Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is a novel zoonotic coronavirus that was identified in 2012. MERS-CoV infection in humans can result in an acute, severe respiratory disease and in some cases multi-organ failure; the global mortality rate is approximately 35 %. The MERS-CoV spike (S) protein is a major target for neutralizing antibodies in infected patients. The MERS-CoV microneutralization test (MNt) is the gold standard method for demonstrating prior infection. However, this method requires the use of live MERS-CoV in biosafety level 3 (BSL-3) containment. The present work describes the generation and validation of S protein-bearing vesicular stomatitis virus (VSV) pseudotype particles (VSV-MERS-CoV-S) in which the VSV glycoprotein G gene has been replaced by the luciferase reporter gene, followed by the establishment of a pseudoparticle-based neutralization test to detect MERS-CoV neutralizing antibodies under BSL-2 conditions. Using a panel of human sera from confirmed MERS-CoV patients, the VSV-MERS-CoV particle neutralization assay produced results that were highly comparable to those of the microneutralization test using live MERS-CoV. The results suggest that the VSV-MERS-CoV-S pseudotype neutralization assay offers a highly specific, sensitive and safer alternative method to detect MERS-CoV neutralizing antibodies in human sera.
Keywords: Middle East respiratory syndrome coronavirus (MERS-CoV), vesicular stomatitis virus, pseudoparticle, luciferase, neutralizing antibodies, microneutralization test (MNt)
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is a zoonotic pathogen that is associated with respiratory virus infections ranging in severity from asymptomatic to severe respiratory illnesses and death. MERS-CoV was first isolated from a 60-year-old patient who died with viral pneumonia and acute renal failure in 2012 in Saudi Arabia [1, 2]. MERS-CoV cases have been identified in 27 countries, although all cases outside of the Arabian Peninsula have been associated with exportations from the Arabian Peninsula [3–7]. As of 28 February 2019, the World Health Organization (WHO) has reported 2374 confirmed cases of MERS-COV infection globally, resulting in 823 deaths (https://www.who.int/emergencies/mers-cov/en/). Genetically, MERS-CoV most closely resembles severe acute respiratory syndrome coronavirus (SARS-CoV), a betacoronavirus known as the first coronavirus to cause severe respiratory infections in humans. SARS-CoV emerged from an animal host in 2002 and caused a worldwide outbreak comprising an estimated 8000 cases with 800 deaths [7–10]. Similar to SARS-CoV, MERS-CoV infections represent zoonotic transmission events [11–13]. Several sero-epidemiology studies have demonstrated that dromedary camels from the Arabian Peninsula and North Africa have high titres of neutralizing antibodies against MERS-CoV, which can be detected in camel sera collected over 30 years [14–17]. In contrast to the SARS outbreak, which was brief, MERS-CoV cases continue to occur 7 years after its identification. The continuous spread of MERS-CoV serves as a constant reminder of the propensity of novel human coronaviruses to emerge from zoonotic reservoirs and cause severe disease in humans, and so continuous serological surveillance efforts remain essential for these viruses.
The risk factors for camel–human transmission, human–human transmission and hospital outbreaks, and the roles of asymptomatic cases in human–human transmission are not fully understood. The clinical symptoms for MERS patients overlap with those for other lower respiratory tract infections, further demonstrating the need to develop low biosafety-level laboratory-based diagnostic assays with high specificity and sensitivity. Furthermore, countermeasures for the treatment or prevention of MERS-CoV infection, including vaccines, are unavailable. Therefore, methods for measuring the potency and breadth of neutralization antibodies against MERS-CoV are essential to address these gaps in our knowledge, as well as to strengthen disease surveillance and disease preparedness platforms.
The spike (S) protein of MERS-CoV is a surface glycoprotein that facilitates receptor binding, membrane fusion and viral entry into host cells via attachment to the cellular receptor dipeptidyl peptidase IV (DPP4), thus playing a pivotal role in MERS-CoV infection of the host cell [18]. The S protein is a major immunogenic component of CoVs and is the major target for neutralizing antibodies [19–22]. Laboratory tests, such as immunofluorescence assays (IFAs) and conventional enzyme-linked immunosorbent assays (ELISAs), are commonly used screening tools for detecting anti-MERS-CoV serum antibodies. However, these assays often yield false-positive reactions due to cross-reactivity with other common human coronaviruses, and so confirmatory tests such as neutralization tests are utilized [23]. The most widely used confirmatory serological assays to detect and measure neutralizing antibody responses to MERS-CoV utilize neutralization of live virus, through either the plaque reduction neutralization test (PRNT) or the microneutralization test (MNt) methods. However, due to the highly pathogenic nature of MERS-CoV and the absence of treatments and vaccines for MERS-CoV infection, PRNT and MNt tests must be conducted under strict bio-containment procedures in a biosafety level 3 (BSL-3) laboratory. This limits epidemiological and pathogenesis studies globally. The development of an alternative method that can be reliably and conveniently used to determine the prevalence of MERS-CoV antibodies at a population level is crucial to prevent the spread of MERS-CoV.
A safe and convenient way to detect neutralizing antibodies against MERS-CoV in serum is the use of viral pseudotypes. Viral pseudotype particles have proven to be very valuable tools for studying virus entry pathways, evaluating the efficacy of vaccines, serological surveillance and gene therapy studies [24–31]. Several reports have utilized MERS-lentivirus pseudotyped viruses in sero-epidemiological studies and basic research investigations [21, 32–36]. Additionally, vesicular stomatitis virus (VSV) pseudotypes with MERS-CoV spike glycoproteins have been utilized to investigate the role of the MERS S protein in virus–receptor-mediated entry, virus/host tropism and drug screening [37–39]. However, systematic comparative equivalency analysis between VSV pseudotypes bearing MERS-CoV spike glycoproteins and the conventional MNt using human sera from confirmed MERS-CoV patients have not been performed.
The purpose of this study was to develop a VSV pseudotype-based neutralization assay to detect MERS-CoV S protein neutralizing antibodies and calculate its equivalence to traditional neutralization assays, therefore circumventing the use of live virus and the need for widely unavailable high-level containment biosafety facilities. Here, we validate a MERS-CoV neutralization assay using a membraned VSV pseudotype system, where its glycoprotein is replaced with a luciferase reporter gene and its membrane is decorated with MERS-CoV S proteins. Additionally, we perform equivalence testing in comparison with the conventional MNt. We demonstrate that these pseudoparticles are neutralized by sera from MERS-CoV-infected patients. Furthermore, most of the patient sera samples have neutralization activities, and these results were consistent with the neutralization activity that was measured by the conventional microneutralization assay using live MERS-CoV. These results demonstrate that the MERS-CoV-S protein pseudotyped VSV particle-based neutralization assay would serve as a safe, reliable and highly specific alternative method to detect MERS-CoV neutralizing antibodies to be used for future sero-epidemiological studies.
Methods
Cells
Baby hamster kidney-21 (BHK-21) cells were cultured and maintained in Dulbecco’s modified essential medium (DMEM) containing 5 % fetal bovine serum (FBS) (Life Technologies/Gibco), and 1× penicillin/streptomycin (p/s) (Sigma) at 37 °C under a 5 % CO2 atmosphere. Vero (African green monkey kidney epithelial) cells were cultured and maintained in DMEM containing 10 % FBS and 1× p/s 37 °C under a 5 % CO2 atmosphere. Different VSV-based pseudotypes bearing either MERS-CoV-S or VSV-G glycoproteins were produced in BHK-21 cells. The pseudoparticle titrations and neutralization assays were carried out in Vero cells.
Serum samples and monoclonal antibody
Pooled normal human serum (pNHS) was purchased from Lee Biosolutions and used as a negative control serum. Human serum from a single patient with laboratory-confirmed MERS-CoV infection was used as the positive control serum. The positive control serum was collected from the first imported case of MERS-CoV in the USA during the case investigation, with a neutralizing titre of 320. A laboratory-confirmed SARS-CoV patient serum sample and a panel of human sera with confirmed high neutralizing antibody titres to human coronaviruses 229E, HKU1, OC43 and NL63 were used in this study to evaluate the VSV-MERS-CoV-S particle-based neutralization assay for potential cross-neutralization. A total of 52 human sera samples from MERS-CoV-infected patients in Saudi Arabia were used to examine equivalences. Anti-VSV-G monoclonal antibody was purchased from Kerafast, Inc.
Production of pseudotyped viruses
A codon-optimized S gene from the MERS-CoV Florida isolate (GenBank accession number: KJ829365.1) was synthesized by GeneScript and sub-cloned into pCAGGS 2.0 eukaryotic expression vector. VSV-MERS-CoV-S pseudoparticles were generated as previously described [24]. Briefly, BHK-21 cells were transfected with pCAGGS plasmid encoding MERS-CoV-FL-S (pCAGGS-MERS-FL-S), VSV-G (pCAGGS-VSV-G), or pCAGGS empty vector (pCAGGS-EV) with Lipofectamine 2000 (Life Technologies). At 24 h post-transfection, cells were infected with pVSVΔG*-G-luciferase at a multiplicity of infection (m.o.i.) of 5. Supernatants containing pseudoparticles were harvested at 24 h post-pVSVΔG*-G-luciferase inoculation and aliquots were stored at −80 °C.
Titration of VSV-MERS-CoV-S pseudoparticles
The pseudoparticle titres were determined in Vero cells. Vero cells were seeded in 96-well black and white tissue culture-treated plates (Perkin-Elmer) and were inoculated with 50 µl of pseudoparticles five fold serially diluted in serum-free DMEM (1× p/s in triplicate in . After 1 h adsorption, the inocula were removed, replaced with fresh DMEM containing 10 % FBS and 1× p/s, and incubated at 37 °C. At 24 h post-inoculation, luciferase activity was determined using the Luciferase Assay kit (Promega, Inc.) according to the manufacturer’s instructions. The titre of the VSV-MERS-CoV-S pseudoparticles was defined as the highest dilution yielding 105 relative luciferase units (RLU). For the titration of VSV-MERS-CoV-S pseudoparticles, the pCAGGS plasmids encoding VSV recombinant G and the empty vector (EV) pCAGGS-EV were used to generate VSV-G pseudoparticles and pseudoparticles without heterologous protein (EV) as positive and negative controls, respectively. Ten fold serial dilutions of VSV pseudotyped with MERS-S (VSV-MERS-S) or negative control pCAGGS empty vector containing no glycoprotein (VSV-EV) were used to infect Vero cells in 96-well plates. RLU were measured 24 h post-infection. The results are expressed as average RLU±standard deviation (sd) of triplicate wells from an experiment repeated three times with similar results. The error bars indicate the sd.
Pseudotype neutralization assay
Briefly, Vero cells were seeded in 96-well black and white tissue culture-treated plates 1 day prior to infection. Human sera were heat-inactivated at 56 °C for 30 min, and diluted as two fold serial dilutions with an initial dilution of 1 : 40 in serum-free DMEM containing 1× p/s. Two hundred microlitres of each serum dilution were mixed thoroughly with 105 RLU-equivalent VSV-MERS-CoV-S pseudoparticles diluted to 200 µl and incubated at 37 °C under a 5 % CO2 atmosphere for 1 h. Positive and negative control sera were included with each assay run. In triplicates, 100 μl of the pseudoparticle–serum mixtures were transferred onto Vero cell monolayers and incubated at 37 °C under a 5 % CO2 atmosphere for 1 h. After adsorption, 100 µl of DMEM containing 10 % FBS p/s was added and incubated at 37 °C and 5 % CO2 for 23 h. At 24 h post-inoculation, luciferase activity was determined using the Luciferase Assay kit according to the manufacturer’s instructions. The results are expressed as the percentage neutralization ±sd of three parallel wells from three independent experiments.
To calculate neutralization, the luciferase activity from each serial diluted human sera sample was normalized to that from the negative control, pNHS. Neutralization was calculated as: (average RLU value of pNHS−average RLU value with test serum/average RLU value of pNHS)×100. Reciprocal endpoint titres were defined as the highest dilution of the test serum sample that decreased RLU values by 80 % or more. Serum samples that failed to show at least an 80 % reduction at the 1 : 40 dilution were considered to be below the limit of detection.
Immunofluorescence assay (IFA)
The slides were first coated with poly-l-lys and seeded with HEK-293T cells 24 h prior to transfection. The cells were transfected with 1 µg of pCAGGS-MERS-CoV-S plasmid DNA or mock transfection. At 24 h post-transfection, the cells’ expression of MERS-CoV-S was detected by convalescent sera from a MERS-CoV patient with a known neutralizing titre against live virus as a positive control (MERS immune sera; 1 : 400), followed by FITC-labelled anti-human IgA/IgG/IgM (1 : 500) (Abcam). DAPI stain was used and cells were imaged using a Zeiss AxioImager microscope at 20× magnification.
Statistical analyses
All graphs were generated with the GraphPad Prism 7 software. All results were calculated and presented as the means±sd obtained from duplicate or triplicate independent experiments. Pearson’s correlation analysis and the Bland–Altman method were used to assess the comparative analyses between the VSV-MERS-CoV-S pseudoparticle neutralization assay and the MNt assay. Statistical analyses were performed using GraphPad Prism 7 software.
The specimens used in this study were determined to be non-human subject research by the Centers for Disease Control and Prevention (CDC) Institutional Review Board (IRB).
Results
Generation and characterization of VSV-MERS-CoV-S pseudoparticles
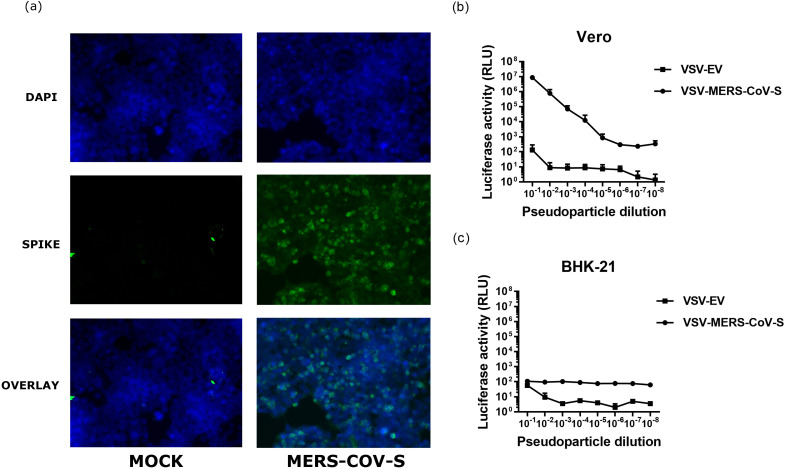
To establish VSV-MERS-CoV-S pseudotype neutralization assays, we generated VSV-MERS-CoV pseudoparticles with surface S protein and a firefly luciferase reporter gene as previously described [24]. The expression of MERS-CoV S proteins was confirmed by immunofluorescence at 24 h post-transfection in HEK293T cells (Fig. 1a). It is known that BHK-21 cells do not have the S-binding MERS-CoV receptor, DPP4, and are therefore not permissible to MERS-CoV infection, but Vero cells do [18, 39]. Efficient packaging of MERS-CoV S on the surface of the VSV pseudoparticles and the ability of the pseudoparticles to enter target cells through receptor binding were confirmed by comparing the titrated inoculation of Vero and BHK-21 cells of negative control VSV pseudoparticles containing no viral glycoproteins on their surface (VSV-EV) with that for VSV pseudotyped with MERS-CoV-S proteins, designated VSV-MERS-S (Fig. 1b and c). As expected, in the Vero cells the luciferase activity was proportional to the dilution of the MERS-S pseudoparticle inoculum (Fig. 1). In comparison to the control pseudoparticles, the MERS-S pseudoparticles demonstrated an approximately six fold increase in luciferase activity (Fig. 1). Neither the MERS-S pseudoparticles nor the control pseudoparticles induced high levels of luciferase activity in BHK-21 cells (Fig. 1), further supporting the notion that BHK-21 cells are resistant to MERS-CoV infection [39]. Based on the RLU values observed at each serial dilution, it was decided to use a 1 : 200 dilution as the input pseudoparticle dose for further experiments for the MERS-CoV S protein pseudotyped VSV particle-based neutralization assay.
Fig. 1.
Generation and characterization of VSV-MERS-CoV-S pseudoparticles. (a) IFA analysis of the expression of MERS-CoV-S protein. HEK-293T cells were transiently transfected with pCAGGS-MERS-CoV-S plasmid DNA or mock transfected. The expression of MERS-CoV-S was detected by convalescent sera from a MERS-CoV patient with a known neutralizing titre against live virus as a positive control (MERS immune sera). (b, c) Titration of VSV-based pseudoparticles on Vero cells (b) and (c) BHK-21 cells. VSV pseudotyped with MERS-S (VSV-MERS-S) or negative control pCAGGS empty vector containing no glycoprotein (VSV-EV) was used to infect Vero cells. The results are expressed as the average relative luciferase units (RLU) ±standard deviation (sd). The error bars indicate the sd.
Optimization of the VSV-MERS-CoV-S pseudoparticle assay
To establish optimal parameters for the MERS-CoV S protein pseudotyped VSV particle-based neutralization assay, the cell density was optimized for maximum luciferase activity. No significant differences in luciferase activities were observed at 103–105 cellsper well, although maximum luciferase activity was reached at 104 cells per well, and so 104 cells per well was selected as the standard assay cell density (Fig. S1a, available in the online version of this article). Next, we investigated the incubation time for optimal luciferase activity. The maximum RLU values peaked at 24 h post-infection and the RLU values decreased with prolonged incubation times (Fig. S1b). Therefore, 24 h post-infection was determined to be the most suitable infection time for MERS-CoV-S pseudoparticles.
Serum neutralization test with MERS-CoV-S pseudoparticles
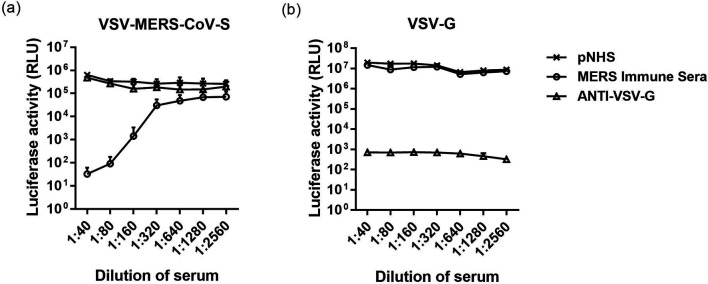
To investigate whether anti-MERS-CoV human sera neutralized VSV-MERS-CoV-S pseudoparticles, VSV-MERS-CoV-S pseudoparticles were preincubated with two fold serial dilutions of pNHS serum (negative control), convalescent sera from a MERS-CoV patient with a known neutralizing titre against live virus as a positive control (MERS immune sera) or anti-VSV-G monoclonal antibody. Luciferase activity was reduced when VSV-MERS-CoV-S was preincubated with positive control serum in a dose-dependent manner (Fig. 2a). In contrast, preincubation with the negative control pNHS or anti-VSV-G monoclonal antibody did not significantly reduce luciferase activity at any dilution factor, thus demonstrating that VSV-MERS-CoV-S pseudoparticles were specifically neutralized by serum from an individual with a confirmed prior MERS-CoV infection (Fig. 2a). As expected, at any dilution factor, preincubation with anti-VSV-G monoclonal antibody efficiently neutralized VSV-ΔG pseudoparticles packaged with VSV-G, although neither the MERS-CoV positive control serum or the negative control pNHS neutralized VSV-ΔG pseudoparticles packaged with VSV-G (Fig. 2b).
Fig. 2.
Neutralization of VSV-based pseudoparticles. (a) VSV-MERS-CoV-S pseudoparticles or (b) VSV-G pseudoparticles were preincubated with equal volumes of two fold serially diluted negative control pooled normal human serum (pNHS), positive control MERS-CoV immune serum or anti-VSV-G monoclonal antibody. The results are expressed as the average relative luciferase units (RLU) ±sd. The error bars indicate the sd.
Establishment of percentage neutralization cutoff for endpoint titration of human sera
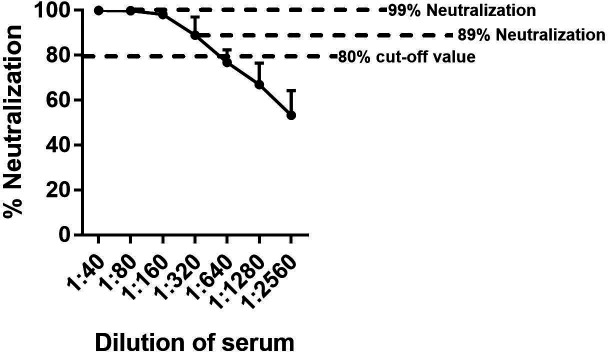
Neutralization may be assessed as the percentage reduction in infectivity at a single dilution. Since our goal is to measure the endpoint neutralization titres of human sera that possibly contain MERS-CoV neutralizing antibodies, we evaluated the percentage neutralization reduction curve of VSV-MERS-CoV-S pseudoparticles when preincubated with a MERS-CoV immune serum with a known live-virus endpoint neutralization titre. The percentage reduction in RLU (% neutralization) was determined by calculating the difference between the average RLU number in triplicate wells with the anti-MERS-CoV serum and the average RLU number in triplicate wells with the negative control pNHS sample, dividing it by the average RLU number in triplicate wells with the negative control pNHS sample and then multiplying the result by 100.
Neutralization of VSV-MERS-CoV-S pseudoparticles was dependent on the serum dilution, reducing RLU between 99 and 53 % at the dilutions tested (Fig. 3). Using a target value of 80 % reduction in RLU in comparison to the pNHS control was within the linear range of reduction. Our standard positive control MERS-CoV immune serum crossed the 80 % reduction line between 320 and 640 dilutions. This same serum has a known endpoint titre of approximately 320 in neutralization assays using live virus (unpublished data), demonstrating relative equivalence in the two assays for this serum.
Fig. 3.
Percentage neutralization of RLU of VSV-MERS-CoV-S pseudoparticles in comparison to pNHS. VSV-MERS-CoV-S pseudoparticles were preincubated with equal volumes of two fold serial dilutions of MERS-CoV immune serum and then inoculated onto Vero cells. The results are normalized to the negative control pNHS serum. The results are expressed as percentage neutralization ±sd. The error bars indicate sd.
Equivalence testing of VSV-MERS-CoV-S pseudoparticle neutralization titration
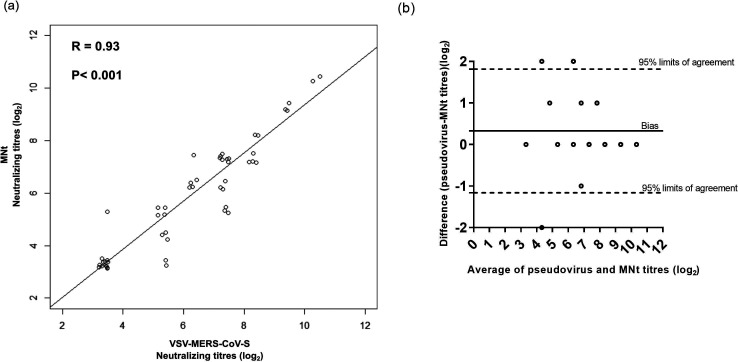
We previously measured the endpoint neutralizing antibody titres of 52 human sera collected from MERS-CoV patients using a live virus MNt. To determine if the VSV-MERS-CoV-S pseudoparticle neutralization test results in equivalent endpoint titres in comparison to the live virus MNt, we compared the endpoint titres from the live virus MNt to the endpoint titre necessary to achieve an 80 % reduction in RLU with VSV-MERS-CoV-S pseudoparticles. Neutralizing antibody titres ranging from 40 to 1280 from 52 human serum samples against MERS-CoV were successfully detected by both neutralization assays (Fig. 4). Sixty-seven per cent (n=35) of the serum samples had the same endpoint neutralization titres as those measured by either technique. We observed that approximately 29 % (n=15) of the serum samples gave higher neutralizing titres in the VSV-MERS-CoV-S pseudoparticle neutralization assay than the conventional MNt. Furthermore, five sera with neutralization titres below the limit of detection by MNt had neutralizing titres that were detectable by the VSV-MERS-CoV-S pseudoparticle neutralization assay, thus demonstrating that the VSV-MERS-CoV-S pseudoparticle neutralization assay may be more sensitive (Fig. 4). The titres from both assays were then analysed statistically. Pearson’s correlation analysis of all data showed a strong correlation between the two neutralization assays, with R=0.9348, P<0.0001, 95 % confidence interval (CI)=0.8886 to 0.9622 (Fig. 4a). Bland–Altman method comparison analysis further demonstrated that these two methods are highly comparable. The average of the logarithmic difference of the two methods was 0.3269 (Fig. 4b). Collectively, these data indicate that the VSV-MERS-CoV-S pseudoparticle neutralization assay can be a reliable alternative to the live virus-based MNt.
Fig. 4.
Correlation analysis between VSV-MERS-CoV-S pseudoparticle and live virus neutralization tests. VSV-MERS-CoV-S pseudoparticles were preincubated with equal volumes of 52 human serum samples in two fold serial dilutions and then inoculated in triplicate onto Vero cells. (a) Pearson correlation analysis of anti-MERS-CoV neutralizing antibody titres measured both by MNt and VSV-MERS-CoV-S pseudoparticle assays. The results on the x-axis are expressed as the log2 inverse dilution that resulted in 80 % or greater reduction in RLU as compared to the negative control pNHS serum. The results on the y-axis are expressed as the log2 inverse dilution that blocked cell death by live MERS-CoV infection. The Pearson’s correlation coefficient (R) value and the P value are depicted for the comparison. (b) Bland–Altman method comparative analysis of anti-MERS-CoV neutralizing antibody titres measured both by the MNt and VSV-MERS-CoV-S pseudoparticle assays. The solid line represents the bias, while the dashed lines represent the upper and lower 95 % confidence limits.
Specificity testing for the VSV-MERS-CoV-S pseudoparticle neutralization assay
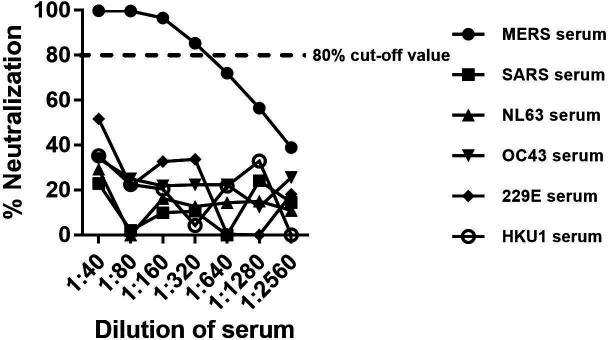
There are six known human coronaviruses that cause respiratory disease in humans. In order to test the specificity of our VSV-MERS-CoV-S pseudoparticles and their potential cross-reactivity with other common human coronaviruses, we tested 40 serum samples from humans with no known anti-MERS-CoV exposure. When using the 80 % reduction in RLU as the cut-off value, none of the serum samples neutralized VSV-MERS-CoV-S pseudoparticles at any dilution factor (Fig. S2). We also tested potential cross-neutralization of VSV-MERS-CoV pseudoparticles with sera from other humans with confirmed infections with the human coronaviruses 229E, HKU1, OC43, NL63 and SARS-CoV. Using the neutralization parameters of 80 % or greater reduction in RLU, we found no cross-neutralization with 229E, HKU1, OC43, NL63 or SARS-CoV anti-sera (Fig. 5). These data confirm the specificity of the VSV-MERS-CoV-S pseudoparticles.
Fig. 5.
Cross-neutralization of VSV-MERS-CoV-S by sera from patients with confirmed human coronavirus infections. VSV-MERS-CoV pseudoparticles were preincubated with equal volumes of two fold serially diluted MERS-CoV immune serum or antisera from the human coronaviruses 229E, HKU1, OC43, NL63 and SARS-CoV and then inoculated in triplicate onto Vero cells for each dilution of serum samples. The results are normalized to the negative control pNHS serum. The results are expressed as percentage neutralization and are the average of two independent experiments.
Discussion
MERS-CoV continues to circulate on the Arabian Peninsula and can cause severe infections, including fatalities. At present, there are no antiviral therapeutics or approved vaccines to combat MERS-CoV infections. The presence of serum neutralizing antibodies ≥20 is a reliable indicator of prior MERS-CoV exposure. Neutralization assays using live viruses are recognized as the gold standard serological assays to confirm the antibody responses to MERS-CoV infection. However, one major obstacle to performing the MNt assay is the requirement for access to high-level biocontainment laboratories, due to the use of live MERS-CoV. Thus, only a limited number of facilities and researchers are able to perform such neutralization assays. Additionally, determining the neutralization of infectious virus in this assay requires the assessment of cytopathic effect in 96-well plates, which is labour-intensive and time-consuming. Furthermore, because of the large viral genome, reverse genetics for MERS-CoV is challenging, and still needs to be used under BSL-3 containment, thereby limiting molecular manipulation and studies. These limitations highlight the need to develop alternative methods for detecting MERS-CoV neutralizing antibodies.
An alternative approach to overcome these shortcomings is the use of viral pseudotypes. Generally, lentiviral and VSV-based pseudotypes are used for the study of enveloped viruses. Several studies have utilized lentiviral and VSV pseudotype systems harbouring MERS-CoV spike glycoproteins to study viral entry, viral tropism and other basic science questions [27, 29, 37–40]. In addition to being utilized to probe basic science questions, pseudoparticle-based neutralization assays have been used for serological surveillance of reservoir species for several other zoonotic viruses in addition to MERS-CoV [17, 33, 41–46]. Studies have employed lentiviral and env-deficient HIV-1 backbone vector pseudotype systems for MERS-CoV seroepidemiological surveillance in humans [32–36]. Using dromedary camel sera, Hemida et al. demonstrated that lentiviral-based MERS-CoV S pseudoparticles detected antibodies specific to MERS-CoV. In our hands, lentiviral particles grew to lower titres than VSV particles, making it challenging to prepare high volumes of stock for use in multiple studies. Standardizing many particles for use in multiple studies is important for making comparisons between studies in this context.
Barlan et al. utilized VSV pseudotyped with MERS-CoV-S and demonstrated that susceptibility to MERS-CoV infection is enhanced by cellular proteases that cleave the S protein [37]. Fukuma et al. demonstrated that infection with VSV-based pseudoparticles bearing truncated C-terminal MERS-CoV-S protein and non-truncated MERS-CoV-S protein was inhibited by rabbit anti-MERS-CoV S serum and that infection was dependent on the expression of DPP4 [40]. This demonstrated that VSV-based MERS-S pseudoparticles can be utilized for neutralization assays. Nonetheless, studies featuring seroepidemiological surveillance in humans utilizing a VSV pseudotype system bearing MERS-CoV spike glycoproteins had not previously been performed. This report describes the first comparison between the neutralization assay using VSV-MERS-CoV-S pseudoparticles and the MNt using a panel of human sera.
This study is limited by only utilizing 52 specimens, which limits its statistical power. Although analysing more specimens might yield a more powerful comparative analysis, the complexities of limited availability, diplomatic issues and biosafety concerns make acquiring MERS-CoV human specimens less feasible. Despite this limitation, statistical analyses comparing MNt and VSV-MER-CoV pseudoparticle neutralization indicate there was a positive correlation between the two methods. Although there was a strong correlation between the two methods, the VSV-MERS-CoV-S pseudoparticle neutralization assay is slightly more sensitive than the conventional MNt. One potential explanation for this is the surface density of S protein. The generation of pseudoparticles by transient transfection in continuous cell lines, rather than producer cells that are more physiologically relevant, may lead to the production of large amounts of virus with less S protein on its surface. Thus, the pseudoparticles may be more susceptible to neutralization because fewer neutralizing antibodies are required to block the entry of MERS-CoV. However, in corroboration with our data, Fukushi et al. previously demonstrated that a neutralization test based on a VSV-based SARS-CoV-S protein bearing pseudoparticles was also more sensitive than the conventional virus neutralization test using live SARS-CoV [47]. In comparison with the MNt method, the pseudoparticle neutralization assay required half as much time to test for MERS-CoV neutralizing antibodies. More importantly, this assay can be performed in a low-biosafety containment laboratory.
In addition to being faster and more sensitive than conventional neutralization assays while maintaining specificity, our VSV-MERS-CoV-S pseudoparticle system offers a convenient approach for examining the antigenic and neutralizing epitope differences among different S proteins from divergent strains of MERS-CoV. The data we have presented here use one S plasmid, but the pseudoparticles can be packaged using the spike sequence from other virus strains. Because S protein is provided in trans in a pCAGGS vector, it can be easily mutated or swapped for any spike protein an investigator is interested in studying. There are existing reverse genetics systems for human coronaviruses, including MERS-CoV, but those systems are more difficult to generate and require BSL-3 facilities [48–50]. Using those full-virus reverse genetics systems are essential for viral replication and pathogenesis studies, but for neutralization studies, the presence of spike alone is sufficient.
MERS convalescent sera did not neutralize VSV particles packaged with VSV-G, demonstrating that neutralization was due to serum antibodies binding specifically to MERS S. One of the main concerns in designing a MERS-CoV neutralization test is the presence of five other known human coronaviruses, four of which cause common infections. It had previously been shown that anti-coronavirus sera cross-neutralize amongst similar coronaviruses [51]. In this study we determined that VSV-MERS-CoV-S pseudoparticles were not neutralized by antisera from alpha- or betacoronaviruses, further demonstrating the assay’s high specificity.
In conclusion, we established a neutralization test using a VSV-based pseudotyped virus possessing MERS-CoV S protein and expressing a luciferase reporter gene. We used the VSV-MERS-CoV-S pseudoparticle neutralization assay at BSL-2 containment, improving the safety of MERS-CoV neutralization assays. Additionally, we found the system to be more rapid and sensitive and easier to use than a conventional microneutralization assay. These characteristics make the VSV-MERS-CoV-S pseudoparticle neutralization assay a more attractive, convenient and suitable assay for MERS-CoV screening and confirmatory serology testing. It offers a safe, rapid alternative method to monitor the potency and breadth of neutralization antibodies against MERS-CoV exposure in future seroepidemiological studies.
Supplementary Data
Funding information
The authors wish to acknowledge financial support from CDC LASSI 2016.
Author contributions
Sandra Lester: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, validation, visualization, writing – original draft, Writing – review and editing.
Jennifer Harcourt: methodology, resources, validation, writing – review and editing.
Michael Whitt: methodology, resources, supervision, writing – review and editing.
Hail M. Al-Abdely: resources, writing – review and editing.
Claire M. Midgley: resources, writing – review and editing.
Abdulrahim M. Alkhamis: resources, writing – review and editing.
Hani A. Aziz Jokhdar: resources, writing – review and editing.
Abdullah M. Assiri: resources, writing – review and editing.
Azaibi Tamin- Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – review & editing.
Natalie Thornburg- Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
Conflicts of interest
The authors declare that there are no conflicts of interest. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
Ethical statement
The specimens used in this study were determined to be non-human subject research by the Centers for Disease Control and Prevention (CDC) Institutional Review Board (IRB).
Footnotes
Abbreviations: BCoV, Betacoronavirus; BHK-21, baby hamster kidney-21; BSL-3, biosafety level 3; CDC, Center for Disease Control and Prevention; CoV, Coronavirus; DMEM, Dulbecco’s Modified Essential Medium; DPP4, dipeptidyl peptidase IV; ELISA, enzyme-linked immunosorbent assay; IFA, immunofluorescence assay; IRB, Institutional Review Board; MERS-CoV, Middle East Respiratory Syndrome Coronavirus; MNt, microneutralization test; PBNA, particle-based neutralization assay; pNHS, Pooled normal human serum; PRNT, plaque reduction neutralization test; RLU, relative luciferase units; S, Spike; SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; SD, standard deviation; VSV, vesicular stomatitis virus; WHO, World Health Organization.
Two supplementary figures are available with the online version of this article.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the middle East, September 2012. Euro Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 3.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kupferschmidt K, diseases E. Emerging diseases. Soaring MERS cases in Saudi Arabia raise alarms. Science. 2014;344:457–458. doi: 10.1126/science.344.6183.457. [DOI] [PubMed] [Google Scholar]
- 5.Min J, Cella E, Ciccozzi M, Pelosi A, Salemi M, et al. The global spread of middle East respiratory syndrome: an analysis fusing traditional epidemiological tracing and molecular phylodynamics. Glob Health Res Policy. 2016;1:14. doi: 10.1186/s41256-016-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JY. An outbreak of middle East respiratory syndrome coronavirus infection in South Korea, 2015. Yonsei Med J. 2015;56:1174–1176. doi: 10.3349/ymj.2015.56.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay IM, Arden KE. Middle East respiratory syndrome: an emerging coronavirus infection tracked by the crowd. Virus Res. 2015;202:60–88. doi: 10.1016/j.virusres.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 9.Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui DS, Memish ZA, Zumla A. Severe acute respiratory syndrome vs. the middle East respiratory syndrome. Curr Opin Pulm Med. 2014;20:233–241. doi: 10.1097/MCP.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 11.Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemida MG, Elmoslemany A, Al-Hizab F, Alnaeem A, Almathen F, et al. Dromedary camels and the transmission of middle East respiratory syndrome coronavirus (MERS-CoV) Transbound Emerg Dis. 2017;64:344–353. doi: 10.1111/tbed.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer B, Müller MA, Corman VM, Reusken CBEM, Ritz D, et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. 2014;20:552–559. doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller MA, Corman VM, Jores J, Meyer B, Younan M, et al. Mers coronavirus neutralizing antibodies in camels, eastern Africa, 1983-1997. Emerg Infect Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemida MG, Perera RA, Al Jassim RA, Kayali G, Siu LY, et al. Seroepidemiology of middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Euro Surveill. 2014;19:20828. doi: 10.2807/1560-7917.ES2014.19.23.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Deng Y, Wen B, Wang H, Meng X, et al. The amino acids 736-761 of the MERS-CoV spike protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. Viral Immunol. 2014;27:543–550. doi: 10.1089/vim.2014.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du L, Yang Y, Zhou Y, Lu L, Li F, et al. Mers-Cov spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Deng Y, Wen B, Wang H, Meng X, et al. The amino acids 736-761 of the MERS-CoV spike protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. Viral Immunol. 2014;27:543–550. doi: 10.1089/vim.2014.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer B, Drosten C, Müller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitt MA. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods. 2010;169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giroglou T, Cinatl J, Rabenau H, Drosten C, Schwalbe H, et al. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J Virol. 2004;78:9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZY, Werner HC, Kong W-pui, Leung K, Traggiai E, et al. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millet JK, Whittaker GR. Host cell entry of middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urbanowicz RA, McClure CP, King B, Mason CP, Ball JK, et al. Novel functional hepatitis C virus glycoprotein isolates identified using an optimized viral pseudotype entry assay. J Gen Virol. 2016;97:2265–2279. doi: 10.1099/jgv.0.000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alharbi NK, Padron-Regalado E, Thompson CP, Kupke A, Wells D, et al. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine. 2017;35:3780–3788. doi: 10.1016/j.vaccine.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molesti E, Wright E, Terregino C, Rahman R, Cattoli G, et al. Multiplex evaluation of influenza neutralizing antibodies with potential applicability to in-field serological studies. J Immunol Res. 2014;2014:1–11. doi: 10.1155/2014/457932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joglekar AV, Sandoval S. Pseudotyped lentiviral vectors: one vector, many guises. Hum Gene Ther Methods. 2017;28:291–301. doi: 10.1089/hgtb.2017.084. [DOI] [PubMed] [Google Scholar]
- 32.Park SW, Perera RAPM, Choe PG, Lau EHY, Choi SJ, et al. Comparison of serological assays in human Middle East respiratory syndrome (MERS)-coronavirus infection. Euro Surveill. 2015;20:30042. doi: 10.2807/1560-7917.ES.2015.20.41.30042. [DOI] [PubMed] [Google Scholar]
- 33.Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574. doi: 10.2807/1560-7917.ES2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 34.Gierer S, Hofmann-Winkler H, Albuali WH, Bertram S, Al-Rubaish AM, et al. Lack of MERS coronavirus neutralizing antibodies in humans, eastern Province, Saudi Arabia. Emerg Infect Dis. 2013;19:2034–2036. doi: 10.3201/eid1912.130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Shi W, Joyce MG, Modjarrad K, Zhang Y, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun. 2015;6 doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang X-C, Agnihothram SS, Jiao Y, Stanhope J, Graham RL, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlan A, Zhao J, Sarkar MK, Li K, McCray PB, et al. Receptor variation and susceptibility to middle East respiratory syndrome coronavirus infection. J Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JE, Li K, Barlan A, Fehr AR, Perlman S, et al. Proteolytic processing of middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci USA. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letko M, Miazgowicz K, McMinn R, Seifert SN, Sola I, et al. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018;24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuma A, Tani H, Taniguchi S, Shimojima M, Saijo M, et al. Inability of rat DPP4 to allow MERS-CoV infection revealed by using a VSV pseudotype bearing truncated MERS-CoV spike protein. Arch Virol. 2015;160:2293–2300. doi: 10.1007/s00705-015-2506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamin A, Harcourt BH, Lo MK, Roth JA, Wolf MC, et al. Development of a neutralization assay for Nipah virus using pseudotype particles. J Virol Methods. 2009;160:1–6. doi: 10.1016/j.jviromet.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molesti E, Cattoli G, Ferrara F, Böttcher-Friebertshäuser E, Terregino C, et al. The production and development of H7 influenza virus pseudotypes for the study of humoral responses against avian viruses. J Mol Genet Med. 2012;7:315–320. [PMC free article] [PubMed] [Google Scholar]
- 43.Molesti E, et al. Comparative serological assays for the study of H5 and H7 avian influenza viruses. Influenza Res Treat, 2013. 2013:p. 286158. doi: 10.1155/2013/286158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu C, Huang Y, Zhang A, Tian D, Wan Y, et al. Safe pseudovirus-based assay for neutralization antibodies against influenza A(H7N9) virus. Emerg Infect Dis. 2013;19:1685–1687. doi: 10.3201/eid1910.130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Qi J, Yuan Y, Xuan Y, Han P, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Du L, Liu C, Wang L, Ma C, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukushi S, Mizutani T, Saijo M, Kurane I, Taguchi F, et al. Evaluation of a novel vesicular stomatitis virus pseudotype-based assay for detection of neutralizing antibody responses to SARS-CoV. J Med Virol. 2006;78:1509–1512. doi: 10.1002/jmv.20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yount B, Curtis KM, Fritz EA, Hensley LE, Jahrling PB, et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, et al. Reverse genetics with a full-length infectious cDNA of the middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muth D, Meyer B, Niemeyer D, Schroeder S, Osterrieder N, et al. Transgene expression in the genome of middle East respiratory syndrome coronavirus based on a novel reverse genetics system utilizing red-mediated recombination cloning. J Gen Virol. 2017;98:2461–2469. doi: 10.1099/jgv.0.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan KH, Chan JFW, Tse H, Chen H, Lau CCY, et al. Cross-Reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013;67:130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.