival curves of p53Mutant/+ mice should be more similar to p53-null mice. One argument against this logic is that p53 mutants retain some activity. This is unlikely, as the p53R172H allele (p53R172H/R172H) in mice is clearly a loss-of-function allele as it can rescue the Mdm2-null phenotype that is p53-dependent (Lang et al. 2004; Terzian et al. 2008). With regards to LOH, lymphomas and carcinomas from both p53R172H and p53R270H heterozygous mice show ∼50% LOH, suggesting again that in these cases the WT p53 protein has activity that is not sufficiently dampened by the presence of the mutant p53 protein and thus has to be removed for tumor growth. These multiple studies indicate that mutant p53 cannot fully inhibit WT p53 function.
Boettcher et al. (2019) also examined DNE for p53R172H and p53R270H in vivo. Mixed chimeric mice were generated by transplanting hematopoietic stem and progenitor cells (HSPCs) of different genotypes: p53+/−, p53−/−, p53R172H/+, and p53R270H/+ into mice and then exposed to sublethal irradiation to assess competitive fitness. p53Mutant/+ cells outcompeted p53+/− cells. However, p53Mutant/+ had a disadvantage over p53-null cells, suggesting that DNE is incomplete in vivo.
In numerous cases, the effects of mutant p53 on WT p53 seem to be tissue and context-specific. For example, whole-body IR exposure of p53R246S/+ mice showed thymocyte cell death similar to p53-null (Lee et al. 2012). However, after exposure to ex vivo radiation, thymus, spleen, and lymph nodes cells had intermediate cell viability outcomes between p53+/− and p53-null cells, while the bone marrow exhibited a cell death phenotype reminiscent of p53+/− cells. In addition, MEFs from p53R246S/+ mice were equally sensitive to IR or nutlin treatment, and had similar growth rates as p53+/− (Lee et al. 2012). Both of these experiments show clear evidence of WT p53 activity in the presence of mutant p53. Thus, these data support the idea that the effect of mutant p53 on WT p53 is partial in some cases, and is highly influenced by the environment. Hence, we propose that mutant p53 can sometimes have an inhibitory effect (IE) that is not defined as DNE, on WT p53 function. Just like the DNE, the IE is dependent on mutant p53 tetramerization and protein levels relative to WT. Mutant p53 is often present at high levels in tumor cells, increasing the likelihood of formation of inactive p53 complexes to exert IE (Fig. 5; Bartek et al. 1990; Alsner et al. 2008). The myriad of proteins and PTMs discussed above may also affect tetramerization, DNA binding, and recruitment of transcriptional machinery and consequently impose an IE. p53-interacting proteins may be present at different levels in different cancers. DNA damage and the signaling cascades that follow, modify WT and mutant p53 (perhaps differentially), and likely also inflict an IE. Data described in Table 2 show that most of the time when DNE is not observed, a mutant p53 IE over WT function is observed in the presence of DNA damage. Certainly, the IE could contribute to the course of disease in cancers that retain WT p53 (such as mucinous ovarian carcinomas) by helping transition from benign or borderline to malignant tumors (Vergara et al. 2010; Semczuk et al. 2017). Importantly, tumors that retain a WT p53 allele may possess other mechanisms, such as increased levels of MDM2, to keep WT p53 inactive.
LOH of TP53 is found in about half of LFS patients (Varley et al. 1997; Alexandrova et al. 2017) and at frequencies close to 80% in sporadic cancers (Parikh et al. 2014). LOH has been interpreted to mean that inactivation of WT TP53 is needed to increase tumor cell fitness and suggests lack of DNE (Varley et al. 1991; Dearth et al. 2007; Alexandrova et al. 2017). Additionally, age of onset, patient survival, and tumor phenotype are used to describe clinical outcomes of mutation “aggressiveness” and should be stronger in mutants with the ability to suppress the remaining WT allele; i.e., DN p53 mutants. In support of this concept, Bougeard et al. (2015) report earlier tumor onset in a French cohort of LFS carriers of tetramerization-intact p53 mutants (e.g., DBD mutants; 21.3 yr) as compared with carriers of tetramerization-deficient mutants (p53 TD or truncation mutants; 28.5 yr). However, Shahbandi and Jackson (2019) more recently report no difference in LOH frequency, tumor phenotype (grade, stage, and size) or patient survival among tetramerization-intact or tetramerization-deficient p53 mutant tumors of the breast, ovary, and lung from the cBioPortal database. Additionally, there is no difference for breast cancer onset in LFS carriers. Boettcher et al. (2019) analyzed a cohort of 164 AML patients with p53 mutations and did not find differences in aggressiveness or survival between carriers of missense (possible DNE) or truncation mutants (no DNE), again suggesting that the DNE does not contribute to outcomes. As discussed previously, differences may be context-specific and stratifying by tumor type could clarify the effect of DNE in clinical outcomes. Furthermore, consistent DNA damage may potentiate the DNE for which stratifying by DNA damage markers, such as microsatellite instability, may be informative.
Conclusions/perspectives
In summary, the p53 TD regulates many aspects of p53 biology such as protein degradation, localization, DNA binding, and target gene specificity. Mutations in the TD occur in cancers because they disrupt p53 tumor suppressor activities. The TD is also at the heart of the DNE of mutant p53 over WT p53, although, as pointed out in this review, even with a functional TD, mutant p53 often does not overwhelm WT activity. Mutant p53 proteins with functional TD often partially inhibit WT p53 activity, a process that we call an inhibitory effect (IE). The DNE clearly occurs in response to DNA damage and is likely dependent on PTMs. Overcoming DNE, or even the IE, would release WT p53 and inhibit tumor growth. A deeper understanding of the regulation of the TD is needed for development of therapeutic options that would release the p53 tumor suppressor from inhibition by mutant p53. In vivo models for monomeric and dimeric p53 proteins could help elucidate regulation of p53 tetramerization with the potential for therapeutic advancement.
Acknowledgments
G.L. is supported by grants from the National Institutes of Health (CA47296 and CA82577). J.G.-A. is a recipient of a Rosalie B. Hite Graduate Fellowship in Cancer Research.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.340976.120.
References
- Alexandrova EM, Mirza SA, Xu S, Schulz-Heddergott R, Marchenko ND, Moll UM. 2017. p53 loss-of-heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain-of-function in vivo. Cell Death Dis 8: e2661 10.1038/cddis.2017.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsner J, Jensen V, Kyndi M, Offersen BV, Vu P, Børresen-Dale AL, Overgaard J. 2008. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta Oncol 47: 600–607. 10.1080/02841860802047411 [DOI] [PubMed] [Google Scholar]
- Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. 2018. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ 25: 104–113. 10.1038/cdd.2017.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurelio ON, Kong XT, Gupta S, Stanbridge EJ. 2000. p53 mutants have selective dominant-negative effects on apoptosis but not growth arrest in human cancer cell lines. Mol Cell Biol 20: 770–778. 10.1128/mcb.20.3.770-778.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Iggo R, Gannon J, Lane DP. 1990. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene 5: 893–899. [PubMed] [Google Scholar]
- Beckerman R, Yoh K, Mattia-Sansobrino M, Zupnick A, Laptenko O, Karni-Schmidt O, Ahn J, Byeon I-J, Keezer S, Prives C. 2016. Lysines in the tetramerization domain of p53 selectively modulate G1 arrest. Cell Cycle 15: 1425–1438. 10.1080/15384101.2016.1170270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H. 2003. Announcing the worldwide Protein Data Bank. Nat Struct Mol Biol 10: 980 10.1038/nsb1203-980 [DOI] [PubMed] [Google Scholar]
- Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, Giacomelli AO, Wong W, Kim J, Chao S, et al. 2019. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365: 599–604. 10.1126/science.aax3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, Olivier M. 2016. TP53 Variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat 37: 865–876. 10.1002/humu.23035 [DOI] [PubMed] [Google Scholar]
- Bougeard G, Renaux-Petel M, Flaman JM, Charbonnier C, Fermey P, Belotti M, Gauthier-Villars M, Stoppa-Lyonnet D, Consolino E, Brugières L, et al. 2015. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol 33: 2345–2352. 10.1200/JCO.2014.59.5728 [DOI] [PubMed] [Google Scholar]
- Brown J, Higo H, McKalip A, Herman B. 1997. Human papillomavirus (HPV) 16 E6 sensitizes cells to atractyloside-induced apoptosis: role of p53, ICE-like proteases and the mitochondrial permeability transition. J Cell Biochem 66: 245–255. [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. 2002. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med 8: 282–288. 10.1038/nm0302-282 [DOI] [PubMed] [Google Scholar]
- Chan WM, Siu WY, Lau A, Poon RY. 2004. How many mutant p53 molecules are needed to inactivate a tetramer? Mol Cell Biol 24: 3536–3551. 10.1128/mcb.24.8.3536-3551.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chène P. 2001. The role of tetramerization in p53 function. Oncogene 20: 2611–2617. 10.1038/sj.onc.1204373 [DOI] [PubMed] [Google Scholar]
- Chène P, Bechter E. 1999a. Cellular characterisation of p53 mutants with a single missense mutation in the β-strand 326-333 and correlation of their cellular activities with in vitro properties. J Mol Biol 288: 891–897. 10.1006/jmbi.1999.2738 [DOI] [PubMed] [Google Scholar]
- Chène P, Bechter E. 1999b. p53 mutants without a functional tetramerisation domain are not oncogenic. J Mol Biol 286: 1269–1274. 10.1006/jmbi.1999.2563 [DOI] [PubMed] [Google Scholar]
- Chène P, Mittl P, Grütter M. 1997. In vitro structure–function analysis of the β-strand 326-333 of human p53. J Mol Biol 273: 873–881. 10.1006/jmbi.1997.1360 [DOI] [PubMed] [Google Scholar]
- Clore GM, Omichinski JG, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn AM. 1994. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science 265: 386–391. 10.1126/science.8023159 [DOI] [PubMed] [Google Scholar]
- Cox ML, Meek DW. 2010. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal 22: 564–571. 10.1016/j.cellsig.2009.11.014 [DOI] [PubMed] [Google Scholar]
- D'Abramo M, Bešker N, Desideri A, Levine AJ, Melino G, Chillemi G. 2016. The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain. Oncogene 35: 3272–3281. 10.1038/onc.2015.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearth LR, Qian H, Wang T, Baroni TE, Zeng J, Chen SW, Yi SY, Brachmann RK. 2007. Inactive full-length p53 mutants lacking dominant wild-type p53 inhibition highlight loss of heterozygosity as an important aspect of p53 status in human cancers. Carcinogenesis 28: 289–298. 10.1093/carcin/bgl132 [DOI] [PubMed] [Google Scholar]
- de Vries A, Flores ER, Miranda B, Hsieh HM, van Oostrom CT, Sage J, Jacks T. 2002. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc Natl Acad Sci 99: 2948–2953. 10.1073/pnas.052713099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49. 10.1038/ng0492-45 [DOI] [PubMed] [Google Scholar]
- Eliyahu D, Goldfinger N, Pinhasi-Kimhi O, Shaulsky G, Skurnik Y, Arai N, Rotter V, Oren M. 1988. Meth A fibrosarcoma cells express two transforming mutant p53 species. Oncogene 3: 313–321. [PubMed] [Google Scholar]
- Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. 1992. Wild-type p53 activates transcription in vitro. Nature 358: 83–86. 10.1038/358083a0 [DOI] [PubMed] [Google Scholar]
- Fischer NW, Prodeus A, Malkin D, Gariépy J. 2016. p53 oligomerization status modulates cell fate decisions between growth, arrest and apoptosis. Cell Cycle 15: 3210–3219. 10.1080/15384101.2016.1241917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer NW, Prodeus A, Tran J, Malkin D, Gariépy J. 2018. Association between the oligomeric status of p53 and clinical outcomes in li-fraumeni syndrome. J Natl Cancer Inst 110: 1418–1421. 10.1093/jnci/djy114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, Ashton AW, Fu W, Mani K, Chin S-F, et al. 2007. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci 104: 20826–20831. 10.1073/pnas.0710017104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebourg T, Sadelain M, Ng YS, Kassel J, Friend SH. 1994. Equal transcription of wild-type and mutant p53 using bicistronic vectors results in the wild-type phenotype. Cancer Res 54: 878–881. [PubMed] [Google Scholar]
- Friedman PN, Chen X, Bargonetti J, Prives C. 1993. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci 90: 3319–3323. 10.1073/pnas.90.8.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon R, Friedler A. 2014. Allosteric modulation of protein oligomerization: an emerging approach to drug design. Front Chem 2: 9 10.3389/fchem.2014.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon R, Brandt T, Sukenik S, Lahav N, Lebendiker M, Shalev DE, Veprintsev D, Friedler A. 2012. Specific recognition of p53 tetramers by peptides derived from p53 interacting proteins. PLoS One 7: e38060 10.1371/journal.pone.0038060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia G, Lahav G. 2014. Constant rate of p53 tetramerization in response to DNA damage controls the p53 response. Mol Syst Biol 10: 753 10.15252/msb.20145168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia G, Guan Y, Shah JV, Lahav G. 2013. Activation and control of p53 tetramerization in individual living cells. Proc Natl Acad Sci 110: 15497–15501. 10.1073/pnas.1311126110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, Howard TP, Takeda DY, Ly SH, Kim E, et al. 2018. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet 50: 1381–1387. 10.1038/s41588-018-0204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo S, Martos V, Santos E, Menendez M, Bo C, Giralt E, de Mendoza J. 2008. Stability and structural recovery of the tetramerization domain of p53-R337H mutant induced by a designed templating ligand. Proc Natl Acad Sci 105: 16426–16431. 10.1073/pnas.0805658105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O, Hall A, Oren M. 1989. Stabilization of the p53 transformation-related protein in mouse fibrosarcoma cell lines: effects of protein sequence and intracellular environment. Mol Cell Biol 9: 3385–3392. 10.1128/MCB.9.8.3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908. 10.1038/sj.onc.1208615 [DOI] [PubMed] [Google Scholar]
- Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower LA. 1995. A mutant p53 transgene accelerates tumour development in heterozygous but not nullizygous p53-deficient mice. Nat Genet 9: 305–311. 10.1038/ng0395-305 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299. 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- Hegi ME, Klein MA, Rüedi D, Chène P, Hamou MF, Aguzzi A. 2000. p53 transdominance but no gain of function in mouse brain tumor model. Cancer Res 60: 3019–3024. [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, Torres-Ramos M, Farrás R, Hay RT, Rodríguez MS. 2010. Oligomerization conditions Mdm2-mediated efficient p53 polyubiquitylation but not its proteasomal degradation. Int J Biochem Cell Biol 42: 725–735. 10.1016/j.biocel.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27. 10.1016/s0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- Imagawa T, Terai T, Yamada Y, Kamada R, Sakaguchi K. 2009. Evaluation of transcriptional activity of p53 in individual living mammalian cells. Anal Biochem 387: 249–256. 10.1016/j.ab.2009.01.030 [DOI] [PubMed] [Google Scholar]
- Itahana Y, Ke H, Zhang Y. 2009. p53 Oligomerization is essential for its C-terminal lysine acetylation. J Biol Chem 284: 5158–5164. 10.1074/jbc.M805696200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. 2005. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65: 10280–10288. 10.1158/0008-5472.CAN-05-2193 [DOI] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. 2008. Arginine methylation regulates the p53 response. Nat Cell Biol 10: 1431–1439. 10.1038/ncb1802 [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP. 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267: 1498–1502. 10.1126/science.7878469 [DOI] [PubMed] [Google Scholar]
- Johnson CR, Morin PE, Arrowsmith CH, Freire E. 1995. Thermodynamic analysis of the structural stability of the tetrameric oligomerization domain of p53 tumor suppressor. Biochemistry 34: 5309–5316. 10.1021/bi00016a002 [DOI] [PubMed] [Google Scholar]
- Kamada R, Nomura T, Anderson CW, Sakaguchi K. 2011. Cancer-associated p53 tetramerization domain mutants: quantitative analysis reveals a low threshold for tumor suppressor inactivation. J Biol Chem 286: 252–258. 10.1074/jbc.M110.174698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada R, Toguchi Y, Nomura T, Imagawa T, Sakaguchi K. 2016. Tetramer formation of tumor suppressor protein p53: structure, function, and applications. Biopolymers 106: 598–612. 10.1002/bip.22772 [DOI] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. 2007. Inhibition of Crm1–p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol 9: 1175–1183. 10.1038/ncb1638 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Lozano G. 1998. Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc Natl Acad Sci 95: 2834–2837. 10.1073/pnas.95.6.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. 2003. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci 100: 8424–8429. 10.1073/pnas.1431692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz C, Low-Calle AM, Choe JH, Laptenko O, Tong D, Joseph-Chowdhury JN, Garofalo F, Zhu Y, Friedler A, Prives C. 2018. Wild-type and cancer-related p53 proteins are preferentially degraded by MDM2 as dimers rather than tetramers. Genes Dev 32: 430–447. 10.1101/gad.304071.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Kato S, Otsuka K, Watanabe G, Kumabe T, Tominaga T, Yoshimoto T, Ishioka C. 2005. The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene 24: 6976–6981. 10.1038/sj.onc.1208839 [DOI] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256: 827–830. 10.1126/science.1589764 [DOI] [PubMed] [Google Scholar]
- Klein MA, Rüedi D, Nozaki M, Dell EW, Diserens AC, Seelentag W, Janzer RC, Aguzzi A, Hegi ME. 2000. Reduced latency but no increased brain tumor penetrance in mice with astrocyte specific expression of a human p53 mutant. Oncogene 19: 5329–5337. 10.1038/sj.onc.1203941 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. 2009. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J Biol Chem 284: 3250–3263. 10.1074/jbc.M805658200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303. 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- Kulikov R, Letienne J, Kaur M, Grossman SR, Arts J, Blattner C. 2010. Mdm2 facilitates the association of p53 with the proteasome. Proc Natl Acad Sci 107: 10038–10043. 10.1073/pnas.0911716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119: 861–872. 10.1016/j.cell.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Lang V, Pallara C, Zabala A, Lobato-Gil S, Lopitz-Otsoa F, Farrás R, Hjerpe R, Torres-Ramos M, Zabaleta L, Blattner C, et al. 2014. Tetramerization-defects of p53 result in aberrant ubiquitylation and transcriptional activity. Mol Oncol 8: 1026–1042. 10.1016/j.molonc.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Shiff I, Freed-Pastor W, Zupnick A, Mattia M, Freulich E, Shamir I, Kadouri N, Kahan T, Manfredi J, et al. 2015. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell 57: 1034–1046. 10.1016/j.molcel.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. 1989. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol Cell Biol 9: 3982–3991. 10.1128/mcb.9.9.3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Sabapathy K. 2008. The R246S hot-spot p53 mutant exerts dominant-negative effects in embryonic stem cells in vitro and in vivo. J Cell Sci 121: 1899–1906. 10.1242/jcs.022822 [DOI] [PubMed] [Google Scholar]
- Lee W, Harvey TS, Yin Y, Yau P, Litchfield D, Arrowsmith CH. 1994. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol 1: 877–890. 10.1038/nsb1294-877 [DOI] [PubMed] [Google Scholar]
- Lee MK, Teoh WW, Phang BH, Tong WM, Wang ZQ, Sabapathy K. 2012. Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell 22: 751–764. 10.1016/j.ccr.2012.10.022 [DOI] [PubMed] [Google Scholar]
- Lomax ME, Barnes DM, Hupp TR, Picksley SM, Camplejohn RS. 1998. Characterization of p53 oligomerization domain mutations isolated from Li-Fraumeni and Li-Fraumeni like family members. Oncogene 17: 643–649. 10.1038/sj.onc.1201974 [DOI] [PubMed] [Google Scholar]
- Lu H, Taya Y, Ikeda M, Levine AJ. 1998. Ultraviolet radiation, but not γ radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc Natl Acad Sci 95: 6399–6402. 10.1073/pnas.95.11.6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui K, Sheikh MS, Huang Y. 2015. Regulation of p53 oligomerization by Ras superfamily protein RBEL1A. Genes Cancer 6: 307–316. 10.18632/genesandcancer.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki CG. 1999. Oligomerization is required for p53 to be efficiently ubiquitinated by MDM2. J Biol Chem 274: 16531–16535. 10.1074/jbc.274.23.16531 [DOI] [PubMed] [Google Scholar]
- Malkin D. 2011. Li-fraumeni syndrome. Genes Cancer 2: 475–484. 10.1177/1947601911413466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF, Nelson CE, Kim DH, Kassel J, Gryka M, Bischoff F, Tainsky M, et al. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250: 1233–1238. 10.1126/science.1978757 [DOI] [PubMed] [Google Scholar]
- Martinez J, Georgoff I, Martinez J, Levine AJ. 1991. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev 5: 151–159. 10.1101/gad.5.2.151 [DOI] [PubMed] [Google Scholar]
- Mateu MG, Fersht AR. 1998. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J 17: 2748–2758. 10.1093/emboj/17.10.2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F, Clark R, Harlow E, Tjian R. 1981. SV40 T antigen binds specifically to a cellular 53 K protein in vitro. Nature 292: 63–65. 10.1038/292063a0 [DOI] [PubMed] [Google Scholar]
- McLure KG, Lee PW. 1998. How p53 binds DNA as a tetramer. EMBO J 17: 3342–3350. 10.1093/emboj/17.12.3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. 2009. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol 1: a000950 10.1101/cshperspect.a000950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Yue J, Liu Z, Shen Z. 2007. Abrogation of the transactivation activity of p53 by BCCIP down-regulation. J Biol Chem 282: 1570–1576. 10.1074/jbc.M607520200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J, Medcalf EA. 1990. Temperature-dependent switching between ‘wild-type’ and ‘mutant’ forms of p53-Val135. J Mol Biol 216: 481–484. 10.1016/0022-2836(90)90371-R [DOI] [PubMed] [Google Scholar]
- Milner J, Medcalf EA. 1991. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65: 765–774. 10.1016/0092-8674(91)90384-B [DOI] [PubMed] [Google Scholar]
- Mittl PR, Chène P, Grütter MG. 1998. Crystallization and structure solution of p53 (residues 326-356) by molecular replacement using an NMR model as template. Acta Crystallogr D Biol Crystallogr 54: 86–89. 10.1107/s0907444997006550 [DOI] [PubMed] [Google Scholar]
- Moll UM, Riou G, Levine AJ. 1992. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci 89: 7262–7266. 10.1073/pnas.89.15.7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. 1995. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci 92: 4407–4411. 10.1073/pnas.92.10.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora P, Carbajo RJ, Pineda-Lucena A, Sánchez del Pino MM, Pérez-Payá E. 2008. Solvent-exposed residues located in the β-sheet modulate the stability of the tetramerization domain of p53–a structural and combinatorial approach. Proteins 71: 1670–1685. 10.1002/prot.21854 [DOI] [PubMed] [Google Scholar]
- Muscolini M, Montagni E, Caristi S, Nomura T, Kamada R, Di Agostino S, Corazzari M, Piacentini M, Blandino G, Costanzo A, et al. 2009. Characterization of a new cancer-associated mutant of p53 with a missense mutation (K351N) in the tetramerization domain. Cell Cycle 8: 3396–3405. 10.4161/cc.8.20.9910 [DOI] [PubMed] [Google Scholar]
- Muscolini M, Montagni E, Palermo V, Di Agostino S, Gu W, Abdelmoula-Souissi S, Mazzoni C, Blandino G, Tuosto L. 2011. The cancer-associated K351N mutation affects the ubiquitination and the translocation to mitochondria of p53 protein. J Biol Chem 286: 39693–39702. 10.1074/jbc.M111.279539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan E, Hirschberg D, Morgner N, Robinson CV, Fersht AR. 2009. Ultraslow oligomerization equilibria of p53 and its implications. Proc Natl Acad Sci 106: 14327–14332. 10.1073/pnas.0907840106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls CD, McLure KG, Shields MA, Lee PW. 2002. Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J Biol Chem 277: 12937–12945. 10.1074/jbc.M108815200 [DOI] [PubMed] [Google Scholar]
- Nomura T, Kamada R, Ito I, Chuman Y, Shimohigashi Y, Sakaguchi K. 2009. Oxidation of methionine residue at hydrophobic core destabilizes p53 tetrameric structure. Biopolymers 91: 78–84. 10.1002/bip.21084 [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119: 847–860. 10.1016/j.cell.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Ono W, Hayashi Y, Yokoyama W, Kuroda T, Kishimoto H, Ito I, Kimura K, Akaogi K, Waku T, Yanagisawa J. 2014. The nucleolar protein Myb-binding protein 1A (MYBBP1A) enhances p53 tetramerization and acetylation in response to nucleolar disruption. J Biol Chem 289: 4928–4940. 10.1074/jbc.M113.474049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero EI, Schüler-Faccini L, Caleffi M, Achatz MI, Olivier M, Martel-Planche G, Marcel V, Aguiar E, Giacomazzi J, Ewald IP, et al. 2008. Detection of R337H, a germline TP53 mutation predisposing to multiple cancers, in asymptomatic women participating in a breast cancer screening program in Southern Brazil. Cancer Lett 261: 21–25. 10.1016/j.canlet.2007.10.044 [DOI] [PubMed] [Google Scholar]
- Parikh N, Hilsenbeck S, Creighton CJ, Dayaram T, Shuck R, Shinbrot E, Xi L, Gibbs RA, Wheeler DA, Donehower LA. 2014. Effects of TP53 mutational status on gene expression patterns across 10 human cancer types. J Pathol 232: 522–533. 10.1002/path.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Li J, Starost MF, Liu C, Zhuang J, Chen J, Achatz MI, Kang J-G, Wang P-y, Savage SA, et al. 2018. Mouse homolog of the human TP53 R337H mutation reveals its role in tumorigenesis. Cancer Res 78: 5375–5383. 10.1158/0008-5472.CAN-18-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Chambers KA, Pabo CO. 1993. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev 7: 2556–2564. 10.1101/gad.7.12b.2556 [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629. 10.1002/humu.20495 [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 2008. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res 36: 5983–5991. 10.1093/nar/gkn598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Huang F, Fersht AR. 2011. Single-molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res 39: 2294–2303. 10.1093/nar/gkq800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro RC, Sandrini F, Figueiredo B, Zambetti GP, Michalkiewicz E, Lafferty AR, DeLacerda L, Rabin M, Cadwell C, Sampaio G, et al. 2001. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc Natl Acad Sci 98: 9330–9335. 10.1073/pnas.161479898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412. 10.1038/nrm2395 [DOI] [PubMed] [Google Scholar]
- Rollenhagen C, Chène P. 1998. Characterization of p53 mutants identified in human tumors with a missense mutation in the tetramerization domain. Int J Cancer 78: 372–376. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Sakamoto H, Lewis MS, Anderson CW, Erickson JW, Appella E, Xie D. 1997. Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry 36: 10117–10124. 10.1021/bi970759w [DOI] [PubMed] [Google Scholar]
- Schumacher B, Mondry J, Thiel P, Weyand M, Ottmann C. 2010. Structure of the p53 C-terminus bound to 14-3-3: implications for stabilization of the p53 tetramer. FEBS Lett 584: 1443–1448. 10.1016/j.febslet.2010.02.065 [DOI] [PubMed] [Google Scholar]
- Semczuk A, Gogacz M, Semczuk-Sikora A, Jóźwik M, Rechberger T. 2017. The putative role of TP53 alterations and p53 expression in borderline ovarian tumors—correlation with clinicopathological features and prognosis: a mini-review. J Cancer 8: 2684–2691. 10.7150/jca.19691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbandi A, Jackson JG. 2019. Analysis across multiple tumor types provides no evidence that mutant p53 exerts dominant negative activity. NPJ Precis Oncol 3: 1 10.1038/s41698-018-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulsky G, Goldfinger N, Peled A, Rotter V. 1991. Involvement of wild-type p53 protein in the cell cycle requires nuclear localization. Cell Growth Differ 2: 661–667. [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 14: 289–300. 10.1101/gad.14.3.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingerland JM, Jenkins JR, Benchimol S. 1993. The transforming and suppressor functions of p53 alleles: effects of mutations that disrupt phosphorylation, oligomerization and nuclear translocation. EMBO J 12: 1029–1037. 10.1002/j.1460-2075.1993.tb05744.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Wang S, Tong YA, Hao ZM, Chang EH. 1993. Dominant negative effect of a germ-line mutant p53: a step fostering tumorigenesis. Cancer Res 53: 4452–4455. [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 18: 1660–1672. 10.1093/emboj/18.6.1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK, Van Pelt C, Terzian T, Lozano G. 2011. Multiple stress signals activate mutant p53 in vivo. Cancer Res 71: 7168–7175. 10.1158/0008-5472.CAN-11-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851. 10.1016/j.molcel.2006.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839. 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. 2008. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 22: 1337–1344. 10.1101/gad.1662908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. 2009a. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem 284: 13804–13811. 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dieck J, Teufel DP, Jaulent AM, Fernandez-Fernandez MR, Rutherford TJ, Wyslouch-Cieszynska A, Fersht AR. 2009b. Posttranslational modifications affect the interaction of S100 proteins with tumor suppressor p53. J Mol Biol 394: 922–930. 10.1016/j.jmb.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Varley JM, Brammar WJ, Lane DP, Swallow JE, Dolan C, Walker RA. 1991. Loss of chromosome 17p13 sequences and mutation of p53 in human breast carcinomas. Oncogene 6: 413–421. [PubMed] [Google Scholar]
- Varley JM, Thorncroft M, McGown G, Appleby J, Kelsey AM, Tricker KJ, Evans DGR, Birch JM. 1997. A detailed study of loss of heterozygosity on chromosome 17 in tumours from Li-Fraumeni patients carrying a mutation to the TP53 gene. Oncogene 14: 865–871. 10.1038/sj.onc.1201041 [DOI] [PubMed] [Google Scholar]
- Vergara D, Tinelli A, Martignago R, Malvasi A, Chiuri VE, Leo G. 2010. Biomolecular pathogenesis of borderline ovarian tumors: focusing target discovery through proteogenomics. Curr Cancer Drug Targets 10: 107–116. 10.2174/156800910790980269 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431. 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- Walerych D, Pruszko M, Zyla L, Wezyk M, Gaweda-Walerych K, Zylicz A. 2018. Wild-type p53 oligomerizes more efficiently than p53 hot-spot mutants and overcomes mutant p53 gain-of-function via a “dominant-positive” mechanism. Oncotarget 9: 32063–32080. 10.18632/oncotarget.25944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Suh YA, Fuller MY, Jackson JG, Xiong S, Terzian T, Quintás-Cardama A, Bankson JA, El-Naggar AK, Lozano G. 2011. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest 121: 893–904. 10.1172/JCI44504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock LJ, Knox A, Mee TR, Raines SA, Milner J. 2008. Influence of tetramerisation on site-specific post-translational modifications of p53: comparison of human and murine p53 tumour suppressor protein. Cancer Biol Ther 7: 1481–1489. 10.4161/cbt.7.9.6473 [DOI] [PubMed] [Google Scholar]
- Wasylishen AR, Lozano G. 2016. Attenuating the p53 pathway in human cancers: many means to the same end. Cold Spring Harb Perspect Med 6: a026211 10.1101/cshperspect.a026211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Fersht AR. 2004. Cooperative binding of tetrameric p53 to DNA. J Mol Biol 341: 1145–1159. 10.1016/j.jmb.2004.06.071 [DOI] [PubMed] [Google Scholar]
- Wijnhoven SW, Speksnijder EN, Liu X, Zwart E, vanOostrom CT, Beems RB, Hoogervorst EM, Schaap MM, Attardi LD, Jacks T, et al. 2007. Dominant-negative but not gain-of-function effects of a p53.R270H mutation in mouse epithelium tissue after DNA damage. Cancer Res 67: 4648–4656. 10.1158/0008-5472.CAN-06-4681 [DOI] [PubMed] [Google Scholar]
- Willis A, Jung EJ, Wakefield T, Chen X. 2004. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 23: 2330–2338. 10.1038/sj.onc.1207396 [DOI] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine J-C, et al. 2011. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 7: 285–295. 10.1038/nchembio.546 [DOI] [PubMed] [Google Scholar]
- Xu J, Zhou X, Wang J, Li Z, Kong X, Qian J, Hu Y, Fang JY. 2013. RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep 3: 1526–1538. 10.1016/j.celrep.2013.04.017 [DOI] [PubMed] [Google Scholar]
- Yakovlev VA, Bayden AS, Graves PR, Kellogg GE, Mikkelsen RB. 2010. Nitration of the tumor suppressor protein p53 at tyrosine 327 promotes p53 oligomerization and activation. Biochemistry 49: 5331–5339. 10.1021/bi100564w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonish-Rouach E, Resnftzky D, Lotem J, Sachs L, Kimchi A, Oren M. 1991. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352: 345–347. 10.1038/352345a0 [DOI] [PubMed] [Google Scholar]