Abstract
We introduce Goslin, a polyglot grammar for common lipid shorthand nomenclatures based on the LIPID MAPS nomenclature and the shorthand nomenclature established by Liebisch and coauthors and used by LipidHome and SwissLipids. Goslin was designed to address the following pressing issues in the lipidomics field: (1) to simplify the implementation of lipid name handling for developers of mass spectrometry-based lipidomics tools, (2) to offer a tool that unifies and normalizes the main existing lipid name dialects enabling a lipidomics analysis in a high-throughput fashion, and (3) to provide a consistent mapping from lipid shorthand names to lipid building blocks and structural properties. We provide implementations of Goslin in four major programming languages, namely, C++, Java, Python 3, and R to kick-start adoption and integration. Further, we set up a web service for users to work with Goslin directly. All implementations are available free of charge under a permissive open source license.
Lipids are, besides DNA/RNA, proteins, and metabolites, the most frequently occurring biomolecules in cells. Several lipid classes have a similar molecular structure. Therefore, lipid names were designed to represent their systematic structure in a clear and concise way to help classify and therefore distinguish lipids by name.1,2 Within the past 2 decades, a systematic shorthand notation for lipids has evolved, spurred by initial standardization approaches within the LIPID MAPS consortium.3,4
With the advent of high-resolution mass spectrometry (MS), as the key tool to investigate lipids, the hierarchical representation of lipids was extended to be able to report multiple levels of structural knowledge.5,6 Since mass spectrometry is currently the dominant technology for the identification and quantification of lipids,7,8 these notations are primarily designed to satisfy the requirements in that application area. Structural knowledge about lipids identified with MS can be represented as a hierarchical tree or table (see Table 1), as introduced by LipidHome9 and later refined by SwissLipids.10
Table 1. Hierarchical Classification of PE(16:1(6Z)/16:0)a.
| level | name | description |
|---|---|---|
| category (LM) | glycerophospholipids | lipid category |
| class (LM) | glycerophosphoethanolamine | lipid class |
| species (SL, LM subclass) | phosphatidylethanolamine (32:1), PE (32:1) | HG, FA summary |
| molecular subspecies (SL) | PE(16:0_16:1) | HG, two FAs |
| structural subspecies (SL) | PE(16:1/16:0) | HG, SN position for FA1 and FA2 |
| isomeric subspecies (SL, LM) | PE(16:1(6Z)/16:0) | HG, SN position, stereo |
Higher levels represent the lipid with less detail and more summarized information, e.g., the top most category level reports that PE(32:1) as a species is a glycerophospholipid. The species level expresses only the head group (HG), and the sum of no. of carbon atoms and no. of double bonds in both fatty acyls (FAs). Intermediate levels report additional details about FA composition (molecular subspecies) and stereospecific numbering (SN) position with respect to the head group (structural subspecies). At the lowest level, the FA composition (no. of carbon atoms, no. of double bonds), their SN position (first is SN1, second is SN2), and the position of the double bond are expressed (6th carbon in SN1 FA, in Z configuration). The “level” column details the mapping of Goslin’s levels to the respective levels in the LIPID MAPS (LM) and SwissLipids hierarchies (SL). Hydroxyl groups have been omitted from this example for brevity, but they are also fully supported by Goslin.
This tree is rooted at the category level (e.g., glycerophospholipids) and descends from class (e.g., glycerophosphoethanolamine) to species (e.g., phosphatidylethanolamine(32:1), PE(32:1)), to molecular subspecies (e.g., PE(16:0_16:1) with determined fatty acyl (FA) lengths and saturation), then to structural subspecies (e.g., PE(16:1/16:0) with defined stereospecific numbering (SN) positions for the FAs), and ultimately to isomeric level (e.g., PE(16:1(6Z)/16:0) or 1-(6Z-hexadecenoyl)-2-hexadecanoyl-sn-glycero-3- phosphoethanolamine). Using the shorthand abbreviations for the lipid classes (e.g., PE for phosphatidylethanolamine or SM for sphingomyelin) together with the shorthand notation for various variable building blocks, such as length of fatty acyl chains and long chain bases as well as the number of double bonds and hydroxylations, lipid names become both manageable and descriptive.
With the current developments in lipidomics and the upcoming high-throughput analyses, an increasing demand for automated computational data handling is on the horizon. Hence, it is absolutely essential that identified lipids are correctly and unambiguously named and stored for follow-up analyses during a computational lipid analysis workflow that may consist of several consecutive tools.
Lipid naming has however evolved into several dialects which complicates the unified computational treatment and parsing of lipid names. For instance, in the literature one may find for essentially the same lipid species any of the following names: Cer(d18:1/16:0), Cer (d18:1/16:0), d18:1/16:0 Cer, Cer 18:1;2/16:0, or CER[N(16)S(18)]. Although some of these lipid names may look very similar or differ perhaps only in one character, a simple string matching fails when these names are not exactly equal.
As a consequence, long and error-prone manual curation is necessary in order to streamline lists of lipid names for their processing in follow-up analysis scripts, workflows, tools, or for their submission to research data repositories. Improper annotation, misidentification, and over-reporting are recognized as some of the most common problems in lipidomics today, thus calling for tailored solutions to help mitigate their impact.11
Methods
To overcome these challenges, we developed the “Grammar of Succinct Lipid Nomenclature” (Goslin). It is designed to act as a library for the development of lipidomics tools providing a standardized data structure for storing structural lipid information. The parsing of lipid names as well as the lipid name generation are the main functions of Goslin. Lipid names may be decomposed into their structural components headgroup (HG), fatty acyls (FA), and long-chain base (LCB), where applicable. We therefore defined a context free grammar12 that defines rules and productions for all structural properties of the lipid nomenclature, including mass spectrometry specific information about unlabeled and heavy isotope labeled species as well as fragments.
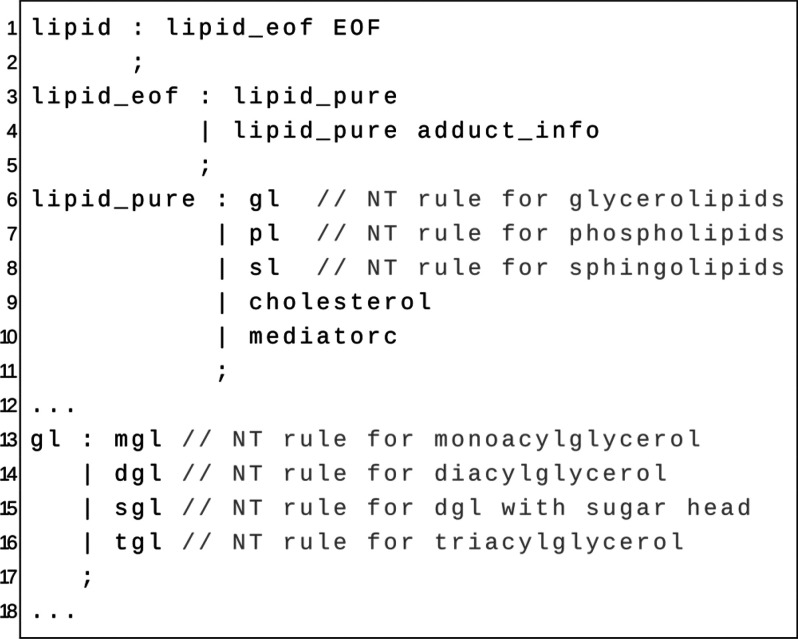
A short extract of that grammar is illustrated in Figure 1. Currently, the grammar covers 289 lipid classes within the seven most occurring lipid categories in eukaryotic organisms, namely fatty acyls, glycerolipids, glycerophospholipids, saccharolipids, sphingolipids, sterol lipids, and polyketides (for a detailed lipid list, see Table S1). To keep all implementations up to date, one authoritative lipid list containing the current lipid name abbreviation including all its synonyms is used by the different implementations.
Figure 1.
Snippet of the Goslin grammar: on the left-hand side of each colon, one nonterminal (NT) rule defines a set of production rules which are either terminal or nonterminal rules. For instance, the production rule lipid_eof defines a lipid name by identifying either a pure lipid name or a pure lipid name followed by an identified scheme of an adduct. The complete grammars are provided in our repositories (see https://github.com/lifs-tools/goslin).
The major advantages of using a grammar rather than a manually coded parser are its flexibility and extensibility. Regular expressions are also not suitable for parsing lipid names, since they are incapable of recognizing nested patterns and can only recognize words from regular languages.12,13 Obviously, lipid names do not exhibit such a regular structure. Changes or updates to the lipid nomenclature can easily be applied to the grammar. Parsers based on these grammars can be generated automatically14 to check whether a string is a valid word within a language defined by the grammar.
Goslin is able to map lipid names to either species level (which includes category and class), subspecies level, to molecular and structural subspecies level, as well as to isomeric subspecies level. Another advantage is its compatibility with lipid nomenclatures from existing publications. For instance, we defined additional grammars capable of parsing the structural lipid names from LIPID MAPS,3,4 SwissLipids,10 and HMDB.15 Variations (e.g., a blank after the headgroup abbreviation or none) are already defined within the grammar and handled automatically without the need for manually coded handling or additional validation. We also consider and handle different common abbreviations of the head groups (e.g., SPH vs Sph or DG vs DAG) and support different strings as separator symbols between multiple fatty acyl chains (either “–”, “_”, or “/” from the literature). Changes of these symbols can easily be updated in the grammar.
Given a lipid name on a certain hierarchy level, the Goslin parser implementation is also able to report the lipid names for all of its parent levels. This feature simplifies several use-cases, such as the computation of the distribution of lipids among the lipid classes or lipid categories. The polyglot approach already covers several existing dialects of the lipid nomenclature and can be extended arbitrarily to cover new lipid categories and classes, head groups, and FA modifications. Tailored event handlers (one for each grammar) transfer all recognized elements of the structural lipid name into a common data structure that reflects the hierarchy similarly to that used in LipidHome9 and SwissLipids.10
Our implementations are written in C++ (version 11 and higher), Java (version 11 and higher), Python (version 3), and R (version 3.6 and higher) and can easily be included into existing lipidomics tools. Detailed descriptions on the integration and usage of the programming libraries are provided in the Supporting Information. Additionally, we provide several instructive code snippets on the usage of Goslin for input and output of lipid names.
Results
We benchmarked our implementations with respect to their execution time for parsing. As a benchmark device, we used an Intel(R) Xeon(R) 2.80 GHz quad core desktop computer with 16 GB RAM. The results are listed in Table 2. All implementations are fast enough to parse lipid name lists of regular experiment sizes (about 1000 lipid names) within a second.
Table 2. Time Performance Benchmark Results of the Different Grammars Applied to Different Lipid Name Test Setsa.
| test set | avg len | C++* | Java* | Python* |
|---|---|---|---|---|
| Goslin (S) | 15.59 | 8237.23 | 7074.57 | 2957.81 |
| Goslin (L) | 37.76 | 2880.18 | 7771.71 | 807.31 |
| LIPID MAPS | 38.57 | 3727.17 | 5498.62 | 996.36 |
| SwissLipids | 36.84 | 3691.4 | 5810.29 | 973.80 |
| HMDB | 32.41 | 4266.21 | 9100.97 | 1280.73 |
All values for the programming languages C++, Java, and Python 3 have the unit parsed lipid names per second*. All test sets contain 10 000 lipid names parsable by their grammar. Here, the test set Goslin (S) contains rather short lipid names on average and Goslin (L) rather long lipid names. The parsing time is dependent on the length of the lipid string. Therefore, the average length (avg len) over all lipids in each data set is reported. Note, that the R implementation utilizes the C++ library internally. Therefore, it is not listed here explicitly.
To provide a user-friendly alternative to the libraries and command line interface (CLI), we developed a Goslin web application (see https://apps.lifs.isas.de/goslin) that offers form-based lipid name translation as well as a REST API for language agnostic, programmatic translation. Lipid names can be directly copied and pasted into the submission form of the web application or can be uploaded as a file, with one lipid name per line. Upon submission, the application parses all lipid names and provides a result list with all identified lipids and additional structural information as well as cross-links to LIPID MAPS and SwissLipids, where applicable.
For lipid names that cannot be parsed, it reports specific errors within the submission form to help the user fix the name. The result list can also be downloaded in a spread-sheet friendly tab-separated value format. For programmatic access, the user can send a web request (HTTPS) to translate his/her lipid list using the provided REST application programming interface (API) by sending a POST request with a JavaScript Object Notation (JSON) body (JSON list of lipid name strings). The response is a JSON list with embedded objects (for usage and tutorials, see the Supporting Information). The REST API is meant to be used by automated workflows, when neither of our offline implementations are suitable or applicable. We also offer instructions to build a Docker image of the web-application for local and customized deployments as well as a Bioconda recipe for the jgoslin CLI16 for inclusion in automated bioinformatics workflows.
Conclusions
With Goslin, we provide another building block toward a more harmonized and interoperable bioinformatics tool landscape for lipidomics. Equipped with multiple grammars, Goslin is able to interpret several lipid name dialects, such as the LIPID MAPS nomenclature, the shorthand nomenclature by Liebisch and co-workers, the SwissLipids nomenclature, and the HMDB nomenclature. Goslin translates them into a normalized, up-to-date nomenclature and structured representation. It therefore may also serve as an educational resource for students and researchers who want to learn more about lipids that are not yet represented in any of the aforementioned lipid databases. Owing to its concept of normalized lipid names and the hierarchical, taxonomic classification, Goslin will be an indispensable tool to integrate lipidomics with other omics and databases.
Currently, we are working on several improvements of the library (1) to offer an implementation of Goslin in additional programming languages (e.g., C#); (2) to improve the performance of the existing implementations; and (3) to add more structural lipid classes and categories into the grammar for a higher coverage of lipids.
We already applied Goslin in other recent projects17−19 and achieved a high-performance boost especially when exchanging our lipid lists with our collaboration partners. The Goslin implementations are freely available at https://github.com/lifs-tools/goslin under the terms of liberal open source licenses.
Acknowledgments
We gratefully acknowledge support by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen, the Regierende Bürgermeister von Berlin–inkl. Wissenschaft und Forschung, and the Bundesministerium für Bildung und Forschung (BMBF) to D.K. (de.NBI/Code 031A534B), N.H. (de.NBI/Code 031L0108A), and R.A. We thank Harald Köfeler and Gerhard Liebisch for the close collaboration within the International Lipidomics Society on lipidomics standardization.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c01690.
Overview of the Goslin libraries and applications with installation instructions and usage examples, the Goslin object model, and the list of supported lipid classes (PDF)
Author Contributions
∥ D.K. and N.H. share first authorship. D.K. and N.H. designed the grammars, N.H. implemented the Java and R libraries and the Goslin web-application. D.K. implemented the C++ and Python versions. B.P. and R.A. contributed their structural and chemical knowledge of lipids. R.A. supervised the project. All authors wrote and approved of the manuscript.
The authors declare no competing financial interest.
Notes
The complete grammars are provided in our repositories (see https://github.com/lifs-tools/goslin). The Goslin implementations are freely available at https://github.com/lifs-tools/goslin under the terms of liberal open source licenses.
Supplementary Material
References
- The nomenclature of lipids. J. Lipid Res. 1967, 8, 523–528. [PubMed] [Google Scholar]
- Chester M. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN) Nomenclature of glycolipids. Eur. J. Biochem. 1998, 257, 293–298. 10.1046/j.1432-1327.1998.2570293.x. [DOI] [PubMed] [Google Scholar]
- Fahy E.; Subramaniam S.; Brown H. A.; Glass C. K.; Merrill A. H. Jr; Murphy R. C.; Raetz C. R. H.; Russell D. W.; Seyama Y.; Shaw W.; Shimizu T.; Spener F.; van Meer G.; VanNieuwenhze M. S.; White S. H.; Witztum J. L.; Dennis E. A. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–862. 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Fahy E.; Subramaniam S.; Murphy R. C.; Nishijima M.; Raetz C. R. H.; Shimizu T.; Spener F.; van Meer G.; Wakelam M. J. O.; Dennis E. A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Vizcaíno J. A.; Köfeler H.; Trötzmüller M.; Griffiths W. J.; Schmitz G.; Spener F.; Wakelam M. J. O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling J. K.; Hermansson M.; Hartler J.; Christiansen K.; Gallego S. F.; Peng B.; Ahrends R.; Ejsing C. S. Proposal for a common nomenclature for fragment ions in mass spectra of lipids. PLoS One 2017, 12, e0188394. 10.1371/journal.pone.0188394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanksby S. J.; Mitchell T. W. Advances in Mass Spectrometry for Lipidomics. Annu. Rev. Anal. Chem. 2010, 3, 433–465. 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- Wenk M. R. Lipidomics: New Tools and Applications. Cell 2010, 143, 888–895. 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Foster J.; Moreno P.; Fabregat A.; Hermjakob H.; Steinbeck C.; Apweiler R.; Wakelam M.; Vizcaino J. LipidHome: A Database of Theoretical Lipids Optimized for High Throughput Mass Spectrometry Lipidomics. PLoS One 2013, 8, e61951. 10.1371/journal.pone.0061951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimo L.; Liechti R.; Hyka-Nouspikel N.; Niknejad A.; Gleizes A.; Götz L.; Kuznetsov D.; David F. P.; van der Goot F. G.; Riezman H.; Bougueleret L.; Xenarios I.; Bridge A. The SwissLipids knowledgebase for lipid biology. Bioinformatics 2015, 31, 2860–2866. 10.1093/bioinformatics/btv285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Ahrends R.; Arita M.; Arita M.; Bowden J. A.; Ejsing C. S.; Griffiths W. J.; Holčapek M.; Köfeler H.; Mitchell T. W.; Wenk M. R.; Ekroos K. Lipidomics needs more standardization. Nat. Metab. 2019, 1, 745–747. 10.1038/s42255-019-0094-z. [DOI] [PubMed] [Google Scholar]
- Chomsky N. Three models for the description of language. IEEE Trans. Inf. Theory 1956, 2, 113–124. 10.1109/TIT.1956.1056813. [DOI] [Google Scholar]
- Kleene S. C.Representation of Events in Nerve Nets and Finite Automata; RAND Corporation, 1951. [Google Scholar]
- Parr T.The Definitive ANTLR 4 Reference; O’Reilly UK Ltd., 2013. [Google Scholar]
- Wishart D. S.; Tzur D.; Knox C.; Eisner R.; Guo A. C.; Young N.; Cheng D.; Jewell K.; Arndt D.; Sawhney S.; Fung C.; Nikolai L.; Lewis M.; Coutouly M. A.; Forsythe I.; Tang P.; Shrivastava S.; Jeroncic K.; Stothard P.; Amegbey G.; Block D.; Hau D. D.; Wagner J.; Miniaci J.; Clements M.; Gebremedhin M.; Guo N.; Zhan Y.; Duggan G. E.; MacInnis G. E.; Weljie A. M.; Dowlatabadi R.; Bamforth F.; Clive D.; Greiner R.; Li L.; Marrie T.; Sykes B. D.; Vogel H. J.; Querengesser L. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B.; Dale R.; Sjödin A.; Chapman B. A.; Rowe J.; Tomkins-Tinch C. H.; Valieris R.; Köster J. The Bioconda Team. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 2018, 15, 475–476. 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B.; Weintraub S.; Coman C.; Ponnaiyan S.; Sharma R.; Tews B.; Winter D.; Ahrends R. A Comprehensive High-Resolution Targeted Workflow for the Deep Profiling of Sphingolipids. Anal. Chem. 2017, 89, 12480–12487. 10.1021/acs.analchem.7b03576. [DOI] [PubMed] [Google Scholar]
- Peng B.; Geue S.; Coman C.; Münzer P.; Kopczynski D.; Has C.; Hoffmann N.; Manke M.-C.; Lang F.; Sickmann A.; Gawaz M.; Borst O.; Ahrends R. Identification of key lipids critical for platelet activation by comprehensive analysis of the platelet lipidome. Blood 2018, 132, e1–e12. 10.1182/blood-2017-12-822890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B.; Kopczynski D.; Pratt B. S.; Ejsing C. S.; Burla B.; Hermansson M.; Benke P. I.; Tan S. H.; Chan M. Y.; Torta F.; Schwudke D.; Meckelmann S. W.; Coman C.; Schmitz O. J.; MacLean B.; Manke M.-C.; Borst O.; Wenk M. R.; Hoffmann N.; Ahrends R. LipidCreator workbench to probe the lipidomic landscape. Nat. Commun. 2020, 11, 2057. 10.1038/s41467-020-15960-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.