Abstract
Klebsiella pneumoniae is recognized as one of the most important healthcare-associated pathogens worldwide due to its tendency to develop antibiotic resistance and cause fatal outcomes. Bacterial identification methods such as culture and biochemical tests are routinely used with limited accuracy in many low- and middle-income countries, including Sudan. The aim of this study was to test the accuracy of identification of K. pneumoniae in Khartoum, Sudan. Two hundred and fifty K. pneumoniae isolates were collected and identified using conventional phenotypic methods, biochemically using API 20E and genotypically by amplification of 16S−23S rDNA and sequencing of rpoB, gapA and pgi. Only 139 (55.6 %) of the isolates were confirmed as K. pneumoniae genotypically by PCR and 44.4 % were identified as non- K. pneumoniae . The results demonstrate that the identification panels used by the hospitals were inaccurately identifying K. pneumonia and led to overestimation of the prevalence of this organism. The current identification methods used in Khartoum hospitals are highly inaccurate, and therefore we recommend the use of a comprehensive biochemical panel or molecular methods, when possible, for accurate identification of K. pneumoniae .
Keywords: Identification methods, Klebsiella pneumoniae, Sudan
Introduction
Klebsiella pneumoniae has been medically recognized as one of the most important opportunistic pathogens, causing worldwide healthcare-associated infections (HAIs) such as pulmonary, urinary tract, blood and soft tissue infections [1]. Moreover, K. pneumoniae has become a clinically important micro-organism, particularly in the last two decades, due to its tendency to develop antibiotic resistance and cause fatal outcomes [2]. It is part of the ESKAPE organism group ( Enterococcus faecium , Staphylococcus aureus , Klebsiella pneumoniae , Acinetobacter baumanii, Pseudomonas aeruginosa and Enterobacter species), which effectively ‘escape’ the effects of antibacterial drugs [3]. The use of limited routine methods (culture and conventional biochemical tests) to isolate pathogenic strains of K. pneumoniae may not be accurate due to the similarity of its biochemical reaction to that of other coliforms, leading to incorrect identification of the organism [4]. Unfortunately, these are the only methods of identification used in the hospital laboratories in Khartoum, Sudan, due to molecular techniques being unavailable as routine methods for identification and the expensive price of analytical profile index (API) kits. The burden of HAIs in general, and K. pneumoniae in particular, is not known in many low- and middle-income countries (LMICs) such as Sudan, due to the lack of adequate diagnostic and research infrastructure. The aim of this study was to determine the accuracy of identification of K. pneumoniae using the limited tests performed at the hospital laboratories.
Methods
Two hundred and fifty isolates identified phenotypically as K. pneumoniae by the hospital laboratories were collected from four different hospitals (Rabat n=98, Souba n=52, Um Durman n=74 and Bahri n=26) in Khartoum state from April 2015 to December 2016. There is no specific algorithm for bacterial identification across most hospitals. Identification of K. pneumoniae from urine and wound swab samples in Khartoum hospitals is based on culture, colony morphology and Gram stain results. Blood and MacConkey agar is used for wound swab cultures, and blood and MacConkey agar or only CLED agar are used for urine samples. Colonies that are mucoid on blood agar, appear as Gram-negative rods under the light microscope after staining, and are lactose-fermenting mucoid colonies in MacConkey’s and CLED agar are identified as K. pneumoniae by the hospital laboratories. In other samples contributing to invasive infections [blood, cerebrospinal fluid (CSF) and pulmonary] a limited number of biochemical tests, such as indole tests, citrate tests, urease tests and Kligler iron agar (KIA) tests, are used subsequent to the identification by colony morphology and Gram staining (Fig. S1, available in the online version of this article). In the current study, API 20E/NE was used as an initial confirmatory test for 79 randomly selected samples . The isolates were cultured in MacConkey agar at 37 °C overnight, and processed as per the manufacturer’s instructions (File. S1). Further identification and confirmation of the all strains was carried out genotypically based on amplification of the 16S−23S rDNA internal transcribed spacer (ITS) of K. pneumoniae as previously described by Yin Liu et al. [5] Briefly, two pairs of K. pneumoniae -specific primers Pf (5ʹ-ATT TGA AGA GGT TGC AAA CGA T-3ʹ)/Pr1(5ʹ-TTC ACT CTG AAG TTT TCT TGT GTT C-3ʹ) and Pf/Pr2(5ʹ-CCG AAG ATG TTT CAC TTC TGA TT-3ʹ)5 were used for the PCR and analysed by gel electrophoresis. The PCR conditions were as follows: 1 µl template DNA (~10 ng, extracted using the Guanidine method) was amplified in a 25 µl containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1 mM each of the four dNTPs, 1 unit Taq DNA polymerase and 1 µM of each primer. The cycling conditions were 10 min at 94 °C followed by 35 cycles of 30 s at 94 °C, 20 s at 57 °C and 20 s at 72 °C, followed by a 10 min hold at 72 °C. The K. pneumoniae isolates produced a 260 bp product with the Pf/Pr2 primer pair, in addition to a 130 bp product with the Pf/Pr1 primer pair. Other Klebsiella spp. (not pneumoniae) produced an amplicon with one primer pair but not with the other. Following identification with the 16S−23S rDNA ITS method, all K. pneumoniae isolates were confirmed by amplification and sequencing of the rpoB (beta-subunit of RNA polymerase), gapA (glyceraldehyde 3-phosphate dehydrogenase) and pgi (phosphoglucose isomerase), all of which are housekeeping genes in the K. pneumoniae genome [6]. The PCR conditions were similar to those used in the 16S−23S rDNA ITS methods, but with annealing temperature of 50 °C. The PCR products of the rpoB, gapA and pgi were purified using the Invitrogen PureLink PCR Purification kit and sequenced. Sequences were analysed using the blast database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm that the sequences were specific to K. pneumoniae . The McNemar test was used to compare the specificity of the identification methods used by hospital laboratories.
Results and Discussion
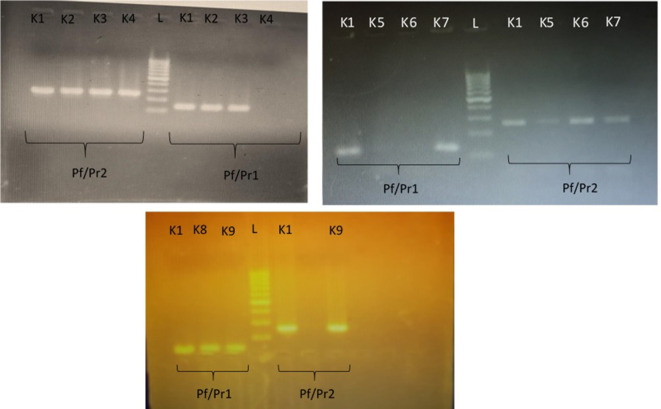
Of the 250 of Gram-negative isolates identified as K. pneumoniae by the clinical laboratory, 139 (55.6 %) were confirmed according to genotypic methods (16S−23S rDNA ITS followed by sequencing of housekeeping genes). The results of API 20E/NE for the 79 tested isolates showed that only 26 isolates were identified as K. pneumoniae (32.9%). The 26 isolates from the API-20NE were further confirmed using the 16S−23S rDNA ITS method and sequencing of the housekeeping genes. K. pneumoniae isolates produced 2=two bands with the primer pairs, as seen in Fig. 1 – a band at 260 bp with the Pf/Pr2 primer pair, in addition to a 130 bp band with the Pf/Pr1 primer pair. The remaining 111 (44.4 %) strains had been wrongly assigned as K. pneumoniae and only produced bands with one but not the other primer pair, as seen in Fig. 1. All 139 K . pneumoniae isolates underwent sequencing of the three housekeeping genes rpoB, gapA and pgi, confirming the accuracy of the 16S−23S rDNA ITS method.
Fig. 1.
PCR amplification of the 16S−23S rDNA ITS of K. pneumoniae . Strain K1 was used as a positive control for all runs. K. pneumoniae isolates (K2, K3, K7 and K9) produced a band at 260 bp with the Pf/Pr2 primer pair, in addition to a 130 bp band with the Pf/Pr1 primer pair. Non- K. pneumoniae isolates (K4, K5 and K6) only produced a 260 bp band with primer pair Pf/Pr2 and failed to produce a band with Pf/Pr1, whereas K8 did not produce a band with Pf/Pr2, but did with Pf/Pr1. L, 100 bp ladder.
Using the McNemar test we found that the specificity of the identification methods used by the hospitals is significantly lower than that of genotypic methods (McNemar chi-squared statistic with Yates correction of 0.5 is 110.002252, P-value is 0.000000).The bacteria studied were isolated from different clinical samples, as shown in Fig. 2 below. Our data show a high rate of misidentification for Gram-negative pathogenic organisms in Khartoum state, which does not give an accurate epidemiological picture, and may be contributing to wrong administration of antibiotics to patients. Identification of K. pneumoniae was only 100 % accurate in cerebrospinal fluid (CSF) samples, as these samples were subjected to different identification methods: culture in different kinds of media and using a panel of biochemical tests (such as citrate tests, urase tests, KIA tests, indole tests, MR tests, VP tests and motility tests). Conversely, accuracy was as low as 43 % for urine samples, where only colony morphology was used as the method of identification. Apart from CSF and nasal swabs, where 100 and 83%, respectively of K. pneumoniae were accurately identified by the laboratories, there was a large percentage of misidentification (39 –57 %) due to the difference in identification methods used for the different clinical samples (invasive vs non-invasive infections) (Table 1).
Fig. 2.
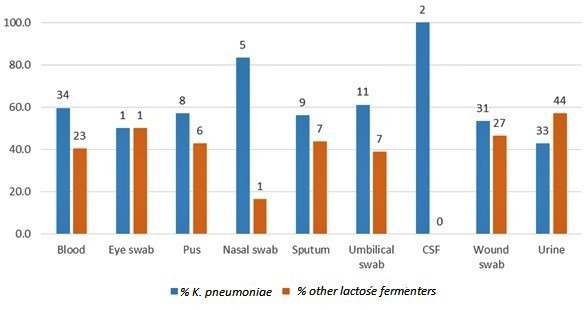
Samples from which K. pneumoniae was isolates. The blue bars indicate the number of isolates identified by the 16S−23S rDNA ITS, whereas the orange bars indicate the number of misidentified organisms.
Table 1.
16S−23S rDNA K. pneumoniae identification versus phenotypic identification
|
Blood |
Eye swab |
Pus |
Nasal swab |
Sputum |
Umbilical swab |
CSF |
Wound swab |
Urine |
|
|---|---|---|---|---|---|---|---|---|---|
|
34 |
1 |
8 |
5 |
9 |
11 |
2 |
31 |
33 |
|
|
Other lactose fermenters |
23 |
1 |
6 |
1 |
7 |
7 |
None |
27 |
44 |
The literature lacks sufficient evidence from Sudan, particularly on the burden of K. pneumoniae in healthcare settings. Hamdan et al. investigated the epidemiology of urinary tract infections from adult diabetic patients in Khartoum [7] and found that K. pneumoniae is the second most predominant cause of infections (after E. coli), causing 23 % of these infections. Here we studied K. pneumoniae isolates collected from various clinical specimens of patients in hospitals, which may explain the high prevalence at 55.6 % of all Gram-negative isolates collected as compared to the study that only studied urinary isolates.
Conventional biochemical tests, if not carefully selected, were shown by several studies to be inadequate in identifying K. pneumoniae [4, 8]. A study by Claus showed that the biochemical tests used to identify bacterial species may not be accurate and need to be tested to see if they are sufficient to discriminate between species [9]. On the other hand, genetic-based identification was shown to be highly accurate. Genes such as 16S−23S rRNA exhibit highly conserved polymorphism within K. pneumoniae clinical isolates, which make them a useful tool for the identification of K. pneumoniae isolates [10, 11]. Furthermore, it has been shown that using the 16S−23S rRNA gene ITS sequences to discriminate Klebsiella species and subspecies was feasible [11]. A study by Ahmed et al. (2015) stressed the need to use molecular biology techniques as essential diagnostic tools in microbiology laboratories [12], and the combination of microbiology and molecular biology led to high sensitivity and specificity. Low contamination levels and high speed have made molecular techniques appealing methods for the diagnosis of many infectious diseases. Molecular methods do, however, require costly equipment and expertise, and may therefore not be available in many LMICs due to lack of available funds.
The microbiological identification methods currently used in hospitals in Khartoum are highly inaccurate and no specific algorithm is used for bacterial identification. To avoid misidentification, we recommend that Khartoum state hospitals review and improve their routine identification methodology for pathogenic organisms to exhibit correct and accurate results. We recommend using molecular identification where possible to obtain the greatest accuracy. However, given the current limitations present in most hospitals (lack of infrastructure, facilities and funding), we would alternatively suggest including an appropriate biochemical testing panel to limit misidentification. The abbreviated algorithm for presumptive identification of Enterobacteriaceae tested by Ng et al. could be a good alternative [13].
Supplementary Data
Funding information
This work was supported by a grant from The Association of Physicians of Great Britain and Ireland.
Author contributions
All authors participated in the design and implementation, analysis and interpretation of the study, and the development of the manuscript. All authors had full access to the data and gave final approval before submission.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This work contains no human or animal data. Institutional approval was obtained from The Sudanese Ministry of Health.
Footnotes
Abbreviations: API, Analytical Profile Index; CSF, Cerebrospinal fluid; ESKAPE, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter species; ITS, Internal Transcribed Spacer; KIA, Kligler Iron Agar; LMIC, Low and Middle Income Countries.
Supplementary material is available with the online version of this article.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 3.Ko W-C, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, et al. Community-Acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen DS, Aucken HM, Abiola T, Podschun R. Recommended test panel for differentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. J Clin Microbiol. 2004;42:3665–3669. doi: 10.1128/JCM.42.8.3665-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ab YL, Wenjie Zheng C, Xia Zhang C. Pcr detection of Klebsiella pneumoniae in infant formula based on 16S–23S internal transcribed spacer. Intl J Food Microbiol. 2008;125:230–235. doi: 10.1016/j.ijfoodmicro.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamdan Z, Kubbara E, Adam AM, Hassan OS, Suliman SO, et al. Urinary tract infection and antimicrobial sensitivity among diabetic patients at Khartoum, Sudan. Ann Clin Microbiol Antimicrob. 2015;78:15–17. doi: 10.1186/s12941-015-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alves MS, Dias RCdaS, de Castro ACD, Riley LW, Moreira BM. Identification of clinical isolates of indole-positive and indole-negative Klebsiella spp. J Clin Microbiol. 2006;44:3640–3646. doi: 10.1128/JCM.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claus H. Optimal selection of biochemical tests to identify microbial species. Zentralblatt für Bakteriologie. 1993;278:522–528. doi: 10.1016/S0934-8840(11)80823-6. [DOI] [PubMed] [Google Scholar]
- 10.Lopes ACS, Rodrigues JF, Clementino MBM, Miranda CAC, Nascimento APA, et al. Application of PCR ribotyping and tDNA-PCR for Klebsiella pneumoniae identification. Mem Inst Oswaldo Cruz. 2007;102:827–832. doi: 10.1590/S0074-02762007005000113. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Cao B, Yu Q, Liu L, Gao Q, et al. Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species. J Clin Microbiol. 2008;46:3555–3563. doi: 10.1128/JCM.00927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed SS, Alp E, Ulu-Kilic A, Doganay M. Establishing molecular microbiology facilities in developing countries. J Infect Public Health. 2015;8:513–525. doi: 10.1016/j.jiph.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 13.Ng LSY, Tan TY, Yeow SCS. A cost-effective method for the presumptive identification of Enterobacteriaceae for diagnostic microbiology laboratories. Pathology. 2010;42:280–283. doi: 10.3109/00313021003631338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.