Abstract
A crucial bacterial strategy to avoid killing by antibiotics is to enter a growth arrested state, yet the molecular mechanisms behind this process remain elusive. The conditional overexpression of mazF, the endoribonuclease toxin of the MazEF toxin–antitoxin system in Staphylococcus aureus, is one approach to induce bacterial growth arrest, but its targets remain largely unknown. We used overexpression of mazF and high-throughput sequence analysis following the exact mapping of non-phosphorylated transcriptome ends (nEMOTE) technique to reveal in vivo toxin cleavage sites on a global scale. We obtained a catalogue of MazF cleavage sites and unearthed an extended MazF cleavage specificity that goes beyond the previously reported one. We correlated transcript cleavage and abundance in a global transcriptomic profiling during mazF overexpression. We observed that MazF affects RNA molecules involved in ribosome biogenesis, cell wall synthesis, cell division and RNA turnover and thus deliver a plausible explanation for how mazF overexpression induces stasis. We hypothesize that autoregulation of MazF occurs by directly modulating the MazEF operon, such as the rsbUVW genes that regulate the sigma factor SigB, including an observed cleavage site on the MazF mRNA that would ultimately play a role in entry and exit from bacterial stasis.
INTRODUCTION
The occurrence of antibiotic resistant bacteria is reaching worrying proportions and it is especially disturbing that the number of new antibiotics that reach the market is diminishing. Understanding how bacteria avoid killing by antibiotics will contribute to future development of new antimicrobial drugs. Notably, one mechanism that was observed decades ago and still remains poorly understood relates to avoidance of killing by antibiotics (1) wherein exposure to lethal concentrations of an antibiotic reveals the survival of a bacterial subpopulation which nevertheless remains genetically susceptible. Upon drug exposure, most of the growing cells in the bacterial population will be rapidly killed by the antibiotic and the remaining fraction of non-growing cells will survive because of a slowed-down metabolism within these cells (2).
Antibiotic tolerance in non-growing cells is explained by the inactive state of antibiotic targets and their associated biological processes and not by heritable genetic mutations of the bacterial population. In fact, upon antibiotic removal, non-growing cells regrow and re-exposure to antibiotics leads again to a rapid killing of the majority of the bacteria population (2–4). The antibiotic tolerance in non-growing bacteria may explain the clinical occurrence of bacterial chronic and relapsing infections, which emphasizes the urgent need to better understand the molecular pathways leading to the development of this state (5).
The bacterial non-growing state has been linked in some studies to expression of toxin–antitoxin systems (TAS) in various organisms that interferes with replication (6,7), ribosomal activity (8), cell wall synthesis (9) and cell division (10). However, the role of TAS on antibiotic tolerance in non-growing bacteria is still debated (11). One of the best characterized TAS inducing bacterial growth arrest is MazEF. Homologs of this type II TAS operon are found in clinically important bacteria such as Escherichia coli, Mycobacterium tuberculosis and Staphylococcus aureus. MazF toxin activity is inhibited by a stoichiometric complex formed with the MazE antitoxin during normal growth conditions. Upon stressed conditions, MazE is degraded by cellular proteases, thereby liberating MazF toxin endoribonucleolytic activity that induces a growth arrest (11). In E. coli and M. tuberculosis, MazF specifically cleaves single-stranded RNA molecules and the identification of specific cleaved targets has led to a model, that MazF activity reduces overall translation and consequently inhibits bacterial growth (12–16). Unless otherwise stated, MazF in this study refers to the S. aureus toxin.
To understand the general function of MazEF in S. aureus, several studies first focused on the regulation of MazEF expression. The mazEF gene is chromosomally encoded and located upstream and co-transcribed with the sigB operon that encodes for the alternative stress sigma factor B (σB) and its regulators RsbUVW. MazEF transcription is activated by sub-inhibitory concentrations of erythromycin and tetracycline and it is regulated negatively and positively by the transcriptional regulator σB and SarA, respectively. Its cognate antitoxin MazE is regulated by proteolysis through ClpCP proteolytic complex primed with the adaptor protein TrfA (17,18) (Figure 5B). In vitro analysis of MazF cleavage activity on an exogeneous transcript, found that MazF toxin is a sequence-specific endoribonuclease that recognizes the pentad sequence U∧ACAU (∧ indicates MazF cleavage site) of mRNAs and cleaves independently of ribosomes (19,20). The MazF cleavage motif was further used to predict MazF target genes. A link between MazEF and virulence was proposed based on the identification and in vitro cleavage of selected virulence genes such as hla (α-hemolysin), spa (Protein A virulence factor), σB and sraP (adhesion factor) (19,21). MazF expression was also shown to inhibit biofilm formation and to increase antibiotic tolerance allowing transition of S. aureus from acute to chronic infections (22).
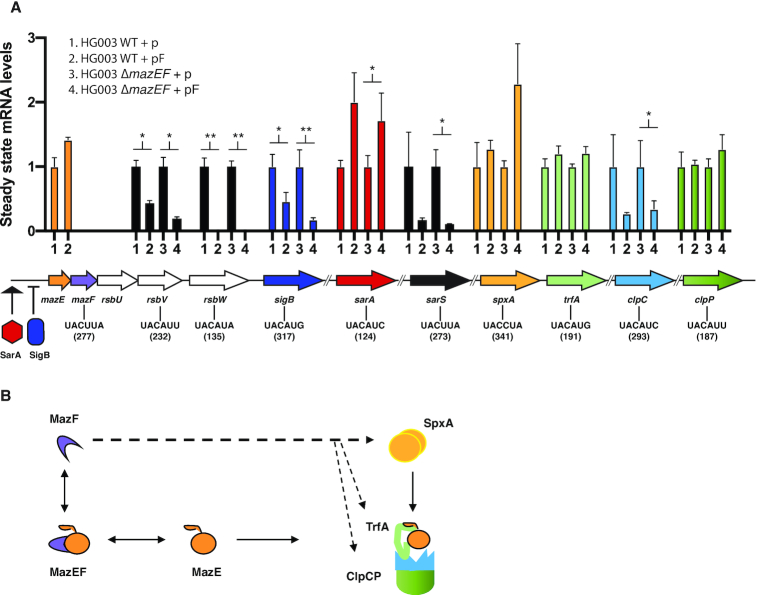
Figure 5.
(A) Steady state transcript levels recorded by qRT-PCR for ten genes in wild-type or mazEF-deleted strains carrying the empty plasmid (p) compared to mazF-plasmid (pF) 10 mpi with ATc. Results are presented as means ± SD of three independent experiments; ** P < 0.01; * P < 0.05 significant differences between the strain carrying the empty plasmid (p) and the strain carrying the mazF-plasmid (pF). The gyrB gene was used to normalize for RNA content. Gene hydrolysis probes hybridize specifically onto the MazF detected site. The MazF recognition sequence and position within the gene is indicated in the gene diagram below. (B) Diagram showing MazF effect on proteins involved in MazE antitoxin proteolysis and containing MazF cleavage motifs. MazF can potentially cleave spxA, trfA, clpC and clpP transcripts (denoted by dashed arrows). SpxA regulates transcription of TrfA adaptor protein, TrfA adaptor in turn is responsible of presenting MazE substrate for degradation to the ClpCP proteolytic system.
In S. aureus, stasis is induced upon overexpression of MazF toxin (21,23), yet whether transcripts are cleaved in vivo to prevent growth and which ones are targeted preferentially is still unknown. Bioinformatic prediction of targets based on the MazF pentad recognition sequence is not feasible since this motif is found across the genome. In this study, we have used a genome-wide high-throughput method to identify bona fide MazF targets in vivo and identified an extended and variable, but robust sequence recognition specificity beyond the canonical UACAU motif for MazF RNA cleavage. The identification of hundreds of precise cleavage sites now provides a snapshot of the extensive collection of genes affected by the MazF endoribonuclease and permits a refined model explaining toxin-mediated growth arrest.
MATERIALS AND METHODS
Induction of mazF and growth arrest
The pRAB11-mazF plasmid (AR1880), expressing mazF under control of an anhydrotetracycline (ATc) inducible promoter was constructed by amplification of S. aureus HG003 mazF using primers carrying BglII and EcoRI restriction sites (Supplementary Table S1). The amplified product was cut with BglII and EcoRI and ligated the into pRAB11 (24). To induce the expression of MazF, ATc was added at a final concentration of 0.2 μM to the cell cultures and incubated for 10–60 min at 37°C with shaking. ATc was freshly prepared and aliquoted since aged ATc was found in pilot experiments to produce toxic breakdown products, which affected interpretation of viability assays.
S. aureus HG003 (RS123) and S. aureus HG003 ΔmazEF (RS125) were electroporated with ATc inducible pRAB11 (RS212, RS191) and pRAB11-mazF (RS124, RS126). Strains carrying pRAB11 were always grown in presence of chloramphenicol (15 μg/ml). Overnight cultures were diluted 1:1000 in 10 ml of Mueller–Hinton broth (Difco) and incubated at 37°C with shaking. At OD600 0.3, ATc was added at a final concentration of 0.2 μM. At each time point (0, 15, 30, 60 min post-induction) the samples were 10-fold serially diluted in 96-well plates using sterile saline solution (0.9% NaCl) and 10 μl aliquots were spot-inoculated in Mueller–Hinton agar plates and incubated at 37°C overnight. Colonies were then counted.
MazF antibody production
The mazE antitoxin gene fused to its cognate toxin gene (i.e. the mazEF genes) was PCR amplified using a synthetic fragment as template (IDT company; Supplementary Table S1). The resulting fragment was digested by NdeI/ScaI and ligated into the expression vector pCWR547 (25) digested by the same restriction enzymes. The resulting plasmids were used to transform competent E. coli strains. All the constructions were verified by PCR and validated by sequencing. His6-SUMO-MazEF proteins were expressed from pCWR547 in E. coli C41 (DE3) strain and purified under standard native conditions using nickel chelate chromatography. The proteins were then excised from a 15% SDS polyacrylamide gel to immunize rabbits (Josman LLC, Napa, CA, USA). Polyclonal antibodies were obtained and used for MazF western blot analysis at a dilution of 1:2500 and revealed by HRP-conjugated goat-anti-rabbit secondary antibodies.
Protein extraction
Cells cultures were grown until exponential phase (O.D600 0.4–0.8). The induction of the MazF expression by ATc was performed for 10–60 min, as needed. Cultures of 10 ml with OD600 of 2–4 were collected and washed three times with 1 ml of phosphate-buffered saline (PBS). Cells were lysed in the presence of lysis buffer (LB) (PBS, 200 μg/ml lysostaphin, 200 μg/ml DNase I, protease inhibitors (Roche) 20 μl per 1 OD unit for 20 min at 37°C). Samples were chilled on ice, and sonicated 10 times with 30-s cycles using Cell Disrupter B-30 (Branson). Extracts were clarified by centrifugation for 10 min at 14 000 × g at 4°C. Total protein concentration was measured in supernatants by the Bradford protein assay. Samples were mixed with Laemmli sample buffer and analyzed by SDS polyacrylamide gel electrophoresis (SDS-PAGE) 16.5% tris–tricine peptide gels (BioRad). Gels were analyzed further by western blot using polyclonal antibodies described above.
RNA extractions for nEMOTE and RNA-seq
Overnight cultures were diluted 1:50 in 20 ml of Mueller–Hinton broth and incubated at 37°C without shaking until OD600 0.3. ATc was added to a final concentration of 0.2 μM and incubated at 37°C during 10 min. Aliquots of 8 ml of bacterial culture were immediately transferred to 40 ml of ice-cold ethanol:acetone (1:1), and centrifugated at 5000 rpm and 4°C during 10 min. The supernatant was discarded, and pellet was washed with 1× TE. The pelleted cells were treated with lysostaphin (200 μg/ml) and RNasin Plus (Promega) in a final volume of 100 μl TE then placed in a heat block at 37°C during 10 min. RNA extractions were done on the same day using the ReliaPrep (Promega) following the manufacturer's instructions. Total RNA was frozen and kept at –80°C.
nEMOTE library preparation and sequencing
RNA was quantified using a Nanodrop 1000 and integrity was assessed with a Bioanalyzer (Agilent Technologies). The nEMOTE protocol was employed as in Kirkpatrick, 2016 (26–28). Briefly, mono-phosphorylated RNA was digested from 8 μg of high-quality total RNA with XRN-1 (New England Biolabs). XRN-1 was removed with a phenol–chloroform–isoamyl alcohol 25:24:1 (Sigma) extraction, and the aqueous phase was recovered using MaXtract High Density (Qiagen) tubes. The recovered RNA was split into two pools, one pool was treated with T4 polynucleotide kinase (+PNK) and the other was subject to the same treatment without PNK (−PNK) and used to establish a background signal. Rp6 RNA oligonucleotide was ligated with T4 RNAse ligase to the 5’-phosphorylated RNA followed by an ethanol precipitation. Reverse transcription was carried out using semi-random primer DROAA and the cDNA was purified using Wizard SV Gel and PCR clean-up system (Supplementary Table S1) (Promega). Second-strand synthesis and barcode incorporation (D6A-D6Q barcodes) were performed using Q5 Hot Start High Fidelity DNA polymerase (New England Biolabs). The PCR products were purified (Qiagen PCR clean-up columns), mixed and size-selected between 300 and 1000 bp on agarose gels. The sample was then quantified by fluorometry using Qubit (Invitrogene) and sequenced in an Illumina HiSeq 2500 sequencer using 50 bp single-read cycle. Two independent replicates were performed.
RNA-seq library preparation and sequencing
Total RNA was quantified using Qubit (Life Technologies) and RNA integrity was assessed with a Bioanalyzer (Agilent Technologies). Total RNA (1 μg) was ribo-depleted with the bacterial Ribo-Zero kit from Illumina. The Truseq total RNA stranded kit from Illumina was used for the library preparation. Library quantity was measured by the Qubit and quality was assessed with a Tapestation on a DNA High sensitivity chip (Agilent Technologies). Libraries were pooled at equimolarity for clustering. Single-read sequencing (100 bases) was performed using the SBS chemistry on an Illumina HiSeq 4000 sequencer.
Bioinformatic analysis of nEMOTE, RNA-seq and annotation of targets
The nEMOTE reads were processed with the R package EMOTE v0.2 (https://github.com/pradosj/EMOTE). This package checks the quality of reads and parses them to extract the mapping sequence as well as the unique molecular identifier (UMI) sequence; it then aligns the reads to the reference genome S. aureus NCTC 8325-4 (with Rbowtie package) and quantifies the number of reads with the unique UMI starting at each genomic position. The genomic positions with less than 4 UMI in both ΔmazEF pRAB11-mazF+PNK replicates used, were not further considered. A beta-binomial model was then used to estimate the probability that +PNK condition is significantly enriched compared to the corresponding −PNK control condition (background noise removal). The P-values obtained for the two replicates are combined with Fisher's method, and the false discovery rate (FDR) computed to correct for multiple testing. To determine confident 5’-OH ends generated by MazF, each genomic position had to match the following criteria: an FDR <0.1 for ΔmazEF pRAB11-mazF condition, and an FDR >0.1 in ΔmazEF condition. Genomic positions mapping onto a rRNA feature were excluded. rRNA cleavage site analysis was performed separately with single rRNA sequences to avoid multiple mapping sites. The MazF cleavages were further annotated with genomic information (DNA sequence around the position, nearest gene), taken from both NCBI and Aureowiki (https://aureowiki.med.uni-greifwald.de/) (29).
The functional characteristics of the MazF-affected genes were obtained by mapping the cleavage sites to genes on the reference genome and performing gene ontology (GO) enrichment analysis using in-house scripts written in R programming language. The script retrieves GO terms annotations from Uniprot for the reference organism STAA8. Then, ancestors of the GO terms were inferred using « is_a » and « part_of » relationships of the ontology go-basic.obo available at http://geneontology.org. The analysis was corrected by identifying the GOs that are enriched in genes containing a MazF cleavage relatively to the 1000 most expressed genes on the genome. A hypergeometric test is performed to determine if a given GO term is enriched in the 260 MazF-affected genes out of the 1000 most expressed genes.
RNA-seq reads 500 bp before and after an nEMOTE-detected site were aligned on the reference genome with ‘bwa mem’ command and ‘samtools’ to generate files in BAM format. The BAM files are further processed to quantify the number of read in genes with the method ‘summarizeOverlaps(mode = ‘IntersectionStrict’,inter.feature = FALSE)’ from R package GenomicAlignments. Quantification around the cleavage sites were obtained with the method ‘coverage()’ of the R package ‘GenomicAlignments’.
Flow cytometry to determine dead and alive bacterial cells
Cell preparation was as above for nEMOTE and RNA-seq protocols. At 10 min postinduction, the cultures were centrifuged, and the pellet was washed three times with 0.2 μm filtered 1× PBS. Cells were standardized to McFarland 0.5 (1.5 × 108 CFU/ml) using a Densimat apparatus (bioMérieux) then diluted 1:100 in 1× PBS. The cells were stained using the BacLight RedoxSensor Green Vitality Kit (Invitrogen) following the manufacturer's instructions. Dead (propidium iodide permeable) and alive cells (positive for bacterial reductase activity) were then counted on a Gallios flow cytometer (Beckman Coulter). Results were analyzed using FlowJo v. 10.6.1 software.
Cell sorting of living bacterial cells
To purify live cells, we used the same cell preparation as above but using a MoFlo Astrios (Beckman–Coulter) cell sorter. Purified live cells were spot inoculated on Mueller Hinton agar plates. In parallel, an equal volume of 2× Mueller–Hinton broth and 10× the ciprofloxacin minimum inhibitory concentration (5 μg/ml) was added to live sorted cells and incubated at 37°C overnight (18 h) without shaking. Cells were then centrifuged at 8000 rpm for 3 min, supernatant was discarded, and cells were washed three times using filtered 1× PBS. Cells were stained again using the BacLight RedoxSensor Green Vitality Kit and dead and live cells were quantified on a Gallios flow cytometer. Results were analyzed using FlowJo v. 10.6.1 software. Significant difference between the strain carrying the empty plasmid (p) and the strain carrying the mazF-plasmid (pF) was analyzed using one-tailed, paired Student's t-test.
RNA extractions for qRT-PCR
Cells (1 ml) were collected at OD600 0.3, centrifuged for 3 min at 8000 rpm, supernatant was discarded, and the pellet was washed with 1 ml 1× TE buffer. The pellet was resuspended in 100 μl lysostaphin buffer composed of 200 μg/ml lysostaphin, 200 μg/ml DNase I, 40 U RNasin Plus Ribonuclease inhibitor (Promega) in 1× TE buffer and placed at 37°C for 10 min. RNA was then extracted using RNeasy Plus Mini Kit (Qiagen) following the manufacturer's instructions and adding two modifications: cell homogenization was performed with QIAshredder (Qiagen) columns after adding RTL Plus lysis buffer and DNase treatment using RNase-Free DNase Set (Qiagen) was applied after the first column wash.
Quantitative reverse transcription PCR (qRT-PCR)
Primers and probes were designed using PrimerExpress software (Applied Biosystems) and obtained from Eurogentec. Absence of DNA contamination in total RNA extractions was assessed by qPCR using 2× Takyon probe assay (Eurogentec). Optimal concentration of primers and TaqMan® hydrolysis probes was assessed by using 4 ng of total RNA per well and a final concentration of primers of probes between 0.05 and 0.2 μM to determine an efficiency of 100% (amplification factor of 2). mRNA levels of each gene were determined by quantitative reverse transcription PCR (qRT-PCR) using the Platinum qRT-PCR ThermoScript One-Step system (Invitrogen). The mRNA levels of each gene were normalized to gyrB RNA levels, which were assayed in each round of qRT-PCR as internal controls. To quantify qRT-PCR data the 2–ΔΔCt method was used (30), where fold change of target gene expression in a target (treated) samples relative to a reference (non-treated) samples was normalized to the reference gene gyrB. Thus, the relative gene expression in non-treated samples was set to 1. The errors for the ΔΔCt were obtained by least square error propagation of the standard deviation for the individual qRT-PCR measurements performed in triplicates. The statistical significance of strain-specific differences in normalized cycle threshold (CT) values of each transcript was evaluated by paired Student's t-test, and data were considered significant when P was <0.05.
RESULTS
Effect of mazF overexpression on S. aureus growth
To identify sites targeted and cleaved by MazF endoribonuclease in vivo, we applied conditional overexpression of MazF toxin in S. aureus. We first confirmed that our MazF expression plasmid induces the characteristic growth defect observed upon mazF overexpression in S. aureus (21). Since MazF toxin activity is mitigated in the presence of MazE antitoxin, we analyzed the effect of mazF overexpression in the presence and absence of MazE antitoxin using wild-type, trfA-deleted (a condition previously shown to stabilize MazE antitoxin by reducing its proteolytic turnover (17)) or mazEF-deleted strains (20). All strains were transformed with a tightly regulated anhydrotetracycline (ATc) inducible plasmid carrying MazF toxin gene (pF) (Supplementary Table S2) and western blot analysis confirmed MazF protein (13.4 kDa) upon ATc induction (Figure 1A). In control strains lacking pF plasmid, MazF was not detected in the mazEF-deleted strain while MazF was detected at low levels in both the wild-type and trfA-deleted strain (as expected easily observed upon over-exposure of the immunoblot detection system, Supplementary Figure S1).
Figure 1.
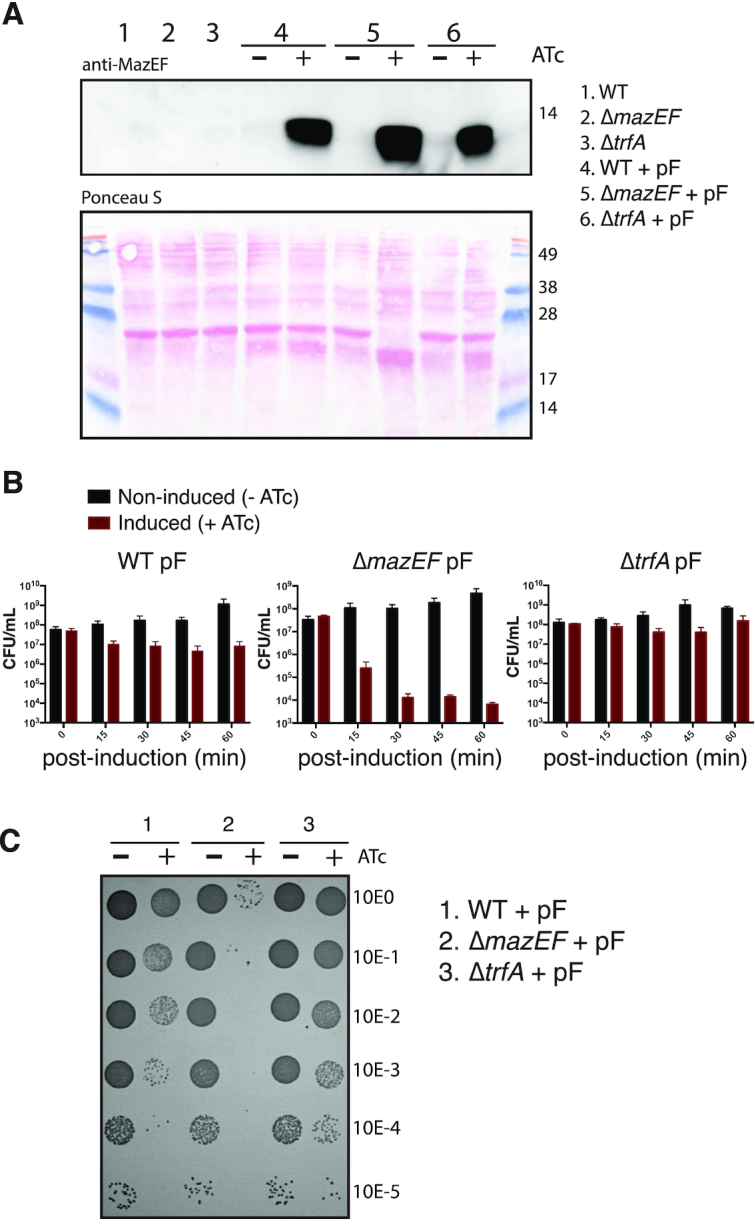
(A) Western blot analysis of MazF protein produced in S. aureus strains with or without the mazF overexpression plasmid (pF). After mazF gene induction with anhydrotetracycline (+ATc) or in non-induced cells (−ATc), total soluble protein extracts from S. aureus strains were loaded in SDS 16.5% polyacrylamide gels and MazF protein (13.4 kDa) was detected using a rabbit-polyclonal anti-MazEF antibody (top) and Ponceau S staining (bottom). A Coomassie Brilliant Blue stain on same samples loaded above is shown in Supplementary Figure S2. (B) Effect of mazF overexpression on S. aureus growth. Colony forming units (CFU) counts at five time points (between 0 and 60 min) after mazF-induced (+ATc) or uninduced (−ATc) S. aureus strains. Data are represented as mean ± SD of three independent experiments. (C) Representative image showing the spot (10 μl) serial dilutions of bacterial cultures on agar plates, after 60 min of mazF-induced (+ATc) and uninduced (−ATc) S. aureus strains. Serial dilutions are indicated at the right margin. CFU counts of S. aureus carrying an empty vector with and without ATc is shown in Supplementary Figure S3.
We observed a decrease in bacterial colony counts (efficiency of plating) at different time points after mazF overexpression (Figure 1B and C). This effect was enhanced in the absence of MazE antitoxin. Stabilization of MazE antitoxin through deletion of the ClpCP adaptor protein TrfA abolished the growth defect caused by the induction of MazF expression and restored colony formation comparable to wild-type control strain (Figure 1B and C). Collectively, these results show that MazF can be conditionally expressed and such cells show reduced ability to form colonies on agar plates in accordance with Fu et al. (21).
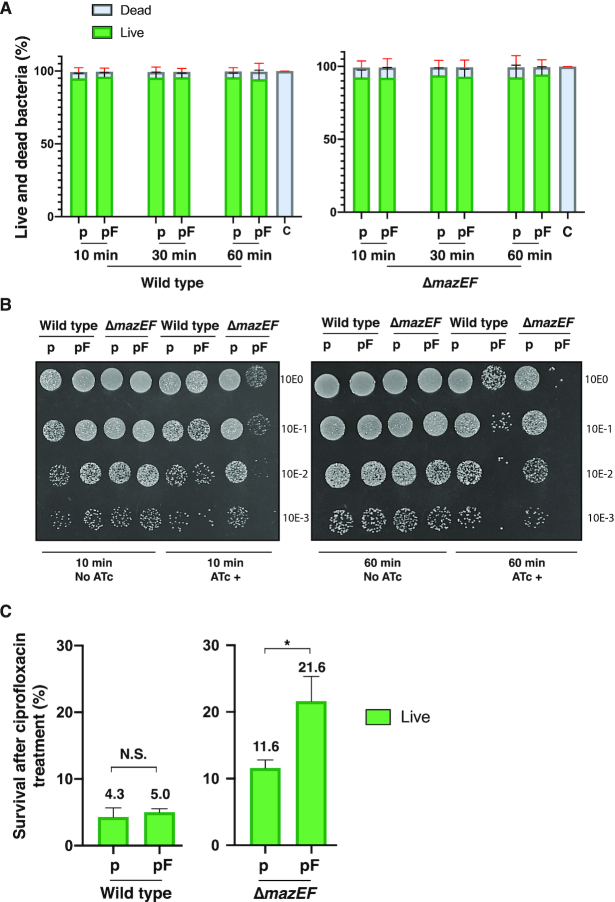
To further analyze the effect of mazF overexpression on growth, bacterial populations with and without mazF induction and in the presence (wild-type) and absence of MazE antitoxin (mazEF-deleted strain) were analyzed by flow cytometry to quantify cell viability. Interestingly, no difference in the percentage of live (RedoxSensorGreen labeled) or dead (propidium iodide labeled) bacteria was observed after 30 or 60 min of MazF induction in either the presence or absence of MazE antitoxin (Figure 2A). In all conditions tested, we counted 50 000 cells and detected overall 95% live and 5% dead bacteria. These live/dead measurements are in sharp contrast with the absence of detectable bacterial colony counts observed after 30 or 60 min of MazF induction (Figure 1C). These observations suggest that after MazF induction, bacteria remain alive but cannot divide, resulting in reduced colony formation observed on agar plates.
Figure 2.
(A) Cell cytometry analysis showing the percentage of live and dead bacteria present after mazF overexpression. S. aureus wild-type and mazEF-deleted strains carrying a control (p) or mazF (pF) plasmid were subject to ATc induction during time. Bacteria were subsequently labeled using RedoxSensorGreen and propidium iodide. Live and dead bacterial counts were determined by fluorescence detection using cell cytometry. Fluorescence detection of heat-killed bacteria was used as a control (c). The percentage of live or dead bacteria from a total of 50 000 bacterial counts is reported. Data is represented as mean ± SD of three independent experiments. (B) Bacterial growth of sorted live bacteria after mazF overexpression. Wild-type and mazEF-deleted strains carrying a control plasmid (p) or a mazF plasmid (pF) were treated or not with ATc. After 10 min (left panel) or 60 min (right panel) of ATc treatment, bacteria were washed, labeled and sorted to obtain 8 × 105 live cells and were serially diluted (top to bottom 100 to 10−3), 10 μl of diluted bacteria were immediately spot-inoculated on Mueller–Hinton agar plates. (C) Wild-type and mazEF-deleted strains carrying a control (p) or a mazF (pF) plasmid were treated with ATc during 10 min. Cell were labeled and sorted and a purified live population of bacteria (8 × 105 cells) was exposed to ciprofloxacin overnight. The remaining bacteria were again labeled, and live cells were counted by cell cytometry. Results are presented as means ± SD of three independent experiments; N.S. is not significant; * P < 0.05 significant differences between the strain carrying the empty (p) or mazF (pF) plasmid.
To further confirm that after MazF induction bacteria were alive but unable to divide, a second analysis was performed by sorting a fixed number of live bacteria (8 × 105) from each condition: MazF induced or uninduced cultures in the presence or absence of MazE antitoxin. Sorted live bacteria were immediately spotted on agar plates (Figure 2B). In the absence of MazE antitoxin, induction of MazF in the ΔmazEF strain, we observed that despite viability measured by FACS analysis, bacteria were unable to divide to form visible bacterial colonies on agar plates. In contrast, live bacteria after MazF induction but carrying MazE antitoxin could divide and give rise to visible bacterial colonies on agar plates. We conclude from these analyses that under our conditions mazF overexpression induced bacterial growth arrest corroborating the characteristic mazF overexpression phenotype in S. aureus.
We next verified that under our conditions of MazF induction, bacteria were indeed in a growth arrested state by evaluating bacterial survival when challenged with the bactericidal antibiotic ciprofloxacin. As antibiotics targeting active growth processes kill less efficiently during growth arrest (31,32), we expect an increased survival to antibiotic treatment upon mazF overexpression. Wild-type and ΔmazEF cells carrying mazF-expression plasmids or control plasmids, were induced with ATc. Ten minutes post induction (mpi), cells were labeled, and the population was sorted to obtain 8 × 105 live bacteria. These live cells were exposed to 10X the ciprofloxacin minimum inhibitory concentration (MIC) overnight. After washing, the remaining bacteria were again labeled, and live and dead cells were determined by flow cytometry. In the presence of MazE antitoxin (wild-type strain background), we recovered ∼5% of the original live population and no significant difference was found between the population carrying an empty plasmid or overexpressing mazF. In contrast, upon mazF overexpression in the absence of the antitoxin (ΔmazEF strain background), we recovered on average nearly twice as many cells (21.6%) of the original population than in the population carrying an empty plasmid (11.6%) (Figure 2C). These results suggest that induction of active MazF in the absence of MazE antitoxin increases bacterial survival towards the antibiotic ciprofloxacin. All together, these results confirm that MazF induction in our experimental conditions induces bacterial growth arrest.
MazF cleavage sequence recognition identified by nEMOTE
To determine the molecular link between mazF overexpression and the observed growth arrest, we identified MazF-cleaved targets on genome-wide scale using nEMOTE (non-phosphorylated exact mapping of transcriptome ends) coupled to high-throughput sequencing analysis. The nEMOTE procedure detects toxin endoribonuclease cleavages that produce 5’-OH transcript-ends in vivo and in the native host. Briefly, total RNA was isolated from mazEF-deleted strain carrying the overexpression plasmid with or without mazF gene following a 10 min treatment with ATc. The nEMOTE protocol was then applied to enrich for 5’-OH transcripts-ends and the resulting library was sequenced. MazF specific cleavage sites were identified by comparing 5’-ends enriched in mazF induced strain compared to the control condition after elimination of background noise using internal nEMOTE controls (see materials and methods).
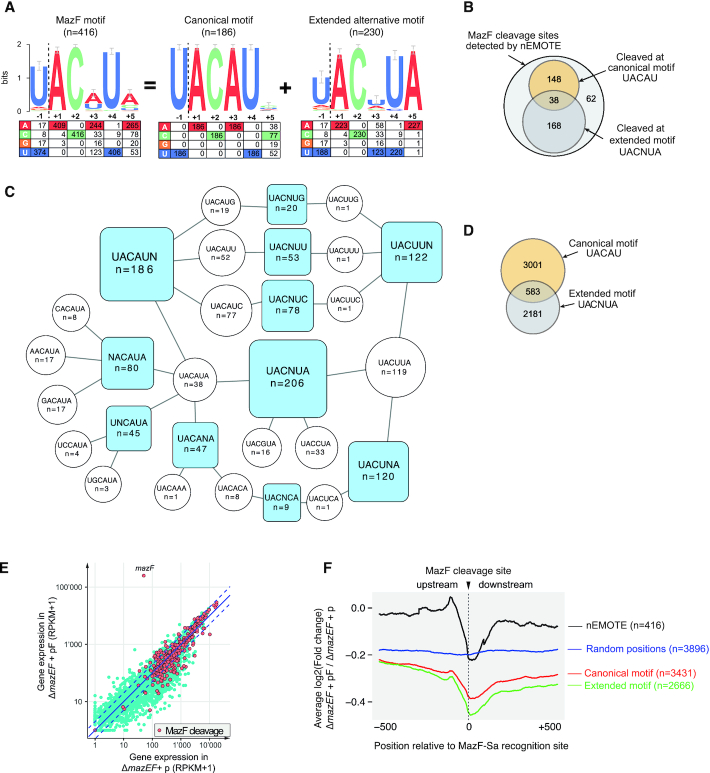
Our nEMOTE analysis detected 416 different RNA sites cleaved by MazF toxin (Supplementary Table S3) that we have precisely mapped on the S. aureus NCTC 8325 genome. The sequence logo plot of the 416 RNA sequences around the detected cleavage sites is shown in Figure 3A. The experiment recovered the MazF canonical sequence recognition U^ACAU previously identified in vitro for coa and spa genes (20). This strict canonical motif explains only 44.7% (n = 186) of all detected MazF cleavages, leaving other cleavage sites showing a variation in the motif at +3 position. The remaining sequences display an alternative extended MazF recognition sequence (motif U∧ACNUA). Compared to the canonical motif, the alternative extended motif shows a decrease in base conservation at positions –1 and +3 and concomitantly, a conserved +5 position emerges (Figure 3A). As shown in Figure 3B, this alternative motif overlaps with the canonical motif because the RNA sequence UACAUA (cleaved n = 38 times) matches both patterns (i.e. UACAU and UACNUA). The extended motif UACNUA was identified 168 times, although other variations were also detected (n = 62). To better understand the exact cleavage sites that compose the motifs and their overlap, we show in Figure 3C motifs with a single variable position (blue squares) linked to the cleavage site sequence, and the number of times observed (white circles), which explain the motif.
Figure 3.
(A) Logo plot obtained from the alignment of the 416 MazF cleavage sites detected by nEMOTE and the base frequency at each position is indicated in the table below. A logo plot with the strict canonical motif (previously identified) was extracted from the 416 MazF cleavage sites revealing a second extended alternative motif. (B) Venn diagram showing the number of MazF cleavage sites detected by nEMOTE. (C) Diagram showing motifs with a single variable position (blue square) and the cleaved sequences composing each blue motif are shown in white circles. (D) Venn diagram showing the number of predicted canonical and extended alternative MazF recognition sequence along the S. aureus transcriptome based on the nEMOTE results. (E) Overlap of RNA-seq global expression profile (blue dots) of mazEF–deleted strain overexpressing mazF (pF) and control (p) and nEMOTE cleavage sites (orange dots). (F) RNA-seq read coverage profiles 500 bp before and after the nEMOTE-detected sites between mazEF-deleted strain overexpressing or not mazF (log2mazEF-pF/mazEF-p). Profiles obtained around the MazF cleaved sites detected by nEMOTE (black line), around nEMOTE predicted canonical (red line) and alternative extended (green line) sites and around random genome positions (blue line). The y-axis positioning of the curves reflects the average impact of MazF activity on the genes.
The MazF cleavage motifs determined by nEMOTE allow prediction of other potential MazF cleavage sites by bioinformatics. Transcriptome-wide scan of the S. aureus 2.4 MB genome for canonical and extended alternative motifs, predicted a total of 3584 and 2181 potential MazF cleavage sites (Figure 3D), but nEMOTE detected 186 and 230 cleaved sites, respectively (Figure 3B). Although potential cleavage sites are widespread throughout the transcriptome, we were able to positively identify just a small fraction of these sites. We did not expect all recognition sequences to be cleaved because other factors involved might impede or determine cleavage at these sites (33). They may be also localized in genes with low expression levels that would avoid capturing by the nEMOTE technique.
RNA-seq expression profile and cleavage sites
To further validate and evaluate the limits of detection of the nEMOTE technique in our experimental conditions, we correlated RNA-seq expression data with the identified cleavage sites. RNA-seq was conducted on ΔmazEF strains carrying mazF overexpression plasmid or the empty vector. A global comparison of gene expression between the two conditions indicates an overall decrease of expression when mazF is overexpressed in the cell (Figure 3E and Supplementary Table S4). By overlapping the expression levels and the 416 nEMOTE cleavage sites we observed that cleavages are mostly detectable for highly expressed genes.
We reasoned that we would detect fewer reads across a MazF cleavage site in the presence of MazF than in its absence. We analyzed the RNA-seq read coverage profiles 500 bp before and after a nEMOTE-detected site between mazEF deleted strain carrying or not mazF overexpression plasmid (log2mazEF-pF/mazEF-p) (see Material and Methods). Aggregated analysis of transcriptomic regions containing MazF sites detected by nEMOTE shows that 500 bp before the cleavage site there is no discernible read count changes between the strain carrying or not the MazF toxin, while a decrease in read counts is observed approaching nEMOTE MazF cleavage site (black line, Figure 3F). In contrast, no decrease was observed in genomic regions without detected cleavage by MazF or lacking predicted MazF cleavage motifs (blue line, Figure 3F). The analysis of regions containing the predicted canonical (red line, Figure 3F) or alternative MazF motifs (green line, Figure 3F) similarly shows a decrease in fragment ratios approaching nEMOTE MazF cleavage site. These results suggest that MazF is also cleaving sites we did not detect by nEMOTE because the expression level was too low or because of biases in labeling the 5′-OH ends in the nEMOTE procedure. RNA-seq data corroborated the identified nEMOTE cleavage sites by clearly showing a marked decrease in number of reads crossing MazF cleavage sites.
MazF cleaves motifs present in coding and non-coding regions
To further characterize MazF toxin cleavage sites, we analyzed the position of nEMOTE-detected cleavages relative to coding sequences (CDSs). As shown in Supplementary Figure S4A, MazF recognizes RNA sequences present in coding (n = 372) or non-coding (n = 44) regions with a higher frequency in coding regions. From the 44 cleavages in non-coding regions, 31 and 13 are located in 5′ promoter or 3′ regions, respectively. Moreover, MazF cleaves both canonical and alternative sites in all three reading frames (In-frame or 0, frame +1 or frame +2). As shown in Supplementary Figure S4B, no preference of cleavage was observed taking into account the relative position of the motif inside a gene. This analysis shows that MazF cleaves inside and outside open reading frames and does not show a reading frame preference.
Global MazF target cleavage and its correlation with S. aureus growth arrest
To understand the molecular link between mazF overexpression and the observed growth defect, we analyzed MazF targets according to their global functional role. An overview of nEMOTE MazF cleaved targets highlights the effect on different mRNAs that are transcribed from essential or non-essential genes corresponding to several metabolic pathways involved in mRNA turnover, translation, transcription, replication, cell division, cell wall metabolism among others (Supplementary Table S3).
The functional characteristics of the MazF-affected genes were obtained by performing gene ontology (GO) enrichment analysis. To avoid the functional analysis to reflect the function of highly expressed genes instead of MazF-affected genes, we limited the analysis to the 1000 most expressed genes, according to the gene-expression given by the ΔmazEF RNA-seq data (measured as read per kilobase pair per million), and assume that the presence of a cleavage is equally likely to be detected in each of them. Out of the 315 genes affected by a MazF cleavage (Supplementary Table S3), 260 (82%) are in the top 1000 most expressed genes. The analysis using gene ontology biological process classifications showed that the most commonly found annotations were metabolic process resulting in cell growth (GO:0008152) and related categories such as generation of precursor metabolites and energy (GO:00006091), ADP metabolic process (GO:0046031) and ATP generation from ADP (GO:0006757). Similarly, molecular function classifications identified affected categories such as RNA binding (GO:0003723), translation factor activity, RNA binding (GO:0008135), endoribonuclease activity (GO:00004521) and translation elongation factor activity (GO:00003746), among others. The complete set of GO enriched categories can be found in Supplementary Table S6.
The increased interest to understand bacterial growth has also pinpointed protein synthesis as an important metabolic pathway to regulate growth (34–36). Interestingly, MazF is known to affect translation-related genes in other species (16). Accordingly, we observed by nEMOTE (Table 1) that in our conditions, MazF cleaves several transcripts involved in protein synthesis. We found transcripts cleaved by MazF at canonical or alternative sequence recognition sites that are present once or multiple times per transcript (Table 1). RNA-seq confirmed the MazF cleavage of translation-related transcripts. Fragment counts between ΔmazEF strain carrying or not mazF overexpression plasmid (log2mazEF-pF/mazEF p), show a clear decrease in fragment ratios detected at or near nEMOTE MazF cleavage sites (Figure 4). More specifically, MazF cleaves transcripts encoding translation initiation factors (IF-1 and IF-2), translation elongation factors (Ts, P, Tu, G), termination factors (peptide release factors 2 and 3), several tRNA synthetases required for amino acid loading on tRNAs, an uracil tRNA methyltransferase required for tRNA maturation and a rlmB methyltransferase affecting the posttranscriptional modification of 23S rRNA essential for peptidyl-tRNA ribosomal recognition. Moreover, as recently observed for E. coli (16), we observed MazF cleavage on several ribosomal protein-related mRNAs (Table 1) that predict altered ribosomal protein production that will disrupt ribosome biogenesis. We identified canonical and alternative cleavage sites in 5S, 16S and 23S rRNAs that would also predict to affect ribosome biogenesis (Supplementary Table S5).
Table 1.
nEMOTE detected cleavage sites in genes involved in protein synthesis
| Gene | Gene number | Gene descriptiona | MazF motif and localizationb | GO categoryc |
|---|---|---|---|---|
| rlmB | SAOUHSC_00513 | 23S rRNA (guanosine(2251)-2′-O)-methyltransferase RlmB | [290/UACUUA] | RNA methylation [GO:0001510] |
| rplK | SAOUHSC_00518 | 50S ribosomal protein L11 | [233/UACUUA] | Structural constituent of ribosome [GO:0003735] |
| rplA | SAOUHSC_00519 | 50S ribosomal protein L1 | []104/UACCUA | Structural constituent of ribosome [GO:0003735] |
| rpsG | SAOUHSC_00528 | 30S ribosomal protein S7 | 5/UACAUU[] | Structural constituent of ribosome [GO:0003735] |
| SAOUHSC_00580 | SAOUHSC_00580 | Hypothetical protein | [9/UACAUU] | n/a |
| prfB | SAOUHSC_00771 | Peptide chain release factor 2 | [492/UACAUU] | n/a |
| trpS | SAOUHSC_00933 | Tryptophanyl-tRNA synthetase | [372/AACAUA] | tRNA aminoacylation for protein Translation [GO:0006418] |
| prfC | SAOUHSC_00956 | Peptide chain release factor 3 | [590/UACAUU] | Regulation of translational Termination [GO:0006449] |
| pheS | SAOUHSC_01092 | Phenylalanyl-tRNA synthetase subunit alpha | [100/UACUUA] | Phenylalanyl-tRNA aminoacylation [GO:0006432] |
| ileS | SAOUHSC_01159 | Isoleucyl-tRNA synthetase | [188/UACAUA] | n/a |
| rpsP | SAOUHSC_01208 | 30S ribosomal protein S16 | [114/UACUUA] | Ribosome [GO:0005840] |
| Gid | SAOUHSC_01223 | tRNA (uracil-5-)-methyltransferase Gid | [526/UACUUA] | tRNA wobble uridine modification [GO:0002098] |
| infB | SAOUHSC_01246 | Translation initiation factor IF-2 | [1257/UACUUA] | Translational initiation [GO:0006413] |
| rpsA | SAOUHSC_01493 | 30S ribosomal protein S1 | [253/UACAUC] | Structural constituent of ribosome [GO:0003735] |
| Efp | SAOUHSC_01625 | Elongation factor P | [310/UACUUA] | translation elongation factor activity [GO:0003746] |
| aspS | SAOUHSC_01737 | Aspartyl-tRNA synthetase | [1362/UACAUC] | Aspartyl-tRNA aminoacylation [GO:0006422] |
| queA | SAOUHSC_01749 | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | [457/UACAUC] | n/a |
| valS | SAOUHSC_01767 | valyl-tRNA synthetase | [229/UACUUA] | Valyl-tRNA aminoacylation [GO:0006438] |
| rpsD | SAOUHSC_01829 | 30S ribosomal protein S4 | [175/UACUUA] | Structural constituent of ribosome [GO:0003735] |
| leuS | SAOUHSC_01875 | Leucyl-tRNA synthetase | [575/UACUUA] | Aminoacyl-tRNA editing activity [GO:0002161] |
| SAOUHSC_02248 | SAOUHSC_02248 | Hypothetical protein | [417/UACAUC] | n/a |
| SAOUHSC_02297 | SAOUHSC_02297 | S1 RNA-binding domain-containing protein | [1662/UACAUC] | Structural constituent of ribosome [GO:0003735] |
| rpsI | SAOUHSC_02477 | 30S ribosomal protein S9 | [314/UACUUA] | Structural constituent of ribosome [GO:0003735] |
| rpsE | SAOUHSC_02494 | 30S ribosomal protein S5 | 2/UACAUG[] | Structural constituent of ribosome [GO:0003735] |
| rplX | SAOUHSC_02501 | 50S ribosomal protein L24 | [2/UGCAUA] | Structural constituent of ribosome [GO:0003735] |
| rplP | SAOUHSC_02505 | 50S ribosomal protein L16 | [169/UACAUG] | Structural constituent of ribosome [GO:0003735] |
| rpsC | SAOUHSC_02506 | 30S ribosomal protein S3 | [570/UACUUA] | Structural constituent of ribosome [GO:0003735] |
| SAOUHSC_02512a | SAOUHSC_02512a | 30S ribosomal protein S10 | [145/UACACA] | n/a |
| SAOUHSC_02519 | SAOUHSC_02519 | Hypothetical protein | [406/UACAUA] | n/a |
| SAOUHSC_02827 | SAOUHSC_02827 | Hypothetical protein | [167/UACUUA] | n/a |
| rpmH | SAOUHSC_03055 | 50S ribosomal protein L34 | [12/UACUUA] | Structural constituent of ribosome [GO:0003735] |
| rpsF | SAOUHSC_00348 | 30S ribosomal protein S6 | [6/AACAUA] [22/UACAUC] | Structural constituent of ribosome [GO:0003735] |
| fusA | SAOUHSC_00529 | Elongation factor G | 2/UACAUG[] [829/UACUUA] | GTPase activity [GO:0003924] |
| SAOUHSC_01091 | SAOUHSC_01091 | SpoU rRNA Methylase family protein | [665/UACCUA] []186/UACUUA | n/a |
| Tsf | SAOUHSC_01234 | Elongation factor Ts | [466/UACUUA] [548/UGCAUA] | Translational elongation [GO:0006414] |
| asnC | SAOUHSC_01471 | Asparaginyl-tRNA synthetase | [789/UACAUC] [559/UACUUA] | Asparaginyl-tRNA aminoacylation [GO:0006421] |
| queG | SAOUHSC_01989 | Hypothetical protein | [867/AACAUA] [839/UACGUA] | Epoxyqueuosine reductase activity [GO:0052693] |
| infA | SAOUHSC_02489 | Translation initiation factor IF-1 | [201/UACUUA] [130/UACAUU] | Translation initiation factor activity [GO:0003743] |
| rplN | SAOUHSC_02502 | 50S ribosomal protein L14 | []15/UACAAA [226/UACAUC] | Structural constituent of ribosome [GO:0003735] |
| cysS | SAOUHSC_00511 | Cysteinyl-tRNA synthetase | [540/UACAUU] [665/UACAUG] [1017/GACAUA] | Cysteinyl-tRNA aminoacylation [GO:0006423] |
| rpsL | SAOUHSC_00527 | 30S ribosomal protein S12 | [185/UACGUA] [235/UACAUC] [251/GACAUA] | Structural constituent of ribosome [GO:0003735] |
| rpsS | SAOUHSC_02508 | 30S ribosomal protein S19 | [234/UACAUU] [137/GACAUA] 25/CACAUA[] | Structural constituent of ribosome [GO:0003735] |
| Tuf | SAOUHSC_00530 | Elongation factor Tu | [591/UACUUA] [595/UACAUU] [743/UACAUG] [928/UACGUA] | Translational elongation [GO:0006414] |
| rplC | SAOUHSC_02512 | 50S ribosomal protein L3 | [324/UACUUU] | Structural constituent of ribosome [GO:0003735] |
| tadA | SAOUHSC_00541 | Hypothetical protein | []8/UACAUA | n/a |
| trnaT | SAOUHSC_T00055 | tRNA-Thr | 7/UACAUA[] | n/a |
| hemA | SAOUHSC_01776 | Glutamyl-tRNA reductase | [455/CACAUA] | Glutamyl-tRNA reductase activity [GO:0008883] |
| SAOUHSC_00526 | SAOUHSC_00526 | 50S ribosomal protein L7Ae-like protein | [90/UACAUC] | Ribosome [GO:0005840] |
| ychF | SAOUHSC_00346 | GTP-dependent nucleic acid-binding protein EngD | [152/UACUUA] [846/AACAUA] | Ribosome binding [GO:0043022] |
| hflX | SAOUHSC_01283 | Hypothetical protein HflX | [422/UACCUA] | Ribosome binding [GO:0043022] |
aGene product as annotated in Aureowiki for NCTC8325 genome
bBrackets represent the ORF; number inside brackets indicate The MazF cleavage position relative to the ATG codon; while brackets before or after the cleavage position indicates cleavage outside the ORF, downstream or upstream respectively.
cComplete gene ontology enrichment analysis in Supplementary Table S6.
Figure 4.
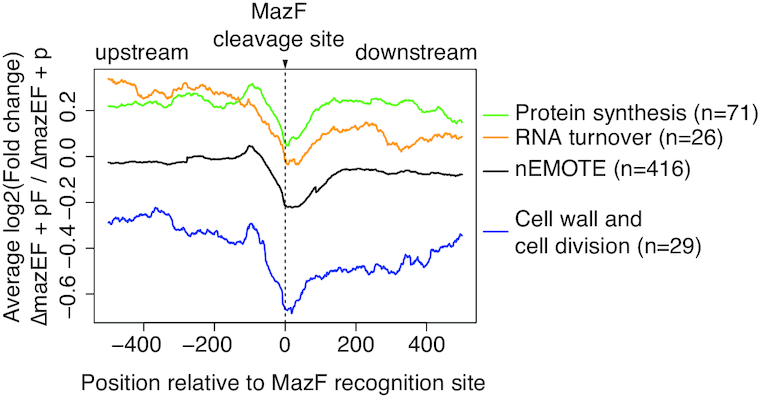
RNA-seq reads coverage profiles 500 bp before and after an nEMOTE-detected cleavage site between mazEF-deleted strain overexpressing or not mazF (log2 mazEF-pF/mazEF-p). Profiles obtained around MazF cleaved sites on protein synthesis genes (green line), RNA turnover genes (yellow line), cell wall and cell division-related genes (blue line). The complete set of nEMOTE-detected sites are shown for comparison (black line).
Another set of important transcripts that we identified in our nEMOTE analysis and confirmed by RNA-seq data are those encoding cell division and cell wall synthesis genes. These two cellular processes drive growth of the cell envelope and thus reproduction. As such, defects in these pathways may lead to growth arrest and a reduction in colony formation. As shown in Table 2 and Figure 4, division-related transcripts of ftsA and ftsZ that are required for Z-ring formation, the pbp1 and pbp2 transcripts, the divIC late division transcript, the mraZ transcript encoding and transcriptional regulator of cell division, the sle1 transcript encoding an autolysin and lytM transcript encoding a cell wall hydrolase are detected as MazF-cleaved messages. Peptidoglycan synthesis or teichoic acid-related transcripts including fmhB, uppS, glmM, femAB, murE and murA2, murI, mprF (fmtC), dltA, oatA, tagB and tagG are detected (37–40). Cell morphology-related mRNAs such as ssaA and alr and the division localized mreD transcripts were found to be cleaved by MazF. The link of mreD cleavage and cell division is unknown as recently it has been shown that mreD has no effect on cell morphology, cell volume or peptidoglycan composition (41).
Table 2.
nEMOTE detected cleavage sites in genes involved in cell wall and cell division
| Gene | Gene number | Gene descriptiona | MazF motif and localizationb | GO categoryc |
|---|---|---|---|---|
| Alr | SAOUHSC_02305 | Alanine racemase | [839/UACCUA] | Alanine racemase activity [GO:0008784] |
| capF | SAOUHSC_00119 | Capsular polysaccharide biosynthesis protein Cap8F | [988/GACAUA] | Catalytic activity [GO:0003824] |
| capJ | SAOUHSC_00123 | Capsular polysaccharide biosynthesis protein Cap5J | [434/UACCUA] | n/a |
| divIC | SAOUHSC_00482 | Hypothetical protein | [313/UACUUA] | n/a |
| dltA | SAOUHSC_00869 | d-Alanine–poly(phosphoribitol) ligase subunit 1 | [1077/UACAUA] | Cell wall organization or biogenesis [GO:0071554] |
| femA | SAOUHSC_01373 | Methicillin resistance factor FemA | [993/UACAUC] | n/a |
| femB | SAOUHSC_01374 | Methicillin resistance factor | [453/UACAUC] | Cell wall organization or biogenesis [GO:0071554] |
| fmhB | SAOUHSC_02527 | Peptidoglycan pentaglycine interpeptide biosynthesis protein FmhB | [952/UACUUA] | Cell wall organization or biogenesis [GO:0071554] |
| fmtC | SAOUHSC_01359 | Hypothetical protein | [2089/UACUUA] | Lysyltransferase activity [GO:0050071] |
| ftsA | SAOUHSC_01149 | Cell division protein | [1257/UACUCA] [1322/AACAUA] | Cell division site [GO:0032153] |
| ftsZ | SAOUHSC_01150 | Cell division protein FtsZ | [477/UACAUU] | Cell division site [GO:0032153] |
| glmM | SAOUHSC_02405 | Phosphoglucosamine mutase | [446/UACAUU] | Cell wall organization or biogenesis [GO:0071554] |
| lytM | SAOUHSC_00248 | Peptidoglycan hydrolase | [237/UACAUU] | Cell wall organization or biogenesis [GO:0071554] |
| mraZ | SAOUHSC_01142 | Cell division protein MraZ transcriptional regulator | [173/UACCUA] | Regulation of biosynthetic process [GO:0009889] |
| mreD | SAOUHSC_01758 | Cell-wall related protein | []30/UACAUA | n/a |
| murA2 | SAOUHSC_02365 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | [470/CACAUA] | Cell wall organization or biogenesis [GO:0071554] |
| murE | SAOUHSC_00954 | UDP-N-acetylmuramoylalanyl-d-glutamate-l-lysine ligase | [1330/GACAUA] | Cell wall organization or biogenesis [GO:0071554] |
| murI | SAOUHSC_01106 | Glutamate racemase | [94/UACUUA] | Cell wall organization or biogenesis [GO:0071554] |
| oatA | SAOUHSC_02885 | O-Acetylation of cell-wall peptidoglycan | [49/UACUUA] | Integral component of membrane [GO:0016021] |
| pbp1 | SAOUHSC_01145 | Penicillin-binding protein 1 | [1095/UACUUA] | Cell wall organization or biogenesis [GO:0071554] |
| pbp2 | SAOUHSC_01467 | Penicillin-binding protein 2 | [1660/UACAUG] [1761/UACUUA] | Cell wall organization or biogenesis [GO:0071554] |
| sle1 | SAOUHSC_00427 | Autolysin | [753/UACUUA] | Cell wall organization or biogenesis [GO:0071554] |
| ssaA | SAOUHSC_02576 | Secretory antigen SsaA, autolysin | [159/UACAUC] | n/a |
| tagB | SAOUHSC_00643 | Teichoic acid biosynthesis protein TagB | [304/UACCUA] [469/UACUUA] | n/a |
| tagG | SAOUHSC_00642 | Teichoic acid biosynthesis protein | [431/UACAUA] | n/a |
| uppS | SAOUHSC_01237 | Undecaprenyl pyrophosphate synthase | [211/UACUUA] | Isoprenoid biosynthetic process [GO:0008299] |
aGene product as annotated in Aureowiki for NCTC8325 genome
bBrackets represent the ORF; number inside brackets indicate the MazF cleavage position relative to the ATG codon; brackets before or after the cleavage position indicates cleavage outside the ORF, downstream or upstream respectively.
cComplete gene ontology enrichment analysis in Supplementary Table S6.
The regulation of mRNA turnover can be an important pathway to modulate the transcriptional demand that must be imposed to establish a bacterial growth stasis. Deletion of mRNA degradation genes has been shown to reduce growth, suggesting that mRNA turnover is an essential process for growth (42–44). From the 31 genes annotated as involved in transcription, MazF was found to cleave 12 important mRNAs coding for both RNA polymerase subunits B and B’ (rpoB and rpoC), termination factor rho together with endo and exoribonucleases such as RNase III (mrnc) and RNase R (rnr) respectively, mRNAs coding for CshA, J1, J2 and RNase Y proteins that are part of the S. aureus RNA degradation machinery (cshA, rnjA, rnjB and rny genes), regulating RNA decay in bacteria (Table 3). We found canonical or alternative sequence recognition sites and the majority are found cleaved inside open reading frames. To visualize the MazF-dependent cleavage of mRNA turnover transcripts, we again used RNA-seq technique as described above. As shown in Figure 4, a decrease in the number of sequenced reads across the MazF cleavage site is observed in cells expressing MazF compared to cells in which MazF is not expressed, showing a MazF-dependent cleavage of selected mRNA turnover genes.
Table 3.
nEMOTE detected cleavage sites in genes involved in RNA turnover
| Gene | Gene number | Gene descriptiona | MazF motif and localizationb | GO categoryc |
|---|---|---|---|---|
| nusG | SAOUHSC_00517 | Transcription antitermination protein | [42/UACAUA] | Regulation of transcription, DNA-templated [GO:0006355] |
| rnr | SAOUHSC_00803 | Ribonuclease R | [1677/UACAUC] | 3′-5′ exonuclease activity [GO:0008408] |
| nusA | SAOUHSC_01243 | Transcription elongation factor NusA | [413/UACUUA] | Transcription regulator activity [GO:0140110] |
| rnjB | SAOUHSC_01252 | RNAse J2 | [936/UACAUU] | Exonuclease activity [GO:0004527] |
| rpoB | SAOUHSC_00524 | DNA-directed RNA polymerase subunit beta | [2697/UACAUU] [2836/UACUUA] | 5′-3′ RNA polymerase activity [GO:0034062] |
| rnjA | SAOUHSC_01035 | RNAse J1 | [1160/UACCUA] [275/UACCUA] | exonuclease activity [GO:0004527] |
| rho | SAOUHSC_02362 | Transcription termination factor Rho | [18/UACAUC] 39/UACUUA[] | DNA-templated transcription, termination [GO:0006353] |
| rny/cvfA | SAOUHSC_01263 | RNAse Y phosphodiesterase | [1004/UACUUA] [1097/UACAUG] [1233/UACAUC] | Hydrolase activity, acting on ester bonds [GO:0016788] |
| rpoC | SAOUHSC_00525 | DNA-directed RNA polymerase subunit beta' | [1205/UACUUA] [1286/UACAUC] [2801/UCCAUA] [3209/UACGUA] | 5′-3′ RNA polymerase activity [GO:0034062] |
| mrnC | SAOUHSC_00512 | Hypothetical protein RNAse III | [131/UACAUC] | Ribonuclease III activity [GO:0004525] |
| cshA | SAOUHSC_02316 | DEAD-box ATP dependent DNA helicase | [972/UACACA] | RNA helicase activity [GO:0003724] |
| chsB | SAOUHSC_01659 | DEAD-box helicase-magnesium homeostasis | [730/UACUUA] | RNA helicase activity [GO:0003724] |
aGene product as annotated in Aureowiki for NCTC8325 genome
bBrackets represent the ORF; number inside brackets indicate the MazF cleavage position relative to the ATG codon; brackets before or after the cleavage position indicates cleavage outside the ORF, downstream or upstream respectively.
cComplete gene ontology enrichment analysis in Supplementary Table S6.
MazF-dependent cleavage of candidate genes potentially modulating MazF toxin
To clarify if MazF can also modulate its own production or activity, we searched for MazF cleavage sites present on transcripts spanning the mazEF locus or that may modulate MazF activity (Table 4 and Figure 5). Transcription of the mazEF locus was previously shown to be negatively and positively regulated by σB and SarA transcriptional regulators, respectively (45). We therefore analyzed in detail the MazF sequence recognition and cleavage in the transcriptomic product of σB operon and sarA genomic regions (Figure 5A). MazF recognition sequences were detected or predicted in sarA, in σB/rsbV/rsbW operon and mazF transcripts while no motif sequence was present in mazE or rsbU transcripts. qRT-PCR was used to confirm MazF cleavage of selected transcripts, using TaqMan® hydrolysis probes hybridizing exactly over the selected MazF recognition sequence. We expected a reduced target-probe hybridization and detection of mRNA levels present upon mazF overexpression due to MazF cleavage. qRT-PCR analyses were normalized using gyrB, which does not have a MazF recognition sequence (21).
Table 4.
MazF cleavage on potential MazEF TAS regulatory circuit
| Gene | Gene number | Gene descriptiona | MazF motif and localizationb | Detection method |
|---|---|---|---|---|
| sigB | SAOUHSC_02298 | RNA polymerase sigma factor SigB | [317/UACAUG] | Predicted, probe |
| rsbW | SAOUHSC_02299 | Serine–protein kinase RsbW | [135/UACAUA] | Predicted, probe |
| rsbV | SAOUHSC_02300 | STAS domain-containing protein | [232/UACAUU] | Probe, nEMOTE |
| rsbU | SAOUHSC_02301 | Phosphatase RsbU | No motif | – |
| mazF | SAOUHSC_02303 | Hypothetical protein | [277/UACUUA] | nEMOTE |
| mazE | SAOUHSC_02304 | Hypothetical protein | No motif | – |
| clpC | SAOUHSC_00505 | Endopeptidase | [293/UACAUC] | Probe, nEMOTE |
| clpP | SAOUHSC_00790 | ATP-dependent Clp protease proteolytic subunit | [187/UACAUU] | Predicted, No experimental detection |
| spxA | SAOUHSC_00934 | Transcriptional regulator Spx | [341/UACCUA] | nEMOTE |
| trfA | SAOUHSC_00935 | Adaptor protein MecA | [191/UACAUG] | Predicted, no experimental detection |
| sarA | SAOUHSC_00620 | Accessory regulator A | [124/UACAUC] | Predicted, no experimental detection |
| sarS | SAOUHSC_00070 | Accessory regulator-like protein | [273/UACUUA] | Probe, nEMOTE |
aGene product as annotated in Aureowiki for NCTC8325 genome
bBrackets represent the ORF; number inside brackets indicate the MazF cleavage position relative to the ATG codon; brackets before or after the cleavage position indicates cleavage outside the ORF, downstream or upstream respectively.
The mRNA levels of selected genes were analyzed upon mazF overexpression in the presence (wild type strain background) or absence (ΔmazEF strain background) of MazE antitoxin (Figure 5A). As expected, mazF overexpression did not affect mRNA levels of mazE gene lacking a MazF recognition sequence. However, >2-fold reduction in mRNA levels was observed upon induction of MazF toxin in the presence or absence of MazE antitoxin for the rsbV, rsbW and sigB transcripts. As previously observed, despite the presence of a canonical motif in sarA mRNA (UACAU motif at position 124), no decrease of sarA mRNA level was observed upon mazF overexpression in either wild type or mazEF-deleted strain backgrounds (21).
The sarA sequence recognition site may potentially be protected by RNA-binding proteins such as CshA or forming a double strand RNA structure precluding its cleavage as previously proposed (33). The nEMOTE detected cleavage of mazF mRNA at the alternative sequence recognition U∧ACUUA at position 277, showing that MazF in S. aureus is prone to autoregulation as recently shown for MazF in E. coli (46). As the same mazF alternative recognition sequence (UACUUA) is found in sarS mRNA at position 273, we predicted a MazF-dependent cleavage of sarS. Both nEMOTE and TaqMan® qRT-PCR analyses show a MazF-dependent cleavage of sarS mRNA (Figure 5A).
MazF toxic activity can be modulated by activation of antitoxin production or stabilization, as antitoxins inherently block toxin activity (47–49). We further hypothesized that production or stabilization of the MazE antitoxin can be achieved through cleavage of trfA, clpP, clpC and spx transcripts. These transcripts encode factors promoting the proteolytic degradation of the MazE antitoxin (Figure 5B and Table 4) and they carry predicted MazF recognition sequence. However, qRT-PCR shows only a decrease of clpC mRNA levels upon mazF overexpression in both strain backgrounds. All together these results show that under our growth conditions of mazF overexpression, transcripts of the σB operon, clpC and mazF itself are targets of MazF cleavage.
DISCUSSION
Controlled overexpression of some toxins in TAS, is one of the experimental tools used to induce a non-growing state and to identify key metabolic pathways potentially involved. This strategy has yielded important insights in several model organisms despite the artificial nature of the toxin induction (14,48,50). In this study, we used conditional overexpression of mazF endoribonuclease toxin to facilitate a global in vivo identification of genes involved in S. aureus growth arrest. Using the nEMOTE transcriptome-wide mapping technique, we provide an extensive catalog of targets, including its own transcript, identified an extended MazF recognition sequence and revealed insights into the MazF cleavage mechanism.
MazF cleavage mechanism
MazF is a toxin present across phylogenetically distant bacterial species including E. coli, M. tuberculosis and S. aureus. In general, MazF was described as a ribosome-independent and sequence specific endoribonuclease recognizing a specific sequence in RNA molecules such as mRNA, tRNA and rRNA (14–16,51–54). The ACA RNA sequence is recognized by E. coli MazF toxin while in M. tuberculosis MazF toxin recognizes sequences such as UGGCU or UUCCU.
In this study, the nEMOTE technique detected the previously recognized MazF sequence specificity (U∧ACAU), showing that nEMOTE is a reliable technique to detect global toxin cleavage sites in vivo. The massive quantity of RNAs analyzed in parallel and in vivo by the nEMOTE procedure has proven advantageous in also identifying a new extended alternative recognition sequence, previously unrecognized by in vitro studies that used single-molecule testing. We could detect that cleavages outside the canonical U^ACAU sequence have an extended recognition specificity. It is interesting to point out that E. coli MazF has also been found to have extended recognition beyond its canonical ACA motif (16). As for E. coli, we found MazF cleavage on transcripts within the coding sequence, but also in the untranslated regions.
Our finding that MazF has different sequence recognition sites might have implications related to cleavage preferences or the need of other factors for full functionality, such as structural or protein factors helping MazF sequence recognition and/or cleavage, as recently suggested in other bacterial species. Indeed, whole-genome studies on E. coli MazF activity have shown that not all ACA sites are cleaved. While reasons for uncleaved E. coli ACA sites remain unknown, studies concerning MazF-mt9 of M. tuberculosis show that the RNA structure is a key element that can determine MazF toxin cleavage, as MazF-mt9 preferentially cleaves tRNA carrying a MazF specific sequence and structure (14). In S. aureus, MazF recognition sites may also be targets of other RNA binding proteins that can promote or inhibit MazF cleavage. We observed that not all MazF recognition sites in highly expressed genes are cleaved and accordingly, S. aureus CshA RNA helicase has been proposed to confer protection to specific mRNAs from MazF cleavage (33). Interestingly, despite the in vivo MazF activity measurement using nEMOTE, this technique identifies the same canonical MazF motif previously observed with classical in vitro techniques, suggesting that at least for cleavage at the MazF canonical recognition site no other factors are apparently required. We do not exclude the possibility of the activation of a second mazF-dependent TAS that could be cleaving RNA, although we do not have evidence that other S. aureus TAS are induced transcriptionally upon mazF overexpression. Further studies will decipher whether structural and/or other proteins and/or RNA post-transcriptional modifications are involved in MazF endoribonuclease activity in S. aureus.
Analysis of the S. aureus genome shows that 700 genes do not carry any MazF cleavage site and that they are essentially short genes (414 bp average length). These genes might have evolved to avoid being cleaved by MazF, and they might be indicative of the core functions that allow the cell to survive in a non-growing state triggered by MazF toxin and/or necessary for regrowth. The functional analysis of the 100 longest genes that do not carry MazF recognition sequence reveals that these genes are implicated in oxidoreductase activity [GO:0016491] and tRNA methylation [GO:0032259]. Whether oxidoreductase and methylation activity are necessary for S. aureus survival during growth arrest remains to be studied. However, defects in methylation-related genes in other bacteria are related to growth defects or shown to be essential for life (55–57).
Identified MazF targets and correlation with growth stasis
The overexpression of MazF used in this study allowed the identification of a thorough set of MazF target transcripts, but currently we can only speculate about the correlation between them and the observed MazF-induced growth stasis phenotype. However, it is worth mentioning that several other studies observed defects in growth upon deletion of gene targets that we also found cleaved by MazF: such as spa (protein A) or the sigB transcriptional regulator using other technical methods (21). We also found transcripts previously related to MazF function such as CshA (33) and others that can be intuitively linked to the MazF induced non-growing state (e.g., cell wall, division or protein translation genes).
As shown by Culviner et al. in E. coli, our analysis identified some MazF targets affecting ribosome biogenesis, however we also observed cleavage of mRNAs involved in protein translation (see Table 1). Intriguingly, we observed cleavage of RNA encoding the HflX GTPase, a factor shown in S. aureus to be involved in disassembly of 100S ribosomes (70S homodimeric) (58). The 100S ribosomes are assembled upon stress conditions and promote bacterial survival by sequestering ribosomes, protecting them from degradation and preventing their use for translation. The 100S ribosome assembly and disassembly is reversible (58,59). Based on our data, the predicted GTPase HflX cleavage by MazF argues for the role of MazF to maintain ribosome dimers, decreasing translation and contributing to growth stasis. This hypothesis is in agreement with our unpublished results, showing that upon mazF overexpression formation of S100 ribosomes is observed.
Identified MazF targets and correlation with regrowth
Genes non-targeted by MazF may be essential for survival under growth arrest and others may be essential to promote regrowth (60–64). Interestingly, among the transcripts not targeted by MazF are the antitoxin mazE (171bp) and the sigB-related rsbU (991bp) transcripts. MazE and RsbU may be seen as key proteins to decrease MazF activity and to promote exit from the MazF-induced growth stasis. RsbU may be necessary to ensure production of MazE antitoxin by affecting the negative regulator SigB. Indeed, SigB is a negative transcriptional regulator of mazEF transcription whose activity is modulated by an upstream pathway involving the RsbU phosphatase (65). RsbU inactivation will result in SigB sequestration by its anti-sigma factor RsbW and predicting a decrease amount of SigB and de-repression of mazEF transcription. The exact mechanism of RsbU-dependent control of mazEF transcription to restore regrowth remains to be determined.
Besides guaranteeing production of MazE antitoxin by maintaining its transcription and avoiding cleavage, antitoxin stabilization or an induced imbalance between MazE and MazF concentrations may be another mechanism to restore growth (47–49). While antitoxin stabilization is modulated by affecting proteolytic systems involved in antitoxin degradation, toxin–antitoxin equilibrium is modulated through autoregulation of mazEF gene expression by transcriptional and post-transcriptional mechanisms as shown in E. coli (46,66). Recently, a mazF gene post-transcriptional autoregulation has been observed, where MazF cleaves its own transcript, producing pulsed MazF levels and affecting bacterial division and a higher antitoxin protein synthesis (46,66).
In S. aureus, ClpCP proteolytic system degrades MazE antitoxin, and interestingly, we identified a MazF motif present and cleaved in clpC transcript. This will predict an increase in stability of MazE antitoxin through disruption of ClpC chaperone. Moreover, our nEMOTE and qRT-PCR techniques detected an alternative extended MazF recognition site cleaved inside the mazF coding sequence. This observation suggests a possible post-transcriptional MazF autoregulation in S. aureus through cleavage of mazF transcript, similar to E. coli. We can propose that decreased mazF transcript and protein levels will produce a toxin–antitoxin imbalance favoring antitoxin concentration needed for regrowth.
The small subset of transcripts lacking a MazF sequence recognition motif argue for their importance for survival and regrowth during and after stasis. A major future task is to identify whether precise targets are linked to the induction of stasis and whether this is entirely stochastic. Evolution must have led to fine tuning to not compromise all cells in order to support the re-establishment of the population.
DATA AVAILABILITY
The data associated with our analyses are available in the GEO data repository accession GSE141855.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Friedrich Götz (Universität Tübingen, Tübingen, Germany) for HG003 mazEF-deleted strain.
Contributor Information
Roberto Sierra, Service of Infectious Diseases, Department of Medical Specialties, Geneva University Hospitals and Medical School, Geneva 1211, Switzerland; Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
Julien Prados, Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
Olesya O Panasenko, Service of Infectious Diseases, Department of Medical Specialties, Geneva University Hospitals and Medical School, Geneva 1211, Switzerland.
Diego O Andrey, Service of Infectious Diseases, Department of Medical Specialties, Geneva University Hospitals and Medical School, Geneva 1211, Switzerland; Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
Betty Fleuchot, Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
Peter Redder, Centre de Biologie Intégrative, Université de Toulouse III, Toulouse 31400, France.
William L Kelley, Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
Patrick H Viollier, Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
Adriana Renzoni, Service of Infectious Diseases, Department of Medical Specialties, Geneva University Hospitals and Medical School, Geneva 1211, Switzerland; Department of Microbiology and Molecular Medicine, University of Geneva, Geneva 1211, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [310030-169404, 310030-166611, CRSII3_160703 to A.R., W.K., P.V.]; Foundation privée [HUG RS1-27 to A.R., P.V.]. Funding for open access charge: Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bigger J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944; 244:497–500. [Google Scholar]
- 2. Dorr T., Vulic M., Lewis K.. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010; 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Claudi B., Sprote P., Chirkova A., Personnic N., Zankl J., Schurmann N., Schmidt A., Bumann D.. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 2014; 158:722–733. [DOI] [PubMed] [Google Scholar]
- 4. Moyed H.S., Bertrand K.P.. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983; 155:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balaban N.Q., Helaine S., Lewis K., Ackermann M., Aldridge B., Andersson D.I., Brynildsen M.P., Bumann D., Camilli A., Collins J.J et al.. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019; 17:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harms A., Stanger F.V., Scheu P.D., de Jong I.G., Goepfert A., Glatter T., Gerdes K., Schirmer T., Dehio C.. Adenylylation of gyrase and Topo IV by FicT toxins disrupts bacterial DNA topology. Cell Rep. 2015; 12:1497–1507. [DOI] [PubMed] [Google Scholar]
- 7. Aakre C.D., Phung T.N., Huang D., Laub M.T.. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell. 2013; 52:617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castro-Roa D., Garcia-Pino A., De Gieter S., van Nuland N.A.J., Loris R., Zenkin N.. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 2013; 9:811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mutschler H., Gebhardt M., Shoeman R.L., Meinhart A.. A novel mechanism of programmed cell death in bacteria by toxin–antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011; 9:e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masuda H., Tan Q., Awano N., Wu K.P., Inouye M.. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012; 84:979–989. [DOI] [PubMed] [Google Scholar]
- 11. Harms A., Brodersen D.E., Mitarai N., Gerdes K.. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell. 2018; 70:768–784. [DOI] [PubMed] [Google Scholar]
- 12. Yamaguchi Y., Park J.H., Inouye M.. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011; 45:61–79. [DOI] [PubMed] [Google Scholar]
- 13. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A.C., Engelberg-Kulka H., Moll I.. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011; 147:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schifano J.M., Cruz J.W., Vvedenskaya I.O., Edifor R., Ouyang M., Husson R.N., Nickels B.E., Woychik N.A.. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2016; 44:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schifano J.M., Vvedenskaya I.O., Knoblauch J.G., Ouyang M., Nickels B.E., Woychik N.A.. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014; 5:3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Culviner P.H., Laub M.T.. Global analysis of the E. coli toxin MazF reveals widespread cleavage of mRNA and the inhibition of rRNA maturation and ribosome biogenesis. Mol. Cell. 2018; 70:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Donegan N.P., Marvin J.S., Cheung A.L.. Role of adaptor TrfA and ClpPC in controlling levels of SsrA-tagged proteins and antitoxins in Staphylococcus aureus. J. Bacteriol. 2014; 196:4140–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donegan N.P., Thompson E.T., Fu Z., Cheung A.L.. Proteolytic regulation of toxin–antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 2010; 192:1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu L., Inoue K., Yoshizumi S., Kobayashi H., Zhang Y., Ouyang M., Kato F., Sugai M., Inouye M.. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009; 191:3248–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schuster C.F., Mechler L., Nolle N., Krismer B., Zelder M.E., Gotz F., Bertram R.. The MazEF toxin-antitoxin system alters the beta-lactam susceptibility of Staphylococcus aureus. PLoS One. 2015; 10:e0126118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu Z., Tamber S., Memmi G., Donegan N.P., Cheung A.L.. Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 2009; 191:2051–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma D., Mandell J.B., Donegan N.P., Cheung A.L., Ma W., Rothenberger S., Shanks R.M.Q., Richardson A.R., Urish K.L.. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio. 2019; 10:e01658-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sierra R., Viollier P., Renzoni A.. Linking toxin–antitoxin systems with phenotypes: a Staphylococcus aureus viewpoint. Biochim. Biophys. Acta. Gene. Regul. Mech. 2019; 1862:742–751. [DOI] [PubMed] [Google Scholar]
- 24. Helle L., Kull M., Mayer S., Marincola G., Zelder M.E., Goerke C., Wolz C., Bertram R.. Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology. 2011; 157:3314–3323. [DOI] [PubMed] [Google Scholar]
- 25. Radhakrishnan S.K., Pritchard S., Viollier P.H.. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev. Cell. 2010; 18:90–101. [DOI] [PubMed] [Google Scholar]
- 26. Kirkpatrick C.L., Martins D., Redder P., Frandi A., Mignolet J., Chapalay J.B., Chambon M., Turcatti G., Viollier P.H.. Growth control switch by a DNA-damage-inducible toxin–antitoxin system in Caulobacter crescentus. Nat. Microbiol. 2016; 1:16008. [DOI] [PubMed] [Google Scholar]
- 27. Redder P. Using EMOTE to map the exact 5′-ends of processed RNA on a transcriptome-wide scale. Methods Mol. Biol. 2015; 1259:69–85. [DOI] [PubMed] [Google Scholar]
- 28. Redder P. Mapping 5′-ends and their phosphorylation state with EMOTE, TSS-EMOTE, and nEMOTE. Methods Enzymol. 2018; 612:361–391. [DOI] [PubMed] [Google Scholar]
- 29. Fuchs S., Mehlan H., Bernhardt J., Hennig A., Michalik S., Surmann K., Pane-Farre J., Giese A., Weiss S., Backert L. et al.. AureoWiki the repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 2018; 308:558–568. [DOI] [PubMed] [Google Scholar]
- 30. Rao X., Huang X., Zhou Z., Lin X.. An improvement of the 2∧(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013; 3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 31. Gefen O., Chekol B., Strahilevitz J., Balaban N.Q.. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci. Rep. 2017; 7:41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brauner A., Fridman O., Gefen O., Balaban N.Q.. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016; 14:320–330. [DOI] [PubMed] [Google Scholar]
- 33. Kim S., Corvaglia A.R., Leo S., Cheung A., Francois P.. Characterization of RNA Helicase CshA and its role in protecting mRNAs and small RNAs of Staphylococcus aureus strain Newman. Infect. Immun. 2016; 84:833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu M., Dai X.. On the intrinsic constraint of bacterial growth rate: M. tuberculosis's view of the protein translation capacity. Crit. Rev. Microbiol. 2018; 44:455–464. [DOI] [PubMed] [Google Scholar]
- 35. Deris J.B., Kim M., Zhang Z., Okano H., Hermsen R., Groisman A., Hwa T.. The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science. 2013; 342:1237435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shahrezaei V., Marguerat S.. Connecting growth with gene expression: of noise and numbers. Curr. Opin. Microbiol. 2015; 25:127–135. [DOI] [PubMed] [Google Scholar]
- 37. Lund V.A., Wacnik K., Turner R.D., Cotterell B.E., Walther C.G., Fenn S.J., Grein F., Wollman A.J., Leake M.C., Olivier N. et al.. Molecular coordination of Staphylococcus aureus cell division. Elife. 2018; 7:e32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monteiro J.M., Fernandes P.B., Vaz F., Pereira A.R., Tavares A.C., Ferreira M.T., Pereira P.M., Veiga H., Kuru E., VanNieuwenhze M.S. et al.. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 2015; 6:8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu W., Missiakas D., Schneewind O.. Septal secretion of protein A in Staphylococcus aureus requires SecA and lipoteichoic acid synthesis. Elife. 2018; 7:e34092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rohrer S., Ehlert K., Tschierske M., Labischinski H., Berger-Bachi B.. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9351–9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tavares A.C., Fernandes P.B., Carballido-Lopez R., Pinho M.G.. MreC and MreD proteins are not required for growth of Staphylococcus aureus. PLoS One. 2015; 10:e0140523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cho K.H. The structure and function of the Gram-positive bacterial RNA degradosome. Front. Microbiol. 2017; 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morrison J.M., Dunman P.M.. The modulation of Staphylococcus aureus mRNA turnover. Future Microbiol. 2011; 6:1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Linder P., Lemeille S., Redder P.. Transcriptome-wide analyses of 5′-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 2014; 10:e1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Donegan N.P., Cheung A.L.. Regulation of the mazEF toxin–antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 2009; 191:2795–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikolic N., Bergmiller T., Vandervelde A., Albanese T.G., Gelens L., Moll I.. Autoregulation of mazEF expression underlies growth heterogeneity in bacterial populations. Nucleic Acids Res. 2018; 46:2918–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Amitai S., Yassin Y., Engelberg-Kulka H.. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 2004; 186:8295–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cho J., Carr A.N., Whitworth L., Johnson B., Wilson K.S.. MazEF toxin–antitoxin proteins alter Escherichia coli cell morphology and infrastructure during persister formation and regrowth. Microbiology. 2017; 163:308–321. [DOI] [PubMed] [Google Scholar]
- 49. Pedersen K., Christensen S.K., Gerdes K.. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 2002; 45:501–510. [DOI] [PubMed] [Google Scholar]
- 50. Mok W.W., Park J.O., Rabinowitz J.D., Brynildsen M.P.. RNA futile cycling in model persisters derived from MazF accumulation. MBio. 2015; 6:e01588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Zhang J., Hoeflich K.P., Ikura M., Qing G., Inouye M.. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003; 12:913–923. [DOI] [PubMed] [Google Scholar]
- 52. Schifano J.M., Edifor R., Sharp J.D., Ouyang M., Konkimalla A., Husson R.N., Woychik N.A.. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schifano J.M., Woychik N.A.. 23S rRNA as an a-Maz-ing new bacterial toxin target. RNA Biol. 2014; 11:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mets T., Lippus M., Schryer D., Liiv A., Kasari V., Paier A., Maivali U., Remme J., Tenson T., Kaldalu N.. Toxins MazF and MqsR cleave Escherichia coli rRNA precursors at multiple sites. RNA Biol. 2017; 14:124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bjork G.R., Jacobsson K., Nilsson K., Johansson M.J., Bystrom A.S., Persson O.P.. A primordial tRNA modification required for the evolution of life. EMBO J. 2001; 20:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gamper H.B., Masuda I., Frenkel-Morgenstern M., Hou Y.M.. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 2015; 6:7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Graille M., Figaro S., Kervestin S., Buckingham R.H., Liger D., Heurgue-Hamard V.. Methylation of class I translation termination factors: structural and functional aspects. Biochimie. 2012; 94:1533–1543. [DOI] [PubMed] [Google Scholar]
- 58. Basu A., Yap M.N.. Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E8165–E8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gohara D.W., Yap M.F.. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr. Genet. 2018; 64:753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joers A., Kaldalu N., Tenson T.. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J. Bacteriol. 2010; 192:3379–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mukamolova G.V., Turapov O., Malkin J., Woltmann G., Barer M.R.. Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum. Am. J. Respir. Crit. Care. Med. 2010; 181:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pinto D., Sao-Jose C., Santos M.A., Chambel L.. Characterization of two resuscitation promoting factors of Listeria monocytogenes. Microbiology. 2013; 159:1390–1401. [DOI] [PubMed] [Google Scholar]
- 63. Rosser A., Stover C., Pareek M., Mukamolova G.V.. Resuscitation-promoting factors are important determinants of the pathophysiology in Mycobacterium tuberculosis infection. Crit. Rev. Microbiol. 2017; 43:621–630. [DOI] [PubMed] [Google Scholar]
- 64. Pascoe B., Dams L., Wilkinson T.S., Harris L.G., Bodger O., Mack D., Davies A.P.. Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS One. 2014; 9:e85998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Giachino P., Engelmann S., Bischoff M.. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 2001; 183:1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nikolic N. Autoregulation of bacterial gene expression: lessons from the MazEF toxin–antitoxin system. Curr. Genet. 2019; 65:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with our analyses are available in the GEO data repository accession GSE141855.