Abstract
SARS-CoV-2 utilizes the IMPα/β1 heterodimer to enter host cell nuclei after gaining cellular access through the ACE2 receptor. Ivermectin has shown antiviral activity by inhibiting the formation of the importin-α (IMPα) and IMPβ1 subunits as well as dissociating the IMPα/β1 heterodimer and has in vitro efficacy against SARS-CoV-2. Plasma and lung ivermectin concentrations vs. time profiles in cattle were used to determine the apparent plasma to lung tissue partition coefficient of ivermectin. This coefficient, together with a simulated geometric mean plasma profile of ivermectin from a published population pharmacokinetic model, was utilized to develop a minimal physiologically-based pharmacokinetic (mPBPK) model. The mPBPK model accurately described the simulated ivermectin plasma concentration profile in humans. The mPBPK model was also used to simulate human lung exposure to ivermectin after 12, 30, and 120 mg oral doses. The simulated ivermectin lung exposures reached a maximum concentration of 772 ng/mL, far less than the estimated 1750 ng/mL IC50 reported for ivermectin against SARS-CoV-2 in vitro. Further studies of ivermectin either reformulated for inhaled delivery or in combination with other antivirals with differing mechanisms of action is needed to assess its therapeutic potential.
Keywords: COVID-19, Importins, Ivermectin, Minimal physiologically-based pharmacokinetic model, SARS-CoV-2, Pharmacokinetics, Pharmacometrics, Physiologically based pharmacokinetic modeling, Kinetics, Pharmacokinetic/pharmacodynamic (PK/PD) modeling
Introduction
The novel SARS-CoV-2 coronavirus causes COVID-19 and has caused a global pandemic resulting in over twenty-four million infections and eight hundred thousand deaths at the time of writing. Healthcare providers and researchers are currently repurposing approved drugs in an attempt to treat COVID-19 patients as urgent efforts to develop effective vaccines and treatments continue.
SARS-CoV-2 causes infection through entering host cells by binding to angiotensin-converting enzyme 2 (ACE2) receptors, which are present in many tissues but predominantly human alveolar epithelial cells.1 , 2 Ivermectin is an approved human and animal anthelmintic which has also shown antiviral properties. Ivermectin inhibits the formation of the importin-α (IMPα) and IMPβ1 subunits as well as causes the dissociation of the formed IMPα/β1 heterodimer.3, 4, 5, 6, 7 The IMPα/β1 heterodimer is thought to be used by the SARS-CoV-2 nucleoprotein to infiltrate the nucleus, thereby delaying cellular proliferation and establishing an optimal environment inside the host cell for virus assembly and replication.8, 9, 10 Previous studies have shown in vitro activity of ivermectin against other RNA viruses such as the Zika, dengue-related West Nile, dengue, human immunodeficiency, and influenza viruses, which also utilize the IMPα/β1 heterodimer to enter host cell nuclei.3 , 7 More recently, ivermectin has demonstrated in vitro activity against SARS-CoV-2 with a reported IC50 value of 1750 ng/mL (2 μM).11
Understanding the pharmacokinetics (PK) and penetration of ivermectin to the site of infection and intended action, the lung in the case of SARS-CoV-2, is an important step in transferring in vitro knowledge to potential in vivo utilization. Therefore, the objective of this study was to develop a minimal physiologically-based pharmacokinetic (mPBPK) model to simulate human lung exposure of ivermectin after oral administration.
Materials and Methods
Ivermectin Pharmacokinetic Data
Ivermectin lung and plasma concentrations in Holstein calves were obtained from studies performed by Lifschitz and colleagues (including unpublished data found in Table S1 of the supplementary material).12 Calves were treated with ivermectin by subcutaneous injection at 0.2 mg/kg using a commercially available formulation for cattle. Lung homogenates from the left lobus cranialis and plasma samples obtained between 1 and 48 days post-treatment were analyzed by high-performance liquid chromatography. Animal procedures were performed according to the Animal Welfare Policy of the Faculty of Veterinary Medicine, Universidad Nacional del Centro de la Provincia de Buenos Aires (UNCPBA), Tandil, Argentina.
Parameter estimates from a phase 1 study assessing the PK of ivermectin in 18 subjects receiving a 0.2 mg/kg dose orally were utilized to simulate a geometric mean plasma concentration profile in humans without residual unexplained variability (RUV) or interindividual variability using Phoenix® WinNonlin (Version 8.2, Certara USA, Inc.; Princeton, NJ).13 The average dose administered in the study, 15 mg, was used for the simulation.
Parameters
Physiologic parameters for cardiac output, lung, and plasma volume were based on population averages.14 , 15 The species-independent apparent lung partition coefficient, Kp, lung/F, was determined by dividing ivermectin lung area under the curve (AUC) by the plasma AUC obtained from the cattle study.12 The absorption rate (Ka), apparent plasma clearance (CLp/F), fraction of cardiac output to other tissues (Fraction), apparent partition coefficient (Kp, other/F), and transit rate (Ktr) for humans were estimated using the matrix exponential solver in Phoenix® WinNonlin (Version 8.2, Certara USA, Inc.). A multiplicative error model was evaluated to describe the RUV of the simulated human plasma data during the mPBPK model development.
Human Lung Exposure Simulation
Human lung and plasma exposure simulations were performed with the final model parameters reported in Table 1 as well as an assumed 20% RUV in R (version 4.0.2) using the mrgsolve package (version 0.10.4) by simulating 1000 patients at observation time intervals of 0.1 h. Therapeutic and supra-therapeutic single doses of ivermectin (12, 30, and 120 mg) were evaluated. The 12 mg dose was simulated to represent the typical single dose of ivermectin administered to humans while the supratherapeutic 30 and 120 mg doses were simulated since these single doses have been shown to be safe with minimal toxicities and exhibiting linear pharmacokinetics.16 A 95% confidence interval of the simulated human plasma and lung PK at each dose was constructed based on the 1000 simulations performed.
Table 1.
Model Predicted Pharmacokinetic Parameter Estimates and Resulting Precision.
| Parameter | Description | Estimate (CV%) | Units |
|---|---|---|---|
| Ka | Absorption rate | 0.14 (2.52) | Hr−1 |
| Vpa | Plasma volume | 315 | L |
| CLp/F | Plasma clearance | 10.85 (3.78) | L/hr |
| VLa | Lung volume | 1.314 | L |
| Kp, lung/Fa | Lung partition coefficient | 2.68 | – |
| Qcoa | Cardiac output | 28215 | L/hr |
| VOthera | Remaining volume | 65.7 | L |
| Kp, other/F | Tissue partition coefficient | 17.32 (5.6) | – |
| Fraction | Fraction of cardiac output | 0.08 (1.18) | – |
| Ktr | Transit rate | 0.36 (3.09) | hr−1 |
Parameter not estimated.
Results
Ivermectin Pharmacokinetic Data
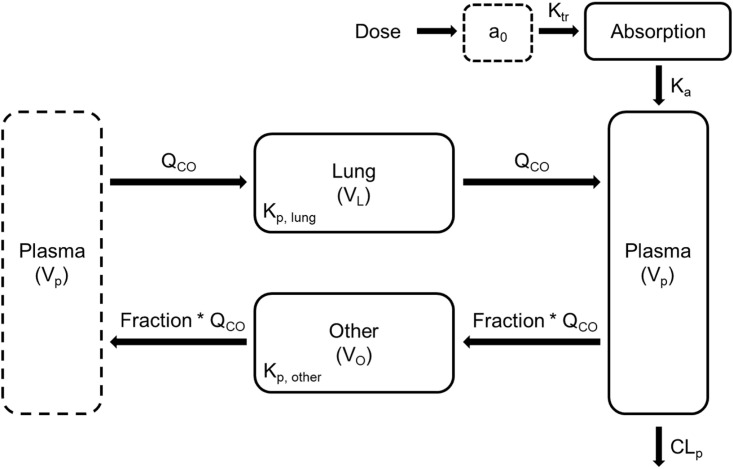
The resulting mPBPK model shown in Fig. 1 adequately captured the simulated geometric mean plasma PK profile in humans (Fig. 2 ).
Fig. 1.
Schematic of the developed mPBPK model for ivermectin following administration of an oral dose in humans (Dose). The plasma compartment (dashed line) on the left is identical to the plasma compartment on the right. The mPBPK model comprises two tissue compartments (Lung and Other) and a plasma compartment (Plasma). The lung receives all of the cardiac output while the other compartment describing the rest of the body receives a fraction of the cardiac output. The delay following oral dosing in humans is described by a transit compartment. Symbols are defined in Table 1.
Fig. 2.
Observations and predictions of the mPBPK model illustrated in Fig. 1. Simulated plasma concentrations from a historical Phase I study of patients receiving a 0.2 mg/kg dose of ivermectin are represented by red circles, and the model prediction is represented by the blue line.
Parameters
Model estimates and precision (CV%) for Ka, CLp/F, Fraction, Kp, other/F, and Ktr are shown in Table 1. Parameter precision around data-informed parameters such as Ka and CLp/F were below 5%. The Kp,other/F estimate of 17.3 conditioned on bioavailability, could be explained by ivermectin's large volume of distribution (estimated to be ~1077 L) as well as the potential poor bioavailability from oral administration of this highly lipophilic drug.13
Model Equations
The following system of ordinary differential equations describes the mPBPK model schematic shown in Fig. 1:
Ivermectin disposition in humans is described by an mPBPK model containing compartments for absorption, plasma, lung, and the rest of the body (Other). The transit compartment (a 0) accounts for the delay in transit through the gastrointestinal tract, and the clearance from plasma is linear. The delay and absorption are described by:
where K tr is the transit rate constant, a 0 is the amount of ivermectin in the transit compartment at a given time, K a is the first-order absorption rate constant, and Dose is the orally administered dose of ivermectin.
Disposition in plasma is described by:
where Lung, Plasma, and Other represent the amount of ivermectin and V L, V p, and V Other represent the physiological volumes of the lung, plasma, and the rest of the body, respectively. Q co is the cardiac output, and Fraction is the fraction of cardiac output that perfuses the rest of the body – the Other compartment. K p,lung /F is the cattle-informed apparent tissue partition coefficient, K p,other /F is the apparent tissue partition coefficient for the rest of the body – the Other compartment – and CL p /F is the apparent clearance of ivermectin from the plasma where F represents bioavailability.
Lung disposition is described by:
Disposition throughout the rest of the body is described by:
Human Lung Exposure Simulation
Human lung exposure simulations following oral administration of 120 mg ivermectin are shown in Fig. 3A while the plasma profile is shown in Fig. 3B. Profiles for human plasma and lung exposure simulations resulting from 12 to 30 mg doses are shown in Figure S1 in the supplementary material. The maximum simulated ivermectin concentrations in plasma and lung were 288 and 772 ng/mL respectively at 5.1 h which is similar to published time to maximum concentration values.17
Fig. 3.
Simulations following oral administration of a single 120 mg ivermectin dose. (A) Median ivermectin concentration in the lung (blue) and 95% CI from simulation of 1000 subjects. The dashed line represents the published in vitro ivermectin IC50 of 1750 ng/mL against SARS-CoV-2. (B) Median ivermectin concentration in the plasma (red) and 95% CI from simulation of 1000 subjects.
Discussion
Interest in the utilization of ivermectin against SARS-CoV-2 as a potential treatment for COVID-19 increased after the demonstration of its in vitro activity.11 To understand the predicted exposure of ivermectin in the lungs, where SARS-CoV-2 causes infection and diffuse alveolar damage, we developed an mPBPK model utilizing the cattle-informed Kp,lung/F parameter as well as the geometric mean plasma profile based on a published population PK model.
The population PK model was selected because it provided a conservative estimate of the mean plasma concentration profile that produced parameters in agreement with a published meta-analysis on ivermectin PK.13 , 17 In our opinion, these plasma data provide a conservative estimate of simulated lung exposure as the mPBPK model was developed based off the geometric mean plasma concentration profile. Our simulated peak lung concentration of 772 ng/mL following a single oral 120 mg dose is well below the reported in vitro IC50 of 1750 ng/mL as seen by other groups.11 , 18 , 19 The mPBPK model allows the simulation of different formulations (e.g., an ethanol-containing solution shown to have approximately twice the systemic availability of ivermectin tablets) as well as the effect of high fat meals to increase lung exposure to ivermectin.17 Reformulation of ivermectin to an inhaled therapy may increase drug exposure in the lungs while minimizing the potential toxicities associated with high plasma concentrations that have not been established as safe.
Ivermectin is a compelling candidate for consideration as part of a combination regimen by making the host cell environment unfavorable for SARS-CoV-2 assembly and replication after IMPα/β1 heterodimer dissociation and inhibition.8, 9, 10 Ivermectin is also lipophilic with a pKa > 8, which has been shown to be beneficial for uptake into the lungs from plasma (Fig. 3).20 We hypothesize that ivermectin in combination with other agents of differing antiviral mechanisms of action could be efficacious and/or reduce the ivermectin exposure necessary to eradicate SARS-CoV-2 through synergistic effects.
Although our model development and approach is unique compared to current literature, our results and conclusions are in agreement with previous analysis in regards to iveremctin's potential as a COVID-19 therapeutic agent.18 , 19 Our efforts are different in that we have provided a framework for mPBPK model development that can be utilized for any future compound thought to be a potential treatment option for COVID-19. Also, by providing the differential equations, future investigators will not be limited by the modeling software/tool available to them and will be able to implement this mPBPK model structure. Our mPBPK model can be applied to the future directions stated by previous investigators through enabling optimization of either systemic or inhalational doses.
The main limitations of our approach are the inability to validate our simulated human lung exposure with any published values as well as not being able to simulate drug accumulation in tissue after multiple dosing. Ivermectin is highly protein bound (~93%), and the proposed simulation represents the total ivermectin concentration.17 , 21 Total ivermectin concentrations were simulated because protein binding in the lung has not been assessed, but it is likely the free drug concentration is significantly lower than the 772 ng/mL peak lung concentration simulated which reinforces the need for an inhalational dose. Ruminants such as cattle have similarly high plasma binding compared to humans (96–99%), and total ivermectin concentration was assessed by Lifschitz and colleagues.12 , 22 Also, because of the similarity in plasma binding of ivermectin between species, the Kp,lung/F was considered species-independent due to the assumed similar impact plasma binding would have on distribution which is an assumption commonly made in physiologically based pharmacokinetic modeling.23 The low parameter estimate of 0.08 for the fraction of cardiac output that perfuses the rest of the body (Fraction) indicates the disposition of ivermectin is perfusion rate-limited. This is a common generic assumption for PBPK models and is also typical of small lipophilic molecules such as ivermectin (logP = 5.83).24
To conclude, we successfully developed an mPBPK model to simulate ivermectin lung exposure in humans after oral dosing. Simulated concentration profiles of single-dose ivermectin administered orally at doses proven to be safe in clinical studies did not achieve lung concentrations above the published in vitro IC50 concentration for SARS-CoV-2. Clinical trials of ivermectin reformulated for inhalational delivery or in vitro studies assessing synergy of ivermectin in combination with antiviral agents of differing mechanisms of action are needed to understand ivermectin's value in the treatment of COVID-19. This approach for model development can be used for current and future COVID-19 therapeutic agents of interest as the repurposing of approved compounds continues.
Footnotes
Funding: The study performed by Lifschitz and colleagues was partially supported by CONICET (Argentina), Universidad Nacional del Centro (Argentina), and Agencia Nacional de Promoción Científica y Tecnológica (PICT 08-00000-00817) Argentina.
Declarations of interest: None.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.xphs.2020.08.024.
Appendix A. Supplementary Data
References
- 1.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jans D.A., Martin A.J., Wagstaff K.M. Inhibitors of nuclear transport. Curr Opin Cell Biol. 2019;58:50–60. doi: 10.1016/j.ceb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Crump A. Ivermectin: enigmatic multifaceted ‘wonder’ drug continues to surprise and exceed expectations. J Antibiot (Tokyo) 2017;70(5):495–505. doi: 10.1038/ja.2017.11. [DOI] [PubMed] [Google Scholar]
- 5.Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443(3):851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tay M.Y., Fraser J.E., Chan W.K. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013;99(3):301–306. doi: 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Yang S.N.Y., Atkinson S.C., Wang C. The broad spectrum antiviral ivermectin targets the host nuclear transport importin alpha/beta1 heterodimer. Antiviral Res. 2020;177:104760. doi: 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- 8.Timani K.A., Liao Q., Ye L. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114(1-2):23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wulan W.N., Heydet D., Walker E.J., Gahan M.E., Ghildyal R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front Microbiol. 2015;6:553. doi: 10.3389/fmicb.2015.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75(1):506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lifschitz A., Virkel G., Sallovitz J. Comparative distribution of ivermectin and doramectin to parasite location tissues in cattle. Vet Parasitol. 2000;87(4):327–338. doi: 10.1016/s0304-4017(99)00175-2. [DOI] [PubMed] [Google Scholar]
- 13.El-Tahtawy A., Glue P., Andrews E.N., Mardekian J., Amsden G.W., Knirsch C.A. The effect of azithromycin on ivermectin pharmacokinetics–a population pharmacokinetic model analysis. PLoS Negl Trop Dis. 2008;2(5):e236. doi: 10.1371/journal.pntd.0000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molina D.K., DiMaio V.J. Normal organ weights in men: part II-the brain, lungs, liver, spleen, and kidneys. Am J Forensic Med Pathol. 2012;33(4):368–372. doi: 10.1097/PAF.0b013e31823d29ad. [DOI] [PubMed] [Google Scholar]
- 15.Boron W.F., Boulpaep E.L. Elsevier; Saint Louis, UNITED STATES: 2016. Medical Physiology E-Book. [Google Scholar]
- 16.Guzzo C.A., Furtek C.I., Porras A.G. Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects. J Clin Pharmacol. 2002;42(10):1122–1133. doi: 10.1177/009127002401382731. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez Canga A., Sahagun Prieto A.M., Diez Liebana M.J., Fernandez Martinez N., Sierra Vega M., Garcia Vieitez J.J. The pharmacokinetics and interactions of ivermectin in humans–a mini-review. AAPS J. 2008;10(1):42–46. doi: 10.1208/s12248-007-9000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmith V.D., Zhou J.J., Lohmer L.R. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin Pharmacol Ther. 2020 doi: 10.1002/cpt.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray M., Rayner C., Noël F., Jans D., Wagstaff K. Ivermectin and COVID-19: a report in Antiviral Research, widespread interest, an FDA warning, two letters to the editor and the authors' responses. Antiviral Res. 2020;178:104805. doi: 10.1016/j.antiviral.2020.104805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boer F. Drug handling by the lungs. Br J Anaesth. 2003;91(1):50–60. doi: 10.1093/bja/aeg117. [DOI] [PubMed] [Google Scholar]
- 21.Klotz U., Ogbuokiri J.E., Okonkwo P.O. Ivermectin binds avidly to plasma proteins. Eur J Clin Pharmacol. 1990;39(6):607–608. doi: 10.1007/BF00316107. [DOI] [PubMed] [Google Scholar]
- 22.Bassissi M.F., Alvinerie M., Lespine A. Macrocyclic lactones: distribution in plasma lipoproteins of several animal species including humans. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138(4):437–444. doi: 10.1016/j.cca.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Cao Y., Jusko W.J. Applications of minimal physiologically-based pharmacokinetic models. J Pharmacokinet Pharmacodyn. 2012;39(6):711–723. doi: 10.1007/s10928-012-9280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones H., Rowland-Yeo K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacometrics Syst Pharmacol. 2013;2(8):e63. doi: 10.1038/psp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.