Abstract
The eukaryotic nucleus is not a homogenous single‐spaced but a highly compartmentalized organelle, partitioned by various types of membraneless structures, including nucleoli, PML bodies, paraspeckles, DNA damage foci and RNA clouds. Over the past few decades, these nuclear structures have been implicated in biological reactions such as gene regulation and DNA damage response and repair, and are thought to provide “microenvironments,” facilitating these reactions in the nucleus. Notably, an altered morphology of these nuclear structures is found in many cancers, which may relate to so‐called “nuclear atypia” in histological examinations. While the diagnostic significance of nuclear atypia has been established, its nature has remained largely enigmatic and awaits characterization. Here, we review the emerging biophysical principles that govern biomolecular condensate assembly in the nucleus, namely, liquid‐liquid phase separation (LLPS), to investigate the nature of the nuclear microenvironment. In the nucleus, LLPS is typically driven by multivalent interactions between proteins with intrinsically disordered regions, and is also facilitated by protein interaction with nucleic acids, including nuclear non–coding RNAs. Importantly, an altered LLPS leads to dysregulation of nuclear events and epigenetics, and often to tumorigenesis and tumor progression. We further note the possibility that LLPS could represent a new therapeutic target for cancer intervention.
Keywords: chromatin structure, intrinsically disordered region/protein , liquid‐liquid phase separation , non–coding RNA , nuclear microenvironment
In this review article, we focus on the emerging biophysical principle that governs biomolecular condensate assembly in the cell nucleus, referred to as biological liquid‐liquid phase separation (LLPS). Altered LLPS leads to dysregulation of nuclear events and epigenetic mechanisms, and, thus, to tumorigenesis and tumor development. We further explore the important possibility that LLPS could be a new therapeutic target for cancer therapy.
![]()
1. INTRODUCTION
Tumor cells exhibit atypical nuclear morphologies that may reflect changes in genomic integrity and nuclear substructures and their functions. The nucleus is compartmentalized, and genomic DNA is surrounded by membraneless structures composed of proteins and RNAs. These include nucleoli, paraspeckles, promyelocytic leukemia (PML) bodies, chromosome territories and DNA damage foci 1 (Figure 1A).
Figure 1.
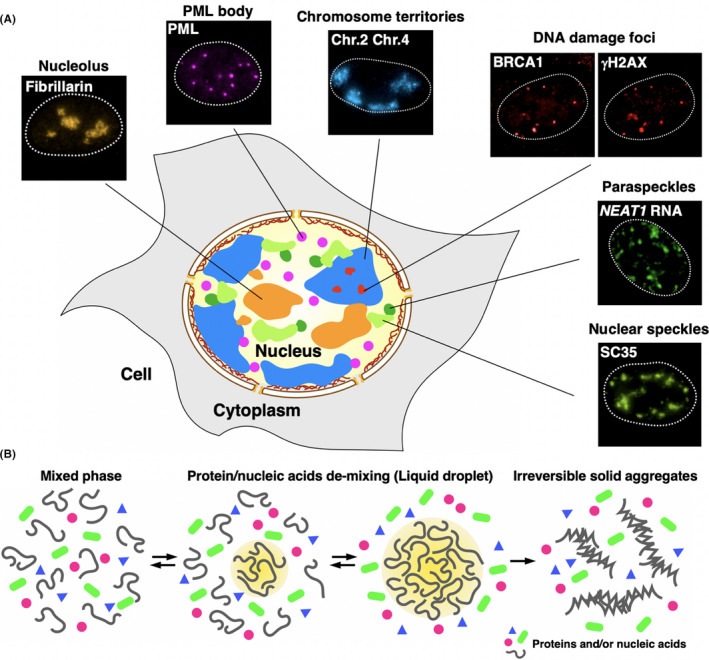
Nuclear microenvironments and principles of their formation. A, The eukaryotic nucleus is highly compartmentalized and contains various membraneless organelles (nuclear bodies/structures). Their immuno‐stained or FISH images are shown. B, Multivalent interactions between proteins and nucleic acids trigger liquid‐liquid phase separation (LLPS). LLPS is reversible (left) and the resultant liquid droplet (yellow‐highlighted area) is dynamic (middle); however, its hyper self‐assembly causes irreversible solid aggregate formation (right)
Over 10 years ago, Zaidi et al proposed the insightful concept that chromatin and nuclear substructures can be considered as “nuclear microenvironments,” which serve as platforms to spatially organize nuclear machineries. Significantly, the composition and organization of nuclear microenvironments are compromised under stress. 2 For example, upon DNA damage, BRCA1/2 (breast cancer‐associated protein 1/2), whose mutations are associated with breast and ovarian cancer, 3 , 4 forms a compartment (Figure 1A) that promotes DNA damage repair and maintenance of genome integrity. 5 Histone proteins in chromatin become phosphorylated at damaged sites, which extend to long megabase‐sized regions, 6 and create nuclear foci (Figure 1A), where numerous DNA damage repair molecules are recruited. Considering nuclear microenvironments, rather than a single biochemical reaction, may be appropriate because a single component cannot confer multilayered cancer‐related changes. From this viewpoint, the discovery and identification of the novel and fundamental properties that create and regulate the nuclear microenvironments have been long awaited.
A series of recent collaborations between cell biologists and biophysicists have robustly revealed that membraneless structures, nuclear bodies or macromolecule condensates are formed through biophysical phenomena referred to as liquid‐liquid phase separation (LLPS). 7 , 8 LLPS is a process in which a single liquid phase composed of mutually soluble components demixes into two or more distinct phases, creating a “liquid droplet” (Figure 1B). Inside cells, LLPS concentrates certain factors at specific places and excludes other factors, creating a heterogenous environment. The molecules inside the droplet are highly mobile, and the droplet itself is also dynamic and reversible. These biophysical properties may bring efficient biological processes and acute cellular responses.
Liquid‐liquid phase separationis thought to be triggered by weak, multivalent interactions between proteins and nucleic acids. 7 , 8 Peptides with intrinsically disordered regions (IDR) in RNA binding proteins (RBPs) tend to undergo LLPS and form liquid droplets or hydrogels in vitro and in vivo. 9 These peptides often contain limited kinds of amino acids, and are referred to as low‐complexity (LC) or prion‐like domains. 10 LLPS is in principle reversible, and the resultant liquid droplets are dynamic and easily undergo fusion and fission. However, liquid droplets composed of specific proteins such as fused in sarcoma (FUS) can also excessively self‐assemble and irreversibly transform into solid aggregates that are known to have a causative role in neurodegenerative diseases 11 (Figure 1B).
In this review, we revisit the concept of nuclear microenvironments with the perspective of LLPS, to provide a new pathological framework for the aberrant chromatin and nuclear structures in cancer. We discuss recent discoveries regarding LLPS in chromatin and nuclear organization, and the role of nuclear ncRNAs as LLPS regulators. We specifically focus on instances in which the dysregulation of the nuclear microenvironment relates to cancer proliferation, progression and recurrence.
2. CHROMATIN ORGANIZATION IS MODULATED BY LLPS
In the eukaryotic nucleus, the genomic DNA is packaged into chromatin. Chromatin organization is a major determinant for nuclear functions such as gene regulation, DNA replication and repair, and chromosome segregation, because chromatin serves as a platform for them. Therefore, chromatin can be considered as one of the components that create a nuclear microenvironment. It is becoming clear that chromatin organization is modulated by LLPS.
2.1. A nucleosome array undergoes LLPS
The nucleosome is the fundamental repeat unit of chromatin, in which 146 bp of DNA are wrapped around an octamer of core histones, H2A, H2B, H3 and H4. 12 DNA has a negatively charged phosphate backbone, while histones are enriched with positively charged amino acids. Thus, the nucleosome is formed by electrostatic interactions between DNA and the histone octamer, which may halve the negative charge of DNA 13 (Figure 2A).
Figure 2.
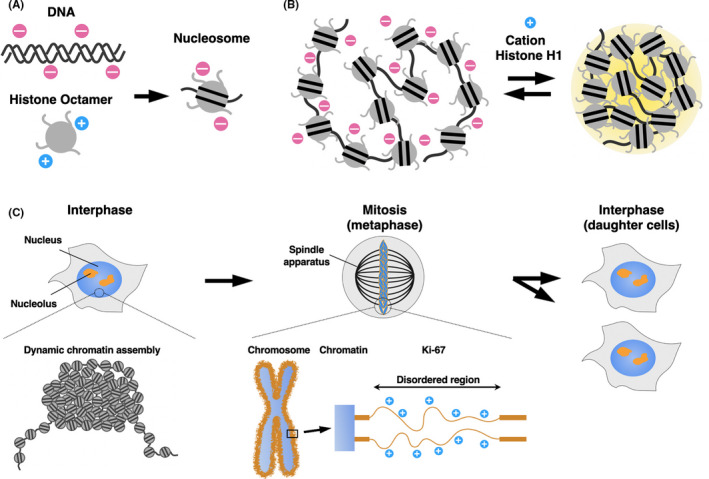
A nucleosome array undergoes LLPS. A, DNA has a negatively charged phosphate backbone, while the core histone proteins are enriched with positively charged amino acids. The nucleosome is formed by electrostatic interactions between DNA and the histone octamer, which may halve the negative charge of DNA. B, A nucleosome array undergoes LLPS upon cation or histone H1 addition in vitro. C, Interphase chromatin forms a large and irregular assembly that behaves like a “liquid droplet” in the nucleus. Metaphase chromosomes are coated with Ki‐67 (orange), which serves as a surfactant
A recent study indicated that nucleosome arrays form liquid droplets through LLPS, at a physiological cation concentration 14 (Figure 2B). Positively charged core histone tails promoted the droplet formation with electrostatic interactions between DNA and the histone octamer. Histone H1 is another histone that binds to a linker region of the nucleosome array. The addition of histone H1 or cations neutralized the repulsion among negatively charged nucleosomes, promoted multivalent interactions among the nucleosomes, and facilitated the droplet formation. Increasing the concentration or number of nucleosomes also promoted LLPS, by increasing the chance of interactions among the multiple nucleosomes. Further analysis revealed that histone acetylation resulted in the dissolution of the nucleosome droplets, but adding the bromodomain‐containing protein 4 (BRD4), which binds to acetylated histone tails, triggered re–phase‐separation of nucleosomes. 14 Droplets of nucleosomes with unmodified histone and newly‐induced droplet of nucleosomes by BRD4 did not coalesce, indicating that distinct chromatin compartments are induced by LLPS with histone modifications and epigenetic “reader” proteins. These findings imply that LLPS is involved in the compartmentalization of nucleosome arrays, by concentrating the components and excluding the arrays with different properties.
2.2. Chromatin structure is regulated by LLPS
Liquid‐liquid phase separation is also involved in higher‐order chromatin structure. Super‐resolution imaging in live cells demonstrated that chromatin represents dynamic and liquid‐like properties in the nucleus, rather than static and physically constrained solid‐phase structures 15 (Figure 2C). This observation inspired us to imagine that on a larger scale, chromatin may also be phase‐separated, and dynamically form liquid droplets.
Compacted and decompacted chromatin structures are known to tightly correlate to gene repression and activation, respectively. Constitutive heterochromatin is cytologically defined as a chromatin segment that is highly condensed throughout the cell cycle, and heterochromatin protein 1 (HP1) is its major component. 16 An in vitro analysis revealed that the Schizosaccaromyces pombe HP1 protein Swi6 interacts with the nucleosome through histone H2B, facilitating LLPS of the nucleosome array. 17 A plausible interpretation of this phenomenon is that the histone octamer conformation is disorganized by Swi6 and buried nucleosome regions become exposed, which newly induces multivalent interactions and triggers LLPS. This may lead to a higher concentration of nucleosomes within the droplets, explaining how the HP1 LLPS facilitates chromatin compaction and concentration, and steric exclusion of transcription factors.
Polymer modelling analyses suggested that a compacted chromatin segment plays a role in “spreading” to large‐scale chromatin structures in a self‐organizing manner. 18 , 19 In this model, an active system to decompact chromatin would also be required to preserve decompacted chromatin structures. Nozawa et al demonstrated that scaffold attachment factor A (SAF‐A)/heterogeneous nuclear ribonucleoprotein U (hnRNP U) decompacts transcriptionally active large‐scale chromatin structures. 20 SAF‐A apparently formed a filament‐shaped oligomer with nuclear RNA, which is required for chromatin decompaction. The SAF‐A/RNA filaments are predicted to biochemically assemble into a nuclear mesh in the nucleus. Nozawa et al suggested that SAF‐A and RNA form a transcriptionally responsive, dynamic nuclear mesh that creates a highly viscous microenvironment that keeps chromatin decompacted. 21 Furthermore, the disruption of nuclear mesh led to chromosomal instability; however, the detailed mechanisms remain to be elucidated. Taken together, it is tempting to speculate that a nuclear SAF‐A/RNA mesh partitions the genome into functionally diverse nuclear microenvironments, an essential process for maintaining genome integrity.
2.3. Mitotic chromosome dynamics is governed by LLPS
Faithful genome segregation during cell division depends on the precise formation of mitotic chromosomes (Figure 2C), involving dynamic structural changes consisting of chromosome condensation, resolution of sister chromatids and individualization of chromosomes as spatially separate bodies. A failure in any of these steps can cause chromosome segregation errors, resulting in genome rearrangements and aneuploidy. Recent reports have suggested that the concept of phase separation could also be applicable to these processes.
Ki‐67 was first identified as a nuclear antigen in Hodgkin lymphoma cells, 22 and is now widely used as a proliferation marker for human tumor cells. Ki‐67 primarily exists in nucleoli during interphase, and becomes re–localized to chromosome surfaces in mitosis. 23 In cells, the depletion of Ki‐67 collapsed chromosomes into a single chromatin mass after mitotic entry. 23 Further analyses using a series of Ki‐67 truncations indicated that size and overall electric charge are important to keep mitotic chromosomes apart from each other. These results suggest that Ki‐67 acts as a steric or electrostatic charge barrier, or surfactant, by phase‐separating individual mitotic chromosome arms.
Aurora B kinase, a component of the chromosomal passenger complex (CPC), accumulates at inner centromeres in mitosis and controls the kinetochore‐microtubule attachments to ensure accurate chromosome segregation through its kinase activity. The accumulation of CPC at inner centromeres is reportedly mediated by LLPS. 24 Significance of the accumulation of CPC in the regulation of the Aurora B kinase activity remains enigmatic. As in the nuclear microenvironment in interphase cells, LLPS is involved in the control of chromosome stability during mitosis, by concentrating appropriate components, excluding unnecessary ones, and conferring dynamics and plasticity.
3. RNA REGULATES NUCLEAR FUNCTIONS THROUGH LLPS
RNA is an important factor in LLPS, and early in vitro experiments showed that RNA binding proteins (RBPs) drive LLPS. 25 , 26 Many non–coding RNAs (ncRNAs) are localized on chromatin, 27 and often occupy a particular nuclear area, forming an “RNA cloud.” ncRNA is a major component of membraneless structures such as the nucleolus and paraspeckles that exhibit biophysical properties as liquid droplets. 25 , 26 , 28 , 29 , 30 The expression and resulting nuclear structure seem physiologically relevant, as ncRNA is frequently dysregulated in cancers. For example, the MALAT1 ncRNA, consisting of nuclear speckles, is highly expressed in lung cancer and breast cancer, and aberrant nuclear speckles with excess amounts of MALAT1 ncRNA are known to be involved in cancer metastasis and oncogene activation during cancer progression. 31 , 32 , 33
3.1. ncRNAs regulate chromatin structure and gene expression
The XIST ncRNA acts in X‐chromosome inactivation in mammalian females. 34 In early development, one of the two X chromosomes in females is suppressed for dosage compensation. In this process, approximately 17 kb of the XIST RNA is produced from a region called the X‐inactivation center in the inactive X chromosome. It then spreads along the entire chromosome, creates a XIST RNA cloud over it, and compacts and represses it. 34 , 35 In the nucleus, the XIST RNA may serve as a platform to create a microenvironment for the repression of genes on the inactive X chromosome. For example, the XIST RNA recruits the SMCHD1‐HBiX1 complex and polycomb repressive complex 2 (PRC2), which contribute to chromosome compaction and gene repression. 36 , 37 ncRNAs may be machineries for concentrating certain factors at specific sites in LLPS. In the human body, the XIST RNA is required for the suppression of leukemia and breast cancers. 38
Of note, ncRNA is not always involved in repressing gene expression. A recent study showed that nuclear ncRNAs, including the XIST RNA, have an ability to destabilize nucleosomes, both in vitro and in vivo. This intrinsic RNA activity may conversely facilitate the access of regulatory factors to chromatin for remodeling. 39 There are also ncRNAs that activate transcription, including HOTTIP, XACT and ELEANORS, which we will discuss later.
3.2. ncRNAs organize and regulate membraneless structures in the nucleus
Various mechanisms for LLPS modulation by RNAs have been proposed (Figure 3). First, the transcription of a specific ncRNA can trigger nuclear body formation (Figure 3A). For example, the nucleolus, the site of ribosome biogenesis, is formed around the ribosomal RNA (rRNA) transcription sites. When the rRNAs are artificially transcribed elsewhere in the chromosome, a nucleolus‐like structure is newly formed at the site. 40 , 41 The paraspeckle is a nuclear body involved in RNA editing and gene regulation, and its formation is dependent on the NEAT1 ncRNA. 42 , 43 When the NEAT1 RNA is transcribed ectopically, other paraspeckle components are recruited, and de novo functional paraspeckle is formed at the site. 44 These observations suggest that nuclear body formation is initially driven by RNA seeding events and followed by the accumulation of RBP; a high local concentration of RBP leads to LLPS.
Figure 3.
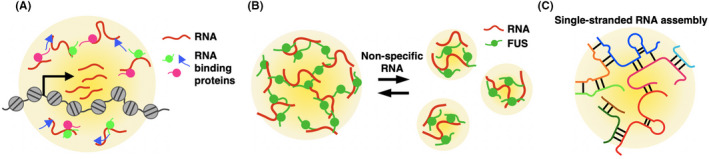
RNAs are regulatory factors for LLPS in the nucleus. A, RNA serves in the “seeding” of membraneless organelles in the nucleus. Many non–coding RNAs (ncRNAs) are localized on chromatin and recruit RNA binding proteins that drive LLPS. B, Non–specific RNAs buffer LLPS of proteins. A specific ncRNA, such as NEAT1, facilitates LLPS of the fused in sarcoma (FUS) protein, while the LLPS of FUS is repressed by an increasing concentration of non–specific RNAs. C, Single‐stranded RNAs containing repetitive sequences tend to self‐assemble through their multivalent base‐pairings
Second, RNA buffers LLPS (Figure 3B). FUS is an LC domain‐containing RBP and forms FUS bodies with the NEAT1 RNA, while FUS bodies are dissolved with an increasing concentration of non–specific RNAs. This suggests that the LLPS mediated by the FUS protein is modulated by both specific (NEAT1 RNA) and non–specific RNAs. 45 An excessive LLPS caused by a reduction of the nuclear RNA levels, genetic ablation of FUS and other RBP, and overmaturation or aging of their liquid droplets may trigger the formation of cytotoxic solid‐like, insoluble assemblies in cells. Therefore, buffering the LLPS level by RNA is important for keeping RBP in a soluble state. 45
Finally, RNA itself also undergoes LLPS 46 (Figure 3C). The expansion of short nucleotide repeats is found in patients with neurological and neuromuscular disorders. A transcript containing such repetitive sequences drives self‐assembly through multivalent base‐pairing with a similar repeat number, resulting in RNA condensates in the nucleus, which potentially disrupt cellular functions. Increasingly, various mechanisms for the regulation of nuclear microenvironments by ncRNAs are being explored.
4. NUCLEAR MICROENVIRONMENTS ARE ALTERED IN CANCERS
In cancer cells, the nuclear morphologies are frequently altered, in a phenomenon referred to as nuclear atypia. 2 For example, changes in composition, number, size and activity on nucleoli are seen, and their diagnostic significance has been proposed 47 , 48 The prominent nucleolus observed in a rare hematological malignancy, blastic plasmacytoid dendritic cell neoplasm, is a compelling case. 49 Another relevant alteration is found in the PML body, in which the level of SUMOylation may underlie LLPS. 50 In acute promyelocytic leukemia (APL), heterozygotic chromosomal translocation between the long arms of chromosome 15 and 17, t(15;17)(q22;q21), produces the PML‐retinoic acid receptor alpha (RARα) fusion protein, leading to PML body disruption and loss of its function. 50 Treatment with all‐trans retinoic acid (ATRA) induces degradation of the fusion protein and recovers the PML body and the cellular functions, which corresponds to an improved outcome of patients. As the integrity of these nuclear structures and thereby the microenvironments are governed by LLPS, a manipulation of LLPS must provide a way to develop novel therapeutics.
4.1. DNA damage foci provide nuclear microenvironments crucial for genome integrity
Defects in DNA repair pathways lead to genomic mutations that can contribute to tumorigenesis or increase cancer aggressiveness. The DNA damage response (DDR) pathway is crucial for the maintenance of genome integrity. The early steps in DDR are the recognition of the DNA lesion and the recruitment of DNA repair proteins to it. Part of the DDR pathway is regulated by LLPS. 51 p53‐binding protein1 (53BP1) is a major player in the DDR pathway and accumulates at DNA lesions, where it creates a nuclear environment called “53BP1 nuclear bodies” to scaffold factors for downstream signal cascades. 52 A live‐cell imaging analysis revealed that the 53BP1 bodies undergo dynamic fusion and fission, a hallmark of LLPS. 51 The p53 tumor suppressor, a transcription factor, and USP28, a deubiquitinase that stabilizes p53, are assembled on the 53BP1 bodies, which are required for the activation of target genes including p21, encoding another tumor suppressor. 53 Therefore, the 53BP1 nuclear body coordinates the DNA damage recognition with gene expression. A super‐resolution microscopic analysis revealed that the 53BP1 nuclear body is colocalized with megabase‐sized chromatin domains called topologically associating domains (TAD), and re–organizes the 3D genome architectures surrounding the DNA damage site, which may be important for reducing the chance of chromosomal translocations. 54 Altogether, the 53BP1 bodies represent the LLPC’s role in DDR: the concentration of proper factors on DNA damage sites and the exclusion of undamaged DNA from damaged DNA.
4.2. Aberrant nuclear microenvironment is created through LLPS in cancers
Gene expression is dysregulated in cancer. RNA polymerase II (RNA pol II) and nascent transcripts are concentrated at discrete sites in the nucleus, the so‐called transcription factories, 55 which can now be explained by LLPS. The C‐terminal domain (CTD) of RNA pol II is a disordered, low‐complexity region that is differentially phosphorylated at transcriptional initiation, elongation, and termination. At transcription initiation, the phosphorylated CTD plays a central role in assembling transcription factors, including the mediator subunit MED1, through LLPS, resulting in a transcription factory or condensate. 56 , 57 , 58 Upon entering the elongation stage, the differential phosphorylation of CTD dissociates RNA pol II from the transcriptional condensate and relocates it to another condensate for splicing factors 59 These findings imply that LLPS is involved in transcription processes, including initiation and the switch to elongation, by concentrating factors and conferring dynamics with phosphorylation.
Super‐enhancers are proposed to be large clusters of enhancers that lead to the highly active transcription of nearby target genes. 60 At each super‐enhancer, RNA pol II, transcription factors, MED1 and BRD4 undergo LLPS and form condensates (Figure 4A). 57 , 61 LLPS thus selectively concentrates factors at specific genomic regions. 61 It was postulated that cancer cells gain large super‐enhancers for driver oncogenes, and, therefore, they are more sensitive to transcriptional inhibitors than normal cells. 61
Figure 4.
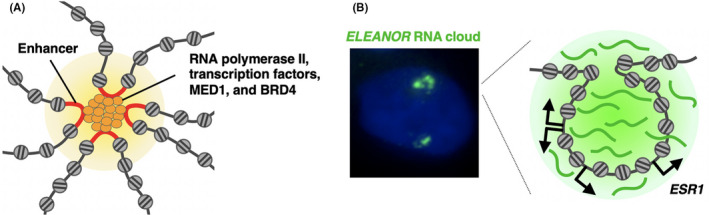
Transcription regulation by membraneless structures in the nucleus. A, A super‐enhancer is a cluster of enhancers that are bound by RNA polymerase II, transcription factors, the mediator subunit MED1 and bromodomain‐containing protein 4 (BRD4). Super‐enhancers are locally concentrated and create a microenvironment for highly active transcription. B, ELEANOR RNA cloud (green) in the nucleus (blue) in a recurrent estrogen receptor‐positive breast cancer model cell (left). ELEANORS activate a large chromatin domain containing multiple breast cancer‐related genes (right)
When estrogen receptor (ER)‐positive breast cancer cells are deprived of estrogen for a long period of time, they acquire estrogen independent proliferation ability. These cells called long‐term estrogen deprivation (LTED) cells recapitulate and, therefore, are a model for recurrence after endocrine therapy. In these cells, a cluster of ncRNAs, named ELEANORS, is produced from the approximately 700 kb chromatin domain containing the estrogen receptor 1 (ESR1) gene, encoding estrogen receptor α. ELEANORS activate the entire domain and lead to the high expression of ESR1 and neighboring genes. ELEANORS form a cloud, providing a nuclear microenvironment for active transcription 62 , 63 , 64 (Figure 4B). ELEANORS also play a role in mediating long‐range chromatin interactions and balancing between cell proliferation and apoptosis. 64
5. CONCLUDING REMARKS
In this review, we have discussed the emerging concept of LLPS in nuclear microenvironment formation, and how it regulates nuclear functions such as gene expression and maintains genome integrity. LLPS is a well‐studied phenomenon in physics and protein chemistry, and now this knowledge can be transferred to understanding the structure and functions of the nucleus. We reconsidered the nuclear atypia from the viewpoint of LLPS, and gained a clearer understanding of genome dysregulation in cancer cells. The nuclear microenvironments that are newly created or disrupted in cancer may potentially be therapeutic targets. In fact, oligonucleotide‐mediated inhibition of the ncRNAs seems promising. 65 , 66 , 67 Searches for small compounds that target LLPS have started for neurodegenerative diseases, 68 in which pathogenic membraneless aggregates are formed. These studies will pave the way for novel cancer therapies.
DISCLOSURE STATEMENT
R‐SN received a research grant from the Uehara Memorial Foundation, the Naito Foundation and the Vehicle Racing Commemorative Foundation. NS received a research grant from DAIZ Inc., the Uehara Memorial Foundation, the Naito Foundation, the Vehicle Racing Commemorative Foundation, the Takeda Science Foundation and the Princess Takamatsu Cancer Research Fund. MT received a research grant from the Vehicle Racing Commemorative Foundation. Other authors declare that they have no conflict of interest.
ACKNOWLEDGMENTS
R‐SN is supported by the Leading Initiative for Excellent Young Researchers, MEXT, Japan. This work was supported in part by JSPS KAKENHI grant numbers JP20H03190 (to R‐SN), JP19K23736 (to TY), JP18K14629 (to MT), JP17H05013 (to HT), JP20K06496 (to HT), JP20H05397 (to HT), JP18H0 (to TH), JP15H05977 (to TH), JP18H05531 (to NS), and JP20H03520 (to NS). This work was supported by grants from the Uehara Memorial Foundation (to R‐SN and NS), the Naito Foundation (to R‐SN and NS), the Vehicle Racing Commemorative Foundation (to R‐SN, MT and NS), the Takeda Science Foundation (to NS), the Princess Takamatsu Cancer Research Fund (to NS) and research grant from DAIZ Inc.
Nozawa R-S, Yamamoto T, Takahashi M, et al. Nuclear microenvironment in cancer: Control through liquid-liquid phase separation. Cancer Sci. 2020;111:3155–3163. 10.1111/cas.14551
Contributor Information
Ryu‐Suke Nozawa, Email: ryusuke.nozawa@jfcr.or.jp.
Noriko Saitoh, Email: noriko.saito@jfcr.or.jp.
REFERENCES
- 1. Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zaidi SK, Young DW, Javed A, et al. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454‐463. [DOI] [PubMed] [Google Scholar]
- 3. Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676‐689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jazaeri AA. Molecular profiles of hereditary epithelial ovarian cancers and their implications for the biology of this disease. Mol Oncol. 2009;3:151‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson SP, Bartek J. The DNA‐damage response in human biology and disease. Nature. 2009;461:1071‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuo LJ, Yang L‐X. Gamma‐H2AX ‐ a novel biomarker for DNA double‐strand breaks. In Vivo. 2008;22:305‐309. [PubMed] [Google Scholar]
- 7. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 9. Lin Y, Protter DSW, Rosen MK, Parker R. Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell. 2015;60:208‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiang S, Kato M, Wu LC, et al. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid‐like droplets, and nuclei. Cell. 2015;163:829‐839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aguzzi A, Altmeyer M. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016;26:547‐558. [DOI] [PubMed] [Google Scholar]
- 12. Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868‐871. [DOI] [PubMed] [Google Scholar]
- 13. Maeshima K, Imai R, Tamura S, Nozaki T. Chromatin as dynamic 10‐nm fibers. Chromosoma. 2014;123:225‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibson BA, Doolittle LK, Schneider MWG, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470‐484.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nozaki T, Imai R, Tanbo M, et al. Dynamic organization of chromatin domains revealed by super‐resolution live‐cell imaging. Mol Cell. 2017;67:282‐293.e287. [DOI] [PubMed] [Google Scholar]
- 16. Allshire RC, Madhani HD. Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol. 2018;19:229‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanulli S, Trnka MJ, Dharmarajan V, et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature. 2019;575:390‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brackley CA, Taylor S, Papantonis A, Cook PR, Marenduzzo D. Nonspecific bridging‐induced attraction drives clustering of DNA‐binding proteins and genome organization. Proc Nat Aca Sci USA. 2013;110:E3605‐E3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erdel F, Rippe K. Formation of chromatin subcompartments by phase separation. Biophys J. 2018;114:2262‐2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nozawa R‐S, Boteva L, Soares DC, et al. SAF‐A regulates interphase chromosome structure through oligomerization with chromatin‐associated RNAs. Cell. 2017;169:1214‐1227.e1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nozawa R‐S, Gilbert N. RNA: nuclear glue for folding the genome. Trends Cell Biol. 2019;29:201‐211. [DOI] [PubMed] [Google Scholar]
- 22. Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13‐20. [DOI] [PubMed] [Google Scholar]
- 23. Cuylen S, Blaukopf C, Politi AZ, et al. Ki‐67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535:308‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trivedi P, Palomba F, Niedzialkowska E, Digman MA, Gratton E, Stukenberg PT. The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat Cell Biol. 2019;21:1127‐1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kato M, Han TW, Xie S, et al. Cell‐free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han TW, Kato M, Xie S, et al. Cell‐free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768‐779. [DOI] [PubMed] [Google Scholar]
- 27. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid‐like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Nat Aca Sci USA. 2011;108:4334‐4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feric M, Vaidya N, Harmon TS, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686‐1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamazaki T, Souquere S, Chujo T, et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70:1038‐1053.e7. [DOI] [PubMed] [Google Scholar]
- 31. Ji P, Diederichs S, Wang W, et al. MALAT‐1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene. 2003;22:8031‐8041. [DOI] [PubMed] [Google Scholar]
- 32. Li Z‐X, Zhu Q‐N, Zhang H‐B, Hu Y, Wang G, Zhu Y‐S. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757‐6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Avner P, Heard E. X‐chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2:59‐67. [DOI] [PubMed] [Google Scholar]
- 35. Senner CE, Brockdorff N. Xist gene regulation at the onset of X inactivation. Curr Opin Genet Dev. 2009;19:122‐126. [DOI] [PubMed] [Google Scholar]
- 36. Nozawa R‐S, Nagao K, Igami K‐T, et al. Human inactive X chromosome is compacted through a PRC2‐independent SMCHD1‐HBiX1 pathway. Nat Struct Mol Biol. 2013;20:566‐573. [DOI] [PubMed] [Google Scholar]
- 37. Jonkers I, Monkhorst K, Rentmeester E, Grootegoed JA, Grosveld F, Gribnau J. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28:5583‐5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Can Res. 2017;77:3965‐3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujita R, Yamamoto T, Arimura Y, et al. Nucleosome destabilization by nuclear non–coding RNAs. Commun Biol. 2020;3:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745‐1763. [DOI] [PubMed] [Google Scholar]
- 41. Oakes ML, Johzuka K, Vu L, Eliason K, Nomura M. Expression of rRNA genes and nucleolus formation at ectopic chromosomal sites in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:6223‐6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirose T, Nakagawa S. Paraspeckles: possible nuclear hubs by the RNA for the RNA. Biomol Concepts. 2012;3:415‐428. [DOI] [PubMed] [Google Scholar]
- 43. Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43:124‐135. [DOI] [PubMed] [Google Scholar]
- 44. Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167‐173. [DOI] [PubMed] [Google Scholar]
- 45. Maharana S, Wang J, Papadopoulos DK, et al. RNA buffers the phase separation behavior of prion‐like RNA binding proteins. Science. 2018;360:918‐921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin Cancer Biol. 2016;37–38:36‐50. [DOI] [PubMed] [Google Scholar]
- 49. Sakamoto K, Katayama R, Asaka R, et al. Recurrent 8q24 rearrangement in blastic plasmacytoid dendritic cell neoplasm: association with immunoblastoid cytomorphology, MYC expression, and drug response. Leukemia. 2018;32:2590‐2603. [DOI] [PubMed] [Google Scholar]
- 50. Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165‐170. [DOI] [PubMed] [Google Scholar]
- 51. Kilic S, Lezaja A, Gatti M, et al. Phase separation of 53BP1 determines liquid‐like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lukas C, Savic V, Bekker‐Jensen S, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243‐253. [DOI] [PubMed] [Google Scholar]
- 53. Cuella‐Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, Chapman JR. 53BP1 integrates DNA repair and p53‐dependent cell fate decisions via distinct mechanisms. Mol Cell. 2016;64:51‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ochs F, Karemore G, Miron E, et al. Stabilization of chromatin topology safeguards genome integrity. Nature. 2019;574:571‐574. [DOI] [PubMed] [Google Scholar]
- 55. Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription "factories' in human nuclei. J Cell Sci. 1996;109:1427‐1436. [DOI] [PubMed] [Google Scholar]
- 56. Boehning M, Dugast‐Darzacq C, Rankovic M, et al. RNA polymerase II clustering through carboxy‐terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833‐840. [DOI] [PubMed] [Google Scholar]
- 57. Cho W‐K, Spille J‐H, Hecht M, et al. Mediator and RNA polymerase II clusters associate in transcription‐dependent condensates. Science. 2018;361:412‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chong S, Dugast‐Darzacq C, Liu Z, et al. Imaging dynamic and selective low‐complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guo YE, Manteiga JC, Henninger JE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sabari BR, Dall’Agnese A, Boija A, et al. Coactivator condensation at super‐enhancers links phase separation and gene control. Science. 2018;361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tomita S, Abdalla MOA, Fujiwara S, et al. A cluster of noncoding RNAs activates the ESR1 locus during breast cancer adaptation. Nat Commun. 2015;6:6966‐7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yamamoto T, Sakamoto C, Tachiwana H, et al. Endocrine therapy‐resistant breast cancer model cells are inhibited by soybean glyceollin I through Eleanor non–coding RNA. Sci Rep. 2018;8:15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Abdalla MOA, Yamamoto T, Maehara K, et al. The Eleanor ncRNAs activate the topological domain of the ESR1 locus to balance against apoptosis. Nat Commun. 2019;10:3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kondo Y, Shinjo K, Katsushima K. Long non–coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927‐1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Misawa A, Takayama KI, Inoue S. Long non–coding RNAs and prostate cancer. Cancer Sci. 2017;108:2107‐2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non–coding RNAs in cancer. Trends Mol Med. 2018;24:257‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wheeler RJ, Lee HO, Poser I, et al. Small molecules for modulating protein driven liquid‐liquid phase separation in treating neurodegenerative disease. bioRxiv. 2019;430:4711. [Google Scholar]


