Abstract
Calcium (Ca2+) is a universal signaling ion, whose major informational role shaped the evolution of signaling pathways, enabling cellular communications and responsiveness to both the intracellular and extracellular environments. Elaborate Ca2+ regulatory networks have been well characterized in eukaryotic cells, where Ca2+ regulates a number of essential cellular processes, ranging from cell division, transport and motility, to apoptosis and pathogenesis. However, in bacteria, the knowledge on Ca2+ signaling is still fragmentary. This is complicated by the large variability of environments that bacteria inhabit with diverse levels of Ca2+. Yet another complication arises when bacterial pathogens invade a host and become exposed to different levels of Ca2+ that (1) are tightly regulated by the host, (2) control host defenses including immune responses to bacterial infections, and (3) become impaired during diseases. The invading pathogens evolved to recognize and respond to the host Ca2+, triggering the molecular mechanisms of adhesion, biofilm formation, host cellular damage, and host-defense resistance, processes enabling the development of persistent infections. In this review, we discuss: (1) Ca2+ as a determinant of a host environment for invading bacterial pathogens, (2) the role of Ca2+ in regulating main events of host colonization and bacterial virulence, and (3) the molecular mechanisms of Ca2+ signaling in bacterial pathogens.
Keywords: Calcium signaling, Calcium channels, Calcium sensors, Toxins, Adhesins, Biofilm, Attachment, Two component regulatory systems, Secretion, Bacterial pathogens
33.1. Elevated External Calcium (Ca2+) Regulates Adaptation of Bacterial Pathogens to Their Host Environment
33.1.1. Host-Associated Ca2+
In order to survive, bacteria must sense the chemical landscape of their environment and respond to it by adjusting their biological activities. Bacterial pathogens have an additional challenge of recognizing the transition between outside and inside the host and efficiently rearranging their gene expression to enable survival in the hostile host. The environment inside the host has a drastically different chemistry regulated by complex signaling systems, including one of the most versatile intracellular messengers, calcium (Ca2+).
Ca2+ signaling has been widely studied in eukaryotes [1, 2]. Ca2+ signaling is based on tightly regulated fluctuations in the levels of the ion in different cellular compartments, that trigger multiple molecular pathways. Whereas the cytoplasmic concentration of free Ca2+ is maintained at high nanomolar level, the extracellular concentration of the ion reaches millimolar levels [3–5] differing between different body fluids, tissues, and organs. Several examples are summarized in Table 33.1.
Table 33.1.
Examples of free Ca2+ levels in human body fluids
| Body fluid | [Ca2+] | References |
|---|---|---|
| Joint fluids | 4 mM | [26] |
| Plasma | 1.3–1.5 mM | [23, 27–29] |
| Serum | 0.7 to 1.4 mM. | [23, 30–34] |
| Saliva (in CF patients) | 0.3 mM (4.8 ± 0.7 mM) | [35–1] |
| Nasal secretions (in CF patients) | 3.1 ± 1.6 mM (4.7 ± 2.2 mM) | [42–4] |
| Sputum (in CF patients) | l.l mM (2.5mM) | [11] |
| Urine | 1.6–5 mM | [45] |
Since Ca2+ signaling regulates most essential cellular processes, slight abnormalities in Ca2+ homeostasis cause diseases or are a result of certain pathologies. For example, in cystic fibrosis (CF) [6, 7], different types of cells, including skin fibroblasts and bronchial epithelium cells, show elevated intracellular Ca2+ pools [8, 9]. In addition, abnormally elevated levels of Ca2+ were registered in multiple body fluids of CF patients (Table 33.1). Further, the elevation of cytosolic Ca2+ concentration was shown to trigger host immune responses against invading pathogens. For example, intestinal epithelial cells infected with Salmonella serotype Typhimurium require an increased cytosolic Ca2+ to express pro-inflammatory chemokine IL-8 [10]. Elevated Ca2+ in CF sputum positively correlates with the release of IL-8 in the necrotic immune cells [11]. As a part of the innate immunity defense, production of antimicrobial peptides (AMPs) by epidermal keratinocytes in response to infection by Pseudomonas aeruginosa, Staphylococcus aureus and other pathogens is induced by elevated levels of Ca2+ [12]. Some of the AMPs, including a family of Ca2+ binding EF-hand S100 family, require Ca2+ for their interactions with targets [13].
Some bacterial pathogens are able to alter the hosts [Ca2+]in levels through activating Ca2+ flux across the plasma membrane and, releasing Ca2+ from the intracellular stores into the cytosol [10, 14–17]. These interactions can be mediated by bacterial surface associated proteins such as PilC of Neisseria meningitidis [17], FliC of P. aeruginosa and Salmonella [18], and FimH of Escherichia coli [19] or by secreted effectors, such as hemolysin A from S. aureus [20], pyocyanin and homoserine lactones from P. aeruginosa and Serratia liquefaciens [21–25]. Such alterations in the host Ca2+ have been shown to facilitate bacterial adherence and subsequent internalization into the host cells.
In plants, Ca2+ is one of the earliest signaling elements that coordinate adaptive immune responses to invading pathogenic bacteria. Cytoplasmic Ca2+ ([Ca2+]cyt) increases in response to infecting pathogens, such as P. syringae [46]. A sustained elevation of [Ca2+]cyt serves as an important early signal, which links the recognition of infection to downstream defenses including generation of reactive oxygen species (ROS) and oxidative burst [47, 48]. The ROS burst may lead to cell death preventing the pathogen establishment inside the plant [49].
Overall, Ca2+ is an essential component of the host environment that both responds to the presence of bacterial pathogens, and regulates specific defense mechanisms. Ca2+ levels in a host may signal to the invading pathogens that they are entering a host and also indicate the status of immune protection in the host. Therefore, recognizing the host Ca2+ level can be beneficial to the invaders and trigger their adaptation to the host environment, and lead to their increased virulence and survival of the pathogen.
33.1.2. Ca2+ Triggers Life Style Switches in Bacterial Pathogens
Bacteria possess efficient regulatory systems that enable their adaptation to continuously changing environments. Regulation of gene expression is key for bacterial survival in a variety of environments. One particularly efficient and complex mechanism of surviving hostile environments is a switch between free-swimming or planktonic lifestyle to sessile life as surface-associated community, called biofilm. This transition is enabled by major molecular rearrangements ultimately enabling increased resistance, cell-cell communication and efficient metabolism [50, 51]. This mechanism is of particularly high importance to extracellular pathogenic bacteria colonizing host surfaces and surviving both host defenses and antimicrobial treatments.
There is a growing body of evidence that Ca2+ plays both a structural and a regulatory role in the transition to surface-associated biofilm lifestyle. Bacterial adhesion is the first step in biofilm formation, and itself is a survival mechanism, as nutrients, for example, tend to accumulate at surfaces [52]. The effect of Ca2+ on adhesion is partially due to electrostatic interactions, but also due to strong interactions of the surfaces with the cell structures, such as pili and fimbriae [53–55], and other macromolecules including teichoic acids, adhesins, lipopolysaccharide (LPS), and extracellular polysaccharides (EPS). It was shown that cell surface properties and their electrostatic interactions with the substratum contribute to Ca2+-enhanced adhesion of non-motile and motile P. aeruginosa [56]. Ca2+- enhanced cell adhesion to diverse host molecules and in vitro substrates, as well as cell-cell aggregation, relies on the presence of type I and type IV pili in a number of pathogens, including Xylella fastidiosa, [53], P. aeruginosa [57], Vibrio vulnificus [58], and N. gonorrhoeae [59]. The Ca2+ regulation of the type IV pilus is determined by its binding to pilus-biogenesis factor, PilY1, enabling pilus extension and retraction [60]. This interaction is also required for the bacterium twitching motility. By interacting with type I pili and fimbriae, Ca2+ modulates invasion of bacterial pathogens, such as E. coli, into host cells [19, 61].
Ca2+ also enhances bacterial adhesion via large cell surface Ca2+-binding adhesins, such as SdrC and SdrD in S. aureus [62, 63] and BapA in Paracoccus denitrificans [64]. The former contain EF hand-like motifs that bind Ca2+ required for protein folding. The latter belongs to repeats-in-toxin (RTX) family, containing multiple nonapeptide Ca2+-binding domains, secreted via Type I Secretion System (TISS), and serving a variety of functions, including cell-cell or cell-surface interactions or contributing to protection against hostile environments by forming bacterial surface (S)-layers (reviewed in [65]). In Listeria monocytogenes, elevated Ca2+ has been reported to stabilize the complex between the adhesin InlA (Internalin A) and the human extracellular E-cadherin domain 1 (hEC1). Once inside the host cell, where Ca2+ concentrations are lower, the InlA-hEC1 complex dissociates, which facilitates the liberation of the bacteria from the host cell membrane into the cytosol [66]. Ca2+ is required for multimerization of large adhesin LapF involved in colonization, microcolony formation, and biofilm maturation of P. putida [67–69].
Due to its interactions with surface proteins and by forming ionic bridges between negatively charged macromolecules, Ca2+ enhances cell aggregation and strengthens biofilm matrixes, including cell aggregation in oral Streptococci [70] and alginate cross-linking of P. aeruginosa biofilm matrix [71]. Ca2+ was also shown to bind extracellular DNA (eDNA), another important component of biofilm matrix, and this thermodynamically favorable binding increases bacterial aggregation in several Gram-positive and Gram-negative species, including S. aureus and P. aeruginosa. The authors concluded that Ca2+ did not affect DNA release [72]. However, this observation is likely species- and strain-specific [73], as Ca2+ was shown to induce production of P. aeruginosa pyocyanin, which promotes DNA release [74]. Furthermore, the presence of Ca2+ increased eDNA release, contributing to biofilm formation in Streptococcus mutans [75]. Ca2+ was also shown to increase the adhesive nature of P. fluorescence biofilm, but reduced its elastic properties [76].
In different bacterial species, elevated Ca2+ either stimulates or reduces biofilm formation. Positive regulation was observed in response to 1–10 mM Ca2+ in Pectobacterium carotovorum [77], Rhizobium leguminosarum [78], Pseudoalteromonas sp. [79], Shewanella oneidensis [80], P. aeruginosa [73], X. fastidiosa [81], V. cholerae [82], and V. fischeri [83]. This regulation was shown to be mediated by diverse mechanisms. For example, elevated Ca2+ activates the transcription of genes responsible for production of surface adhesins and EPS: alginate in P. aeruginosa [73] and P. syringae [84]; symbiosis polysaccharide (syp) or cellulose in V. fisheri. Ca2+-dependent hemophilic interactions of surface-associated adhesion SdrC promotes biofilm formation in S. aureus [62].
Negative regulation of biofilm by elevated Ca2+ was reported in S. aureus [85] and V. cholerae [86]. In V. cholerae, this regulation is mediated by negatively regulated two-component system CarSR and vps (Vibrio polysaccharide) genes. However, V. cholerae also produces Vps-independent biofilm, which is preferred under high Ca2+ sea water conditions, where Ca2+ interacts directly with the O-antigen polysaccharide [87]. S. aureus produces several surface adhesins, such as clumping factors A and B (ClfA and ClfB) [88, 89] and biofilm-associated protein (Bap) [85]. These proteins contain Ca2+-binding EF-hand-like motifs, and binding the ion inhibits their role in cell adhesion and biofilm formation. A point mutation in protease aureolysin (aur) gene in one of S. aureus strains led to increased activity of ClfB, required for biofilm growth under Ca2+-depleted conditions [90].
Some factors contributing to biofilm formation are known to be regulated by cyclic-di-GMP (c-di-GMP) (reviewed in [91]) and quorum sensing (QS) (reviewed in [92]). This raises the possibility of interconnections between c-di-GMP, QS and Ca2+ regulatory networks that warrant further studies.
33.1.3. Virulence Factors Regulated by Ca2+
Factors that enable pathogenic bacteria to cause diseases can be broadly grouped into several categories, such as penetration, colonization, damage of host cells, evasion of host defenses, and proliferation, all ultimately contributing to the developing infections. Colonization requires pathogens to establish interactions with host tissues by producing extracellular or cell-associated molecules. It may also involve communication between invaders themselves or those with commensals. The relationship between some of these factors and Ca2+ is discussed above. Here we outline virulence factors attributed to more invasive host-pathogen interactions that are directly or indirectly regulated by Ca2+.
Bacterial invasion is commonly facilitated by the production and secretion of molecules that cause enzymatic or non-enzymatic damage to the host cells [93, 94]. A number of secreted enzymes are known to be regulated by Ca2+ in bacterial pathogens. For example, in P. aeruginosa, Ca2+ promotes the production of extracellular proteases LasB, LasA, PrpL (protease IV), and AprA [73, 95–97]. In the case of elastase LasB, Ca2+ not only increases the production of the protein, but modulates its processing, export, stability and enzymatic activity [95, 98, 99]. The enzymatic activity covers a wide repertoire of substrates, including elastin, collagen, and human immunoglobulins, underlining the significance of the protein and its Ca2+ regulation in P. aeruginosa pathogenicity. The alkaline protease A (AprA) binds Ca2+ through its C-terminal RTX domain, enabling folding of both C- and N-terminal proteolytic domains, which is required for stable conformation and enzymatic activity of the protease [73, 80, 96]. Both AprA and LasB are capable of degrading exogenous flagellin monomers under Ca2+-replete condition, which prevents flagellin-mediated immune recognition and killing of P. aeruginosa via complement-mediated phagocytosis [99, 100]. The Ca2+-enhanced production of the two proteases with anti-flagellin activity provides a robust strategy for P. aeruginosa to escape the detection by the complement system.
Our earlier studies showed that production of pyocyanin, the extracellular redox cycling compound and a virulence factor of P. aeruginosa [101, 102] is up-regulated in response to elevated Ca2+ [73]. Pyocyanin is found in pulmonary fluids of CF patients and shown to disrupt Ca2+ homeostasis of the host epithelial cells [21, 103], potentially contributing to a further increase of extracellular host Ca2+ and therefore induction of Ca2+-regulated virulence.
Toxins represent one of the most powerful strategies of bacterial pathogens to conquer a host. Ca2+ modulates the production, secretion, and function of several toxins in a number of pathogens. In E. coli, Ca2+ is required for Hemolysin A (HlyA) binding to erythrocytes [104]. Binding Ca2+ causes conformational change in the toxin increasing its surface hydrophobicity and promoting the irreversible binding to the lipid bilayer of erythrocytes. This interaction preludes and ensures the lytic effect [105]. In V. cholerae, Ca2+ enhances bile salt-dependent activation of virulence. The mechanism relies on Ca2+ promoting the bile salt-induced activation of transmembrane virulence regulator TcpP, which then induces the production of major virulence factors, including toxin-coregulated pilus (TCP) [106]. The presence of Ca2+ has been reported to be essential for the toxicity of anthrax-edema toxin (composed of protective antigen and edema factor) produced by Bacillus anthracis [107]. The edema factor has adenylate cyclase activity synthesizing cAMP. Once in the host cytosol, the edema factor produces cAMP, which causes a rapid influx of Ca2+. The accumulation of cAMP in the cytosol requires the presence of extracellular Ca2+. As a potent inhibitor of immune response, accumulated cAMP leads to suppression of proinflammatory cytokines, phagocytosis and bactericidal activity of leukocytes thereby facilitating the survival of bacteria in the host [107, 108].
On the other hand, elevated host Ca2+ may have a negative regulatory effect on virulence and thus contribute to host defenses. One example is a cell wall degrading enzyme endopolygalacturonase (PehA) that is down-regulated by high (10–30 mM) levels of Ca2+ in a plant pathogen Pectobacterium carotovorum. This prevents the pathogen from infecting the plant [109].
Overall, Ca2+ regulates many virulence factors of invading bacterial pathogens, which stresses the importance of a detailed understanding of Ca2+ regulatory pathways in these pathogens.
33.1.4. Ca2+-Regulated Secretion Systems
Most bacteria can respond to and manipulate their environment through the secretion of extracellular proteins. Bacterial secreted proteins are often involved in breakdown of macromolecules, such as polysaccharides or polypeptides to simple sugars or amino acids that the bacteria can take up and utilize as carbon, nitrogen, and energy sources. Secreted proteins may also act as virulence factors, as in the case for the proteases described above, LasA, LasB, PrpL, and AprA, which modulate immune effectors and degrade elastic tissues [73, 95, 96, 99, 100]. Pathogenic bacteria also use protein secretion to kill other cells, including eukaryotic cells [110] and, in some cases, competing bacteria within biofilm communities [111]. Extracellular Ca2+ concentration plays a direct or indirect signaling role in many of the bacterial protein secretion systems.
Bacteria use at least six different strategies to secrete proteins (termed: T1SS to T6SS) reviewed in [112]. The T1SS transports specific proteins directly from the cytoplasm to the extracellular medium, with no apparent periplasmic intermediate. The Ca2+ requiring protease, AprA [113], is secreted by the T1SS, composed of three components, AprDEF, which include cytoplasmic ATPase, an inner membrane protein component, and an outer membrane protein [114]. These proteins form a molecular complex, dedicated to the export of AprA [113]. AprA accumulates in the biofilms of P. aeruginosa under Ca2+-replete conditions, but not under Ca2+ − limiting conditions [73]. The other protease virulence factors described above that require Ca2+ for activity or structural integrity, LasA, LasB, and the PrpL [98], are secreted by the T2SS [115–117]. The T2SS is a general pathway for secretion of a variety of extracellular proteins. In the T2SS, proteins with N-terminal export signal peptides, are first exported to the periplasm by either the Sec export machinery or the twin-arginine translocation (TAT) system [118, 119]. Sec, exports proteins in an unfolded state, then folds the proteins into their three dimensional confirmation in the periplasm, with the help of proteins such as disulfide bond isomerase, DsbA. The TAT system exports folded proteins into the periplasm. Once in the periplasm, proteins are secreted across the outer member via the secretion apparatus. For example, in P. aeruginosa the secretion apparatus is composed of the Xcp proteins (XcpA and XcpP-Z) or the homologous system, Hxc (composed of HxcP-Z) [112]. In addition to secretion of enzymes into the extracellular medium, the T2SS also plays a role in generation of certain types of pili, the Type IV pilus, which plays role in bacterial attachment and movement along surface. In twitching motility, bacteria move along surfaces by extension and retraction of the pili, through polymerization and depolymerization of the pilin subunits. Some bacteria, including E. coli and several Vibrio spps, requires Ca2+ for structural integrity of the major pseudopilin subunit, GspG [120].
The type III secretion system (T3SS), encoded on pathogenicity islands of many pathogenic bacteria, delivers effector protein toxins directly into the cytosol of eukaryotic cells during infection. The toxins, including enzymes such as ADP-ribosyltransferases, phospholipases, or adenylate cyclases, disrupt such host cell activities as actin remodeling, and gene regulation [112]. Perhaps the most interesting role of Ca2+ in secretion of bacterial virulence factors is its direct role in expression regulation (activation or repression) of the genes encoding the secretion apparatuses. It has been known for many years that expression of the T3SS genes is induced by either host-cell contact or by chelation of Ca2+ from the medium (low [Ca2+]) [121, 122]. The T3SS forms a complex needle-like structure that is related to the bacterial flagella basal body [123]. The T3SS includes inner and outer membrane ring structures, and cytoplasmic protein components that dock to the inner membrane ring. The needle-like structure protrudes from the basal body, punctures the host cell membrane, and secretes toxins directly into the host cells. For this reason, the T3SS has also been termed the injectosome.
Regulation of expression of the T3SS gene clusters by host cell contact has been well characterized in P. aeruginosa [124, 125]. Transcription of the T3SS in P. aeruginosa is controlled by the transcriptional activator, ExsA, an AraC/XylS-type regulator. ExsA is inhibited by a cascade of protein-protein interactions that prevent ExsA binding to the DNA. The cascade involves interactions of ExsA, ExsD, ExsC, and ExsE. Expression of the T3SS genes is induced when ExsE is translocated from the cell through the T3SS, ultimately titrating the anti-activator, ExsD away from the ExsA and allowing transcription. If translocation of ExsE through the T3SS is functional (e.g. during host cell contact or at low Ca2+), transcription of the TS33 genes is activated. If ExsE builds up in the cell due to lack of host cell contact, then further transcription of the TS33 genes is inhibited.
Another role of Ca2+ in regulation of T3SS was recently shown to involve the Ca2+-sensor protein, LadS [126]. LadS is a hybrid membrane-bound sensor, containing both a histidine kinase domain and a periplasmic Ca2+-binding DISMED2 domain. Broder et al. [126] mutated potential Ca2+-binding residues in P. aeruginosa and found that the resulted T3SS gene expression became insensitive to Ca2+ conditions. Ca2+ binding to the LadS DISMED2 domain is the first step in a regulatory cascade that responds to external Ca2+. The cascade involves two component system GacCS, two small regulatory RNAs, RsmY and RsmZ, and the RNA binding protein, RsmA. Ultimately, binding of RsmA to specific mRNA sequences results in gene regulation at the translational level [126].
Assembly of the T3SS is a dynamic process that responds to external Ca2+. Using Yersinia entercolitica, Diepold et al. [127] tagged components of the T3SS with fluorescent reporter proteins, so that they could image the membrane and cytosolic components. Using Fluorescence correlation spectroscopy, they calculated the diffusion rates of the T3SS cytoplasmic components under inducing (low Ca2+) and non-inducing (high Ca2+) conditions, as they assembled. They found that the rate of diffusion of the cytoplasmic components changed with the external Ca2+ concentration and proposed this as a novel mechanism for the role of Ca2+ in regulation of T3SS assembly.
The switch between expression of T3SS (low Ca2+) to more recently discovered type T6SS [128] may correlate with the switch from acute to chronic infections of P. aeruginosa [126]. The T6SS also uses direct injection of effector proteins into other cells, which may be host cells, or other competing bacteria [111]. However, rather than being evolutionarily related to the flagella basal body, the T6SS appears related to bacteriophage tail-associated proteins [129]. The tail-spike is used to puncture the membranes of cells, and the effector molecules at the tip of the spike are injected directly into the cytosol. Regulation of expression of T6SS is not well characterized and the role of Ca2+ in regulation of T6SS is not well known. However, Allsopp et al. [130] using a transposon mutagenesis screening approach identified the RNA binding protein, RsmA as a primary component involved in the regulation of all the three T6SS gene operons of P. aeruginosa. Therefore, a common link in the inverse regulation of T3SS and T6SS involves the RNA binding protein RsmA.
33.2. Molecular Mechanisms of Ca2+ Responses in Pathogens
33.2.1. Two Component Systems
As discussed above, Ca2+ levels differ within a host, fluctuate during disease progression, and thus form a complex signaling landscape for invading pathogens. Therefore, sensing host Ca2+ is an important task enabling pathogens to efficiently adjust to the host environment by triggering the expression of genes responsible for virulence and resistance. Bacteria accomplish this in part by employing two-component regulatory systems (TCS). A traditional TCS consists of a sensor kinase and a response regulator. The sensor kinase is usually embedded into the inner membrane with the sensor region often facing the periplasm. Upon signal binding, the sensor kinase auto-phosphorylates followed by phosphorylating the response regulator, typically regulating transcription of a set of target genes [131–133].
Several Ca2+ sensing TCSs have been identified. Some of them are positively and some are negatively regulated by elevated Ca2+ (Table 33.2). To test whether the relationship with the ion is defined by the recognition sequence within the TCS sensor, we aligned the sensor sequences of the TCS experimentally shown to be regulated by Ca2+. Based on the Clustal Omega alignment, a maximum likelihood tree was constructed using MEGA4 algorithm [134, 135] (Fig.33.1). The distinct grouping of positively and negatively regulated sensors supports the idea of sequence-dependent relationship with Ca2+.
Table 33.2.
Sensors from two-component regulatory systems (TCS) regulated by Ca2+
| Name | GenBank ID | Stimuli | Ca2+-dependent regulatory targets | Regulated Phenotype | Organism | References |
|---|---|---|---|---|---|---|
| Positively Regulated by Elevated Ca2+ | ||||||
| CarS | AAG06044.1 | Ca2+ | carP, carO | Virulence | P. aeruginosa | [153] |
| CvsS | AAO56858.1 | Ca2+ | hspR, algU | Biofilm formation, cellulose production, virulence, T3SS | P. syringae | [84] |
| AtoS | AEH68827.1 | Ca2+ SpermidineHis-tamineAcetoacetate | Ato operon | Synthesis of PHB-PP, motility | E. coli | [154–157] |
| VicK | PNM00564.1 | Ca2+, Mn2+, sucrose | atlA operon | eDNA release, response to host immunity, attachment, and biofilm formation | S. mutans | [91, 158, 159] |
| Negatively Regulated by Elevated Ca2+ | ||||||
| PhoQ | AIB53821.1 | Mg2+,Ca2+, acetate | LPS modification | E. coli | [137, 147] | |
| PhoQ | P0DM80.1 | Mg2+,Ca2+ | LPS modification, SOS response | S. enterica | [136, 145, 160] | |
| PhoQ | AEU17992.1 | Mg2+,Ca2+ | LPS modification | P. aeruginosa | [137, 141, 143] | |
| PehS | BAJ11971.1 | Ca2+ | pehA | Virulence | P. carotovorum | [148] |
| CiaH | P0A4I6.1 | Ca2+ | ciaX | Ca2+ mediated cell functions and biofilm production | S. pneumoniae | [161] |
| CarS | 2,614,773 | Ca2+ | vps | Vps-dependent biofilms, antibiotic resistance | V. cholerae | [86, 162] |
Fig. 33.1. Molecular Phylogenetic analysis of Ca2+regulated TCS.
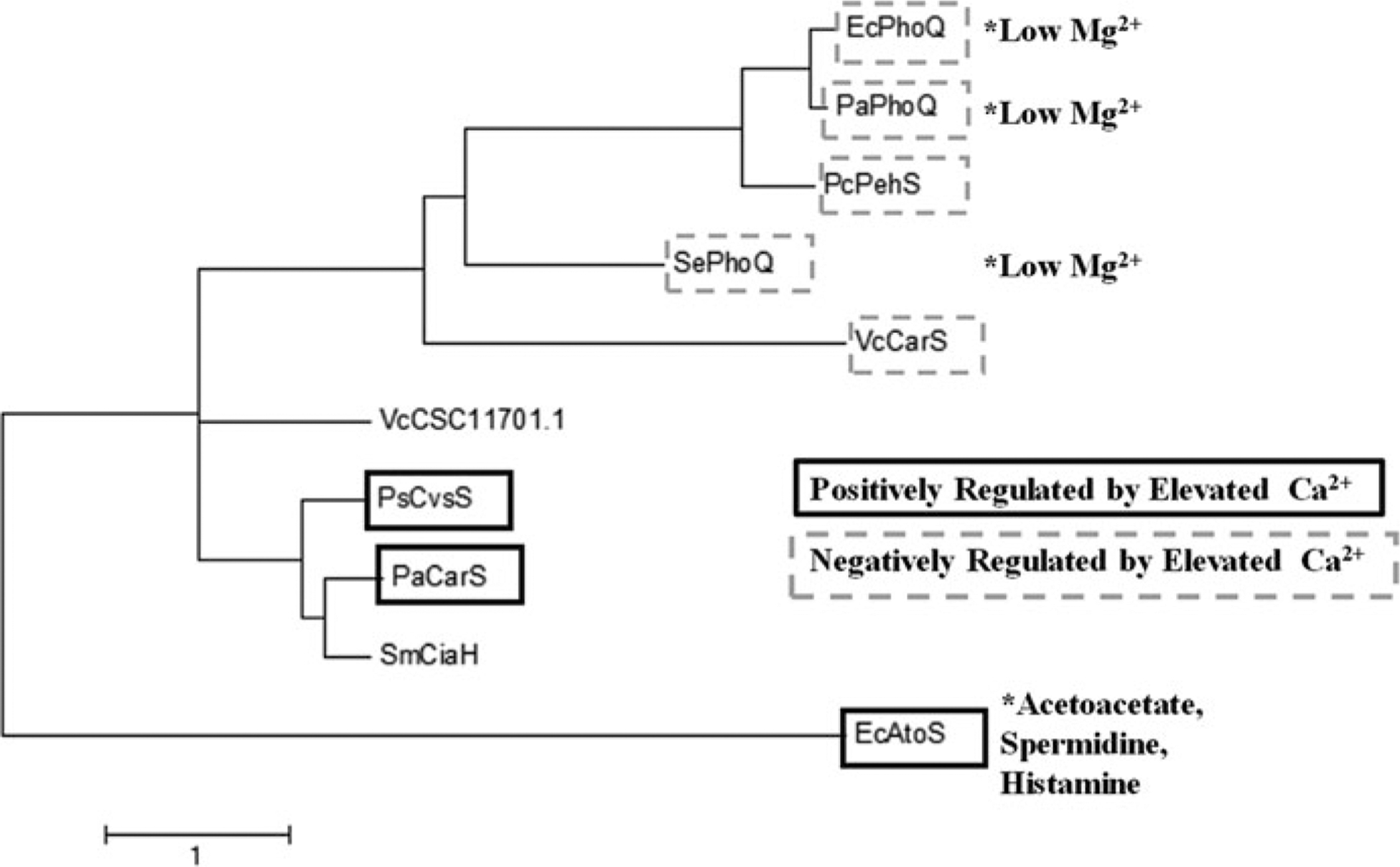
To analyze the sequences of Ca2+ regulated TCS sensors, we first determined the sensor regions by selecting the periplasmic loop of the proteins based on TMHMM analyses. After the 11 sequences were aligned, all positions containing gaps and missing data were eliminated. The phylogenetic relationship of the sensor regions was inferred by using the Maximum Likelihood algorithm based on the JTT matrix-based model [1] in MEGA7 [2]. The tree with the highest log likelihood (−2667.30) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site
The TCSs that are negatively regulated by Ca2+ have been studied in more details. PhoPQ is a well-characterized TCS, found in a variety of Gram-negative pathogens, such as P. aeruginosa, E. coli, Yersinia pestis, Shigella flexneri and S. enterica [82, 131, 136–138]. PhoPQ systems regulate multiple virulence factors, resistance and motility. The PhoPQ regulon includes the SOS response of S. enterica [139], and the arn operon of P. aeruginosa, which is responsible for lipid A modifications and increased resistance to antimicrobial peptides [138, 140–144]. Interestingly, PhoPQ systems are also repressed by elevated Mg2+, and two-distinct Mg2+ and Ca2+ binding regions were identified in PhoQ sensor of S. enterica [136]. These regions are conserved in other PhoQ homologs [145, 146]. While the PhoPQ systems share overall sequence similarly, they differ in their responses. Thus, mutating Ca2+-binding residues in PhoQ have less of an impact on transcriptional regulation in E. coli than in S. enterica [146]. PhoQ interactions with ligands also differ in different species: Mg2+ binds to PhoQ dimer causing destabilization and preventing signal transduction in P. aeruginosa PhoQ (PaPhoQ), but not in E. coli PhoQ (EcPhoQ) [137]. Finally, PhoQ may respond to additional ligands such as acetate in the E. coli PhoQ [147]. The TCS most closely grouped with PhoPQ is PehSR from a plant pathogen Pectobacterium carotovorum (Fig.33.1). This system regulates the production of a secreted endopolygalacturonase, PehA which is required for initial invasion of the pathogen into a host [109, 148–150]. Homologs of PehA, although not yet characterized, have been identified in another plant pathogen Erwinia chrysanthemi [151, 152].
Another TCS negatively regulated by elevated Ca2+ is CarSR from V. cholera [86]. CarSR regulates Vibrio polysaccharide (vps), the main matrix component of vps-dependent biofilms. Although elevated Ca2+ decreases the formation of vps- dependent biofilms, it increases the formation of vps-independent biofilm in V. cholerae, as well as production of bile-salt-dependent virulence factors required for colonization of the gut [82, 163, 164]. The mechanisms for this positive Ca2+ regulation of V. cholerae virulence are not known and potentially involve an alternative TCS that is positively regulated by Ca2+. To predict such a TCS, we performed BLASTP alignment of the sensor region from the positively regulated by Ca2+ P. aeruginosa CarS against the V. cholerae genome and identified a putative Ca2+ responsive sensor protein CSC11701.1. The close grouping of its sensor sequence with P. aeruginosa CarS and P. syringae CvsS supports its potentially positive regulation by Ca2+ (Fig.33.1).
An atypical TCS, CiaHR, was identified in Streptococcus mutans. CiaHR con- tains a third component, CiaX, a small protein, which upon binding Ca2+, interacts with the sensor portion of CiaH and prevents the phosphor-relay. The system was shown to regulate antibiotic resistance, biofilm formation, eDNA uptake, as well as stress response [161, 165–167]. Interestingly, CiaH grouped closely with the positively regulated sensors, indicating a possibility that CiaH itself may be regulated by Ca2+. In addition, S. mutans biofilm formation and eDNA release can be positively regulated in response to Ca2+ via the TCS VicKR [91, 159, 168, 169]. However, VicK is more distantly related to the positively regulated Ca2+ sensors, which could be attributed to its potential to respond to other stimuli (Fig.33.1).
The TCS that are positively regulated by elevated Ca2+ were shown to be involved in regulating virulence and resistance factors in response to Ca2+. Earlier, our group identified a TCS that is positively regulated by Ca2+ in P. aeruginosa. It was named it Calcium Regulated Senor/ Regulator, CarSR [153]. This system regulates at least two identified targets, CarO and CarP, involved in Ca2+–induced production of virulence factors pyocyanin and pyoverdine and contributes to the regulation of the intracellular Ca2+ homeostasis and tolerance to elevated Ca2+. Recently, another Ca2+-induced TCS was found in plant pathogen P. syringae, named Calcium, Virulence, and Swarming Sensor and Regulator, CvsSR. CvsSR is required for P. syringae pathogenicity in plants by enhancing transcription of genes for T3SS and small RNAs while repressing alginate and flagella biosynthesis [84]. In E. coli, synthesis of the polyhydoxybutyrate polyphosphate (PHB-PP) Ca2+ channels is positively regulated by the AtoSC TCS. These non-proteinaceous Ca2+ channels play a role in eDNA uptake and folding of the outer membrane protein OmpA [154, 170, 171]. In addition, AtoSC regulates other processes, such as motility in response to acetoacetate, histamine, and spermidine [155–157]. This may be reflected in its distant grouping from CarSR and CvsSR (Fig.33.1).
In summary, bacterial pathogens utilize multiple TCS to recognize changes in the environmental Ca2+ and adjust their transcriptional activity. These systems are versatile as they evolved to enable bacterial adaptations to multi-variant environments. Understanding the regulation by TCS is challenged by several factors: the presence of multiple systems in one organism, sensors responding to different stimuli, and additional components involved in signal recognition [172–175]. Different TCS may also cross-talk enabling multiple signals to control similar responses, as in the case of SypFG, proposed to mediate Ca2+ induction of biofilm formation in V. fischeri [83]. The sensing portion of SypF, however, was not required for Ca2+ induction, and involved an alternative sensor kinase RscS phosphorylating SypF in response to Ca2+ [83]. A much better understanding of the TCS signaling networks triggered by Ca2+ is needed to fully appreciate their role in Ca2+-mediated communication between invading pathogens and their hosts.
33.2.2. Ca2+ Sensors
In eukaryotes, members of the calmodulin superfamily are the best studied Ca2+ sensors [176, 177]. Calmodulin (CaM) contains two canonical EF-hand motifs coordinating Ca2+ binding [178, 179]. Upon binding Ca2+, CaM undergoes conformational changes enabling binding and activating diverse target proteins [180]. Therefore, searches for components of Ca2+ signaling networks in bacteria have focused on proteins with EF hands [181]. In addition, proteins that play roles in translocating or buffering Ca2+ have been studied [182–188]. A number of calmodulin-like proteins have been reported based on their sequence and structure similarity to CaM and binding to anti-calmodulin antibodies. Several of these Ca2+ sensors are summarized in Table 33.3. The first proposed bacterial Ca2+ sensor was CasA, from Rhizobium etli. CasA has three pairs of EF hand domains, similarly to eukaryotic calbindin and calretinin [189]. CasA mediates Ca2+-dependent symbiotic relationship between R. etli and its plant host. Our group identified a homolog of CasA in P. aeruginosa and named it EfhP (EF-hand protein) [190]. EfhP is required for maintaining Ca2+in homeostasis and involved in Ca2+ regulation of P. aeruginosa virulence. Our current studies verified the ability of this protein to selectively bind Ca2+ and undergo Ca2+–dependent conformational changes supporting its role as a Ca2+ sensor (Kayastha et al. in preparation). Another EF hand protein, proposed to function as a Ca2+-sensor, is CabD from Streptomyces coelicolor. CabD contributes to Ca2+ regulation of aerial mycelium formation. CabD has high- and low-affinity Ca2+-binding sites, suggesting their distinct roles in mediating Ca2+-regulatory and buffering roles, respectively [177].
Table 33.3.
Ca2+ Sensors
| Protein name/GenBank ID | Organism | Function/Properties | Domain | References |
|---|---|---|---|---|
| EfhP | P. aeruginosa | Regulates Ca2+ induced virulence and intracellular Ca2+ homeostasis | One pair of EF hands | Kayastha et al. (in preparation) |
| AAG07494.1 | ||||
| CasA | R. etli | Mediates Ca2+ dependent symbiosis with leguminous host | Three pairs of EF hands | [189] |
| AF288533 | ||||
| CabD | S. coelicolor | Affects formation of aerial mycelium | Two pairs of EF hands | [177] |
| 3AKA_A | ||||
| Protein S | M. xanthus | Required for assembly of spore coat | Beta-gamma crystalline fold | [191] |
| WP_020477824 | ||||
| CAMLP | M. tuberculosis | Activates NAD kinase and phosphodiesterase upon Ca2+ binding | Single EF hand motif | [192, 193] |
| NP_215727 | ||||
| CAMLP | M. smegmatis | Activates phosphodiesterase | Single EF hand motif | [194] |
| AY319523.1 | ||||
| CALP | B. subtilis | Activates phosphodiesterase in Ca2+ dependent manner | [195] | |
| YP_004243569 | ||||
| CLP | B. pertussis | Activates adenylate cyclase in Ca2+ dependent manner | [196] | |
| LadS | P. aeruginosa | Ca2+ dependent phosphor-relay to GacSA | Histidine kinase, 7 transmembrane, DISMED2 | [197, 198] |
| AAG07361 |
CaM-like proteins have been reported in Mycobacteria and are suggested to play a role in sensing Ca2+ [199]. The CaM-like protein, Rv1211 of M. tuberculosis binds Ca2+ through its single EF hand domain and stimulates the activities of NAD kinase and phosphodiesterase (PDE), targets that are similar to those of eukaryotic CaM. Reduced expression of this protein has been shown to impair M. tuberculosis growth and survival in macrophages, suggesting its importance during infection [192, 193]. A homologous CaM-like protein from M. smegmatis has been shown to stimulate phosphodiesterase activity and regulate the metabolism of phospholipids [200] supporting the role of this protein as a Ca2+ sensor.
Protein S from Myxococcus xanthus is a member of ßγ-crystalline family. This protein shares structural similarity to CaM and binds Ca2+, which is required for the protein assembly on the surface of the spores [201]. A more recent report showed that the protein’s Ca2+-binding site forms a high charge density pocket similar to that in calsequestrin and human Hsp70. However it is still not clear whether the Ca2+-induced conformational changes in protein S play roles in signaling events [191].
Recently, a hybrid histidine kinase LadS was shown to trigger a Ca2+-induced switching between acute and chronic type of virulence in P. aeruginosa [198]. As discussed above, LadS belongs to a unique class of bacterial sensors that possess histidine kinase, seven transmembrane, and the periplasmic DISMED2 domain. The latter was shown to bind Ca2+ via Asp80 and Asp90 residues and activate the kinase activity leading to phosphorelay cascade [197, 198]. In contrast to typical sensors of TCS that phosphorylate partnered response regulators, LadS phosphorylates the TCS GacSA and thus activates GacS/Rsm pathway responsible for the global regulation of chronic infection-type virulence and tolerance [88]. In silico analysis of the sequence conservation of LadS showed that this protein is unique to Pseudomonas. Interestingly, during searches for homologous DISMED2 domains among selected bacterial pathogens, we identified several proteins including putative alkaline phosphatase synthesis sensors CJK91172.1 and CJK40304.1, and a membrane protein with GGDEF domain, CJK88170.1, in S. pneumoniae. The GGDEF domain is known to act as a diguanylate cyclase responsible for synthesis of cyclic-di-GMP, a second messenger mainly regulating biofilm formation [202]. In addition to DISMED2 domain, these proteins contain the two residues (Asp80 and Asp90) required for Ca2+ recognition, suggesting they may sense and respond to Ca2+.
Pathogenic bacteria possess Ca2+-sensing proteins that play an important role in modulating their pathogenicity. However, despite evidence of the significance of Ca2+ regulation in bacterial physiology and virulence, the knowledge on Ca2+ sensors and regulators in bacteria is still limited. Further studies are needed to determine whether these proteins sense the changes in the intracellular Ca2+ and thus enable Ca2+ to serve as second messenger.
33.3. Components of Intracellular Ca2+ Signaling in Pathogenic Bacteria Mediating Ca2+ Regulation of Virulence
A number of studies have shown that bacterial metabolic processes respond to elevated Ca2+ (reviewed in [203–205]). Some of these processes are regulated by extracellular Ca2+ levels and are mediated by TCS. However, other processes may respond to the transient changes in the intracellular Ca2+ (Ca2+in), thus implicating Ca2+ as a second messenger. Although the latter still requires experimental proof, here we discuss the components of Ca2+in signaling network that have been shown to play a role in Ca2+ regulation, supporting the idea of a regulatory role of Ca2+in in bacteria.
Similarly to eukaryotic cells, bacteria maintain their basal [Ca2+]in at high nanomolar level and generate transient changes in [Ca2+]in in response to diverse stimuli [206–210]. These stimuli include a variety of extracellular factors, such as Ca2+ex, pH, mechanical stimulation, intermediates of carbohydrate metabolism, and oxidative stress; all factors potentially to be encountered upon entering a host [206, 208, 210–213]. A number of proteins have been shown to contribute to the maintenance of Ca2+in homeostasis and to the generation of Ca2+in fluctuations [203, 206]. However, it is still not clear whether bacteria have an intracellular source of Ca2+ for these fluctuations (e.g. a compartment for storing and releasing Ca2+ into the cytoplasm) or if they rely on influx of extracellular Ca2+. E. coli accumulates Ca2+ in the periplasm to millimolar levels when grown in the presence of millimolar extracellular Ca2+ [207]. It is possible that the periplasmic Ca2+ may be released into the cytoplasm and used for generating intracellular Ca2+ transients. However, further mechanistic studies are imperative.
33.3.1. Ca2+ Channels
Several types of Ca2+ transporters have been identified in bacteria. The transporters contribute to the regulation of virulence and host-pathogen interactions. First, the poly-β-hydroxybutyrate polyphosphate (PHB-PP) in E. coli forms non-proteinaceous Ca2+ channels and translocates Ca2+ into the cytoplasm [214]. In addition, PHB-PP channels are required for Ca2+-dependent genetic competence, which plays a key role in uptake of foreign DNA, eventually enhancing bacterial adaptation to the host environment and resistance [215]. The production of the corresponding PHB-PP synthases was shown to be induced by elevated extracellular Ca2+ and several other stimuli via Ca2+-dependent TCS AtoSC [154].
Another type of Ca2+ channels, a pH-dependent Ca2+ leak channel, YetJ was identified in B. subtilis [216]. This protein contains a BAX-1 inhibitor domain homologous to the one in a Ca2+ leak channel found in the endoplasmic reticulum (ER) membrane. Eukaryotic channels containing transmembrane BAX inhibitor-1 motif (TMBIM) mediate Ca2+in homeostasis and apoptosis [217]. Interestingly, this highly conserved domain has been identified in a number of bacterial proteins. Initially, this domain was identified in E. coli protein YccA, and was shown to play a role in biofilm maturation [99]. While more research is needed to determine the roles of YetJ and YccA in B. subtilis and E. coli physiology, our group recently identified another homolog of the channel in P. aeruginosa [Guragain et al. in preparation]. We named it CalC for Ca2+ Leak Channel and determined that the protein is responsible for generating transient changes in [Ca2+] in the P. aeruginosa cytoplasm in response to extracellular Ca2+. Transcriptional profiling of the mutant strain with disrupted calC revealed that the responses to elevated Ca2+ were impaired, particularly for genes encoding virulence factors and biofilm determinants [Guragain et al. in preparation]. This work provides experimental proof of the regulatory role of Ca2+in transients in bacterial responses to Ca2+. Furthermore, homology searches identified a number of putative BAX-1 Ca2+ leak channels in bacterial pathogens including S. pneumoniae, P. carotovorum, Coexiella burnetti, S. enterica, and H. pylori, indicating the conserved nature of the Ca2+ leak channel in bacterial pathogens, and possibly suggesting a role in the pathogenic life style.
Mechanosensitive channels (MSC) are large, non-selective channels that usually allow the passage of ions in response to mechanical or osmotic stress (reviewed in [205, 218, 219]). MSC are found in a variety of different bacteria including human and plant pathogens [220–222]. In B. subtilis, MSC SpoVAC releases Ca-dipicolinic acid complex, which is required for spore formation [223]. Mechanical stress in E. coli was shown to cause an increase in [Ca2+]in leading to altered gene expression [212]. However, the MSC, MscL in this organism did not impact Ca2+ in homeostasis [224], raising a possibility of an alternative Ca2+ channel responding to mechanical stress. Our studies with P. aeruginosa identified several Ca2+ transporters including a putative MSC encoded by PA4614, which contributed to the restoration of the [Ca2+]in basal level and the regulation of Ca2+-induced swarming motility [206].
33.3.2. Ca2+ Pumps
Elevated levels of free Ca2+in can be toxic to bacterial cells, and the recovery to the basal [Ca2+]in is critical for re-sensitizing cells for the next wave of [Ca2+]in signaling. Therefore, the mechanisms of Ca2+ efflux are of high importance for cellular survival and for Ca2+in regulation. Underlining their physiological significance, multiple families of efflux transporters have evolved and been shown to play a role in bacterial physiology and virulence (reviewed in [203]). The first group includes two types (P and F) of ATPases that couple Ca2+ export to ATP hydrolysis [60, 225, 226]. These proteins are highly conserved and were identified in diverse bacterial pathogens. In addition to translocating Ca2+, or likely because of it, some of these proteins are important in diverse bacterial processes related to survival in a host. For example, CaxP plays a role in host colonization by S. pneumoniae [227], CtpE of M. smegmatis contributes to cell surface integrity [228], and PA3920 and PA2435 of P. aeruginosa mediate Ca2+ regulation of swarming motility [206]. The second group includes ion exchange transporters coupling Ca2+ export to co-transport of other ions. Although many of these transporters have been identified in bacterial pathogens (reviewed in [203]), there is little evidence yet about their role in virulence.
33.3.3. Predicting Novel Components of Ca2+ Signaling Network
To expand our knowledge on the components of Ca2+in signaling network in pathogenic bacteria, we aimed to predict novel Ca2+-recognizing or translocating proteins based on their homology to well-characterized components of eukaryotic Ca2+ network. For this, we selected well-characterized eukaryotic Ca2+-binding proteins, whose homologs in bacteria have not been reported. All sequence alignments were carried out using NCBI BLASTP [76, 229]. CarR, a Ca2+ sensor that is a G protein-coupled receptor, plays an essential role in fluctuating intracellular Ca2+ homeostasis in response to minute changes in [Ca2+]ex and other stimuli [230–234]. The protein contains three cooperative Ca2+ binding sites. To our surprise, four (underlined) out of five (in bold) residues required for Ca2+ binding (RXXEXXEEAEERD) were found in a large number of bacterial proteins involved in a variety of life-sustaining functions, including recruitment of replisome in S. aureus [235], cell division protein FtsA in E. persicina [236], putative Rhs toxin and DNA recombination regulation system in E. coli [237–239], and putative pili assembly gene in Enterobacter cloacae. Another eukaryotic Ca2+ signaling protein is RyR1, which is a Ca2+ channel known to release Ca2+ stored in the sarcoplasmic reticulum into the cytoplasm [240]. Sequence homology searches against bacterial genomes identified only a small fragment of the protein as aligning with bacterial proteins. This region happened to be located within the lining of the pore required for Ca2+ sensitivity [240–242]. Four (underlined) out of five residues E3893, H3895, E3967, Q3970, T5001 required for Ca2+ binding were found with similar spacing in several putative ABC transporters of pathogenic bacteria, including S. aureus and S. pneumoniae. We also detected these residues in putative bestrophin transporters of many bacterial species including P. aeruginosa and E. coli, in an orphan transcriptional regulator unique to a P. aeruginosa clinical isolate, and in several enzymes including a putative purine phosphatase of P. aeruginosa. The discovery of a putative Ca2+ binding site in bacterial bestrophin channels is particularly interesting, since human bestrophin is a Ca2+-gated potassium channel [243]. However, the only characterized bacterial bestrophin channel (in K. pneumoniae) was shown to not require Ca2+ for its function [244] nor did it contain the Ca2+-binding site found in the human bestrophin. This raises a possibility of two types of bestrophin channels in bacteria, Ca2+-dependent and Ca2+-independent. Interestingly, the eukaryotic bestrophin has been demonstrated to possess multiple splicing variants: with and without Ca2+ binding region [245]. Overall, these findings suggest that a significantly greater number of bacterial processes are likely regulated by Ca2+ than already known. The fact that most of these predicted Ca2+- binding proteins were detected in bacterial pathogens suggests the importance of Ca2+ regulation in their physiology and, possibly, interactions with hosts, whose vital processes are regulated by Ca2+.
33.4. Concluding Remarks
Bacteria have very dynamic and complex responses to Ca2+. Over the past 10 years, the evidence that bacteria utilize Ca2+ for signaling has grown, yet important pieces are still missing. An area that needs study is on intracellular Ca2+ signaling. While a number of Ca2+ sensors and Ca2+-dependent regulatory systems have been shown to regulate essential functions, most of the findings are of correlative nature with no direct experimental evidence linking the changes in the intracellular Ca2+ to the regulation of transcription or translation. Even less is known about how the amplitude and the frequency of intracellular Ca2+ signals modulate the response. An interesting aspect is the conservation of many Ca2+-binding domains in eukaryotes and bacteria indicating an evolutionary lineage between Ca2+ signaling networks in these domains of life. One technical problem, that is worth mentioning, is the disregard for the presence of Ca2+ in commonly used rich bacteriological growth media, such as LB or BHI. Consequently, Ca2+ regulation of the resultant bacterial phenotypes may be underestimated. Overall, Ca2+ signaling in bacteria is an exciting and quickly developing field, which is providing not only the fundamental understanding of bacterial life and evolution but also generating insights into the regulation of bacterial pathogenicity.
References
- 1.Edel KH, Kudla J (2015) Increasing complexity and versatility: how the calcium signaling toolkit was shaped during plant land colonization. Cell Calcium 57(3):231–246 [DOI] [PubMed] [Google Scholar]
- 2.Permyakov EA, Kretsinger RH (2009) Cell signaling, beyond cytosolic calcium in eukaryotes. J Inorg Biochem 103(1):77–86 [DOI] [PubMed] [Google Scholar]
- 3.Clapham DE (1995) Calcium signaling. Cell 80(2):259–268 [DOI] [PubMed] [Google Scholar]
- 4.Bush DS, Jones RL (1990) Measuring intracellular Ca2+ levels in plant cells using the fluorescent probes, Indo-1 and Fura-2: progress and prospects. Plant Physiol 93(3):841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose J et al. (2011) Calcium efflux systems in stress signaling and adaptation in plants. Front Plant Sci 2:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Von Ruecker AA, Bertele R, Harms HK (1984) Calcium metabolism and cystic fibrosis: mitochondrial abnormalities suggest a modification of the mitochondrial membrane. Pediatr Res 18(7):594. [DOI] [PubMed] [Google Scholar]
- 7.Aris RM et al. (1999) Altered calcium homeostasis in adults with cystic fibrosis. Osteoporos Int 10(2):102–108 [DOI] [PubMed] [Google Scholar]
- 8.Ceder O, Roomans G, Hösli P (1982) Increased calcium content in cultured fibroblasts from trisomy patients: comparison with cystic fibrosis fibroblasts. Scan Electron Microsc 1982(Pt 2):723–730 [PubMed] [Google Scholar]
- 9.Roomans GM (1986) Calcium and cystic fibrosis. Scan Electron Microsc 1986(Pt 1):165–178 [PubMed] [Google Scholar]
- 10.Gewirtz AT et al. (2000) Salmonella typhimurium induces epithelial IL-8 expression via Ca2+−mediated activation of the NF-κB pathway. J Clin Invest 105(1):79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith DJ et al. (2014) Elevated metal concentrations in the CF airway correlate with cellular injury and disease severity. J Cyst Fibros 13(3):289–295 [DOI] [PubMed] [Google Scholar]
- 12.Büchau AS, Gallo RL (2007) Innate immunity and antimicrobial defense systems in psoriasis. Clin Dermatol 25(6):616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donato R (2001) S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol 33(7):637–668 [DOI] [PubMed] [Google Scholar]
- 14.Pace J, Hayman MJ, Galán JE (1993) Signal transduction and invasion of epithelial cells by S. Typhimurium. Cell 72(4):505–514 [DOI] [PubMed] [Google Scholar]
- 15.Gekara NO et al. (2007) The multiple mechanisms of Ca2+ signalling by listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Cell Microbiol 9(8):2008–2021 [DOI] [PubMed] [Google Scholar]
- 16.Hu Y et al. (2011) Structures of Anabaena calcium-binding protein CcbP INSIGHTS INTO CA2+ SIGNALING DURING HETEROCYST DIFFERENTIATION. J Biol Chem 286(14):12381–12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asmat TM et al. (2011) Streptococcus pneumoniae infection of host epithelial cells via polymeric immunoglobulin receptor transiently induces calcium release from intracellular stores. J Biol Chem 286(20):17861–17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nhieu GT et al. (2004) Calcium signalling during cell interactions with bacterial pathogens. Biol Cell 96(1):93–101 [DOI] [PubMed] [Google Scholar]
- 19.Khan NA et al. (2007) FimH-mediated Escherichia coli K1 invasion of human brain microvascular endothelial cells. Cell Microbiol 9(1):169–178 [DOI] [PubMed] [Google Scholar]
- 20.Eichstaedt S et al. (2009) Effects of Staphylococcus aureus-hemolysin a on calcium signalling in immortalized human airway epithelial cells. Cell Calcium 45(2):165–176 [DOI] [PubMed] [Google Scholar]
- 21.Denning GM et al. (1998) Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am J Phys Lung Cell Mol Phys 274(6):L893–L900 [DOI] [PubMed] [Google Scholar]
- 22.Schwarzer C et al. (2010) Pseudomonas aeruginosa homoserine lactone activates store-operated cAMP and cystic fibrosis transmembrane regulator-dependent cl– secretion by human airway epithelia. J Biol Chem 285(45):34850–34863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsen S, Kordel J (1994) Calcium in biological systems. University Science Books, Mill Valley, CA, p 107 [Google Scholar]
- 24.Vikström E et al. (2010) Role of calcium signalling and phosphorylations in disruption of the epithelial junctions by Pseudomonas aeruginosa quorum sensing molecule. Eur J Cell Biol 89(8):584–597 [DOI] [PubMed] [Google Scholar]
- 25.Werthén M, Lundgren T (2001) Intracellular Ca2+ mobilization and kinase activity during acylated homoserine lactone-dependent quorum sensing in Serratia liquefaciens. J Biol Chem 276(9):6468–6472 [DOI] [PubMed] [Google Scholar]
- 26.Maroudas A (1979) Physicochemical properties of articular cartilage In: Freeman MAS(ed) Adult articular cartilage. Pitman Medical, Tunbridge Wells, pp 215–290 [Google Scholar]
- 27.Prohaska C, Pomazal K, Steffan I (2000) Determination of ca, mg, Fe, cu, and Zn in blood fractions and whole blood of humans by ICP-OES. Fresenius J Anal Chem 367(5):479–484 [DOI] [PubMed] [Google Scholar]
- 28.Baker S, Worthley L (2002) The essentials of calcium, magnesium and phosphate metabolism: part I. Physiology. Critical care and. Resuscitation 4:301–306 [PubMed] [Google Scholar]
- 29.Oreskes I et al. (1968) Measurement of ionized calcium in human plasma with a calcium selective electrode. Clin Chim Acta 21(3):303–313 [DOI] [PubMed] [Google Scholar]
- 30.Sava L et al. (2005) Serum calcium measurement: total versus free (ionized) calcium. Indian J Clin Biochem 20(2):158–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore EW (1970) Ionized calcium in normal serum, ultrafiltrates, and whole blood determined by ion-exchange electrodes. J Clin Invest 49(2):318–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz H, McConville B, Christopherson E (1971) Serum ionized calcium by specific ion electrode. Clin Chim Acta 31(1):97–107 [DOI] [PubMed] [Google Scholar]
- 33.Reinhart RA (1988) Magnesium metabolism: a review with special reference to the relationship between intracellular content and serum levels. Arch Intern Med 148(11):2415–2420 [DOI] [PubMed] [Google Scholar]
- 34.Fanconi A, Rose GA (1958) The ionized, complexed, and protein-bound fractions of calcium in plasma: an investigation of patients with various diseases which affect calcium metabolism, with an additional study of the role of calcium ions in the prevention of tetany. Q J Med 27:463–494 [PubMed] [Google Scholar]
- 35.Fiyaz M et al. (2013) Association of salivary calcium, phosphate, pH and flow rate on oral health: a study on 90 subjects. J Ind Soc Periodontol 17(4):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blomfield J, Warton KL, Brown J (1973) Flow rate and inorganic components of submandibular saliva in cystic fibrosis. Arch Dis Child 48(4):267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chernick WS, Barbero GJ, Parkins FM (1961) Studies on submaxillary saliva in cystic fibrosis. J Pediatr 59(6):890–898 [DOI] [PubMed] [Google Scholar]
- 38.Marmar J, Barbero GJ, Sibinga MS (1966) The pattern of parotid gland secretion in cystic fibrosis of the pancreas. Gastroenterology 50(4):551–556 [PubMed] [Google Scholar]
- 39.Blomfield J et al. (1976) Parotid gland function in children with cystic fibrosis and child control subjects. Pediatr Res 10(6):574. [DOI] [PubMed] [Google Scholar]
- 40.Moreira A et al. (2009) Flow rate, pH and calcium concentration of saliva of children and adolescents with type 1 diabetes mellitus. Braz J Med Biol Res 42(8):707–711 [DOI] [PubMed] [Google Scholar]
- 41.Agha-Hosseini F, Dizgah IM, Amirkhani S (2006) The composition of unstimulated whole saliva of healthy dental students. J Contemp Dent Pract 7(2):104–111 [PubMed] [Google Scholar]
- 42.Halmerbauer G et al. (2000) The relationship of eosinophil granule proteins to ions in the sputum of patients with cystic fibrosis. Clin Exp Allergy 30(12):1771–1776 [DOI] [PubMed] [Google Scholar]
- 43.Lorin MI, Gaerlan PF, Mandel ID (1972) Quantitative composition of nasal secretions in normal subjects. J Lab Clin Med 80(2):275–281 [PubMed] [Google Scholar]
- 44.Lorin M et al. (1976) Composition of nasal secretion in patients with cystic fibrosis. J Lab Clin Med 88(1):114–117 [PubMed] [Google Scholar]
- 45.Taylor EN, Curhan GC (2007) Differences in 24-hour urine composition between black and white women. J Am Soc Nephrol 18(2):654–659 [DOI] [PubMed] [Google Scholar]
- 46.Nemchinov LG, Shabala L, Shabala S (2008) Calcium efflux as a component of the hypersensitive response of Nicotiana benthamiana to Pseudomonas syringae. Plant Cell Physiol 49(1):40–46 [DOI] [PubMed] [Google Scholar]
- 47.Grant M et al. (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23(4):441–450 [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Du L, Poovaiah B (2014) Calcium signaling and biotic defense responses in plants. Plant Signal Behav 9(11):e973818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma W, Berkowitz GA (2007) The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol 9(11):2571–2585 [DOI] [PubMed] [Google Scholar]
- 50.Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2(2):95–108 [DOI] [PubMed] [Google Scholar]
- 51.Sauer K (2003) The genomics and proteomics of biofilm formation. Genome Biol 4(6):219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Loosdrecht MC et al. (1990) Influence of interfaces on microbial activity. Microbiol Rev 54(1):75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruz LF, Cobine PA, De La Fuente L (2012) Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl Environ Microbiol 78(5):1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romantschuk M (1992) Attachment of plant pathogenic bacteria to plant surfaces. Annu Rev Phytopathol 30(1):225–243 [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi T et al. (2009) Gene cloning and characterization of Streptococcus intermedius fimbriae involved in saliva-mediated aggregation. Res Microbiol 160(10):809–816 [DOI] [PubMed] [Google Scholar]
- 56.Kerchove AJ, Elimelech M (2008) Calcium and magnesium cations enhance the adhesion of motile and nonmotile pseudomonas aeruginosa on alginate films. Langmuir 24(7):3392–3399 [DOI] [PubMed] [Google Scholar]
- 57.Johnson MD et al. (2011) Pseudomonas aeruginosa PilY1 binds integrin in an RGD-and calcium-dependent manner. PLoS One 6(12):e29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams TC, Ayrapetyan M, Oliver JD (2015) Molecular and physical factors that influence attachment of Vibrio vulnificus to chitin. Appl Environ Microbiol 81(18):6158–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Y et al. (2013) Mutation of the conserved calcium-binding motif in Neisseria gonorrhoeae PilC1 impacts adhesion but not piliation. Infect Immun 81(11):4280–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orans J et al. (2010) Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc Natl Acad Sci U S A 107(3):1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eto DS et al. (2008) Clathrin, AP-2, and the NPXY-binding subset of alternate endo-cytic adaptors facilitate FimH-mediated bacterial invasion of host cells. Cell Microbiol 10(12):2553–2567 [DOI] [PubMed] [Google Scholar]
- 62.Barbu EM et al. (2014) SdrC induces staphylococcal biofilm formation through a homophilic interaction. Mol Microbiol 94(1):172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Josefsson E et al. (1998) The binding of calcium to the B-repeat segment of SdrD, a cell surface protein of Staphylococcus aureus. J Biol Chem 273(47):31145–31152 [DOI] [PubMed] [Google Scholar]
- 64.Kumar S, Spiro S (2017) Environmental and genetic determinants of biofilm formation in Paracoccus denitrificans. mSphere 2(5):e00350–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linhartova I et al. (2010) RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev 34(6):1076–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niemann HH, Schubert W-D, Heinz DW (2004) Adhesins and invasins of pathogenic bacteria: a structural view. Microbes Infect 6(1):101–112 [DOI] [PubMed] [Google Scholar]
- 67.Martínez-Gil M et al. (2012) Calcium causes multimerization of the large adhesin LapF and modulates biofilm formation by Pseudomonas putida. J Bacteriol 194(24):6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M (2010) LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol 77(3):549–561 [DOI] [PubMed] [Google Scholar]
- 69.Espinosa-Urgel M, Salido A, Ramos J-L (2000) Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol 182(9):2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rose RK (2000) The role of calcium in oral streptococcal aggregation and the implications for biofilm formation and retention. BBA-Gen Subjects 1475(1):76–82 [DOI] [PubMed] [Google Scholar]
- 71.Korstgens V et al. (2001) Influence of calcium ions on the mechanical properties of a model biofilm of mucoid Pseudomonas aeruginosa. Water Sci Technol 43(6):49–57 [PubMed] [Google Scholar]
- 72.Das T et al. (2014) Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS One 9(3):e91935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarkisova S et al. (2005) Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J Bacteriol 187(13):4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das T, Manefield M (2012) Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One 7(10):e46718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jung CJ et al. (2017) AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun 85(9):pii: e00252–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Safari A et al. (2014) The significance of calcium ions on Pseudomonas fluorescens biofilms - a structural and mechanical study. Biofouling 30(7):859–869 [DOI] [PubMed] [Google Scholar]
- 77.Haque MM et al. (2017) CytR homolog of Pectobacterium carotovorum subsp. carotovorum controls air-liquid biofilm formation by regulating multiple genes involved in cellulose production, c-di-GMP signaling, motility, and type III secretion system in response to nutritional and environmental signals. Front Microbiol 8:972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vozza NF et al. (2016) A rhizobium leguminosarum CHDL- (cadherin-like-) Lectin participates in assembly and remodeling of the biofilm matrix. Front Microbiol 7:1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patrauchan MA et al. (2005) Calcium influences cellular and extracellular product formation during biofilm-associated growth of a marine Pseudoalteromonas sp. Microbiology-Sgm 151:2885–2897 [DOI] [PubMed] [Google Scholar]
- 80.Theunissen S et al. (2010) The 285 kDa bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res Microbiol 161(2):144–152 [DOI] [PubMed] [Google Scholar]
- 81.Parker JK et al. (2016) Calcium transcriptionally regulates the biofilm machinery of Xylella fastidiosa to promote continued biofilm development in batch cultures. Environ Microbiol 18(5):1620–1634 [DOI] [PubMed] [Google Scholar]
- 82.Hay AJ et al. (2017) Calcium enhances bile salt-dependent virulence activation in Vibrio cholerae. Infect Immun 85(1):pii: e00707–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tischler AH et al. (2018) Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J Bacteriol 200(15):pii: e00016–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fishman MR et al. (2018) Ca2+-induced two-component system CvsSR regulates the type III secretion system and the Extracytoplasmic function sigma factor AlgU in Pseudomonas syringae pv. Tomato DC3000. J Bacteriol 200(5):e00538–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arrizubieta MJ et al. (2004) Calcium inhibits bap-dependent multicellular behavior in Staphylococcus aureus. J Bacteriol 186(22):7490–7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bilecen K, Yildiz FH (2009) Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11(8):2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watnick PI et al. (2001) The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol 39(2):223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Connell DP et al. (1998) The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+−dependent inhibitory site. J Biol Chem 273(12):6821–6829 [DOI] [PubMed] [Google Scholar]
- 89.Eidhin DN et al. (1998) Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol 30(2):245–257 [DOI] [PubMed] [Google Scholar]
- 90.Abraham NM, Jefferson KK (2012) Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology 158(Pt 6):1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Romling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77(1):1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Whiteley M, Diggle SP, Greenberg EP (2017) Progress in and promise of bacterial quorum sensing research. Nature 551(7680):313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilson J, Schurr M, LeBlanc C (2002) Mechanisms of bacterial pathogenicity. Postgrad Med J 78:216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ribet D, Cossart P (2015) How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect 17(3):173–183 [DOI] [PubMed] [Google Scholar]
- 95.Olson JC, Ohman DE (1992) 1992, Efficient production and processing of elastase and LasA by Pseudomonas aeruginosa require zinc and calcium ions. J Bacteriol 174:4140–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang L, Conway JF, Thibodeau PH (2012) Calcium-induced folding and stabilization of the Pseudomonas aeruginosa alkaline protease. J Biol Chem 287(6):4311–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marquart ME et al. (2005) Calcium and magnesium enhance the production of Pseudomonas aeruginosa protease IV, a corneal virulence factor. Med Microbiol Immunol 194(1–2):39–45 [DOI] [PubMed] [Google Scholar]
- 98.Thayer M, Flaherty KM, McKay DB (1991) Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-a resolution. J Biol Chem 266(5):2864–2871 [DOI] [PubMed] [Google Scholar]
- 99.Casilag F et al. (2016) The LasB elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect Immun 84(1):162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laarman AJ et al. (2012) Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol 188(1):386–393 [DOI] [PubMed] [Google Scholar]
- 101.Hall S et al. (2016) Cellular effects of pyocyanin, a secreted virulence factor of Pseudomonas aeruginosa. Toxins 8(8):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rada B, Leto TL (2011) The redox-active Pseudomonas virulence factor pyocyanin induces formation of neutrophil extracellular traps. FASEB J 25(1 Supplement):360.1–360.1 [Google Scholar]
- 103.Ran H, Hassett DJ, Lau GW (2003) Human targets of Pseudomonas aeruginosa pyocyanin. Proc Natl Acad Sci 100(24):14315–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boehm DF (1990) Welch R a, and Snyder IS, Calcium Is Required for Binding of Escherichia Coli Hemolysin (HlyA) to Erythrocyte Membranes. Infect Immun 58(6):1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bakas L et al. (1998) Calcium-dependent conformation of E. coli α-haemolysin. Implications for the mechanism of membrane insertion and lysis. Biochim Biophys Acta 1368(2):225–234 [DOI] [PubMed] [Google Scholar]
- 106.Hay AJ et al. (2017) Calcium enhances bile salt-dependent virulence activation in Vibrio cholerae. Infect Immun 85(1):e00707–e00716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumar P, Ahuja N, Bhatnagar R (2002) Anthrax edema toxin requires influx of calcium for inducing cyclic AMP toxicity in target cells. Infect Immun 70(9):4997–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Serezani CH et al. (2008) Cyclic AMP: master regulator of innate immune cell function. Am J Respir Cell Mol Biol 39(2):127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Flego D et al. (1997) Control of virulence gene expression by plant calcium in the phy-topathogen Erwinia carotovora. Mol Microbiol 25(5):831–838 [DOI] [PubMed] [Google Scholar]
- 110.Deng W et al. (2017) Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15(6):323–337 [DOI] [PubMed] [Google Scholar]
- 111.Ho BT, Dong TG, Mekalanos JJ (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15(1):9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Filloux A (2011) Protein secretion Systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol 2:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baumann U et al. (1993) Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J 12(9):3357–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Duong F et al. (1992) Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene 121:47–54 [DOI] [PubMed] [Google Scholar]
- 115.Kessler E et al. (1993) Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268(10):7503–7508 [PubMed] [Google Scholar]
- 116.Thayer MM, Flaherty KM, McKay DB (1991) Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-Å resolution. J Biol Chem 266(5):2864–2871 [DOI] [PubMed] [Google Scholar]
- 117.Wilderman PJ et al. (2001) Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect Immun 69(9):5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Palmer T, Berks BC (2012) The twin-arginine translocation (tat) protein export pathway. Nat Rev Microbiol 10(7):483–496 [DOI] [PubMed] [Google Scholar]
- 119.Papanikou E, Karamanou S, Economou A (2007) Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol 5(11):839–851 [DOI] [PubMed] [Google Scholar]
- 120.Korotkov KV et al. (2009) Calcium is essential for the major pseudopilin in the type 2 secretion system. J Biol Chem 284(38):25466–25470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dasgupta N et al. (2006) Transcriptional induction of the Pseudomonas aeruginosa type III secretion system by low Ca2+ and host cell contact proceeds through two distinct signaling pathways. Infect Immun 74(6):3334–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vallis AJ et al. (1999) Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun 67(2):914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schraidt O, Marlovits TC (2011) Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331(6021):1192–1195 [DOI] [PubMed] [Google Scholar]
- 124.Vakulskas CA, Brutinel ED, Yahr TL (2010) ExsA recruits RNA polymerase to an extended −10 promoter by contacting region 4.2 of sigma-70. J Bacteriol 192(14):3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brutinel ED, Vakulskas CA, Yahr TL (2010) ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA binding activity. J Bacteriol 192(6):1479–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Broder UN, Jaeger T, Jenal U (2016) LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol 2:16184. [DOI] [PubMed] [Google Scholar]
- 127.Diepold A et al. (2017) A dynamic and adaptive network of cytosolic interactions governs protein export by the T3SS injectisome. Nat Commun 8:15940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mougous JD et al. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312(5779):1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leiman PG et al. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106(11):4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Allsopp LP et al. (2017) RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114(29):7707–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.He X, Wang S (2014) DNA consensus sequence motif for binding response regulator PhoP, a virulence regulator of Mycobacterium tuberculosis. Biochemistry 53(51):8008–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kreamer NN, Costa F, Newman DK (2015) The ferrous iron-responsive BqsRS two-component system activates genes that promote cationic stress tolerance. MBio 6(2):e02549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schaaf S, Bott M (2007) Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J Bacteriol 189(14):5002–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sievers F et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol 7:539–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tamura K et al. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599 [DOI] [PubMed] [Google Scholar]
- 136.Vescovi EG et al. (1997) Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J Biol Chem 272(3):1440–1443 [DOI] [PubMed] [Google Scholar]
- 137.Lesley JA, Waldburger CD (2001) Comparison of the Pseudomonas aeruginosa and Escherichia coli PhoQ sensor domains: evidence for distinct mechanisms of signal detection. J Biol Chem 276(33):30827–30833 [DOI] [PubMed] [Google Scholar]
- 138.Cai X et al. (2011) The effect of the potential PhoQ histidine kinase inhibitors on Shigella flexneri virulence. PLoS One 6(8):e23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tu X et al. (2006) The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc Natl Acad Sci 103(36):13503–13508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bishop RE et al. (2004) Enzymology of lipid a palmitoylation in bacterial outer membranes. J Endotoxin Res 10(2):107–112 [DOI] [PubMed] [Google Scholar]
- 141.Macfarlane EL et al. (1999) PhoP–PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol 34(2):305–316 [DOI] [PubMed] [Google Scholar]
- 142.Shin D et al. (2006) A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314(5805):1607–1609 [DOI] [PubMed] [Google Scholar]
- 143.Macfarlane EL, Kwasnicka A, Hancock RE (2000) Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146(10):2543–2554 [DOI] [PubMed] [Google Scholar]
- 144.Gooderham WJ et al. (2009) The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155(3):699–711 [DOI] [PubMed] [Google Scholar]
- 145.Chamnongpol S, Cromie M, Groisman EA (2003) Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J Mol Biol 325(4):795–807 [DOI] [PubMed] [Google Scholar]
- 146.Regelmann AG et al. (2002) Mutational analysis of the Escherichia coli PhoQ sensor kinase: differences with the Salmonella enterica serovar typhimurium PhoQ protein and in the mechanism of Mg2+ and Ca2+ sensing. J Bacteriol 184(19):5468–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lesley JA, Waldburger CD (2003) Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme a. J Bacteriol 185(8):2563–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Flego D et al. (2000) A two-component regulatory system, pehR-pehS, controls Endopoly-galacturonase production and virulence in the plant pathogen Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact 13(4):447–455 [DOI] [PubMed] [Google Scholar]
- 149.Kariola T et al. (2003) Erwinia carotovora subsp carotovora and Erwinia-derived elicitors HrpN and PehA trigger distinct but interacting defense responses and cell death in Arabidop-sis. Mol Plant-Microbe Interact 16(3):179–187 [DOI] [PubMed] [Google Scholar]
- 150.Palomaki T et al. (2002) A putative three-dimensional targeting motif of polygalacturonase (PehA), a protein secreted through the type II (GSP) pathway in Erwinia carotovora. Mol Microbiol 43(3):585–596 [DOI] [PubMed] [Google Scholar]
- 151.Kaneshiro WS et al. (2008) Characterization of Erwinia chrysanthemi from a bacterial heart rot of pineapple outbreak in Hawaii. Plant Dis 92(10):1444–1450 [DOI] [PubMed] [Google Scholar]
- 152.Hugouvieux-Cotte-Pattat N, Shevchik VE, Nasser W (2002) PehN, a Polygalacturonase homologue with a low hydrolase activity, is Coregulated with the other Erwinia chrysanthemi Polygalacturonases. J Bacteriol 184(10):2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Guragain M et al. (2016) The Pseudomonas aeruginosa PAO1 two-component regulator CarSR regulates calcium homeostasis and calcium-induced virulence factor production through its regulatory targets CarO and CarP. J Bacteriol 198(6):951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Theodorou MC et al. (2006) Involvement of the AtoS-AtoC signal transduction system in poly-(R)-3-hydroxybutyrate biosynthesis in Escherichia coli. Biochim Biophys Acta 1760(6):896–906 [DOI] [PubMed] [Google Scholar]
- 155.Theodorou MC, Theodorou EC, Kyriakidis DA (2012) Involvement of AtoSC two-component system in Escherichia coli flagellar regulon. Amino Acids 43(2):833–844 [DOI] [PubMed] [Google Scholar]
- 156.Theodorou MC et al. (2007) Spermidine triggering effect to the signal transduction through the AtoS-AtoC/Az two-component system in Escherichia coli. Biochim Biophys Acta 1770(8):1104–1114 [DOI] [PubMed] [Google Scholar]
- 157.Theodorou EC, Theodorou MC, Kyriakidis DA (2013) Regulation of poly-(R)-(3-hydroxybutyrate-co-3-hydroxyvalerate) biosynthesis by the AtoSCDAEB regulon in phaCAB(+) Escherichia coli. Appl Microbiol Biotechnol 97(12):5259–5274 [DOI] [PubMed] [Google Scholar]
- 158.Jung C-J et al. (2017) AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun 85(9):e00252–e00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Downey JS et al. (2014) In vitro manganese-dependent cross-talk between Streptococcus mutans VicK and GcrR: implications for overlapping stress response pathways. PLoS One 9(12):e115975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kawasaki K (2012) Complexity of lipopolysaccharide modifications in Salmonella enterica: its effects on endotoxin activity, membrane permeability, and resistance to antimicrobial peptides. Food Res Int 45(2):493–501 [Google Scholar]
- 161.He X et al. (2008) The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol Microbiol 70(1):112–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bilecen K et al. (2015) Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect Immun 83(3):1199–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kierek K, Watnick PI (2003) The Vibrio cholerae O139 O-antigen polysaccharide is essential for ca2+−dependent biofilm development in sea water. Proc Natl Acad Sci 100(24):14357–14362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kierek K, Watnick PI (2003) Environmental determinants of Vibrio cholerae biofilm development. Appl Environ Microbiol 69(9):5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Giammarinaro P, Sicard M, Gasc AM (1999) Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology 145(Pt 8):1859–1869 [DOI] [PubMed] [Google Scholar]
- 166.Ibrahim YM et al. (2004) Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J Bacteriol 186(16):5258–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu C et al. (2010) Regulation of ciaXRH operon expression and identification of the CiaR regulon in Streptococcus mutans. J Bacteriol 192(18):4669–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jung CJ et al. (2017) AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun 85(9):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Senadheera MD et al. (2005) A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 187(12):4064–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reusch R (2013) The role of short-chain conjugated poly-(R)-3-Hydroxybutyrate (cPHB) in protein folding. Int J Mol Sci 14(6):10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Theodorou MC, Tiligada E, Kyriakidis DA (2009) Extracellular Ca2+ transients affect poly-(R)-3-hydroxybutyrate regulation by the AtoS-AtoC system in Escherichia coli. Biochem J 417(3):667–672 [DOI] [PubMed] [Google Scholar]
- 172.Yamamoto K et al. (2005) Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem 280(2):1448–1456 [DOI] [PubMed] [Google Scholar]
- 173.Rodrigue A et al. (2000) Cell signalling by oligosaccharides. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol 8(11):498–504 [DOI] [PubMed] [Google Scholar]
- 174.Siryaporn A, Goulian M (2008) Cross-talk suppression between the CpxA–CpxR and EnvZ–OmpR two-component systems in E. Coli. Mol Microbiol 70(2):494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wanner BL (1992) Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol 174(7):2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chin D, Means AR (2000) Calmodulin: a prototypical calcium sensor. Trends Cell Biol 10(8):322–328 [DOI] [PubMed] [Google Scholar]
- 177.Zhao X et al. (2010) Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein. Protein Cell 1(8):771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang M et al. (2012) Structural basis for calmodulin as a dynamic calcium sensor. Structure 20(5):911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ikura M (1996) Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci 21(1):14–17 [PubMed] [Google Scholar]
- 180.Crivici A, Ikura M (1995) Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct 24(1):85–116 [DOI] [PubMed] [Google Scholar]
- 181.Zhou Y et al. (2006) Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins 65(3):643–655 [DOI] [PubMed] [Google Scholar]
- 182.Inouye S, Franceschini T, Inouye M (1983) Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci 80(22):6829–6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Iwasa Y et al. (1981) Calmodulin-like activity in the soluble fraction of Escherichia coli. Biochem Biophys Res Commun 98(3):656–660 [DOI] [PubMed] [Google Scholar]
- 184.Kerson GW, Miernyk JA, Budd K (1984) Evidence for the occurrence of, and possible physiological role for, cyanobacterial calmodulin. Plant Physiol 75(1):222–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Leadlay PF, Roberts G, Walker JE (1984) Isolation of a novel calcium-binding protein from streptomyces erythreus. FEBS Lett 178(1):157–160 [Google Scholar]
- 186.Swan D et al. (1989) Cloning, characterization, and heterologous expression of the Saccha-ropolyspora erythraea (Streptomyces erythraeus) gene encoding an EF-hand calcium-binding protein. J Bacteriol 171(10):5614–5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Swan DG et al. (1987) A bacterial calcium-binding protein homologous to calmodulin. Nature 329(6134):84. [DOI] [PubMed] [Google Scholar]
- 188.Yonekawa T, Ohnishi Y, Horinouchi S (2005) A calmodulin-like protein in the bacterial genus Streptomyces. FEMS Microbiol Lett 244(2):315–321 [DOI] [PubMed] [Google Scholar]
- 189.Xi C et al. (2000) Symbiosis-specific expression of rhizobium etli casA encoding a secreted calmodulin-related protein. Proc Natl Acad Sci 97(20):11114–11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sarkisova SA et al. (2014) A Pseudomonas aeruginosa EF-hand protein, EfhP (PA4107), modulates stress responses and virulence at high calcium concentration. PLoS One 9(6):e98985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Scholl ZN et al. (2016) Single-molecule force spectroscopy reveals the calcium dependence of the alternative conformations in the native state of a βγ-Crystallin protein. J Biol Chem 291(35):18263–18275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Koul S, et al. , A novel calcium binding protein in Mycobacterium tuberculosis—potential target for trifluoperazine. 2009 [PubMed] [Google Scholar]
- 193.Advani MJ, Rajagopalan M, Reddy PH (2014) Calmodulin-like protein from M. Tuberculosis H37Rv is required during infection. Sci Rep 4:6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Reddy PT et al. (2003) Cloning and expression of the gene for a novel protein from Mycobacterium smegmatis with functional similarity to eukaryotic calmodulin. J Bacteriol 185(17):5263–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fry I, Becker-Hapak M, Hageman J (1991) Purification and properties of an intracellular calmodulinlike protein from Bacillus subtilis cells. J Bacteriol 173(8):2506–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nagai M et al. (1994) Purification and characterization of Bordetella calmodulin-like protein. FEMS Microbiol Lett 116(2):169–174 [DOI] [PubMed] [Google Scholar]
- 197.Vincent F et al. (2010) Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site. Environ Microbiol 12(6):1775–1786 [DOI] [PubMed] [Google Scholar]
- 198.Broder UN, Jaeger T, Jenal U (2017) LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol 2(1):16184. [DOI] [PubMed] [Google Scholar]
- 199.Falah A et al. (1988) On the presence of calmodulin-like protein in mycobacteria. FEMS Microbiol Lett 56(1):89–93 [Google Scholar]
- 200.Burra SS et al. (1991) Calmodulin-like protein and the phospholipids of Mycobacterium smegmatis. FEMS Microbiol Lett 80(2–3):189–194 [DOI] [PubMed] [Google Scholar]
- 201.Teintze M, Inouye M, Inouye S (1988) Characterization of calcium-binding sites in development-specific protein S of Myxococcus xanthus using site-specific mutagenesis. J Biol Chem 263(3):1199–1203 [PubMed] [Google Scholar]
- 202.Ryjenkov DA et al. (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187(5):1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dominguez DC, Guragain M, Patrauchan M (2015) Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 57(3):151–165 [DOI] [PubMed] [Google Scholar]
- 204.Dominguez DC (2004) Calcium signalling in bacteria. Mol Microbiol 54(2):291–297 [DOI] [PubMed] [Google Scholar]
- 205.Booth IR et al. (2015) The evolution of bacterial mechanosensitive channels. Cell Calcium 57(3):140–150 [DOI] [PubMed] [Google Scholar]
- 206.Guragain M et al. (2013) Calcium homeostasis in Pseudomonas aeruginosa requires multiple transporters and modulates swarming motility. Cell Calcium 54(5):350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jones HE, Holland IB, Campbell AK (2002) Direct measurement of free Ca2+ shows different regulation of Ca2+ between the periplasm and the cytosol of Escherichia coli. Cell Calcium 32(4):183–192 [DOI] [PubMed] [Google Scholar]
- 208.Naseem R et al. (2008) pH and monovalent cations regulate cytosolic free Ca2+ in E. Coli. Biochim Biophys Acta Biomembr 1778(6):1415–1422 [DOI] [PubMed] [Google Scholar]
- 209.Knight MR et al. (1991) Recombinant aequorin as a probe for cytosolic free Ca2+ in Escherichia coli. FEBS Lett 282(2):405–408 [DOI] [PubMed] [Google Scholar]
- 210.Jones HE et al. (1999) Slow changes in cytosolic free Ca2+ inEscherichia colihighlight two putative influx mechanisms in response to changes in extracellular calcium. Cell Calcium 25(3):265–274 [DOI] [PubMed] [Google Scholar]
- 211.Campbell AK et al. (2007) Fermentation product butane 2, 3-diol induces Ca2+ transients in E. Coli through activation of lanthanum-sensitive Ca2+ channels. Cell Calcium 41(2):97–106 [DOI] [PubMed] [Google Scholar]
- 212.Bruni GN et al. (2017) Voltage-gated calcium flux mediates Escherichia coli mechanosensation. Proc Natl Acad Sci 114(35):9445–9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Herbaud M-L et al. (1998) Calcium signalling in Bacillus subtilis. Biochim Biophys Acta 1448(2):212–226 [DOI] [PubMed] [Google Scholar]
- 214.Pavlov E et al. (2005) A high-conductance mode of a poly-3-hydroxybutyrate/calcium/polyphosphate channel isolated from competent Escherichia coli cells. FEBS Lett 579(23):5187–5192 [DOI] [PubMed] [Google Scholar]
- 215.Huang R, Reusch RN (1995) Genetic competence in Escherichia coli requires poly-beta-hydroxybutyrate/calcium polyphosphate membrane complexes and certain divalent cations. J Bacteriol 177(2):486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chang Y et al. (2014) Structural basis for a pH-sensitive calcium leak across membranes. Science 344(6188):1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu Q (2017) TMBIM-mediated Ca2+ homeostasis and cell death. BBA-Mol Cell Res 1864(6):850–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Haswell ES, Phillips R, Rees DC (2011) Mechanosensitive channels: what can they do and how do they do it? Structure 19(10):1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Martinac B (2004) Mechanosensitive ion channels: molecules of mechanotransduction. J Cell Sci 117(12):2449–2460 [DOI] [PubMed] [Google Scholar]
- 220.Moe PC, Blount P, Kung C (1998) Functional and structural conservation in the mechanosensitive channel MscL implicates elements crucial for mechanosensation. Mol Microbiol 28(3):583–592 [DOI] [PubMed] [Google Scholar]
- 221.Chang G et al. (1998) Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science 282(5397):2220–2226 [DOI] [PubMed] [Google Scholar]
- 222.Blount P et al. (1996) Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J 15(18):4798–4805 [PMC free article] [PubMed] [Google Scholar]
- 223.Velasquez J et al. (2014) Bacillus subtilis spore protein SpoVAC functions as a mechanosensitive channel. Mol Microbiol 92(4):813–823 [DOI] [PubMed] [Google Scholar]
- 224.Cox CD et al. (2013) Selectivity mechanism of the mechanosensitive channel MscS revealed by probing channel subconducting states. Nat Commun 4:2137. [DOI] [PubMed] [Google Scholar]
- 225.Kühlbrandt W (2004) Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol 5:282. [DOI] [PubMed] [Google Scholar]
- 226.Rensing C et al. (2000) CopA: an Escherichia coli cu(I)-translocating P-type ATPase. Proc Natl Acad Sci 97(2):652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rosch JW et al. (2008) Calcium efflux is essential for bacterial survival in the eukaryotic host. Mol Microbiol 70(2):435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gupta HK, Shrivastava S, Sharma R (2017) A novel calcium uptake transporter of unchar-acterized P-type ATPase family supplies calcium for cell surface integrity in Mycobacterium smegmatis. MBio 8(5):e01388–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.States DJ, Gish W (1994) Combined use of sequence similarity and codon bias for coding region identification. J Comput Biol 1(1):39–50 [DOI] [PubMed] [Google Scholar]
- 230.Huang Y et al. (2009) Multiple Ca2+ binding sites in the extracellular domain of Ca2+-sensing receptor corresponding to cooperative Ca2+ response. Biochemistry 48(2):388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hendy GN et al. (2000) Mutations of the calcium-sensing receptor (CASR) in familial hypocalciuric hypercalcemia, neonatal severe hyperparathyroidism, and autosomal dominant hypocalcemia. Hum Mutat 16(4):281–296 [DOI] [PubMed] [Google Scholar]
- 232.Holstein DM et al. (2004) Calcium-sensing receptor-mediated ERK1/2 activation requires Gαi2 coupling and dynamin-independent receptor internalization. J Biol Chem 279(11):10060–10069 [DOI] [PubMed] [Google Scholar]
- 233.Di Mise A et al. (2018) Activation of calcium-sensing receptor increases intracellular calcium and decreases cAMP and mTOR in PKD1 deficient cells. Sci Rep 8(1):5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Smith KA et al. (2016) Calcium-sensing receptor regulates cytosolic [Ca2+] and plays a major role in the development of pulmonary hypertension. Front Physiol 7:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Huang Y-H et al. (2016) Characterization of Staphylococcus aureus Primosomal DnaD protein: highly conserved C-terminal region is crucial for ssDNA and PriA helicase binding but not for DnaA protein-binding and self-Tetramerization. PLoS One 11(6):e0157593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mosyak L et al. (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J 19(13):3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Koskiniemi S et al. (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110(17):7032–7037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Drees JC et al. (2006) Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J Biol Chem 281(8):4708–4717 [DOI] [PubMed] [Google Scholar]
- 239.Briggs GS et al. (2007) Ring structure of the Escherichia coli DNA-binding protein RdgC associated with recombination and replication fork repair. J Biol Chem 282(17):12353–12357 [DOI] [PubMed] [Google Scholar]
- 240.Hernández-Ochoa EO et al. (2015) Critical role of intracellular RyR1 calcium release channels in skeletal muscle function and disease. Front Physiol 6:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Xu L et al. (2018) G4941K substitution in the pore-lining S6 helix of the skeletal muscle ryanodine receptor increases RyR1 sensitivity to cytosolic and luminal Ca2+. J Biol Chem 293(6):2015–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Clarke OB, Hendrickson WA (2016) Structures of the colossal RyR1 calcium Release Channel. Curr Opin Struct Biol 39:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kane Dickson V, Pedi L, Long SB (2014) Structure and insights into the function of a Ca2+- activated Cl(−) channel. Nature 516(7530):213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yang T et al. (2014) Structure and selectivity in bestrophin ion channels. Science 346(6207):355–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kuo YH et al. (2014) Effects of alternative splicing on the function of bestrophin-1 calcium-activated chloride channels. Biochem J 458:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]


