Abstract
Melanoma cells utilize multiple mechanisms to exit the primary tumor mass, invade the surroundings and subsequently distant tissues. We have previously reported that the expression of the RNA editing enzyme ADAR1 (adenosine deaminase acting on RNA) is downregulated in metastatic melanoma, which facilitates proliferation and invasion. Here we show that ADAR1 controls melanoma invasiveness by regulating ITGB3 expression via miR-30a and miR-30d. ADAR1 overexpression or knockdown leads to an increase or decrease, respectively, in the expression of both microRNAs. The effect is independent of RNA-editing. Dual luciferase assays show that both microRNAs directly regulate the expression of the ITGB3 integrin. Overexpression of the miR-30a or miR-30d lead to a decrease in ITGB3 and a resultant decreased invasive and metastatic capacities. Neutralization of the endogenous miR-30a or miR-30d leads to the opposite effect. The microRNAs regulate ITGB3 levels probably through a post-transcriptional effect, as both mRNA and protein levels of ITGB3 are affected. These results further expand our knowledge on the ADAR1-ITGB3 network and its central role in acquisition of the invasive phenotype of metastatic melanoma.
Keywords: Melanoma, invasion, metastasis, integrin, ITGB3, RNA editing
Introduction
Malignant melanoma is an aggressive tumor with a high metastatic potential as a very early event. Thus, understanding the acquisition of invasive behavior is of clear importance. It was previously shown that the transition from radial growth phase (RGP) to the vertical growth phase (VGP) enhances the metastatic potential [1]. One of the most important proteins associated with melanoma metastatic potential is beta-3 integrin (ITGB3) [1-3]. Together with the αV subunit, it forms the heterodimeric adhesion receptor vitronectin. It is a major cell-extracellular matrix (ECM) mediator that binds a range of ligands containing the amino acid sequence RGD, mainly collagen, laminin and fibronectin. Changes in the cytoskeleton organization and altered contacts with the ECM are required for increasing cell motility and intravasation [4,5]. Upregulation of αVβ3 expression has been associated with malignant potential. It was previously shown that the transcription factors SP1 [6], FoxC2 [7], and CDK11P58 [8] are involved in the regulation of ITGB3 expression. Additional studies show that miRNAs [9-16] and other regulatory elements, such as protein kinase C [17], activated RAF-MEK-ERK signaling [18], and CCND1b [19] as putative regulators of ITGB3 expression.
Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional mechanism through which RNA sequences are directly altered by members of the family of adenosine deaminases that act on RNA (ADARs) enzymes, namely ADAR1 and ADAR2 [20]. ADARs can affect multiple cellular functions by causing amino acid substitutions [21], changes in splicing, RNA stabilization, and nuclear retention [22], as well through general control of non-coding RNAs biogenesis and their target gene specificity [23,24]. ADARs can also operate in an RNA editing-independent manner by affecting processing of miRNA [25-27], formation of protein-protein complexes [28], and decreasing protein kinase activities [29,30]. We have previously shown that ADAR1 expression is downregulated along melanoma progression, particularly during the metastatic transition [26], thereby enhancing proliferation [26], resistance to tumor infiltrating lymphocytes [31], and invasiveness [32]. Downregulation of ADAR1 was also found to increase melanoma metastasis in vivo [33]. Network analysis identified ITGB3 as a prime member in the ADAR1-related network. Indeed, ADAR1 regulates melanoma cell invasion by controlling ITGB3 expression independently of RNA editing, at the transcriptional and post-transcriptional levels via miR-22 and PAX6 transcription factor, respectively [32].
Here we further expand the ADAR1-ITGB3-Invasion network. We identify miR-30a and miR-30d as ADAR1-regulated microRNAs, and define their role as direct regulators of ITGB3 expression and thereby of melanoma cell invasion.
Methods
Cells and antibodies
In this study the following cell lines were used: 624mel and 526mel (obtained from Dr. Steve Rosenberg, NCI), 003mel was generated and authenticated in our laboratories [31], and WM-115, WM-266-4, SKmel-5, SKmel-28, A375, MeWo, 526mel, and HEK293T were obtained from ATCC. Cells were maintained in RPMI-1640 supplemented with 10% heat-inactivated FBS, 1 mM L-glutamine, 1 mM non-essential amino acids, 1 mM pyruvate and penicillin-streptomycin (all from Biological Industries, Israel). Stably transfected cell lines were cultured with 1 μg/ml puromycin (Calbiochem) or 2 mg/ml G418 (Alexis Biochemicals). The following antibodies were used: Mouse anti human-ITGB3 FITC (DAKO), Mouse anti human-Isotype Control IgG1 FITC (DAKO), Rabbit anti human ADAR1 (Sigma-Aldrich), mouse anti human β-actin (MP Biochemicals).
Cohort of melanoma patients
Formalin-fixed paraffin-embedded blocks of melanoma metastases obtained from 36 metastatic melanoma patients were used to explore miR and ITGB3 expression, and investigate the correlations with overall survival. Written informed consent was obtained based on approval of Israel Ministry of Health protocol 3518/2004.
RNA isolation and reverse transcription
Total RNA from cell lines was isolated using Tri Reagent (Sigma) extraction method. Briefly, the cells pellet first homogenized in Trizol, and then 0.2 ml chloroform per ml Tri reagent was added, samples were centrifuged and the aqueous phase collected. Then 0.5 ml isopropanol per ml Tri reagent was added and the sample was again centrifuged. After discarding the supernatant, the pellet was re-suspended in 75% ethanol, centrifuged and re-suspended in RNase free water. Integrity of the RNA was determined by spectrophotometry and electrophoresis.
Prior to each biopsy block sectioning, the microtome was meticulously cleaned using a 70% ethanol solution followed by RNAase AWAY™ spray (Thermo Fischer, USA), and the water in the water plate were replaced. All tumor biopsy blocks were sectioned 10 times, each section was 6 μm thick. An expert pathologist reviewed the H&E slides of each of the tumor biopsies to enable gross dissection of the tumor area into sterile 1.5 mL vials. miRNA were extracted with miRNeasy (Qiagen) and total RNA was extracted with RNeasy (Qiagen).
The cDNA pools were generated with a Transcriptor high fidelity transcriptor kit (Roche) using random hexamer primers or Universal cDNA synthesis kit Exiqon® microRNA cdna kit (Exiqon).
Real-time quantitative PCR analysis
Primers (Sigma-Aldrich) were designed according to Primer-Express® software guidelines (Applied Biosystems). Forward and reverse primers were designed from different exons to eliminate possible DNA contamination [26]. miRNA expression was tested with custom Exiqon® primers (Exiqon). The real-time PCR (quantitative PCR, qPCR) reactions were normalized to GAPDH or U6 endogenous control. Fold of expression was calculated with the accepted ΔΔCt method, as reported previously [26].
Expression constructs and stable transfections
The expression systems used in this work were pQCXIP.puro and psiCheck2 (Promega). The various primers that were designed for cloning and introduction of mutations are described in Supplementary Table 1. Transfections were performed with Turbofect® (Fermentas) according to manufacturer’s instructions. Retroviral transductions were performed as previously described [26]. Site directed mutagenesis was performed using QuickChange® kit (Stratagene) according to manufacturer’s instructions.
Anti-miR, oligos and transient transfection
Anti-miR-30a or anti-miR-30d oligos were used along with proper negative control (Dharmacon). The various oligos were transiently transfected (20 nM) with JetPrime® (polyplus) in 96 well microplates, and the cells were tested for miRNA and protein expression 48 hours post transfection.
Invasion assay
Melanoma cells (2×105) were seeded into the upper wells of Transwell invasion system (44) onto Matrigel (BD Biosciences) coated ThinCerts® PET membranes containing 8-μm pores (Greiner-bio-one) in RPMI 1640 with 0.1% fetal bovine seru at invaded each membrane was measured by XTT staining as previously described. Percent of invasion was calculated as: (Total number of invading cells)/(Total number of seeded cells) ×100. The values were adjusted to the relative growth ratio of cells within 24 hrs evaluated by Net proliferation (standardized XTT), as previously described [34].
Flow cytometry
Staining for extracellular antigens was performed on 1×105 cells with the appropriate fluorochrome-conjugated antibodies diluted in FACS medium (PBS, 0.02% sodium azide, 0.5% BSA) on ice for 30 min. Following incubation, cells were centrifuged (5 min, 500 g, 4°C), washed and re-suspended in 200 µl FACS medium and collected for FACS analysis. All experiments were performed using a FACSCalibur® instrument (BD Biosciences) and data analysis using FlowJo® software (Tree Star Inc.).
Western blot
Lysates of 5×106 cells were washed with PBS and lysed in RIPA (Sigma Aldrich) lysis buffer and protease inhibitor cocktail (Roche) on ice for 20 min. Insoluble material was removed by centrifugation at 14,000 rpm for 10 min at 4°C. Protein concentration was measured using Pierce™ BCA protein kit (Thermo Scientific). Proteins were separated by 10-12% SDS-PAGE, transferred onto Nitrocellulose membranes and incubated with specific antibodies. The antigen-antibody complexes were visualized by standard ECL (Enhanced Chemiluminescence) reaction (Biological-Industries).
Luciferase reporter assay
HEK293T cells were co-transfected with 1 μg of psiCheck2-ITGB3 3’UTR (UTR), different psiCheck2-ITGB3 mutated 3’UTR seed sequences (UTR-MUT-30A, UTR-MUT-30D) corresponding to the miRs binding site(s) or psiCheck2-empty vector (No UTR), and 0.1 μg of the pQCXIP-miRs-30a, -30d or pQCXIP-empty vector (Mock) as control. Cells were harvested 48 hours post transfection and assayed with Dual Luciferase Reporter Assay System® (Promega) according to the manufacturer’s instructions.
In vivo
Melanoma stable transfectants (Mock, miR-30a or miR-30d) were labeled with Calcein Green (Thermo-Fisher) and 293 cells were labeled with Calcein Red (Thermo-Fisher) according to the manufacturer’s instructions, and mixed in a 1:1 ratio. A total of 4×106 cells were injected to the tail vein of each SCID/NOD mouse, 6-8 weeks of age. Lungs were harvested after 12 h and were rendered into single cell suspension using mechanic mincing and enzymatic digestion using Collagenase IV and DNAse I [35]. Single cell suspension was then analyzed by flow cytometry for the ratio between the green melanoma cells and the red 293 cells that were used as an internal control. Animal experiments were performed in compliance with the Animal Welfare Act.
Statistical analysis
Data were analyzed using the unpaired two-tailed Student’s t test or one-way ANOVA. Correlations were examined with Pearson’s correlation test. Two tailed P value ≤0.05 was considered significant.
Results
Expression of miR-30a, miR-30d and ITGB3 in melanoma
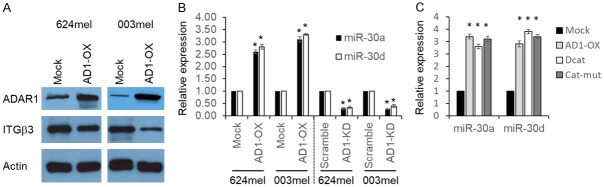
High throughput analysis of microRNA expression in 624mel cells following ADAR1 knockdown was performed and published previously [26]. Crossing this list together a list of miRNAs predicted to target ITGB3 3’UTR, based on TargetScan 7.2 [36] identified miR-30a and miR-30d as putative candidates as both ADAR1-controlled and ITGB3 regulators. We have previously generated melanoma cell lines with ADAR1 overexpression (624/AD1-OX, 003/AD1-OX) or knockdown (624/AD1-KD, 003/AD1-KD). Transfectants with empty vector (624/Mock, 003/Mock) or scrambled sequences (624/Scramble, 003/Scramble), respectively, serve as negative controls [31,32]. We have previously shown that knockdown of ADAR1 leads to an increase in ITGB3 at the protein level [32]. Here we show that overexpression of ADAR1 led to a decrease in ITGB3 at the protein level, as confirmed in western blot (Figure 1A). Significant and consistent alterations in both microRNAs were validated using qPCR, as ADAR1 overexpression increased the expression levels of both microRNAs, whereas ADAR1 knockdown led to the opposite effects (Figure 1).
Figure 1.
ADAR1 controls the expression of miR-30a and miR-30d in an RNA editing independent manner. A. Shows the effect of ADAR1 overexpression on ITGB3 expression. ADAR1 was stably overexpression (AD1-OX) in the indicated cell lines. Mock overexpression (Mock) served as negative control. A representative Western Blot analysis of ADAR1, ITGB3 and Actin is shown. B. Shows the effect of ADAR1 on miR-30a and miR-30d expression. ADAR1 was stably overexpression (AD1-OX) or knocked down (AD1-KD) in the indicated cell lines. Mock overexpression (Mock) or scrambled sequence (Scramble) served as the respective negative controls. miRs expression levels relative to the appropriate negative control were determined by real time PCR. C. Shows that ADAR1 controls miR-30a and miR-30d in an RNA editing independent manner. Overexpression of ADAR1 without the catalytic domain (DCat) or with a neutralizing mutation within the catalytic domain (Cat-mut) was compared to Mock and AD1-OX. Figure shows the average ± SE of three experiments on independent RNA purifications, each performed in triplicates. Student’s T-test was used to determine statistical significance. *denotes P<0.05.
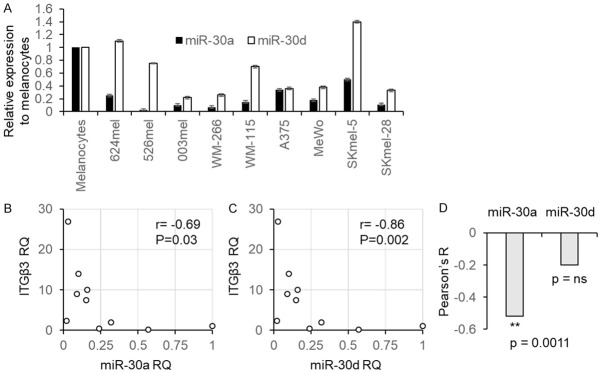
The expression of miR-30a, miR-30d and ITGB3 was investigated in 9 melanoma lines and in normal melanocytes as a reference. The expression levels of miR-30a and miR-30d were significantly lower than in the normal melanocytes in 9/9 and 7/9 of the melanoma lines tested, respectively (Figure 2A). This is in accordance with the lower ADAR1 levels in metastatic melanoma lines as compared to normal melanocytes we reported in the past [26]. A statistically significant, strong negative correlation, was observed between each of the microRNAs and ITGB3 at the RNA level (Figure 2B, 2C). We further tested in 36 specimens derived from metastatic melanoma patients the expression levels of miR-30a, miR-30d, and of ITGB3 using real time PCR. miR-30a was negatively correlated with ITGB3 in a highly significant manner (r=-0.519, P=0.0011), but not miR-30d (Figure 2D).
Figure 2.
miR-30a and miR-30d are inversely correlated with ITGB3. Normalized miR-30a, miR-30d and ITGB3 expression levels were determined in 10 melanoma cell lines, with the expression levels among melanocytes serving as reference level. A. Shows the expression of miR-30a and miR-30d; B. Shows the correlation of miR-30a and ITGB3; C. Shows the correlation of miR-30d and ITGB3. D. Shows the Peasron correlation R of ITGB3 with miR-30a or miR-30d in a cohort of 36 metastatic melanoma patients. Correlation was calculated using Pearson test; the mean ± SE of three experiments on independent RNA purifications, each performed in triplicates. ns denotes non-significant.
ITGB3 is directly regulated by miR-30a and miR-30d
A portion of ITGB3 3’UTR containing the putative binding sites for miR-30a and miR-30d was cloned upstream to Renilla luciferase in a dual luciferase reporting psiCheck2 system. The putative binding sites were altered with three point mutations (UTR-MUT) for each miR. miRs were cloned into the pQCXIP expression vector. Empty psiCheck2 (NO-UTR) and pQCXIP (Mock) served as negative controls. The various constructs were co-transfected into the easily transfectable HEK 293T cells. The luciferase signal of cells co-transfected with both empty vectors served as point of reference. Forced expression of miRs-30a and -30d with the UTR construct significantly inhibited the luciferase signal while the inhibitory effect was abolished when the UTR-MUT construct was tested (Figure 3). These results show that both microRNAs directly regulate the 3’UTR of ITGB3.
Figure 3.

ITGB3 expression is directly controlled by miR-30a and miR-30d. Figure shows the results of dual luciferase assays. UTRs (UTR-30A or UTR-30D) and MUT-UTRs (UTR-MUT-30A, UTR-MUT-30D) denote ITGB3 3’UTR segments containing the reference sequence or mutated sequence in the binding site of the respective miR. Each tested microRNA is indicated above the respective UTRs. The results are presented as normalized dual luciferase activity, and represent the mean ± SE of three biologically independent experiments, each performed in triplicates. Asterisks represent P values: **P<0.01 (2-tailed t-test).
Functional effect of miR-30a and miR-30d
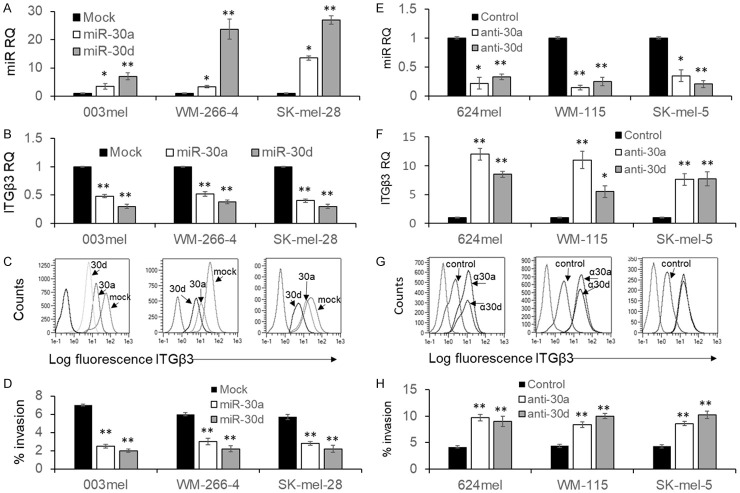
The effect of miR-30a and miR-30d on ITGB3 expression and subsequent function was tested in six melanoma cell models. Each of the microRNAs was overexpressed in the melanoma cell lines 003mel, WM-266-4 and SKmel-28 (Figure 4A). These melanoma lines expressed miR-30a and miR-30d at the lowest levels (Figure 2A). In all three melanoma lines, miR-30d was more pronouncedly overexpressed than miR-30a (Figure 4A). In agreement with the luciferase experiments (Figure 3), in all three melanoma cell lines, both microRNAs caused downregulation of ITGB3 at the mRNA level (Figure 4B) and at the protein level (Figure 4C). The downregulation was greater with miR-30d, in correlation to its stronger overexpression. These results indicate that the regulation of ITGB3 by miR-30a and miR-30d occurs at the RNA level. Importantly, a significant reduction in the invasiveness through matrigel of was observed in all three melanoma lines tested with both microRNAs (Figure 4D).
Figure 4.
miR-30a or miR-30d control ITGB3 expression and melanoma cell invasion. (A) Overexpression of miR-30a and miR-30d in 3 indicated melanoma cell lines confirmed in real time PCR, as compared to Mock transfection; (B) Expression levels of ITGB3 mRNA determined with real time PCR or (C) protein determined with flow cytometry in each of the stably transfected constructs in the indicated cell lines; (D) Percent invasion in each of the stably transfected constructs in the indicated cell lines. (E) Expression of miR-30a and miR-30d in 3 indicated melanoma cell lines after transfection of the respective anti-miR, as determined by real time PCR. Control sequence served as reference; (F) Expression levels of ITGB3 mRNA determined with real time PCR or (G) protein determined with flow cytometry in each of the transfected anti-miRs in the indicated cell lines; (H) Percent invasion in each of the transfected anti-miRs in the indicated cell lines. The results represent the mean ± SE of three biologically independent experiments, each performed in triplicates. Asterisks represent P values: *P<0.05, **P<0.01 (2-tailed t-test). Flow cytometry in (C, G) shows one representative experiment out of three performed.
To further ratify these results in an even more physiologic intervention, the effect of neutralization of miR-30a and miR-30d was investigated. For that purpose, another set of three melanoma lines with the highest expression of the microRNAs was selected, including 624mel, WM-115 and SKmel-5 (Figure 2A). Introduction of anti-miR-30a and anti-miR-30d significantly reduced the endogenous corresponding microRNA levels (Figure 4E). The reduction of each microRNA led to a significant increase in ITGB3 expression in all three melanoma lines at the mRNA level (Figure 4F) and at the protein level (Figure 4G). Accordingly, the invasiveness through matrigel of all melanoma lines was significantly increased with both microRNAs (Figure 4H). These collective observations confirm the regulation of ITGB3 by both microRNAs at the RNA level, and their functional importance on melanoma cell invasiveness.
Finally, the effect of miR-30a and miR-30d on metastasis was evaluated in vivo. Melanoma cell lines 003mel or WM-266-4 stably transfected with Mock, miR-30a or miR-30d used in Figure 4 were labeled with green calcein, and were mixed with 293 cells labeled with red calcein. A 1:1 mixture of the cells was injected into the tail vein of SCID/NOD mice, and the lungs were harvested 12 h afterwards. The 293 served as an internal control. Remarkably, stable transfection with miR-30a or miR-30d significantly reduced the melanoma/293 ratio, in line with the results presented above. Due to the short time frame, these differences are less likely to be accounted to differences in proliferation.
Discussion
ADAR1 plays an important role in melanoma cell invasion and in-vivo metastasis [32,33]. ITGB3 is centrally involved in melanoma progression [32] and metastatic potential [1-3]. We have previously demonstrated robust network interactions between ADAR1 and ITGB3, occurring at both the transcriptional and post-transcription levels, as demonstrated in multiple melanoma cell lines and clinical samples [32]. The existence of multiple regulatory mechanisms on ITGB3 expression via ADAR1 attests for its importance. Here we further uncover this network by pointing on miR-30a and miR-30d as ADAR1-controlled microRNAs, which play a direct role in controlling ITGB3 expression and invasion phenotype.
The role of miR-30a in inhibition melanoma cell invasion was recently described, by targeting E-Cadherin [37] or SOX4 [38]. A similar role for miR-30a was recently described in renal cell carcinoma by targeting ATG12 [39], in lung squamous cell carcinoma by targeting FoxD1 [40] and in lung adenocarcinoma by targeting CNPY2 [41]. The role of miR-30d in inhibition of invasion was demonstrated in breast carcinoma by targeting KLF11 [42], in non-small cell lung carcinoma by targeting NFIB [43] and in colorectal cancer by targeting LRH-1 [44]. There is hardly any published information on the role of miR-30d in melanoma. Here we expand the knowledge on miR-30a or miR-30d by describing their regulation via the ADAR1 enzyme. ADAR1 overexpression or knockdown results in upregulation or downregulation, respectively, of both miR-30a and miR-30d (Figure 1). Moreover, the regulation of miR-30a and miR-30d by ADAR1 is independent of RNA-editing (Figure 1), which is also consistent with our previous findings that ADAR1 controls the biogenesis of microRNAs in a similar manner [26]. We show that both miR-30a and miR-30d inhibit the invasion of melanoma cells (Figure 4) and their metastatic potential (Figure 5), by directly targeting the central ITGB3 integrin (Figure 3). The role of ITGB3 in melanoma cell invasion was previously shown by using blocking monoclonal antibodies [32]. These results complement the understanding on how miR-30 family controls the invasiveness of melanoma cells. ITGB3 expression is controlled by miR-30a and miR-30d directly (Figure 3), probably at the mRNA level, as both mRNA and protein levels are affected (Figure 4). We provide substantial evidence in several melanoma cell lines by overexpression of microRNAs (Figure 4) or by targeting endogenous microRNAs with anti-miRs (Figure 4). A significant, inverse correlation between each of these microRNAs and ITGB3 was indeed demonstrated in multiple melanoma cell lines and in a cohort of patients (Figure 2).
Figure 5.
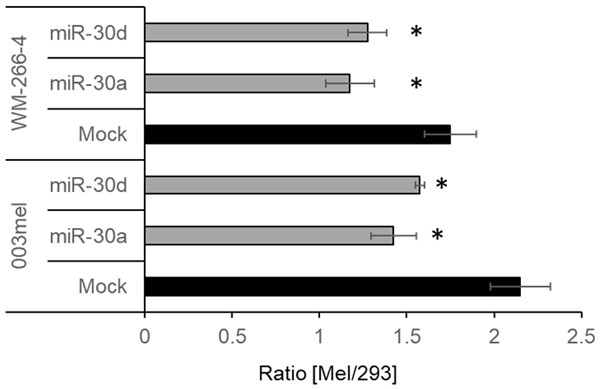
miR-30a and miR-30d decrease experimental lung metastasis. The indicated melanoma cell lines transfected with Mock, miR-30a or miR-30d were each injected to the tail vein of SCID/NOD mice in a 1:1 ratio with 293 cells. Melanoma cells were labeled with red calcein and 293 with green calcein. 12 h after injection, lungs were harvested, rendered into a single cell suspension and analyzed by flow cytometry. Figure shows the average melanoma/293 ratio of 3 independent experiments, each included 4 mice per group. Error bars indicate SE. *denotes P<0.05 using one-way ANOVA.
These collective results are in line with our past reports on the inhibitory role of invasion [32] and proliferation [26] of melanoma cells by ADAR1. Indeed, ADAR1 leads to increased miR-30a and miR-30d expression (Figure 1), which down-modulates ITGB3 and thereby decreases melanoma cell invasiveness (Figure 4). On the other end, ADAR1 downregulation leads to decreased miR-30a and miR-30d expression (Figure 1), which up-modulates ITGB3 and thereby enhances melanoma cell invasiveness (Figure 5). The latter corresponds to the frequently observed ADAR1 downregulation occurring at the transition from primary to metastatic melanoma [26]. As all components demonstrated here, including ADAR1, miR-30a/d and ITGB3, are expressed in multiple types of cancer, it is reasonable to assume that this mechanism plays an important role in multiple malignancies.
In conclusion, here we further uncover the ADAR1-ITGB3 network by pointing on miR-30a and miR-30d as ADAR1-controlled microRNAs, which play a direct role in the post-transcriptional expression control of ITGB3 and of the invasive melanoma cell phenotype. The results of this study could suggest on miR-30a or miR-30d as promising therapeutic agents, capable of reducing the metastatic potential of melanoma cells. Investigation of the therapeutic value of these microRNAs is warranted.
Acknowledgements
This work was supported by research grant ISF (Israel Science Foundation) 1925/15 and by Samueli Foundation Grant for Integrative Immuno-Oncology. Special thanks for the Lemelbaum Family, Aronson Foundation and Allen Berg Fund for Excellence in Immuno-Oncology for their generous support.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Gaggioli C, Sahai E. Melanoma invasion-current knowledge and future directions. Pigment Cell Res. 2007;20:161–72. doi: 10.1111/j.1600-0749.2007.00378.x. [DOI] [PubMed] [Google Scholar]
- 2.Haass NK, Smalley KS, Li L, Herlyn M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18:150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 3.Orgaz JL, Sanz-Moreno V. Emerging molecular targets in melanoma invasion and metastasis. Pigment Cell Melanoma Res. 2013;26:39–57. doi: 10.1111/pcmr.12041. [DOI] [PubMed] [Google Scholar]
- 4.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinon P, Wehrle-Haller B. Integrins: versatile receptors controlling melanocyte adhesion, migration and proliferation. Pigment Cell Melanoma Res. 2011;24:282–94. doi: 10.1111/j.1755-148X.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Wilhide CC, Dang C, Li L, Li SX, Villa-Garcia M, Bray PF. Human integrin β3 gene expression: evidence for a megakaryocytic cell-specific cis-acting element. Blood. 1998;92:2777–2790. [PubMed] [Google Scholar]
- 7.Hayashi H, Sano H, Seo S, Kume T. The Foxc2 transcription factor regulates angiogenesis via induction of integrin β3 expression. J Biol Chem. 2008;283:23791–800. doi: 10.1074/jbc.M800190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi Y, Huang S, Wang L, Zhou R, Wang L, Xiao X, Li D, Cai Y, Zhou X, Wu J. CDK11(p58) inhibits ERα-positive breast cancer invasion by targeting integrin β3 via the repression of ERα signaling. BMC Cancer. 2014;14:577. doi: 10.1186/1471-2407-14-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller DW, Bosserhoff AK. Integrin [beta] 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Ni R, Huang Y, Wang J. miR-98 targets ITGB3 to inhibit proliferation, migration, and invasion of non-small-cell lung cancer. Onco Targets Ther. 2015;8:2689–97. doi: 10.2147/OTT.S90998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang C, Zhang H, Guo Y, Hong Y, Liu Y, Xue Y. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol Biol Rep. 2014;41:2521–7. doi: 10.1007/s11033-014-3110-0. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Liu B, Lin Z, Yao Y, Chen Y, Li Y, Chen J, Yu D, Tang Z, Wang B, Zeng S, Fan S, Wang Y, Li Y, Song E, Li J. MiR-320a acts as a prognostic factor and inhibits metastasis of salivary adenoid cystic carcinoma by targeting ITGB3. Mol Cancer. 2015;14:96. doi: 10.1186/s12943-015-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang SY, Choi SA, Lee JY, Park AK, Wang KC, Phi JH, Koh EJ, Park WY, Park SH, Hwang DW, Jung HW, Kim SK. miR-192 suppresses leptomeningeal dissemination of medulloblastoma by modulating cell proliferation and anchoring through the regulation of DHFR, integrins, and CD47. Oncotarget. 2015;6:43712–43730. doi: 10.18632/oncotarget.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, Zheng Q, Ma Y, Zhang J, Wu N, Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 16.Wei W, Yang Y, Cai J, Cui K, Li R, Wang H, Shang X, Wei D. MiR-30a-5p suppresses tumor metastasis of human colorectal cancer by targeting ITGB3. Cell Physiol Biochem. 2016;39:1165–76. doi: 10.1159/000447823. [DOI] [PubMed] [Google Scholar]
- 17.Symonds JM, Ohm AM, Tan AC, Reyland ME. PKCδ regulates integrin α V β 3 expression and transformed growth of K-ras dependent lung cancer cells. Oncotarget. 2016;7:17905–19. doi: 10.18632/oncotarget.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woods D, Cherwinski H, Venetsanakos E, Bhat A, Gysin S, Humbert M, Bray PF, Saylor VL, McMahon M. Induction of β3-integrin gene expression by sustained activation of the ras-regulated raf-MEK-extracellular signal-regulated kinase signaling pathway. Mol Cell Biol. 2001;21:3192–205. doi: 10.1128/MCB.21.9.3192-3205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu FH, Luo LQ, Liu Y, Zhan QX, Luo C, Luo J, Zhang GM, Feng ZH. Cyclin D1b splice variant promotes αvβ3-mediated adhesion and invasive migration of breast cancer cells. Cancer Lett. 2014;355:159–167. doi: 10.1016/j.canlet.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 20.Zinshteyn B, Nishikura K. Adenosine-to-inosine RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2009;1:202–209. doi: 10.1002/wsbm.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–4. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 22.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27–R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heale BS, Eulalio A, Schulte L, Vogel J, O’Connell MA. Analysis of A to I editing of miRNA in macrophages exposed to salmonella. RNA Biol. 2010;7:621–627. doi: 10.4161/rna.7.5.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemlich Y, Greenberg E, Ortenberg R, Besser MJ, Barshack I, Jacob-Hirsch J, Jacoby E, Eyal E, Rivkin L, Prieto VG, Chakravarti N, Duncan LM, Kallenberg DM, Galun E, Bennett DC, Amariglio N, Bar-Eli M, Schachter J, Rechavi G, Markel G. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest. 2013;123:2703–18. doi: 10.1172/JCI62980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie Y, Ding L, Kao PN, Braun R, Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Samuel CE. Adenosine Deaminase ADAR1 increases gene expression at the translational level by decreasing protein Kinase PKR-dependent eIF-2[alpha] phosphorylation. J Mol Biol. 2009;393:777–87. doi: 10.1016/j.jmb.2009.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clerzius G, Gélinas JF, Daher A, Bonnet M, Meurs EF, Gatignol A. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol. 2009;83:10119–28. doi: 10.1128/JVI.02457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galore-Haskel G, Nemlich Y, Greenberg E, Ashkenazi S, Hakim M, Itzhaki O, Shoshani N, Shapira-Fromer R, Ben-Ami E, Ofek E, Anafi L, Besser MJ, Schachter J, Markel G. A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme. Oncotarget. 2015;6:28999–29015. doi: 10.18632/oncotarget.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemlich Y, Baruch EN, Besser MJ, Shoshan E, Bar-Eli M, Anafi L, Barshack I, Schachter J, Ortenberg R, Markel G. ADAR1-mediated regulation of melanoma invasion. Nat Commun. 2018;9:2154. doi: 10.1038/s41467-018-04600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, Velazquez-Torres G, Nip KM, Zhu K, Brooks D, Jones SJ, Birol I, Mosqueda M, Wen YY, Eterovic AK, Sood AK, Hwu P, Gershenwald JE, Robertson AG, Calin GA, Markel G, Fidler IJ, Bar-Eli M. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 2015;17:311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenberg E, Hershkovitz L, Itzhaki O, Hajdu S, Nemlich Y, Ortenberg R, Gefen N, Edry L, Modai S, Keisari Y, Besser MJ, Schachter J, Shomron N, Markel G. Regulation of cancer aggressive features in melanoma cells by microRNAs. PLoS One. 2011;6:e18936. doi: 10.1371/journal.pone.0018936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortenberg R, Sapir Y, Raz L, Hershkovitz L, Ben Arav A, Sapoznik S, Barshack I, Avivi C, Berkun Y, Besser MJ, Ben-Moshe T, Schachter J, Markel G. Novel immunotherapy for malignant melanoma with a monoclonal antibody that blocks CEACAM1 homophilic interactions. Mol Cancer Ther. 2012;11:1300–10. doi: 10.1158/1535-7163.MCT-11-0526. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 37.Noori J, Javanmard SH, Sharifi M. The role of microRNA-30a and downstream snail1 on the growth and metastasis of melanoma tumor. Iran J Basic Med Sci. 2019;22:534–540. doi: 10.22038/IJBMS.2019.32317.7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu EB, Sun XY, Li JP, Zhang C. miR-30a-5p inhibits the proliferation, migration and invasion of melanoma cells by targeting SOX4. Mol Med Rep. 2018;18:2492–2498. doi: 10.3892/mmr.2018.9166. [DOI] [PubMed] [Google Scholar]
- 39.Chen YH, Zhou JL, Wu XR, Huang JW, Chen W, Liu DM, Zhang J, Huang YR, Xue W. miR-30a-3p inhibits renal cancer cell invasion and metastasis through targeting ATG12. Transl Androl Urol. 2020;9:646–653. doi: 10.21037/tau.2019.12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CH, Tang JH, Xu S, Zhang WX, Jiang HL. miR-30a-5p inhibits proliferation and migration of lung squamous cell carcinoma cells by targeting FOXD1. Bio Res Int. 2020;2020:2547902. doi: 10.1155/2020/2547902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Wang HT, Kanmangne D, Li R, Qian ZQ, Xia XY, Wang XJ, Wang T. miR-30a-3p suppresses the proliferation and migration of lung adenocarcinoma cells by downregulating CNPY2. Oncol Rep. 2020;43:646–654. doi: 10.3892/or.2019.7423. [DOI] [PubMed] [Google Scholar]
- 42.Han ML, Wang YM, Guo GC, Li L, Dou DW, Ge X, Lv PW, Wang F, Gu YT. microRNA-30d mediated breast cancer invasion, migration, and EMT by targeting KLF11 and activating STAT3 pathway. J Cell Biochem. 2018;119:8138–8145. doi: 10.1002/jcb.26767. [DOI] [PubMed] [Google Scholar]
- 43.Wu YB, Zhang JN, Hou SZ, Cheng ZM, Yuan MX. Non-small cell lung cancer: miR-30d suppresses tumor invasion and migration by directly targeting NFIB. Biotechnol Lett. 2017;39:1827–1834. doi: 10.1007/s10529-017-2428-9. [DOI] [PubMed] [Google Scholar]
- 44.Yan LK, Qiu J, Yao JF. Downregulation of microRNA-30d promotes cell proliferation and invasion by targeting LRH-1 in colorectal carcinoma. Int J Mol Med. 2017;39:1371–1380. doi: 10.3892/ijmm.2017.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.