Abstract
Epigenetic processes are essential for normal development and the maintenance of tissue-specific gene expression in mammals. Changes in gene expression and malignant cellular transformation can result from disruption of epigenetic mechanisms, and global disruption in the epigenetic landscape is a key feature of cancer. The study of epigenetics in cancer has revealed that human cancer cells harbor both genetic alterations and epigenetic abnormalities that interplay at all stages of cancer development. Unlike genetic mutations, epigenetic aberrations are potentially reversible through epigenetic therapy, providing a therapeutically relevant treatment option. Histone methyltransferase inhibitors are emerging as an epigenetic therapy approach with great promise in the field of clinical oncology. The recent accelerated approval of the enhancer of zeste homolog 2 (EZH2; also known as histone-lysine N-methyltransferase EZH2) inhibitor tazemetostat for metastatic or locally advanced epithelioid sarcoma marks the first approval of such a compound for the treatment of cancer. Many other histone methyltransferase inhibitors are currently in development, some of which are being tested in clinical studies. This review focuses on histone methyltransferase inhibitors, highlighting their potential in the treatment of cancer. We also discuss the role for such epigenetic drugs in overcoming epigenetically driven drug resistance mechanisms, and their value in combination with other therapeutic approaches such as immunotherapy.
Keywords: Cancer, Epigenetics, Histone methyltransferase, Oncology
Key Summary Points
| Global disruptions in the epigenetic landscape are a key feature of cancer. |
| Unlike genetic mutations, epigenetic aberrations are potentially reversible through therapy with epigenetic modifiers. |
| In a rapidly evolving field of research, histone methyltransferases have been targeted with a range of small-molecule inhibitors. |
| Histone methyltransferase inhibitors are in development for the treatment of both solid and hematologic malignancies. |
| The success in this field is highlighted by the clinical development of molecules and the recent approval of the methyltransferase inhibitor tazemetostat (EZH2 inhibitor). |
Introduction
In eukaryotic cell nuclei, DNA is packaged and organized in a DNA–protein complex called chromatin. The basic component of chromatin is the nucleosome, consisting of approximately 147 bp of DNA and core histones (H2A, H2B, H3, and H4) [1–4]. Nucleosomes associated with active genes are more accessible than those with inactive genes, suggesting that the degree of chromatin packaging correlates with gene activity [5].
In contrast to inherited genetic variation that remains constant over the lifetime of an individual, epigenetics differ as disease changes over time. This provides the opportunity to employ epigenetic changes as biomarkers for disease and targets for disease modification in many fields of healthcare [6]. Unlike genetic mutations, epigenetic aberrations are potentially reversible. Thus, targeting relevant epigenetic factors with small molecules might provide an efficient approach to “fix” dysregulated gene/chromosome-regulatory systems resulting from aberrant epigenetic profiles. Epigenetic mechanisms are important for the regulation of cell proliferation, differentiation, survival, and other cellular processes [7]. Epigenetic dysregulation results in changes to the transcriptional landscape, affecting multiple pathways which might contribute to tumor pathogenesis [8]. Here we provide an overview of epigenetic modification with relevance to cancer treatment, focusing on histone methyltransferases. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Histone Methyltransferase Inhibitors
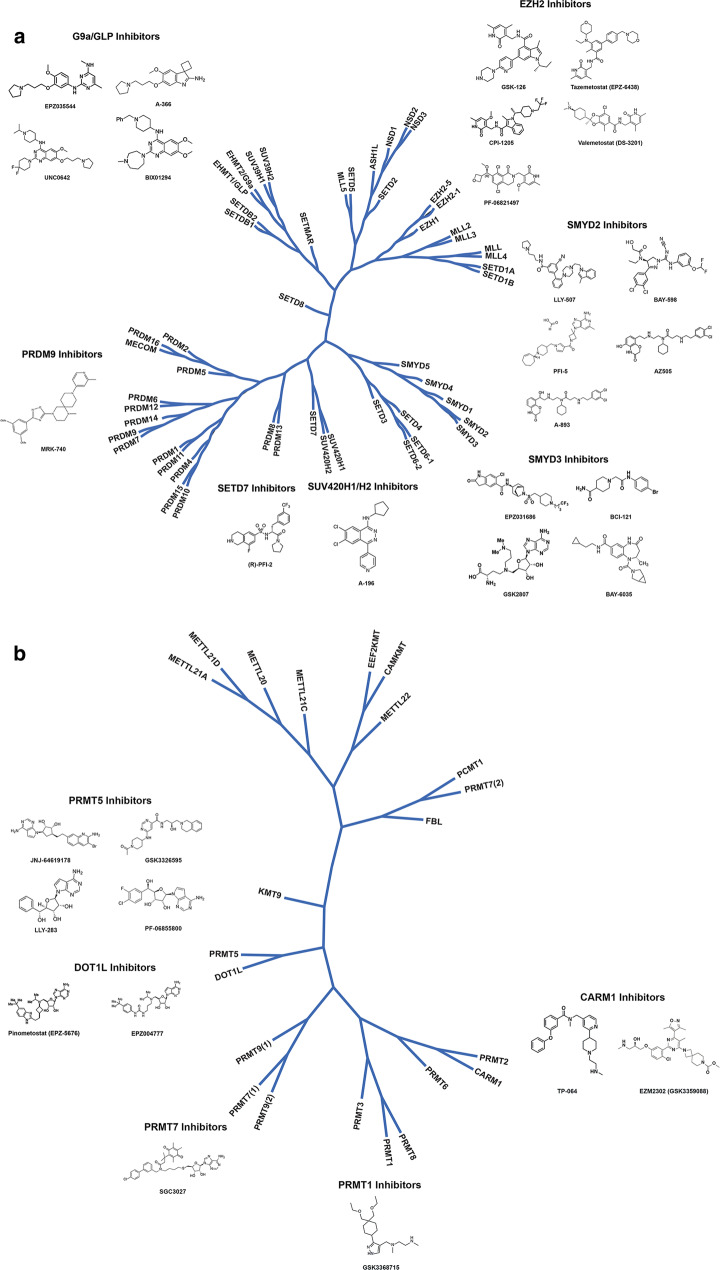
According to the histone code hypothesis [9], transcription is in part regulated by methylation of histone proteins resulting in recruitment of other proteins through the specific recognition of modified histone proteins [9]. Lysine as well as arginine residues of the N-terminal tail can be methylated [9]. Lysine methylation has three methyl additions (mono-, di-, and trimethylation). Arginine methylation has two methyl additions (mono- and di-), with dimethylation occurring on either single (asymmetric) or two (symmetric) nitrogen atoms [10]. Key lysine and arginine methyltransferases under investigation for cancer treatment include enhancer of zeste homolog 2 (EZH2), G9a, disruptor of telomeric silencing 1-like protein (DOT1L), and protein arginine methyltransferases (PRMT) 1 and 5. Figure 1 illustrates a phylogenetic tree of protein methyltransferases and the publicly available chemical structures for some of the associated inhibitors.
Fig. 1.
Phylogenetic tree of protein methyltransferases and molecules in development
Genome-wide studies have revealed that different levels of histone methylation of H3K4, which can be mediated by distinct histone methyltransferases [11, 12], have been linked to specific gene activities: H3K4 monomethylation, together with H3K27 acetylation, marks enhancer activity, whereas H3K4 trimethylation and dimethylation are associated with the transcription start site of active genes [11, 13, 14]. It is also known that specialized proteins such as Jumonji domain-containing 2A (JMJD2A), p53-binding protein 1 (53BP1), SAGA-associated factor 29 (SGF29), and others interact with a broad spectrum of histone methylation marks to facilitate DNA damage repair or regulate transcription [15].
Histone methylation was initially considered relatively stable compared with histone acetylation, but it can also be removed by histone demethylases, e.g., lysine-specific demethylase 1A (LSD1 or KDM1A), the lysine demethylase for H3K4 and H3K9 [16, 17]. The Jumonji C [JmjC] domain-containing protein UTX [ubiquitously transcribed tetratricopeptide repeat, X chromosome], as well as the related JMJD3 [Jumonji domain containing 3], specifically remove methyl marks on H3K27 [18]. Interplay of these epigenetic mechanisms regulates tissue-specific gene expression patterns and preserves cellular identity. For example, histone methyltransferases (G9a, Suv39h1, and PRMT5) recruit DNA methyltransferases (DNMTs) to specific genomic loci to promote DNA methylation [19–21]. On the other hand, DNMTs can recruit histone deacetylases (HDACs) and other effector methyl-binding proteins to promote gene silencing, highlighting the linkage between DNA methylation and histone modification machinery critical for maintaining the cellular epigenetic landscape.
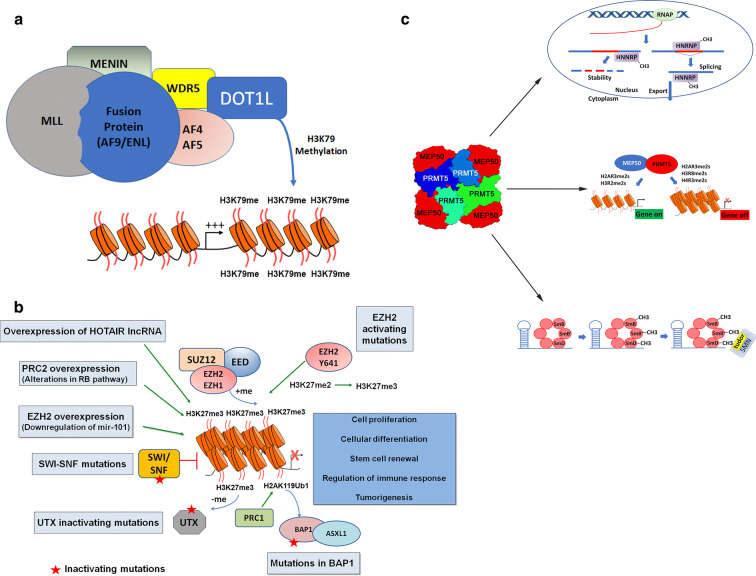
In an area that has evolved at a rapid pace in recent years, histone methyltransferases have been targeted with a range of small-molecule inhibitors. The success in this field is highlighted by the clinical and translational development of molecules discussed in the following paragraphs. Figure 2 is a graphical representation of the major complexes covered in this review (polycomb repressive complex 2 [PRC2], EZH2, DOT1L, and PRMT5).
Fig. 2.
Graphical representation of the mechanisms of selected methyltransferases targeted by protein methyltransferase inhibitors. a In acute myeloid leukemia, chromosomal translocations result in formation of oncogenic fusion proteins that combine the chromatin targeting domains of MLL to the protein recruitment domains of fusion partners including AF9 and ENL. These fusion partners initiate the recruitment of DOT1L H3K79 histone methyltransferase function to genes that govern cell proliferation and cell differentiation. At these genes, DOT1L enzyme function increases transcriptional output of these genes, resulting in oncogenic programs. b EZH2 or the related protein EZH1 comprise the polycomb repressive complex 2 with partner proteins EED and SUZ12. In normal development and tissue regulation, PRC2 regulates gene expression programs governing cell proliferation, cellular differentiation, stem cell renewal, and immune regulation. In cancer, activation of PRC2 through gain-of-function mutations in EZH2 or overexpression of EZH2 alter the normal regulation of PRC2 function leading to uncontrolled cell growth, survival, and immune evasion. Cancers harboring mutations in epigenetic regulators including SWI/SNF, EED, and BAP1 that antagonize EZH2 and PRC2 function also create opportunities for targeting EZH2 as a synthetic lethal vulnerability in cancer. c PRMT5 and MEP50 form a heterooctomer complex catalyzing the symmetric dimethylation of proteins involved in RNA transcription, processing, metabolism, and splicing. PRMT5 works both in the regulation of gene expression through histone methylation as well as alternative splicing of pre-mRNA through methylation of splicing proteins. Both types of regulation impact gene expression and pre-mRNA processing and thus can be considered “epigenetic”. The first part of c demonstrates the role of PRMT5 in the methylation of HNRNP proteins. These proteins are involved in the regulation of mRNA stability, splicing, and nuclear export. PRMT5 adds symmetric dimethylation to arginine residues on hnRNP proteins, hnRNPA1 and hnRNAH1. The middle of c shows PRMT5 methylation of histone proteins. This type of methylation can activate or repress gene expression depending on the arginine residue that is modified. Symmetric dimethylation of histone arginine residues by PRMT5 on H2AR3, H3R2, H3R8, and H4R3 has been demonstrated to impact both transcriptional activation and repression in a context-specific manner. The third part of c involves the methylation of Sm proteins by PRMT5. PRMT5 directly methylates Sm proteins including SmB, SmB′, and SmD. This methylation is critical for forming key components of the spliceosome through the recruitment of SMN proteins and RNAs for assembly and biogenesis of snRNP core particles required for pre-mRNA splicing that form snRNP complexes
EZH2
EZH2 is a histone lysine methyltransferase that forms the catalytic subunit of the PRC2 complex, also including EED, SUZ12, and RbAp46. It primarily catalyzes H3K27 methylation which results in gene silencing [22]. It is important in the control of genes involved in the regulation of cell fate decisions, such as self-renewal and differentiation. EZH2 has also been associated with repression of tumor suppressor and epithelial-mesenchymal transition (EMT)-related genes such as p16INK4a and E-cadherin [23]. Apart from its transcriptional repressor activity on chromatin, EZH2 has also been reported to catalyze certain non-histone substrates, including signal transducer and activation of transcription 3 (STAT3), GATA4, androgen receptor (AR), and retinoic acid-related orphan nuclear receptor α [24–27].
Alterations in EZH2 are associated with cancer development and progression. Gain-of-function point mutations at Y641 and A677 in EZH2, which lead to activation of methyltransferase activity and increased H3K27 trimethylation, have been observed in follicular lymphoma (FL) and diffuse large B cell lymphoma (DLBCL), and associated with disease progression [28–30]. Increased EZH2 activity in such tumors has been associated with suppression of differentiation. EZH2 is overexpressed in solid tumors, including breast, bladder, endometrial, liver, ovarian, prostate, small cell lung cancer (SCLC), melanoma, glioblastoma, and pediatric glioma, as well as lymphomas, correlating with disease progression and poorer prognosis [31–37].
Upregulated EZH2 activity in SCLC, associated with frequent inactivation of the retinoblastoma (RB1) tumor suppression gene, promotes tumor growth and resistance to standard-of-care chemotherapy, resulting in poor overall survival [38]. EZH2 activity has been associated with tumor progression in prostate cancer, and EZH2 inhibition resulted in antitumor effects both in in vitro and in in vivo prostate cancer preclinical models. Moreover, EZH2 inhibitors synergize with the androgen receptor antagonist enzalutamide in inhibiting growth of adenocarcinoma castration-resistant prostate cancer (CRPC) cells [39–41]. EZH2 activity has also been associated with induction of cellular reprogramming and trans-differentiation of prostate tumors into enzalutamide-resistant neuroendocrine prostate cancer and EZH2 inhibition re-sensitized such NEPC tumors to enzalutamide [42, 43].
In certain cancers, epigenetic factors that interact with EZH2 to regulate cellular transcriptional programs have been shown to harbor mutations. Mutation or loss of INI1 (SMARCB1) and SMARCA4 subunits of the SWI/SNF complex are associated with malignant rhabdoid tumor and ovarian small cell carcinoma of the hypercalcemic type [44–47]. Mutations of BRCA-associated protein 1 (BAP1), a deubiquitinating enzyme and a subunit of the polycomb repressive complex containing H2A deubiquitinase activity (PR-DUB) [48], is associated with mesothelioma, uveal melanoma, cutaneous melanoma, renal cell carcinoma, and other cancers [48]. Loss of function mutations in UTX, an H3K27 histone demethylase that antagonizes EZH2 activity, has been reported in multiple myeloma, bladder, medulloblastoma, esophageal, and pancreatic cancers [49, 50]. Preclinical studies imply that such cancer cells are driven by EZH2 activity, with EZH2 inhibition resulting in reduced tumor growth [51–54], prompting their study in the clinical setting.
A number of selective SAM-competitive EZH2 inhibitors have been developed that inhibit both the mutant form of EZH2 as well as the wild-type form [55]. These small molecules mainly inhibit H3K27 di- and trimethylation, leading to reactivation of EZH2 target genes. Six SAM-competitive molecules, CPI-0209, CPI-1205, tazemetostat (EPZ-6438), GSK2816126, PF-06821497, and DS-3201, a dual inhibitor of EZH1 and EZH2, have entered into clinical trials. The clinical trial of GSK2816126 has since been terminated because of modest anticancer activity at tolerable doses and dosing limitations of the drug [56]. Selective SAM-competitive EZH2 inhibitors presently in clinical trials are shown in Table 1.
Table 1.
SAM-competitive EZH2 inhibitors in clinical trials
| Molecule | Cancer indications in clinical studies | Clinical phase | ClinicalTrials.gov identifier |
|---|---|---|---|
| CPI-1205 | B cell lymphoma | 1 | NCT02395601 |
| Solid tumor | 1/2 | NCT03525795a | |
| mCRPC | 1/2 | NCT03480646 | |
| CPI-0209 | Solid tumor | 1/2 | NCT04104776 |
| DS-3201 | AML or ALL | 1 | NCT03110354 |
| SCLC | 1/2 | NCT03879798 | |
| r/r adult T cell lymphoma | 2 | NCT04102150 | |
| Tazemetostat | r/r B cell NHL | 2 | NCT03456726 |
| Malignant mesothelioma | 2 | NCT02860286 | |
| r/r INI1-negative tumors or synovial sarcoma | 1 | NCT02601937 | |
| r/r FL and DLBCL | 1 | NCT02220842b | |
| r/r B cell NHL | 1 | NCT03009344 | |
| B cell NHL, FL, DLBCL, and solid tumor | 1/2 | NCT01897571 | |
| PF-06821497 | SCLC, r/r CRPC, and r/r FL | 1 | NCT03460977 |
ALL acute lymphoblastic leukemia, AML acute myeloid lymphoma, CRPC castration-resistant prostate cancer, DLBCL diffuse large B cell lymphoma, FL follicular lymphoma, mCRPC metastatic castration-resistant prostate cancer, NHL non-Hodgkin’s lymphoma, NPC nasopharyngeal carcinoma, PD-1 programmed cell death-1, r/r relapsed/refractory, SAM S-adenosyl-methionine, SCLC small cell lung cancer
aIn combination with anti-CTLA-4
bIn combination with anti-PD1 and anti-CD20
Tazemetostat, an orally bioavailable potent and selective SAM-competitive EZH2 inhibitor, was recently approved by the US Food and Drug Administration for epithelioid sarcoma [57], and is currently the most investigated agent in the clinical setting, with antitumor properties demonstrated in preclinical models [58, 59]. A phase 2 study of tazemetostat in patients with relapsed/refractory (r/r) malignant mesothelioma with BAP1 inactivation (NCT02860286) has also reported favorable safety and tolerability, and promising antitumor activity [60]. Tazemetostat is being tested in an ongoing phase 1/2 clinical trial in patients with r/r B cell non-Hodgkin’s lymphoma (NHL) or advanced solid tumors (NCT01897571). The study reported objective responses of 38% (n = 8/21) in patients with B cell NHL and 5% (n = 2/43) in patients with solid tumors, with favorable safety profiles [61]. Notably, durable objective response or prolonged stable disease was observed in 38% (n = 5/13) of patients with INI-negative or SMARCA4-negative solid tumors, whereas responses were not observed in patients with wild-type INI1 or wild-type SMARCA4 proteins [61].
A phase 2 trial of tazemetostat in patients with r/r FL or r/r DLBCL reported clinical activity and durable response with favorable safety profiles in interim analyses. Efficacy for tazemetostat was observed in lymphoma with EZH2 mutations as well as wild-type EZH2. In 39 evaluable patients with r/r FL with an activating EZH2 mutation, the objective response rate (ORR) was 74% with a complete response (CR) rate of 10%. In 53 evaluable EZH2 wild-type patients, the ORR was 34%, with a CR rate of 6% [62]. In patients with r/r DLBCL, the ORR was 17% in both mutant and wild-type subgroups, and a substantially longer duration of response was seen in the mutant arm [63]. A recent update reported an ORR of 77% for 43 patients with EZH2 mutations [64]. The accelerated approval of tazemetostat in epithelioid sarcoma was based on efficacy data from a single-arm cohort (Cohort 5) of a multicenter trial (NCT02601950). In this phase 2 trial, tazemetostat was tested across various cohorts of patients with INI1 deficiency with variable responses. Not all patients with tumors with INI1 loss responded to therapy, but the epithelioid sarcoma cohort provided evidence for the efficacy of the drug in this setting [65]. Although tazemetostat was tested in other tumors, so far no predictors of response have been identified, even in tumors with INI1 loss. Although a predictive biomarker for response to EZH2 inhibition is lacking, some data support lower SMARCA2 expression as a biomarker in SWI/SNF mutant cancers [66].
PF-06821497, a potent and selective EZH2 inhibitor with a Ki of less than 0.1 nM against both wild-type and mutant Y641N-EZH2, induced robust tumor growth inhibition in a dose-dependent manner in DLBCL xenograft models in vivo [67]. This molecule is currently under evaluation in a multicenter, phase 1 study (NCT03460977) in patients with CRPC (in combination with enzalutamide), SCLC (in combination with cisplatin or carboplatin plus etoposide), and FL.
GSK2816126 is a highly selective, SAM-competitive inhibitor of EZH2 methyltransferase activity [68], and was the first commercial EZH2 inhibitor, opening the door for other compounds developed in this space. GSK2816126 efficiently inhibits the growth of EZH2-activating mutant DLBCL cells both in vitro and in vivo [68]; however, a phase 1 study of patients with hematologic malignancies (DLBCL, FL, NKL, and malignant mesothelioma) and solid tumors [69] was terminated because of the lack of clinical activity, potentially due to poor pharmacokinetic properties.
An ongoing phase 1 study of CPI-1205, a potent, selective SAM-competitive inhibitor [70], in patients with B cell lymphoma reported favorable toxicity, with evidence of antitumor activity and target engagement [71]. In patients with metastatic CRPC, an ongoing randomized phase 1b/2 study (ProSTAR) of CPI-1205 in combination with either abiraterone or enzalutamide reported that CPI-1205 was well tolerated, with a promising pharmacokinetic/pharmacodynamic profile and clinical activity in both arms in second-line treatment [72, 73]. CPI-0209 is a follow-up compound to CPI-1205 with better potency and pharmacokinetics.
Although targeting EZH2 appears to be a promising strategy to treat many types of cancers, when KARPAS422 lymphoma cells were treated with the EZH2 inhibitor El1 for a prolonged duration, resistant cells carrying the secondary EZH2 mutations Y111L and Y661D emerged [74]. Those mutant cells were not only resistant to El1 but also to EPZ-6438, which shares the pyridine scaffold with EI1 [74]. Although emergence of such resistant mutations has not yet been observed in the clinic, considering that most potent EZH2 inhibitors contain a common 2-pyridine ring [19, 55], it would be desirable to inhibit EZH2 activity through mechanisms other than a SAM-competitive inhibition. In this context, growing interest in targeting the embryonic ectoderm development (EED) component of the PRC2 complex has evolved as an alternative strategy to inhibit EZH2 [75–79]. For example, Novartis has reported a number of EED binders in development that inhibit basal PRC2 activity, including MAK683, which is an allosteric inhibitor that binds EED and currently in a clinical trial for DLBCL and advanced solid tumors [77].
Some cancers depend on both EZH1 and EZH2 for progression; for example, preclinical studies have shown that the sole inhibition of EZH2 did not suppress MLL-AF9-mediated acute myeloid leukemia (AML), but simultaneous inhibition of EZH2 and EZH1 produced complete suppression [80, 81], and other preclinical studies have shown greater antitumor efficacy for EZH1/2 dual inhibitors compared with EZH2 selective inhibition [82]. The superior antitumor activity of dual inhibition was observed in vitro and in vivo against DLBCL cells harboring gain-of-function mutations in EZH2 [82]. One of these previously reported EZH1/2 dual inhibitors, DS-3201, has since shown clinical activity in patients with NHL, although further clinical data are required to assess if dual inhibition provides a better therapeutic index [83]. Overall, EZH2 inhibitors are emerging as a novel therapeutic option based on their promising clinical data in patients with NHL, especially FL, while their clinical activity in solid tumors is still under investigation. Since oncogenic activity of EZH2 is context-dependent, identification of biomarkers for response along with rational combination approaches might be critical for their successful application in solid tumors. Inactivating EZH2 mutations are also observed in certain tumors, e.g., myelodysplastic syndrome, myelofibrosis, myeloproliferative neoplasm, chronic myelomonocytic leukemia, and T cell acute lymphoblastic leukemia (ALL), so it might play a tumor suppressor role in certain contexts [84–86]. Thus it is important to understand the diverse role of EZH2 in cancer to determine the optimal use of EZH2 inhibitors in the clinic.
DOT1L
The H3K79 methyltransferase DOT1L is involved in gene activation [87, 88]. DOT1L interaction with fusion partners of mixed lineage leukemia (MLL) results in inappropriate DOT1L recruitment and aberrant expression of target genes, leading to MLL-rearranged leukemogenesis [89, 90]. MLL fusion partners AF9, ENL, and AF4 account for over two-thirds of MLL rearrangements [91]. Three compounds (EPZ004777, EPZ-5676 [pinometostat], and SGC0496) have been examined as specific DOT1L inhibitors [92]. Although EPZ004777 demonstrated robust activity in a mouse model of MLL as well as in cultured cells, its pharmacokinetic properties were unsuited to clinical application [92]. EPZ-5676, a DOT1L inhibitor related to EPZ004777, demonstrated better potency and selectivity compared with EPZ00477 in a rat xenograft model of MLL-rearranged leukemia, with durable complete tumor regression [93].
A phase 1 study of EPZ-5676 (pinometostat) in patients with r/r leukemia involving MLL translocation (NCT01684150) showed that EPZ-5676 was well tolerated with 2 of 51 patients experiencing complete remission [94]. A phase 1 study in children with r/r MLL-rearranged acute leukemia (NCT02141828) demonstrated an acceptable safety profile, though no objective response occurred [95].
The modest clinical response to DOT1L inhibition might be explained by the possibility that residual DOT1L methyltransferase activity is sufficient for cell viability. Further clinical studies, including the ongoing phase 1/2 trial examining EPZ-5676 in combination with chemotherapy agents (NCT03724084) or EPZ-5676 in combination with the DNMT inhibitor azacitidine (NCT03701295), will help evaluate the applicability of DOT1L inhibitors for MLL-rearranged leukemia.
Protein Arginine Methyltransferases
Arginine methylation, mediated by the nine members of the PRMT family, is an important epigenetic mechanism for regulating transcription. The PRMTs are classified as type I, type II, or type III on the basis of the type of methylation made to the target substrates. All three types of PRMT enzymes are capable of catalyzing arginine monomethylation, whereas type I PRMTs catalyze asymmetric dimethylation (me2a) and type II enzymes catalyze symmetric dimethylation (me2s) [96, 97]. PRMTs methylate a variety of substrates, including histones and many non-histone proteins important in cancer signaling pathways.
Coactivator-associated arginine methyltransferase 1 (CARM1 [PRMT4]) is overexpressed in AML and breast, prostate, and liver cancers and is an important regulator of cell differentiation [98–101]. CARM1 asymmetrically dimethylates H3 residues R17 and R26, and also modifies many non-histone proteins. These include members of MLL, SWI/SNF, and mediator complexes [102], RNA-binding proteins, and some splicing factors [103]. CARM1 has been implicated in both pluripotency [104, 105] and cellular differentiation in multiple lineages [106–108]. It blocks myeloid differentiation of human hematopoietic stem/progenitor cells and is downregulated during myeloid differentiation. CARM1 inhibitors have been reported in preclinical studies from Epizyme and Takeda, but these have yet to enter the clinic [109, 110]. For example, depletion of CARM1 reduced the leukemic cell burden in an AML mouse model [98] and pharmacologic inhibition with the CARM1 inhibitor EZM2302 increased survival of AML-transplanted mice [111]. EZM2302 also demonstrated antiproliferative activity of lymphoma cells in vitro and inhibited growth of multiple myeloma tumors in a mouse xenograft model [109].
PRMT5 is overexpressed or upregulated in leukemia, lymphoma, and solid tumors, including glioblastoma and lung cancers [96, 112]. It is the major type II enzyme catalyzing symmetric arginine dimethylation of proteins involved in pathways known to be dysregulated in cancer, including transcription, cell signaling, mRNA translation, DNA damage, receptor trafficking, protein stability, and pre-mRNA splicing [113]. In a colony-forming assay, PRMT5 inactivation inhibited the abilities of cyclin D1, c-MYC, NOTCH1, and MLL-AF9 to induce neoplastic growth [114]. Loss of PRMT5 in primary glioblastoma cells and cell lines resulted in cell-cycle arrest, p53-independent apoptosis, and reduced migration capacity [115]. Genetic knockdown in non-small cell lung carcinoma (NSCLC) cells demonstrated antiproliferative activity that was not rescued by catalytically inactive protein, and it reduced tumor growth in an orthotopic lung cancer mouse model [116]. Taken together, these results suggest that PRMT5 is a potential therapeutic target for many types of cancers.
Preclinical studies have shown the potential for pharmacologic inhibition of PRMT5 in the treatment of cancer. EPZ015666 (GSK3235025), a potent orally bioavailable PRMT5 inhibitor, demonstrated antiproliferation and antitumor activity in vitro and in vivo in mantle cell lymphoma [117] and glioblastoma [118]. The orally bioavailable PRMT5 inhibitor JNJ-64619178 demonstrated tumor regression in a SCLC xenograft model [119] and has since entered clinical development (NCT03573310). PF-06939999 is a potent and selective orally bioavailable PRMT5 inhibitor with antitumor activity in leukemia and pancreatic cancer xenografts [120]. LLY-283 is a potent and selective PRMT5 inhibitor with demonstrated antitumor effects in a mouse AML xenograft tumor model [121]. CTx-034, developed by a partnership between Merck and Cancer Research Technology (CRT), is an orally bioavailable and potent PRMT5 inhibitor. In mice, administration of CTx-034 was associated with a dose-dependent reduction of the H4R3me2s mark, without a significant reduction in body weight or hematologic changes [122]. Thus far, three PRMT5 inhibitors, GSK3326595 (EPZ015938), JNJ-6461978, and PF-06939999, have entered into clinical studies in patients with hematologic malignancies and solid tumors. A presentation at the European Society for Medical Oncology congress reported data from the METEOR-1 study (NCT02783300) for GSK3326595 in advanced solid tumors [123]. Patients (n = 54) received doses of 12.5 mg to 600 mg once daily, and from 50 mg to 200 mg twice daily, with a median time on treatment of 1.8 months (range 1 day to 18.7 months). Grade 3 and 4 adverse events were reported for 35% of patients, and stable disease was reported for 19 patients. Partial responses were observed in patients with human papillomavirus–positive cervical cancer (1 response/1 subject) and adenoid cystic carcinoma (ACC) (3 responses/14 subjects); the responses in ACC were observed at the 200-mg dose. A summary of PRMT inhibitors currently in clinical trials is shown in Table 2.
Table 2.
PRMT inhibitors in clinical trials
| Molecules | Cancer indications in clinical studies | Clinical phase | Identifier |
|---|---|---|---|
| PRMT1 inhibitor | |||
| GSK3368715 | r/r DLBCL, r/r solid tumors | 1 | NCT03666988 |
| PRMT5 inhibitor | |||
| GSK3326595 | MDS, AML | 1/2 | NCT03614728a |
| r/r solid tumors, NHL | 1 | NCT02783300 | |
| JNJ-64619178 | r/r B cell NHL, advanced solid tumors | 1 | NCT03573310 |
| PF-06939999 | Advanced or metastatic solid tumors | 1 | NCT03854227 |
AML acute myeloid lymphoma, DLBCL diffuse large B cell lymphoma, MDS myelodysplastic syndrome, NHL non-Hodgkin’s lymphoma, PRMT protein arginine methyltransferases, r/r relapsed/refractory
aIn combination with azacitidine and best available care
Research in this field has uncovered potential genetic biomarkers of PRMT5 response. Homozygous deletion of the methylthioadenosine phosphorylase (MTAP) gene occurs in many cancer cells because of its location in proximity to CDKN2A, a tumor-suppressor gene often deleted in cancers. Viability of MTAP-null lines was hampered by shRNA-based inhibition of PRMT5 and its cofactor WDR77 compared with MTAP-positive cells [124], though treatment with the inhibitor EPZ015666 did not demonstrate a significant difference in sensitivity between MTAP-null or wild-type cell lines [125, 126]. Similarly, the TMPRSS2:ERG fusion gene is commonly found in AR-positive prostate cancer, and PRMT5 shRNA robustly inhibited the growth of AR-positive prostate cancer cells carrying the ERG fusion gene, whereas no or only minor effect of growth inhibition was observed in ERG fusion-negative cells [127]. GSK3235025-sensitive glioblastoma cell lines demonstrated an altered expression ratio of two PRMT5 cofactors, CLNS1A and RIOK1 [118, 128]. Additionally, AML cell lines with mutations in splicing factors (SF3B1, SRSF2, and U2AF1) demonstrated enhanced sensitivity to GSK591, the first chemical probe of PRMT5 co-developed by Epizyme and GlaxoSmithKline [117, 129]. These results suggest potential biomarkers for preselecting patients who may respond to a PRMT5 inhibitor in clinical studies, though more research is warranted to further refine the responder hypothesis.
PRMT1 is responsible for up to 85% of protein arginine methylation reactions [130] and modifies H4R3, which recruits coactivator p300/CBP-associated factor complex, leading to additional histone acetylation [97, 130]. PRMT1 also methylates many non-histone proteins, including BRACA1, EGFR [112], and Ash2L, a component of H3K4 methyltransferase complexes [131]. Overexpression or upregulation of PRMT1 and its isoforms has been shown in breast cancer, ALL, NSCLC, lung cancer, and glioma [112]. PRMT1 plays an important role in hematologic malignancies [103] and solid tumors [132, 133], although just one PRMT1 inhibitor (GSK3368715) is currently in development. A recent publication reported strong antiproliferative activity for this molecule across a range of solid tumor and hematological models, and synergy with PRMT5 inhibition [134]. Deletion of the MTAP gene, which results in accumulation of the metabolite 2-methylthioadenosine, increased sensitivity to GSK3368715 in cell lines [134]. This finding suggests that MTAP status may be a useful biomarker for selecting patients that may respond best to GSK3368715. Recently, GSK3368715 entered phase 1 clinical trials for hematologic malignancies or solid tumors with increased incidence of MTAP deletion (NCT03666988). Table 3 summarizes various methyltransferase inhibitors under clinical investigation.
Table 3.
Overview of methyltransferase inhibitors under clinical investigation
| Target class | Clinical compounds | Manufacturers |
|---|---|---|
| Protein lysine methyltransferase inhibitors | ||
| DOT1L inhibitor | Pinometostat (EPZ-5676) | Epizyme |
| EZH2 inhibitors | Tazemetostat, CPI-0209, CPI-1205, PF-06821497, DS-3201, MAK683 | Epizyme, Constellation Pharma, Pfizer, Daiichi Sankyo, Novartis |
| G9a inhibitor | EZM8266 | Epizyme |
| Protein arginine methyltransferase inhibitors | ||
| PRMT5 inhibitors | GSK3326595, JNJ-64619178, PF-06939999 | GlaxoSmithKline, Johnson & Johnson, Pfizer |
| PRMT1 inhibitor | GSK3368715 | GlaxoSmithKline |
Emerging targets
G9a
G9a (EHMT2) belongs to the Suv39h family, and was initially identified as a histone methyltransferase that mediates H3K9 and H3K27 methylation in vitro [135]. Subsequent in vivo studies suggest that G9a is a major enzyme for mono- and dimethylation of H3 in euchromatic regions [136–138] and involved in trimethylation of H3K9 [139]. A structurally related methyltransferase, G9a-like protein (GLP/EHMT1), shares the same histone substrate specificities [140–142]. Like many other methyltransferases, G9a (EHMT2) modifies a number of non-histones, including p53 [143]. G9a and GLP (EHMT1) are involved in MYC-dependent repression, and G9a inhibition suppresses MYC-dependent tumor growth [144].
G9a is dysregulated in a variety of cancers, including leukemia and ovarian, lung, breast, and other cancers [145]. Pharmacologic inhibition of G9a with BIX-01294, the first-identified G9a inhibitor [146], repressed tumor growth in mouse xenograft models of hepatocellular carcinoma, oral squamous cell carcinoma, and breast cancer [147–149], although cellular toxicity [146] prevented clinical application. Subsequently, several derivatives of BIX-01294 were developed. UNC0638, a potent, selective inhibitor of G9a and glucagon-like peptide-1 (GLP-1), demonstrated a lower toxicity compared with BIX-01294 [150], with an antitumor effect in mouse xenograft models of pancreatic cancer and NSCLC [151, 152]. A-366, a G9a inhibitor structurally unrelated to BIX-01294, demonstrated a 10-fold higher potency for G9a compared with GLP-1 [153], and a lower cytotoxicity compared with other G9a inhibitors, with equivalent cellular activity on H3K9 dimethylation. A recently published report identified G9a as a possible new target for the treatment of bladder cancer [154]. Sustained responses in a mouse model of metastatic bladder cancer were observed with G9a/DNMT inhibition in combination with programmed death ligand 1 (PD-L1) blockade. The study also reported increased G9a levels in bladder cancer patients resistant to PD-L1 inhibition, suggesting potential clinical relevance of this combination approach [154].
Mixed Lineage Leukemia Protein Family
MLL1 is commonly associated with chromosomal translocations in childhood leukemias [155, 156]. In recent years, tumor exome sequencing studies have shown that MLL3 and MLL2 are mutated in a significant proportion of malignancies, particularly solid tumors. The MLL family (MLL1-4) encodes histone methyltransferases for H3K4 that play crucial roles in the regulation of development-related genes, including Hox loci. One of the functions of the MLL family of proteins is as a catalytic subunit of the MLL/COMPASS complex, responsible for mono-, di-, and trimethylation of H3K4 [157]. Although small-molecule inhibitors of the MLL complex catalytic function have not been developed, small-molecule protein–protein interaction inhibitors have been identified for proteins that comprise the MLL/COMPASS complex. Among COMPASS components, menin and WDR5 are of particular interest. Menin is a tumor-suppressor protein that is part of the MLL1/COMPASS or MLL2/COMPASS complex [157] that plays a critical role in H3K4 methylation of Hox loci [158]. WDR5 is a common component among all of MLL/COMPASS complexes, and interacts with a number of transcription factors, cofactors, and non-coding RNAs [159]. Like menin, WDR5 is essential for MLL’s methyltransferase activities.
The MLL family and their interaction partners are frequently dysregulated in numerous types of cancers. Menin plays important roles in MLL fusion-dependent proliferation, differentiation block, and leukemic transformation [160–162]. WDR5 is overexpressed in solid tumors and leukemia [163–165] and, in patients with leukemia, high WDR5 expression levels are associated with those with high-risk disease [166]. Both menin and WDR5 function as recruiters of MLL fusions to chromatin, and play central roles in leukemic transformation [167, 168], and are thus potential therapeutic targets, with preclinical research underway mostly in MLL-rearranged leukemia.
SMYD2 and SMYD3
A recent report found abundant expression of SMYD2 to be an independent biomarker for poor prognosis of patients with hepatocellular carcinoma, which might imply that SMYD2 overexpression is associated with a more aggressive phenotype [169]. Potent and selective SMYD2 inhibitors have been developed to interrogate this potentially important anticancer approach [170–172].
Expressed at high levels in a number of cancers, SMYD3 is a lysine methyltransferase associated with a poor prognosis [173–181]. A causative association between cancer development and SMYD3 function has been shown in Ras-driven lung and pancreatic adenocarcinomas [182]. Although SMYD3 may be involved in heart development [183] and early development [184], whole-body or organ-specific SMYD3 deletion in mice did not result in aberrant pathology [185]. On the basis of these observations, if developed for the treatment of cancer, one can speculate that SMYD3 inhibition may have few side effects [186–188].
Although there has been extensive preclinical development of small-molecule SMYD3 inhibitors, no clinical trials with these agents are in progress at the time of this writing.
SETD7
Increasing evidence suggests that SET domain-containing (lysine methyltransferase) 7 (SETD7) plays a critical role in a number of physiological and pathological processes, such as metabolism, immunity, and cancer [189]. Histone and non-histone substrates for SETD7 are involved in various distinct cellular operations, including regulation of the cell cycle, response to DNA damage, RNA polymerase II-dependent gene transcription, cell differentiation, and chromatin modulation [12, 190, 191]. Like most lysine methyltransferases, SETD7 contains a SET domain that is responsible for the transfer of a methyl group to a lysine residue [190, 192, 193]. Recently, SETD7 was identified as a potential therapeutic target to inhibit intestinal tumorigenesis, as it is required for Wnt-driven intestinal tumorigenesis and regeneration [194].
SETD8
Current evidence suggests that SETD8 is the only methyltransferase known to catalyze the monomethylation of histone H4 at lysine 20 [195]. Overexpression of SETD8 is observed in some cancers, including pancreatic and bladder cancers, NSCLC, SCLC, chronic myelogenous leukemia, and hepatocellular carcinoma [196]. Various inhibitors of SETD8 have been developed but none have entered into clinical trials at the time of this writing [197, 198].
Drug Resistance, Epigenetic Plasticity, and Anticancer Immunity
In 1976, Nowell proposed a well-known tumor evolution model, where the acquired genetic instability combined with clonal selection of tumor cells results in resistance to therapeutic intervention [199]. Inherent variability of cancer cells provides them with the plasticity to survive, even when under the treatment of efficient drugs [200]. Considering a much higher rate of epigenetic “errors” compared with genetic mutations, epigenetic regulators may play an important role in generating new cancer stem/progenitor-like cells as well as in developing cancer heterogeneity, both of which are fundamental mechanisms of developing drug resistance [8, 201].
A certain type of drug resistance caused by epigenetic dysregulation can be reversed by low doses of epigenetic modifiers [201]. Silencing through hypermethylation of the Schlafen 11 (SLFN11) gene is linked to resistance to platinum drugs [38, 202] and treatment of class I (romidepsin, entinostat), but not class II (ricolinostat) HDAC inhibitors increased SLFN11 expression [203]. A recent study found that combining an EZH2 inhibitor with standard cytotoxic therapies prevented emergence of acquired resistance and improved chemotherapeutic efficacy in chemoresistant and chemosensitive models of SCLC via upregulation of SLFN11 [38]. These findings suggest the possibility of applying epigenetic modifiers to evade drug resistance, and the study of various drug combinations is warranted in this context.
In addition to tumor intrinsic resistance mechanisms, epigenetic changes in cancer cells have also been associated with a variety of mechanisms mediating escape from immune surveillance [204–208]. These mechanisms include downregulation of cellular antigen presentation machinery, reduced expression of tumor-associated antigens, inhibition of proinflammatory cytokines, and altered expression of checkpoint proteins [209–213]. Epigenetic mechanisms have also been associated with altered function of immune cells and hence reduced antitumor immunity [205, 206].
Epigenetic reprogramming conferred by EZH2 dysregulation has been identified as a major mechanism of transcriptional repression of genes that confer immunogenicity to tumor cells. This is best exemplified by the role of EZH2 in transcriptional repression of genes involved in antigen-presenting pathways in cancer, and a pivotal role for EZH2-mediated H3K27me3 repressive marks in the maintenance of MHC-I silencing in MHC-I-deficient cancers was recently reported [209]. Interestingly, in neuroendocrine tumors including SCLC and neuroblastoma, EZH2 function appears to highjack a conserved developmental program in neural progenitor cells to potentially confer immune privilege via MHC-I downregulation. Genetic inhibition of EZH2 in a genetically engineered mouse model of SCLC led to MHC-I upregulation and immune-mediated tumor regression [209].
Wang et al. highlighted the critical role of EZH2 activity in mediating Treg cell function in anticancer immunity [214]. Selective upregulation of EZH2 was observed in tumor-infiltrating Treg cells but not in effector T cells and Treg cells in peripheral blood. Genetic deletion or pharmacological inhibition of EZH2 in vitro or in vivo destabilized FOXp3 expression in Treg cells thus promoting immune-mediated rejection of tumors in mouse syngeneic models [214]. These results were further confirmed in a study demonstrating that genetic depletion of EZH2 in Treg cells or EZH2 catalytic inhibition using CPI-1205 elicited phenotypic and functional alterations to Treg cells, along with enhanced activation of effector T cells leading to robust antitumor immunity [215]. However, EZH2 inhibition has also been associated with reduced long-term survival of effector T cells [216, 217], along with induction of immunosuppressive myeloid-derived suppressor cells [218], highlighting the need for further evaluation of its diverse role in mediating immune cell function. Collectively, EZH2-mediated epigenetic programs are essential for the lineage commitment as well as the functional immune suppressive activity of Treg cells, along with tumor escape from immune surveillance via downregulation of the antigen presentation pathway, providing a rationale for exploring combination therapies of EZH2 inhibitors with immune checkpoint inhibitors.
Future Perspective and Challenges
Although a number of epigenetic modifiers have demonstrated promising results in preclinical studies, progressing them for solid tumors has been challenging, and careful consideration of drug combinations may affect their future clinical utility in this respect. A lack of predictive biomarkers has also made progress difficult. However, some traction is being made. For example, recent data suggest that SMARCA2 expression could be predictive for EZH2-targeted approaches in the context of SWI/SNF mutant cancers [66]. It was shown that EZH2 inhibition is effective in SMARCA4 mutant cancers that concurrently transcriptionally silence the paralog helicase SMARCA2. Cellular sensitivity to EZH2 inhibition in SMARCA4 mutant cancer models was associated with lower expression of SMARCA2, independent of tissue derivation.
The combination of epigenetic modifiers with immunotherapy is evolving as another area of great interest, with numerous clinical trials underway in both solid and liquid tumors across a range of epigenetic targets. Other approaches under clinical investigation include combination with treatments such as standard chemotherapy, radiotherapy, angiogenesis inhibition, epidermal growth factor receptor inhibition, and hormone therapy. Methyltransferase inhibitors in combination with checkpoint blockade can play an important role in inhibiting the suppressive function of Treg cells [215], for example, and counter resistance mechanisms that limit the effectiveness of checkpoint inhibitors. As mentioned above, a number of EZH2 inhibitors have now entered into clinical trials, including both CPI-205 and tazemetostat in combination with checkpoint blockade.
Conclusions
The modulation of epigenetic targets in the treatment of cancer is proving to be a rich area for research, with a plethora of novel epigenetic modifiers in development, some of which are showing early signs of activity in clinical trials. Histone methyltransferase inhibitors are a promising new approach for targeting epigenetic dysregulation in cancer and the recent approval of tazemetostat leads the way for future advances in this field. However, the most optimal approach to using such drugs, including patient selection, the identification of biomarkers, as well as the correct drug combinations, is an area of active research with still many unanswered questions. The outcomes of various ongoing clinical trials will no doubt provide the answers to some of these questions and pave the way to providing novel treatments for patients with an unmet need.
Acknowledgements
Funding
This review, the journal’s Rapid Service and Open Access Fees were funded by Pfizer Inc.
Medical Writing Assistance
Medical writing support was provided by David Sunter, PhD, CMPP™ of Engage Scientific Solutions and was funded by Pfizer Inc.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Gabriel G. Malouf is a consultant for Pfizer, Ipsen, BMS, Astellas; he has received travel support from Pfizer, Ipsen, BMS, MSD and Astellas; Hope S. Rugo has received research support to the University of California San Francisco from Immunomedics, Daiichi Sankyo, Eisai, Eli Lilly, Genentech, including F. Hoffmann-La Roche, Ltd, MacroGenics, Merck, Novartis, OBI, and Pfizer, and travel support from Daiichi Sankyo, Genentech, including F. Hoffmann-La Roche, Ltd, MacroGenics, Mylan, Novartis, Pfizer, and PUMA; Kristen Jensen-Pergakes, Ira Jacobs, Shikhar Sharma, Frank Scappaticci, and Thomas A. Paul are employees of Pfizer and hold Pfizer stock.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
Digital Features
To view digital features for this article go to 10.6084/m9.figshare.12249719.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. [DOI] [PubMed]
- 2.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–113. [DOI] [PubMed]
- 3.McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 5.Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozek LS, Dolinoy DC, Sartor MA, Omenn GS. Epigenetics: relevance and implications for public health. Annu Rev Public Health. 2014;35:105–122. doi: 10.1146/annurev-publhealth-032013-182513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaffidi P, Misteli T. Cancer epigenetics: from disruption of differentiation programs to the emergence of cancer stem cells. Cold Spring Harb Symp Quant Biol. 2010;75:251–258. doi: 10.1101/sqb.2010.75.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 10.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Local A, Huang H, Albuquerque CP, et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat Genet. 2018;50:73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011;6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 14.Koch CM, Andrews RM, Flicek P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Wang GG. Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem Sci. 2013;38:546–555. doi: 10.1016/j.tibs.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 18.Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Q, Rank G, Tan YT, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fioravanti R, Stazi G, Zwergel C, Valente S, Mai A. Six years (2012–2018) of researches on catalytic EZH2 inhibitors: the boom of the 2-pyridone compounds. Chem Rec. 2018;18:1818–1832. doi: 10.1002/tcr.201800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JM, Lee JS, Kim H, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He A, Shen X, Ma Q, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCabe MT, Graves AP, Ganji G, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci USA. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yap DB, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 32.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 34.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato T, Kaneda A, Tsuji S, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammad F, Weissmann S, Leblanc B, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 38.Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryant RJ, Cross NA, Eaton CL, Hamdy FC, Cunliffe VT. EZH2 promotes proliferation and invasiveness of prostate cancer cells. Prostate. 2007;67:547–556. doi: 10.1002/pros.20550. [DOI] [PubMed] [Google Scholar]
- 40.Karanikolas BD, Figueiredo ML, Wu L. Comprehensive evaluation of the role of EZH2 in the growth, invasion, and aggression of a panel of prostate cancer cell lines. Prostate. 2010;70:675–688. doi: 10.1002/pros.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao L, Tien JC, Vo J, et al. Epigenetic reprogramming with antisense oligonucleotides enhances the effectiveness of androgen receptor inhibition in castration-resistant prostate cancer. Cancer Res. 2018;78:5731–5740. doi: 10.1158/0008-5472.CAN-18-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ku SY, Rosario S, Wang Y, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dardenne E, Beltran H, Benelli M, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim KH, Roberts CW. Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genet. 2014;207:365–372. doi: 10.1016/j.cancergen.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30:356–363. doi: 10.1016/j.tig.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jelinic P, Mueller JJ, Olvera N, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witkowski L, Carrot-Zhang J, Albrecht S, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–443. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- 48.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones DT, Jager N, Kool M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knutson SK, Warholic NM, Wigle TJ, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci USA. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan-Penebre E, Armstrong K, Drew A, et al. Selective killing of SMARCA2- and SMARCA4-deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of EZH2: in vitro and in vivo preclinical models. Mol Cancer Ther. 2017;16:850–860. doi: 10.1158/1535-7163.MCT-16-0678. [DOI] [PubMed] [Google Scholar]
- 54.LaFave LM, Beguelin W, Koche R, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21:1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stazi G, Zwergel C, Mai A, Valente S. EZH2 inhibitors: a patent review (2014–2016) Expert Opin Ther Pat. 2017;27:797–813. doi: 10.1080/13543776.2017.1316976. [DOI] [PubMed] [Google Scholar]
- 56.Yap TA, Winter JN, Giulino-Roth L, et al. Phase 1 study of the novel enhancer of zeste homolog 2 (EZH2) inhibitor GSK2816126 in patients with advanced hematological and solid tumors. Clin Cancer Res. 2019;25:7331–7339. doi: 10.1158/1078-0432.CCR-18-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.US Food and Drug Administration. FDA approves first treatment option specifically for patients with epithelioid sarcoma, a rare soft tissue cancer. 2020. https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-option-specifically-patients-epithelioid-sarcoma-rare-soft-tissue. Accessed Mar 5, 2020.
- 58.Knutson SK, Kawano S, Minoshima Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 59.Kawano S, Grassian AR, Tsuda M, et al. Preclinical evidence of anti-tumor activity induced by EZH2 inhibition in human models of synovial sarcoma. PLoS One. 2016;11:e0158888. doi: 10.1371/journal.pone.0158888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zauderer MG, Szlosarek P, Le Moulec S, et al. Phase 2, multicenter study of the EZH2 inhibitor tazemetostat as monotherapy in adults with relapsed or refractory (R/R) malignant mesothelioma (MM) with BAP1 inactivation. J Clin Oncol. 2018;36(15_suppl):8515.
- 61.Italiano A, Soria JC, Toulmonde M, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 62.Morschhauser F, Tilly H, Chaidos A, et al. Interim update from a phase 2 multicenter study of tazemetostat, an EZH2 inhibitor, in patients with relapsed or refractory follicular lymphoma. Hematol Oncol. 2019;37:154–156. [Google Scholar]
- 63.Ribrag V, Morschhauser F, McKay P, et al. Interim results from an ongoing phase 2 multicenter study of tazemetostat, an EZH2 inhibitor, in patients with relapsed or refractory (R/R) diffuse large B-cell lymphoma (DLBCL) Blood. 2018;132:4196. [Google Scholar]
- 64.Biospace. Epizyme reports updated data from phase 2 trial of tazemetostat in patients with relapsed or refractory follicular lymphoma. 2019. https://www.biospace.com/article/releases/epizyme-reports-updated-data-from-phase-2-trial-of-tazemetostat-in-patients-with-relapsed-or-refractory-follicular-lymphoma/. Accessed Mar 5, 2020.
- 65.US Food and Drug Administration. FDA approves tazemetostat for advanced epithelioid sarcoma. 2020. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-tazemetostat-advanced-epithelioid-sarcoma. Accessed Mar 5, 2020.
- 66.Januario T, Ye X, Bainer R, et al. PRC2-mediated repression of SMARCA2 predicts EZH2 inhibitor activity in SWI/SNF mutant tumors. Proc Natl Acad Sci USA. 2017;114:12249–12254. doi: 10.1073/pnas.1703966114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kung PP, Bingham P, Brooun A, et al. Optimization of orally bioavailable enhancer of zeste homolog 2 (EZH2) inhibitors using ligand and property-based design strategies: identification of development candidate (R)-5,8-dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497) J Med Chem. 2018;61:650–665. doi: 10.1021/acs.jmedchem.7b01375. [DOI] [PubMed] [Google Scholar]
- 68.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 69.Yap TA, Johnson PWM, Winter J, et al. A phase I, open-label study of GSK2816126, an enhancer of zeste homolog 2 (EZH2) inhibitor, in patients with relapsed/refractory diffuse large B-cell lymphoma (DLBCL), transformed follicular lymphoma (tFL), other non-Hodgkin’s lymphomas (NHL), multiple myeloma (MM) and solid tumor. J Clin Oncol. 2016;34(15 suppl):TPS2595-TPS.
- 70.Vaswani RG, Gehling VS, Dakin LA, et al. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J Med Chem. 2016;59:9928–9941. doi: 10.1021/acs.jmedchem.6b01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harb W, Abramson J, Lunning M, et al. A phase 1 study of CPI-1205, a small molecule inhibitor of EZH2, preliminary safety in patients with B-cell lymphomas. Ann Oncol. 2018;29(Suppl 3):iii7–iii9. 10.1093/annonc/mdy048.001.
- 72.Taplin M-E, Hussain A, Shore ND, et al. A phase 1b/2 study of CPI-1205, a small molecule inhibitor of EZH2, combined with enzalutamide (E) or abiraterone/prednisone (A/P) in patients with metastatic castration resistant prostate cancer (mCRPC). J Clin Oncol. 2018;36(6 suppl):TPS398-TPS.
- 73.Constellation Pharmaceuticals. Constellation Pharmaceuticals presents results from phase 1b portion of ProSTAR clinical trial of CPI-1205 at AACR Meeting [press release]. 2019. https://www.globenewswire.com/news-release/2019/04/01/1791015/0/en/Constellation-Pharmaceuticals-Presents-Results-from-Phase-1b-Portion-of-ProSTAR-Clinical-Trial-of-CPI-1205-at-AACR-Meeting.html. Accessed July 29, 2019.
- 74.Gibaja V, Shen F, Harari J, et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene. 2016;35:558–566. doi: 10.1038/onc.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi W, Zhao K, Gu J, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol. 2017;13:381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 76.He Y, Selvaraju S, Curtin ML, et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat Chem Biol. 2017;13:389–395. doi: 10.1038/nchembio.2306. [DOI] [PubMed] [Google Scholar]
- 77.Li L, Zhang H, Zhang M, et al. Discovery and molecular basis of a diverse set of polycomb repressive complex 2 inhibitors recognition by EED. PLoS One. 2017;12:e0169855. doi: 10.1371/journal.pone.0169855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lingel A, Sendzik M, Huang Y, et al. Structure-guided design of EED binders allosterically inhibiting the epigenetic polycomb repressive complex 2 (PRC2) methyltransferase. J Med Chem. 2017;60:415–427. doi: 10.1021/acs.jmedchem.6b01473. [DOI] [PubMed] [Google Scholar]
- 79.Huang Y, Zhang J, Yu Z, et al. Discovery of first-in-class, potent, and orally bioavailable embryonic ectoderm development (EED) inhibitor with robust anticancer efficacy. J Med Chem. 2017;60:2215–2226. doi: 10.1021/acs.jmedchem.6b01576. [DOI] [PubMed] [Google Scholar]
- 80.Neff T, Sinha AU, Kluk MJ, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci USA. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim W, Bird GH, Neff T, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Honma D, Kanno O, Watanabe J, et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 2017;108:2069–2078. doi: 10.1111/cas.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Honma D, Nosaka E, Shiroishi M, et al. DS-3201, a potent EZH1/2 dual inhibitor, demonstrates antitumor activity against non-Hodgkin lymphoma (NHL) regardless of EZH2 mutation. Blood. 2018;132(suppl 1):2217. [Google Scholar]
- 84.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 85.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 86.Ntziachristos P, Tsirigos A, Van Vlierberghe P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 88.Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 89.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 90.Chang MJ, Wu H, Achille NJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 92.McLean CM, Karemaker ID, van Leeuwen F. The emerging roles of DOT1L in leukemia and normal development. Leukemia. 2014;28:2131–2138. doi: 10.1038/leu.2014.169. [DOI] [PubMed] [Google Scholar]
- 93.Daigle SR, Olhava EJ, Therkelsen CA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stein EM, Garcia-Manero G, Rizzieri DA, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131:2661–2669. doi: 10.1182/blood-2017-12-818948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shukla N, Wetmore C, O’Brien MM, et al. Final report of phase 1 study of the DOT1L inhibitor, pinometostat (EPZ-5676), in children with relapsed or refractory MLL-r acute leukemia. Blood. 2016;128(22):2780.
- 96.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 97.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vu LP, Perna F, Wang L, et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 2013;5:1625–1638. doi: 10.1016/j.celrep.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng H, Qin Y, Fan H, et al. Overexpression of CARM1 in breast cancer is correlated with poorly characterized clinicopathologic parameters and molecular subtypes. Diagn Pathol. 2013;8:129. doi: 10.1186/1746-1596-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hong H, Kao C, Jeng M-H, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–89. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 101.Osada S, Suzuki S, Yoshimi C, et al. Elevated expression of coactivator-associated arginine methyltransferase 1 is associated with early hepatocarcinogenesis. Oncol Rep. 2013;30:1669–1674. doi: 10.3892/or.2013.2651. [DOI] [PubMed] [Google Scholar]
- 102.Cheng D, Vemulapalli V, Lu Y, et al. CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci Alliance. 2018;1:e201800117. doi: 10.26508/lsa.201800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Greenblatt SM, Liu F, Nimer SD. Arginine methyltransferases in normal and malignant hematopoiesis. Exp Hematol. 2016;44:435–441. doi: 10.1016/j.exphem.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 104.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu Q, Bruce AW, Jedrusik A, et al. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 107.Kim J, Lee J, Yadav N, et al. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279:25339–25344. doi: 10.1074/jbc.M402544200. [DOI] [PubMed] [Google Scholar]
- 108.Yadav N, Cheng D, Richard S, et al. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9:193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Drew AE, Moradei O, Jacques SL, et al. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep. 2017;7:17993. doi: 10.1038/s41598-017-18446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakayama K, Szewczyk MM, Dela Sena C, et al. TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget. 2018;9:18480–18493. doi: 10.18632/oncotarget.24883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Greenblatt SM, Man N, Hamard PJ, et al. CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis. Cancer Cell. 2018;33(1111–27):e5. doi: 10.1016/j.ccell.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poulard C, Corbo L, Le Romancer M. Protein arginine methylation/demethylation and cancer. Oncotarget. 2016;7:67532–67550. doi: 10.18632/oncotarget.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Y, Chitnis N, Nakagawa H, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5:288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan F, Alinari L, Lustberg ME, et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74:1752–1765. doi: 10.1158/0008-5472.CAN-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gu Z, Gao S, Zhang F, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J. 2012;446:235–241. doi: 10.1042/BJ20120768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chan-Penebre E, Kuplast KG, Majer CR, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 118.Braun CJ, Stanciu M, Boutz PL, et al. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell. 2017;32(411):26.e11. doi: 10.1016/j.ccell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brehmer D, Wu T, Mannens G, et al. Abstract DDT02-04: a novel PRMT5 inhibitor with potent in vitro and in vivo activity in preclinical lung cancer models. Cancer Res. 2017;77:DDT02-4.
- 120.Patman R. Design and synthesis of carbonucleoside inhibitors of PRMT5 for the treatment of cancer: a desymmetrization approach. In: Medicinal Chemistry Gordon Research Conference; 7 Aug 2019; New London.
- 121.Bonday ZQ, Cortez GS, Grogan MJ, et al. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med Chem Lett. 2018;9:612–617. doi: 10.1021/acsmedchemlett.8b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Falk H, Foitzik RC, Allan E, et al. Abstract 5371: PRMT5 inhibitors as novel treatment for cancers. Cancer Res. 2015;75:5371. [Google Scholar]
- 123.Siu LL, Rasco DW, Vinay SP, et al. METEOR-1: a phase I study of GSK3326595, a first-in-class protein arginine methyltransferase 5 (PRMT5) inhibitor, in advanced solid tumours. Ann Oncol. 2019;30(Suppl 5):v159–v193. 10.1093/annonc/mdz244.
- 124.Kryukov GV, Wilson FH, Ruth JR, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mavrakis KJ, McDonald ER, Schlabach MR, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 126.Marjon K, Cameron MJ, Quang P, et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 127.Mounir Z, Korn JM, Westerling T, et al. ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the androgen receptor. Elife. 2016;5:e13964. [DOI] [PMC free article] [PubMed]
- 128.Guderian G, Peter C, Wiesner J, et al. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J Biol Chem. 2011;286:1976–1986. doi: 10.1074/jbc.M110.148486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fong JY, Pignata L, Goy PA, et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell. 2019;36(194–209):e9. doi: 10.1016/j.ccell.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li X, Wang C, Jiang H, Luo C. A patent review of arginine methyltransferase inhibitors (2010–2018) Expert Opin Ther Pat. 2019;29:97–114. doi: 10.1080/13543776.2019.1567711. [DOI] [PubMed] [Google Scholar]
- 131.Butler JS, Zurita-Lopez CI, Clarke SG, Bedford MT, Dent SY. Protein-arginine methyltransferase 1 (PRMT1) methylates Ash2L, a shared component of mammalian histone H3K4 methyltransferase complexes. J Biol Chem. 2011;286:12234–12244. doi: 10.1074/jbc.M110.202416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Avasarala S, Van Scoyk M, Karuppusamy Rathinam MK, et al. PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J Biol Chem. 2015;290:13479–13489. doi: 10.1074/jbc.M114.636050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liao H-W, Hsu J-M, Xia W, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125:4529–4543. doi: 10.1172/JCI82826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fedoriw A, Rajapurkar SR, O’Brien S, et al. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell. 2019;36(100):14.e25. doi: 10.1016/j.ccell.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 135.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 136.Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Peters AH, Kubicek S, Mechtler K, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 138.Rice JC, Briggs SD, Ueberheide B, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 139.Yokochi T, Poduch K, Ryba T, et al. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 141.Tachibana M, Ueda J, Fukuda M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weiss T, Hergeth S, Zeissler U, et al. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenet Chromatin. 2010;3:7. doi: 10.1186/1756-8935-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tu WB, Shiah YJ, Lourenco C, et al. MYC interacts with the G9a histone methyltransferase to drive transcriptional repression and tumorigenesis. Cancer Cell. 2018;34(579):95.e8. doi: 10.1016/j.ccell.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 145.Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol. 2015;6:487. doi: 10.3389/fimmu.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kubicek S, O’Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 147.Yokoyama M, Chiba T, Zen Y, et al. Histone lysine methyltransferase G9a is a novel epigenetic target for the treatment of hepatocellular carcinoma. Oncotarget. 2017;8:21315–21326. doi: 10.18632/oncotarget.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dong C, Wu Y, Yao J, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ren A, Qiu Y, Cui H, Fu G. Inhibition of H3K9 methyltransferase G9a induces autophagy and apoptosis in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2015;459:10–17. doi: 10.1016/j.bbrc.2015.01.068. [DOI] [PubMed] [Google Scholar]
- 150.Vedadi M, Barsyte-Lovejoy D, Liu F, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pan MR, Hsu MC, Luo CW, Chen LT, Shan YS, Hung WC. The histone methyltransferase G9a as a therapeutic target to override gemcitabine resistance in pancreatic cancer. Oncotarget. 2016;7:61136–61151. doi: 10.18632/oncotarget.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang K, Wang J, Yang L, et al. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1alpha and APC2 gene expression in non-small cell lung cancer. Mol Cancer. 2018;17:153. doi: 10.1186/s12943-018-0896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sweis RF, Pliushchev M, Brown PJ, et al. Discovery and development of potent and selective inhibitors of histone methyltransferase G9a. ACS Med Chem Lett. 2014;5:205–209. doi: 10.1021/ml400496h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Segovia C, San Jose-Eneriz E, Munera-Maravilla E, et al. Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat Med. 2019;25:1073–1081. doi: 10.1038/s41591-019-0499-y. [DOI] [PubMed] [Google Scholar]
- 155.Ford DJ, Dingwall AK. The cancer COMPASS: navigating the functions of MLL complexes in cancer. Cancer Genet. 2015;208:178–191. doi: 10.1016/j.cancergen.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 156.Ziemin-van der Poel S, McCabe NR, Gill HJ, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–739. [DOI] [PMC free article] [PubMed]
- 157.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang P, Lin C, Smith ER, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lu K, Tao H, Si X, Chen Q. The histone H3 lysine 4 presenter WDR5 as an oncogenic protein and novel epigenetic target in cancer. Front Oncol. 2018;8:502. doi: 10.3389/fonc.2018.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 161.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen YX, Yan J, Keeshan K, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci USA. 2006;103:1018–1023. doi: 10.1073/pnas.0510347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen X, Xie W, Gu P, et al. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci Rep. 2015;5:8293. doi: 10.1038/srep08293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun Y, Bell JL, Carter D, et al. WDR5 supports an N-Myc transcriptional complex that drives a protumorigenic gene expression signature in neuroblastoma. Cancer Res. 2015;75:5143–5154. doi: 10.1158/0008-5472.CAN-15-0423. [DOI] [PubMed] [Google Scholar]
- 165.Kim JY, Banerjee T, Vinckevicius A, et al. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol Cell. 2014;54:613–625. doi: 10.1016/j.molcel.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ge Z, Song EJ, Kawasawa YI, Li J, Dovat S, Song C. WDR5 high expression and its effect on tumorigenesis in leukemia. Oncotarget. 2016;7:37740–37754. doi: 10.18632/oncotarget.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu J, Li L, Xiong J, et al. MLL1 and MLL1 fusion proteins have distinct functions in regulating leukemic transcription program. Cell Discov. 2016;2:16008. doi: 10.1038/celldisc.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zuo SR, Zuo XC, He Y, et al. Positive expression of SMYD2 is associated with poor prognosis in patients with primary hepatocellular carcinoma. J Cancer. 2018;9:321–330. doi: 10.7150/jca.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cowen SD, Russell D, Dakin LA, et al. Design, synthesis, and biological activity of substrate competitive SMYD2 inhibitors. J Med Chem. 2016;59:11079–11097. doi: 10.1021/acs.jmedchem.6b01303. [DOI] [PubMed] [Google Scholar]
- 171.Eggert E, Hillig RC, Koehr S, et al. Discovery and characterization of a highly potent and selective aminopyrazoline-based in vivo probe (BAY-598) for the protein lysine methyltransferase SMYD2. J Med Chem. 2016;59:4578–4600. doi: 10.1021/acs.jmedchem.5b01890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nguyen H, Allali-Hassani A, Antonysamy S, et al. LLY-507, a cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J Biol Chem. 2015;290:13641–13653. doi: 10.1074/jbc.M114.626861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vieira FQ, Costa-Pinheiro P, Almeida-Rios D, et al. SMYD3 contributes to a more aggressive phenotype of prostate cancer and targets cyclin D2 through H4K20me3. Oncotarget. 2015;6:13644–13657. doi: 10.18632/oncotarget.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu Y, Liu H, Luo X, Deng J, Pan Y, Liang H. Overexpression of SMYD3 and matrix metalloproteinase-9 are associated with poor prognosis of patients with gastric cancer. Tumour Biol. 2015;36:4377–4386. doi: 10.1007/s13277-015-3077-z. [DOI] [PubMed] [Google Scholar]
- 175.Vieira FQ, Costa-Pinheiro P, Ramalho-Carvalho J, et al. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr Relat Cancer. 2014;21:51–61. doi: 10.1530/ERC-13-0375. [DOI] [PubMed] [Google Scholar]
- 176.Sponziello M, Durante C, Boichard A, et al. Epigenetic-related gene expression profile in medullary thyroid cancer revealed the overexpression of the histone methyltransferases EZH2 and SMYD3 in aggressive tumours. Mol Cell Endocrinol. 2014;392:8–13. doi: 10.1016/j.mce.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 177.Liu C, Wang C, Wang K, et al. SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J Natl Cancer Inst. 2013;105:1719–1728. doi: 10.1093/jnci/djt304. [DOI] [PubMed] [Google Scholar]
- 178.Van Aller GS, Reynoird N, Barbash O, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hamamoto R, Furukawa Y, Morita M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 180.Hamamoto R, Silva FP, Tsuge M, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu Y, Luo X, Deng J, Pan Y, Zhang L, Liang H. SMYD3 overexpression was a risk factor in the biological behavior and prognosis of gastric carcinoma. Tumour Biol. 2015;36:2685–2694. doi: 10.1007/s13277-014-2891-z. [DOI] [PubMed] [Google Scholar]
- 182.Mazur PK, Reynoird N, Khatri P, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fujii T, Tsunesumi S, Yamaguchi K, Watanabe S, Furukawa Y. Smyd3 is required for the development of cardiac and skeletal muscle in zebrafish. PLoS One. 2011;6:e23491. doi: 10.1371/journal.pone.0023491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T, Imai H, Minami N. Histone methyltransferase Smyd3 regulates early embryonic lineage commitment in mice. Reproduction. 2015;150:21–30. doi: 10.1530/REP-15-0019. [DOI] [PubMed] [Google Scholar]
- 185.Sarris ME, Moulos P, Haroniti A, Giakountis A, Talianidis I. SMYD3 is a transcriptional potentiator of multiple cancer-promoting genes and required for liver and colon cancer development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 186.Mitchell LH, Boriack-Sjodin PA, Smith S, et al. Novel oxindole sulfonamides and sulfamides: EPZ031686, the first orally bioavailable small molecule SMYD3 inhibitor. ACS Med Chem Lett. 2016;7:134–138. doi: 10.1021/acsmedchemlett.5b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Van Aller GS, Graves AP, Elkins PA, et al. Structure-based design of a novel SMYD3 inhibitor that bridges the SAM-and MEKK2-binding pockets. Structure. 2016;24:774–781. doi: 10.1016/j.str.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 188.Judge RA, Zhu H, Upadhyay AK, et al. Turning a substrate peptide into a potent inhibitor for the histone methyltransferase SETD8. ACS Med Chem Lett. 2016;7:1102–1106. doi: 10.1021/acsmedchemlett.6b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Batista IAA, Helguero LA. Biological processes and signal transduction pathways regulated by the protein methyltransferase SETD7 and their significance in cancer. Signal Transduct Target Ther. 2018;3:19. doi: 10.1038/s41392-018-0017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pradhan S, Chin HG, Esteve PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tao Y, Neppl RL, Huang ZP, et al. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3:39. doi: 10.1186/2045-3701-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Oudhoff MJ, Braam MJS, Freeman SA, et al. SETD7 controls intestinal regeneration and tumorigenesis by regulating Wnt/beta-catenin and Hippo/YAP signaling. Dev Cell. 2016;37:47–57. doi: 10.1016/j.devcel.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 195.Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Takawa M, Cho HS, Hayami S, et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012;72:3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- 197.Milite C, Feoli A, Viviano M, et al. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin Epigenet. 2016;8:102. doi: 10.1186/s13148-016-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ma A, Yu W, Li F, et al. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase SETD8. J Med Chem. 2014;57:6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 200.Davies AE, Albeck JG. Microenvironmental signals and biochemical information processing: cooperative determinants of intratumoral plasticity and heterogeneity. Front Cell Dev Biol. 2018;6:44. doi: 10.3389/fcell.2018.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nogales V, Reinhold WC, Varma S, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084–3097. doi: 10.18632/oncotarget.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Murai J, Tang SW, Leo E, et al. SLFN11 blocks stressed replication forks independently of ATR. Mol Cell. 2018;69(371):84.e6. doi: 10.1016/j.molcel.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Scott-Browne JP, Lopez-Moyado IF, Trifari S, et al. Dynamic changes in chromatin accessibility occur in CD8(+) T cells responding to viral infection. Immunity. 2016;45:1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sen DR, Kaminski J, Barnitz RA, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Philip M, Fairchild L, Sun L, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Corces MR, Granja JM, Shams S, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362(6413):eaav1898. [DOI] [PMC free article] [PubMed]
- 208.Loo Yau H, Ettayebi I, De Carvalho DD. The cancer epigenome: exploiting its vulnerabilities for immunotherapy. Trends Cell Biol. 2019;29:31–43. doi: 10.1016/j.tcb.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 209.Burr ML, Sparbier CE, Chan KL, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019;36(385):401.e8. doi: 10.1016/j.ccell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Peng D, Kryczek I, Nagarsheth N, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.McGranahan N, Rosenthal R, Hiley CT, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171(1259):71.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sade-Feldman M, Jiao YJ, Chen JH, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8:1136. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gounder MM, Zhu G, Roshal L, et al. Immunologic correlates of the abscopal effect in a SMARCB1/INI1-negative poorly differentiated chordoma after EZH2 inhibition and radiotherapy. Clin Cancer Res. 2019;25:2064–2071. doi: 10.1158/1078-0432.CCR-18-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang X, Brea LT, Yu J. Immune modulatory functions of EZH2 in the tumor microenvironment: implications in cancer immunotherapy. Am J Clin Exp Urol. 2019;7:85–91. [PMC free article] [PubMed] [Google Scholar]
- 215.Goswami S, Apostolou I, Zhang J, et al. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J Clin Invest. 2018;128:3813–3818. doi: 10.1172/JCI99760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.He S, Wang J, Kato K, et al. Inhibition of histone methylation arrests ongoing graft-versus-host disease in mice by selectively inducing apoptosis of alloreactive effector T cells. Blood. 2012;119:1274–1282. doi: 10.1182/blood-2011-06-364422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang Y, Kinkel S, Maksimovic J, et al. The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood. 2014;124:737–749. doi: 10.1182/blood-2013-12-544106. [DOI] [PubMed] [Google Scholar]
- 218.Huang S, Wang Z, Zhou J, et al. EZH2 inhibitor GSK126 suppresses antitumor immunity by driving production of myeloid-derived suppressor cells. Cancer Res. 2019;79:2009. doi: 10.1158/0008-5472.CAN-18-2395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.