Abstract
Coordinated regulation of ribosomal RNA (rRNA) synthesis and ribosomal protein gene (RPG) transcription by eukaryotic RNA polymerases (RNAP) is a key requirement for growth control. Although evidence for balance between RNPI-dependent 35S rRNA production and RNAPII-mediated RPG transcription have been described, the molecular basis is still obscure. Here, we found that Rph1 modulates the transcription status of both rRNAs and RPGs in yeast. We show that Rph1 widely associates with RNAPI and RNAPII-transcribed genes. Deletion of RPH1 remarkably alleviates cell slow growth caused by TORC1 inhibition via derepression of rRNA and RPG transcription under nutrient stress conditions. Mechanistically, Rim15 kinase phosphorylates Rph1 upon rapamycin treatment. Phosphorylation-mimetic mutant of Rph1 exhibited more resistance to rapamycin treatment, decreased association with ribosome-related genes, and faster cell growth compared to the wild-type, indicating that Rph1 dissociation from chromatin ensures cell survival upon nutrient stress. Our results uncover the role of Rph1 in coordination of RNA polymerases-mediated transcription to control cell growth under nutrient stress conditions.
INTRODUCTION
Ribosomes provide the basis for cell mass accumulation and protein production, which drives cell growth. Ribosome biogenesis is an intricate process that requires coordinated and balanced production of processed rRNAs, ribosomal proteins (RPs) and ribosome biogenesis-related (Ribi) factors. In rapidly growing yeast cells around 60% of total cellular RNAs including rRNAs are transcribed by RNAPI, and nearly 50% of all RNAPII initiation events occur on ribosomal protein genes (RPGs) (1). Tightly coordination of transcription of rRNAs and RPGs is important for regulation of ribosome biogenesis and cell growth in optimal growth condition as so in response to nutrient stress conditions (2). To ensure efficient ribosome biogenesis, an equimolar production of the different ribosomal components is necessary. The imbalanced production of rRNAs and RPs would lead to impairment of cell growth and G1 cell-cycle arrest in many eukaryotes (3).
The growth-promoting TORC1 (target of rapamycin 1) kinase is a central regulator of both rRNAs and RPGs at the transcriptional level across many species. Under optimal growth conditions, in the yeast Saccharomyces cerevisiae, TORC1 is active and simultaneously stimulates both rRNA and RPG transcription (2). TORC1 inactivation with rapamycin leads to a rapid repression of transcription of both rRNA and RPG to protect cells against proteotoxic stress (4).
Several factors in yeast have been implicated in coordinating the transcription of RPGs and rRNAs, which was initially suggested by Warner group (5). Previous studies found that the essential protein Ifh1 is recruited by a constitutively promoter-bound partner protein Fhl1 to many RPG promoters and activates their expression (6–8). However, they noticed that Ifh1 was concomitantly localized in the nucleolus in a CURI complex formed by CK2 and rRNA processing factors Utp22 and Rrp7, suggesting its coupling role in the production of rRNAs and RPGs (5). Recently, Utp22 was identified to control Ifh1 binding affinity to RPG promoters, thereby coordinating transcription of RPGs and that of rRNAs (9). In addition, Hmo1, was reported to be present in both the rDNA transcription unit and some RPG promoters and involved in TORC1-dependent coregulation of rRNA and RPG transcription (2,10–11). In higher eukaryotes, a line of studies also demonstrated coordinated transcriptional regulation of ribosome components. For example, the drosophila transcriptional factor TIF-IA, a homolog of yeast Rrn3 that is capable of activation of RNAPI, can stimulate rRNA synthesis and feed-forward couple RPG transcription (12). Interesting, the circadian clock controls transcription factors and clock-dependent rhythmic activation of signaling pathways to regulate transcription of RPGs and rRNAs (13). In human cells, the DDX21 RNA helicase could coordinate transcription of rRNAs and RPGs, as well as small nucleolar RNAs (snoRNA) (14). Although many evidence prove that co-activation of RPG and rRNA transcription is exerted under optimal growth conditions, little is known whether repressive factors exist and how they coregulate transcription of those genes to protect extravagant growth.
Rph1 has been characterized as a demethylase that removes histone H3 Lys36 (H3K36) di- and trimethylation in budding yeast (15–17). Rph1 contains a JmjN domain in the N-terminus, which is required for its demethylase activity and a zinc finger (ZF) domain in the C-terminus, which is responsible for DNA binding (15). Several studies including ours demonstrated Rph1 as a negative regulator to control expression of a subset of genes (related to DNA damage, oxidative stress or autophagy) under optimal conditions, whereas dissociation of Rph1 from chromatin increased expression of those aforementioned genes upon DNA damage stress or nitrogen starvation (18–20). Of note, we observed that overexpression of Rph1 retards cell growth, but the molecular mechanism is unclear.
In this work, using unbiased genome-wide analysis we uncovered Rph1 largely occupied the transcribed regions of rDNAs and almost all of RPGs as well as most regions for transcription of snoRNAs. Increased amounts of Rph1 led to decreased expression of ribosomal genes, thereby inhibiting cell growth. In contrast, deletion of RPH1 remarkably increased RNAPII occupancy on RPGs and Ribi genes to facilitate cell growth under rapamycin treatment. We further showed that a derepressive role of Rph1 on gene transcription depends on Rim15-mediated phosphorylation. Inhibition of TOR by rapamycin triggers Rph1 hyperphosphorylation and releases it from transcriptional regions of rDNA and RPG, which ensures cell survival under such harsh conditions. Our data discover a repressor that dynamically regulate transcription of ribosomal genes to control cell growth.
MATERIALS AND METHODS
Yeast strains and media
All yeast strains used in this study are listed in the Supplementary Table S1. Gene disruption and integrated tagging were performed using standard polymerase chain reaction (PCR)-based strategy as described previously (21). The Tet-tor2 strain was obtained from Yeast TET Promoters Hughes (yTHC) kit, and TOR2 gene was silenced by addition of doxycycline (TargetMol, Cat# T1140). The generated strains were verified by PCR and western blot analysis. Media used in this study: YPD medium (1% yeast extract, 2% peptone and 2% dextrose), or SC dropout medium (0.67% yeast nitrogen base without amino acids, supplemented with amino acids and 2% glucose) with appropriate supplements.
Cell cultures and drug treatment
Yeast cells were grown to OD600 0.6–0.8 at 30°C in YPD, synthetic complete (SC) or synthetic minimal (SD) medium with necessary additives unless otherwise indicated. Yeast cells were treated with 100 nM rapamycin for 2 h unless otherwise indicated. Cells were serially diluted as indicated and then spotted onto YPD, SC or SD plates with indicated drugs, respectively. Plates were incubated at 30°C and imaged at 2–5 days.
Whole yeast cell extracts preparation, western blot and antibodies
NaOH-based protocols were used to lyse yeast cells for western blots as described before (21). Briefly, 5-ml mid-log cultures of yeast were quenched in 250 μl of 2 M NaOH with 8% mercaptoethanol and incubated on ice for 5 min. Cell pellets were resuspended gently 250 μl of Buffer A (40 mM HEPES-KOH, pH 7.5, 350 mM NaCl, 0.1% Tween 20, 10% glycerol, 1 × protease inhibitors cocktails) and pelleted. Appropriate 2 × SDS-sample buffers were added into different samples according to the weight of cell pellets and resuspended. After boiling, the same amount of proteins was separated using sodium dodecyl sulphate-polyacrylamide gelelectrophoresis (SDS-PAGE) gels. Western blot analyses were performed as described previously (21). The following antibodies were used: Primary: α-Rph1 (home made, 1:5000), α-G6PDH (Sigma A9521 1:20,000), α-H3K36me3 (Abclonal A2366, 1:3000), α-H3 (Active Motif 39164, 1:10,000), α-Rps6 (Abclonal A6058, 1:1000), α-phosph-Rps6 S235/236 (Abclonal AP0538 1:1000), α-Flag (MBL PM020, 1:5000), α-GFP (Sungen KM8009, 1:3000); Secondary: α-rabbit IgG/HRP (Jackson ImmunoResearch, 715–035-150, 1:10,000), α-mouse IgG/HRP (Jackson ImmunoResearch 115–035-003,1:10,000).
Phos-tag western blots
The phos-tag gels were prepared followed as the product manual (APExBIO F4002) for protein phosphorylation analysis. Briefly, 6% (w/v) acrylamide SDS-PAGE gel containing 10–20 μM of PBR-A (Phos Binding Reagent Acrylamide) was made. Cell extracts were prepared in 2 × SDS-sample buffers.
RNA extraction and real-time quantitative PCR (RT-qPCR) analysis
A total of 30 ml culture of either wild-type (WT) or rph1Δ yeast cells were grown in YPD to OD600 = 0.8 and then treated with rapamycin (100 nM, Sangon Biotech A606203) or dimethyl sulfoxide (DMSO) for 2 h. Yeast cells were collected by spinning down with 3000 rpm for 5 min, then cell pellets were washed with 10-ml sterile water twice. Yeast cell samples were resuspended with 0.5 ml TRIzol (Invitrogen) and snap-frozen in liquid nitrogen and stored at −80°C freezer < 1 week. Total RNAs were extracted using TRIzol and further purified via chloroform extraction methods. The purity and yield of RNAs were examined by measuring the A260/A280 ratio using NanoDrop One Sepctrometer (Gene Company). RNA integrity was examined by agarose electrophoresis. After drying, an aliquot of 1 μg total RNA from each sample was subjected to reverse transcription using the M-MLV Reverse Transcriptase (ThermoFisher Scientific 28025021) according to the manufacturer's protocol. NCBI Primer-BLAST online tool was used to search a specific primer set with the amplification product length of 100–200 bp. qPCR experiments were carried out as the manufacturer's protocol suggested except each reaction volume with 10 μl (Yeasen 11201ES08). The Cq values of all amplification curves were less than 28 and the melt curves of each primer set only contain a single peak. Three technical replicates data were analyzed using Bio-Rad CFX Manager software (version 3.1) with normalized mode (ΔΔCq). qPCR primer sequences were listed in Supplementary Table S2.
RNA-seq analysis
RNA-seq libraries were prepared by Berry Genomics Company (Beijing) and paired end sequencing was performed on Illumina NovaSeq 6000. FastQC was used to do quality control on raw sequence data. Low quality reads and adaptor contamination were removed by Trimmomatic. The filtered reads were mapped to SacCer3 using STAR v2.5.3a and quantified using featureCounts v1.6.3. Reads count normalization and differential gene expression analysis was performed using DESeq2 v1.24.0. Gene ontology (GO) was performed by DAVID v6.8. R version 3.6.0 was used for some custom analysis. For Heatmap, we use R package edgeR to calculate counts per million (CPM) of each gene, then log(CPM) value was used to make heatmap by using R package ComplexHeatmap. The ggplot2 package was used to plot a boxplot of gene expression.
Chromatin immunoprecipitation (ChIP) and ChIP-seq analyses
Yeast cells were grown in YPD to OD600 = 0.8 and then treated with rapamycin (100 nM) or dimethyl sulfoxide (DMSO) for 2 h before harvest. Cells were crosslinked with 1% formaldehyde for 15 min at room temperature and then quenched with glycine for 5 min. Cell pellets were washed twice with sterile cold water, then snap-frozen in liquid nitrogen and stored at −80°C until use. Frozen pellets were lysed in 0.3 ml ChIP lysis (140 mM NaCl) buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 0.1% deoxycholate, 1 mM PMSF and protease inhibitor cocktail from Bimake Company, Cat# B14002). After beads beating, chromatin was sonicated for 20–25 cycles (high output, 30 s on, 30 s off) using the Bioruptor (Diagenode) to produce fragments of ∼300 bp. Cell lysates was centrifuged at 16,000 × g for 15 min at 4°C to pellet debris. Immunoprecipitation was performed overnight at 4°C with 10 μl Flag M2 resin (Sigma A2220). For RNAPII ChIP assay, Immunoprecipitation was performed overnight at 4°C with 10 μg RNAPII antibody (Santa Cruz, SC-56767), then 10 μl Protein G beads were added and incubated for an additional 2 h. Beads were then washed once with ChIP lysis (140 mM NaCl) buffer for 10 min, once with ChIP lysis (500 mM NaCl) buffer (50 mM HEPES-KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 1 mM PMSF) for 20 min and once with LiCl/NP40 buffer (10 mM Tris–HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na Deoxycholate, 1 mM EDTA) for 10 min. Elution was performed with two consecutive 15 min incubations with 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature. After reverse crosslinking and proteinase K digestion, DNA was then column purified using the Universal DNA Purification Kit (TIANGEN). DNA Concentration was quantified using Equalbit dsDNA HS Assay Kit (Vazyme EQ111).
The ChIP assays were performed as described previously using 10 μl IgG Sepharose beads (GE Healthcare 17–0618-05) or 10 μl Flag M2 resin for each sample. Quantitative PCR (Yeasen 11201ES08) was performed as above, and primer sets are listed in the Supplementary Table S2. The ChIP-seq libraries were prepared using VAHTS™ Universal DNA Library Prep Kit for Illumina® V3 (Vazyme ND607). The quality and quantity of constructed libraries were determined using Fragment Analyzer (Advanced Analytical). Paired end sequencing was performed on Illumina NovaSeq 6000.
FastQC v0.11.8 was used to do quality control on raw sequence data. Low quality reads and adaptor contamination were removed by Trimmomatic v0.38. The filtered reads were aligned against SacCer3 genome using Bowtie2 v2.3.4.3 with default parameters. The mapped reads was then converted to BigWig files and visualized using the IGV.
MACS2 v2.2.1 with a P-value threshold of 1e − 5 was used to call ChIP peaks over ‘Input’ background. We used bedtools v2.27.1 to get ChIP peaks related genes. GO was performed by DAVID v6.8 online server. The line plot of ChIP-Seq data was performed by ngsplot v2.63.
Quantification and statistical analysis
Unless otherwise stated, western blot data were quantified using ImageJ software to measure the relative intensity of each band. All quantification data were presented as the mean ± SD (standard deviation) from at least three independent experiments. Statistical differences were determined by two-tailed unpaired t-test, and a P-value of less than 0.05, 0.01, 0.001 or 0.0001 was considered statistically significant and marked as ‘*’, ‘**’, ‘***’, ‘****’, respectively. ‘n.s.’ indicates ‘not significant’.
RESULTS
Cells lacking Rph1 are resistant to the nutrient stress
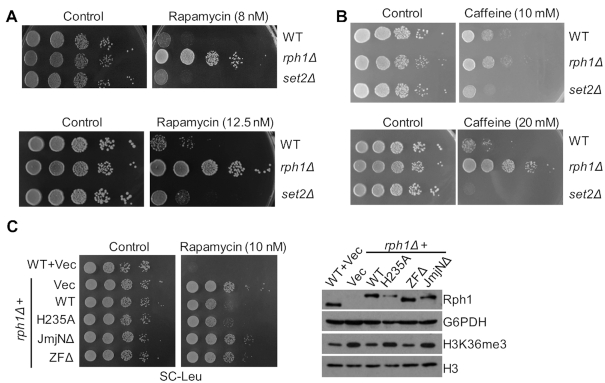
Previous studies have indicated that Rph1 is involved in nitrogen starvation-stressed autophagy process (19). We wondered if Rph1 might be also important for the regulation of nutrient stress pathway. To test this, the growth rates of various cells treated with rapamycin (a potent and specific TORC1 inhibitor) or caffeine (a drug inhibits Tor1, Tor2 and MAP kinases) were monitored. Since Set2 is the histone H3K36 methyltransferase and it has an opposite biochemical function of the H3K36 demethylase Rph1, we chose set2Δ strain as a control (22). Consistent with previous results (23), set2Δ cells are sensitive to rapamycin or caffeine compared to the WT cells (Figure 1A and B). In contrast, rph1Δ cells from two different genetic backgrounds showed much faster growth phenotypes under various doses of drug treatment (Figure 1A and B; Supplementary Figure S1A). Of note, rph1Δ cells but not WT cells can survive under a high concentration of rapamycin treatment (1 μM), indicating a repressive role of Rph1 in nutrient-stress response pathway (Supplementary Figure S1B).
Figure 1.
Cells lacking Rph1 are resistant to nutrient stress. (A andB) The indicated strains were spotted on YPD plates with or without indicated doses of rapamycin or caffeine. (C) WT strain with empty vector (WT + Vec) or rph1Δ strain expressing endogenous promoter-driven WT RPH1 allele (rph1Δ + WT) or various mutation alleles were plated with or without rapamycin (left panel). The expression levels of the indicated strains were examined by immunoblotting with an anti-Rph1 antibody (right panel).
To confirm this, either WT cells expressing empty vector or rph1Δ cells expressing various Rph1 constructs under the control of an endogenous promoter were treated with rapamycin, and their sensitivities toward rapamycin were examined. As expected, rph1Δ cells expressing the WT construct, but not a construct lacking JmjN domain (JmjNΔ) of Rph1 showed a decreased H3K36me3 level (15) and a slow-growth rate even under a low concentration of rapamycin treatment (Figure 1C and Supplementary Figure S1C). Interestingly, unlike expressing the WT Rph1 allele, rph1Δ cells expressing a construct lacking zinc figure domain (ZFΔ) exhibited a demethylase activity toward H3K36me3 (15) but failed to retard cell growth (Figure 1C). Surprisingly, cells overexpressing a catalytic-dead mutant of Rph1 (H235A) showed no change of H3K36me3 but a similar slow-growth phenotype relative to that of WT cells, suggesting that its role is independent of the demethylase activity (Supplementary Figure S1C). Moreover, overexpression of Gis1, a paralog of Rph1 in budding yeast, also did not retard cell growth (Supplementary Figure S1C). Since the effect of Rph1 on cell growth is independent of its demethylase activity and irrelevant to Gis1, we will only focus on Rph1 itself in response to nutrient stress in the following experiments.
RPH1 genetically interacts with TORC1 signaling pathway
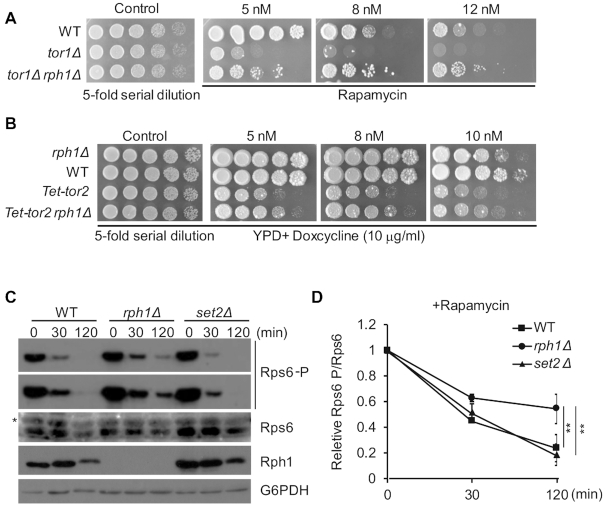
Rapamycin directly inhibits the Target of Rapamycin Complex 1 (TORC1), and Rph1 was identified as an effector of TORC1 (24,25). To test whether RPH1 genetically interacts with the TORC1 pathway, we generated rph1Δ with either a deletion of TOR1 or Tet-promoter driven TOR2 gene (Tet-tor2, TOR2 is essential in budding yeast), and plated these strains on rapamycin. Expectedly, deletion of TOR1 alone or inhibition of TOR2 expression by addition of doxycycline exhibited a synthetic growth defect in the presence of rapamycin. Conversely, double-mutant strains bearing rph1Δtor1Δ or rph1ΔTet-tor2 partially rescued the slow-growth phenotype in the presence of the same concentration of rapamycin (Figure 2A and B).
Figure 2.
RPH1 is genetically involved in the TORC1 pathway. (A) The indicated strains were spotted on YPD plates with low dose of rapamycin. Pictures were taken after grown 2 or 4 days. (B) Tet-off system derived yeast strains were spotted on plates with the indicated doses of rapamycin in the presence of doxycycline. (C) Log-phase cells were cultured under rapamycin treatment, and were isolated at the indicated times and subjected to immunoblot with different antibodies. G6PDH served as a loading control. Asterisk indicates a non-specific band. (D) The phosphorylated Rps6 levels relative to total Rps6 levels in panel C were plotted with the indicated time points and then normalized to the time 0 point of individual strain by densitometric analysis using ImageJ software. The error bars represent mean ± SD from three independent biological repeats. t-test, ∗∗P < 0.01.
To validate Rph1 is indeed involved in the TORC1 regulatory pathway, we examined the protein and phosphorylation levels of ribosomal protein S6 (Rps6), a downstream target of TORC1, which is dephosphorylated with the TORC1 inhibition (26). With rapamycin inhibition of TORC1, Rph1 levels were downregulated due to inactivation of ribosome biogenesis. Notably, the levels of Rps6 gradually decreased with a similar trend in either WT, set2Δ or rph1Δ cells. However, the phosphorylation level of Rps6 diminished much lower in rph1Δ cells compared with WT or set2Δ cells (Figure 2C). More strikingly, even treated with rapamycin for 120 min, nearly half amount of phosphorylated Rps6 was evident, suggesting TORC1 activity was still present in the rph1Δ cells even under nutrient stress conditions (Figure 2D). Altogether, we conclude that Rph1 may regulate cell growth through a TORC1-mediated pathway.
Rph1 negatively regulates transcription of ribosomal genes under non-optimal growth conditions
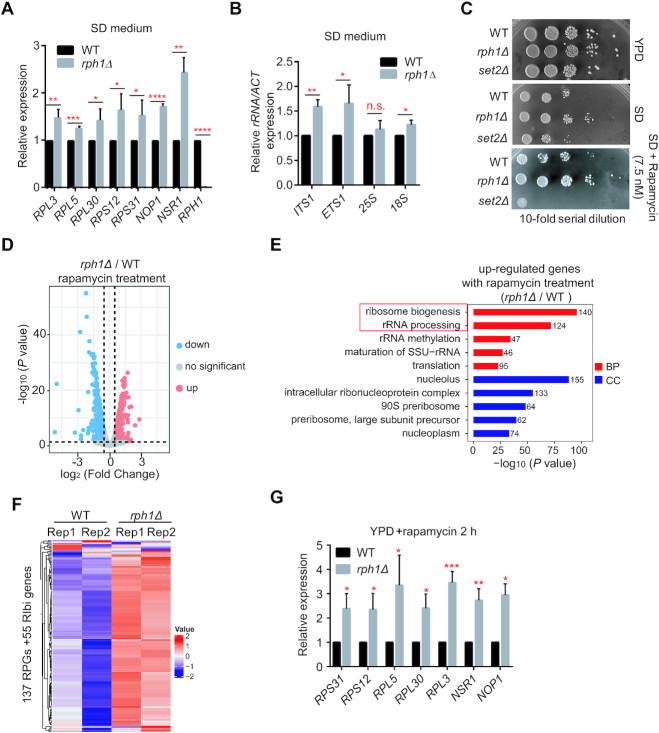
Cell growth potential is governed by ribosomal gene expression, while TORC1 controls rDNA activation and cell growth (10). To figure out whether Rph1 affects cell growth through regulating ribosomal gene expression, RNA-sequencing (RNA-seq) assay was performed and global gene transcriptional levels in rph1Δ and WT cells under both nutrient-rich and nutrient-stress conditions were examined. We identified 537 differentially expressed genes (DEGs), including 357 upregulated genes and 180 downregulated genes in nutrient-rich medium (Supplementary Figure S2A). Agreed with previous study, GO analyses did show that upregulated gene clusters were enriched in autophagy events, but not enriched in ribosome biogenesis events (Supplementary Figure S2B) (19). Consistently, RNA-seq analysis did not exhibit increased RPGs transcription between rph1Δ and WT cells in nutrient-rich condition, which was validated by RT-qPCR analyses (Supplementary Figure S2C and D). However, when Rph1 constructs driven by different promoters were overexpressed in cells that expressed different levels of Rph1 protein, the cells expressing higher levels of Rph1 were associated with slower growth (Supplementary Figure S3A and B). The slow-growth phenotype with higher expression of Rph1 was concomitant with lower transcriptional levels of RPGs as well as rRNA co-transcribed sequences (Supplementary Figure S3C and D). Measurement of expression of 35S rRNA co-transcribed sequence (ETS1) or 18S rDNA co-transcribed sequence (ITS1) has been utilized to quantify difference of nascent rRNA levels (27).
Next, we tested whether Rph1 could regulate transcription of ribosomal genes under non-optimal growth conditions, for instance, in nutrient-limited SD medium or in rapamycin stress conditions. Overnight cultures of WT or rph1Δ cells grown in SD medium were diluted into fresh SD medium and continued growing to middle logarithm phase. Cells were harvested and subjected to RNA extraction. RT-qPCR analysis indicated that rph1Δ cells showed a significant increase of transcription levels of RPGs and 35S rRNAs compared to WT cells (Figure 3A and B). Accordingly, spot assays showed that, under both nutrient limited and rapamycin treatment conditions, rph1Δ cells exhibited faster growth phenotypes, whereas set2Δ cells have slower growth phenotype compared to WT cells (Figure 3C). Moreover, under a nutrient stress conditions with 2 h rapamycin treatment, we found that DEGs in rph1Δ cells were significantly changed (662 upregulated genes and 572 downregulated genes, Figure 3D). In agreement with faster cell growth phenotype in rph1Δ cells upon nutrient stress, GO analyses demonstrated upregulated genes were predominantly correlated with ribosome biogenesis and rRNA processing, implicating its roles in these two regulatory pathways (Figure 3E). Indeed, expression levels of most of 137 RP genes and 55 Rph1-occupied Ribi genes were significantly enhanced in rapamycin treated rph1Δ cells compared to WT cells, and the high-throughput data were validated by RT-qPCR analyses (Figure 3F and G). Altogether, these results strongly suggested that Rph1, as a transcription repressor, negatively regulate expression of RPGs and rRNA synthesis.
Figure 3.
Rph1 negatively regulates transcription of ribosomal genes under nutrient-limited and nutrient stress conditions. (A andB) Expression of representative RPGs (A) or the indicated 35S rRNAs loci (B) from WT and rph1Δ cells cultured in SD medium was examined by RT-qPCR analyses. (C) The indicated strains were spotted onto YPD or SD plates with or without rapamycin. (D) Volcano plots indicating the DEGs from overlapped two biological repeats in rph1Δ cells relative to WT cells with rapamycin treatment for 2 h. The vertical dashed gray lines in the plot represent log2 normalized fold changes of ±0.5. The horizontal dashed gray line represents an adjusted P value of 0.05. (log2(FC) > 0.5, P < 0.05). (E) GO analysis showed the top 5 enriched upregulated gene clusters based on DEGs in panel D by different pathways. BP: biological processes; CC: cellular components. (F) Expression heatmap of all RPGs and Rph1-enriched 55 Ribi genes in either rph1Δ or WT cells with rapamycin treatment. (G) Expression of representative RPGs and Ribi genes was validated by RT-qPCR. Data are represented as mean ± SD from three biological replicates. t-test, ∗P < 0.05; ∗∗P < 0.01; ***P < 0.001, **** P < 0.0001, n.s. ‘not significant’.
Rph1 occupies RNAPI-transcribed rDNA regions and RNAPII-transcribed RP and Ribi genes under nutrient-rich conditions
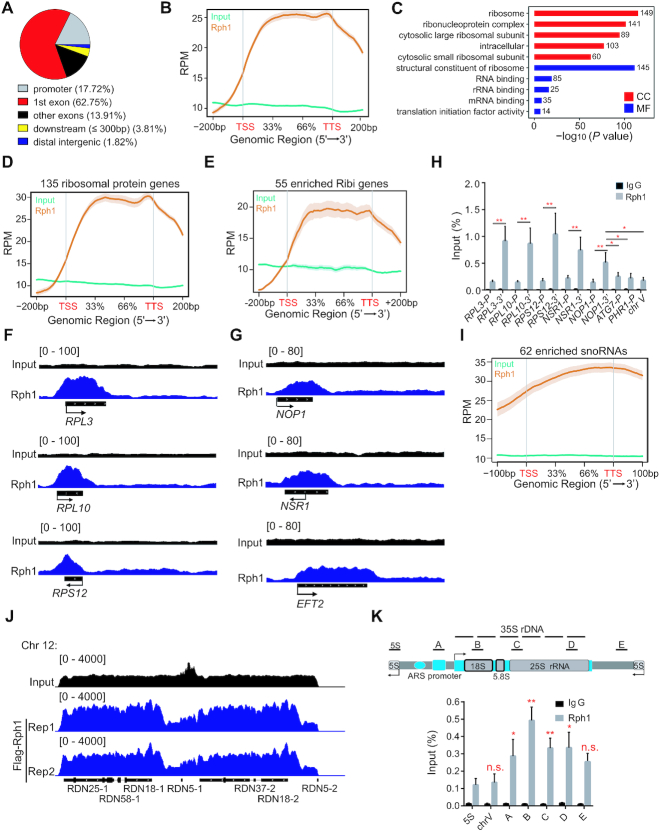
To investigate the potential mechanism of Rph1 regulating ribosome biogenesis, we determined the chromatin landscape of Rph1-bound genomic locations using chromatin immunoprecipitation sequencing (ChIP-seq). Total 689 overlapped peaks from two biological repeats showed that majority of which (94.38%) located at both exons and promoters (Figure 4A and Supplementary Figure S4A). Notably, Rph1 prefers to occupy the transcriptional regions (Figure 4B). GO analyses of Rph1-bound regions revealed a specific and highly significant association with several regulatory arms of the ribosomal pathways (Figure 4C). Among them, 135 from total 137 RPGs and 55 from ∼200 Ribi genes were occupied by Rph1 (Figure 4D and E; Supplementary Figure S4B). We further observed that Rph1 binding was evident at the transcribed regions of genes encoding components of both the 40S (for example, RPS12) and 60S (RPL3 and RPL10) subunits, as well as Ribi genes required for both pre-rRNA processing and ribosome biogenesis (NOP1, NSR1 and EFT2) (Figure 4F and G). Intriguingly, we noticed that Rph1 binds the transcribed regions much more abundant (2- to 5-fold increased) than the promoters of any genes, which was confirmed by ChIP following by qPCR analysis (ChIP-qPCR) (Figure 4H). In addition, it has been known that in budding yeast, the majority of snoRNAs are involved in rRNA processing, which are required for production of ribosome and cell growth (14,28). We found that Rph1 binds more than 80% of snoRNA (62 from 77 known sonRNAs) (Figure 4I). Rph1 enrichment on the transcriptional regions of genes for RP and snoRNAs was reproducible in dataset from previous results (GEO-NCBI: GSE 121635) (Supplementary Figure S4C and D). More interestingly, Genome browser tracks showed that Rph1 also extensively bound the rDNA genomic locus, particularly enriched over the 35S rDNA transcription units, but not on the RNAPIII-transcribed 5S rDNA units, compared to the non-targeted silenced chromosome V (Figure 4J and K). Altogether, we conclude that Rph1 may control ribosome biogenesis through physically interacting with both RPG and rDNA locus.
Figure 4.
Genomic enrichment of Rph1 in budding yeast. (A) Genome-wide distribution of Rph1 ChIP-seq peaks. The numbers represent the percentage of peaks in every genomic region. (B) Enrichment of Rph1 ChIP-seq signals on different regions of gene body. RPM: Read count Per Million mapped reads (thereafter as below). (C) GO analyses show the top 5 of Rph1-enriched peaks by different pathways. CC: cellular components; MF: molecular function. The numbers shown in the plot indicated the classified gene numbers. (D andE) Distributions of Rph1-enriched signals on RPGs (D) and on Ribi genes (E). TSS: transcription start sites; TTS: transcription termination sites. (F andG) Genome browser view of Rph1 enrichment from two biological repeats at representative RPGs (panel F) and Ribi genes (panel G), respectively. (H) Rph1 enrichment at various representative RPGs was confirmed by ChIP-qPCR. IgG ChIP served as control. (I) Distributions of Rph1-enriched signals on snoRNAs. (J) Genome browser view of Rph1 enrichment in the representative rDNA regions of chromosome 12. (K) Representative of one rDNA repeat with the position of primers used (upper panel). ChIP-qPCR validation of Rph1 enrichment over the rDNA locus (bottom panel). In panel H and K, qPCR data are represented as mean ± SD from three biological replicates. t-test, ∗P < 0.05; ∗∗P < 0.01; *** P < 0.001, n.s. ‘not significant’.
Deletion of Rph1 increased expression of ribosomal genes mainly via increasing RNAPII occupancy and facilitating RNAPI transcription process under nutrient stress conditions
We attempted to investigate the correlation of ribosome genomic enrichment of Rph1 with ribosomal gene expression. To test this, the divergence of genome-wide Rph1 enrichment was examined in cells treated with or without rapamycin by ChIP-seq. As expected, most of Rph1 proteins detached from RPGs and snoRNA regions (Supplementary Figure S5A and B), and almost all of Rph1 was released from rDNA regions after normalized to the Input (Supplementary Figure S5C). Intriguingly, the majority of Rph1 was still associated with the Ribi genes even under rapamycin treatment (Supplementary Figure S5D). These results indicated that Rph1 dissociates from ribosomal genes under rapamycin treatment condition.
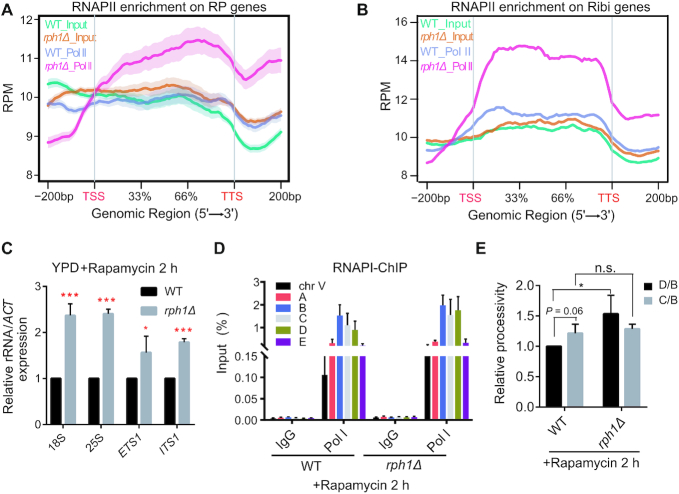
Next, we want to know how Rph1 regulates transcriptional levels of RPGs and rRNAs under nutrient stress conditions. We hypothesized that increased transcriptional levels of RPGs and Ribi genes in rph1Δ cells resulted from increased RNAPII tethering, To test this idea, ChIP-seq analysis was performed under rapamycin stress conditions. As expected, engagement of RPGs and Ribi gene loci by RNAPII was remarkedly increased in rph1Δ cells compared to WT cells (Figure 5A and B), accompanied with elevated expression of RPGs and Ribi genes (Figure 3G), establishing the correlation of increased chromatin occupancy of RNAPII with loss of RPH1. Since rRNA and RPG transcription are tightly co-regulated, we further examine whether RNAPI-transcribed rRNAs were coordinately upregulated in rph1Δ cells. Upon rapamycin treatment, obvious elevated levels of 18S and 25S rRNA, as well as increased levels of ETS1 and ITS1, were detected in rph1Δ cells compared to WT cells, confirming its coordinated role of Rph1 in rRNA synthesis (Figure 5C). Intriguingly, we did not observe changes in association of RNAPI with rDNA repeats in rph1Δ cells regardless of rapamycin treatment (Supplementary Figure S5A and Figure 5D), suggesting that other mechanisms may assist regulation of rRNA transcription.
Figure 5.
Deletion of Rph1 increased RNAPII occupancy and facilitated RNAPI transcription with rapamycin treatment. (A and B) Distribution of RNAPII ChIP-seq signals on gene bodies of RPGs (A) and Ribi genes (B) in WT and rph1Δ cells with rapamycin treatment. (C) Expression of the indicated rRNA regions in WT and rph1Δ cells. (D) ChIP-qPCR analysis were performed to examine RNAPI (Rpa190-Flag) enrichment over different rDNA loci in WT or rph1Δ cells. IgG ChIP served as a negative control. (E) Processivity assay of RNAPI transcription were performed in the rph1Δ cells relative to WT. The IP/Input value for the middle gene body (the C locus) or the 3′ end (the D locus) was divided by the value for the 5′ end (the B locus), and the ratio (‘processivity’) for rph1Δ was normalized to that for WT. qPCR data are represented as mean ± SD from three biological replicates. t-test, ∗P < 0.05; ∗∗P < 0.01; *** P < 0.001, n.s. ‘not significant’.
Since the transcription factor Rrn3 is required for activation of the 35S rRNA transcription via formation of the RNAPI–Rrn3 complex at the rDNA promoters, we wonder if increased rRNA transcription in rph1Δ cells is related with Rrn3 (29). Therefore, WT and rph1Δ strains bearing an integrated C-terminal 3xFlag-tagged Rrn3 were genereated, and deletion of RPH1 did not affect Rrn3 protein levels (Supplementary Figure S6B). ChIP-qPCR analyses showed that, in both treated and untreated conditions, the associated levels of Rrn3 with the promoters of 35S rDNAs are comparable in both WT and rph1Δ cells, although the binding affinities are strikingly reduced upon rapamycin treatment (Supplementary Figure S6C and D). These data rule out the possibility that increased rRNA transcription in rph1Δ cells results from increased Rrn3 association with chromatin. Another possibility is that deletion of RPH1 may affect the elongation step in rRNA transcription. To do this, the processivity assay of RNAPI transcription was performed by comparing the association of RNAPI with the middle gene body (the C locus) or the 3′ end (the D locus) of the 35S rDNA loci to that at the 5′ end (the B locus) using ChIP analysis. This method has been used to measure its ability of RNAPI to stay engaged in a template from initiation to termination of transcription (30). Interestingly, we observed a similar transcription pattern in the processivity of RNAPI without rapamycin treatment (Supplementary Figure S6E), but an 1.5-fold increase in rph1Δ cells compared to WT cells with rapamycin treatment (Figure 5E), which suggests that Rph1 dissociation from chromatin may affect RNAPI elongation process under nutrient stress conditions. Overall, the data indicated that Rph1 regulates transcription of ribosomal genes through affecting occupancy or capacity of RNA polymerases onto chromatin.
Rim15-dependent Rph1 phosphorylation released its repression on ribosome genes
Next, we attempted to investigate the mechanism of upregulation of ribosomal gene expression in rph1Δ cells under nutrient stress conditions. Previous study has indicated that Rph1 became hyperphosphorylated upon rapamycin treatment (25). In addition, the kinase Rim15 could translocate into the nucleus and mediate Rph1 phosphorylation upon nitrogen starvation, which releases repressive role of Rph1 on ATG gene expression (19). Moreover, one of the key effectors functioning downstream of the TORC1 pathway, Sch9, antagonizes the nuclear accumulation of Rim15 in optimal growth conditions (31–33). The evidence above strongly suggest that activated Rim15 may also mediate Rph1 phosphorylation under rapamycin stress conditions. To get a higher resolution of the phosphorylation status of Rph1, phos-tag, a molecule that binds specifically to phosphorylated ions was used. With addition of phos-tag, integrated Rph1–3×Flag showed a significant shift of high molecular weight bands in WT cells, but showed a strong block of shift in rim15Δ cells upon rapamycin treatment (Figure 6A and Supplementary Figure S7A). Importantly, deletion of RPH1 in rim15Δ cells remarkedly rescued rapamycin-sensitive phenotype caused by RIM15 loss, indicating a genetic interaction of RPH1 and RIM15 (Figure 6B).
Figure 6.
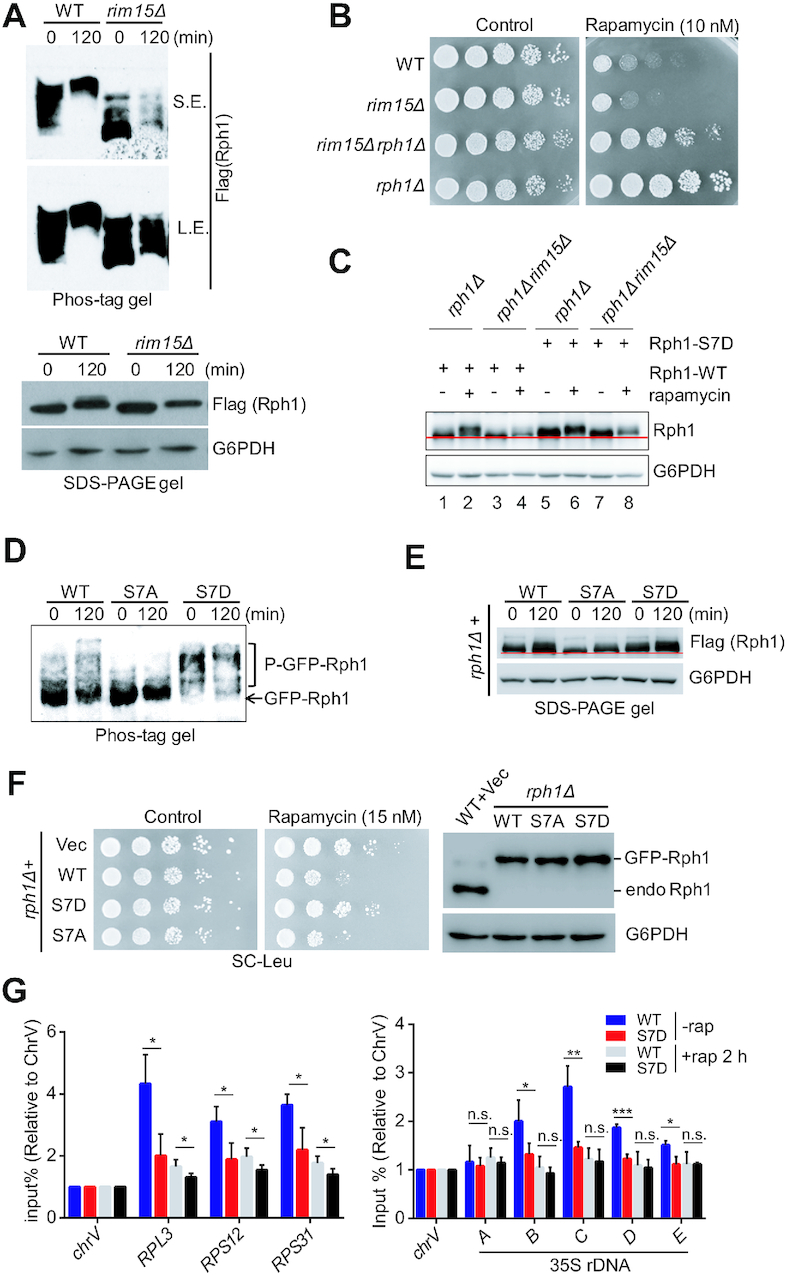
Rim15-dependent Rph1 phosphorylation alleviates cellular sensitivity to rapamycin stress. (A) The phosphorylation states of Rph1-Flag in indicated strains upon rapamycin treatment were examined using phos-tag gel (top panel) or SDS-PAGE gel (bottom panel). (B) The indicated strains were spotted on YPD plates with or without rapamycin. (C) Mobility shift of the indicated cells bearing either WT or S7D Rph1 under rapamycin treatment was examined using regular SDS-PAGE gel. (D andE) The phosphorylation states of GFP-Rph1 or Flag-Rph1 bearing either the WT or mutations driven by endogenous RPH1 promoter upon rapamycin treatment were examined using a phos-tag gel (panel D) or SDS-PAGE gel (panel E), respectively. (F) Rapamycin sensitivity of various Rph1 constructs were monitored by a spot assay (left panel), and protein levels of GFP-Rph1 in the indicated cells were examined by immunoblotting (right panel). (G) ChIP-qPCR analysis showed relative abundance of WT or S7D Rph1 in different RP gene loci (left panel) and different regions of 35S rDNA (right panel) with or without rapamycin treatment. qPCR data are represented as mean ± SD from at least three biological replicates. t-test, ∗P < 0.05; ∗∗P < 0.01; ***P < 0.001, n.s. ‘not significant’
To identify potential Rim15-mediated phosphorylation sites of Rph1, nine phosphorylation sites reported in previous studies will be further analyzed (Supplementary Figure S7B). The reason why we chose those sites as they became phosphorylated only under nutrient stress conditions (25,34). Each serine residue of phosphorylated site on Rph1 was mutated to alanine (A) or aspartic acid (D) to represent phospho-defective mutant or phospho-mimetic mutant, respectively. We then examined growth phenotypes of rph1Δ cells bearing individual Rph1 mutants that are under the control of an endogenous promoter. Compared to cells bearing a WT Rph1 construct, these phospho-defective mutants showed a similar sensitivity of rapamycin (Supplementary Figure S7C); whereas seven of the nine phsopho-mimetic mutants showed resistance to rapamycin except S561D and S575D mutants (Supplementary Figure S7D). It has been reported that S561 and S575 are two phosphorylation sites of CDC28, a cyclin-dependent kinase, which probably are not phosphorylated sites of Rim15 (35). Hence, Flag-tagged or GFP-tagged Rph1 constructs bearing S7A mutation (S412/425/426/429/430/434/557A) or S7D mutation (S412/425/426/429/430/434/557D) were generated, respectively. We confirmed these seven serine residues can be phosphorylated by Rim15, as WT Rph1 protein showed a faster mobility in rim15Δ cells compared to WT cells upon rapamycin treatment (Figure 6C, lane 3 versus lane 4), whereas S7D mutant did not show an obvious shift regardless of in the presence or absence of RIM15 (Figure 6C, lane 6 versus lane 8). As expected, even with rapamycin treatment, S7A mutant severely attenuated phosphorylation of Rph1 despite of bearing a Flag tag or a GFP tag, examined by either phos-tag gel or regular SDS-PAGE gel (Figure 6D and E). In stark contrast, S7D mutant exhibited dramatic phosphorylation states no matter with or without rapamycin treatment (Figure 6D and E). Importantly, although S7D mutant expressed equally to the WT or S7A mutant of Rph1, S7D mutant showed a rapamycin-resistance growth phenotype comparable to rph1Δ strain (Figure 6F). In addition, overexpression of S7A mutant under the control of an endogenous promoter led to a slow-growth phenotype even in nutrient-rich condition, suggesting its predominant repressive role in ribosome biogenesis (Supplementary Figure S7E). Conversely, we validated that S7D mutant binds both RP genes and 35S rDNA locus much less than WT Rph1 does in optimal growth conditions, and decreased association with these regions upon nutrient stress by ChIP-qPCR analysis (Figure 6G). These results demonstrate that Rph1 phosphorylation is required for its release from ribosome genomic locus, thereby derepressing ribosomal gene expression under nutrient-stress conditions.
DISCUSSION
In this work, we present evidence that Rph1 is a cofactor that represses transcription of both RPGs and 35S rRNAs. Under optimal growth conditions, the association of unphosphorylated Rph1 with both RNAPI and RNAPII transcribed genes may provide a simple yet highly efficient mechanism to integrate transcription of RPGs and 35S rRNAs for ribosome biogenesis. In this scenario, RNA polymerases, orchestrated with key transcriptional factors, including Ifh1 and Rrn3, maximally drive the transcription elongation process for production of RPs and rRNAs. Therefore, it is difficult to detect transcriptional changes of RPGs and rRNAs in rph1Δ cells. In contrast, under nutrient limited conditions, lack of Rph1 coordinately enhances RPG transcription and 35S rRNA synthesis to promote cell growth (Figure 3). More importantly, under nutrient stress conditions, phosphorylated Rph1 is released from transcribed regions of RPGs and 35S rRNAs, which allows small amounts of ribosomal genes can be transcribed and synthesis, thereby maintaining cell long-term survival even lacking assistance of activators. Without Rph1, about 1.5-fold increase of expression levels of RPGs and 35S rRNAs was detected, which significantly accelerates cell growth even under such nutrient stress conditions (Figure 7). Our data make us propose a model in which chromatin engagement of Rph1 provide a sensing system to adjust production of ribosomes properly in response to different nutrient context.
Figure 7.
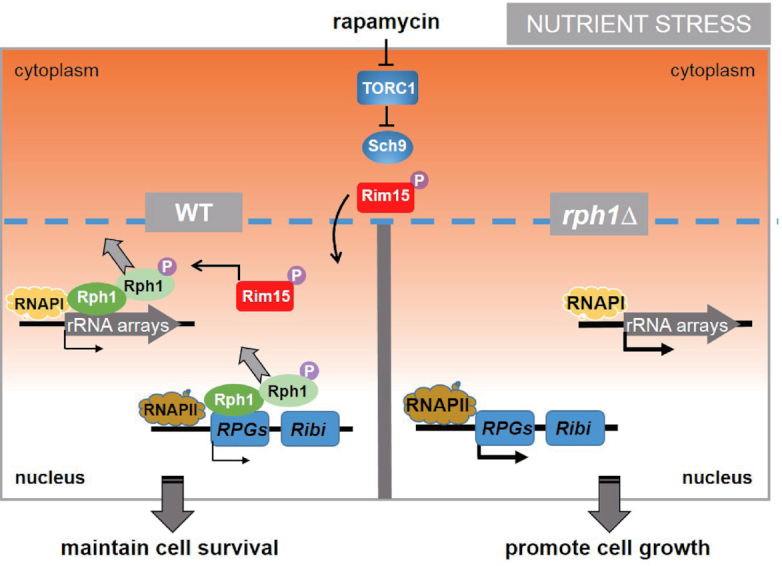
A proposed model shows that Rph1 coordinates transcription of RPGs and rRNAs to control cell growth under nutrient stress conditions. With nutrient stress to inactivate TORC1 and Sch9, phosphorylated Rim15 is translocated to nucleus and catalyzes Rph1 phosphorylation, which releases the majority of Rph1 from transcribed regions of RPGs and 35S rDNAs, thereby maintaining long-term cell survival. Under the same stress condition, complete absence of RPH1 leads to about 1.5-fold increase of expression levels of RPGs and 35S rRNAs compared to WT cells, which further promotes ribosome biogenesis and cell growth.
Our model is supported by several lines of evidence that correlate Rph1 levels with ribosome biogenesis and cell growth. First, Rph1 binds to the majority transcriptional regions of rRNAs and RPGs that are required for production of ribosome assembly and biogenesis. With rapamycin treatment, Rph1 occupancy in RPG loci was decreased concomitantly with loss of Rph1 in rDNA regions (Supplementary Figure S5). Second, Rph1 genetically interacts with the TORC1 pathway, which has been extensively proven to control ribosome biogenesis and cell growth. Deletion of RPH1 in TOR1-deficient cells significantly rescues cell sick-growth phenotype, which strongly supports that Rph1 is involved in ribosome biogenesis (Figure 2). Third, Rph1 levels are oppositely correlated with transcription levels of rRNAs and RPGs as well as cell growth rate under nutrient limited and nutrient stress conditions (Figures 3 and 5; Supplementary Figure S3). Lastly, phosphorylation-mimetic Rph1 S7D mutant possesses less binding affinity of genomes and faster cell growth phenotype (Figure 6). Combined these, we conclude that chromatin-occupied Rph1 levels are important for proper ribosome biogenesis. Intriguingly, RNAPI abundance on rRNA regions were not affected by Rph1 level, which seems to contradict the point that Rph1 ordinates RNAPI and RNAPII-mediated transcription. However, we found that Rph1 is associated with the majority of snoRNAs, which suggests that Rph1 may regulate snoRNAs to affect RNAPI-mediated rRNA processing. Indeed, the processivity assay indicated that Rph1 may affect RNAPI elongation process to regulate transcription levels of RNAPI-mediated rRNAs with rapamycin treatment (Figure 5E). Thus, our data support that Rph1 is involved in ribosome biogenesis and cell growth, mainly by coregulation of transcription of rRNAs and RPGs.
Although our data suggested that Rph1 may function to repress transcription of RPGs and rRNAs by largely occupying the transcribed regions of 35 rDNAs and almost all of RPGs under optimal growth conditions, we surprisingly did not observe increased transcription of those genes as well as accelerated cell growth phenotype. Instead, we observed slightly decreased transcription of RPGs and a little slower growth phenotype in rph1Δ cells compared to WT when cells were cultured in YPD liquid medium (Supplementary Figure S2D), which is consistent with previous observations (15). It has been reported that aberrant ribosome biogenesis led to excess RPs production, which reduced the growth of yeast cells (36). Since Rph1 occupied both RPGs and rDNA loci, we speculate that deletion of RPH1 might cause abnormal production or synthetic imbalance in RPs and rRNAs, which may be proteotoxic to the cell and conversely retards normal cellular growth. Why such repression is necessary under optimal growth conditions? We considered several interpretations. First, in a rapidly growing yeast cell, Rph1 could coordinate regulation of the rRNA genes and 137 RP genes by co-occupancy that is essential for the economy of the cell. Second, Rph1 maintains proteostasis by repressing excessive ribosomal biogenesis. Third, it has been suggested that moderate silencing of rRNA transcription is critical for genome stability. Repression of transcription of ribosomal genes by Rph1 may be necessary for restrict uncontrolled cell proliferation in budding yeast. For multicellular eukaryotes, it has been extensively reported that hyperactivation of ribosome synthesis abnormally accelerates protein synthesis and induces instability of rDNA, thereby promoting cancer cell proliferation and metastasis. For example, many tumor suppressors, such as pRB (retinoblastoma-associated protein), p53, and ARF (alternative reading frame) can repress rRNA transcription through directly interaction with rDNA or transcription apparatus (37,38). Whether and how Rph1 acts such a repressive role needs to be further explored.
Several studies have indicated that Rph1 represses expression of many ATG genes as well as DNA damage genes under optimal growth conditions via binding their promoters (18–19,39). Surprisingly, our genome-wide ChIP-seq analyses illustrated that Rph1 vastly associates with transcribed regions of rDNA and RPGs, instead of the promoters of previous reported genes, suggesting its role in transcriptional elongation but not transcriptionall initiation. These results are also consistent with the fact that localization of H3K36me3 at gene coding region and rDNA repeats, reminiscent of its demethylase character. Although by analyzing public database we did observe a slight enhancement of H3K36me3 levels at those transcriptional regions upon Rph1 deletion (data not shown), expression of the inactive mutant of Rph1 exhibited a slow growth defect with nutrient stress, similar to the WT allele (Figure 1C and Supplementary Figure S1C). Therefore, we believe that the impact of Rph1 on cell growth is specific through regulating transcriptional elongation of ribosomal genes.
Besides RNAPI and RNAPII transcribed genes, RNAPIII transcribed tRNA genes are also involved in ribosome biogenesis. With nutrient stress, transcription of this class of genes was repressed along with other RPGs and rRNAs. In our analysis, we did not observe any enrichment of Rph1 on these tRNA genes, reinforcing Rph1’s binding specificity. Of note, previous studies have demonstrated Maf1 as a general and direct repressor of yeast RNAPIII transcription (40,41), which utilizes similar inhibitory actions of Rph1. For example, both Maf1 and Rph1 only associated with RNA polymerases on chromatin in a dephosphorylated state. Engagement of chromatin allows them to repress associated gene transcription, respectively. Once they are in a phosphorylated state, both protein would dissociate from chromatin. Different from that Rph1 was dissociated from chromatin, Maf1 was stimulated to associate with chromatin as well as disengaged RNAPIII with rapamycin treatment (40). Whether these two parallel signaling pathways act together is particularly of interest.
Understanding how distinct transcriptional machineries involved in cell growth are tightly coordinated to sense nutrient alternations becomes a fundamental but challenging question. Previous studies have demonstrated that two rRNA processing factors, including DDX2 in mammal and Utp22 in yeast, could coordinate rRNA production and RPGs transcription. DDX2 promotes rRNA transcription, processing and modification. At the meantime, DDX2 also facilitates transcription of RPGs by association with promoters of RNAPII-transcribed genes encoding RPs and snoRNAs. Utp22 may act as a negative role in this coupling event. With long-term growth inhibition, rRNA-released Utp22 titrates the transcription activator Ifh1 from RPG promoters thereby maintaining an inhibitory state of ribosome biosynthesis. Both cases revealed how rRNA synthesis guides RPG transcription. In this work, we identified Rph1, which has been found in the RNAPII elongation complex by in vitro reconstitution assay (42), could sense RPG transcription to guide rRNA production from an opposite direction. Our work may help to further decipher the elegant but complex crosstalk machinery between RNAPI and RNAPII and more transcriptional networks in higher eukaryotes.
DATA AVAILABILITY
Raw data have been deposited in the GEO database with accession number GSE141034.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Yu Zhou and his lab members for helping bioinformatics analysis, Drs. Yang Lu and Shanshan Li for sharing strain and reagents. The numerical calculation in this work have been done on the supercomputing system in the Supercomputing Center of Wuhan University.
Notes
Present address: Feng Li, Division of Molecular and Cellular Biology, National Institutes of Health, Bethesda, MA 20892, USA.
Contributor Information
Wen-Jie Shu, Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072 China.
Runfa Chen, Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072 China.
Zhao-Hong Yin, Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072 China.
Feng Li, Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072 China.
Heng Zhang, Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, 3888 Chenhua Road, Shanghai, 201062, China.
Hai-Ning Du, Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, Hubei 430072 China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2019YFA0802501]; Major State Basic Research Development Program of China [2013CB910700]; National Natural Science Foundation of China [31271369, 31770843, 31971231]. Funding for open access charge: National Natural Science Foundation of China [31770843].
Conflict of interest statement. None declared.
REFERENCES
- 1. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 2. Lempiainen H., Shore D.. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009; 21:855–863. [DOI] [PubMed] [Google Scholar]
- 3. de la Cruz J., Gomez-Herreros F., Rodriguez-Galan O., Begley V., de la Cruz Munoz-Centeno M., Chavez S.. Feedback regulation of ribosome assembly. Curr. Genet. 2018; 64:393–404. [DOI] [PubMed] [Google Scholar]
- 4. Albert B., Kos-Braun I.C., Henras A.K., Dez C., Rueda M.P., Zhang X., Gadal O., Kos M., Shore D.. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife. 2019; 8:e45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rudra D., Mallick J., Zhao Y., Warner J.R.. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 2007; 27:4815–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin D.E., Soulard A., Hall M.N.. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004; 119:969–979. [DOI] [PubMed] [Google Scholar]
- 7. Schawalder S.B., Kabani M., Howald I., Choudhury U., Werner M., Shore D.. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004; 432:1058–1061. [DOI] [PubMed] [Google Scholar]
- 8. Wade J.T., Hall D.B., Struhl K.. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004; 432:1054–1058. [DOI] [PubMed] [Google Scholar]
- 9. Albert B., Knight B., Merwin J., Martin V., Ottoz D., Gloor Y., Bruzzone M.J., Rudner A., Shore D.. A molecular titration system coordinates ribosomal protein gene transcription with ribosomal RNA synthesis. Mol. Cell. 2016; 64:720–733. [DOI] [PubMed] [Google Scholar]
- 10. Berger A.B., Decourty L., Badis G., Nehrbass U., Jacquier A., Gadal O.. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell. Biol. 2007; 27:8015–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hall D.B., Wade J.T., Struhl K.. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006; 26:3672–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grewal S.S., Evans J.R., Edgar B.A.. Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J. Cell Biol. 2007; 179:1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jouffe C., Cretenet G., Symul L., Martin E., Atger F., Naef F., Gachon F.. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013; 11:e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calo E., Flynn R.A., Martin L., Spitale R.C., Chang H.Y., Wysocka J.. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015; 518:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klose R.J., Gardner K.E., Liang G., Erdjument-Bromage H., Tempst P., Zhang Y.. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007; 27:3951–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tu S., Bulloch E.M., Yang L., Ren C., Huang W.C., Hsu P.H., Chen C.H., Liao C.L., Yu H.M., Lo W.S. et al.. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol. Chem. 2007; 282:14262–14271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim T., Buratowski S.. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J Biol. Chem. 2007; 282:20827–20835. [DOI] [PubMed] [Google Scholar]
- 18. Liang C.Y., Wang L.C., Lo W.S.. Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 2013; 24:3251–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernard A., Jin M., Gonzalez-Rodriguez P., Fullgrabe J., Delorme-Axford E., Backues S.K., Joseph B., Klionsky D.J.. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 2015; 25:546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li F., Zheng L.D., Chen X., Zhao X., Briggs S.D., Du H.N.. Gcn5-mediated Rph1 acetylation regulates its autophagic degradation under DNA damage stress. Nucleic Acids Res. 2017; 45:5183–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du H.N., Fingerman I.M., Briggs S.D.. Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Genes Dev. 2008; 22:2786–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F. et al.. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002; 22:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDaniel S.L., Hepperla A.J., Huang J., Dronamraju R., Adams A.T., Kulkarni V.G., Davis I.J., Strahl B.D.. H3K36 methylation regulates nutrient stress response in Saccharomyces cerevisiae by enforcing transcriptional fidelity. Cell Rep. 2017; 19:2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wullschleger S., Loewith R., Hall M.N.. TOR signaling in growth and metabolism. Cell. 2006; 124:471–484. [DOI] [PubMed] [Google Scholar]
- 25. Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R.. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009; 23:1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez A., Shimobayashi M., Eisenberg T., Merle D.A., Pendl T., Hall M.N., Moustafa T.. TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae. PLoS One. 2015; 10:e0120250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laribee R.N., Hosni-Ahmed A., Workman J.J., Chen H.. Ccr4-not regulates RNA polymerase I transcription and couples nutrient signaling to the control of ribosomal RNA biogenesis. PLoS Genet. 2015; 11:e1005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dieci G., Preti M., Montanini B.. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009; 94:83–88. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto R.T., Nogi Y., Dodd J.A., Nomura M.. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 1996; 15:3964–3973. [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider D.A., French S.L., Osheim Y.N., Bailey A.O., Vu L., Dodd J., Yates J.R., Beyer A.L., Nomura M.. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. PNAS. 2006; 103:12707–12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., Winderickx J., De Virgilio C.. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell. 2003; 12:1607–1613. [DOI] [PubMed] [Google Scholar]
- 32. Wanke V., Cameroni E., Uotila A., Piccolis M., Urban J., Loewith R., De Virgilio C.. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 2008; 69:277–285. [DOI] [PubMed] [Google Scholar]
- 33. Wanke V., Pedruzzi I., Cameroni E., Dubouloz F., De Virgilio C.. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 2005; 24:4271–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye C., Sutter B.M., Wang Y., Kuang Z., Zhao X., Yu Y., Tu B.P.. Demethylation of the protein phosphatase PP2A promotes demethylation of histones to enable their function as a methyl group sink. Mol. Cell. 2019; 73:1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holt L.J., Tuch B.B., Villen J., Johnson A.D., Gygi S.P., Morgan D.O.. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009; 325:1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tye B.W., Commins N., Ryazanova L.V., Wuhr M., Springer M., Pincus D., Churchman L.S.. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. Elife. 2019; 8:e43002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hein N., Hannan K.M., George A.J., Sanij E., Hannan R.D.. The nucleolus: an emerging target for cancer therapy. Trends Mol. Med. 2013; 19:643–654. [DOI] [PubMed] [Google Scholar]
- 38. Bywater M.J., Pearson R.B., McArthur G.A., Hannan R.D.. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer. 2013; 13:299–314. [DOI] [PubMed] [Google Scholar]
- 39. Liang C.Y., Hsu P.H., Chou D.F., Pan C.Y., Wang L.C., Huang W.C., Tsai M.D., Lo W.S.. The histone H3K36 demethylase Rph1/KDM4 regulates the expression of the photoreactivation gene PHR1. Nucleic Acids Res. 2011; 39:4151–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts D.N., Wilson B., Huff J.T., Stewart A.J., Cairns B.R.. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell. 2006; 22:633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oficjalska-Pham D., Harismendy O., Smagowicz W.J., Gonzalez de Peredo A., Boguta M., Sentenac A., Lefebvre O.. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell. 2006; 22:623–632. [DOI] [PubMed] [Google Scholar]
- 42. Joo Y.J., Ficarro S.B., Chun Y., Marto J.A., Buratowski S.. In vitro analysis of RNA polymerase II elongation complex dynamics. Genes Dev. 2019; 33:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been deposited in the GEO database with accession number GSE141034.