Abstract
Genomic aberrations of the PTEN tumour suppressor gene are among the most common in prostate cancer. Inactivation of the PTEN gene by deletion or mutation is identified in ~20% of primary prostate tumour samples at radical prostatectomy and as many as 50% of castration resistant tumours. Loss of PTEN function leads to activation of the PI3K–AKT pathway and is strongly associated with adverse oncological outcomes, making PTEN a potentially useful genomic marker to distinguish indolent from aggressive disease in patients with clinically localized tumours. At the other end of the disease spectrum, therapeutic compounds targeting nodes in the PI3K/AKT/mTOR signalling pathway are being tested in clinical trials for patients with metastatic castration-resistant prostate cancer (CRPC). Knowledge of PTEN status might be helpful to identify patients who are more likely to benefit from these therapies. To enable the use of PTEN status as a prognostic and predictive biomarker, analytically validated assays have been developed for reliable and reproducible detection of PTEN loss in tumour tissue and in blood liquid biopsies. Use of clinical-grade assays in tumour tissue have shown a robust correlation between loss of PTEN and of its protein as well as a strong association between PTEN loss and adverse pathological features and oncological outcomes. In advanced disease, assessing PTEN status in liquid biopsies shows promise in predicting response to targeted therapy. Finally, studies have shown that PTEN might have additional functions that are independent of the PI3K/AKT pathway, including those affecting tumour growth through modulation of the immune response and tumour microenvironment.
Prostate cancer is the most commonly diagnosed cancer and the third leading cause of cancer-related death in US men1. However, the vast majority of patients diagnosed with prostate cancer will not die from the disease1. This conundrum results in a need to improve clinical risk stratification to identify which patients can be safely treated by active surveillance AS), which patients can be cured by therapies directed solely at the prostate, and which require the integration of systemic therapy. Although clinicopathological variables are useful for risk stratification, as many as 30% of men considered to be eligible for active surveillance based on these variables are found to already harbour or progress to advanced disease and require intervention2, highlighting the unmet need for more informative determinants of prognosis. Next-generation sequencing has elucidated many molecular alterations and molecular subclasses in prostate cancer; however, aside from inhibition of the androgen receptor (AR) signalling axis, few molecular drivers of the disease have been established. The PTEN (phosphatase and tensin homolog on chromosome 10) tumour suppressor and the PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase) signalling axis it restrains are among the most commonly altered pathways in primary prostate cancer3,4. Furthermore, animal models and correlative biomarker studies in humans have overwhelmingly nominated PTEN loss as a critical pathway to disease progression in hormone naive and castration-resistant prostate cancer (CRPC). In CRPC, improved mechanistic understanding and identification of the oncogenic drivers of tumour growth and reciprocal feedback in this signalling pathway has led to the design of biologically informed studies in which patient benefit is being demonstrated. In this setting, PTEN might be an important biomarker to predict the likelihood of therapeutic response. Here, we review the biology of the PTEN tumour suppressor and its relationship to prostate cancer. We further discuss the validation and clinical utility of PTEN as a prognostic biomarker in localized prostate cancer and consider progress towards realizing the potential of PTEN as a predictive biomarker in advanced metastatic CRPC.
Biology of the PTEN tumour suppressor
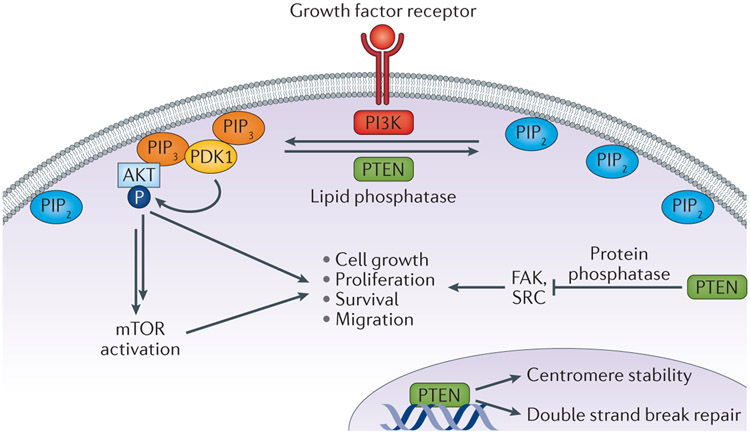
PTEN acts as dual-specificity phosphatase, converting phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3 or PIP3] into phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2 or PIP2] 5. In this manner, PTEN functions as a direct antagonist of the activity of class I PI3K, a family of enzymes which convert PIP2 to PIP3, resulting in the activation of the downstream AKT and mTOR (mammalian target of rapamycin) signalling cascades (Figure 1). Loss and/or inactivating mutation of PTEN results in unopposed activity of PI3Ks and accumulation of PIP3 on the cell membrane, which leads to recruitment and activation of proteins containing pleckstrin homology (PH) domains, including the kinase PDK1 and its substrate AKT. Active phosphorylated AKT modulates a number of downstream targets, including mTOR signalling, which have key roles in the regulation of apoptosis, cell cycle progression, cellular proliferation, metabolism, differentiation, and invasion6. In its role as a lipid phosphatase, PTEN modulates an enormous number of cellular processes, including cell polarity and motility, cellular senescence, and modulation of the tumour microenvironment6. In addition, the phosphatase activity of PTEN seems to mediate aspects of both innate and adaptive immunity7,8.
Figure 1 ∣. The diverse cellular roles of PTEN.
PTEN acts as lipid phosphatase, converting phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3 or PIP3] into phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2 or PIP2]. In this capacity, PTEN antagonizes the function of Class I PI3K activity, which converts PIP2 to PIP3. This lipid phosphatase activity of PTEN suppresses the activation of the downstream oncogenic AKT and mTOR signalling cascades. However, PTEN also has several other noncanonical functions, including weak protein phosphatase activity with known kinase substrates such as FAK and SRC. Finally, PTEN probably functions in the nucleus in a PI3K-independent manner to promote chromosome stability and DNA repair .
Beyond its role as a lipid phosphatase, PTEN has established protein phosphatase activity, leading to functions that are independent of PI3K-AKT signalling. Study of PTEN mutations that have lost lipid phosphatase but retain protein phosphatase activity in cell lines have revealed numerous novel protein substrates of PTEN, and diverse functions in regulating cell adhesions via the focal adhesion kinase FAK9 and the non-receptor tyrosine kinase SRC10. This protein phosphatase activity might modulate many of the nuclear functions of PTEN including cell cycle regulation11, as nuclear PIP3 pools are relatively insensitive to PTEN lipid phosphatase12. In addition, ample evidence supports a role for PTEN in the nucleus that is entirely PI3K-independent. PTEN localizes to centromeres and PTEN mutations that disrupt this interaction lead to centromeric instability. PTEN-null cells demonstrate spontaneous DNA double-strand breaks, probably owing to PTEN-mediated regulation of RAD51 expression13,14 SUMOylation might regulate nuclear localization of PTEN and contribute to its role in DNA damage repair14. These studies have formed the basis for clinical trials evaluating poly (ADP-ribose) polymerase (PARP) inhibitors to treat prostate cancer with PTEN loss.
Characteristics of PTEN inactivation
PTEN genomic deletion in prostate cancer was first identified almost two decades ago15-17, and subsequent sequencing studies have demonstrated that PTEN is the most commonly lost tumour suppressor gene in primary disease3,18,19. The vast majority of prostate tumours with PTEN loss inactivate PTEN by genomic deletion3,18,19 (Figure 2). However, depending on the type of cohort examined and the assay used to determine PTEN status, the reported rate of PTEN gene deletions in prostate cancer varies (Table 1). This variation is probably partly due to the fact that the frequency of PTEN deletion is highly correlated with increasing Gleason score and tumour stage20-22. However, the fact that few analytically validated assays have been performed to determine PTEN status has also contributed to this variability, which is a key consideration to keep in mind when reviewing the data.
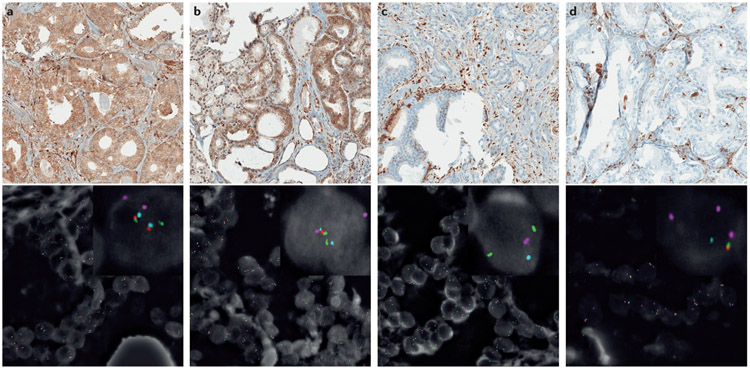
Figure 2 ∣. Prostate cancer samples with variable PTEN protein expression by IHC and corresponding PTEN FISH.
a ∣ Prostate tumour with intact PTEN identified with IHC with two intact PTEN alleles detected using FISH. PTEN IHC demonstrates intact PTEN (top) and four-colour FISH image from an adjacent section (bottom) shows two intact PTEN alleles (red) with two intact copies of flanking genes, WAPAL (green) and FAS (aqua) as well as chromosome 10 centromeres (pink). b ∣ Prostate tumour showing PTEN expression using IHC with hemizygous PTEN deletion using FISH. PTEN IHC demonstrates intact PTEN protein (top), with four-colour FISH image (bottom) from an adjacent section showing a hemizygous PTEN deletion with loss of one PTEN gene (one red signal). As both centromeres (pink) and the WAPAL (green) and FAS (aqua) probes that flank either side of PTEN are retained, this hemizygous deletion is likely to be interstitial and restricted to the PTEN region.c ∣ Prostate tumour showing absence of PTEN expression by IHC with homozygous PTEN gene deletion detected in intraductal tumour by FISH. PTEN IHC image (top) shows loss of PTEN in tumour glands. Four-colour FISH image from an adjacent section (bottom) shows a homozygous deletion with loss of both PTEN genes (red). The retention of the centromeres (pink) and both WAPAL genes (green), but the presence of only one copy of FAS (aqua) indicates that one of the deletions involved both PTEN and FAS. d ∣ Prostate tumour showing PTEN protein loss by IHC with hemizygous PTEN gene deletion by FISH. PTEN IHC demonstrates complete PTEN loss (top), with four-colour FISH image (bottom) from an adjacent section showing a hemizygous PTEN deletion with loss of one PTEN gene (red) along with flanking genes WAPAL (green) and FAS (aqua). This tumour demonstrates complex hemizygous PTEN deletion, in which PTEN is deleted along with adjacent genes (WAPAL and FAS) located on both sides of PTEN.
Table 1 ∣.
Studies of PTEN status in contemporary prostate cancer cohorts
| Technique | Tissue type | PTEN loss % | Study |
|---|---|---|---|
| PTEN deletion* (2 colour FISH) | Incidental tumour (TURP) | 17% (56/322) | Reid et al., 201099 |
| PTEN deletion (4 colour FISH) | Prostate cancer CRPC | 37.5% (112/298) 62% (20/32) |
Yoshimoto et al., 2012118 |
| PTEN deletions (2 colour FISH) | Prostate cancer | 20% (458/2266) | Krohn et al., 201221 |
| PTEN mutation (sequence) | Prostate cancer mCRPC | 10.1% (1/11) 8% (4/50) |
Grasso et al., 201239 |
| PTEN Copy number loss (arrays) | Same cohort | 46% (5/11) 40% (20/50) |
Grasso et al., 201239 |
| PTEN Immunohistochemistry† | Prostate cancer CRPC mCRPC | 15% (42/282) 45% (55/122) 61% (19/31) |
Leinonen et al., 2013115 |
| PTEN deletions (2 colour FISH) | Incidental tumour (TURP) | 16% (104/643) | Cuzick et al., 2013168 |
| PTEN Immunohistochemistry | Same cohort | 18% (119/675) | Cuzick et al., 2013168 |
| PTEN copy number and mutation (sequence) | mCRPC | 40.7% (61/150) | Robinson et al. 201540 |
| PTEN mutation (sequence) | Prostate cancer | 2% (7/333) | TCGA et al., 20153 |
| PTEN Copy number loss (arrays) | Same cohort | 15% (50/333) | TCGA et al., 20153 |
| PTEN Immunohistochemistry | Prostate cancer | 16% (166/1044) | Ahearn et al., 201537 |
| PTEN deletion (4 colour FISH) | Prostate cancer | 18% (112/612) | Troyer et al., 201522 |
| PTEN Immunohistochemistry | Same cohort | 22% (158/731) | Lotan et al., 201636,62 |
| PTEN Immunohistochemistry | Prostate cancer Same cohort as 21 | 24.2% (1890/7813) | Lotan et al., 2017169 |
total number of PTEN deletions (homozygous and hemizygous deletions combined).
only cases with absence of PTEN protein are shown
In early studies using microsatellite analysis, loss of heterozygosity (LOH) at the PTEN locus was reported in 10–55% of primary and advanced tumours from surgical cohorts 15,17,23-26. In studies using fluorescence in situ hybridization (FISH), loss of at least one PTEN allele has been reported in up to 68% of primary tumours from various historical surgical cohorts20,27-35. Subsequent studies have been reporting PTEN deletion in around 15–20% of surgically treated men3,22,36,37(Table 1). Consistent with the strong correlation with tumour stage, PTEN loss is more common in prostate cancer metastases than in primary tumours, with most studies reporting rates of loss near 40%4,17,33,38,39 (Table 1). Data from a CRPC cohort published in 2015 showed deep (likely homozygous) deletions in ~30% of patients, with truncating mutations and gene fusions in an additional 10% 40. Racial ancestry might also affect the frequency of PTEN loss. Primary prostate tumours arising in African American men have lower rates of PTEN loss than tumours arising in matched patients of European American ancestry41-44. However the association of PTEN loss with poor prognosis seems to be independent of racial ancestry42.
Although biallelic deletion is the most common reason for PTEN inactivation, it can also be silenced by alternative genetic and epigenetic mechanisms in a minority of cases. Genomic rearrangements have been reported to involve and inactivate PTEN and nearby genes45,46,47. The frequency with which PTEN is inactivated by mutations or methylation seems to be quite low, at <10% of cases (Table 1). Results of early Sanger sequencing studies reported a high rate of mutations in the PTEN promoter region, but some of these studies were confounded by the existence of a PTEN pseudogene (PTENP1) that harbours a high rate of such alterations48,49. Data from exon sequencing studies have shown PTEN mutation rates hovering around 5% in primary tumours, of which many have hemizygous deletions involving the second allele18,19,39,40,50. The majority of mutations are truncating, with relatively few missense mutations. Epigenetic inactivation, in which loss of PTEN protein is a result of promoter methylation, has been described in other tumour types such as breast cancer51,52 . Few contemporary studies have investigated PTEN hypermethylation in primary prostate cancers, and most have reported negative results53.
Other potential PTEN inactivation mechanisms include microRNA (miRNA) and noncoding RNA (ncRNA). Several mRNAs, including the PTEN pseudogene PTENP1, might have growth-suppressive and tumour-suppressive properties and can act as competing endogenous RNAs (ceRNAs) to microRNAs that can regulate PTEN levels54. PTEN-targeting microRNAs are aberrantly overexpressed in human prostate tumours and are capable of initiating prostate tumorigenesis in vitro and in vivo55. Finally, PTEN is post-translationally regulated by phosphorylation, ubiquitylation, oxidation, acetylation, proteosomal degradation, and subcellular localization, and by its interactions with other proteins56. Among these inactivation mechanisms, post-translational modifications such as phosphorylation and ubiquitination have been shown to decrease PTEN protein levels, whereas oxidation and acetylation reduce PTEN activity57. However, the frequency with which such inactivation events occur in human prostate tumours remains unclear.
Most evidence indicates that PTEN inactivation occurs in primary tumours before they metastasize. Sequencing studies have demonstrated that PTEN deletion typically occurs identically in at least a subset of tumour cells from the primary and all or most sampled metastases38,58,59. However, in contrast to TMPRSS2-ERG rearrangements (the most common gene rearrangement in prostate cancer), PTEN deletion is frequently heterogeneous within the primary tumour, suggesting that it usually occurs after ERG rearrangement34,60,61 (Figure 3). This heterogeneity, occurring in up to 40% of primary tumours with PTEN loss36,37,62, is an important challenge for detection of PTEN status in diagnostic biopsy specimens. The relatively late loss of PTEN in primary tumours is consistent with low rates of PTEN loss observed in isolated prostatic intraepithelial neoplasia (PIN), which is widely believed to represent a precursor to invasive prostate cancer63-65.
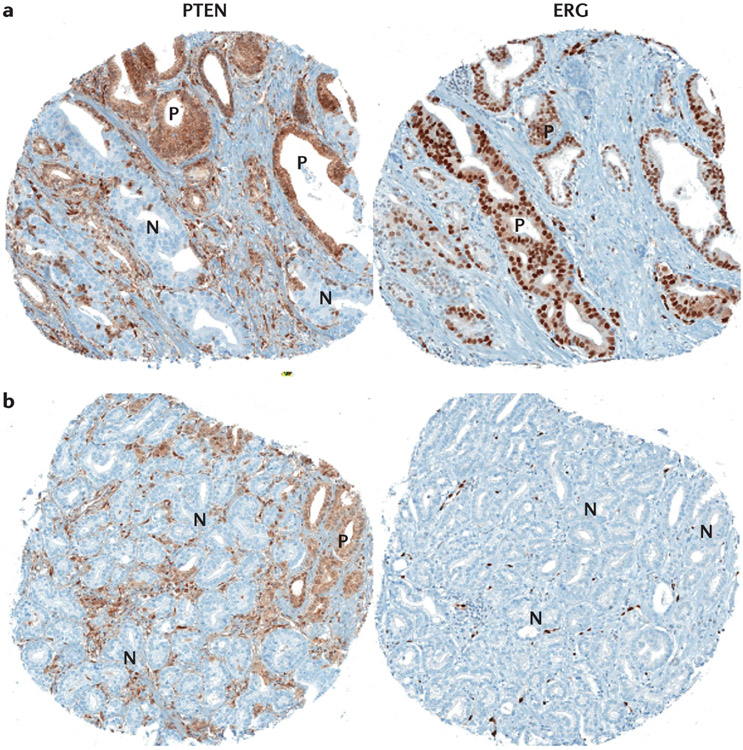
Figure 3 ∣. Heterogeneous immunohistochemical expression of ERG and PTEN in prostate tumours.
a ∣ Heterogeneous PTEN loss is observed in some tumour glands (N), with intact staining in other tumour glands (P) whereas the same areas are uniformly positive for ERG expression indicating that clonal ERG genomic rearrangement is probably present with subsequent subclonal PTEN loss. b ∣ Heterogeneous PTEN loss is seen in some tumour glands (N), with intact staining in other tumour glands (P), whereas the same areas are uniformly negative for ERG expression (ERG genomic rearrangement is absent).
PTEN interactions during tumorigenesis
Mouse models have been useful to elucidate interactions between PTEN loss and other genes and signalling pathways commonly altered in human prostate cancer. In mice, monoallelic Pten loss (Pten+/−) is sufficient to induce PIN, but does not lead to invasive prostatic carcinoma66-69. By contrast, biallelic ablation of pten in mice (pten−/−) results in relatively slow progression to locally microinvasive disease and, much more rarely to micrometastatic prostate carcinoma, the kinetics of which apparently depend, at least in part, on the background strain of mouse used 66,70,71. In mice, evidence supports the role of pten gene dosage in prostatic tumorigenesis66; however, whether this role of PTEN applies in human prostate cancer remains unclear. Patients with PTEN hemizygous tumours have intermediate outcomes between those with wild-type PTEN and those with biallelic loss; however, this effect could be due to inactivating epigenetic or point mutations of the second allele, which are not detectable by FISH36. Mouse prostate tumours with Pten loss are notably less sensitive to castration than those without Pten loss, suggesting a link between PTEN loss and castration resistance72-74. In mouse models, the mechanism of this effect seems to be downregulation of AR levels owing to reciprocal feedback between PI3K activation and AR75; some evidence of this feedback has also been observed in human prostate cancers75-77. These studies have been the basis for numerous ongoing clinical trials testing combinations of ADT and PI3K inhibitors.
Additional genomic aberrations enhance prostate tumorigenesis in the Pten-null mouse prostate78. ERG expression and Pten loss synergize to accelerate prostate carcinogenesis in mouse models79,80. In the context of Pten loss, ERG expression restores AR transcription in mouse models and human samples, providing a potential mechanism for the common co-occurrence of these two alterations81. However, human data have generally not supported the hypothesis that PTEN deletion and ERG rearrangement synergize in terms of poor oncological outcomes (see below). Concomitant loss of Pten and Tp53 results in aggressive tumour growth in the mouse prostate, which is not observed with Tp53 deficiency alone71,82,83. Loss of Tp53 might be associated with LOH of Pten, which facilitates hormone-refractory disease and bypasses cellular senescence71. Combined deletion of Pten, Rb1, and Tp53 in the mouse prostate is associated with development of metastatic tumours demonstrating lineage plasticity84, and human tumours with loss of these three tumour suppressor genes are extremely aggressive and frequently show neuroendocrine differentiation85,86. Similarly, combined loss of Pten and overexpression of c-MYC synergize to result in highly penetrant and aggressive androgen-insensitive tumours with a high rate of metastasis not observed with either aberration on its own87. Mice with c-MYC overexpression in addition to Pten and Tp53 loss also exhibit aggressive tumours with frequent local metastasis71,82.
PTEN loss is typically mutually exclusive with several other genomic alterations in human prostate cancer, including SPOP mutation and CHD1 loss, and study of these alterations in mouse and xenograft models has helped to improve understanding of the biology that underlies these findings. Recurrent SPOP mutations are the most common mutations in primary prostate cancer, occurring in close to 10% of cases. Although SPOP mutation alone is insufficient to drive tumorigenesis in mouse models, it is sufficient to activate PI3K/mTOR signalling on its own and uncouples the negative feedback between PI3K and AR signalling in human prostate organoids88. Similarly, CHD1, a chromatin helicase DNA-binding factor, is rarely lost in PTEN-null tumours, and combined loss of these tumour suppressors is synthetic lethal in vitro and in vivo owing to PTEN-deficiency-induced stabilization of CHD1, which is required for transcription of NF-κB target genes.89
PTEN as a prognostic biomarker
The introduction and widespread use of serum PSA in the late 1980s led to a considerable increase in prostate cancer incidence, and resulted in the overtreatment of many clinically insignificant prostate tumours90. However, distinguishing indolent from aggressive prostate tumours remains a challenge. Determination of which patients are eligible for active surveillance is variable between institutions and is largely based on biopsy pathology variables (Gleason score, number of biopsy cores, maximum percentage of core involvement) and serum PSA levels91. However, even in the most restrictive protocols, as many as 30% of men selected for active surveillance programmes using these parameters will be found to already harbour or to progress to higher grade disease and require intervention2. Those men who demonstrated more aggressive disease within 1–2 years of initial diagnosis were probably misclassified, owing to problems with blind biopsy sampling, and improved targeting of biopsies using MRI guidance has shown promise for risk stratification in this context92,93. Among biopsy parameters, Gleason score is the most prognostic and changes to the Gleason grading system have further refined it94. Gleason score describes the degree of morphological differentiation in the tumour based on histological examination of tumour architecture. When determined on biopsy, the final Gleason score comprises the score attributed to the most common architectural pattern or grade (ranging from grade 3 to grade 5) and the second-most-common or highest grade pattern94. Updates to approaches to tumour scoring in the past few years mean that Gleason scores have been categorized into five discrete and highly prognostic grade groups 95. However, despite these updates, the limit of what information can be inferred using tumour morphology is reaching its limit and validated, tissue-based prognostic biomarkers to identify potentially aggressive prostate tumours are needed.
Although several RNA-based commercial assays have shown potential utility in this context96, DNA-based biomarkers might have the advantage of being more stable, and, therefore, less prone to variation in preanalytic parameters such as tissue fixation conditions and tissue age. Among prognostic DNA biomarkers that have emerged from the sequencing of thousands of prostate cancers, PTEN gene loss is arguably one of the most promising. PTEN inactivation in prostate tumours is a nearly universal and highly replicable finding and is associated with adverse oncological outcomes such as increased tumour grade and stage, earlier biochemical recurrence after radical prostatectomy, metastasis, prostate-cancer-specific death, and androgen-independent progression20,22,27,32,36,37,60,97. A meta-analysis of seven previously published studies confirmed a strong correlation of PTEN genomic deletion with increased Gleason score or increased likelihood of extraprostatic extension in patients with surgically treated localized prostate cancer98. PTEN loss is clearly associated with an increased risk of biochemical recurrence after prostatectomy in several large studies21,22,36. Perhaps most importantly, PTEN was found to be an independent prognostic indicator of prostate-cancer-specific death in patients treated both conservatively or surgically37,99.
By taking advantage of the close association between PTEN loss and increasing Gleason score, PTEN might be useful as a prognostic biomarker in localized prostate cancer. In this context, testing PTEN status could potentially improve on current risk stratification protocols when Gleason score is inaccurate, particularly in low-risk and low–intermediate-risk groups, in which clinical and pathological risk stratification can be inaccurate. PTEN inactivation is generally about twice as common in Gleason score 7 (Grade Group 2–3) disease than in Gleason score 6 (Grade Group 1) at radical prostatectomy when the true Gleason score is known36,37. Accordingly, PTEN loss in Gleason pattern 3 tissue sampled from Gleason score 7 (Grade Group 2/3) tumours is substantially more frequent than PTEN loss in pattern 3 tissue sampled from Gleason score 6 (Grade Group 1) tumours100. Consistent with these data, PTEN loss in Gleason 6 (Grade Group 1) biopsies predicts upgrading to Gleason 7 (Grade Group 2/3) or higher in the radical prostatectomy specimen101,102. In Gleason 3+4=7 (Grade Group 2) tumour biopsies, PTEN loss is independently associated with a twofold increase in risk of extraprostatic extension after prostatectomy103. The first study of the utility of PTEN in an active surveillance cohort of Gleason 6 (Grade Group 1) tumours at biopsy, showed that PTEN loss was associated with an increased risk of subsequent biopsy upgrading, discontinuation of active surveillance and adverse histopathological features at radical prostatectomy104. Finally, several studies examining PTEN status in biopsies have used more concrete oncological end points such as metastasis or death. In one small study, PTEN loss in the biopsy sample predicted increased risk of CRPC, metastasis, and prostate-cancer-specific mortality in surgically treated patients105.
Until further studies are published using metastasis or death as clinical end points, the available data support the use of PTEN loss as an early marker of aggressive prostate cancer in clinical biopsy samples that are determined to be grade group 1 or potentially low-volume grade group 2. In these cases, PTEN status could substantially improve the prognostic information contained in the Gleason grade and might improve stratification of patient therapy – for example the fact that patients with PTEN loss might be inappropriate for active surveillance protocols (Figure 4). Like many molecular markers, the high frequency of PTEN loss heterogeneity in the setting of primary tumours can make it challenging to accurately assess PTEN status in a small biopsy sample. At least one study has suggested that PTEN testing in a minimum of two cores with cancer foci is necessary to accurately assess PTEN status in the context of random needle biopsies.106 In the future, improved prostate imaging modalities, such as multiparametric MRI, might also help to ensure sampling of the dominant tumour nodule, in which PTEN status might be most clinically relevant.
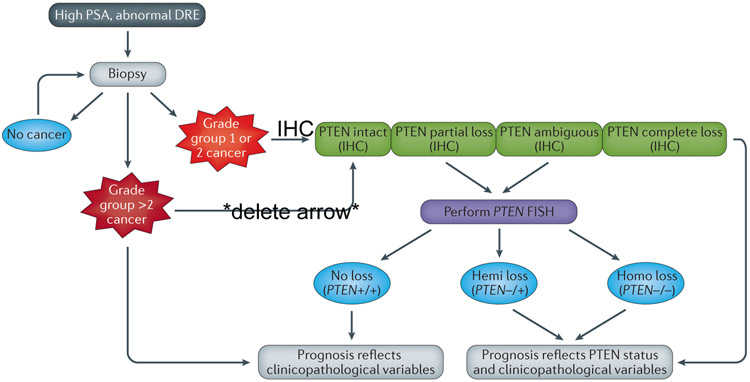
Figure 4 ∣. Algorithm for when to determine PTEN status on diagnostic biopsy material using IHC and FISH.
a ∣ Prostate biopsy is indicated to confirm or exclude cancer in patients with elevated serum PSA and/or abnormal digital rectal exam (DRE). Patients with persistent PSA elevation might require repeat biopsy.. b) If cancer is detected and is low-to-intermediate risk (Grade group 1 or 2; Gleason score 3+3 or 3+4), PTEN status can be ascertained using immunohistochemical (IHC) staining. If PTEN protein expression is intact, the prognosis reflects the patient’s clinicopathological variables (e.g. Grade Group, PSA, age, DRE). If IHC demonstrates complete PTEN loss, biomarker status should be considered in the patient’s prognosis along with clinicopathological variables and FISH is not necessary. However, if PTEN loss is incomplete or ambiguous (e.g. negative PTEN staining in cancer glands along with negative PTEN expression in internal control, such as benign glands and/or stroma), FISH is recommended and bears similar connotations to PTEN loss determined by IHC. c ∣ If cancer is detected and is high risk (Grade group 3, 4 and 5; Gleason score 4+3 and 4+4, ≥4+5), PTEN status does not need to be determined, because prognosis will be strongly driven by clinicopathological variables.
Another emerging area in which PTEN could be used as a biomarker is in the context of differential diagnosis of intraepithelial lesions in prostate biopsies. Intraductal carcinoma of the prostate and high-grade PIN are the two main intraepithelial neoplastic lesions occurring in the prostate; they exist along a morphological spectrum107. PIN is frequently an isolated finding, occurring in biopsies without invasive carcinoma, and is generally not associated with an increased risk of cancer diagnosis on a subsequent biopsy108,109. By contrast, the presence of intraductal carcinoma on needle biopsy is associated with underlying high-grade invasive carcinoma in the prostate >90% of cases110,111. In keeping with its association with underlying high-grade invasive carcinoma, intraductal carcinoma of the prostate commonly shows PTEN loss, whereas this is extraordinarily uncommon in PIN 63-65,112. Thus, PTEN status assessment can help to distinguish these lesions and to clarify the patient prognosis in the setting of intraepithelial proliferations113.
PTEN in combination with ERG
PTEN loss is enriched 2–5-fold among localized tumours with ERG gene rearrangement compared with those without this alteration30,33,36,37,60,79,80. By itself, ERG gene rearrangement does not portend an altered prognosis in most surgical cohorts114. Dissecting the interaction of PTEN and ERG with respect to oncological outcomes in prostate cancer has been complex. Based on animal models demonstrating synergy between PTEN and ERG for tumour progression, and early studies of the interaction of PTEN and ERG with respect to biochemical recurrence, patients with combined ERG rearrangement and PTEN inactivation were initially thought likely to have the worst prognosis of all patient groups30,115,116. However, additional, larger studies using biochemical recurrence as an outcome measure have found that patients with PTEN loss did similarly poorly, regardless of ERG status21,35,36. When lethal prostate cancer, rather than the surrogate outcome measure of biochemical recurrence is used, it seems that tumours with PTEN loss that lack ERG rearrangement have the worst outcomes, after either surgical or conservative therapies37,99. In fact, only the subset of tumours with PTEN loss that lack ERG rearrangement have worse oncological outcomes than those tumours with PTEN intact, suggesting that the two alterations should be assayed together to improve prognostication. This discrepancy between results from studies examining biochemical recurrence versus lethal prostate cancer might be due to an interaction of PTEN/ERG status with treatments received after biochemical recurrence, such as radiation or hormone therapy. An additional study has shown that PTEN loss combined with high immunohistochemical expression of AR, especially in ERG-negative cancers, predicted initiation of secondary treatments, shortened disease-specific survival time, and stratified Gleason score 7 (Grade group 2/3) patients into different prognostic groups117. Cumulatively, these results suggest that PTEN loss is most closely associated with lethal prostate cancer among patients whose tumours have not rearranged ERG. Thus, triaging patients by combining PTEN status with other prognostic biomarkers at the time of biopsy might also help to stratify patients with localized disease into active surveillance versus definitive therapy cohorts.
Assays to detect PTEN loss
Although the potential utility of PTEN as a biomarker in localized disease — with or without other molecular markers — is well established, very few assays have been analytically validated for clinical use in this setting. Analytic validation is an absolute requirement for any molecular biomarker that is to be used in clinical decision making. In clinical pathology laboratories, FISH and immunohistochemistry (IHC) have predominantly been used to assess patient samples for changes in PTEN copy number and protein expression (Figure 2). FISH is a quantitative and highly specific method for determination of gene copy number within interphase cells in tissue sections27. Probe design, sample preparation, hybridization protocols, and signal scoring criteria are critically important factors for quality assurance. PTEN FISH assays can be compromized by a high background level of signal losses associated with tangential sectioning of nuclei during slide preparation. A number of new probe designs (typically using two PTEN-flanking probes for WAPAL and FAS in addition to the PTEN probe and the centromere control probe) have been shown to increase the accuracy of FISH deletion assays, so that true chromosomal deletions can be readily distinguished from the false signal losses caused by sectioning artefacts118,119.
Compared with IHC, one advantage of FISH is that hemizygous and homozygous gene loss can both be detected with high sensitivity. As might be expected from gene dosage effects in mice66, hemizygous PTEN loss is more weakly associated with adverse outcomes than homozygous loss21,22. However, the relatively high cost of FISH probes, the complexity of standardized scoring protocols and the challenge of integrating FISH into clinical pathology laboratory workflows have prompted researchers to develop a less expensive and time-consuming clinical grade immunohistochemical assay to detect PTEN loss36,97. In addition, as PTEN loss is commonly subclonal and/or focal in primary prostate tumours34,61,77,97, detection of PTEN deletion — especially focal losses — by FISH can be technically challenging and is more feasible using IHC than FISH. IHC protocols have been successfully validated on the Ventana Benchmark platform in a Clinical Laboratory Improvement Amendments (CLIA)-certified lab with high interobserver reproducibility in the scoring system36,37. Correlation of PTEN gene loss by FISH and protein loss by IHC is quite high36,97,120,121, but some discordance can result from the fact that PTEN loss can theoretically be due to very small genomic deletions or transcriptional or post-transcriptional regulation, in which case FISH might show a false negative result. Interestingly, heterogeneous or partial PTEN protein loss was a weaker prognostic indicator than homogeneous or complete loss, using either biochemical recurrence or lethal prostate cancer as the outcome variable, but the reasons for this effect remain unclear36,37. Compared with FISH, IHC is not as sensitive in detecting hemizygous PTEN loss36; however, whether this discrepancy adds prognostic information remains unclear22. Overall, the most cost-effective and time-effective protocol to screen for PTEN loss in human prostate tissue should include initial screening by IHC, followed by FISH analysis in cases that are ambiguous or indeterminate by IHC (generally comprising <5% of cases) and potentially in cases with heterogeneous loss of PTEN by IHC, in which FISH can add prognostic information36 (Figure 4).
As sensitive and noninvasive measurement of DNA biomarkers in blood and urine becomes increasingly feasible with advanced sequencing technologies, interest in detecting PTEN deletion in bodily fluids is growing. Such tests could be particularly useful in patients with advanced metastatic prostate cancer, in which PTEN might serve as predictive biomarker. One relevant method is detection of PTEN status in circulating tumour cells (CTCs). EpCAM and cytokeratin-based enrichment protocols for CTCs are currently the only FDA-approved platform for CTC analysis (CellSearch), but other methodologies are gaining traction. PTEN status can be evaluated in CTCs by FISH using an enrichment-free platform, and is both highly concordant with PTEN status in matched fresh-frozen tissues.122 PTEN loss associated with poorer survival in patients with metastatic CRPC (mCRPC) Cell-free DNA in blood might also prove useful to assess PTEN status123, although studies to correlate blood PTEN with tissue PTEN status are still ongoing. Urine is another accessible sample type that might be suitable for interrogation of PTEN status via cell-free DNA124. Most of these studies remain focused on biomarker development, but additional thorough analytical validation studies will be required before fluid-based DNA biomarker measurements can be used clinically125. Ongoing clinical trials incorporating CTCs and cell-free DNA measurements will add to our understanding of the potential use of these assays, though many of these trials are focused on androgen receptor splice variants rather than PTEN 126-129. One ongoing trial creates a multi-institutional database of CTC DNA for chromosomal gains/losses and RNA for androgen receptor splice variants in patients with metastatic castration resistant prostate cancer before and after treatment with abiraterone, enzalutamide, or taxane-based chemotherapy with the hope of identifying markers of therapy-resistance130.
PTEN-targeted therapies in mCRPC
The potential role of PTEN as a predictive biomarker in the context of mCRPC is under examination. Consistent with mouse models predicting the association of PTEN inactivation with development of CRPC, PTEN loss has been associated with decreased response to novel AR-targeted therapies, including abiraterone131. However, the most exciting context for PTEN as a predictive biomarker has been in the setting of therapies targeted to PI3K/AKT/mTOR signaling. PTEN loss is associated with unimpeded PI3K and downstream signalling, so the initial outlook for the efficacy of PI3K/AKT/mTOR inhibitors in prostate cancer was optimistic (Figure 5). However, despite promising performance in preclinical models, the clinical efficacy of these drugs as single agents in CRPC has been uniformly low132,133. Preclinical models have suggested that single-agent therapy with inhibitors targeting the PI3K/AKT/mTOR pathway might activate AR signalling via compensatory crosstalk; thus, co-targeting the AR and the PI3K pathways might be a more effective therapeutic approach than using single-agent therapies 75,134-136. However, early clinical trial data with PI3K inhibitors and novel androgen-targeted therapies do not look uniformly promising in unselected patients. The trial of BKM-120 — a pan-PI3K inhibitor — with enzalutamide did not show efficacy137, whereas BEZ235 — a dual PI3K/mTOR inhibitor — in combination with abiraterone, was poorly tolerated138. Further trials of abiraterone in combination with either BKM-120 or BEZ235 are not planned139. One trial of everolimus (RAD001, an mTOR inhibitor) and bicalutamide warrants further investigation, although the treatment-associated toxic effects were somewhat limiting140. Additional trials of novel AR-signalling inhibitors and mTORC1 inhibition using everolimus are currently in progress141,142. The hope is that the results from these trials will be similar to the breast cancer BOLERO-2 trial, in which a combination of hormonal therapy and mTORC1 inhibition was quite promising143. Other trials testing the efficacy of PI3K/AKT/mTOR inhibition with additional agents in the context of AR axis signalling suppression are currently underway or soon to be reported, including a phase 1b trial of enzalutamide with the mTOR kinase inhibitor CC-115144 and a phase 2 study of the PI3K/mTOR inhibitor GDC-0980 with abiraterone145.
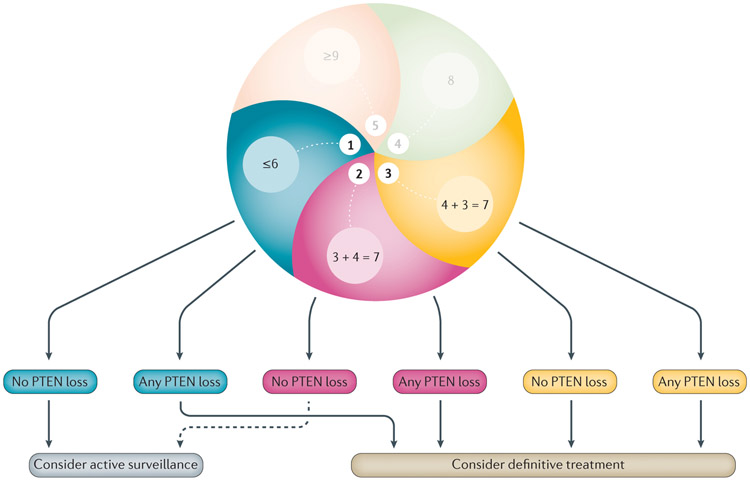
Figure 5 ∣. Proposed management options using clinicopathological variables at biopsy and PTEN status.
Patients with Grade Group 1 (GG1) cancer and no loss of PTEN on biopsy (by IHC and/or FISH) should be considered for active surveillance. Patients with GG1 cancer with PTEN loss should be considered for definitive treatment using radiotherapy or prostatectomy. In some clinical contexts, patients with GG2 cancer with no loss of PTEN could be considered for active surveillance, particularly if they have low-volume disease and a low percentage of Gleason pattern 4 (indicated by dashed arrow). Patients with GG2 tumours with PTEN loss or tumours >GG2 should be considered for definitive treatment in most cases.
One limitation of the aforementioned studies is that they have generally been conducted on unselected and heterogeneous patient populations, some with and some without PTEN loss. This limitation is, in part, due to the historical lack of uniformly accepted and highly analytically validated assays to measure PTEN loss. Given the relatively high frequency of PTEN loss in the CRPC setting, that a therapeutic benefit would be seen even in unselected patients would seem likely. However, subset analyses stratified by PTEN status might be required to identify benefit for agents targeting the PI3K/AKT/mTOR pathway. For example, one trial of a novel ATP-competitive AKT inhibitor (ipatasertib or GDC-0068) with abiraterone showed some responses, which occurred much more frequently in patients harbouring PTEN loss (identified by IHC)145,146. This promising result suggests that PTEN status, if measured using an analytically validated test, could indeed ultimately become a predictive biomarker in terms of selection of patients for treatment with AKT inhibitors. These data, combined with the ease of PTEN status testing by analytically validated IHC or FISH, emphasize the need for additional trials selected for tumours with PTEN loss to improve efficacy assessment of combined AR-targeted and PI3K-targeted, AKT-targeted, or mTOR-targeted therapies.
Another issue with early trials of pan-PI3K inhibitors is that the toxic effects associated with their use are often limiting. Given that evidence for nonredundant roles of PI3K isoforms in different tumour types is increasing, isoform-specific inhibitors of PI3K might have a role in prostate cancer treatment. PI3Ks consist of a catalytic subunit (p110α, p110β, or p110δ) complexed to a regulatory subunit (p85)147. Both the p110α and p110β isoforms are ubiquitously expressed; however, in the setting of PTEN loss, preclinical data suggest that PI3Kα activity is relatively suppressed and tumour cells rely more heavily on the p110β isoform136. Accordingly, ablation of p110β, but not p110α, was sufficient to impede tumorigenesis in the PTEN-null mouse prostate148. However, even PI3Kβ inhibition activates AR activity in preclinical models136; thus, PI3Kβ-selective inhibitors combined with AR axis inhibition are currently being tested for their ability to suppress the reciprocal feedback activation of both pathways149,150.
Additional novel strategies are being evaluated in preclinical studies. One potential therapeutic option is combination therapy of PI3K inhibitors with PARP (Poly [ADP-ribose] polymerase) inhibitors. PARP is an enzyme that modifies DNA at single strand breaks, leading to the recruitment of DNA repair effectors151. Prostate tumour cells deficient in normal DNA repair mechanisms (such as tumours with BRCA2 loss) are highly sensitive to PARP inhibition152. Furthermore, nuclear PTEN might also have a role in DNA repair13,14, raising the question of whether combined PARP and PI3K inhibition might be efficacious. Preclinical prostate cancer models with combined pten/tp53 loss show some response to these combination therapies153,154. Finally, emerging evidence suggests that an alternative variant of PTEN known as PTEN-Long, may be secreted by cells155. The secreted protein can be taken up by other cells, including those that have lost endogenous PTEN activity. When mice with xenograft tumours lacking PTEN were treated with PTEN-Long, their tumours stayed stable and some even regressed155. The findings suggest that, in principle, recombinant secreted PTEN could itself be a novel treatment approach to tumours with PTEN mutations or deletions156. Ultimately, these studies emphasize the importance of understanding PTEN regulatory mechanisms and function for the design of novel therapeutic strategies in advanced prostate cancer.
PTEN loss and immune microenvironment modulation
Prostate cancer is a slow-growing disease making it an ideal tumour for future immunotherapy. This longer disease course provides a considerable time period during which novel therapeutics could be applied to trigger an antitumour immune response157. Emerging data suggest that, in addition to its established role as a tumour suppressor, PTEN loss itself might be an immunosuppressive event8. Successful immunotherapy in prostate cancer will depend on fully understanding the tumour microenvironment and the inflammatory mechanisms that enable the tumour to evade immune responses, as well as identification of actionable targets to enhance immune attack on tumour cells.
The development of chronic prostatic inflammation is often accompanied by histological lesions that seem to be precursors to prostate cancer158, and histological evidence shows that prostate tumours are often infiltrated by immune cells such as CD4+ and CD8+ T-cells, natural killer (NK) cells and antigen-presenting cells (dendritic cells and macrophages)159. The ratios of each subtype are associated with different prognoses. For example, infiltrating NK cells are associated with good prognosis and are thought to provide a strong antitumour response160, whereas CD4+ cells, which are regulatory T cells (Treg cells), suppress immune responses and are associated with a poor prognosis161.
Studies in melanoma, one of the first tumours to show a clinical benefit with immunotherapy, have shown that PTEN loss correlates with a reduction in T cell inflammatory responses and worse outcomes with anti-PD-1 immunotherapy162. PTEN has also been shown to directly regulate interferon response signalling pathways and, in studies using oncolytic viruses, loss of PTEN has a crucial role in mediating antiviral innate immunity7. For example, PTEN-deficient cancer cells have muted type I interferon responses and are more sensitive to viral infections than cells with intact PTEN7,163. Such alterations to IFN regulation signalling pathways by PTEN are likely to have protumorigenic effects in addition to the effects on the innate antiviral immune system. These findings might explain why PTEN-deficient tumour cells are more permissive to IFN-sensitive oncolytic viruses, and are an important consideration for future oncolytic therapy trials targeting PTEN-deficient prostate cancers164.
The role of PTEN in the tumour immune response is likely to act through activation of the signal transducer and activator of transcription (STAT) protein family in prostate cancer165. PTEN dephosphorylates interferon regulatory factor 3 (IRF3) and increases its nuclear translocation, leading to increased expression of IFN1 response genes7. By contrast, mouse embryonic fibroblast cells with Pten knockout have disrupted nuclear import, decreased activity of IRF3 and have reduced type I IFN response. Downstream targets of IRF3 include IFNα and IFNβ28, both of which activate STAT1 and STAT3 transcription factors. STAT proteins are key to both type I and type II interferon responses, such as the induction of chemokines that recruit immune cells into the tissue microenvironment166.
Understanding the dynamic interaction between PTEN-deficient tumours and the immune signalling that takes place in the tumour microenvironment is important for developing effective immunotherapies. For example, Pten-null mouse models secrete immunosuppressive senescence-associated cytokines into the tumour microenvironment167. Interestingly, pharmacological inhibition of the Jak2/Stat3 pathway can reactivate the senescence-associated cytokine network, leading to an antitumour immune response that enhances sensitivity to chemotherapy167. These data suggest that if the immune surveillance of senescent PTEN-null tumours is suppressed, specific pharmacological interventions might be able to restore immunogenicity to tumours.
The emerging relationship between PTEN and the immune system is complex and covers both protumorigenic and antitumourigenic cascades that depend on cellular phenotypes, combinations of these phenotypes, and the tumour microenvironment. Further studies are needed to exploit PTEN-dependent changes, such as reduced type I interferon response and cytokine signalling to the tumour microenvironment, and to develop effective immunotherapy in prostate cancer.
Conclusions
Detection of PTEN loss in prostate cancer has tremendous potential to enhance our understanding of the biology of the disease and improve patient care. As the most commonly lost tumour suppressor in primary prostate cancer, PTEN loss is one of few prognostic biomarkers that is reproducibly associated with poor outcomes in patients with the disease. Easily and inexpensively measured using analytically validated assays, PTEN status determination in diagnostic biopsies might improve patient selection for active surveillance and can identify patients at increased risk for disease progression who could benefit from intensive definitive therapies. In advanced metastatic prostate cancer, PTEN status can be measured in liquid biopsies. At this end of the disease spectrum, further refinements in targeting PI3K–AKT–mTOR signalling, most likely in combination with AR signalling, might be effective in subsets of patients with PTEN-deficient tumours. Finally, emerging evidence suggests that PTEN status influences immune response to tumour progression and has a role in predicting which patients will respond to promising immunotherapies. Although we are clearly in the early days of molecular classification of prostate cancer and its application to clinical care, as a biomarker, PTEN is here to stay.
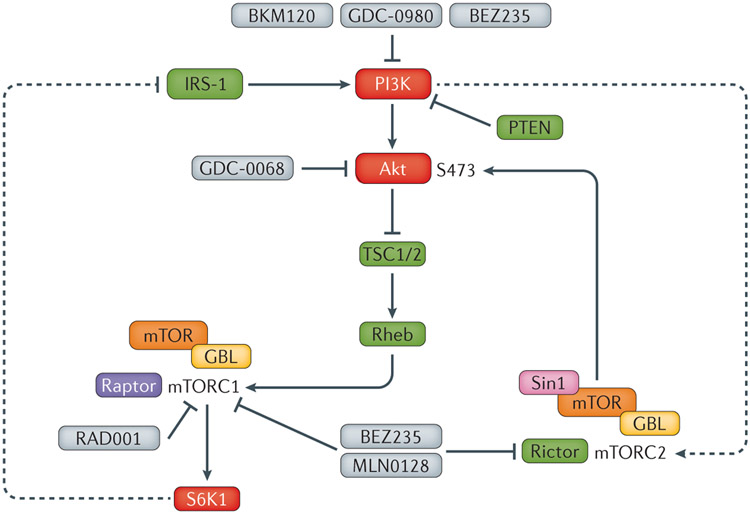
Figure 6 ∣. Selected drugs in clinical trials targeting the PI3K/AKT pathway that have been used in combination with androgen deprivation therapy.
PI3K inhibitors (GDC-0980, BKM120), combined PI3K/mTOR inhibitors (BEZ235), mTORC1 inhibitors (RAD001) and mTOR kinase inhibitors (CC-115) have been used in clinical trials in combination with novel and conventional androgen deprivation therapies. Whereas some trials have been largely negative (BKM-120137) or poorly tolerated (BEZ235138), others are promising and suggest that PTEN status might be a useful predictive biomarker for response in some contexts (eg, GDC-0068 or ipatasertib146).
Key points.
Large-scale next-generation genetic analyses of prostate cancer emphasize the frequent occurrence and importance of focal genomic deletions inactivating PTEN
PTEN loss in radical prostatectomy samples is often concurrent with genomic rearrangements involving the ETS family transcription factors
PTEN loss is reproducibly associated with adverse oncological outcomes by itself or in combination with other biomarkers, and helps distinguish indolent tumours from those likely to progress.
PTEN might be a useful prognostic biomarker to distinguish potentially aggressive grade group 1 or 2 tumours, which might make patients poor candidates for active surveillance programmes
Robust clinical assays using immunohistochemistry and FISH have been developed to reproducibly measure PTEN protein and gene loss using diagnostic tissue biopsies and circulating tumour cells from plasma and cell-free DNA
PTEN loss is associated with suppression of androgen receptor (AR) transcriptional output and PI3K inhibitors activate AR signaling, suggesting potential efficacy of combination therapies targeting the PI3K and AR signaling pathways
Emerging studies indicate that PTEN loss is associated with alterations to cellular interferon responses in the tumour microenvironment — tumours with loss of PTEN are more likely to have an immunosuppressive microenvironment, suggesting that advanced prostate cancers with PTEN loss might be amenable to immune-based therapies
Acknowledgements
Funding for this research was provided in part by a Transformative Impact Award from the CDMRP-PCRP (W81XWH-13-2-0070, H.I.S. and T.L.L.). T.L.L. was additionally supported by the NIH/NCI Cancer Center Support Grant P30 CA006973 and the Patrick Walsh Prostate Cancer Research Fund. H.I.S. was additionally supported by NIH/NCI Prostate SPORE Grant P50-CA92629, NIH/NCI Cancer Center Support Grant P30 CA008748, and the Prostate Cancer Foundation. T.J. and D.M.B. were funded by Prostate Cancer Canada and the Movember Foundation (Grant #T2014-01-PRONTO). T.J. was supported by Transformative Pathology Fellowship funded by Ontario Institute for Cancer Research through funding provided by the Government of Ontario.
Footnotes
Competing interests statement
T.L.L. has received research support from Ventana Medical Systems. D.M.B. has received financial support from Myriad Genetics and Metamark Genetics.
References
- 1.Siegel RL, Miller KD & Jemal A Cancer Statistics, 2017. CA Cancer J Clin 67, 7–30, doi: 10.3322/caac.21387 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Tosoian JJ et al. Intermediate and Longer-Term Outcomes From a Prospective Active-Surveillance Program for Favorable-Risk Prostate Cancer. J Clin Oncol 33, 3379–3385, doi: 10.1200/JCO.2015.62.5764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025, doi: 10.1016/j.cell.2015.10.025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22, doi: 10.1016/j.ccr.2010.05.026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maehama T & Dixon JE The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273, 13375–13378 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Song MS, Salmena L & Pandolfi PP The functions and regulation of the PTEN tumour suppressor. Nature reviews. Molecular cell biology 13, 283–296, doi: 10.1038/nrm3330 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Li S et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol 17, 241–249, doi: 10.1038/ni.3311 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Chen L & Guo D The functions of tumor suppressor PTEN in innate and adaptive immunity. Cell Mol Immunol 14, 581–589, doi: 10.1038/cmi.2017.30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura M et al. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 280, 1614–1617 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Zhang S et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med 17, 461–469, doi: 10.1038/nm.2309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng LP, Brown JL & Eng C PTEN coordinates G(1) arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Hum Mol Genet 10, 599–604 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Lindsay Y et al. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J Cell Sci 119, 5160–5168, doi: 10.1242/jcs.000133 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Shen WH et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128, 157–170, doi: 10.1016/j.cell.2006.11.042 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Bassi C et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341, 395–399, doi: 10.1126/science.1236188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns P et al. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res 57, 4997–5000 (1997). [PubMed] [Google Scholar]
- 16.Steck PA et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 15, 356–362, doi: 10.1038/ng0497-356 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res 58, 204–209 (1998). [PubMed] [Google Scholar]
- 18.Berger MF et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220, doi: 10.1038/nature09744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbieri CE et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44, 685–689, doi: 10.1038/ng.2279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimoto M et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 97, 678–685, doi: 10.1038/sj.bjc.6603924 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krohn A et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol 181, 401–412, doi: 10.1016/j.ajpath.2012.04.026 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Troyer DA et al. A multicenter study shows PTEN deletion is strongly associated with seminal vesicle involvement and extracapsular extension in localized prostate cancer. Prostate 75, 1206–1215, doi: 10.1002/pros.23003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feilotter HE, Nagai MA, Boag AH, Eng C & Mulligan LM Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene 16, 1743–1748, doi: 10.1038/sj.onc.1200205 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Pesche S et al. PTEN/MMAC1/TEP1 involvement in primary prostate cancers. Oncogene 16, 2879–2883, doi: 10.1038/sj.onc.1202081 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Wang SI, Parsons R & Ittmann M Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res 4, 811–815 (1998). [PubMed] [Google Scholar]
- 26.Whang YE et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A 95, 5246–5250, doi:DOI 10.1073/pnas.95.9.5246 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimoto M et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet 169, 128–137, doi: 10.1016/j.cancergencyto.2006.04.003 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Verhagen PC et al. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol 208, 699–707, doi: 10.1002/path.1929 (2006). [DOI] [PubMed] [Google Scholar]
- 29.McCall P, Witton CJ, Grimsley S, Nielsen KV & Edwards J Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer 99, 1296–1301, doi: 10.1038/sj.bjc.6604680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimoto M et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol 21, 1451–1460, doi: 10.1038/modpathol.2008.96 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Attard G et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res 69, 2912–2918, doi: 10.1158/0008-5472.CAN-08-3667 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Sircar K et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol 218, 505–513, doi: 10.1002/path.2559 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Han B et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol 22, 1083–1093, doi: 10.1038/modpathol.2009.69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krohn A et al. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol 27, 1612–1620, doi: 10.1038/modpathol.2014.70 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Steurer S et al. TMPRSS2-ERG fusions are strongly linked to young patient age in low-grade prostate cancer. Eur Urol 66, 978–981, doi: 10.1016/j.eururo.2014.06.027 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Lotan TL et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol 29, 904–914, doi: 10.1038/modpathol.2016.88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahearn TU et al. A Prospective Investigation of PTEN Loss and ERG Expression in Lethal Prostate Cancer. J Natl Cancer Inst 108, doi: 10.1093/jnci/djv346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 15, 559–565, doi: 10.1038/nm.1944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grasso CS et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243, doi: 10.1038/nature11125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson D et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228, doi: 10.1016/j.cell.2015.05.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khani F et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res 20, 4925–4934, doi: 10.1158/1078-0432.CCR-13-2265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tosoian JJ et al. Prevalence and Prognostic Significance of PTEN Loss in African-American and European-American Men Undergoing Radical Prostatectomy. Eur Urol 71, 697–700, doi: 10.1016/j.eururo.2016.07.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindquist KJ et al. Mutational Landscape of Aggressive Prostate Tumors in African American Men. Cancer Res 76, 1860–1868, doi: 10.1158/0008-5472.CAN-15-1787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang FW et al. Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function ERF Mutations. Cancer Discov 7, 973–983, doi: 10.1158/2159-8290.CD-16-0960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid AH et al. Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol 25, 902–910, doi: 10.1038/modpathol.2011.207 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Murphy SJ et al. Integrated analysis of the genomic instability of PTEN in clinically insignificant and significant prostate cancer. Mod Pathol 29, 143–156, doi: 10.1038/modpathol.2015.136 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Ibeawuchi C et al. Exploring prostate cancer genome reveals simultaneous losses of PTEN, FAS and PAPSS2 in patients with PSA recurrence after radical prostatectomy. International journal of molecular sciences 16, 3856–3869, doi: 10.3390/ijms16023856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whang YE, Wu XY & Sawyers CL Identification of a pseudogene that can masquerade as a mutant allele of the PTEN/MMAC1 tumor suppressor gene. J Natl Cancer Inst 90, 859–861, doi:DOI 10.1093/jnci/90.11.859 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Zysman MA, Chapman WB & Bapat B Considerations When Analyzing the Methylation Status of PTEN Tumor Suppressor Gene. The American Journal of Pathology 160, 795–800, doi: 10.1016/s0002-9440(10)64902-4 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beltran H et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol 63, 920–926, doi: 10.1016/j.eururo.2012.08.053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bermudez Brito M, Goulielmaki E & Papakonstanti EA Focus on PTEN Regulation. Frontiers in oncology 5, 166, doi: 10.3389/fonc.2015.00166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia JM et al. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chromosomes Cancer 41, 117–124, doi: 10.1002/gcc.20062 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Konishi N et al. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol 160, 1207–1214, doi: 10.1016/S0002-9440(10)62547-3 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tay Y et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357, doi: 10.1016/j.cell.2011.09.029 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poliseno L et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 3, ra29, doi: 10.1126/scisignal.2000594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leslie NR & Foti M Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol Sci 32, 131–140, doi: 10.1016/j.tips.2010.12.005 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Salmena L, Carracedo A & Pandolfi PP Tenets of PTEN tumor suppression. Cell 133, 403–414, doi: 10.1016/j.cell.2008.04.013 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Gundem G et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357, doi: 10.1038/nature14347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haffner MC et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest 123, 4918–4922, doi: 10.1172/JCI70354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bismar TA et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int 107, 477–485, doi: 10.1111/j.1464-410X.2010.09470.x (2011). [DOI] [PubMed] [Google Scholar]
- 61.Gumuskaya B et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis 16, 209–215, doi: 10.1038/pcan.2013.8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lotan TL et al. PTEN Loss as Determined by Clinical-grade Immunohistochemistry Assay Is Associated with Worse Recurrence-free Survival in Prostate Cancer. Eur Urol Focus 2, 180–188, doi: 10.1016/j.euf.2015.07.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lotan TL et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol 26, 587–603, doi: 10.1038/modpathol.2012.201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morais CL et al. Utility of PTEN and ERG immunostaining for distinguishing high-grade PIN from intraductal carcinoma of the prostate on needle biopsy. Am J Surg Pathol 39, 169–178, doi: 10.1097/PAS.0000000000000348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morais CL et al. ERG and PTEN status of isolated high-grade PIN occurring in cystoprostatectomy specimens without invasive prostatic adenocarcinoma. Hum Pathol 55, 117–125, doi: 10.1016/j.humpath.2016.04.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trotman LC et al. Pten dose dictates cancer progression in the prostate. PLoS Biol 1, E59, doi: 10.1371/journal.pbio.0000059 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Cristofano A, Pesce B, Cordon-Cardo C & Pandolfi PP Pten is essential for embryonic development and tumour suppression. Nat Genet 19, 348–355, doi: 10.1038/1235 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Stambolic V et al. High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/− mice. Cancer Res 60, 3605–3611 (2000). [PubMed] [Google Scholar]
- 69.Podsypanina K et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A 96, 1563–1568 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Chen Z et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730, doi: 10.1038/nature03918 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen MM & Abate-Shen C Pten inactivation and the emergence of androgen-independent prostate cancer. Cancer Res 67, 6535–6538, doi: 10.1158/0008-5472.CAN-07-1271 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Jiao J et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res 67, 6083–6091, doi: 10.1158/0008-5472.CAN-06-4202 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Mulholland DJ et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19, 792–804, doi: 10.1016/j.ccr.2011.05.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carver BS et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586, doi: 10.1016/j.ccr.2011.04.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choucair K et al. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. BMC cancer 12, 543, doi: 10.1186/1471-2407-12-543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bismar TA et al. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. Histopathology 60, 645–652, doi: 10.1111/j.1365-2559.2011.04116.x (2012). [DOI] [PubMed] [Google Scholar]
- 78.Grabowska MM et al. Mouse models of prostate cancer: picking the best model for the question. Cancer Metastasis Rev 33, 377–397, doi: 10.1007/s10555-013-9487-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carver BS et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 41, 619–624, doi: 10.1038/ng.370 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.King JC et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet 41, 524–526, doi: 10.1038/ng.371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Y et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 19, 1023–1029, doi: 10.1038/nm.3216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J et al. A mouse model of heterogeneous, c-MYC-initiated prostate cancer with loss of Pten and p53. Oncogene 31, 322–332, doi: 10.1038/onc.2011.236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Couto SS et al. Simultaneous haploinsufficiency of Pten and Trp53 tumor suppressor genes accelerates tumorigenesis in a mouse model of prostate cancer. Differentiation 77, 103–111, doi: 10.1016/j.diff.2008.09.010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ku SY et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83, doi: 10.1126/science.aah4199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan HL et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res 20, 890–903, doi: 10.1158/1078-0432.CCR-13-1982 [doi] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aparicio AM et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin Cancer Res 22, 1520–1530, doi: 10.1158/1078-0432.CCR-15-1259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hubbard GK et al. Combined MYC Activation and Pten Loss Are Sufficient to Create Genomic Instability and Lethal Metastatic Prostate Cancer. Cancer Res 76, 283–292, doi: 10.1158/0008-5472.CAN-14-3280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blattner M et al. SPOP Mutation Drives Prostate Tumorigenesis In Vivo through Coordinate Regulation of PI3K/mTOR and AR Signaling. Cancer Cell 31, 436–451, doi: 10.1016/j.ccell.2017.02.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao D et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542, 484–488, doi: 10.1038/nature21357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moschini M et al. Low-risk Prostate Cancer: Identification, Management, and Outcomes. Eur Urol 72, 238–249, doi: 10.1016/j.eururo.2017.03.009 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Bruinsma SM et al. Active surveillance for prostate cancer: a narrative review of clinical guidelines. Nature reviews. Urology 13, 151–167, doi: 10.1038/nrurol.2015.313 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Barrett T & Haider MA The Emerging Role of MRI in Prostate Cancer Active Surveillance and Ongoing Challenges. AJR. American journal of roentgenology 208, 131–139, doi: 10.2214/AJR.16.16355 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Ma TM et al. The Role of Multiparametric Magnetic Resonance Imaging/Ultrasound Fusion Biopsy in Active Surveillance. Eur Urol 71, 174–180, doi: 10.1016/j.eururo.2016.05.021 (2017). [DOI] [PubMed] [Google Scholar]
- 94.Epstein JI et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 40, 244–252, doi: 10.1097/PAS.0000000000000530 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Epstein JI et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol 69, 428–435, doi: 10.1016/j.eururo.2015.06.046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ross AE, D’Amico AV & Freedland SJ Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer Prostatic Dis 19, 1–6, doi: 10.1038/pcan.2015.31 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Lotan TL et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res 17, 6563–6573, doi: 10.1158/1078-0432.CCR-11-1244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y & Dai B PTEN genomic deletion defines favorable prognostic biomarkers in localized prostate cancer: a systematic review and meta-analysis. Int J Clin Exp Med 8, 5430–5437 (2015). [PMC free article] [PubMed] [Google Scholar]
- 99.Reid AH et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer 102, 678–684, doi: 10.1038/sj.bjc.6605554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trock BJ et al. PTEN loss and chromosome 8 alterations in Gleason grade 3 prostate cancer cores predicts the presence of un-sampled grade 4 tumor: implications for active surveillance. Mod Pathol 29, 764–771, doi: 10.1038/modpathol.2016.63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lotan TL et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol 28, 128–137, doi: 10.1038/modpathol.2014.85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Picanco-Albuquerque CG et al. In prostate cancer needle biopsies, detections of PTEN loss by fluorescence in situ hybridization (FISH) and by immunohistochemistry (IHC) are concordant and show consistent association with upgrading. Virchows Arch 468, 607–617, doi: 10.1007/s00428-016-1904-2 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Guedes LB, Tosoian JJ, Hicks J, Ross AE & Lotan TL PTEN Loss in Gleason Score 3 + 4 = 7 Prostate Biopsies is Associated with Nonorgan Confined Disease at Radical Prostatectomy. J Urol 197, 1054–1059, doi: 10.1016/j.juro.2016.09.084 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Lokman U, Erickson AM, Vasarainen H, Rannikko AS & Mirtti T PTEN Loss but Not ERG Expression in Diagnostic Biopsies Is Associated with Increased Risk of Progression and Adverse Surgical Findings in Men with Prostate Cancer on Active Surveillance. Eur Urol Focus, doi: 10.1016/j.euf.2017.03.004 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Mithal P et al. PTEN loss in biopsy tissue predicts poor clinical outcomes in prostate cancer. International journal of urology : official journal of the Japanese Urological Association 21, 1209–1214, doi: 10.1111/iju.12571 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Shah RB, Bentley J, Jeffery Z & DeMarzo AM Heterogeneity of PTEN and ERG expression in prostate cancer on core needle biopsies: implications for cancer risk stratification and biomarker sampling. Hum Pathol 46, 698–706, doi: 10.1016/j.humpath.2015.01.008 (2015). [DOI] [PubMed] [Google Scholar]
- 107.Wobker SE & Epstein JI Differential Diagnosis of Intraductal Lesions of the Prostate. Am J Surg Pathol 40, e67–82, doi: 10.1097/PAS.0000000000000609 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Epstein JI & Herawi M Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care. J Urol 175, 820–834, doi: 10.1016/S0022-5347(05)00337-X (2006). [DOI] [PubMed] [Google Scholar]
- 109.Tosoian JJ, Alam R, Ball MW, Carter HB & Epstein JI Managing high-grade prostatic intraepithelial neoplasia (HGPIN) and atypical glands on prostate biopsy. Nature reviews. Urology 15, 55–66, doi: 10.1038/nrurol.2017.134 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Guo CC & Epstein JI Intraductal carcinoma of the prostate on needle biopsy: Histologic features and clinical significance. Mod Pathol 19, 1528–1535, doi: 10.1038/modpathol.3800702 (2006). [DOI] [PubMed] [Google Scholar]
- 111.Robinson BD & Epstein JI Intraductal carcinoma of the prostate without invasive carcinoma on needle biopsy: emphasis on radical prostatectomy findings. J Urol 184, 1328–1333, doi: 10.1016/j.juro.2010.06.017 (2010). [DOI] [PubMed] [Google Scholar]
- 112.Hickman RA et al. Atypical Intraductal Cribriform Proliferations of the Prostate Exhibit Similar Molecular and Clinicopathologic Characteristics as Intraductal Carcinoma of the Prostate. Am J Surg Pathol 41, 550–556, doi: 10.1097/PAS.0000000000000794 (2017). [DOI] [PubMed] [Google Scholar]
- 113.De Marzo AM, Haffner MC, Lotan TL, Yegnasubramanian S & Nelson WG Premalignancy in Prostate Cancer: Rethinking What we Know. Cancer prevention research 9, 648–656, doi: 10.1158/1940-6207.CAPR-15-0431 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Pettersson A et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev 21, 1497–1509, doi: 10.1158/1055-9965.EPI-12-0042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Leinonen KA et al. Loss of PTEN Is Associated with Aggressive Behavior in ERG-Positive Prostate Cancer. Cancer Epidemiol Biomarkers Prev 22, 2333–2344, doi: 10.1158/1055-9965.Epi-13-0333-T (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fontugne J et al. Recurrent prostate cancer genomic alterations predict response to brachytherapy treatment. Cancer Epidemiol Biomarkers Prev 23, 594–600, doi: 10.1158/1055-9965.EPI-13-1180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lahdensuo K et al. Loss of PTEN expression in ERG-negative prostate cancer predicts secondary therapies and leads to shorter disease-specific survival time after radical prostatectomy. Mod Pathol 29, 1565–1574, doi: 10.1038/modpathol.2016.154 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Yoshimoto M et al. PTEN genomic deletions that characterize aggressive prostate cancer originate close to segmental duplications. Genes Chromosomes Cancer 51, 149–160, doi: 10.1002/gcc.20939 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Yoshimoto M et al. Abstract 63: Incorporation of flanking probes reduces truncation losses for fluorescence in situ hybridization analysis of recurrent genomic deletions in tumor sections. Cancer Res 73, 63–63, doi: 10.1158/1538-7445.am2013-63 (2014). [DOI] [Google Scholar]
- 120.Sangale Z et al. A robust immunohistochemical assay for detecting PTEN expression in human tumors. Applied immunohistochemistry & molecular morphology : AIMM 19, 173–183, doi: 10.1097/PAI.0b013e3181f1da13 (2011). [DOI] [PubMed] [Google Scholar]
- 121.Chaux A et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol 25, 1543–1549, doi: 10.1038/modpathol.2012.104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Punnoose EA et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br J Cancer 113, 1225–1233, doi: 10.1038/bjc.2015.332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wyatt AW et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. JAMA oncology 2, 1598–1606, doi: 10.1001/jamaoncol.2016.0494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xia Y et al. Copy number variations in urine cell free DNA as biomarkers in advanced prostate cancer. Oncotarget 7, 35818–35831, doi: 10.18632/oncotarget.9027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wyatt AW et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 109, doi: 10.1093/jnci/djx118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.NCT03236688. Detection of ARv7 in the Plasma of Men With Advanced Metastatic Castrate Resistant Prostate Cancer (MCRP).
- 127.NCT02438007. A Study of Galeterone Compared to Enzalutamide In Men Expressing Androgen Receptor Splice Variant-7 mRNA (AR-V7) Metastatic CRPC.
- 128.NCT03050866. Cabazitaxel in mCRPC Patients With AR-V7 Positive Circulating Tumor Cells (CTCs).
- 129.NCT02853097. Cell-Free DNA and RNA in Blood fromMetastatic Prostate Cancer Patients.
- 130.NCT02269982. Prospective CiRculating prOstate Cancer Predictors in HighEr Risk mCRPC studY.
- 131.Ferraldeschi R et al. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur Urol 67, 795–802, doi: 10.1016/j.eururo.2014.10.027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Edlind MP & Hsieh AC PI3K-AKT-mTOR signaling in prostate cancer progression and androgen deprivation therapy resistance. Asian journal of andrology 16, 378–386, doi: 10.4103/1008-682X.122876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dillon LM & Miller TW Therapeutic targeting of cancers with loss of PTEN function. Curr Drug Targets 15, 65–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thomas C et al. Synergistic targeting of PI3K/AKT pathway and androgen receptor axis significantly delays castration-resistant prostate cancer progression in vivo. Mol Cancer Ther 12, 2342–2355, doi: 10.1158/1535-7163.MCT-13-0032 (2013). [DOI] [PubMed] [Google Scholar]
- 135.Marques RB et al. High Efficacy of Combination Therapy Using PI3K/AKT Inhibitors with Androgen Deprivation in Prostate Cancer Preclinical Models. Eur Urol 67, 1177–1185, doi: 10.1016/j.eururo.2014.08.053 (2015). [DOI] [PubMed] [Google Scholar]
- 136.Schwartz S et al. Feedback suppression of PI3Kalpha signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kbeta. Cancer Cell 27, 109–122, doi: 10.1016/j.ccell.2014.11.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Armstrong AJ et al. Phase II trial of the PI3 kinase inhibitor buparlisib (BKM-120) with or without enzalutamide in men with metastatic castration resistant prostate cancer. European journal of cancer 81, 228–236, doi: 10.1016/j.ejca.2017.02.030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wei XX et al. A Phase I Study of Abiraterone Acetate Combined with BEZ235, a Dual PI3K/mTOR Inhibitor, in Metastatic Castration Resistant Prostate Cancer. The oncologist 22, 503–e543, doi: 10.1634/theoncologist.2016-0432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Massard C et al. Phase Ib dose-finding study of abiraterone acetate plus buparlisib (BKM120) or dactolisib (BEZ235) in patients with castration-resistant prostate cancer. European journal of cancer 76, 36–44, doi: 10.1016/j.ejca.2017.01.024 (2017). [DOI] [PubMed] [Google Scholar]
- 140.Chow H et al. A phase 2 clinical trial of everolimus plus bicalutamide for castration-resistant prostate cancer. Cancer 122, 1897–1904, doi: 10.1002/cncr.29927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.NCT02106507. ARN 509 Plus Everolimus in Men With Progressive Metastatic Castration-Resistant Prostate Cancer After Treatment With Abiraterone Acetate.
- 142.NCT02125084. Enzalutamide Plus Everolimus in Men With Metastatic Castrate-Resistant Prostate Cancer.
- 143.Baselga J et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 366, 520–529, doi: 10.1056/NEJMoa1109653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.NCT02833883. Use of an Experimental Drug, CC-115, With Enzalutamide in Men With Castration-Resistant Prostate Cancer.
- 145.NCT01485861. Study of Ipatasertib or GDC-0980 With Abiraterone Acetate Versus Coralie in Participants With Castration-Resistant Prostate Cancer Previously Treated With Docetaxel Chemotherapy.
- 146.de Bono JS et al. PTEN loss as a predictive biomarker for the Akt inhibitor ipatasertib combined with abiraterone acetate in patients with metastatic castration-resistant prostate cancer (mCRPC). Annals of Oncology 27, doi: 10.1093/annonc/mdw372.2 (2016). [DOI] [Google Scholar]
- 147.Fruman DA et al. The PI3K Pathway in Human Disease. Cell 170, 605–635, doi: 10.1016/j.cell.2017.07.029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jia S et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature 454, 776–779, doi: 10.1038/nature07091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.NCT01884285 . AZD8186 First Time In Patient Ascending Dose Study.
- 150.NCT02215096. Dose-finding Study of GSK2636771 When Administered in Combination With Enzalutamide in Male Subjects With Metastatic Castration-Resistant Prostate Cancer.
- 151.Mateo J et al. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur Urol 71, 417–425, doi: 10.1016/j.eururo.2016.08.037 (2017). [DOI] [PubMed] [Google Scholar]
- 152.Mateo J et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 373, 1697–1708, doi: 10.1056/NEJMoa1506859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gonzalez-Billalabeitia E et al. Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer Discov 4, 896–904, doi: 10.1158/2159-8290.CD-13-0230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.van de Ven AL et al. Nanoformulation of Olaparib Amplifies PARP Inhibition and Sensitizes PTEN/TP53-Deficient Prostate Cancer to Radiation. Mol Cancer Ther 16, 1279–1289, doi: 10.1158/1535-7163.MCT-16-0740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hopkins BD et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341, 399–402, doi: 10.1126/science.1234907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang H et al. Relevance and therapeutic possibility of PTEN-long in renal cell carcinoma. PLoS One 10, e114250, doi: 10.1371/journal.pone.0114250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Drake CG Immunotherapy for prostate cancer: an emerging treatment modality. The Urologic clinics of North America 37, 121–129, Table of Contents, doi: 10.1016/j.ucl.2009.11.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.De Marzo AM et al. Inflammation in prostate carcinogenesis. Nature reviews. Cancer 7, 256–269, doi: 10.1038/nrc2090 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Strasner A & Karin M Immune Infiltration and Prostate Cancer. Frontiers in oncology 5, 128, doi: 10.3389/fonc.2015.00128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gannon PO et al. Characterization of the intra-prostatic immune cell infiltration in androgen-deprived prostate cancer patients. J Immunol Methods 348, 9–17, doi: 10.1016/j.jim.2009.06.004 (2009). [DOI] [PubMed] [Google Scholar]
- 161.Si TG, Wang JP & Guo Z Analysis of circulating regulatory T cells (CD4+CD25+CD127-) after cryosurgery in prostate cancer. Asian journal of andrology 15, 461–465, doi: 10.1038/aja.2013.22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peng W et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 6, 202–216, doi: 10.1158/2159-8290.CD-15-0283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Moussavi M et al. Oncolysis of prostate cancers induced by vesicular stomatitis virus in PTEN knockout mice. Cancer Res 70, 1367–1376, doi: 10.1158/0008-5472.CAN-09-2377 (2010). [DOI] [PubMed] [Google Scholar]
- 164.Champion BR, Fisher K & Seymour L A PTENtial cause for the selectivity of oncolytic viruses? Nat Immunol 17, 225–226, doi: 10.1038/ni.3394 (2016). [DOI] [PubMed] [Google Scholar]
- 165.Pencik J et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun 6, 7736, doi: 10.1038/ncomms8736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu H, Pardoll D & Jove R STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews. Cancer 9, 798–809, doi: 10.1038/nrc2734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Toso A et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell reports 9, 75–89, doi: 10.1016/j.celrep.2014.08.044 (2014). [DOI] [PubMed] [Google Scholar]
- 168.Cuzick J et al. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer 108, 2582–2589, doi: 10.1038/bjc.2013.248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lotan TL et al. PTEN loss detection in prostate cancer: comparison of PTEN immunohistochemistry and PTEN FISH in a large retrospective prostatectomy cohort. Oncotarget 8, 65566–65576, doi: 10.18632/oncotarget.19217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]