Abstract
Bioluminescence has long been used to image biological processes in vivo. This technology features luciferase enzymes and luciferin small molecules that produce visible light. Bioluminescent photons can be detected in tissues and live organisms, enabling sensitive and noninvasive readouts on physiological function. Traditional applications have focused on tracking cells and gene expression patterns, but new probes are pushing the frontiers of what can be visualized. The past few years have also seen the merger of bioluminescence with optogenetic platforms. Luciferase-luciferin reactions can drive light-activatable proteins, ultimately triggering signal transduction and other downstream events. This review highlights these and other recent advances in bioluminescence technology, with an emphasis on tool development. We showcase how new luciferins and engineered luciferases are expanding the scope of optical imaging. We also highlight how bioluminescent systems are being leveraged not just for sensing—but also controlling—biological processes.
Graphical Abstract
eTOC
Love and Prescher review the discovery and optimization of bioluminescent probes. Such probes have found widespread use in biological imaging, and in recent years, have been used in combination with optogenetic switches.
Introduction:
Bioluminescent probes have been used for decades to report on biological functions (Kaskova et al., 2016; Yao et al., 2018; Yeh and Ai, 2019). These tools emit photons that can illuminate events in cells, tissues, and even live organisms (James and Gambhir, 2012) Bioluminescent light derives from a chemical reaction between luciferase enzymes and luciferin small molecules (Paley and Prescher, 2014). Dozens of luciferase-luciferin pairs from the natural world have been coopted for imaging in heterologous organisms (Oba et al., 2017; Tsarkova et al., 2016; Viviani, 2002; Widder, 2010). The probes employ similar mechanisms to produce photons, but they differ in the wavelengths produced and other optical properties (Kaskova et al., 2016). Bioluminescent light can escape tissue, although it is subject to the same scattering and absorption phenomena as other optical probes (Contag and Bachmann, 2002). However, since bioluminescence does not require excitation light, it can circumvent the high background signals (via autofluorescence) commonly associated with fluorescent proteins in tissues and opaque animals (Contag and Bachmann, 2002). As a consequence, luciferase imaging is often a go-to technique for sensitive imaging in rodents and other preclinical organisms (Contag, 2007; Mezzanotte et al., 2017; Xu et al., 2016). Bioluminescence is particularly well suited for long-term studies, as the probes are largely nontoxic and not susceptible to photobleaching. Indeed, luciferase-luciferin pairs have been widely used for examining cell trafficking patterns and proliferation in vivo (Minn et al., 2005; Prescher and Contag, 2010; Santos et al., 2009). Numerous other applications are also possible, owing to the sensitivity, broad dynamic range, and user-friendly features of bioluminescence.
More biological discoveries are on the horizon, thanks to recent achievements in bioluminescent probe design. Over the past few years, novel luciferases and luciferins have been identified in the natural world and coopted for imaging new targets (Yeh and Ai, 2019). Well-established bioluminescent tools have also been tuned to emit different colors and report on cellular metabolites, among other functions (Rathbun and Prescher, 2017). Additional engineering efforts have provided collections of bioluminescent probes with unique reactivity profiles (Yao et al., 2018). These tools can illuminate multiple targets in cells, tissues, and even whole animals. The expansion of the bioluminescent toolkit mirrors the development of other optical probes. Studies of fluorescent protein structure and function, in particular, paved the way for dozens of new reporters (Rodriguez et al., 2017). Many were engineered to exhibit distinct emission profiles (Heim et al., 1994; Shaner et al., 2004), improved photon outputs (Shaner et al., 2013; Shaner et al., 2008), or light-induced conformational changes (Lukyanov et al., 2005), among other properties (Davidson and Campbell, 2009; Duwé and Dedecker, 2019).
Beyond imaging, bioluminescent light can be harnessed to manipulate biological systems. Luciferases are known to participate in a variety of energy transfer reactions in nature (Titushin et al., 2011). Such processes can potentially be coopted to control both intra- and intercellular functions. In recent years, researchers have integrated luciferases with a variety of light-responsive proteins and molecules, including retinal chromophores and photolabile groups (Berglund et al., 2013; Lindberg et al., 2018). These light-sensitive tools can be employed in a variety of mammalian cells and tissues (Fenno et al., 2011; Pudasaini et al., 2015). When illuminated, the biomolecules undergo conformational changes or cleavage reactions, ultimately triggering gene expression and other downstream functions (Bardhan and Deiters, 2019; Fenno et al., 2011). The necessary excitation light is typically delivered via UV bulbs or LED arrays. Bioluminescent photons can take the place of these external sources, providing a means to activate networks noninvasively, via simple addition of a small molecule. While such bioluminescent optogenetic platforms are only in their infancy, they hold much promise for controlling biological activities in the brain and other deep tissues – locales that are typically refractory to external light and require implantation of fiber optic cables (Doronina-Amitonova et al., 2013).
This review highlights recent advances in bioluminescent probe development that are driving new directions in biomedical research. We begin with a basic overview on bioluminescent platforms, then describe the discovery and characterization of new light-emitting systems. We further highlight synthetic efforts that have afforded novel, bioavailable luciferins with unique photophysical properties. We also discuss parallel advances in enzyme engineering that have provided a broad set of luciferases with improved kinetic and spectral emission profiles. Many of these luciferases are also selective for unique luciferin architectures and can be used for multi-parameter imaging. The last section focuses on bioluminescent probes used for applications beyond imaging, including optogenetics. These platforms leverage luciferase-luciferin reactions to selectively activate proteins or molecular bonds, enabling precise chemical control over biological processes. Collectively, bioluminescent probes are not only pushing the boundaries of what can be “seen” in living systems, but also what can be controlled.
How nature produces light
Bioluminescence has been observed in organisms across many domains of life (Lloyd, 1983; Widder, 2010). The natural glow is used for a variety of functions (Haddock et al., 2009; Wood et al., 1989a), including mating and defense (Hastings, 1983). Bioluminescence is strikingly prevalent in the deep ocean where sunlight is scarce and luciferase-luciferin pairs can provide a source of photons. In fact, thousands of creatures have evolved to emit blue light, as these wavelengths travel farther in ocean water compared to other colors (Martini and Haddock, 2017). Among the most recognizable bioluminescent sea creatures is Oplophorus gracilirostris, a shrimp that spews a bright blue cloud of luminescent liquid to distract predators (Inouye and Sasaki, 2007). Other marine organisms glow due to the presence of symbiotic bioluminescent bacteria (Nealson and Hastings, 1979). The microbes use the host for survival and, in return, provide a shimmer of light to attract prey or camouflage the host (Ruby and Lee, 1998). Still other ocean dwellers produce photons using luciferases (or luciferase-like photoproteins) in combination with fluorescent proteins. A famous example is Aequorea victoria, a jellyfish that emits green light via energy transfer between a luciferase-like photoprotein (aequorin) and green fluorescent protein (GFP, Shimomura et al., 1962, 1963). On land, insects such as beetles and fireflies have evolved to emit yellow-green light. The well-known North American firefly, Photinus pyralis, uses this light to communicate and attract mates (Lloyd, 1983). Other terrestrial bioluminescent organisms include glowing fungi (Oliveira et al., 2015) and earthworms (Petushkov et al., 2014). The light-emitting mechanisms for these species have only recently been elucidated.
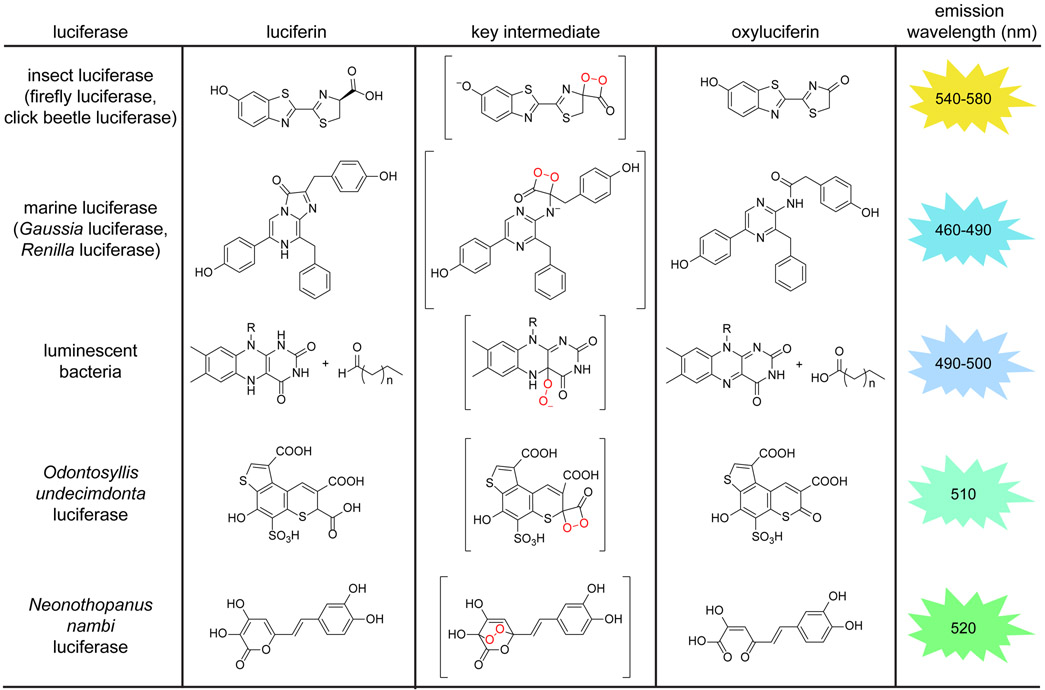
The diversity of bioluminescent organisms contrasts with the surprisingly small number of mechanisms known for light production (Kaskova et al., 2016). All bioluminescent creatures use molecular oxygen and luciferase enzymes to catalyze light emission. The enzyme structures and luciferin substrates vary (Figure 1), although the reactions proceed through similar types of intermediates. The reaction pathways generate excited-state oxyluciferins; relaxation of these molecules to the ground state results in photon emission. Luciferins are “push-pull” chromophores that behave similarly to fluorescent molecules, requiring electron-donating groups and conjugated pi systems for robust light production. The optical properties of bioluminescent reactions are also influenced by the luciferase microenvironment (McElroy et al., 1965). Modulating either the luciferase enzyme or the luciferin small molecule can alter bioluminescent colors and other optical features (Branchini et al., 2010; Hall et al., 2018; Kuchimaru et al., 2016; Rumyantsev et al., 2016).
Figure 1. Bioluminescence in the wild.
Examples of naturally occurring luciferase enzymes and luciferin small molecules. All luciferases oxidize their complementary luciferins to yield photons of light, although unique chemistries are employed. Most bioluminescent probes emit light in the blue-green region. (*Emission wavelengths reported at room temperature.)
Despite the prevalence of bioluminescent systems, few have been widely applied in vivo. The color of light produced, reporter stabilities, and probe bioavailabilities are often sub-optimal for imaging in tissues (Contag and Bachmann, 2002). Perhaps the most widely used luciferin for imaging in vivo is d-luciferin (Yeh and Ai, 2019). This substrate is oxidized by firefly luciferase (Fluc) and related insect luciferases. The mechanism likely proceeds via single electron transfer (Branchini et al., 2015), producing a high-energy dioxetanone intermediate. Elimination of CO2 affords the oxidized product and a photon of yellow-green light (in the case of Fluc) (Kaskova et al., 2016). Different colors are produced when d-luciferin is processed by other enzymes. For example, luciferases from the click beetle generate light ranging from green to red (Wood et al., 1989b), underscoring the role of the enzyme microenvironment in modulating emission color. It should also be noted that insect luciferases require ATP to activate the luciferin substrate (Inouye, 2010). This requirement has limited the use of Fluc and related luciferases in extracellular spaces and other areas devoid of large ATP stores.
Similar to insects, most marine luciferases catalyze light emission via dioxetanone intermediates (Markova et al., 2019). Some of the most popular include Renilla luciferase (Rluc) and Gaussia luciferase (Gluc). The substrate for these (and nearly all) ocean luciferases is the pyrazinone molecule, coelenterazine (CTZ) (Haddock et al., 2009). CTZ, unlike d-luciferin, does not require pre-activation with ATP. Additionally, bioluminescent reactions with CTZ provide primarily blue light (Jiang et al., 2016). The distinct emission spectra and substrate profiles of marine and insect luciferases enables them to be used in tandem for imaging multiple targets (Paley and Prescher, 2014). Multi-component imaging with d-luciferin and CTZ in vivo, though, remains challenging. One substrate is typically injected at a time, and signal from the first luciferin must clear prior to administration of the second. The blue light produced by marine luciferases is also poorly tissue penetrant, limiting detection in some cases. Further, CTZ is quite electron rich and prone to non-specific oxidation, resulting in high background emission (Jiang et al., 2016).
Like marine luciferases, bioluminescent bacteria emit blue photons. However, the light-emitting mechanisms are quite distinct. Bacterial bioluminescence is mediated by a suite of enzymes encoded by the lux operon (Brodl et al., 2018). Two of the operon genes give rise to a functional luciferase, while the others encode a fatty acid reductase complex and a flavin reductase. The luciferase oxidizes long-chain aldehydes using molecular oxygen and reduced flavin mononucleotides. During the reaction sequence, a flavin conjugate is promoted to the excited state; relaxation to the ground state provides light. Importantly, all the machinery necessary for aldehyde oxidation (and thus bioluminescence) is encoded in lux (Brodl et al., 2018). The entire operon can be expressed in heterologous organisms, resulting in a continuous glow (Meighen, 1993) with no exogenous substrate required. The lux operon has been used for decades to examine the spread and life cycle of numerous pathogens in vivo (Contag et al., 1995; Guijarro and Méndez, 2020; Kadurugamuwa et al., 2003; Na et al., 2019; Sweeney et al., 2019; Wiles et al., 2004). The operon has yet to be routinely applied in mammalian cells, though, owing to aldehyde toxicity and low photon outputs. The Sayler group recently introduced the lux genes into a panel of cancer cell lines (Xu et al., 2014). The requisite enzymes were encoded on a single plasmid, separated by self-cleaving elements. The cells exhibited autoluminescence, but light emission was weak. Photon outputs were increased when the expression levels of key enzymes were modulated. Recent engineering efforts have further improved the bacterial bioluminescence platform for imaging in both bacteria (Gregor et al., 2018) and mammalian cells (Gregor et al., 2019).
Contrasting with bacterial bioluminescence, the biosynthetic pathways for d-luciferin and CTZ remain unknown. As a consequence, these substrates must be supplied exogenously in experiments involving common luciferases (Fluc, Rluc, etc.). There remains intense interest in elucidating the origins of d-luciferin and CTZ, to eventually circumvent the need for substrate delivery (Adams Jr and Miller, 2020). Firefly feeding studies with putative precursors suggested that d-luciferin can result from 1,4-hydroquinone condensation with two cysteine residues (Oba et al., 2014). Unfortunately, the exact enzymes involved in the transformation were not determined. In related work, Weng and coworkers undertook large-scale sequencing efforts to probe d-luciferin biosynthesis. The genomes of fireflies and other insects were determined, and transcriptomic analyses revealed candidate biosynthetic genes (Fallon et al., 2018). To date, though, the definitive pathway has not been revealed. Genome mining has also been used to probe CTZ biosynthesis. This luciferin is hypothesized to arise from a tripeptide comprising phenylalanine and two tyrosine residues (i.e., the “FYY” motif) (Francis et al., 2015). A cyclization-dehydration sequence could potentially give rise to a light-emitting substrate. The FYY motif is present in the genomes of many bioluminescent ctenophores, but conspicuously absent from non-luminescent relatives (Francis et al., 2015). Feeding studies with bioluminescent copepods also suggest that CTZ can be produced de novo from phenylalanine and tyrosine (Oba et al., 2009; Tessler et al., 2018), although the precise mechanism is unknown.
Despite the inherent challenges, two new luciferin architectures and their associated biosynthetic pathways have recently been solved. The first luciferin derives from the marine polychaetes, Odontosyllis undecimdonta (Kotlobay et al., 2019). The Yampolsky and Tsarkova groups elucidated the structure using a combination of mass spectrometry, NMR, and X-ray analyses. The tricyclic luciferin is hypothesized to undergo two separate oxidative pathways—one that results in light emission from canonical luciferase activity. Yampolsky and coworkers also uncovered a second luciferin and novel light-emitting mechanism in the bioluminescent fungus, Neonothopanus nambi. The fungus uses 3-hydroxyhispidin as a light-emitting substrate, converting it to caffeylpyruvic acid via a unique endoperoxide intermediate (Kotlobay et al., 2018). Excitingly, the genes involved in 3-hydroxyhispidin biosynthesis were also elucidated. The entire luciferin pathway can thus be expressed in target cells along with the requisite luciferase, enabling light production without the need for exogenous substrate delivery. The fungal bioluminescence system was recently introduced into tobacco and other plants, producing autoluminescent species (Khakhar et al., 2020; Mitiouchkina et al., 2020). Light emission was used to monitor hormone fluxes as well as diurnal flowing patterns (Khakhar et al., 2020). These examples highlight the potential for using fungal bioluminescence in heterologous organisms, and several more applications are anticipated in the near future.
Using nature’s light to “see” biology
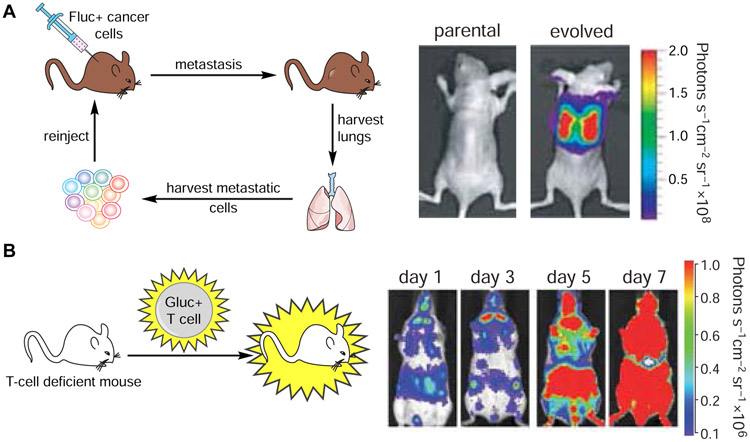
Although several bioluminescent systems exist in the wild, only a handful have been widely used for imaging in vivo. Firefly luciferase (Fluc) remains one of the most popular, as it emits a larger fraction of red, tissue-penetrant photons compared to other naturally occurring luciferases (Zhao et al., 2005). The Fluc-d-luciferin pair also possesses other desirable features, including low background, excellent bioavailability, and easy handling. Historically, Fluc has been used to image cell motility and proliferation, along with gene expression, in live animals. The luciferase is simply expressed in target cells of interest. When d-luciferin is administered, the compound can permeate the cells and light is produced. Photon intensities correlate with the amount of luciferase present, enabling biological features to be easily tracked. Seminal studies with Fluc-d-luciferin include tracking metastatic cancers (Jenkins et al., 2003; Scatena et al., 2004), infection (Luker et al., 2002; Schoggins et al., 2012), and other pathologies (Creusot et al., 2008; Swijnenburg et al., 2008) (Figure 2A).
Figure 2. Noninvasive imaging in vivo with bioluminescent probes.
Luciferase-luciferin pairs have been routinely used to track cell migration and proliferation in vivo. (A) Imaging cancer cell proliferation and metastases. Mice were injected with Fluc-expressing MDA-MB-231 cells. Metastatic cells were harvested from lungs and re-injected into recipient mice. These cells exhibited a higher degree of metastatic outgrowth than the parental tumor. Figure reprinted with permission from Minn, et al., 2005 (B) Imaging T cell trafficking. Gluc-expressing T cells were injected into immunodeficient mice. T cell proliferation and homing were visualized over time via bioluminescence. Figure reprinted with permission from Santos et al., 2009.
Beyond tracking cells and gene products, bioluminescence can be used to report on enzyme activities and small molecule metabolites in vivo. Fluc itself has been engineered to be responsive to certain analytes (Smirnova and Ugarova, 2017). Many analogs of d-luciferin have also been tailored to report on cellular targets (Li et al., 2013; Porterfield and Prescher, 2015). These latter probes typically comprise “cages”, motifs that block Fluc processing and/or light emission. The cages are removed in response to complementary enzymes or metabolites. Once liberated, the luciferin can be used by Fluc to produce photons. Light emission thus correlates with the enzyme or analyte of interest. Promega Corporation has generated a wide variety of caged molecules, including probes for oxidase (Valley et al., 2006; Zhou et al., 2006b) and phosphatase (Zhou et al., 2008) activities, among others (O’Brien et al., 2005; Zhou et al., 2006a). Such molecules have been routinely used to assay enzyme function in lysates, cells, and tissues. Some have further enabled high-throughput screens for inhibitors (Fan and Wood, 2007). Caged probes have also been applied in rodent models. In a recent example, the Miller group designed a caged luciferin to report on fatty acid amide hydrolase (FAAH) (Mofford et al., 2015), a popular therapeutic target. FAAH is expressed in brain tissue and controls the lifetime of key fatty acid metabolites (Blankman and Cravatt, 2013). To detect FAAH activity in live mice, a caged luciferin comprising an amide group (instead of the key carboxylate in d-luciferin) was synthesized. The amide probe is not emissive with Fluc. In the presence of FAAH, though, the amide is hydrolyzed, restoring a functional carboxylate and thus light emission in the presence of Fluc. Miller and coworkers used the probe to visualize FAAH activity and screen drug inhibitors in mice. In related work, the Chang group generated a caged luciferin that was responsive to hydrogen peroxide in vivo (Van de Bittner et al., 2010). This probe comprises a 6’ aryl boronic acid moiety, which blocks production of a light-emitting luciferin. The cage is removed in the presence of cellular hydrogen peroxide, restoring the necessary 6’-OH group and thus light emission.
Blue-light emitting luciferases, including lux-expressing bacteria and marine luciferase-luciferin pairs, have also been co-opted for imaging in vivo. While they emit less tissue-penetrant photons than Fluc, these bioluminescent systems typically exhibit faster turnover kinetics and are thus “brighter” reporters (Bhaumik and Gambhir, 2002; Tannous et al., 2005). Rluc and Gluc also require no additional cofactors, aside from molecular oxygen (Kaskova et al., 2016). Thus, they are well suited for use in the extracellular space where ATP stores are limited. Rluc and Gluc have been used to examine a wide range of biological functions, including T-cell tracking in subcutaneous tumors (Figure 2B) (Jones et al., 2015; Santos et al., 2009). Other recent examples include monitoring stress granule formation in viruses and the dynamics of biofilm formation in vitro (Bonenfant et al., 2019; Charoenviriyakul et al., 2018; Gutierrez Jauregui et al., 2019).
Moving beyond the all-natural
Despite the versatility and ubiquity of bioluminescence in biomedical imaging, limitations remain. As noted earlier, the blue light emission from many luciferases precludes their widespread use in some in vivo environments. Blue photons are prone to absorption by hemoglobin and other abundant chromophores in mammals (Raghuram et al., 2020; Yeh et al., 2019; Zhao et al., 2005). CTZ is also prone to autoxidation and can be toxic to cells (Jiang et al., 2016; Shipunova et al., 2018). While d-luciferin is less susceptible to off-target reactions, the turnover rate for insect luciferases is relatively slow, resulting in suboptimal photon outputs. Large amounts of d-luciferin are also required for in vivo experiments. Additionally, only a fraction of the photons produced by the Fluc-d-luciferin reaction effectively escape tissue. More sensitive imaging could be achieved with bioluminescent tools capable of producing larger numbers of red photons. There are also only a handful of luciferins that have been optimized for use in live animals, and most cannot be easily used in tandem. As a result, multi-component imaging in vivo with bioluminescent reporters remains challenging.
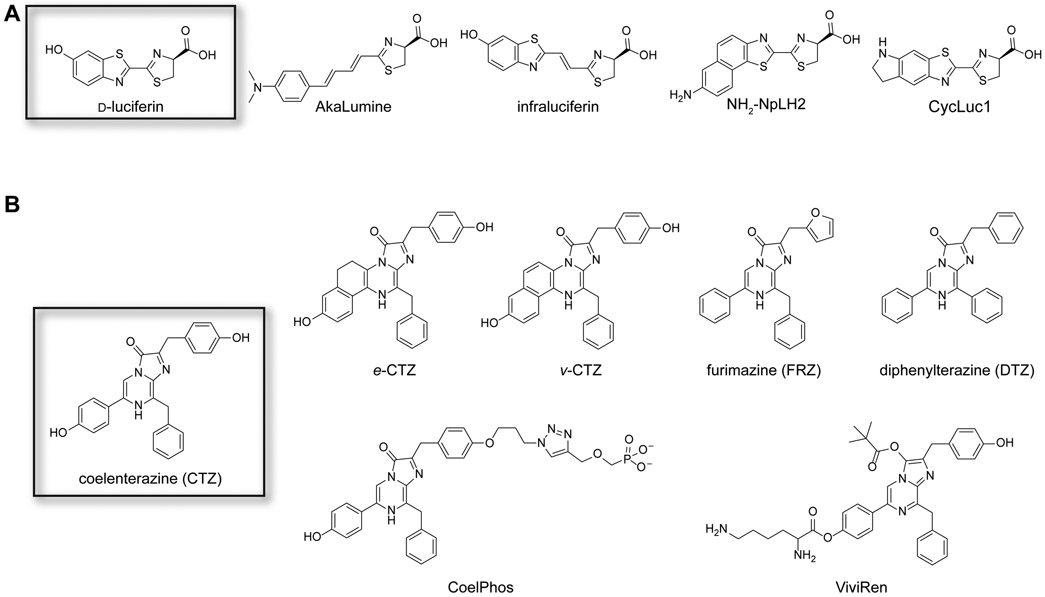
Voids in the bioluminescent toolbox have been addressed with artificial enzymes and substrates. Many recent efforts have focused on expanding the set of viable luminophores. Several groups have generated more stable and/or red-shifted insect luciferins, some of which are pictured in Figure 3A. For example, the Niwa group crafted a collection of red-emitting probes by extending the pi-conjugation of d-luciferin (Iwano et al., 2013). The most extended analog, termed AkaLumine, exhibited a ~100-nm red shift with Fluc compared to the native substrate d-luciferin. The intensity of light emission was low, suggesting that the substrate was poorly processed by Fluc. Some activity was recouped by engineering the luciferase enzyme (Iwano et al., 2018). Similar results were observed with infra-luciferin, a d-luciferin analog comprising an extra vinyl unit between the benzothiazole-thiazole ring juncture (Jathoul et al., 2015). Red-shifted emission has also been achieved with other pi-extended versions of d-luciferin, including naphthyl derivatives (e.g., NH2-NpLH2 and OH-NpLH2) (Hall et al., 2018). These analogs release 650-750 nm photons with click beetle red luciferase and are excellent candidates for deep tissue imaging.
Figure 3. Synthetic luciferins.
(A) Representative analogs of D-luciferin. Some of these molecules exhibit preferential reactivity with mutant luciferases, red-shifted emission, and/or improved bioavailability in organisms. (B) Representative analogs of coelenterazine. Some of these molecules exhibit unique reactivities with mutant luciferases, improved stability and solubility, and/or altered wavelengths of emission.
The Miller group reported a series of cyclic alkylaminoluciferin (CycLuc) analogs that exhibited both red-shifted emission and improved bioavailability (Mofford et al., 2014a; Reddy et al., 2010). CycLuc1, in particular, enabled more sensitive imaging in rodent models when compared to conventional doses of d-luciferin (Evans et al., 2014). CycLuc1 comprises a fused pyrrolidine in place of the hydroxy substituent, a structural perturbation that shifts its emission to ~600 nm (Reddy et al., 2010). The photon output from the Fluc-CycLuc1 reaction was low compared to the native enzyme-substrate pair, but activity was recovered when mutant enzymes were employed (Harwood et al., 2011). Excitingly, synthetic CycLucs were also able to reveal luciferase-like enzymes in non-luminescent organisms (Mofford et al., 2014b). Additional examples of “latent” luciferase activity are likely to be uncovered as new luciferin probes are developed.
Ongoing efforts to produce d-luciferin analogs are benefitting from streamlined and divergent synthetic routes (Kitada et al., 2018; McCutcheon et al., 2015; Sharma et al., 2017). Several of these methods have been used to produce sterically and electronically modified scaffolds (Williams and Prescher, 2019). Importantly, many analogs exhibit preferential activity with certain luciferase enzymes (Jones et al., 2017; Steinhardt et al., 2016; Steinhardt et al., 2017; Zhang et al., 2018). These unique patterns of reactivity can be used to discriminate luciferases in cells and whole organisms, providing an avenue for multi-component imaging that does not rely on color resolution (Rathbun et al., 2017).
A variety of CTZ analogs have also been produced in recent years (Figure 3B). Many efforts have focused on tuning the luciferin architecture to improve brightness, red shift the emission, and mitigate autoxidation. Some of the earliest reported CTZ analogs (e.g., e-CTZ and v-CTZ) included ring fusions at C6. When e-CTZ was paired with Rluc, the luminescence output increased seven-fold compared to CTZ (Inouye and Shimomura, 1997). When v-CTZ was paired with an engineered Rluc variant (Rluc 8.6), the emission spectrum was considerably red-shifted (emission max = 588 nm) (Loening et al., 2007). CTZ has also been modified at other positions to create designer luciferins (Jiang et al., 2016). One example includes CoelPhos, a CTZ analog comprising a phosphorylated appendage at C2 (Lindberg et al., 2013). CoelPhos has limited cell permeability and is thus well suited for visualizing biological events in the extracellular space. Another synthetic analog, ViviRen, is equipped with ester caging groups that temporarily block emission (Yeh and Ai, 2019). These cages can be removed by cellular esterases, releasing a viable luciferin and restoring bioluminescence. ViviRen enables more sustained light emission in vivo and is less susceptible to autoxidation than CTZ. These features can be advantageous for imaging in brain and other tissues (Otto-Duessel et al., 2006). Similar to d-luciferin, there have also been efforts to develop more modular syntheses of CTZ analogs (Coutant et al., 2019; Vece and Vuocolo, 2015). Such chemistries are expanding the number of custom luciferins for imaging applications.
Additional improvements to bioluminescent probes have come from modulating the other half of the light-emitting pair, the luciferase. Luciferases with faster kinetics and altered binding pockets have been developed in recent years (Yao et al., 2018; Yeh and Ai, 2019). For example, as mentioned earlier, the Miyawaki group engineered Fluc to process the pi-extended analog AkaLumine. The resulting enzyme, Akaluc, contains 28 mutations and enables more sensitive imaging than Fluc/d-luciferin in thick tissue and preclinical models (Iwano et al., 2018). Akaluc was transduced in the right striatum of a marmoset brain and visualized with AkaLumine. Real-time imaging was possible for over a year – a stunning achievement in bioluminescence technology. Related engineering efforts from Promega Corporation have provided mutant click beetle luciferases that can efficiently process naphthyl-modified luciferins (Hall et al., 2018). The best mutant showed improved activity not only with the naphthyl analogs, but also with d-luciferin. These bright and red-emitting bioluminescent probes will enable ever-more sensitive imaging applications. Mutant luciferases have also been identified that can selectively process infra-luciferin and d-luciferin, enabling two-component imaging in cells and in vivo (Stowe et al., 2019).
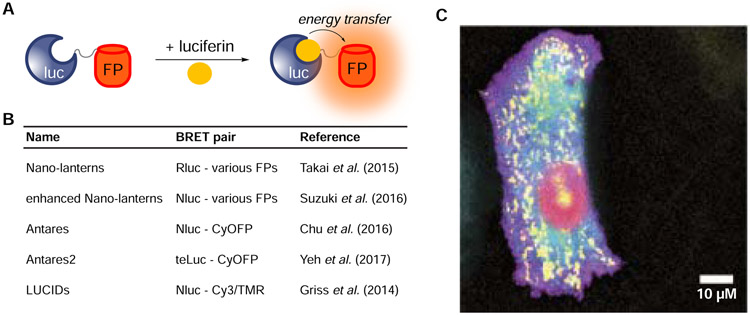
Several engineering efforts have been directed at marine luciferases to provide more desirable colors of light for imaging in thick tissues. Most efforts have capitalized on bioluminescence resonance energy transfer (BRET, Figure 4A), wherein luciferases are tethered to fluorescent proteins that harbor red-shifted emission spectra (Figure 4B, Yeh and Ai, 2019). In some notable examples, Nagai and coworkers fused Rluc (or an engineered variant) to a panel of fluorescent protein acceptors (Saito et al., 2012; Takai et al., 2015). Several BRET reporters were produced that exhibited a range of colors. One of the brightest conjugates, Rluc tethered to Venus fluorescent protein (yellow Nano-lantern, YNL), produced red-shifted light and ten-fold more photons than Rluc alone (Takai et al., 2015). YNL was further engineered to report on cellular Ca2+ levels. The BRET sensor was produced via insertion of the Ca2+-binding domains calmodulin and M13 within the middle of the Rluc sequence. In the presence of calcium, the Rluc fragments can recombine to restore bioluminescence. The yellow emission of YNL enabled the Ca2+ sensor to be applied in whole animals (Saito et al., 2012). Additional BRET probes have been produced by pairing marine luciferases with near-infrared emitting fluorescent proteins (Rumyantsev et al., 2016) and quantum dots (Hasegawa et al., 2013) and can be coopted for numerous sensing applications.
Figure 4. Multicolor imaging with engineered BRET reporters.
(A) A variety of luciferase-fluorescent protein (FP) fusions have been generated. Resonance energy transfer can provide a range of bioluminescent colors. (B) Example BRET reporters. These constructs comprise luciferase donors with either fluorescent protein or small molecule acceptors. (C) A panel of enhanced Nano- lanterns was expressed in HeLa cells, enabling real-time BRET imaging of cellular organelles. Reprinted with permission from Suzuki et al., 2016.
Many recent bioluminescent applications have featured NanoLuc (Nluc). Nluc is an engineered variant of the luciferase from the deep-sea shrimp Oplophorus gracilirostris (Hall et al., 2012). This enzyme was engineered in combination with the native luciferin, CTZ. The goal was to identify a luciferase that yielded bright luminescence with a substrate that produced less background signal. Toward this end, a variety of CTZ analogs were produced that were more stable and less prone to autoxidation. These molecules were used in screens to identify suitable luciferases. The “winning” pair, NanoLuc and furimazine (FRZ), exhibited robust photon emission with reduced background. This pair also produced “glow”-type kinetics, enabling more sustained imaging in biological environments. Bioluminescent reactions that provide steady light outputs for >30 minutes (Andreu et al., 2010; Fraga, 2008) are popular choices for use in rodent models, where capturing peak light outputs can be challening. Many marine luciferases, by contrast, exhibit “flash” type kinetics. Such reporters produce strong, yet short-lived, (seconds to minutes) signals (Bhaumik and Gambhir, 2002; Tannous et al., 2005). The half-life of luminescent signal from Nluc-FRZ under standard imaging protocols is >2 h, a marked improvement over traditional marine luciferase reporters. The photon output is also ~150-fold higher than Rluc or Fluc under similar conditions, making Nluc well suited for long-term imaging applications. Further, Nluc is also remarkably stable at high temperatures and over a range of pH values, making it a “go-to” reporter in the field. Indeed, Nluc and related variants have been used in numerous applications over the past five years, including pathogen tracking (Azevedo et al., 2014; Sasaki et al., 2018), drug screening (Omachi et al., 2018), and biosensing (Aird et al., 2020; Chang et al., 2020). Nluc is also easily resolved from Fluc and other insect luciferases in vitro, based on its unique emission spectrum and substrate profile. Combinations of these reporters have recently been used to monitor multiple signaling pathways at once (Sarrion-Perdigones et al., 2019).
While Nluc has enabled numerous pursuits in vitro and in cellulo, this luciferase has been somewhat limited in vivo (Yeh et al., 2017). The emission maximum of the Nluc-FRZ reaction (460 nm) is slightly blue-shifted from Rluc and Gluc (Hall et al., 2012). To push the wavelength to more a tissue-penetrant regime, researchers have paired Nluc with red-emitting fluorescent proteins. In one example, the Nagai group fused Nluc to tdTomato to create a red Nano-lantern (termed ReNL) (Suzuki et al., 2016). ReNL has an emission maximum of 590 nm, a 130 nm shift from Nluc itself. ReNL is part of a broader series of “enhanced” Nano-lanterns (eNLs) that comprise Nluc and well-known fluorescent proteins (Figure 4C). The fluorescent proteins were selected to provide high quantum yields and a range of colors. Several of the resulting BRET reporters, including the cyan enhanced Nano-lantern (CeNL) and the green enhanced Nano-lantern (GeNL), were found to emit more photons than the parental luciferase. The palette of eNLs has been co-opted for a variety of applications, including multi-spectral imaging of cellular organelles and calcium sensing (Farhana et al., 2019; Suzuki et al., 2016).
Related work from the Lin group provided Antares, an orange/red-emitting BRET reporter for sensitive imaging in vivo (Chu et al., 2016). Antares comprises Nluc flanked by two CyOFP1 fluorescent proteins. CyOFP1 is well suited to be a BRET acceptor with Nluc, as its absorption spectrum overlaps with the Nluc emission profile. CyOFP1 also exhibits a high quantum yield and a large Stokes shift, emitting a large portion of light above 600 nm. Such wavelengths are desirable for imaging in rodents and opaque tissue. Indeed, when a vector encoding Antares was hydrodynamically injected into mice, robust signal was observed in liver tissue. Notably, the signal was 2.6-fold higher than for traditional Fluc/d-luciferin imaging under the same conditions.
Further improvements to Antares were reported by the Ai group, in an effort to identify more robust and red-shifted imaging agents. This group generated libraries of Nluc mutants and screened them with a synthetic CTZ analog (diphenylterazine, DTZ). One of the “winners” (termed teLuc) catalyzed light emission with DTZ, exhibiting a maximum emission of 502 nm, a 60 nm shift from the Nluc-FRZ reaction. DTZ and teLuc enabled more sensitive imaging in thick tissue and mouse models compared to Nluc/FRZ and other CTZ-utilizing luciferases (Yeh et al., 2017). The Ai group also fused CyOFP1 proteins to teLuc to create a new BRET construct termed Antares2. Antares2 emits 3.8-fold more photons above 600 nm when compared to Antares itself, enabling even more sensitive imaging in rodent models.
In addition to classic cell tracking studies, Antares and Antares2 can report on other biological processes in vivo. In a seminal example, the Lin group engineered Antares to report on Ca2+ levels. Calmodulin and M13 were inserted into the Nluc region of Antares, which reduced photon output from the luciferase (Oh et al., 2019). Ca2+ binding to calmodulin triggered a conformational change in the sensor, restoring photon output. The bioluminescent probe was used to study calcium dynamics in different lobes of a mouse liver. The remarkable sensitivity and dynamic range of the sensor bode well for future studies of calcium dynamics in vivo, including in difficult-to-image areas like the brain. A related sensor based on Nluc/Antares has also been applied to study GPCR signaling (Oishi et al., 2020), and additional applications for metabolite and biomolecule detection are anticipated.
Similar to fluorescent protein conjugates, Nluc-fluorescent small molecule probes have proven useful sensors. In recent work, the Johnsson group developed a suite of BRET probes combining SNAP-tags and HaloTags with a circularly permuted version of Nluc (Hiblot et al., 2017). The SNAP-/HaloTag labels were used to append a broad range of fluorophores to the Nluc construct. The fusions were engineered such that the small molecules were positioned close to Nluc, optimizing energy transfer between the luciferase and acceptor. The resulting palette of probes was applied to image cellular organelles. Nluc-HaloTag conjugates have also been evaluated with JaneliaFluor dyes (Thirukkumaran et al., 2020). The best BRET combination enabled single-cell microscopic imaging with exposure times of <1 s.
Another set of BRET sensors was developed by the Johnsson group for measuring drug levels in small volumes (Griss et al., 2014). The probes, termed LUCiferase-based Indicators of Drugs (LUCIDs), comprise engineered Nluc-SNAPtag fusions and a receptor for the drug of interest. The SNAPtag facilitates installation of the requisite fluorophore, along with a competing ligand for the receptor. In the absence of drug, the competing ligand binds the receptor, enabling BRET between Nluc and the fluorophore. In the presence of drug, the ligand is released, and BRET is reduced. Such ratiometric readouts enabled sensitive detection of methotrexate and cyclosporin, among other drugs. The modular design of LUCIDs enables them to be applied to many other target analytes. Most recently, this concept was adapted for sensing NADH (Yu et al., 2019) and NADPH (Yu et al., 2018) in patient blood samples. The luminescent readout was on par with the most sensitive conventional methods for metabolite detection, including LC-MS. Importantly, the assays required just a drop of blood and a cell phone camera for detection, showcasing the simplicity and broad applicability of Nluc-based sensors for clinical diagnoses.
Using nature’s light to drive biological functions
Recent advances in bioluminescence technology have harnessed luciferases not just for visualization, but also to activate biological processes. Inspiration has come from the field of optogenetics, where photons from LEDs and other exogenous sources are used to trigger photoresponsive proteins (and often downstream gene expression) (Fenno et al., 2011). Light delivery is quite facile for experiments involving cultured cells or resected tissue. Light delivery in vivo, though, can be cumbersome, often requiring surgical implantation of optical fibers (Doronina-Amitonova et al., 2013; Häusser, 2014). The powerful light sources used can also heat surrounding tissue and perturb key measurements (Owen et al., 2019). Luciferases have attracted attention as “cold” light sources to drive optogenetic switches. Activation can be achieved by simple injection of the luciferin, a quick and routine procedure. Bioluminescent probes thus offer the advantage of being “chemogenetic”, in addition to optogenetic (Atasoy and Sternson, 2017). Light can be effectively “dosed” with added amounts of luciferin, enabling a tuned response. Bioluminescence is also functional in deep tissue, where activation of photoresponsive proteins has been historically difficult.
The initial targets for bioluminescence-driven optogenetics have been channelrhodopsins and LOV domains, proteins comprising well-known light-harvesting chromophores (Fenno et al., 2011; van der Horst and Hellingwerf, 2004) (Figure 5). The most common channelrhodopsin used in optogenetics derives from algae and features a light-sensitive retinal chromophore (Boyden et al., 2005). Retinal absorption of blue light triggers a conformational change, ultimately enabling cation flux into the cell (Sineshchekov et al., 2009). The light-responsive feature of channelrhodopsin has been coopted to control neurons in a variety of settings (Boyden et al., 2005). The gene encoding channelrhodopsin can be expressed in mammalian cells, and upon irradiation with blue light, neurons are triggered to “fire” action potentials.
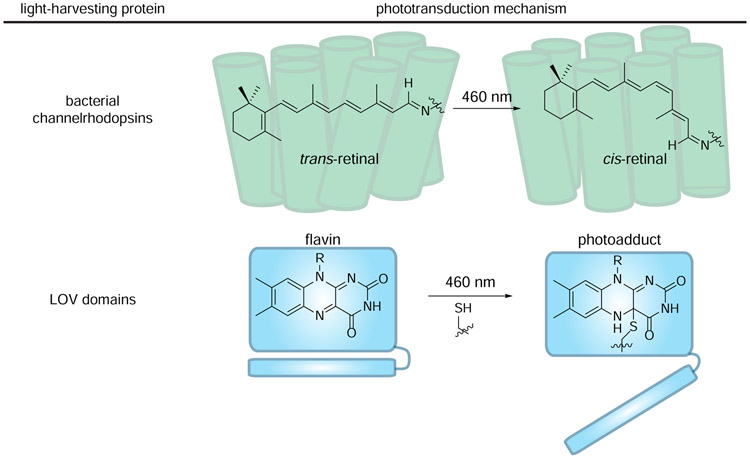
Figure 5. Light-harvesting proteins use unique chromophores to convert light energy to chemical energy.
(top) Channelrhodopsins comprise covalently bound retinal chromophores. Retinal isomerizes when illuminated with blue light, driving the channel pore to open. (bottom) LOV domains comprise noncovalently bound flavins chromophores. When excited with 460 nm light, flavin can be trapped by a nearby cysteine. The resulting conformational change initiates signal transduction.
Flavin-binding proteins have been similarly targeted for bioluminescent optogenetics. Flavins play diverse roles in nature, ranging from catalyzing oxidation-reduction reactions to absorbing light in photocascades (Hemmerich and Nagelschneider, 1970). The most well-known flavin-binding proteins for optogenetics are the LOV domains, the Blue-Light-Using-FAD proteins (BLUFs), and cryptochromes (CRYs) (Losi and Gärtner, 2012). Many organisms use these domains to sense environmental conditions and regulate phototropism (Christie, 2007). Our discussion will focus on LOV domains, as they have been the initial targets of luciferase pairing. LOV domains are found in diverse organisms (Glantz et al., 2016) and play roles in circadian rhythms (Imaizumi et al., 2003) and stress responses (Möglich and Moffat, 2007), among other functions (Huala et al., 1997). These proteins are equipped with a key cysteine residue near the bound flavin (Figure 5). Upon excitation, the cysteine bites into the C4 position of the flavin ring (Christie, 2007; Conrad et al., 2014). Formation of this covalent adduct causes a conformational change in the protein, initiating the phototransduction cascade. Similar to channelrhodopsins, LOV domains have been broadly employed to turn-on gene expression with external light. Both classes of optogenetic probes – and how they have been merged with bioluminescence – are described in more detail below.
Channelrhodopsins.
The first widely used protein for optogenetics, channelrhodopsin (ChR), belongs to a family of ion channels commonly found in light-sensitive algae (Zhang et al., 2011). Like all opsins, ChR binds the chromophore retinal, a vitamin A derivative (Sineshchekov et al., 2009). This polyene is covalently bound to the channel via a Schiff-base linkage. Upon photon absorption, the all-trans form of retinal isomerizes to the 13-cis configuration. This isomerization changes the conformation of ChR, triggering opening of the pore for cation entrance and downstream activation (Figure 5) (Deisseroth and Hegemann, 2017). Known ChRs absorb blue or green light and are thus good targets for photoactivation by the marine-derived luciferases Gluc, Rluc, and Nluc (Deisseroth and Hegemann, 2017).
The Hochgeschwender group took advantage of the spectral overlap between Gluc and ChR to create a “luminopsin” probe (Figure 6A). Gluc is a bright, blue-emitting luciferase that does not require ATP or other cofactors to produce light, rendering it functional in the extracellular space. The researchers hypothesized that luminopsins could produce action potentials when expressed in neurons (Berglund et al., 2013). The first generation comprised Gluc fused to the N-terminus of channelrhodopsin-2 from Chlamydomonas (LMO1, Figure 6A). YFP was fused to the C-terminus to verify channel localization. Upon administration of CTZ, LMO1 action potential firing was observed (Figure 6B). After this initial report, other luminopsins were developed to potentiate nerve cell firing or inhibition (Berglund et al., 2016; Berglund et al., 2020; Park et al., 2020). Inhibitory opsins block cation passage upon light stimulation, thus blocking action potential firing. Therefore, depending on the desired application, CTZ administration can either activate or prevent neuronal firing.
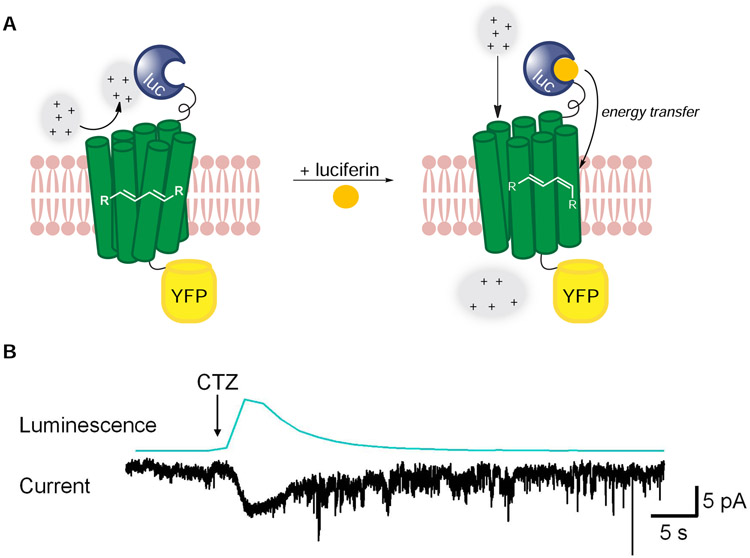
Figure 6. Luciferase-rhodopsin fusions enable spatiotemporal control over ion channel function.
(A) The first-generation luminopsin (LMO1) featured Gluc (blue) tethered to channelrhodopsin2 (ChR2, green). CTZ application potentiates energy transfer, driving retinal isomerization and channel opening. (B) LMO1 was expressed in HEK cells. CTZ application generated photocurrents. Reprinted from Berglund, et al. 2013.
Luminopsins circumvent the need for external light delivery and are applicable in deep tissue. Thus, they can provide a way to non-invasively modulate brain activity. Luminopsins were evaluated in this context using an established mouse behavior assay (Kilpatrick et al., 1982). This assay involves GABAergic neurons in the substantia nigra. When these neurons are activated in one hemisphere of the brain, mice circle ipsilaterally. By contrast, when these neurons are silenced, mice circle in the opposite direction. Mouse GABAergic neurons were targeted with vectors encoding an activating luminopsin (LMO3) or an inhibitory luminopsin (iLMO) (Berglund et al., 2016). Upon administration of CTZ, mice expressing LMO3 circled primarily ipsilaterally. By contrast, mice expressing iLMO circled contralaterally. This experiment demonstrated the utility of luminopsins for controlling select populations of neurons in living animals, via application of a small molecule. Following this work, the Hochgeschwender group explored new combinations of engineered Gluc enzymes and various rhodopsins (Berglund et al., 2020; Park et al., 2020). Some of the resulting luminopsins comprised GlucM23 fused to more light-sensitive activating and inhibiting channelrhodopsins. Others included Gluc fused to ChR2 harboring step-function mutations. Such probes are expanding the optogenetic toolbox for controlling neuron functions (Petersen et al., 2019; Zenchak et al., 2020).
LOV domains.
Flavins represent another major class of light-harvesting species for bioluminescent optogenetics. These molecules come in a variety of flavors, but all possess a tricyclic isoalloxazine core (Hemmerich and Nagelschneider, 1970). Biologically, flavin exists in three main forms: riboflavin (the free vitamin), flavin adenine dinucleotide (FAD), and flavin mononucleotide (FMN). FAD and FMN comprise the majority of flavin-binding proteins, and these molecules are bound via pi-stacking and/or hydrogen bonding interactions (Losi and Gärtner, 2008). Flavin-dependent light-harvesting proteins absorb maximally at 460 nm (Losi and Gärtner, 2008) and have been used as optogenetic platforms (Christie et al., 2012). All absorb light and drive conformational changes in protein structure. We focus exclusively on LOV domains in this section, as they have been targeted with luciferase light sources. As noted earlier, such absorption leads to covalent trapping by neighboring cysteine residues, altering the overall structure of the domain and triggering downstream events. Excited-state flavins can also produce singlet oxygen, and this feature has been similarly exploited in conjunction with bioluminescence.
Some of the earliest examples of harnessing bioluminescence for non-imaging purposes involved LOV domains. For example, Nluc was fused to miniSOG (mini-singlet oxygen generator (Shramova et al., 2016), a LOV domain that was engineered to favor singlet oxygen production over photoadduct formation upon blue light illumination (Shu et al., 2011). miniSOG lacks the key cysteine residue for conventional LOV domain function. Nluc and miniSOG have ideal spectral overlap and, thus, a fusion construct enables cytotoxic singlet oxygen production “on demand”. When Nluc-miniSOG was expressed in cells, cytotoxicity was observed upon administration of FRZ (Proshkina et al., 2018). Whether or not cells were killed due to singlet oxygen alone, though, remains unresolved (Pimenta et al., 2013; Ruiz-González et al., 2013; Shipunova et al., 2018).
The Ting group capitalized on the conformational switch observed when conventional LOV domains absorb light. Photoadduct formation drives opening of the J helix (Konold et al., 2016). This motif can be engineered to contain proteolytic cut sites or other recognition elements. Thus, upon blue light illumination, the J helix is exposed and available to mediate other biological processes (Kim et al., 2017; Sanchez and Ting, 2020; Wang et al., 2017). The Ting group used an engineered LOV domain in conjunction with Nluc to drive downstream gene expression (Kim et al., 2019). The strategy built off of their previously reported SPARK (Specific Protein Association tool giving transcriptional Readout with fast Kinetics) system to probe protein-protein interactions (Kim et al., 2017). SPARK comprised a transcription factor (Gal4) bound to the membrane via the β-adrenergic receptor (β2AR). This GPCR was tethered to a LOV domain with a TEV protease-cleavage site buried in the J helix (Kim et al., 2017). Upon blue-light activation, the J helix swung open to reveal the cleavage site. SPARK thus required two inputs: (1) external light to activate a LOV domain and (2) protein association (arrestin-β2AR) to drive signal to trigger transcription. SPARK decreased the background of conventional transcription-based detection methods of protein-protein interactions. However, high expression of the TEV-arrestin resulted in unintended cleavage of the protease cut site and artificial signal production.
To tune proteolysis more precisely, SPARK was modified to respond to luciferase-derived light. Nluc was expressed between arrestin and the TEV protease, providing SPARK2. When arrestin binds β2AR, Nluc comes into close proximity to the LOV domain. In the presence of FRZ, bioluminescent light can drive J helix release. Subsequent cleavage by TEV protease liberates the Gal4 transcription factor. Energy transfer between Nluc and LOV domain is highly distance dependent, resulting in reduced background signal and more sensitive readouts on protein-protein interactions. The Ting group was further able to show bioluminescent light activation in a transcellular sense. In this scenario, Nluc was fused to neurexin and ICAM in the extracellular space. These latter proteins drive neuron-neuron interactions. Upon cell contact and FRZ administration, Nluc on one cell activated a photoresponsive version of β2AR (Figure 7A) on a second cell, driving SPARK2. Transcription was modest, likely due to low levels of bioluminescent light. Additional enhancements can likely be achieved using engineered luciferases with higher photon outputs.
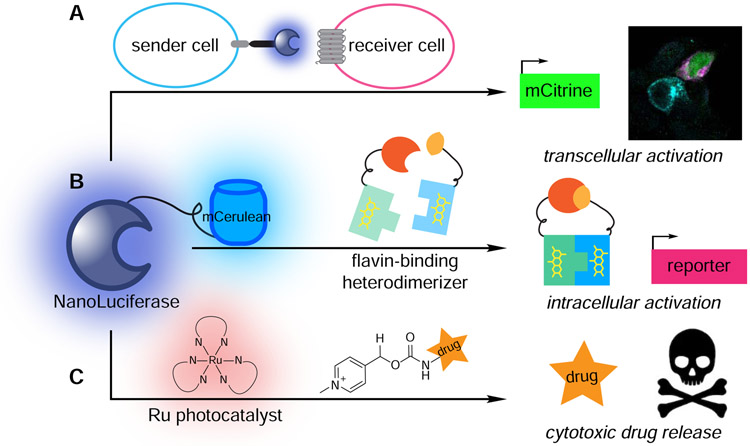
Figure 7. Recent examples of harnessing bioluminescent photons to drive biological processes.
(A) Transcellular activation of gene expression. Nluc expressed in “sender” cells (blue) activated light-sensitive proteins in “receiver” cells (pink), resulting in mCitrine expression (green). (B) Intracellular activation of gene expression. Energy transfer from a Nluc-mCerulean conjugate to a flavin-binding protein facilitated heterodimerization and downstream transcription of reporter genes. (C) Cytotoxic drug release. Energy transfer from Nluc to a ruthenium photocatalyst facilitated bond cleavage reactions and cargo release.
One approach to achieve higher power output involves BRET constructs (Parag-Sharma et al., 2020). As noted in the earlier discussion of Nano-lanterns, higher quantum yields from luciferase-fluorescent protein fusions have been observed (Suzuki et al., 2016). The Amelio group capitalized on this feature by developing a Nluc-mCerulean BRET construct known as LumiFluor. LumiFluor produces three-fold higher power outputs than Nluc alone. At physiologically relevant levels, LumiFluor can provide ~mW/m2 power levels, on par with what is necessary to activate many blue-light sensitive proteins. Indeed, LumiFluor was used in conjunction with flavin-binding heterodimerizers to drive downstream gene expression (Figure 7B). One of the most exciting results was activation of a light-inducible dCas9 system, suggesting genetic manipulation in vivo can be controlled via simple administration of luciferin. Further engineering of the LumiFluor platform will likely expand the number of potential applications.
Bioluminescent light has been used to drive processes beyond conventional optogenetics, including photouncaging reactions. Photolysis has long been used to release drugs and other payloads under spatiotemporal control in cells and tissues (Ellis-Davies, 2007). For example, nitrobenzyl photocages have been extensively employed for targeted activation of gene transcription (Lu et al., 2012), local release of anticancer agents (Dcona et al., 2012), and targeted deposition of imaging probes (Yang et al., 2017). Traditional applications require exogenous UV light, which can damage cells and surrounding tissue. Recent work has focused on developing photocages that are activated with less harmful wavelengths of light, including BODIPY (Peterson et al., 2018) and cyanine scaffolds (Gorka et al., 2018). Bioluminescent activation offers an alternative mechanism for caged molecule release. For example, Nluc can be used to activate ruthenium (Ru) photocatalysts (Lindberg et al., 2018). Such catalysts can cleave pyridinium linkers on caged molecules. Importantly, the photocatalysts are also minimally toxic in living systems, and have good spectral overlap with Nluc. The Winssinger group showed that Nluc undergoes energy transfer with the Ru molecule when in close proximity (Figure 7C). To properly localize the reagents, they used a SNAP-tag conjugate. Incubation with FRZ initiated bioluminescent light production and release of pyridinium-caged materials, including anti-cancer agents. A similar “bioluminolysis” strategy was employed with coumarin-based photocages (Chang et al., 2019), although no catalyst was required. Simple application of a small molecule luciferin could drive drug release. While much remains to be learned about the limits of luciferase-driven reactions, these examples suggest that bioluminescence can be harnessed to manipulate biological systems.
Conclusions and future directions
Bioluminescent tools have been widely applied in recent years – both for imaging and as light sources to activate photocascades. All applications rely on luciferase-luciferin pairs with appropriate emission spectra and robust photon outputs. Synthetic efforts have provided luciferins with altered colors, improved bioavailability, and distinct architectures. Concurrently, dozens of luciferases have been engineered to further modulate photon outputs and selectively process designer substrates. Additional work to uncover novel luciferase-luciferin pairs in nature has further expanded the imaging toolbox and deepened our understanding of how bioluminescence works.
Future imaging experiments will benefit from continued substrate optimization and enzyme engineering. More red-shifted probes will enhance imaging in thicker tissue. Higher photon outputs will also enable fewer cells to be more reliably visualized. With improved tools, it may soon be possible to perform single cell imaging in deep tissues in vivo. Additional substrate-specific luciferases will bolster efforts to track multiple cell types and other biological features. This work will continue to benefit from robust synthetic methods to produce collections of luciferins. Molecules deviating from canonical luciferin architectures are also viable targets. Such chemiluminescent substrates can produce light with alternative oxidases or other cellular enzymes, comprising a distinct class of light-emitting probes (Gross et al., 2009; Hananya and Shabat, 2017; Hananya and Shabat, 2019).
Future optogenetics experiments will benefit from a combination of analytical work and engineering. Much remains to be learned about the mechanisms driving bioluminescent photocascades and the limits of such activation. Signal from bioluminescent-driven systems thus far has been minimal, likely due to the low photon outputs. Such challenges can likely be addressed with additional enzyme engineering and substrate tuning. Synthetic chemistry efforts are likely to yield new luciferins with improved photophysical properties and bioavailability. Most applications to date have relied on CTZ/FRZ, molecules with some liabilities owing to cytotoxicity and optical interference with common reporters. With new probes in hand, we anticipate other photoresponsive proteins and photocages will be pursued in conjunction with luciferases. Such tools will continue to push the boundaries of what is possible with bioluminescent technologies.
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R01 GM107630 to J.A.P.). A.C.L was supported by the BEST IGERT program (National Science Foundation DGE-1144901). We would like to thank members of the Prescher lab for helpful discussions and edits.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams ST Jr, and Miller SC (2020). Enzymatic promiscuity and the evolution of bioluminescence. FEBS J. 287, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird EJ, Tompkins KJ, Ramirez MP, and Gordon WR (2020). Enhanced molecular tension sensor based on bioluminescence resonance energy transfer (BRET). ACS Sens. 5, 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S (2010). Optimisation of bioluminescent reporters for use with mycobacteria. PLoS one 5, e10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, and Sternson SM (2017). Chemogenetic tools for causal cellular and neuronal biology. Physiol. Rev 98, 391–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo MF, Nie CQ, Elsworth B, Charnaud SC, Sanders PR, Crabb BS, and Gilson PR (2014). Plasmodium falciparum transfected with ultra bright NanoLuc luciferase offers high sensitivity detection for the screening of growth and cellular trafficking inhibitors. PLoS One 9, e112571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardhan A, and Deiters A (2019). Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. in Struc. Biol 57, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund K, Birkner E, Augustine GJ, and Hochgeschwender U (2013). Light-emitting channelrhodopsins for combined optogenetic and chemical-genetic control of neurons. PLoS One 8, e59759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund K, Clissold K, Li HE, Wen L, Park SY, Gleixner J, Klein ME, Lu D, Barter JW, Rossi MA, et al. (2016). Luminopsins integrate opto- and chemogenetics by using physical and biological light sources for opsin activation. Proc. Natl. Acad. Sci. USA 113, e358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund K, Fernandez AM, Gutekunst C-AN, Hochgeschwender U, and Gross RE (2020). Step-function luminopsins for bimodal prolonged neuromodulation. J. Neurosci. Res 98, 422–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S, and Gambhir SS (2002). Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc. Natl. Acad. Sci. USA 99, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankman JL, and Cravatt BF (2013). Chemical probes of endocannabinoid metabolism. Pharmacol. Rev 65, 849–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonenfant G, Williams N, Netzband R, Schwarz MC, Evans MJ, and Pager CT (2019). Zika virus subverts stress granules to promote and restrict viral gene expression. J. Virol 93, e00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Branchini BR, Ablamsky DM, Davis AL, Southworth TL, Butler B, Fan F, Jathoul AP, and Pule MA (2010). Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal. Biochem 396, 290–297. [DOI] [PubMed] [Google Scholar]
- Branchini BR, Behney CE, Southworth TL, Fontaine DM, Gulick AM, Vinyard DJ, and Brudvig GW (2015). Experimental support for a single electron-transfer oxidation mechanism in firefly bioluminescence. J. Am. Chem. Soc 137, 7592–7595. [DOI] [PubMed] [Google Scholar]
- Brodl E, Winkler A, and Macheroux P (2018). Molecular mechanisms of bacterial bioluminescence. Comput. Struct. Biotech 16, 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Kim KT, Lindberg E, and Winssinger N (2020). Smartphone DNA or RNA sensing using semisynthetic luciferase-based logic device. aCs Sens. 5, 807–813. [DOI] [PubMed] [Google Scholar]
- Chang D, Lindberg E, Feng S, Angerani S, Riezman H, and Winssinger N (2019). Luciferase-induced photouncaging: Bioluminolysis. Angew. Chem. Int. Ed. Engl 58, 16033–16037. [DOI] [PubMed] [Google Scholar]
- Chang D, Lindberg E, and Winssinger N (2017). Critical analysis of rate constants and turnover frequency in nucleic acid-templated reactions: reaching terminal velocity. J. Am. Chem. Soc 139, 1444–1447. [DOI] [PubMed] [Google Scholar]
- Charoenviriyakul C, Takahashi Y, Morishita M, Nishikawa M, and Takakura Y (2018). Role of extracellular vesicle surface proteins in the pharmacokinetics of extracellular vesicles. Mol. Pharm 15, 1073–1080. [DOI] [PubMed] [Google Scholar]
- Chen S-F, Navizet I, Roca-Sanjuán D, Lindh R, Liu Y-J, and Ferré N (2012). Chemiluminescence of coelenterazine and fluorescence of coelenteramide: A systematic theoretical study. J. Chem. Theory Comput 8, 2796–2807. [DOI] [PubMed] [Google Scholar]
- Christie JM (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol 58, 21–45. [DOI] [PubMed] [Google Scholar]
- Christie JM, Gawthorne J, Young G, Fraser NJ, and Roe AJ (2012). LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol. Plant 5, 533–544. [DOI] [PubMed] [Google Scholar]
- Chu J, Oh Y, Sens A, Ataie N, Dana H, Macklin JJ, Laviv T, Welf ES, Dean KM, Zhang F, et al. (2016). A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol 34, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KS, Manahan CC, and Crane BR (2014). Photochemistry of flavoprotein light sensors. Nat. Chem. Biol 10, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag CH (2007). In Vivo Pathology: Seeing with molecular specificity and cellular resolution in the living body. Annu. Rev.Pathol 2, 277–305. [DOI] [PubMed] [Google Scholar]
- Contag CH, and Bachmann MH (2002). Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng 4, 235–260. [DOI] [PubMed] [Google Scholar]
- Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, and Benaron DA (1995). Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol 18, 593–603. [DOI] [PubMed] [Google Scholar]
- Coutant EP, Goyard S, Hervin V, Gagnot G, Baatallah R, Jacob Y, Rose T, and Janin YL (2019). Gram-scale synthesis of luciferins derived from coelenterazine and original insights into their bioluminescence properties. Org. Biomol. Chem 17, 3709–3713. [DOI] [PubMed] [Google Scholar]
- Creusot RJ, Yaghoubi SS, Kodama K, Dang DN, Dang VH, Breckpot K, Thielemans K, Gambhir SS, and Fathman CG (2008). Tissue-targeted therapy of autoimmune diabetes using dendritic cells transduced to express IL-4 in NOD mice. J. Clin. Immunol 127, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MW, and Campbell RE (2009). Engineered fluorescent proteins: innovations and applications. Nat. Methods 6, 713–717. [DOI] [PubMed] [Google Scholar]
- Dcona MM, Mitra D, Goehe RW, Gewirtz DA, Lebman DA, and Hartman MCT (2012). Photocaged permeability: a new strategy for controlled drug release. Chem. Commun 48, 4755–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, and Hegemann P (2017). The form and function of channelrhodopsin. Science 357, 5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina-Amitonova LV, Fedotov IV, Ivashkina OI, Zots MA, Fedotov AB, Anokhin KV, and Zheltikov AM (2013). Implantable fiber-optic interface for parallel multisite long-term optical dynamic brain interrogation in freely moving mice. Sci. Rep 3, 3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwé S, and Dedecker P (2019). Optimizing the fluorescent protein toolbox and its use. Curr. Opin. Biotechnol 58, 183–191. [DOI] [PubMed] [Google Scholar]
- Ellis-Davies GCR (2007). Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods 4, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MS, Chaurette JP, Adams ST, Reddy GR, Paley MA, Aronin N, Prescher JA, and Miller SC (2014). A synthetic luciferin improves bioluminescence imaging in live mice. Nat. Methods 11, 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon TR, Lower SE, Chang C-H, Bessho-Uehara M, Martin GJ, Bewick AJ, Behringer M, Debat HJ, Wong I, Day JC, et al. (2018). Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife 7, e36495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhana I, Hossain MN, Suzuki K, Matsuda T, and Nagai T (2019). Genetically encoded fluorescence/bioluminescence bimodal indicators for Ca2+ imaging. ACS Sens. 4, 1825–1834. [DOI] [PubMed] [Google Scholar]
- Fan F, and Wood KV (2007). Bioluminescent assays for high-throughput screening. Assay Drug Dev. Techn 5, 127–136. [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, and Deisseroth K (2011). The development and application of optogenetics. Annu. Rev. Neurosci 34, 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga H (2008). Firefly luminescence: A historical perspective and recent developments. Photochem. Photobiol. Sci 7, 146–158. [DOI] [PubMed] [Google Scholar]
- Francis WR, Shaner NC, Christianson LM, Powers ML, and Haddock SHD (2015). Occurrence of isopenicillin-N-synthase homologs in bioluminescent Ctenophores and implications for coelenterazine biosynthesis. PLoS One 10, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorka AP, Yamamoto T, Zhu J, and Schnermann MJ (2018). Cyanine photocages enable spatial control of inducible Cre-mediated recombination. ChemBioChem 19, 1239–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor C, Gwosch KC, Sahl SJ, and Hell SW (2018). Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc. Natl. Acad. Sci. USA 115, 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor C, Pape JK, Gwosch KC, Gilat T, Sahl SJ, and Hell SW (2019). Autonomous bioluminescence imaging of single mammalian cells with the bacterial bioluminescence system. Proc. Natl. Acad. Sci. USA 116, 26491–26496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss R, Schena A, Reymond L, Patiny L, Werner D, Tinberg CE, Baker D, and Johnsson K (2014). Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring. Nat. Chem. Biol 10, 598–603. [DOI] [PubMed] [Google Scholar]
- Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, Ratner L, and Piwnica-Worms D (2009). Bioluminescence imaging of myeloperoxidase activity in vivo. Nat. Med 15, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro JA, and Méndez J (2020). In vivo bioluminescent imaging of Yersinia ruckeri pathogenesis in fish In Bioluminescent Imaging: Methods and Protocols, Ripp S, ed. (New York, NY: Springer US; ), pp. 69–80. [DOI] [PubMed] [Google Scholar]
- Gutierrez Jauregui R, Fleige H, Bubke A, Rohde M, Weiss S, and Forster R (2019). IL-1β promotes Staphylococcus aureus biofilms on implants in vivo. Front. Immunol 10, 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock SHD, Moline MA, and Case JF (2009). Bioluminescence in the sea. Annu. Rev. Mar. Sci 2, 443–493. [DOI] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, and Wood KV (2012). Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol 7, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Woodroofe CC, Wood MG, Que I, van’t Root M, Ridwan Y, Shi C, Kirkland TA, Encell LP, Wood KV, Lowik C, Mezzanotte L (2018). Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hananya N and Shabat D (2017). A glowing trajectory between bio- and chemiluminescence: from luciferin-based Probes to triggerable dioxetanes. Angew Chem Int Ed. 56, 16454–16463. [DOI] [PubMed] [Google Scholar]
- Hananya N and Shabat D (2019). Recent advances and challenges in luminescent imaging: bright outlook for chemiluminescence of dioxetanes in water. ACS Cent Sci. 5, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood KR, Mofford DM, Reddy GR, and Miller SC (2011). Identification of mutant firefly luciferases that efficiently utilize aminoluciferins. Chem. Biol 18, 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Tsukasaki Y, Ohyanagi T, and Jin T (2013). Bioluminescence resonance energy transfer coupled near-infrared quantum dots using GST-tagged luciferase for in vivo imaging. Chem. Commun 49, 228–230. [DOI] [PubMed] [Google Scholar]
- Hastings JW (1983). Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. J. Mol. Evol 19, 309–321. [DOI] [PubMed] [Google Scholar]
- Häusser M (2014). Optogenetics: the age of light. Nat. Methods 11, 1012–1014. [DOI] [PubMed] [Google Scholar]
- Heim R, Prasher DC, and Tsien RY (1994). Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91, 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, and Nagelschneider G (1970). Chemistry and molecular biology of flavins and flavoproteins. FEBS Lett. 8, 69–83. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han I-S, Larsen E, and Briggs WR (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278, 2120. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, and Kay SA (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. [DOI] [PubMed] [Google Scholar]
- Inouye S (2010). Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell. Mol. Life Sci 67, 387–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S, and Sasaki S (2007). Overexpression, purification and characterization of the catalytic component of Oplophorus luciferase in the deep-sea shrimp, Oplophorus gracilirostris. Protein Expr. Purif 56, 261–268. [DOI] [PubMed] [Google Scholar]
- Inouye S, and Shimomura O (1997). The use of Renilla luciferase, Oplophorus luciferase, and Apoaequorin as bioluminescent reporter protein in the presence of coelenterazine analogues as substrate. Biochem. Biophys. Res. Commun 233, 349–353. [DOI] [PubMed] [Google Scholar]
- Iwano S, Obata R, Miura C, Kiyama M, Hama K, Nakamura M, Amano Y, Kojima S, Hirano T, Maki S, and Niwa H (2013). Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron 69, 3847–3856. [Google Scholar]
- Iwano S, Sugiyama M, Hama H, Watakabe A, Hasegawa N, Kuchimaru T, Tanaka KZ, Takahashi M, Ishida Y, Hata J, Shimozono S, Namiki K, Fukano T, Kiyama M, Okano H, Kizaka-Kondoh S, McHugh TJ, Yamamori T, Hioki H, Maki S, Miyawaki A (2018). Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 359, 935–939. [DOI] [PubMed] [Google Scholar]
- James ML, and Gambhir SS (2012). A molecular imaging primer: modalities, imaging agents, and applications. Physiol. Rev 92, 897–965. [DOI] [PubMed] [Google Scholar]
- Jathoul AP, Grounds H, Anderson JC, and Pule MA (2015). A dual-color far-red to near-infrared firefly luciferin analogue designed for multiparametric bioluminescence imaging. Angew. Chem. Int. Ed. Engl 54, 1698–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins DE, Yu S-F, Hornig YS, Purchio T, and Contag PR (2003). In vivo monitoring of tumor relapse and metastasis using bioluminescent PC-3M-luc-C6 cells in murine models of human prostate cancer. Clin. Exp. Metastas 20, 745–756. [DOI] [PubMed] [Google Scholar]
- Jiang T, Du L, and Li M (2016). Lighting up bioluminescence with coelenterazine: strategies and applications. Photochem. Photobiol. Sci 15, 466–480. [DOI] [PubMed] [Google Scholar]
- Hiblot J, Yu Q, Sabbadini M, Reymond L, Xue L, Sallin O, Schena A, Hill N, Griss R, and Johnsson K (2017). Luciferases with tunable emission wavelengths. Angew. Chem. Int. Ed. Engl 56, 14556–14560. [DOI] [PubMed] [Google Scholar]
- Jones KA, Li DJ, Hui E, Sellmyer MA, and Prescher JA (2015). Visualizing cell proximity with genetically encoded bioluminescent reporters. ACS Chem. Biol 10, 933–938. [DOI] [PubMed] [Google Scholar]
- Jones KA, Porterfield WB, Rathbun CM, McCutcheon DC, Paley MA, and Prescher JA (2017). Orthogonal luciferase–luciferin pairs for bioluminescence imaging. J. Am. Chem. Soc 139, 2351–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Sin L, Albert E, Yu J, Francis K, DeBoer M, Rubin M, Bellinger-Kawahara C, Parr JTR, and Contag PR (2003). Direct continuous method for monitoring biofilm infection in a mouse model. Infect. Immun 71, 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaskova ZM, Tsarkova AS, and Yampolsky IV (2016). 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev 45, 6048–6077. [DOI] [PubMed] [Google Scholar]
- Khakhar A, Starker CG, Chamness JC, Lee N, Stokke S, Wang C, Swanson R, Rizvi F, Imaizumi T, and Voytas DF (2020). Building customizable auto-luminescent luciferase-based reporters in plants. eLife 9, e52786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Cho KF, Kim MW, and Ting AY (2019). Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. eLife 8, e43826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MW, Wang W, Sanchez MI, Coukos R, von Zastrow M, and Ting AY (2017). Time-gated detection of protein-protein interactions with transcriptional readout. eLife 6, e30233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada N, Saitoh T, Ikeda Y, Iwano S, Obata R, Niwa H, Hirano T, Miyawaki A, Suzuki K, Nishiyama S, and Maki SA (2018). Toward bioluminescence in the near-infrared region: Tuning the emission wavelength of firefly luciferin analogues by allyl substitution. Tetrahedron Lett. 59, 1087–1090. [Google Scholar]
- Konold PE, Mathes T, Weeiβorn J, Groot ML, Hegemann P, and Kennis JTM (2016). Unfolding of the C-terminal Jα helix in the LOV2 photoreceptor domain observed by time-resolved vibrational spectroscopy. J. Phys. Chem. Lett 7, 3472–3476. [DOI] [PubMed] [Google Scholar]
- Kotlobay AA, Dubinnyi MA, Purtov KV, Guglya EB, Rodionova NS, Petushkov VN, Bolt YV, Kublitski VS, Kaskova ZM, Ziganshin RH, Nelyubina YV, Dorovatovskii PV, Eliseev IE, Branchini BR Bourenkov G, Ivanov IA, Oba Y, Yampolsky IV, and Tsarkova AS (2019). Bioluminescence chemistry of fireworm Odontosyllis. Proc. Natl. Acad. Sci. USA 116, 18911–18916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlobay AA, Sarkisyan KS, Mokrushina YA, Marcet-Houben M, Serebrovskaya EO, Markina NM, Gonzalez Somermeyer L, Gorokhovatsky AY, Vvedensky A, Purtov KV, Petushkov VN, Rodionova NS, Chepurnyh TV, Fakhraurova LI, Guglya EB, Ziganshin R, Tsarkova AS, Kaskova ZM, Shender V, Abakumov M, Abakumova TO, Povolotskaya IS, Eroshkin FM, Zaraisky AG, Mishin AS, Dolgov SV, Mitiouchkina TY, Kopantzev EP, Waldenmaier HE, Oliveira AG, Oba Y, Barsova E, Bogdanova EA, Gabaldon T, Stevani CV, Lukyanova S, Smirnov IV, Gitelson JI, Kondrashov FA, and Yampolsky IV (2018). Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. USA 115, 12728–12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchimaru T, Iwano S, Kiyama M, Mitsumata S, Kadonosono T, Niwa H, Maki S, and Kizaka-Kondoh S (2016). A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging. Nat. Commun 7, 11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen L, Du L, and Li M (2013). Cage the firefly luciferin! – a strategy for developing bioluminescent probes. Chem. Soc. Rev 42, 662–676. [DOI] [PubMed] [Google Scholar]
- Lindberg E, Angerani S, Anzola M, and Winssinger N (2018). Luciferase-induced photoreductive uncaging of small-molecule effectors. Nat. Commun 9, 3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg E, Mizukami S, Ibata K, Fukano T, Miyawaki A, and Kikuchi K (2013). Development of cell-impermeable coelenterazine derivatives. Chem. Sci 4, 4395–4400. [DOI] [PubMed] [Google Scholar]
- Lloyd JE (1983). Bioluminescence and communication in insects. Annu. Rev. Entomol 28, 131–160. [Google Scholar]
- Loening AM, Wu AM, and Gambhir SS (2007). Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods 4, 641–643. [DOI] [PubMed] [Google Scholar]
- Losi A, and Gärtner W (2008). Bacterial bilin- and flavin-binding photoreceptors. Photochem. Photobiol. Sci 7, 1168–1178. [DOI] [PubMed] [Google Scholar]
- Losi A, and Gärtner W (2012). The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu. Rev. Plant Biol 63, 49–72. [DOI] [PubMed] [Google Scholar]
- Lu X, Agasti SS, Vinegoni C, Waterman P, DePinho RA, and Weissleder R (2012). Optochemogenetics (OCG) allows more precise control of genetic engineering in mice with CreER regulators. Bioconjugate Chem. 23, 1945–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker GD, Bardill JP, Prior JL, Pica CM, Piwnica-Worms D, and Leib DA (2002). Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J. Virol 76, 12149–12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukyanov KA, Chudakov DM, Lukyanov S, and Verkhusha VV (2005). Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol 6, 885–890. [DOI] [PubMed] [Google Scholar]
- Markova SV, Larionova MD, and Vysotski ES (2019). Shining light on the secreted luciferases of marine copepods: current knowledge and applications. Photochem. Photobiol 95, 705–721. [DOI] [PubMed] [Google Scholar]
- Martini S, and Haddock SHD (2017). Quantification of bioluminescence from the surface to the deep sea demonstrates its predominance as an ecological trait. Sci. Rep 7, 45750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon DC, Porterfield WB, and Prescher JA (2015). Rapid and scalable assembly of firefly luciferase substrates. Org. Biomol. Chem 13, 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy WD, Seliger HH, and Deluca M (1965). Enzyme catalysis and color of light in bioluminescent reactions In Evolving Genes and Proteins, Bryson V, and Vogel HJ, eds. (Academic Press; ), pp. 319–340. [Google Scholar]
- Meighen EA (1993). Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7, 1016–1022. [DOI] [PubMed] [Google Scholar]
- Mezzanotte L, van’t Root M, Karatas H, Goun EA, and Löwik CWGM (2017). In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol. 35, 640–652. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, and Massagué J (2005). Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitiouchkina T, Mishin AS, Somermeyer LG, Markina NM, Chepurnyh TV, Guglya EB, Karataeva TA, Palkina KA, Shakhova ES, Fakhranurova LI, Chekova SV, Tsarkova AS, Golubev YV, Negrebetsky VV, Dolgushin SA, Shalaev PV, Shlykov D, Melnik OA, Shipunova VO, Deyev SM, Bubyrev AI, Pushin AS, Choob VV, Dolgov SV, Kondrashov FA, Yampolsky IV, and Sarkisyan KS (2020). Plants with genetically encoded autoluminescence. Nat. Biotechnol in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofford DM, Adams ST, Reddy GSKK, Reddy GR, and Miller SC (2015). Luciferin amides enable in vivo bioluminescence detection of endogenous fatty acid amide hydrolase activity. J. Am. Chem. Soc 137, 8684–8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofford DM, Reddy GR, and Miller SC (2014a). Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred dubstrates over d-luciferin. J. Am. Chem. Soc 136, 13277–13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofford DM, Reddy GR, and Miller SC (2014b). Latent luciferase activity in the fruit fly revealed by a synthetic luciferin. Proc. Natl. Acad. Sci 111, 4443–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, and Moffat K (2007). Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol 373, 112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na SH, Oh MH, Jeon H, Lee Y-K, Lee B, Shin M, and Lee JC (2019). Imaging of bioluminescent Acinetobacter baumannii in a mouse pneumonia model. Microb. Pathog 137, 103784. [DOI] [PubMed] [Google Scholar]
- Nealson KH, and Hastings JW (1979). Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev 43, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba Y, Kato S. i., Ojika M, and Inouye S (2009). Biosynthesis of coelenterazine in the deep-sea copepod, Metridia pacifica. Biochem. Biophys. Res. Commun 390, 684–688. [DOI] [PubMed] [Google Scholar]
- Oba Y, Stevani CV, Oliveira AG, Tsarkova AS, Chepurnykh TV, and Yampolsky IV (2017). Selected least studied but not forgotten bioluminescent systems. Photochem. Photobiol 93, 405–415. [DOI] [PubMed] [Google Scholar]
- Oba Y, Yoshida N, Kanie S, Ojika M, and Inouye S (2014). Biosynthesis of firefly luciferin in adult lantern: decarboxylation of L-cysteine is a key step for benzothiazole ring formation in firefly luciferin synthesis. PloS One 8, e84023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MA, Daily WJ, Hesselberth PE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, and Wood KV (2005). Homogeneous, bioluminescent protease assays: caspase-3 as a model. J. Biomol. Screen 10, 137–148. [DOI] [PubMed] [Google Scholar]
- Oh Y, Park Y, Cho JH, Wu H, Paulk NK, Liu LX, Kim N, Kay MA, Wu JC, and Lin MZ (2019). An orange calcium-modulated bioluminescent indicator for non-invasive activity imaging. Nat. Chem. Biol 15, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi A, Dam J, and Jockers R (2020). β-Arrestin-2 BRET biosensors detect different β-arrestin-2 conformations in interaction with GPCRs. ACS Sens. 5, 57–64. [DOI] [PubMed] [Google Scholar]
- Oliveira AG, Stevani CV, Waldenmaier HE, Viviani V, Emerson JM, Loros JJ, and Dunlap JC (2015). Circadian control sheds light on fungal bioluminescence. Curr. Biol 25, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omachi K, Kamura M, Teramoto K, Kojima H, Yokota T, Kaseda S, Kuwazuru J, Fukuda R, Koyama K, Matsuyama S, Motomura K, Shuto T, Suico MA, Kai H (2018). A split-luciferase-based trimer formation assay as a high-throughput screening platform for therapeutics in Alport Syndrome. Cell Chem. Biol 25, 634–643. [DOI] [PubMed] [Google Scholar]
- Otto-Duessel M, Khankaldyyan V, Gonzalez-Gomez I, Jensen MC, Laug WE, and Rosol M (2006). In vivo testing of Renilla luciferase substrate analogs in an orthotopic murine model of human glioblastoma. Mol. Imaging 5, 57–64. [PubMed] [Google Scholar]
- Owen SF, Liu MH, and Kreitzer AC (2019). Thermal constraints on in vivo optogenetic manipulations. Nat. Neurosci 22, 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley MA, and Prescher JA (2014). Bioluminescence: a versatile technique for imaging cellular and molecular features. MedChemComm 5, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parag-Sharma K, O’Banion CP, Henry EC, Musicant AM, Cleveland JL, Lawrence DS, and Amelio AL (2020). Engineered BRET-based biologic light sources enable spatiotemporal control over diverse optogenetic systems. ACS Synth. Biol 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Song S-H, Palmateer B, Pal A, Petersen ED, Shall GP, Welchko RM, Ibata K, Miyawaki A, Augustine GJ, and Hochgeschwender U (2020). Novel luciferase–opsin combinations for improved luminopsins. J. Neurosci. Res 98, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen ED, Sharkey ED, Pal A, Shafau LO, Zenchak JR, Peña AJ, Aggarwal A, Prakash M, and Hochgeschwender U (2019). Restoring function after severe spinal cord injury through bioluminescence-driven optogenetics. bioRxiv, 710194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JA, Wijesooriya C, Gehrmann EJ, Mahoney KM, Goswami PP, Albright TR, Syed A, Dutton AS, Smith EA, and Winter AH (2018). Family of BODIPY photocages cleaved by single photons of visible/near-infrared light. J. Am. Chem. Soc 140, 7343–7346. [DOI] [PubMed] [Google Scholar]
- Petushkov VN, Dubinnyi MA, Tsarkova AS, Rodionova NS, Baranov MS, Kublitski VS, Shimomura O, and Yampolsky IV (2014). A novel type of luciferin from the Siberian luminous warthworm Fridericia heliota: structure elucidation by spectral studies and total synthesis. Angew. Chem. Int. Ed. Engl 53, 5566–5568. [DOI] [PubMed] [Google Scholar]
- Pimenta FM, Jensen RL, Breitenbach T, Etzerodt M, and Ogilby PR (2013). Oxygen-dependent photochemistry and photophysics of "miniSOG," a protein-encased flavin. Photochem. photobiol 89, 1116–1126. [DOI] [PubMed] [Google Scholar]
- Porterfield WB, and Prescher JA (2015). Tools for visualizing cell–cell ‘interactomes’. Curr. Opin. Chem. Biol 24, 121–130. [DOI] [PubMed] [Google Scholar]
- Prescher JA, and Contag CH (2010). Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol 14, 80–89. [DOI] [PubMed] [Google Scholar]
- Proshkina GM, Shramova EI, Shilova ON, Ryabova AV, and Deyev SM (2018). Phototoxicity of flavoprotein miniSOG induced by bioluminescence resonance energy transfer in genetically encoded system NanoLuc-miniSOG is comparable with its LED-excited phototoxicity. J. Photochem. Photobiol. B 188, 107–115. [DOI] [PubMed] [Google Scholar]
- Pudasaini A, El-Arab KK, and Zoltowski BD (2015). LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Front. Mol. Biosci 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram A, Ye F, Adams JK, Shaner N, Robinson JT, and Veeraraghavan A (2020). Determining the depth limit of bioluminescent sources in scattering media. bioRxiv, 044982. [Google Scholar]
- Rathbun CM and Prescher JA (2017). Bioluminescent probes for imaging biology beyond the culture dish. Biochemistry 56, 5178–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun CM, Porterfield WB, Jones KA, Sagoe MJ, Reyes MR, Hua CT, and Prescher JA (2017). Parallel screening for rapid identification of orthogonal bioluminescent tools. ACS Cent. Sci 3, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GR, Thompson WC, and Miller SC (2010). Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J. Am. Chem. Soc 132, 13586–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, and Tsien RY (2017). The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci 42, 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, and Lee K-H (1998). The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol 64, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-González R, Cortajarena AL, Mejias SH, Agut M, Nonell S, and Flors C (2013). Singlet oxygen generation by the genetically encoded tag miniSOG. J. Am. Chem. Soc 135, 9564–9567. [DOI] [PubMed] [Google Scholar]
- Rumyantsev KA, Turoverov KK, and Verkhusha VV (2016). Near-infrared bioluminescent proteins for two-color multimodal imaging. Sci. Rep 6, 36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Chang YF, Horikawa K, Hatsugai N, Higuchi Y, Hashida M, Yoshida Y, Matsuda T, Arai Y, and Nagai T (2012). Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun 3, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MI, and Ting AY (2020). Directed evolution improves the catalytic efficiency of TEV protease. Nat. Methods 17, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos EB, Yeh R, Lee J, Nikhamin Y, Punzalan B, Punzalan B, Perle KL, Larson SM, Sadelain M, and Brentjens RJ (2009). Sensitive in vivo imaging of T cells using a membrane-bound Gaussia princeps luciferase. Nat. Med 15, 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Chang L, Gonzalez Y, Gallego-Flores T, Young DW, and Venken KJT (2019). Examining multiple cellular pathways at once using multiplex hextuple luciferase assaying. Nat Commun. 10, 5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Anindita PD, Phongphaew W, Carr M, Kobayashi S, Orba Y, and Sawa H (2018). Development of a rapid and quantitative method for the analysis of viral entry and release using a NanoLuc luciferase complementation assay. Virus Res. 243, 69–74. [DOI] [PubMed] [Google Scholar]
- Scatena CD, Hepner MA, Oei YA, Dusich JM, Yu S-F, Purchio T, Contag PR, and Jenkins DE (2004). Imaging of bioluminescent LNCaP-luc-M6 Tumors: A new animal model for the study of metastatic human prostate cancer. Prostate 59, 292–303. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A, and Rice CM (2012). Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc. Natl. Acad. Sci. USA 109, 14610–14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, and Tsien RY (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, and Tsien RY (2008). Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Adams ST, Liebmann KL, and Miller SC (2017). Rapid access to a broad range of 6′-substituted firefly luciferin analogues reveals surprising emitters and inhibitors. Org. Lett 19, 5836–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, and Saiga Y (1962). Extraction, purification and properties of Aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol 59, 223–239. [DOI] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, and Saiga Y (1963). Further data on the bioluminescent protein, aequorin. J. Cell. Comp. Physiol 62, 1–8. [DOI] [PubMed] [Google Scholar]
- Shipunova VO, Shilova ON, Shramova EI, Deyev SM, and Proshkina GM (2018). A highly specific substrate for NanoLUC luciferase furimazine is toxic in vitro and in vivo. Russ. J. Bioorg. Chem 44, 225–228. [Google Scholar]
- Shramova E, Proshkina G, Chumakov S, Khodarovich Y, and Deyev S (2016). Flavoprotein miniSOG cytotoxisity can be induced by bioluminescence resonance energy transfer. Acta Naturae 8, 118–123. [PMC free article] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, and Tsien RY (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov OA, Govorunova EG, and Spudich JL (2009). Photosensory functions of channelrhodopsins in native algal cells. Photochem. Photobiol 85, 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova DV, and Ugarova NN (2017). Firefly luciferase-based fusion proteins and their applications in bioanalysis. Photochem. Photobiol 93, 436–447. [DOI] [PubMed] [Google Scholar]
- Steinhardt RC, O'Neill JM, Rathbun CM, McCutcheon DC, Paley MA, and Prescher JA (2016). Design and synthesis of an alkynyl luciferin analogue for bioluminescence imaging. Chem. Eur. J 22, 3671–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt RC, Rathbun CM, Krull BT, Yu JM, Yang Y, Nguyen BD, Kwon J, McCutcheon DC, Jones KA, Furche F, and Prescher JA (2017). Brominated luciferins are versatile bioluminescent probes. ChemBioChem 18, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe CL, Burley TA, Allan H, Vinci M, Kramer-Marek G, Ciobota DM, Parkinson GN, Southworth TL, Agliardi G, Hotblack A, Lythgoe MF, Branchini BR, Kalber TL, Anderson JC, and Pule MA (2019). Near-infrared dual bioluminescence imaging in mouse models of cancer using infraluciferin. eLife 8, e45801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Nakano M, and Nagai T (2016). Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nat. Commun 7, 13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney E, Lovering AM, Bowker KE, MacGowan AP, and Nelson SM (2019). An in vitro biofilm model of Staphylococcus aureus infection of bone. Lett. Appl. Microbiol 68, 294–302. [DOI] [PubMed] [Google Scholar]
- Swijnenburg R-J, Schrepfer S, Govaert JA, Cao F, Ransohoff K, Sheikh AY, Haddad M, Connolly AJ, Davis MM, Robbins RC, and Wu JC (2008). Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc. Natl. Acad. Sci. USA 105, 12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai A, Nakano M, Saito K, Haruno R, Watanabe TM, Ohyanagi T, Jin T, Okada Y, and Nagai T (2015). Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. USA 112, 4352–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA, Kim D-E, Fernandez JL, Weissleder R, and Breakefield XO (2005). Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in Vivo. Mol. Ther 11, 435–443. [DOI] [PubMed] [Google Scholar]
- Tessler M, Gaffney JP, Crawford JM, Trautman E, Gujarati NA, Alatalo P, Pieribone VA, and Gruber DF (2018). Luciferin production and luciferase transcription in the bioluminescent copepod Metridia lucens. PeerJ 6, e5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirukkumaran OM, Wang C, Asouzu NJ, Fron E, Rocha S, Hofkens J, Lavis LD, and Mizuno H (2020). Improved HaloTag ligand enables BRET imaging with NanoLuc. Front. Chem 7, 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titushin MS, Feng Y, Lee J, Vysotski ES, and Liu Z-J (2011). Protein-protein complexation in bioluminescence. Protein Cell. 2, 957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarkova AS, Kaskova ZM, and Yampolsky IV (2016). A tale of two luciferins: fungal and earthworm new bioluminescent systems. Acc. Chem. Res 49, 2372–2380. [DOI] [PubMed] [Google Scholar]
- Valley MP, Zhou W, Hawkins EM, Shultz J, Cali JJ, Worzella T, Bernad L, Good T, Good D, Riss TL, et al. (2006). A bioluminescent assay for monoamine oxidase activity. Anal. Biochem 359, 238–246. [DOI] [PubMed] [Google Scholar]
- Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, and Chang CJ (2010). In vivo imaging of hydrogen peroxide production in a murine tumor model with a chemoselective bioluminescent reporter. Proc. Natl. Acad Sci. USA 107, 21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst MA, and Hellingwerf KJ (2004). Photoreceptor Proteins, “Star Actors of Modern Times”: A review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res 37, 13–20. [DOI] [PubMed] [Google Scholar]
- Vece V, and Vuocolo G (2015). Multicomponent synthesis of novel coelenterazine derivatives substituted at the C-3 position. Tetrahedron 71, 8781–8785. [Google Scholar]
- Viviani VR (2002). The origin, diversity, and structure function relationships of insect luciferases. Cell. Mol. Life Sci 59, 1833–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wildes CP, Pattarabanjird T, Sanchez MI, Glober GF, Matthews GA, Tye KM, and Ting AY (2017). A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol 35, 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widder EA (2010). Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science 328, 704. [DOI] [PubMed] [Google Scholar]
- Wiles S, Clare S, Harker J, Huett A, Young D, Dougan G, and Frankel G (2004). Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacterrodentium. Cell. Microbiol 6, 963–972. [DOI] [PubMed] [Google Scholar]
- Williams SJ, and Prescher JA (2019). Building biological flashlights: orthogonal luciferases and luciferins for in vivo imaging. Acc. Chem. Res 52, 3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KV, Lam YA, and McElroy WD (1989a). Introduction to beetle luciferases and their applications. J. Biolumin. Chemilumin 4, 289–301. [DOI] [PubMed] [Google Scholar]
- Wood KV, Lam YA, Seliger HH, and McElroy WD (1989b). Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science 244, 700. [DOI] [PubMed] [Google Scholar]
- Xu T, Close D, Handagama W, Marr E, Sayler G, and Ripp S (2016). The expanding toolbox of in vivo bioluminescent imaging. Front. Oncol 6, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Ripp S, Sayler GS, and Close DM (2014). Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS One 9, e96347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mu J, and Xing B (2017). Photoactivated drug delivery and bioimaging. Wiley Interdiscip. Rev. Nanomen. Nanobiotechnol 9, e1408. [DOI] [PubMed] [Google Scholar]
- Yao Z, Zhang BS, and Prescher JA (2018). Advances in bioluminescence imaging: new probes from old recipes. Curr. Opin. Chem. Biol 45, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H-W, and Ai H-W (2019). Development and applications of bioluminescent and chemiluminescent reporters and biosensors. Annu. Rev. Anal. Chem 12, 129–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H-W, Karmach O, Ji A, Carter D, Martins-Green MM, and Ai H-W (2017). Red- shifted luciferase-luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 14, 971–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H-W, Wu T, Chen M, and Ai H-W (2019). Identification of factors complicating bioluminescence imaging. Biochemistry 58, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Pourmandi N, Xue L, Gondrand C, Fabritz S, Bardy D, Patiny L, Katsyuba E, Auwerx J, and Johnsson K (2019). A biosensor for measuring NAD+ levels at the point of care. Nat. Metab 1, 1219–1225. [DOI] [PubMed] [Google Scholar]
- Yu Q, Xue L, Hiblot J, Griss R, Fabritz S, Roux C, Binz P-A, Haas D, Okun JG, and Johnsson K (2018). Semisynthetic sensor proteins enable metabolic assays at the point of care. Science 361, 1122–1126. [DOI] [PubMed] [Google Scholar]
- Zenchak JR, Palmateer B, Dorka N, Brown TM, Wagner L-M, Medendorp WE, Petersen ED, Prakash M, and Hochgeschwender U (2020). Bioluminescence-driven optogenetic activation of transplanted neural precursor cells improves motor deficits in a Parkinson's disease mouse model. J. Neurosci. Res 98, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BS, Jones KA, McCutcheon DC, and Prescher JA (2018). Pyridone luciferins and mutant luciferases for bioluminescence imaging. ChemBioChem 19, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, Magnuson J, Hegemann P, Deisseroth K (2011). The microbial opsin family of optogenetic tools. Cell 147, 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, and Contag CH (2005). Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J. Biomed. Opt 10, 41210. [DOI] [PubMed] [Google Scholar]
- Zhou W, Andrews C, Liu J, Shultz JW, Valley MP, Cali JJ, Hawkins EM, Klaubert DH, Bulleit RF, and Wood KV (2008). Self-cleavable bioluminogenic luciferin phosphates as alkaline phosphatase reporters. ChemBioChem 9, 714–718. [DOI] [PubMed] [Google Scholar]
- Zhou W, Shultz JW, Murphy N, Hawkins EM, Bernad L, Good T, Moothart L, Frackman S, Klaubert DH, Bulleit RF, and Wood KV (2006a). Electrophilic aromatic substituted luciferins as bioluminescent probes for glutathione S-transferase assays. Chem. Commun, 4620–4622. [DOI] [PubMed] [Google Scholar]
- Zhou W, Valley MP, Shultz J, Hawkins EM, Bernad L, Good T, Good D, Riss TL, Klaubert DH, and Wood KV (2006b). New bioluminogenic substrates for monoamine oxidase assays. J. Am. Chem. Soc 128, 3122–3123. [DOI] [PubMed] [Google Scholar]