Histone turnover during gene transcription is mediated by regulatory proteins, not by the transcription machinery.
Abstract
Transcription in eukaryotes correlates with major chromatin changes, including the replacement of old nucleosomal histones by new histones at the promoters of genes. The role of these histone exchange events in transcription remains unclear. In particular, the causal relationship between histone exchange and activator binding, preinitiation complex (PIC) assembly, and/or subsequent transcription remains unclear. Here, we provide evidence that histone exchange at gene promoters is not simply a consequence of PIC assembly or transcription but instead is mediated by activators. We further show that not all activators up-regulate gene expression by inducing histone turnover. Thus, histone exchange does not simply correlate with transcriptional activity, but instead reflects the mode of action of the activator. Last, we show that histone turnover is not only associated with activator function but also plays a role in transcriptional repression at the histone loci.
INTRODUCTION
Transcription in eukaryotes is associated with major changes in the chromatin structure. One important event is the replication-independent replacement of “old” nucleosomal histones by “new” histones [for a review, see (1)]. This process, referred to as histone exchange or histone turnover, is largely conserved among eukaryotes (2, 3). In metazoans, histone exchange is highest at the promoters and enhancers of active genes (4, 5). In yeast, histones H2A and H2B that flank the central H3/H4 tetramer in the nucleosome particle exchange genome wide and independently of transcription (6). However, exchange of H3 and H4, which reflects disruption and turnover of the entire nucleosome, is mainly restricted to transcriptionally active promoters where it generally correlates with gene transcription, although not necessarily linearly (5, 7). Histone exchange is regulated by adenosine 5′-triphosphate–dependent chromatin remodeling complexes and histone chaperones. Some of these components are recruited to gene promoters by activators or travel with the elongating RNA polymerase, while others act globally across the genome (1, 8). They are implicated in either retaining the original histones or in promoting histone eviction, histone incorporation, or both (9).
The role of histone exchange in regulated gene transcription is not yet fully established. In particular, the causal relationship, if any, between histone eviction and deposition at promoters and transcription preinitiation complex (PIC) assembly remains unclear. Nucleosomes may be evicted as a passive consequence of PIC assembly, which is highly dynamic (10–12), thus potentially providing the grounds for incorporation of new histones originating from the free pool. Alternatively, histone eviction and deposition may precede and thus regulate PIC formation (13, 14). This may be due to the nucleosome-destabilizing property of promoter sequences (15) or the consequence of an activator-dependent event that leads to histone eviction and incorporation. In this scenario, the continuous exchange of histones may transiently expose promoter sites, thereby facilitating association of the transcription machinery while maintaining proper chromatin structure [for reviews, see (1, 16)].
Here, we show that, unexpectedly, histone exchange largely persists genome wide in the absence of PIC formation and, hence, transcription. The same is true at inducible promoters, where histone exchange requires the presence of the cognate activator but not the PIC. We provide direct evidence that histone turnover involves the activation domain (AD) and is not merely due to competition with the activator for binding to DNA. We further show that Rap1 defines a distinct class of transcriptional activators that stimulate gene expression without inducing histone turnover. Last, we provide evidence that HIR (histone regulatory)–mediated repression of the histone loci involves the continuous incorporation of new histones to maintain a repressive nucleosomal environment.
RESULTS
Histone exchange is largely maintained in the absence of PIC formation and transcription
As a first step toward determining what causes histones to turn over, we set up conditions to monitor histone exchange in the absence of PIC formation and, hence, transcription. For this, we inhibited PIC assembly by rapidly depleting TATA box–binding protein (TBP) from the nucleus using the rapamycin-mediated anchor-away approach (17) (Fig. 1A, upper left panel). Previous work has shown that the PIC is very dynamic and that nuclear depletion of TBP, which is essential for PIC assembly at all genes (18), results in its rapid dissociation and that of the PIC from promoters (10, 11, 19). To monitor histone exchange, we expressed an influenza hemagglutinin (HA) epitope–tagged version of histone H3 (H3HA) from a galactose-inducible promoter. To avoid replication-coupled histone incorporation, cells were first arrested at G1 with alpha factor. H3HA was then induced by galactose, and rapamycin was added to deplete TBP (Fig. 1A, upper middle and right panels; see fig. S5 for experimental overview). As a control, H3HA expression was induced without TBP depletion by replacing rapamycin with glucose to limit galactose induction of H3HA when TBP remains nuclear. TBP occupancy and H3HA incorporation into chromatin, which indirectly measures histone exchange, were assessed at various time points by quantitative chromatin immunoprecipitation (ChIP; Fig. 1A, lower panels). Figure 1 shows that under normal conditions, TBP promoter occupancy is high and H3HA rapidly accumulates at the actively transcribed ACT1 and RPL28 genes, whereas low levels of TBP and H3HA are detected at the silent STE3 gene (Fig. 1A, lower panels, green lines). Addition of rapamycin results in a rapid (i.e., within 10 min) loss of TBP from the ACT1 and RPL28 promoters, but does not affect H3HA incorporation at the ACT1 promoter at later time points, thus pointing to a PIC-independent event (Fig. 1A, lower panels, red lines). However, it leads to decreased incorporation at the 3′ end of the RPL28 gene (Fig. 1A, lower right panels; see below for further explanation). Thus, the TBP depletion strategy provides a suitable means for measuring changes in histone exchange in the absence of PIC formation.
Fig. 1. Genome-wide histone exchange in the absence of TBP.
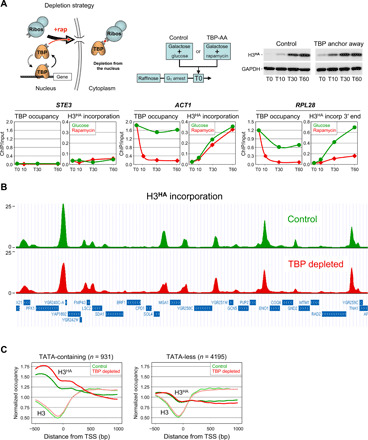
(A) Upper left panel: the TBP anchor-away assay. Upon addition of rapamycin (+rap; red dot), TBP fused to the rapamycin-binding domain FRB is rapidly exported out of the nucleus through its interaction with an RP (ribos) bearing the complementary FKBP rapamycin-binding domain [for details, see (18)]. Upper middle panel: experimental approach. Cells from a TBP anchor-away strain carrying a galactose-inducible H3HA construct were grown in raffinose and arrested in G1 by alpha factor. Galactose was then added at T0 to induce H3HA expression, together with rapamycin to deplete TBPFRB from the nucleus [TBP–anchor away (AA)]. As a control, rapamycin was replaced by glucose to limit galactose induction of H3HA when TBP remains nuclear (control). The right panel is a Western blot analysis for H3HA expression under the indicated conditions. Lower panels: TBP promoter occupancy and H3HA incorporation under control (glucose, green lines) or TBP depletion (rapamycin, red lines) conditions were monitored at the indicated genes and time points (minutes) by quantitative ChIP using antibodies against TBP or HA. Results are expressed as percentage of input DNA recovered. Note that H3HA incorporation at RPL28 was measured at the 3′ gene end. (B) ChIP-seq analysis of H3HA incorporation under control (green) or TBP depletion (red) conditions at T30 after galactose induction. Shown is a representative region in chromosome VII (965,000 to 1,015,000). The H3HA ChIP-seq reads were normalized to histone H3 ChIP-seq reads and are expressed in arbitrary units. Shown is one of two independent experiments. (C) Metagene profiles of H3HA incorporation and H3 occupancy under control (green) or TBP depletion (red) conditions at SAGA-dependent, TATA-containing genes (left panel) and at TFIID-dependent, TATA-less genes (56). Plots show a 1.5-kb region aligned at the transcription start site (TSS) and average H3HA or H3 enrichment for the indicated conditions. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To investigate the role of the PIC and transcription in histone exchange on a genomic scale, we used ChIP sequencing (ChIP-seq). Measurements were performed just before and at an early time point (30 min) after induction of H3HA expression to reduce the risk of reaching a plateau at highly dynamic sites. To normalize for nucleosome occupancy, we also carried out ChIP-seq for histone H3. We performed the experiment twice using independent biological samples and obtained very similar results (fig. S1A). Under control conditions, H3HA typically incorporates within intergenic regions, particularly over promoter regions, as reported (Fig. 1B and fig. S1B) (20, 21). Consistent with previous studies (20, 22), histone exchange is associated more with SAGA (Spt-Ada-Gcn5 acetyltransferase)–dependent, TATA-containing promoters than with TFIID (transcription factor II D)–dependent, TATA-less promoters (Fig. 1C and fig. S1C). Only rare protein-coding genes, exemplified by FIG2, show high histone turnover over the gene body (fig. S1D). Notably, H3HA incorporation largely persists and may even modestly increase at TATA-containing promoters, following nuclear depletion of TBP (Fig. 1, B and C, and fig. S1, B and C). Only a few genomic regions show markedly reduced histone turnover. These include the 3′ end of a few RNA polymerase II (Pol II)–transcribed genes, as illustrated by the EMC33 and RPG1 genes (vertical arrow in fig. S1B) and some transfer RNA (tRNA) and small nuclear RNA (snRNA) gene clusters (fig. S1E). Intriguingly, dynamic sites often show an increase in nucleosome occupancy upon TBP depletion (fig. S1B, lower panels). However, increased nucleosome occupancy upon TBP dissociation also occurs in the absence of H3HA incorporation (fig. S1E), suggesting that the two events are independent. Together, these results suggest that histone exchange at most genomic sites is not simply a consequence of PIC formation or transcription.
Gal4 mediates histone exchange at galactose-inducible genes
The finding that histones globally keep exchanging at dynamic promoters in the absence of a PIC points to a role for regulatory proteins or some intrinsic nucleosome-destabilizing promoter sequences (15). To distinguish between these possibilities, we concentrated on inducible genes up-regulated by well-characterized activators. We monitored H3HA incorporation under noninducing conditions or under inducing conditions in the presence or absence of TBP or the cognate activator protein. We first focused on galactose 4 (Gal4), a well-known activator that stimulates transcription of several galactose metabolic genes, including GAL1, GAL7, and GAL10, when galactose is used as the carbon source. Our ChIP-seq experiment (Fig. 1) performed under galactose conditions, when Gal4 is active, indicates that the GAL gene promoters are dynamic and that H3HA accumulates at similar levels under control conditions and following TBP anchor away (fig. S6A). This was confirmed by quantitative ChIP in an independent experiment performed under similar conditions, in which loss of TBP was inferred from the decrease in steady-state mRNA levels (Fig. 2, A and B). Consistent with the ChIP-seq data, H3HA incorporation remains unaffected upon rapamycin-mediated TBP depletion both at the GAL genes tested and at the HXK1 and PYK1 genes that we used as controls (Fig. 2B). Thus, histone turnover at Gal4-activated genes also persists in the absence of a PIC.
Fig. 2. Histone exchange at Gal4-regulated genes in the presence or absence of TBP or the activator Gal4.
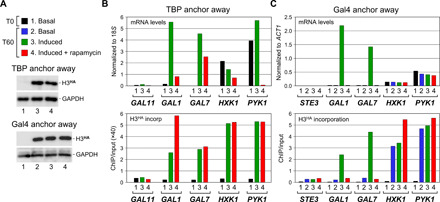
(A and B) Histone turnover in the absence of TBP. mRNA (upper panel) and H3HA incorporation (lower panel) levels for the indicated control, and Gal4-regulated GAL1 and GAL7 genes were monitored by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and quantitative ChIP under the same experimental conditions, as in Fig. 1A. Measurements were made just before (T0) and at 60 min (T60) after galactose addition or not. See fig. S5 for experimental scheme. The PYK1 mRNA signals were divided by 30× to facilitate comparison. GAL11 is a lowly transcribed gene that shows no histone exchange (41). Shown in (A) is the sample color code and a Western blot analysis for H3HA expression. (C) Histone turnover in the absence of Gal4. Cells from a Gal4 anchor-away strain carrying a cadmium-inducible H3HA allele were grown in raffinose and arrested in G1 by alpha factor. Cadmium was added at T0 to induce expression of H3HA. Cells were then maintained under basal (i.e., raffinose) conditions, or galactose was added 15 min later without or with prior treatment with rapamycin to deplete Gal4FRB from the nucleus. mRNA (upper panel) and H3HA incorporation (lower panel) levels at the indicated genes were monitored as in (B). On the left is a Western blot analysis for H3HA expression. See figs. S5 and S6 for experimental diagrams and biological replicates of this and all subsequent figures.
To assess the role of Gal4, we constructed a Gal4 anchor-away strain expressing H3HA independently of the carbon source from a cadmium-inducible promoter. Under noninducing (i.e., basal) conditions, mRNA levels and H3HA incorporation at the Gal4-regulated GAL1 and GAL7 promoters are low, comparable to those detected at the silent STE3 gene (Fig. 2C). Upon galactose induction, GAL1 and GAL7 mRNA levels strongly increase, as expected. This is accompanied by a marked increase in H3HA incorporation at these genes to reach levels similar to those observed at the highly transcribed PYK1 gene. This, however, does not occur when rapamycin is added to deplete Gal4 before galactose induction, despite having similar H3HA expression levels and incorporation at PYK1 (Fig. 2C). Thus, histone turnover at galactose-regulated promoters takes place only under inducing conditions and is mediated by the Gal4 activator.
Gal4 has been reported to recruit the Gcn5 histone acetyl transferase–containing SAGA complex. SAGA acetylates histone H3 lysines 9 and 14 (H3K9/14ac), two modification marks that typically accumulate at active promoters (23). Consistently, galactose activation of the GAL1 and GAL7 genes is associated with increased H3K9ac at their promoters (fig. S2A). This is true under both control and TBP depletion conditions, thus pointing to a Gal4 function (fig. S2A). To assess the role of these modifications in histone exchange, we converted H3 K9 and K14 into arginines (K9/14R). Incorporation of an HA-tagged histone H3(K9/14R) modification mutant into a wild-type chromatin background occurs normally (fig. S2B, upper panels), as does incorporation of wild-type H3 into chromatin containing the histone H3 mutant (fig. S2B, lower panels). The same is true for H3K4, which is commonly trimethylated at active promoters (23, 24). Thus, these promoter-specific histone modifications do not play a major role in Gal4-mediated histone exchange at the active GAL genes.
Msn2/4 mediates histone exchange at heat shock–inducible genes
To investigate whether other activators trigger promoter histone turnover, we turned to Msn2 and Msn4. These two redundant transcriptional activators are well known to up-regulate transcription of a number of target genes in response to various stresses such as heat shock (25). We measured mRNA levels and H3HA incorporation at two such genes, HSP12 and CTT1, and as a control at the constitutively active ACT1 and silent STE3 genes, either under basal conditions or upon heat shock following TBP depletion or not (see the experimental scheme in fig. S5). Figure 3 shows that HSP12 and CTT1 are strongly induced by heat shock (Fig. 3B, upper panel). This correlates with a marked increase in H3HA incorporation (Fig. 3, A and B). Depletion of TBP before heat shock totally abrogates HSP12 and CTT1 gene inductions. However, H3HA is unaffected, thus pointing again to a PIC-independent event (Fig. 3B).
Fig. 3. Histone exchange at Msn2/4-dependent heat shock–inducible genes.
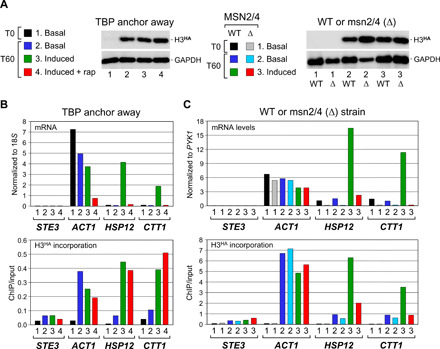
(A and B) Histone turnover in the absence of TBP. mRNA (upper panel) and H3HA incorporation (lower panel) levels under control or TBP anchor-away conditions were measured, as in Fig. 2, except that, where indicated (induced), transcription of the Msn2/4-dependent HSP12 and CTT1 genes was induced by heat shock at 37°C starting 10 min after galactose induction of H3HA expression. The HSP12 mRNA signals were divided by 5 to facilitate comparison. See fig. S5 for experimental scheme. Shown in (A) is the sample color code and a Western blot analysis for H3HA expression. WT, wild type. (C) Histone turnover in the absence of Msn2/4. mRNA levels (upper panel) and H3HA incorporation (lower panel) were monitored at the same genes in a wild-type strain and in a strain deleted for MSN2 and MSN4 [msn2/4 (Δ)]. H3HA induction and heat shock were as in (B). The CTT1 mRNA signals were multiplied by 5 to facilitate comparison. The sample color code and a Western blot analysis for H3HA expression are shown above in (A).
To examine the role of the Msn2/4 activators, we used a strain deleted for both the MSN2 and MSN4 genes (25). While increased transcription and concomitant H3HA incorporation at HSP12 and CTT1 upon heat shock are evident in the parental wild-type strain, much lower levels are found in the mutant strain under the same conditions (Fig. 3, A and C). Thus, as for the Gal4-regulated genes, histone turnover at Msn2/4-regulated genes is low under noninducing conditions, and it increases upon heat shock in an Msn2/4-dependent manner.
Ace1 mediates histone exchange at the copper-inducible CUP1 gene
To further substantiate the role of activators in promoter histone turnover, we considered a third activator, Ace1 (activator of CUP1 expression). In the presence of copper, Ace1 binds upstream to and activates transcription of the CUP1 gene, which encodes a metallothionein that protects cells from copper toxicity (26). We investigated the contribution of the PIC and of Ace1 to histone exchange at the CUP1 promoter as before, using TBP and Ace1 anchor-away strains expressing H3HA from a galactose-inducible promoter (Fig. 4A and fig. S5). Figure 4 shows that CUP1 expression and H3HA incorporation are low in both strains under noninducing conditions (Fig. 4, B and C). Upon copper induction, H3HA incorporation markedly increases to reach levels comparable to those measured at the highly dynamic ACT1 or ADH1 promoters (Fig. 4, B and C). H3HA incorporation remains high following TBP depletion, indicating a PIC-independent event, but drops back to basal levels upon Ace1 anchor away (Fig. 4, B and C, lower panels). By contrast, no changes are observed at the control ACT1 and ADH1 genes and, unexpectedly, at the SOD1 gene reported to be also regulated by Ace1 (27). Thus, as for the Gal4- and Msn2/4-regulated genes, histone turnover at the CUP1 gene requires the presence of the activator but not the PIC.
Fig. 4. Histone exchange at the Ace1-dependent, copper-inducible CUP1 gene is mediated by the Ace1 AD.
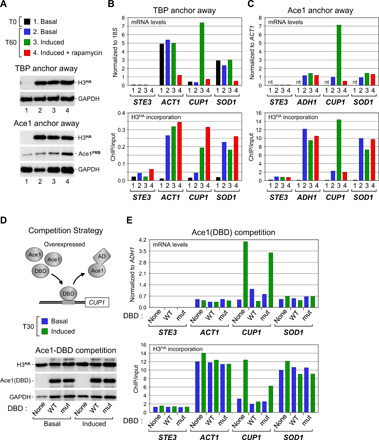
(A and B) Histone turnover in the absence of TBP. Similar experiment as before except that, where indicated (induced), Ace1 binding to DNA was induced by adding copper to the medium. (C) Histone turnover in the absence of Ace1. Similar experiment but after rapamycin-mediated nuclear depletion of Ace1 instead of TBP. nt, not tested. See fig. S5 for experimental details. (D) Upper: Competition strategy to assess the relative contribution of the Ace1 DBD and AD in histone exchange. See fig. S5 for experimental scheme and text for details. Lower: Western blot analysis for expression of the Ace1 truncations produced as GFP fusions from a galactose-inducible expression vector (28). (E) mRNA levels and H3HA incorporation at the Ace1-regulated CUP1 and indicated control genes under basal or inducing conditions in control cells (none) and in cells overexpressing the wild-type (WT) Ace1 DBD or a point mutant (mut) with attenuated DNA binding affinity (28).
To assess the relative contribution of the Ace1 DNA-binding domain (DBD) and AD to histone turnover at CUP1, we used a competition strategy (Fig. 4D). It has been shown that overexpression of a transcriptionally inactive, truncated Ace1 protein that retains the copper-dependent DBD but lacks the AD blocks activation of CUP1 by displacing endogenous Ace1 from its binding site (28). Consistently, we find that copper-induced CUP1 expression is reduced to basal levels in cells overexpressing the wild-type Ace1 DBD, whereas a point mutant with attenuated DNA binding activity is without effect (Fig. 4E, upper panel) (28). Notably, this decrease in CUP1 expression is accompanied by a corresponding decrease in H3HA incorporation (Fig. 4E, lower panel). Thus, histone turnover at the active CUP1 promoter is not due to Ace1 binding to its cognate DNA element, but instead reflects a function of the Ace1 AD. The same must be true for Gal4, since Gal4 is already bound in an inactive form to its regulatory element under noninducing conditions when histones do not exchange (Fig. 2C) (29).
Rap1 activates gene transcription without inducing histone exchange
Inspection of our ChIP-seq data reveals that a number of highly active genes exhibit unexpectedly low histone turnover at their promoters. These include ribosomal protein (RP) genes, which are typically regulated by the activator Rap1 and are among the most heavily transcribed yeast genes (30, 31). Metagene analysis confirmed the low level of H3HA incorporation at RP gene promoters, both in the presence or absence of TBP (fig. S3A). The same was observed by quantitative ChIP at two well-known Rap1-regulated RP genes, RPS13 and RPL30 (Fig. 5, A and B). Low histone exchange was also detected at Rap1-dependent, non-RP genes (fig. S3B). Thus, a lack of histone exchange is commonly observed at Rap1-dependent genes and is not due to the PIC-blocking access to the promoter. To examine the impact of Rap1 on histone exchange, we used the auxin [indole-3-acetic acid (IAA)]–induced degradation system (fig. S3C). Unexpectedly, while remaining largely unchanged at all other genes tested, H3HA incorporation increases at the RPL30 gene following Rap1 degradation (Fig. 5, A and C, and fig. S3B). Thus, if anything, Rap1 prevents histones from exchanging at these promoters.
Fig. 5. Rap1 activates gene transcription without inducing histone exchange.
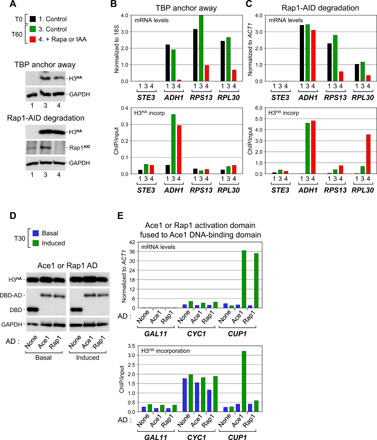
(A and B) mRNA and H3HA incorporation levels at the Rap1-regulated RPS13 and RPL30 genes and at the inactive STE3 and active ADH1 control genes under normal conditions or following TBP anchor away. (C) Same but following auxin (IAA)–mediated degradation of Rap1 fused to auxin-inducible degron (AID; Rap1AID). See fig. S5 for experimental details. (D and E) Activation of CUP1 by the Ace1 AD and Rap1 AD. Cells depleted for endogenous Ace1 (see fig. S3D) were made to express the Ace1 DBD alone (none) or fused to either the Ace1 or Rap1 AD. Gene expression and H3HA incorporation were monitored, as before.
To exclude that the lack of histone exchange is a specific feature of the RPS13 and RPL30 promoters, we tested Rap1 at the highly dynamic CUP1 promoter (Fig. 4). For this, and to exclude any possible effect of the Rap1 DBD, we fused the Rap1 AD to the DBD of Ace1, the natural copper-dependent CUP1 activator. The resulting Rap1-Ace1 fusion and, as a control, native Ace1 and a truncation containing only the DBD were tested in the absence or presence of copper to induce Ace1 binding to DNA. While the Ace1 DBD alone has no activity, as expected, full-length Ace1 and the Rap1-Ace1 fusion produced in similar amounts activate comparably CUP1 transcription in response to copper (Fig. 5, D and E, upper panel). Yet notably, a marked increase in H3HA incorporation is only observed with Ace1 (Fig. 5E, lower panel). This excludes a promoter effect and provides further evidence that histone exchange at the activated CUP1 promoter is not due to Ace1 binding to its cognate DNA element. Thus, Rap1 defines another class of activators that stimulate transcription without triggering histone exchange.
HIR-dependent nucleosome incorporation and occupancy at the histone loci
Another unexpected finding from our ChIP-seq analysis is the massive histone turnover observed at the histone loci regulatory regions. Yeast contains four histone loci, each containing two divergently transcribed genes separated by a central regulatory region. The HHT1-HHF1 and HHT2-HHF2 loci encode H3 and H4, whereas the HTA1-HTB1 and HTA2-HTB2 loci encode H2A and H2B. These loci are highly expressed only during S phase due to both activation and repression mechanisms. Three of the four gene pairs contain a negative element (NEG) in their regulatory region to which the HIR histone chaperone complex is recruited to silence histone gene expression outside of the S phase (32, 33). Figure 6 shows that all three NEG-containing loci accumulate much higher levels of H3HA than seen at the highly dynamic ADH1 gene promoter, regardless of whether TBP is depleted or not (Fig. 6A, upper panels). This is not the case at the HTA2-HTB2 locus that lacks a NEG element (Fig. 6A, upper right panel). This points to a role for the HIR complex in histone exchange at these loci. To examine this possibility, we monitored H3HA incorporation in cells deleted for HPC2, a subunit of the HIR complex involved in its recruitment to the NEG-containing histone loci (32). Notably, while having little effect on induction of H3HA expression (Fig. 6A, lower right panel) and incorporation at ADH1 and the HTA2-HTB2 loci (Fig. 6A, upper left and right panels), HIR inactivation almost completely abrogates H3HA incorporation at the NEG-containing loci (Fig. 6A, lower panels, black bars). This event is accompanied by a drastic decrease in H3 occupancy (Fig. 6A, gray bars). This suggests that the HIR complex silences histone gene transcription through a mechanism that involves incorporation of new histones.
Fig. 6. HIR-mediated histone incorporation at the NEG-containing histone loci.
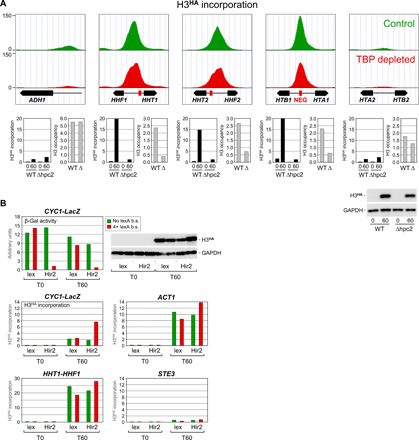
(A) Upper panels: H3HA incorporation under control (green) or TBP depletion (red) conditions at the four histone loci and at the highly transcribed ADH1 gene. Data are presented as in Fig. 1B but using a different scale on the y axis. Lower panels: H3HA incorporation (black bars) and H3 occupancy (gray bars) in wild-type cells or in cells carrying a deletion (Δ) of the gene encoding the Hpc2 subunit of the HIR complex. See fig. S5 for experimental detail. H3 occupancy levels are relative to those measured within a subtelomeric region on chromosome VI, which was set to 10. Note that the H3 occupancy results for ADH1 are presented on a different scale. Lower right: Western blot analysis of H3HA expression. (B) Transcriptional repression by artificial recruitment of the HIR complex. Upper left panel: The activity of a CYC1 promoter–driven β-galactosidase reporter gene (CYC1-LacZ) carrying (red) or not (green) four upstream LexA-binding sites was monitored in G1-arrested cells expressing LexA alone or LexA fused to the Hir2 subunit of the HIR complex (36). See text and fig. S5 for experimental details. Lower panels: H3HA incorporation within the upstream promoter regions of the indicated genes. Note the different scales on the y axis. b.s., binding sites.
The HIR complex has been shown to repress histone genes via recruitment of the RSC (remodel the structure of chromatin) chromatin-remodeling complex (34). We therefore examined whether RSC is involved in histone exchange at the histone loci. We found that RSC inactivation has no effect on H3HA incorporation at the HTA1-HTB1 histone locus (fig. S4, upper panels). Unexpectedly, however, the TBP-dependent exchange observed at the 3′ ends of some transcribed genes decreases, and this occurs without changes in PIC formation or transcription (fig. S4) (35).
To further examine the functional link between HIR-mediated histone incorporation and repression, we used an artificial recruitment strategy (36). In this assay, the Hir2 subunit of the HIR complex is fused to the LexA DBD and tested for its effect on transcription of a reporter gene carrying either no or four LexA-binding sites upstream of the natural CYC1 promoter and regulatory elements. As reported previously, LexA-Hir2, but not LexA alone, strongly represses activity of only the LexA-containing reporter (Fig. 6B, upper left panel) (36). ChIP analysis for H3HA reveals that the decrease in reporter gene transcription correlates with an increase in histone incorporation at the CYC1 promoter region (Fig. 6B, middle left panel). The effect is specific; no major change in H3HA incorporation occurred at the reporter lacking LexA-binding sites, at the endogenous HHF1-HHT1 locus, or at the active ACT1 or silent STE3 genes. Furthermore, LexA alone had no effect, pointing to a HIR activity (Fig. 6B, middle and lower panels). Thus, artificial recruitment of the HIR complex to a transcriptionally active test promoter leads to increased incorporation of new histones and concomitant transcriptional repression. This is the opposite of what is observed at inducible promoters, where H3HA incorporation increases with transcriptional activity. Together, these data suggest that chromatin within the NEG-containing histone regulatory regions is intrinsically unstable and that the HIR complex acts to maintain a repressive chromatin structure by continuously incorporating new histones.
DISCUSSION
Activator-mediated histone exchange
A major finding of the present study is that histone exchange is largely maintained across the genome in the absence of TBP and, thus, PIC assembly. This was unexpected since the commonly accepted view is that promoter nucleosomes and the PIC, which assembles and disassembles in a highly dynamic fashion (10, 11), are in competition with each other (37, 38). Thus, histone exchange is not a consequence of, but instead precedes and perhaps facilitates, PIC assembly and transcription. This points to a role for regulatory proteins or some intrinsic nucleosome-destabilizing property of promoter sequences (15). Consistent with the former possibility, we show that histone turnover at inducible promoters regulated by three unrelated activators, Gal4, Msn2/4, and Ace1, occurs only under activating conditions and requires the presence of the cognate activator, but not the PIC. For Ace1 and Gal4, we provide direct evidence that this effect is mediated by the AD. Thus, the three activators function by a mechanism that ultimately results in histone exchange. It is likely that the same is true for other inducible genes and for a number of the many constitutively active promoters showing TBP-independent histone exchange.
TBP-dependent histone exchange
Our ChIP-seq analysis revealed that only a few sites show TBP-dependent histone exchange across the genome. These include tRNA and snRNA gene clusters and are likely due to these genes being highly transcribed. Intriguingly, TBP-dependent exchange is also observed at the 3′ ends of a few RNA Pol II–transcribed genes (Fig. 1A and fig. S1B). Examination of few such genes that are facing each other in a tail-to-tail orientation suggests that the phenomenon implicates the RSC complex and is not due to TBP binding at the 3′ regions of these genes (fig. S4). This points to exchange at these regions being transcription dependent. However, the mechanism remains elusive.
Histone exchange and histone modifications
Indirect evidence points to a potential causal relationship between the two covalent histone modification marks H3K9ac and H3K4me3, which typically accumulate at the promoters of transcribed genes and histone turnover (39). For example, increased H3K9ac and H3K4me3 levels at active promoters correlate with higher H3.3 turnover rates in mammalian cells (5, 40). Consistently, we found that activation of GAL genes by galactose is associated with both increased H3K9ac and increased histone exchange at the promoters (fig. S2A). This occurs independently of TBP, thus pointing to a Gal4 activator function. However, the use of histone lysine-to-arginine mutants revealed that H3K4me and H3K9ac do not play a critical role in either eviction of nucleosomal histones or deposition of new histones at the GAL promoters, in agreement with previous data on other genes (fig. S2B) (41). Thus, histone exchange at GAL promoters is not regulated by these histone modifications.
The histone loci: Repression by incorporation of new histones
Another important finding that emerges from our ChIP-seq analysis is that the three NEG-containing histone loci show, by far, the highest levels of H3HA incorporation genome wide. The NEG element has been shown to recruit the HIR complex to repress histone gene transcription outside of S phase (33, 34, 42). In vitro, the HIR complex can deposit histones onto DNA in a replication-independent manner, indicating that it functions as a nucleosome assembly factor (43, 44). Consistently, HIR inactivation in yeast results in a loss of nucleosomes over the NEG-containing histone regulatory regions (45). Here, we confirm these findings and further show that this is due to a defect in the incorporation of new histones. Furthermore, we show that artificial recruitment of the HIR complex to a test reporter results in increased H3HA incorporation and reduced reporter gene expression (Fig. 6) (36). Thus, the HIR complex appears to act as a sensor of free histones outside of S phase and to repress histone gene transcription by continuously promoting the assembly of new nucleosomes.
Rap1: Activation without histone exchange
Whereas activation by Gal4, Msn2/4, and Ace1 triggers histone exchange, activation by Rap1 does not at the genes we tested (Fig. 5). This is not due to Rap1 being a weak activator; Rap1 binds to and activates many RP genes, which are among the most heavily transcribed yeast genes (30, 31). Furthermore, fusing the Rap1 AD to the Ace1 DBD results in a protein that is as powerful as native Ace1 in stimulating CUP1 gene transcription. However, only Ace1 triggers histone exchange. This excludes that the lack of histone exchange at the natural Rap1-regulated RPS13 and RPL30 genes is due to a core promoter-specific effect. Instead, the results point to a specific feature of the Rap1 AD. Thus, Rap1 defines a distinct class of transcriptional activators that stimulate gene expression without inducing histone turnover. Rap1 has been reported to activate transcription by recruiting the TBP-containing general transcription factor TFIID to the promoter to facilitate PIC assembly (19, 46, 47). The absence of histone exchange at Rap1-regulated promoters therefore provides additional evidence that histones are exchanging independently of the PIC. It is also consistent with previous findings, suggesting that nucleosome eviction is not a prerequisite for PIC assembly (12, 48).
Overall, our data suggest two classes of activators. One class, exemplified by Gal4, Msn2/4, and Ace1, activates transcription by a mechanism that likely involves the recruitment of chromatin remodeling factors [for reviews, see (49, 50)] and that ultimately results in increased histone exchange at the promoter. The second class, illustrated by Rap1, activates transcription without triggering histone exchange. A prediction from this interpretation is that histone exchange should correlate more tightly with activator type than with transcription rates, as commonly reported.
MATERIALS AND METHODS
Strains and plasmids
A full list of the yeast strains used in this study is available in table S1. Plasmids and oligonucleotides are listed in tables S2 and S3, respectively. To allow efficient G1 arrest by alpha factor, all strains are MATa and defective for the secreted BAR1-encoded protease that cleaves alpha factor. Genes were deleted or modified to encode C-terminally tagged proteins using one-step polymerase chain reaction (PCR) replacement. In Fig. 5 (D and E), the Ace1 (residues 124 to 225) and Rap1 (residues 601 to 695) ADs were fused at the C terminus of the Ace1 DBD by PCR amplification from genomic DNA and in-frame insertion into p1883 at the unique Cla1 site between the Ace1 DBD and green fluorescent protein (GFP; see tables S2 and S3). In Figs. 2C and 6A (lower panels), H3HA was expressed from a cadmium-inducible promoter by replacing the GAL1 promoter in the original construct (10) by the PCR-amplified MET3 promoter (nucleotides −780 to −7 relative to the AUG start codon) using Kpn1 and Msc1 restriction sites. All other constructs have been described. Details of the plasmid constructions are available upon request.
Growth conditions
Cells were grown overnight at 30°C (unless otherwise indicated) to an optical density at 600 nm (OD600) of 0.5 to 0.7 in casamino acid medium supplemented with 2% raffinose and 0.1% glucose and lacking adenine, tryptophan, and/or uracil for plasmid selection. Methionine (0.2 mM final) was included when H3HA expression was from the MET3 promoter. Cells were then arrested in G1 by treatment with alpha factor (600 ng/ml; Primm Srl, Italy) for 3 hours. Expression of H3HA was induced at T0 (minutes) by directly adding galactose or cadmium sulfate to a final concentration of 2% and 0.5 mM, respectively. Where indicated, rapamycin (LC laboratories) was added at the same time to a final concentration of 4 μg/ml to induce nuclear depletion of the FRB-tagged proteins (i.e., TBP, Gal4, or Ace1). In the TBP anchor-away experiments, glucose (0.5% final) was added to the control cultures (without rapamycin) to limit the induction of H3HA. Degradation of RAP1 fused to the auxin-inducible degron was triggered by addition of 3-indole acetic acid (Sigma-Aldrich) at 500 μM final concentration to cell cultures arrested in G1 at an OD600 not exceeding 0.3 to 0.4. Culture aliquots were pelleted at the indicated time points (see experimental schemes in fig. S5) and processed for Western blot, reverse transcription PCR (RT-PCR), and ChIP analyses.
RNA isolation and reverse transcription
RNA was extracted from 10 ml of yeast cells grown as described above using the hot phenol method. After treatment with RQ1 ribonuclease (RNase)–free deoxyribonuclease (Promega) and heat inactivation, a fraction of the RNA (500 ng) was reverse transcribed with M-MLV (Moloney murine leukemia virus) reverse transcriptase (Promega) and oligo d(T)15 primer in the presence of the RNase inhibitor RNasin (80 U/ml; Fermentas) according to the manufacturer’s instructions. Real-time PCR quantification was carried out as described below using 1/50 of the complementary DNA product and primer pairs specific for the indicated genes (see table S3). The values were normalized to those obtained for the 18S ribosomal RNA or a control mRNA, as indicated in the figures to account for differences in total RNA across samples.
ChIP and real-time PCR quantification
A detailed protocol for ChIP and quantitative PCR (qPCR) analysis can be found at www.mimo.unige.ch/STRUBIN_LABb.htm. Chromatin prepared from about 2 × 108 yeast cells was immunoprecipitated with mouse monoclonal anti-HA antibodies (2 μg; 16B12, Covance), rabbit polyclonal anti-TBP antibodies (2 μg; from Laurie Stargell, Colorado State University), rabbit polyclonal anti-H3 antibodies (1 μg; 1791, Abcam), or rabbit polyclonal anti–acetyl-histone H3 (Lys9; 2 μg; 07-352, Millipore). The recovered DNA and two standard dilutions of the input DNA were quantified in duplicate by real-time PCR using the KAPA SYBR FAST qPCR Kit Master Mix (2×) Universal (Kapa Biosystems) and the Bio-Rad CFX96 Real-time PCR System. Sequences of the oligonucleotide primers are found in table S3. The immunoprecipitation value for a given region was calculated as the ratio between the immunoprecipitation signal and the respective input DNA signal to correct for variation between different samples and primer pairs. All data are representative of at least two completely independent experiments. Independent biological replicates can be found in fig. S6.
Western blotting
Protein samples were prepared by boiling the pelleted cells in radioimmunoprecipitation assay buffer, as described (41). The membranes were probed with 1:5000 mouse monoclonal anti-HA (16B12; Covance), 1:5000 rabbit polyclonal anti-FRB (from David Shore, University of Geneva), 1:5000 rabbit polyclonal anti-RAP1 (from David Shore, University of Geneva), 1:10,000 rabbit polyclonal anti–glyceraldehyde-3-phosphate dehydrogenase (A9521; Sigma-Aldrich), and 1:5000 mouse monoclonal anti-GFP antibodies (11814460001, Roche). Horseradish peroxidase–conjugated goat anti-mouse (Bio-Rad) or anti-rabbit (A8275, Sigma-Aldrich) immunoglobulin G were used at 1:10,000 as secondary antibodies.
Next-generation sequencing library construction
ChIP-seq was performed using chromatin prepared from approximately 2 × 109 yeast cells (10-fold more than for conventional ChIP). Immunoprecipitation was with 20 μg of anti-HA antibodies (16B12, Covance) or 10 μg of anti-H3 antibodies (1791, Abcam). After decross-linking and DNA purification, samples were treated with RNase A (20 mg/ml; Sigma-Aldrich) for 1 hour at 37°C in 1× NEB (New England Biolabs) buffer 2 followed by a 45-min calf intestinal phosphatase (NEB) treatment. DNA was extracted with phenol:chloroform:isoamyl alcohol and ethanol precipitated with GlycoBlue. DNA ends were polished with End-it (Epicentre) at room temperature for 1 hour. AMPure XP beads (Beckman-Coultier) were used to perform a double size selection by first adding 0.5× beads to deplete longer fragments from the supernatant, which was then applied to 1.3× more beads to size-select upward of 100 base pair (bp). A-tailing was performed on the beads with Klenow exo− polymerase (Epicentre). Cleanup was performed with the addition of 1.8× ABR (Ampure XP Bead Resuspension) buffer (15% polyethylene glycol and 2.5 M NaCl) to the beads, which were then washed with 70% ethanol. DNA ligation (Fast-Link, Epicentre) to NEXTFlex (Bioo Scientific) multiplexed adapters was performed on the beads. Cleanup was performed, as above, with the exception of an additional ethanol wash and elution of the DNA from the beads with 39 μl of water. Test 18- and 20-cycle PCR reactions were performed with 10 μl each, with a view to reduce the number of amplification cycles. The remainder of the ligated material was PCR amplified under optimum conditions. Amplified material was cleaned up with AMPure beads, as above. StrataClone was used to check the quality of the libraries. Samples were mixed in equimolar amounts (determined by KAPA Library Quantitation) and submitted to the UMass Deep Sequencing Core for HiSeq 100SR, with multiplex indexing. In the replicate experiment, ChIP-seq libraries were generated using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs) and submitted to the iGE3 Deep Sequencing Core for HiSeq 100PE.
Data analysis
Sequence data were aligned to SacCer3 via bowtie2 (51), with default settings. To visualize occupancy on the genome browser, Homer’s makeTagDirectory and makeUCSCfile (52) codes were used to generate bigWig files, which were each normalized to 10 million. The cross-correlation of Watson and Crick reads was used to infer the average fragment length, and reads were extended by this value. Genome-wide coverage was calculated with bedtools (53), and counts were averaged in 10-bp bins surrounding −500 to +1000 bp of the edge of the +1 nucleosome (54) and normalized to coverage over genes. Average nucleosome profiles for sets of genes were created in ggplot2 (55). In fig. S1C, TATA-containing and TATA-less genes were selected, as defined by (56). RP and non-RP genes used for fig. S3A are defined in table S4.
Supplementary Material
Acknowledgments
We are most grateful to D. Shore for providing us with the yeast strains YJB25 (RAP1-AID) and STH1-mAID and with anti-FRB and anti-RAP1 antibodies; to J. Labbare for the msn2msn4∆ and isogenic control strains; to M. A. Osley for plasmids pBTM116, pBTM116-HIR2, pAJ1, and pAJ1621; to D. Winge for plasmids p793, p1883, and p1885; to L. Stargell for anti-TBP antibodies; and to F. Stutz for helpful discussions and critical reading of the manuscript. Funding: This work was supported by grants from the Swiss National Science Foundation 310030-149626 and 310030-175781 to M.S. and by the Canton of Geneva. Author contributions: S.K., P.F., and M.S. designed the experiments. S.K. and P.F. performed all Western blotting, RT-qPCR, and ChIP experiments. P.F. and A.L.H. performed ChIP-seq. A.L.H., J.S., and O.J.R. analyzed genomic datasets. S.K. and M.S. wrote the manuscript, with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The ChIP-seq datasets have been deposited in the NCBI Gene Expression Omnibus under accession GSE143305. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/36/eabb0333/DC1
REFERENCES AND NOTES
- 1.Venkatesh S., Workman J. L., Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Das C., Tyler J. K., Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta 1819, 332–342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talbert P. B., Henikoff S., Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Deaton A. M., Gómez-Rodríguez M., Mieczkowski J., Tolstorukov M. Y., Kundu S., Sadreyev R. I., Jansen L. E., Kingston R. E., Enhancer regions show high histone H3.3 turnover that changes during differentiation. eLife 5, e15316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraushaar D. C., Jin W., Maunakea A., Abraham B., Ha M., Zhao K., Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 14, R121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamai A., Imoberdorf R. M., Strubin M., Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Weiner A., Hsieh T.-H. S., Appleboim A., Chen H. V., Rahat A., Amit I., Rando O. J., Friedman N., High-resolution chromatin dynamics during a yeast stress response. Mol. Cell 58, 371–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai W. K. M., Pugh B. F., Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimaldi Y., Ferrari P., Strubin M., Independent RNA polymerase II preinitiation complex dynamics and nucleosome turnover at promoter sites in vivo. Genome Res. 24, 117–124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Werven F. J., van Teeffelen H. A. A. M., Holstege F. C. P., Timmers H. T. M., Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat. Struct. Mol. Biol. 16, 1043–1048 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Zanton S. J., Pugh B. F., Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 20, 2250–2265 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubik S., Bruzzone M. J., Jacquet P., Falcone J.-L., Rougemont J., Shore D., Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol. Cell 60, 422–434 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Rhee H. S., Pugh B. F., Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekinger E. A., Moqtaderi Z., Struhl K., Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18, 735–748 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Haruki H., Nishikawa J., Laemmli U. K., The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31, 925–932 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Vannini A., Cramer P., Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Zaidi H. A., Auble D. T., Bekiranov S., RNA synthesis is associated with multiple TBP-chromatin binding events. Sci. Rep. 7, 39631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dion M. F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O. J., Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Rufiange A., Jacques P.-É., Bhat W., Robert F., Nourani A., Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 Acetylation and Asf1. Mol. Cell 27, 393–405 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Tirosh I., Barkai N., Two strategies for gene regulation by promoter nucleosomes. Genome Res. 18, 1084–1091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., Friedman N., Rando O. J., Single-nucleosome mapping of histone modifications in S. cerevisiae. PLOS Biol. 3, e328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe F. S., Fischl H., Murray S. C., Mellor J., Is H3K4me3 instructive for transcription activation? Bioessays 39, e201600095 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Boy-Marcotte E., Lagniel G., Perrot M., Bussereau F., Boudsocq A., Jacquet M., Labarre J., The heat shock response in yeast: Differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33, 274–283 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Fürst P., Hu S., Hackett R., Hamer D., Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell 55, 705–717 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Gralla E. B., Thiele D. J., Silar P., Valentine J. S., ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc. Natl. Acad. Sci. U.S.A. 88, 8558–8562 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller G., Bird A., Winge D. R., Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae. Eukaryot. Cell 4, 1863–1871 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalley K., Johnston S. A., Kodadek T., Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442, 1054–1057 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Lieb J. D., Liu X., Botstein D., Brown P. O., Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat. Genet. 28, 327–334 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Yarragudi A., Parfrey L. W., Morse R. H., Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 35, 193–202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin A. D., Vishnoi N., Prochasson P., A global requirement for the HIR complex in the assembly of chromatin. Biochim. Biophys. Acta 1819, 264–276 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Kurat C. F., Recht J., Radovani E., Durbic T., Andrews B., Fillingham J., Regulation of histone gene transcription in yeast. Cell. Mol. Life Sci. 71, 599–613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriksson P. R., Ganguli D., Nagarajavel V., Clark D. J., Regulation of histone gene expression in budding yeast. Genetics 191, 7–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubik S., O’Duibhir E., de Jonge W. J., Mattarocci S., Albert B., Falcone J.-L., Bruzzone M. J., Holstege F. C. P., Shore D., Sequence-directed action of RSC remodeler and general regulatory factors modulates +1 nucleosome position to facilitate transcription. Mol. Cell 71, 89–102.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Spector M. S., Raff A., DeSilva H., Lee K., Osley M. A., Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17, 545–552 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adkins M. W., Tyler J. K., Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21, 405–416 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Struhl K., Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98, 1–4 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Zentner G. E., Henikoff S., Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 20, 259–266 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Chow C.-M., Georgiou A., Szutorisz H., Maia e Silva A., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N., Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 6, 354–360 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrari P., Strubin M., Uncoupling histone turnover from transcription-associated histone H3 modifications. Nucleic Acids Res. 43, 3972–3985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osley M. A., Gould J., Kim S., Kane M., Hereford L., Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell 45, 537–544 (1986). [DOI] [PubMed] [Google Scholar]
- 43.Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., Yates J. R., Kaufman P. D., Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15, 2044 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prochasson P., Florens L., Swanson S. K., Washburn M. P., Workman J. L., The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19, 2534–2539 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fillingham J., Kainth P., Lambert J.-P., van Bakel H., Tsui K., Peña-Castillo L., Nislow C., Figeys D., Hughes T. R., Greenblatt J., Andrews B. J., Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35, 340–351 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Garbett K. A., Tripathi M. K., Cencki B., Layer J. H., Weil P. A., Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell. Biol. 27, 297–311 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mencía M., Moqtaderi Z., Geisberg J. V., Kuras L., Struhl K., Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9, 823–833 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Zhang H., Gao L., Anandhakumar J., Gross D. S., Uncoupling transcription from covalent histone modification. PLOS Genet. 10, e1004202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becker P. B., Workman J. L., Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 5, a017905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Z., Chen K., Xia Z., Chavez M., Pal S., Seol J.-H., Chen C.-C., Li W., Tyler J. K., Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 28, 396–408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsankov A. M., Thompson D. A., Socha A., Regev A., Rando O. J., The role of nucleosome positioning in the evolution of gene regulation. PLOS Biol. 8, e1000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2016).
- 56.Basehoar A. D., Zanton S. J., Pugh B. F., Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699–709 (2004). [DOI] [PubMed] [Google Scholar]
- 57.A. Kassambara, ggpubr: ’ggplot2’ Based Publication Ready Plots. R package version 0.2.5 (2020).
- 58.Knight B., Kubik S., Ghosh B., Bruzzone M. J., Geertz M., Martin V., Dénervaud N., Jacquet P., Ozkan B., Rougemont J., Maerkl S. J., Naef F., Shore D., Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev. 28, 1695–1709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Candelli T., Challal D., Briand J.-B., Boulay J., Porrua O., Colin J., Libri D., High-resolution transcription maps reveal the widespread impact of roadblock termination in yeast. EMBO J. 37, e97490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D., Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68, 709–719 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/36/eabb0333/DC1


