Abstract
Glioblastoma (GBM) is the most lethal primary brain tumor of the central nervous system in adults. Despite advances in surgical and medical neuro-oncology, the median survival is about 15 months. For this reason, initial diagnosis, prognosis, and targeted therapy of GBM represent very attractive areas of study. Aptamers are short three-dimensional structures of single-stranded nucleic acids (RNA or DNA), identified by an in vitro process, named systematic evolution of ligands by exponential enrichment (SELEX), starting from a partially random oligonucleotide library. They bind to a molecular target with high affinity and specificity and can be easily modified to optimize binding affinity and selectivity. Thanks to their properties (low immunogenicity and toxicity, long stability, and low production variability), a large number of aptamers have been selected against GBM biomarkers and provide specific imaging agents and therapeutics to improve the diagnosis and treatment of GBM. However, the use of aptamers in GBM diagnosis and treatment still represents an underdeveloped topic, mainly due to limited literature in the research world. On these bases, we performed a systematic review aimed at summarizing current knowledge on the new promising DNA and RNA aptamer-based molecules for GBM diagnosis and treatment. Thirty-eight studies from 2000 were included and investigated. Seventeen involved the use of aptamers for GBM diagnosis and 21 for GBM therapy. Our findings showed that a number of DNA and RNA aptamers are promising diagnostic and therapeutic tools for GBM management.
Keywords: aptamer, nucleic acid, glioblastoma, diagnosis, therapy
1. Introduction
Glioblastoma (GBM) is classified by the World Health Organization as grade IV astrocytoma and is the most common and fatal primary brain tumor of the central nervous system (CNS) in adults [1,2]. Despite advances in surgical and medical neuro-oncology, median survival is only 15 months after the first diagnosis and with standard surgery and chemoradiation [3,4,5,6,7]. For this reason, initial diagnosis and targeted therapy of GBM represent very attractive areas of study.
Currently, diagnostic methods are based on initial neurological examination and imaging tests (magnetic resonance imaging, MRI, and computerized tomography, CT) to evidence brain abnormality. Nevertheless, there are many limitations in standard GBM diagnostic methods: in fact, distinguishing glioma grades or discriminating GBM from other intracranial mass lesions remain very difficult or even impossible by conventional techniques [8]. Moreover, often only surgical biopsy can definitely confirm the diagnosis. In addition to these factors, the fact that GBM symptoms overlap with those of other brain disorders makes diagnosis even more difficult, with GBM in certain cases being misdiagnosed as other CNS pathologies (sinusitis, tension headache, myasthenia gravis, or demyelinating disease) [9].
After initial diagnosis, current clinical treatment for GBM requires a multidisciplinary approach, consisting in initial tumor resection followed by concurrent radiotherapy and chemotherapy. However, despite maximal safe surgical resection and multimodality therapy, about 70% of these tumors invariably recur, with standards of care at recurrence far less well defined than in the newly diagnosed setting [7,10,11]. Radiotherapy and chemotherapy in GBM are affected by de novo or acquired resistance, reasons why surgery and temozolomide (TMZ) treatment before or during standard care therapy can increase patient survival by only around 15 months. Therefore, novel drugs able to inhibit GBM cell growth and not subjected to the side effects of the current therapies are needed [6].
Given the above-mentioned limitations and the lack of standardization related to GBM diagnosis and treatment, the need to develop new diagnostic and therapeutic strategies for GBM diagnosis and treatment, also involving targeted imaging and drug delivery platforms, has led to the investigation of aptamers. Indeed, these molecules possess many features making them ideal novel imaging and therapeutic agents for diagnosis and treatment of GBM [12].
To date, nucleic acid aptamers have attracted growing interest as biosensors and diagnostic and therapeutic elements for tumor imaging due to the following properties: (i) chemical synthesis allowing for low batch variation, (ii) an ability to recognize targets with high specificity and affinity, (iii) a stability at high or low temperatures and critical pH values, and (iv) a small size that allows good tissue penetration and rapid clearance [13,14]. Further, aptamers can be internalized into cells to deliver payloads [15]. Moreover, very recent studies on aptamers demonstrate a capacity to cross the blood brain barrier (BBB), considered the major obstacle for innovative GBM approaches [16]. Thanks to these attractive characteristics, these innovative molecules have been studied so far to improve the quality and sensitivity of imaging techniques for diagnostic purposes [17] and to explore new therapeutic approaches for GBM management [18].
Concerning the use of aptamers in GBM therapy, various aptamers have been tested as therapeutic agents. Several studies investigated aptamer-based drugs that bind to and inhibit specific proteins, while others were on aptamers as systems for drug delivery specifically to the tumor cells. However, despite their promise, the use of aptamers in GBM diagnosis and treatment still represents an underdeveloped topic, mainly due to limited literature in the research world. In this scenario, the aim of this systematic review is to collect and summarize the current state of the art on the role of aptamers in GBM, focusing on their use in diagnostic and therapeutic applications.
2. Materials and Methods
2.1. Search Strategy and Selection Criteria
A systematic search for all published studies concerning the application of aptamers for GBM management was conducted. The most relevant scientific electronic databases (PubMed, Cochrane Library, MEDLINE, ScienceDirect, Google Scholar) were comprehensively explored and used to build the search. Only studies published since 2000 were selected. The search strategy included the key terms listed in S1.
The literature search was restricted to English language publications. Two reviewers, after having independently screened identified titles and abstracts, assessed the full text of the original articles involving aptamers used as probe or therapeutic tools for GBM management. For articles meeting these criteria with full text available, the following further selection criteria were used: articles were excluded if they also involved any type of brain tumor other than GBM and if they were off-topic after investigating the full text. Moreover, we only included articles demonstrating the utility of aptamers for human applications. The entire flow and results of the literature research were finally checked by a third researcher.
2.2. Data Extraction and Collection
After the above-mentioned selection procedure, selected articles that met the inclusion criteria were analyzed by two reviewers, and data useful for conducting the systematic review were extracted and collected in a pre-designed sheet. Extracted data included the following: study characteristics (first author name, year of publication, and method of study, namely in vivo and/or in vitro), name of the aptamer investigated, nucleic acid sequence, aptamer target, diagnostic or therapeutic application depending on if the study used aptamers for GBM diagnosis or therapy, and dissociation constant (Kd).
2.3. Planning and Conducting the Review
The articles were classified according to the purpose they had, namely if they concerned application for GBM diagnosis or therapy. This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (See S2 for PRISMA Checklist).
3. Results
3.1. Study Selection
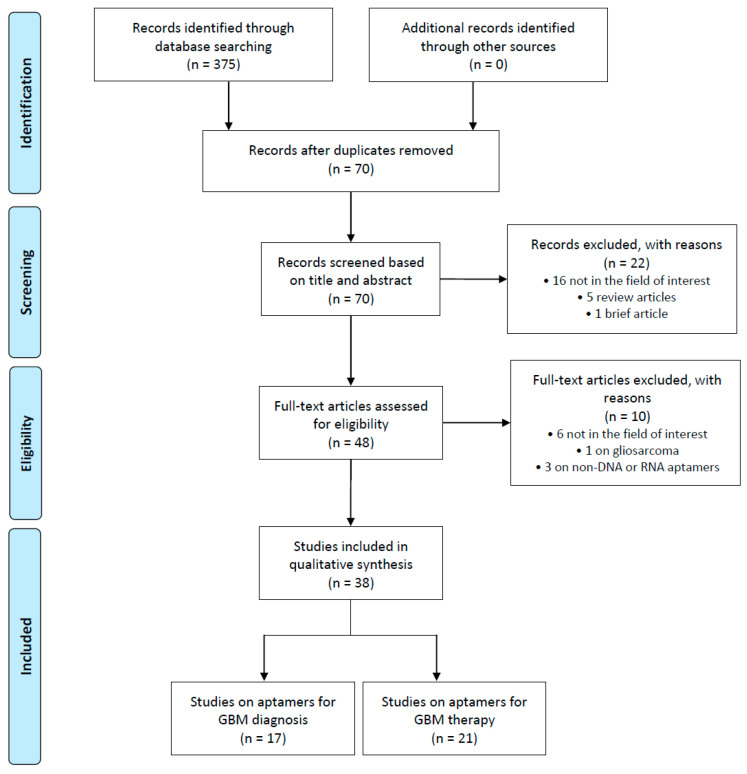
A total of 375 articles was retrieved from the PubMed, Google Scholar, Web of Science, and Science Direct databases. After the removal of 305 duplicate articles, we performed a screening based on titles and abstracts of the remaining 70 articles. Twenty-two records in this step were excluded for the following reasons: 5 were review articles, 1 was a brief article, and the remaining 16 were off-topic and only mentioned the words “aptamers” and/or “GBM” in the abstract. Screening by titles and abstracts yielded 49 potentially eligible articles, which were evaluated by their full text. Of these articles, 1 record was excluded because it involved gliosarcoma, 3 were excluded because they investigated non-DNA or RNA aptamers, and 6 were excluded because they were off-topic and did not investigate the role of aptamers in GBM diagnosis or treatment. Finally, 38 records were included for qualitative synthesis. The PRISMA flow diagram of included studies according to the inclusion and exclusion criteria is reported in Figure 1, and their characteristics are summarized in Table 1 and Table 2.
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram.
Table 1.
Characteristics of included studies on diagnosis applications of aptamers in glioblastoma (GBM). Kd = dissociation constant.
| Author | Year | Aptamer | Nucleic Acid | Target | Tested | Kd | Conjugated Molecule | Diagnostic Application |
|---|---|---|---|---|---|---|---|---|
| Affinito et al. [19] | 2019 | A40s | 2′-fluoropyrimidine- RNA | In vitro; Ex vivo | 41.92 nM | miRNA; anti-miRNA; Cy5, Alexa488 | Confocal fluorescence microscope, histochemistry | |
| Fechter et al. [20] | 2019 | H02 | 2′-fluoropyrimidine- RNA | Integrin α5β1 | In vitro; Ex vivo | 72–277.8 nM | Cy5, Alexa564 | Confocal fluorescence microscope, SPR |
| Wu et al. [21] | 2018 | WYZ-41a WYZ-50a | DNA | A172 cells | In vitro | 75.27–168.56 nM | Cy5, FITC | Fluorescence microscope |
| Hasan et al. [22] | 2018 | Anti-EGFR | 2′-fluoropyrimidine- RNA | EGFRvIII | In vitro | Dynamic morphology | ||
| Mahmood et al. [23] | 2015 | Anti-EGFR | 2′-fluoropyrimidine- RNA | EGFRvIII | In vitro | Dynamic morphology | ||
| Alibolandi et al. [24] | 2014 | AS1411 | 2′-fluoropyrimidine- RNA | Nucleolin | In vitro | CdTe QDs | Fluorescence microscope | |
| Wu et al. [25] | 2014 | U2, U8, U19, U31 | DNA | EGFRvIII | in vivo; Ex vivo | 3.37–16.78 nM | 188RE | SPECT |
| Tan et al. [26] | 2013 | 32, 41, 43, 47 | DNA | EGFRvIII | In vitro | 0.62–37.57 nM | FITC, Biotin | Confocal fluorescence microscope, ELISA |
| Kim B. et al. [27] | 2013 | Anti-VEGFR2 | DNA | VEGFR2 | In vivo | 0.12 nM | carboxylated magnetic nanocrystal | MRI |
| Kim Y. et al. [28] | 2013 | A1, A2, A3, A4, A5 | DNA | TICs | In vitro | 0.12–3.75 nM | Cy3 | Fluorescence microscope |
| Li et al. [29] | 2013 | GB-10 | DNA | Tenascin-C | in vitro | 110 µM | NHS–PEG18–aldehyde | MR-FS |
| Kang et al. [30] | 2012 | GM128, GM131 | DNA | U118-MG cells | In vitro; Ex vivo | 20–37 nM | FAM | Confocal fluorescence microscope |
| Wan et al. [31] | 2011 | Anti-EGFR | 2′-fluoropyrimidine- RNA | EGFR and EGFRvIII | In vitro | Microfluidic device | ||
| Bayrac et al. [32] | 2011 | GMT3, GMT5, GMT9 | DNA | A172 cells | In vitro | 75.27–168.56 nM | Biotin | Fluorescence microscope |
| Wan et al. [33] | 2010 | Anti-EGFR | 2′-fluoropyrimidine- RNA | EGFR and EGFRvIII | In vitro | 2.4 nM | FAM | Fluorescence microscopy |
| Hicke et al. [34] | 2006 | TTA1 | 2′-fluoropyrimidine and 2′-OH purine RNA | Tenascin-C | In vivo | Rhodamine-RED, technetium-99m | Fluorescence microscopy, scintigraphy | |
| Daniels et al. [35] | 2003 | GB-10 | DNA | Tenascin-C | In vitro | 150 nM | Biotin | ELISA, SPR |
| Hicke et al. [36] | 2001 | TTA1 | 2′-fluoropyrimidine and 2′-OH purine RNA | Tenascin-C | In vivo | 3 nM | Phosphorus-32 | SPR |
Table 2.
Characteristics of included studies on therapeutic applications of aptamers in GBM. Kd = dissociation constant.
| Author | Year | Aptamer | Nucleic Acid | Target | Tested | Kd | Conjugated Molecule | Therapeutic Application |
|---|---|---|---|---|---|---|---|---|
| Peng et al. [37] | 2020 | U2 | DNA | EGFRvIII | In vitro; In vivo | Gold nanoparticle (AuNPs) | Growth inhibition in vitro and prolongs the survival time in vivo | |
| Affinito et al. [38] | 2020 | A40s | 2′-fluoropyrimidine- RNA | EphA2 | In vitro | 0.76 ± 0.2641 nM | Growth inhibition, stemness, and migration of GSCs | |
| Fu et al. [39] | 2019 | GS24 | DNA | Transferrin receptor | In vitro; In vivo | tFNA-TMZ | ||
| Wang et al. [40] | 2019 | CL-4RNV616 | 20-OMe/DNA mixmer | EGFR | In vitro | 18.24 nM | Growth inhibition and apoptosis induction | |
| Shi et al. [41] | 2019 | GMT8 and Gint4.T | DNA; 2′-fluoropyrimidine- RNA | Unknown; PDGFRβ | In vitro | tFNA-paclitaxel | Growth inhibition, migration, and invasion and apoptosis induction of GBM cells. | |
| Yoon et al. [42] | 2019 | PDR3 | 2′-fluoropyrimidine- RNA | PDGFRα | In vitro | 0.25 nM | STAT3 siRNA | Growth inhibition, apoptosis induction. |
| Wei et al. [43] | 2019 | 4-1BB-OPN | 2′-O-methylation for all C and U nucleotides | OPN and 4-1BB | In vivo | Immunostimolatory effect and increases survival rate | ||
| Zhang et al. [44] | 2018 | U2 | DNA | EGFRvIII | In vitro; In vivo | 6.27 nM | RE188 | Growth inhibition, radiosensitivity, and radiotherapy |
| Esposito et al. [45] | 2018 | Gint4.T | 2′-fluoropyrimidine- RNA | PDGFRβ | In vitro; In vivo | STAT3 siRNA | Growth inhibition and migration in vitro and inhibition of tumor growth and angiogenesis in vivo | |
| Bayrac et al. [46] | 2018 | GMT-3 | DNA | A-172 cell line | In vitro | Doxorubicin (DOX) | Cytotoxic effects | |
| Luo et al. [47] | 2017 | AS1411 | DNA | Nucleolin | In vitro; In vivo | poly (l-γ-glutamyl-glutamine)-paclitaxel (PGG-PTX) | Pro-apoptotic effect, increases median survival time and cell apoptosis in vivo | |
| Esposito et al. [48] | 2016 | GL21.T; Gint4.T | 2′-fluoropyrimidine- RNA | Axl; PDGFRβ | In vitro | miR-137; antimiR-10b | Inhibition of GSC propagation | |
| Amero et al. [49] | 2016 | GL43.T | 2′-fluoropyrimidine- RNA | EphB3 and EphB2 | In vitro | 433.5 nM | Cell migration | |
| Camorani et al. [50] | 2015 | CL4; Gint4.T | 2′-fluoropyrimidine- RNA | EGFRvIII; PDGFRβ | In vitro | Growth migration and invasion inhibition | ||
| Zhang et al. [51] | 2014 | 32-biotin (BA) | DNA | EGFRvIII | In vitro | c-METsiRNA | Apoptosis induction and growth inhibition | |
| Camorani et al. [52] | 2014 | Gint4.T | 2′-fluoropyrimidine- RNA | PDGFRβ | In vitro; In vivo | 9.6 nM | Growth inhibition in vitro and in vivo. | |
| Wan et al. [53] | 2013 | anti-EGFR | 2′-fluoropyrimidine- RNA | EGFR | In vitro | 2.4 nM | Growth and motility inhibition | |
| Gao et al. [54] | 2012 | GMT8 | DNA | A-172 cell line | In vitro; In vivo | docetaxel-loaded ApNP | Apoptosis induction and growth inhibition | |
| Verhoeff et al. [55] | 2009 | Pegaptanib | 2′-O-methyl purine/2′fluoro pyrimidine with two 2′-ribo purines conjugated to 40 kDa PEG, 3′ inverted dT | VEGF | In vivo | 200 pM | Decreases tumor blood vessel density | |
| Liu et al. [56] | 2009 | E21 | 2′-fluoropyrimidine- RNA | EGFRvIII | In vitro | 33 nM | Apoptosis induction (after transfection) |
3.2. Aptamers in GBM Diagnosis
Seventeen articles investigated aptamers in imaging and diagnostic systems for GBM. Their characteristics are summarized in Table 1. According to the targets, we distinguish two groups: the first aimed at visualizing Tenascin-C-positive cells, the second at detecting epidermal growth factor receptor- (and its variant III-) expressing cells. In addition, single articles were found for the following GBM-associated proteins: nucleolin, vascular endothelial growth factor receptor (VEGF), and integrin α5β1; and for the following GBM cell lines: T98G, U118-MG, A172, and GBM-initiating cells (also known as GBM stem cells).
3.3. Aptamers as Diagnostic
The first aptamers used in GBM detection tools, and thus potentially applicable for GBM diagnosis, were aimed at identifying GBM cells by means of the extracellular matrix glycoprotein Tenascin-C (TN). As early as 2001, Hicke BJ et al. used TN-expressing U251 cells and purified TN to obtain TTA1, an RNA aptamer (modified with f2′-F-pyrimidine and 2′-OH purine nucleotides), specific to the tumor-associated protein TN. TTA1 proved to bind TN with a very high affinity (Kd, 3 × 10−9 M) [36]. Moreover, the group evaluated the binding via surface plasmon resonance (SPR), a technique detecting circulating cancer biomarkers usable as key actors of liquid biopsy for GBM diagnosis [57]. A few years later, the same group used scintigraphy to assess the distribution in mice of the technetium-99m-labeled aptamer, showing the potential clinical application of radiolabeled TTA1 [34]. In 2003, the same target was used by Gold et al. to select a DNA aptamer, GBI-10, in the U251 cell line. They demonstrate the selectivity of GBI-10 for GBM cells, with the aptamer-TN interaction assessed via enzyme-linked immunosorbent assay (ELISA) and biosensor analysis with SPR [35]. Thereafter, Li Y et al. emphasize the potential diagnostic application of GBI-10 with a molecular recognition force spectroscopy (MR-FS) study investigating dynamic aptamer-target interactions [29].
More recently, interest has been mainly focused on EGFR and on its receptor variant III, EGFRvIII, which is associated with GBM aggressiveness. Over the past few years, DNA and RNA aptamers have been generated and used for EGFR-mediated detection of GBM. In 2010, Iqbal et al. isolated an anti-EGFR RNA aptamer by iterative selection using the purified human protein [33]. The authors proved not only the ability of the aptamer to bind wild-type and mutant EGFR with high affinity (Kd = 2.4 nM), but also demonstrated the aptamer’s capacity to detect and capture murine and human GBM cells when it is immobilized on a glass substrate, so it is possible to use the technique for early detection and the monitoring of residual disease [33]. In fact, one year later, the same group designed a flow-through lab-on-chip device that took advantage of the surface-bound aptamer’s affinity for EGFR, the biomarker overexpressed in GBM, to demonstrate that a microfluidic-based approach can be used to detect and isolate GBM cells [31]. The group then advanced the diagnostic use of anti-EGFR aptamers with two subsequent articles on the tracking of the differential dynamics of GBM cell morphology on aptamer-grafted substrates, and on the analysis of dynamic morphology in computational single-cell metrics to detect and recognize tumor cells [22,29].
Working on the same target, Tan et al. and Wu et al. performed two-cell-systematic- evolution- of-ligands-by-exponential-enrichment (SELEX) selections on U87-EGFRvIII cells in order to specifically recognize receptor variant III. Both groups isolated DNA aptamers that proved to be usable in GBM diagnosis. In fact, Tan et al. demonstrated, in vitro, the imaging of different selected FITC-labelled aptamers in U87-mutated cells [26]. Furthermore, the authors measured the binding affinity of the aptamers by biotin-avidin ELISA (BA-ELISA) assay: all the sequences analyzed had a Kd less than 100 nmol/L. By contrast, Wu et al. used imaging with single-photon emission computed tomography (SPECT), in vivo and ex vivo, with tumor-bearing mice [25]. This nuclear medicine imaging technique is similar to scintigraphy, the conventional diagnostic radioisotope-based approach, but has the ability to provide 3D details and, therefore, is useful for solid tumors like GBM.
Despite a greater focus on these two targets, there are many other groups concentrating their forces on detecting GBM by targeting other tumor-related proteins. Indeed, Hadizadeh et al. utilized the ssDNA aptamer AS1411, which targets nucleolin, to generate AS1411-cadmium telluride (CdTe) quantum dots (QDs) [58], which are usable as versatile fluorescent probes and sensors in several applications. The authors prepared CdTe QDs with four different colors through a microwave-assisted reduction method, and then assessed, with a fluorescence microscope, the aptamer-QD conjugates. They demonstrated the potential of the conjugates as nanoprobes for GBM imaging in vitro [24]. By contrast, Kim et al. generated an aptamer-based diagnostic nanoprobe by conjugating an anti-VEGFR2 DNA aptamer with magnetic nanocrystals. In vivo tests in an orthotopic GBM mouse model showed a high magnetic resonance (MR) imaging sensitivity of the tumor vasculature [27]. Moreover, in the last year, Choulier et al. combined protein- and cell-SELEX to isolate RNA aptamers that selectively bind the integrin α5β1, an αβ heterodimeric cell surface receptor associated with tumor angiogenesis and GBM aggressiveness (Kd in the nM range). The authors prove the diagnostic potential of the labeled aptamer, using it in a fluorescence-based assay on cell lines and in histo-fluorescence assays on patient-derived xenografts (PDXs) [20].
Other groups, such as Kang et al. and Bayrac et al., have used aptamers in microscopy imaging. The former identified high affinity ssDNA aptamers for GBM with the use of U118-MG cells: confocal images show cell and human tumor tissue section staining after FAM- or Cy5-labeled aptamers treatment, respectively [30]. The latter selected GBM-targeting ssDNA aptamers with the use of A172 cells and investigated cell detection via streptavidin-PE staining of the biotinylated aptamer [32]. Both groups have not further investigated the aptamers’ targets in subsequent studies. Similarly, in 2018, Yang et al. selected two aptamers targeting T98G cells—WYZ-41a and WYZ-50a—and, without discovering the targets, proved their diagnostic applicability. Indeed, both aptamers detected their target cells in complex mixtures, such as undiluted fetal bovine serum (FBS) or cerebral spinal fluid (CSF), demonstrating the power of aptamers in improving GBM diagnosis through liquid biopsy [21].
Finally, two groups recently selected aptamers against a specific subgroup of GBM cells, brain tumor-initiating cells (TICs) and GBM stem cells (GSCs), responsible for GBM metastasis and relapse. Rich et al. selected a pool of DNA aptamers recognizing TICs with a very low dissociation constant (Kd between 0.12 and 3.75 nM); binding was proved by aptamer-Cy3-based fluorescence [28]. Some years later, Affinito et al., in 2019, similarly selected the GBM-specific 2′-F-RNA aptamer A40s, using an RNA library on primary human GSCs. The authors demonstrated A40s-mediated detection in GBM cells and in human GSCs tissue sections [19]. Here too, the aptamer proved to have a high affinity for target cells in a low nanomolar range (Kd of 41.92 nM). Since stem cells have a role in metastasis and chemoresistance, these aptamers could be applied in the clinical setting for the early detection of new metastatic niches and for the monitoring of GBM treatment.
Thus, many studies have been carried out to take advantage of the unique characteristics of aptamers to improve the quality and sensitivity of conventional imaging techniques and enhance GBM diagnosis.
3.4. Aptamers in GBM Therapy
Twenty-one studies investigated the role of DNA and RNA aptamers in targeted therapy of GBM. Their characteristics are summarized in Table 2. Among the selected studies, 11 investigated aptamer-based therapy with binding inhibiting a specific protein target, and 10 investigated the therapeutic effect of aptamer-based conjugates specifically delivering molecules (non-coding RNA, nanoparticles, chemotherapeutics) to tumor cells after ligand recognition.
3.5. Aptamers as Inhibitors
Here we review the therapeutic effect of DNA and RNA aptamers developed as promising new drugs for GBM in the field of targeted therapy. Eleven articles on the therapeutic targets specifically recognized by the aptamers were discussed. We found that half of these studies aimed to block EGFR or its mutated form (EGFRVIII), 3 studies aimed to block the Eph receptor family, whereas fewer investigated aptamers recognizing VEGF, platelet-derived growth factor receptor (PDGFR), or osteopontin (OPN) (one paper for each).
Numerous studies were aimed at selecting inhibitors, including aptamers, for EGFR because it is a relevant feature in primary GBM (it is overexpressed in 60% of tumors) and its mutation correlates with poor prognosis and tumor aggressiveness [59,60]. In this regard, several aptamers were selected and characterized and all of them produced a good reduction in cell proliferation in vitro.
In 2012, Wan et al. developed a chip to observe cell behavior in vitro after treatment with a 2′-fluoropyrimidine-containing RNA aptamer against EGFR. They found that the anti-EGFR aptamer reduces migration and proliferation of GBM cells by blocking EGFR phosphorylation [53].
Very recently, Wang et al. selected and characterized an aptamer against EGFR named CL-4RNV616, containing 20-O-Methyl RNA and DNA nucleotides. CL-4RNV616 was tested not only on U87MG glioblastoma cells but also on Huh-7 liver cancer and MDA-MB-231 breast cancer. The authors demonstrated that CL-4RNV616 binds and inhibits EGFR, has high stability in human serum, and increases apoptosis in cancer cells; in breast cancer, biopsy-based immunostaining demonstrated high EGFR, but no data were given on GBM tissues [40].
Other studies selected and characterized aptamers targeting a variant of EGFR named EGFRvIII, which correlates with EGFRwt amplification in clinical GBM samples. EGFRvIII is constitutively active thanks to a particular deletion and is found in 25% of tumors, but instability makes it a difficult target.
Liu et al. described, for the first time, an aptamer for EGFRvIII with a low dissociation constant (Kd). Unluckily, the aptamer selected in Escherichia coli for EGFRvIII was not able to bind the human EGFRvIII protein, the reason why the aptamer was not investigated further [56]. However, two promising aptamers for human EGFRvIII are (i) CL4, a 2′-fluoropyrimidine-containing RNA aptamer which binds wild-type and mutant forms of EGFR [50], and (ii) U2, a DNA aptamer specific to the above-mentioned EGFR variant [44].
In vitro, CL4 proved to be a promising inhibitor. In fact, the authors nicely demonstrate that CL4 inhibits GMB growth as well as or better than the current EGFR inhibitor (gefitinib), representing, therefore, an attractive alternative for GBM management. Moreover, solid evidence demonstrates that EGFR inhibitors make the growth and survival of GBM cells dependent on PDGFRβ signaling, the reason why being in order to improve the therapeutic effect of CL4. Camorani et al. combined CL4 with a second aptamer, named Gint4, which binds and inhibits the phosphorylation of PDGFRβ [50]. Gint4 was previously characterized in vitro and in vivo, with the authors demonstrating a drastic inhibition of cell viability and tumor growth [52]. The combined therapy resulted in a stronger reduction of GBM aggressiveness when compared to TMZ or approved PDGFRβ and EGFR inhibitors (imatinib, gefitinib, and cetuximab) [50]. Instead, the DNA aptamer U2 inhibits tumor growth and acts as a molecular imaging probe in vitro and in vivo [56].
Other promising therapeutic targets for GBM are represented by Eph receptors, known to be involved in tumor invasion and metastasis [61]. Within the family, EphA2 is highly expressed in GBM, but it is not detectable in the normal brain [62]. It correlates with neovascularization [63], proliferation, pathological grade, and patient survival [64,65,66]. Moreover, EphA2 expression increases in GBM stem cells (GSCs), a subpopulation of GBM cells resistant to conventional therapies, and being co-expressed with other stem cell markers, such as CD133 and integrin alpha 7, it represents an important molecular feature of GBM stemness. These well-characterized findings make EphA2 a candidate for efficacious targeted therapy for GBM: in fact, Affinito et al. very recently selected and characterized 2′-fluoropyrimidine-containing RNA aptamers (40L and its truncated form, A40s). As a first attempt, the authors described an innovative cell-SELEX approach aimed at identifying RNA aptamers binding primary GSCs. 40L and A40s recognize not only primary GSCs but also human GBM tissues positive for the GBM stemness marker CD133. Interestingly, A40s binds primary GSCs in the nanomolar range (about 42 nM), and the authors demonstrate that A40s can be used to deliver secondary molecules, for example, microRNAs [19]. The same authors in 2020 published a second paper in which A40s is fully characterized in vitro for its therapeutic effect thanks to the discovery of the aptamer’s target, EphA2.
As expected, considering the relationship between EphA2 expression and GBM progression, the inhibition of EphA2 mediated by A40s treatment reduces self-renewal of GSCs and GBM tumor propagation. A40s is stabile in human serum (~24 h) and crosses the blood brain barrier (BBB), reaching the brain in healthy mice after systemic administration [38]. Although the data support that A40s has potential applicability as a therapeutic tool to block the GSC population and, thus, GBM recurrence, the authors neither investigated in-depth A40s affinity for the other members of the Eph family nor the therapeutic effect of the aptamer in vivo.
EphB3 and EphB2 are reported to be overexpressed in GBM cell lines and to have a pro-tumoral role promoting migration and invasion in cancer cells. GL43.T is an RNA aptamer modified with 2′-fluoro pyrimidine (2′-F-Py) that binds EphB3/EphB2. It blocks cell proliferation and antagonizes migration in GBM cells. The binding affinity for the EphB3 receptor is in the high nM range (433.5 nM–136.6 µM), but the authors do not give any information on the affinity of GL43.T for EphB2 or other Eph receptors [49].
Other approaches have used the bispecific aptamer named 4-1BB–OPN. The authors concomitantly targeted OPN and the costimulatory receptors on CD8+ T cells by using two different aptamers, respectively OPN-aptamer and 4-1BB, attached by an RNA linker sequence. 4-1BB–OPN was tested in vivo in mice with established GL261 GBM cells: the authors solidly demonstrated that the treatment drastically increases survival time by 68%, so is a promising approach to potentiate naturally occurring antitumor immunity via tumor targeting [43].
Another promising biomarker for recurrent GBM is the mediator of angiogenesis, VEGF. The alternative splicing of VEGF mRNA produces four principal isoforms (containing 121, 165, 189, and 206 amino acids), among which VEGF165 is the predominant one. Pegaptanib, approved by the US FDA in December 2004 for treatment of age-related macular degeneration (AMD), is an RNA aptamer targeting VEGF165. Considering that Bevacizumab (a monoclonal antibody against VEGF) was approved for GBM treatment, Verhoeff et al. investigated the effect of aptamer-based anti-VEGF treatment in combination with irradiation in an orthotopic mouse model of GBM. Their results demonstrated that the treatment suppresses invasive growth, increasing progression-free survival (PFS) in vivo [55].
3.6. Aptamers as Carriers of Therapeutics
Here, we fully investigate aptamers as main components of targeted delivery systems in GBM. The ten selected articles can be divided into two major groups: those on aptamers as carriers of nanoparticles (five articles) and those on aptamers as carriers of ncRNAs (four articles). Only one article concerned aptamers as carriers of chemotherapeutic agents.
Regarding the first group, Gao et al. published in 2012 the first studies on aptamers as carriers of nanoparticles in GBM. The authors describe ApNP, a conjugate obtained from nanoparticles of MPEG-PCL functionalized with GMT8, a DNA aptamer binding GBM cells. In order to induce GBM cell death, Docetaxel (DTX), an inhibitor of microtubule depolymerization causing mitotic arrest, was used alone as a control or loaded in ApNP (DTX-loaded ApNP). The results show that DTX-ApNP not only induces cell apoptosis and inhibits tumor spheroid growth in vitro, but that it targets GBM tumor mass, prolonging the survival of GBM-bearing mice compared to controls [54].
GMT8 was also tested in association with the PDGFRβ-binding RNA aptamer Gint4.T to deliver tFNA, a three-dimensional (3D) tetrahedral framework nucleic acid to GBM cells. tFNA is a self-assembly of four single-stranded DNAs (ssDNAs) named S1–S4 and represents a promising targeted approach for the delivery of oligonucleotides or drugs. Firstly, the nanoconjugate, named GTG, was characterized in vitro as a delivery tool in two different GBM cell lines (U87MG and bEnd.3). The authors nicely demonstrated that GTG is efficiently internalized in GBM cells and is able to cross the BBB in an in vitro model [41]. When GTG was loaded with paclitaxel (PTX), named GPC-PTX, to investigate anti-GBM efficacy, they found a reduction in cell proliferation and apoptosis.
Fu et al. used the same approach with tFNA functionalized with aptamers for the specific delivery of therapeutic agents in order to kill GBM cells. In this case, tFNA nanoparticles were loaded with TMZ. To confer GBM tissue specificity, ssDNAs named S2 and S3 were elongated with AS1411 and GS24 aptamers, respectively. AS1411, which binds nucleolin and induces tumor cell apoptosis, and GS24, which binds the transferrin receptor (TRF), helped to pass the BBB. The authors demonstrated in vitro that the lethality of tFNA-TMZ was higher than TMZ alone. Furthermore, thanks to GS24, tFNA crossed the BBB in vivo and stayed in brain vessels for 1 h, suggesting that tFNA might be a favorable delivery vehicle [39].
The application of AS1411 as a nanoparticle delivery tool was previously investigated by Luo et al. In this case, poly (L-γ-glutamyl-glutamine) loaded with paclitaxel (PGG-PTX) was functionalized with AS1411. The nanoconjugate exhibited cytotoxicity in vitro on 2D cultures and 3D tumor spheroids. The authors also studied tissue distribution and therapeutic effects of AS1411-PGG-PTX in vivo. They proved that the conjugate’s tissue distribution was related to tumor localization and that the median survival of mice treated with AS1411-PGG-PTX was higher compared to controls [47].
Peng et al. assessed the proprieties of U2-AuNP, a novel brain-targeting complex. As previously reported, the aptamer U2 binds EGFRVIII. U2, thanks to a thiol group, was conjugated to a gold nanoparticle (AuNPs) through an Au-S bond. U2-AuNP inhibited the activation of EGFRvIII, affected U87-EGFRvIII cell proliferation and invasion, and increased the survival of GBM-bearing mice [37].
In the second group of articles, aptamers are used as carriers of ncRNA (miRNA, anti-miR, siRNA). In the first published study, investigating the delivery of small interfering RNAs (siRNAs), the authors characterized the DNA aptamer 32 as a carrier for a c-Met siRNA binding EGFRvIII. Aptamer 32, as well as the c-Met siRNA, were biotinylated (Ba), and both moieties incubated with streptavidin containing four binding sites to form a stable conjugate. The conjugate was characterized in vitro, demonstrating that the Ba aptamer is an efficient delivery tool for c-Met siRNA into U87-EGFRvIII cells, offering a combined treatment regimen for patients with high expression of c-Met and EGFRvIII [44].
The next two studies investigated aptamer-mediated delivery of siRNA targeting STAT3, the role of which as a key regulator in GBM has been demonstrated. In the first, the STAT3 siRNA was conjugated to PDR3, an anti-PDGFRα RNA aptamer. The conjugate strengthens the effect of the aptamer alone: in fact, the authors firstly demonstrated the effect of PDR3 on apoptosis and then confirmed that PDR3-siSTAT3 mediates strong reduction in cell viability in vitro [42]. In the second, Esposito et al. characterized a modified 2′F-Py nuclease-resistant RNA conjugate in vitro and in vivo. In that study, Gint4.T (an anti-PDGFRβ) was conjugated via a sticky bridge approach to ansiRNA specifically targeting STAT3. The conjugate reduced STAT3 mRNA levels and, accordingly, in vitro viability and migration of GBM cells. Gint4.T AsiC proved to lead to a selective functional effect expressed only by PDGFRβ-positive cells. Furthermore, systemic administration of the conjugate in vivo led to reduction of tumor mass and inhibition of neovascularization [45].
The same group designed two different conjugates to deliver miRNA/antimiRNA to glioblastoma stem-like cells simultaneously. In particular, GL21.T (an anti-AXL aptamer) was conjugated to miR-137, and Gint4.T (an anti-PDGFRβ aptamer) to anti-miR-10b. By combining the therapeutic potential of the miRNA-based approach and RTKs, the authors found a drastic inhibition of self-renewal and migration GSCs [48]. The other approach delivered a chemotherapeutic agent via a specific aptamer: the ssDNA aptamer GMT-3 was conjugated to the anticarcinogenic drug Doxorubicin (DOX), known to intercalate into DNA strands by noncovalent interaction. The authors demonstrated in GBM cells that GMT-3 delivers DOX well to GBM cell lines, with a selective release of the cytotoxic drug in targeted cells, overcoming the cytotoxic effects of the chemotherapeutic on normal cells [46].
4. Discussion
In this systematic review, we highlight the potential value of the RNA and DNA aptamers in the management of GBM. Aptamers have been shown to be useful tools in diagnostic and therapeutic approaches in several diseases [67,68]; an important milestone was achieved in December 2004, when the US FDA approved the first RNA aptamer for the treatment of neovascular age-related macular degeneration (AMD), named Pegaptanib (Macugen).
To date several studies have investigated the diagnostic role and clinical value of aptamers in GBM, a pathology in which the standard approaches of diagnosis and treatment are still limited [69].
In the literature, there are many data collected on aptamers in GBM. One promising clinical trial started in 2019, which uses a pegylated structured L-oligoribonucleotide aptamer (Spiegelmer) that binds and neutralizes CXCL12, named Olaptesed Pegol (NOX-A12) (NCT04121455). Nevertheless, the benefits of using aptamers as diagnostic and therapeutic agents remains unclear. We have carried out a systematic review examining all studies on RNA and DNA aptamers found to be promising as diagnostic and/or therapeutic molecules published since 2000. This qualitative analysis has involved 38 studies: the results and conclusions of the selected studies reveal the great potential of aptamers in the management of GBM. Despite this, the actual use of aptamers is generally still in its infancy.
In particular, there is a more promising group of aptamers: those described for the development of new treatments for GBM; the clinical usefulness of the other categories of aptamers remains to be clarified since they have been only poorly characterized. With regard to GBM treatment, the best-characterized aptamers inhibiting cancer cell proliferation and with good affinity for their targets are Gint4 and U2. The former binds PDGFRβ and has been studied by different groups. Selected against GBM cells, it was initially studied for its great potential as an inhibitor of cell proliferation. It was then further characterized (i) as a carrier in combination with a second aptamer to increase the therapeutic effect on GBM cells, and (ii) as a carrier of ncRNA (siRNA, antimiR) and PTX-loaded tFNA. In addition, Gint4 binds and functions as a carrier against GSCs, suggesting a potential use in combination with chemotherapeutic agents. Very recently, Cerchia et al. published a short communication in which they demonstrated the ability of GINT4 to cross the BBB in orthotropic GBM mice, confirming its great potential for new therapeutic approaches in GBM. The latter binds EGFRVIII and can be used as a therapeutic nanoparticle carrier and as a diagnostic probe, making it a promising tool in GBM management.
Compared to the above, there are fewer studies on other aptamers in the literature, so it was even more complicated to evaluate the ability of those molecules to help in the management of GBM. One way to assess the value of each individual aptamer may be to look for the year of discovery. In fact, it should be noted that some aptamers, listed in Table 1, were discovered some years ago and since then have not been studied further. It can be assumed that they have been abandoned. However, other aptamers, such as A40s, were first published only last year, so it is likely that further studies are underway, and their value will be assessed in the coming years.
Currently, research in the advancement of GBM therapy is focused on tumor-targeted administration. To date, several aptamers with interesting in vitro and in vivo results have been characterized as carriers of a therapeutic load. Among this group, AS1411 seems to be a promising molecule [57]. AS1411 is a DNA aptamer targeting nucleolin, and is now in clinical trials on advanced solid tumors and acute myeloid leukemia (NCT00512083 phase II, completed; NCT00881244 phase I, completed). Other aptamers, such as GMT8, are good candidates for targeted administration approaches because they have great binding capacity and specificity for GBM, and should be further investigated to determine targets.
In parallel, in the diagnostic technology landscape, aptamer immobilization to develop microfluidic devices and dynamic morphology studies using anti-EGFR aptamers seem to be very promising and practically applicable for GBM diagnosis [21,31]. Concerning the possible aptamer manipulations in this field, the most attractive modifications involve aptamer-isotope conjugations, which have demonstrated a great potential the molecular imaging laboratory, like MRI and SPECT. These diagnostic techniques have been demonstrated to be improved with the use of aptamers.
In this scenario, it would be desirable to reach a common view on the use of aptamers in standard GBM diagnosis and therapy. With our systematic review, we have summarized the available data, highlighting the high value of these molecules. However, despite the interesting characteristics of aptamers, several challenges, either intrinsic to the nature of these molecules (RNA, DNA) or linked to GBM, still need to be overcome. While the limits of aptamers are mostly related to their sensitivity to body-fluid nucleases and CpG toxicity, intrinsic limits in the management of GBM are related to the BBB, which limits the number of drugs that can reach the tumor. Future studies should be aimed at improving the stability of aptamers, discovering aptamers that cross the BBB, and testing aptamers in animal models for preclinical testing.
5. Conclusions
Although the potential role of aptamers in the management of GBM is only just emerging, giving a clear direction to the development of promising aptamers for the clinic is an important objective. In this regard, our review first describes the DNA and RNA aptamers that have been studied as diagnostic and therapeutic tools for GBM, and then highlights the most promising aptamers and the challenges that need to be met for their use in clinical practice.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/8/2173/s1. S1. Key terms used in literature search; S2. PRISMA Checklist.
Author Contributions
Conceptualization, S.N. and V.B.; methodology, V.B.; validation, S.N., A.A., and V.B.; formal analysis, S.N.; investigation, S.N. and A.A.; writing—original draft preparation, S.N. and A.A.; writing—review and editing, V.B., C.C., and G.C.; visualization, S.N.; supervision, M.S., C.C. and G.C. All authors discussed the results and commented on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by “Ricerca Corrente” Grant from Italian Ministry of Health (IRCCS SDN) to (S.N. and C.C.), Associazione Italiana Ricercasul Cancro (AIRC) (IG 2016 N. 18473, to G.C.), H2020-MSCA-RISE-2017, CANCER 777682, SATIN grant 2018-2020 (to G.C.), H2020-MSCA-RISE-2019 cONCReTE 872391 (to G.C.), H2020-MSCA-RISE-2019 PRISAR2 872860 (to G.C.), MSCA IF n. 891551 Gl.EXO (to A.A.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Shergalis A., Bankhead A., Luesakul U., Muangsin N., Neamati N. Current Challenges and Opportunities in Treating Glioblastoma. Pharm. Rev. 2018;70:412–445. doi: 10.1124/pr.117.014944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang R.Y., Neagu M.R., Reardon D.A., Wen P.Y. Pitfalls in the Neuroimaging of Glioblastoma in the Era of Antiangiogenic and Immuno/Targeted Therapy—Detecting Illusive Disease, Defining Response. Front. Neurol. 2015;6 doi: 10.3389/fneur.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander B.M., Cloughesy T.F. Adult Glioblastoma. J. Clin. Oncol. 2017;35:2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y.-H., Wang Z.-F., Pan Z.-Y., Péus D., Delgado-Fernandez J., Pallud J., Li Z.-Q. A Meta-Analysis of Survival Outcomes Following Reoperation in Recurrent Glioblastoma: Time to Consider the Timing of Reoperation. Front. Neurol. 2019;10:286. doi: 10.3389/fneur.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossignol J., Srinageshwar B., Dunbar G.L. Current Therapeutic Strategies for Glioblastoma. Brain Sci. 2019;10:15. doi: 10.3390/brainsci10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016;20:S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope W.B., Brandal G. Conventional and advanced magnetic resonance imaging in patients with high-grade glioma. Q. J. Nucl. Med. Mol. Imaging. 2018;62:239–253. doi: 10.23736/S1824-4785.18.03086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakhan S.E., Harle L. Difficult diagnosis of brainstem glioblastoma multiforme in a woman: A case report and review of the literature. J. Med. Case Rep. 2009;3:87. doi: 10.1186/1752-1947-3-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallick S., Benson R., Hakim A., Rath G.K. Management of glioblastoma after recurrence: A changing paradigm. J. Egypt. Natl. Cancer Inst. 2016;28:199–210. doi: 10.1016/j.jnci.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 11.The German Glioma Network. Seystahl K., Hentschel B., Loew S., Gramatzki D., Felsberg J., Herrlinger U., Westphal M., Schackert G., Thon N., et al. Bevacizumab versus alkylating chemotherapy in recurrent glioblastoma. J. Cancer Res. Clin. Oncol. 2020;146:659–670. doi: 10.1007/s00432-019-03086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hays E., Duan W., Shigdar S. Aptamers and Glioblastoma: Their Potential Use for Imaging and Therapeutic Applications. Int. J. Mol. Sci. 2017;18:2576. doi: 10.3390/ijms18122576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Methods in molecular biology . In: Nucleic Acid Aptamers: Selection, Characterization, and Application. Mayer G., editor. Humana Press; New York, NY, USA: 2016. [Google Scholar]
- 14.Fu Z., Xiang J. Aptamers, the Nucleic Acid Antibodies, in Cancer Therapy. Int. J. Mol. Sci. 2020;21:2793. doi: 10.3390/ijms21082793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan X., Jia F., Wang P., Zhang K. Nucleic acid-based drug delivery strategies. J. Control. Release. 2020;323:240–252. doi: 10.1016/j.jconrel.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monaco I., Camorani S., Colecchia D., Locatelli E., Calandro P., Oudin A., Niclou S., Arra C., Chiariello M., Cerchia L., et al. Aptamer Functionalization of Nanosystems for Glioblastoma Targeting through the Blood–Brain Barrier. J. Med. Chem. 2017;60:4510–4516. doi: 10.1021/acs.jmedchem.7b00527. [DOI] [PubMed] [Google Scholar]
- 17.Gijs M., Aerts A., Impens N., Baatout S., Luxen A. Aptamers as radiopharmaceuticals for nuclear imaging and therapy. Nucl. Med. Biol. 2016;43:253–271. doi: 10.1016/j.nucmedbio.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y., Yang D., Schluesener H.J., Zhang Z. Advances in SELEX and application of aptamers in the central nervous system. Biomol. Eng. 2007;24:583–592. doi: 10.1016/j.bioeng.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Affinito A., Quintavalle C., Esposito C.L., Roscigno G., Vilardo C., Nuzzo S., Ricci-Vitiani L., De Luca G., Pallini R., Kichkailo A.S., et al. The Discovery of RNA Aptamers that Selectively Bind Glioblastoma Stem Cells. Mol. Ther. Nucleic Acids. 2019;18:99–109. doi: 10.1016/j.omtn.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fechter P., Cruz Da Silva E., Mercier M.-C., Noulet F., Etienne-Seloum N., Guenot D., Lehmann M., Vauchelles R., Martin S., Lelong-Rebel I., et al. RNA Aptamers Targeting Integrin α5β1 as Probes for Cyto- and Histofluorescence in Glioblastoma. Mol. Ther. Nucleic Acids. 2019;17:63–77. doi: 10.1016/j.omtn.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q., Wang Y., Wang H., Wu L., Zhang H., Song Y., Zhu Z., Kang D., Yang C. DNA aptamers from whole-cell SELEX as new diagnostic agents against glioblastoma multiforme cells. Analyst. 2018;143:2267–2275. doi: 10.1039/C8AN00271A. [DOI] [PubMed] [Google Scholar]
- 22.Hasan M.R., Hassan N., Khan R., Kim Y.-T., Iqbal S.M. Classification of cancer cells using computational analysis of dynamic morphology. Comput. Methods Programs Biomed. 2018;156:105–112. doi: 10.1016/j.cmpb.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Mahmood M.A.I., Hasan M.R., Khan U.J.M., Allen P.B., Kim Y., Ellington A.D., Iqbal S.M. One-step tumor detection from dynamic morphology tracking on aptamer-grafted surfaces. Technology. 2015;03:194–200. doi: 10.1142/S2339547815500089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alibolandi M., Abnous K., Ramezani M., Hosseinkhani H., Hadizadeh F. Synthesis of AS1411-Aptamer-Conjugated CdTe Quantum Dots with High Fluorescence Strength for Probe Labeling Tumor Cells. J. Fluoresc. 2014;24:1519–1529. doi: 10.1007/s10895-014-1437-5. [DOI] [PubMed] [Google Scholar]
- 25.Wu X., Liang H., Tan Y., Yuan C., Li S., Li X., Li G., Shi Y., Zhang X. Cell-SELEX Aptamer for Highly Specific Radionuclide Molecular Imaging of Glioblastoma In Vivo. PLoS ONE. 2014;9:e90752. doi: 10.1371/journal.pone.0090752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y., Shi Y., Wu X., Liang H., Gao Y., Li S., Zhang X., Wang F., Gao T. DNA aptamers that target human glioblastoma multiforme cells overexpressing epidermal growth factor receptor variant III in vitro. Acta Pharm. Sin. 2013;34:1491–1498. doi: 10.1038/aps.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim B., Yang J., Hwang M., Choi J., Kim H.-O., Jang E., Lee J., Ryu S.-H., Suh J.-S., Huh Y.-M., et al. Aptamer-modified magnetic nanoprobe for molecular MR imaging of VEGFR2 on angiogenic vasculature. Nanoscale Res. Lett. 2013;8:399. doi: 10.1186/1556-276X-8-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y., Wu Q., Hamerlik P., Hitomi M., Sloan A.E., Barnett G.H., Weil R.J., Leahy P., Hjelmeland A.B., Rich J.N. Aptamer Identification of Brain Tumor-Initiating Cells. Cancer Res. 2013;73:4923–4936. doi: 10.1158/0008-5472.CAN-12-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Qiao H., Yan W., Zhang J., Xing C., Wang H., Zhang B., Tang J. Molecular recognition force spectroscopy study of the dynamic interaction between aptamer GBI-10 and extracellular matrix protein tenascin-C on human glioblastoma cell: Molecular Recognition Based on AFM. J. Mol. Recognit. 2013;26:46–50. doi: 10.1002/jmr.2242. [DOI] [PubMed] [Google Scholar]
- 30.Kang D., Wang J., Zhang W., Song Y., Li X., Zou Y., Zhu M., Zhu Z., Chen F., Yang C.J. Selection of DNA Aptamers against Glioblastoma Cells with High Affinity and Specificity. PLoS ONE. 2012;7:e42731. doi: 10.1371/journal.pone.0042731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan Y., Tan J., Asghar W., Kim Y., Liu Y., Iqbal S.M. Velocity Effect on Aptamer-Based Circulating Tumor Cell Isolation in Microfluidic Devices. J. Phys. Chem. B. 2011;115:13891–13896. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 32.Bayrac A.T., Sefah K., Parekh P., Bayrac C., Gulbakan B., Oktem H.A., Tan W. In Vitro Selection of DNA Aptamers to Glioblastoma Multiforme. ACS Chem. Neurosci. 2011;2:175–181. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y., Kim Y., Li N., Cho S.K., Bachoo R., Ellington A.D., Iqbal S.M. Surface-Immobilized Aptamers for Cancer Cell Isolation and Microscopic Cytology. Cancer Res. 2010;70:9371–9380. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 34.Hicke B.J., Stephens A.W., Gould T., Chang Y.-F., Lynott C.K., Heil J., Borkowski S., Hilger C.-S., Cook G., Warren S., et al. Tumor targeting by an aptamer. J. Nucl. Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 35.Daniels D.A., Chen H., Hicke B.J., Swiderek K.M., Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hicke B.J., Marion C., Chang Y.-F., Gould T., Lynott C.K., Parma D., Schmidt P.G., Warren S. Tenascin-C Aptamers Are Generated Using Tumor Cells and Purified Protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 37.Peng L., Liang Y., Zhong X., Liang Z., Tian Y., Li S., Liang J., Wang R., Zhong Y., Shi Y., et al. Aptamer-Conjugated Gold Nanoparticles Targeting Epidermal Growth Factor Receptor Variant III for the Treatment of Glioblastoma. Int. J. Nanomed. 2020;15:1363–1372. doi: 10.2147/IJN.S238206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Affinito A., Quintavalle C., Esposito C.L., Roscigno G., Giordano C., Nuzzo S., Ricci-Vitiani L., Scognamiglio I., Minic Z., Pallini R., et al. Targeting Ephrin Receptor Tyrosine Kinase A2 with a Selective Aptamer for Glioblastoma Stem Cells. Mol. Ther. Nucleic Acids. 2020;20:176–185. doi: 10.1016/j.omtn.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu W., You C., Ma L., Li H., Ju Y., Guo X., Shi S., Zhang T., Zhou R., Lin Y. Enhanced Efficacy of Temozolomide Loaded by a Tetrahedral Framework DNA Nanoparticle in the Therapy for Glioblastoma. ACS Appl. Mater. Interfaces. 2019;11:39525–39533. doi: 10.1021/acsami.9b13829. [DOI] [PubMed] [Google Scholar]
- 40.Wang T., Philippovich S., Mao J., Veedu R.N. Efficient Epidermal Growth Factor Receptor Targeting Oligonucleotide as a Potential Molecule for Targeted Cancer Therapy. Int. J. Mol. Sci. 2019;20:4700. doi: 10.3390/ijms20194700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi S., Fu W., Lin S., Tian T., Li S., Shao X., Zhang Y., Zhang T., Tang Z., Zhou Y., et al. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier. Nanomed. Nanotechnol. Biol. Med. 2019;21:102061. doi: 10.1016/j.nano.2019.102061. [DOI] [PubMed] [Google Scholar]
- 42.Yoon S., Wu X., Armstrong B., Habib N., Rossi J.J. An RNA Aptamer Targeting the Receptor Tyrosine Kinase PDGFRα Induces Anti-tumor Effects through STAT3 and p53 in Glioblastoma. Mol. Ther. Nucleic Acids. 2019;14:131–141. doi: 10.1016/j.omtn.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei J., Marisetty A., Schrand B., Gabrusiewicz K., Hashimoto Y., Ott M., Grami Z., Kong L.-Y., Ling X., Caruso H., et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2018;129:137–149. doi: 10.1172/JCI121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., Peng L., Liang Z., Kou Z., Chen Y., Shi G., Li X., Liang Y., Wang F., Shi Y. Effects of Aptamer to U87-EGFRvIII Cells on the Proliferation, Radiosensitivity, and Radiotherapy of Glioblastoma Cells. Mol. Ther. Nucleic Acids. 2018;10:438–449. doi: 10.1016/j.omtn.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito C.L., Nuzzo S., Catuogno S., Romano S., de Nigris F., de Franciscis V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther. Nucleic Acids. 2018;10:398–411. doi: 10.1016/j.omtn.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayraç A.T., Akça O.E., Eyidoğan F.İ., Öktem H.A. Target-specific delivery of doxorubicin to human glioblastoma cell line via ssDNA aptamer. J. Biosci. 2018;43:97–104. doi: 10.1007/s12038-018-9733-x. [DOI] [PubMed] [Google Scholar]
- 47.Luo Z., Yan Z., Jin K., Pang Q., Jiang T., Lu H., Liu X., Pang Z., Yu L., Jiang X. Precise glioblastoma targeting by AS1411 aptamer-functionalized poly (l-γ-glutamylglutamine)–paclitaxel nanoconjugates. J. Colloid Interface Sci. 2017;490:783–796. doi: 10.1016/j.jcis.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Esposito C.L., Nuzzo S., Kumar S.A., Rienzo A., Lawrence C.L., Pallini R., Shaw L., Alder J.E., Ricci-Vitiani L., Catuogno S., et al. A combined microRNA-based targeted therapeutic approach to eradicate glioblastoma stem-like cells. J. Control. Release. 2016;238:43–57. doi: 10.1016/j.jconrel.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 49.Amero P., Esposito C.L., Rienzo A., Moscato F., Catuogno S., de Franciscis V. Identification of an Interfering Ligand Aptamer for EphB2/3 Receptors. Nucleic Acid Ther. 2016;26:102–110. doi: 10.1089/nat.2015.0580. [DOI] [PubMed] [Google Scholar]
- 50.Camorani S., Crescenzi E., Colecchia D., Carpentieri A., Amoresano A., Fedele M., Chiariello M., Cerchia L. Aptamer targeting EGFRvIII mutant hampers its constitutive autophosphorylation and affects migration, invasion and proliferation of glioblastoma cells. Oncotarget. 2015;6:37570–37587. doi: 10.18632/oncotarget.6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X., Liang H., Tan Y., Wu X., Li S., Shi Y. A U87-EGFRvIII cell-specific aptamer mediates small interfering RNA delivery. Biomed. Rep. 2014;2:495–499. doi: 10.3892/br.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camorani S., Esposito C.L., Rienzo A., Catuogno S., Iaboni M., Condorelli G., de Franciscis V., Cerchia L. Inhibition of Receptor Signaling and of Glioblastoma-derived Tumor Growth by a Novel PDGFRβ Aptamer. Mol. Ther. 2014;22:828–841. doi: 10.1038/mt.2013.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan Y., Tamuly D., Allen P.B., Kim Y., Bachoo R., Ellington A.D., Iqbal S.M. Proliferation and migration of tumor cells in tapered channels. BioMed Microdevices. 2013;15:635–643. doi: 10.1007/s10544-012-9721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao H., Qian J., Yang Z., Pang Z., Xi Z., Cao S., Wang Y., Pan S., Zhang S., Wang W., et al. Whole-cell SELEX aptamer-functionalised poly(ethyleneglycol)-poly(ε-caprolactone) nanoparticles for enhanced targeted glioblastoma therapy. Biomaterials. 2012;33:6264–6272. doi: 10.1016/j.biomaterials.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 55.Verhoeff J.J.C., Stalpers L.J.A., Claes A., Hovinga K.E., Musters G.D., Peter Vandertop W., Richel D.J., Leenders W.P.J., van Furth W.R. Tumour control by whole brain irradiation of anti-VEGF-treated mice bearing intracerebral glioma. Eur. J. Cancer. 2009;45:3074–3080. doi: 10.1016/j.ejca.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y., Kuan C.-T., Mi J., Zhang X., Clary B.M., Bigner D.D., Sullenger B.A. Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol. Chem. 2009;390:137–144. doi: 10.1515/BC.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellassai N., D’Agata R., Jungbluth V., Spoto G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-invasive Cancer Diagnosis. Front. Chem. 2019;7:570. doi: 10.3389/fchem.2019.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mongelard F., Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. 2010;12:107–114. [PubMed] [Google Scholar]
- 59.Flynn J.F., Wong C., Wu J.M. Anti-EGFR Therapy: Mechanism and Advances in Clinical Efficacy in Breast Cancer. J. Oncol. 2009;2009:16. doi: 10.1155/2009/526963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harari P.M. Epidermal growth factor receptor inhibition strategies in oncology. Endocr. Relat. Cancer. 2004;11:689–708. doi: 10.1677/erc.1.00600. [DOI] [PubMed] [Google Scholar]
- 61.Day B.W., Stringer B.W., Boyd A.W. Eph receptors as therapeutic targets in glioblastoma. Br. J. Cancer. 2014;111:1255–1261. doi: 10.1038/bjc.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wykosky J. EphA2 as a Novel Molecular Marker and Target in Glioblastoma Multiforme. Mol. Cancer Res. 2005;3:541–551. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 63.Wu N., Zhao X., Liu M., Liu H., Yao W., Zhang Y., Cao S., Lin X. Role of MicroRNA-26b in Glioma Development and Its Mediated Regulation on EphA2. PLoS ONE. 2011;6:e16264. doi: 10.1371/journal.pone.0016264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X., Wang Y., Wang Y., Zhen H., Yang H., Fei Z., Zhang J., Liu W., Wang Y., Zhang X. Expression of EphA2 in Human Astrocytic Tumors: Correlation with Pathologic Grade, Proliferation and Apoptosis. Tumor Biol. 2007;28:165–172. doi: 10.1159/000103010. [DOI] [PubMed] [Google Scholar]
- 65.Liu F., Park P.J., Lai W., Maher E., Chakravarti A., Durso L., Jiang X., Yu Y., Brosius A., Thomas M., et al. A Genome-Wide Screen Reveals Functional Gene Clusters in the Cancer Genome and Identifies EphA2 as a Mitogen in Glioblastoma. Cancer Res. 2006;66:10815–10823. doi: 10.1158/0008-5472.CAN-06-1408. [DOI] [PubMed] [Google Scholar]
- 66.Wang L.-F., Fokas E., Bieker M., Rose F., Rexin P., Zhu Y., Pagenstecher A., Engenhart-Cabillic R., An H.-X. Increased expression of EphA2 correlates with adverse outcome in primary and recurrent glioblastoma multiforme patients. Oncol. Rep. 2008;19:151–156. doi: 10.3892/or.19.1.151. [DOI] [PubMed] [Google Scholar]
- 67.Kaur H., Bruno J.G., Kumar A., Sharma T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics. 2018;8:4016–4032. doi: 10.7150/thno.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santosh B., Yadava P.K. Nucleic Acid Aptamers: Research Tools in Disease Diagnostics and Therapeutics. BioMed Res. Int. 2014;2014:13. doi: 10.1155/2014/540451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain K.K. A Critical Overview of Targeted Therapies for Glioblastoma. Front. Oncol. 2018;8:419. doi: 10.3389/fonc.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.