Abstract
The imbalance between the proliferation and apoptosis of B-cell precursors is an important contributor to the pathogenesis of B-cell precursor acute lymphoblastic leukemia (BCP-ALL), while its specific regulatory mechanism remains perplexing. This study aimed to expound the underlying mechanism of the proliferation and apoptosis of BCP-ALL cells from the perspective of non-coding RNA. In this study, long non-coding RNA colorectal neoplasia differentially expressed (LncRNA CRNDE) was upregulated in the bone marrow of BCP-ALL patients and BCP-ALL cell lines (NALM-6 and RS4;11). Functionally, LncRNA CRNDE knockdown restrained cell proliferation and boosted cell apoptosis in NALM-6 and RS4;11 cells. The subsequent investigation confirmed that LncRNA CRNDE bound to miR-345-5p and negatively regulated miR-345-5p expression. The overexpression of miR-345-5p suppressed cell proliferation and boosted cell apoptosis in NALM-6 and RS4;11 cells. Further experiments revealed that miR-345-5p downregulated cyclic AMP response element-binding protein (CREB) expression by targeting its mRNA directly. CREB overexpression reversed the effect of miR-345-5p mimic on cell proliferation and apoptosis in NALM-6 and RS4;11 cells. Finally, in vivo experiments showed that LncRNA CRNDE knockdown prolonged the survival of mice xenotransplanted with NALM-6 cells. In conclusion, LncRNA CRNDE upregulated CREB expression by suppressing miR-345-5p, thus promoting cell proliferation and reducing cell apoptosis in BCP-ALL.
Keywords: B-cell precursor acute lymphoblastic leukemia, cyclic AMP-binding protein, long non-coding RNA colorectal neoplasia differentially expressed, miR-345-5p
INTRODUCTION
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is a malignancy characterized by the aberrant accumulation of immature clonal B-cell precursors in the bone marrow (BM) (Zhuang et al., 2014). Despite the advances in treating BCP-ALL, the outcomes of BCP-ALL patients remain unsatisfactory (Jacobson et al., 2016). Buske et al. (1997) reported that the imbalance between the proliferation and apoptosis of B-cell precursors is an important contributor to the pathogenesis of BCP-ALL. Therefore, identifying the specific regulatory mechanism of the proliferation and apoptosis of B-cell precursors is important for the development of novel therapeutic approaches for BCP-ALL treatment.
Long non-coding RNA colorectal neoplasia differentially expressed (LncRNA CRNDE) was first found to be activated in colorectal neoplasia (Graham et al., 2011), and it is involved in various cancers by regulating cell proliferation and apoptosis (Hu et al., 2017; Liu et al., 2017). By conducting a transcriptome analysis, Lajoie et al. (2017) found that LncRNA CRNDE was upregulated in the blood samples of BCP-ALL patients. Our preliminary experimental results showed that compared with the healthy controls, the BM of BCP-ALL patients exhibited a higher level of LncRNA CRNDE expression, indicating that LncRNA CRNDE could play a role in BCP-ALL progression.
Increasing evidence has confirmed that LncRNAs can act as competing endogenous RNAs (ceRNAs) that bind to miRNAs and remove their suppressive effect on mRNA expression, thus regulating the various biological processes in disease (Cesana et al., 2011). Han et al. (2019) demonstrated that in liver cancer cells, LncRNA CRNDE released a high-mobility group protein A2 expression by sponging miR-33a, proving the ability of LncRNA CRNDE to function as a ceRNA. Using Bioinformatics prediction software (LncBase Predicted v.2), we found potential binding sites between LncRNA CRNDE and several miRNAs (miR-345-5p, miR-515-5p, miR-4282, miR-543, and miR-4301). Interestingly, these miRNAs have been reported to be tumor suppressor genes because of their inhibitory effect on cell proliferation, invasion, and migration in various cancers (Avval et al., 2019; Chen et al., 2016; Kang et al., 2016; Liu et al., 2019; Zhang et al., 2019).
Inspired by previous studies and the forecast of Bioinformatics software, we speculate that LncRNA CRNDE may regulate the proliferation and apoptosis of BCP-ALL cells by sponging the target miRNA, thus contributing to the development of BCP-ALL. In this study, the BM samples of patients with BCP-ALL and BCP-ALL cell lines were used to determine the role of LncRNA CRNDE in BCP-ALL and expound its regulation mechanism in light of the ceRNA mechanism.
MATERIALS AND METHODS
Human study cohort
BM biopsies from 26 BCP-ALL patients were collected. The BM biopsies from 15 patients who presented with unexplained thrombocytosis or anemia but with no hematologic malignancy or an autoimmune disease found during diagnostic procedure and follow-up were collected as the controls. This study was approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University (No. 2019-KY-194). Each subject signed an informed consent form.
Cell culture and cell transfection
The primary normal precursor B-cells were isolated from healthy controls using fluorescence-activated cell sorting on a FACSVantage (BD, USA) (Buske et al., 1997). The BCP-ALL cell lines (NALM-6, RS4;11, CEMO-1, CCRF-SB, and SUP-B15) were obtained from the American Type Culture Collection (USA). The cells were maintained in the RPMI 1640-Glutamax-I medium (Thermo Fischer Scientific, USA) containing 10% fetal calf serum (Gibco, USA), penicillin (100 U/ml; Invitrogen, USA), and streptomycin (100 mg/ml; Invitrogen) with 5% CO2 at 37°C.
The RNAi vectors (shRNA-CRNDE and miR-345-5p inhibitor), overexpression vectors (miR-345-5p mimic and pcDNA-CREB), and relative negative controls (shRNA, inhibitor-NC, mimic-NC, and pcDNA) were synthesized by RiboBio (China) and transfected into cells using Lipofectamine 3000 (Invitrogen).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the cells or BM tissues of mice using the TURBO DNA-free Kit (Thermo Fischer Scientific). After determining their concentration and purity, the RNA samples were reversely transcripted into cDNA. The qRT-PCR assay was conducted in the ABI 7500 Real-Time PCR System (Applied Biosystems, USA). The sequences of the primers are shown in Table 1. U6 or GAPDH was used as an endogenous control.
Table 1.
Sequences of primers and shRNAs
| Gene name | Sequence |
|---|---|
| LncRNA CRNDE | Forward: 5′-GAGGACGTGCTGGGGCT-3′ |
| Reverse: 5′-CTGAGTCCATGTCCCGAATC-3′ | |
| CREB | Forward: 5′-TGCCACATTAGCCCAGGTA-3′ |
| Reverse: 5′-GCTGTATTGCTCCTCCCT-3′ | |
| GAPDH | Forward: 5′-GGTGGCAGAGGCCTTTG-3′ |
| Reverse: 5′-TGCCCATTTAGCATCTCCTT-3′ | |
| miR-345-5p | Forward: 5′-GCTGACTCCTAGTCCA-3′ |
| Reverse: 5′-TGGTGTCGTGGAGTCG-3-3′ | |
| U6 | Forward: 5′-CGCTTCGGCAGCACATATAC-3′ |
| Reverse: 5′-TTCACGAATTTGCGTGTCAT-3′ | |
| shRNA-NC | 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| shRNA1-CRNDE | 5′-GAGUGCUAGUUCUCUUGUATT-3′ |
| shRNA2-CRNDE | 5′-GGATGCTGTCAGCTAAGTTCA-3′ |
| shRNA3-CRNDE | 5′-GUCACGCAGAAGAAGGUUATT-3′ |
Measurement of cell viability, apoptosis, and proliferation
Cell viability was assessed by methyl thiazolyl tetrazolium (MTT) assay. The cells seeded in 96-well plates were reacted with MTT (10 μl/well; Cwbio, China) at 37°C in the dark. Four hours later, the formazan crystals at the bottom of each well were dissolved by the addition of dimethyl sulfoxide (150 μl/well; Cwbio) at 37°C for 15 min. The absorbance intensity was measured at 490 nm.
Cell apoptosis was measured by the Annexin V-FITC Apoptosis Detection Kit (Univ-bio, China) using a flow cytometer (FACScan; BD).
Cell proliferation was detected using the bromodeoxyuridine (BrdU) incorporation assay kit (Sigma-Aldrich, USA). Briefly, the cells were incubated with BrdU, blocking buffer, a primary antibody against BrdU, secondary antibody, and DAPI. The BrdU-positive cells were counted using a fluorescence microscope (Nikon, Japan).
Western blot
The protein levels of cleaved caspase 3 (cl-caspase 3), total caspase 3 (T-caspase 3), B-cell lymphoma-2 (Bcl-2), Bcl-2-associated X (Bax), cleaved poly ADP-ribose polymerase (cl-PARP), and cyclic AMP response element-binding protein (CREB) were determined by Western blot. The primary antibodies used were as follows: anti-cl-caspase 3 (ab49822, 1:500; Abcam, UK), anti-T-caspase 3 (ab13847, 1:500; Abcam), anti-Bcl-2 (ab182858, 1:2,000; Abcam), anti-Bax (ab32503, 1:1,000; Abcam), anti-cl-PARP (ab32064, 1:2,000; Abcam), anti-CREB (ab32515, 1:500; Abcam), and anti-GAPDH (ab9485, 1:2,500; Abcam).
Dual-luciferase reporter assay
The binding sites between LncRNA CRNDE and miR-345-5p were predicted by LncBase Predicted version 2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted). LncRNA CRNDE wild-type (CRNDE-WT) and LncRNA CRNDE mutant-type (CRNDE-Mut) were synthesized by GenePharma (China) and inserted into pmirGLO plasmids, respectively. Then, 0.5 μg plasmid and 20 nM miR-345-5p mimic or miR-345-5p inhibitor or their negative controls (mimic-NC or inhibitor-NC) were cotransfected in well-grown NALM-6 cells. Forty-eight hours after transfection, the cells were lysed, and the luciferase activities were measured with the Dual-Luciferase Reporter Assay Kit (Promega, China) following the manufacturer’s protocol.
The binding sites between miR-345-5p and CREB mRNA were predicted by Targetscan (http://www.targetscan.org/vert_72/). A full-length of the wild-type CREB mRNA (CREB-WT) and a full-length of the CREB mRNA with a mutating miR-345-5p binding site (CREB-Mut) were synthesized by GenePharma. The luciferase activities were measured as described above. The elevated renilla luciferase activities of CRNDE-WT, CRNDE-MUT, CREB-WT, and CREB-MUT plasmids indicated that the plasmids were transfected into the NALM-6 cells successfully (Supplementary Fig. S1).
RNA pull-down assay
The combination of LncRNA CRNDE and miR-345-5p was determined by the RNA pull-down assay. The biotin-labeled LncRNA CRNDE (Bio-CRNDE) and its negative control (Bio-NC) were provided by RiboBio. The NALM-6 cells (1.5 × 107) were collected and lysed using a 100 μl specific lysis buffer (Thermo Fischer Scientific). The lysate was incubated with 50 pmol Bio-CRNDE and 50 μl streptavidin agarose magnetic beads for 1 h at 4°C. miR-345-5p in the complex pulled down by Bio-CRNDE was detected by qRT-PCR. The complex pulled down by Bio-NC was served as the negative control.
Mouse xenograft models
Lentivirus (LV)-shRNA-CRNDE and its negative control (LV-shRNA) were provided by RiboBio. NOD/SCID mice (4-6 weeks old; Shanghai SLAC Laboratory Animal, China) were divided into the LV-shRNA-CRNDE group and the LV-shRNA group. In the LV-shRNA-CRNDE group (n = 10), 1 × 106 NALM-6 cells transfected with LV-shRNA-CRNDE were diluted in 100 μl of PBS and injected into mice through the tail vein. In the LV-shRNA group (n = 10), 1 × 106 NALM-6 cells transfected with LV-shRNA were diluted in 100 μl of PBS and injected into the same position of mice. The survival rates of mice from day 0 to day 100 were analyzed by the Kaplan–Meier analysis, and statistical analysis was performed using the log-rank test. The mice were sacrificed on day 100, and BM samples were collected from each mouse for qRT-PCR and Western blot. All experiments were approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University (No. 2020-KS-HNSR227).
RESULTS
The effect of LncRNA CRNDE knockdown on BCP-ALL cell lines
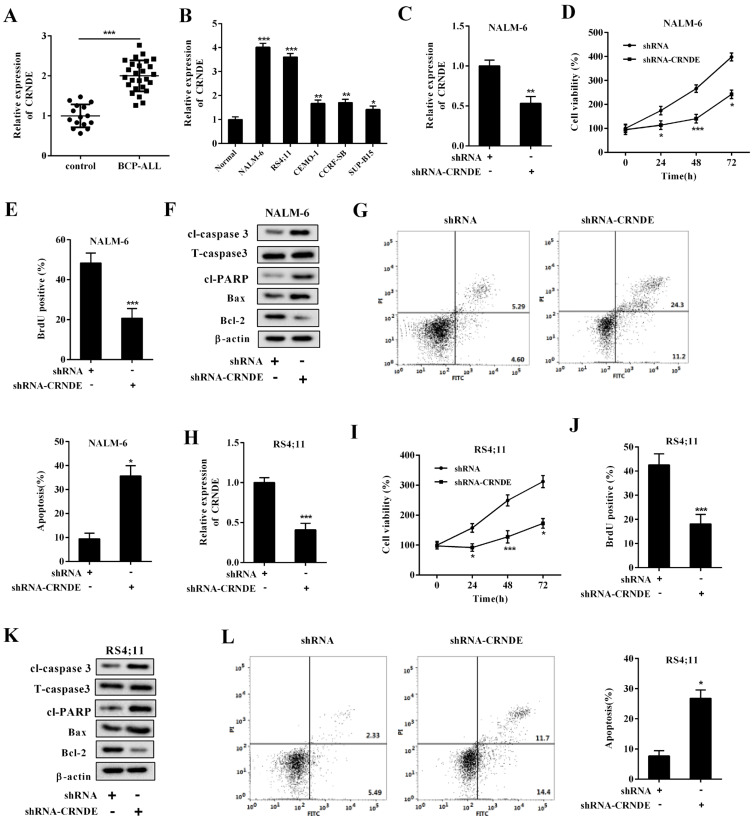
As shown in Fig. 1A, which compared the BM samples collected from the controls, the expression level of LncRNA CRNDE strongly increased in the BM samples collected from the BCP-ALL patients. Similarly, compared with the normal precursor B-cell line, the expression level of LncRNA CRNDE was upregulated in the BCP-ALL cell lines (NALM-6, RS4;11, CEMO-1, CCRF-SB, and SUP-B15) (Fig. 1B). Given that the BCP-ALL cell lines NALM-6 and RS4;11 showed higher expression levels of LncRNA CRNDE than others, we chose them to conduct the succeeding experiments.
Fig. 1. The effect of LncRNA CRNDE knockdown on the BCP-ALL cell lines.
The expression level of LncRNA CRNDE was measured in the (A) BM samples collected from patients with B-cell precursor acute lymphoblastic leukemia (BCP-ALL, n = 26) and controls (n = 15) and in (B) normal primary precursor B-cells (Normal) and BCP-ALL cell lines (NALM-6, RS4;11, CEMO-1, CCRF-SB, and SUP-B15) using qRT-PCR. Then, NALM-6 and RS4;11 cells were transfected with shRNA-CRNDE or its negative control (shRNA). (C and H) The expression level of LncRNA CRNDE was measured using qRT-PCR in NALM-6 cells and RS4;11 cells, respectively. (D and I) Cell viability was measured using the MTT assay in NALM-6 cells and RS4;11 cells, respectively. (E and J) Cell proliferation was measured using the bromodeoxyuridine (BrdU) assay in NALM-6 cells and RS4;11 cells, respectively. (F and K) The protein levels of cleaved (cl-) caspase 3, total (T) caspase 3, cleaved poly ADP-ribose polymerase (cl-PARP), B-cell lymphoma-2 (Bcl-2), and Bcl-2-associated X (Bax) were measured using Western blot in NALM-6 cells and RS4;11 cells, respectively. Β-actin was used as an internal control. (G and L) Cell apoptosis was measured using flow cytometry in NALM-6 cells and RS4;11 cells, respectively. *P < 0.05, **P < 0.01, ***P < 0.001 vs control or Normal or shRNA.
To test the effect of LncRNA CRNDE on the BCP-ALL cells, we silenced the LncRNA CRNDE expression by transfecting shRNA-CRNDE into the BCP-ALL cell lines NALM-6 and RS4;11 (Figs. 1C and 1H). As shown in Supplementary Fig. S2, compared with shRNA2 and 3, shRNA1 (shRNA-CRNDE) most effectively suppressed the LncRNA CRNDE expression in both NALM-6 and RS4;11 cells. Therefore, we selected shRNA1 (shRNA-CRNDE) for the succeeding experiments. Interestingly, the abrogation of the LncRNA CRNDE expression resulted in decreased cell viability (Figs. 1D and 1I), cell proliferation (Figs. 1E and 1J), and protein levels of Bcl-2 (an anti-apoptotic protein [Hassan et al., 2014]; Figs. 1F and 1K) in the BCP-ALL cell lines. In addition, the protein levels of pro-apoptotic proteins (cl-caspase 3 and Bax (Gu et al., 2018); Figs. 1F and 1K) and the number of apoptotic cells (Figs. 1G and 1L) were increased in the BCP-ALL cell lines transfected with shRNA-CRNDE.
Regulation of LncRNA CRNDE on miR-345-5p
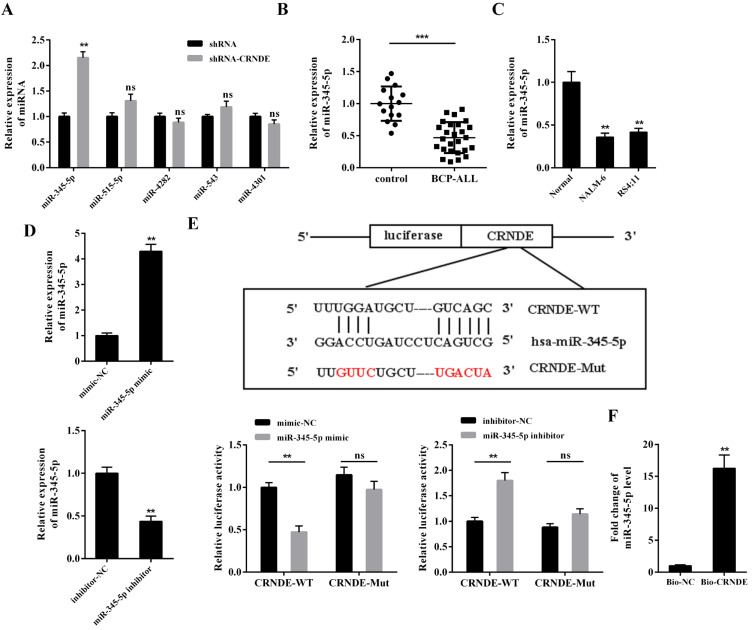
Utilizing the prediction of Bioinformatics software LncBase v.2, we found that five miRNAs (miR-345-5p, miR-515-5p, miR-4282, miR-543, and miR-4301) had potential binding sites with LncRNA CRNDE (Supplementary Fig. S3, Fig. 2E). The results in Fig. 2A showed that the silence of LncRNA CRNDE notably raised the miR-345-5p expression while not affecting the expression levels of other miRNAs in the NALM-6 cells. Compared with the BM samples of the controls and the primary normal precursor B-cells, the BM samples of the BCP-ALL patients and BCP-ALL cell lines exhibited a lower expression level of miR-345-5p (Figs. 2B and 2C). The luciferase activity of CRNDE-WT was significantly reduced by miR-345-5p mimic and raised by miR-345-5p inhibitor, whereas the luciferase activity of CRNDE-Mut was not significantly affected by either of them (Figs. 2D and 2E). Furthermore, the results of the RNA pull-down assay showed that miR-345-5p was more abundant in the Bio-CRNDE pull-down complex than in the Bio-NC pull-down complex (Fig. 2F). Taken together, these findings hinted that LncRNA CRNDE could bind to miR-345-5p and negatively regulate its expression in BCP-ALL cells.
Fig. 2. Regulation of LncRNA CRNDE on miR-345-5p.
(A) NALM-6 cells were transfected with shRNA or shRNA-CRNDE. Forty-eight hours later, the cells were harvested, and the expression levels of miR-345-5p, miR-515-5p, miR-4282, miR-543, and miR-4301 were measured by qRT-PCR. **P < 0.01 vs shRNA. ns, not significant. (B) The expression level of miR-345-5p was measured in the BM samples collected from patients with BCP-ALL (n = 26) and healthy controls (n = 15) and from (C) primary normal precursor B-cells and BCP-ALL cell lines by qRT-PCR. ***P < 0.001 vs control, **P < 0.01 vs Normal. (D) The transfection efficiencies of miR-345-5p mimic and miR-345-5p inhibitor were measured by qRT-PCR. **P < 0.01 vs mimic-NC or inhibitor-NC. (E) Top: the potential binding sites between LncRNA CRNDE and miR-345-5p. Bottom: NALM-6 cells were transfected with a dual-luciferase reporter vector containing either the LncRNA CRNDE WT or the LncRNA CRNDE Mut in the presence of miR-345-5p mimic or miR-345-5p inhibitor or their negative controls (mimic-NC or inhibitor-NC). Forty-eight hours later, the luciferase reporter activities were detected by dual-luciferase reporter gene assay. **P < 0.01 vs mimic-NC or inhibitor-NC. (F) RNA pull-down with biotin-labeled (Bio)-CRNDE was used to assess the interaction between LncRNA CRNDE and miR-345-5p in NALM-6 cells. The expression level of miR-345-5p was detected by qRT-PCR. **P < 0.01 vs Bio-NC.
The regulatory effect of LncRNA CRNDE on the BCP-ALL cell lines mediated by miR-345-5p
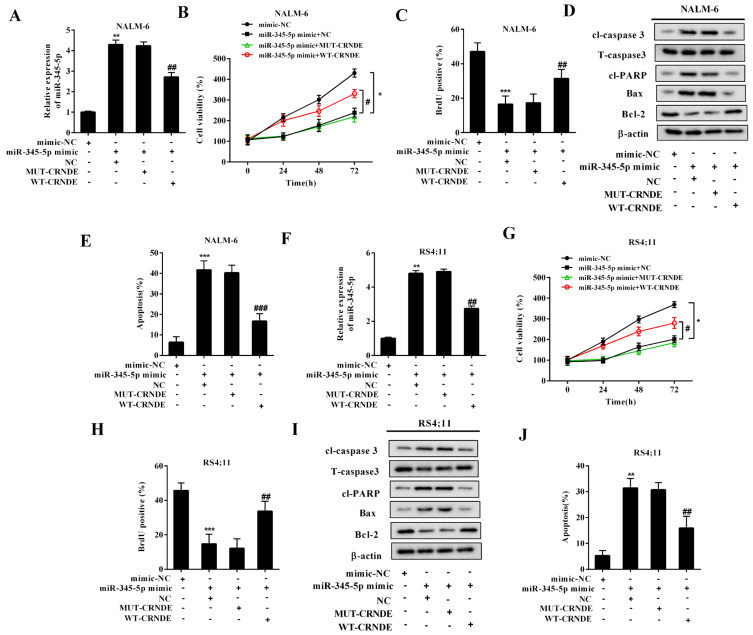
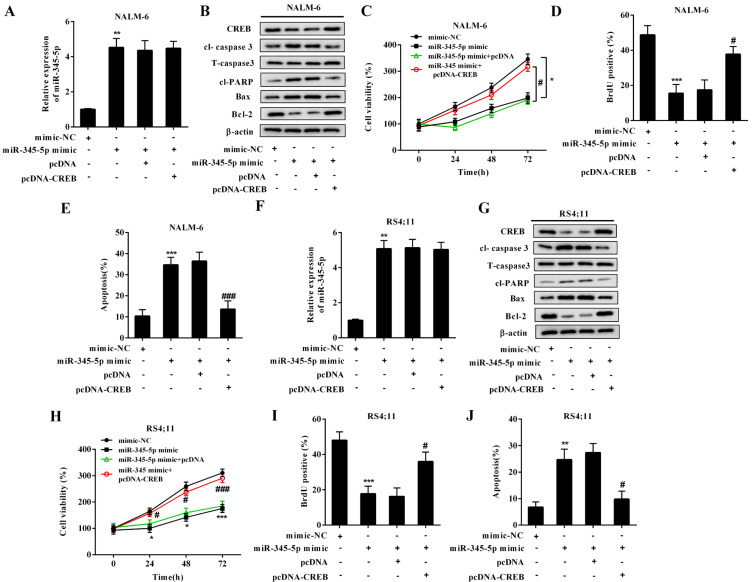
To verify whether miR-345-5p was involved in the regulatory effect of LncRNA CRNDE on the proliferation and apoptosis of BCP-ALL cell lines, the following experiments were conducted. The BCP-ALL cell lines (NALM-6 and RS4;11) were transfected with miR-345-5p mimic, and this transfection effectively raised the expression levels of miR-345-5p (Figs. 3A and 3F). In response to the miR-345-5p overexpression, the viability (Figs. 3B and 3G) and the proliferation (Figs. 3C and 3H) of the BCP-ALL cell lines were decreased. The protein levels of cl-caspase 3, Bax, and cl-PARP were upregulated, whereas the protein level of Bcl-2 was decreased in the miR-345-5p-overexpressed BCP-ALL cell lines (Figs. 3D and 3I), indicating that the overexpression of miR-345-5p boosted cell apoptosis. Accordingly, the results of flow cytometry showed that the miR-345-5p-overexpressed BCP-ALL cell lines exhibited a higher apoptosis rate than the BCP-ALL cell lines transfected with the mimic-NC (Figs. 3E and 3J). Then, the BCP-ALL cell lines were cotransfected with miR-345-5p mimic + WT-CRNDE/MUT-CRNDE to explore whether LncRNA CRNDE could partly reverse the effect of miR-345-5p mimic. As shown in Figs. 3A and 3F, compared with NC, the transfection of WT-CRNDE effectively downregulated the miR-345-5p expression in the BCP-ALL cell lines in the presence of miR-345-5p mimic. As expected, WT-CRNDE upregulated the cell viability (Figs. 3B and 3G), increased the cell proliferation (Figs. 3C and 3H), and facilitated the cell apoptosis (Figs. 3D, 3E, 3I, and 3J) in the miR-345-5p mimic-transfected BCP-ALL cell lines, indicating that WT-CRNDE reserved the effect of miR-345-5p mimic on the cells by downregulating the miR-345-5p expression.
Fig. 3. LncRNA CRNDE promoted cell proliferation and decreased cell apoptosis by downregulating the miR-345-5p expression in the BCP-ALL cell lines.
The BCP-ALL cell lines (NALM-6 and RS4;11) were divided into four groups: mimic-NC, miR-345-5p mimic + WT-CRNDE, miR-345-5p mimic + negative control (MUT-CRNDE), and miR-345-5p mimic + blank control (NC). (A and F) The expression level of miR-345-5p was measured by qRT-PCR. (B and G) Cell viability was measured using MTT assay. *P < 0.05, # P < 0.05. (C and H) Cell proliferation was measured using the BrdU assay. (D and I) The protein levels of cl-caspase 3, T- caspase 3, cl-PARP, Bax, and Bcl-2 were measured by Western blot. Β-actin was used as an internal control. (E and J) Cell apoptosis was measured using flow cytometry. **P < 0.01, ***P < 0.001 vs mimic-NC. ## P < 0.01, ### P < 0.001 vs miR-345-5p mimic + NC.
miR-345-5p negatively regulated CREB in BCP-ALL cell lines
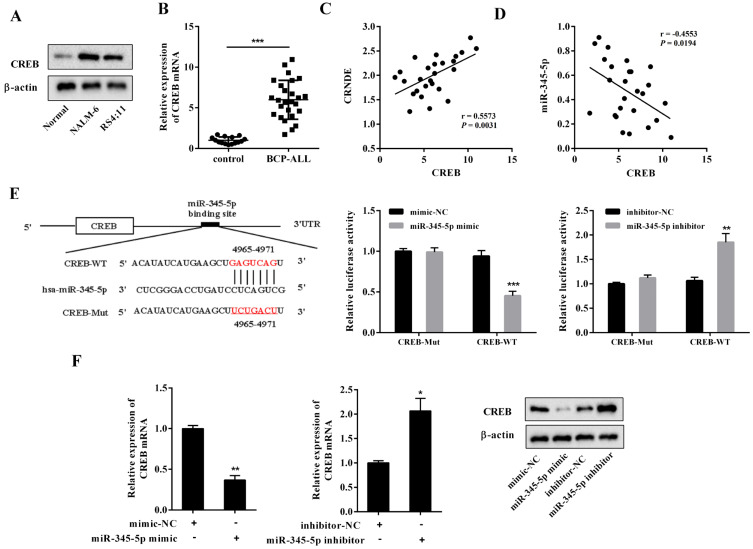
A recent study showed that CREB, a cellular transcription factor, promoted BCP-ALL progression and that the interference of CREB induced the apoptosis of BCP-ALL cell lines by activating apoptosis-associated caspases (Shabestari et al., 2017). In the present study, CREB was upregulated in both the BCP-ALL cell lines and BM samples of patients with BCP-ALL (Figs. 4A and 4B). The correlation plots further showed that the mRNA level of CREB was positively correlated with the LncRNA CRNDE expression (Fig. 4C) and inversely correlated with the miR-345-5p expression (Fig. 4D) in the BM samples of patients with BCP-ALL. Using the Bioinformatics software Targetscan, we found that miR-345-5p had putative binding sites with CREB (Fig. 4E). To explore the effect of miR-345-5p on CREB, dual-luciferase reporter assays were conducted. As shown in Fig. 4E, the luciferase activity of CREB-WT was significantly decreased in the NALM-6 cells transfected with miR-345-5p mimic, and increased in the NALM-6 cell transfected with miR-345 inhibitor. Neither miR-345-5p mimic nor miR-345-5p inhibitor affected the luciferase activity of CREB-Mut. Then, the NALM-6 cells were transfected with miR-345-5p mimic or miR-345-5p inhibitor. The results in Fig. 4F showed that the mRNA and protein levels of CREB were downregulated by miR-345-5p mimic and upregulated by miR-345-5p inhibitor. These data demonstrate that miR-345-5p negatively regulated the CREB expression by binding to its mRNA.
Fig. 4. miR-345-5p negatively regulated CREB in the BCP-ALL cell lines.
(A) The CREB expression was measured by Western blot in the primary normal precursor B-cells and BCP-ALL cell lines. (B) The CREB expression was measured by qRT-PCR in the BM samples collected from patients with BCP-ALL (n = 26) and healthy controls (n = 15). (C) Correlation plot of the CREB mRNA level (x-axis) and LncRNA CRNDE expression (y-axis) in the BM samples of patients with BCP-ALL. (D) Correlation plot of the CREB mRNA level (x-axis) and miR-345-5p expression (y-axis) in the BM samples of patients with BCP-ALL. (E) The putative binding sites between miR-345-5p and CREB were forecasted by Bioinformatics software. NALM-6 cells were transfected with a dual-luciferase reporter vector containing either the CREB Mut or the CREB WT in the presence of miR-345-5p mimic or miR-345-5p inhibitor or their negative controls (mimic-NC or inhibitor-NC). Forty-eight hours later, the luciferase reporter activities were detected by dual-luciferase reporter gene assay. (F) NALM-6 cells were transfected with miR-345-5p mimic or miR-345-5p inhibitor or mimic-NC or inhibitor-NC. Forty-eight hours later, the cells were harvested. The mRNA level of CREB was measured by qRT-PCR, and the protein level of CREB was measured by Western blot. *P < 0.05, **P < 0.01, ***P < 0.001 vs control or mimic-NC or inhibitor-NC.
The CREB expression mediated the modulatory effect of miR-345-5p on the BCP-ALL cell lines
To confirm whether the regulatory effects of miR-345-5p on cell proliferation and apoptosis were dependent on CREB, NALM-6 cells were transfected with miR-345-5p mimic/mimic-NC or cotransfected with miR-345-5p mimic + pcDNA-CREB/miR-345-5p mimic + pcDNA. In response to the miR-345-5p mimic transfection, miR-345-5p was overexpressed (Fig. 5A), the protein levels of CREB and anti-apoptotic protein were decreased (Fig. 5B), the protein levels of pro-apoptotic proteins were increased (Fig. 5B), cell viability was declined (Fig. 5C), cell proliferation was suppressed (Fig. 5D), and the number of apoptotic cells was increased (Fig. 5E). By contrast, in NALM-6 cells cotransfected with miR-345-5p mimic + pcDNA-CREB, pcDNA-CREB reserved the inhibitory effect of miR-345-5p mimic on cell proliferation (Fig. 5D) and the promoting effect of miR-345-5p mimic on cell apoptosis (Figs. 5B and 5E). To further verify our findings, we repeated the above experiments in RS4;11 cells and obtained similar results (Figs. 5F-5J), indicating that the miR-345-5p overexpression inhibited cell proliferation and induced cell apoptosis by negatively regulating the CREB expression.
Fig. 5. miR-345-5p overexpression inhibited cell proliferation and induced cell apoptosis by downregulating the CREB expression in the BCP-ALL cell lines.
The BCP-ALL cell lines (NALM-6 and RS4;11) were divided into four groups: mimic-NC, miR-345-5p mimic, miR-345-5p mimic + pcDNA-CREB, and miR-345-5p mimic + the negative control of pcDNA-CREB (pcDNA). (A and F) The expression level of miR-345-5p was measured by qRT-PCR. (B and G) The protein levels of CREB, cl-caspase 3, T-caspase 3, cl-PARP, Bax, and Bcl-2 were measured by Western blot. Β-actin was used as an internal control. (C and H) Cell viability was measured using MTT assay. (D and I) Cell proliferation was measured using the BrdU assay. (E and J) Cell apoptosis was measured using flow cytometry. **P < 0.01, ***P < 0.001 vs mimic-NC. # P < 0.05, ### P < 0.001 vs miR-345-5p mimic + pcDNA.
LncRNA CRNDE knockdown prolonged survival of mice xenotransplanted with NALM-6 cells
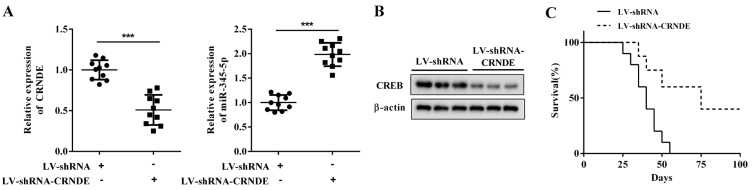
To determine whether LncRNA CRNDE knockdown could slow down the BCP-ALL progression in an animal model, 1 × 106 NALM-6 cells transfected with LV-shRNA-CRNDE or LV-shRNA were injected into NOD/SCID mice through the tail vein. As illustrated in Fig. 6C, the Kaplan-Meier analysis showed that the administration of LV-shRNA-CRNDE prolonged the survival of NOD/SCID mice xenotransplanted with NALM-6 cells. Meanwhile, LV-shRNA-CRNDE downregulated the expression levels of LncRNA CRNDE and CREB and upregulated the miR-345-5p expression in the BM samples of mice (Figs. 6A and 6B), indicating that miR-345-5p and CREB were also involved in the treatment effect of LV-shRNA-CRNDE on NOD/SCID mice xenotransplanted with NALM-6 cells.
Fig. 6. LncRNA CRNDE knockdown prolonged the survival of mice xenotransplanted with NALM-6 cells.
NOD/SCID mice were divided into the lentivirus (LV)-shRNA-CRNDE group and the LV-shRNA group. In the LV-shRNA-CRNDE group (n = 10), 1 × 106 NALM-6 cells transfected with LV-shRNA-CRNDE were diluted in 100 μl of PBS and injected into mice through the tail vein. In the LV-shRNA group (n = 10), 1 × 106 NALM-6 cells transfected with LV-shRNA were diluted in 100 μl of PBS and injected into the same position of mice. One hundred days later, the BM samples were collected from each mouse. (A) The expression levels of LncRNA CRNDE and miR-345-5p were measured by qRT-PCR. (B) The protein level of CREB was measured by Western blot. (C) The survival rates of mice were assessed by Kaplan–Meier analysis. ***P < 0.001 vs LV-shRNA.
DISCUSSION
Although much effort has been made to comprehend the pathogenesis of BCP-ALL, the precise regulatory mechanisms that lead to the misbalance between the proliferation and apoptosis of B-cell precursors are not well understood. Thus, the study identified a new regulatory pathway for cell proliferation and apoptosis in BCP-ALL cells involved in LncRNA CRNDE, miR-345-5p, and CREB. Briefly, we verified that the LncRNA CRNDE knockdown downregulated the CREB expression by targeting miR-345-5p, thus promoting cell apoptosis and reducing cell proliferation in BCP-ALL cells.
Using integrative bioinformatics analysis, James et al. (2019) found that 1,235 LncRNAs were aberrantly dysregulated in the BM samples of BCP-ALL patients and that 942 LncRNAs were closely related to the relapse of BCP-ALL, highlighting the role of LncRNAs in BCP-ALL. However, the specific regulatory mechanisms of LncRNAs in BCP-ALL are still unexplored. In our study, we found that LncRNA CRNDE was upregulated in the BM samples from BCP-ALL patients and BCP-ALL cell lines. The LncRNA CRNDE knockdown significantly declined cell viability and proliferation, elevated the protein levels of pro-apoptotic proteins cleaved caspase 3 and Bax, and increased the number of apoptotic cells in the BCP-ALL cell lines, indicating that LncRNA CRNDE knockdown restrained cell proliferation and boosted cell apoptosis in BCP-ALL. Furthermore, the in vivo experiments showed that the LncRNA CRNDE knockdown efficiently prolonged the overall survival of mice xenotransplanted with NALM-6 cells, confirming the therapeutic effect of the LncRNA CRNDE knockdown on BCP-ALL for the first time.
The CeRNAs hypothesis was proposed by Harvard researchers in 2011. The hypothesis states that various types of RNA (e.g., LncRNA and circRNA) can completely bind to the same miRNA, thus decreasing the number of miRNAs available to target mRNAs and abolishing the downstream effects of these miRNAs on the target mRNAs (Salmena et al., 2011). In our study, a clear increase in miR-345-5p expression was observed in the BCP-ALL cell lines transfected with shRNA-CRNDE. The following dual-luciferase reporter gene assay and RNA pull-down assay showed a combination of LncRNA CRNDE and miR-345-5p. The WT-CRNDE eliminated the regulatory effect of miR-345-5p mimic on cells, increased cell proliferation, and inhibited cell apoptosis in the BCP-ALL cell lines. Subsequent experiments proved that the regulatory effect of miR-345-5p on cell proliferation and apoptosis relied on its negative regulation of the CREB expression. These findings confirmed that LncRNA CRNDE could function as a ceRNA for miR-345-5p and remove its inhibitory effect on CREB.
CREB is a cellular transcription factor that can directly regulate multiple genes involved in cell proliferation and differentiation (Johannessen et al., 2004). A recent study showed that CREB could regulate hematopoiesis and contribute to the leukemia phenotype (Shima and Kitabayashi, 2011). Cho et al. (2011) reported that enriched CREB could be detected in the majority of BCP-ALL primary samples. Shabestari et al. (2017) found that CREB knockdown induced the apoptosis of BCP-ALL cell lines by activating apoptosis-associated caspases, suggesting that CREB is a potential target for the treatment of BCP-ALL. In accordance with previous studies, we found that the CREB overexpression could distinctly raise cell viability, reduce pro-apoptosis protein expression, and decrease the number of apoptotic cells in BCP-ALL cell lines, further proving the promoting effect of CREB on BCP-ALL progression.
In conclusion, this study elucidated the essential role of the LncRNA CRNDE/miR-345-5p/CREB axis in the modulation of cell proliferation and apoptosis in BCP-ALL. Although the experiment has many aspects that require improvement, we still hope this study can provide a new perspective for the clinical treatment of BCP-ALL.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grant No. 81700138 to Weimin Wang) and the Young Foundation of the First Affiliated Hospital of Zhengzhou University (to Weimin Wang).
Footnotes
AUTHOR CONTRIBUTIONS
Z.J. and F.G. participated in conceptualization, methodology, and project administration. W.W. participated in data curation and writing–original draft preparation. F.W. and P.M. participated in collection of clinical samples. S.G. and X.L. participated in partial animal experiments. L.C. participated in data analysis. L.S. and H.S. participated in complete examination of manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Avval A.J., Majd A., Gholipour N., Noghabi K.A., Ohradanova-Repic A., Ahangari G. An inventive report of inducing apoptosis in non-small cell lung cancer (NSCLC) cell lines by transfection of miR-430. 2019 doi: 10.2174/1871520619666190416114145. [DOI] [PubMed] [Google Scholar]
- Buske C., Becker D., Feuring-Buske M., Hannig H., Wulf G., Schafer C., Hiddemann W., Wormann B. TGF-beta inhibits growth and induces apoptosis in leukemic B cell precursors. Leukemia. 1997;11:386–392. doi: 10.1038/sj.leu.2400586. [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.G., Zhou W., Han T., Du S.Q., Li Z.H., Zhang Z., Shan G.Y., Kong C.Z. MiR-345 suppresses proliferation, migration and invasion by targeting Smad1 in human prostate cancer. J. Cancer Res. Clin. Oncol. 2016;142:213–224. doi: 10.1007/s00432-015-2016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.C., Mitton B., Sakamoto K.M. CREB and leukemogenesis. Crit. Rev. Oncog. 2011;16:37–46. doi: 10.1615/CritRevOncog.v16.i1-2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L.D., Pedersen S.K., Brown G.S., Ho T., Kassir Z., Moynihan A.T., Vizgoft E.K., Dunne R., Pimlott L., Young G.P., et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y.Y., Chen M.H., May B.H., Liao X.Z., Liu J.H., Tao L.T., Man-Yuen Sze D., Zhang A.L., Mo S.L. Matrine induces apoptosis in multiple colorectal cancer cell lines in vitro and inhibits tumour growth with minimum side effects in vivo via Bcl-2 and caspase-3. Phytomedicine. 2018;51:214–225. doi: 10.1016/j.phymed.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Han S., Han B., Li Z., Sun D. Downregulation of long noncoding RNA CRNDE suppresses drug resistance of liver cancer cells by increasing microRNA-33a expression and decreasing HMGA2 expression. Cell Cycle. 2019;18:2524–2537. doi: 10.1080/15384101.2019.1652035. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hassan M., Watari H., AbuAlmaaty A., Ohba Y., Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res. Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hu C.E., Du P.Z., Zhang H.D., Huang G.J. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145. Cell. Physiol. Biochem. 2017;42:13–21. doi: 10.1159/000477107. [DOI] [PubMed] [Google Scholar]
- Jacobson S., Tedder M., Eggert J. Adult acute lymphoblastic leukemia: a genetic overview and application to clinical practice. Clin. J. Oncol. Nurs. 2016;20:E147–E154. doi: 10.1188/16.CJON.E147-E154. [DOI] [PubMed] [Google Scholar]
- James A.R., Schroeder M.P., Neumann M., Bastian L., Eckert C., Gökbuget N., Tanchez J.O., Schlee C., Isaakidis K., Schwartz S., et al. Long non-coding RNAs defining major subtypes of B cell precursor acute lymphoblastic leukemia. J. Hematol. Oncol. 2019;12:8. doi: 10.1186/s13045-018-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M., Delghandi M.P., Moens U. What turns CREB on? Cell. Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kang X., Wang M., Wang H., Shen X., Guan W. MiR-4282 suppresses proliferation and mobility of human colorectal carcinoma cells by targeting semaphorin 3E. Panminerva Med. 2016;58:197–205. [PubMed] [Google Scholar]
- Lajoie M., Drouin S., Caron M., St-Onge P., Ouimet M., Gioia R., Lafond M.H., Vidal R., Richer C., Oualkacha K., et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS One. 2017;12:e0174124. doi: 10.1371/journal.pone.0174124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gan L., Zhang J. miR-543 inhibites cervical cancer growth and metastasis by targeting TRPM7. Chem. Biol. Interact. 2019;302:83–92. doi: 10.1016/j.cbi.2019.01.036. [DOI] [PubMed] [Google Scholar]
- Liu X.X., Xiong H.P., Huang J.S., Qi K., Xu J.J. Highly expressed long non-coding RNA CRNDE promotes cell proliferation through PI3K/AKT signalling in non-small cell lung carcinoma. Clin. Exp. Pharmacol. Physiol. 2017;44:895–902. doi: 10.1111/1440-1681.12780. [DOI] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353-358. 2011 doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabestari R.M., Safa M., Alikarami F., Banan M., Kazemi A. CREB knockdown inhibits growth and induces apoptosis in human pre-B acute lymphoblastic leukemia cells through inhibition of prosurvival signals. Biomed. Pharmacother. 2017;87:274–279. doi: 10.1016/j.biopha.2016.12.070. [DOI] [PubMed] [Google Scholar]
- Shima Y., Kitabayashi I. Deregulated transcription factors in leukemia. Int. J. Hematol. 2011;94:134–141. doi: 10.1007/s12185-011-0905-9. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhou J., Xue D., Li Z., Liu Y., Dong L. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int. J. Biol. Macromol. 2019;129:227–232. doi: 10.1016/j.ijbiomac.2019.01.127. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Li D., Fu J., Shi Q., Lu Y., Ju X. Overexpression of AIOLOS inhibits cell proliferation and suppresses apoptosis in Nalm-6 cells. Oncol. Rep. 2014;31:1183–1190. doi: 10.3892/or.2013.2964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.