Abstract
In addition to the paternal genome, spermatozoa carry several intrinsic factors, including organelles (e.g., centrioles and mitochondria) and molecules (e.g., proteins and RNAs), which are involved in important steps of reproductive biology such as spermatogenesis, sperm maturation, oocyte fertilization and embryo development. These factors constitute potential biomarkers of “viable sperm” and male fertility status and may become major assets for diagnosing instances of idiopathic male infertility in both humans and livestock animals. A better understanding of the mechanism of action of these sperm intrinsic factors in the regulation of reproductive and developmental processes still presents a major challenge that must be addressed. This review assembles the main data regarding morpho-functional and intrinsic sperm features that are associated with male infertility, with a particular focus on microRNA (miRNA) molecules.
Keywords: infertility, diagnosis, spermatozoa, semen, intrinsic factors, ncRNAs, human, cattle
Introduction
Male fertility potential relies on several physical, endocrine, and genetic factors, which underlie the production of fully functional spermatozoa as well as their successful arrival at the site of fertilization. Impairment of these male reproductive functions accounts for half of the infertility issues that couples face during their reproductive age (Agarwal et al., 2015; Zegers-Hochschild et al., 2017). While male infertility diagnosis is mainly based on physical characteristics, hormonal analysis, and use of the traditional spermogram, its etiology remains unexplained in more than 30% of patients (Irvine, 1998; Agarwal et al., 2015). Thus, the design of novel customized diagnostic tools to fully assess male fertility potential and male infertility etiology is a necessity. Furthermore, an improved method to assess male fertility potential would be a major asset to the livestock industry, since this parameter constitutes a serious economic challenge in this field. Research performed on sperm samples from humans, livestock animals and rodents recently questioned the definition of the sperm cell as a simple carrier of the paternal genome. From this, it now appears that the spermatozoon is a well-differentiated cell that carries a widely diverse signature of specific organelles and molecules, that could vary according to male fertility potential, and be transferred to the oocyte at the time of fertilization. This contemporary view of the sperm cell opens potential new avenues concerning the development of a novel generation of diagnostic tools. In this review, we will delineate structural, morpho-functional and intrinsic sperm features, and comment on their contribution to the post-fertilization processes. The detection of sperm microRNAs (miRNAs) as a predictor of male fertility potential will be discussed with regard to its possible applications in both clinic and industry.
“Healthy Sperm” Concept: Structural, Morpho-Functional and Intrinsic Features
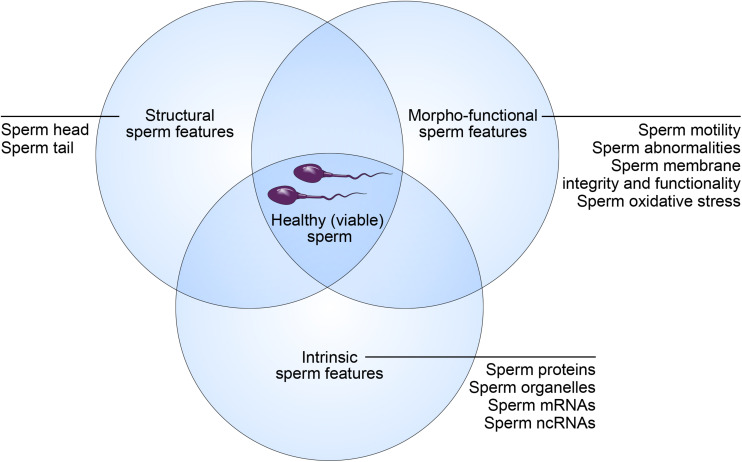
While sperm quality is a key determinant of male fertility potential, the discrimination between a high- and low-quality sperm sample is challenging. Indeed, this relies on a broad spectrum of sperm features (Amann and Hammerstedt, 1993). According to this concept, “healthy spermatozoa” also referred to as “viable spermatozoa” should possess the ability to reach the fertilization site, bind to and fertilize the oocytes, and properly contribute to initiation of early embryo development (Krawetz, 2005). These abilities are strictly dependent on specific structural, morpho-functional, and intrinsic features (Figure 1).
FIGURE 1.
Concept of “healthy (viable) sperm.” Male fertility potential relies on structural, morpho-functional, and intrinsic sperm features that shape equally the concept of “healthy (viable) sperm.”
Structural Sperm Features
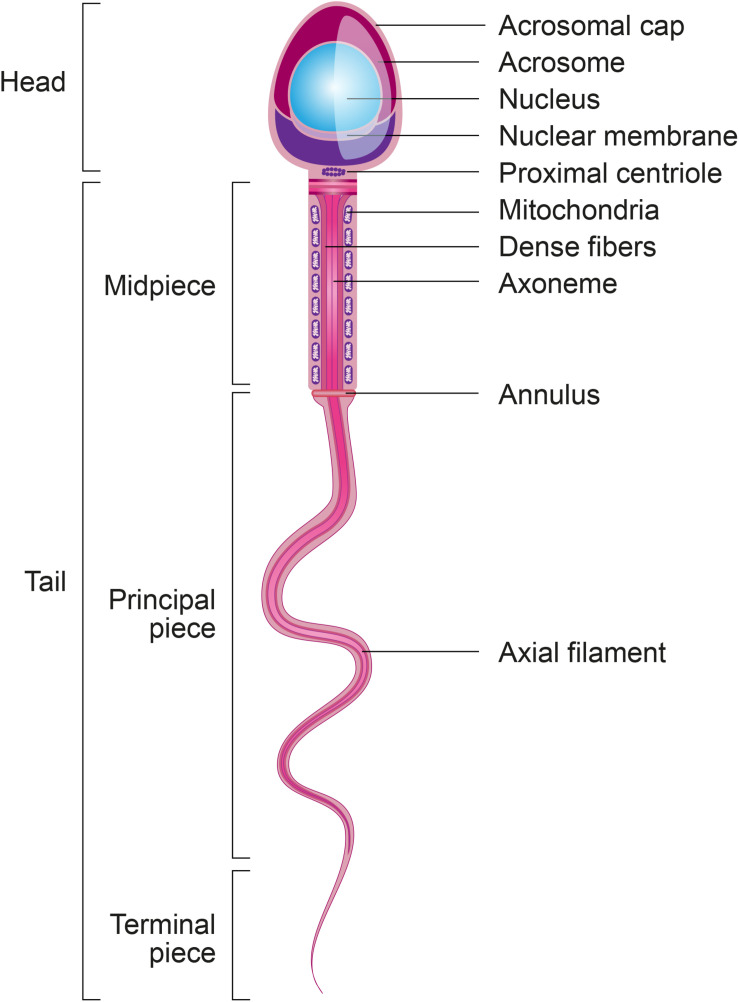
Spermatozoa are the most differentiated cells of the organism and display particular features related to their main function: the delivery of paternal DNA to the oocyte (Krawetz, 2005). In this regard, spermatozoa are composed of two main parts: a head to carry and protect the genome, and a tail or flagellum to propel the cell to the site of fertilization (Figure 2).
FIGURE 2.
Structural sperm features. Spermatozoa are composed of two main parts: head and tail (or flagellum). The sperm head is constituted basically by the acrosome and nucleus. The sperm tail includes: the neck that contains mainly the proximal centriole; the midpiece which is composed by mitochondria, outer dense fibers (ODF) and axoneme; principal piece containing the fibrous sheath and axoneme; and terminal piece.
Within the sperm head is a limited quantity of cytoplasm, highly condensed DNA, and a well-delimited acrosome. A substantial portion of the cytoplasm is lost during the final steps of spermatogenesis, specifically during spermiogenesis, the process during which round spermatids differentiate into elongated spermatids and then spermatozoa (Gadea et al., 2013). The remaining cytoplasm from intercellular bridges, called the cytoplasmic droplet, is lost during sperm transit through the epididymis (Cooper and Yeung, 2003). During spermiogenesis, the sequential replacement of histones by transitional proteins and then by protamines within sperm chromatin triggers genome condensation (Zini and Agarwal, 2011; Gadea et al., 2013). The acrosome, located at the top of the head, contains specific enzymes that promote specific functions trailing sperm capacitation and acrosome reaction: (1) exposure of acrosome zona pellucida binding proteins during sperm capacitation; (2) sperm ability to cross cumulus cells that surrounds the oocytes, (3) sperm binding to zona pellucida and acrosome reaction, and (4) migration of IZUMO1 protein from the outer acrosomal membrane to the equatorial segment of sperm surface to ensure its binding to JUNO receptor on the oocyte (Satouh et al., 2012; Bianchi et al., 2014). Since acrosome reacted sperm retain the ability to penetrate the zona pellucida, the previous paradigm supporting the role of enzymes released during acrosome reaction in the digestion of the zona pellucida has been revisited in different species (Hirohashi and Yanagimachi, 2018).
The sperm tail (or flagellum) includes the neck, midpiece, principal piece and terminal piece. The sperm centrioles are important to early embryo development and are localized in the sperm neck (Avidor-Reiss and Fishman, 2019). The axoneme, located internally along the entire flagellum, is composed of nine peripheral doublets and two central single microtubules (9 + 2 structure) integrated by the intraflagellar transport (IFT) system. Surrounding the axoneme, there is the outer dense fibers (ODF) and mitochondria in the sperm midpiece and the fibrous sheath (FS), formed by nine bundles of fibers of different lengths, in the principal piece (Inaba, 2003). Depending on species, approximately 22–75 mitochondria are present in the midpiece to produce enough energy necessary for a spermatozoon to transit along the female reproductive tract and to reach the fertilization site in the oviduct (Song and Lewis, 2008). All of these sperm structural characteristics are essential to ensure the ability of spermatozoa to cross the muco-cervical and uterine barriers and reach the oviduct, where they bind to and penetrate the oocyte to deliver their DNA content.
Morpho-Functional Sperm Features
Morpho-functional sperm attributes are capable of modulating male fertility potential and are tightly related to the structural sperm features. Parameters such as sperm motility/kinetics, morphological abnormalities, integrity of plasma and acrosome membranes, mitochondrial activity production of reactive oxygen species (ROS), DNA fragmentation and capacitation status are included in this group and usually are associated with male fertility (Table 1 and Figure 3). Thus, assessment of these parameters is essential to determine male fertility potential: evaluation of a higher number of morpho-functional attributes establishes the highest relationship with male fertility (Amann and Hammerstedt, 1993). A high proportion of morphological abnormalities – referred to as teratozoospermia – including, e.g., spermatozoa with large, small or piriform heads as well as coiled-tails, is associated with reproductive dysfunctions (Gillan et al., 2008). This common cause of male infertility is routinely assessed by light microscopic analysis of semen in fertility clinics.
TABLE 1.
Correlation between sperm morpho-functional attributes and male fertility.
| Sperm morpho-functional attribute | Definition | Required equipment | Correlation with fertility | References |
| Sperm subjective motility | Maximum percentage of sperm cells with movement | Light field optical microscope, preferably with phase contrast | r = 0.53 (P < 0.01) (160 to 90-day non-return) | Correa et al., 1997 |
| r = 0.67 (P = 0.03) (156-day non-return) | Gillan et al., 2008 | |||
| Computer assisted sperm analysis (CASA) | Percentage of sperm with motility and measurement of different sperm kinetic features | Light field optical microscope, with phase contrast coupled to a computer | WOB2: r = 0.57 (P < 0.05) (156-day non-return) | Morrell et al., 2018 |
| r = 0.67 (P = 0.03) (156-day non-return) | Gillan et al., 2008 | |||
| VSL3: r = −0.34 (P < 0.05) (156-day non-return) | Sellem et al., 2015 | |||
| ALH4 and progressive motility presented r2 of 0.68 | Farrell et al., 1998 | |||
| ALH4, BCF5, LIN6, VAP7 and VSL3 presented r2 of 0.98 | Farrell et al., 1998 | |||
| Sperm morphological abnormalities | Percentage of cells that present with form defects, often called abnormal cell percentage (Blom, 1973) | Light field optical microscope preferably with differential interference contrast | r = −0.59 (P = 0.01) (160 to 90-day non-return) | Correa et al., 1997 |
| r = −0.76 (P < 0.05) (156-day non-return) | Gillan et al., 2008 | |||
| r = −0.62 (P < 0.05) (156-day non-return) | Morrell et al., 2018 | |||
| IPIAH8 | Percentage of IPIAH8 sperm | Fluorescence microscopy or flow cytometry | High IPIAH8 (44.5%): 64.7% of pregnancy rate9 Low IPIAH8 (8.5%): 36.2% of pregnancy rate9 | Oliveira et al., 2014 |
| ROS | Percentage of sperm cells producing ROS | Fluorescence microscopy or flow cytometry | High ROS (10.63%): low fertility rates Low ROS (6.11%): high fertility rates | Celeghini et al., 2019 |
| DNA fragmentation | Percentage of sperm cells with DNA fragmentation | Fluorescence microscopy or flow cytometry | r = −0.56 (P < 0.01) | Morrell et al., 2018 |
| Capacitation status | Percentage of acrosome-reacted spermatozoa | Fluorescence microscopy or flow cytometry | r = 0.37 (P < 0.05) (1litter size) | Kwon et al., 2015 |
1Fertility measurement; 2wobble (%); 3progressive velocity (μm/s); 4lateral head amplitude (μm); 5beat cross frequency (Hz); 6linearity (%); 7path velocity (μm/s); 8spermatozoa with intact plasma and acrosome membranes and high mitochondrial membrane potential; 9ultrassonography exam 30 days after insemination.
FIGURE 3.
Morpho-functional sperm features. Schematic figure representing the spermatozoa with satisfactory (left) and unsatisfactory (right) morpho-functional features. Sperm acrosome membrane integrity, sperm plasma membrane integrity, sperm DNA integrity, low quantity of ROS, sperm mitochondrial membrane high activity, high sperm motility and normal sperm morphology characterize the satisfactory morpho-functional sperm features. Sperm acrosome membrane damage, sperm plasma membrane damage, sperm DNA fragmentation, high quantity of ROS, sperm mitochondrial membrane low activity, low sperm motility and abnormal sperm morphology characterize the unsatisfactory morpho-functional sperm features.
Several other techniques are currently available to assess sperm morpho-functional attributes. Sperm motility, kinetics, vigor and hyperactivation levels, are morpho-functional features classically assessed by light microscopy or computer-assisted sperm analysis (i.e., CASA) (Vincent et al., 2012). In addition, since sperm motility requires a substantial amount of energy produced by mitochondrial activity (Piomboni et al., 2012), high fertility samples usually present with high mitochondrial membrane potential. This feature can be assessed by fluorescence microscopy and flow cytometric approaches mainly using the JC-1 fluorescent probe, which forms J-aggregates when mitochondria are active and polarized; in this situation, more JC-1 monomers enter the spermatozoa and aggregate to generate red fluorescence (Gillan et al., 2005; Celeghini et al., 2007). In addition to energy production, mitochondria also produce large quantities of ROS that promote oxidative stress and damage to sperm membranes and DNA. Therefore, ROS quantification in sperm cells is also an important morpho-functional feature that can be assessed by fluorescence microscopy, flow cytometry and molecular/biochemical assays using fluorescent probes. Such assays include BODIPY, which measures the susceptibility of sperm to lipid peroxidation, CellROX Deep Red®, which stains ROS present in sperm cells, and measurement of malondialdehyde concentration (produced during lipid peroxidation) using the TBARS (Thiobarbituric acid reactive substances) assay (Aitken et al., 2007; Celeghini et al., 2019). In addition, since damage to the sperm membranes and DNA impairs sperm capacitation and early embryo development, both plasma and acrosome membrane as well as DNA integrity should be evaluated by fluorescence microscopy or flow cytometric approaches. Propidium iodide fluorescent probes and fluorescent lectin from Pisum sativum can be used to assess plasma and acrosome membrane damage (Celeghini et al., 2007), respectively, as well as SCSA® (Sperm Chromatin Structure Assay) to detect the susceptibility of sperm to DNA fragmentation (Evenson and Wixon, 2006). Post-sperm capacitation status could be assessed using the combination of Hoechst 33258/chlortetracycline fluorescence probes and is proposed to predict litter size and fertility success in pigs (Kwon et al., 2015).
While the assessment of these features is essential to determine male fertility potential, based on data from human clinics, about 30% of sperm samples presenting with satisfactory morpho-functional features (Figure 3) are still unable to fertilize the oocytes or to trigger early embryo development. Recent advances in analyses of sperm molecular and subcellular content suggested that intrinsic factors may participate to early embryo development and, consequently, could be potential targets for the assessment of male fertility potential.
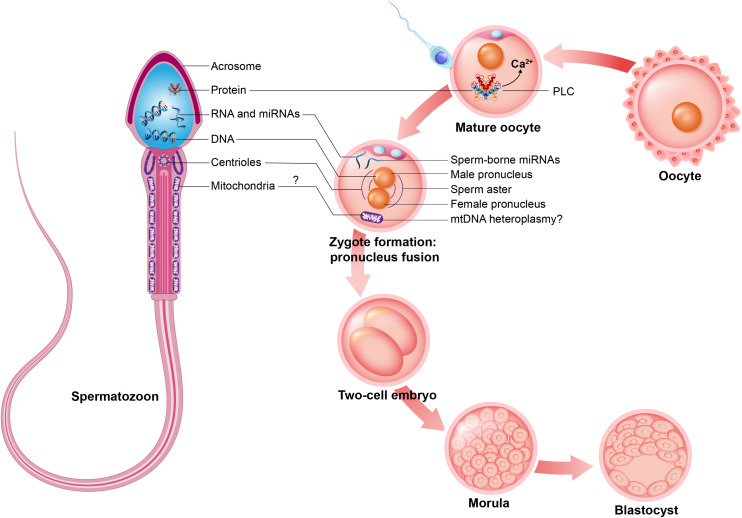
Intrinsic Sperm Features
In addition to the paternal DNA, the spermatozoon carries numerous molecules and organelles, which constitute a growing focus of interest in fundamental research. Among these intrinsic factors, sperm centrioles, sperm mitochondria, sperm proteins, and sperm RNAs are particularly interesting with respect to their potential contribution to the post-fertilization process (Figure 4). As shown in Figure 2, sperm centrioles are localized in the sperm neck while sperm mitochondria are arranged in the midpiece. Since mature spermatozoa possess a minimal amount of cytoplasm and a nucleus with highly condensed DNA that prevents transcription, the presence of proteins and RNAs in sperm cells is limited. However, several proteins and ribonucleic acid molecules are likely acquired by the maturing spermatozoa during spermatogenesis and their transit through the epididymis and post-ejaculatory journey (Boerke et al., 2007).
FIGURE 4.
Intrinsic sperm features. Spermatozoon contributes early embryo development during mature oocyte fertilization, zygote formation and embryo cleavage potentially with intrinsic sperm features such as: sperm proteins (e.g., PLCζ, to promote oocyte activation); sperm RNAs and microRNAs (miRNAs); sperm DNA, to generate male pronucleus; sperm centrioles, to form sperm aster; and may contribute sperm mitochondria, promoting mtDNA heteroplasmy.
RNAs Species Are Major Sperm Intrinsic Factors
microRNAs and Other Sperm RNA Species
Sperm RNAs were first described in the 1970s in murine and bovine spermatozoa (Betlach and Erickson, 1973; Paul and Duerksen, 1975). Given the limited quantity of cytoplasm and high level of sperm DNA condensation, this finding has been questioned as RNAs were potentially providing from mitochondria or contamination from somatic cells (Krawetz, 2005). In that regards, the roles and the origin of sperm RNAs remain to be determined (Ostermeier et al., 2004; Krawetz, 2005).
Four different RNA species can be distinguished in spermatozoa (Boerke et al., 2007): (1) mRNAs that are remnants from the spermatogenesis process with no known function in the oocyte (e.g., protamine-2 encoding RNA, Ziyyat, 2001); (2) mRNAs that originate from spermatogenesis with a potential function in the oocytes (e.g., PLCζ encoding RNA, Saunders et al., 2002); (3) mRNAs classified as “foreign” RNAs as a result of their acquisition by sperm during passage through the epididymis and by the contact with the fluids coming from the accessory sexual glands (e.g., clusterin encoding RNA, Hermo et al., 1994); and (4) non-coding RNAs (ncRNAs) that are acquired during the late steps of spermatogenesis or during post-testicular sperm maturation with a potential function at the time of fertilization (e.g., miRNAs and tRNA derived fragments, Sharma et al., 2016; Yuan et al., 2016). Among RNA species present in spermatozoa, ribosomal RNAs and mitochondrial RNAs (mtRNAs) are the most abundant, followed by ncRNAs. In contrast to somatic cells that comprise approximately 10 pg of RNA, mature spermatozoa carry around a 1,000-fold lower RNA content (Odia et al., 2018).
In addition to mRNAs, other sperm-borne RNA species, including miRNAs have been associated with sperm cell fertility status and early embryo development (Liu et al., 2012). The central dogma of molecular biology supports that genes are transcribed in the form of mRNAs, which in turn are translated into proteins, with the exception of structural RNAs such as ribosomal RNA (Mattick, 2001). Advancements in molecular biology and studies on the human genome have revealed that only 2% of all genes actually encode proteins (Mattick, 2001). In addition, many genomic sequences that were first described as “junk DNA” due to the fact that they did not encode proteins and had no apparent function, now appear to play an important role as post-transcriptional modulators. In this context, a large group of RNAs called non-coding RNAs (ncRNAs) has been identified in spermatozoa, including miRNAs (Wightman et al., 1993).
MicroRNAs are ∼18–22-nucleotide sequences that modulate gene expression following their pairing with the untranslated region (3′UTR) of target transcripts (Ambros, 2004; Bartel, 2004). The biosynthesis of miRNAs is performed in the cell nucleus with the generation of primary miRNA transcripts (pri-miRNA) and then precursor miRNAs (pre-miRNA), which are transported to the cytoplasm. In the cytoplasm, the hairpin structure is cleaved by Dicer, resulting in small double-stranded transcripts. The miRNA double strand is then transferred to Argonaut proteins, which bind to the mature miRNA also called guide-miRNA. This complex promotes gene silencing through translational inhibition or cleavage of mRNA (Ambros, 2004; Bartel, 2004). Such cleavage is common in plants and occurs when the pairing between the miRNA and 3′UTR region is perfect. On the other hand, an imperfect pairing implies the blockade of translation machinery on that particular mRNA without degradation—the most common process in animals (Bartel, 2004, 2018).
Of interest is that miRNAs either act in the cell where they are transcribed or in target cells through an intercellular communication mechanism (Raposo and Stoorvogel, 2013). Extracellular vesicles are important mediators of this communication. Exosomes comprise a population of small (40–160 nm) extracellular vesicles derived from intracellular multivesicular bodies carrying diverse bioactive molecules, including miRNAs, mRNAs and proteins (Raposo and Stoorvogel, 2013). Once exosomes are released into the extracellular environment, they can be internalized by recipient target cells (Raposo and Stoorvogel, 2013). Exosomes are released from different internal organs of the male reproductive tract, including the epididymis (epididymosomes) and prostate (prostasomes) (Sullivan and Saez, 2013). Epididymosomes play a key role during cellular communication that promotes sperm maturation during spermatozoa passage into the microenvironment of the epididymis (Belleannée, 2015).
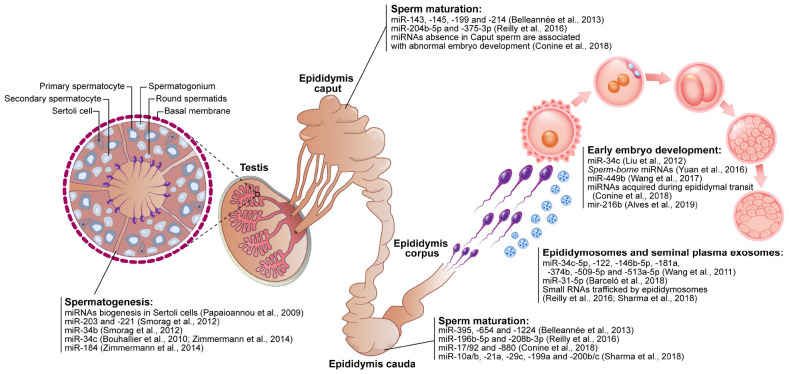
In general, miRNAs are post-transcriptional regulators that play important roles in physiological processes and their impairment results in different pathologies. These molecules have been shown to have great importance in the regulation of spermatogenesis (Papaioannou et al., 2009; Kotaja, 2014) and sperm maturation during sperm passage through the epididymis (Björkgren et al., 2012; Jerczynski et al., 2016; Sipilä and Björkgren, 2016) as shown in Figure 5. With the knowledge that miRNAs are very stable, conserved bioactive molecules derived from the different internal organs of the male reproductive tract, they could be considered potent fertility biomarkers. In light of these findings, sperm cells have gained the status of gametes that are not only responsible for transporting genetic material, but also as contributors to the activation of the oocyte and the cellular structure and molecular components of the zygote as shown in Figure 5.
FIGURE 5.
Schematic figure demonstrating the contribution of microRNAs (miRNAs) in the reproductive events. The sperm-related-miRNAs molecules display functions in spermatogenesis (testis), sperm maturation (epididymis caput, corpus, and cauda), sperm and seminal plasma interaction (e.g., epididymosomes) as well as modulating early embryo development.
miRNAs and Spermatogenesis
Spermatogenesis is a well-organized process that culminates in the production of sperm cells. Conditional knock-out of the miRNA processing enzyme Dicer1 in Sertoli cells triggers male infertility due to the absence of sperm in the lumen of the seminiferous tubules and spermatogenesis disorders (Papaioannou et al., 2009; Zimmermann et al., 2014), suggesting that the biogenesis of small non-coding RNA is important to spermatogenesis. Indeed, each cell type of the seminiferous tubule has a different miRNA profile that plays an important role in proliferation and differentiation (Hayashi et al., 2008; Smorag et al., 2012). In pigs, miR-26a has been shown to inhibit proliferation and to promote apoptosis in Sertoli cells, thus impairing sperm production (Ran et al., 2018). In mice, miR−221 and miR−290 regulate the proliferation of spermatogonia and primary spermatocytes (Smorag et al., 2012). In addition, miR−203 modulates spermatocyte meiosis and the miR−34 family regulates the formation of spermatids (Smorag et al., 2012). In humans, pigs and mice, the miR−34 family is also important in the regulation of spermatogenesis (Bouhallier et al., 2010; Zimmermann et al., 2014). Thus, miRNAs such as miR−26a, miR−221, miR−290, miR−203, and the miR−34 have been proposed to contribute to spermatozoa production (Figure 5), as their dysregulation results in disturbances in spermatogenesis and consequently a decrease in male fertility potential.
miRNAs and Sperm Maturation
Once produced in the testis, spermatozoa must pass through the epididymis to acquire their motility and oocyte-fertilizing ability. During their passage through the different epididymal segments, spermatozoa come into contact with different repertoires of ncRNAs, transcripts and proteins that are released from the epithelium of the epididymis mostly via epididymosomes (Sullivan et al., 2007; Belleannée et al., 2013; Sullivan, 2015; Reilly et al., 2016; Sharma et al., 2018). The transfer of proteins and other contents from epididymosomes to spermatozoa has been proposed to occur via the dynamin 1 mechanoenzyme following the tethering of epididymosomes to specific sperm receptors located in the post-acrosomal region and the fusion of epididymosomes with the sperm membrane (Zhou et al., 2019). In mice, sperm miRNAs present in the sperm are altered along the epididymis (Figure 5), showing that the sperm miRNA content is dynamic even following spermatogenesis (Nixon et al., 2015).
Conditional knock-out mice for the Dicer enzyme in principal cells of the epididymis dysregulated the differentiation process (Björkgren et al., 2012). Thus, biogenesis of ncRNAs seems to be important in the regulation of epididymal epithelium, sperm maturation and fertility. While miR-10a/b, −21a, −29c, −196b−5p, −199a, −200b/c, and −208b−3p accumulate in spermatozoa during passage through the epididymis, miR−204b−5p and miR−375−3p are more abundant in spermatozoa from the caput and corpus of the epididymis (Reilly et al., 2016; Sharma et al., 2018). Epididymosomes also display different miRNA profiles and traffic small RNAs to spermatozoa (Sharma et al., 2018), including miR−143, −145, −199, and −214 that are more abundant in epididymosomes present in the caput of the bovine epididymis, and miR−395, −654, and −1224 that are more abundant in epididymosomes from the cauda (Belleannée et al., 2013). In men with reproductive abnormalities, miRNAs present in seminal plasma (miR−34c−5p, −122, −146b−5p, −181a, −374b, −509−5p, and −513a−5p) decreased in abundance in men presenting with azoospermia (absence of spermatozoa in the ejaculate) and increased in abundance in men presenting with asthenozoospermia (decreased sperm motility) (Wang et al., 2011). Although studies have shown the importance of miRNAs along the epididymis, further work is required to show how miRNAs are able to regulate sperm maturation during passage of spermatozoa through the epididymis and how regulation of motility acquisition and the ability of the sperm to become fertile occur.
Contribution of Sperm Intrinsic Factors to Post-Fertilization Processes
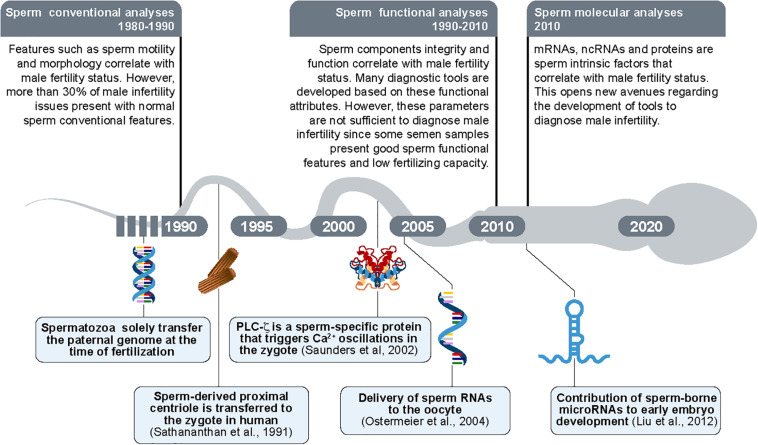
For many years it was postulated that sperm cells had the exclusive function of transporting genetic material to the oocyte. However, at the end of the 20th century, this sole contribution was challenged by the fact that specific sperm intrinsic factors, including sperm centrioles, sperm proteins, ribonucleic acids and mitochondrial DNA (mtDNA) could contribute to fertilization and/or early embryo development (Figure 6).
FIGURE 6.
Timeline of the new findings regarding sperm morpho-functional and intrinsic features. The paternal DNA was considered as the sole intrinsic sperm feature transferred from spermatozoon to oocyte until 1990s. In parallel, the sperm evaluation was limited to sperm conventional analyses (sperm motility and sperm morphology/abnormalities). Proximal centrioles were then shown to be transmitted by sperm to the oocytes during fertilization for the first time in 1991 (Sathananthan et al., 1991). The sperm-borne PLCζ protein was shown as a promotor of oocyte activation in 2002 (Saunders et al., 2002). The delivery of RNAs molecules was then revealed as transferred from sperm to the oocyte in 2004 (Ostermeier et al., 2004). In parallel, the sperm evaluation was updated to sperm functional analyses (e.g., sperm plasma membrane integrity). In 2010s, small sperm-borne RNAs molecules (microRNAs/miRNAs) were shown as important to early embryo development (Liu et al., 2012) potentially constituting a new group of sperm analyses composed by evaluation of molecular targets.
Contribution of the Sperm Centriole to Embryo Development
Centrioles are cytoplasmic structures involved in cell division and in the formation of cilia and flagella (Avidor-Reiss and Fishman, 2019). While spermatozoa present with centrioles arranged in the neck of their tail (Figure 2), mature oocytes lack centrioles (Simerly et al., 2018). Therefore, sperm-derived centrioles ensure the formation of sperm aster and centrosome in the fertilized oocyte, which bring the female and male pronuclei into close proximity and promote syngamy (Simerly et al., 1995; Schatten and Sun, 2009), as shown in Figure 4. Thus, centrioles are organelles that ensure proper embryonic development. In addition, the proper formation of the sperm aster is an important factor related to male fertility. For instance, large and well organized sperm aster formations in human spermatozoa correlate with samples that present with high fertility, while small and disorganized sperm aster formations correlate with low fertility samples (Navara et al., 1997). Therefore, beyond the transmission of the centrioles by the sperm cells, the quality of the sperm-borne centrioles is also important to embryo development. It has recently been shown that sperm from non-rodent mammals are responsible for transmitting a second atypical centriole to oocyte (Fishman et al., 2018). This centriole potentially forms the sperm aster and centrosome in the oocytes and, together with the first centriole, supports the fusion between the female and male pronucleus.
The Questioned Contribution of Sperm Mitochondrial DNA to Embryo Paternal Inheritance
Mitochondria are peculiar organelles that possess a specific DNA type: mtDNA. This organelle is essentially responsible for energy production, which is important to the cell’s activity. During spermatogenesis, sperm mitochondria change from a conventional to a condensed form that ensures more efficient energy production (Amaral et al., 2013). While some mitochondria are lost during spermatogenesis, between 22 and 75 mitochondria remain in the midpiece of the mature spermatozoon depending on species (Figure 2) (Bahr and Engler, 1970; Song and Lewis, 2008). Although spermatic mitochondria also contain mtDNA, the current paradigm is that the embryo exclusively possesses maternal mitochondria and, consequently, mitochondrial diseases are only inherited from the mother (Hecht et al., 1984). However, the inheritance of paternal mtDNA was suggested following the discovery of a high level of mtDNA heteroplasmy, i.e., mix of maternal and paternal mitochondrial genome, in mouse (Gyllensten et al., 1991) and human (17 individuals) (Luo et al., 2018) as schematized in Figure 4. These findings open new research avenues to better understand mitochondrial disease inheritance. However, further studies are necessary to confirm this potential paradigm shift.
Contribution of Sperm Proteins to Oocyte Activation
While mature spermatozoa are transcriptionally inactive upon their entrance to the epididymis, their proteome is dynamic due to the acquisition of proteins from the surrounding fluid, the cleavage of others from the sperm surface, and the rearrangement of acrosomal proteins during sperm capacitation. These proteins ensure diverse functions at different stages including: sperm maturation, e.g., the glioma pathogenesis-related 1-like protein 1 (GLiPr1L1) that is acquired by spermatozoa and participates in the binding of sperm cells to zona pellucida (Caballero et al., 2012); tagging of sperm sub-populations to ensure heterogeneity, e.g., the sperm binding protein 1 (ELSPBP1) that discriminates dead spermatozoa (D’Amours et al., 2012); and sperm ability to bind to the oocyte, e.g., Spermadhesin AWN (AWN) that promotes sperm binding to oocyte’s zona pellucida during fertilization (Kwon et al., 2014).
In addition to these proteins, sperm cells also acquire proteins during the final steps of spermatogenesis that are important to oocyte activation, e.g., the sperm-specific WW domain-binding protein (PAWP) and phospholipase C zeta (PLCζ) (Saunders et al., 2002; Wu et al., 2007). Among these spermatic proteins, PLCζ is an important player during fertilization, since it triggers intracellular calcium impulses required for oocyte activation (Saunders et al., 2002; Kashir et al., 2010). As a result of these impulses, and once fertilized, the metaphase II mature oocyte is able to resume and progress through meiotic division. This ensures the formation of female and male pronuclei, followed by syngamy (Figure 4). The presence of these proteins in spermatozoa positively correlates with male fertility (Saunders et al., 2002; Aarabi et al., 2014).
Contribution of Sperm RNA Species to Embryo Development
Sperm RNAs
The detection of sperm-borne RNAs, including clusterin and AKAP4, in embryos raised questions regarding their potential contribution to the molecular regulation of embryonic development (Ostermeier et al., 2004). The discovery of zygote transcripts originating exclusively from spermatozoa opened new avenues into the investigation of sperm RNAs and their importance to embryo development (Ostermeier et al., 2005; Jodar et al., 2013). Most sperm RNAs are transcribed and then stored or inactivated in spermatozoa during spermiogenesis. During this stage of spermatogenesis, histones are replaced by protamines in sperm DNA, changing the shape of nucleosomes to a toroid structure with high sperm nuclear condensation culminating in gene expression silencing. However, 1% of histones in mice and 10–15% of histones in humans and cattle remain in the form of nucleosomes in spermatozoa (Samans et al., 2014). The remaining nucleosomes in human and bovine spermatozoa are located in similar promoter regions, indicating transcriptional conservation of specific genes related to early embryonic development (Samans et al., 2014). Even if the presence of RNAs is now accepted in spermatozoa, there is no evidence showing that RNAs can be transcribed from DNA in mature spermatozoa. Most of the RNAs present in sperm cells are thus potentially acquired during spermatogenesis as well as during passage through the epididymis.
Even though sperm cells possess limited quantities of sperm RNAs, sperm-borne RNAs have been associated with many relevant reproductive processes and several correlate with fertility potential in the bull, including AK1 (adenylate kinase 1; R2 = 0.90), IB (integrin β5, R2 = 0.95), NGF (R2 = 0.47), TIMP (tissue inhibitor of metalloproteinases, R2 = 0.39), SNRPN (small nuclear ribonucleoprotein polypeptide N, R2 = 0.71) and PLCζ (phospholipase C, R2 = 0.69) (Kasimanickam et al., 2012). Similarly, the presence of 415 transcripts out of 24,000, were found in different proportions between bovine spermatozoa presenting with high or low fertility potential (Feugang et al., 2010). In human sperm cells, ANXA2 (Annexin A2), BRD2 (Bromodomain Containing 2), OAZ3 (Ornithine Decarboxylase Antizyme 3), PRM1 (Protamine 1) and PRM2 (Protamine 2) transcripts were present in lower amounts in sperm cells with low motility (Jodar et al., 2013).
Sperm miRNAs
Until embryonic gene activation, i.e., when the embryo begins to transcribe its newly formed genome, embryo development relies on the expression of maternal transcripts originating from the oocyte and, to a lesser extent, from spermatozoa. MicroRNAs are also delivered by gametes to the embryos. Until activation of the embryonic genome the abundance of miRNA transcripts present in the embryo decreases until embryonic gene activation begins (Tang et al., 2007). Murine embryos produced from Dicer enzyme knock-out oocytes fail to undergo the first cell division, demonstrating the importance of the miRNAs or other ncRNAs contributed by the female gamete during early embryonic development (Tang et al., 2007). Similarly, murine embryos generated with normal oocytes and Dicer or Drosha enzyme knock-out spermatozoa showed decreased development rates, which recovered following the replacement of miRNAs (Yuan et al., 2016). In addition, the inhibition of miR-34c, which is exclusively delivered by sperm to the embryo in mice (Liu et al., 2012), but not in cattle (Tscherner et al., 2014), prevents the first cleavage (Liu et al., 2012). Sperm-borne miR−449b overexpression in cloned embryos is associated with enhanced cleavage rates at the 8-cell stage and lower rates of apoptosis in the blastocyst (Wang et al., 2017). Sperm-borne miR−216b was recently shown to be present at higher abundance in spermatozoa from low fertility bulls and is associated with a reduced first cleavage rate as well as a reduced number of cells in blastocysts (Alves et al., 2019). In parallel, the RNAs that are acquired by spermatozoa during epididymal transit were shown to be essential for embryo implantation and development in mice (Conine et al., 2018, 2019). Thus, despite the decrease in miRNA levels until activation of the embryonic genome, proper embryo development potentially relies on miRNAs delivered by the gametes (Figure 5).
Contribution of Small Non-coding RNAs to Transgenerational Inheritance of Traits
The transgenerational inheritance of traits concerns the transmission of characteristics from the parents to the offspring, i.e., heritable phenotype, without altering the DNA sequence (Weinhold, 2006). Following advances in the field of epigenetics, i.e., the study of inheritance that is not based on DNA sequence, but on how the DNA sequence is utilized, miRNAs transmitted by spermatozoa at the time of fertilization are considered epigenetic factors that have some effects on the offspring.
Environmental conditions such as toxicant exposure (Schuster et al., 2016), mental stress (Rodgers et al., 2013), alcohol consumption (Rompala et al., 2018), and low protein or high fat diet (Sharma et al., 2016) are considered principal modulators of epigenetic markers by promoting alterations in the sperm “epigenome.” Rodgers et al. (2015) identified nine miRNAs that had altered abundance in the sperm cells from mice submitted to chronic stress. Surprisingly, the injection of these nine miRNAs into embryos of non-stressed parents recapitulated the effects observed in parents undergoing chronic stress (Rodgers et al., 2015). In a similar manner, regulation of another group of ncRNAs in sperm cells, the tRNA-derived fragments, was dependent on the diet of the sire and this influenced the offspring phenotype (Sharma et al., 2016). These studies show that sperm-borne ncRNAs potentially own epigenetic influence. However, further studies are necessary in order to explain the mechanism of action of these molecules in the offspring.
Linking miRNAs to Male (In)Fertility Status
Since miRNAs play an important role in the physiological processes related to spermatogenesis, sperm maturation and early embryonic development (Figure 5), these molecules have the potential to modulate sperm morpho-functional features as well as male fertility ability. In that regards, different miRNA profiles have been associated with different sperm quality levels and fertility phenotypes from animal models and human clinical samples as listed in Table 2.
TABLE 2.
Sperm miRNAs associated with different infertility issues in porcine, human, bovine and mouse.
| miRNAs | Abundance | Phenotype associate | Specie | References |
| let-7a | Down-regulated | Low abnormalities | Porcine | Curry et al., 2011 |
| let-7a-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| let-7d | Down-regulated | Low abnormalities/High motility | Porcine | Curry et al., 2011 |
| let-7e | Down-regulated | Low abnormalities/High motility | Porcine | Curry et al., 2011 |
| let-7f-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-9-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-15a | Down-regulated | High fertility | Bovine | Menezes et al., 2019 |
| miR-15b | Up-regulated | Low abnormalities | Porcine | Curry et al., 2011 |
| miR-15b | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-16 | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-17-5p | Up-regulated | High motility sperm | Bovine | Capra et al., 2017 |
| miR-19a | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-19b-3p | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-20a-5p | Up-regulated | High motility sperm | Bovine | Capra et al., 2017 |
| miR-22 | Down-regulated | Low abnormalities | Porcine | Curry et al., 2011 |
| miR-22-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-26a-5p | Up-regulated | High motility sperm | Bovine | Capra et al., 2017 |
| miR-27a-5p | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-29b | Down-regulated | High fertility | Bovine | Menezes et al., 2019 |
| miR-30b-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-30d-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-33b | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-34a-5p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-34b | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-34c | Up-regulated | High fertility | Mouse | Liu et al., 2012 |
| miR-34c-5p | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-34c-3p | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-93-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-103a-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-122 | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-122-5p | Down-regulated | High motility sperm | Bovine | Capra et al., 2017 |
| miR-126-5p | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-127 | Down-regulated | Capacitated sperm | Porcine | Li et al., 2018 |
| miR-130b-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-132-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-145-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-148b-3p | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-149-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-151-3p | Up-regulated | Capacitated sperm | Porcine | Li et al., 2018 |
| miR-152 | Down-regulated | Capacitated sperm | Porcine | Li et al., 2018 |
| miR-181a-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-184 | Down-regulated | High motility sperm | Bovine | Capra et al., 2017 |
| miR-193b-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-205 | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-208a | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-212-3p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-216b | Down-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-222-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-296-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-320a | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-324-3p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-339a | Down-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-339-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-340-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-346 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-365a-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-432-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-449a | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-483-5p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-486-5p | Down-regulated | High motility sperm | Bovine | Capra et al., 2017 |
| miR-487a | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-491-5p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-500 | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-502-5p | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-505 | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-517-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-518d-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-518f-3p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-520c-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-520d-3p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-526b-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-532 | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-542-5p | Up-regulated | High fertility | Bovine | Alves et al., 2019 |
| miR-543 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-552 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-564 | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-573 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-596 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-622 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-636 | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-644a | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-659-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-708-5p | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-744-5p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-766-3p | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-935 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-942 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-1249 | Down-regulated | High fertility | Bovine | Fagerlind et al., 2015 |
| miR-1254 | Up-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-1285 | Down-regulated | Capacitated sperm | Porcine | Li et al., 2018 |
| miR-1296 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-1298 | Down-regulated | High fertility | Human | Salas-Huetos et al., 2016 |
| miR-1973 | Up-regulated | Asthenozoospermic | Human | Abu-Halima et al., 2013 |
| miR-1973 | Down-regulated | Oligoasthenozoospermic | Human | Abu-Halima et al., 2013 |
Functional studies have been carried out in knock-out mouse models to determine the contribution of individual miRNAs to male fertility (Comazzetto et al., 2014; Wu et al., 2014). For instance, simultaneous deletion of miR34b/c and miR−449 triggered male infertility following the impairment of spermatogenesis, motile ciliogenesis, and sperm maturation (Comazzetto et al., 2014; Wu et al., 2014). While targeted deletion is essential to unravel the function of specific miRNAs in vivo, phenotypic effects are rarely observed after invalidation of a single miRNA-encoding gene due to compensatory functions of related miRNAs. These functional approaches were complemented by several correlative studies performed in large mammals and humans.
Within livestock animals, bulls that presented satisfactory sperm quality and history of distinct fertility (high vs. low fertility) displayed different profiles of sperm miRNAs that are summarized in Table 2 (Fagerlind et al., 2015; Alves et al., 2019; Menezes et al., 2019). Similarly, spermatozoa from boar featuring low motility and a high percentage of abnormalities, displayed a higher abundance of the miRNAs: let−7a, let−7d, let−7e, and miR−22; and lower abundance of miR−15b (Curry et al., 2011) compared with spermatozoa presenting with normal features. In addition, Capra et al. (2017) sorted spermatozoa according to their motility and showed that miR−17−5p, −20a−5p, −26a−5p, −122−5p, −184, and −486−5p were differently displayed between the two subpopulations (Capra et al., 2017). Capacitated and non-capacitated spermatozoa also appeared to show different miRNAs profiles. For instance, Li et al. (2018) showed that miR−127, −151−3p, −152, −1285 were differently expressed in in natura (fresh) compared to capacitated boar sperm (Li et al., 2018). Based on that sperm miRNAs features, variation in expression of miRNAs associated with economically important infertility issues might open avenues to the development of selection programs in livestock animals. Targeting seminal miRNAs candidates as non-invasive biomarkers for male infertility could thus have profound economic impact on livestock industry.
In humans, sperm miRNA profiles from fertile and infertile men accompanied or not by morpho-functional sperm alterations have been extensively investigated. In that regards, men with reproductive disorders presented different sperm miRNA profiles compared with normospermic control individuals: asthenozoospermia (low sperm motility) was associated with higher abundance of 50 sperm miRNAs, among then hsa-miR−34b, −122, and −1973, and lower abundance of 27 sperm miRNAs; oligoasthenozoospermia (low sperm concentration and low sperm motility) was associated with higher abundance of 42 miRNAs (hsa-miR−15b, −16, −19a, −34b, −34c−5p, −122, −449a e −1973 were the most abundant) and lower abundance of 44 miRNAs (Abu-Halima et al., 2013); teratozoospermia (abnormal sperm morphology) was associated with the downregulation of six miRNAs (hsa-miR−151−5p, −935, −125a−3p, −132−5p, −320b, −195−5p) (Salas-Huetos et al., 2015). On the other hand, the sperm miRNA profile of men who presented normal semen quality (normozoospermic), but had a different fertility performance (fertile vs. infertile) showed that 57 miRNAs were presented at a different abundance between the groups. Among these miRNAs miR-15b−5p and 34a−5p, were related to pathways linked to embryonic development and chromatin remodeling (Salas-Huetos et al., 2016). Correlations were also observed between sperm miRNA content and assisted reproductive technology (ART) outcomes. For instance, the study from Cui et al. (2015) indicated that the proportion of good quality embryos obtained after 3 days post-ICSI positively correlated with the amount of hsa-miR−34c found in spermatozoa (Cui et al., 2015). While several confounding factors might be considered before drawing extrapolative conclusions, some spermatic miRNAs may thus become predictive biomarker of defective spermatozoa and ART outcome. Tentative explanations have been proposed regarding the molecular mechanism involved in these correlative observations between sperm miRNA profiles and fertility levels. For instance, hsa-miR−27a that is increased in asthenoteratozoospermic patients regulates the post-transcriptional expression of the Cysteine-Rich Secretory Protein2 (CRISP2), which encodes for a protein playing a role in sperm motility, acrosome reaction and gamete fusion (Zhou et al., 2017).
Overall, the different studies performed on human clinical samples as well as on animal models have shown that different profiles of sperm miRNA content can be observed according to sperm morpho-functional features and fertility status. Extracellular miRNAs correlated with male infertility issues are also retrieved in seminal plasma (Barceló et al., 2018), which encompasses secretions from the different internal organs of the male reproductive tract. While both spermatic and extra-cellular RNA could be used as molecular targets for the non-invasive diagnosis of male infertility and reproductive system diseases, a better understanding of miRNA-mediated regulation of reproductive functions is needed.
Final Considerations
In summary, sperm cells should display specific factors to fulfill the “healthy sperm” concept, including proper morpho-functional features in addition to intrinsic factors. While sperm cell morpho-functional attributes are not always sufficient to predict male fertility potential, spermatozoa carry different factors, including organelles (centrioles, mitochondria) and molecules (proteins, RNAs, ncRNAs), which are involved in important steps of reproductive biology, e.g., spermatogenesis, sperm maturation, fertilization and embryo development. These factors constitute potential biomarkers of “healthy sperm” and male fertility status and may become major assets for diagnosing instances of idiopathic male infertility in both humans and livestock animals. A better understanding of the mechanism of action of these sperm intrinsic factors in the regulation of reproductive and developmental processes still presents a major challenge that must be addressed.
Author Contributions
CB, MA, and EC wrote the manuscript and designed the figures. All the authors revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. We are grateful to the São Paulo Research Foundation (FAPESP) and the Natural Sciences and Engineering Research Council of Canada for financing the studies by the authors described in this review (MA was funded by FAPESP Grants #2015/09154-6 and #2016/23243-4. EC was funded by FAPESP Grant #2016/05395-1. CB was the recipient of an NSERC discovery grant).
References
- Aarabi M., Balakier H., Bashar S., Moskovtsev S. I., Sutovsky P., Librach C. L., et al. (2014). Sperm content of postacrosomal WW binding protein is related to fertilization outcomes in patients undergoing assisted reproductive technology. Fertil. Steril. 102 440–447. 10.1016/j.fertnstert.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Abu-Halima M., Hammadeh M., Schmitt J., Leidinger P., Keller A., Meese E., et al. (2013). Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 99 1249.e16–1255.e16. 10.1016/j.fertnstert.2012.11.054 [DOI] [PubMed] [Google Scholar]
- Agarwal A., Mulgund A., Hamada A., Chyatte M. R. (2015). A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 13 1–9. 10.1186/s12958-015-0032-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken R. J., Wingate J. K., De Iuliis G. N., McLaughlin E. A. (2007). Analysis of lipid peroxidation in human spermatozoa using BODIPY C11. MHR Basic Sci. Reprod. Med. 13 203–211. 10.1093/molehr/gal119 [DOI] [PubMed] [Google Scholar]
- Alves M. B. R., de Arruda R. P., De Bem T. H. C., Florez-Rodriguez S. A., Sá Filho M. F., de Belleannée C., et al. (2019). Sperm-borne miR-216b modulates cell proliferation during early embryo development via K-RAS. Sci. Rep. 9 1–14. 10.1038/s41598-019-46775-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. P., Hammerstedt R. H. (1993). In vitro evaluation of sperm quality: an opinion. J. Androl. 14 397–406. 10.1002/j.1939-4640.1993.tb03247.x [DOI] [PubMed] [Google Scholar]
- Amaral A., Lourenço B., Marques M., Ramalho-Santos J. (2013). Mitochondria functionality and sperm quality. Reproduction 146 163–174. 10.1530/REP-13-0178 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2004). The functions of animal microRNAs. Nature 431 350–355. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., Fishman E. L. (2019). It takes two (centrioles) to tango. Reproduction 157 R33–R51. 10.1530/REP-18-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr G. F., Engler W. F. (1970). Considerations of volume, mass, DNA, and arrangement of mitochondria in the midpiece of bull spermatozoa. Exp. Cell Res. 60 338–340. 10.1016/0014-4827(70)90526-4 [DOI] [PubMed] [Google Scholar]
- Barceló M., Mata A., Bassas L., Larriba S. (2018). Exosomal microRNAs in seminal plasma are markers of the origin of azoospermia and can predict the presence of sperm in testicular tissue. Hum. Reprod. 33 1087–1098. 10.1093/humrep/dey072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2018). Metazoan MicroRNAs. Cell 173 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannée C. (2015). Extracellular microRNAs from the epididymis as potential mediators of cell-to-cell communication. Asian J. Androl. 17 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannée C., Calvo É, Caballero J., Sullivan R. (2013). Epididymosomes convey different repertoires of microRNAs throughout the bovine epididymis. Biol. Reprod. 89:30. 10.1095/biolreprod.113.110486 [DOI] [PubMed] [Google Scholar]
- Betlach C., Erickson R. (1973). A unique RNA species from maturing mouse spermatozoa. Nature 242 114–115. 10.1038/242114a0 [DOI] [PubMed] [Google Scholar]
- Bianchi E., Doe B., Goulding D., Wright G. J. (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508 483–487. 10.1038/nature13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkgren I., Saastamoinen L., Krutskikh A., Huhtaniemi I., Poutanen M., Sipilä P. (2012). Dicer1 ablation in the mouse epididymis causes dedifferentiation of the epithelium and imbalance in sex steroid signaling. PLoS One 7:e38457. 10.1371/journal.pone.0038457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom E. (1973). The ultrastructure of some characteristic sperm defects and a proposal for a new classification of the bull spermiogram. Nord. Vet. Med. 25 383–391. [PubMed] [Google Scholar]
- Boerke A., Dieleman S. J., Gadella B. M. (2007). A possible role for sperm RNA in early embryo development. Theriogenology 68(Suppl. 1) S147–S155. 10.1016/j.theriogenology.2007.05.058 [DOI] [PubMed] [Google Scholar]
- Bouhallier F., Allioli N., Lavial F., Chalmel F., Perrard M.-H., Durand P., et al. (2010). Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16 720–731. 10.1261/rna.1963810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero J., Frenette G., Amours O., Belleannée C., Lacroix-Pepin N., Robert C., et al. (2012). Bovine sperm raft membrane associated Glioma Pathogenesis-Related 1-like protein 1 (GliPr1L1) is modified during the epididymal transit and is potentially involved in sperm binding to the zona pellucida. J. Cell. Physiol. 227 3876–3886. 10.1002/jcp.24099 [DOI] [PubMed] [Google Scholar]
- Capra E., Turri F., Lazzari B., Cremonesi P., Gliozzi T. M., Fojadelli I., et al. (2017). Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between High- and Low-motile sperm populations. BMC Genomics 18:14. 10.1186/s12864-016-3394-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeghini E. C. C., Alves M. B. R., de Arruda R. P., de Rezende G. M., Florez-Rodriguez S. A., de Sá Filho M. F. (2019). Efficiency of CellROX deep red ® and CellROX orange ® fluorescent probes in identifying reactive oxygen species in sperm samples from high and low fertility bulls. Anim. Biotechnol. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Celeghini E. C. C., de Arruda R. P., de Andrade A. F. C., Nascimento J., Raphael C. F. (2007). Practical techniques for bovine sperm simultaneous fluorimetric assessment of plasma, acrosomal and mitochondrial membranes. Reprod. Domest. Anim. 42 479–488. 10.1111/j.1439-0531.2006.00810.x [DOI] [PubMed] [Google Scholar]
- Comazzetto S., Di Giacomo M., Rasmussen K. D., Much C., Azzi C., Perlas E., et al. (2014). Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci. PLoS Genet. 10:e1004597. 10.1371/journal.pgen.1004597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Sun F., Song L., Rivera-Pérez J. A., Rando O. J. (2018). Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell 46 470.e3–480.e3. 10.1016/j.devcel.2018.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Sun F., Song L., Rivera-Pérez J. A., Rando O. J. (2019). MicroRNAs absent in caput sperm are required for normal embryonic development. Dev. Cell 50 7–8. 10.1016/j.devcel.2019.06.007 [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Yeung C.-H. (2003). Acquisition of volume regulatory response of sperm upon maturation in the epididymis and the role of the cytoplasmic droplet. Microsc. Res. Tech. 61 28–38. 10.1002/jemt.10314 [DOI] [PubMed] [Google Scholar]
- Correa J. R., Pace M. M., Zavos P. M. (1997). Relationships among frozen-thawed sperm characteristics assessed via the routine semen analysis, sperm functional tests and fertility of bulls in an artificial insemination program. Theriogenology 48 721–731. 10.1016/S0093-691X(97)00296-3 [DOI] [PubMed] [Google Scholar]
- Cui L., Fang L., Shi B., Qiu S., Ye Y. (2015). Spermatozoa micro ribonucleic acid–34c level is correlated with intracytoplasmic sperm injection outcomes. Fertil. Steril. 104 312.e1–317.e1. 10.1016/j.fertnstert.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Curry E., Safranski T. J., Pratt S. L. (2011). Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology 76 1532–1539. 10.1016/j.theriogenology.2011.06.025 [DOI] [PubMed] [Google Scholar]
- D’Amours O., Frenette G., Bordeleau L.-J., Allard N., Leclerc P., Blondin P., et al. (2012). Epididymosomes transfer epididymal sperm binding protein 1 (ELSPBP1) to dead spermatozoa during epididymal transit in bovine. Biol. Reprod. 87 1–11. 10.1095/biolreprod.112.100990 [DOI] [PubMed] [Google Scholar]
- Evenson D. P., Wixon R. (2006). Clinical aspects of sperm DNA fragmentation detection and male infertility. Theriogenology 65 979–991. 10.1016/j.theriogenology.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Fagerlind M., Stalhammar H., Olsson B., Klinga-Levan K. (2015). Expression of miRNAs in bull spermatozoa correlates with fertility rates. Reprod. Domest. Anim. 50 587–594. 10.1111/rda.12531 [DOI] [PubMed] [Google Scholar]
- Farrell P. B., Presicce G. A., Brockett C. C., Foote R. H. (1998). Quantification of bull sperm characteristics measured by computer-assisted sperm analysis (CASA) and the relationship to fertility. Theriogenology 49 871–879. 10.1016/S0093-691X(98)00036-3 [DOI] [PubMed] [Google Scholar]
- Feugang J. M., Rodriguez-Osorio N., Kaya A., Wang H., Page G., Ostermeier G. C., et al. (2010). Transcriptome analysis of bull spermatozoa: implications for male fertility. Reprod. Biomed. Online 21 312–324. 10.1016/j.rbmo.2010.06.022 [DOI] [PubMed] [Google Scholar]
- Fishman E. L., Jo K., Nguyen Q. P. H., Kong D., Royfman R., Cekic A. R., et al. (2018). A novel atypical sperm centriole is functional during human fertilization. Nat. Commun. 9:2210. 10.1038/s41467-018-04678-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea J., Parrington J., Kashir J., Coward K. (2013). “The male reproductive tract and spermatogenesis,” in Textbook of Clinical Embryology, eds Coward K., Wells D. (Cambridge: Cambridge University Press; ), 18–26. 10.1017/cbo9781139192736.005 [DOI] [Google Scholar]
- Gillan L., Evans G., Maxwell W. M. C. (2005). Flow cytometric evaluation of sperm parameters in relation to fertility potential. Theriogenology 63 445–457. 10.1016/j.theriogenology.2004.09.024 [DOI] [PubMed] [Google Scholar]
- Gillan L., Kroetsch T., Chis Maxwell W. M., Evans G. (2008). Assessment of in vitro sperm characteristics in relation to fertility in dairy bulls. Anim. Reprod. Sci. 103 201–214. 10.1016/j.anireprosci.2006.12.010 [DOI] [PubMed] [Google Scholar]
- Gyllensten U., Wharton D., Josefsson A., Wilson A. C. (1991). Paternal inheritance of mitochondrial DNA in mice. Nature 352 255–257. 10.1038/352255a0 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Chuva de Sousa Lopes S. M., Kaneda M., Tang F., Hajkova P., Lao K., et al. (2008). MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3:e0001738. 10.1371/journal.pone.0001738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht N. B., Liem H., Kleene K. C., Distel R. J., Ho S. (1984). Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev. Biol. 102 452–461. 10.1016/0012-1606(84)90210-0 [DOI] [PubMed] [Google Scholar]
- Hermo L., Oka R., Morales C. R. (1994). Secretion and endocytosis in the male reproductive tract: a role in sperm maturation. Int. Rev. Cytol. 154 106–189. 10.1016/S0074-7696(08)62199-3 [DOI] [PubMed] [Google Scholar]
- Hirohashi N., Yanagimachi R. (2018). Sperm acrosome reaction: its site and role in fertilization. Biol. Reprod. 99 127–133. 10.1093/biolre/ioy045 [DOI] [PubMed] [Google Scholar]
- Inaba K. (2003). Molecular architecture of the sperm flagella: molecules for motility and signaling. Zool. Sci. 20 1043–1056. 10.2108/zsj.20.1043 [DOI] [PubMed] [Google Scholar]
- Irvine D. S. (1998). Epidemiology and aetiology of male infertility. Hum. Reprod. 13 33–38. 10.1093/humrep/13.1.33 [DOI] [PubMed] [Google Scholar]
- Jerczynski O., Lacroix-Pépin N., Boilard E., Calvo E., Bernet A., Fortier M. A., et al. (2016). Role of Dicer1-dependent factors in the paracrine regulation of epididymal gene expression. PLoS One 11:e0163876. 10.1371/journal.pone.0163876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M., Selvaraju S., Sendler E., Diamond M. P., Krawetz S. A. (2013). The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 19 604–624. 10.1093/humupd/dmt031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashir J., Heindryckx B., Jones C., De Sutter P., Parrington J., Coward K. (2010). Oocyte activation, phospholipase C zeta and human infertility. Hum. Reprod. Update 16 690–703. 10.1093/humupd/dmq018 [DOI] [PubMed] [Google Scholar]
- Kasimanickam V., Kasimanickam R., Arangasamy A., Saberivand A., Stevenson J. S., Kastelic J. P. (2012). Association between mRNA abundance of functional sperm function proteins and fertility of Holstein bulls. Theriogenology 78 2007–2019. 10.1016/j.theriogenology.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Kotaja N. (2014). MicroRNAs and spermatogenesis. Fertil. Steril. 101 1552–1562. 10.1016/j.fertnstert.2014.04.025 [DOI] [PubMed] [Google Scholar]
- Krawetz S. A. (2005). Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 6 633–642. 10.1038/nrg1654 [DOI] [PubMed] [Google Scholar]
- Kwon W.-S., Rahman M., Lee J.-S., Kim J., Yoon S.-J., Park Y.-J., et al. (2014). A comprehensive proteomic approach to identifying capacitation related proteins in boar spermatozoa. BMC Genomics 15:897. 10.1186/1471-2164-15-897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon W.-S., Rahman M. S., Lee J.-S., You Y.-A., Pang M.-G. (2015). Improving litter size by boar spermatozoa: application of combined H33258/CTC staining in field trial with artificial insemination. Andrology 3 552–557. 10.1111/andr.12020 [DOI] [PubMed] [Google Scholar]
- Li Y., Li R. H., Ran M. X., Zhang Y., Liang K., Ren Y. N., et al. (2018). High throughput small RNA and transcriptome sequencing reveal capacitation-related microRNAs and mRNA in boar sperm. BMC Genomics 19:1–12. 10.1186/s12864-018-5132-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Pang R. T. K., Chiu P. C. N., Wong B. P. C., Lao K., Lee K. (2012). Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. U.S.A. 109 490–494. 10.1073/pnas.1110368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Valencia C. A., Zhang J., Lee N.-C., Slone J., Gui B., et al. (2018). Biparental Inheritance of Mitochondrial DNA in Humans. Proc. Natl. Acad. Sci. U.S.A. 115 13039–13044. 10.1073/pnas.1810946115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. (2001). Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2 986–991. 10.1093/embo-reports/kve230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes E. S. B., Badial P. R., El Debaky H., Husna A. U., Ugur M. R., Kaya A., et al. (2019). Sperm miR-15a and miR-29b are associated with bull fertility. Andrologia 52:e13412. 10.1111/and.13412 [DOI] [PubMed] [Google Scholar]
- Morrell J. M., Valeanu A. S., Lundeheim N., Johannisson A. (2018). Sperm quality in frozen beef and dairy bull semen. Acta Vet. Scand. 60 1–10. 10.1186/s13028-018-0396-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navara C. S., Hewitson L. C., Simerly C. R., Sutovsky P., Schatten G. (1997). The implications of a paternally derived centrosome during human fertilization: consequences for reproduction and the treatment of male factor infertility. Am. J. Reprod. Immunol. 37 39–49. 10.1111/j.1600-0897.1997.tb00191.x [DOI] [PubMed] [Google Scholar]
- Nixon B., Stanger S. J., Mihalas B. P., Reilly J. N., Anderson A. L., Tyagi S., et al. (2015). The MicroRNA signature of mouse spermatozoa is substantially modified during epididymal maturation. Biol. Reprod. 93 1–20. 10.1095/biolreprod.115.132209 [DOI] [PubMed] [Google Scholar]
- Odia M., Swanson G., Krawetz S. A. (2018). A history of why fathers’ RNA matters. Biol. Reprod. 99 147–159. 10.1093/biolre/ioy007 [DOI] [PubMed] [Google Scholar]
- Oliveira B. M., Arruda R. P., Thomé H. E., Maturana Filho M., Oliveira G., Guimarães C., et al. (2014). Fertility and uterine hemodynamic in cows after artificial insemination with semen assessed by fluorescent probes. Theriogenology 82 767–772. 10.1016/j.theriogenology.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Ostermeier G. C., Goodrich R. J., Diamond M. P., Dix D. J., Krawetz S. A. (2005). Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil. Steril. 83 1687–1694. 10.1016/j.fertnstert.2004.12.046 [DOI] [PubMed] [Google Scholar]
- Ostermeier G. C., Miller D., Huntriss J. D., Diamond M. P., Krawetz S. A. (2004). Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429:2603. 10.1038/nature02602 [DOI] [PubMed] [Google Scholar]
- Papaioannou M. D., Pitetti J. L., Ro S., Park C., Aubry F., Schaad O., et al. (2009). Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 326 250–259. 10.1016/j.ydbio.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J., Duerksen J. D. (1975). Chromatin-associated RNA content of heterochromatin and euchromatin. Mol. Cell. Biochem. 9 9–16. 10.1007/BF01731728 [DOI] [PubMed] [Google Scholar]
- Piomboni P., Focarelli R., Stendardi A., Ferramosca A., Zara V. (2012). The role of mitochondria in energy production for human sperm motility. Int. J. Androl. 35 109–124. 10.1111/j.1365-2605.2011.01218.x [DOI] [PubMed] [Google Scholar]
- Ran M., Weng B., Cao R., Li Z., Peng F., Luo H., et al. (2018). miR-26a inhibits proliferation and promotes apoptosis in porcine immature Sertoli cells by targeting the PAK2 gene. Reprod. Domest. Anim. 53 1375–1385. 10.1111/rda.13254 [DOI] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200 373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly J. N., McLaughlin E. A., Stanger S. J., Anderson A. L., Hutcheon K., Church K., et al. (2016). Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci. Rep. 6 1–15. 10.1038/srep31794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers A. B., Morgan C. P., Bronson S. L., Revello S., Bale T. L. (2013). Paternal stress exposure alters sperm MicroRNA content and reprograms offspring HPA stress axis regulation. J. Neurosci. 33 9003–9012. 10.1523/JNEUROSCI.0914-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers A. B., Morgan C. P., Leu N. A., Bale T. L. (2015). Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. U.S.A. 112 13699–13704. 10.1073/pnas.1508347112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompala G. R., Simons A., Kihle B., Homanics G. E. (2018). Paternal preconception chronic variable stress confers attenuated ethanol drinking behavior selectively to male offspring in a pre-stress environment dependent manner. Front. Behav. Neurosci. 12:257. 10.3389/fnbeh.2018.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Huetos A., Blanco J., Vidal F., Godo A., Grossmann M., Pons M. C., et al. (2015). Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil. Steril. 104 591–601. 10.1016/j.fertnstert.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Salas-Huetos A., Blanco J., Vidal F., Grossmann M., Pons M. C., Garrido N., et al. (2016). Sperm from normozoospermic fertile and infertile individuals convey a distinct miRNA cargo. Andrology 4 1–9. 10.1111/andr.12276 [DOI] [PubMed] [Google Scholar]
- Samans B., Yang Y., Krebs S., Sarode G. V., Blum H., Reichenbach M., et al. (2014). Uniformity of nucleosome preservation pattern in mammalian sperm and Its connection to repetitive DNA elements. Dev. Cell 30 23–35. 10.1016/j.devcel.2014.05.023 [DOI] [PubMed] [Google Scholar]
- Sathananthan A. H., Kola I., Osborne J., Trounson A., Ng S. C., Bongso A., et al. (1991). Centrioles in the beginning of human development. Proc. Natl. Acad. Sci. U.S.A. 88 4806–4810. 10.1073/pnas.88.11.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satouh Y., Inoue N., Ikawa M., Okabe M. (2012). Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J. Cell Sci. 125 4985–4990. 10.1242/jcs.100867 [DOI] [PubMed] [Google Scholar]
- Saunders C. M., Larman M. G., Parrington J., Cox L. J., Royse J., Blayney L. M., et al. (2002). PLC ζ: a sperm-specific trigger of Ca 2 + oscillations in eggs and embryo development. Development 3544 3533–3544. [DOI] [PubMed] [Google Scholar]
- Schatten H., Sun Q.-Y. (2009). The role of centrosomes in mammalian fertilization and its significance for ICSI. Mol. Hum. Reprod. 15 531–538. 10.1093/molehr/gap049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A., Skinner M. K., Yan W. (2016). Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ. Epigenet. 2:dvw001. 10.1093/eep/dvw001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellem E., Broekhuijse M. L. W. J., Chevrier L., Camugli S., Schmitt E., Schibler L., et al. (2015). Use of combinations of in vitro quality assessments to predict fertility of bovine semen. Theriogenology 84 1447.e5–1454.e5. 10.1016/j.theriogenology.2015.07.035 [DOI] [PubMed] [Google Scholar]
- Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351 391–396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Sun F., Conine C. C., Reichholf B., Kukreja S., Herzog V. A., et al. (2018). Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46 481–494. 10.1016/j.devcel.2018.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerly C., Manil-Ségalen M., Castro C., Hartnett C., Kong D., Verlhac M. H., et al. (2018). Separation and loss of centrioles from primordidal germ cells to mature oocytes in the mouse. Sci. Rep. 8 1–17. 10.1038/s41598-018-31222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerly C., Wu G.-J., Zoran S., Ord T., Rawlins R., Jones J., et al. (1995). The paternal inheritance of the centrosome, the cell’s microtubule-organizing center, in humans, and the implications for infertility. Nat. Med. 1 47–52. 10.1038/nm0195-47 [DOI] [PubMed] [Google Scholar]
- Sipilä P., Björkgren I. (2016). Segment-specific regulation of epididymal gene expression. Reproduction 152 R91–R99. 10.1530/REP-15-0533 [DOI] [PubMed] [Google Scholar]
- Smorag L., Zheng Y., Nolte J., Zechner U., Engel W., Pantakani D. V. K. (2012). MicroRNA signature in various cell types of mouse spermatogenesis: evidence for stage-specifically expressed miRNA-221, -203 and -34b-5p mediated spermatogenesis regulation. Biol. Cell 104 677–692. 10.1111/boc.201200014 [DOI] [PubMed] [Google Scholar]
- Song G. J., Lewis V. (2008). Mitochondrial DNA integrity and copy number in sperm from infertile men. Fertil. Steril. 90 2238–2244. 10.1016/j.fertnstert.2007.10.059 [DOI] [PubMed] [Google Scholar]
- Sullivan R. (2015). Epididymosomes: a heterogeneous population of microvesicles with multiple functions in sperm maturation and storage. Asian J. Androl. 17 726–729. 10.4103/1008-682X.155255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R., Frenette G., Girouard J. (2007). Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J. Androl. 9 483–491. 10.1111/j.1745-7262.2007.00281.x [DOI] [PubMed] [Google Scholar]
- Sullivan R., Saez F. (2013). Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction 146 R21–35. 10.1530/REP-13-0058 [DOI] [PubMed] [Google Scholar]
- Tang F., Kaneda M., O’Carroll D., Hajkova P., Barton S. C., Sun Y. A., et al. (2007). Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 21 644–648. 10.1101/gad.418707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherner A., Gilchrist G., Smith N., Blondin P., Gillis D., LaMarre J. (2014). MicroRNA-34 family expression in bovine gametes and preimplantation embryos. Reprod. Biol. Endocrinol. 12:85. 10.1186/1477-7827-12-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P., Underwood S. L., Dolbec C., Bouchard N., Kroetsch T., Blondin P. (2012). Bovine semen quality control in artificial insemination centers. Anim. Reprod. 9 153–165. [Google Scholar]
- Wang C., Yang C., Chen X., Yao B., Yang C., Zhu C., et al. (2011). Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin. Chem. 57 1722–1731. 10.1373/clinchem.2011.169714 [DOI] [PubMed] [Google Scholar]
- Wang M., Gao Y., Qu P., Qing S., Qiao F., Zhang Y., et al. (2017). Sperm-borne miR-449b influences cleavage, epigenetic reprogramming and apoptosis of SCNT embryos in bovine. Sci. Rep. 7 1–12. 10.1038/s41598-017-13899-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold B. (2006). Epigenetics: the science of change. Environ. Health Perspect. 114 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (1993). Posttranscriptional regulation of the heterochronic gene lin- 14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 855–862. 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Wu A. T. H., Sutovsky P., Manandhar G., Xu W., Katayama M., Day B. N., et al. (2007). PAWP, a sperm-specific WW domain-binding protein, promotes meiotic resumption and pronuclear development during fertilization. J. Biol. Chem. 282 12164–12175. 10.1074/jbc.M609132200 [DOI] [PubMed] [Google Scholar]
- Wu J., Bao J., Kim M., Yuan S., Tang C., Zheng H., et al. (2014). Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 111 E2851–E2857. 10.1073/pnas.1407777111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Schuster A., Tang C., Yu T., Ortogero N., Bao J., et al. (2016). Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143 635–647. 10.1242/dev.131755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers-Hochschild F., Adamson G. D., Dyer S., Racowsky C., De Mouzon J., Sokol R., et al. (2017). The international glossary on infertility and fertility care, 2017. Hum. Reprod. 32 1786–1801. 10.1093/humrep/dex234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-H., Zhou Q.-Z., Yang J.-K., Lyu X.-M., Bian J., Guo W.-B., et al. (2017). MicroRNA-27a-mediated repression of cysteine-rich secretory protein 2 translation in asthenoteratozoospermic patients. Asian J. Androl. 19:591. 10.4103/1008-682X.185001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Stanger S. J., Anderson A. L., Bernstein I. R., De Iuliis G. N., McCluskey A., et al. (2019). Mechanisms of tethering and cargo transfer during epididymosome-sperm interactions. BMC Biol. 17:1–18. 10.1186/s12915-019-0653-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C., Romero Y., Warnefors M., Bilican A., Borel C., Smith L. B., et al. (2014). Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS One 9:e107023. 10.1371/journal.pone.0107023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zini A., Agarwal A. (2011). Sperm Chromatin. New York, NY: Springer. [Google Scholar]
- Ziyyat A. (2001). Differential gene expression in pre-implantation embryos from mouse oocytes injected with round spermatids or spermatozoa. Hum. Reprod. 16 1449–1456. 10.1093/humrep/16.7.1449 [DOI] [PubMed] [Google Scholar]