Abstract
Background
Francisella tularensis is a rare zoonotic bacterium that spreads sporadically by various routes, including infected arthropod bites, ingestion of contaminated water and inhalation of contaminated dust. However, its occurrence in postoperative chest infection has never been reported. Pathogen isolation, serology and molecular detection methods are commonly used for the diagnosis of tularaemia.
Case presentation
We present the first case report of the isolation of F. tularensis from a patient with a chest infection (a boy in his teens) following cardiac surgery for closure of a ventral septal defect. It was isolated on blood and chocolate agar on the third day after the subculture of drain fluid collected in a blood culture bottle incubated in Bact T/Alert 3-D (bioMerieux, France). The organism was identified as F. tularensis by Vitek GN ID Cards (Vitek 2 Compact, bioMerieux, France). The patient made a smooth recovery with antibiotic therapy.
Conclusion
F. tularensis can cause post-operative infection, especially in patients with a rural background.
Keywords: Francisella tularensis, Zoonosis, infection, bacterium
Introduction
Tularaemia is a bacterial zoonotic disease caused by Francisella tularensis , which is a Gram-negative coccobacillus. The organism is thought to be maintained in the environment principally by various terrestrial and aquatic mammals, such as ground squirrels, rabbits, hares, voles, muskrats, water rats and other rodents [1]. It is highly infectious and may be transmitted to humans by a number of different routes. It is commonly spread by handling infected animals, ingestion of contaminated food or water, inhalation of infective aerosols, and bites from ticks and insects [2]. F. tularensis has three biovars: biovar A (biovar tularensis), which is highly virulent and is mostly found in North America and exceptionally in Europe [3]; biovar B, which is not as virulent as biovar A and has been traced in Europe, Asia and (rarely) North America; and biovar novicida, which was identified recently [3, 4].
Very few classical virulence factors have been identified for this pathogen. The bacterial capsule is not required for survival after phagocytosis of bacteria by polymorphonuclear leukocytes [5]. The lipopolysaccharide (LPS) of F. tularensis does not have the properties of a classical endotoxin. Moreover, it does not induce the release of interleukin-1 from mononuclear cells [6]. Phase variation of LPS and lipid A in F. tularensis has been observed. Probing the genome sequence data will help to identify putative virulence factors, such as adhesions [7].
The American Society for Microbiology has recommended biosafety level (BSL) 2 practices for dealing with clinically suspected specimens from patients of F. tularensis [8]. The diagnostic methods that are currently used in the developed world include pathogen isolation, molecular confirmation and serology [9]. Since culture procedures pose a risk to laboratory staff, serological tests can be carried out on nonvaccinated laboratory staff [10]. Antibodies against F. tularensis appear about 2 weeks after the onset of the disease. These can be detected in serum by either agglutination or by enzyme-linked immunosorbent assay (ELISA) [11]. Direct fluorescent antibody (DFA) assays can be carried in a reference laboratory to confirm the identification of the isolate as F. tularensis [12]. PCR-based methods have been evaluated for their potential to identify F. tularensis and to discriminate between its different subspecies. However, its highly infectious nature and the lack of a standardized PCR protocol makes the diagnosis difficult to confirm [13]. Real-time PCR assays have also been developed for the classification of F. tularensis subspecies [9]. Long random-sequence oligonucleotide primers and primers specific for repetitive extragenic palindromic (REP) sequences and enterobacterial repetitive intragenic consensus (ERIC) sequences are also being studied [14].
F. tularensis has been reported in humans and animals in every state of the USA except Hawaii [8]. Little is known about the distribution of F. tularensis in Asia except that it is present in the former USSR, eastern Turkey, China and Japan [15]. It was first reported in India in 2015 in blood cultures of a febrile patient [16]. The first case of tularaemia infection in humans was reported in 1980 in Marivan, in the Kurdistan Province of Iran [15]. However, there is a scarcity of data concerning F. tularensis in Pakistan. The only available study was carried out on F. tularensis -contaminated soil using PCR with the support of Pennsylvania State University, USA [17].
We did not find any report of F. tularensis being identified in clinical specimens from Pakistan [17]. We present the first case report of chest infection due to F. tularensis , which led to pleural and pericardial effusion in a cardiac patient who had been operated on for closure of a ventricular septal defect (VSD).
Patient introduction
A boy in his teens who had been diagnosed with VSD was operated on to effect its closure at Rawalpindi Institute of Cardiology (RIC), Rawalpindi, Pakistan in 2018. The patient was a resident of a rural area of Gujar Khan in Rawalpindi district in the province of Punjab, Pakistan. He had a history of close contact with birds at home, and cattle and farm animals in his neighbourhood. Well water was used for both bathing and drinking. The well was uncovered and did not have a surrounding parapet. The patient was discharged 6 days after surgery. However, the patient presented again to RIC 2 weeks later with fever and fullness in the right side of his chest. The wound was found to be infected with reddish exudate and was debrided. Chest X-rays showed right-sided pleural and pericardial effusion (Fig. 1). The pleural fluid was drained and found to be haemorrhagic, and its culture sensitivity was advised on. Laboratory investigations revealed a white blood cell count (WBC) of 15×109 l−1 with 75 % polymorphs, a haemoglobin (Hb) level of 9.1 g dl−1 and a platelet level of 179×109 l−1. The C-reactive protein (CRP) level was 36 g dl−1 and the erythrocyte sedimentation rate (ESR) 46 mm fell at the end of the first hour. The patient’s liver, renal and coagulation profiles were within normal limits. The serological test for Brucella antibodies was negative. A culture swab taken from the operation site did not yield any growth. The patient was treated with injections of ceftiaxone (1 g twice daily). However, the fever did not settle even after 5 days, although the chest wound was healing and granulating and the blood cultures were negative to date.
Fig. 1.
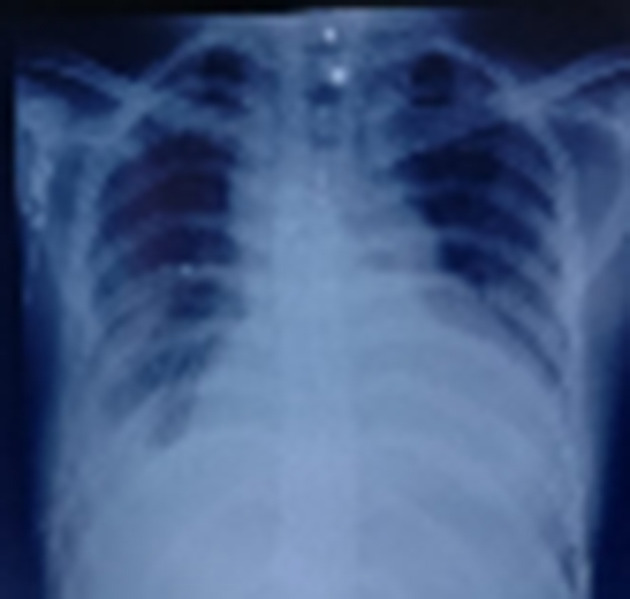
Chest X-ray showing right-sided pleural effusion with a chest tube and globular cardiac silhouette due to pericardial effusion.
Methods
The drain fluid from the right-sided pleural space was inoculated in Bact T/Alert 3-D (bioMerieux, France). After 4 days of incubation at 37 °C, the system showed a positive signal. Fluid was sub-cultured on blood and chocolate agar, and MacConkey agar. Biochemical tests such as a catalase test, an oxidase test, growth on urea and motility were performed using standard microbiological methods [18].
The saline suspension of the isolate was prepared to obtain a microbial concentration of 108 colony-forming units (c.f.u.) ml−1. The bacterial suspension was inoculated in Vitek GN ID Cards and was identified by the Vitek-2 Compact (bioMerieux, France) [19, 20].
The isolate and its subsequent subcultures were identified using Vitek GN ID Cards in the Vitek-2 Compact (bioMerieux, France). The antibiotic susceptibility of the isolate was determined as recommended in the Clinical and Laboratory Standards Institute (CLSI) guidelines (M45A2) [21].
Results
After 48 h of incubation at 37 °C, the subcultures showed a lawn of small non-haemolytic greyish white colonies with more than 105 c.f.u. ml−1 on the blood and chocolate agar (Fig. 2). However, no growth was found on the MacConkey agar plate.
Fig. 2.
Growth of F. tularensis on blood and chocolate agar.
Gram staining of the colonies showed small, faintly staining, Gram-negative cocobacilli (Fig. 3). The biochemical characteristics of the culture isolate are shown (Table 1). The organism was identified by Vitek 2 Compact as F. tularensis (Figs 4 and 5). It was susceptible to gentamycin, tobramycin, tetracycline and ciprofloxacin. Intravenous treatment with tobramycin was initiated and the patient became afebrile in the next 24 h. The patient improved dramatically after receiving antibiotics.
Fig. 3.
Gram staining of F. tularensis .
Table 1.
Biochemical characteristics of the culture isolate
|
Gram stain |
Catalase test |
Oxidase test |
Glucose |
Growth in urea |
Motility |
|---|---|---|---|---|---|
|
Weakly staining Gram-negative cocobacilli |
Negative |
Negative |
Fermented |
Negative |
Negative |
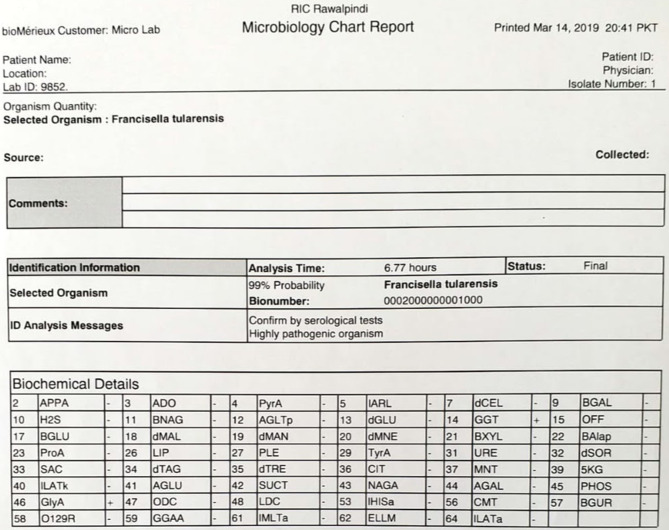
Fig. 4.
Vitek 2 Compact identification print-out from 14 March 2019.
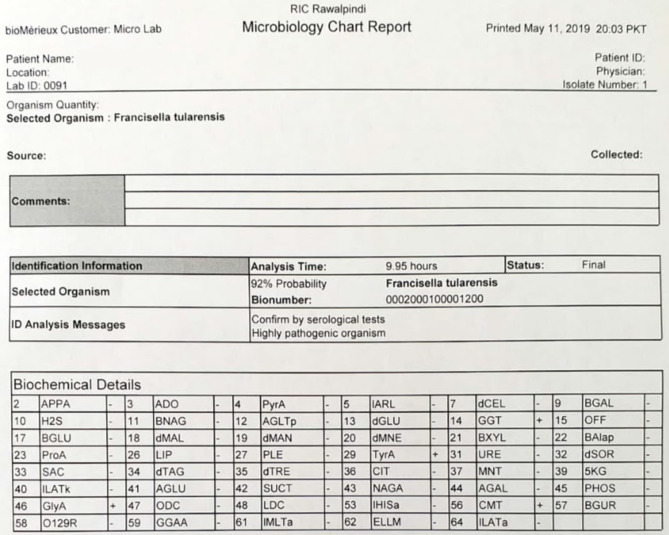
Fig. 5.
Vitek 2 Compact identification print-out from 11 May 2019.
Outcome
The patient improved clinically. There was resolution of the chest infection and pleural effusion in the follow-up chest X-rays (Fig. 6). The other haematological and biochemical profiles also became normal. The chest tube was removed as the drain became dry. The patient was discharged in good condition after 10 days of treatment.
Fig. 6.
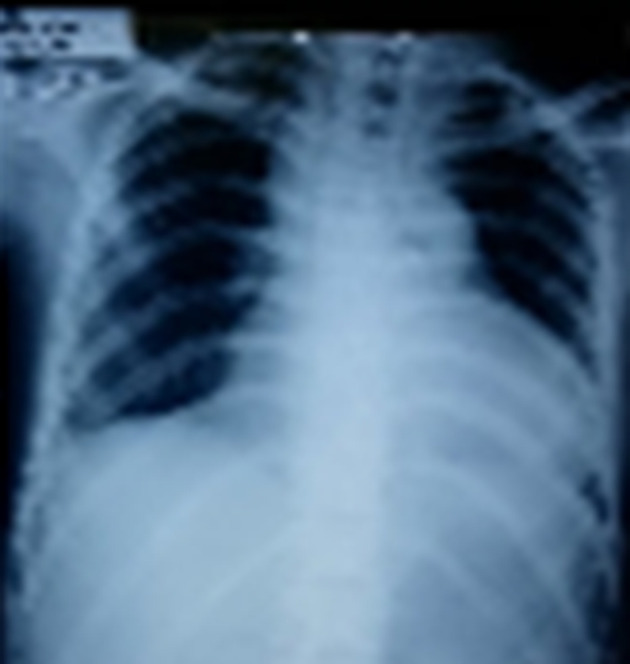
Chest X-rays showing resolution of pleural effusion.
Discussion
Most infections with F. tularensis have been reported from North America, Europe and northern Asia [8–12]. We report a novel and contemporary case of a post-operative chest infection with F. tularensis in a boy operated on for treatment of VSD. To the best of our knowledge, this is the first report of F. tularensis isolation in Pakistan and the first case of post-cardiac surgery chest infection due to the organism worldwide.
F. tularensis is a Gram-negative coccobacillus that causes tularaemia, a zoonotic disease primarily observed in the Northern Hemisphere. It is acquired by direct contact with infected animals or following the ingestion of contaminated water and food or the inhalation of contaminated aerosols [2]. The clinical presentation of the disease depends on the route of transmission. Symptoms develop within 3–5 days of infection and include fever with chills, sore throat, headache and generalized body aches. The clinical significance of tularaemia is unknown, but it can be fatal in patients with other comorbid illnesses, such as diabetes mellitus and pneumonia [9]. Our patient was operated on for VSD closure but developed a post-operative chest infection with right-sided pleural effusion 2 weeks after discharge from the hospital (Fig. 1). He had a history of consumption of open well water for bathing purposes only. Cattle and farm animals in his neighborhood with associated tick, deer fly or mosquito bites are another possible route of transmission. The source of infection in our patient is probably environmental in origin.
The first case of F. tularensis from India was reported in a febrile patient with positive blood culture that was similarly identified by Vitek-2 [16]. A rare case report of pneumonia due to F. tularensis was reported from Turkey in an elderly lady with a history of contact with infected animals [22]. There were also both birds and farm animals in our patient’s neighborhood.
The organism is rarely isolated from blood culture directly, but has been identified with the development of sensitive blood culture systems [23]. Our patient also had bloody pleural fluid that yielded greyish white non-haemolytic colonies on blood and chocolate agar, with no growth on MacConkey agar. These were negative for oxidase, urease, motility and fermentation of lactose and other sugars. Similar findings have been reported in a study from Sudan, where F. tularensis was isolated from blood culture in brain heart infusion [24]. A similar case of F. tularensis isolation has been reported from haemorrhagic pleural effusion and blood specimen in a middle aged man from Norway with sepsis [25].
F. tularensis is known as a potential bioterrorism agent due to its high virulence and low infective dose [23]. Tularaemia is the most commonly reported laboratory-acquired bacterial infection. Culture techniques are usually avoided for diagnosis because of the high virulence of F. tularensis infections, especially in areas where biovar A is highly prevalent [9].
F. tularensis is a fastidious aerobic organism that is usually slow to grow and difficult to culture due to its requirement for cysteine for enhanced growth [23]. In our patient the drain fluid was inoculated in blood culture bottles and incubated in Bact T/Alert 3-D (bioMerieux, France) that showed signal positivity on the fourth day. The fluid from signal-positive culture bottles showed minimal growth of small, non-haemolytic, greyish white colonies on subculture after 48 h on chocolate and blood agar (Fig. 2). Gram staining of the colonies showed small, faintly staining, Gram-negative cocobacilli. (Fig. 3). F. tularensis is relatively inert biochemically, with only a few sugars (glucose, maltose, sucrose and glycerol) being utilized. F. tularensis does not reduce nitrate. It is oxidase- and urease-negative and is weakly catalase-positive [15, 18].
The phenotypic and biochemical characteristics of our patient’s isolate were characteristic for F. tularensis (Table 1). The isolate and its subsequent subcultures were identified by Vitek-2 as F. tularensis (Figs 4 and 5).
Serological-based tests can be used in setups that lack the recommended biosafety level facilities for bacterial culture. The detection of serum antibodies is most frequently achieved by agglutination or by ELISA. An immunochromatographic handheld assay can detect polyclonal and a monoclonal antibody to the LPS of F. tularensis [23]. However, these tests are not readily available in our country.
Fluorescence-labelled antibodies can be used to detect F. tularensis in clinical specimens. PCR-based assays can be used to identify F. tularensis DNA in human skin lesions or in the blood of infected mice [12]. These procedures permit a more rapid diagnosis, but are usually performed in reference laboratories. Only one PCR-based study on F. tularensis -contaminated soil has been reported from Pakistan, and no study on human infection has so far been reported [17]. Long random-sequence oligonucleotide primers and primers specific for REP and ERIC sequences are also being evaluated in the developed world [23].
Thus, in most clinical laboratories a diagnosis of tularaemia relies on serological tests, except in patients suspected of tularaemia with possible bacteraemia, where blood cultures can yield F. tularensis [5, 6]. In our country, no study has been carried out to isolate F. tularensis from clinical specimens [17]. This is either due to the rarity of the disease or because there is often a lack of diagnostic facilities in our country. Confirmation of the isolate with antigen detection or molecular methods could not be performed due to the lack of these facilities in our country and due to failure to transport it, since the organism is a bioterrorism agent.
Conclusion
We report the first case of F. tularensis isolated from the haemorrhagic pleural fluid of a post-cardiac surgery patient from a rural background in Pakistan. The specimens in clinically suspected cases can yield F. tularensis on enriched media on subculture from infected fluid. Moreover, there is a dire need for diagnostic facilities to identify the pathogen in our country for early detection and treatment of clinically suspected patients. Until that time vigilance is required by clinicians and microbiologists to identify suspected cases.
Data bibliography
Funding information
The authors received no specific grant from any funding agency.
Author contributions
Dr Arif Maqsood Ali:Conception and design, Drafting the article or revising itcritically, Final approval; Muhammad Noor ul Amin: Conception anddesign, Drafting the article or revising it critically, Final approval; Dr Shazia Arif: Conception and design, Drafting thearticle or revising it critically, Final approval.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Written consent was obtained from the patient for publishing the data.
Footnotes
Abbreviations: BSL, biosafety level; CRP C, reactive protien; DFA, direct fluorescent antibody; ELISA, enzyme linked immunosorbant assay; ERIC, enterobacterial repetitive intragenic consensus; ESR, erythrocyte sedimentation rate; F. tularensis , Francisella tularensis ; Hb, haemoglobin; LPS, lipopolysacharide; REP, repetitive extragenic palindromic; RIC, Rawalpindi Institute of Cardiology; VSD, ventricular septal defect; WBC, white blood cell count.
References
- 1.Oyston PC, Sjöstedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis . Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 2.Kılıç S. A general overview of Francisella tularensis and the epidemiology of tularemia in Turkey. Flora. 2010;15:37–58. [Google Scholar]
- 3.Oyston PC. Francisella tularensis: unravelling the secrets of an intracellular pathogen. J Med Microbiol. 2008;57:921–930. doi: 10.1099/jmm.0.2008/000653-0. [DOI] [PubMed] [Google Scholar]
- 4.McCoy GW, Chapin CW. Bacterium tularense, the cause of a plague-like disease of rodents. Public Health Bull. 1912;53:17–23. [Google Scholar]
- 5.Sandström G, Löfgren S, Tärnvik A. A capsule-deficient mutant of Francisella tularensis LVS exhibits enhanced sensitivity to killing by serum but diminished sensitivity to killing by polymorphonuclear leukocytes. Infect Immun. 1988;56:1194–1202. doi: 10.1128/iai.56.5.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandström G, Sjöstedt A, Johansson T, Kuoppa K, Williams JC. Immunogenicity and toxicity of lipopolysaccharide from Francisella tularensis LVS. FEMS Microbiol Immunol. 1992;5:201–210. doi: 10.1111/j.1574-6968.1992.tb05902.x. [DOI] [PubMed] [Google Scholar]
- 7.Cowley SC, Myltseva SV, Nano FE. Phase variation in Francisella tularensis affecting intracellular growth, lipopolysaccharide antigenicity and nitric oxide production. Mol Microbiol. 1996;20:867–874. doi: 10.1111/j.1365-2958.1996.tb02524.x. [DOI] [PubMed] [Google Scholar]
- 8.Stidham RA, Freeman DB, von Tersch RL, Sullivan PJ, Tostenson SD. Epidemiological review of Francisella tularensis: a case study in the complications of dual diagnoses. PLoS Currents. 2018;10 doi: 10.1371/currents.outbreaks.8eb0b55f377abc2d250314bbb8fc9d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization Epidemic and Pandemic Alert and Response . WHO Guidelines on Tularemia. Geneva, Switzerland: World Health Organization; 2007. [Google Scholar]
- 10.Burke DS. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J Infect Dis. 1977;135:55–60. doi: 10.1093/infdis/135.1.55. [DOI] [PubMed] [Google Scholar]
- 11.Koskela P, Salminen A. Humoral immunity against Francisella tularensis after natural infection. J Clin Microbiol. 1985;22:973–979. doi: 10.1128/jcm.22.6.973-979.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morse S, Henkel R. Francisella tularensis: understanding reported occupational exposures and laboratory methods used for the identification of Francisella tularensis . Applied Biosafety. 2018;23:11–18. doi: 10.1177/1535676017746695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma N, Hotta A, Yamamoto Y, Fujita O, Uda A, et al. Detection of Francisella tularensis-specific antibodies in patients with tularemia by a novel competitive enzyme-linked immunosorbent assay. Clin Vaccine Immunol. 2013;20:9–16. doi: 10.1128/CVI.00516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Puente-Redondo VA, del Blanco NG, Gutiérrez-Martín CB, García-Peña FJ, Rodríguez Ferri EF, Ferri ER. Comparison of different PCR approaches for typing of Francisella tularensis strains. J Clin Microbiol. 2000;38:1016–1022. doi: 10.1128/jcm.38.3.1016-1022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arata A, Chamsa H, Farhang-Azad A, Mescerjakova O, Neronov V, et al. First detection of tularaemia in domestic and wild mammals in Iran. Bull World Health Organ. 1973;49:597. [PMC free article] [PubMed] [Google Scholar]
- 16.Nirkhiwale SD, Gehlot GS, Bandi AK, Jasani AN. Isolation of Francisella tularensis from blood culture. Indian J Med Microbiol. 2015;33:329. doi: 10.4103/0255-0857.154901. [DOI] [PubMed] [Google Scholar]
- 17.Winn WC. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. Lippincott Williams & Wilkins; 2017. [Google Scholar]
- 18.Pincus DH. Encyclopedia of Rapid Microbiological Methods. Bethesda, MD: Parenteral Drug Association; 2006. Microbial identification using the bioMérieux Vitek® 2 system. [Google Scholar]
- 19.Puttaswamy S, Gupta SK, Regunath H, Smith LP, Sengupta S. A comprehensive review of the present and future antibiotic susceptibility Testing (AST) systems. Arch Clin Microbiol. 2018;09 doi: 10.4172/1989-8436.100083. [DOI] [Google Scholar]
- 20.Wayne PA. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. approved guideline—. Document M45-A2. 2010. [Google Scholar]
- 21.Agca M, Duman D, Sulu E, Ozbaki F, Barkay O, et al. Rare cause of Pleuropnemonia: tularemia disease. J Coll Physicians Surg Pak. 2017;27:574–576. [PubMed] [Google Scholar]
- 22.Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohamed SE, Mubarak AI, Alfarooq LO. Francisella tularensis bacteremia: a case report from Sudan. Case Rep Infect Dis. 2012;2012:1–2. doi: 10.1155/2012/405737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haristoy X, Lozniewski A, Tram C, Simeon D, Bevanger L, et al. Francisella tularensis bacteremia. J Clin Microbiol. 2003;41:2774–2776. doi: 10.1128/JCM.41.6.2774-2776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muhammad J, Rabbani M, Shabbir MZ, Muhammad K, Ghori MT, et al. Cross sectional study and risk factors analysis of Francisella tularensis in soil samples in Punjab Province of Pakistan. Front Cell Infect Microbiol. 2019;9:89. doi: 10.3389/fcimb.2019.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]