Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently causing the respiratory illness termed as the coronavirus disease 2019 or the COVID-19 pandemic. Indeed, the significant increase in deaths in the current days due to influenza around the world started in 1889 is a continued public health threat because of its intermittent style of pandemic outbreaks. An array of research on the influenza viruses has been conducted especially pointing on (1) the development of the anti-viral drugs and the design of probable vaccines on trial basis, (2) the biochemical and genetic aspects underlying the viral pathogenicity, (3) the viral epidemiology, and on (4) the protective immunity against the influenza viruses. Current review briefly discussed the epidemic/ pandemic history of influenza and correlated with the current epidemiology, the possible preventive measures that may be taken by the public health professionals as well as to increase the protective awareness among the general people. The viral reassortments during the initiation of pandemics have also been focused based on the previous literatures.
Keywords: Influenza viruses, COVID-19, Pandemic, History, Public health
Introduction
Disease outbreaks, epidemics and pandemics (like the ongoing COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 or the SARS-CoV-2; and the Spanish influenza H1N1 in 1918–1920) are very well known terms for the epidemiologists as well as for the other health professionals [1, 2]. A disease outbreak, both regional and seasonal, is known to take place when a disease propagates into a relatively large numbers of people within a community to several countries, lasting from days to years. In contrast to an outbreak, an epidemic is known to occur when an infectious disease tends to spread more quickly among a huge number of populations, as can be exemplified by the acute respiratory syndrome (SARS) epidemic in 2003, causing 774 deaths worldwide [2]. A pandemic is simply known as global disease like the Spanish influenza H1N1 (causing around 50 million people in 1918–1920), Hong Kong influenza H3N2, causing approximately 1 million people in 1968, Asian influenza H2N2 (causing nearly 2 million deaths in 1957–1958), or the Russian influenza H2N2 (causing more than 1 million deaths in 1889–1892) [1–5]. The ongoing pandemic by the SARS-CoV-2, which started in late 2019, has appeared no less dreadful than the Spanish influenza in terms of morbidity and mortality [4]. COVID-19 is already well known as the life threatening infectious respiratory disease which actually shares the same routes and means of transmission as influenza; and hence influenza can be correlated with COVID-19 disease which has been further confirmed by the case of a patient co-infected with SARS-CoV-2 and influenza A virus [6]. Therefore, measures taken for the influenza (and other infectious respiratory diseases) prevention and control may also serve effectively to combat the COVID-19 pandemic [4]. The influenza pandemic is of significant public health importance since it is frequent with the rise of a new subtype of a virus which in turn weakens or escapes the host immunity completely [3, 4]. Moreover, the virus is easily transmissible from human to human through the daily unhygienic activities like irresponsible and close contact coughing and sneezing [3–6].
The 1918 influenza pandemic, the most severe one in the history, occurred in three waves: first at Spring 1918, second one at Fall 1918 (the death cases were the highest), and the third wave at the Winter 1918 and the Spring at 1919, suggesting that a pandemic can dreadfully harm the global human lives especially regarding morbidity and mortality [5]. Along with a "very high alert” for the pandemic announced by the regulatory bodies, scientists usually concentrate on the transmission of viruses, viral pathogenesis, the associated host immunity, effectual diagnosis of the disease, and the preventive therapeutic strategies including the possible vaccine development [7]. Although a scientific discussion usually focuses primarily on the biomedical research advances along with the disease mitigation possibilities, the control of any pandemic largely requires the preventive realization within the broader demographic, socioeconomic, cultural, and even the political context at all levels regardless the local, regional, national or international levels [6]. Indeed, responding to any pandemic with the highest level of emergency is of prime significance to limit the viral transmission and eventually to maintain the global public health sustainability. Such responses may include medical interventions, appropriate drug designing and vaccine development, research on the host immune responses during viral pathogenesis and the evolution of virus, detecting virus transmission, rapid and accurate diagnosis, and various preventive measures along with the public awareness, etc.[1, 6–9]. The current review briefly highlighted on the major pandemic outbreaks in a simplistic way to communicate such importance of the technical knowhow in order to attain the common knowledge to be safe during a pandemic situation.
Russian influenza (La Grippe) H2N2 (1889–1892)
Russian Flu, the first true epidemic cases observed in May 1889 (in Bukhara, Turkestan; in Athabasca, Canada, and in Greenland), was another outbreak caused by the Influenza A virus subtype H2N2; however, recently it is thought that the subtype was actually H3N8 [10]. The flu specifically spread into the urban areas prior to propagate into the whole world, ultimately causing over a million of deaths [10]. Only a small fraction of the population got affected during the first three months; however, after that four‐fifths of the people were noticed to be severely affected without any early symptoms of influenza [5, 8]. Briefly it is to be noted that the mortality rate was higher among the elderly people in 1889–1890 pandemic whereas this pattern was dominantly noticed among the young people during the 1918–1919 pandemic outbreaks [4, 8–11]. There is an interesting factor to note that influenza was attributed to a Bacillus species, and it was not shown to be caused by a virus until the 1930s [11, 12]. Moreover, a hypothesis was drawn that young adults could have increased risk of dying from the Spanish flu in 1918–1920 (the H1N1 subtype) if they had been previously infected by the Russian flu (1889–1890) [12].
Viral reassortment and pathogenicity
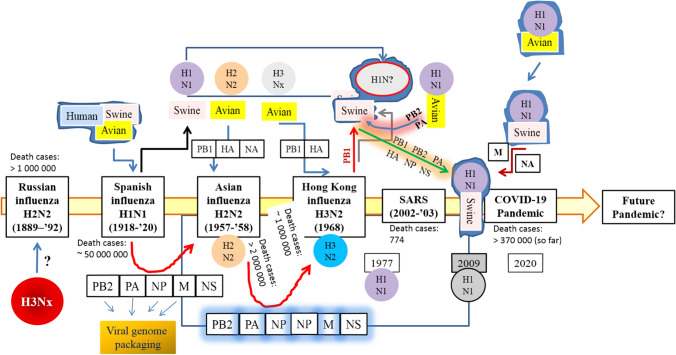
Interestingly, it has been recently inferred that the causative virus of the Russian influenza could be of the H3Nx subtype which circulated up to the 1918 influenza pandemic outbreak [9]. The logic behind such inference is that the causative virus of the 1918 H1N1 pandemic remained undefined in an aspect that whether the virus originated from the multiple reassortment events or not [9]. The reassortment event could be held between the avian-, swine- and human influenza viruses, or it could be introduced by a direct zoonotic transmission by the vectors stated above [9, 11–14]. As a known information, it’s clear that the influenza pandemics occurs when the hemagglutinin (HA, or H) with or without neuraminidase (NA, or N) subtypes of influenza viruses infect humans (with little or no immunity) with the subsequent follow up of person to person transmission. So far 18 HA subtypes (H1 to H18) and 11 NA subtypes (N1 to N11) have been recognized [13]. Besides the reassortment events as stated above, another interpretation on the viral pathogenecity underlies the point mutations within the set of genes encoding the corresponding amino acids required for antigenic expressions. Eventually such mutations augment the enhanced rate of replacement of NA and HA (possibly by the positive selection pressure) which are already known as the antigenic cluster of the viral subtype [15]. Such an event leads to parallel replacement of the antigens or even may drive the current antigens back to their ancestral forms which go a long way to impart the viral adaptation and survival within the host inactivating the protective immunity [15]. A simple illustration of the viral reassortment between the influenza viruses is shown in Fig. 1.
Fig. 1.
Simple illustration of viral reassortment between the influenza viruses. After the 1918–1920 Spanish influenza outbreak the 1918 A/H1N1 subtype (thought to be generated from manifold reassortments between avian-, swine-, and human borne viruses) was likely to circulate continuously and thereby triggering the subsequent epidemics [20]. In 1957 the A/H1N1 subtype was depicted to be reassorted with an avian A/H2N2 virus, which circulated until 1968, and reassorted again with the avian H3Nx virus [37]. In 1977 the A1/H1N1 virus was reestablished among the humans (contained the M gene segment of 1918 origin, encoding the matrix protein, M1 and M2, grossly required for the viral pathogenecity) and was co-circulated with H3N2 viruses till 2009 [20, 36, 38, 39] when it was thought to be swapped by another A/H1N1 subtype, H1N1pdm2009 [38] as a result of the numerous reassortments between avian-, swine-, and human influenza viruses yy[20, 36–38]. This is to be noted that such viral reaasortments have not been shown between the SARS or SARS-CoV2 in this illustration. PB2 Polymerase basic protein 2, involved in transcription initiation and cap stealing mechanism; PA polymerase acidic protein, involved in viral RNA transcription and replication; NP nuceloprotein, involved in the encapsulation of the viral genome for RNA transcription, replication and packaging; M matrix protein, involved in the formation of the viral envelope; NS non-structural protein, involved in the inhibition of host immune responses; PB1: Polymerase basic protein 1, a determinant of the influenza virus virulence; HA hemagglutinin, a homotrimeric glycoprotein found on the surface of influenza viruses which is integral to its infectivity, and is involved in the viral attachment and acts as a membrane fusion protein.; NA neuraminidase, involved in cleaving sialic acid groups from glycoproteins and are required for influenza virus replication; i.e., it is essential for release of progeny virus particles from the surface of an infected cell
Spanish influenza H1N1 in 1918–1920
As stated above, the Spanish influenza pandemic was in three consecutive waves: spring 1918, autumn 1918, and winter 1918–19, responsible for killing approximately 50 million people worldwide with abnormally severe clinical manifestations among the healthy and young adults [5, 9, 16, 17]. Moreover, lack of cellular immunity as well as the pre-existing virus-specific and/or cross-reactive antibodies, individual genetic makeup like the single-nucleotide polymorphisms (SNPs), co-infections with bacterial pathogens, and with the measles and malaria causing agents; and finally the malnutrition or obesity has been found to contribute to the high mortality due to the 1918 H1N1 virus infection [9]. Though the 1918 influenza pandemic is known as the “Spanish Flue”; in real the 1918 influenza virus didn’t originate in Spain; rather it originated in the Midwest of the United States of America and then France, from where the viral transmission spread throughout Europe and within the rest of the world [9, 17, 18]. So, the geographic origins of the 1918 virus remained obscure, and eventually the viral vector (animal reservoir) became controversial. Being a segmented virus, influenza virus was capable of undergoing the reassortment process which is known to occur when two influenza virus strains co-infect the same cell, thereby triggering the evolution of a novel “reassortant” virus consisting of an unusual assemblage of genes [9]. Such an interesting mechanism between the avian- and the human influenza viruses later gave rise to the 1957 and 1968 influenza pandemics [9, 19].
Demolition of protective immunity during viral pathogenesis
Some of the viral factors including haemagglutinin (HA) and the polymerase gene segments are stringently known to trigger the abnormal increase in the pro-inflammatory cytokines and chemokines, for example: Interleukin (IL)-2, 5, 6 and 8, the interferon (IFN)-inducible protein (IP-10), macrophage inflammatory protein 1α (MIP1α), MIP1β and monocyte chemoattractant protein-1 (MCP1), tumor necrosis factor alpha (TNF-α), etc. in course of the viral pathogenesis during a pandemic outbreak [9]. However, a dysregulated pro-inflammatory response was noticed to be triggered by the 1918 influenza virus, which in turn imparted severe lung lesions among the infected patients [20]. Besides, the lack of pre-existing antibodies (virus-specific or cross-reactive) as well as the shortage of the pre-existing influenza virus specific CD8+ T cells among the young population was another prime factor for the viral pathogenecity [9, 21]. This is to be noted that the protective influenza virus-specific antibody responses may be long-lived [9, 21, 22]. However, pointing out the virally infected persons as well applying the proper anti-viral therapy is always essential to limit the dreadful propagation of the disease. During the years 1918–1920, there were no certain drugs that could be effectively implanted for the elimination of virus. On the contrary, in the current days, several anti-viral drugs like the neuraminidase inhibitors oseltamivir and zanamivir are employed prophylactically in the outbreak situations [9, 23].
Regarding the preparation or development of Influenza virus vaccines, a major shortfall underlies the fact that antibodies elicited by the seasonal influenza vaccines cannot impart protection in the case of an antigenically distinct influenza virus of a novel subtype [24]. More specifically, during the interpandemic periods, the antigenic drift (mutations in the genes of influenza viruses) variants may arise which in turn created difficulties in the development of a corresponding vaccine [24]. The mutations in the genes lead to changes in the HA and NA surface proteins (serving as antigens) which are to be recognized by the immune system with the concomitant triggering of the protective immunity and the production of specific antibodies blocking the infection. One potential strategy to counter this problem has been noticed when a trivalent hemagglutinin DNA prime-trivalent inactivated influenza vaccine (IIV3) boost regimen (the 2012/13 seasonal trivalent HA DNA vaccine; i.e., VRC-FLUDNA063-00-VP) was developed to cure the children suffering from seasonal influenza in the US [25]. In healthy adults, such DNA influenza vaccines against the emerging subtypes of avian origin (including H5 and H7) administered as a prime injection prior to an inactivated boost was observed to increase the overall antibody titers [25].
Asian influenza H2N2 in 1957–1958
The H2N2 subtype influenza A virus (consisting of the genes from an H2N2 subtype encoding H2 hemagglutinin and N2 neuraminidase) that originated from an avian influenza A virus emerged in East Asia, which triggered another pandemic outbreak known as Asian Flu [11]. The first reported case was Singapore in February 1957, Hong Kong in April 1957 and in the coastal cities of the USA in summer 1957, with the final fatality exerted by more than one million deaths worldwide [11]. However, according to Chowell and clleagues (in 2017), the first local outbreak originated in continental China during February‐March 1957, and then the virus reached Hong Kong and other parts of Asia within a few weeks; and within July of that year, the pandemic reached Europe and South American countries of the Pacific coast, New Zealand, and South Africa. In the USA, the sporadic outbreaks were reported during June‐August 1957 [26]. Moreover, the mortality burden of the 1957 influenza pandemic in 39 countries was assessed with the WHO associated adequate data since such pandemic appeared as the second one of this century after the Spanish flue which had no mathematical measurement of morbidity and mortality except the gross calculations of death cases [27]. Interestingly, the study conducted by Viboud and colleagues (in 2016) covering the full range of global extrapolations of mortality revealed the plausible estimates of the 1957–1959 pandemic burden were in between 0.7 million–2.1 million excess deaths [27].
During a pandemic, a rigorous question arises in the scientists’ mind whether humans may possess the cross-subtype immunity in order to develop an appropriate vaccine [28]. Such a puzzle was nearly resolved through the analysis of the archival records of the Cleveland Family Study before and during the 1957 pandemic; i.e., during the shift from subtype H1N1 to H2N2 to investigate the development of the heterosubtypic immunity. The study revealed that only 5.6% of the adults having symptomatic influenza A, developed the infection earlier during the pandemic, despite living in households with participants who had influenza; whereas the infection frequency was much higher (55.2%) in the children having the symptomatic influenza A. This finding clearly showed the accumulation of the heterosubtypic immunity during a pandemic which may draw sufficient attention for the development of vaccines [29].
Hong Kong influenza H3N2 in 1968
The H1N1 subtype influenza A virus causing the 1918 pandemic outbreak caused the seasonal epidemics until 1957 when a new influenza A virus of the H2N2 subtype emerged [28]. This was referred to as the “Asian” influenza virus, causing the second pandemic after the Spanish flue as stated above [29]. The H2N2 kept continuing the dissemination of the seasonal influenza from 1957 to 1967; and in 1968 the third pandemic appeared with the new H3N2 subtype [29, 30]. The H2N2 subtype, causing the pandemic in 1957, indeed continued to disseminate in humans till 1968 [28–31]. The antigenic diversity of human H2N2 viruses isolated between the intermittent 12 years (from 1957 to 1968) has been well characterized; and two amino acid substitutions, T128D and N139K, located in the head domain of the H2 hemagglutinin (HA) molecule, have been shown to be the functional determinants of the antigenic change during the evolution of H2N2 subtype [29].
The 1968 influenza A virus of H3N2 subtype, with the especial feature of antigenic drift, consisted of two genes from an avian influenza A virus: a new H3 hemagglutinin (HA), and the N2 neuraminidase (NA) from the 1957 H2N2 subtype [24]. As stated earlier, the estimated number of deaths was approximately 1 million worldwide with higher death rate among the elderly people (those who were aged 65 on an average). It has been reported that the neuraminidase (NA)-specific immunity provides protection against influenza, and antibodies against HA and NA are likely to fortify the protection [27–30]. Like HA, NA may achieve point mutations during the passage of the viruses within the host, which in turn results in the alteration in the viral antigenicity; i.e., the antigenic drift which is very likely to reduce NA-antibody-based protection against the viral infection as revealed from the in vivo studies [30]. It is to be noted that the World Health Organization (WHO) selects the influenza candidate vaccine strains based only on the HA antigenicity and not that of NA. Thus the continued surveillance of the NA of circulating influenza viruses and the subsequent of NA antigenicity to optimize the influenza vaccines has been suggested [30].
Severe acute respiratory syndrome (SARS) in 2002–2003
SARS, the severe acute respiratory syndrome, caused by a novel coronavirus (termed as SARS-CoV, causing a massive epidemic outbreak among 24 countries resulting in 774 deaths out of 8000 infected cases), is another highly infectious and lethal atypical form of pneumonia, originating in China too; and spreading from Southeast-Asia to over all five continents [32]. The SARS-CoV was found to be transmitted from bats to small carnivores, and then from them to humans [33]. Since the mutations of the viral receptor genes (more specifically, the point mutations and recombination of genomes among different species/ strains) were responsible for its pathogenesis and fatality, the SARS pandemics is thought be the unique model of an emerging disease [32–34]. In 2012, some incidences of novel coronavirus infection were reported in Arabian Peninsula with pneumonia and acute kidney injury; and the virus was named Middle East respiratory syndrome coronavirus (MERS-CoV) which was thought to be propagated from camels and bats [32].
This is to be noted that four coronaviruses (HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1) appeared to be endemic (i.e., an infectious microorganism that is restricted to a specific locality or region) so far and mainly associated with the mild respiratory illnesses; whereas the other two coronaviruses: SARS-CoV and the MERS-CoV) are widely known as emerging infections triggering severe respiratory syndrome worldwide [32, 33]. Indeed, the current COVID-19 pandemic is the third serious Coronavirus outbreak within the last 20 years, following SARS in 2002–2003 and MERS in 2012 [32–34]. SARS patients were found to suffer from acute renal impairment and proteinuria together with hematological abnormalities like thrombocytopenia and lymphopenia [34–36].
In contrast the SARS-CoV-2 is principally causing headache, fever, sore throat, cough, acute respiratory distress syndrome, acute cardiac injury, upper airway congestion, pharyngalgia, myalgia, and diarrhea, anemia, and even the secondary infections [37]. However, the onset of acute respiratory distress syndrome (ARDS) by three β-coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV is common [32]. Finally, it is to be stated that though several candidate vaccines against SARS-CoV have been manufactured and tested in small scale; however, unfortunately still there is no FDA approved vaccine against SARS [34]. Interestingly, the etiological agent that caused 50 million deaths in 1918–919 was unknown at that very time (will be discussed later); but since that old time the world has experienced three additional influenza pandemics: the 1957 “Asian” influenza pandemic, the 1968 “Hong Kong” influenza pandemic (described in the descendent paragraphs) and the currently ongoing COVID-19 pandemic [1–4, 9, 16, 17]. It is to be pondered that both the ongoing infection SARS-CoV-2 and the previous SARS-CoV infections are associated with the elevation of the pro-inflammatory cytokines and chemokines (cytokine storm) while the pathogenicity of MERS-CoV is based on its interferon (IFN) antagonist proteins [32].
Coronavirus disease in 2019–2020 (COVID-19) by SARS-CoV-2
The pandemic of the Coronavirus Disease 2019 (COVID-19) resulting in novel coronavirus-infected pneumonia (NCIP) is the ongoing pandemics around the world which originated in China [38]. COVID-19 has already caused more than 3,500,000 deaths with approximately 5,500,000 infected cases in more than 200 countries with its utmost severity in the USA, Italy, Spain, Germany, France, UK and Iran [39, 40]. A significant genomic similarity with Severe Acute Respiratory Syndrome (SARS, a pandemics occurred in 2002) was noticed [32, 38, 40]. This virus represents severe risks not only in the complex humanitarian crises lacking the adequate health infrastructure but also; surprisingly its severity is being noted in the highly developed countries. However, this is worth to note that the poor monitoring, displacement, crowded housing, malnutrition, inadequate water, sanitation, and hygiene (WASH) tools may provoke the viral fatality [41]. Coronaviruses are already known as the enveloped single-stranded zoonotic RNA viruses causing the symptoms nearly similar to those as seen during the common cold or more severe respiratory syndromes [42]. Besides the running pandemic outbreak by the COVID-19, Coronaviruses have been known to cause two large-scale pandemics along the last two decades: SARS and MERS (Middle East respiratory syndrome coronavirus) [43]. Together with an array of published articles on coronaviruses, the China CDC (Centers for Disease Control and Prevention) published a guideline for the public awareness on combating COVID-19 pandemic [38].
Viral pathogenesis and the host immunity escaping strategies
Dissemination of the COVID-19 is mediated by via cough or sneeze droplets or direct contact within human-to-human [37]. The viral particles are thought to bind to the angiotensin-converting enzyme 2 receptor in humans thereby rendering the infected individual prone to the respiratory disorders [31]. The coronavirus pneumonia develops within maximum 7 days with the early symptoms of fever, headache, cough, congestion, pharyngalgia, etc.[38–42]. The sequential secondary bacterial infections are not surprising during the viral infections. Suppression of the intricate innate antiviral responses induces the viral infection. The SARS-CoV-2 RNA encodes at least two proteases possessing the capacity of cleaving the cells of the innate immunity [42, 43]. The viruses may replicate extensively in the cytoplasm by modifying the intracellular membranes to form specific replication organelles which protect the recognition of viral RNA by the innate immune sensors [38, 40]. The viral endoribonuclease (RNase) activity is another means of excaping the innate immunity. Besides, the viruses can modify their RNAs to keep protected the recognition by the foreign RNA sensors of the host immune cells [35, 39]. Moreover, the receptor-binding domain (RBD) of viral spike can mediate the viral entry. Interestingly, a monoclonal antibody (MAb) can bind to the cell surface of the immunolglobulin G (IgG) Fc receptor which in turn mediates the viral entry into the host as also happens during the MERS-CoV infection [32, 38, 40]. Indeed, using the spike (S) protein, both the SARS-CoV-2 and SARS-CoV bind to the angiotensin-converting enzyme 2 (ACE 2) receptor of the host whereas MERS-CoV utilizes the host receptor Dipeptidyl peptidase 4 (DPP4); and it’s to be noted that SARS-CoV-2 possesses a longer spike protein than that of SARS-CoV; and moreover, the SARS-CoV-2 entry depends on the non-neutralizing antibodies while in case of SARS-CoV, the neutralizing antibodies block the viral entry whereas the MERS-CoV entry principally depends on the host natural killer (NK) cell inactivation [31].
Preventive strategies and public awareness
The 1918 H1N1 subtype pandemic followed the H2N2 pandemic in 1957; and the third pandemic was caused by H3N2 subtype in 1968. An integrated interplay between viral genome, host immunity including the dysregulated pro-inflammatory response, and societal factors are usually responsible for morbidity and mortality due to any epidemic- or pandemic outbreak [9, 19]. As discussed earlier, the high pathogenicity exerted by the 1918 influenza virus was due to the proteins encoded by the viral haemagglutinin (HA) and polymerase gene segments [9, 19]. Lack of appropriate humoral immunity; i.e., the pre-existing virus-specific and/or cross-reactive antibodies and the shortage in the cellular immunity can be another promoting factor for the onset of viral infection [20]. Thus the anti-HA antibody, anti-NA antibody, and cell-mediated immunity are very important for the protective immunity against influenza [31]. Besides, (1) the genetic factors such as single-nucleotide polymorphisms (SNPs), (2) the co-infections with other bacterial pathogens, (3) malnutrition, (4) obesity, (5) population demographics, (6) antibiotic resistance, (7) climate change, etc. may influence the dreadfulness of influenza virus infections [11].
Quick identification of suspect cases is the major step to limit the spread of any transmissible virus. The identification based on the molecular biological tools like the sequencing and annotation of the viral genome as well as the development of rapid diagnostic tests may facilitate the isolation of the infected persons to ensure others’ safety [3, 41]. In the current time, the diagnosis of viral strains responsible for influenza has spectacularly improved both in terms of accuracy, speed and sensitivity by means of an array of molecular diagnostic techniques. Moreover, the risk assessment of the pandemic viruses has also improved through the screening of the viral genome within the infected individuals. Monitoring the possible mutations within the viral genome is also possible.
The Public Health Emergency of International Concern (PHEIC) along with the World Health Organization (WHO) and the CDC (Centers for Disease Control and Prevention) is already working on the pandemic cases with set goals to maintain the global public health sustainability. Respiratory hygiene (maintained by hand hygiene, safe cough practice, and social distancing) is the most effectual public health intercession as a preventive strategy against the spread of respiratory viruses [41]. During the SARS pandemic in 2003 the need for the quick detection of the nucleic acid sequence evolved which is major breakthrough in the diagnosis of viral infections [35]. Also, a forceful home quarantine strategy was launched which is currently being practiced to combat the transmission of COVID-19 [35]. Moreover, as stated earlier, the CDC guidelines described very well about the method of wearing face masks, hand washing style, protective measures while residing at home or in public gathering, different disinfection and sanitation methods, etc.[42]. Such strategies would further boost the medical observation at home as well as go a long way to eliminate panic among the general population during the pandemics [42].
Concluding remarks
An array of informative reports on especially the pathogenicity and epidemiology on the pandemics of influenza viruses are now a days well available in a huge scientific and social database. Such information indeed, from the Spanish flu pandemic in 1918 to the current COVID-19 pandemic in 2020, have been really helpful to develop different approaches prevent the circulation of the different influenza subtypes and the dreadful β-coronaviruses; as well as to treat the virally infected patients. Both pharmaceutical and non-pharmaceutical interventions have been developed including the development of drug/vaccine development, implementation of quarantine measures, wearing protective cloths with glove and masks, limiting the global travelling, maintain proper hygiene, etc. The medical interventions employed different anti-viral drugs and tried for the development of vaccines; however, no appropriate vaccine has been developed till date to circumvent these respiratory viruses. Rather, proper medical nursing seemed to be useful along with ventilation. To counteract the antigenic drift as stated earlier, increased surveillance is needed along with the novel strategies to develop the vaccine composition, production, formulation, presentation, and delivery.
The policy makers working especially in WHO and CDC, the public health experts, the specialist scientists and other health professionals have long been working to protect human civilization from these dreadful virus outbreaks. Current review is apparently devoid of new information on influenza virus from the molecular point of virological research; nevertheless, here some points have been addressed which might be useful for gaining general knowledge and the protective against the viral fatality by a broad range of population. For the epidemiologists working the densely populated developing or under developed countries, the information (based on the published epidemiological and medical reports) especially regarding the viral transmission, the time wave pattern of viral infection, the possible failures in developing the appropriate vaccines and the unfortunate events of neutralizing human immune systems by the viruses (through the antigenic drift and/ or the viral reassortment) could be of great importance for outbreak predictions, pandemic planning and possible public awareness.
Acknowledgement
We thank the authors whose articles have been cited during the preparation of this review paper.
Compliance with ethical standards
Conflict of interest
None declared.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kępińska AP, Iyegbe CO, Vernon AC, Yolken R, Murray RM, Pollak TA. Schizophrenia and influenza at the centenary of the 1918–1919 Spanish influenza pandemic: mechanisms of psychosis risk. Front Psychiatry. 2020;11:72. doi: 10.3389/fpsyt.2020.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003; 348(20):1953–66. 10.1056/nejmoa030781 [DOI] [PubMed]
- 3.Leung Y, To M, Lam T, Yau S, Leung O, Chuang S. Epidemiology of human influenza A(H7N9) infection in Hong Kong. J Microbiol Immunol Infect. 2017;50(2):183–188. doi: 10.1016/j.jmii.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Lin Q, Zhao S, Gao D, Lou Y, Yang S, Musa SS, Wang MH, Cai Y, Wang W, Yang L, He D. A conceptual model for the coronavirus disease 2019 (COVID-19) outbreak in Wuhan, China with individual reaction and governmental action. Int J Infect Dis. 2020;93:211–216. doi: 10.1016/j.ijid.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC (Centers for Disease Control and Prevention). 1918 Pandemic influenza: three waves. https://www.cdc.gov/flu/pandemic-resources/1918-commemoration/three-waves.htm accessed on May 30, 2020.
- 6.Wu D, Lu J, Ma X, Liu Q, Wang D, Gu Y, Li Y, He W. Coinfection of influenza virus and severe acute respiratory syndrome Coronavirus 2 (SARS-COV-2) Ped Infect Dis J. 2020 doi: 10.1097/INF.0000000000002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel OA, Manicassamy B. Broadly protective strategies against influenza viruses: universal vaccines and therapeutics. Front Microbiol. 2020;7(11):135. doi: 10.3389/fmicb.2020.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing ET. La Grippe or Russian influenza: mortality statistics during the 1890 epidemic in Indiana. Influenza Other Respir Viruses. 2019;13(3):279–287. doi: 10.1111/irv.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Short KR, Kedzierska K, van de Sandt CE. Back to the future: lessons learned from the 1918 influenza pandemic. Front Cell Infect Microbiol. 2018;8:343. doi: 10.3389/fcimb.2018.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D, Lu J, Liu Y, Zhang Z, Luo L. Positive effects of COVID-19 control measures on influenza prevention. Int J Infect Dis. 2020;S1201–9712(20):30225–30233. doi: 10.1016/j.ijid.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC (Centers for Disease Control and Prevention). 1957–1958 Pandemic (H2N2 virus). https://www.cdc.gov/flu/pandemic-resources/1968-pandemic.html. Accessed on May 30, 2020.
- 12.Bernd Heinrich. Profiling a pandemic. Natural History. https://www.naturalhistorymag.com/features/243463/profiling-a-pandemic. Accessed on April 4, 2020.
- 13.van de Sandt CE, Kreijtz JH, Geelhoed-Mieras MM, Nieuwkoop NJ, Spronken MI, van de Vijver DA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Differential recognition of influenza a viruses by M158–66 epitope-specific CD8+ T cells is determined by extraepitopic amino acid residues. J Virol. 2016;90(2):1009–1022. doi: 10.1128/jvi.02439-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenberger JK. The origin and virulence of the 1918 "Spanish" influenza virus. Proc Am Philos Soc. 2006;150(1):86–112. doi: 10.2807/esw.10.42.02812-en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neverov AD, Lezhnina KV, Kondrashov AS, Bazykin GA. Intrasubtype reassortments cause adaptive amino acid replacements in H3N2 influenza genes. PLoS Genet. 2014;10(1):e1004037. doi: 10.1371/journal.pgen.1004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray CJ, Lopez AD, Chin B, Feehan D, Hill KH. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: a quantitative analysis. Lancet. 2006;368(9554):2211–2218. doi: 10.1016/s0140-6736(06)69895-4. [DOI] [PubMed] [Google Scholar]
- 17.Collins SD. Age and sex incidence of influenza and pneumonia morbidity and mortality in the epidemic of 1928–29 with comparative data for the epidemic of 1918–19: based on surveys of families in certain localities in the United States following the epidemics. Public Health Rep. 1931;46:1909–1937. doi: 10.2307/4580139. [DOI] [Google Scholar]
- 18.Barry JM, Viboud C, Simonsen L. Cross-protection between successive waves of the 1918–1919 influenza pandemic: epidemiological evidence from US Army camps and from Britain. J Infect Dis. 2008;198(10):1427–1434. doi: 10.1086/592454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schäfer JR, Kawaoka Y, Bean WJ, Süss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194(2):781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 20.Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, Suzuki H, Nishimura H, Mitamura K, Sugaya N, Usui T, Murata T, Maeda Y, Watanabe S, Suresh M, Suzuki T, Suzuki Y, Feldmann H, Kawaoka Y. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431(7009):703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed R, Oldstone MBA, Palese P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol. 2007;8(11):1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed C, Katz JM. Serological surveys for 2009 pandemic influenza A H1N1. Lancet. 2010;375(9720):1062–1063. doi: 10.1016/s0140-6736(09)62194-2. [DOI] [PubMed] [Google Scholar]
- 23.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, García-Sastre A. Influenza. Nat Rev Dis Primers. 2018 doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Jong JC, Rimmelzwaan GF, Fouchier RAM, Osterhaus ADME. Influenza virus: a master of metamorphosis. J Infect. 2000;40(3):218–228. doi: 10.1053/jinf.2000.0652. [DOI] [PubMed] [Google Scholar]
- 25.Houser KV, Yamshchikov GV, Bellamy AR, May J, Enama ME, Sarwar U, Larkin B, Bailer RT, Koup R, Paskel M, Subbarao K, Anderson E, Bernstein DI, Creech B, Keyserling H, Spearman P, Wright PF, Graham BS, Ledgerwood JE; VRC 702 Study Team. DNA vaccine priming for seasonal influenza vaccine in children and adolescents 6 to 17 years of age: A phase 1 randomized clinical trial. PLoS One. 2018;13(11):e0206837. 10.1371/journal.pone.0206837 [DOI] [PMC free article] [PubMed]
- 26.Chowell G, Simonsen L, Fuentes R, Flores J, Miller MA, Viboud C. Severe mortality impact of the 1957 influenza pandemic in Chile. Influenza Other Respir Viruses. 2017;11(3):230–239. doi: 10.1111/irv.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viboud C, Simonsen L, Fuentes R, Flores J, Miller MA, Chowell G. Global mortality impact of the 1957–1959 influenza pandemic. J Infect Dis. 2016;213(5):738–745. doi: 10.1093/infdis/jiv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 Pandemic of 1957: an experiment of nature. J Infect Dis. 2006;193(1):49–53. doi: 10.1086/498980. [DOI] [PubMed] [Google Scholar]
- 29.Linster M, Schrauwen EJA, van der Vliet S, Burke DF, Lexmond P, Bestebroer TM, Smith DJ, Herfst S, Koel BF, Fouchier RAM. The Molecular basis for antigenic drift of human A/H2N2 influenza viruses. J Virol. 2019;93(8):e01907–e1918. doi: 10.1128/JVI.01907-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan H, Gao J, Yang H, Yang S, Harvey R, Chen YQ, Zheng NY, Chang J, Carney PJ, Li X, Plant E, Jiang L, Couzens L, Wang C, Strohmeier S, Wu WW, Shen RF, Krammer F, Cipollo JF, Wilson PC, Stevens J, Wan XF, Eichelberger MC, Ye Z. The neuraminidase of A (H3N2) influenza viruses circulating since 2016 is antigenically distinct from the A/Hong Kong/4801/2014 vaccine strain. Nat Microbiol. 2019;4(12):2216–2225. doi: 10.1038/s41564-019-0522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babu TM, Perera RAPM, Wu JT, Fitzgerald T, Nolan C, Cowling BJ, Krauss S, Treanor JJ, Peiris M. Population serologic immunity to human and avian H2N2 viruses in the United States and Hong Kong for pandemic risk assessment. J Infect Dis. 2018;218(7):1054–1060. doi: 10.1093/infdis/jiy291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan PK, Chan MC. Tracing the SARS-coronavirus. J Thorac Dis. 2013;5(Suppl 2):S118–S121. doi: 10.3978/j.issn.2072-1439.2013.06.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Peng F, Wang R, Guan K, Jiang T, Xu G, Chang C. SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2003;2020:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu KH, Tsang WK, Tang CS, Lam MF, Lai FM, To KF, Fung KS, Tang HL, Yan WW, Chan HWH, Lai TST, Tong KL, Lai KN. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67(2):698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Booth CM. Clinical features and short-term outcomes of 144 patients with SARS in the Greater Toronto Area. JAMA. 2003;289(21):2801. doi: 10.1001/jama.289.21.joc30885. [DOI] [PubMed] [Google Scholar]
- 37.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chafekar A, Fielding BC. MERS-CoV: understanding the latest human coronavirus threat. Viruses. 2018;10(2):93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO (World Health Organization). Coronavirus diseases (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed on 29 May 2020.
- 40.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4(12):951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole DN, Escudero DJ, Gostin LO, Leblang D, Talbot EA. Responding to the COVID-19 pandemic in complex humanitarian crises. Int J Equity Health. 2020;19(1):41. doi: 10.1186/s12939-020-01162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, Sun C, Sylvia S, Rozelle S, Raat H, Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hörmansdorfer S, Campe H, Sing A. SARS—Pandemic and emerging DISEASE. Jl Für Verbraucherschutz Und Lebensmittelsicherheit. 2008;3(4):417–420. doi: 10.1007/s00003-008-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]