ABSTRACT
Polo-like kinase 1 (Plk1) is a cell cycle kinase essential for mitosis progression, but also important for checkpoint recovery and adaptation in response to DNA damage and replication stress. However, although Plk1 is expressed in S phase, little is known about its function during unperturbed DNA replication. Using Xenopus laevis egg extracts, mimicking early embryonic replication, we demonstrate that Plk1 is simultaneously recruited to chromatin with pre-replication proteins where it accumulates throughout S phase. Further, we found that chromatin-bound Plk1 is phosphorylated on its activating site T201, which appears to be sensitive to dephosphorylation by protein phosphatase 2A. Extracts immunodepleted of Plk1 showed a decrease in DNA replication, rescued by wild type recombinant Plk1. Inversely, modest Plk1 overexpression accelerated DNA replication. Plk1 depletion led to an increase in Chk1 phosphorylation and to a decrease in Cdk2 activity, which strongly suggests that Plk1 could inhibit the ATR/Chk1-dependent intra-S phase checkpoint during normal S phase. In addition, we observed that phosphorylated Plk1 levels are high during the rapid, early cell cycles of Xenopus development but decrease after the mid-blastula transition when the cell cycle and the replication program slow down along with more active checkpoints. These data shed new light on the role of Plk1 as a positive regulating factor for DNA replication in early, rapidly dividing embryos.
KEYWORDS: DNA replication, Polo-like kinase 1 (Plk1), checkpoint, Xenopus
Introduction
To maintain genome stability, eukaryotic DNA replication must be strictly controlled in space and time during S phase [1]. In higher eukaryotes, DNA replication starts from several thousand replication origins, each activated at different times during S phase. Assembly of the pre-replicative complex (pre-RC) during G1 phase at origins is initiated by the binding of the origin recognition complex (ORC) to DNA sequences, this in turn recruits Cdc6, Cdt1 and the MCM 2–7 complex. The pre-RCs are subsequently activated at the G1/S phase transition by Cyclin- and Dbf4-dependent kinases (respectively, CDKs and DDKs). CDKs and DDKs function to recruit additional factors that activate the functional helicase MCM-GINS-Cdc45 and start DNA synthesis at the origins. When replication forks stall or DNA is damaged during S phase, the replication checkpoint is activated. In the yeast S. cerevisiae, this checkpoint depends on Mec1 and Rad53 which stabilize stalled replication forks [2,3] and prevent or delay firing of late origins [4,5]. In mammalian culture cells and in the Xenopus in vitro system, forks stalled by the DNA polymerase inhibitor aphidicolin cause the helicase to uncouple from polymerase activities. This generates the signal to activate ATR [6,7], that phosphorylates and activates the kinase Chk1 [8,9]. Chk1 in turn phosphorylates the phosphatase Cdc25, leading to its degradation [10]. Cdc25 is required for Cyclin-Cdk2 activation [11]. Replication stress can be induced by exogenous agents, like aphidicolin, or endogenous sources like spontaneous DNA damage, difficult to replicate structures or depletion of limiting replication factors [12]. We and others underlined the role of ATR and Chk1 in regulating origin firing during normal S phase in the absence of induced fork stalling and DNA damage in Xenopus [13–16] and in mammalian cells [17,18].
In order to resume the cell cycle after checkpoint activation, adaptation or recovery mechanisms have been described in yeast, Xenopus and mammalian cells [19]. The Serine-Threonine kinase, Polo-like kinase 1 (Plk1) plays a critical role in these pathways during G2/M transition and mitosis [20]. It was first shown in the Xenopus in vitro system that, after prolonged replication fork stalling, ATR phosphorylates the mediator Claspin, creating a docking site for Plk1 through its polo-box domain (PBD) [21]. Plk1 then phosphorylates Claspin, which leads to its dissociation from chromatin and the inactivation of Chk1. This succession of events abrogates the checkpoint and restores G2/M progression of the cell cycle. Plk1 abundance and activity are highly regulated during the cell cycle via expression, stability and post-translational modifications. In mammalian cells, the Plk1 protein level is low in G1 phase, increases during S phase, remains high through G2-M phases as does its kinase activity [22–24]. At the end of mitosis, Plk1 is rapidly degraded by the proteasome after poly-ubiquitination [25]. In the Xenopus in vitro system, Plk1 is not degraded between M and S phases [26], but its activity is regulated during the cell cycle by its spatio-temporal phosphorylation [27]. The phosphorylation of Plk1 in its T loop dissociates the N-terminal kinase domain from the C-terminal Polo-box domain (PBD), switching the protein conformation to an open and active form. The key activation site, highly conserved from yeast to humans, is located on T201 in Xenopus (equivalent to T210 in human) [28]. A polo-like kinase kinase (Plkk) has been identified, which phosphorylates and activates Plk1 in Xenopus oocytes at the G2/M transition [29]. However, protein kinase A seems to be the main T201 phosphorylating kinase in vitro and the S340 phosphorylation would be responsible for electrophoretic mobility retardation [27]. In human cells, Plk1 is phosphorylated in G2 by Bora/Aurora A [30,31] and other PAK kinases [32] on its activating equivalent T210 site. Upon DNA damage or replication stress protein, protein phosphatase 2A (PP2A-B55α) has been shown to bind to and dephosphorylate Plk1 during G2 [33]. Plk1 is also methylated on K209 upon DNA damage [34] in S, G2 and M phases, which antagonizes T210 phosphorylation in G2/M. The localization of Plk1 to the nucleus during S and G2 phase is due to a bipartite nuclear localization sequence (NLS) [35], which is further promoted by its sumoylation at G2/M [36]. Plk1 is frequently overexpressed in human cancers (reviewed in [37]), but its contribution to tumor development is unclear. The overexpression of Plk1 in mammalian culture cells [38,39] or in mice models [40] results in the misregulation of G2 phase or mitotic events. Plk1 knockdown is lethal during early development in Xenopus [41] and mice [42] with embryos arresting due to severe defects in mitosis.
Consequently, most studies of Plk1 focused on its role during G2 and M, however, new functions of Plk1 have recently emerged during S phase upon replication stress and DNA damage [43]. Especially, Plk1 interaction with MCM7 during S phase was shown to increase in response to double strand breaks in a human cell culture line [44]. Likewise, in the Xenopus in vitro system, it was shown that Plk1 recruitment to aphidicolin induced stalled forks via its PBD domain is dependent on MCM2 phosphorylation by ATR. In this context, Plk1 depletion leads to a decrease of DNA synthesis and a more active intra-S phase checkpoint [45]. Other studies in mammalian cells reported that Plk1 phosphorylates Hbo1 (Histone acetyltransferase binding to Orc1) [46] and Orc2 under conditions of replication stress [47], both necessary for MCM recruitment, underlining a possible role for Plk1 in pre-RC assembly. Further, Plk1 depletion in human cancer cells leads to a decrease of DNA synthesis and to DNA damage at the G1/S border due to disrupted binding of the MCM complex [48]. Plk1 is also important for replication fork stability upon stress via its interaction with BRCA2 [49] and FANCM [50].
However, how Plk1 is regulated during unperturbed S phase and how its deregulation could affect the DNA replication program in the absence of exogenous replication stress is unknown. In order to study Plk1 during an unperturbed S phase, we took advantage of the Xenopus in vitro system, in which the replication components are easily manipulated without being impaired by other phases of the cell cycle. We show here that active, phosphorylated (T201) Plk1 is recruited to chromatin concomitant with, but independently of pre-RC assembly in the absence of replication stress. Plk1 depletion results in a decrease of DNA replication during early and mid S phase whereas addition of recombinant wild type Plk1 accelerates DNA replication. Our experiments show that Plk1 depletion increases Chk1 phosphorylation and decreases Cdk2 activity but does not affect pre-RC assembly. We provide here for the first time evidences for an important role of Plk1 as a positive regulator for the DNA replication program in the absence of induced replication stress.
Materials and methods
Reagents and antibodies
Rubratoxin A was purchased from Cayman Chemical (19605). Unless otherwise specified, reagents were from Sigma-Aldrich, France. Antibodies directed against the following proteins were used: human S345-P-Chk1 (1/1000, Cell Signaling Technology 2341); human T210-P-Plk1 (1/1000, Abcam ab39068), homologous to the Xenopus T201 site; XCdk2 (1/1000, gift from Catherine Bonne-Andrea); hCdc45 (1/1000, Santa Cruz Biotechnology sc-20685), hMCM2 (1/2000, Bethyl Laboratories A300-191A); Tubulin (1/10000, Sigma T5168); XChk1 (1/500 [16]); Phospho-S139 Histone H2AX (1/1000, Millipore 05–636); Histone H3 (1/1000, Abcam ab1791); XCyclin E1 (1/2000, European Xenopus Resource center/JL. Maller); XMCM7, XOrc1 and XOrc2 (1/1000, gifts from R. A. Laskey). Recombinant His-geminin, GST- p27, GST-p21 were prepared as described, respectively [51–53].
Recombinant Xenopus Plk1 and Plk1 antibody
Recombinant His-tagged Xenopus Plk1 was cloned in pFastBac1vector, expressed in the baculovirus Bac-to-Bac expression system (Invitrogen), purified using Nickel-Sepharose beads (Amersham Bioscience) as described by the supplier then dialyzed over night against 25 mM Hepes pH 7.8, 250 mM NaCl, 5 mM imidazole, 5% glycerol, 7.5 mM MgCl2, 1 mM DTT, 1 mM EDTA, its kinase activity was assayed in vitro using casein as substrate. Purified recombinant His-tagged Plk1 was then used as an antigen to raise antibodies in rabbits at a commercial facility (Covalab, Villeurbanne, France). The anti-Plk1 serum was further purified on protein A Sepharose (GE Healthcare) according to the manufacturer’s protocol.
Replication of sperm nuclei in Xenopus egg extracts
Replication competent extracts from unfertilized Xenopus eggs and sperm nuclei from testis of male frogs were prepared as described [54]. Egg extracts were used fresh unless stated otherwise. Sperm nuclei (2000 nuclei/µl or 7000 nuclei/µl) were incubated in untreated, mock or Plk1 depleted extracts in the presence of cycloheximide (250 µg/ml), energy mix (7.5 mM creatine phosphate, 1 mM ATP, 0.1 mM EGTA, pH 7.7, 1 mM MgCl2). Replication was allowed to continue for indicated time. In vitro fertilization of Xenopus eggs with sperm was performed according to standard techniques [55], and developmental stages of embryos were determined according to Nieuwkoop and Faber [56]. Our institutional Animal Care and Use Committee (IACUC), namely Paris Center and South number 59, approved the study and the protocols herein (approvals number 2012–0062 and 2012–0063), following the French and the European laws on animal experimentation.
Immunodepletions
Anti-Xenopus Plk1 serum, anti-Orc2 serum, pre-immune serum or rabbit IgG were incubated overnight at 4°C with protein A Sepharose beads (GE Healthcare). For Orc2, beads were washed with EB buffer (50 mM Hepes-KOH, pH 7.4, 50 mM KCl, 5 mM MgCl2) without DTT and briefly with a small volume of fresh extract to eliminate buffer and incubated twice 30 min at 4°C with egg extract (volume ratio 1:2) under rotation. For Plk1, anti-XPlk1 coupled beads were washed with EB buffer without DTT and incubated in extracts 1 hour at 4°C (volume ratio 1:3).
Neutral and alkaline agarose gel electrophoresis
Sperm nuclei were incubated in fresh extracts complemented with indicated reagents and one-fiftieth volume of [α-32P] dCTP (3000 Ci/mmol). DNA was recovered after DNAzol® treatment (Invitrogen protocol) followed by ethanol precipitation, separated on 1.1% alkaline agarose gels, and analyzed as described [57]. From one extract to another, the replication extent (percent of replication) differs at a specific time point, because each egg extract replicates nuclei with its own replication kinetics. In order to compare different independent experiments, performed using different egg extracts, the data points of each control sample were independently fitted to a logistic curve and scaled by the inferred maximum incorporation value to 0–100 %. To include statistics, the scaled data points were grouped into 4 bins (0–20% = early; 21–60% = mid; 61–90% = late; 91–100% = very late S phase); mean and standard deviation were calculated for each bin and the t-test was used to assess statistically significant differences between the data in each bin.
Western blot analysis
For analysis of whole extract samples, replication reactions were stopped at indicated times by the addition of SDS sample buffer. For analysis of nuclei, reactions were diluted into a 20-fold volume of nuclear isolation buffer (NIBS) (50 mM Hepes, pH 7.5, 150 mM NaCl, 2 mM MgCl2, protease inhibitors, phosphatase inhibitors, 10% sucrose) and nuclei were pelleted through a 20% sucrose cushion in NIBS buffer at 4000 g, 5 min, 4°C. The purification was repeated and the pellet was dissolved in SDS sample buffer. For analysis of chromatin-bound proteins we used a slightly modified protocol from [58]. Briefly, reactions were diluted into a 13-fold volume of ELB buffer (10 mM Hepes pH 7.5, 50 mM KCl, 2.5 mM MgCl2) containing 1 mM DTT, 0.2% Triton X100, protease inhibitors and phosphatase inhibitors; chromatin was recovered through a 500 mM sucrose cushion in ELB buffer, at 6780 g, 50 sec, 4°C. Interphase was washed twice with 200 µl ELB, 250 mM sucrose and resuspended in SDS sample buffer. For detection of Histone H2A.X phosphorylation (adapted from [59]), 50 µl of mock-depleted or Plk1 depleted interphase extract were incubated with 7000 nuclei/µl for 90 min at 23°C. Chromatin was isolated as described above, however, in the continuous presence of 10 mM NaF, 1 mM Na Vanadate and spun at 6000 g for 20 min through the 500 mM sucrose ELB cushion. Positive and negative controls were prepared by incubating frozen extracts containing 7000 nuclei/µl with either 150 ng/µl EcoRI-linearized pGEX plasmid or 2 µM Okadaic Acid (OA) [60] or their respective vehicles.
Proteins were subjected to 12% or 15% SDS-polyacrylamide gels [61]. To increase the separation of Plk1 and Chk1, western blots were performed using 8.5% acrylamide gels prepared with a 120:1 acrylamide:bisacrylamide ratio [23]. Immunoblots were carried out by standard methods after semi-dry transfer to P-Immobilon (Millipore). Immunodetection was performed using appropriate horseradish peroxidase-labeled secondary antibodies (1/10000), followed by chemiluminescence using Super Signal West Pico or Femto Chemiluminescence Kit (Pierce), then imaged and quantified using ChemiDoc Imaging System (Bio-Rad). For immuno-depleted or immuno-precipitated samples, horseradish peroxidase-labeled protein A (1/20000, Invitrogen 101023) was used for immunodetection to minimize denatured IgG chains recognition.
Cdk2 immunoprecipitation and kinase assays
Anti-Xenopus Cdk2 antibody or mock rabbit IgG were coupled to Protein A Sepharose as described above and washed in dilution buffer (50 mM Hepes/KOH pH 8.0, 50 mM KCl, 20 mM K2HPO4/KH2PO4 pH 8). Replication reactions (50 µl) containing 2000 nuclei/µl were stopped after 45 min with 5 volumes dilution buffer supplemented with protease and phosphatase inhibitors then overlaid on 150 µl dilution buffer containing 30% sucrose and centrifuged at 5000 g for 5 min. The pellet was resuspended in 200 µl dilution buffer supplemented with 0.2% Triton X100 to extract nuclear proteins, incubated 10 min on ice and centrifuged 14000 rpm for 5 min. The supernatant was incubated with Cdk2 or mock coupled beads at 4°C for 2 h. Beads were washed three times in dilution buffer with 0.2% Triton X100, once in dilution buffer without Triton X100 and finally in EB buffer. H1 histone kinase assays in duplicates were performed using 10 µl beads, 0.1 µCi γ32P-ATP, 50 µM ATP and 0.5 µg histone H1 for 30 min at 30°C. Reactions were stopped with SDS sample buffer, proteins were separated by SDS gel electrophoresis. Gels were dried and bands were quantified on a phosphorimager Typhoon Trio (GE Healthcare).
Results
Plk1 recruitment to chromatin coincides with Orc-complex recruitment during pre-RC assembly
Previous studies highlighted the role of Plk1 in the checkpoint recovery or adaptation pathway in the presence of exogenous DNA damage, fork stalling and minimal licensing [21,45]. Based on numerical simulations of experimental data, our previous study suggested a mechanism that locally inhibits the replication checkpoint [16] for the regulation of origin firing in the Xenopus in vitro system in the absence of exogenous replication stress. Since Plk1 was a good candidate to be part of this mechanism, we investigated its role during unperturbed S phase using a specific anti-Xenopus Plk1 antibody, which was raised against wild-type recombinant Plk1 (Suppl. Figure 1). Xenopus Plk1 levels are constant in M and S phase egg extracts [26] and its activity is regulated by posttranslational modification and localization [27], but when Plk1 is recruited to chromatin during normal S phase, was unknown. Permeabilized sperm nuclei were incubated in S phase egg extract, then purified chromatin fractions were collected starting from pre-RC assembly and continuing further after the start of DNA replication. Upon addition of sperm DNA to S phase egg extracts, the nuclear membrane is formed, chromatin is assembled and replication proteins are imported and recruited on chromatin [54]. Western blot analysis (Figure 1a) revealed that the recruitment of Plk1 to chromatin coincides with the recruitment of the ORC complex (Orc1), but was slightly before the MCM complex (MCM2) and well before the start of DNA synthesis marked by the presence of the sliding clamp PCNA on chromatin. Plk1 further accumulates on chromatin as S phase progresses. Therefore, our results show that in the absence of exogenous stress, during normal DNA replication, Plk1 is recruited to chromatin simultaneous to the pre-RC proteins, in the Xenopus in vitro system.
Figure 1.
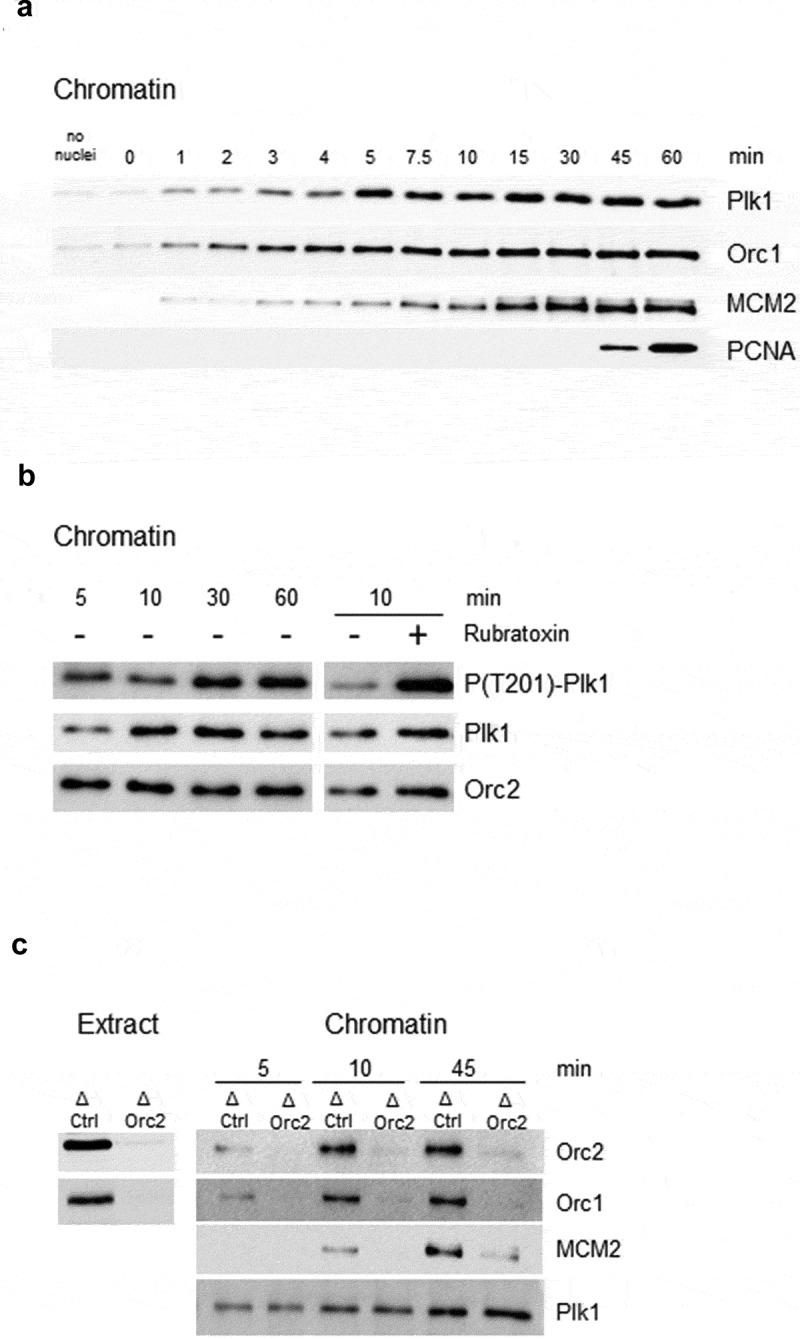
Early chromatin recruitment of T201-phosphorylated Plk1 during normal S phase is independent of pre-RC assembly. A Sperm nuclei (2000 n/µl) were incubated at indicated times in thawed S phase egg extract, chromatin was purified and analyzed by western blot with indicated antibodies. B Chromatin fractions as in A, but in the presence of PP2A inhibitor rubratoxin A (1.5 µM), Orc 2 as loading control. C sperm nuclei (2000 n/µl) were incubated in control IgG (ΔCtrl) depleted or Orc2 (ΔOrc2) depleted fresh egg extract, chromatin was purified at different times during S phase and proteins were analyzed by western blotting with indicated antibodies.
Plk1 is phosphorylated on chromatin at T201 and is dephosphorylated in a PP2A dependent manner
In Xenopus, Plk1 is phosphorylated and activated at S128 and T201 during mitosis [27,28,41] but the activity of mammalian as well as Xenopus Plk1 mainly depend on T210 phosphorylation (equivalent to T201 in Xenopus) [27,62]. Using a specific anti-phospho (T201) Plk1 antibody, we found that Plk1 is recognized by this antibody in S phase extracts (Suppl. Figure 2) and in different S phase chromatin fractions (Figure 1b and Suppl. Figure 3). The phosphorylation at T201 on chromatin increased during S phase together with Plk1 levels which shows that at least part of chromatin-bound Plk1 is in its T201 phosphorylated, active form. Since this phosphorylation is higher in M phase than in S phase (Suppl. Figure 2), we reasoned that the activity of Plk1 must be regulated by a specific phosphatase during S phase. Therefore, the effect of the inhibition of a protein phosphatase, PP2A, by rubratoxin A [63] on Plk1 phosphorylation was tested. Rubratoxin A added to sperm nuclei replicating in egg extract strongly inhibited DNA replication, consistent with the effect previously obtained using okadaic acid, another PP2A inhibitor [64,65]. We found, that the inhibition of PP2A with rubratoxin A strongly increased Plk1 phosphorylation in chromatin fractions (Figure 1b). PP2A inhibition by okadaic acid also strongly increased the phosphorylation of Plk1 in interphase extracts (Suppl. Figure 4).
We were intrigued by the early recruitment of Plk1 to chromatin during unperturbed replication, which coincided with the recruitment of the Orc complex. Therefore, we checked whether Orc2 would be necessary for the early recruitment of Plk1 to chromatin in the Xenopus in vitro system. Orc2 was immunodepleted from egg extract (Figure 1c), sperm nuclei were incubated in mock and Orc2 depleted extracts for different times and the chromatin recruitment of Plk1 was analyzed (Figure 1c). It was shown that immunoprecipitation of Orc2 co-precipitates the whole Orc complex along with the MCM complex and abolishes MCM complex recruitment and DNA replication in the in vitro system [66–68]. We found that Plk1 was still recruited to chromatin to the same extent in Orc2/Orc1-depleted replication reactions compared to control depleted reactions. Inhibiting the initiation of DNA replication by a Cdk2 inhibitor p21 or by geminin did not inhibit Plk1 recruitment, consistent with the above described Orc2 depletion experiments, but the inhibition of nuclear transport by WGA (wheat germ agglutinin) strongly decreased chromatin bound Plk1 (suppl. Figure 5). In conclusion, during unperturbed S phase in Xenopus extracts, Plk1 is recruited to chromatin independently of pre-RC assembly and DNA replication; the chromatin bound Plk1 is phosphorylated on its activating site T201 and the dephosphorylation depends on PP2A.
Plk1 depletion decreases DNA replication and results in an increase of Chk1 S344 phosphorylation and a decrease in Cdk2 kinase activity in the absence of replication stress
Based on these observations we investigated whether Plk1 could play a role in regulating DNA replication in the absence of exogenous replication stress. Plk1 was immunodepleted from interphase egg extracts (Figure 2a) and sperm nuclei were incubated in the presence of α32P-dCTP in mock or Plk1 depleted extracts and in Plk1 depleted extracts to which recombinant Plk1 had been added back. Replication reactions were stopped at indicated times during S phase and nascent strand synthesis was monitored by alkaline gel electrophoresis (Figure 2b), then quantified (Figure 2c). We observed that Plk1 depletion slowed down DNA replication and that adding back the wild type recombinant Plk1, partially rescued DNA replication with an increasing potency over time in this experiment (Figure 2c). The experiment was repeated six times using different extracts and normalized for early, middle, late and very late S phase time points (Figure 2d). The decrease of DNA replication after Plk1 depletion was significant and most pronounced during early and mid S phase (65 and 59% decrease, respectively) and the rescue with recombinant Plk1 was best in mid to late S phase. This result clearly shows that Plk1 plays a positive role in regulating DNA replication in the Xenopus in vitro system during normal S phase.
Figure 2.
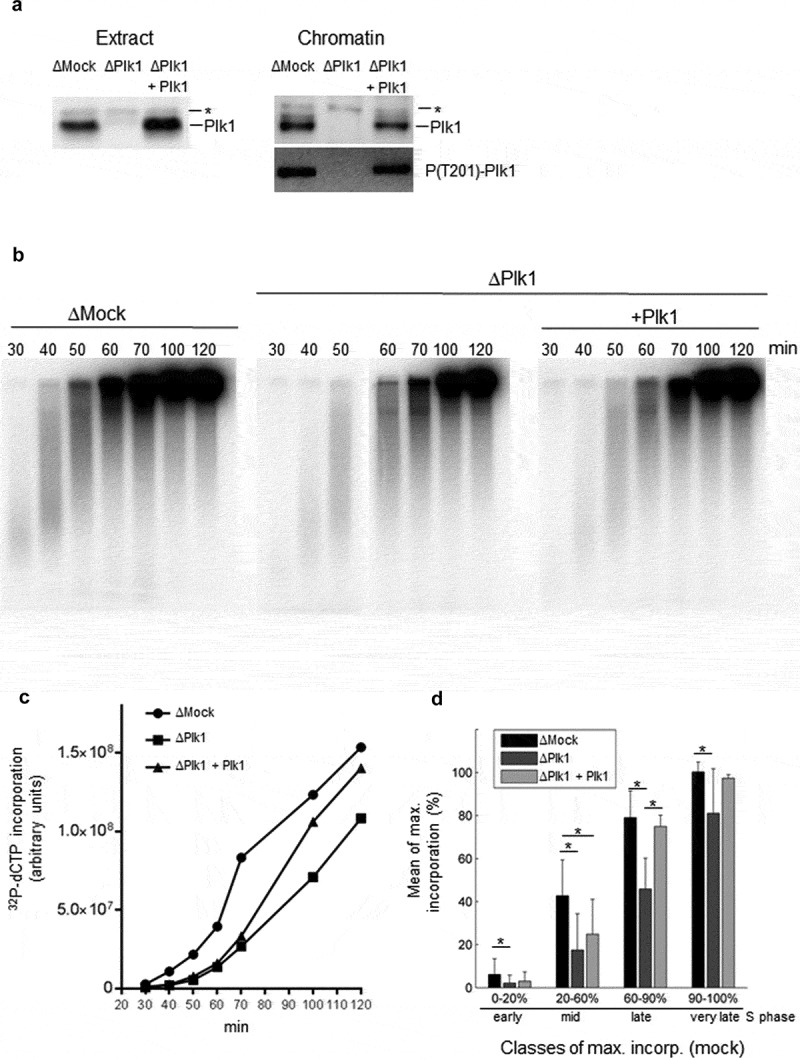
Depletion of Plk1 in the Xenopus in vitro system decreases DNA replication in the absence of stress. A Western blot analysis after mock depletion (ΔMock), Plk1 depletion (ΔPlk1), Plk1 depletion plus recombinant WT Plk1 (150 nM) (ΔPlk1+ Plk1); Left: S phase egg extracts; Right: Chromatin fractions, * nonspecific band. B Sperm nuclei (2000 n/µl) were incubated in ΔMock, in ΔPlk1 depleted extracts or in ΔPlk1+ Plk1 extracts in the presence of α32P-dCTP, reactions were stopped at indicated times, DNA was purified and separated by alkaline gel electrophoresis. C Quantification of α32P-dCTP incorporation in B (arbitrary units), D Mean percentages of maximal incorporation in six independent experiments, * statistically different, P < 0.05, t-test.
We have previously shown that the Chk1/ATR dependent replication checkpoint controls the replication program in the absence of exogenous replication stress [16]. Since Plk1 depletion in stress conditions (presence of aphidicolin) leads to an increase of activated P-(S344)-Chk1 levels in the Xenopus in vitro system [45], we investigated whether this would also be the case in the absence of replication stress. Replication reactions were performed in mock and Plk1 depleted extracts using a high concentration of sperm nuclei (7000 nuclei/µl) and the activation of Chk1 was monitored at several time points all over S phase in the presence and absence of aphidicolin. Consistent with the former study, in the presence of aphidicolin, we observed an increase of normalized P-Chk1 levels after depletion of Plk1 in comparison to mock-depleted replication reactions (Suppl. Fig. 6 A, B). Interestingly, in the absence of aphidicolin an increase of P-Chk1 (Figure 3a,b) was also detected all over S phase, but which was most elevated during early and mid S phase. This is entirely consistent with the highest inhibition of DNA replication during this period (see Figure 2d) after Plk1 depletion. The mean increase of P-Chk1 after Plk1 depletion was about 2-fold throughout S phase in four independent experiments (Figure 3c). No P-Chk1 signal was detected in the absence of sperm nuclei after Plk1 depletion (Suppl. Fig. 6 C). We were not able to detect a significant decrease of P-Chk1 signal by replenishment with the recombinant protein after Plk1 depletion (Suppl. Fig. 6D). This observation could be explained by the partial, delayed rescue on DNA replication we observed after Plk1 depletion and add back (Figure 2d). When active, Chk1 phosphorylates and inactivates Cdc25 that can no longer dephosphorylate and activate Cdk2. Therefore, in order to confirm whether Plk1 depletion inhibits the Chk1/Cdc25/Cdk2 axis of the checkpoint cascade, Cdk2 activity was directly measured in nuclei incubated in mock and Plk1 depleted extracts. Immunoprecipitation experiments were performed using protein A beads coupled to anti-Cdk2 or to control antibodies in mock and Plk1 depleted nuclear extracts (Figure 4a). Cdk2 activities of immunoprecipitates were then measured using H1 kinase assays in triplicates in two independent experiments (Figure 4b). Our results showed that Plk1 depletion leads to a mean 37% reduction of Cdk2 activity in replicating nuclei in the absence of replication stress (Figure 4c,d). Studies in mammalian cells reported that Plk1 is required for the replication licensing step [46–48], but we could not detect a decrease of the pre-RC loading on chromatin after Plk1 depletion in the Xenopus system (Suppl. Fig. 7). We conclude that Plk1 depletion slows down DNA replication through a more active Chk1-dependent replication checkpoint and by a direct or indirect inhibition of Cdk2 activity. This observed increase in checkpoint activity could be due to a direct interaction of Plk1 and the actors of the checkpoint cascade or due to an indirect effect of Plk1 on genome stability, leading to double strand breaks (DSBs). We therefore monitored the presence of phosphorylated H2AX (γH2AX) on chromatin as a known DSB marker after mock or Plk1 depletion (Figure 4e,f). We could not detect an increase of γH2AX levels on chromatin after Plk1 depletion compared to mock depletion when compared to induced DSBs after addition of EcoRI-cut plasmid or the addition of okadaic acid [60]. Therefore, we conclude that Plk1 depletion did not increase DNA double strand breaks and genome instability under normal replication conditions.
Figure 3.
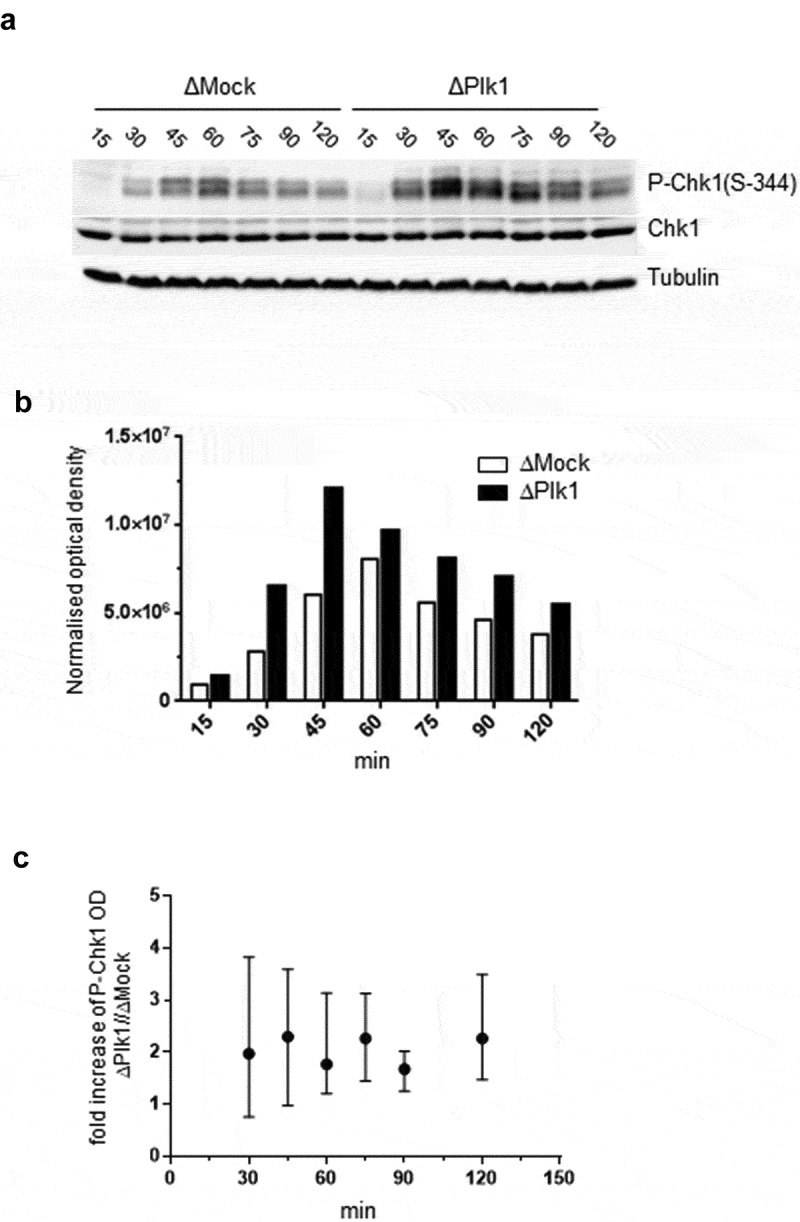
Depletion of Plk1 increases S344-phosphorylation of Chk1 in the absence of replication stress. A Sperm nuclei (7000 n/µl) were incubated in Mock (ΔMock), Plk1 (ΔPlk1) depleted egg extracts and aliquots were taken from the whole replication reactions at indicated time then processed for western blotting (no aphidicolin). B quantifications of P-Chk1 bands (optical density, OD) normalized to tubulin signal from experiment A. C Mean increase of P-Chk1 signal (OD) after Plk1 depletion with range from four independent experiments.
Figure 4.
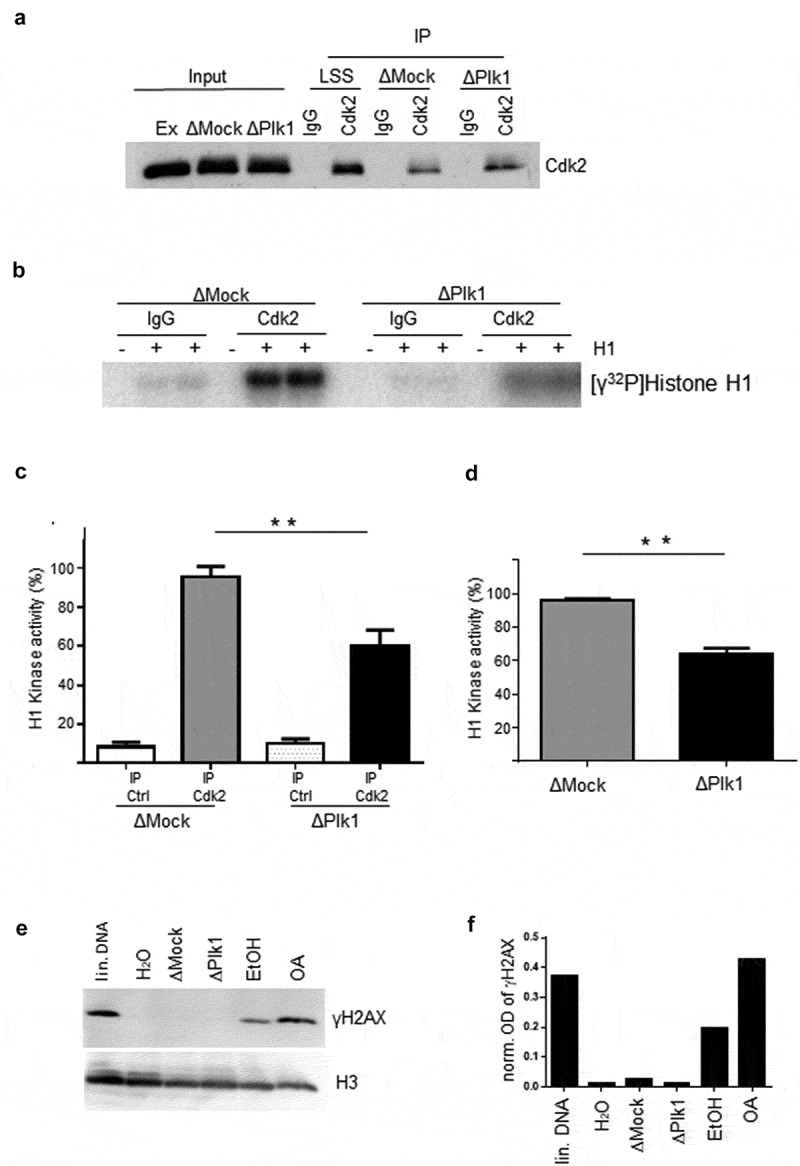
Cdk2 activity is decreased in replicating nuclei after Plk1 depletion A Sperm nuclei (2000 n/µl) were incubated in mock (ΔMock) or Plk1(ΔPlk1) depleted egg extract for 60 min, nuclear extracts were prepared and incubated with control (IP ctrl) or Cdk2 (IP Cdk2) antibody coupled beads. B control or Cdk2 coupled beads from Mock (ΔMock) or Plk1 (ΔPlk1) depleted nuclear extracts were incubated in kinase reactions (triplicates) in the presence of γ32P-ATP and Histone H1 as substrate, proteins were separated on SDS-PAGE and revealed by autoradiography. C Rel. quantification of B as % of maximum H1 kinase activity (mock as 100%). D, Mean H1 kinase activity from Cdk2 coupled beads incubated in mock (ΔMock) or Plk1 (ΔPlk1) depleted nuclear extracts of two independent experiments, * statistically different, P < 0.001, t-test. E Sperm nuclei (7000 n/µl) were incubated for 90 min in Mock (ΔMock), Plk1 (ΔPlk1) depleted egg extracts or in egg extracts containing or not 150 ng/µl EcoRI-linearized pGEX plasmid (lin. DNA)/water (H2O) or in egg extracts containing or not 2 µM Okadaic Acid (OA)/Ethanol (EtOH). Chromatin was purified and processed for western blotting using anti-phospho-Histone H2AX (γH2AX), then anti-Histone H3 antibodies. F quantifications of γH2AX bands (optical density, OD) normalized to H3 signal of experiment E.
Twofold overexpression of Plk1 accelerates DNA replication
In order to better distinguish between a direct or indirect role of Plk1 in checkpoint regulation, the effect of a modest overexpression of Plk1 was investigated. Endogenous Plk1 was measured at 150 nM by SDS-PAGE in interphase egg extracts (Suppl. Fig. 8 A, B). Purified recombinant wild type Xenopus Plk1 was added to replication reactions to obtain a twofold overexpression (300 nM) (Figure 5a, upper panel). Consistent with the increase observed in the cytosol, the recruitment of both, Plk1 and its phosphorylated form, to chromatin (Figure 5a, lower panel) was increased after overexpression in comparison to the control. The effect of this Plk1 overexpression on DNA replication was monitored by nascent strand analysis in three independent experiments in the presence of α32P-dCTP (Figure 5b). We observed that overexpression of Plk1 increased mean DNA replication in a reproducible manner 1.3 to 1.5 fold depending on S phase progression in the absence of exogenous stress, which is consistent with a mean 1.7 fold decrease of the normalized P-Chk1 signal after addition of recombinant Plk1 at all time-points during S phase from different experiments (Figure 5c,d). This result strongly suggests that Plk1 directly inhibits the replication checkpoint in the absence of exogenous replication stress, but it does not exclude other additional mechanisms.
Figure 5.
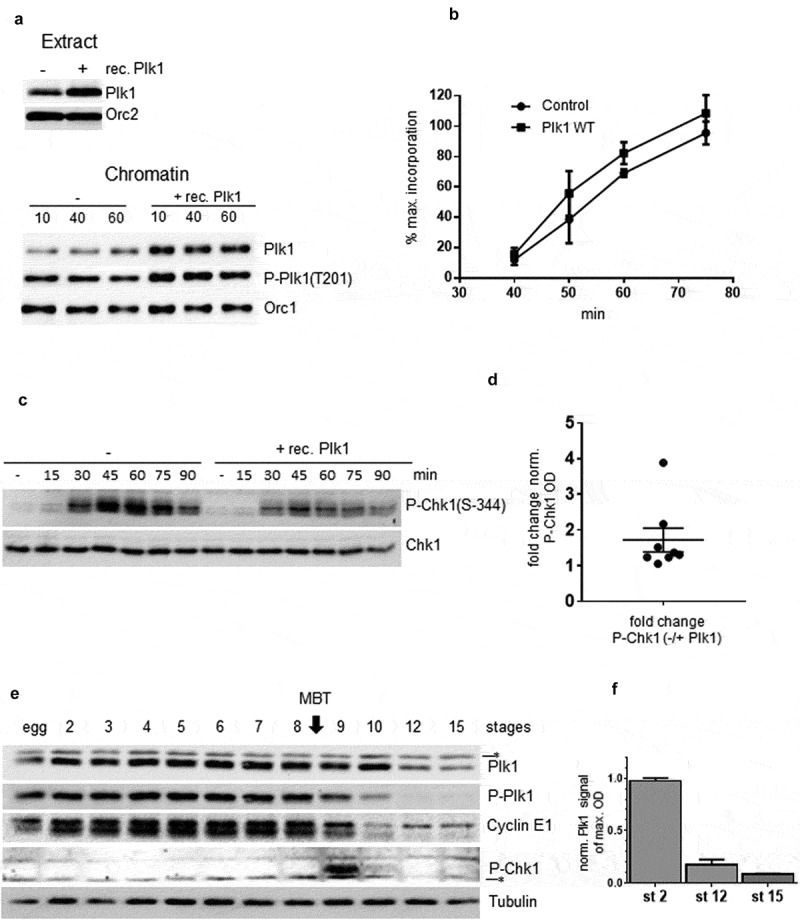
Accelerating effect of Plk1 overexpression on DNA replication and changes in Plk1 abundance during early Xenopus development A Western blot analysis of Plk1 in egg extract (upper) or in chromatin fractions (lower) in control or after Plk1 addition. B Sperm nuclei (2000 n/µl) were incubated in the presence of α32P-dCTP and recombinant Plk1 and reactions were stopped at indicated time, then DNA was purified and analyzed by alkaline gel electrophoresis, mean of percent of maximal incorporation from three independent experiments. C Western blot analysis of P-Chk1 and Chk1 at different time (min) during S phase with (+) or without (-) addition of recombinant Plk1 addition in whole extract. D quantification of 8 different P-Chk1 optical densities of western blots from two independent S phase kinetics experiments normalized with Chk1, points indicating fold change of P-Chk1 from control over Plk1 overexpression samples, with mean (1.722) and standard error of mean 0.33 (SEM). E Xenopus embryos were harvested after in vitro fertilization at indicated stages and whole embryo lysates were analyzed by western blot with Plk1, P-Plk1(T201), cyclin E1, P-Chk1(S344) and tubulin antibodies. F Quantification of western blot, optical densities (OD) of Plk1 normalized to tubulin, at stage 2, 12 and 15 from three different in vitro fertilizations, different egg batches from three female frogs.
Phosphorylated Plk1 levels are high during the first rapid cell cycles and decline after the mid-blastula transition
Plk1 levels have been shown to be similar during S and M phases in Xenopus egg extracts at least during the first cell cycle [26] whereas it is degraded in anaphase in differentiated mammalian cells [25]. In order to understand when and how Plk1 regulation changes during early Xenopus development, Xenopus eggs were fertilized in vitro and embryos harvested at indicated stages, then Plk1 from whole embryo extracts was analyzed by western blotting (Figure 5e). We found that Plk1 levels remained constantly high during the first cell divisions before the onset of zygotic transcription, but were decreased to 20% at stage 12 after the midblastula transition (MBT), then to less than 10% after stage 15 (figure 5f). This observation was reproducible with two other independent series of early embryos (Suppl. Figure 9). Interestingly, the phosphorylation of Plk1 at T201 started to decrease earlier, at stage 9 after 12–13 cell cycles, shortly after the MBT at a time when Cyclin E1 levels decline and Chk1 is transiently phosphorylated. In conclusion, Plk1 is present at constant levels with a steady phosphorylation at T201 in early embryos until the transient activation of Chk1 marking late MBT. Starting from this time, T201 phosphorylation of Plk1, followed by Plk1 protein levels rapidly decline.
Discussion
Polo-like kinase 1 has been shown to play a crucial role in maintaining genome integrity because it is essential for sister chromatid segregation, but also because it is involved in the DNA damage recovery pathway during G2 phase. However, its role during S phase remains poorly understood. In the present study, we used Xenopus’ synchronous in vitro replication system to focus on the role of Plk1 during S phase, thus avoiding side effects of passing through other phases of the cell cycle when Plk1 is up or downregulated. Our study clearly shows that Plk1 positively regulates DNA replication during normal S phase in Xenopus. First, Plk1 is present and phosphorylated on its known activating site T201 on chromatin before the initiation of DNA replication. Second, Plk1 depletion slows down DNA replication whereas Plk1 overexpression accelerates it. Third, Plk1 depletion leads to an increase of Chk1 phosphorylation and a decrease of Cdk2 kinase activity, which supports the idea that Plk1 could inhibit the ATR/Chk1-dependent replication checkpoint during S phase in the absence of any external replication stress.
Plk1 depletion induced a reproducible decrease of DNA synthesis, mainly during early-to mid S phase in a detailed kinetic analysis. Adding back the wild type recombinant Plk1 was sufficient to rescue DNA replication showing that the effect is due to Plk1 and not to co-immunoprecipitated proteins. The added back Plk1 was recruited on chromatin and phosphorylated at the same time and to similar extents than the endogenous Plk1. We always observed that in rescue experiments DNA synthesis recovered with a certain delay compared to the control which could be due to missing or delayed posttranslational modifications of the recombinant Plk1 aside of the detected T201 phosphorylation. Indeed, it has recently been reported that sumoylation could stabilize Plk1 and promote its nuclear import [36]. A previous study did not show any effect of Plk1 depletion on DNA synthesis in the absence of replication stress or on fully licensed chromatin [45], however, DNA replication was only analyzed at the end of S phase when the Plk1 depletion effect was lower. This latter observation leads us to the assumption that the absence of Plk1 could be compensated later in S phase to ensure timely DNA replication and could not change the overall S phase length.
We show that the recruitment of Plk1 on chromatin takes place prior to initiation of DNA replication, at the same time than the recruitment of the Orc complex and before the recruitment of the MCM complex. However, immunodepletion of Orc2 did not prevent Plk1 recruitment which demonstrates that the recruitment Plk1 is independent of the Orc complex as well as the MCM complex and DNA replication itself. It has been reported that when replication stress is induced by aphidicolin, Plk1 is recruited to chromatin after the phosphorylation of MCM2 by ATR in Xenopus [45]. Other studies in human cells show interactions between Plk1 and MCM7 [44]. Our result does not exclude the possibility that Plk1 already on chromatin could focus on sites of stalled replication forks with ongoing DNA synthesis but how Plk1 is recruited to chromatin in the Xenopus in vitro system in the first place remains unknown. Plk1 is known to interact in mitosis with other chromatin bound factors such as topoisomerase II [69] and cohesins [70] but whether these or other chromatin factors are necessary for Plk1 recruitment in interphase would need further investigation.
The activity of Plk1 is fine-tuned by phosphorylation and dephosphorylation. We show that chromatin bound Plk1 is phosphorylated at its major activating site T201 within the T-loop domain. Several mitotic kinases have been shown to phosphorylate Plk1 during G2 phase or mitosis. In Xenopus oocytes, a Polo-like kinase kinase (Plkk) phosphorylates and activates Plk1 at the G2/M transition [29], whereas PKA can phosphorylate Plk1 T201 in vitro [27]. In human cells, during the G2 to M phase transition, Plk1 is phosphorylated in G2 by Bora/Aurora A [30,31] and other PAK kinases [32] on T210 equivalent to Xenopus T201. Whether these kinases are also active during interphase is unknown but, in our hands, the inhibition of Aurora A/B or PKA using specific inhibitors (respectively ZM 447439 and PKI) did not affect replication nor decrease the phosphorylation of Plk1. The inhibition of the S phase kinase Cdk2 by p27 did only slightly, and therefore probably indirectly, decrease Plk1 phosphorylation. Plk1 has been shown to interact with PP2A in G2 phase and mitosis during DNA damage recovery [33,71]. We show here that PP2A inhibition by rubratoxin or okadaic acid strongly increased Plk1 phosphorylation in S phase. PP2A activity therefore out passes the activity of a Plk1 activating kinase during S phase. Moreover, this demonstrates the existence of a phosphorylation turn-over on Plk1-T201 during S phase. This delicate S phase phosphorylation balance must then be reversed when entering mitosis, where Plk1 activity reaches its maximum (Suppl. Figure 2) and when PP2A is inhibited [72]. PP2A inhibition or depletion inhibits DNA replication [64,65] to a greater extent than the Plk1 depletion alone, reflecting the fact that PP2A also targets many other DNA replication proteins such as Cdc6 [73], DNA unwinding element-binding protein (DUE-B) [74] or RPA [75]. However, our results provide evidence that PP2A might be involved in a tight regulation of chromatin bound Plk1 during normal S phase.
Plk1 is a major player of the DNA damage recovery or adaptation pathway in G2/mitosis and has been shown to inhibit the DNA damage checkpoint. We demonstrate here that Plk1 depletion during unperturbed S phase leads to an increase of S344 phosphorylation of Chk1 and a decrease of Cdk2 activity, showing that the replication checkpoint activity increased, even in the absence of external stress. Since Plk1 overexpression leads to opposite effects, this strongly argues in favor of Plk1 directly inhibiting the ATR/Chk1/Cdc25 axis, although we cannot exclude that other replication factors could be additionally targeted. Consistent with an earlier study [45], we could not detect any increase of DNA double strand breaks after Plk1 depletion under normal replication conditions. In fact, DSBs were only described under low, limiting MCM conditions, but not in normal replication conditions after Plk1 depletion in replicating nuclei in Xenopus [45]. This observation further argues in favor of Plk1 directly targeting the checkpoint pathway or replication proteins during normal S phase. It has been shown that Plk1 phosphorylates and inhibits the Chk1 mediator protein Claspin only after prolonged exposure to aphidicolin in Xenopus [21,45]. We and others have shown that the ATR/Chk1 dependent replication checkpoint is activated in Xenopus and mammalian cells in the absence of external stress [13,14,16,17]. This indicates that replication fork stalling occurs during normal S phase, probably due to difficulties in replicating certain regions, limiting replication factors or spontaneous DNA damage [12]. Therefore, during normal S phase, Plk1 could inhibit the transiently activated replication checkpoint in order to promote S phase progression. Previous studies in mammalian cancer cell lines showed that Plk1 depletion affects S phase entry and progression via the inhibition of the recruitment of the MCM complex [46–48]. However, we could not detect any effect of Plk1 depletion on pre-RC assembly nor a visible delay in the entry of S phase (see appearance of first nascent strands in Figure 2b and in Suppl. Fig. 10) in the Xenopus system. It is possible that this regulation would not be conserved in Xenopus embryos whose short cycles are devoid of G1 and G2 phases.
Plk1 overexpression in cultured cells [38–40] disturbs G2 or mitosis events, impairing the close examination of this overexpression on DNA replication. Using S phase extracts, we were able to show that a moderate Plk1 overexpression accelerates DNA replication. It would be interesting to investigate whether excess of Plk1 also confers a selective advantage to rapidly cycling cancer cells. To follow up on this idea, we looked at Plk1 expression during the first embryonic divisions, before midblastula transition (MBT) when the cell cycle is deprived of G1and G2 phases and lasts only 30 min. We found that the level of Plk1 is high during the first rapid cell cycles of development, then suddenly declines at the time when the zygotic transcription resumes and when the cell cycle and the DNA replication program slows down [76]. This could be the consequence of the switch from a stabilized pre-MBT form of Plk1 to the cyclic (degradation/accumulation) control of Plk1 at later stages of development similar to that of cell culture lines [22–25]. It is therefore tempting to speculate that steady high levels of Plk1 during the first cleavages contribute to quick S and M phases in early Xenopus embryos because adaptation or recovery pathways, which include Plk1, would override checkpoint responses.
In early Xenopus embryos, S phase is brief and replication initiates without any apparent sequence specificity [77]. To better understand the mechanisms that ensure complete DNA replication, we proposed a numerical model for the control of DNA replication in Xenopus [78]. Numerical simulations of initiation frequencies are consistent with a global inhibition of replication clusters (group of origins) by Chk1 but need to be combined with a local repression of Chk1 action close to activated origins to fit replication frequencies [16]. The present study now clearly shows that Plk1 positively regulates normal S phase progression, most probably through the inhibition of the replication checkpoint, which would make Plk1 a good candidate for such a local inhibition in the proposed mechanism of our recent numerical model. Further studies including DNA combing experiments followed by the analysis of initiation frequencies after Plk1 depletion or overexpression will tell whether Plk1 can fulfill this role.
Supplementary Material
Acknowledgments
We thank the anonymous referees for their constructive comments. We thank B. Miroux for critical reading of the manuscript, Odile Bronchain for Xenopus embryos, P. Drevet (I2BC) for the production of recombinant Plk1 baculovirus, C. Bonne-Andrea for XCdk2 and R.A. Laskey for XOrc1, XOrc2, XMCM7 antibody, Carl Mann for γ-H2A.X antibody. Anti-Xenopus cyclin E1, developed by JL. Maller were obtained from the European Xenopus Resource Centre, curated with funding from the Wellcome Trust/BBSRC and maintained by the University of Portsmouth, School of Biological Sciences.
Funding Statement
D.C., O.H. and K.M. are supported by the Centre National de Recherche Scientifique (CNRS) and the department of genome biology of I2BC, D.C is supported by a PhD fellowship of IdEX Paris-Saclay University, A.G. is supported by the Fondation de la Recherche Medicale (FRM, DEI20151234404), Institut National du Cancer (INCa, PLBIO16-302) and Commissariat à l’Energie Atomique (CEA).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data of this article can be accessed here.
References
- [1].Ganier O, Prorok P, Akerman I, et al. Metazoan DNA replication origins. Curr Opin Cell Biol. 2019;58:134–141. [DOI] [PubMed] [Google Scholar]
- [2].Lopes M, Cotta-Ramusino C, Pellicioli A, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. [DOI] [PubMed] [Google Scholar]
- [3].Tercero JA, Diffley JF.. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. [DOI] [PubMed] [Google Scholar]
- [4].Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. [DOI] [PubMed] [Google Scholar]
- [5].Shirahige K, Hori Y, Shiraishi K, et al. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. [DOI] [PubMed] [Google Scholar]
- [6].Byun TS, Pacek M, Yee M, et al. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].MacDougall CA, Byun TS, Van C, et al. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guo Z, Kumagai A, Wang SX, et al. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jin J, Shirogane T, Xu L, et al. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Donzelli M, Draetta GF. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003;4:671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2013;16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Marheineke K, Hyrien O. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J Biol Chem. 2004;279:28071–28081. [DOI] [PubMed] [Google Scholar]
- [14].Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. [DOI] [PubMed] [Google Scholar]
- [15].Guo C, Kumagai A, Schlacher K, et al. Interaction of Chk1 with treslin negatively regulates the initiation of chromosomal DNA replication. Mol Cell. 2015;57:492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Platel M, Goldar A, Wiggins JM, et al. Tight Chk1 levels control replication cluster activation in Xenopus. PloS One. 2015;10:e0129090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Maya-Mendoza A, Petermann E, Gillespie DAF, et al. Chk1 regulates the density of active replication origins during the vertebrate S phase. Embo J. 2007;26:2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Michelena J, Gatti M, Teloni F, et al. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J Cell Biol. 2019;218:2865–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shaltiel IA, Krenning L, Bruinsma W, et al. The same, only different - DNA damage checkpoints and their reversal throughout the cell cycle. J Cell Sci. 2015;128:607–620. [DOI] [PubMed] [Google Scholar]
- [20].Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. [DOI] [PubMed] [Google Scholar]
- [21].Yoo HY, Kumagai A, Shevchenko A, et al. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004;117:575–588. [DOI] [PubMed] [Google Scholar]
- [22].Golsteyn RM. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hamanaka R, Smith MR, O’Connor PM, et al. Polo-like kinase is a cell cycle-regulated kinase activated during mitosis. J Biol Chem. 1995;270:21086–21091. [DOI] [PubMed] [Google Scholar]
- [24].Lee KS, Yuan YL, Kuriyama R, et al. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Descombes P, Nigg EA. The polo-like kinase Plx1 is required for M phase exit and destruction of mitotic regulators in Xenopus egg extracts. Embo J. 1998;17:1328–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kelm O. Cell cycle-regulated phosphorylation of the Xenopus Polo-like Kinase Plx1. J Biol Chem. 2002;277:25247–25256. [DOI] [PubMed] [Google Scholar]
- [28].Qian Y-W, Erikson E, Maller JL. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol Cell Biol. 1999;19:8625–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qian Y. Purification and cloning of a protein kinase that phosphorylates and activates the Polo-like Kinase Plx1. Science. 1998;282:1701–1704. [DOI] [PubMed] [Google Scholar]
- [30].Macůrek L, Lindqvist A, Lim D, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. [DOI] [PubMed] [Google Scholar]
- [31].Seki A, Coppinger JA, Jang C-Y, et al. Bora and the kinase aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ji J-H, Hwang H-I, Lee H-J, et al. Purification and proteomic identification of putative upstream regulators of polo-like kinase-1 from mitotic cell extracts. FEBS Lett. 2010;584:4299–4305. [DOI] [PubMed] [Google Scholar]
- [33].Wang L, Guo Q, Fisher LA, et al. Regulation of polo-like kinase 1 by DNA damage and PP2A/B55α. Cell Cycle. 2015;14:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li W, Wang H-Y, Zhao X, et al. A methylation-phosphorylation switch determines Plk1 kinase activity and function in DNA damage repair. Sci Adv. 2019;5:eaau7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Taniguchi E, Toyoshima-Morimoto F, Nishida E. Nuclear translocation of Plk1 mediated by its bipartite nuclear localization signal. J Biol Chem. 2002;277:48884–48888. [DOI] [PubMed] [Google Scholar]
- [36].Wen D, Wu J, Wang L, et al. SUMOylation promotes nuclear import and stabilization of polo-like kinase 1 to support its mitotic function. Cell Rep. 2017;21:2147–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–660. [DOI] [PubMed] [Google Scholar]
- [38].Smith MR, Wilson ML, Hamanaka R, et al. Malignant transformation of mammalian cells initiated by constitutive expression of thePolo-like Kinase1. Biochem Biophys Res Commun. 1997;234:397–405. [DOI] [PubMed] [Google Scholar]
- [39].Mundt KE, Golsteyn RM, Lane HA, et al. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239:377–385. [DOI] [PubMed] [Google Scholar]
- [40].de Cárcer G, Venkateswaran SV, Salgueiro L, et al. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat Commun. 2018;9:3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Qian Y-W, Erikson E, Li C, et al. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol Cell Biol. 1998;18:4262–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lu L-Y, Wood JL, Minter-Dykhouse K, et al. Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol Cell Biol. 2008;28:6870–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Song B, Liu XS, Liu X. Polo-like kinase 1 (Plk1): an unexpected player in DNA replication. Cell Div. 2012;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tsvetkov L, Stern DF. Interaction of chromatin-associated Plk1 and Mcm7. J Biol Chem. 2005;280:11943–11947. [DOI] [PubMed] [Google Scholar]
- [45].Trenz K, Errico A, Costanzo V. Plx1 is required for chromosomal DNA replication under stressful conditions. Embo J. 2008;27:876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu Z-Q LX. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci. 2008;105:1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Song B, Liu XS, Davis K, et al. Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol Cell Biol. 2011;31:4844–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yim H, Erikson RL. Polo-like kinase 1 depletion induces DNA damage in early S prior to caspase activation. Mol Cell Biol. 2009;29:2609–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yata K, Bleuyard J-Y, Nakato R, et al. BRCA2 coordinates the activities of cell-cycle kinases to promote genome stability. Cell Rep. 2014;7:1547–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. Embo J. 2010;29:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. [DOI] [PubMed] [Google Scholar]
- [52].Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. [DOI] [PubMed] [Google Scholar]
- [53].Frank-Vaillant M, Jessus C, Ozon R, et al. Two distinct mechanisms control the accumulation of cyclin B1 and Mos in Xenopus oocytes in response to progesterone. Mol Biol Cell. 1999;10:3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. [DOI] [PubMed] [Google Scholar]
- [55].Sive HL, Grainger RM, Harland RM. Xenopus laevis in vitro fertilization and natural mating methods. CSH Protoc. 2007;2007:pdb.prot4737. [DOI] [PubMed] [Google Scholar]
- [56].Nieuwkoop PD, Faber J, editors. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Pub; 1994. [Google Scholar]
- [57].Marheineke K, Hyrien O. Aphidicolin triggers a block to replication origin firing in Xenopus egg extracts. J Biol Chem. 2001;276:17092–17100. [DOI] [PubMed] [Google Scholar]
- [58].Räschle M, Knipscheer P, Enoiu M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Costanzo V, Shechter D, Lupardus PJ, et al. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. [DOI] [PubMed] [Google Scholar]
- [60].Peng A, Lewellyn AL, Schiemann WP, et al. Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Curr Biol. 2010;20:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. [DOI] [PubMed] [Google Scholar]
- [62].Jang Y-J, Ma S, Terada Y, et al. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002;277:44115–44120. [DOI] [PubMed] [Google Scholar]
- [63].Wada S, Usami I, Umezawa Y, et al. Rubratoxin A specifically and potently inhibits protein phosphatase 2A and suppresses cancer metastasis. Cancer Sci. 2010;101:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chou DM, Petersen P, Walter JC, et al. Protein phosphatase 2A regulates binding of Cdc45 to the prereplication complex. J Biol Chem. 2002;277:40520–40527. [DOI] [PubMed] [Google Scholar]
- [65].Murphy CM, Michael WM. Control of DNA replication by the nucleus/cytoplasm ratio in xenopus. J Biol Chem. 2013;288:29382–29393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rowles A, Chong JP, Brown L, et al. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. [DOI] [PubMed] [Google Scholar]
- [67].Carpenter PB, Dunphy WG. Identification of a novel 81-kDa component of the Xenopus origin recognition complex. J Biol Chem. 1998;273:24891–24897. [DOI] [PubMed] [Google Scholar]
- [68].Romanowski P, Madine MA, Rowles A, et al. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol CB. 1996;6:1416–1425. [DOI] [PubMed] [Google Scholar]
- [69].Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J Biol Chem. 2008;283:6209–6221. [DOI] [PubMed] [Google Scholar]
- [70].Sumara I, Vorlaufer E, Stukenberg PT, et al. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. [DOI] [PubMed] [Google Scholar]
- [71].Kim S-Y, Hyun S-Y, Jang Y-J. Dephosphorylation of Plk1 occurs through PP2A-B55/ENSA/Greatwall pathway during mitotic DNA damage recovery. Cell Cycle. 2019:18(10)1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Castro A, Lorca T. Greatwall kinase at a glance. J Cell Sci. 2018;131(20). [DOI] [PubMed] [Google Scholar]
- [73].Yan Z, Fedorov SA, Mumby MC, et al. PR48, a novel regulatory subunit of protein phosphatase 2A, interacts with Cdc6 and modulates DNA replication in human cells. Mol Cell Biol. 2000;20:1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Gao Y, Yao J, Poudel S, et al. Protein phosphatase 2A and Cdc7 kinase regulate the DNA unwinding element-binding protein in replication initiation. J Biol Chem. 2014;289:35987–36000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang F, Zhu S, Fisher LA, et al. Protein interactomes of protein phosphatase 2A B55 regulatory subunits reveal B55-mediated regulation of replication protein A under replication stress. Sci Rep. 2018;8:2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Platel M, Narassimprakash H, Ciardo D, et al. Genome wide decrease of DNA replication eye density at the midblastula transition of Xenopus laevis. Cell Cycle. 2019;18:1458–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hyrien O, Méchali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. Embo J. 1993;12:4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Goldar A, Labit H, Marheineke K, et al. A dynamic stochastic model for DNA replication initiation in early embryos. PloS One. 2008;3:e2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


