Abstract
Protein secretion mechanisms are essential for the virulence of most bacterial pathogens. Typhoid toxin is an essential virulence factor for Salmonella Typhi, the cause of typhoid fever in humans. This toxin is unique in that it is only produced within mammalian cells, and it must be trafficked to the extracellular space prior to intoxicating target cells. An essential and poorly understood aspect of this transport pathway is the secretion of typhoid toxin from the bacterium into the S. Typhi-containing vacuole. We show here that typhoid toxin secretion requires its translocation to the trans side of the peptidoglycan layer at the bacterial poles for subsequent release through the outer membrane. This translocation process depends on a specialized muramidase, whose activity requires the localized editing of peptidoglycan by a specific LD-transpeptidase. These studies describe a protein export mechanism that is likely conserved in other bacterial species.
Keywords: protein secretion, muramidases, peptidoglycan remodeling, bacterial envelope, typhoid fever, Salmonella Typhi, L-D transpeptidases
Bacterial pathogens have evolved sophisticated protein secretion systems that can direct proteins to the extracellular environment or even directly into eukaryotic cells 1-4. Protein transport across the bacterial envelope is particularly challenging in Gram-negative bacteria, as proteins must move through at least three barriers, the inner membrane, the peptidoglycan layer, and the outer membrane. The Gram-negative bacterial pathogen Salmonella enterica serovar Typhi (S. Typhi) is the cause of typhoid fever, a systemic disease of humans that remains a global health concern 5-9. Typhoid toxin is an essential virulence factor of this pathogen and it is believed to be largely responsible for the acute, life-threatening symptoms of typhoid fever 10-12. This toxin, which is encoded within a discrete genomic islet (Supplementary Fig. 1), has a unique A2B5 architecture with two covalently linked enzymatic “A” subunits, PltA and CdtB, associated to a homopentameric “B” subunit made up of PltB 11. A unique feature of typhoid toxin is that it is exclusively expressed by intracellularly localized bacteria 10,13,14. This unique pattern of gene expression is the result of a silencing and counter-silencing transcriptional regulatory mechanism through the opposing actions of the PhoP/PhoQ two-component regulatory system and the histone-like protein H-NS 14. Once synthesized and exported to the periplasmic space by the sec-dependent pathway, the toxin is assembled into a 115-kDa multimeric complex, which is secreted into the lumen of the S. Typhi-containing vacuole. The toxin is then packaged into vesicle carrier intermediates, which transport it to the extracellular space from where it can reach its targets 10,15. The mechanism by which the toxin is secreted from the bacteria into the lumen of its enclosing phagocytic vacuole is not understood, although it is known to require ttsA, which is located within the same locus as the typhoid toxin genes (Supplementary Fig. 1), and encodes a homolog of bacteriophage N-acetyl-β-D-muramidases 16. Here we show that TtsA mediates the translocation of typhoid toxin from the cis to the trans side of the peptidoglycan layer, positioning the toxin within a compartment from where it can be rapidly released by membrane active compounds encountered by S. Typhi during infection. We found that TtsA activity requires the editing of the peptidoglycan at the bacterial poles by a specific LD-transpeptidase, which is essential for toxin secretion. This study describes a protein export mechanism, which has evolved to ensure the release of a toxin in the appropriate environment.
Results
TtsA-dependent translocation and agonist-mediated release of typhoid toxin after S. Typhi growth in vitro.
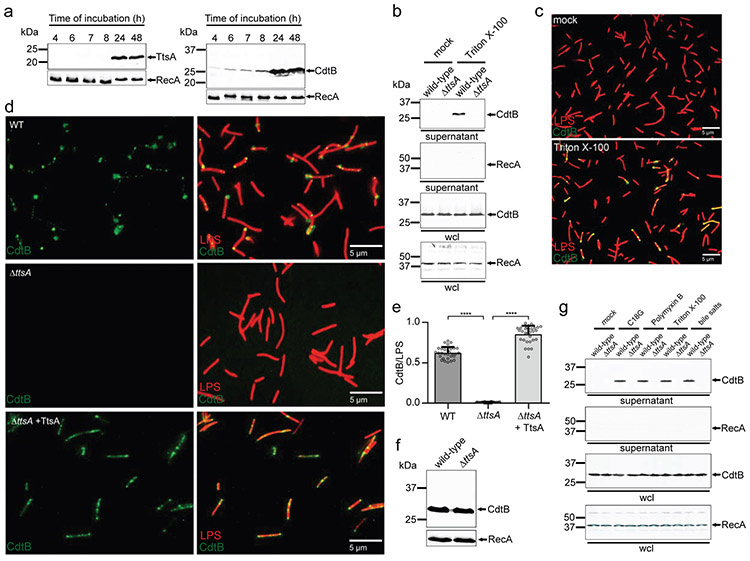
Since typhoid toxin is exclusively produced within infected cells, the visualization of vesicle carrier intermediates has been the only surrogate assay available to study typhoid toxin secretion (Supplementary Fig. 2) 16, an assay poorly suited for mechanistic studies. We have recently unraveled the regulatory network that controls the exclusive intracellular expression of typhoid toxin 14, allowing us to identify in vitro culture conditions that closely mimic those encountered by bacteria within mammalian cells 17 and that are therefore permissive for typhoid toxin and TtsA expression at levels equivalent to those in infected cells (Fig. 1a and Supplementary Fig. 2) 14,16,18. Using these growth conditions (henceforth referred to as “typhoid toxin inducing media” or TTIM) we examined the TtsA-dependent secretion of typhoid toxin into culture supernatants monitored through the detection of its CdtB subunit. Surprisingly, we did not detect typhoid toxin in culture supernatants or on the surface of bacteria grown under these conditions, although we readily detected it in whole bacterial lysates (Fig. 1b and 1c). However, we found that the addition of a very low amount of detergent to bacterial cells, which did not affect viability (Supplementary Fig. 3), resulted in the detection of typhoid toxin both in the supernatants (Fig. 1b) and on the bacterial cells (Fig. 1c). The detection of the toxin in these locations was strictly dependent on TtsA function since it was not detected on S. Typhi ΔttsA bacterial cells (Fig. 1d and 1e, Supplementary Data set 1) or supernatants (Fig. 1b) after its growth in TTIM, even though the toxin levels of expression in the ΔttsA mutant and wild-type strains were indistinguishable (Fig. 1f). Intracellular bacteria, particularly those that like S. Typhi inhabit lysosome-related-organelles 18, are exposed to a myriad of membrane disrupting compounds such as antimicrobial peptides, or membrane active proteins such as those of the saposin or perforin families 19-21. S. Typhi in particular, is known to colonize the gallbladder and to form biofilms in gallbladder stones 22,23, which would readily expose it to bile salts. We hypothesized that, similarly to Triton X-100, these compounds could act as stimulators of toxin release. In fact, we have previously shown that sub-inhibitory concentrations of antimicrobial peptides can stimulate typhoid toxin expression 14. We therefore tested whether the addition of antimicrobial peptides or bile salts could trigger typhoid toxin release to cell-free supernatants after S. Typhi growth in TTIM. Western blot analysis indicated that addition of the antimicrobial peptides C18G, polymyxin B, or bile salts at concentrations that did not inhibit S. Typhi growth (Supplementary Fig. 3) stimulated the rapid release of up to ~20% of the total typhoid toxin pool in wild-type S. Typhi but not in its isogenic ΔttsA mutant grown under the same conditions (Fig. 1g). We also utilized LC-MS/MS to examine the cell-free supernatants of wild type and ΔttsA S. Typhi grown in TTIM after stimulation with the same agonists. We found that addition of antimicrobial peptides C18G, polymyxin B, or bile salts resulted in the selective release of typhoid toxin into the cell-free supernatants of wild type bacteria as no other resident periplasmic proteins were detected by our analysis (Supplementary Data Set 6). Consistent with the Western blot analysis, typhoid toxin was not detected by LC-MS/MS in cell-free supernatants of the S. Typhi ΔttsA mutant strain under any condition. These results indicate that the TtsA-mediated translocation of typhoid toxin through the peptidoglycan (PG) layer must position the toxin at a subcellular site from where it can be subsequently released upon reception of the appropriate stimuli.
Figure 1. TtsA-dependent translocation and agonist-mediated release of typhoid toxin in vitro.
(a) Expression of TtsA and typhoid toxin in typhoid toxin inducing media (TTIM). S. Typhi carrying chromosomally encoded FLAG-tagged CdtB (as a surrogate for typhoid toxin) or TtsA were grown in TTIM and the expression of TtsA and CdtBin equal number of bacterial cells was monitored over time by Western blot analysis. The amount of RecA (loading control) was analyzed on a separate Western blot, (b-e) Outer membrane disruption results in the TtsA-dependent detection of typhoid toxin in culture supernatants and on bacterial cells. Wild-type and ΔttsA S. Typhi (CdtB-FLAG) were grown in TTIM for 24 hs, pelleted and washed twice with PBS, and subsequently treated with Triton X-100 (0.1 %). Bacterial whole cell lysates (wcl) and filtered culture supernatants were then analyzed by Western blotting for the presence of CdtB and RecA (as a cell lysis and loading control) (b). Alternatively, wild-type S. Typhi was grown 24 hs in TTIM, fixed, treated with Triton X-100 (0.1%) or PBS (mock), and then stained with mouse anti-FLAG (to stain CdtB) (green) and rabbit anti S. Typhi LPS (to visualize bacterial cells) (red) (c). (d and e) The indicated strains grown in the same manner were fixed, treated with Triton X-100 (0.1 %) and stained as indicated above. The average ratios of typhoid toxin positive cells (green) vs total cells (LPS, red) ± standard deviation are shown (**** p < 0.0001, two-sided Student’s t Test) (Supplementary data set 1) (e). The levels of CdtB in the strains used in panel (c) were determined by Western blot analysis using RecA as a loading control analyzed on a separate Western blot (f). (g) Antimicrobial peptides trigger the release of typhoid toxin to the culture supernatants. Wild-type and ΔttsA S. Typhi strains (CdtB-FLAG) CdtB were grown 24 hs in TTIM, pelleted and washed twice with PBS and subsequently treated with sub-inhibitory concentrations of the antimicrobial peptide C18G (2.5μg/ml) for 30 min, Polymyxin B (0.1μg/ml), Triton X-100 (0.1%), or bile salts (0.05 %). Filtered supernatants and whole cell lysates were analyzed by Western blotting for the presence of CdtB and RecA (as a cell lysis and loading control). All data in (a-g) were derived from at least three independent experiments.
TtsA mediates the transport of typhoid toxin to the trans side of the peptidoglycan layer.
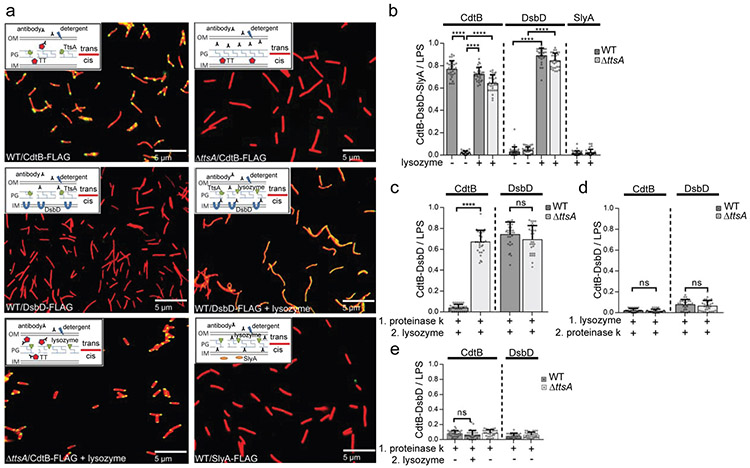
To characterize the subcellular compartment from which typhoid toxin is released to the extracellular space, we investigated its subcellular location in bacterial cells grown in vitro using immunofluorescence microscopy and different membrane permeabilization protocols. We found that, in contrast to wild type, typhoid toxin could not be detected on the S. Typhi ΔttsA mutant cells even after treatment with higher concentration of Triton X-100 (1%) than that required for its release from wild-type bacteria (0.1 %) (Fig. 2a and 2b). Under these conditions, the inner-membrane-anchored periplasmic protein DsbD epitope tagged at its periplasmic C-terminus 24 could not be detected by antibody staining in wild-type S. Typhi (Fig. 2a and 2b). This observation indicates that in the absence of TtsA, typhoid toxin, like DsbD, is located on the cis side of the PG layer inaccessible to antibody molecules. Consistent with this hypothesis, disruption of the PG layer by lysozyme treatment resulted in the detection on the bacterial cell of both, typhoid toxin and DsbD, in the presence or in the absence of TtsA (Fig. 2a and 2b and Supplementary data set 2). These experimental conditions did not result in the detection of a cytoplasmic control protein (Fig. 2a and 2b and Supplementary Fig. 4) indicating that although the treatment disrupted the outer membrane and PG layer, it did not allow access of the antibody to the bacterial cytoplasm, a measure of an intact inner membrane. These results indicate that TtsA allows the translocation of typhoid toxin to the trans side of the PG layer, a location where it becomes accessible to antibody molecules after a minor disruption of the outer membrane. In the absence of TtsA, typhoid toxin remains in the cis side of the PG layer where it is inaccessible to antibody molecules even after disruption of the outer membrane. To further probe this hypothesis, we carried out proteinase K susceptibility studies after different permeabilization protocols. We found that although proteinase K treatment of wild-type S. Typhi cells readily eliminated the detection of typhoid toxin, it did not affect the detection of the control protein DsbD, or typhoid toxin in a ΔttsA mutant S. Typhi (Fig. 2c and Supplementary data set 2). However, disruption of the PG layer by lysozyme pre-treatment completely abolished the detection of DsbD and typhoid toxin in both wild-type or ΔttsA S. Typhi after addition of proteinase K (Fig. 2d and Supplementary data set 2). Taken together, these results demonstrate that: 1) typhoid toxin resides in a compartment different from the compartment harboring the canonical periplasmic protein DsbD; 2) these compartments are separated by the PG layer; and 3) typhoid toxin resides in the trans side of the PG layer. Notably, in wild-type bacteria, proteinase K treatment without addition of lysozyme resulted in the removal of the entire population of typhoid toxin (Fig. 2e and Supplementary data set 2) indicating that the bulk of typhoid toxin molecules are located on the trans side of the PG layer, thus arguing that the TtsA-dependent toxin translocation process must be highly efficient.
Figure 2. TtsA mediates the transport of typhoid toxin to the trans side of the peptidoglycan layer.
(a-e) Wild-type and ΔttsA S. Typhi carrying chromosomally-encoded FLAG-tagged CdtB (as a surrogate for typhoid toxin), DsbD, or SlyA (as indicated) were grown in TTIM for 24 hs, fixed, mock treated or treated with lysozyme for 20 min (to permeabilize the PG layer), and stained as in Fig. 1 (a). To probe the TtsA-dependent translocation of typhoid toxin from the cis to the trans side of the PG layer, bacteria (wild-type and ΔttsA mutant) were first treated with proteinase K for 30 min, followed by lysozyme or mock treatment for 20 min (c and e), or first treated with lysozyme for 20 min, followed by proteinase K treatment for 30 min (d). In all cases, the number of bacterial cells positive for CdtB, DsbD, or SlyA signals (green), was quantified relative to the total number of bacteria analyzed (LPS, red signal). The average ratios of green positive cells to red total cells ± standard deviation from 3 independent experiments are shown. In all cases, a total of 30 images were collected from which 100 randomly selected bacteria (total number) per image were analyzed, resulting in 3,000 bacterial cells analyzed per bacterial strain or condition (**** p < 0.0001, (c) ns p = 0.131, (d) ns p = 0.2078, p = 0.28, (e) ns p = 0.2646, two-sided Student’s t Test) (Supplementary data set 2) (b-e).
Typhoid toxin and TtsA localize to the bacterial poles.
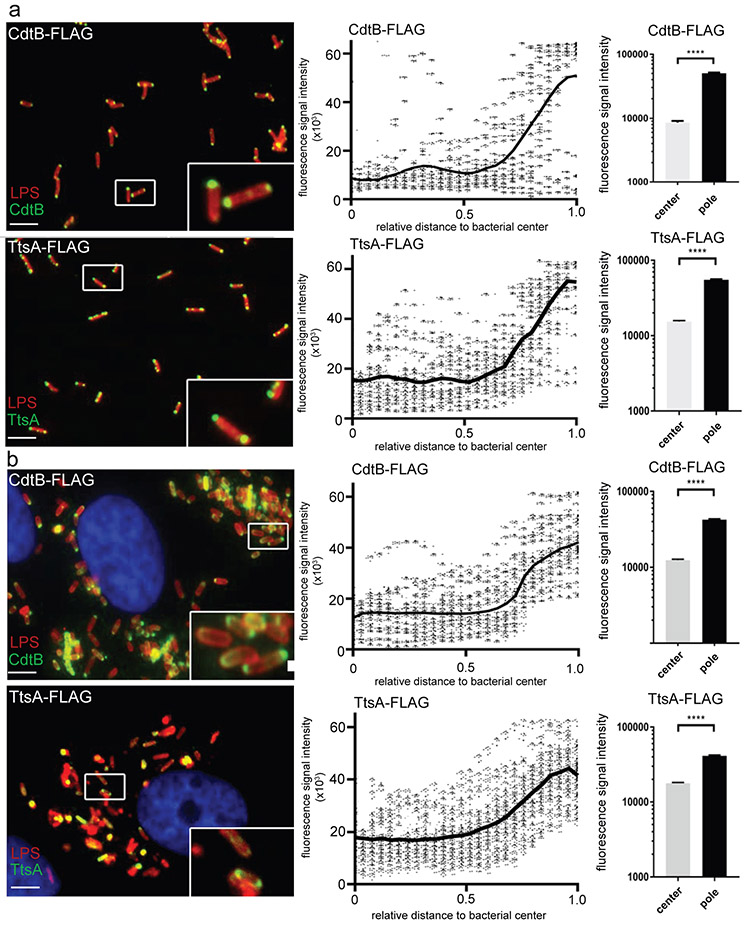
During execution of these experiments, we noticed that typhoid toxin was preferentially detected at the bacterial poles. We therefore quantified the distribution of the typhoid toxin components CdtB and PltB using immunofluorescence microscopy and image analysis. We found a strong polar localization for both proteins (Fig. 3a, Supplementary Fig. 5 and Supplementary data set 3). Furthermore, the polar localization was also observed in intracellular bacteria after infection of cultured epithelial cells (Fig. 3b, Supplementary Fig. 5 and Supplementary data set 3). The localization of typhoid toxin at the bacterial poles was confirmed by live imaging of GFP-tagged CdtB (Supplementary Fig. 6) and by immunoelectron microscopy both in bacteria grown in TTIM, as well as in bacteria within cultured epithelial cells (Supplementary Fig. 7). We also examined the localization of TtsA both after growth in vitro and after bacterial infection in vivo. We found that, like typhoid toxin, TtsA is also localized at the bacterial poles (Fig. 3a and 3b and Supplementary data set 3). These results indicate that typhoid toxin is likely translocated at the bacterial poles and that the activity of TtsA may be selectively exerted at a discrete site of the bacterial envelope.
Figure 3. Typhoid toxin and TtsA localize to the bacterial poles.
(a and b) Typhoid toxin and TtsA localize to the bacterial poles after in vitro growth or cell infection. S. Typhi strains carrying chromosomally-encoded FLAG-tagged CdtB (as a surrogate for typhoid toxin) or TtsA were grown for 24 hours in TTIM, fixed, and stained with a mouse antibody directed to the FLAG-epitope (green) (to visualize CdtB or TtsA, as indicated) and a rabbit antibody directed to S. Typhi LPS (red) (a). Alternatively, the same S. Typhi strains were grown in LB (non inducing media), applied to cultured Henle-407 human epithelial cells, and 24 hours post infection the infected cells were stained with an antibody directed to the FLAG-epitope (green) to visualize CdtB or TtsA (as indicated), a rabbit antibody directed to S. Typhi LPS (red), and DAPI for DNA detection (blue) (b) (scale bar: 5 μm). The scatter blot diagrams of each panel show fluorescence signal intensities for the corresponding FLAG-tagged protein distributed along the axes of individual bacterial cells. Twenty six measuring points were defined from the center (0) to each of the poles (1.0) of the bacterial cells and the distribution of the fluorescence intensity along the axes of bacteria were analyzed with the MicrobeJ plug-in of ImageJ (https://imagej.nih.gov/ij). The black line depicts the average of the intensities measured at each of the 26 measuring points. Data in each panel are from 1,800 individual measurements at each of the measuring points from 900 bacteria analyzed in opposite directions from the center. The bar graphs show the quantification of the signal intensities at the measuring point furthest from the center (1.0), and at the center of each bacterium (0) for each condition. Numbers represent the mean ± SEM from 1,800 measurements (**** p < 0.0001, two-sided Student’s t Test) (Supplementary data set 3).
TtsA exerts its activity at the bacterial poles.
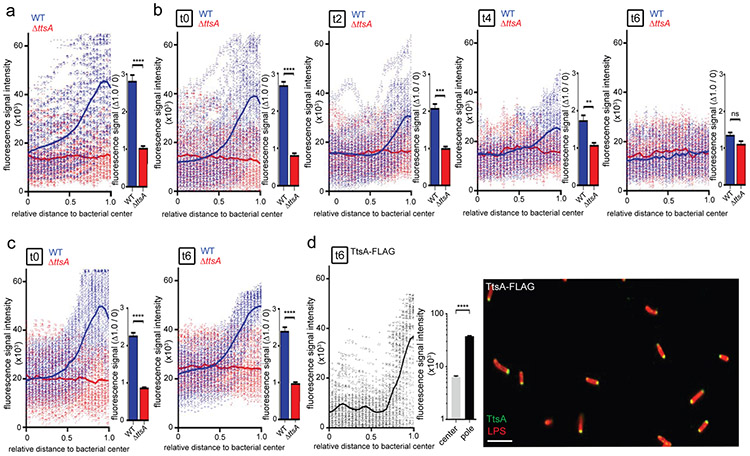
The polar localization of TtsA suggested that its activity might be exerted at the bacterial poles, in which case TtsA-dependent PG remodeling should be detectable at these sites. To test this hypothesis we labeled the S. Typhi PG using an alkyne-modified D-alanine and an azide-containing fluorophore that can be linked by click chemistry 25. The PG structure features linear glycan strands connected by short peptides, which contain D-alanine residues26-28. Consequently, metabolic incorporation of alkyne-modified D-alanine can be used to label remodeling PG 25,29,3. We found that after a short (5 min) alkyne-D-alanine labeling pulse in exponentially growing S. Typhi, labeled PG could be readily visualized at the cell division septum (Supplementary Fig. 8). As expected, a long (60 min) labeling pulse led to PG labeling all around the entire body of the bacteria (Supplementary Fig. 8). These results confirmed that this approach can be used to detect PG remodeling in S. Typhi, and therefore we used it to label PG after growth to stationary phase in TTIM, conditions that lead to TtsA and typhoid toxin expression. We found that, in contrast to what we observed after logarithmic growth in LB broth, when S. Typhi was grown to stationary phase in TTIM for 24 hs followed by an additional 4 hs of growth in the presence of alkyne-D-alanine, PG labeling was preponderantly detected at the poles of the bacteria (Fig. 4a, Supplementary Fig. 9, and Supplementary data set 4). Importantly, in the S. Typhi ΔttsA mutant we observed markedly reduced labeling, which was distributed throughout the body of the bacteria rather than concentrated at the poles (Fig. 4a, Supplementary Fig. 9, and Supplementary data set 4). These results indicate that when S. Typhi is grown under conditions that lead to typhoid toxin expression, TtsA exerts its activity on PG located at the bacterial poles, resulting in PG remodeling likely as a result of the repair process that may follow its PG hydrolytic action.
Figure 4. TtsA exerts its activity at the bacterial poles.
(a) S. Typhi wild-type and the isogenic ΔttsA mutant were grown in TTIM for 24 hs, and metabolically labeled by the addition of alkyne-D-alanine (2mM) for 240 minutes. Fixed bacteria were treated with an azido-AF488 fluorophore (10μM), which was linked by click chemistry to alkyne-D-alanine that had been incorporated into to the PG layer, (b and c) Growth-phase dependency of the PG remodeling at the bacterial poles. S. Typhi wild type and the isogenic ΔttsA mutant were grown 24 hs in TTIM, and subsequently labeled metabolically by the addition of alkyne-D-alanine (2mM) for 240 minutes. Bacteria cultures were then split into TTIM (mock) (c) or into TTIM containing 1 % casamino acids (both conditions in the presence of alkyne-D-alanine) (b), which allows bacteria to re-enter the cell cycle (see Supplementary Fig. 10). At the indicated time points bacteria were harvested and PG was labeled as indicated above. The scatter plots in (a-c) show the results of the line scan analysis of fluorescence intensity along the axes of individual bacterial cells as described in Fig. 3. The blue or red line depicts the average fluorescence from each measured point. The bar graph show the average ratios of the signal intensities at the measuring point furthest from the center (1.0) to the measuring point at the center of each bacterium (0) ((** p < 0.01; *** p < 0.001; **** p < 0.0001, n. s. differences not statistically significant, p = 0.0545, two-sided Student’s t Test) (Supplementary data set 4). (d) TtsA localization after re-growth of S. Typhi (TtsA-FLAG) in casamino acid-supplemented TTIM as described above. Bacteria were fixed, treated with lysozyme (50 μg/ml) and Triton X-100 (0.3 %). TtsA was visualized by fluorescence microscopy as described in Fig. 3 (scale bar: 5 μm). The scatter plot shows the line scan analysis of fluorescence intensity along the axes of individual bacterial cells as described in Fig. 3. The bar graph shows the quantification of the signal intensities at the measuring point furthest from the center (1.0), and at the center of each bacterium (0). Numbers represent the mean ± SEM from 1,800 measurements (**** p < 0.0001, two-sided Student’s t Test) (Supplementary data set 4).
To gain insight into the mechanisms that may restrict the activity of TtsA at the poles we examined the fate of TtsA and peptidoglycan remodeling at the poles of bacteria that had been allowed to re-enter the cell cycle (conditions not permissive for TtsA activity), after growth to stationary phase (conditions permissive for TtsA expression and activity). We grew S. Typhi to stationary phase in TTIM and labeled the bacterial cultures with alkyne-D-alanine as described above. After 4 hs of growth in the presence of alkyne-D-alanine, we split the cultures and added either casamino acids, which stimulate re-entry of the bacteria into the logarithmic phase of growth (see Supplementary Fig. 10), or eluent (mock) and further grew the cultures for various times (always in the presence of alkyne-D-alanine). We found that, in the presence of casamino acids the alkyne-D-alanine-dependent fluorescent signal at the poles was significantly diminished over time (Fig. 4b and Supplementary data set 4), even though under these conditions TtsA remained at the poles (Fig. 4d, Supplementary Fig. 11, and Supplementary data set 4). In contrast, in cultures without casamino acids, the fluorescent signal was readily detected at the poles (Fig. 4c). Taken together these results indicate that the presence of TtsA at the bacterial poles is not sufficient to induce polar PG remodeling and that a specific PG structure may be required for TtsA to exert its activity.
YcbB-dependent peptidoglycan editing is required for typhoid toxin translocation across the bacterial envelope.
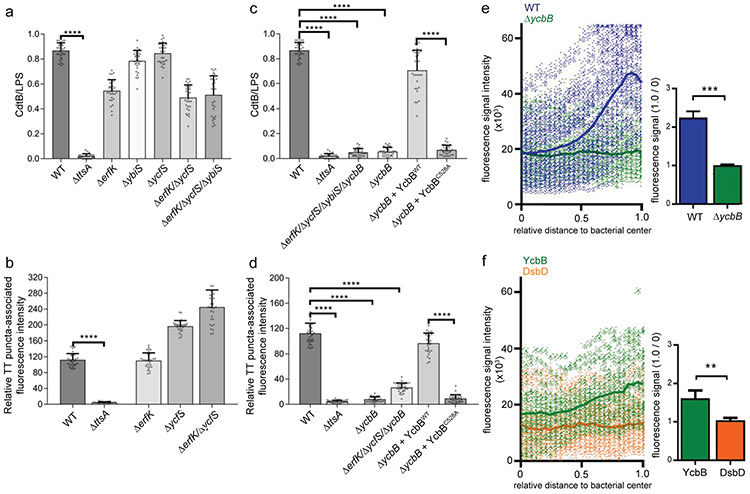
The glycan strands that form the PG layer are connected by short 6-9 amino acid peptides to maintain the rigidity of the cell wall 26-28. The peptides contain L- and D-amino acids and in Gram-negative bacterial species like S. Typhi, the nascent peptide sequence is L-Ala-D-Glu-m-Dap (i. e. meso-diaminopimelic acid)-D-Ala-D-Ala. Some of the peptides are crosslinked by transpeptidase enzymes, including the so-called penicillin-binding proteins (PBPs), which produce the most commonly observed cross-links between the carboxyl group of D-Ala at position 4 of one peptide to the ε-amino group of the m-Dap residue at position 3 of another (the so called 4–3 or DD-cross-links) 31. Other minor cross-links between two m-Dap residues of adjacent stem peptides (the so called 3–3 or LD cross-links) are also observed, which in exponentially growing cells amount to only ~2% of all cross links but that in stationary phase, or in intracellular bacteria can account for as much as ~20% of all cross-links 31-33. These cross-links are catalyzed by LD-transpeptidases, which are structurally and functionally unrelated to PBPs 34,35. Unlike DD-transpeptidases, LD-transpeptidases are non-essential for growth. The activities of PG hydrolases are often dependent on specific modifications of the PG chain imparted by various enzymes including transpeptidases 36. Our observation that the TtsA activity may depend on specific structural features of the PG subunits prompted us to investigate the potential contribution of LD-transpeptidases to typhoid toxin secretion. S. Typhi encodes four LD-transpeptidases: YbiS, ErfK, YcfS, which cross-link the PG to Braun's lipoprotein 34, and YcbB, which catalyzes the formation of a direct link between two m-DAP residues 35. A homolog of ynhG, which encodes another enzyme with this activity, is a pseudogene in S. Typhi. We found that introduction of deletion mutations in ybiS, erfK, ycfS, alone or in combination, had no effect on typhoid toxin secretion (Fig. 5a, 5b, and Supplementary data set 5). However, introduction of a deletion in ycbB completely abolished typhoid toxin secretion, both in vitro (Fig. 5c and Supplementary data set 5) and after bacterial infection of cultured cells (Fig. 5d and Supplementary data set 5), although the introduction of any of the mutations did not affect typhoid toxin expression (Supplementary Fig. 12). The contribution of YcbB to typhoid toxin secretion was dependent on its catalytic activity since expression of the catalytic mutant YcbBC528A was unable to complement a ΔycbB S. Typhi mutant strain (Fig. 5c and 5d and Supplementary data set 5). These results suggest that YcbB imparts localized PG modifications that are essential for TtsA activity. Consistent with this hypothesis, we found that the TtsA-dependent polar remodeling of the peptidoglycan was strictly dependent on YcbB (Fig. 5E, Supplementary Fig. 13, and supplementary data set 5), and that YcbB itself was enriched at the bacterial poles (Fig. 5f and Supplementary data set 5).
Figure 5. YcbB-dependent peptidoglycan editing is required for typhoid toxin translocation across the bacterial envelope.
(a-d) Contribution of LD-transpeptidases to typhoid toxin translocation. S. Typhi (CdtB-FLAG) strains carrying the indicated mutations were grown for 24 hs in TTIM, fixed, and stained as indicated in Fig. 3. The average ratios of the green/red fluorescence intensity ± standard deviation are shown (**** p < 0.0001, two-sided Student’s t Test) (Supplementary data set 5) (a and c). Alternatively, Henle-407 cells were infected with the indicated S. Typhi strains and 24 hs post infection the infected cells were fixed and stained as indicated in Fig. 3. The quantification of the fluorescence intensity of typhoid toxin-associated fluorescent puncta in infected cells is shown (b and d). Values represent relative fluorescence intensity and are the mean ± standard deviation (****: p < 0.0001, two-sided Student’s t test (Supplementary data set 5). (e) YcbB-dependent polar remodeling of the peptidoglycan layer. S. Typhi wild-type or the isogenic ΔycbB mutant were grown in TTIM for 24 hs, and metabolically labeled by the addition of alkyne-D-alanine (2mM) for 240 minutes as indicated in Fig. 4. The scatter plot shows the results of the line scan analysis of fluorescence intensity along the axes of individual bacterial cells for wild-type (blue) and the ΔycbB (green) S. Typhi strains carried out as described in Fig. 2. The blue or green line depicts the average of the intensities measured at each of the measuring points. Bar graph shows the average ratios of the signal intensities measured at the point furthest from the center (1.0) to the signal intensities measured at the center (0) of each bacterium. Numbers represent the mean ± standard deviations from 1,800 measurements (*** p < 0.001, Student’s t Test) (Supplementary data set 5). (f) YcbB is enriched at the bacterial poles. S. Typhi (YccB-FLAG) strains expressing FLAG-tagged YcbB (green) or DsbD (orange) were grown for 24 hours in TTIM, fixed, and stained with a mouse antibody directed to the FLAG-epitope. The scatter plot shows the fluorescence signal intensities and average intensities (lines) for the indicated proteins as described in Fig. 3. The bar graph shows the average ratios of the signal intensities measured at the point furthest from the center (1.0) to the signal intensities measured center (0) of each bacterium. Numbers represent the mean ± standard deviation from 1,800 measurements (** p < 0.01, two-sided Student’s t Test) (Supplementary data set 5). All data in (a-f) were derived from at least three independent experiments.
YcbB-edited peptidoglycan is a substrate for TtsA activity.
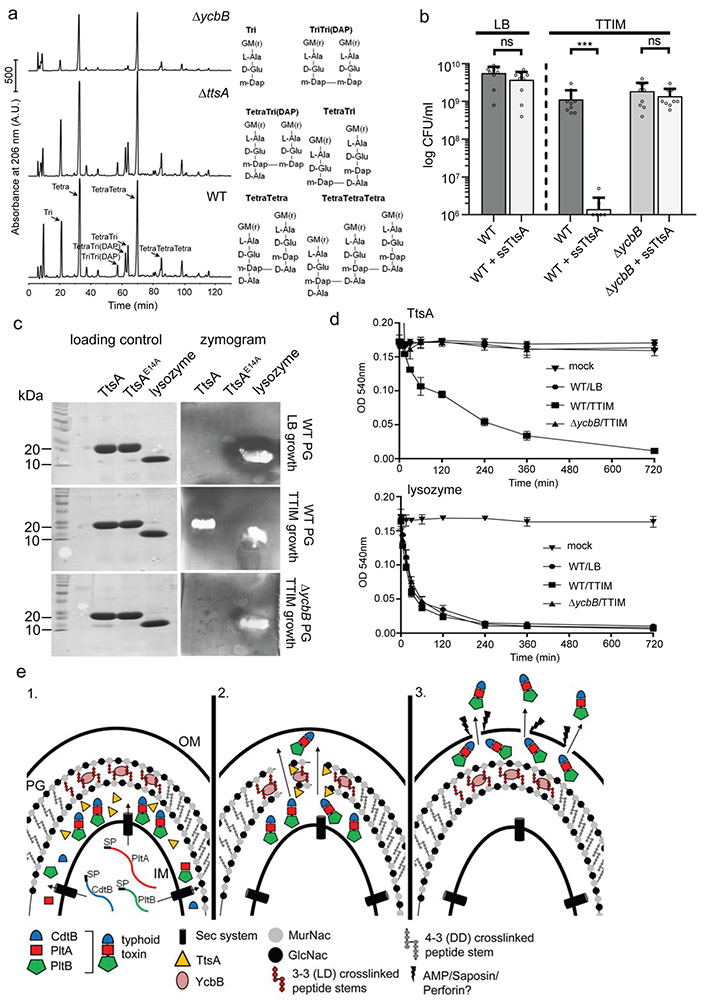
To further explore the mechanisms for TtsA specificity, we compared the PG structures of wild-type and ΔycbB S. Typhi strains grown in TTIM, which is permissive for TtsA activity. We found that while the PG of wild-type S. Typhi grown under these conditions exhibited a significant amount of tripeptides and 3-3 (mDAP-mDAP) cross-links, these cross-links were not detect in the ΔycbB mutant (Fig. 6a and Table S1). Consistent with these observations we found that expression of a form of TtsA containing a Sec-dependent secretion signal (ssTtsA) in S. Typhi (for its efficient delivery into the periplasm) led to a significant reduction in CFU (presumably due to deleterious effects affecting viability or growth) after growth in TTIM, which leads to a PG structure containing significant amount of 3-3 cross-links (Fig. 6b and Supplementary Fig. 14). In contrast, such an effect was not observed when ssTtsA was expressed in a ΔycbB mutant grown under the same conditions, or in wild-type S. Typhi grown in LB broth (Fig. 6b and Supplementary Fig. 14), a condition that leads to a reduced amount of 3-3 cross-links 33. These results indicate that TtsA cannot exert its toxic effect in PG that is not rich in 3-3 crosslinks. We then tested the in vitro enzymatic activity of purified TtsA on PG obtained from wild-type or ΔycbB S. Typhi strains grown under different conditions and using two different assays. We found that while TtsA was able to hydrolyze S. Typhi PG from wild-type bacteria grown in TTIM, no TtsA-mediated hydrolysis was detected in PG from wild-type bacteria grown in LB to logarithmic phase (conditions incompatible with TtsA activity), or in PG from the ΔycbB mutant grown under any condition (Fig. 6c and 6d), while lysozyme was able to hydrolyze all PG preparations (Fig. 6c and 6d). Taken together these results indicate that the TtsA activity is strictly exerted on YcbB-edited PG at the bacterial poles and that it most likely requires 3-3 cross-linked PG to exert its hydrolytic activity.
Figure 6. TtsA activity requires YcbB-mediated peptidoglycan editing.
(a) Muropeptide profiles of different S. Typhi strains showing the YcbB-dependent PG editing. The indicated S. Typhi strains were grown 24 hs in TTIM, PG was isolated and analyzed by HPLC-MS. The ycbB mutant shows a significant decrease in the monomeric disaccharide tripeptide (Tri) and the 3-3 cross-linked muropeptides TriTri(Dap) and TetraTri(Dap). (b) YcbB-dependent growth arrest after sec-dependent TtsA translocation to the periplasmic space. S. Typhi wild type and the ΔycbB isogenic mutant strain, both carrying a plasmid expressing ttsA containing a sec-secretion signal (sec-TtsA) under the control of an arabinose-inducible promoter, were grown in LB or TTIM media containing 0.001 % arabinose, and the number of CFU was determined after 24 hs of growth. Values represent the mean +/− standard deviation (*** p < 0.001, n. s. difference not statistically significant, p = 0.1367 and p = 0.316, two-sided Student’s t test). (c and d) The TtsA muramidase activity is dependent on YcbB. Peptidoglycan was isolated from wild-type S. Typhi grown in either LB, or TTIM, or from the ΔycbB S. Typhi mutant grown in TTIM, as indicated. In-gel digestion zymograms were performed with equal amounts of purified wild-type TtsA, the catalytic mutant TtsAE14A, or lysozyme (shown in a coomassie stained PAGE, left panels) (c). Alternatively, the activity of purified TtsA and lysozyme (as a control) was evaluated using a turbidimetric assay using purified peptidoglycan as indicated above (d). Graphs show the mean turbidity (measured at OD540 nm) ± standard deviation. All data in (a-d) were derived from at least three independent experiments. (e) Model for the typhoid toxin secretion mechanism. The different subunits of typhoid toxin (PltB, PltA, and CdtB) are secreted to the cis side of the periplasm by the sec pathway, where they assemble into the holotoxin complex (1). The muramidase TtsA introduces a fenestration in the YcbB-modified peptidoglycan layer at the bacterial poles allowing the passage of typhoid toxin to the trans side of the periplasmic space, positioning the toxin in close proximity to the outer membrane (2) from where it is released to the exterior upon minor disruptions to the outer membrane caused by various agonists encountered by S. Typhi during infection (3).
Discussion
Gram-negative bacteria have evolved multiple secretion mechanisms of varying complexities to transport proteins across the bacterial envelope 1-4. While some protein secretion machines can move their substrates through all the layers of the bacterial envelope in one step, others operate by engaging the substrates in the bacterial periplasm after their translocation by the Sec machinery. We have described here a mechanism of protein translocation across the bacterial envelope responsible for the export of S. Typhi’s typhoid toxin (Fig. 6e). Through this mechanism the individual subunits of the toxin are first secreted to the periplasm by the Sec machinery, where they assemble into the holotoxin complex. The holotoxin is then translocated across the PG layer to a compartment on the trans side of the PG layer. We propose that translocation of the toxin to this compartment positions it so that it can be released in the presence of specific agonists with the capacity to infringe minor disruption to the bacterial outer membrane. The specific nature of such agonist(s) is not known and may vary depending on the specific environments in which S. Typhi resides during infection. However, we have shown here that sub-inhibitory amounts of antimicrobial peptides or bile salts, which are readily encountered by S. Typhi during infection, are capable of efficiently triggering the release of the toxin. Therefore, it is likely that these compounds may be functionally relevant triggers of typhoid toxin release during S. Typhi infection. Of note, it has been reported that the release of a Zymomonas mobilis extracellular levansucrase, a process that depends on ZlyS 37,38, a homolog of TtsA, can be also stimulated by the presence of membrane active compounds 39.
We demonstrated that typhoid toxin translocation occurs at the bacterial poles and depends on the activity of TtsA, a specialized muramidase, which is also located at the bacterial poles. The activity of TtsA requires the specific edition of peptidoglycan by the LD-transpeptidase YcbB, which catalyzes the so-called 3-3 (or LD) cross-links. Consistent with this observation, no typhoid toxin secretion or translocation to the trans side of the PG layer was observed in the absence of YcbB. This is remarkable in that it has been reported that, at least in E. coli, the vast majority (up to 98%) of the PG crosslinks are between D-Ala and meso-Dap, the so-called 4-3 (or DD) crosslinks. However, these observations have been made with bacteria grown exponentially in rich medium, conditions that differ substantially from those encountered by intracellular bacteria (or bacteria grown in TTIM medium). In fact, there is evidence suggesting that the PG structure of intracellularly localized Salmonella may differ from that of bacteria grown exponentially in LB broth 33,40. Our results also suggest that the PG structure at the poles in bacteria grown under TTIM conditions may differ from the structure elsewhere in the bacterial body. The specialized location of typhoid toxin in the trans side of the PG at the bacterial poles also raises the intriguing possibility that in some specific environments such as the Salmonella containing vacuole the bacterial envelope itself, and in particular the architecture of the periplasmic space, may be distinct. Most knowledge about bacterial physiology or the architecture of its envelope is largely derived from studies of bacteria grown in artificial rich media. However, throughout their entire life cycle bacterial pathogens inhabit environments that differ substantially from those of the media routinely used in the laboratory. Consequently, more studies are required to understand the composition and architecture of the bacterial envelope under environmental conditions that mimic those encountered by bacterial pathogens.
The protein export mechanism described here differs significantly in its evolutionary design from other two-step protein secretion systems that mediate the transport of Sec-translocated substrates 2-4. Indeed, all two-step protein secretion systems known today engage their substrates in what would be considered the cis side of the PG in the periplasmic space and utilize various specific outer-membrane protein channels to move their substrates through the outer membrane. It is intriguing that all the necessary elements to build the different secretion machines are always encoded within a common gene cluster, which sometimes although not always, also encodes the cognate secretion substrates. In contrast, other than ttsA, no other genes required for typhoid toxin secretion are present within the pathogenicity islet that harbors ttsA and the typhoid toxin genes. Furthermore, despite close scrutiny, we have been unable to identify additional S. Typhi genes required for typhoid toxin secretion. Periplasmic proteins can be released to the extracellular space in outer membrane vesicles (OMVs) that are shed from bacterial cells 41,42. However, we have been unable to detect outer membrane vesicles containing typhoid toxin in S. Typhi grown under the conditions used in these studies or in intracellularly localized S. Typhi. Furthermore, in infected cells, the packaging of typhoid toxin into vesicle carrier intermediates requires the interaction of PltB with a glycan receptor on the luminal side of the membrane of the Salmonella- containing vacuole 15. The topology of potential toxin containing OMVs would therefore be incompatible with this packaging mechanism. Consequently, it is unlikely that OMVs play a role in the release of typhoid toxin from S. Typhi cells.
Phylogenetic and genetic analyses 16 indicate that TtsA appears to be a recent exaptation of related enzymes that are utilized by bacteriophages to exit from infected bacterial cells 43. Given the presence of homologous muramidases encoded in close proximity to bacterial toxins or large extracellular enzymes in other bacterial genomes 16,37,38,44, we predict that this protein secretion mechanism is likely to be conserved in other bacteria. We hypothesize that this recently evolved export system provide bacterial pathogens with the unique advantage of releasing pre-synthesized toxins upon the reception of environmental cues (e. g. antimicrobial peptides) whose presence itself may define the very specific environment in which the toxin must exert its function.
Methods
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table S2. All S. Typhi strains are derived from the clinical isolate ISP2825 45. All in frame deletions or insertions into the S. Typhi chromosome were generated by standard recombinant DNA and allelic exchange procedures using the E. coli β-2163 Δnic35 as the conjugative donor strain 46 and the R6K-derived suicide vector pSB890 as previously described 47. For S. Typhi ΔttsA complementation studies, we used plasmid pSB3783, which encodes an arabinose-inducible promoter and is derived from plasmid pBAD24 48. For TtsA expression for protein purification, we used the expression plasmid pET28b (Invitrogen) in E. coli strain BL21. All plasmids used in this study were constructed using the Gibson assembly cloning strategy 49. All generated plasmids and strains used in this study have been verified by nucleotide sequencing.
Bacterial and eukaryotic cell culture.
S. Typhi strains were routinely cultured in L-broth (LB) on a rotation wheel at 37°C. For in vitro typhoid toxin secretion assays, bacteria were sub-cultured in a chemically defined medium, which induces typhoid toxin expression and therefore is referred to as TTIM for typhoid toxin induction medium. This defined medium was adapted from previous studies 14,17 and its composition is as follows: K2SO4 (0.5 mM), KH2PO4 (1 mM), (NH4)2SO4 (7.5 mM), Tris Base (50mM), Bis Tris (50 mM), casamino acids (0.1 %), KCL (5 mM), cysteine (50 μg/ml), tryptophan (50 μg/ml), glycerol (32.5 mM) and magnesium (15 μM). For host cell infection assays, bacteria were sub-cultured in LB containing 0.3 M NaCl, to stimulate expression of the SPI-1 type III secretion system 50. When appropriate, ampicillin (100 μg/ml), Kanamycin (50 μg/ml) and tetracycline (10 μg/ml) were added to the bacterial cultures. The Henle-407 human intestinal epithelial cells (obtained from the Roy Curtiss III collection in 1987) were grown in Dulbecco's modified Eagle medium (DMEM, Gibco) supplemented with 10 % bovine calf serum (BCS, Gemini Bioproducts) at 37°C with 5 % CO2 in a humidified incubator. Cells were routinely tested for the presence of mycoplasma by a standard PCR method. The cells were also frequently checked for their morphological features, growth characteristics, and functionalities, but were not authenticated by short tandem repeat (STR) profiling.
In vitro typhoid toxin expression and secretion assay.
To detect typhoid toxin and TtsA expression in S. Typhi strains carrying chromosomally encoded 3xFLAG-epitope-tagged versions of CdtB (a typhoid toxin subunit) or TtsA, the different strains were grown overnight in LB, washed twice with 1 x PBS (without MgCl supplement, Difco) and sub-cultured in TTIM after a 1:50 dilution. At the indicated time points, an equal number of bacteria (standardized by CFU) were harvested and bacterial pellets were resuspended in Laemmli buffer and boiled for 5 minutes. The expression profile of TtsA and CdtB was determined by Western blot analysis using a mouse monoclonal antibody directed to the FLAG-epitope (Invitrogen). As a loading control, samples were analyzed on a separate Western blot with a polyclonal rabbit antibody directed to RecA.
To detect secreted typhoid toxin in bacterial supernatants, S. Typhi wild type and the isogenic ΔttsA mutant strains, both with chromosomally-encoded 3xFLAG-tagged CdtB, were grown 24 hs in TTIM to induce typhoid toxin and TtsA expression. Bacterial cultures were then mock treated for 30 min, or treated with Triton X-100 (0.1 %) (Sigma-Aldrich) for 15 min, bile salts (0.05 %) (Sigma-Aldrich) for 15 min, polymyxin B (0.1 μg/ml) (Sigma-Aldrich) for 10 min, or C18G antimicrobial peptide (2.5μg/ml) (AnaSpec Inc.) for 30 min. Bacterial pellets and trichloroacetic acid (TCA)-precipitated supernatants (overnight at 4°C) were resuspended in Laemmli buffer and boiled for 5 minutes. The presence of typhoid toxin (CdtB subunit) in the supernatants and in the pellets was determined by Western blot analyses as indicated above. Cytoplasmic RecA served as a negative control for cell lyses in the supernatant analyses and as loading controls for the whole cell lysates. Detection of secreted proteins in the bacterial supernatants by LC-MS/MS analysis after TCA precipitation (overnight at 4°C) was carried out as previously described 51.
Immunofluorescence microscopy of bacteria grown in vitro.
The indicated S. Typhi strains were grown in TTIM for 24 hs and bacteria were then washed with PBS and fixed using 4 % paraformaldehyde (PFA) for 15 min. No fixation or other treatments were performed for the visualization of S. Typhi strains chromosomally encoding CdtB-sfGFP and plasmid-born mCherry. When indicated, fixed bacteria were treated with lysozyme (Sigma Aldrich) (50 μg/ml) for 20 min in Tris-buffer (pH 8) at 37°C. Bacteria were then washed and mounted on poly-(D)-lysine (Sigma-Aldrich) coated glass coverslips. The coverslips with attached bacteria were washed with PBS and, if applicable, incubated with 1 % Triton X-100 in PBS for 30 min, washed again, and incubated overnight at 4°C with primary anti-FLAG M2 mouse monoclonal antibody (Sigma) (1:10,000) and anti-Salmonella O poly A-1 & Vi rabbit antiserum (Becton, Dickinson & Co.) (1:10,000) in PBS, containing 1 % bovine serum albumin (BSA) and 0.01 % Triton X-100. After removal of the primary antibody, the coverslips were washed 6 times with 1 x PBS and incubated with secondary Alexa-Fluor 488-conjugated anti-mouse and Alexa-Fluor 594-conjugated anti-rabbit (Invitrogen) (1:2,000) antibodies in PBS, containing 1% bovine serum albumin (BSA) and 0.01 % Triton X-100 for 60 min at RT protected from light. Mounted samples (ProLong Gold antifade, Molecular Probes) were visualized in an Eclipse TE2000-U (Nikon) microscope equipped with an Andor Zyla 5.5 sCMOS camera driven by Micromanager software (https://www.micro-manager.org).
S. Typhi cultured cell infection assays.
The different S. Typhi strains were grown overnight and then sub-cultured (1:50) in fresh LB containing 0.3 M NaCl, to stimulate expression of the SPI-1 type III secretion system 50. When bacteria reached an OD600 Of 0.9, cultured epithelial Henle-407 cells were infected for 1 hour in Hank’s balanced salt solution (HBSS, Gibco) at a multiplicity of infection (MOI) of 30. Cells were then washed twice with HBSS and further incubated with culture medium supplemented with gentamicin (100 μg/ml) for 1 hr to kill remaining extracellular bacteria, and further incubated in culture medium containing gentamicin (5 μg/ml) to avoid cycles of reinfection.
Immunofluorescence microscopy of infected cultured epithelial cells.
Twenty-four hours after infection with the indicated S. Typhi strains, Henle-407 cells cultured on glass coverslips were rinsed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. No fixation or other treatments were performed prior to the visualization of cultured cells infected with S. Typhi strains expressing CdtB-sfGFP and mCherry. Samples were incubated in 50 mM NH4Cl in PBS for 10 min and subsequently blocked in 1% bovine serum albumin and 0.1 % Triton X-100 in PBS for 30 minutes. Coverslips were washed and incubated overnight at 4°C with primary anti-FLAG M2 mouse monoclonal antibody (Sigma) (1:10,000) and anti-Salmonella O poly A-1 & Vi rabbit antiserum (Becton, Dickinson & Co.) (1:10,000) in PBS, containing 1 % bovine serum albumin (BSA) and 0.01 % Triton X-100. Coverslips were then washed 6 times with 1 x PBS and incubated for 60 min at RT (protected from light) with secondary Alexa-Fluor 488-conjugated anti-mouse and Alexa-Fluor 594-conjugated anti-rabbit (Invitrogen) (1:2,000) antibodies and 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS, containing 1 % bovine serum albumin (BSA) and 0.01 % Triton X-100. Mounted samples (ProLong Gold antifade, Molecular Probes) were visualized in an Eclipse TE2000-U (Nikon) microscope equipped with an Andor Zyla 5.5 sCMOS camera driven by Micromanager software (https://www.micro-manager.org).
Quantification of fluorescence-positive bacteria after growth in vitro and within infected cells.
Bacterial associated fluorescence signals from FLAG-tagged CdtB (to visualize typhoid toxin), DsbD, or SlyA were quantified by fluorescence microscopy using the open source software ImageJ with MicrobeJ plug-in (http://rsbweb.nih.gov/ij/) 52. Images were captured from randomly selected fields in both, the red (LPS signal to identify all bacterial cells) and green (to identify CdtB- ,DsbD-, or SlyA-positive bacteria) channels. The number of bacterial cells positive for green fluorescence signals (CdtB, DsbD or SlyA) was quantified relative to the total number of bacterial cells analyzed (LPS signal, red). In total 30 images were collected from which 100 randomly selected bacteria per image were analyzed resulting in 3,000 bacterial cells analyzed per bacterial strain and/or condition.
Quantification of typhoid toxin-associated transport intermediates.
The typhoid toxin vesicular transport intermediates were quantified by fluorescent microscopy using the open source software ImageJ (http://rsbweb.nih.gov/ij/) as described previously 15. Briefly, images were captured from randomly-selected fields. Images from the LPS stain were used to identify the area corresponding to the bacterial cell body and this area was used to obtain the bacterial-associated typhoid toxin fluorescence signal, which was substracted from the typhoid toxin-associated fluorescence. The remaining fluorescence was considered to be associated with typhoid toxin carrier intermediates. Finally, the intensity of fluorescence associated with toxin carriers in each field was normalized using the fluorescence-associated with typhoid toxin in bacterial cells in the same field.
Immunoelectron microscopy.
A S. Typhi strain carrying chromosomally-encoded encoded FLAG-tagged CdtB was grown in TTMI for 24 hs, fixed with 4 % PFA, washed, and bacterial pellets were quickly frozen in liquid nitrogen. Alternatively, the same bacterial strain was grown in LB as indicated above to infect Henle 407 human epithelial cells, and 24 hs after infection, infected cells were fixed with 4% PFA, washed, and infected cell pellets were quickly frozen in liquid nitrogen. Samples were then processed for immunoelectron microscopy as previously described 53 using a mouse primary antibody (1:500), directed to the FLAG-epitopes and a secondary immunogold (8 nm)-labeled anti-mouse antibody (1:150).
Proteinase K protection assay.
S. Typhi wild-type and ΔttsA mutant strains expressing chromosomally encoded FLAG-tagged CdtB or DsbD were grown in TTIM for 24 hs, washed with 1 x PBS and subjected to two alternative procedures:
Procedure 1:
Bacterial cells were treated with proteinase K (100 μg/ml) at 37°C for 30 min in Tris buffer (pH 8.0), washed thoroughly 4 times with 1 x PBS to remove any remaining proteinase K, fixed with 4 % PFA for 15 min at RT, and treated with lysozyme (50 μg/ml) for 20 min at 37°C in Tris buffer (pH 8.0).
Procedure 2:
Bacterial cells were fixed with 4% PFA for 15 min at RT, washed twice with 1 x PBS, followed by mock treatment or treatment with lysozyme (50 μg/ml) for 20 min at 37°C in Tris buffer (pH 8.0), washed twice with 1 x PBS, followed by mock treatment or treatment with proteinase K (100 μg/ml) for 30 min at 37°C in Tris buffer pH 8.0
All samples were subsequently treated with 0.3 % Triton X-100 for 30 min at RT and mounted on (D)-polylysine coated coverslips. Fluorescence microscopy and quantifications of tagged-protein-associated fluorescence signals (green) normalized to the LPS signals (red) were performed as described above.
Fluorescence labeling of peptidoglycan.
S. Typhi wild type, ΔttsA and ΔycbB mutant strains were grown in TTIM for 24 hs, and subsequently incubated in TTIM containing alkyne-D-alanine (2 mM) (Boao Pharma, Boston) for 4 hs. To analyze peptidoglycan remodeling in bacteria that had been allowed to re-enter the cell cycle after growth to stationary phase, S. Typhi strains were grown to stationary phase in TTIM and labeled with alkyne-D-alanine as described above. Bacterial cultures were then split, casamino acids or eluent were added, and cultures were further grown for the indicated times (always in the presence of alkyne-D-alanine). In all cases, after the various growth conditions bacteria were washed with PBS and fixed for 15 min in 4% paraformaldehyde (PFA) at room temperature. Fixed bacteria were resuspended in Click-iT Cell Reaction Buffer (Invitrogen) and copper-catalyzed click-chemistry was performed in the dark at room temperature for 30 min using the Click-iT Cell Reaction Kit (Invitrogen) with 10 μM azido-Alexa-Fluoro488 fluorophore (Invitrogen). Bacteria were then washed three times and attached to poly-(D) lysine coated coverslips. If applicable bacterial cells were counterstained for LPS (red) with primary anti-Salmonella O poly A-1 & Vi rabbit antiserum (Becton, Dickinson & Co.) (1:10,000) and secondary anti-rabbit Alexa-Fluor594 (Invitrogen) (1:2,000) antibody.
Fluorescence distribution analyses.
The line scan analysis of the distribution of fluorescence signal intensities of labeled peptidoglycan or stained bacteria along the medial axes of the bacterial cell bodies was carried out using the MicrobeJ plug-in 52 of ImageJ (https://imagej.nih.gov/ij/index.html) software. Random fluorescence microscopy images were obtained and bacterial cells were defined as regions of interest (ROI) by using LPS (red) counterstaining. The center within each ROI was identified and 26 measuring points along the medial axes of the bacteria from the center (0) to each of the poles (1.0) were automatically selected on the peptidoglycan or labeled antibody fluorescence channels (green). For the peptidoglycan-associated fluorescence distribution analysis, the fluorescence signal associated with non-TtsA-dependent PG labeling was substracted .
TtsA protein purification.
The coding sequence for ttsA was amplified from S. Typhi strain ISP2825 and cloned by Gibson cloning strategy 49 into the expression vector pET28a+ (Novagen) resulting in N-terminal his-epitope tagged TtsA. Expression and purification of TtsAWT and catalytic mutant TtsAE14A were carried out as previously described 11. Escherichia coli strain BL21 carrying the different plasmids were grown in LB containing kanamycin (50 μg/ml) to an OD600 nm of 0.6–0.7 at 37°C. Expression of TtsA was subsequently induced by the addition of 0.5 mM IPTG, and induced cultures were incubated overnight at 25°C. Bacterial cells were pelleted by centrifugation, resuspended in lysis buffer [Tris-HCl (150 mM, pH 8.0), NaCl (100 mM), imidazol (10 mM), lysozyme (100 μg/ml), DNAse (100 μg/ml), saturated PMSF] and lysed with a French press. Lysates were subsequently pelleted (20,000g 1h, 4°C), and affinity-purified using a Nickel-resin (Qiagen) column. The eluates were diluted in 20 mM Tris-HCl, pH 8.0 buffer and loaded onto a Hi Trap Q ion-exchange column. Fractions from the ion-exchange chromatography were monitored on SDS–PAGE, concentrated, and further purified by using a Superdex 200 column. Final fractions were examined for purity on a 12 % SDS–PAGE.
Peptidoglycan hydrolysis assays.
S. Typhi wild-type and ΔycbB mutant strains were grown in TTIM for 24 hs or, when indicated, in LB to logarithmic growth phase (OD600 0.5), and peptidoglycan was then isolated as previously described 54. Purified peptidoglycan preparations were resuspended in MilliQ water containing 0.01 % NaN3 and stored at 4°C until their use in the different assays. Muramidase activity of purified TtsA was detected by in gel digestion (zymographic assay) or by a turbidimetry assay. For the zymogram assay, purified TtsA (10 μg), its catalytic mutant TtsAE14A (10 μg), or lysozyme (10 μg) were separated on a 12 % low SDS (0.01 %) polyacrylamide gels containing 0.1 % (w/v) purified S. Typhi peptidoglycan. Protein samples were not boiled and the sample loading buffer contained 0.1 % coomassie blue G-250 with 150 mM 6-aminocaproic acid (ACA) but no SDS. Following electrophoresis, the gels were split in half. One half was used as protein loading control and was stained with coomassie (0.5 %). The other half was used for the zymogram assay. In this case the gels were rinsed and soaked in water twice for 5 min at room temperature, incubated twice for 1 hr in washing buffer [100 mM Tris–HCl (pH 8.0), 1% (v/v) Triton X-100] at room temperature with gentle agitation, rinsed and soaked twice with water for 5 min, and subsequently incubated overnight at 37°C in enzyme activity buffer [20 mM Tris-HCL (pH6.8) and 50 mM NaCl] with gentle agitation. Gels were stained for peptidoglycan with 0.1% (w/v) methylene blue in 0.01% (w/v) KOH, and after de-staining with water, the unstained bands due to local degradation of peptidoglycan appeared as a clear zone in a blue background of stained peptidoglycan. The turbidimetric assay was performed with the same highly purified peptidoglycan preparation as used for the zymographic assay. Isolated peptidoglycan was diluted in enzyme activity buffer [20 mM Tris-HCL (pH6.8), 50 mM NaCl] to an OD540 of ~0.175. Purified TtsA, TtsAE14A or lysozyme (5 μg of each) were added to the buffer and optical densities measurements at 540nm were performed at the indicated time points.
Bacterial growth arrest assay.
S. Typhi wild-type and ΔycbB mutant strains, expressing plasmid-born TtsA containing a N-terminal Sec translocation signal sequence from DsbA under the control of an arabinose inducible promoter, were grown overnight in LB medium supplemented with 5 μg/ml tetracycline. Overnight grown bacteria were subcultured (1:50) in TTIM or LB and grown to an OD600 of 0.3, at which point 0.001 % arabinose was added to the bacterial cultures to induce the expression of TtsA, and further incubated for 20 hs. Colony forming units (CFUs) were then determined by plating bacterial dilutions on LB agar plates.
HPLC-MS analyses of isolated S. Typhi peptidoglycan.
PG was isolated and analyzed according to a previously described procedure 55.
Supplementary Material
Acknowledgements
We thank Hesper Rego for useful discussions and members of the Galán laboratory for critical review of the manuscript. T.G. was supported in part by a Postdoctoral Fellowship (GE 2653/1-1) from the Deutsche Forschungsgemeinschaft (German Research Foundation). This work was supported by National Institute of Allergy and Infectious Diseases grant AI079022 (to J.E.G.) and the UK Medical Research Council grant MR/N002679/1 (to W.V.).
Footnotes
Data availability statement
All data generated or analyzed during this study are included in this published article (and its supplementary information).
Competing interests
The authors declare no competing interests
REFERENCES
- 1.Galán J & Waksman G Protein-Injection Machines in Bacteria. Cell 172, 1306–1318(2018. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa T et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 13, 343–359 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Green E & Mecsas J Bacterial Secretion Systems: An Overview. Microbiol Spectr. 4, doi: 10.1128/microbiolspec.VMBF-0012-2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koster M, Bitter W & Tommassen J Protein secretion mechanisms in Gram-negative bacteria. Int. J. Med. Microbiol 290, 325–331 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Parry C, Hien TT, Dougan G, White N & Farrar J Typhoid fever. N Engl J Med. 347, 1770–1782 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Crump J & Mintz E Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 50, 241–246 (2010. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raffatellu M, Wilson R, Winter S & Bäumler A Clinical pathogenesis of typhoid fever. J Infect Dev Ctries 2, 260–266 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Wain J, Hendriksen R, Mikoleit M, Keddy K & Ochiai R Typhoid fever. Lancet 385, 1136–1145 (2015. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougan G & Baker S Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol. 68, 317–336 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Spano S, Ugalde JE & Galan JE Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3, 30–38, doi: 10.1016/j.chom.2007.11.001 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Song J, Gao X & Galan JE Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature 499, 350–354, doi: 10.1038/nature12377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan JE Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc Natl Acad Sci U S A 113, 6338–6344, doi: 10.1073/pnas.l606335113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haghjoo E & Galan JE Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 101, 4614–4619, doi: 10.1073/pnas.0400932101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowler C & Galán J Decoding a Salmonella Typhi Regulatory Network that Controls Typhoid Toxin Expression within Human Cells. Cell Host Microbe 23, 65–76(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang S, Song J & Galán J Receptor-Mediated Sorting of Typhoid Toxin during Its Export from Salmonella Typhi-Infected Cells. Cell Host Microbe. 20, 682–689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodak H & Galán J Salmonella Typhi homolog of bacteriophage muramidases controls Typhoid toxin secretion. EMBO Reports 14, 95–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beuzón C, Banks G, Deiwick J, Hensel M & Holden D pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 33, 806–816 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Spanò S & Galán J A Rab32-dependent pathway contributes to Salmonella typhi host restriction. Science 338, 960–963 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darmoise A, Maschmeyer P & Winau F The immunological functions of saposins. Adv Immunol. 105, 25–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCormack R et al. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. Elife 4, e06508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prost L, Sanowar S & Miller S Salmonella sensing of anti-microbial mechanisms to promote survival within macrophages. Immunol Rev. 219, 55–65(2007). [DOI] [PubMed] [Google Scholar]
- 22.Di Domenico E, Cavallo I, Pontone M, Toma L & Ensoli F Biofilm Producing Salmonella Typhi: Chronic Colonization and Development of Gallbladder Cancer. Int J Mol Sci. 18, E1887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunn J et al. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 22, 648–655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porat A, Cho S & Beckwith J The unusual transmembrane electron transporter DsbD and its homologues: a bacterial family of disulfide reductases. Res Microbiol. 155, 617–622 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Siegrist M et al. (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 8, 500–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner R, Vollmer W & Foster S Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 91, 862–874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egan A, Biboy J, van't Veer I, Breukink E & Vollmer W Activities and regulation of peptidoglycan synthases. Philos Trans R Soc Lond B Biol Sci. 370, 20150031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vollmer W, Blanot D & de Pedro MA Peptidoglycan structure and architecture. FEMS Microbiol Rev. 32, 149–167 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Cameron T, Anderson-Furgeson J, Zupan J, Zik JJ1, & Zambryski P Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. MBio 5, e01219–01214 (2014. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuru E, Tekkam S, Hall E, Brun Y & Van Nieuwenhze M Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc. 10, 33–52 (2015. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glauner B, Holtje JV & Schwarz U The composition of the murein of Escherichia coli. J. Biol. Chem 263, 10088–10095 (1988). [PubMed] [Google Scholar]
- 32.Holtje JV Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62, 181–203 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintela J, de Pedro M, Zöllner P, Allmaier G & Garcia-del Portillo F Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol Microbiol. 23, 693–704 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Magnet S et al. Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol 189, 3927–3931 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnet S, Dubost L, Marie A, Arthur M & Gutmann L Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol 190, 4782–4785 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcorlo M, Martinez-Caballero S, Molina R & Hermoso JA Carbohydrate recognition and lysis by bacterial peptidoglycan hydrolases. Current Opinons in Structural Biology 44, 87–100 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Kondo Y et al. Cloning and characterization of a pair of genes that stimulate the production and secretion of Zymomonas mobilis extracellular levansucrase and invertase. Biosci Biotechnol Biochem. 58, 526–530. (1994). [DOI] [PubMed] [Google Scholar]
- 38.Oda Y, Yanase H, Kato N & Tonomura K Liberation of Sucrose-Hydrolyzing Enzymes from Cells by the zliS Gene Product That Mediates Protein Secretion in Zymomonas mobilis. J. Fermentation and Bioengineering 77,419–422(1994). [Google Scholar]
- 39.Takeda K et al. The effect of amphiphilic compounds on the secretion of levansucrase by Zymomonas mobilis. Process Biochemistry 40, 3723–3731 (2005). [Google Scholar]
- 40.Rico-Pérez G et al. A novel peptidoglycan D,L-endopeptidase induced by Salmonella inside eukaryotic cells contributes to virulence. Mol Microbiol. 99, 546–556 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Takeda K, Guerrero-Mandujano A, Hernández-Cortez C, Ibarra J & Castro-Escarpulli G The outer membrane vesicles: Secretion system type zero. Traffic 18, 425–432 (2017. ). [DOI] [PubMed] [Google Scholar]
- 42.Schwechheimer C & Kuehn M Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 13, :605–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang I, Smith D & Young R Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 54, 799–825 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Hamilton J et al. A holin and an endopeptidase are essential for chitinolytic protein secretion in Serratia marcescens. J. Cell Biol 207, 615–626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galan JE & Curtiss R 3rd. Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun 59,2901–2908(1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demarre G et al. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol 156, 245–255 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Kaniga K, Bossio JC & Galan JE The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol 13, 555–568 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Guzman LM, Belin D, Carson MJ & Beckwith J Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6, 343–345, doi: 10.1038/nmeth.1318 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Galán JE & Curtiss R Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun 58, 1879–1885 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Gao B, Novik V & Galan JE Quantitative Proteomics of Intracellular Campylobacter jejuni Reveals Metabolic Reprogramming. PLoS Pathog 8, e1002562, doi: 10.1371/journal.ppat.1002562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ducret A, Quardokus EM & Brun YV MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature microbiology 1, 16077, doi: 10.1038/nmicrobiol.2016.77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann J, Pypaert M, Taguchi T, Malsam J & Warren G Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, 848–851 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Heidrich C et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Molecular microbiology 41, 167–178 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Glauner B Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem 172, 451–464 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.