Abstract
Tuberculosis (TB), a chronic disease caused by Mycobacterium tuberculosis (Mtb), is a global health issue across the world. Pakistan ranks fifth among the countries, which are facing, a significantly great number of mortalities and morbidities due to TB. Unfortunately, all previously reported treatments are not successful for the eradication of TB. Here in this study, we report an emerging treatment option for this disease. We have applied immunoinformatics to predict highly conserved B and T-cell epitopes from Mtb, showing significant binding affinities to the frequent HLA alleles in the Pakistani population. A total of ten highly referenced and experimentally validated epitopes were selected from the Immune Epitope Database (IEDB), followed by their conservancy analysis using weblogos. The consensus sequences and variants derived from these sequences were examined, for their binding affinities, with prevalent HLA alleles of Pakistan. Moreover, the antigenic and allergenic natures of these peptides were also evaluated via Vaxijen and AllerTOP, respectively. Consequently, all potentially allergenic and non-antigenic, peptide fragments, were excluded from the analysis. Among all putative epitopes, three CD8 + T-cell epitopes were selected, as ideal vaccine candidates and, population coverage analysis revealed that the combination of these three peptides was covering, 67.28% Pakistani Asian and 57.15% mixed Pakistani populations. Likewise, eleven linear and six conformational or discontinuous B-cell epitopes were also marked as potential vaccine candidates based on their prediction score, non-allergenic nature, and antigenic properties. These epitopes, however, need the final validation via wet-lab studies. After their approval, these epitopes would be effective candidates for the future designing of epitope-based vaccines against Mtb infections in Pakistan.
Electronic supplementary material
The online version of this article (10.1007/s10989-020-10111-w) contains supplementary material, which is available to authorized users.
Keywords: Epitope-based vaccines, Mycobacterium tuberculosis, Human leukocyte antigen, Pakistani population
Introduction
Tuberculosis (TB) is a serious health concern globally, caused by the infections of Mycobacterium tuberculosis (Mtb) (Ullah et al. 2016). Mtb is responsible for the infections of lungs as well as many other organs including, lymph nodes, abdomen, gastrointestinal, and bones (Sunnetcioglu et al. 2015). Mtb-based infections are, therefore, described as pulmonary or extrapulmonary tuberculosis (Campbell and Bah-Sow 2006). The infectious droplets released during speaking, coughing, and sneezing, are responsible for the transmission of Mtb infections among individuals (Glaziou et al. 2015). The general symptoms associated with TB include weight loss, fever, fatigue, loss of appetite, and sweat, while 90% of the patients acquire latent infections, with no obvious symptom of the disease (Kaufmann et al. 2010; Knechel 2009). According to the world health organization (WHO), in 2019, approximately 10.4 million people were infected with TB worldwide, of which 1.3 million died (Annabel et al. 2019). Pakistan stands fifth in position among the most affected country with TB, with an estimated 510,000 cases per year and a mortality rate of 23 deaths per 100,000 individuals (NTP 2016). The death rate could increase significantly if these infections are left untreated.
Till now, several treatment options are available to control the Mtb infections, including, anti-TB drugs and the Bacille Calmette-Guerin (BCG) vaccine (Iseman 2002). BCG is used to control severe pulmonary tuberculosis in children, with 60 to 80% efficiency, however, it is not as efficient for individuals with disease recurrence (Roy et al. 2014). There is evidence of multidrug resistance (MDR) in Mtb strains, suggesting even more aggravated infections, which would be difficult to control using available treatments (Munir et al. 2018). Thus, it is the need of time to present alternative treatment options for these infections. Currently, epitope-based vaccines are proved efficient in different infections and, therefore, are substituting trivial vaccines (Li et al. 2014). In this strategy, only small immunogenic fragments, epitopes, from the full-length sequence of the antigenic proteins, are considered as potential vaccine candidates (Sette and Fikes 2003). These vaccines elicit the immune response, only against the immunogenic fragments, thus minimizing the chances of non-specific immune responses (Tissot et al. 2010). In vaccine designing, computational tools have gained notable attention, as they help to identify highly immunogenic and non-allergenic epitopes, as suitable vaccine candidates (María et al. 2017). Furthermore, the 3D structure analysis and the assessment of the binding affinities of putative epitopes with HLA alleles are also possible. Another important feature is the population coverage analysis, which helps to screen the potential epitopes for population-specific vaccines (Denisova et al. 2010). Additionally, these methods are cost-effective, less laborious, and are relatively safe compared to traditional methods (Tomar and De 2010).
This study aims to use multiple in-silico tools for the identification of putative B and T-cell epitopes, from the pathogenic proteins of the Mtb, followed by their conservancy analysis, allergenic and antigenic predictions. The antigenic and non-allergenic epitopes restricting with the prevalent HLA alleles of Pakistan were selected, and their population coverage was measured. Finally, through structural modelling, the position of epitopes on the corresponding antigenic proteins were evaluated. The selected peptide would help in the future vaccine designing for tuberculosis.
Materials and Methods
Figure 1 represents all steps followed for the screening of epitope-based vaccine candidates specific to control Mycobacterium tuberculosis in Pakistani populations.
Fig. 1.
The stepwise approach for the in-silico design of epitope-based vaccine candidates for Mycobacterium tuberculosis in Pakistani populations
T Cell Epitope Selection
T-cell epitopes are immunogenic peptides fragments capable of producing a specific immune response. These peptides are, therefore, important candidates for epitope-based vaccines (Weber et al. 2009). The highly referenced and experimentally validated MHC class-I and class-II, T-cell epitopes of Mycobacterium tuberculosis were retrieved using the online Immune Epitope Database (IEDB) (https://www.iedb.org). IEDB contains comprehensive information on epitopes and offers multiple tools for epitope prediction and analysis (Vita et al. 2015).
Sequence Retrieval
The sequences of all antigenic proteins containing the selected epitopes were obtained from the National Center for Biotechnology Information, NCBI (https://www.ncbi.nlm.nih.gov), with the accession IDs of NP_216402.1 (acyltransferase/ mycolyltransferase Ag85B), P9WNK7.1 (6 kDa early secretory antigenic target), NP_218280.1 (Lipoprotein LpqH), NP_214802.1 (ESAT-6-like protein EsxH), NP_216547.1 (Alpha-crystallin), P9WGU1.1 (Phosphate Binding Protein Pst1) and NP_218391.1 (ESAT-6-like protein EsxB) (Benson et al. 1993). Moreover, the Basic Local Alignment Search Tool, BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), was used to search all homologous sequences from the strains of Mycobacterium tuberculosis(Altschul et al. 1990).
Consensus Sequence Prediction
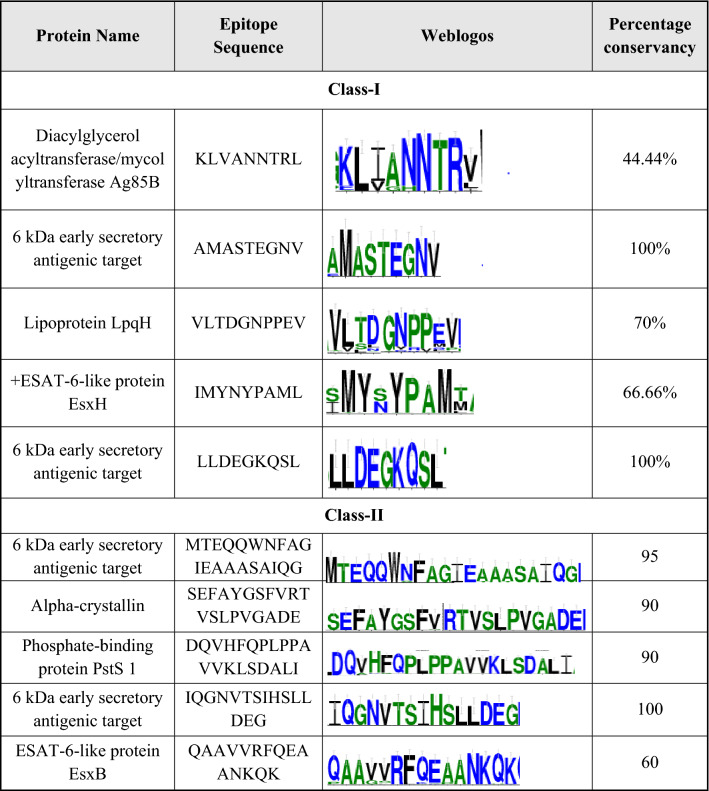
Multiple Sequence Alignment (MSA) performed via BioEdit v.7.2.6, was submitted to Weblogo3 software (https://weblogo.threeplusone.com) to obtain the consensus sequences (Table 2). Weblogo3 generates sequence logos in the form of stacks of letters. The height of each letter stack represents the conservancy of corresponding amino acids in the sequence (Crooks et al. 2004).
Table 2.
Weblogos and the conservancy analysis of the class-I and class-II epitopes via weblogo3
Retrieval of Common HLA Alleles from Pakistani Populations
The Allele Frequency Net Database (AFND) (https://www.allelefrequencies.net), was used to screen the prevalent HLA class-I and class-II alleles from Pakistan at the threshold of 0.01 (Table 3).AFND is an online database that contains information about alleles and their frequencies present in different populations concerning geographical regions (Gonzalez-Galarza et al. 2010).
Table 3.
Prevalent class-I and class-II HLA alleles in Pakistani populations retrieved through AFND
| Most prevalent class-I and class-II HLA alleles in Pakistani population | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| HLA Class-I restricted alleles | HLA Class-II restricted alleles | ||||||||
| Class A | Frequency | Class B | Frequency | Class C | Frequency | DQB1 | Frequency | DRB1 | Frequency |
| A*11:01 | 1.094 | B*51 | 1.125 | C*04 | 1.167 | DQB1*02 | 2.055 | DRB1*03 | 1.552 |
| A*33:03 | 0.658 | B*35 | 1.112 | C*07 | 1.025 | DQB1*05 | 1.699 | DRB1*15 | 1.099 |
| A*02:01 | 0.602 | B*08 | 0.628 | C*15 | 0.905 | DQB1*06 | 1.33 | DRB1*11 | 0.947 |
| A*24:02 | 0.573 | B*40 | 0.537 | C*07:02 | 0.822 | DQB1*03 | 0.962 | DRB1*01 | 0.747 |
| A*26:01 | 0.524 | B*58 | 0.414 | C*12 | 0.666 | DQB1*03:01 | 0.878 | DRB1*07 | 0.624 |
| A*01:01 | 0.491 | B*13 | 0.316 | C*06:02 | 0.62 | DQB1*03:02 | 0.406 | DRB1*13 | 0.585 |
| A*03:01 | 0.456 | B*15 | 0.286 | C*07:01 | 0.5 | DQB1*06:01 | 0.13 | DRB1*16 | 0.354 |
| A*32:01 | 0.445 | B*52 | 0.278 | C*12:03 | 0.492 | DQB1*04 | 0.127 | DRB1*14 | 0.335 |
| A*02:06 | 0.389 | B*44 | 0.263 | C*06 | 0.483 | DQB1*03:05 | 0.066 | DRB1*04 | 0.285 |
| A*30:01 | 0.335 | B*07 | 0.245 | C*03:02 | 0.454 | DQB1*03:03:02 | 0.062 | DRB1*10:01 | 0.251 |
| A*11 | 0.321 | B*27 | 0.193 | C*12:02 | 0.381 | DQB1*06:02 | 0.058 | DRB1*10 | 0.221 |
| A*02 | 0.29 | B*18 | 0.165 | C*14 | 0.361 | DRB1*08 | 0.139 | ||
| A*68:01:02 | 0.281 | B*53 | 0.14 | C*03 | 0.316 | DRB1*13:02 | 0.034 | ||
| A*24 | 0.268 | B*37 | 0.139 | C*01 | 0.218 | DRB1*12 | 0.02 | ||
| A*33 | 0.255 | B*38 | 0.133 | C*02 | 0.17 | DRB1*13:01 | 0.011 | ||
| A*01 | 0.187 | B*41 | 0.127 | C*16 | 0.143 | ||||
| A*03 | 0.187 | B*50 | 0.124 | C*08:02 | 0.142 | ||||
| A*26 | 0.18 | B*14:01 | 0.117 | C*16:02 | 0.136 | ||||
| A*31:01:02 | 0.177 | B*14:02 | 0.104 | C*05 | 0.112 | ||||
| A*33:01 | 0.163 | C*17 | 0.104 | ||||||
| A*68:01 | 0.151 | C*08:01 | 0.065 | ||||||
| A*02:11 | 0.095 | C*03:03 | 0.059 | ||||||
| A*30:02 | 0.09 | C*08 | 0.05 | ||||||
| A*29:01 | 0.089 | C*16:01 | 0.05 | ||||||
| A*03:02 | 0.084 | ||||||||
Binding Affinity Prediction
T-cell epitopes present on the surface of immune cells complexed with MHC molecules. Among these, CD4 + T-cells target MHC class-I, while CD8 + T-cells to the MHC class-II. The binding affinities of T-cell epitopes, with corresponding MHC molecules, are, therefore, the crucial determinant of elicitation of the efficient immune response (Cunha-Neto 1999). MHC-I (https://tools.iedb.org/mhci) and MHC-II (https://tools.iedb.org/mhcii) binding prediction tools from IEDB, were used for the evaluation of the binding affinities, of selected epitopes and their consensus variants with the prevalent class-I and class-II HLA alleles of Pakistan. For the prediction of binding affinities with MHC class-I HLA alleles, a total of two algorithms, ANN and netMHCpan, were considered, as ANN algorithm, was not trained for A*33:03, C*03:02, C*12:02, C*08:01 and C*16:01 alleles (Hoof et al. 2009; Nielsen and Andreatta 2017; Nielsen et al. 2003). Likewise, for MHC-II binding prediction, NN-align, and netMHCIIpan, were used. NN-align is trained on experimental data, while the netMHCIIpan algorithm is based on the artificial neural network method and trained on five groups of datasets of protein and peptide sequences based on the common motif clustering (Karosiene et al. 2013; Nielsen and Lund 2009). Binding affinities of DQB1*06:01, DQB1*03:03:02, DQB1*03:05, DRB1*10:01, DRB1*13:01 and DRB1*13:02, were predicted by netMHCIIpan. The results of binding affinities were expressed, in the form of IC50nM (the maximal half inhibitory concentration), where IC50 < 50 nM indicates significantly high binding, values in between 50-500 nM means intermediate affinity, while 500-5000 nM exhibits very low binding affinities. (Koşaloğlu-Yalçın et al. 2018) (Additional files 1 and 2).
Allergenicity Prediction
Allergenicity of selected epitopes and their consensus variants was predicted, by web-based server AllerTop v.2.0 (https://www.ddg-pharmfac.net/AllerTOP/index.html). AllerTop uses the auto cross-covariance (ACC) method, developed by wold et al. (Wold et al. 1993). The server is trained, on several known allergens and non-allergens from different species (Dimitrov et al. 2013).
Antigenicity Prediction
For the confirmation of the immunogenic character of all peptide fragments, online Vaxijen v.2.0 (https://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), was used. Vaxijen is based on the alignment independent method, which predicts antigenicity, using physicochemical properties and ACC methods (Doytchinova and Flower 2007). For antigenicity assessment, peptide fragments with a threshold greater than 0.5, were marked as potentially antigenic.
Population Coverage Calculation
Following antigenicity and allergenicity predictions, all antigenic and non-allergenic T-cell epitopes, having good binding affinities for the prevalent class-I and class-II HLA alleles of Pakistan, were considered for the population coverage analysis, using IEDB population coverage tool (https://tools.iedb.org/population).
Structural modelling and validation of Mtb proteins
In addition to the T-cells, B-cells are also crucial for the maximum elicitation of the immune response, the screening of B-cell epitopes is, therefore, equally essential in vaccine designing (Mauri and Bosma 2012). For B-cell epitope predictions, three proteins from Mtb including, ESAT-6 like protein EsxB, Lipoprotein LpqH and, Phosphate binding protein PstS 1, were selected. These three proteins were considered, as most of the referenced epitopes, in IEDB and literature against Mtb, were from these selected proteins (Converse and Cox 2005; Peirs et al. 2005; Sánchez et al. 2012). As the 3D models of these proteins, were required for the prediction of conformational B-cell epitopes, we firstly performed structural modelling, via Iterative Threading ASSembly Refinement (I-TASSER), followed by model validations, (Yang et al. 2015; Zheng et al. 2019) using Structural Analysis and Verification Server, SAVES version 5 (https://servicesn.mbi.ucla.edu/SAVES/). The models were validated based on Verify3D, PROCHECK, and ERRAT (Colovos and Yeates 1993; Eisenberg et al. 1997; Laskowski et al. 1993).
Prediction of Linear and Conformational B Cell Epitopes
IEDB ElliPro tool (https://tools.iedb.org/ellipro/), was used to predict linear and conformational B-cell, using three selected proteins of the Mtb as templates (Ponomarenko et al. 2008). In Ellipro, the minimum gap between residues was selected as 0.5, while the maximum allowed distance was 6.0A. For the visualization of 3D structures and to assess their orientations, DiscoveryStudio was used (Gazi et al. 2016; Gurung et al. 2012). As for T-cell epitopes, the allergenic and antigenic natures of B-cell epitopes were also determined using the AllerTop and Vaxijen servers, respectively, with the same parameters, as mentioned previously, for T-cell epitopes.
Results
Selection of T-cell Epitopes
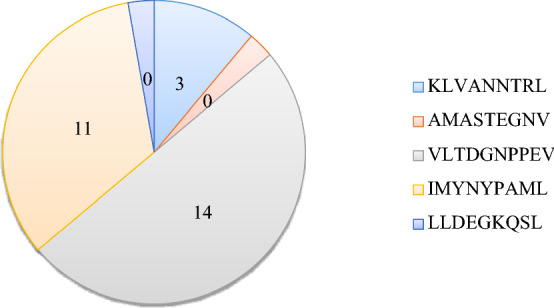
A total of ten highly referenced and experimentally validated epitope sequences were selected, from IEDB, as the potential immunogenic peptide for Mtb. Among these ten epitopes, five epitopes were from class-I, and five from class-II (Table 1). From these ten epitope sequences, weblogos were generated, to assess the conservancy of these epitopes among different strains (Table 2). In the case of the class-I epitope, the three variants were derived, from the first epitope KLVANNTRL, based on its weblogo. Likewise, fourteen variants from the third epitope, VLTDGNPPEV and, eleven variants from the fourth epitope, IMYNYPAML, were derived considering their web logos. The second and fifth epitopes with 100% conservancy, produce no variants and their original sequences, were analyzed (Additional file 1).
Table 1.
Highly referenced and, experimentally validated class-I and class-II epitopes retrieved through IEDB
| Sr.no | IEDB epitope ID | Protein name | Sequence | Restriction | Number of references listed at IEDB |
|---|---|---|---|---|---|
| Class I | |||||
| 1 | 32,213 | Diacylglycerol acyltransferase/mycolyltransferase Ag85B | KLVANNTRL | HLA-A*02:01 | 7 |
| 2 | 3064 | 6 kDa early secretory antigenic target | AMASTEGNV | HLA-A*02:01 | 6 |
| 3 | 69,798 | Lipoprotein LpqH | VLTDGNPPEV | HLA-A*02:01 | 6 |
| 4 | 27,586 | ESAT-6-like protein EsxH | IMYNYPAML | HLA-A*02:01 | 5 |
| 5 | 37,146 | 6 kDa early secretory antigenic target | LLDEGKQSL | HLA-A*02:01 | 5 |
| Class II | |||||
| 1 | 42,797 | 6 kDa early secretory antigenic target | MTEQQWNFAGIEAAASAIQG | HLA-DRB1*04 | 7 |
| 2 | 57,417 | Alpha-crystallin | SEFAYGSFVRTVSLPVGADE | HLA-DRB1*01:01 | 7 |
| 3 | 9924 | Phosphate-binding protein PstS 1 | DQVHFQPLPPAVVKLSDALI | HLA-DRB1*01 | 5 |
| 4 | 161,641 | 6 kDa early secretory antigenic target | IQGNVTSIHSLLDEG | HLA-DRB1*15:01 | 5 |
| 5 | 161,666 | ESAT-6-like protein EsxB | QAAVVRFQEAANKQK | HLA-DRB5*01:01 | 5 |
Similarly, for class-II epitopes, in addition to original epitope sequences, sequence variants were also derived. The first, second, and third epitopes (MTEQQWNFAGIEAAASAIQG, SEFAYGSFVRTVSLPVGADE, and DQVHFQPLPPAVVKLSDALI) produced six sequence variants each, while the fifth epitope, QAAVVRFQEAANKQK, produced one variant. The fourth class-II epitope, however, did not produce any variant (Additional file 2). Thus, a total of thirty-two class-I and twenty-one class-II, peptide fragments were considered for further analysis (Figs. 2 and 3).
Fig. 2.
Number of variants derived from the five MHC class-I epitopes retrieved through IEDB (Additional file 1)
Fig. 3.
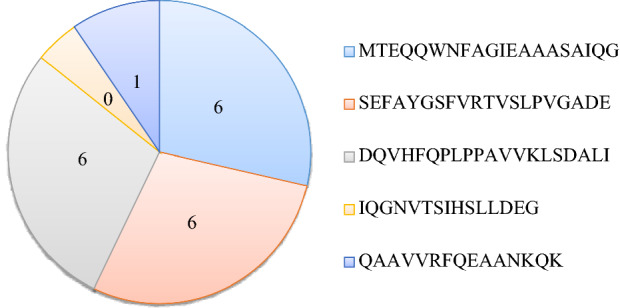
Number of variants derived from the five MHC class-II epitopes retrieved through IEDB (Additional file 2)
Prediction of prevalent HLA alleles from Pakistan
AFND reveals the prevalence of ninety-four, common HLA alleles in Pakistani populations, with sixty-eight alleles, belonging to class-I, and twenty-six to class-II. Among sixty-eight class-I HLA alleles, twenty-five belongs to HLA-A, nineteen to HLA-B, and twenty-four to the HLA-C category. Likewise, eleven class-II specific HLA alleles belong to DQB1 and fifteen to DRB1 (Additional File 3, Table 3).
Binding Affinity Prediction
The MHC binding prediction tools, from IEDB, were used to calculate the bindings of selected class-I and class-II epitopes as well as the variants derived from them, with the prevalent class-I and class-II HLA alleles of Pakistani populations. For MHC-I binding predictions, ANN and netMHCpan algorithms calculated the 736 interactions, of all selected class-I epitopes and their variants, with the prevalent class-I HLA alleles. Of these 736 interactions, twenty-seven interactions were the strong interactions with the IC50 value of less than 50 nM. (Additional File1, Fig. 4). Likewise, NNalign and netMHCIIpan algorithms, calculated total 189 interactions for class-II epitopes, and their variants with the class-II HLA alleles and thirty-five interactions were the strong interactions having IC50 threshold of less than 50 nM (Additional File 2, Fig. 4).
Fig. 4.
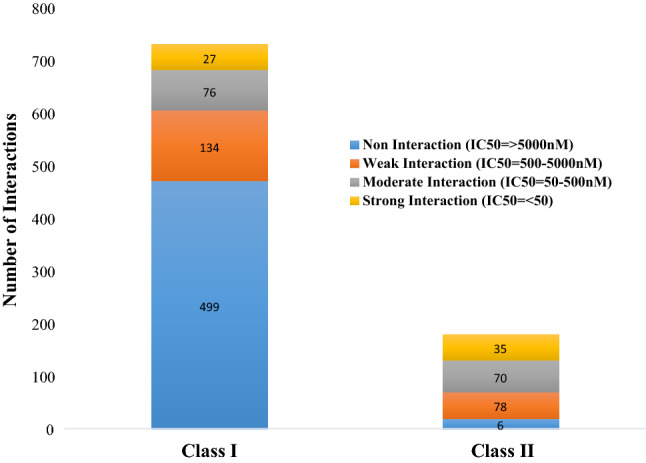
Chart summarizing the interactions between selected class-I and class-II epitopes as well as their variants with the prevalent HLA alleles of Pakistan
Allergenicity Assessment
AllerTop v.2.0 marked six of thirty-two epitopes from class-I, and eight of twenty-one epitopes from class-II, as non-allergens. The remaining class-I and class-II, epitopes were reported, as the potential allergens (Figs. 5 and 6 and Additional files 1 and 2).
Fig. 5.
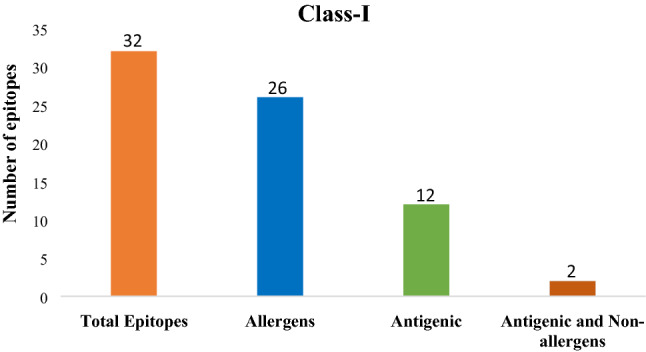
Epitope statistics: Number of antigenic and non-allergenic class-I peptide fragments, selected after the exclusion of allergenic and non-antigenic peptides
Fig. 6.
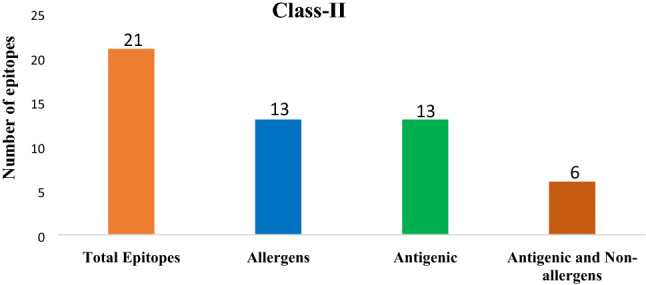
Epitope statistics: Number of antigenic and non-allergenic class-II peptide fragments, selected after the exclusion of allergenic and non-antigenic peptides
Antigenicity of Epitopes
Vaxijen v.2.0 server showed that twelve of thirty-two class-I epitopes and, thirteen of twenty-one class-II epitopes, were possessing antigenic nature (Figs. 5 and 6 and Additional files 1 and 2).
Population Coverage
For population coverage analysis, class-I and class-II epitopes, with antigenic as well as non-allergenic natures and having strong binding affinities, with prevalent HLA alleles in Pakistan were considered relevant. Based on these selection parameters, one class-I epitope (LTDGNPPEV) from lipoprotein LpqH and two class-II epitopes, one from the Phosphate-binding protein PstS1 (QVHFQPLPPAVVKLS) and second from ESAT-6-like protein EsxB (QAAGVRFQEAANKQK) were selected. Population coverage analysis revealed that the mixture of these three epitopes covers 67.28% of Pakistani Asian (Table 4) and 57.15% of Pakistani mixed populations (Table 5).
Table 4.
Population coverage for Pakistani Asian population
| Epitope | Coverage | HLA allele (genotypic frequency (%)) | Total HLA hits | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class I and II | HLA-A*01:01 (4.99) | HLA-A*02:01 (8.68) | HLA-DQB1*03:01 (11.63) | HLA-DQB1*06:01 (13.00) | HLA-DQB1*06:02 (5.80) | HLA-DRB1*10:01 (3.40) | HLA-DRB1*13:01 (1.10) | HLA-DRB1*13:02 (0.73) | ||
| Epitope #1: QVHFQPLPPAVVKLS | 48.98% | − | − | + | + | − | + | + | + | 5 |
| Epitope #2: QAAGVRFQEAANKQK | 31.82% | − | − | + | − | + | − | − | − | 2 |
| Epitope #3: LTDGNPPEV | 24.73% | + | + | − | − | − | − | − | − | 2 |
| Epitope set | 67.28% | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 |
Table 5.
Population coverage for Pakistani mixed population
| Epitope | Coverage | HLA allele (genotypic frequency (%)) | Total HLA hits | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class I and II | HLA-A*01:01 (13.05) | HLA-A*02:01 (7.60) | HLA-DQB1*03:01 (14.54) | HLA-DQB1*06:01 (0.0) | HLA-DQB1*06:02 (0.0) | HLA-DRB1*10:01 (4.00) | HLA-DRB1*13:01 (0.0) | HLA-DRB1*13:02 (0.00) | ||
| Epitope #1: QVHFQPLPPAVVKLS | 32.69% | − | − | + | + | − | + | + | + | 5 |
| Epitope #2: QAAGVRFQEAANKQK | 26.97% | − | − | + | − | + | − | − | − | 2 |
| Epitope #3: LTDGNPPEV | 36.34% | + | + | − | − | − | − | − | − | 2 |
| Epitope set | 57.15% | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 |
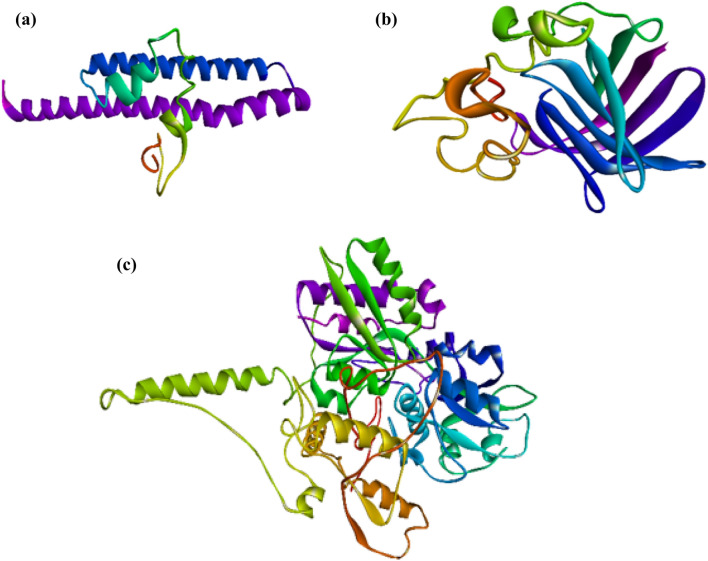
3D structure modelling and validation
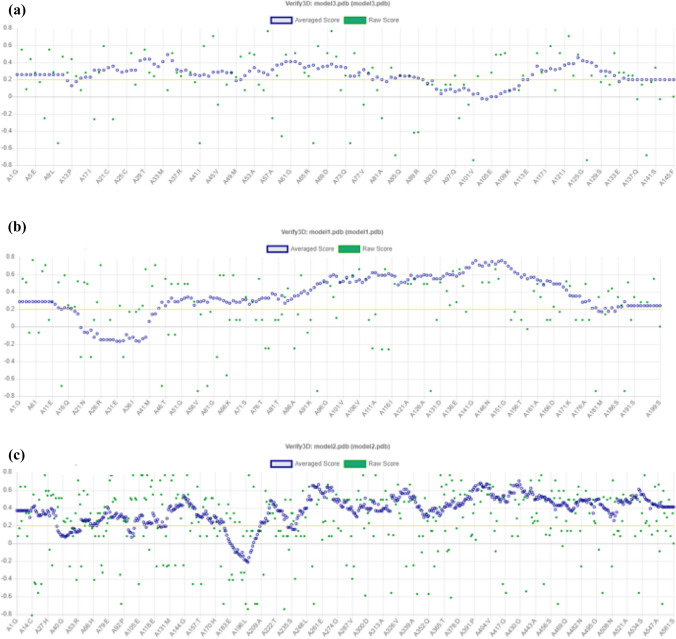
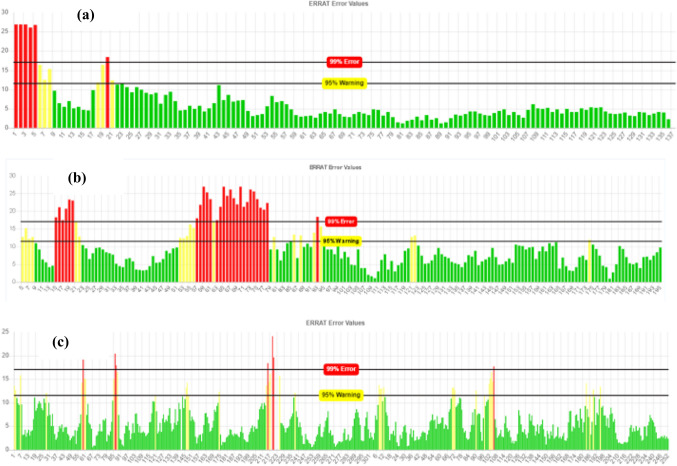
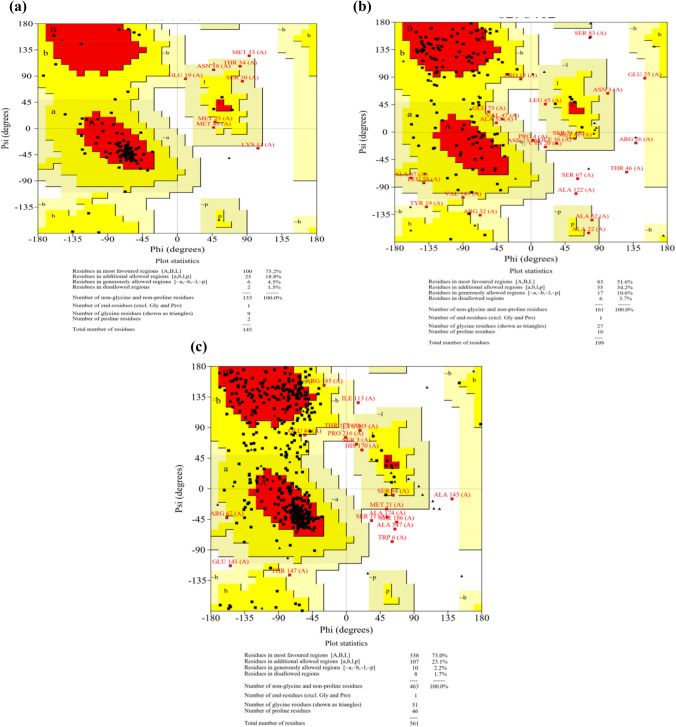
The 3D structure of three proteins (ESAT-6 like protein EsxB, Lipoprotein LpqH, and Phosphate binding protein PstS 1), containing finally selected epitopes, were modelled using I-TASSER (Fig. 7a–c). After structural modelling, Verify 3D, ERRAT, and PROCHECK from SAVES server were used for the model validations (Mahmood et al. 2020). The verify3D evaluates these models by determining the compatibility of models with amino acid sequences and assigned the structural scores for the alpha, beta, loop, polar, and non-polar to these sequences. The score ranges from − 1 (poor score) to + 1 (good score). 81.38% of the sequence in ESAT 6 like protein EsxB (Fig. 8a), 84.92% in lipoprotein LpqH (Fig. 8b) and, 87.34% in Phosphate binding protein PstS1 (Fig. 8c) were having the verify-3D score > 0.2, ensuring good quality of the protein models. The non-bonded interactions of atoms of these three protein models were checked using ERRAT. The overall quality factor of the three models was greater than 70Ao (Fig. 9a–c). Finally, the stereochemical properties of protein models were, verified by Ramachandran plots generated by the PROCHECK server, which shows 75.2% residues of ESAT-6 like protein EsxB (Fig. 10a), 51.6% residues of Lipoprotein LpqH (Fig. 10b) and 73.0% residues of Phosphate binding protein PstS 1 (Fig. 10c) were in the most favourable regions, while 18.8, 34.2 and 23.1% residues from these three proteins, respectively, were in the allowed regions.
Fig. 7.
Three-dimensional structure of proteins obtained from I-TASSER a ESAT-6 like protein EsxB b Lipoprotein LpqH c Phosphate binding protein PstS 1
Fig. 8.
Model quality analysis through VERIFY3D a ESAT-6 like protein EsxB b Lipoprotein LpqH c Phosphate binding protein PstS 1
Fig. 9.
Non-bonded interaction of atoms checked by ERRAT a ESAT-6 like protein EsxB b Lipoprotein LpqH c Phosphate binding protein PstS 1
Fig. 10.
a–c Ramachandran plots generated by PROCHECK for stereochemical analysis of protein models
Linear and Conformational B Cell Epitope Prediction
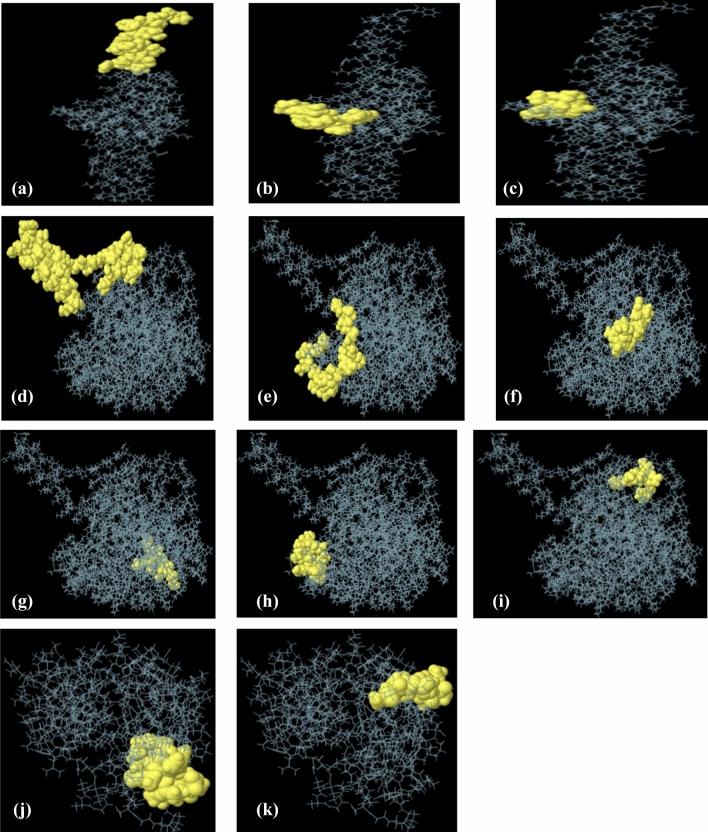
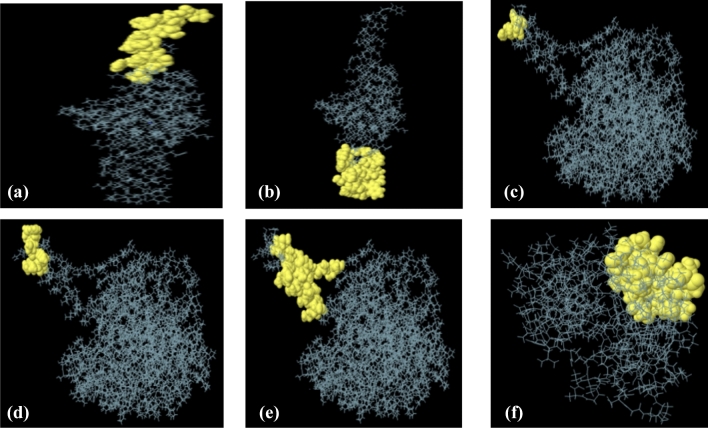
IEDB Ellipro tool identified linear and conformational B-cell epitopes with default epitope prediction parameters. A total of twenty-nine linear epitopes, eighteen from Phosphate binding protein PstS 1, four from ESAT 6 like protein EsxB and, seven from lipoprotein LpqH, were selected with a protrusion index (PI) score in between 0.8 and 0.5. Of these, three epitopes from ESAT 6 like protein EsxB, six from phosphate-binding protein PstS1 and, two from lipoprotein LpqH, were found non-allergenic and antigenic (Table 6). Moreover, the position of these eleven linear B-cell epitopes on corresponding protein models, are also presented in Fig. 11. In addition to linear B-cell epitopes, a total of seventeen conformational epitopes, from three proteins (Phosphate binding protein PstS 1, ESAT 6 like protein EsxB and lipoprotein LpqH), were also predicted using the IEDB Ellipro tool. The epitopes having prediction scores, between 0.704 and 0.992, were taken into considerations. The higher score designates the high solvent accessibility (Mishra et al. 2018). The information about the amino acid residues, length, prediction score, and the position of selected conformational epitopes, are mentioned in (Table 7 and Fig. 12).
Table 6.
Linear B cell epitopes of Mycobacterium tuberculosis
| Protein name | Epitope sequence | Allergenicity | Antigenicity | No. of residues | Score | 3D structure |
|---|---|---|---|---|---|---|
| ESAT-6-like protein EsxB | SRADEEQQQALSSQMGF | Non-allergen | Antigen | 17 | 0.832 | Figure 11 a |
| IKEPRTEINES | Non-allergen | Antigen | 11 | 0.767 | Figure 11 b | |
| GTNPESA | Non-Allergen | Antigen | 7 | 0.646 | Figure 11 c | |
| Phosphate-binding protein PstS 1 | PRTEINANTIGENNSHRTPANFLAGSPRECRSRMKIRLHTLLAVLTAAPLLL | Non-allergen | Antigen | 52 | 0.836 | Figure 11 d |
| KPPSGSPETGAGAGTVATTPASSPVT | Non-allergen | Antigen | 26 | 0.763 | Figure 11 e | |
| YCLIPPRTEINA | Non-allergen | Antigen | 12 | 0.712 | Figure 11 f | |
| SQRGLGEAQ | Non-allergen | Antigen | 9 | 0.601 | Figure 11 g | |
| QKDAATAQTLQA | Non-allergen | Antigen | 12 | 0.602 | Figure 11 h | |
| PRTEINSH | Non-allergen | Antigen | 8 | 0.596 | Figure 11 i | |
| Lipoprotein LpqH | SGTGQGNASATKDGSH | Non-allergen | Antigen | 16 | 0.742 | Figure 11 j |
| DGKDQNV | Non-allergen | Antigen | 7 | 0.549 | Figure 11 k |
Fig. 11.
Three-dimensional presentation of linear epitopes (a–k). Yellow surface represented the epitopes while grey color represented the protein
Table 7.
Conformational B cell epitopes of Mycobacterium tuberculosis
| Protein name | Residues | No. of residues | Score | 3D structure |
|---|---|---|---|---|
| ESAT-6-like protein EsxB | A:V126, A:S129, A:R130, A:A131, A:D132, A:E133, A:E134, A:Q135, A:Q137, A:A138, A:L139, A:S140, A:S141, A:Q142, A:M143, A:G144, A:F145 | 17 | 0.814 | Figure 12a |
| A:E78, A:S79, A:T80, A:A81, A:G82, A:S83, A:L84, A:Q85, A:G86, A:Q87, A:W88, A:R89, A:G90, A:A91, A:A92, A:G93, A:T94, A:A95, A:A96, A:Q97, A:A98, A:V101, A:R102 | 23 | 0.755 | Figure 12b | |
| Phosphate-binding protein PstS 1 | A:R182, A:E183, A:R187 | 3 | 0.992 | Figure 12c |
| A:C184, A:R185, A:S186, A:K189, A:I190, A:R191 | 6 | 0.988 | Figure 12d | |
| A:T172, A:P173, A:A174, A:N175, A:F176, A:L177, A:A178, A:G179, A:S180, A:P181, A:M188, A:L192, A:H193, A:T194, A:L195, A:L196, A:A197, A:V198, A:L199, A:T200, A:A201, A:A202, A:P203, A:L204, A:L205, A:L206 | 26 | 0.877 | Figure 12e | |
| Lipoprotein LpqH | A:D95, A:G96, A:K97, A:V145, A:N146, A:G147, A:T177, A:G178, A:V179, A:D180, A:M181, A:A182, A:N183, A:P184, A:M185, A:S186, A:P187, A:V188, A:N189, A:K190, A:S191 | 21 | 0.704 | Figure 12f |
Fig. 12.
Three-dimensional presentation of conformational epitopes (a–f). Yellow surface represented the epitopes while grey color represented the protein
Discussion
Recently, the rapidly mutating genome of Mycobacterium tuberculosis and the chances of disease reoccurrence have made almost all available treatment options for this pathogen inefficient (Ford et al. 2013). Moreover, with the emergence of multidrug-resistant TB strains has further curtailed the efficacy of these treatment options (Falzon et al. 2013). This catastrophe has transformed the research trends toward designing the epitope-based vaccines, which are the most recent and reliable method in vaccine designing. The epitope-based vaccines are advantageous over trivial vaccines because of their ability to aptly combat mutating pathogens and, producing prominent immune responses (Oyarzún and Kobe 2016). Moreover, by directing the immune response against highly immunogenic components of the pathogen, these vaccines, guarantee an ideal immune elicitation (Purcell et al. 2007). Additionally, these vaccines are safe, efficient, long-lasting, and more economical. Despite these obvious benefits, the epitope prediction, their contribution to immune elicitation, and experimental validations are quite laborious (Sette and Fikes 2003). The advancements in the field of bioinformatics have proved helpful for vaccine development (Barh et al. 2010). In-silico approaches have helped in the screening of putative epitopes from the pathogenic proteins of different organisms. It is also possible to distinguish significantly allergenic vaccine targets likely to induce allergenic reactions in hosts. During immune elicitation, antigen-presenting cells (APCs) display the epitopes on their surface in complex with the MHC molecules. It is, therefore, essential to screen prevalent HLA alleles in the population of interest and to assess the binding affinities of putative epitopes with these alleles (Braciale et al. 1987).
These strategies help in designing population-specific vaccines and have been successful for various pathogens (Ashraf et al. 2016; Chakraborty et al. 2010; Khan et al. 2015; Mahmood et al. 2019; Moradi et al. 2017). In the recent past, a few studies on epitope-based vaccine designing for Mtb have come forward, Mistry and Flower, in 2017, designed an epitope ensemble against TB, however, they did not mention the allergenic natures as well as the population specificity for this epitope ensemble (Mistry and Flower 2017). In Rai et al. 2018 reported a bi-epitope vaccine, namely L4.8, comprising of MHC-I and MHC-II binding peptides of active (TB10.4) and latent (Acr1) stages of Mtb antigens, respectively. L4.8 significantly elicited CD8 + and CD4 + T cells and was claimed for better protection against Mtb infection than BCG. It is, however, not approved clinically and needs validation on model organisms (Rai et al. 2018). In 2019, an immunoinformatics based study from Elhag Mustafa et al. reported four immunogenic peptides, from the conserved regions of the PPE65 family protein of Mycobacterium tuberculosis, along with the population coverage of these epitopes. However, allergenic assessment of the selected peptides was missing. Moreover, the study has ignored the highly referenced pathogenic proteins of tuberculosis from, which experimentally validated peptides, are listed in IEDB (Elhag et al. 2019). Herein, we present a computational strategy, to report the significantly immunogenic and non-allergenic B and T-cell epitopes restricting to the common HLA alleles of Pakistani populations, as the potential vaccine candidates for these populations. The study starts by selecting ten experimentally validated and highly referenced MHC-I and MHC-II epitopes from the Immune Epitope Database (IEDB) (Table 1). Considering the conservancy of ten selected peptides via weblogo3, all possible variants of original peptide fragments were derived and analyzed together with the epitopes (Table 2). The conservancy analysis is important because it considers not only the original epitope sequence but also the all possible variants that could derive from the parent epitope, in any strain of the pathogen. Thus, it helps in the broad-level screening of all possible peptide fragments for their possible antigenic potential and immune elicitation. The interactions between MHC-I and MHC-II epitopes with the class-I and class-II HLA alleles are essential for the antigen presentations and, therefore, for the elicitation of the immune response (Arlehamn et al. 2014; Ivanyi 2014). These alleles are highly polymorphic and vary among the populations, and the assessments of prevalent HLA alleles from the population of interest are significantly important while designing population-based vaccines (Shahsavar et al. 2017). AFND marked sixty-eight class-I and twenty-six class-II HLA alleles as the significantly prevalent in Pakistani populations (Additional file 3). The peptide fragments showing binding affinities to the selected class-I and class-II HLA alleles were selected at IC50 value < 50 nM (Additional files 1 and 2).
The immunogenicity and allergenicity assessments, respectively, via Vaxijen and AllerTop, helped in the selection of one class-I, LTDGNPPEV, and two class-II epitopes, QVHFQPLPPAVVKLS and QAAGVRFQEAANKQK as ideal vaccine candidates. The three peptide fragments were immunogenic, non-allergenic, and restricting to the broad range of the HLA alleles of the Pakistani populations. The combination of these three epitopes was covering 67.28% of Pakistani Asian and 57.15% of mixed Pakistani populations (Tables 4 and 5). The low population coverage, in this case, maybe because of the limited information on Pakistani HLA available in AFND. The information about the subtypes of most HLA alleles was missing for Pakistani populations.
In the adaptive immune system, humoral, or antibody-based response forms the first line of defence against most of the pathogens. Antibodies can recognize the linear or conformational B-cell epitopes and initiate an immune response against them (Ahmad et al. 2016). Thus, in addition to the T-cell epitopes, the screening of linear as well as conformational B-cell epitopes and their characterization is precisely important in vaccine designing. The three immunogenic proteins from Mtb including, ESAT-6 like protein EsxB, Lipoprotein LpqH and Phosphate binding protein PstS 1 were selected for the prediction of B-cell epitopes based on extensive literature review as well as high references in IEDB for these proteins (Birhanu et al. 2019; Mistry and Flower 2017; Monterrubio-López and Ribas-Aparicio 2015; Mustafa 2013; Zhang et al. 2009). After 3D structural modelling and validation, the protein models were used for predicting B-cell epitopes. A total of eleven linear and six conformational B-cell epitopes retrieved through IEDB, ElliPro tool possess the antigenic and non-allergic properties similar to T-cell epitopes and selected. This analysis suggests that the identified B and T-cell epitopes and are likely to elicit a maximum immune response against Mtb infections in Pakistani populations without any allergenic side effects.
Conclusion
In this study, we report B and T-cell epitopes, which are significantly antigenic, non-allergenic, and restrict to the majority of the Pakistani HLA alleles, therefore, can be used to design an epitope-based vaccine against these populations. These epitopes need further experimental validation through wet-lab practices.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We like to acknowledge Mr Aadil Hussain for his help in graphics editing. Authors are also thankful for the online available open access databases which were used in this research. The research was self-supported by the authors.
Compliance with Ethical Standards
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Malik Siddique Mahmood, Email: Siddique.phd.ibb@pu.edu.pk.
Duaa Bin-T-Abid, Email: duaabintabid@gmail.com.
Saba Irshad, Email: saba.ibb@pu.edu.pk.
Hina Batool, Email: hina.batool@umt.edu.pk.
References
- Ahmad TA, Eweida AE, Sheweita SA. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinol. 2016;5:71–83. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Annabel B, Anna D, Hannah M. Global tuberculosis report 2019. Geneva: World Health Organization; 2019. [Google Scholar]
- Arlehamn CSL, Lewinsohn D, Sette A, Lewinsohn D. Antigens for CD4 and CD8 T cells in tuberculosis. Cold Spring Harb Perspect Med. 2014;4:a018465. doi: 10.1101/cshperspect.a018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf NM, Bilal M, Mahmood MS, Hussain A, Mehboob MZ. In-silico analysis of putative HCV epitopes against Pakistani human leukocyte antigen background: an approach towards development of future vaccines for Pakistani population Infection. Genet Evolut. 2016;43:58–66. doi: 10.1016/j.meegid.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Barh D, Misra AN, Kumar A, Vasco A. A novel strategy of epitope design in Neisseria gonorrhoeae. Bioinformation. 2010;5:77. doi: 10.6026/97320630005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D, Lipman DJ, Ostell J. GenBank. Nucleic Acids Res. 1993;21:2963–2965. doi: 10.1093/nar/21.13.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birhanu AG, et al. Ample glycosylation in membrane and cell envelope proteins may explain the phenotypic diversity and virulence in the Mycobacterium tuberculosis complex. Sci Rep. 2019;9:1–15. doi: 10.1038/s41598-019-39654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braciale TJ, Morrison LA, Sweetser MT, Sambrook J, Gething MJ, Braciale VL. Antigen presentation pathways to class I and class II MHC-restricted T lymphocytes. Immunol Rev. 1987;98:95–114. doi: 10.1111/j.1600-065x.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Campbell IA, Bah-Sow O. Pulmonary tuberculosis: diagnosis and treatment BMJ. BMJ. 2006;332:1194. doi: 10.1136/bmj.332.7551.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, et al. A computational approach for identification of epitopes in dengue virus envelope protein: a step towards designing a universal dengue vaccine targeting endemic regions. In silico Biol. 2010;10:235–246. doi: 10.3233/ISB-2010-0435. [DOI] [PubMed] [Google Scholar]
- Colovos C, Yeates T. ERRAT: an empirical atom-based method for validating protein structures. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse SE, Cox JS. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol. 2005;187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Neto E. MHC-restricted antigen presentation and recognition: constraints on gene, recombinant and peptide vaccines in humans. Braz J Med Biol Res. 1999;32:199. doi: 10.1590/s0100-879x1999000200008. [DOI] [PubMed] [Google Scholar]
- Denisova GF, Denisov DA, Bramson JL. Applying bioinformatics for antibody epitope prediction using affinity-selected mimotopes–relevance for vaccine design. Immunome Res. 2010;6:1–8. doi: 10.1186/1745-7580-6-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Flower DR, Doytchinova I. AllerTOP-a server for in silico prediction of allergens. BMC Bioinform. 2013;6:1–9. doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Lüthy R, Bowie JU. [20] VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Elhag M, Sati AOM, Saadaldin MM, Hassan MA. Immunoinformatics prediction of epitope based peptide vaccine against mycobacterium tuberculosis PPE65 family protein. bioRxiv. 2019 doi: 10.1101/755983. [DOI] [Google Scholar]
- Falzon D, et al. Resistance to fluoroquinolones and second-line injectable drugs: impact on multidrug-resistant TB outcomes. Eur Respir J. 2013;42:156–168. doi: 10.1183/09031936.00134712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CB, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazi MA, et al. Functional, structural and epitopic prediction of hypothetical proteins of Mycobacterium tuberculosis H37Rv: an in silico approach for prioritizing the targets. Gene. 2016;591:442–455. doi: 10.1016/j.gene.2016.06.057. [DOI] [PubMed] [Google Scholar]
- Glaziou P, Sismanidis C, Floyd K, Raviglione M. Global epidemiology of tuberculosis. Cold Spring Harb Perspect Med. 2015;5:a017798. doi: 10.1101/cshperspect.a017798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2010;39:D913–D919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung RB, Purdie AC, Begg DJ, Whittington RJ. In silico identification of epitopes in Mycobacterium avium subsp. paratuberculosis proteins that were upregulated under stress conditions. Clin Vaccine Immunol. 2012;19:855–864. doi: 10.1128/CVI.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseman M. Tuberculosis therapy: past, present and future. Eur Respir J. 2002;20:87S–94s. doi: 10.1183/09031936.02.00309102. [DOI] [PubMed] [Google Scholar]
- Ivanyi J. Function and potentials of M. tuberculosis epitopes. Front Immunol. 2014;5:107. doi: 10.3389/fimmu.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M. NetMHCIIpan-3. 0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 2013;65:711–724. doi: 10.1007/s00251-013-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Hussey G, Lambert P-H. New vaccines for tuberculosis. Lancet. 2010;375:2110–2119. doi: 10.1016/S0140-6736(10)60393-5. [DOI] [PubMed] [Google Scholar]
- Khan M, Hossain M, Rakib-Uz-Zaman S, Morshed M. Epitope-based peptide vaccine design and target site depiction against Ebola viruses: an immunoinformatics study. Scand J Immunol. 2015;82:25–34. doi: 10.1111/sji.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knechel NA. Tuberculosis: pathophysiology, clinical features, and diagnosis. Critic Care Nurse. 2009;29:34–43. doi: 10.4037/ccn2009968. [DOI] [PubMed] [Google Scholar]
- Koşaloğlu-Yalçın Z, et al. Predicting T cell recognition of MHC class I restricted neoepitopes. Oncoimmunology. 2018;7:e1492508. doi: 10.1080/2162402X.2018.1492508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK. Peptide vaccine: progress and challenges. Vaccines. 2014;2:515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood MS, Asad-Ullah M, Batool H, Batool S, Ashraf NM. Prediction of epitopes of Neisseria Gonorrhoeae against USA human leukocyte antigen background: an immunoinformatic approach towards development of future vaccines for USA population. Mol Cell Probes. 2019;43:40–44. doi: 10.1016/j.mcp.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Mahmood MS, Irshad S, Butt TA, Batool H, Batool S, Ashraf NM. In-silico analysis of deleterious missense SNPs of human TYR gene associated with oculocutaneous albinism type 1 (OCA1) Meta Gene. 2020;24:100674. [Google Scholar]
- María R, Arturo C, Alicia JA, Paulina M, Gerardo AO. The impact of bioinformatics on vaccine design and development. Vaccines. 2017 doi: 10.5772/intechopen.69273. [DOI] [Google Scholar]
- Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- Mishra A, Ranganathan S, Jayaram B, Sattar A. Role of solvent accessibility for aggregation-prone patches in protein folding. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-31289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Flower DR. Designing epitope ensemble vaccines against TB by selection: prioritizing antigens using predicted immunogenicity. Bioinformation. 2017;13:220. doi: 10.6026/97320630013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterrubio-López GP, Ribas-Aparicio RM. Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology. BioMed Res Int. 2015 doi: 10.1155/2015/483150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi J, Tabrizi M, Izad M, Mosavari N, Feizabadi MM. Designing a novel multi-epitope DNA-based vaccine against tuberculosis: in silico approach. Jundishapur J Microbiol. 2017 doi: 10.5812/jjm.43950. [DOI] [Google Scholar]
- Munir MK, Rehman S, Iqbal R. Meeting the challenge, making a difference: multidrug resistance tuberculosis in Pakistan. Pakistan J Med Res. 2018;57:1–2. [Google Scholar]
- Mustafa A. Diagnostic and vaccine potentials of ESAT-6 family proteins encoded by M. tuberculosis genomic regions absent in M. bovis. BCG Mycobact Dis J Mycobact Dis. 2013;3:129. [Google Scholar]
- Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Andreatta M. NNAlign: a platform to construct and evaluate artificial neural network models of receptor–ligand interactions. Nucleic Acids Res. 2017;45:W344–W349. doi: 10.1093/nar/gkx276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP NTcP (2016) Annual report
- Oyarzún P, Kobe B. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum Vaccines Immunother. 2016;12:763–767. doi: 10.1080/21645515.2015.1094595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs P, et al. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect Immun. 2005;73:1898–1902. doi: 10.1128/IAI.73.3.1898-1902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J, Bui H-H, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6:404. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- Rai PK, et al. A lipidated bi-epitope vaccine comprising of MHC-I and MHC-II binder peptides elicits protective CD4 T cell and CD8 T cell immunity against Mycobacterium tuberculosis. J Transl Med. 2018;16:1–12. doi: 10.1186/s12967-018-1653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, et al. Effect of BCG vaccination against Mycobacterium tuberculosis infection in children: systematic review and meta-analysis. BMJ. 2014;349:g4643. doi: 10.1136/bmj.g4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez A, Espinosa P, García T, Mancilla R. The 19 kDa Mycobacterium tuberculosis lipoprotein (LpqH) induces macrophage apoptosis through extrinsic and intrinsic pathways: a role for the mitochondrial apoptosis-inducing factor. Clin Dev Immunol. 2012;2012:950503. doi: 10.1155/2012/950503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Fikes J. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr Opin Immunol. 2003;15:461–470. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- Shahsavar F, Varzi A-M, Ahmadi SAY. A genomic study on distribution of human leukocyte antigen (HLA)-A and HLA-B alleles in Lak population of Iran. Genomics Data. 2017;11:3–6. doi: 10.1016/j.gdata.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnetcioglu A, Sunnetcioglu M, Binici I, Baran AI, Karahocagil MK, Saydan MR. Comparative analysis of pulmonary and extrapulmonary tuberculosis of 411 cases. Ann Clin Microbiol Antimicrob. 2015;14:34. doi: 10.1186/s12941-015-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot AC, et al. Versatile virus-like particle carrier for epitope based vaccines. PLoS ONE. 2010;5:e9809. doi: 10.1371/journal.pone.0009809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar N, De RK. Immunoinformatics: an integrated scenario. Immunology. 2010;131:153–168. doi: 10.1111/j.1365-2567.2010.03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I, Javaid A, Tahir Z, Ullah O, Shah AA, Hasan F, Ayub N. Pattern of drug resistance and risk factors associated with development of drug resistant Mycobacterium tuberculosis in Pakistan. PLoS ONE. 2016;11:e0147529. doi: 10.1371/journal.pone.0147529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CA, Mehta PJ, Ardito M, Moise L, Martin B, De Groot AS. T cell epitope: friend or foe? Immunogenicity of biologics in context. Adv Drug Deliv Rev. 2009;61:965–976. doi: 10.1016/j.addr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Jonsson J, Sjörström M, Sandberg M, Rännar S. DNA and peptide sequences and chemical processes multivariately modelled by principal component analysis and partial least-squares projections to latent structures. Anal Chim Acta. 1993;277:239–253. [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12:7. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. Screening and assessing 11 Mycobacterium tuberculosis proteins as potential serodiagnostical markers for discriminating TB patients from BCG vaccinees. Genomics Proteomics Bioinform. 2009;7:107–115. doi: 10.1016/S1672-0229(08)60039-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, et al. Detecting distant-homology protein structures by aligning deep neural-network based contact maps. PLoS Comput Biol. 2019;15:e1007411. doi: 10.1371/journal.pcbi.1007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.