Abstract
Key discoveries in Drosophila have shaped our understanding of cellular “enhancers.” With a special focus on the fly, this chapter surveys properties of these adaptable cis-regulatory elements, whose actions are critical for the complex spatial/temporal transcriptional regulation of gene expression in metazoa. The powerful combination of genetics, molecular biology, and genomics available in Drosophila has provided an arena in which the developmental role of enhancers can be explored. Enhancers are characterized by diverse low- or high-throughput assays, which are challenging to interpret, as not all of these methods of identifying enhancers produce concordant results. As a model metazoan, the fly offers important advantages to comprehensive analysis of the central functions that enhancers play in gene expression, and their critical role in mediating the production of phenotypes from genotype and environmental inputs. A major challenge moving forward will be obtaining a quantitative understanding of how these cis-regulatory elements operate in development and disease.
Keywords: embryo patterning, enhancer, gene regulation, transcription, transcription factor, FlyBook
THE body plans of complex eukaryotes are composed of hundreds of distinct cell types, but with few exceptions, all cells in an organism contain identical genomic sequences. What makes cell types different from each other is that each one expresses a unique combination of messenger RNAs (mRNAs) and structural RNAs. During development, two major processes, asymmetric cell division and cell–cell signaling, drive cell differentiation and embryo patterning. Asymmetric cell division leads to the unequal distribution of cytoplasmic determinants in each daughter cell; signals from sending cells are received by other cells, and relayed to second messengers in the cytoplasm. Both processes trigger changes in the combinations, concentrations, or activities of transcription factors (TFs) in the nucleus, which activate and/or repress transcription of downstream target genes by binding specifically to regulatory DNA. Some target genes encode other TFs or signaling molecules that stimulate neighboring cells, contributing to a network that regulates cellular differentiation and organizes distinct differentiation pathways in precise regions of the developing embryo over time.
Research on Drosophila melanogaster has provided insights into the molecular workings that unfold the genetic code during the process of embryo development. Here, we focus on enhancers, specific regulatory elements capable of influencing RNA polymerase II-dependent gene expression in a distance- and orientation-independent manner (Banerji et al. 1981; Moreau et al. 1981). The interactions among TFs on enhancers dictate the regulatory potential of these elements, which is realized by contacts between enhancers and the general transcriptional machinery found at transcriptional start sites (TSSs).
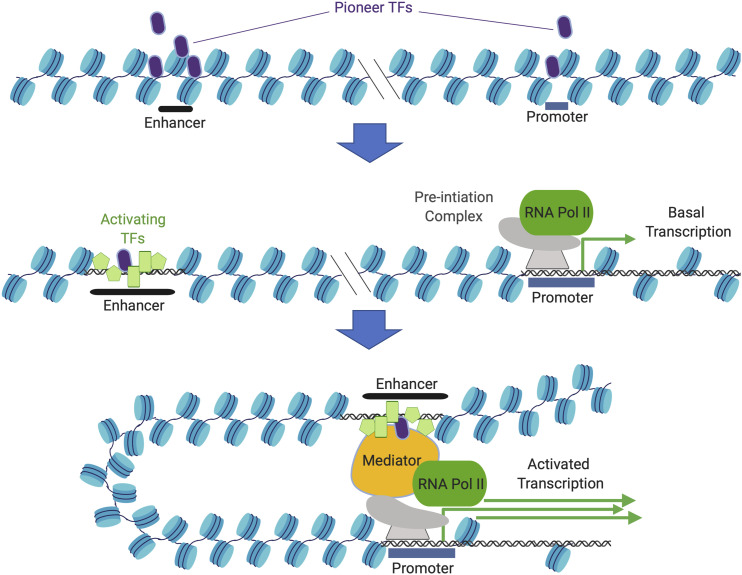
Enhancer activity is controlled at three general levels (Figure 1). First, if the enhancer lies in a region of compacted chromatin, the region must be converted to a less-compacted or open state, which can involve the action of so-called “pioneer” TFs [reviewed in Zaret and Mango (2016)]. Unlike many TFs that are targeted to nucleosome-free areas, pioneer factors can bind to motifs even when they are wrapped on a nucleosome. Second, primed by pioneer factors, enhancers (and many promoters) are bound by additional sequence-specific TFs that are critical for the execution of their functions [reviewed in Peter (2015)]. The TSS is the location of the basal promoter, which determines directionality and marks where transcription starts [reviewed in Vo Ngoc et al. (2019)]. In some cases, even before a gene is expressed, the basal promoter is occupied by RNA polymerase II and proteins of the general transcriptional machinery [reviewed in Gaertner and Zeitlinger (2014) and Core and Adelman (2019)]. This “preinitiation complex” (PIC) can include an RNA polymerase II that has not yet begun transcription and has not acquired specific phosphorylation marks on the C-terminus (poised), or has produced a short transcript and then arrested (paused or stalled) (Rougvie and Lis 1988; Radonjic et al. 2005). Third, enhancers bound by sequence-specific TFs are sometimes capable of activating target genes in a “hard-wired” mode, without requiring the selective activation of facultative signaling systems. Alternatively, some enhancers require further signaling to permit the binding of relevant transcriptional cofactors to engage the transcriptional machinery (Barolo and Posakony 2002).
Figure 1.
Developmental regulation of gene expression by transcriptional enhancers. (A) Enhancer and promoter sequences are compacted in chromatin prior to gene activity. Pioneer factors can interact with the locus, changing its accessibility for binding by activating TFs. (B) Partially or fully assembled sets of TFs can associate with the enhancer and the promoter. In cell types in which the enhancer is actively repressed, specific TFs may remodel chromatin structure to reduce but not eliminate access. Bifunctional TFs may be alternatively associated with corepressors or coactivators depending on the signaling state of the cell. (C) In the active state, the enhancer may physically associate with the promoter, triggering formation of a preinitiation complex and/or promoting the release of paused transcriptional complexes. Created with Biorender.com. RNA Pol II, RNA polymerase II; TF, transcription factor.
An active enhancer can strongly increase the production of the associated full-length mRNA. Because enhancers and promoters can be separated by many kilobases along the genomic sequence, DNA-looping mechanisms have been proposed to explain how they can physically interact in the nucleus [reviewed in Schoenfelder and Fraser (2019)]; these interactions may be on the same strand (cis), although interactions between an enhancer on one allele and a promoter on the other have been observed (Lewis 1954). This process is called transvection, and is especially prevalent in Drosophila, in which autosomes are paired during interphase (Morris et al. 1998). Single-molecule assays have shown that enhancers can affect the activity of a promoter in several ways, including increasing the frequency of the promoter’s switch to an “on” state, increasing the length of time that the promoter stays “on,” and increasing the rate of polymerase initiation while the promoter is activated [reviewed in Nicolas et al. (2017)].
The focus of this chapter will be on enhancers and the sequence-specific TFs that bind them, with a strong emphasis on the role of Drosophila as an experimental model for understanding how these interactions contribute to embryo development. Three characteristics of Drosophila make it especially well suited for studying enhancer-mediated mechanisms. First, from extensive genetic analysis, a broad set of mutants unlocked the key elements of enhancers and their cognate TFs. Specific regulatory mutations of key patterning genes turned out to affect loci containing enhancers, mirroring the classic lac operator mutants that were instrumental for Jacob and Monod (1961). Further, genetic screens in the second half of the 20th century identified > 50 loci involved in patterning the major axes of the Drosophila body plan (Lewis 1978; Kaufman et al. 1980; Lewis et al. 1980; Nüsslein-Volhard and Wieschaus 1980; Kornberg 1981). When cloned, the great majority of these genes were found to encode TFs, including the Hox proteins, and components of signaling pathways, including Wingless (Wg) and Hedgehog (Hh). Second, enhancer activities are easily studied in early embryogenesis, which involves a series of 14 synchronized nuclear divisions that generate a syncytium of ∼6000 tightly packed nuclei in a two-dimensional space near the cortical region of the embryo (Foe and Alberts 1983). This special developmental stage provides an unparalleled platform for visualizing mRNA and protein expression patterns as they form (see Box 1). Finally, many technical advances, most notably the use of P-elements to generate animals with single-copy, intact transgenes (Spradling and Rubin 1982), and recombination-based methods to insert transgenes into specific genomic landing sites (Golic et al. 1997; Groth et al. 2004; Bateman et al. 2006), were pioneered in Drosophila. These methods allow the manipulation of any cis-regulatory element, and any trans-acting factor, in specific developmental settings. Together, these advantages have allowed Drosophila researchers to make unparalleled progress toward understanding how enhancers respond to specific concentrations of TFs, integrate the binding activities of multiple TFs, and form the spatial and temporal patterns of transcription that foreshadow the Drosophila body plan.
Box 1 Assaying enhancer activity in cells and in vivo
Reporter Gene Assays in Cultured Cells
These assays are extremely efficient for the rapid assessment of enhancers that are normally active in the transfected cells. Inputs of specific factors can be effectively assessed by deletion or mutation of their binding sites, while factors normally absent can be introduced with a low background from the endogenous genes. In classic experiments, reporter gene outputs were quantified by measuring the radioactive products of enzymes such as chloramphenicol acetyl transferase. These assays were largely replaced by reporter genes that express enzymes such as luciferase, which produce fluorescent products.
Reporter Gene Assays in vivo (Fixed Specimens and Tissues)
The use of P-elements and recombination systems have allowed fly researchers to introduce single-copy reporter genes into the genome, which allows them to be studied in a more biologically relevant trans-acting environment. Initial assays of lacZ reporter genes used antibodies to detect LacZ protein or antisense RNA probes to detect the lacZ mRNA. Foundational experiments, including the identification of HOX and pair-rule gene expression patterns, relied on radiolabeled antisense probes hybridized to sectioned embryos, which took weeks to image. Refinements in in situ hybridization using nonradioactive probes allowed this method to become widespread. The development of sensitive fluorescent reporters, including the expression of GFP and derivatives, has greatly improved the sensitivity of in vivo reporter assays. High-resolution imaging combined with in situ hybridization has provided additional avenues for quantitative assessment of gene expression. Single molecular fluorescence in situ hybridization (FISH) techniques are sensitive enough to measure single mRNA transcripts in the cytoplasm of fixed samples. By multiplexing probes with distinct wavelengths, many individual mRNA species can be visualized in single cells of fixed specimens or tissues.
Live Imaging Assays in vivo
A decisive advance in following transcriptional control came about with the use of in vivo live imaging techniques. These approaches rely on visualizing nascent transcripts as they are produced in the nucleus. Briefly, RNA sequences that form hairpins are inserted into the transcription units of reporter genes. These hairpins represent high-affinity binding sites for bacteriophage capsid proteins such as MS2, which can be expressed ubiquitously in Drosophila as a fusion to GFP. When the hairpin-containing mRNA is produced in the nucleus, MS2-GFP protein binds to nascent transcripts, appearing as bright puncta. As the mRNAs are processed and exported to the cytoplasm, the puncta disappear. Thus, the GFP signals serve as a proxy for timing and quantifying immediate transcriptional activity. These methods have in recent years allowed researchers to measure transcription rates in live embryos, which has contributed greatly to our understanding of the dynamics involved in enhancer-mediated transcription.
Discovery of Eukaryote Enhancers
Studies of eukaryotic transcriptional regulation were built on foundations laid years before, when bacteria and phages were used to identify the molecules and processes of the Central Dogma. In the 30 years following the Second World War, genetic and biochemical approaches in bacteria combined to identify RNA polymerase [reviewed in Hurwitz (2005)], DNA elements (promoters) that help position and regulate its activity (Pribnow 1975), and activator proteins (σ factors) that directly recruit RNA polymerase to promoters (Busby and Ebright 1999). Influenced by these early studies, an analogous foray with mammalian viruses was deemed the most efficient route to elucidating transcriptional mechanisms in eukaryotes. Fractionation of crude extracts led to the isolation of protein components, and highly sensitive radiolabeled substrates allowed accurate identification of minute amounts of correctly initiated products (Reinberg et al. 1987). Viral transcripts were produced in vitro and compared with those from infected cells (Dignam et al. 1983). Endogenous cellular transcripts expressed at high levels were also studied, and the TATA box was discovered to be proximal to the histone TSS, similar to the −10 TATA sequence from bacterial genes (Grosschedl et al. 1981). This discovery motivated the search for eukaryotic specificity factors analogous to bacterial σ factors; one of the first discovered eukaryotic TFs, the “specificity protein” Sp1, was shown to be just such a factor because it could stimulate in vitro transcription from promoters bearing GC-rich Sp1-binding sites (Dynan and Tjian 1983).
By the mid-1980s, a basic framework for the specificity of eukaryotic transcription was established. RNA polymerase II was shown to interact with basal promoter complexes that establish the position of transcription initiation, much like the σ factor-containing bacterial enzyme (Reinberg et al. 1987; Helmann and Chamberlin 1988). Sequence-specific activators bound in close proximity to the promoter boosted the efficiency of this process. Studies in the budding yeast Saccharomyces cerevisiae, which has a very compact genome, showed that the regulatory sequences of most genes were located within the 1-kbp region 5′ of the basal promoter (Strathern et al. 1981; Struhl 1982). In parallel, Walter Schaffner at the University of Zurich and Pierre Chambon at the Centre National de la Recherche Scientifique showed that the simian vacuolating virus 40 (SV40) virus genome contains regulatory segments that greatly increase the expression of the rabbit β-globin gene in HeLa cells [Banerji et al. 1981; Moreau et al. 1981; reviewed in Schaffner (2015)]. These segments functioned when physically separated by > 1 kbp from the β-globin promoter, and when placed 5′ or 3′ of the β-globin transcription unit. Thus the SV40 sequences were the first identified enhancers. Originally, these sequences were thought to be unique to viruses, but the subsequent identification of endogenous cellular enhancers in an intron of the IgH gene (Banerji et al. 1983; Gillies et al. 1983) suggested that enhancers in higher eukaryotes function at long distances from their TSSs. In mammals, enhancers can lie great distances (even > 1 Mbp) from their target promoters (Lettice et al. 2002; Sagai et al. 2004), and action from a distance seems to be the rule in such highly dispersed genomes.
Defining Enhancers in Flies
The ∼140-Mbp Drosophila genome contains ∼15,000 genes with an average size of ∼10 kbp, and is thus intermediate in gene density between the compact yeast genome (6000 genes in 13 Mbp) and highly dispersed genomes of mammals (> 20,000 genes dispersed in a 3-Gbp genome). Certain classes of genes in Drosophila, including widely expressed “housekeeping” genes such as those encoding ribosomal proteins (Baumann and Gilmour 2017) and genes associated with terminal differentiation (Michiels et al. 1989; Papatsenko et al. 2001), tend to feature promoter-proximal regulatory sequences. In contrast, genomic comparisons of fly and worm genes have demonstrated that average intergenic spacing is particularly large for genes encoding TFs and signaling molecules, suggesting that much DNA is dedicated to distal enhancers for the regulation of these classes of genes (Nelson et al. 2004). For example, the segmentation genes that establish the body plans of insects are expressed in complex temporal and spatial patterns (Akam 1987; Ingham 1988). Individual parts of these patterns are regulated by modular enhancers that can be located several tens of kilobases away from their associated TSSs. When tested in reporter genes, these enhancers function at a distance and when placed 5′ or 3′ of a TSS, and thus fulfill the classical enhancer definition.
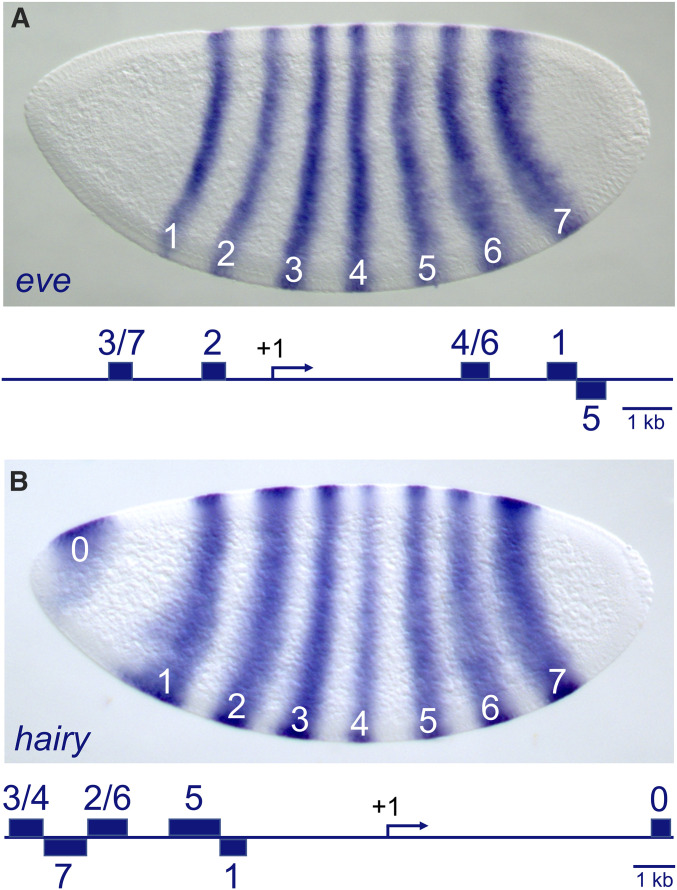
Among the best-characterized enhancers are those that control the expression of the pair-rule genes, which are expressed in patterns of seven stripes in blastoderm-stage embryos (e.g., Figure 2). These genes were originally grouped together based on their mutant phenotypes, which exhibit reiterated deletions in every other segment along the anterior–posterior (AP) axis of the first-instar larva (Nüsslein-Volhard and Wieschaus 1980). For example, embryos bearing mutations in the pair-rule hairy (h) gene fail to form the anterior region of the even-numbered segments (numbers 2, 4, 6, etc.), while the odd-numbered segments are unaffected (Ish-Horowicz et al. 1985). Intriguingly, an analysis of h deletion mutants with breakpoints at different positions upstream of the TSS showed that defects in individual even-numbered segments were always associated with the absence of specific genomic regions (Howard et al. 1988). These observations suggested that h expression in different segment primordia is controlled by specific genomic regions, each of which responds to a different set of positional cues. By cloning these regions into reporter genes and truncating fragments that contain stripe-forming activities, modular enhancers were identified for most of the h stripes (Figure 2; Hooper et al. 1989; Howard and Struhl 1990) (Pankratz et al. 1990; Riddihough and Ish-Horowicz 1991). Similar reporter gene studies of the pair-rule gene even-skipped (eve) identified stripe-specific enhancers in regions upstream and downstream of the eve transcription unit (Goto et al. 1989; Harding et al. 1989; Stanojevic et al. 1991; Small et al. 1996; Fujioka et al. 1999). Indeed, all pair-rule genes analyzed so far contain at least one stripe-specific enhancer (Yu and Pick 1995; Klingler et al. 1996; Schroeder et al. 2011). These early studies emphasized the one-enhancer/one-regulatory-pattern idea; however, many enhancers are reused in multiple developmental windows, sometimes binding to the same TFs or alternatively recruiting different stage-specific factors (Preger-Ben Noon et al. 2018).
Figure 2.
Enhancer-mediated expression of the pair-rule genes even-skipped (eve) and hairy. Embryos were stained by in situ hybridization to detect eve (A) and hairy (B) messenger RNAs. Both genes are expressed in striped patterns that help organize the segmented body of the Drosophila embryo. The genetic loci for eve and hairy are shown as schematics below their expression patterns. Transcription start sites are indicated by the +1 arrows, and modular enhancer sequences are shown as blue rectangles. Individual enhancers contain unique combinations of transcription factor-binding sites (e.g., see Figure 5), and independently direct the expression of one or two stripes. In situ images courtesy of Michael Zhang and Pinar Onal.
Genome-Wide Identification of Enhancers in Drosophila
The first Drosophila enhancers were studied using reporter genes and in vitro DNA-binding assays. As data accumulated, it became clear that enhancers share features that could be harnessed to efficiently identify similar elements. Most enhancers consist of discrete, uninterrupted segments of DNA ranging in size from several hundred to 1000 bp. These segments contain a high density of accessible binding sites for TFs compared to the whole genome, and computer algorithms were developed to search for clusters of sites in genomic DNA, which led to the discovery of many novel enhancers (Chen et al. 1995; Berman et al. 2002; Halfon et al. 2002; Markstein et al. 2002; Papatsenko et al. 2002; Rajewsky et al. 2002). Enhancers, like other functional regions of the genome, are likely to be conserved during evolution, and this property was used to improve the predictive ability of site cluster-finding algorithms (Bergman et al. 2002; Emberly et al. 2003; Berman et al. 2004). In addition, while sequences of enhancers can be extensively altered over evolutionary time (Hare et al. 2008; Swanson et al. 2010), the enrichment of conserved motifs within an enhancer region can reveal the functional element, even over great evolutionary distances, such as Drosophila to mosquito (Kantorovitz et al. 2009). Specific molecular approaches have also accelerated the discovery of enhancers; the use of chromatin immunoprecipitation (ChIP) and DNA Adenine Methyltransferase Identification (DAM-ID) methods has enabled researchers to isolate chromatin fragments that are directly bound by a given TF (Gilmour and Lis 1984; van Steensel et al. 2001; Li et al. 2008). A variety of methods [nascent transcript mapping, ChIP-sequencing (Seq) with antibodies against histone modifications, DNAse-Seq, Mnase-Seq, FAIRE (formaldehyde-assisted isolation of regulatory elements)-Seq, and ATAC-Seq] have been used to make genome-wide maps of accessible, active regions of chromatin. These methods have demonstrated that enhancer activation can be correlated with a loosening of chromatin (Thomas et al. 2011; Cusanovich et al. 2018), the appearance of specific histone modifications (Bonn et al. 2012), and to some degree low levels of transcription of the enhancer sequences themselves [enhancer RNAs (eRNAs)] (Mikhaylichenko et al. 2018).
Several studies have examined the utility of employing chromatin accessibility and modifications to identify regulatory elements critical for tissue-specific gene expression in Drosophila. However, accessibility alone may not provide a strong prediction of gene expression. For instance, patterns of open chromatin among distinct imaginal discs of the same developmental stage are similar, despite their different expression patterns (McKay and Lieb 2013). These global patterns do change with developmental time. A more differentiated picture emerges from fine-scale cell-type analysis; while open chromatin segments themselves are not very predictive of enhancers driving gene expression in the Drosophila embryo CNS midline cells, regions showing tissue-specific enrichment in the FAIRE-Seq signal were strongly enriched in midline enhancers (Pearson et al. 2016). Similarly, DNA elements differentially enriched for greater ATAC-Seq accessibility in specific segments of the Drosophila blastoderm embryo were highly indicative of active enhancers for pair-rule genes (Bozek et al. 2019). Similar trends emerge from single-cell assessment of chromatin from three distinct embryonic stages (Cusanovich et al. 2018).
Two recent studies used unbiased high-throughput methods to identify enhancers and estimate how much of the noncoding genome is dedicated to enhancer-like activities. The first analyzed a collection of 7705 transgenic lines, each of which contained a randomly chosen 2-kbp fragment of unique noncoding sequence (Kvon et al. 2014). Embryos were collected throughout embryogenesis (0–24 hr after egg laying) for each line and assayed by in situ hybridization to detect reporter gene RNA at any time during this period. Remarkably, nearly one-half (3557) showed some patterned reporter gene expression, showing that the fly genome is densely populated with regulatory elements. Because the collection covers roughly 10% of the unique noncoding regions of the genome, the authors estimate that there are between 50,000 and 100,000 enhancers involved in the process of embryogenesis. In a second study from the same laboratory, a high-throughput method [self-transcribing active regulatory region (STARR)-Seq] was used to screen random fragments from the genome for enhancer activity in cultured cells (Arnold et al. 2013). Briefly, randomly chosen genomic DNA fragments were cloned into two plasmid vectors designed to transcribe an open reading frame and the cloned regions themselves if the elements had enhancer-like activity. After transfection of the library into cultured cells, RNA-Seq was performed, which led to the identification of thousands more fragments that showed positive regulatory activity in cells. The key advantage to STARR-seq is that one can in theory scan the entire genome for regulatory elements, although this approach is limited to testing one cell type at a time. Stark and colleagues further showed that this method produces very different results depending on the basal promoter used in the library vector, indicating that false negatives are likely (Zabidi et al. 2015). Unlike the in vivo setting, signaling-dependent enhancers can be identified only if the relevant signal, such as a hormone, is known and can be added in vitro (Shlyueva et al. 2014).
In the 35 years since the discovery of the first Drosophila enhancers, > 1100 published studies have identified > 24,000 enhancers that are active in vivo or in cultured cells [http://redfly.ccr.buffalo.edu (Halfon et al. 2008)]. Remarkably, the activities of almost 14,000 enhancers have been validated by reporter gene assays in transgenic embryos.
Limitations of Reporter Genes and Complementary Methods
Reporter gene assays (Box 1) have been widely used for the study of enhancer function, but they have significant limitations. For example, reporters in cultured cells only respond to TFs expressed by those cells; thus, they cannot mimic the diversity of cell types encountered in vivo. Furthermore, transient transfection assays introduce variable numbers of transgenes into each cell, with an uncharacterized chromatin state, which significantly complicates any quantification of the results. Also, false-positive results may stem from the relaxed accessibility of regulatory sites on these transfected genes. Other limitations arise from the design of most reporter genes, and apply to both cell culture and in vivo assays. For example, most studies place the enhancer adjacent to the basal promoter, which may introduce chromatin displacement or steric effects not present in the endogenous gene.
These limitations are readily seen in studies of single-copy reporter genes that are stably integrated into the genome and analyzed in vivo. While many reporter genes drive expression patterns that are indistinguishable from those produced by their associated genes, this is not always the case. Such discrepancies can be explained in several ways. First, because reporter genes are inserted into nonendogenous genomic regions, they may be subjected to position effects from regulatory regions surrounding the insertion site. This is a serious concern, even when transgenes are inserted by recombination into commonly used landing sites. For example, Kvon et al. took 78 different enhancers that showed positive activities when inserted into a landing site on chromosome 2, and inserted them into a second landing site on chromosome 3 (Kvon et al. 2014). Only 47 (60%) showed identical patterns at both locations. The rest drove patterns that were weaker/negative (12 fragments) or spatially different (19 fragments) when inserted into the chromosome 3 landing site. Second, fragments tested for enhancer-like activities in reporter genes are tested in isolation, and may lack adjacent TF sites that ensure full activity, boundary elements, and/or polycomb response elements. Third, the identification of a minimal sequence with enhancer activity may lead to the false conclusion that this is the main source of that activity. Recent studies have identified an increasing number of genes that contain multiple “shadow” enhancers that drive very similar expression patterns (Zuo et al. 1991; Hong et al. 2008a; Perry et al. 2010, 2011; Fujioka and Jaynes 2012; Cannavò et al. 2016). Multiple enhancers may work together to create more robust expression patterns or refine each other’s expression patterns through synergistic or antagonistic interactions (Perry et al. 2012; Bothma et al. 2015). They may also provide a redundant system that permits one enhancer to evolve, while the other maintains critical patterning activities (Hong et al. 2008a).
A complementary approach to reporter gene assays is to delete them from larger genomic fragments that more accurately reflect the endogenous chromatin environment. These experiments test whether a specific sequence is necessary for gene expression and function. For very large genes (up to 200 kbp in length), researchers have constructed artificial chromosomes in bacteria (O’Connor et al. 1989) or yeast (Murray and Szostak 1983; Mouse Genome Sequencing Consortium et al. 2002). For smaller genes (up to 20 kb), these experiments are performed using traditional transgenes. For example, the eve locus is contained in a 16-kbp genomic sequence that can rescue the eve null phenotype (Fujioka et al. 1999). In the first experiment of its kind, the 480-bp minimal eve stripe 2 enhancer was deleted from the rescue transgene to test whether it is required for gene function (Ludwig et al. 2005). This deletion dramatically reduced stripe 2 expression and changed the expression pattern of the downstream gene engrailed, causing a lethal phenotype. Unexpectedly, the eve expression pattern driven by the 480-bp deletion construct retained residual expression at the stripe 2 position, indicating that additional sequences outside this minimal element contain spatial patterning information.
The most powerful way to test enhancer requirement is to delete it or mutate it in the context of the endogenous locus. Traditional genetic screens have identified mutations that disrupt or delete enhancers, as mentioned above for hairy, while more directed approaches have employed homologous recombination strategies in Drosophila (Rong and Golic 2000). More recent clustered regularly interspaced short palindromic repeats (CRISPR)/CAS9 approaches have greatly facilitated such studies, allowing the engineering of precise mutations in endogenous genes [reviewed in Bier et al. (2018)]. Significantly, while such mutations provide the most physiologically relevant information, there is a strong possibility of false-negative results because of enhancer redundancy. For instance, mutations of the complex enhancers regulating shaven-baby cause phenotypic changes only when assayed in specific genetic backgrounds, or in heat-stressed conditions (Tsai et al. 2019). Also, genomic perturbations may impact gene expression in unexpected ways. For example, deleting or mutating enhancers contained within introns can disrupt the function of an unannotated exon or interfere with mRNA splicing [reviewed in Catarino and Stark (2018)].
Finally, population sequence variations have been recently used to study enhancer function. The impact of such population variation on transcription is measured by expression Quantitative Trait Locus (eQTL) tests, in which transcriptomic and genomic data are combined for distinct Drosophila lines. Genetic variants associated with higher or lower expression can be mapped to relevant regulatory regions, although the specific changes that impact function are not necessarily known, due to linkage disequilibrium (Huang et al. 2015; Cannavò et al. 2017).
TFs Bind to Specific Sequences (Binding Sites) in Enhancers
Enhancers function as templates for TF binding; thus, it is critical to consider the structures and functions of the TFs themselves. As with enhancers, studies in Drosophila were critical for discovering sequence-specific DNA-binding TFs. A prime example is the homeodomain (HD), a 60-amino acid domain that was discovered by comparing the coding sequences of several TFs [Antennapedia (Antp), Ultrabithorax (Ubx), and Fushi-tarazu (Ftz)] involved in embryonic patterning (McGinnis et al. 1984b; Scott and Weiner 1984). Structurally, HDs form three α-helices, one of which interacts directly with DNA base pairs in the major groove (Otting et al. 1990), and this structure is conserved throughout metazoans (McGinnis et al. 1984a). Remarkably, two of the helices are similar to a conserved helix-turn-helix (HTH) motif in the Cro repressor of bacteriophage λ and two Escherichia coli proteins, the catabolite gene activator protein and the lac repressor (Qian et al. 1989). All three HTH proteins were shown to autoregulate by binding to specific DNA sequences in the operator regions of their own genes (Anderson et al. 1981; McKay and Steitz 1981; Matthews et al. 1982; Sauer et al. 1982), and similar activities were proposed for the HDs in Drosophila, but potential target genes for the Drosophila HD proteins were not known at the time. However, using a pull-down assay with the HD of Engrailed (En), another HD protein involved in embryo segmentation (Kornberg 1981), Desplan and co-workers showed that the En HD binds specifically to sequences in bacteriophage λ, and to sequences located upstream of ftz and en itself (Desplan et al. 1985). These studies defined the first sequences that HD proteins bind, and led to thousands of studies on the DNA-binding activities of hundreds of Drosophila TFs.
The Drosophila genome encodes > 700 TFs with characterized DNA-binding domains (Hammonds et al. 2013). In addition to the HD, with its HTH structure, other common DNA-binding domains include the basic helix-loop-helix, the basic leucine zipper, the winged helix, the high-mobility group (HMG), and the zinc finger (ZF). Crystal structures reveal the importance of ionic interactions with the DNA backbone as well as sequence-specific hydrogen bonding with bases, generally but not always in the major groove. Developmental and physiological changes of state are often driven by changes in expression or activity of these TFs, thus a major focus of Drosophila studies has been on understanding the roles of individual TFs in specific biological processes, which includes knowing about the DNA directly bound by these factors.
Despite the potential for TFs to select targets in vivo based on preferences for specific motifs, there is not always a high correlation between predicted binding based on in silico or in vitro assays, and in vivo binding assayed by ChIP assays (Pique-Regi et al. 2011; Cheng et al. 2013). Thus, experimental determinations are essential to understand the specificity of action of these proteins on enhancers. Early techniques relied on direct in vitro protein-DNA studies of discrete elements likely to contain regulatory sites. Electrophoretic mobility shift assays (Fried and Crothers 1981) and DNAseI protection (footprint) assays were then used to define the exact sequences (binding sites) preferred by a given TF (Galas and Schmitz 1978). By aligning sequences from multiple bound fragments, it was possible to infer the binding specificity of the TF from a relatively small number of footprinted sites. However, some TFs bind to multiple related sequences with similar affinities, which made it impossible to assign a simple consensus. To address this issue, Stormo invented the position weight matrix (PWM) (Stormo et al. 1982). In a simplified example of a PWM, binding sites are aligned as precisely as possible, and a matrix is made that lists the probability of having a specific base (A, C, G, or T) at each position, which can be represented in logo form (Hertz and Stormo 1999). The PWM can then be used to score the potential binding activity for any sequence. In general, there is a reasonable correlation between PWM score and affinity-binding constants as measured in vitro by quantitative gel shift or surface plasmon resonance (Majka and Speck 2007), although other factors influence in vivo occupancy.
These in vitro studies of TF-binding preference have been greatly expanded by a number of more comprehensive experimental approaches, including protein-binding microarrays (PBMs), in which all possible 8-bp DNA sequences (8-mers) are arrayed on a glass slide and probed with a fluorescently labeled DNA-binding domain (Berger et al. 2006). PBM experiments are particularly powerful because they generate quantitative binding information for every possible DNA sequence. Another in vitro approach [systematic evolution of ligands by exponential enrichment (SELEX)] uses a resin-bound DNA-binding domain to iteratively pull down specific sequences from a mixture of randomized oligos, followed by deep sequencing after every round of pull down (Riley et al. 2014; Rastogi et al. 2018). An alternative in vivo system involves expression of the TF in bacteria, where binding is measured by the ability to recruit bacterial RNA polymerase to a library of promoters containing randomized DNA sequences (Bulyk 2005) (Noyes et al. 2008b). A recently developed highly quantitative assay permits the assessment of TF–DNA interactions in solution, using fluorescence anisotropy in a high-throughput system (Jung et al. 2018).
TF–enhancer interactions can also be studied in more physiologically relevant contexts using a number of techniques, including chromatin ChIP and DAM-ID. ChIP involves fixing cells or tissues with formaldehyde to stabilize TF–DNA interactions (Gilmour and Lis 1984), while DAM-ID involves in vivo expression of a protein fusion between a DNA-binding domain and a bacterial methyl transferase to methylate DNA where the fusion protein binds (van Steensel and Henikoff 2000). In both cases, the DNA is sheared, and immunoprecipitation is used to isolate TF-bound or methylated fragments, respectively. High-throughput sequencing reveals regions contacted by the TF, and computational analysis yields overrepresented sequences that represent candidate binding motifs (Li et al. 2008). This is a very powerful method, but reproducibility of ChIP experiments is often low, varying by antibody, experimenter, and laboratory, and cross-linking over extended periods of time can lead to many false positives (Teytelman et al. 2013). Thus validation steps are essential, although often neglected. These include the use of independent antibodies that recognize distinct epitopes, depletion of the TF as a negative control, and, of course, biological replicates. A more recent variant of ChIP, Cut and Run, involves the interaction of the antibody to a TF with intact chromatin in the nucleus, followed by binding of an antibody-binding MNase fusion protein, to release specifically bound DNA segments for sequencing (Skene and Henikoff 2017). This method appears to be more sensitive than ChIP, as in principle only specifically bound DNA is isolated, reducing the background. These genome-wide approaches show that DNA sequence is an important factor, but is not sufficient for determining in vivo binding preferences. As discussed earlier, the availability of open regulatory regions is required for TF accessibility (Kaplan et al. 2011), as is the presence of neighboring cooperatively acting factors. In some cases, no consensus motifs are found in regions that are strongly bound in vivo, suggesting that binding is dictated strictly by protein–protein interactions, by a so-called “TF collective” model (Junion et al. 2012).
Even in cases where binding sites for a specific TF are present in a functioning enhancer, defining which sites are functional in vivo can be very challenging. One reasonable prediction is that sites that bind with high affinity in vitro are more likely to be functional in vivo, but this is not a general rule. For example, several studies have shown that intermediate-affinity sites are critical for enhancer function (Parker et al. 2011; Crocker et al. 2015; Farley et al. 2015; Datta et al. 2018). Also, it is clear that many enhancers contain multiple copies of binding sites for individual TFs. In some cases, cooperativity between sites for the same factor has been shown to be important for the activity of a specific TF (Lebrecht et al. 2005). Cooperative binding would make an enhancer more strongly affected by the absolute TF concentration in the nucleus, and is probably determined by site spacing and the relative orientation of adjacent sites, but we still have little understanding of the rules of cooperativity.
Most DNA-binding domains make specific contacts with 3–6 bp of DNA in isolation, but because TFs can contain multiple DNA-binding domains, and function as dimers and other complexes, the sequences that mediate binding and function can range from 6 to 20 bp in length, which greatly increases the sequence specificity required for binding the correct enhancers in vivo. For example, all HD-containing proteins in the Ubx and Antp complexes bind in vitro to very similar sequences with low complexity (4 bp), but each protein binds and activates distinct sets of target genes in vivo. Factors affecting in vivo binding specificity include subtle binding motif preferences (Noyes et al. 2008a), clustering of multiple low-affinity sites (Crocker et al. 2015), and interactions with cofactors that bind sequences adjacent to the 4-bp site directly contacted by the HD. The importance of cofactors was shown by a series of SELEX-Seq experiments that compared the in vitro binding activities of all eight Hox proteins when expressed as trimeric fusions with their cofactors Extradenticle and Homothorax (Slattery et al. 2011). These fusions bound to specific motifs that were on average 9 bp in length, compared with 4 bp for the unfused HDs.
The complementary in vivo and in vitro approaches described above are used to identify the sequence motifs and enhancers bound by a known TF. There are also situations in which a genomic sequence is known to have enhancer activity, but the TFs that regulate it are unknown. Aside from consulting existing DNA-binding databases, which do not include all TFs in all types of cells, there are several methods to directly identify candidate TFs using DNA sequences. First, tandem copies of the DNA sequence (the bait) can be attached to a chromatographic resin, and used to affinity purify proteins in vitro from nuclear extracts (Kadonaga and Tjian 1986). Second, in a method called a “one-hybrid screen,” several tandem copies of the bait sequence can be placed upstream of a selectable marker gene in yeast or bacteria, and a complementary DNA library containing fusions to a strong transcriptional activation domain is then transformed into the cells (Ouwerkerk and Meijer 2001). Proteins containing domains that bind to the bait sequence will activate the selectable marker and be identified by sequencing the clones harbored by surviving strains. Most recently, specific regulatory sequences have been purified directly from living tissues and bound proteins identified using sensitive mass spectrometry methods [reviewed in Wierer and Mann (2016)].
TFs Contain Effector Domains that Mediate Activation and Repression
In addition to DNA-binding domains, TFs commonly contain effector (activation and/or repression) domains that interact with components of the basal transcription machinery, scaffolding proteins, or chromatin-modifying enzymes to activate and repress genes [reviewed in Frietze and Farnham (2011)]. Among the first and best-characterized activation domains are those from the yeast Gal4 and GCN4 factors, and the human Sp1 protein (Gill and Ptashne 1987; Hope et al. 1988; Courey et al. 1989). When fused to a protein fragment containing only a DNA-binding domain, these domains can mediate activation in vitro and in vivo. A number of activation domains have been shown to directly contact general TFs including TATA-box-binding protein-associated factors (TAFs) from the TFIID complex that binds the TATA box, as well as Mediator, a mega-Dalton complex that directly contacts and regulates RNA polymerase II (Goodrich et al. 1993; Gill et al. 1994). Activation is also achieved by TF interactions with scaffolding proteins and chromatin-modifying or -remodeling enzymes.
Some enhancers, such as those found in viruses, contain only activator binding sites, but most developmentally regulated enhancers are also bound by TFs that repress activation. Mechanistically, most repressors inhibit transcription through chromatin-mediated pathways (see below) and in Drosophila, several effector domains that mediate repression have been discovered. These regions of the TFs include motifs capable of directly interacting with non-DNA-binding corepressors. Hairy, Engrailed, and Sloppy-paired contain short hydrophobic sequences that recruit the corepressor Groucho (Gro) (Fisher et al. 1996; Tolkunova et al. 1998; Andrioli et al. 2004; Jennings et al. 2006); other motifs are present in early-acting repressor proteins that recruit the corepressor dCtBP (Arnosti et al. 1996b; Nibu et al. 1998; Struffi 2004). Both CtBP and Gro corepressors recruit histone deacetylases and demethylases, which can compact chromatin and erase important marks recognized by chromatin regulatory proteins (Sundqvist et al. 1998; Chen et al. 1999).
Finally, in some cases, a single TF can mediate activation or repression depending on whether it interacts with coactivators or corepressors. For example, many signal-transduction systems terminate in TF binding to enhancers that mediate the cellular signal [reviewed in Barolo and Posakony (2002)]. Such enhancers bear similarities in many systems: weak activators bind constitutively, but in the absence of a signal, a repressor complex built upon a dual-output TF suppresses their ability to activate. Tissue-specific signals remodel the repressor complex into an activator complex, providing a new stimulatory output as well as relieving the inhibition of the general activators. This overall architecture provides a greater dynamic range in signaling than would be possible with a single recruited activator.
Mechanisms of Enhancer-Mediated Activation and Repression
Enhancers have been traditionally classified by functional properties in cell- and organism-based assays, as well as in cell-free in vitro studies, and more recently by similarities in the chromatin properties associated with these regulatory elements. In general, most enhancers have the potential to activate transcription in a specific setting. However, there are a large variety of proteins that associate with enhancers, and it is not unreasonable to ask whether their biochemical properties are similar, or whether they impact gene expression through diverse mechanisms.
Common properties shared by many enhancers are the presence of multiple binding sites for TFs, which may function at different levels in the transition of an enhancer from a compacted chromatin state to a fully active state (Figure 1). Recent work in a number of systems suggests that pioneer TFs may function to loosen chromatin, making it possible for other sequence-specific TF “settlers” to bind (Zaret and Mango 2016). The best candidate for a pioneer TF in Drosophila is the ubiquitous maternal factor Zelda (Zld), which is a key activator of the zygotic genome in early development (Liang et al. 2008). Zld was originally identified as a mutation [vielfaeltig, loosely translated as “manifold” (i.e., in defects)] because of its effects on many different embryonic processes (Staudt et al. 2006). Zld is a large ZF protein that binds to a consensus motif found in many developmental enhancers (ten Bosch et al. 2006; De Renzis et al. 2007; Li et al. 2008; Harrison et al. 2011; Nien et al. 2011; Satija and Bradley 2012). Loss of Zld reduces the binding efficiency of Bicoid (Bcd), Dorsal (Dl), and Twist, TFs that pattern the AP and dorsal–ventral (DV) axes of the embryo, consistent with its role as a pioneer factor (Yanez-Cuna et al. 2012; Foo et al. 2014; Xu et al. 2014).
How pioneer TFs function at the molecular level is still not clear, but they may directly or indirectly recruit chromatin-modifying and -remodeling complexes (Li et al. 2014; Schulz et al. 2015; Sun et al. 2015), and these activities may be shared with settler TFs. Once bound, TFs can also directly contact components of the basal machinery, including subunits of Mediator, as well as TAFs (Wright et al. 2006; Vojnic et al. 2011). An additional activity described for activation domains is the recruitment of the pTEF-b kinase, which phosphorylates the C-terminal domain of RNA polymerase II, as well as recruitment or regulation of pause-regulating factors DSIF and NELF [Bieniasz et al. 1999; Fujita et al. 2008; reviewed in Core and Adelman (2019)]. The phosphorylation of RNA polymerase II physically rearranges the structure of the PIC, permitting transition from promoter assembly to RNA polymerase II escape (Joo et al. 2019). TFs such as HMG-domain proteins, which have little DNA-binding specificity, can also interact with accessible enhancers. These TFs can function as architectural elements that interact with DNA-bound TFs to stabilize overall complex formation (Ellwood et al. 2000).
In the biochemical approaches employed to study these interactions, it is difficult to determine whether activator proteins bound to distally-located enhancers physically interact in vivo with promoter-localized Mediator or specific basal factors. However, most models of enhancer-bound activators, supported by chromatin conformation capture data, as well as fluorescent imaging studies, suggest these proteins are physically engaged with the basal machinery through enhancer–promoter looping [reviewed in Schoenfelder and Fraser (2019)]. Visualization of initiating transcripts via “tagging” of primary transcripts with aptamers that bind to modified GFP proteins provides an additional direct readout of transient promoter bursts [reviewed in George et al. (2018)]. The frequency of bursting is correlated with the proximity of distally located enhancers, strongly supporting the idea that distally bound factors interact directly with basal promoter-bound transcriptional machinery. An exciting new dimension of these studies relating to enhancer–promoter contacts comes from a realization that within the nucleus, unstructured protein domains of many components of the transcriptional machinery might self-associate into membraneless compartments (condensates) [Hnisz et al. 2017; Shrinivas et al. 2019; see also Peng and Weber (2019)]. The physical nature of these condensates is poorly understood, but the correlation between liquid–liquid-phase separation formation in vitro and activity in vivo indicates that such nonwell-mixed compositions may play major roles in defining enhancer–promoter interactions. In the Drosophila embryo, such membraneless compartments may potentiate the recruitment of primary patterning proteins Bcd and Dl by the Zld pioneer factor (Mir et al. 2018; Yamada et al. 2019). These compartments may underlie local subnuclear concentrations of TFs that appear to attract or at least activate enhancers containing TF binding sites of low or high affinity (Tsai et al. 2017, 2019).
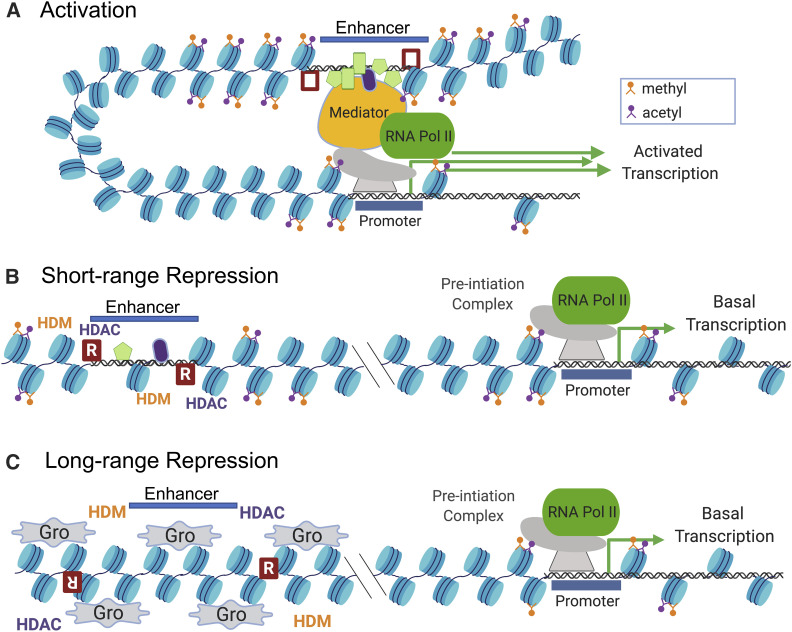
As mentioned above, most developmentally regulated enhancers also contain binding sites for repressors. Repressor sites can overlap with activator sites; in these situations, repression occurs via competition with the activator for binding. However, repressors need not overlap with activator sites, and most repressors function by generating more compacted chromatin environments that are inimical for activator binding (Li and Arnosti 2011; Kok et al. 2015). Such mechanisms have been divided into two classes: short- and long-range (Figure 3). Well-characterized short-range repressors include the gap proteins Kruppel (Kr), Giant (Gt), and Knirps, which form boundaries of stripes driven by eve enhancers (Arnosti et al. 1996b; Gray and Levine 1996; Hewitt et al. 1999), and Snail, which acts on Dl/Twist-activated neurectodermal enhancers (Gray et al. 1994). For these repressors, the exact position of the binding site is critical: moving the site > 100 bp away from the nearest activator site can severely reduce the ability of the repressor to function.
Figure 3.
Mechanisms of enhancer-mediated transcriptional repression. In nuclei containing mostly activators (A), activator binding recruits histone methyl transferases and histone acetyl transferases (HMTs and HATs, both not shown), which add methyl and acetyl groups to nucleosomes at the enhancer and promoter. Direct contacts (possibly by looping) are established, which substantially increase expression levels. When repressors are present in sufficient numbers, they bind to sites close to the enhancer (open red boxes in A; filled boxes in B and C). In short-range mechanisms (B), sequence-specific repressors (R) (and their associated corepressors, not shown) recruit histone demethylases (HDMs) and histone deacetylases (HDACs), which remove methyl and acetyl groups, and prevent activator binding and loop formation. In long-range repression, corepressors such as Groucho (Gro) spread along the chromatin, recruiting HDMs and HDACs that remove activating histone modifications in a broad region of chromatin. Created with Biorender.com.
Chromatin studies have shown that the nucleosomes associated with active enhancers contain high levels of acetylation and methylation on specific lysine residues of histone tails [reviewed in Allis and Jenuwein (2016)]. Short-range repressors such as Knirps remove these modifications, compacting the overall structure at a local level (Figure 3B). In contrast, long-range repressors such as Hairy have been shown to inhibit distal activator sites through generation of large-scale deacetylated and demethylated domains of chromatin [Figure 3C; Barolo and Levine 1997; Kok et al. 2015]. Interestingly, the impact of Hairy on chromatin accessibility appears to be weaker than that of the short-range repressor Knirps, indicating that losses of acetyl and methyl marks are not always directly connected to overall compaction, although this distinction has not been thoroughly explored (Li and Arnosti 2011).
Repression is considered to be associated with promoter silencing, and classical repressors such as those discussed above do effectively turn expression down to undetectable levels when expressed at peak levels (Surkova et al. 2008). The significance of partial repression of target genes in regions of low repressor concentration is generally not understood; if developmental switches operate with strong thresholds, reducing target gene activity below a specific level may be adequate for regulation. Another type of repression, “soft repression,” has been less well studied. Soft repression complexes reduce transcription levels, but never completely abolish them (Wei et al. 2016). The broadly expressed Drosophila insulin receptor (InR) gene appears to operate by this mechanism, in which promoter-proximal retinoblastoma corepressors can influence overall expression up to twofold, but constitutive enhancers located in the gene body maintain activity in all cell types at a minimum level (Wei et al. 2016).
Notably, the inhibitory effects of the best-studied Drosophila repressors appear to be largely restricted to individual enhancers, so that silencing of one enhancer does not impact other nearby enhancers, unless the elements are artificially close together (Small et al. 1993; Gray et al. 1994). The sophisticated switching activity of enhancers driving separate eve stripes depends on independence of biochemical reactions on each element. The limited size of the block of chromatin where histone modifications are changed by short-range repressors appears to explain how enhancers can function in this autonomous fashion (Li and Arnosti 2011; Kok et al. 2015).
An important property of most cellular enhancers, as noted above in the description of pair-rule gene enhancers, is that their action is additive, meaning that they act in a modular fashion. This independence of action is not absolute; for instance, promoter output is saturable, such that addition of more enhancers will eventually bring diminishing returns (Bothma et al. 2015). Such observations stem from biophysical properties of promoter activity; rather than showing smooth and continuous outputs, most cellular promoters show stochastic properties, whereby even when a gene is being expressed, over short time spans the average rate of initiation changes, and even comes to a complete stop. Such “bursting” is a function of the core promoter structure as well as the types and concentrations of activators operating on the promoter [reviewed in Nicolas et al. (2017)]. From the kinetics of looping observed in vivo, which generally involves enhancer–promoter complexes with a stability of minutes, it is likely that saturation of a promoter occurs when the “off” state of the promoter is reduced to a minimum (Bothma et al. 2015). Recent studies of Notch-dependent transcription in the embryo indicate that when signaling is at a maximum, Notch-dependent promoters hold their “on” state for longer periods, while the average length of the “off” state is unchanged (Falo-Sanjuan et al. 2019). This contrasts with other enhancers, in which transcriptional activation is associated with a shorter “off” state, or with average higher transcriptional initiation frequency during those periods when the promoter is on (Fukaya et al. 2016).
Enhancers Function as Responsive Combinatorial Switches
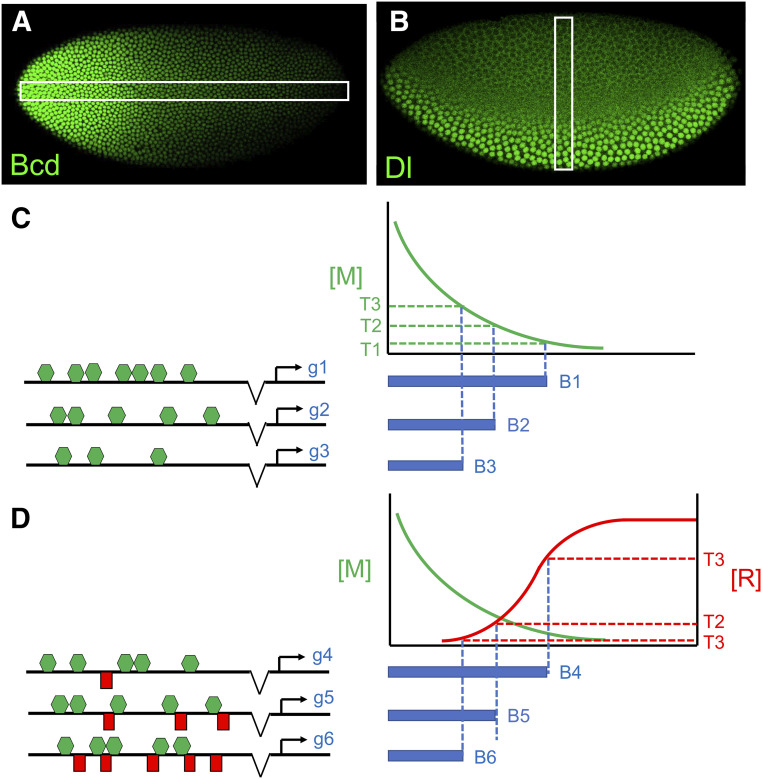
Developmentally regulated enhancers can contain dozens of binding sites for individual TFs, including pioneer factors that increase chromatin accessibility. Once accessible, the specific activity of each enhancer (when and where it activates transcription) is determined by its complement of binding sites and the combination and concentration of TFs in each cell. Cell type-specific TFs often control batteries of genes through independent enhancers, and different target genes can exhibit unique temporal or spatial expression patterns. In its simplest form, an enhancer might contain only sites for activator TFs, where gene activation is enabled only in nuclei with sufficiently high concentrations of those factors. If enhancers associated with different target genes contain different numbers of binding sites, or sites with different binding affinities, then each enhancer would have its own concentration threshold for activation, in a model referred to as the “differential affinity hypothesis” (Figure 4C; Driever et al. 1989).
Figure 4.
Morphogen-mediated patterning mechanisms. (A and B) Immunofluorescence detection of the Bicoid (A) and Dorsal (B) transcription factors (TFs), which are distributed in concentration gradients and function as morphogens in the early fly embryo. (C and D) Two models for target-gene patterning. (C) The differential affinity model. Three hypothetical target genes (g1–g3) are shown. Each gene contains an enhancer with a different number of binding sites (green hexagons) for a TF that functions as an activating morphogen (M). The blue bar to the right of each gene represents its expression pattern, and each gene makes a threshold-dependent expression boundary at a position determined by the number of binding sites in its enhancer. All three enhancers are activated in regions with high levels of the morphogen. Enhancers containing more binding sites are bound by M and activated in regions with lower levels of morphogen. Differences in binding site affinity (not shown here) can also determine binding sensitivity and boundary positioning. (D) A combinatorial model that integrates opposing gradients of an activator (M) and a repressor (R). In this model, the enhancers associated with three genes (g4–g6) contain the same number (and affinity) of activator sites, but different numbers of repressor sites. In this model, boundary positions are determined by threshold concentrations of the repressor. Embryo images in (A) and (B) are courtesy of Pinar Onal and Christine Rushlow, respectively.
According to this hypothesis, enhancers containing fewer binding sites and/or lower-affinity sites would be activated only in regions that contain high levels of activator proteins, making boundaries of target gene expression that lie close to the source of the gradient (Figure 4C; Driever et al. 1989). Enhancers containing more and/or higher-affinity sites would have lower activation thresholds, and make boundaries that lie farther from the gradient source. This hypothesis has been tested extensively in the early Drosophila embryo, where long-range nuclear gradients of the activator TFs Bcd and Dl are thought to act as morphogens (Wolpert 1971) that organize the AP and DV axes of the embryo (Figure 4, A and B). Bcd and Dl both activate dozens of target genes in domains with on/off expression boundaries at different positions within their gradients (Driever and Nüsslein-Volhard 1988; Roth et al. 1989; Rushlow et al. 1989; Struhl et al. 1989; Stathopoulos et al. 2002; Chen et al. 2012).
For Bcd, which has > 50 confirmed target genes, several studies argue against the strict interpretation of the differential affinity hypothesis. First, there is no significant correlation between expression boundary position and aggregate Bcd-binding strength as estimated by site affinity, site number, or Bcd ChIP-peak height of the 66 known Bcd-dependent enhancers (Ochoa-Espinosa et al. 2005; Xu et al. 2014; Hannon et al. 2017). The simplest explanation for this is that Bcd does not work alone in the activation of its target genes, and two cofactors, the pioneer factor Zld and Hunchback, bind to most Bcd target enhancers and help activate them (Simpson-Brose et al. 1994; Porcher et al. 2010; Xu et al. 2014; Hannon et al. 2017; Mir et al. 2018). Also, the boundary positions of most Bcd target genes are set by one or more repressor TFs expressed in gradients that are spatially opposed to the Bcd gradient (Figure 4D; Lohr et al. 2009; Chen et al. 2012). Almost all confirmed Bcd-dependent enhancers contain binding sites for at least one of these repressors, suggesting that repressor-mediated antagonism of Bcd-dependent activation is a key mechanism for setting the boundaries of most Bcd target genes.
For Dl, several genes expressed only in regions with high Dl levels were found to have enhancers with low-affinity sites, while others expressed in regions with lower Dl concentrations have enhancers with higher-affinity sites (Jiang and Levine 1993; Hong et al. 2008b). These results are consistent with the differential affinity hypothesis. However, Dl-dependent enhancers are also bound by Zld and two other coactivators (Twist and Grainyhead), both of which increase the apparent sensitivity to Dl binding (Jiang and Levine 1993; Garcia and Stathopoulos 2011; Foo et al. 2014; Yamada et al. 2019), and there is evidence that at least one repressor antagonizes the positive effect of Zld on Dl-mediated activation (Ozdemir et al. 2014).
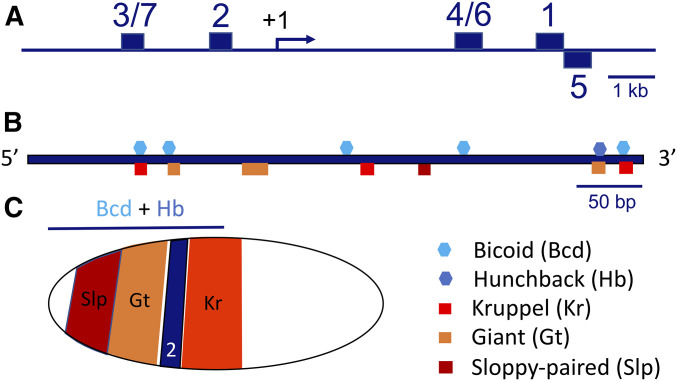
Studies of many fly enhancers show that, in general, they are regulated by multiple TFs, and the on/off state of each enhancer is controlled by the stochiometric balance between activators and repressors in each nucleus (Pankratz et al. 1990; Guss et al. 2001; Rushlow et al. 2001; Swanson et al. 2010). A classic example is the enhancer that drives the expression of eve stripe 2 (Figure 5, B and C). This enhancer has been studied intensely as a 480-bp minimal fragment that autonomously drives reporter gene expression (Small et al. 1991, 1992; Arnosti et al. 1996a; Andrioli et al. 2002), but sequences outside this core element also contribute to the regulation of the stripe (Janssens et al. 2006; Crocker and Stern 2017; Barr et al. 2019). The stripe 2 enhancer contains multiple binding sites for the activator TFs Bcd, Hb, and possibly Zld, which could potentially activate the stripe throughout the anterior half of the embryo (Figure 5C). However, the enhancer drives only a narrow stripe of expression because it contains binding sites for three repressor proteins [Sloppy-paired 1 (Slp1), Gt, and Kr], which are located in regions of the embryo that lie anterior and posterior to the stripe (Stanojevic et al. 1991; Small et al. 1992; Arnosti et al. 1996a; Andrioli et al. 2002). Mutations in the activator sites reduce stripe expression levels, while mutations in repressor sites cause ectopic activation in regions occupied by the repressors. These data strongly suggest that this enhancer acts as a switch that integrates the effects of multiple factors to generate an on/off expression pattern that appears as a precise stripe.
Figure 5.
Regulation of the eve stripe 2 enhancer. (A) Schematic representation of the eve locus (A) shows the position of the stripe 2 enhancer upstream of the transcription start site (+1). (B) The stripe 2 enhancer (B) contains at least 13 binding sites for five different transcription factors, including Bicoid (Bcd) and Hunchback (Hb), which activate transcription, and Sloppy-paired (Slp), Giant (Gt), and Kruppel (Kr), which function as repressors. (C) Schematic representation of an early embryo (anterior to the left, dorsal up) showing the expression patterns of the regulators of eve 2. Bcd and Hb are distributed throughout the anterior half of the embryo (denoted by the horizontal line above the embryo), while the repressors are expressed in discrete domains along the anterior–posterior axis. The enhancer is activated in a stripe of cells that contain high levels of Bcd and Hb proteins, and very low levels of repressors. Repression by Gt and Kr form the anterior and posterior boundaries of the stripe, respectively.
Understanding how all the binding sites in an enhancer contribute to its function in vivo is still a major challenge. The underlying biophysical interactions that drive transcriptional events are still incompletely understood, and models that emphasize different aspects of the critical events help define computational needs and drive experimental design. At one end of a spectrum of ideas, each binding site functions independently, and the overall activity of the enhancer can be deduced by summing the activities of individual sites (Arnosti and Kulkarni 2005). The molecular model associated with this picture is that individual stabilizing interactions between specific portions of the basal machinery and activators are dynamic, so that the PIC is repeatedly contacted by different stimulatory surfaces that are in close spatial proximity due to their grouping on the enhancer DNA. This idea has been called the “billboard” model, which proposes that each site is important, but that the exact placement of sites along the length of the enhancer is not critical. Strong support for the billboard model comes from studies of enhancer evolution. For example, several studies have shown that the spacing between critical binding sites in the eve stripe 2 enhancer have dramatically changed during insect evolution (Ludwig and Kreitman 1995; Hare et al. 2008). Despite these changes, the eve 2 enhancers from these species drive a stripe of expression when tested by reporter gene assays in Drosophila.
At the opposite end of the spectrum from the billboard model is the “enhanceosome” model, which suggests that spacing between sites is critical for enhancer function. This model is largely based on studies of the IFN-β enhancer, which fails to function if the spacing between its activator sites is perturbed (Kim and Maniatis 1997). For most developmentally regulated enhancers, some flexibility in spacing is permitted if the full length of the enhancer is considered. However, within this more flexible framework, critical spacing requirements between individual pairs of sites do exist. For example, within the eve 2 enhancer, some regions show no evolutionary variation in the spacing between adjacent binding sites (Ludwig and Kreitman 1995). This constant spacing may be required for cooperative binding events between TFs. A third model, the TF collective model, emphasizes the importance of indirect enhancer–TF interactions mediated by protein–protein interactions, allowing some factors to regulate an enhancer relatively independently of DNA motifs (Junion et al. 2012). These models are not mutually exclusive, but highlight particular aspects of the complex biochemical interactions that can take place between TFs on an enhancer (Park et al. 2019).
A major, mostly unrealized goal of studying enhancers is to use their DNA sequences to directly predict their associated in vivo expression patterns. Some progress has been made toward this goal for groups of enhancers that pattern the early embryo along the AP (Janssens et al. 2006; Segal et al. 2008; Markstein et al. 2004; He et al. 2010) and DV axes (Markstein et al. 2004; Zinzen et al. 2006), and that organize the presumptive mesoderm into a number of different muscle cell types (Zinzen et al. 2009; Wilczynski et al. 2012). In a study of AP patterning, Segal and co-workers used the expression patterns of eight TFs, along with a thermodynamic model to estimate their binding activities, to establish the parameters of a model to optimally compute the known expression patterns of a training set of 44 well-characterized enhancers (Segal et al. 2008). The model they obtained could indeed predict patterns that appeared similar to the known patterns of most of the training-set enhancers, and also did reasonably well in predicting patterns for test enhancers not included in the training set, although several test enhancers were shorter delineations of those in the training set, and thus not truly independent. While not perfect, this study identified enhancers whose predicted and observed patterns are quite similar, which supports the idea that most of the patterning inputs for those enhancers have been identified. In contrast, enhancers that showed striking differences between their predicted and observed expression patterns suggest that factors critical for their regulation are still unknown.
More significant progress in predicting patterns from sequences has also come from two studies of mesoderm patterning in flies. In the first (Zinzen et al. 2009), a genome-wide atlas of in vivo binding occupancy was generated for five different TFs that are active in specific cell types at different stages of mesodermal development. Filtering the data in the binding atlas identified a large number of novel mesoderm-specific enhancers: 35 of 36 tested fragments drove mesoderm-specific expression. The in vivo occupancy data and previously known expression patterns of 310 enhancers were combined to develop a machine-learning algorithm, which indicated five distinct classes, each of which showed a high correlation between binding activities and expression patterns. Support vector machines were then developed to distinguish members within a single class from those outside and applied to > 8000 predicted enhancers. At least six members of each class were tested by reporter genes, and completely or partially correct patterns were predicted for > 85% of the tested fragments. In the second study (Wilczynski et al. 2012), a Bayesian model was used to predict the expression patterns of genes based only on genome-wide binding data for the same five TFs, and genome-wide maps of the positions of insulator elements and regions with specific marks of open chromatin, which assisted greatly in associating individual enhancers with specific transcription units. The approach was quite successful in predicting the temporal and spatial expression patterns of 600 newly annotated genes, and validated by 20 reporter genes that showed expression patterns that were accurately predicted in time (95%) and space (50%). Altogether, these pioneering studies suggest that predicting temporal and spatial expressions directly from DNA sequence is an attainable goal, and that success in other systems will depend in large part on gathering complete data sets for the binding activities of the major TFs that regulate the activities of that system. However, interpretation of transcriptional outputs directly from DNA sequences may be complicated by the fact that in many loci, similar transcriptional activities are encoded in multiple “shadow enhancers.” These redundant or partially redundant enhancers can be programmed by an identical set of TFs, but this is not always observed (Frankel et al. 2010; Staller et al. 2015; Wunderlich et al. 2015).
Transcription of Enhancers (eRNAs)
Early studies of immunoglobulin enhancers in mammalian cells showed that transcription of noncoding RNA was detectable even before promoter activity commenced (Yancopoulos and Alt 1985); such transcription was suggested to promote access of the recombination machinery, prior to initiation of mRNA production (Cobb et al. 2006). Genome-wide analysis of eukaryotic transcription often focuses on cytoplasmic mRNA, and emphasizes steady-state levels of transcripts. However, approaches that detect production of RNA directly on the chromatin, as well as measurements made in the absence of nuclear exosome (nuclease complex) activity have revealed an entirely different scenario. In both Drosophila and in mammals, regulatory regions can be very actively transcribed, similarly to promoters [reviewed in Arnold et al. (2019)]. Such eRNAs are generally short and highly unstable, and for the most part, we do not know if the production of these RNAs is critical for gene expression, or rather reflects a side reaction on enhancers that is an consequence of RNA polymerase II firing from promoters, or off-target effects of TFs that are tolerated by the system.
A comprehensive survey of such nonpromoter-associated transcripts in Drosophila S2 cells demonstrated that there are strong correlations between the presence of a regulatory region, DNAseI accessibility, and enhancer activity. However, there are many exceptions to these correlations (Henriques et al. 2018). This study furthermore established the presence of basal promoter-like motifs directly at the site of initiation within enhancers, and showed that factors regulating pausing at the promoter also impact pausing at the enhancer. A similar survey of noncoding, unstable transcription in Drosophila embryos revealed that, unlike many mammalian enhancers, Drosophila regulatory regions are often asymmetrically transcribed. Therefore, divergent transcription appears not to be a suitable mark for enhancer region discovery in Drosophila, because the classical differential between TSSs (predominantly transcribed over gene) and enhancers (symmetrical bidirectional level of initiation) is less pronounced (Mikhaylichenko et al. 2018). In this study, putative enhancers were assessed for promoter activity, and promoters assessed for activation at a distance, as for an enhancer. The authors suggest that the partial overlap of these activities indicates that there is a continuum between regulatory elements that function as enhancers and promoters, rather than two wholly discrete biochemical entities. The extent of distal activation potential can be measured in a specific context (Arnold et al. 2013); however, it is possible that in another context (distance, different target promoter, etc.) the element may be more or less active.
Specificity of Enhancer–Promoter Interactions
It is critical that enhancers and promoters interact specifically to avoid improper regulation of neighboring genes. With tens of thousands of transcription units, enhancers, and promoters, there are multiple mechanisms that ensure that the correct interactions are made [reviewed in van Arensbergen et al. (2014) and Zabidi and Stark (2016)]. Physical proximity plays an important role; in a survey of enhancers identified in transgenic assays, of 482 validated enhancer-gene pairs in Drosophila (Kvon et al. 2014), 88% of the enhancers were located within introns or in regions that lie immediately 5′ or 3′ of the associated transcription units, and the remaining 12% appeared to act on a promoter separated by one or more intervening genes. Physical interactions between TF-bound enhancers and promoters can be identified by modified chromatin conformation capture assays [Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET)] (Fullwood et al. 2009), in which transient enhancer–promoter interactions are fixed by treatment with formaldehyde, followed by immunoprecipitation with antibodies to specific TFs or RNA polymerase II. Finally, the hybridization of probes specific to promoter and distal enhancer regions has been used to detect transient enhancer–promoter interactions, which appear as a superposition of probe-labeled spots in a micrograph (Chen et al. 2018). High-throughput “painting” of chromosomes using this methodology has been used to measure the architecture of enhancer–promoter interactions on the BX-C HOX locus at single-cell resolution in the developing embryo (Mateo et al. 2019).
These data suggest that transcriptional regulation in Drosophila is dominated by local interactions between enhancers and promoters, and a number of mechanisms exist to ensure correct enhancer–promoter associations. At the highest level, each individual chromosome is divided transcriptional neighborhoods called topologically associating domains (TADs) (Dixon et al. 2012). Genes contained within a TAD are thought to be coordinately regulated, and regulatory interactions between genes in adjacent TADs are prevented by stable nucleoprotein complexes located at TAD boundaries. Early studies on the HSP70 locus in flies were important for defining higher-order chromosome domains (Udvardy et al. 1985; Kellum and Schedl 1991), and more recent chromatin conformation capture assays (Lieberman-Aiden et al. 2009) have identified TADs genome-wide (Hou et al. 2012; Sexton et al. 2012). Nucleoprotein complexes at the TAD boundaries include the DNA-binding proteins suppressor of Hairy Wing [su(Hw)], Zw5, BEAF-32, and Drosophila CTCF, which interact with CP190 and mod(mdg)4 boundary element factors to prevent enhancer–promoter interactions (Geyer and Corces 1992; Roseman et al. 1993; Bushey et al. 2009). Interestingly, CTCF is highly conserved in mammals, and plays a similar role in defining chromatin domains through its interaction with cohesin complexes.
The best-characterized insulator-binding protein in Drosophila is su(Hw), a ZF-containing transcriptional repressor that functions with its partner protein mod(mdg4). Studies from many laboratories have shown that insertion of su(Hw)-binding sites between enhancers and promoters can prevent enhancer-mediated transcription (Geyer and Corces 1992; Dorsett 1993; Cai and Levine 1995). At the genome-wide level, antibodies to su(Hw) and mod(mdg4) have shown that both proteins bind to hundreds of loci in polytene chromosomes, suggesting that su(Hw)-mediated insulation is important for the regulation of many genes. However, when the same antibodies were used on diploid cells, only 20–25 intensely stained foci appeared, suggesting that su(Hw)-bound loci (possibly domain boundaries) had coalesced into a few regions within the nucleus, which were named “insulator bodies” (Gerasimova et al. 2000).
Taken together, these results strongly support the idea that chromosomes are organized into a series of loops, and that interactions between regulatory elements and genes on different loops are prevented by insulator bodies. Consistent with this, insulators are underrepresented in regions between dedicated enhancer–promoter pairs compared to the regions between genes (Kvon et al. 2014). However, the relationship between TAD boundaries and DNA elements that act as insulators is still not clear. Recent gene-specific and genome-wide studies testing the roles of TADs in gene expression have revealed that these structural domains are not absolute, or even critical, determinants of enhancer–promoter specificity. Gross chromosomal structural rearrangements associated with X-ray treatment, represented by Drosophila balancer chromosomes, disrupt many TADs, but in most cases there are minimal changes in gene expression (Ghavi-Helm et al. 2019). Similar findings in mammals suggest that TADs likely exert their effects in more subtle ways, with certain exceptional cases where TAD disruption produces major phenotypes (Rodríguez-Carballo et al. 2017; Schwarzer et al. 2017).
For enhancers located in densely packed regions of the genome, additional layers of regulation are required to ensure enhancer–promoter specificity (Corbin and Maniatis 1989; Li and Noll 1994). For example, the dpp gene contains a number of enhancers required for its expression in the wing imaginal disc (Merli et al. 1996). These enhancers are contained in a genomic interval that lies between 20 and 35 kb downstream of the dpp promoter. Two other genes, Slh and oaf, lie closer to the dpp enhancers, but are not expressed in wing discs at all. In an ingenious experiment, Merli et al. (1996) used homologous recombination-mediated repair of a P-element excision to precisely replace the native oaf promoter with the hsp70 promoter, which responds to dpp wing enhancer activity. This replacement caused the activation of the oaf gene in a dpp-like pattern, showing convincingly that basal promoters contain sequences that can affect promoter usage. A further demonstration of enhancer–promoter specificity came from studies of an “enhancer trap” construct, which was inserted into random genomic locations where it could be influenced by local enhancers (Butler and Kadonaga 2001). The enhancer trap construct contained two promoters, one with and one without the Downstream Promoter Element (DPE), which is critical for TFIID binding to the +30 region. Controlled recombination events were used to separately remove each of the promoters, and several insertions were identified that contained enhancer activity that selectively activated the DPE-containing basal promoter.
Several other studies presented evidence that specific interactions (tethering) between enhancers and promoters are important for endogenous gene regulation. (Calhoun et al. 2002; Kwon et al. 2009; Swanson et al. 2010). In another approach, dual reporter genes were used to artificially place enhancers between two promoters. These experiments showed that some enhancers preferentially activate one basal promoter, while others can activate both, but at lower overall levels (e.g., Ohtsuki et al. 1998).
Defining the promoter sequences controlling promoter choice in vivo is still a challenge. One intriguing idea is that the specific TFs bound to an enhancer prefer promoters containing specific sequence motifs, such as TATA, the initiator, the DPE, or combinations of these motifs [Butler and Kadonaga 2001; see also Vo Ngoc et al. (2019)]. As examples, a TATA-containing promoter was shown to be preferentially activated by the IAB5 enhancer (Calhoun and Levine 2003), and two TFs, Caudal and Dl, were shown to prefer promoters containing DPE elements (Juven-Gershon et al. 2008).
It is hard to imagine that enhancer–promoter specificity in vivo is primarily controlled by simple sequence motifs, as basal promoter elements exhibit a complex diversity of sequences. Certain combinations of sequence motifs may provide more specific “codes” that can be recognized by enhancer-bound protein complexes (Rach et al. 2009). This idea was tested by studying Bcd-dependent activation of hunchback (hb), which contains two promoters: one that responds to Bcd and one that does not (Ling et al. 2019). Mutation analysis showed that the active promoter contains two important motifs, TATA and a binding site for the ubiquitous TF Zld, which is thought to act as a pioneer TF. Because Zld also binds to the Bcd-dependent enhancers, changes in chromatin might be synchronized to increase the probability that they will interact with the correct promoter. If the combination of two motifs identified in the active hb promoter represents a general code for Bcd-dependent activation, one might expect to see them overrepresented in the promoters of other known Bcd target genes. Unfortunately, this does not seem to be the case. A scan of 25 known Bcd-dependent promoters showed that most do not contain either motif, and only two contain both Zld and TATA (Ling et al. 2019).
More general surveys of enhancer–promoter specificity have revealed additional factors that play into regulation. Enhancer-mediated activation of specific promoters might involve the recruitment of different cofactors that create physical contacts between TF-bound enhancers and components of the basal machinery (Hsu et al. 2008; Stampfel et al. 2015). To assess the significance of the diverse types of transcriptional coactivators involved in enhancer activity, Haberle et al. fused 23 different cofactors to the Gal4-DNA-binding domain, and challenged these fusions with 72,000 candidate promoters in a high-throughput assay in cultured cells (Haberle et al. 2019). Promoters with similar cofactor-driven activities were found to lie in five major clusters, each with a particular signature of overrepresented sequence motifs including TATA, specific DPE variants, etc. Some of these cofactor-mediated activities were assayed by the expression of specific genes in loss-of-function mutants for the candidate cofactor. Although these mutants induce pleiotropic effects, the results support the idea that transcriptional cofactors possess distinct biochemical potentials that differentially impact different basal promoters, pointing to a molecular mechanism for enhancer-preferred promoter activation.
At the molecular level, homotypic interactions between enhancer- and promoter-bound factors may link the two regulatory elements together, as demonstrated in mammalian cells for the YY1 TF (Sigova et al. 2015). In addition, cohesin complexes involved in chromosome mechanics may play important roles. Cohesins are ring-like structures important for sister chromatid cohesion during mitosis and meiosis. The involvement of cohesins in enhancer–promoter interactions was first discovered in pioneering Drosophila genetic experiments from the Dorsett laboratory, where mutations in the Nipped-B cohesin-loader gene were found to interfere with the regulation of the cut gene by its distal enhancer (Rollins et al. 1999). The involvement of cohesins is likely to be conserved, based on recent studies in mammals, which uncovered multiple roles for cohesins in establishing TADs and mediating enhancer–promoter interactions (Kagey et al. 2010; Fudenberg et al. 2016; Schwarzer et al. 2017).
In summary, many factors (positions of insulators, tethering mechanisms, chromatin accessibility, distance along the DNA, and cofactor usage) appear to determine which promoters are activated by individual enhancers in different cellular contexts. Each of these factors alone can be shown to control promoter choice in a reporter gene, but in endogenous loci, where multiple enhancers and promoters may lie in close proximity, complex interplays between them may be necessary to ensure appropriate enhancer–promoter interactions.
Challenges for the Future
Transgenesis and high-throughput sequencing technology coupled to genomics has transformed our understanding of cellular enhancers by permitting the enumeration of possible regulatory sequences, and providing a pathway to validate the activity of identified regulatory regions. As with protein-coding regions, the elucidation of an enhancer regulatory “parts list” is by no means enough for us to understand the biological function of these elements. Several challenges will require integrated approaches to make progress. First, the finding that many parts of the genome have enhancer-like properties does not mean that all of these elements are functional; such false-positive sites of TF binding and chromatin opening may represent the inevitable side reactions that stem from having large genomes regulated from distal areas by transcriptional regulators with low specificity. If a side reaction occurs to produce an enhancer-like region of no regulatory importance, it may not be selected against. Comparative evolutionary studies would be expected to be able to measure a lack of positive selection, but the flexible architecture of enhancers means that their sequences show much more variation, even under strong functional selection. In addition, for many of the enumerations of enhancers using genomic techniques, highly cell-specific enhancers would not be identified if they were active only in a small proportion of the total cell population. Single-cell methods may provide an inroad to identify such elements, although the coverage on a cell-by-cell basis is very low.
Many putative cis-regulatory regions have been identified through analysis of populations, using genome-wide association study and eQTL tests. In a few meritorious studies, powerful genomic and gene editing methods showed the actual regulatory significance of such variants for transcriptional regulation, but in large part, most of the extant data are merely correlative, and in some cases linking single-nucleotide polymorphisms to the nearest gene provides a false picture of where the enhancers are actually impacting gene expression (Claussnitzer et al. 2015). Chromatin conformation data can be helpful in connecting the regions of interest to target genes, although such interactions must be measured in the relevant cell type.
The demonstration that enhancers can act in a modular manner galvanized the field and permitted a reductionistic analysis of enhancers in developmental biology. However, we are finding individual cases where enhancers do not act in a strictly additive fashion, and it remains to be determined whether nonadditivity involves enhancer–enhancer, enhancer–promoter, or other types of interactions. The idea that the modules identified are identical to the biological control element has also been challenged by findings that an enhancer’s activity may be dispersed over a large region, not amenable to simple dissection, CRISPR disruption, or even ChIP analysis. The possible involvement of discrete binding sites that are necessary but not sufficient for wild-type activity was raised by pioneering computational work of Reinitz and colleagues, but remains to be tested on a genome-wide basis (Janssens et al. 2006).
The idea of a genome as a hard-wired computer program that can unfold through developmental time was the basis of biochemical and molecular studies of the sea urchin by Eric Davidson (Peter et al. 2012), and a similar picture from Drosophila research has been extremely powerful in elucidating conserved metazoan regulatory pathways. However, we have made little progress in converting a simple Boolean model for action of enhancer-based regulatory circuits into quantitative, dynamic models. The impacts of genetic background effects and environmental signals are not at all captured by these simplistic models, and it is likely that quantitative assessments of enhancers will be needed to properly evaluate the impacts of these important influences on phenotype. Such quantitative approaches are likely to integrate data from widely different fields, including biophysical studies, population genetic work, and genome-wide measurements using ever more-powerful single-cell data sets. Because of the deep resources available to current researchers, these are fields in which Drosophila is poised to make unique contributions. Forty years after the initial characterization of enhancers, one of the unique properties of higher eukaryotic genomes, we are now in a position to fully recognize how these powerful transcriptional regulators impact development and disease.
Acknowledgments
We thank all our Drosophila colleagues who have contributed to understanding of how genes are regulated at the transcriptional level, especially Michael Levine, one of the pioneers who developed the Drosophila embryo as a preeminent system for enhancer studies, and D.N.A. would like to acknowledge Walter Schaffner’s input and continued enthusiasm. This work was supported by National Institutes of Health grants GM-51946 to S.S. and GM-124137 to D.N.A.
Footnotes
Communicating editor: B. Oliver
Literature Cited
- Akam M., 1987. The molecular basis for metameric pattern in the Drosophila embryo. Development 101: 1–22. [PubMed] [Google Scholar]
- Allis C. D., and Jenuwein T., 2016. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17: 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., and Matthews B. W., 1981. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature 290: 754–758. 10.1038/290754a0 [DOI] [PubMed] [Google Scholar]
- Andrioli L. P., Vasisht V., Theodosopoulou E., Oberstein A., and Small S., 2002. Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development 129: 4931–4940. [DOI] [PubMed] [Google Scholar]
- Andrioli L. P., Oberstein A. L., Corado M. S., Yu D., and Small S., 2004. Groucho-dependent repression by Sloppy-paired 1 differentially positions anterior pair-rule stripes in the Drosophila embryo. Dev. Biol. 276: 541–551. 10.1016/j.ydbio.2004.09.025 [DOI] [PubMed] [Google Scholar]
- Arnold C. D., Gerlach D., Stelzer C., Boryn L. M., Rath M. et al. , 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077. “https://urldefense.proofpoint.com/v2/url?u=https-3A__doi.org_10.1126_science.1232542&d=DwMGaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=rN_e1qtolURbJFd28oFUKQ&m=Ff1RySX7X2udg8wAzZUvbETopoTfIFepxOINDUOzuKg&s=BO8VyrseICn0-H1fXpuGyq4wjHjEJ2H2ay3eCzuHHm4&e=” 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Arnold P. R., Wells A. D., and Li X. C., 2019. Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front. Cell Dev. Biol. 7: 377 10.3389/fcell.2019.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti D. N., and Kulkarni M. M., 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94: 890–898. 10.1002/jcb.20352 [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Barolo S., Levine M., and Small S., 1996a The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122: 205–214. [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Gray S., Barolo S., Zhou J., and Levine M., 1996b The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 15: 3659–3666. 10.1002/j.1460-2075.1996.tb00735.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., and Schaffner W., 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308. 10.1016/0092-8674(81)90413-X [DOI] [PubMed] [Google Scholar]
- Banerji J., Olson L., and Schaffner W., 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33: 729–740. 10.1016/0092-8674(83)90015-6 [DOI] [PubMed] [Google Scholar]
- Barolo S., and Levine M., 1997. Hairy mediates dominant repression in the Drosophila embryo. EMBO J. 16: 2883–2891. 10.1093/emboj/16.10.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S., and Posakony J. W., 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16: 1167–1181. 10.1101/gad.976502 [DOI] [PubMed] [Google Scholar]
- Barr K., Reinitz J., and Radulescu O., 2019. An in silico analysis of robust but fragile gene regulation links enhancer length to robustness. PLOS Comput. Biol. 15: e1007497 10.1371/journal.pcbi.1007497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Lee A. M., and Wu C. T., 2006. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173: 769–777. 10.1534/genetics.106.056945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann D. G., and Gilmour D. S., 2017. A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes. Nucleic Acids Res. 45: 10481–10491. 10.1093/nar/gkx676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. F., Philippakis A. A., Qureshi A. M., He F. S., Estep P. W. 3rd et al. , 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 24: 1429–1435. 10.1038/nbt1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, C. M., B. D. Pfeiffer, D. E. Rincón-Limas, R. A. Hoskins, A. Gnirke et al., 2002 Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol 3: RESEARCH0086. doi: 10.1186/gb-2002-3-12-research0086 10.1186/gb-2002-3-12-research0086 [DOI] [PMC free article] [PubMed]
- Berman B. P., Nibu Y., Pfeiffer B. D., Tomancak P., Celniker S. E. et al. , 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 757–762. 10.1073/pnas.231608898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman B. P., Pfeiffer B. D., Laverty T. R., Salzberg S. L., Rubin G. M. et al. , 2004. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5: R61 10.1186/gb-2004-5-9-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P. D., Grdina T. A., Bogerd H. P., and Cullen B. R., 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 96: 7791–7796. 10.1073/pnas.96.14.7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Harrison M. M., O’Connor-Giles K. M., and Wildonger J., 2018. Advances in engineering the fly genome with the CRISPR-Cas system. Genetics 208: 1–18. 10.1534/genetics.117.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S., Zinzen R. P., Girardot C., Gustafson E. H., Perez-Gonzalez A. et al. , 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44: 148–156. 10.1038/ng.1064 [DOI] [PubMed] [Google Scholar]
- Bothma J. P., Garcia H. G., Ng S., Perry M. W., Gregor T. et al. , 2015. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife 4: e07956 10.7554/eLife.07956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek M., Cortini R., Storti A. E., Unnerstall U., Gaul U. et al. , 2019. ATAC-seq reveals regional differences in enhancer accessibility during the establishment of spatial coordinates in the Drosophila blastoderm. Genome Res. 29: 771–783. 10.1101/gr.242362.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M. L., 2005. Discovering DNA regulatory elements with bacteria. Nat. Biotechnol. 23: 942–944. 10.1038/nbt0805-942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S., and Ebright R. H., 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293: 199–213. 10.1006/jmbi.1999.3161 [DOI] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E., and Corces V. G., 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23: 1338–1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. E., and Kadonaga J. T., 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15: 2515–2519. 10.1101/gad.924301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., and Levine M., 1995. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376: 533–536. 10.1038/376533a0 [DOI] [PubMed] [Google Scholar]
- Calhoun V. C., and Levine M., 2003. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 100: 9878–9883. 10.1073/pnas.1233791100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V. C., Stathopoulos A., and Levine M., 2002. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 99: 9243–9247. 10.1073/pnas.142291299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E., Khoueiry P., Garfield D. A., Geeleher P., Zichner T. et al. , 2016. Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 26: 38–51. 10.1016/j.cub.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E., Koelling N., Harnett D., Garfield D., Casale F. P. et al. , 2017. Genetic variants regulating expression levels and isoform diversity during embryogenesis. Nature 541: 402–406. 10.1038/nature20802 [DOI] [PubMed] [Google Scholar]
- Catarino R. R., and Stark A., 2018. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 32: 202–223. 10.1101/gad.310367.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Fernandez J., Mische S., and Courey A. J., 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13: 2218–2230. 10.1101/gad.13.17.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xu Z., Mei C., Yu D., and Small S., 2012. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149: 618–629. 10.1016/j.cell.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Levo M., Barinov L., Fujioka M., Jaynes J. B. et al. , 2018. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 50: 1296–1303. 10.1038/s41588-018-0175-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. K., Hertz G. Z., and Stormo G. D., 1995. MATRIX SEARCH 1.0: a computer program that scans DNA sequences for transcriptional elements using a database of weight matrices. Comput. Appl. Biosci. 11: 563–566. [DOI] [PubMed] [Google Scholar]
- Cheng Q., Kazemian M., Pham H., Blatti C., Celniker S. E. et al. , 2013. Computational identification of diverse mechanisms underlying transcription factor-DNA occupancy. PLoS Genet. 9: e1003571 10.1371/journal.pgen.1003571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M., Dankel S. N., Kim K. H., Quon G., Meuleman W. et al. , 2015. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 373: 895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R. M., Oestreich K. J., Osipovich O. A., and Oltz E. M., 2006. Accessibility control of V(D)J recombination. Adv. Immunol. 91: 45–109. 10.1016/S0065-2776(06)91002-5 [DOI] [PubMed] [Google Scholar]
- Corbin V., and Maniatis T., 1989. The role of specific enhancer-promoter interactions in the Drosophila Adh promoter switch. Genes Dev. 3: 2191–2120. 10.1101/gad.3.12b.2191 [DOI] [PubMed] [Google Scholar]
- Core L., and Adelman K., 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33: 960–982. 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Holtzman D. A., Jackson S. P., and Tjian R., 1989. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59: 827–836. 10.1016/0092-8674(89)90606-5 [DOI] [PubMed] [Google Scholar]
- Crocker J., and Stern D. L., 2017. Functional regulatory evolution outside of the minimal even-skipped stripe 2 enhancer. Development 144: 3095–3101. 10.1242/dev.149427 [DOI] [PubMed] [Google Scholar]
- Crocker J., Abe N., Rinaldi L., McGregor A. P., Frankel N. et al. , 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160: 191–203. 10.1016/j.cell.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich D. A., Reddington J. P., Garfield D. A., Daza R. M., Aghamirzaie D. et al. , 2018. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555: 538–542. 10.1038/nature25981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R. R., Ling J., Kurland J., Ren X., Xu Z. et al. , 2018. A feed-forward relay integrates the regulatory activities of Bicoid and Orthodenticle via sequential binding to suboptimal sites. Genes Dev. 32: 723–736. 10.1101/gad.311985.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S., Elemento O., Tavazoie S., and Wieschaus E. F., 2007. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5: e117 (erratum: PLoS Biol. 5: e213). 10.1371/journal.pbio.0050117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan C., Theis J., and O’Farrell P. H., 1985. The Drosophila developmental gene, engrailed, encodes a sequence- specific DNA binding activity. Nature 318: 630–635. 10.1038/318630a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., and Roeder R. G., 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y. et al. , 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D., 1993. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics 134: 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., and Nüsslein-Volhard C., 1988. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54: 95–104. 10.1016/0092-8674(88)90183-3 [DOI] [PubMed] [Google Scholar]
- Driever W., Thoma G., and Nüsslein-Volhard C., 1989. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature 340: 363–367. 10.1038/340363a0 [DOI] [PubMed] [Google Scholar]
- Dynan W. S., and Tjian R., 1983. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 32: 669–680. 10.1016/0092-8674(83)90053-3 [DOI] [PubMed] [Google Scholar]
- Ellwood K. B., Yen Y. M., Johnson R. C., and Carey M., 2000. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol. 20: 4359–4370. 10.1128/MCB.20.12.4359-4370.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberly E., Rajewsky N., and Siggia E. D., 2003. Conservation of regulatory elements between two species of Drosophila. BMC Bioinformatics 4: 57 10.1186/1471-2105-4-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falo-Sanjuan J., Lammers N. C., Garcia H. G. and Bray S. J., 2019. Enhancer priming enables fast and sustained transcriptional responses to Notch signaling. Dev. Cell 50: 411–425.e8. 10.1016/j.devcel.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley E. K., Olson K. M., Zhang W., Brandt A. J., Rokhsar D. S. et al. , 2015. Suboptimization of developmental enhancers. Science 350: 325–328. 10.1126/science.aac6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A. L., Ohsako S., and Caudy M., 1996. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16: 2670–2677. 10.1128/MCB.16.6.2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., and Alberts B. M., 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61: 31–70. [DOI] [PubMed] [Google Scholar]
- Foo S. M., Sun Y., Lim B., Ziukaite R., O’Brien K. et al. , 2014. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr. Biol. 24: 1341–1346. 10.1016/j.cub.2014.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N., Davis G. K., Vargas D., Wang S., Payre F. et al. , 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493. 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., and Crothers D. M., 1981. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9: 6505–6525. 10.1093/nar/9.23.6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S., and Farnham P. J., 2011. Transcription factor effector domains. Subcell. Biochem. 52: 261–277. 10.1007/978-90-481-9069-0_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N. et al. , 2016. Formation of chromosomal domains by loop extrusion. Cell Rep. 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., and Jaynes J. B., 2012. Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev. Biol. 362: 309–319. 10.1016/j.ydbio.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Emi-Sarker Y., Yusibova G. L., Goto T., and Jaynes J. B., 1999. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126: 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Ryser S., Piuz I., and Schlegel W., 2008. Up-regulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Mol. Cell. Biol. 28: 1630–1643. 10.1128/MCB.01767-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T., Lim B., and Levine M., 2016. Enhancer control of transcriptional bursting. Cell 166: 358–368. 10.1016/j.cell.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H. et al. , 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462: 58–64. 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B., and Zeitlinger J., 2014. RNA polymerase II pausing during development. Development 141: 1179–1183. 10.1242/dev.088492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., and Schmitz A., 1978. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5: 3157–3170. 10.1093/nar/5.9.3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., and Stathopoulos A., 2011. Lateral gene expression in Drosophila early embryos is supported by Grainyhead-mediated activation and tiers of dorsally-localized repression. PLoS One 6: e29172 10.1371/journal.pone.0029172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L., Indig F. E., Abdelmohsen K., and Gorospe M., 2018. Intracellular RNA-tracking methods. Open Biol. 8: 180104. 10.1098/rsob.180104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T. I., Byrd K., and Corces V. G., 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6: 1025–1035. 10.1016/S1097-2765(00)00101-5 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., and Corces V. G., 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. 10.1101/gad.6.10.1865 [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Jankowski A., Meiers S., Viales R. R., Korbel J. O. et al. , 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51: 1272–1282. 10.1038/s41588-019-0462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., and Ptashne M., 1987. Mutants of GAL4 protein altered in an activation function. Cell 51: 121–126. 10.1016/0092-8674(87)90016-X [DOI] [PubMed] [Google Scholar]
- Gill G., Pascal E., Tseng Z. H., and Tjian R., 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91: 192–196. 10.1073/pnas.91.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., and Tonegawa S., 1983. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33: 717–728. 10.1016/0092-8674(83)90014-4 [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., and Lis J. T., 1984. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc. Natl. Acad. Sci. USA 81: 4275–4279. 10.1073/pnas.81.14.4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic M. M., Rong Y. S., Petersen R. B., Lindquist S. L., and Golic K. G., 1997. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 25: 3665–3671. 10.1093/nar/25.18.3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Hoey T., Thut C. J., Admon A., and Tjian R., 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75: 519–530. 10.1016/0092-8674(93)90386-5 [DOI] [PubMed] [Google Scholar]
- Goto T., Macdonald P., and Maniatis T., 1989. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57: 413–422. 10.1016/0092-8674(89)90916-1 [DOI] [PubMed] [Google Scholar]
- Gray S., and Levine M., 1996. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev. 10: 700–710. 10.1101/gad.10.6.700 [DOI] [PubMed] [Google Scholar]
- Gray S., Szymanski P., and Levine M., 1994. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 8: 1829–1838. 10.1101/gad.8.15.1829 [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Wasylyk B., Chambon P., and Birnstiel M. L., 1981. Point mutation in the TATA box curtails expression of sea urchin H2A histone gene in vivo. Nature 294: 178–180. 10.1038/294178a0 [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., and Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss K. A., Nelson C. E., Hudson A., Kraus M. E., and Carroll S. B., 2001. Control of a genetic regulatory network by a selector gene. Science 292: 1164–1167. 10.1126/science.1058312 [DOI] [PubMed] [Google Scholar]
- Haberle V., Arnold C. D., Pagani M., Rath M., Schernhuber K. et al. , 2019. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570: 122–126. 10.1038/s41586-019-1210-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M. S., Grad Y., Church G. M., and Michelson A. M., 2002. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 12: 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M. S., Gallo S. M., and Bergman C. M., 2008. REDfly 2.0: an integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res. 36: D594–D598. 10.1093/nar/gkm876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds A. S., Bristow C. A., Fisher W. W., Weiszmann R., Wu S. et al. , 2013. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14: R140 10.1186/gb-2013-14-12-r140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon C. E., Blythe S. A., and Wieschaus E. F., 2017. Concentration dependent chromatin states induced by the bicoid morphogen gradient. Elife 6: e28275 10.7554/eLife.28275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K., Hoey T., Warrior R., and Levine M., 1989. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 8: 1205–1212. 10.1002/j.1460-2075.1989.tb03493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare E. E., Peterson B. K., Iyer V. N., Meier R., and Eisen M. B., 2008. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 4: e1000106 10.1371/journal.pgen.1000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Li X. Y., Kaplan T., Botchan M. R., and Eisen M. B., 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7: e1002266 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Samee M. A. H., Blatti C., and Sinha S., 2010. Thermodynamics-based models of transcriptional regulation by enhancers: the roles of synergistic activation, cooperative binding and short-range repression. PLoS Comput. Biol. 6: e1000935 10.1371/journal.pcbi.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., and Chamberlin M. J., 1988. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 57: 839–872. 10.1146/annurev.bi.57.070188.004203 [DOI] [PubMed] [Google Scholar]
- Henriques T., Scruggs B. S., Inouye M. O., Muse G. W., Williams L. H. et al. , 2018. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32: 26–41. 10.1101/gad.309351.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G. Z., and Stormo G. D., 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15: 563–577. 10.1093/bioinformatics/15.7.563 [DOI] [PubMed] [Google Scholar]
- Hewitt G. F., Strunk B. S., Margulies C., Priputin T., Wang X. D. et al. , 1999. Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development 126: 1201–1210. [DOI] [PubMed] [Google Scholar]
- Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., and Sharp P. A., 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. W., Hendrix D. A., and Levine M. S., 2008a Shadow enhancers as a source of evolutionary novelty. Science 321: 1314 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. W., Hendrix D. A., Papatsenko D., and Levine M. S., 2008b How the Dorsal gradient works: insights from postgenome technologies. Proc. Natl. Acad. Sci. USA 105: 20072–20076. 10.1073/pnas.0806476105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper K. L., Parkhurst S. M., and Ish-Horowicz D., 1989. Spatial control of hairy protein expression during embryogenesis. Development 107: 489–504. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., and Struhl K., 1988. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature 333: 635–640. 10.1038/333635a0 [DOI] [PubMed] [Google Scholar]
- Hou C., Li L., Qin Z. S., and Corces V. G., 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48: 471–484. 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. R., and Struhl G., 1990. Decoding positional information: regulation of the pair-rule gene hairy. Development 110: 1223–1231. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham P., and Rushlow C., 1988. Region-specific alleles of the Drosophila segmentation gene hairy. Genes Dev. 2: 1037–1046. 10.1101/gad.2.8.1037 [DOI] [PubMed] [Google Scholar]
- Hsu J. Y., Juven-Gershon T., Marr M. T. 2nd, Wright K. J., Tjian R. et al. , 2008. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent vs. TATA-dependent transcription. Genes Dev. 22: 2353–2358. 10.1101/gad.1681808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F. et al. , 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: E6010–E6019. 10.1073/pnas.1519159112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz J., 2005. The discovery of RNA polymerase. J. Biol. Chem. 280: 42477–42485. 10.1074/jbc.X500006200 [DOI] [PubMed] [Google Scholar]
- Ingham P. W., 1988. The molecular genetics of embryonic pattern formation in Drosophila. Nature 335: 25–34. (erratum Nature 335: 744). 10.1038/335025a0 [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Howard K. R., Pinchin S. M., and Ingham P. W., 1985. Molecular and genetic analysis of the hairy locus in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 50: 135–144. 10.1101/SQB.1985.050.01.019 [DOI] [PubMed] [Google Scholar]
- Jacob F., and Monod J., 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3: 318–356. 10.1016/S0022-2836(61)80072-7 [DOI] [PubMed] [Google Scholar]
- Janssens H., Hou S., Jaeger J., Kim A. R., Myasnikova E. et al. , 2006. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat. Genet. 38: 1159–1165. 10.1038/ng1886 [DOI] [PubMed] [Google Scholar]
- Jennings B. H., Pickles L. M., Wainwright S. M., Roe S. M., Pearl L. H. et al. , 2006. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell 22: 645–655. 10.1016/j.molcel.2006.04.024 [DOI] [PubMed] [Google Scholar]
- Jiang J., and Levine M., 1993. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72: 741–752. 10.1016/0092-8674(93)90402-C [DOI] [PubMed] [Google Scholar]
- Joo Y. J., Ficarro S. B., Chun Y., Marto J. A., and Buratowski S., 2019. In vitro analysis of RNA polymerase II elongation complex dynamics. Genes Dev. 33: 578–589. 10.1101/gad.324202.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Bandilla P., von Reutern M., Schnepf M., Rieder S. et al. , 2018. True equilibrium measurement of transcription factor-DNA binding affinities using automated polarization microscopy. Nat. Commun. 9: 1605 [corrigenda: Nat. Commun. 10: 689 (2019)]. 10.1038/s41467-018-03977-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G., Spivakov M., Girardot C., Braun M., Gustafson E. H. et al. , 2012. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148: 473–486. 10.1016/j.cell.2012.01.030 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T., Hsu J. Y., and Kadonaga J. T., 2008. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 22: 2823–2830. 10.1101/gad.1698108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., and Tjian R., 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 83: 5889–5893. 10.1073/pnas.83.16.5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A. et al. , 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [corrigenda: Nature 472: 247 (2011)]. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantorovitz M. R., Kazemian M., Kinston S., Miranda-Saavedra D., Zhu Q. et al. , 2009. Motif-blind, genome-wide discovery of cis-regulatory modules in Drosophila and mouse. Dev. Cell 17: 568–579. 10.1016/j.devcel.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T., Li X. Y., Sabo P. J., Thomas S., Stamatoyannopoulos J. A. et al. , 2011. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 7: e1001290 10.1371/journal.pgen.1001290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Lewis R., and Wakimoto B., 1980. Cytogenetic analysis of chromosome 3 in DROSOPHILA MELANOGASTER: the homoeotic gene complex in polytene chromosome interval 84a-B. Genetics 94: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., and Schedl P., 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. 10.1016/0092-8674(91)90318-S [DOI] [PubMed] [Google Scholar]
- Kim T. K., and Maniatis T., 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell 1: 119–129. 10.1016/S1097-2765(00)80013-1 [DOI] [PubMed] [Google Scholar]
- Klingler M., Soong J., Butler B., and Gergen J. P., 1996. Disperse vs. compact elements for the regulation of runt stripes in Drosophila. Dev. Biol. 177: 73–84. 10.1006/dbio.1996.0146 [DOI] [PubMed] [Google Scholar]
- Kok K., Ay A., Li L. M., and Arnosti D. N., 2015. Genome-wide errant targeting by Hairy. Elife 4: e06394 10.7554/eLife.06394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T., 1981. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proc. Natl. Acad. Sci. USA 78: 1095–1099. 10.1073/pnas.78.2.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E. Z., Kazmar T., Stampfel G., Yanez-Cuna J. O., Pagani M. et al. , 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. 10.1038/nature13395 [DOI] [PubMed] [Google Scholar]
- Kwon D., Mucci D., Langlais K. K., Americo J. L., DeVido S. K. et al. , 2009. Enhancer-promoter communication at the Drosophila engrailed locus. Development 136: 3067–3075. 10.1242/dev.036426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrecht D., Foehr M., Smith E., Lopes F. J., Vanario-Alonso C. E. et al. , 2005. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc. Natl. Acad. Sci. USA 102: 13176–13181. 10.1073/pnas.0506462102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L. A., Horikoshi T., Heaney S. J., van Baren M. J., van der Linde H. C. et al. , 2002. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl. Acad. Sci. USA 99: 7548–7553. 10.1073/pnas.112212199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239. 10.1086/281833 [DOI] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lewis R. A., Wakimoto B. T., Denell R. E., and Kaufman T. C., 1980. Genetic analysis of the antennapedia gene complex (ant-C) and adjacent chromosomal regions of DROSOPHILA MELANOGASTER. II. Polytene chromosome segments 84a–84B1,2. Genetics 95: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. M., and Arnosti D. N., 2011. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr. Biol. 21: 406–412. 10.1016/j.cub.2011.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., and Noll M., 1994. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 13: 400–406. 10.1002/j.1460-2075.1994.tb06274.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Y., MacArthur S., Bourgon R., Nix D., Pollard D. A. et al. , 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6: e27 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Y., Harrison M. M., Villalta J. E., Kaplan T., and Eisen M. B., 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3: e03737 10.7554/eLife.03737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H. L., Nien C. Y., Liu H. Y., Metzstein M. M., Kirov N. et al. , 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403. 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T. et al. , 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., Umezawa K. Y., Scott T. and Small S., 2019. Bicoid-dependent activation of the target gene hunchback requires a two-motif sequence code in a specific basal promoter. Mol. Cell 75: 1178–1187.e4. 10.1016/j.molcel.2019.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr U., Chung H. R., Beller M., and Jackle H., 2009. Antagonistic action of Bicoid and the repressor Capicua determines the spatial limits of Drosophila head gene expression domains. Proc. Natl. Acad. Sci. USA 106: 21695–21700. 10.1073/pnas.0910225106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. Z., and Kreitman M., 1995. Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol. Biol. Evol. 12: 1002–1011. [DOI] [PubMed] [Google Scholar]
- Ludwig M. Z., Palsson A., Alekseeva E., Bergman C. M., Nathan J. et al. , 2005. Functional evolution of a cis-regulatory module. PLoS Biol. 3: e93 10.1371/journal.pbio.0030093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J., and Speck C., 2007. Analysis of protein-DNA interactions using surface plasmon resonance. Adv. Biochem. Eng. Biotechnol. 104: 13–36. [PubMed] [Google Scholar]
- Markstein M., Markstein P., Markstein V., and Levine M. S., 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99: 763–768. 10.1073/pnas.012591199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Zinzen R., Markstein P., Yee K. P., Erives A. et al. , 2004. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131: 2387–2394. 10.1242/dev.01124 [DOI] [PubMed] [Google Scholar]
- Mateo L. J., Murphy S. E., Hafner A., Cinquini I. S., Walker C. A. et al. , 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., and Takeda Y., 1982. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc. Natl. Acad. Sci. USA 79: 1428–1432. 10.1073/pnas.79.5.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., and Gehring W. J., 1984a A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37: 403–408. 10.1016/0092-8674(84)90370-2 [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., and Gehring W. J., 1984b A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308: 428–433. 10.1038/308428a0 [DOI] [PubMed] [Google Scholar]
- McKay D. J., and Lieb J. D., 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27: 306–318. 10.1016/j.devcel.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., and Steitz T. A., 1981. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature 290: 744–749. 10.1038/290744a0 [DOI] [PubMed] [Google Scholar]
- Merli C., Bergstrom D. E., Cygan J. A., and Blackman R. K., 1996. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 10: 1260–1270. 10.1101/gad.10.10.1260 [DOI] [PubMed] [Google Scholar]
- Michiels F., Gasch A., Kaltschmidt B., and Renkawitz-Pohl R., 1989. A 14 bp promoter element directs the testis specificity of the Drosophila beta 2 tubulin gene. EMBO J. 8: 1559–1565. 10.1002/j.1460-2075.1989.tb03540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylichenko O., Bondarenko V., Harnett D., Schor I. E., Males M. et al. , 2018. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32: 42–57. 10.1101/gad.308619.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M., Stadler M. R., Ortiz S. A., Hannon C. E., Harrison M. M. et al. , 2018. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. Elife 7: e40497 10.7554/eLife.40497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium; Waterston R. H., Lindblad-Toh K., Birney E., Rogers J. et al. , 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P. et al. , 1981. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 9: 6047–6068. 10.1093/nar/9.22.6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Chen J. L., Geyer P. K., and Wu C. T., 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95: 10740–10745. 10.1073/pnas.95.18.10740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., and Szostak J. W., 1983. Construction of artificial chromosomes in yeast. Nature 305: 189–193. 10.1038/305189a0 [DOI] [PubMed] [Google Scholar]
- Nelson C. E., Hersh B. M., and Carroll S. B., 2004. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 5: R25 10.1186/gb-2004-5-4-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y., Zhang H., Bajor E., Barolo S., Small S. et al. , 1998. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 17: 7009–7020. 10.1093/emboj/17.23.7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas D., Phillips N. E., and Naef F., 2017. What shapes eukaryotic transcriptional bursting? Mol. Biosyst. 13: 1280–1290. 10.1039/C7MB00154A [DOI] [PubMed] [Google Scholar]
- Nien C. Y., Liang H. L., Butcher S., Sun Y., Fu S. et al. , 2011. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7: e1002339 10.1371/journal.pgen.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B., Christensen R. G., Wakabayashi A., Stormo G. D., Brodsky M. H. et al. , 2008a Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289. 10.1016/j.cell.2008.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B., Meng X., Wakabayashi A., Sinha S., Brodsky M. H. et al. , 2008b A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 36: 2547–2560. 10.1093/nar/gkn048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- O’Connor M., Peifer M., and Bender W., 1989. Construction of large DNA segments in Escherichia coli. Science 244: 1307–1312. 10.1126/science.2660262 [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A., Yucel G., Kaplan L., Pare A., Pura N. et al. , 2005. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc. Natl. Acad. Sci. USA 102: 4960–4965. 10.1073/pnas.0500373102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine M., and Cai H. N., 1998. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12: 547–556. 10.1101/gad.12.4.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Muller M., Affolter M. et al. , 1990. Protein–DNA contacts in the structure of a homeodomain–DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 9: 3085–3092. 10.1002/j.1460-2075.1990.tb07505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk P. B., and Meijer A. H., 2001. Yeast one-hybrid screening for DNA-protein interactions. Curr. Protoc. Mol. Biol. Chapter 12: Unit 12 12 10.1002/0471142727.mb1212s55 [DOI] [PubMed] [Google Scholar]
- Ozdemir A., Ma L., White K. P., and Stathopoulos A., 2014. Su(H)-mediated repression positions gene boundaries along the dorsal-ventral axis of Drosophila embryos. Dev. Cell 31: 100–113. 10.1016/j.devcel.2014.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz M. J., Seifert E., Gerwin N., Billi B., Nauber U. et al. , 1990. Gradients of Kruppel and knirps gene products direct pair-rule gene stripe patterning in the posterior region of the Drosophila embryo. Cell 61: 309–317. 10.1016/0092-8674(90)90811-R [DOI] [PubMed] [Google Scholar]
- Papatsenko D., Nazina A., and Desplan C., 2001. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech. Dev. 101: 143–153. 10.1016/S0925-4773(00)00581-5 [DOI] [PubMed] [Google Scholar]
- Papatsenko D. A., Makeev V. J., Lifanov A. P., Regnier M., Nazina A. G. et al. , 2002. Extraction of functional binding sites from unique regulatory regions: the Drosophila early developmental enhancers. Genome Res. 12: 470–481. 10.1101/gr.212502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Estrada J., Johnson G., Vincent B. J., Ricci-Tam C. et al. , 2019. Dissecting the sharp response of a canonical developmental enhancer reveals multiple sources of cooperativity. Elife 8: e41266 10.7554/eLife.41266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. S., White M. A., Ramos A. I., Cohen B. A., and Barolo S., 2011. The cis-regulatory logic of Hedgehog gradient responses: key roles for gli binding affinity, competition, and cooperativity. Sci. Signal. 4: ra38 10.1126/scisignal.2002077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. C., McKay D. J., Lieb J. D., and Crews S. T., 2016. Chromatin profiling of Drosophila CNS subpopulations identifies active transcriptional enhancers. Development 143: 3723–3732. 10.1242/dev.136895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A., and Weber S. C., 2019. Evidence for and against liquid-liquid phase separation in the nucleus. Noncoding RNA 5: 50 10.3390/ncrna5040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Bothma J. P., and Levine M., 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20: 1562–1567. 10.1016/j.cub.2010.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., and Levine M., 2011. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 108: 13570–13575. 10.1073/pnas.1109873108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Bothma J. P., Luu R. D., and Levine M., 2012. Precision of hunchback expression in the Drosophila embryo. Curr. Biol. 22: 2247–2252. 10.1016/j.cub.2012.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I. S., and Davidson E. F., 2015. Genomic Control Processes: Development and Evolution. Academic Press, Cambridge, MA. [Google Scholar]
- Peter I. S., Faure E., and Davidson E. H., 2012. Predictive computation of genomic logic processing functions in embryonic development. Proc. Natl. Acad. Sci. USA 109: 16434–16442. 10.1073/pnas.1207852109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R., Degner J. F., Pai A. A., Gaffney D. J., Gilad Y. et al. , 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 21: 447–455. 10.1101/gr.112623.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher A., Abu-Arish A., Huart S., Roelens B., Fradin C. et al. , 2010. The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development 137: 2795–2804. 10.1242/dev.051300 [DOI] [PubMed] [Google Scholar]
- Preger-Ben Noon E., Sabaris G., Ortiz D. M., Sager J., Liebowitz A. et al. , 2018. Comprehensive analysis of a cis-regulatory region reveals pleiotropy in enhancer function. Cell Rep. 22: 3021–3031. 10.1016/j.celrep.2018.02.073 [DOI] [PubMed] [Google Scholar]
- Pribnow D., 1975. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc. Natl. Acad. Sci. USA 72: 784–788. 10.1073/pnas.72.3.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Muller M., Gehring W. J. et al. , 1989. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell 59: 573–580. 10.1016/0092-8674(89)90040-8 [DOI] [PubMed] [Google Scholar]
- Rach E. A., Yuan H. Y., Majoros W. H., Tomancak P., and Ohler U., 2009. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 10: R73 10.1186/gb-2009-10-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonjic M., Andrau J. C., Lijnzaad P., Kemmeren P., Kockelkorn T. T. et al. , 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 18: 171–183. 10.1016/j.molcel.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Rajewsky N., Vergassola M., Gaul U., and Siggia E. D., 2002. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics 3: 30 10.1186/1471-2105-3-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi C., Rube H. T., Kribelbauer J. F., Crocker J., Loker R. E. et al. , 2018. Accurate and sensitive quantification of protein-DNA binding affinity. Proc. Natl. Acad. Sci. USA 115: E3692–E3701. 10.1073/pnas.1714376115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., and Roeder R. G., 1987. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J. Biol. Chem. 262: 3322–3330. [PubMed] [Google Scholar]
- Riddihough G., and Ish-Horowicz D., 1991. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 5: 840–854. 10.1101/gad.5.5.840 [DOI] [PubMed] [Google Scholar]
- Riley T. R., Slattery M., Abe N., Rastogi C., Liu D. et al. , 2014. SELEX-seq: a method for characterizing the complete repertoire of binding site preferences for transcription factor complexes. Methods Mol. Biol. 1196: 255–278. 10.1007/978-1-4939-1242-1_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carballo E., Lopez-Delisle L., Zhan Y., Fabre P. J., Beccari L. et al. , 2017. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31: 2264–2281. 10.1101/gad.307769.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins R. A., Morcillo P., and Dorsett D., 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152: 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., and Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. 10.1126/science.288.5473.2013 [DOI] [PubMed] [Google Scholar]
- Roseman R. R., Pirrotta V., and Geyer P. K., 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12: 435–442. 10.1002/j.1460-2075.1993.tb05675.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., Stein D., and Nüsslein-Volhard C., 1989. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59: 1189–1202. 10.1016/0092-8674(89)90774-5 [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., and Lis J. T., 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804. 10.1016/S0092-8674(88)91087-2 [DOI] [PubMed] [Google Scholar]
- Rushlow C. A., Han K., Manley J. L., and Levine M., 1989. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell 59: 1165–1177. 10.1016/0092-8674(89)90772-1 [DOI] [PubMed] [Google Scholar]
- Rushlow C., Colosimo P. F., Lin M. C., Xu M., and Kirov N., 2001. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 15: 340–351. 10.1101/gad.861401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T., Masuya H., Tamura M., Shimizu K., Yada Y. et al. , 2004. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh). Mamm. Genome 15: 23–34. 10.1007/s00335-033-2317-5 [DOI] [PubMed] [Google Scholar]
- Satija R., and Bradley R. K., 2012. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 22: 656–665. 10.1101/gr.130682.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., and Pabo C. O., 1982. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature 298: 447–451. 10.1038/298447a0 [DOI] [PubMed] [Google Scholar]
- Schaffner W., 2015. Enhancers, enhancers - from their discovery to today’s universe of transcription enhancers. Biol. Chem. 396: 311–327. 10.1515/hsz-2014-0303 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S., and Fraser P., 2019. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20: 437–455. 10.1038/s41576-019-0128-0 [DOI] [PubMed] [Google Scholar]
- Schroeder M. D., Greer C., and Gaul U., 2011. How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development 138: 3067–3078. 10.1242/dev.062141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz K. N., Bondra E. R., Moshe A., Villalta J. E., Lieb J. D. et al. , 2015. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25: 1715–1726. 10.1101/gr.192682.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G. et al. , 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., and Weiner A. J., 1984. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Natl. Acad. Sci. USA 81: 4115–4119. 10.1073/pnas.81.13.4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Raveh-Sadka T., Schroeder M., Unnerstall U., and Gaul U., 2008. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540. 10.1038/nature06496 [DOI] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B. et al. , 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shlyueva D., Stelzer C., Gerlach D., Yanez-Cuna J. O., Rath M. et al. , 2014. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol. Cell 54: 180–192. 10.1016/j.molcel.2014.02.026 [DOI] [PubMed] [Google Scholar]
- Shrinivas K., Sabari B. R., Coffey E. L., Klein I. A., Boija A. et al. , 2019. Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75: 549–561.e7. 10.1016/j.molcel.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova A. A., Abraham B. J., Ji X., Molinie B., Hannett N. M. et al. , 2015. Transcription factor trapping by RNA in gene regulatory elements. Science 350: 978–981. 10.1126/science.aad3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Brose M., Treisman J., and Desplan C., 1994. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell 78: 855–865. 10.1016/S0092-8674(94)90622-X [DOI] [PubMed] [Google Scholar]
- Skene P. J., and Henikoff S., 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6: e21856 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P. et al. , 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. 10.1016/j.cell.2011.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Kraut R., Hoey T., Warrior R., and Levine M., 1991. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 5: 827–839. 10.1101/gad.5.5.827 [DOI] [PubMed] [Google Scholar]
- Small S., Blair A., and Levine M., 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11: 4047–4057. 10.1002/j.1460-2075.1992.tb05498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Arnosti D. N., and Levine M., 1993. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119: 762–772. [PubMed] [Google Scholar]
- Small S., Blair A., and Levine M., 1996. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 175: 314–324. 10.1006/dbio.1996.0117 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., and Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. 10.1126/science.6289435 [DOI] [PubMed] [Google Scholar]
- Staller M. V., Vincent B. J., Bragdon M. D., Lydiard-Martin T., Wunderlich Z. et al. , 2015. Shadow enhancers enable Hunchback bifunctionality in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 112: 785–790. 10.1073/pnas.1413877112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfel G., Kazmar T., Frank O., Wienerroither S., Reiter F. et al. , 2015. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature 528: 147–151. 10.1038/nature15545 [DOI] [PubMed] [Google Scholar]
- Stanojevic D., Small S., and Levine M., 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254: 1385–1387. 10.1126/science.1683715 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Van Drenth M., Erives A., Markstein M., and Levine M., 2002. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111: 687–701. 10.1016/S0092-8674(02)01087-5 [DOI] [PubMed] [Google Scholar]
- Staudt N., Fellert S., Chung H. R., Jackle H., and Vorbruggen G., 2006. Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol. Biol. Cell 17: 2356–2365. 10.1091/mbc.e05-11-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., and Ehrenfeucht A., 1982. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 10: 2997–3011. 10.1093/nar/10.9.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J., Hicks J., and Herskowitz I., 1981. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J. Mol. Biol. 147: 357–372. 10.1016/0022-2836(81)90488-5 [DOI] [PubMed] [Google Scholar]
- Struffi, P., 2004 Transcriptional repression mediated by the Drosophila Knirps protein: contributions of CtBP and Rpd3. Ph.D. Thesis, Michigan State University, East Lansing. [Google Scholar]
- Struhl K., 1982. Regulatory sites for his3 gene expression in yeast. Nature 300: 285–286. 10.1038/300284a0 [DOI] [PubMed] [Google Scholar]
- Struhl G., Struhl K., and Macdonald P. M., 1989. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57: 1259–1273. 10.1016/0092-8674(89)90062-7 [DOI] [PubMed] [Google Scholar]
- Sun Y., Nien C. Y., Chen K., Liu H. Y., Johnston J. et al. , 2015. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25: 1703–1714. 10.1101/gr.192542.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A., Sollerbrant K., and Svensson C., 1998. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 429: 183–188. 10.1016/S0014-5793(98)00588-2 [DOI] [PubMed] [Google Scholar]
- Surkova S., Kosman D., Kozlov K., Manu E. Myasnikova et al. , 2008. Characterization of the Drosophila segment determination morphome. Dev. Biol. 313: 844–862. 10.1016/j.ydbio.2007.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson C. I., Evans N. C., and Barolo S., 2010. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18: 359–370. 10.1016/j.devcel.2009.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch J. R., Benavides J. A., and Cline T. W., 2006. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133: 1967–1977. 10.1242/dev.02373 [DOI] [PubMed] [Google Scholar]
- Teytelman L., Thurtle D. M., Rine J., and van Oudenaarden A., 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA 110: 18602–18607. 10.1073/pnas.1316064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Li X. Y., Sabo P. J., Sandstrom R., Thurman R. E. et al. , 2011. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 12: R43 10.1186/gb-2011-12-5-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunova E. N., Fujioka M., Kobayashi M., Deka D., and Jaynes J. B., 1998. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell. Biol. 18: 2804–2814. 10.1128/MCB.18.5.2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A., Muthusamy A. K., Alves M. R., Lavis L. D., Singer R. H. et al. , 2017. Nuclear microenvironments modulate transcription from low-affinity enhancers. Elife 6: e28975 10.7554/eLife.28975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A., Alves M. R., and Crocker J., 2019. Multi-enhancer transcriptional hubs confer phenotypic robustness. Elife 8: e45325 10.7554/eLife.45325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A., Maine E., and Schedl P., 1985. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185: 341–358. 10.1016/0022-2836(85)90408-5 [DOI] [PubMed] [Google Scholar]
- van Arensbergen J., van Steensel B., and Bussemaker H. J., 2014. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 24: 695–702. 10.1016/j.tcb.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., and Henikoff S., 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18: 424–428. 10.1038/74487 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., and Henikoff S., 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27: 304–308. 10.1038/85871 [DOI] [PubMed] [Google Scholar]
- Vo Ngoc L., Kassavetis G. A., and Kadonaga J. T., 2019. The RNA Polymerase II Core Promoter in Drosophila. Genetics 212: 13–24. 10.1534/genetics.119.302021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojnic E., Mourao A., Seizl M., Simon B., Wenzeck L. et al. , 2011. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 18: 404–409. 10.1038/nsmb.1997 [DOI] [PubMed] [Google Scholar]
- Wei Y., Gokhale R. H., Sonnenschein A., Montgomery K. M., Ingersoll A. et al. , 2016. Complex cis-regulatory landscape of the insulin receptor gene underlies the broad expression of a central signaling regulator. Development 143: 3591–3603. 10.1242/dev.138073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierer M., and Mann M., 2016. Proteomics to study DNA-bound and chromatin-associated gene regulatory complexes. Hum. Mol. Genet. 25: R106–R114. 10.1093/hmg/ddw208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B., Liu Y. H., Yeo Z. X., and Furlong E. E., 2012. Predicting spatial and temporal gene expression using an integrative model of transcription factor occupancy and chromatin state. PLoS Comput. Biol. 8: e1002798 10.1371/journal.pcbi.1002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L., 1971. Positional information and pattern formation. Curr. Top. Dev. Biol. 6: 183–224. 10.1016/S0070-2153(08)60641-9 [DOI] [PubMed] [Google Scholar]
- Wright K. J., Marr M. T. 2nd, and Tjian R., 2006. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. USA 103: 12347–12352. 10.1073/pnas.0605499103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z., Bragdon M. D., Vincent B. J., White J. A., Estrada J. et al. , 2015. Krüppel expression levels are maintained through compensatory evolution of shadow enhancers. Cell Rep. 12: 1740–1747 [corrigenda: Cell Rep. 14: 3030 (2016)]. 10.1016/j.celrep.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Chen H., Ling J., Yu D., Struffi P. et al. , 2014. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 28: 608–621. 10.1101/gad.234534.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Whitney P. H., Huang S.-K., Eck E. C., Garcia H. G. et al. , 2019. The Drosophila pioneer factor Zelda modulates the nuclear microenvironment of a dorsal target enhancer to potentiate transcriptional output. Curr. Biol. 29: 1387–1393.e5. 10.1016/j.cub.2019.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D., and Alt F. W., 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40: 271–281. 10.1016/0092-8674(85)90141-2 [DOI] [PubMed] [Google Scholar]
- Yanez-Cuna J. O., Dinh H. Q., Kvon E. Z., Shlyueva D., and Stark A., 2012. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 22: 2018–2030. 10.1101/gr.132811.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., and Pick L., 1995. Non-periodic cues generate seven ftz stripes in the Drosophila embryo. Mech. Dev. 50: 163–175. 10.1016/0925-4773(94)00333-I [DOI] [PubMed] [Google Scholar]
- Zabidi M. A., and Stark A., 2016. Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet. 32: 801–814. 10.1016/j.tig.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi M. A., Arnold C. D., Schernhuber K., Pagani M., Rath M. et al. , 2015. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. 10.1038/nature13994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., and Mango S. E., 2016. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37: 76–81. 10.1016/j.gde.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R. P., Senger K., Levine M., and Papatsenko D., 2006. Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 16: 1358–1365. 10.1016/j.cub.2006.05.044 [DOI] [PubMed] [Google Scholar]
- Zinzen R. P., Girardot C., Gagneur J., Braun M., and Furlong E. E., 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70. 10.1038/nature08531 [DOI] [PubMed] [Google Scholar]
- Zuo P., Stanojevic D., Colgan J., Han K., Levine M. et al. , 1991. Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev. 5: 254–264. 10.1101/gad.5.2.254 [DOI] [PubMed] [Google Scholar]