Abstract
Halitosis is a common ailment concerning 15% to 60% of the human population. Halitosis can be divided into extra-oral halitosis (EOH) and intra-oral halitosis (IOH). The IOH is formed by volatile compounds, which are produced mainly by anaerobic bacteria. To these odorous substances belong volatile sulfur compounds (VSCs), aromatic compounds, amines, short-chain fatty or organic acids, alcohols, aliphatic compounds, aldehydes, and ketones. The most important VSCs are hydrogen sulfide, dimethyl sulfide, dimethyl disulfide, and methyl mercaptan. VSCs can be toxic for human cells even at low concentrations. The oral bacteria most related to halitosis are Actinomyces spp., Bacteroides spp., Dialister spp., Eubacterium spp., Fusobacterium spp., Leptotrichia spp., Peptostreptococcus spp., Porphyromonas spp., Prevotella spp., Selenomonas spp., Solobacterium spp., Tannerella forsythia, and Veillonella spp. Most bacteria that cause halitosis are responsible for periodontitis, but they can also affect the development of oral and digestive tract cancers. Malodorous agents responsible for carcinogenesis are hydrogen sulfide and acetaldehyde.
Keywords: halitosis, malodor, volatile sulfur compounds, hydrogen sulfide, microbiota, Fusobacterium, Porphyromonas, Prevotella, periodontitis, carcinogenesis
1. Introduction
Halitosis is a common problem that manifests as an unpleasant and disgusting odor emanating from the mouth [1]. Malodor is mainly caused by putrefactive actions of microorganisms on endogenous or exogenous proteins and peptides. Oral malodor is an embarrassing condition that affects a large percentage of the human population. This condition often results in nervousness, humiliation, and social difficulties, such as the inability to approach people and speak to them [2,3,4,5,6]. Halitosis experiences from about 15% to 60% of the human population worldwide [7,8,9,10,11,12]. Halitosis can be divided into extra-oral halitosis (EOH) and intra-oral halitosis (IOH) [2,3,5].
The factors that increase the likelihood of halitosis include periodontal diseases, dry mouth, smoking, alcohol consumption, dietary habits, diabetes, and obesity. Halitosis can also be affected by the general hygiene of the body (i.e., dehydration, starvation, and high physical exertion), advanced age, bleeding gums, decreased brushing frequency, but also by stress [3,13,14,15,16]. Produced during stress, catecholamines and cortisol increased hydrogen sulfide production by sub-gingival anaerobic bacteria [17]. The medications which can cause extra-oral halitosis were categorized into 10 groups: acid reducers, aminothiols, anticholinergics, antidepressants, antifungals, antihistamines and steroids, antispasmodics, chemotherapeutic agents, dietary supplements, and organosulfur substances [18].
More and more patients are struggling with bad breath and report this problem to their primary care practitioner for diagnosis and management [19,20]. However, many physicians, dentists, and biologists have insufficient knowledge regarding the cause and biochemistry of this disease.
In this review, we focused on intra-oral halitosis, regardless of classification.
2. Classifications of Halitosis
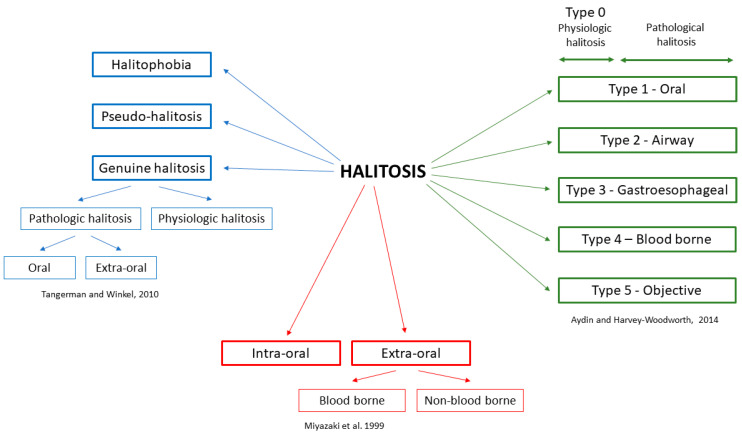
In the literature, mainly three classifications of halitosis are used, described by Miyazaki et al., 1999 [21], Tangerman and Winkel in 2010 [22], and Aydin and Harvey-Woodworth in 2014 [23] (Figure 1).
Figure 1.
Miyazaki et al. divided halitosis as intra-oral (IOH) and extra-oral (EOH) [21]. Extra-oral halitosis can be of bloodborne or non-bloodborne origin and covers about 5–10% of all halitosis [22]. Bloodborne-related causes include diabetes metabolic disorders, kidney and liver diseases, and certain drugs and food. Non-bloodborne-related causes include respiratory and gastrointestinal diseases. Meanwhile, pathological conditions in the oral cavity are responsible for 80–90% of IOH [2,3,25]. Both aerobic and anaerobic bacteria can be responsible for IOH. These microorganisms tend to produce foul-smelling, sulfur-containing gases called volatile sulfur compounds (VSCs) [23,26].
In the classification of Tangerman and Winkel [22], halitosis is classified as genuine and delusional. Delusional halitosis (monosymptomatic hypochondriasis; imaginary halitosis) is a condition in which patients believe that their breath is smelly and offensive. The social pressure of having fresh smelling breath increases the number of people that are preoccupied with this condition. However, the perception of oral malodor does not always reflect actual clinical oral malodor [27]. Self-perceived halitosis was found to be more prevalent amongst males, particularly smokers, compared to females. However, there are no statistical differences when comparing with different age groups [28]. Genuine halitosis is further subdivided into physiological and pathological halitosis. Physiological halitosis (foul morning breath, morning halitosis) is caused by saliva retention, as well as the putrefaction of entrapped food particles. Meanwhile, intra- and extra-oral causes are responsible for pathological halitosis [3,4,19].
Aydin and Harvey-Woodworth divided pathologic halitosis into five types: Type 1 (oral), Type 2 (airway), Type 3 (gastroesophageal), Type 4 (blood-borne) and Type 5 (subjective). Moreover, it is Type 0 halitosis (physiologic odor), which can be a connection of the physiologic contributions of oral, airway, gastroesophageal, blood-borne, and subjective halitosis. Any combination of the above types can be present in every healthy person [23].
3. Volatile Compounds
Halitosis is formed by volatile compounds, which are produced mainly by bacteria in the oral cavity. In the oral cavity, nearly 700 different compounds have been detected [29]. To these volatile substances belong sulfur compounds, aromatic compounds, amines, short-chain fatty or organic acids, alcohols, aliphatic compounds, aldehydes, and ketones (Table 1) [25,30,31,32,33]. It is considered that hydrogen sulfide, methyl mercaptan, and dimethyl sulfide are the main volatile compounds in IOH [34,35,36,37]. In many studies, the measurement of malodor substances concerns only volatile sulfur compounds (VSCs). The most commonly used are VSC monitors, such as the Halimeter (Interscan, Chatsworth, USA) [11,36,38,39,40,41]. This method has a significant disadvantage because the measure of dimethyl sulfide is not exact [42]. Moreover, the presence of alcohols, phenyl compounds, and polyamines can interfere with readings [16,43]. For this reason, in the assessment of IOH, other substances are often not taken into account. However, they can have an equally important role. It is confirmed by studies using gas chromatography-mass spectrometry [29,32,44]. In the paper of Monedeiro et al., in the persons with IOH, 85 volatiles, were detected, and the most predominant classes of malodor compounds were alcohols and ketones. In this group, in comparison to healthy persons, an increased number of volatile sulfur compounds and esters was observed. Simultaneously, authors found ten VSCs substances: methyl thioacetate, dimethyl disulfide, dimethyl trisulfide, dimethyl tetrasulfide, dimethyl pentasulfide, dimethyl sulfone, allyl thiocyanate, allyl isothiocyanate, S-methyl pentanethioate, and thiolan-2-one [44]. In other studies, in halitosis patients, the 30 most abundant volatile compounds in the oral cavity belonged to alkanes or alkane derivatives, therein methyl benzene, tetramethyl butane, and ethanol [45]. Dadamio et al. reported VSC and amines (such as putrescine, cadaverine, and trimethylamine) as the most abundant organic compounds in IOH patients [46].
Table 1.
| Group of Compounds | Compound Name | Chemical Formula | Chemical Structure | Odor Threshold (ppm) [49,50,51,52] |
Toxicity in Rats LD50 (mg/kg) |
|---|---|---|---|---|---|
| Volatile sulfur compounds (VSC) | Hydrogen sulfide | H2S |
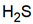
|
0.00004 | 15 [53] |
| Methyl mercaptan | CH4S |
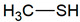
|
5.1 × 10−13 | 61 (unspecified mammal species) [54] | |
| Dimethyl sulfide | C2H6S |
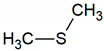
|
0.00012 | 3300 [54,55] | |
| Dimethyl disulfide | C2H6S2 |
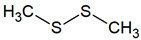
|
0.00029 | 190 [54] | |
| Dimethyl trisulfide | C2H6S3 |
|
no data | no data | |
| Allyl methyl sulfide | C4H8S |
|
0.00014 | no data | |
| Aromatic compounds | Pyridine | C5H5N |
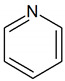
|
0.01 | 360–891 [54,55] |
| Picoline | C6H7N |
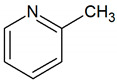
|
0.0026 | 200–790 [54,55] | |
| Indole | C8H7N |
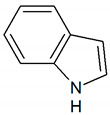
|
0.0003 | 1000 [54,55] | |
| Skatole | C9H9N |
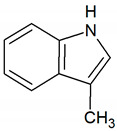
|
0.0000056 | 3450 [54,55] | |
| Amines | Ammonia | H3N |
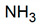
|
0.043 | 350 [56] |
| Urea | CH4N2O |
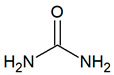
|
no data | 567–8471 [54,55] | |
| Methylamine | CH5N |
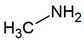
|
0.00075 | 100 [54,55] | |
| Dimethylamine | C2H7N |
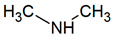
|
0.00076 | 698 [54,55,57] | |
| Trimethylamine | C3H9N |
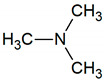
|
0.00002 | 500–535 [54,55] | |
| Putrescine | C4H12N |
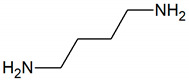
|
no data | 463–2000 [54,55,58] | |
| Cadaverine | C5H14N |
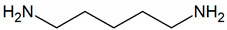
|
no data | 2000 [58] | |
| Short/medium fatty or organic acids | Acetic acid | C2H4O2 |
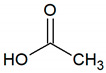
|
0.0004 | 3310 [54,55] |
| Propionic acid | C3H6O2 |
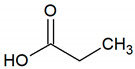
|
0.00099 | 2600–3500 [54,55] | |
| Butyric acid | C4H8O2 |
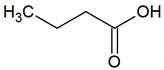
|
0.001 | 1500–2000 [54,55] | |
| Valeric acid | C5H10O |
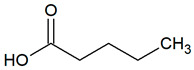
|
0.000037 | 2000–4600 [59] | |
| Isovaleric acid | C5H10O |
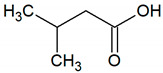
|
0.000078 | 2 [54] | |
| Alcohols | Methanol | CH4O |
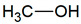
|
3.05 | 2131–7529 [54,55] |
| Ethanol | C2H6O |
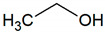
|
0.09 | 1440–7060 [54,55] | |
| Propanol | C3H8O |
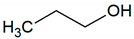
|
0.031 | 590–2200 [54,55] | |
| Aliphatic compounds | Cyclopropane | C3H6 |
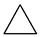
|
no data | no data |
| Cyclobutane | C4H8 |
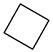
|
no data | no data | |
| Pentane | C5H12 |
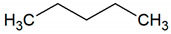
|
1.29 | 400–>2000 [54,55] | |
| Aldehydes and ketones | Acetaldehyde | C2H4O |
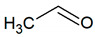
|
0.0015 | 640–1930 [54,55] |
| Acetone | C3H6O |
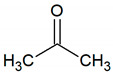
|
0.4 | 5500–5800 [54,55,57] | |
| Acetophenone | C8H8O |
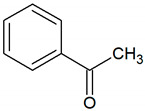
|
0.00024 | 815–2650 [54,55] | |
| Benzophenone | C13H10O |
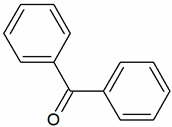
|
no data | >10,000 [54,55] |
In Table 1, among others, values of odor thresholds are presented. Amid VSCs, which are the most often described compounds in IOH, the lowest value of odor threshold has methyl mercaptan, followed by hydrogen sulfide and dimethyl sulfide. This means that these substances are mainly responsible for the unpleasant smell in the mouth. Besides, methyl mercaptan is felt in much lower concentrations than the other compounds.
In the oral cavity, the most relevant anatomical part related to IOH is the tongue. The tongue-associated microbiota produce malodorous compounds and fatty acids. The VSCs are the most essential substances responsible for malodor. They are products of metabolism of sulfur amino acids: methionine, cysteine, and homocysteine in the Gram-negative anaerobic bacteria [25,30,47,60]. Hydrogen sulfide and mercaptans are the principal end products [38]. In healthy volunteers, the concentration of H2S in saliva was within a range of 1.641–7.124 μM [61]. In other studies, the mean amount of H2S in the saliva of healthy persons was 0.5 ng/10 mL, whereas in patients with IOH it was 6.7 ng/10 mL [62]. Gram-positive bacteria can support Gram-negative anaerobic bacteria in the production of VSC. They cut off sugar chains from glycoproteins and provide proteins that are necessary for proteolytic processes [60]. Streptococcus salivarius has an impact on the deglycosylation of salivary glycoproteins, mainly mucins, which can next be degraded to VSC by Porphyromonas gingivalis [63]. In turn, Solobacterium moorei is associated with the production of VSC through β-galactosidase activity and the degradation of glycoproteins [60,64].
The essential VSCs are hydrogen sulfide, dimethyl sulfide, dimethyl disulfide, and methyl mercaptan [25,30] (Table 1). These are produced mostly by anaerobic bacteria. The increased production of malodorous gases occurs mainly in tongue coating, and diseases such as gingivitis and periodontitis and, to a less extent, in pericoronitis, oral ulcers, periodontal abscesses, and herpetic gingivitis [65]. Other volatile organoleptic compounds, such as indole, skatole, amines, and ammonia, are produced by the putrefaction of non-sulfur containing amino acids (i.e., tryptophan, lysine and ornithine). Studies have shown that volatile sulfur compounds are the major contributors to bad breath. Hydrogen sulfide, methyl mercaptan and, to a lesser extent, dimethyl sulfide, represent 90% of the volatile sulfur compounds in halitosis [2,27].
Volatile sulfur compounds can be toxic for human cells even at low concentrations. They contain thiols (-SH groups) that interact with other proteins and support the negative interaction of bacterial antigens and enzymes. The result of this effect is chronic inflammation, periodontal gingivitis, and periodontitis [66]. In human gingival fibroblasts, H2S activates the mitochondrial pathway of apoptosis [67]. The H2S is a known genotoxic agent, which has an impact on genomic instability and cumulative mutations [68]. In studies on rats, it was demonstrated that hydrogen sulfide leads to ultrastructural changes in epithelial cells and periodontal destruction [69]. Increased amounts of H2S by the activation of proliferation, migration, and invasion can also lead to carcinogenesis [70,71]. Fusobacterium nucleatum and Porphyromonas gingivalis belong to the most essential carcinogenic oral bacteria producing VSCs [70,72]. Cancerogenic is also acetaldehyde produced from ethanol by mucosal epithelial cells or oral microflora, e.g., Candida albicans, Candida non-albicans, Neisseria sp., and Streptococcus sp. Acetaldehyde binds to DNA and leads to the formation of DNA adducts, point mutations, and DNA cross-linking [73,74].
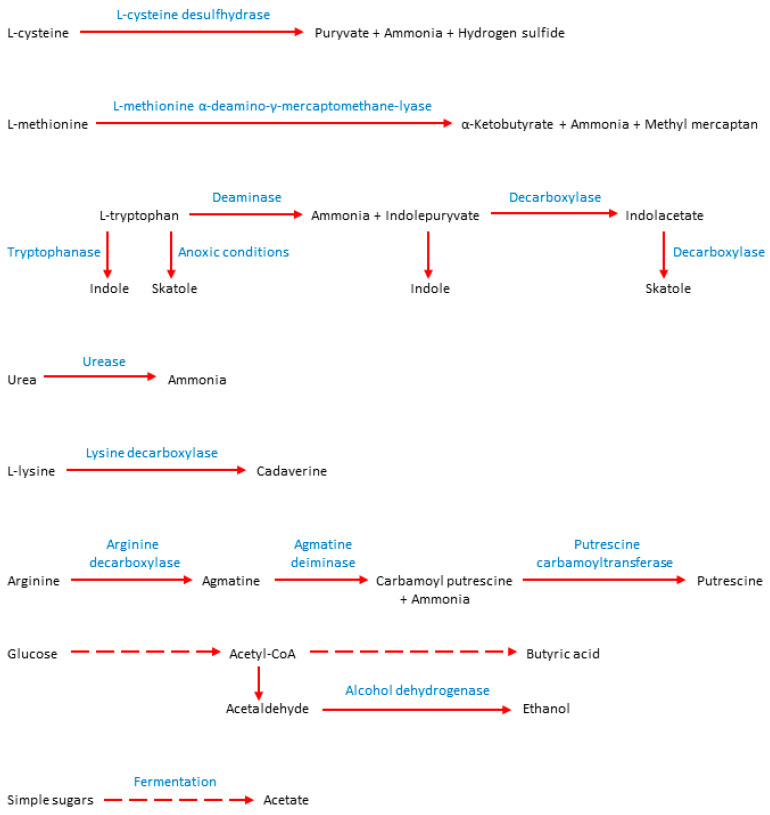
Other important substances causing IOH are diamines, such as putrescine and cadaverine. Both compounds are produced from amino acids, putrescine from arginine, and cadaverine from L-lysine [75,76] (Figure 2). Both diamines are associated with the putrefaction of food by bacteria occurring in the dental plaque and severe periodontitis [77].
Figure 2.
Simplified ways of bacterial production of selected odorous compounds [30,75,76,96,97,98,99,100,101,102].
Gram-negative bacteria, mostly Enterobacteriaceae, which can colonize the oral cavity and dentures, produce urease that hydrolyzes urea into carbon dioxide and ammonia [78]. Escherichia coli can form ammonia from cysteine using cysteine desulfhydrase [79] or reduce nitrates to ammonia [73]. Major contributors to trimethylamine production are gut bacteria, which can be inhabitants of the oral cavity, such genera as Anaerococcus, Clostridium, Collinsella, Desulfovibrio, Lactobacillus, E. coli, Citrobacter, Edwardsiella, Providencia, and Proteus [74,80,81,82,83,84].
Indole and skatole are produced in high amounts by intra-oral, Gram-positive Streptococcus milleri, and anaerobic Gram-negative bacteria such as Porphyromonas intermedia, Fusobacterium nucleatum, and Porphyromonas gingivalis. Small amounts of both aromatic compounds produced Aggregatibacter aphrophilus, Staphylococcus epidermidis, and Streptococcus sanguis [85].
4. Microbiota Responsible for Intra-Oral Halitosis
The human oral cavity microbiota is an ecosystem consisting of various symbiotic microbes. There is a relationship between the global composition of indigenous bacterial populations and human health [86,87]. The oral microbiota is truly diverse and consists of 50–100 billion bacteria. There are about 700 taxa, of which one-third cannot be grown in vitro [88,89]. A vast range of microorganisms inhabit the human oral cavity, including bacteria, fungi, viruses, and protozoa [90,91]. The basic oral microbiota consists of phyla, such as Firmicutes, Proteobacteria, Fusobacteria, Bacteroidetes, and Actinobacteria. The most dominant genera are Streptococcus, Veillonella, Gemella, Granulicatella, Neisseria, Haemophilus, Selenomonas, Fusobacterium, Leptotrichia, Prevotella, Porphyromonas, and Lachnoanaerobaculum. Lots of current findings reported that oral bacteria can be biomarkers that differentiate healthy and pathological conditions within the oral cavity. The oral microbiota research is used as a diagnostic and prognostic tool in the aspect of human health. In the human body, the oral cavity is the second site, after the colon, containing the largest diversity of microbial populations [92]. Simultaneously, changes in the gut microbiota are reflected in the oral microbiota, and the microbial communities of the oral cavity and gastrointestinal tract are predictive of each other [93,94,95].
The oral bacteria that are most likely to produce hydrogen sulfide from L-cysteine or serum are Bacteroides spp., Eubacterium spp., Fusobacterium spp., Peptostreptococcus spp., Porphyromonas spp., Selenomonas spp., Tannerella forsythia, and Veillonella spp. Another essential component of VSC is methyl mercaptan produced from L-methionine or serum. It is a metabolic product mainly derived from Bacteroides spp., Eubacterium spp., Fusobacterium spp., Porphyromonas spp., and Treponema denticola [30,96] (Table 2).
Table 2.
| Chemical Compound | Bacteria |
|---|---|
| Hydrogen sulfide from L-cysteine | Bacteroides intermedius, Bacteroides spp., Capnocytophaga ochracea, Centipeda periodontii, Eikenella corrodens, Eubacterium brachy, E. limosum, Eubacterium spp., Fusobacterium alocis, F. nucleatum, F. periodonticum, F. sulei, Peptostreptococcus anaerobius, P. micros, P. prevotii, Porphyromonas endodontalis, Propionibacterium propionicum, Selenomonas artemidis, S. dianae, S. flueggei, S. infelix, S. noxia, S. sputigena, Tannerella forsythia, Veillonella dispar, V. parvula |
| Methyl mercaptan from L-methionine | Bacteroides spp., Eubacterium spp., F. nucleatum, F. periodonticum, Porphyromonas endodontalis |
| Hydrogen sulfide from serum | Bacteroides gracilis, B. intermedius, B. loescheii, B. oralis, Eubacterium lentum, Eubacterium spp., F. nucleatum, Mitsuokella dentalis, Peptostreptococcus magnus, P. micros, P. prevotii, P. propionicum, Porphyromonas gingivalis, T. forsythia, Treponema denticola, V. parvula |
| Methyl mercaptan from serum | P. endodontalis, P. gingivalis, T. denticola |
Ye at al.’s studies showed a correlation between high H2S and CH4S levels and the growth of microorganisms such as Prevotella spp., Peptostreptococcus spp., Eubacterium nodatum, and Alloprevotella spp. Comparing the study and control group, the authors noted significantly higher concentrations of all compounds (total VSC, H2S, CH4S, and C2H6S) in the malodor group [103]. The most active producers of hydrogen sulfide are Gram-negative anaerobes Prophyromonas gingivalis, Treponema denticola, and Tannerella forsythia (red complex). Furthermore, the red complex microorganisms are associated with periodontal disease. Hydrogen sulfide and methyl mercaptan are produced in large quantities in periodontal inflammations [104,105,106]. During periodontitis, Porphyromonas spp., Prevotella spp., and Treponema denticola may play the most crucial role in providing amino acids to other anaerobic bacteria. Through this process, anaerobes acquire the opportunity to produce H2S and CH4S [60] (Figure 2). In the studies of Takeshita et al., the producers of hydrogen sulfide in saliva were bacteria from the genera Neisseria, Fusobacterium, Porphyromonas, and SR1. In contrast, producers of the methyl mercaptan are representatives of the genera Prevotella, Veillonella, Atopobium, Megasphaera, and Selenomonas [107]. Significant contributors to methyl mercaptan production are also gut bacteria, which can be inhabitants of the oral cavity, such as E. coli, Citrobacter spp., and Proteus spp. [48,84].
Many studies showed that bacterial diversity in the group of patients with IOH is much higher than in the control group. Furthermore, many publications draw attention to the correlation between halitosis and individual microorganisms. The relationship between tongue bacterial composition structure and VSC gases is also mentioned by many authors [3,108]. Many oral bacteria that cause IOH contain similar enzymes. These enzymes are proteins encoded by related genes (megL, lcs, mgl) in the genomes of various bacterial species. The main enzymes are methionine γ-lyase, L-cysteine desulfhydrase, and L-methionine α-deamino-γ-mercaptomethane-lyase [109].
Veloso et al. mentioned that in 85% of the patients IOH is caused by Gram-negative bacteria [6]. According to Wei et al., the oral microbiota responsible for IOH includes a wide range of microbial communities, including 13 phyla, 23 classes, 37 orders, 134 genera, 266 species, and 349 operational taxonomic units. The largest percentage amongst the oral cavity microorganisms are genera, like Prevotella, Alloprevotella, Leptotrichia, Peptostreptococcus, and Stomatobaculum. These bacteria present a higher percentage of occurrence in the sample of patients with IOH than in the control samples from healthy patients [103]. In turn, the presence of bacteria, such as Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria, was demonstrated in both the samples from examined and control groups. Firmicutes was the most abundant phylum in saliva samples from both groups [110,111].
The composition of the tongue microbiota has an essential influence on IOH. The most common molecular technique for testing and evaluating an oral cavity microbiome is the sequencing [5,107,112,113]. Seerangaiyan et al. published a review in 2017, in which they showed the composition of the bacteria of Aggregatibacter, Campylobacter, Capnocytophaga, Clostridiales, Leptotrichia, Parvimonas, Peptostreptococcus, Peptococcus, Prevotella, Selenomonas, Dialister, Tannerella, and Treponema in the group of patients with IOH. Using the amplification of 16S rRNA, the researchers also demonstrated a high prevalence of Solobacterium moorei strains in the IOH group. By testing the control group, significant differences were found in both groups of healthy and sick people. Furthermore, using polymerase chain reactions (PCRs), Seerangaiyan et al. showed the positive correlation of Leptotrichia spp. and Prevotella spp. to oral malodor severity, contrary to Haemophilus spp., Gemella spp. and Rothia spp. [5].
Patients with IOH have a specific biofilm on the dorsal part of the tongue. Bernardi et al. stated that this biofilm consists of a significant proportion of Fusobacterium nucleatum and Streptococcus spp. The occurrence of these two types of bacteria in patients with IOH was completely related. According to the authors, these microorganisms contribute significantly to IOH and can be treated as treatment targets [114]. In other research, Bernardi and partners showed that Actinomyces graevenitzii and Veillonella rogosae were closely related to the occurrence of IOH in a group of volunteers. Also, Streptococcus mitis/oralis, S. pseudopneumoniae, and S. infantis, as well as Prevotella spp. were detected often in malodor patients. Moreover, following the earlier findings, the researchers’ results revealed the presence of Actinomyces odontolyticus, Solobacterium moorei, Prevotella melaninogenica, Fusobacterium periodonticum, and Tannerella forsythia in IOH patients. Furthermore, microorganisms such as Streptococcus parasanguinis, S. salivarius, Veillonella spp., and Rothia mucilaginosa dominated in the oral microbiota of healthy people [112].
Yitzhaki et al. noticed the connection between IOH and wearing dentures. The unpleasant odor was organoleptically assessed and the oral microbiome was analyzed using Next Generation Sequencing 16S rDNA technology. Researchers have identified bacterial taxa, including nine phyla, 29 genera, and 117 species. The samples taken from patients with IOH showed the dominance of the phyla Firmicutes and Fusobacteria and the genera Leptotrichia, Atopobium, Megasphaera, Oribacterium, and Campylobacter. The analyses revealed a significant diversity of the oral microbiota among samples from IOH patients wearing alveolar dentures and significant differences in comparison to the control group [113].
The use of tobacco also has a huge impact on the oral microbiota diversity. After examining a group of smokers and non-smokers, researchers reported that in both groups, most of the oral microbiota were Gram-negative bacterial strains. Simultaneously, Klebsiella pneumoniae dominated in smokers’ saliva and Pseudomonas aeruginosa in non-smokers’ saliva samples. An essential finding of the research was also that the Candida species accounted for the largest percentage of microbes amongst smokers with halitosis [97]. Al-Zyound et al. performed tests showing an increased level of three bacterial genera in smokers: Streptococcus, Prevotella, and Veillonella. Researchers provided evidence that tobacco smoking has a direct effect on the oral microbiota. They also suggested that after smoking cessation, it is possible to return to the standard composition of the oral cavity microbiota [115].
Wu et al. noticed significant changes in the oral microbiota that occurred amongst obese people suffering from malodor. The Prevotella, Granulicatella, Peptostreptococcus, Solobacterium, Catonella, and Mogibacterium were more abundant genera in the obesity group than in healthy persons [116].
Halitosis has often been reported amongst the symptoms related to Helicobacter pylori infection and gastroesophageal reflux disease. Anbari et al. made the observations that the incidence of malodor amongst Helicobacter pylori-positive patients was 74% [2]. However, Tagerman et al. disagreed about a possible relationship between Helicobacter pylori infection and objective halitosis [22].
It is difficult to identify bacteria that promote malodor in children. The most common groups of oral bacteria in children with IOH are Veillonella spp., Prevotella spp., Fusobacterium spp. However, there is no difference in the abundance of these microorganisms in children with IOH and those without [110].
In Table 3, results of studies concerning microbiota associated with IOH are presented. Summarizing the table, the oral bacteria that are most related to IOH are Actinomyces spp., Bacteroides spp., Dialister spp., Eubacterium spp., Fusobacterium spp., Leptotrichia spp., Peptostreptococcus spp., Porphyromonas spp., Prevotella spp., Selenomonas spp., Solobacterium spp., Tannerella forsythia, and Veillonella spp.
Table 3.
Results of studies concerning bacteria associated with intra-oral halitosis (IOH).
| Bacteria Related to Intra-Oral Halitosis | Studied Population | Study Method | Reference |
|---|---|---|---|
| Bacteroides gracilis, B. intermedius, B. loescheii, B. oralis, Capnocytophaga ochracea, Centipeda periodontii, Eikenella corrodens, Eubacterium brachy, E. lentum, E. limosum, Fusobacterium alocis, F. nucleatum, F. periodonticum, F. sulei, Mitsuokella dentalis, Peptostreptococcus anaerobius, P. magnus, P. micros, P. prevotii, Porphyromonas endodontalis, P. gingivalis, Propionibacterium propionicum, Selenomonas artemidis, S. dianae, S. flueggei, S. infelix, S. noxia, S. sputigena, Tannerella forsythia, Treponema denticola, Veillonella dispar, V. parvula | 9 persons | Bacterial culture | [96] |
| Fusobacterium sp., P. gingivalis, Prevotella intermedia | 16 IOH adults or children | Bacterial culture | [117] |
| Campylobacter rectus, F. nucleatum, P. micros, P. gingivalis, P. intermedia, T. forsythia | 40 IOH patients | Anaerobic culture | [118] |
| Fusobacterium sp., P. gingivalis, P. intermedia, T. forsythia | 20 IOH adults | Anaerobic culture | [119] |
| P. gingivalis, P. intermedia, P. melaninogenica, P. nigrescens, Streptococcus constellatus, T. forsythia, T. denticola, V. parvula | 10 adult persons | checkerboard DNA-DNA hybridization technique | [120] |
| Actinomyces israelii, A. neuii, A. odontolyticus, Aggregatibacter actinomycetemcomitans (serotype a), Atopobium parvulum, Prevotella bivia, P. disiens, P. nigrescens, Pseudomonas aeruginosa, Staphylococcus epidermis, S. constellatus, Streptococcus mitis, T. forsythia, V. parvula | 21 IOH adults | Checkerboard DNA-DNA hybridization |
[121] |
| F. nucleatum, P. gingivalis, T. forsythia | 30 adults | PCR | [122] |
| P. gingivalis, P. intermedia, T. forsythia | 101 IOH adults | PCR | [123] |
| P. gingivalis, P. intermedia, P. nigrescens, T. forsythia, T. denticola | 29 IOH patients and 10 healthy adults | Real-time PCR | [124] |
| F. nucleatum, Solobacterium moorei, T. forsythia | 78 adult males | Quantitative real-time PCR | [35] |
| A. actinomycetemcomitans, F. nucleatum, P. gingivalis, P. intermedia, T. denticola | 31 IOH patients and 31 healthy adults | 16S rDNA-directed PCR | [125] |
| Atopobium sp., Dialister sp., Eubacterium sp., Fusobacterium nucleatum, Leptotrichia sp., Megasphaera sp., Neisseria sp., Parvimonas sp., Peptococcus sp., Peptostreptococcus sp., P. gingivalis, P. endodontalis, Prevotella sp., Selenomonas sp., Solobacterium sp., SR1 sp., Veillonella sp. | 30 IOH patients and 13 healthy persons | PCR and sequencing | [107] |
| A. odontolyticus, F. periodonticum, Leptotrichia sp., Okadaella gastrococcus, Prevotella melaninogenica, S. moorei, T. forsythia | 6 IOH patients and 6 healthy adults | PCR and sequencing | [112] |
| phyla Firmicutes and Fusobacteria, genera Atopobium, Campylobacter, Leptotrichia, Megasphaera, Oribacterium | 26 full dentures patients | PCR and sequencing | [113] |
| A. odontolyticus, Atopobium parvulum, Lysobacter-type species, Porphyromonas sp., P. melaninogenica, P. pallens, P. veroralis, Streptococcus salivarius, S. mitis, S. oralis, V. parvula | 20 IOH patients and 12 healthy adults | PCR and DNA sequencing | [126] |
| Eubacterium sp., Dialister sp., Granulicatella elegans, Porphyromonas sp., P. intermedia, Staphylococcus warneri, S. moorei | 8 IOH patients and 5 healthy adults | PCR and DNA sequencing | [127] |
| Aggregatibacter sp., A. segnis, Campylobacter sp., Capnocytophaga sp., Clostridiales, Dialister sp., Leptotrichia sp., Parvimonas sp., Peptostreptococcus sp., Peptococcus sp., Prevotella sp., Selenomonas sp., SR1, Tannerella sp., TM7-3, Treponema sp. | 16 IOH patients and 10 healthy adults | 16S rRNA sequencing | [5] |
| Prevotella sp., Leptotrichia sp., Actinomyces sp., Porphyromonas sp., Selenomonas sp., Selenomonas noxia, Capnocytophaga ochracea | 5 IOH children and 5 healthy | 16S rRNA sequencing | [128] |
| A. parvulum, Eubacterium sulci, F. periodonticum, Dialister sp., S. moorei, Streptococcus sp., TM7-8, | 6 IOH patients and 5 healthy adults | 16S rRNA sequencing | [129] |
| A. odontolyticus, Hemophilus parainfluenzae, Gemella sp., Leptotrichia wadei, Prevotella tannerae, Streptococcus sp., | 29 adults | 16S rDNA amplicon sequencing | [130] |
| Actinomyces sp., Prevotella sp., Veillonella sp. | 10 adults | 16S rRNA gene sequencing | [131] |
| Aggregatibacter sp., Anaerovorax sp., Bacteroidales, Butyrivibrio sp., Dialister sp., Eikenella sp., Mogibacterium sp., Moraxella sp., Peptococcus sp., Peptostreptococcaceae, RF39, Tannerella sp., Treponema sp., Veillonellaceae | 40 IOH adults | 16S rRNA sequencing | [132] |
| Streptococcus halitosis sp. nov. strain VT-4 | - | 16S rRNA sequencing | [133] |
5. Conclusions
The IOH is formed by volatile compounds, among which volatile sulfur compounds (VSCs), such as hydrogen sulfide, dimethyl sulfide, dimethyl disulfide, and methyl mercaptan, are predominant. VSCs are produced mainly by anaerobic bacteria belonging to genera Actinomyces, Bacteroides, Dialister, Eubacterium, Fusobacterium, Leptotrichia, Peptostreptococcus, Porphyromonas, Prevotella, Selenomonas, Solobacterium, Tannerella, and Veillonella. A combination of different microbial techniques is recommended to analyze the etiological microflora associated with IOH. Increased knowledge of the microbiota of the oral cavity and especially tongue biofilm is essential for further research to develop new halitosis therapy strategies.
Author Contributions
Conceptualization, K.H. and T.M.K.; data curation, K.H., M.M.J., Z.Ł.B. and T.M.K.; writing—original draft preparation, K.H., M.M.J. and T.M.K.; writing—review and editing, K.H., Z.Ł.B. and T.M.K.; visualization, T.M.K.; supervision, T.M.K. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the Chancellor of the Faculty of Pharmacy, PUMS, Lucjusz Zaprutko.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pieniążek A., Pietrzak M. Halitoza-etiologia, metody diagnostyki i leczenie. Halitosis-etiology, methods of diagnosis and treatment. J. Health Study Med. 2017;2:101–122. [Google Scholar]
- 2.Anbari F., Ashouri Moghaddam A., Sabeti E., Khodabakhshi A. Halitosis: Helicobacter pylori or oral factors. Helicobacter. 2019;24:e12556. doi: 10.1111/hel.12556. [DOI] [PubMed] [Google Scholar]
- 3.De Geest S., Laleman I., Teughels W., Dekeyser C., Quirynen M. Periodontal diseases as a source of halitosis: A review of the evidence and treatment approaches for dentists and dental hygienists. Periodontology 2000. 2016;71:213–227. doi: 10.1111/prd.12111. [DOI] [PubMed] [Google Scholar]
- 4.Kapoor U., Sharma G., Juneja M., Nagpal A. Halitosis: Current concepts on etiology, diagnosis and management. Eur. J. Dent. 2016;10:292–300. doi: 10.4103/1305-7456.178294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seerangaiyan K., van Winkelhoff A.J., Harmsen H.J.M., Rossen J.W.A., Winkel E.G. The tongue microbiome in healthy subjects and patients with intra-oral halitosis. J. Breath. Res. 2017;11:036010. doi: 10.1088/1752-7163/aa7c24. [DOI] [PubMed] [Google Scholar]
- 6.Veloso D.J., Abrão F., Martins C.H.G., Bronzato J.D., Gomes B.P.F.A., Higino J.S., Sampaio F.C. Potential antibacterial and anti-halitosis activity of medicinal plants against oral bacteria. Arch. Oral Biol. 2020;110:104585. doi: 10.1016/j.archoralbio.2019.104585. [DOI] [PubMed] [Google Scholar]
- 7.Seemann R., Filippi A., Michaelis S., Lauterbach S., John H.-D., Huismann J. Duration of effect of the mouthwash CB12 for the treatment of intra-oral halitosis: A double-blind, randomised, controlled trial. J. Breath Res. 2016;10:036002. doi: 10.1088/1752-7155/10/3/036002. [DOI] [PubMed] [Google Scholar]
- 8.Kumbargere Nagraj S., Eachempati P., Uma E., Singh V.P., Ismail N.M., Varghese E. Interventions for managing halitosis. Cochrane Database Syst. Rev. 2019;12:CD012213. doi: 10.1002/14651858.CD012213.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadanovsky P., Carvalho L.B.M., Ponce de Leon A. Oral malodour and its association with age and sex in a general population in Brazil. Oral Dis. 2007;13:105–109. doi: 10.1111/j.1601-0825.2006.01257.x. [DOI] [PubMed] [Google Scholar]
- 10.Nalçaci R., Dülgergil T., Oba A.A., Gelgör I.E. Prevalence of breath malodour in 7–11-year-old children living in Middle Anatolia, Turkey. Community Dent. Health. 2008;25:173–177. [PubMed] [Google Scholar]
- 11.Bornstein M.M., Kislig K., Hoti B.B., Seemann R., Lussi A. Prevalence of halitosis in the population of the city of Bern, Switzerland: A study comparing self-reported and clinical data. Eur. J. Oral Sci. 2009;117:261–267. doi: 10.1111/j.1600-0722.2009.00630.x. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama S., Ohnuki M., Shinada K., Ueno M., Wright F.A.C., Kawaguchi Y. Oral malodor and related factors in Japanese senior high school students. J. Sch. Health. 2010;80:346–352. doi: 10.1111/j.1746-1561.2010.00512.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen X., Zhang Y., Lu H.-X., Feng X.-P. Factors Associated with Halitosis in White-Collar Employees in Shanghai, China. PLoS ONE. 2016;11:e0155592. doi: 10.1371/journal.pone.0155592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyanova L. Stress hormone epinephrine (adrenaline) and norepinephrine (noradrenaline) effects on the anaerobic bacteria. Anaerobe. 2017;44:13–19. doi: 10.1016/j.anaerobe.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 15.De Lima P.O., Nani B.D., Rolim G.S., Groppo F.C., Franz-Montan M., Alves De Moraes A.B., Cogo-Müller K., Marcondes F.K. Effects of academic stress on the levels of oral volatile sulfur compounds, halitosis-related bacteria and stress biomarkers of healthy female undergraduate students. J. Breath Res. 2020;14:036005. doi: 10.1088/1752-7163/ab944d. [DOI] [PubMed] [Google Scholar]
- 16.Wu J., Cannon R.D., Ji P., Farella M., Mei L. Halitosis: Prevalence, risk factors, sources, measurement and treatment—A review of the literature. Aust. Dent. J. 2020;65:4–11. doi: 10.1111/adj.12725. [DOI] [PubMed] [Google Scholar]
- 17.Calil C.M., Oliveira G.M., Cogo K., Pereira A.C., Marcondes F.K., Groppo F.C. Effects of stress hormones on the production of volatile sulfur compounds by periodontopathogenic bacteria. Braz. Oral Res. 2014;28 doi: 10.1590/1807-3107BOR-2014.vol28.0008. [DOI] [PubMed] [Google Scholar]
- 18.Mortazavi H., Rahbani Nobar B., Shafiei S. Drug-related Halitosis: A Systematic Review. Oral Health Prev. Dent. 2020;18:399–407. doi: 10.3290/j.ohpd.a44679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bicak D.A. A Current Approach to Halitosis and Oral Malodor—A Mini Review. Open Dent. J. 2018;12:322–330. doi: 10.2174/1874210601812010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gokdogan O., Catli T., Ileri F. Halitosis in otorhinolaryngology practice. Iran. J. Otorhinolaryngol. 2015;27:145–153. [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki H., Arao M., Okamura K., Kawaguchi Y., Toyofuku A., Hoshi K., Yaegaki K. Tentative classification of halitosis and its treatment needs. Niigata Dent. J. 1999;32:7–11. [Google Scholar]
- 22.Tangerman A., Winkel E.G. Extra-oral halitosis: An overview. J. Breath Res. 2010;4:017003. doi: 10.1088/1752-7155/4/1/017003. [DOI] [PubMed] [Google Scholar]
- 23.Aydin M., Harvey-Woodworth C.N. Halitosis: A new definition and classification. Br. Dent. J. 2014;217 doi: 10.1038/sj.bdj.2014.552. [DOI] [PubMed] [Google Scholar]
- 24.Seemann R., Conceicao M.D., Filippi A., Greenman J., Lenton P., Nachnani S., Quirynen M., Roldan S., Schulze H., Sterer N., et al. Halitosis management by the general dental practitioner—Results of an international consensus workshop. J. Breath Res. 2014;8:017101. doi: 10.1088/1752-7155/8/1/017101. [DOI] [PubMed] [Google Scholar]
- 25.Koczorowski R., Karpiński T.M. Halitosis—Problem społeczny. Halitosis—A social problem. Now. Lek. 2001;70:657–664. [Google Scholar]
- 26.Tangerman A., Winkel E.G. Volatile Sulfur Compounds as the Cause of Bad Breath: A Review. Phosphorus Sulfur Silicon Relat. Elem. 2013;188:396–402. doi: 10.1080/10426507.2012.736894. [DOI] [Google Scholar]
- 27.Rani H., Ueno M., Zaitsu T., Furukawa S., Kawaguchi Y. Factors associated with clinical and perceived oral malodor among dental students. J. Med. Dent. Sci. 2015;62:33–41. doi: 10.11480/620202. [DOI] [PubMed] [Google Scholar]
- 28.AlSadhan S.A. Self-perceived halitosis and related factors among adults residing in Riyadh, Saudi Arabia. A cross sectional study. Saudi Dent. J. 2016;28:118–123. doi: 10.1016/j.sdentj.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van den Velde S., van Steenberghe D., Van Hee P., Quirynen M. Detection of odorous compounds in breath. J. Dent. Res. 2009;88:285–289. doi: 10.1177/0022034508329741. [DOI] [PubMed] [Google Scholar]
- 30.Aylıkcı B.U., Colak H. Halitosis: From diagnosis to management. J. Nat. Sci. Biol. Med. 2013;4:14–23. doi: 10.4103/0976-9668.107255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loesche W.J., Kazor C. Microbiology and treatment of halitosis. Periodontology 2000. 2002;28:256–279. doi: 10.1034/j.1600-0757.2002.280111.x. [DOI] [PubMed] [Google Scholar]
- 32.van den Velde S., Quirynen M., van Hee P., van Steenberghe D. Halitosis associated volatiles in breath of healthy subjects. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007;853:54–61. doi: 10.1016/j.jchromb.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 33.Madhushankari G.S., Yamunadevi A., Selvamani M., Mohan Kumar K.P., Basandi P.S. Halitosis—An overview: Part-I—Classification, etiology and pathophysiology of halitosis. J. Pharm. Bioallied Sci. 2015;7:S339–S343. doi: 10.4103/0975-7406.163441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi K.Y., Lee B.S., Kim J.H., Kim J.J., Jang Y., Choi J.W., Lee D.J. Assessment of Volatile Sulfur Compounds in Adult and Pediatric Chronic Tonsillitis Patients Receiving Tonsillectomy. Clin. Exp. Otorhinolaryngol. 2018;11:210–215. doi: 10.21053/ceo.2017.01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nani B.D., de Lima P.O., Marcondes F.K., Groppo F.C., Rolim G.S., de Moraes A.B.A., Cogo-Müller K., Franz-Montan M. Changes in salivary microbiota increase volatile sulfur compounds production in healthy male subjects with academic-related chronic stress. PLoS ONE. 2017;12:e0173686. doi: 10.1371/journal.pone.0173686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laleman I., Dekeyser C., Wylleman A., Teughels W., Quirynen M. The OralChromaTM CHM-2: A comparison with the OralChromaTM CHM-1. Clin. Oral Investig. 2020 doi: 10.1007/s00784-019-03148-9. [DOI] [PubMed] [Google Scholar]
- 37.Dinc M.E., Altundag A., Dizdar D., Avincsal M.O., Sahin E., Ulusoy S., Paltura C. An objective assessment of halitosis in children with adenoid vegetation during pre- and post-operative period. Int. J. Pediatr. Otorhinolaryngol. 2016;88:47–51. doi: 10.1016/j.ijporl.2016.06.042. [DOI] [PubMed] [Google Scholar]
- 38.He M., Lu H., Cao J., Zhang Y., Wong M.C.M., Fan J., Ye W. Psychological characteristics of Chinese patients with genuine halitosis. Oral Dis. 2020 doi: 10.1111/odi.13376. [DOI] [PubMed] [Google Scholar]
- 39.Wang F., Qiao W., Bao B., Wang S., Regenstein J., Shi Y., Wu W., Ma M. Effect of IgY on Periodontitis and Halitosis Induced by Fusobacterium nucleatum. J. Microbiol. Biotechnol. 2019;29:311–320. doi: 10.4014/jmb.1810.10044. [DOI] [PubMed] [Google Scholar]
- 40.Sezen Erhamza T., Ozdıler F.E. Effect of rapid maxillary expansion on halitosis. Am. J. Orthod. Dentofac. Orthop. 2018;154:702–707. doi: 10.1016/j.ajodo.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Interscan Welcomes You to the Official Halimeter® Website. [(accessed on 9 July 2020)]; Available online: https://www.halimeter.com/
- 42.van den Broek A.M.W.T., Feenstra L., de Baat C. A review of the current literature on aetiology and measurement methods of halitosis. J. Dent. 2007;35:627–635. doi: 10.1016/j.jdent.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Armstrong B.L., Sensat M.L., Stoltenberg J.L. Halitosis: A review of current literature. J. Dent. Hyg. 2010;84:65–74. [PubMed] [Google Scholar]
- 44.Monedeiro F., Milanowski M., Ratiu I.-A., Zmysłowski H., Ligor T., Buszewski B. VOC Profiles of Saliva in Assessment of Halitosis and Submandibular Abscesses Using HS-SPME-GC/MS Technique. Molecules. 2019;24:2977. doi: 10.3390/molecules24162977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips M., Cataneo R.N., Greenberg J., Munawar M., Nachnani S., Samtani S. Pilot study of a breath test for volatile organic compounds associated with oral malodor: Evidence for the role of oxidative stress. Oral Dis. 2005;11(Suppl. S1):32–34. doi: 10.1111/j.1601-0825.2005.01085.x. [DOI] [PubMed] [Google Scholar]
- 46.Dadamio J., Van Tornout M., Van den Velde S., Federico R., Dekeyser C., Quirynen M. A novel and visual test for oral malodour: First observations. J. Breath Res. 2011;5:046003. doi: 10.1088/1752-7155/5/4/046003. [DOI] [PubMed] [Google Scholar]
- 47.Koczorowski R., Karpiński T.M., Hofman J. Badanie zależności między halitosis a chorobami przyzębia. A study of the relationship between halitosis and periodontal diseases. Dent. Forum. 2004;30:51–56. [Google Scholar]
- 48.Mogilnicka I., Bogucki P., Ufnal M. Microbiota and Malodor-Etiology and Management. Int. J. Mol. Sci. 2020;21:2886. doi: 10.3390/ijms21082886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murnane S.S., Lehocky A.H., Owens P.D. Odor Thresholds for Chemicals with Established Occupational Health Standards (2nd Edition)—Knovel. American Industrial Hygiene Association; Falls Church, VA, USA: 2013. [Google Scholar]
- 50.Odorous Substances (Osmogenes) and Odor Thresholds. [(accessed on 29 July 2020)]; Available online: https://www.lenntech.com/table.htm.
- 51.Leonardos G., Kendall D., Barnard N. Odor Threshold Determinations of 53 Odorant Chemicals. J. Air Pollut. Control. Assoc. 1969;19:91–95. doi: 10.1080/00022470.1969.10466465. [DOI] [Google Scholar]
- 52.Nagata Y. Measurement of Odor Threshold by Triangle Odor Bag Method. Office of Odor, Noise and Vibration, Environmental Management Bureau, Ministry of Environment; Tokyo, Japan: 2003. pp. 118–127. [Google Scholar]
- 53.Warenycia M.W., Goodwin L.R., Benishin C.G., Reiffenstein R.J., Francom D.M., Taylor J.D., Dieken F.P. Acute hydrogen sulfide poisoning. Demonstration of selective uptake of sulfide by the brainstem by measurement of brain sulfide levels. Biochem. Pharm. 1989;38:973–981. doi: 10.1016/0006-2952(89)90288-8. [DOI] [PubMed] [Google Scholar]
- 54.PubChem. [(accessed on 12 July 2020)]; Available online: https://pubchem.ncbi.nlm.nih.gov/
- 55.ChemIDplus Advanced Chemical Information with Searchable Synonyms, Structures, and Formulas. [(accessed on 12 July 2020)]; Available online: https://chem.nlm.nih.gov/chemidplus/
- 56.Safety Data Sheet. Material Name: Ammonia, Anhydrous. Mathesson Tri-Gas; Basking Ridge, NJ, USA: 2009. 11p [Google Scholar]
- 57.CDC—Index of Chemicals—NIOSH Publications and Products. [(accessed on 12 July 2020)]; Available online: https://www.cdc.gov/niosh/idlh/intridl4.html.
- 58.Til H.P., Falke H.E., Prinsen M.K., Willems M.I. Acute and subacute toxicity of tyramine, spermidine, spermine, putrescine and cadaverine in rats. Food Chem. Toxicol. 1997;35:337–348. doi: 10.1016/S0278-6915(97)00121-X. [DOI] [PubMed] [Google Scholar]
- 59.Safety Data Sheet Valeric Acid—Perstorp. [(accessed on 12 July 2020)]; Available online: https://www.perstorp.com/-/media/files/perstorp/msds/valeric%20acid/msds_valeric%20acid_engeu-10857.pdf.
- 60.Suzuki N., Yoneda M., Takeshita T., Hirofuji T., Hanioka T. Induction and inhibition of oral malodor. Mol. Oral Microbiol. 2019;34:85–96. doi: 10.1111/omi.12259. [DOI] [PubMed] [Google Scholar]
- 61.Zaorska E., Konop M., Ostaszewski R., Koszelewski D., Ufnal M. Salivary Hydrogen Sulfide Measured with a New Highly Sensitive Self-Immolative Coumarin-Based Fluorescent Probe. Molecules. 2018;23:2241. doi: 10.3390/molecules23092241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sopapornamorn P., Ueno M., Shinada K., Yanagishita M., Kawaguchi Y. Relationship between total salivary protein content and volatile sulfur compounds levels in malodor patients. Oral. Surg. Oral. Med. Oral Pathol. Oral Radiol. Endodontol. 2007;103:655–660. doi: 10.1016/j.tripleo.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 63.Sterer N., Rosenberg M. Streptococcus salivarius promotes mucin putrefaction and malodor production by Porphyromonas gingivalis. J. Dent. Res. 2006;85:910–914. doi: 10.1177/154405910608501007. [DOI] [PubMed] [Google Scholar]
- 64.Tanabe S., Grenier D. Characterization of volatile sulfur compound production by Solobacterium moorei. Arch. Oral Biol. 2012;57:1639–1643. doi: 10.1016/j.archoralbio.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 65.Quirynen M., Dadamio J., van den Velde S., Smit M.D., Dekeyser C., Tornout M.V., Vandekerckhove B. Characteristics of 2000 patients who visited a halitosis clinic. J. Clin. Periodontol. 2009;36:970–975. doi: 10.1111/j.1600-051X.2009.01478.x. [DOI] [PubMed] [Google Scholar]
- 66.Milella L. The Negative Effects of Volatile Sulphur Compounds. J. Vet. Dent. 2015;32:99–102. doi: 10.1177/089875641503200203. [DOI] [PubMed] [Google Scholar]
- 67.Fujimura M., Calenic B., Yaegaki K., Murata T., Ii H., Imai T., Sato T., Izumi Y. Oral malodorous compound activates mitochondrial pathway inducing apoptosis in human gingival fibroblasts. Clin. Oral Investig. 2010;14:367–373. doi: 10.1007/s00784-009-0301-5. [DOI] [PubMed] [Google Scholar]
- 68.Attene-Ramos M.S., Wagner E.D., Plewa M.J., Gaskins H.R. Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 69.Yalçın Yeler D., Aydın M., Hocaoğlu P.T., Koraltan M., Özdemir H., Kotil T., Gül M. Ultrastructural changes in epithelial cells of rats exposed to low concentration of hydrogen sulfide for 50 days. Ultrastruct. Pathol. 2016;40:351–357. doi: 10.1080/01913123.2016.1234530. [DOI] [PubMed] [Google Scholar]
- 70.Karpiński T.M. Role of Oral Microbiota in Cancer Development. Microorganisms. 2019;7:20. doi: 10.3390/microorganisms7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S., Bian H., Li X., Wu H., Bi Q., Yan Y., Wang Y. Hydrogen sulfide promotes cell proliferation of oral cancer through activation of the COX2/AKT/ERK1/2 axis. Oncol. Rep. 2016;35:2825–2832. doi: 10.3892/or.2016.4691. [DOI] [PubMed] [Google Scholar]
- 72.Karpiński T.M. The Microbiota and Pancreatic Cancer. Gastroenterol. Clin. N. Am. 2019;48:447–464. doi: 10.1016/j.gtc.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Tiso M., Schechter A.N. Nitrate reduction to nitrite, nitric oxide and ammonia by gut bacteria under physiological conditions. PLoS ONE. 2015;10:e0119712. doi: 10.1371/journal.pone.0119712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ufnal M., Zadlo A., Ostaszewski R. TMAO: A small molecule of great expectations. Nutrition. 2015;31:1317–1323. doi: 10.1016/j.nut.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 75.Ma W., Chen K., Li Y., Hao N., Wang X., Ouyang P. Advances in Cadaverine Bacterial Production and Its Applications. Engineering. 2017;3:308–317. doi: 10.1016/J.ENG.2017.03.012. [DOI] [Google Scholar]
- 76.Liu Y.-L., Nascimento M., Burne R.A. Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int. J. Oral Sci. 2012;4:135–140. doi: 10.1038/ijos.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sakanaka A., Kuboniwa M., Hashino E., Bamba T., Fukusaki E., Amano A. Distinct signatures of dental plaque metabolic byproducts dictated by periodontal inflammatory status. Sci. Rep. 2017;7:42818. doi: 10.1038/srep42818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J., Lkhagva E., Chung H.-J., Kim H.-J., Hong S.-T. The Pharmabiotic Approach to Treat Hyperammonemia. Nutrients. 2018;10:140. doi: 10.3390/nu10020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Awano N., Wada M., Mori H., Nakamori S., Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol. 2005;71:4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romano K.A., Vivas E.I., Amador-Noguez D., Rey F.E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio. 2015;6:e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velasquez M.T., Ramezani A., Manal A., Raj D.S. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins. 2016;8:326. doi: 10.3390/toxins8110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rath S., Heidrich B., Pieper D.H., Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome. 2017;5:54. doi: 10.1186/s40168-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z., Tang W.H.W., Buffa J.A., Fu X., Britt E.B., Koeth R.A., Levison B.S., Fan Y., Wu Y., Hazen S.L. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 2014;35:904–910. doi: 10.1093/eurheartj/ehu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He X., Slupsky C.M. Metabolic fingerprint of dimethyl sulfone (DMSO2) in microbial-mammalian co-metabolism. J. Proteome Res. 2014;13:5281–5292. doi: 10.1021/pr500629t. [DOI] [PubMed] [Google Scholar]
- 85.Codipilly D., Kleinberg I. Generation of indole/skatole during malodor formation in the salivary sediment model system and initial examination of the oral bacteria involved. J. Breath Res. 2008;2:017017. doi: 10.1088/1752-7155/2/1/017017. [DOI] [PubMed] [Google Scholar]
- 86.Garg N., Luzzatto-Knaan T., Melnik A.V., Caraballo-Rodríguez A.M., Floros D.J., Petras D., Gregor R., Dorrestein P.C., Phelan V.V. Natural products as mediators of disease. Nat. Prod. Rep. 2017;34:194–219. doi: 10.1039/C6NP00063K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takeshita T., Suzuki N., Nakano Y., Shimazaki Y., Yoneda M., Hirofuji T., Yamashita Y. Relationship between oral malodor and the global composition of indigenous bacterial populations in saliva. Appl. Environ. Microbiol. 2010;76:2806–2814. doi: 10.1128/AEM.02304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.HOMD: Human Oral Microbiome Database. [(accessed on 19 June 2020)]; Available online: http://www.homd.org/
- 89.Krishnan K., Chen T., Paster B.J. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017;23:276–286. doi: 10.1111/odi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baker J.L., Bor B., Agnello M., Shi W., He X. Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y., Wang X., Li H., Ni C., Du Z., Yan F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018;99:883–893. doi: 10.1016/j.biopha.2018.01.146. [DOI] [PubMed] [Google Scholar]
- 92.Mok S.F., Karuthan C., Cheah Y.K., Ngeow W.C., Rosnah Z., Yap S.F., Ong H.K.A. The oral microbiome community variations associated with normal, potentially malignant disorders and malignant lesions of the oral cavity. Malays J. Pathol. 2017;39:1–15. [PubMed] [Google Scholar]
- 93.Bashan A., Gibson T.E., Friedman J., Carey V.J., Weiss S.T., Hohmann E.L., Liu Y.-Y. Universality of human microbial dynamics. Nature. 2016;534:259–262. doi: 10.1038/nature18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rautava J., Pinnell L.J., Vong L., Akseer N., Assa A., Sherman P.M. Oral microbiome composition changes in mouse models of colitis. J. Gastroenterol. Hepatol. 2015;30:521–527. doi: 10.1111/jgh.12713. [DOI] [PubMed] [Google Scholar]
- 96.Persson S., Edlund M.B., Claesson R., Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol. Immunol. 1990;5:195–201. doi: 10.1111/j.1399-302X.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 97.Ma Q., Zhang X., Qu Y. Biodegradation and Biotransformation of Indole: Advances and Perspectives. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deslandes B., Gariépy C., Houde A. Review of microbiological and biochemical effects of skatole on animal production. Livestock Prod. Sci. 2001;71:193–200. doi: 10.1016/S0301-6226(01)00189-0. [DOI] [Google Scholar]
- 99.Formolo M. The Microbial Production of Methane and Other Volatile Hydrocarbons. In: Timmis K.N., editor. Handbook of Hydrocarbon and Lipid Microbiology. Springer; Berlin/Heidelberg, Germany: 2010. pp. 113–126. [Google Scholar]
- 100.Lan W., Yang C. Ruminal methane production: Associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci. Total Environ. 2019;654:1270–1283. doi: 10.1016/j.scitotenv.2018.11.180. [DOI] [PubMed] [Google Scholar]
- 101.Huang J., Tang W., Zhu S., Du M. Biosynthesis of butyric acid by Clostridium tyrobutyricum. Prep. Biochem. Biotechnol. 2018;48:427–434. doi: 10.1080/10826068.2018.1452257. [DOI] [PubMed] [Google Scholar]
- 102.Lynch K.M., Zannini E., Wilkinson S., Daenen L., Arendt E.K. Physiology of Acetic Acid Bacteria and Their Role in Vinegar and Fermented Beverages. Compr. Rev. Food Sci. Food Saf. 2019;18:587–625. doi: 10.1111/1541-4337.12440. [DOI] [PubMed] [Google Scholar]
- 103.Ye W., Zhang Y., He M., Zhu C., Feng X.-P. Relationship of tongue coating microbiome on volatile sulfur compounds in healthy and halitosis adults. J. Breath Res. 2019;14:016005. doi: 10.1088/1752-7163/ab47b4. [DOI] [PubMed] [Google Scholar]
- 104.Pham T.A.V., Ueno M., Zaitsu T., Takehara S., Shinada K., Lam P.H., Kawaguchi Y. Clinical trial of oral malodor treatment in patients with periodontal diseases. J. Periodont. Res. 2011;46:722–729. doi: 10.1111/j.1600-0765.2011.01395.x. [DOI] [PubMed] [Google Scholar]
- 105.Niederauer A.J., Guimarães R.A., Oliveira K.L., Pires A.R., Demasi A.P., Ferreira H.H., Sperandio M., Napimoga M.H., Peruzzo D.C. H2S in periodontal immuneinflammatory response and bone loss: A study in rats. Acta Odontol. Latinoam. 2019;32:164–171. [PubMed] [Google Scholar]
- 106.Silva M.F., Nascimento G.G., Leite F.R.M., Horta B.L., Demarco F.F. Periodontitis and self-reported halitosis among young adults from the 1982 Pelotas Birth Cohort. Oral Dis. 2020;26:843–846. doi: 10.1111/odi.13286. [DOI] [PubMed] [Google Scholar]
- 107.Takeshita T., Suzuki N., Nakano Y., Yasui M., Yoneda M., Shimazaki Y., Hirofuji T., Yamashita Y. Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci. Rep. 2012;2:215. doi: 10.1038/srep00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Demmer R.T., Jacobs D.R., Singh R., Zuk A., Rosenbaum M., Papapanou P.N., Desvarieux M. Periodontal Bacteria and Prediabetes Prevalence in ORIGINS: The Oral Infections, Glucose Intolerance and Insulin Resistance Study. J. Dent. Res. 2015;94:201S–211S. doi: 10.1177/0022034515590369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang K.Y. Future Management and Possible Treatment of Halitosis. [(accessed on 21 June 2020)]; Available online: https://munin.uit.no/handle/10037/7626.
- 110.Ileri Keceli T., Gulmez D., Dolgun A., Tekcicek M. The relationship between tongue brushing and halitosis in children: A randomized controlled trial. Oral Dis. 2015;21:66–73. doi: 10.1111/odi.12210. [DOI] [PubMed] [Google Scholar]
- 111.Nakano Y., Suzuki N., Kuwata F. Predicting oral malodour based on the microbiota in saliva samples using a deep learning approach. BMC Oral Health. 2018;18:128. doi: 10.1186/s12903-018-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bernardi S., Karygianni L., Filippi A., Anderson A.C., Zürcher A., Hellwig E., Vach K., Macchiarelli G., Al-Ahmad A. Combining culture and culture-independent methods reveals new microbial composition of halitosis patients’ tongue biofilm. Microbiologyopen. 2020;9:e958. doi: 10.1002/mbo3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yitzhaki S., Reshef L., Gophna U., Rosenberg M., Sterer N. Microbiome associated with denture malodour. J. Breath Res. 2018;12:027103. doi: 10.1088/1752-7163/aa95e0. [DOI] [PubMed] [Google Scholar]
- 114.Bernardi S., Continenza M.A., Al-Ahmad A., Karygianni L., Follo M., Filippi A., Macchiarelli G. Streptococcus spp. and Fusobacterium nucleatum in tongue dorsum biofilm from halitosis patients: A fluorescence in situ hybridization (FISH) and confocal laser scanning microscopy (CLSM) study. New Microbiol. 2019;42:108–113. [PubMed] [Google Scholar]
- 115.Al-Zyoud W., Hajjo R., Abu-Siniyeh A., Hajjaj S. Salivary Microbiome and Cigarette Smoking: A First of Its Kind Investigation in Jordan. Int. J. Environ. Res. Public Health. 2019;17:256. doi: 10.3390/ijerph17010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu Y., Chi X., Zhang Q., Chen F., Deng X. Characterization of the salivary microbiome in people with obesity. PeerJ. 2018;6:e4458. doi: 10.7717/peerj.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Boever E.H., Loesche W.J. Assessing the contribution of anaerobic microflora of the tongue to oral malodor. J. Am. Dent. Assoc. 1995;126:1384–1393. doi: 10.14219/jada.archive.1995.0049. [DOI] [PubMed] [Google Scholar]
- 118.Roldán S., Winkel E.G., Herrera D., Sanz M., Van Winkelhoff A.J. The effects of a new mouthrinse containing chlorhexidine, cetylpyridinium chloride and zinc lactate on the microflora of oral halitosis patients: A dual-centre, double-blind placebo-controlled study. J. Clin. Periodontol. 2003;30:427–434. doi: 10.1034/j.1600-051X.2003.20004.x. [DOI] [PubMed] [Google Scholar]
- 119.Grover H.S., Blaggana A., Jain Y., Saini N. Detection and measurement of oral malodor in chronic periodontitis patients and its correlation with levels of select oral anaerobes in subgingival plaque. Contemp. Clin. Dent. 2015;6:S181–S187. doi: 10.4103/0976-237X.166825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Faveri M., Feres M., Shibli J.A., Hayacibara R.F., Hayacibara M.M., de Figueiredo L.C. Microbiota of the dorsum of the tongue after plaque accumulation: An experimental study in humans. J. Periodontol. 2006;77:1539–1546. doi: 10.1902/jop.2006.050366. [DOI] [PubMed] [Google Scholar]
- 121.Ademovski S.E., Persson G.R., Winkel E., Tangerman A., Lingström P., Renvert S. The short-term treatment effects on the microbiota at the dorsum of the tongue in intra-oral halitosis patients—A randomized clinical trial. Clin. Oral Investig. 2013;17:463–473. doi: 10.1007/s00784-012-0728-y. [DOI] [PubMed] [Google Scholar]
- 122.Kamaraj D.R., Bhushan K.S., Laxman V.K., Mathew J. Detection of odoriferous subgingival and tongue microbiota in diabetic and nondiabetic patients with oral malodor using polymerase chain reaction. Indian J. Dent. Res. 2011;22:260–265. doi: 10.4103/0970-9290.84301. [DOI] [PubMed] [Google Scholar]
- 123.Awano S., Gohara K., Kurihara E., Ansai T., Takehara T. The relationship between the presence of periodontopathogenic bacteria in saliva and halitosis. Int. Dent. J. 2002;52(Suppl. S3):212–216. doi: 10.1002/j.1875-595X.2002.tb00927.x. [DOI] [PubMed] [Google Scholar]
- 124.Tanaka M., Yamamoto Y., Kuboniwa M., Nonaka A., Nishida N., Maeda K., Kataoka K., Nagata H., Shizukuishi S. Contribution of periodontal pathogens on tongue dorsa analyzed with real-time PCR to oral malodor. Microbes Infect. 2004;6:1078–1083. doi: 10.1016/j.micinf.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 125.Adedapo A.H., Kolude B., Dada-Adegbola H.O., Lawoyin J.O., Adeola H.A. Targeted polymerase chain reaction-based expression of putative halitogenic bacteria and volatile sulphur compound analysis among halitosis patients at a tertiary hospital in Nigeria. Odontology. 2020;108:450–461. doi: 10.1007/s10266-019-00467-x. [DOI] [PubMed] [Google Scholar]
- 126.Riggio M.P., Lennon A., Rolph H.J., Hodge P.J., Donaldson A., Maxwell A.J., Bagg J. Molecular identification of bacteria on the tongue dorsum of subjects with and without halitosis. Oral Dis. 2008;14:251–258. doi: 10.1111/j.1601-0825.2007.01371.x. [DOI] [PubMed] [Google Scholar]
- 127.Haraszthy V.I., Zambon J.J., Sreenivasan P.K., Zambon M.M., Gerber D., Rego R., Parker C. Identification of oral bacterial species associated with halitosis. J. Am. Dent. Assoc. 2007;138:1113–1120. doi: 10.14219/jada.archive.2007.0325. [DOI] [PubMed] [Google Scholar]
- 128.Ren W., Zhang Q., Liu X., Zheng S., Ma L., Chen F., Xu T., Xu B. Supragingival Plaque Microbial Community Analysis of Children with Halitosis. J. Microbiol. Biotechnol. 2016;26:2141–2147. doi: 10.4014/jmb.1605.05012. [DOI] [PubMed] [Google Scholar]
- 129.Kazor C.E., Mitchell P.M., Lee A.M., Stokes L.N., Loesche W.J., Dewhirst F.E., Paster B.J. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang F., Huang S., He T., Catrenich C., Teng F., Bo C., Chen J., Liu J., Li J., Song Y., et al. Microbial basis of oral malodor development in humans. J. Dent. Res. 2013;92:1106–1112. doi: 10.1177/0022034513507065. [DOI] [PubMed] [Google Scholar]
- 131.Washio J., Sato T., Koseki T., Takahashi N. Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour. J. Med. Microbiol. 2005;54:889–895. doi: 10.1099/jmm.0.46118-0. [DOI] [PubMed] [Google Scholar]
- 132.Oshiro A., Zaitsu T., Ueno M., Kawaguchi Y. Characterization of oral bacteria in the tongue coating of patients with halitosis using 16S rRNA analysis. Acta Odontol. Scand. 2020:1–6. doi: 10.1080/00016357.2020.1754459. [DOI] [PubMed] [Google Scholar]
- 133.Tetz G., Vikina D., Brown S., Zappile P., Dolgalev I., Tsirigos A., Heguy A., Tetz V. Draft Genome Sequence of Streptococcus halitosis sp. nov., Isolated from the Dorsal Surface of the Tongue of a Patient with Halitosis. Microbiol. Resour. Announc. 2019;8:e01704-18. doi: 10.1128/MRA.01704-18. [DOI] [PMC free article] [PubMed] [Google Scholar]