SUMMARY
The liver communicates with the intestine via the portal vein, biliary system, and mediators in the circulation. Microbes in the intestine maintain liver homeostasis but can also serve as a source of pathogens and molecules that contribute to fatty liver diseases. We review changes in the gut microbiota that can promote development or progression of alcohol-associated and non-alcoholic fatty liver disease—the most common chronic liver diseases in Western countries. We discuss how microbes and their products contribute to liver disease pathogenesis, putative microbial biomarkers of disease, and potential treatment approaches based on manipulation of the gut microbiota. Increasing our understanding of interactions between the intestinal microbiome and liver might help us identify patients with specific disease subtypes and select specific microbiota-based therapies.
INTRODUCTION
The human gastrointestinal tract contains trillions of microbes that facilitate digestion and nutrient absorption, affect host metabolism, shape immunity, and are involved in drug metabolism. Beyond bacteria, immense populations of viruses and fungi, as well as archaea, reside in the gut—they interact with each other and with human cells in a complex and only marginally understood manner.
The liver receives portal vein blood drained from the gastrointestinal tract and is therefore the first organ exposed to gut-derived microbes and their components and metabolites. On the other hand, the liver affects the intestine by producing bile, which is secreted into the biliary system and directly released into the small intestine. Although commensal gut microbes help maintain liver homeostasis (Mazagova et al., 2015), they can also produce factors that cause liver damage under pathologic conditions (Duan et al., 2019; Llopis et al., 2016).
Alcohol-associated liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD) are global health burdens with growing incidence. With an overall prevalence of 6% and 25%, respectively, ALD and NAFLD are the most common chronic liver diseases in Western countries (Xiao et al., 2019). ALD has become the leading cause for liver transplantation in the United States (Lee et al., 2019), whereas non-alcoholic steatohepatitis (NASH) has become the second-leading liver disease among patients awaiting liver transplantation (Wong et al., 2015).
NAFLD and ALD share histopathology characteristics, ranging from benign simple steatosis to steatohepatitis and to more advanced disease, including advanced fibrosis and cirrhosis, which can ultimately lead to hepatocellular carcinoma, liver failure, and death.
Only 10%–20% of patients with alcohol-associated or non-alcoholic fatty liver disease develop progressive liver disease (Loomba and Adams, 2019; Parker et al., 2019). Therefore, other factors, which might be behavioral, environmental, and/or genetic, affect progression. Changes in gut microbiota composition, microbial metabolism, and gut barrier function are additional co-factors that can contribute to progression of ALD and NAFLD. Transplanting feces from alcoholic hepatitis patients into germ-free mice, exacerbates ethanol-induced liver disease. Liver damage was reversible when feces from alcoholics without alcoholic hepatitis were re-transplanted (Llopis et al., 2016). Germ-free mice are protected from obesity, and fecal transfer from obese mice into germ-free mice resulted in greater body fat gain than fecal transplant from lean mice (Bäckhed et al., 2004; Turnbaugh et al., 2006). Although these studies suggest an important role of the gut microbiota in modulating alcohol-associated and metabolic diseases, studying the microbiome in the controlled setting of preclinical models has several limitations and causal relationships are elusive in the very complex human ecological situation.
In this review, we elucidate the role of the gut microbiome in ALD and NAFLD. We focus on pathogenic pathways in humans, the potential to use the gut microbiome as diagnostic tool, and on strategies to alter the intestinal microbiome for treatment of liver disease.
PATHOGENESIS
Fatty liver development
Increased and chronic ethanol intake is the main cause of alcohol-associated fatty liver disease. Ethanol is converted in the liver into acetaldehyde by alcohol dehydrogenases and the microsomal ethanol-oxidizing system. Highly toxic acetaldehyde is further metabolized to acetate, which is then quickly released into the bloodstream, where it can be used as biological fuel for energy production. In addition to the toxic properties of acetaldehyde, hepatic ethanol metabolism results in the generation of highly reactive molecular fragments that create an oxidative stress environment and contribute to liver damage. Ethanol affects gene regulation in hepatocytes, resulting in fat accumulation by promoting lipogenesis and inhibiting beta-oxidation and lipolysis.
Overnutrition, physical inactivity and adiposity are the most important contributors to NAFLD. Accumulation of fat in the liver is a consequence of an increased availability of free fatty acids derived from adipose tissue, which contributes up to 60% of hepatic fat content, while hepatic de novo lipogenesis, is responsible for up to 30% of hepatic fat, with added contribution from dietary lipid intake (Donnelly et al., 2005). An interplay between adipose tissue inflammation, adipocyte lipolysis, increased free serum fatty acids, impaired adiponectin signaling, and increased hepatic lipogenesis contributes to NAFLD. These features are interrelated with insulin resistance, a critical pathophysiological factor in NAFLD, that worsens adipocyte dysfunction and allows hepatic de novo lipogenesis to continue (Figure 1). A consecutive inflammatory cascade contributes to hepatocyte mitochondrial dysfunction, endoplasmatic reticulum stress and generation or reactive oxygen species, ultimately leading to hepatic inflammation, cell death and fibrosis (Buzzetti et al., 2016). In this vicious cycle, gut microbial components modulate disease at the level of the intestine, liver and systemic circulation.
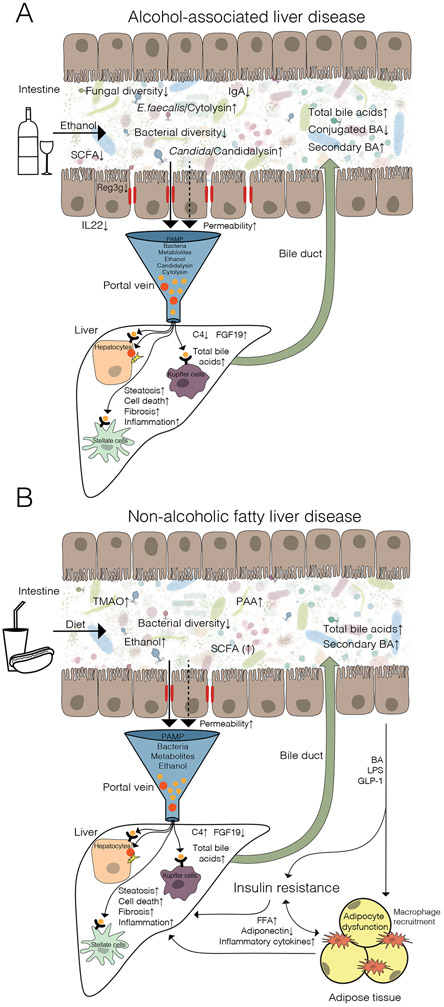
Figure 1. Dysregulated interactions between the intestine and liver in patients with fatty liver disease.
Alterations in the gut microbiota in (A) alcohol-associated liver disease and (B) non-alcoholic fatty liver disease. Gut-derived pathogen-associated molecular patterns (PAMP) that reach the liver through the portal vein are recognized by pattern recognition receptors such as toll-like receptors. The activation ultimately results in steatosis, cell death, inflammation, and fibrosis. Alternatively, direct hepatocyte damage can be induced. Bile acids (BA), produced by the liver, are secreted in the intestine via the bile duct. In NAFLD, insulin resistance, macrophage recruitment and adipocyte dysfunction can be modulated by intestinal derived BA signaling, lipopolysaccharide (LPS) delivery and glucagon like peptide-1 (GLP-1), which in turn affect NAFLD development. C4, 7-alpha-hydroxy-4-cholesten-3-one; IL-22, interleukin-22; REG3G, regenerating islet derived protein 3 gamma; IgA, immunoglobulin A; SCFAs, short-chain fatty acids; TMAO, trimethylamine N-oxide; PAA, phenylacetic acid; FXR, farnesoid X receptor; FGF19, fibroblast growth factor 19.
Changes in the gut microbiome
Microbiota research in liver diseases has largely focused on intestinal bacteria. Alcohol intake is associated with reduced intestinal bacterial diversity and changes in the composition of the microbiome, with a depletion of bacteria that are considered beneficial, and an increase of pathobionts, with a shift towards gram-negative bacteria (Duan et al., 2019; Smirnova et al., 2020). In addition to changes in the bacterial microbiome, alcohol-related liver disease is associated with reduced diversity of intestinal fungi and overgrowth of Candida spp. (Chu et al., 2020; Lang et al., 2020b; Yang et al., 2017).
Several studies have described the composition of the bacterial microbiome in fecal samples from patients with NAFLD, but results are inconsistent and contradictory. The most consistent changes observed in patients with NAFLD compared with healthy individuals, or in patients with mild vs more severe NAFLD, were increased relative abundances of Proteobacteria, Enterobacteriaceae, Escherichia and Dorea, and decreased abundances of Ruminococcaceae, Anaerospacter, Coprococcus, Eubacterium, Faecalibacterium, and Prevotella (Aron-Wisnewsky et al., 2020). Intestinal viruses, which are the most abundant microbes in the gut, have not been characterized in patients with any liver disease.
Certain aspects of gut microbiota signatures associated with ALD and NAFLD show remarkable similarities to changes observed in patients with inflammatory bowel disease (IBD) (Zuo and Ng, 2018). The increase in gram-negative bacteria described in ALD and NAFLD, as well as other diseases (including IBD and colorectal cancer), represents a source of lipopolysaccharide (LPS), a major component of the outer membrane of gram-negative bacteria. LPS can cause a local intestinal immune response, activate the inflammasome in the liver and recruit macrophages to adipose tissue (Caesar et al., 2012). Other bacteria, such as Ruminococcaceae, Eubacterium or Faecalibacterium, that are depleted in NAFLD and ALD and other diseases, are important short-chain fatty acid producers, and might therefore mediate colonic inflammation, gut barrier dysfunction and energy harvest.
Gut barrier dysfunction and microbe translocation
The intestinal gut barrier provides the first line of protection against pathogens. The physical and biochemical barrier includes a thick mucus layer and cellular tight junctions, which firmly link intestinal epithelial cells, and antimicrobial proteins. The immune system component of the barrier includes secretory immunoglobulin A, B and T cells, dendritic cells, and neutrophils. This multilayer defense prevents transfer of pathobionts, pathogens, and microbial products to the circulation and other organs.
Translocation of microbial components
Chronic alcohol intake can disrupt the gut barrier integrity. Approximately 50% of heavy drinkers have evidence of intestinal dysbiosis, which is associated with gut barrier dysfunction. Short-term abstinence largely restores intestinal eubiosis and reduces intestinal permeability (Leclercq et al., 2014). Microbial components, such as LPS, can translocate from the intestinal lumen through the portal vein to the liver. They are recognized by pattern recognition receptors such as toll-like receptors (TLRs) in the liver, resulting in hepatic inflammation, hepatocyte damage, and liver fibrosis (Figure 1). Similar pathways are believed to contribute to development of NAFLD, but instead of alcohol intake, other stimuli such as dietary factors contribute to intestinal inflammation and gut barrier disruption. Inflammasome activation, in particular the NOD-like receptor protein 3 (NLRP3) inflammasome has been identified as another trigger for liver inflammation responding to pathogen-associated molecular patterns (PAMPs) such as LPS. NLRP3 activation in the liver leads to activation of caspase 1 and the production of interleukin-1ß and several other inflammatory cytokines, ultimately resulting in programmed cell death, inflammation and fibrosis. NLRP3 is significantly upregulated in the liver of patients with NASH compared with simple steatosis (Wree et al., 2014), and liver inflammasome components correlate with the activity of alcohol-associated liver disease (Voican et al., 2015). On the other hand, NLRP6 and NLRP3 inflammasome deficiency modulates the gut microbiota, which causes gut barrier dysfunction and exacerbates steatohepatitis in mice (Henao-Mejia et al., 2012).
Similar to early-stage ALD, the role of gut barrier disruption in NAFLD development is unclear—only a subset of patients with NAFLD has increased intestinal permeability (Chu et al., 2019). TLR4 activation was reported to contribute to liver disease in mouse models of NAFLD, but inhibition of TLR4 with JKB-121 in a phase 2, randomized, placebo-controlled, double-blind trial in NASH patients (NCT02442687) had discouraging results. Gut barrier disruption in combination with TLR4 or inflammasome activation might contribute to a subset of cases of ALD and NAFLD. It is tempting to speculate that increased intestinal permeability is a factor that contributes to disease progression, but further studies are necessary to elucidate this association.
Translocation of viable bacteria
Not only bacterial products, but viable bacteria can translocate across the intestinal barrier. Findings from preclinical models support the contribution of translocated viable bacteria to ethanol-induced liver disease (Duan et al., 2019; Llorente et al., 2017; Wang et al., 2016). The space between intestinal epithelial cells with disrupted tight junctions is too narrow to allow passage of intact bacteria (Turner, 2009), and the molecular mechanisms of bacterial translocation across the intestinal epithelial cell barrier are not fully understood. This process can contribute to development of disease, but also occurs under healthy conditions, in which commensal bacteria are transported to mesenteric lymph nodes by dendritic cells, induce a local immune response, and are eliminated by the innate immune system.
Patients with advanced stages of chronic liver fibrosis or with acute hepatitis have been reported to have deficits in innate and adaptive immune responses. These deficits can lead to the systemic invasion of gut pathobionts beyond mesenteric lymph nodes, resulting in a systemic inflammatory response and infections—particularly those caused by aerobic, gram-negative bacteria. However, further studies are needed to determine the contribution of translocated viable bacteria to progression of early stages of liver disease in patients.
Bacterial metabolites
Bile acids
Changes in microbe-derived metabolites have been implicated in pathogenesis of fatty liver diseases. Bile acids are not only important for the emulsification of dietary lipids, they also act as signaling molecules through binding to host nuclear and G-protein-coupled receptors. Binding to these receptors impacts several host metabolic functions including regulation of glucose homeostasis and lipid metabolism. Primary bile acids are synthesized and conjugated in hepatocytes and reach the intestine through secretion in the biliary tract. Primary, conjugated, and hydrophilic bile acids can be reabsorbed from the terminal ileum. Microbial bile salt hydrolases (BSH) deconjugate primary bile acids, which prevents their reabsorption. Many bacterial species can deconjugate bile acids, including Clostridium, Lactobacillus, Bifidobacterium, Eubacterium, Escherichia and Bacteroides (Wahlström et al., 2016). A small fraction of unabsorbed bile acids enter the distal gastrointestinal tract, where they undergo formation to secondary bile acids through bacterial hydroxylation, epimerization, esterification and desulfation (Molinero et al., 2019). Generation of secondary bile acids is a microbial collaborative act, whereas the 7α/ß-dehydroxylation pathway, known to be performed by bacterial species belonging to Clostridium, Lachnospiraceae and Eggerthella, is one major relevant pathway in humans (Heinken et al., 2019). Hepatic bile acid synthesis is regulated by a negative feedback mechanism that includes activation of the farnesoid X receptor (FXR), which is expressed in many tissues but has been mostly studied in intestinal and liver cells. Activation of FXR in the intestine, predominantly by primary bile acids, leads to the transcription of fibroblast growth factor 19 (FGF19), which reaches the liver through the portal vein. In the liver, FGF19 suppresses expression of CYP7A1 and thereby decreases bile acid synthesis. Takeda G protein-coupled receptor 5 (TGR5) is another bile acid responsive receptor, expressed on several cell types such as Kupffer cells, immune cells and adipose tissue, and is involved in important metabolic processes (Arora and Bäckhed, 2016).
Although intestinal bacteria control the rate-limiting step in production of secondary bile acids, bile acids themselves affect the composition of the gut microbiota (Lorenzo-Zúñiga et al., 2003). Increasing bacterial BSH activity in mouse intestine, changed the bile acid pool and had a beneficial impact on host metabolism and adiposity (Joyce et al., 2014).
Ethanol intake leads to quantitative and qualitative changes in bile acid homeostasis. Chronic administration of ethanol to mice caused increased expression of genes that control bile acid biosynthesis in the liver and increased plasma and fecal levels of unconjugated bile acids. In humans, secondary bile acids were increased in fecal and serum samples from active consumers of alcohol (Kakiyama et al., 2014). Bile acid homeostasis and FGF19 signaling are dysregulated in patients with alcoholic hepatitis (Brandl et al., 2018), which is the most severe form of ALD and characterized by severe hepatic cholestasis. Unconjugated and conjugated bile acids were increased in serum samples from patients with alcoholic hepatitis, but in contrast to findings from studies of mice, de novo bile acid biosynthesis was decreased, based on serum levels of 7-alpha-hydroxy-4-cholesten-3-one (C4) (Brandl et al., 2018).
FXR signaling is involved in regulation of glucose and lipid metabolism, and also increases insulin sensitivity (Zhang et al., 2006), which is reduced in patients with NAFLD. FXR is activated by the primary bile acid chenodeoxycholic acid (CDCA) but inhibited by the secondary bile acid deoxycholic acid (DCA) (Parks et al., 1999). Similar to ALD, NAFLD is associated with changes in intestinal and circulatory bile acid composition. Serum samples from patients with NASH have higher levels of DCA and lower levels of CDCA compared with healthy individuals (controls). Liver tissues from patients with NASH have increased expression of genes involved in bile acid synthesis and reduced expression of FGF19, compared with healthy controls (Jiao et al., 2018). Pediatric patients with NASH have decreased serum levels of FGF19 and reduced levels of FXR in the liver (Nobili et al., 2018). Fecal levels of bile acids are increased in patients with NASH patients compared with controls, and serum levels of C4 are also increased (Mouzaki et al., 2016). Patients with NAFLD and higher stages of liver fibrosis have increased serum levels of bile acids (Caussy et al., 2019).
These findings indicate that bile acid and FXR dysregulation is a metabolic feature of patients with ALD or NAFLD. Although bile acid metabolism is regulated by gut microbes, it is not clear to what extent alterations in the intestinal microbiota of patients contribute to bile acid dysregulation, or if bile acids affect the function and composition of the intestinal microbiome.
Short-chain fatty acids
Bacterial fermentation of non-digestible carbohydrates results in production of short-chain fatty acids (SFCAs)—butyrate, propionate, and acetate are the most abundant in the intestine. SCFAs have beneficial effects, providing energy sources for enterocytes and colonocytes, helping maintain the gut barrier, and inhibiting proliferation of hepatic cells. SCFAs also have anti-inflammatory effects, increase satiety, and decrease food intake (Schwenger et al., 2019). The most abundant SFCA-producing bacteria belong to Lachnospiriaceae and Ruminococcacecae. Within these families, Eubacterium rectale, Faecalibacterium prausnitzii and several Roseburia species as well as Anaerostipes are known butyrate producers (Baxter et al., 2019).
Chronic alcohol use is associated with reduced fecal levels of SCFAs (Couch et al., 2015). Fecal samples from patients with alcoholic hepatitis contain lower concentrations of SCFAs and fewer SCFA-producing bacteria than samples from heavy drinkers (controls) (Smirnova et al., 2020). In patients with cirrhosis, low circulating levels of butyrate are associated with markers of inflammation, serum endotoxin, and more advanced liver disease (Juanola et al., 2019).
In contrast, higher concentrations of SCFAs have been associated with weight gain in mouse models of obesity (Turnbaugh et al., 2006) and with overweight in humans (Schwiertz et al., 2010). A study of a small number of patients with NASH found higher levels of SCFAs and a higher abundance of SCFA-producing bacteria in fecal samples from patients with NASH than controls. The increase in SCFAs was associated with immune features of disease progression (Rau et al., 2018).
Increased energy harvest
The gut microbiota is essential for efficient nutrient harvest from polysaccharide rich diet. Early mouse studies suggested an increased capacity to harvest energy from diet in the obese mouse microbiota through increased generation of SCFA (Turnbaugh et al., 2006), or increased luminal absorption of monosaccharides with resulting de novo lipogenesis (Bäckhed et al., 2004). A later study with 12 lean and 9 obese individuals could not confirm these results. Although rapid changes in the gut microbiota were observed following changes in the caloric intake, there were no differences in nutrient absorption at baseline between healthy and obese subjects, as measured with bomb calorimetry of fecal contents (Jumpertz et al., 2011). Overall, the role of potentially increased energy harvest by the gut microbiota in humans requires additional studies.
Ethanol
Microbial fermentation in the gut can lead to endogenous alcohol production. Even in the absence of ethanol supplementation, increased ethanol has been measured in the breath of obese, but not of lean mice, and can be decreased by antibiotic administration (Cope et al., 2000). Pediatric patients with NASH had increased blood levels of ethanol related to endogenous ethanol production by gut bacteria (Zhu et al., 2013). A recent study identified high ethanol-producing strains of K. pneumoniae in fecal samples from a patient with NASH who had autobrewery syndrome, which is characterized by extensive production of ethanol by intestinal microbes. This results in high blood concentrations of alcohol in the absence of alcohol consumption. Transfer of fecal microbes from patients with NASH into intestines of mice results in liver damage in the recipient mice. The severity of liver damage can be reduced by eliminating the K. pneumoniae strains that produce high levels of ethanol. Furthermore, weight loss in patients with NASH was associated with a significant decrease in the ability of intestinal microbes to produce ethanol (Yuan et al., 2019). Increased endogenous ethanol production by the microbiota in the absence of exogenous alcohol intake can cause the development and progression of NAFLD. This might contribute to the similar histopathologic features of ALD and NAFLD.
Virulence factors
Studies investigating associations of the gut microbiota and liver diseases have predominantly focused on diversity, relative abundance, and metabolic profiles of specific microorganisms. However, little is known about the specific features of microbes believed to be responsible for the disease development and progression. Virulence factors are proteins or peptides encoded by microbial genes that help the organisms colonize the intestine or mediate disease. A recent study found that cytolysin, a virulence factor expressed by E. faecalis, causes direct lysis of hepatocytes and liver damage, likely through its ability to form pores. The presence of cytolysin, but not the abundance of E. faecalis, was highly associated with worse clinical outcomes and increased mortality in patients with alcoholic hepatitis. This means that liver disease might be mediated by microbial expression of specific virulence factors. Cytolysin is specifically associated with alcoholic hepatitis—it was not associated with the presence or progression of NAFLD (Lang et al., 2020a).
THE GUT MICROBIOME AS DIAGNOSTIC TOOL
Although it is unclear whether changes in the gut microbiota of patients with ALD or NAFLD cause disease development and progression or result from it, features of the gut microbiota might serve as diagnostic or prognostic markers. Cytolysin, a two-subunit toxin that is expressed and secreted by E. faecalis, which has been mechanistically linked to liver damage in patients with ALD, had better performance in identification of people who died within 90 days than other commonly used prognostic scoring systems, such as the model for end-stage liver disease score (Duan et al., 2019). This study was performed in an international multicenter cohort of patients with alcoholic hepatitis, but the finding requires validation in an independent and larger cohort.
A set of gut microbial markers derived from metagenomic sequencing, together with age and body mass index, is able to identify patients with NAFLD who have advanced fibrosis with high accuracy (Loomba et al., 2017). This is important because liver biopsy is still required for grading and staging of patients with NAFLD—non-invasive markers of risk of development of liver-related complications are urgently needed. Other potential biomarkers for NAFLD include microbe-derived metabolites, such as metabolites from aromatic amino acid and branched-chain amino acid metabolism (Caussy et al., 2018; Hoyles et al., 2018).
These markers and scoring system must be validated in independent cohorts of patients with NAFLD and controls, of different geographical and therefore dietary, ethnic, and environmental background. Furthermore, their superiority to other clinical or imaging-based methods of evaluation, or whether these types of analyses are easier to perform, has not been shown.
MICROBE-BASED TREATMENT OF LIVER DISEASES
Bugs as drugs
If changes in the intestinal microbiome can cause fatty liver diseases, strategies to alter the composition of the gut microbiome in an additive way might be used as treatments (Mimee et al., 2016). One approach is fecal microbiota transplantation (FMT), which involves transplant of entire microbiota communities. Probiotics (living bacteria), prebiotics (groups of nutrients that promote expansion of specific bacteria), and synbiotics (combination of probiotics and prebiotics) are first-generation microbe-based therapies; these might add or nurture beneficial bacterial strains to the commensal microbiota. Beneficial bacteria are thought to either replenish diminished commensal bacteria or provide resistance against colonization by pathogens and pathobionts. Engineered bacteria have been classified as next-generation microbiome therapeutics (Mimee et al., 2016). These bacteria are precisely designed to either produce a beneficial metabolite or metabolize toxic products (Figure 2).
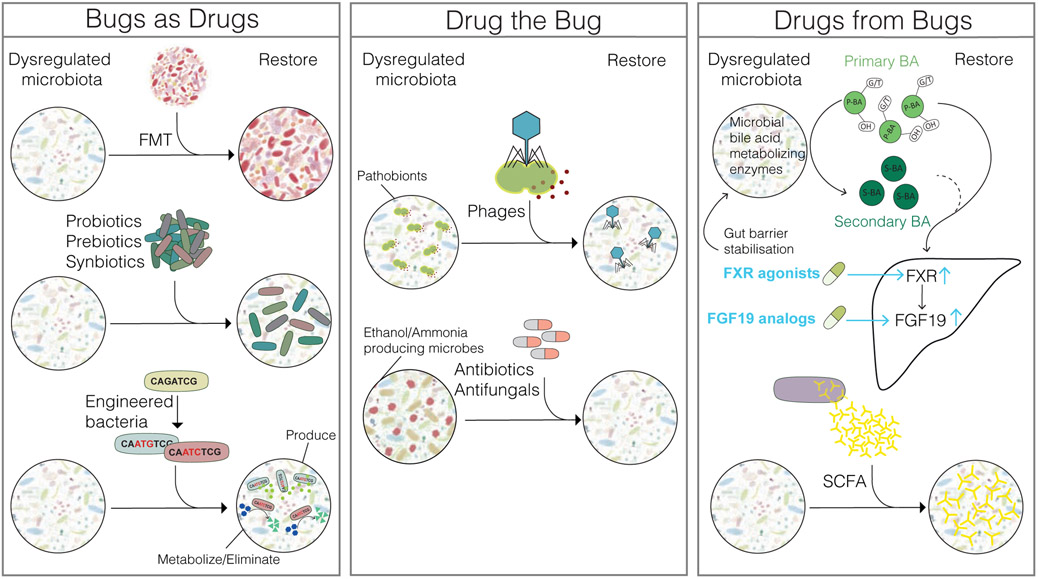
Figure 2. Targeting the gut microbiota for treatment of liver disease (bugs as drugs).
Additive approaches include fecal microbiota transfer (FMT), transplanting entire microbe communities; and administration of probiotics (living bacteria), prebiotics (group of nutrients that enhance growth of specific bacteria), synbiotics (combination of probiotics and prebiotics), and engineered bacteria, which can produce beneficial metabolites or metabolize toxins into non-toxic products (drug the bug). Specific pathogens or pathobionts can be eliminated using bacteriophages (phages) targeting specific bacterial strains. Pathogenic microbes, such as those with the ability to generate high levels of ethanol or ammonia could be targeted with antibiotics or antifungals (drugs from bugs). Supplementation of microbe-derived molecules or their derivatives such as farnesoid X receptor (FXR) agonists, FGF19 analogs, or short-chain fatty acids (SCFA), can replace lost metabolic activity of microbes. Microbial bile acid metabolizing enzymes include bile salt hydrolases that catalzye deconjugation, enzymes for hydroxylation, epimerization, esterification and desulfation, which eventually leads to the transformation of primary bile acids (P-BA) to secondary bile acids (S-BA). G/T represents glycine/taurine conjugates, OH represents hydroxy groups.
FMT
FMT is a radical approach of changing the composition of an individual’s gut microbiome. FMT has been investigated in cirrhotic patients with recurrent hepatic encephalopathy and alcoholic hepatitis with overall promising results (Table 1). There have been no published reports of the effects of FMT in patients with NASH. In FMT studies including patients with metabolic syndrome and obesity, FMT led to improved peripheral insulin sensitivity in some studies, however, effects were mild and no significant changes were observed related to other metabolic parameters and body mass index (Table 1). In addition to recently raised safety concerns (DeFilipp et al., 2019), there are questions about whether FMT alone, without other treatments such as lifestyle interventions, will have long and lasting effects—particularly for patients with NAFLD, characterized by disease progression over years and multifactorial pathogenesis.
Table 1.
Selected studies targeting the gut-liver axis
| Type | Study subjects | Intervention details |
Key outcomes | Safety | Reference |
|---|---|---|---|---|---|
| FMT | n=20 cirrhotic patients (mainly due to chronic hepatitis C virus and/or alcohol use disorder) with MELD <17 and recurrent HE | Single FMT enema following antibiotic treatment vs. SOC | FMT well tolerated, significantly less patients receiving FMT developed HE. FMT increased microbial diversity and increased beneficial bacterial taxa | Less SAEs in the FMT group compared with SOC, no complications related to FMT | (Bajaj et al., 2019) |
| n=8 severe AH patients, n=18 historical controls with severe AH, both groups steroid-ineligible | Daily FMT via naso-jejunal tube for 7 days | 87.5% of patients who received FMT survived for 1 year, compared with 33.5% of matched controls | Excessive flatulence was a complaint of 50% of patients with FMT | (Philips et al., 2017) | |
| n=51 patients with severe AH | n=8 corticosteroids, n=17 nutritional support only, n=10 pentoxifylline, n=16 FMT (daily via naso-jejunal tube for 7 days) | Proportions of patients surviving at the end of 3 months in the steroids, nutrition, pentoxifylline, and FMT group were 38%, 29%, 30%, and 75%, respectively | Not reported | (Philips et al., 2018) | |
| n=38 patients with metabolic syndrome | Single allogenic FMT via naso-jejunal tube from lean donor vs. autologous FMT | No metabolic changes at 18 weeks after FMT, insulin sensitivity at 6 weeks after allogenic FMT was significantly improved | No SAEs in both groups | (Kootte et al., 2017) | |
| n=18 patients with metabolic syndrome | Single allogenic FMT via naso-jejunal tube from lean donor vs. autologous FMT | Significantly improved peripheral insulin sensitivity 6 weeks after FMT | No SAEs in both groups | (Vrieze et al., 2012) | |
| n=22 obese subjects without diabetes, NASH or metabolic syndrome | Randomized, double-blind study, FMT by capsules from a lean donor or placebo capsules | No significant changes in BMI at 12-week follow-up | No adverse events in both groups | (Allegretti et al., 2020) | |
| n=24 obese subjects with mild to moderate insulin resistance | Randomized, double-blind study, weekly FMT by capsules vs. placebo capsules for 6 weeks | No significant differences between groups for insulin sensitivity, body mass index and fat mass | No significant differences in adverse events, no FMT related SAEs | (Yu et al., 2020) | |
| Next-generation engineered bacteria | Mouse models of hyperammonemia | Oral gavage of engineered Escherichia coli Nissle strains (SYNB1020) | Significant lowering of blood ammonia levels | (Kurtz et al., 2019) | |
| n=23 patients with cirrhosis | Escherichia coli Nissle strains (SYNB1020) for 6 days vs. placebo | No evidence of blood ammonia lowering or changes in other exploratory endpoints relative to placebo | No adverse events reported | (Synlogic, 2019) | |
| Experimental alcoholic liver disease (mice) | Oral gavage of engineered Lactobacillus reuteri-producing IL-22 vs. non-engineered Lactobacillus reuteri | Reduced bacterial translocation to the liver by increasing the expression of REG3G in the intestine, reduced ethanol-induced liver damage | (Hendrikx et al., 2019) | ||
| Bacteriophages | Humanized mouse models of ethanol-induced liver damage | Oral gavage of bacteriophages targeting cytolysin-positive E. faecalis vs. control phages | Bacteriophage administration reduced the severity of ethanol-induced liver disease | (Duan et al., 2019) | |
| Targeting the bile acid pathway | Mouse models of ethanol-induced liver damage | Non-tumorigenic variant of FGF19 vs. vehicle | Improved gut barrier function and reduced liver damage | (Hartmann et al., 2018) | |
| n=19 AH patients | Obeticholic acid (OCA) vs. placebo | Decrease in MELD score from baseline to Day 42 of −3.4 in the OCA arm, and −2.2 in the placebo arm (P-Value of 0.62) | No AEs related to study drug but discontinued due to safety concerns in patients with advanced liver disease | Clinicaltrials.gov identifier: NCT02039219 | |
| n=82 biopsy-proven NASH | Randomized, double-blind, placebo-controlled, phase 2 study, NGM282 (engineered FGF19 analogue) vs. placebo | Significant reduction in histological features, such as the NAFLD activity score and liver fibrosis after 12 weeks of treatment | AEs included diarrhea, nausea, frequent bowel movements, and abdominal pain. No SAE related to study drug | (Harrison et al., 2018; Harrison et al., 2019) | |
| n= 931 biopsy-proven NASH | Interim analysis of a randomized, double-blind, placebo-controlled study, OCA vs. Placebo | OCA 25 mg significantly improved fibrosis and key components of NASH disease activity after 18 months of treatment | Most common adverse event were pruritus and increases in LDL cholesterol related to OCA | (Younossi et al., 2019) | |
| SCFAs | Mouse models of ethanol-induced liver damage | Administration of tributyrin (ester composed of butyrate and glycerol) | Gut barrier stabilization, reduced liver injury,however, no effect on liver injury from long-term ethanol administration | (Cresci et al., 2014). | |
| n=60 overweight subjects | Randomized, controlled study, inulin-propionate ester vs. inulin-control administration over 24 weeks | Significantly reduced weight gain, intra-abdominal adipose tissue distribution, hepatic lipid content in the Inulin-propionate ester group | No increased AEs in the inulin-propionate ester group | (Chambers et al., 2015) | |
| n=10 patients with metabolic syndrome, n=9 healthy | Oral administration of sodium butyrate daily for 4 weeks | No effect on plasma and fecal butyrate levels after treatment. Significantly improved insulin sensitivity only in healthy lean subjects | Not reported | (Bouter et al., 2018) |
Bold font in the second column highlights the studied organism or disease. AEs, adverse events, AH, alcoholic hepatitis; BMI, body mass index; FMT, fecal microbiota transfer; HE, hepatic encephalopathy; OCA, obeticholic acid; REG3G, antimicrobial molecule regenerating islet derived 3 gamma; SAEs, severe adverse events; SCFAs, short-chain fatty acids; SOC, standard of care
First-generation biotics
Several studies have investigated the effect of probiotics, prebiotics, or synbiotics in patients with chronic liver diseases (for a review, see (Schwenger et al., 2019). Administration of probiotics, prebiotics, or synbiotics to patients with ALD, cirrhosis, and hepatic encephalopathy led to improvements in direct and indirect markers of disease severity (Schwenger et al., 2019). A meta-analysis of data from 1252 participants in 21 trials concluded that administration of probiotics or synbiotics to patients with NAFLD was associated with significant reductions in levels of alanine aminotransferase, in liver stiffness measurements, and in hepatic steatosis graded by ultrasound (Sharpton et al., 2019). However, due to heterogeneity among studies and small sample sizes in individual trials, no clear recommendation can be made for the use of probiotics, prebiotics, or synbiotics in patients with chronic liver diseases.
Next-generation engineered bacteria
Administration of ammonia consuming engineered bacteria has been tested as treatment for hepatic encephalopathy after demonstrating the potential to lower blood ammonia level in mouse models of hyperammonemia. However, further drug development was discontinued because blood levels of ammonia could not be lowered in patients with cirrhosis (Table 1). Another potential application of engineered bacteria is increasing interleukin 22 (IL-22), a cytokine that signals to epithelial cells in several organs, including the intestine and the liver (Hendrikx and Schnabl, 2019). In parallel to studies in mice, using engineered Lactobacillus reuteri-producing IL-22 (Table 1), the IL-22 agonist F-652 was evaluated in a phase 2a open-label study of patients with alcoholic hepatitis. F-652 was safe and resulted in reduced serum and plasma markers of inflammation and high rates of survival (Arab et al., 2019). Overall, engineered microbes might be used to restore homeostatic functions to the gut microbiome and improve liver-related outcomes.
Drug the bug
Bacteria, pathobionts, or pathogens can also be harmful and promote liver disease progression. However, bacteria can be targeted and eliminated with subtractive treatment approaches (Mimee et al., 2016). Currently used approaches with antibiotics are not specific and can have side effects because they also eliminate beneficial commensals. Highly specific treatment approaches with bacteriophages (phages) have been developed and can effectively edit the gut microbiota. Even more sophisticated approaches could target specific microbial enzymes to change synthesis of metabolites. One preclinical example demonstrated that a specific chemical inhibitor targeting microbial trimethylamine lyase decreases trimethylamine N-oxide (TMAO) production, which has been implicated in pathogenesis of cardiovascular diseases (Wang et al., 2015) (Figure 2).
Antibiotics
Patients with advanced liver diseases are frequently given antibiotics due to an increased susceptibility to pathobiont invasion caused by immune dysfunction, or to reduce episodes of hepatic encephalopathy. Due to the relative non-specificity of most antibiotics and the risk for emergence of multidrug-resistant bacteria, it is unlikely that management of patients with ALD or NAFLD without cirrhosis will involve long-term administration of antibiotics.
Bacteriophages
Bacteriophages (phages) are viruses that infect bacteria, replicate inside them, and kill them. Phage-based therapies have been predominantly studied to treat infectious diseases caused by multidrug-resistant bacteria. Administration of phages against cytolytic E. faecalis to gnotobiotic mice that received transplantations of feces from patients with alcoholic hepatitis who carried cytolysin-positive E. faecalis reduced the severity of ethanol-induced liver disease in the mice (Duan et al., 2019). This study provided the first evidence that strategies to target single gut pathobionts might be developed for treatment of liver disease, but testing in humans, and addressing bacterial resistance against phages, is required.
Drugs from bugs
Microbe-derived metabolites (also called postbiotics) and their related signaling pathways, in humans or in other microbes, might be used in treatment of ALD and NAFLD (Mimee et al., 2016). Supplementation with these molecules or their derivatives could replace some metabolic activities of lost or reduced microbes (Figure 2).
Targeting the bile acid pathway
Given the associations between dysregulation of bile acid homeostasis and severity of ALD and NAFLD, it might be effective to target the bile acid and FXR pathway. This has been successfully proven in mouse models of ethanol-induced liver damage but studies in humans are needed (Table 1).
Targeting the bile acid pathway, e.g. with FGF19 analogues and FXR agonists such as obeticholic acid (OCA) is one of the most promising approaches for treatment of NAFLD (Table 1). OCA leads to alterations in the human gut microbiome (Friedman et al., 2018) and preclinical studies found that OCA prevents disruption of the gut vascular barrier (Mouries et al., 2019). Therefore, the positive effects of OCA could be mediated, in part, by modulating the intestinal microbiome or gut barrier integrity. On the other hand, patients might respond in different ways to OCA or other drugs that affect interactions between the intestine and liver, depending on the composition of the gut microbiome, presence of gut bacterial overgrowth, and gut barrier dysfunction.
SCFAs
Overall, there are many questions about the relationship between SCFAs and ALD and NAFLD and data on using SCFAs as therapeutic approach in these patients is limited (Table 1). Trials should investigate the effects of increasing intestinal levels of SCFAs, via direct supplementation or administration of SCFA-producing bacteria, in patients with these liver diseases.
CHALLENGES AND FUTURE PERSPECTIVES
It is important to study the gut microbiome in humans, because rodents develop only some features of human specific alterations in the gut microbiome. The microbiota of mice can be humanized, by transplantation of fecal material into intestines of gnotobiotic mice; this allows for studies of relationships between human microbes and specific diseases, but this model has several important limitations. Most importantly, some human taxa cannot colonize mice, and the microbial communities differ substantially between recipient mice and their human donors (Walter et al., 2020).
Studies of the human microbiome have predominantly relied on cross-sectional, observational designs—an important shortcoming. The composition of the gut microbiome responds rapidly to variations in nutrition, lifestyle, medications, and environmental conditions (Gupta et al., 2017). Most studies rely on a single snapshot of gut microbiota composition, so it is not surprising that there are issues in reproducibility. These issues have limited the utility of gut microbes or microbial profiles as prognostic or diagnostic markers. Furthermore, patients with fatty liver often have co-morbidities that can bias results—identified gut microbiota signatures could be affected by obesity or type 2 diabetes, or medications used. Longitudinal sample collection and better-matched controls are needed to establish associations between gut microbiota signatures and specific disorders. Studies that monitor gut microbiota dynamics over time, in large numbers of patients with different features, and in response to different events, are needed to determine the mechanisms by which the intestinal microbiota contributes to or responds to liver disease (Poyet et al., 2019).
In addition to studies of the intestinal bacteria associated with liver diseases, studies are needed of other microbes, such as fungi, viruses, and archaea, as well as the interactions among all gut microbes. There is evidence that microbes behave differently in different gut environmental conditions, so microbiota research might shift from taxonomic descriptions toward functional studies of interactions among human cells, microbes, and their metabolic pathways. Further research is needed to determine the presence and abundance of small metabolites and the expression of microbial virulence factors in network analyses. All of these aspects will hopefully lead to a better understanding of the dynamic interactions between the gut microbiome and liver diseases.
One of the biggest challenges will be to categorize patients with ALD or NAFLD into subgroups and to identify those associated with specific microbiome alterations. We have made progress in developing methods to edit the gut microbiota, such as with phages, and introduce engineered bacteria that can restore a specific deficiency or supplement depleted microbial-derived metabolites as postbiotics. Despite these technological advances, current trial designs are still one size fits all. As we have shown in this review, not all patients have gut barrier dysfunction—a drug that restores gut barrier function will fail if participants are not selected based on biomarkers of increased intestinal permeability. Microbiota-based treatments must be carefully selected based on the specific alteration in a patient’s intestine or microbiome.
Individualized treatment approaches can now target specific strains of bacteria to increase the efficacy of medical treatments and decrease side effects. It is unlikely that patients with NAFLD all have increased endogenous ethanol production or increased bile acid synthesis. Therefore, it will be important to not only report the mean values of test results for the entire patient population, but to assign patients to subgroups based on microbial, genetic, and metabolic features—or a combination of these (Figure 3). These markers might help identify patients at risk for progression of ALD and NAFLD and select treatment approaches.
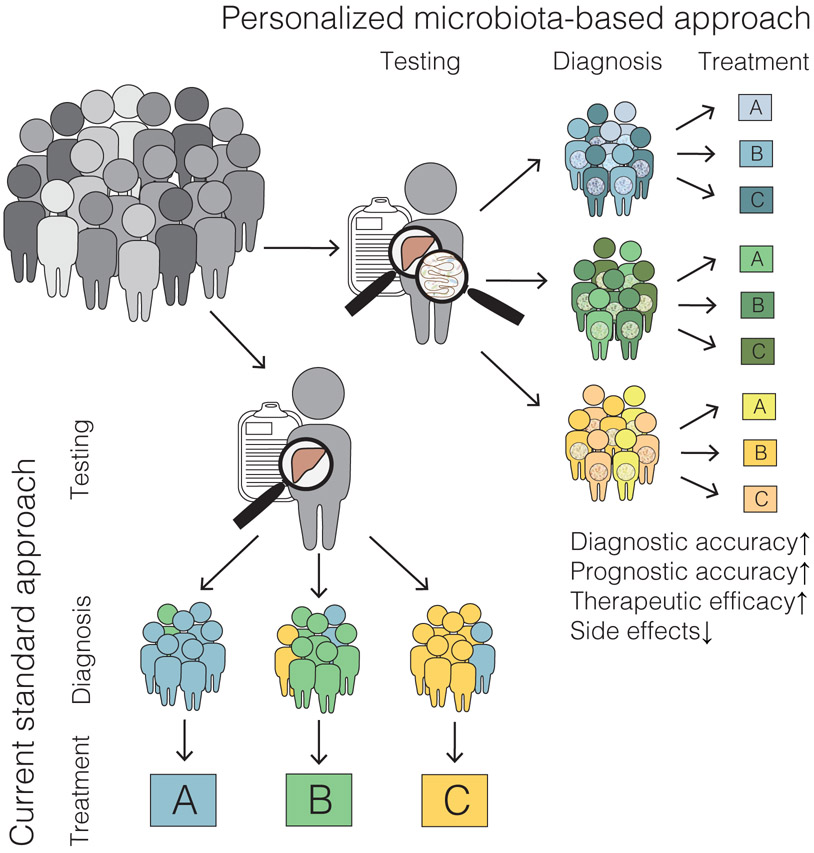
Figure 3. Precision therapies for fatty liver disease.
Microbiome studies might identify new pathways and factors that contribute to progression of fatty liver diseases; these might be used to assign patients to subgroups and identify those associated with specific alterations to the gut microbiota. Integration of this knowledge into diagnostic analyses might increase diagnostic and prognostic accuracy, and lead to personalized microbiota-based therapeutics. These approaches could increase therapeutic efficacy and reduce treatment-associated side effects, compared with standard approaches.
Understanding the intimate, essential, and sometimes toxic relationship among gut microbes and humans has tremendous potential for development of new non-invasive diagnostic and prognostic approaches as well as for development of personalized microbiota-based treatments. It appears, though, that there is much more that we don’t know than what we truly understand.
Fatty liver diseases are the most common chronic liver diseases in Western countries. The liver is exposed to components of the gut microbiota which can modulate disease. This review discusses our current understanding of the gut-liver crosstalk and implications for microbe-based biomarkers and treatment approaches.
ACKNOWLEDGEMENTS
This work was supported by a DFG fellowship (LA 4286/1-1) (to S.L.), NIH grants R01 AA020703, R01 AA24726, U01 AA026939 and by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.) and services provided by NIH-funded centers P30 DK120515 and P50 AA011999.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics and Patara Pharmaceuticals. B.S.’s institution UC San Diego has received research support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics. S.L. and B.S. have patents related to this work (PCT/US2019/024703 and US62/946,182).
REFERENCES
- Allegretti JR, Kassam Z, Mullish BH, Chiang A, Carrellas M, Hurtado J, Marchesi JR, McDonald JAK, Pechlivanis A, Barker GF, et al. (2020). Effects of Fecal Microbiota Transplantation With Oral Capsules in Obese Patients. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 18(4), 855–863.e852. [DOI] [PubMed] [Google Scholar]
- Arab JP, Sehrawat TS, Simonetto DA, Verma VK, Feng D, Tang T, Dreyer K, Yan X, Daley WL, Sanyal A, et al. (2019). An Open Label, Dose Escalation Study To Assess The Safety And Efficacy Of IL-22 Agonist F-652 In Patients With Alcoholic Hepatitis. Hepatology. 10.1002/hep.31046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, Nieuwdorp M, and Clément K (2020). Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nature reviews. Gastroenterology & hepatology. 10.1038/s41575-41020-40269-41579. [DOI] [PubMed] [Google Scholar]
- Arora T, and Bäckhed F (2016). The gut microbiota and metabolic disease: current understanding and future perspectives. J Intern Med. 280(4), 339–349. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, and Gordon JI (2004). The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America. 101(44), 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj JS, Salzman NH, Acharya C, Sterling RK, White MB, Gavis EA, Fagan A, Hayward M, Holtz ML, Matherly S, et al. (2019). Fecal Microbial Transplant Capsules Are Safe in Hepatic Encephalopathy: A Phase 1, Randomized, Placebo-Controlled Trial. Hepatology. 70(5), 1690–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, and Schmidt TM (2019). Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio. 10(1), e02566–02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouter K, Bakker GJ, Levin E, Hartstra AV, Kootte RS, Udayappan SD, Katiraei S, Bahler L, Gilijamse PW, Tremaroli V, et al. (2018). Differential metabolic effects of oral butyrate treatment in lean versus metabolic syndrome subjects. Clinical and translational gastroenterology. 9(5), 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K, Hartmann P, Jih LJ, Pizzo DP, Argemi J, Ventura-Cots M, Coulter S, Liddle C, Ling L, Rossi SJ, et al. (2018). Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol. 69(2), 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzetti E, Pinzani M, and Tsochatzis EA (2016). The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism: clinical and experimental. 65(8), 1038–1048. [DOI] [PubMed] [Google Scholar]
- Caesar R, Reigstad CS, Bäckhed HK, Reinhardt C, Ketonen M, Lundén G, Cani PD, and Bäckhed F (2012). Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 61(12), 1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Hsu C, Lo MT, Liu A, Bettencourt R, Ajmera VH, Bassirian S, Hooker J, Sy E, Richards L, et al. (2018). Link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology. 68(3), 918–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Hsu C, Singh S, Bassirian S, Kolar J, Faulkner C, Sinha N, Bettencourt R, Gara N, Valasek MA, et al. (2019). Serum bile acid patterns are associated with the presence of NAFLD in twins, and dose-dependent changes with increase in fibrosis stage in patients with biopsy-proven NAFLD. Aliment Pharmacol Ther. 49(2), 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, MacDougall K, Preston T, Tedford C, Finlayson GS, et al. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 64(11), 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Duan Y, Lang S, Jiang L, Wang Y, Llorente C, Liu J, Mogavero S, Bosques-Padilla F, Abraldes JG, et al. (2020). The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J Hepatol. 72(3), 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Duan Y, Yang L, and Schnabl B (2019). Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 68(2), 359–370. [DOI] [PubMed] [Google Scholar]
- Cope K, Risby T, and Diehl AM (2000). Increased gastrointestinal ethanol production in obese mice: implications for fatty liver disease pathogenesis. Gastroenterology. 119(5), 1340–1347. [DOI] [PubMed] [Google Scholar]
- Couch RD, Dailey A, Zaidi F, Navarro K, Forsyth CB, Mutlu E, Engen PA, and Keshavarzian A (2015). Alcohol induced alterations to the human fecal VOC metabolome. PLoS One. 10(3), e0119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresci GA, Bush K, and Nagy LE (2014). Tributyrin supplementation protects mice from acute ethanol-induced gut injury. Alcoholism, clinical and experimental research. 38(6), 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen YB, and Hohmann EL (2019). Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. The New England journal of medicine. 381(21), 2043–2050. [DOI] [PubMed] [Google Scholar]
- Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, and Parks EJ (2005). Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. The Journal of clinical investigation. 115(5), 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al. (2019). Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 575(7783):505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman ES, Li Y, Shen TD, Jiang J, Chau L, Adorini L, Babakhani F, Edwards J, Shapiro D, Zhao C, et al. (2018). FXR-Dependent Modulation of the Human Small Intestinal Microbiome by the Bile Acid Derivative Obeticholic Acid. Gastroenterology. 155(6), 1741–1752.e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VK, Paul S, and Dutta C (2017). Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front Microbiol. 8, 1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SA, Rinella ME, Abdelmalek MF, Trotter JF, Paredes AH, Arnold HL, Kugelmas M, Bashir MR, Jaros MJ, Ling L, et al. (2018). NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (London, England). 391(10126), 1174–1185. [DOI] [PubMed] [Google Scholar]
- Harrison SA, Rossi SJ, Paredes AH, Trotter JF, Bashir MR, Guy CD, Banerjee R, Jaros MJ, Owers S, Baxter BA, et al. (2019). NGM282 Improves Liver Fibrosis and Histology in 12 Weeks in Patients With Nonalcoholic Steatohepatitis. Hepatology. 10.1002/hep.30590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, Llorente C, Wang L, Alnouti Y, Fouts DE, Starkel P, et al. (2018). Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 67(6), 2150–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, and Thiele I (2019). Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. 7(1), 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. (2012). Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 482(7384), 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, Starkel P, Ho SB, Gao B, Fiehn O, et al. (2019). Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 68(8), 1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrikx T, and Schnabl B (2019). Indoles: metabolites produced by intestinal bacteria capable of controlling liver disease manifestation. J Intern Med. 286(1), 32–40. [DOI] [PubMed] [Google Scholar]
- Hoyles L, Fernandez-Real JM, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, et al. (2018). Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nature medicine. 24(7), 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ, et al. (2018). Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 67(10), 1881–1891. [DOI] [PubMed] [Google Scholar]
- Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, and Gahan CGM (2014). Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proceedings of the National Academy of Sciences. 111(20), 7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanola O, Ferrusquia-Acosta J, Garcia-Villalba R, Zapater P, Magaz M, Marin A, Olivas P, Baiges A, Bellot P, Turon F, et al. (2019). Circulating levels of butyrate are inversely related to portal hypertension, endotoxemia, and systemic inflammation in patients with cirrhosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 33(10), 11595–11605. [DOI] [PubMed] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, and Krakoff J (2011). Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. The American journal of clinical nutrition. 94(1), 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, Takei H, Nittono H, Ridlon JM, Fuchs M, et al. (2014). Colonic inflammation and secondary bile acids in alcoholic cirrhosis. American journal of physiology. Gastrointestinal and liver physiology. 306(11), G929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, Hermes G, Bouter KE, Koopen AM, Holst JJ, et al. (2017). Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell metabolism. 26(4), 611–619.e616. [DOI] [PubMed] [Google Scholar]
- Kurtz CB, Millet YA, Puurunen MK, Perreault M, Charbonneau MR, Isabella VM, Kotula JW, Antipov E, Dagon Y, Denney WS, et al. (2019). An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Science translational medicine. 11(475). [DOI] [PubMed] [Google Scholar]
- Lang S, Demir M, Duan Y, Martin A, and Schnabl B (2020a). Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver International. 40(4), 860–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S, Duan Y, Liu J, Torralba MG, Kuelbs C, Ventura-Cots M, Abraldes JG, Bosques-Padilla F, Verna EC, Brown RS Jr., et al. (2020b). Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatology. 71(2), 522–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, Windey K, Tremaroli V, Backhed F, Verbeke K, et al. (2014). Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proceedings of the National Academy of Sciences of the United States of America. 111(42), E4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BP, Vittinghoff E, Dodge JL, Cullaro G, and Terrault NA (2019). National Trends and Long-term Outcomes of Liver Transplant for Alcohol-Associated Liver Disease in the United States. JAMA Internal Medicine. 179(3), 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, Puchois V, Martin JC, Lepage P, Le Roy T, et al. (2016). Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 65(5), 830–839. [DOI] [PubMed] [Google Scholar]
- Llorente C, Jepsen P, Inamine T, Wang L, Bluemel S, Wang HJ, Loomba R, Bajaj JS, Schubert ML, Sikaroodi M, et al. (2017). Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat Commun. 8(1), 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomba R, and Adams LA (2019). The 20% Rule of NASH Progression: The Natural History of Advanced Fibrosis and Cirrhosis Caused by NASH. Hepatology (Baltimore, Md.). 70(6), 1885–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, et al. (2017). Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell metabolism. 25(5), 1054–1062.e1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Zúñiga V, Bartolí R, Planas R, Hofmann AF, Viñado B, Hagey LR, Hernández JM, Mañé J, Alvarez MA, Ausina V, et al. (2003). Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology (Baltimore, Md.). 37(3), 551–557. [DOI] [PubMed] [Google Scholar]
- Mazagova M, Wang L, Anfora AT, Wissmueller M, Lesley SA, Miyamoto Y, Eckmann L, Dhungana S, Pathmasiri W, Sumner S, et al. (2015). Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 29(3), 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimee M, Citorik RJ, and Lu TK (2016). Microbiome therapeutics - Advances and challenges. Adv Drug Deliv Rev. 105(Pt A), 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero N, Ruiz L, Sánchez B, Margolles A, and Delgado S (2019). Intestinal Bacteria Interplay With Bile and Cholesterol Metabolism: Implications on Host Physiology. Front Physiol. 10(185). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouries J, Brescia P, Silvestri A, Spadoni I, Sorribas M, Wiest R, Mileti E, Galbiati M, Invernizzi P, Adorini L, et al. (2019). Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol. 71(6), 1216–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, Fung S, Fischer SE, McGilvray IG, and Allard JP (2016). Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One. 11(5), e0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobili V, Alisi A, Mosca A, Della Corte C, Veraldi S, De Vito R, De Stefanis C, D'Oria V, Jahnel J, Zohrer E, et al. (2018). Hepatic farnesoid X receptor protein level and circulating fibroblast growth factor 19 concentration in children with NAFLD. Liver Int. 38(2), 342–349. [DOI] [PubMed] [Google Scholar]
- Parker R, Aithal GP, Becker U, Gleeson D, Masson S, Wyatt JI, and Rowe IA (2019). Natural history of histologically proven alcohol-related liver disease: A systematic review. Journal of hepatology. 71(3), 586–593. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, et al. (1999). Bile acids: natural ligands for an orphan nuclear receptor. Science (New York, N.Y.). 284(5418), 1365–1368. [DOI] [PubMed] [Google Scholar]
- Philips CA, Pande A, Shasthry SM, Jamwal KD, Khillan V, Chandel SS, Kumar G, Sharma MK, Maiwall R, Jindal A, et al. (2017). Healthy Donor Fecal Microbiota Transplantation in Steroid-Ineligible Severe Alcoholic Hepatitis: A Pilot Study. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 15(4), 600–602. [DOI] [PubMed] [Google Scholar]
- Philips CA, Phadke N, Ganesan K, Ranade S, and Augustine P (2018). Corticosteroids, nutrition, pentoxifylline, or fecal microbiota transplantation for severe alcoholic hepatitis. Indian J Gastroenterol. 37(3), 215–225. [DOI] [PubMed] [Google Scholar]
- Poyet M, Groussin M, Gibbons SM, Avila-Pacheco J, Jiang X, Kearney SM, Perrotta AR, Berdy B, Zhao S, Lieberman TD, et al. (2019). A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nature medicine. 25(9), 1442–1452. [DOI] [PubMed] [Google Scholar]
- Rau M, Rehman A, Dittrich M, Groen AK, Hermanns HM, Seyfried F, Beyersdorf N, Dandekar T, Rosenstiel P, and Geier A (2018). Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J. 6(10), 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenger KJ, Clermont-Dejean N, and Allard JP (2019). The role of the gut microbiome in chronic liver disease: the clinical evidence revised. JHEP Rep. 1(3), 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, and Hardt PD (2010). Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring, Md.). 18(1), 190–195. [DOI] [PubMed] [Google Scholar]
- Sharpton SR, Maraj B, Harding-Theobald E, Vittinghoff E, and Terrault NA (2019). Gut microbiome-targeted therapies in nonalcoholic fatty liver disease: a systematic review, meta-analysis, and meta-regression. The American journal of clinical nutrition. 110(1), 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Puri P, Muthiah MD, Daitya K, Brown R, Chalasani N, Liangpunsakul S, Shah VH, Gelow K, Siddiqui MS, et al. (2020). Fecal microbiome distinguishes alcohol consumption from alcoholic hepatitis but does not discriminate disease severity. Hepatology. 10.1002/hep.31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synlogic (2019). https://investor.synlogictx.com/news-releases/news-release-details/synlogic-discontinues-development-synb1020-treat-hyperammonemia.
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, and Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444(7122), 1027–1031. [DOI] [PubMed] [Google Scholar]
- Turner JR (2009). Intestinal mucosal barrier function in health and disease. Nature reviews. Immunology. 9(11), 799–809. [DOI] [PubMed] [Google Scholar]
- Voican CS, Njiké-Nakseu M, Boujedidi H, Barri-Ova N, Bouchet-Delbos L, Agostini H, Maitre S, Prévot S, Cassard-Doulcier A-M, Naveau S, et al. (2015). Alcohol withdrawal alleviates adipose tissue inflammation in patients with alcoholic liver disease. Liver International. 35(3), 967–978. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 143(4), 913–916.e917. [DOI] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall HU, and Backhed F (2016). Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell metabolism. 24(1), 41–50. [DOI] [PubMed] [Google Scholar]
- Walter J, Armet AM, Finlay BB, and Shanahan F (2020). Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell. 180(2), 221–232. [DOI] [PubMed] [Google Scholar]
- Wang L, Fouts DE, Starkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho SB, Brenner DA, et al. (2016). Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell host & microbe. 19(2), 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, and Ahmed A (2015). Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 148(3), 547–555. [DOI] [PubMed] [Google Scholar]
- Wree A, McGeough MD, Peña CA, Schlattjan M, Li H, Inzaugarat ME, Messer K, Canbay A, Hoffman HM, and Feldstein AE (2014). NLRP3 inflammasome activation is required for fibrosis development in NAFLD. Journal of Molecular Medicine. 92(10), 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Wang F, Wong N-K, He J, Zhang R, Sun R, Xu Y, Liu Y, Li W, Koike K, et al. (2019). Global liver disease burdens and research trends: Analysis from a Chinese perspective. Journal of Hepatology. 71(1), 212–221. [DOI] [PubMed] [Google Scholar]
- Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, et al. (2017). Intestinal fungi contribute to development of alcoholic liver disease. The Journal of clinical investigation. 127(7), 2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi ZM, Ratziu V, Loomba R, Rinella M, Anstee QM, Goodman Z, Bedossa P, Geier A, Beckebaum S, Newsome PN, et al. (2019). Obeticholic acid for the treatment of non-alcoholic steatohepatitis: interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet (London, England). 394(10215), 2184–2196. [DOI] [PubMed] [Google Scholar]
- Yu EW, Gao L, Stastka P, Cheney MC, Mahabamunuge J, Torres Soto M, Ford CB, Bryant JA, Henn MR, and Hohmann EL (2020). Fecal microbiota transplantation for the improvement of metabolism in obesity: The FMT-TRIM double-blind placebo-controlled pilot trial. PLoS medicine. 17(3), e1003051–e1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, Zhao X, Li N, Li S, Xue G, et al. (2019). Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell metabolism. 30(4), 675–688.e677. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, and Edwards PA (2006). Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 103(4), 1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, and Gill SR (2013). Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 57(2), 601–609. [DOI] [PubMed] [Google Scholar]
- Zuo T, and Ng SC (2018). The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front Microbiol. 9, 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]