Abstract
FLT3 receptor is an important therapeutic target in acute myeloid leukemia due to high incidence of mutations associated with poor clinical outcome. Targeted therapies against the FLT3 receptor, including small-molecule FLT3 tyrosine kinase inhibitors (TKIs) and anti-FLT3 antibodies, have demonstrated promising preclinical and even clinical efficacy. Yet, even with the current FDA approval for two FLT3 inhibitors, these modalities were unable to cure AML or significantly extend the lives of patients with a common mutation called FLT3-ITD. While FLT3 is viable target, the approaches to inhibit its activity were inadequate. To develop a new modality for targeting FLT3, our team engineered an α-FLT3-A192 fusion protein composed of a single chain variable fragment antibody conjugated with an elastin-like polypeptide. These fusion proteins assemble into multi-valent nanoparticles with excellent stability and pharmacokinetic properties as well as in vitro and in vivo pharmacological activity in cellular and xenograft murine models of AML. In conclusion, α-FLT3-A192 fusions appear to be a viable new modality for targeting FLT3 in AML and warrant further preclinical development to bring it into the clinic.
Keywords: FLT3, AML, ELP, therapeutic target
Introduction
Acute myeloid leukemia (AML) is a hematological malignancy characterized by a block in differentiation and uncontrolled proliferation of immature myeloid cells called blasts [1]. Clinical outcome of patients with AML remains poor, with less than 30% of patients surviving 5 years [2]. Mutations within the FMS-like tyrosine kinase 3 (FLT3) occur frequently in AML (~ 30% of cases) and are associated with higher incidence of relapse and shorter overall survival [3]. In particular the FLT3-internal tandem duplication (FLT3-ITD) mutation occurs in the juxtamembrane (JM) domain of FLT3, which leads to constitutive activation of the FLT3 kinase and downstream JAK/STAT5, Raf/MEK/ERK, and PI3K/Akt. All of these pathways promote leukemia growth and cell survival [4, 5].
Multiple tyrosine kinase inhibitors (TKI) (midostaurin, gilteritinib, and sorafenib) with activity in FLT3 are under clinical evaluation [6–8]. Although these TKIs have promising activity in patients with relapsed or refractory AML or in combination with chemotherapy as first line therapy, they remain limited mainly by acquired resistance. Evolving of secondary mutations in FLT3, upregulation of the FLT3-WT receptor, and the FL ligands, in addition to activation of other kinase survival pathways are mechanisms that contribute to the development of TKI resistance in treated patients [9, 10]. Other emerging therapies gaining significance are target-specific antibodies. Due to their high binding specificity and affinity, antibodies against FLT3 could serve as an attractive approach to target FLT3.
Single-chain antibody fragments (scFv) are about 30 kDa and consist of variable regions of heavy (VH) and light (VL) chains, which are joined by a flexible peptide linker. scFvs are as specific as full-length monoclonal antibodies yet possess lower immunogenicity and are more easily manipulated through recombinant protein engineering. Unfortunately, scFvs have short circulating half-lives in humans, likely as a result of glomerular filtration in the kidneys [11]. When compared to monoclonal antibodies that require expression in mammalian cells [12], scFvs can be more economically produced in Escherichia Coli [13]. However, purification of scFvs from E. coli has been challenging because they lack the stability of monoclonal antibodies and tend to aggregate. In some cases, scFvs can be identified as inclusion bodies in the periplasm of E. coli [14, 15]. Inclusion body purification requires specialized steps of solubilization and protein refolding [16], which lowers protein yield [17]. To address these challenges, we developed anti-FLT3 scFv fused with an elastin-like polypeptide (ELP), A192. ELPs are genetically engineered protein polymers that consist of the amino acid sequence (VPGXG)n, which is derived from human tropoelastin. “X” represents a guest amino acid, and “n” indicates the number of the pentameric repeats. As ELP sequences are derived from human tropoelastin, some ELPs including A192 appear to be biocompatible and biodegradable [18], and in fact, one of ELP fusions, Vasomara™, has successfully entered clinical trials by applying ELP technology [19]. ELPs undergo reversible phase separation above a tunable transition temperature, which is determined by X and n. This phase separation can be induced with mild salt or heat, and high purity ELP fusions can be obtained without chromatography [20]. Our team showed that fusion of an scFv to A192 generates a soluble bioactive nanoparticle [21]. In that study, a CD20 targeted scFv was shown to assemble into worm-shaped nanoparticles, which efficiently induced apoptosis in models of B-cell lymphoma [21]. In this study we now report the development of a new nanoparticle that targets the FLT3 receptor, has an extended pharmacokinetic half-life, and demonstrates excellent pharmacological activity, a first of its kind therapy for AML.
Methods
α-FLT3-A192 cloning, expression and purification
The amino acid sequence for heavy and light variable fragment encoding α-FLT3 scFv (Supplemental methods and Fig. S1) was obtained from IMC-EB10, one of the anti-FLT3 monoclonal antibodies patented by US20090297529A1, and the monoclonal antibody binds domain 4 of FLT3 and cross-reacts with mouse FLT3 [22]. The gene was fused to the amino terminus of an ELP called A192, in the pET-25b(+) vector, encoding α-FLT3-A192. For cloning, an α-FLT3 scFv gene was purchased (Integrated DNA Technologies, IA, USA) and ligated into an empty pET25b(+) vector cut using NdeI and BamHI restriction enzymes (New England Biolabs, MA, USA). Next, BseRI and BssHII restriction enzymes (New England Biolabs, MA, USA) were used to digest pET25b(+)-α-FLT3 scFv and pET25b(+)-A192 to construct pET25b(+)-α-FLT3-A192. To confirm the correct ligation product, a diagnostic DNA digestion was performed using NdeI and BamHI restriction enzymes. The digestion was observed after electrophoresis on a 1% agarose gel. Further, the plasmid was sent for DNA sequencing with the primer specific for T7 promoter (TAATACGACTCACTATAGGG) and T7 terminator (GCTAGTTATTGCTCAGCGG), which confirmed in-frame insertion into the pET-25b(+) vector. To purify the fusion protein, Clearcoli® BL21 (DE) Electrocompetent Cells (60810, Lucigen, WI, USA) were transformed with the α-FLT3-A192 plasmid. After transformation, colonies were picked and cultured in 60 ml of Terrific Broth with Glycerol (TB) (C8153, CulGeneX, CA, USA) with 100 μg/ml of carbenicillin at 37°C for 16–18 hours. ~10 mL of Clearcoli BL21 culture was added to an autoclaved 1L of TB with 100 μg/ml carbenicillin to culture a total of 6L of bacteria at 37 °C until the optical density at 600 nm (OD600) reached between 0.6 and 0.8. Once the measured OD600 reached 0.6, 400 μl of 1M isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to each 1L flask to bring the final concentration to 400 μM IPTG. IPTG induction was allowed to proceed overnight at room temperature. The next day, bacteria were recovered by centrifugation at 4,000 rpm for 15 minutes, and the supernatant was discarded. Each pellet was resuspended with 30 ml of cold PBS, vortexed, and disrupted using a probe-tip Misonix sonicator S-4000 (Misonix, NY, USA). After sonication, 0.5 % of polyethyleneimine (PEI) was added to each cell lysate, and the cell lysate was incubated on ice for 5 minutes. The cell lysate was centrifuged at 13,000 rpm for 15 minutes at 4 °C. The supernatant was collected, and solid NaCl was added to the supernatant to reach a final concentration of 2 M. Each supernatant was placed in the water bath at 37 °C until phase separation was observed. The supernatant was recovered by centrifugation at 4,000 rpm for 15 minutes at 37 °C. After centrifugation, the supernatant was discarded, and the pellet was resolubilized with cold PBS on ice. The resolubilized pellet was centrifuged at 13,000 rpm for 15 minutes at 4 °C, and the supernatant was collected. This process, hot and cold centrifugation, was repeated three times with a decreasing volume of PBS and NaCl concentration to obtain pure α-FLT3-A192.
α-FLT3 scFv cloning, expression and purification
To purify α-FLT3 scFv, the gene was inserted into pFUSE vector with a C-terminal His6 tag, and the gene was expressed in Expi293F cells (Thermo Fisher, MA, USA). The expressed protein was purified by using immobilized metal affinity chromatography (IMAC) using Ni-NTA beads (Thermo Fisher, MA, USA) followed by a Superdex 75 10/300 GL column (GE Healthcare Life Sciences, MA, USA) (Supplemental Information).
α-FLT3-A192 and α-FLT3 scFv protein concentration measurements
To measure the concentration of α-FLT3-A192, the fusion protein was denatured with 6 M guanidine hydrochloride to disrupt the assembly of nanoparticles. The protein concentration was measured using the following equation:
| (1) |
Where A280 is absorbance at 280 nm, A350 is absorbance at 350 nm, ε is the molecular extinction coefficient at 280 nm, and l is the path length (cm). To obtain ε, the following equation was employed [23]:
| (2) |
Using the above equation, ε of α-FLT3 scFv was estimated to be 40,130 L mol−1 cm−1 assuming all pairs of cysteine residues are oxidized to form cysteine. ε of A192 was estimated to be 1,490 L mol−1 cm−1 as it has only one tyrosine. Therefore, ε of α-FLT3-A192 was estimated to be 41,620 L mol−1 cm−1. The ε of α-FLT3 scFv expressed and purified from Expi293F cells was estimated to be 44,600 L mol−1 cm−1 assuming all pairs of cysteine residues are oxidized. The optical absorbance at 280 and 350 nm was measured with a NanoDrop 2000 (ThermoFisher Scientific Inc., MA, USA), which has a path length of 0.1 cm.
α-FLT3-A192 purity and transition temperature analysis
The purity of α-FLT3-A192 was measured using ImageJ (NIH, MD, USA). To run SDS-PAGE, 10 μg and 20 μg of α-FLT3-A192 were loaded to a 4–20% precast SDS-PAGE gel (4561095, BioRad Laboratories, CA, USA). The gel was stained with GelCode™ Blue Safe Protein Stain (24596, Thermo Fisher, MA, USA). The SDS-PAGE gel was imaged with a ChemiDoc Touch Image System (Bio-Rad Laboratories, CA, USA). Using ImageJ, a whole lane was plotted to obtain the area under each peak. The following equation was used to get the purity of the protein:
| (3) |
Apeak is the area under the peak of the interest, and Atotal peak is the area under all peaks observed on the SDS-PAGE gel. The transition temperature (Tt) was measured using Beckman Coulter DU 800 UV/Vis spectrometer (Beckman Coulter, CA, USA). To measure the transition temperature, four different concentrations of α-FLT3-A192, 3.125, 6.25, 12.5, and 25 μM were prepared. Then, the samples were heated at a rate of 1°C/min. starting from 20 °C to 85 °C. To find the transition temperature, OD350 was measured every 18 seconds. Tt was determined where the maximum first derivative of the OD350 with respect to temperature occurred. This data was fit to the following equation:
| (4) |
Where b is the y-intercept representing the extrapolated Tt at a concentration of 1 μM, m is a slope representing the change in °C per 10-fold change in ELP concentration, CELP.
α-FLT3-A192 protein refolding
To maximize the biological activity, the fusion protein was refolded (Fig. S2). The same volume of 8 M urea buffer with 10 mM β-mercaptoethanol (BME) as the volume of the fusion protein in PBS was added. After mixing the 8 M urea buffer, the fusion protein was dialyzed to 3 M urea buffer with 2 mM glutathione (GSH) and 0.4 mM oxidized glutathione (GSSH) for 24 hours at 4 °C. Then, the protein was dialyzed to 1 M urea buffer with 2 mM GSH and 0.4 mM GSSH, 0.5 M urea buffer, 0 M urea buffer, and then 2 times of PBS for 24 hours for each step at 4 °C. To measure the endotoxin level of the refolded α-FLT3-A192 and A192, three dilutions (10−3, 10−4, 105), were used to prepare protein samples. The chromogenic endotoxin testing kit, Pyrochrome® (Associates of Cape Cod, Inc., MA, USA), was used to quantify the endotoxin burden of the fusion protein and A192 (Supplemental method).
Measurements of the hydrodynamic radius of α-FLT3-A192 and colloidal stability
To measure the hydrodynamic radius and the stability of α-FLT3-A192, dynamic light scattering (DLS) was employed. To run DLS, 25 μM of α-FLT3-A192 was prepared in PBS, filtered with a 0.22 μm filter, and 60 μl of the protein was added to each well of a 384-well plate. To prevent evaporation of the protein, 15 μl of mineral oil was added to each sample. The hydrodynamic radius was measured with a DynaPro Plate Reader II (Wyatt Technology, CA, USA) at 37 °C. The hydrodynamic radius of A192 was also measured to observe how the fusion of α-FLT3-A192 changes the hydrodynamic radius. To measure the stability of the fusion protein, the plate was incubated at 37 °C, and every 24 hours, the hydrodynamic radius of α-FLT3-A192 was measured until 72 hours.
Measurement of absolute size and weight of α-FLT3-A192 nanoparticles
To measure the absolute molecular mass of α-FLT3-A192 nanoparticles, size exclusion chromatography-multi-angle light scattering (SEC-MALS) was employed. To run SEC-MALS, 10 μM of α-FLT3-A192 was prepared, and the protein was filtered with a 0.22 μm filter. A Shodex protein KW-804 (8.0mmI.D.×300mm) (Showa Denko America, NY, USA) was equilibrated with PBS. The fusion protein elusion was observed with three detectors, UV 210 mm detector (SYCLC-1200) (Agilent Technologies, CA, USA), multi-angle static light scattering detector (DAWN HELEOS) (Wyatt Technology, CA), USA, and differential refractometer (Optilab rEX) (Wyatt Technology, CA, USA). The data were analyzed by ASTRA 6 software.
α-FLT3-A192 binding study
To determine the binding specificity of α-FLT3-A192 to cells overexpressing FLT3 receptors, the fusion protein was labeled with 5(6)-Carboxytetramethylrhodamine succinimidyl ester (NHSrhodamine) (46406, Thermo Fisher, MA, USA) and free dye was removed by Zeba™ Spin Desalting Columns (Thermo Fisher, MA, USA). Optical density was used to quantify the rhodamine concentration for labeled ELPs using the following equation:
| (5) |
Where CELP is the concentration of rhodamine-labeled proteins, A555 is absorbance at 555 nm, εRhodamine is molecular extinction coefficient of NHS-rhodamine, which is 80,000 L mol−1 cm−1. Based on these measurements, the degree of labeling was ~1.2, ~1.6 rhodamine per A192, α-FLT3-A192 respectively. MOLM-13, MV4–1, and U937 cells were treated with 1 and 10 μM of the rhodamine-labeled protein and incubated for 30 minutes at 4 °C. As a negative control, 25 μM of rhodamine-labeled A192 was used. Cells were washed twice with PBS and transferred to glass-bottom MatTek plates (P35G-0.170–14-C, MatTek Corporation, MA, USA) for live cell imaging. Images were captured using ZEISS LSM 880 Confocal Laser Scanning Microscope (Zeiss, Germany) using a 63× objective. Images were quantified with ImageJ to compare the intensity levels among the different cell lines and treatment doses. For the competitive binding assays, 0.5×105 MOLM-13 and MV4–11 cells were pre-incubated with 0.4μg IgG (Santa Cruz, Cat # 2025) or anti-FLT3mAb (Thermo Fisher, Cat # 17135182) at 0.1, 0.2, and 0.4 μg in 100 μl PBS for 30 mins on ice. Cells were then washed and treated with rhodamine-labeled α-FLT3-A192 (10 μM) for 30 mins on ice. Unbound α-FLT3-A192 was washed away and binding of α-FLT3A192 was analyzed by measuring the shift in the mean fluorescent intensity (MFI) by flow cytometry.
Pharmacokinetic (PK) study of α-FLT3-A192 and α-FLT3 scFv
To compare the PK parameters of the α-FLT3-A192 Nanoparticles to those of free α-FLT3 scFv both formulations were NHS-rhodamine labeled and administered IV to ~20 g mice, injected into tail vein of female 4 to 6-week-old NOD-scid /Il2rg−/− (NSG) mice (n=5). The same concentration of α-FL3-A192 with the one in vivo study was used, and as the yield of α-FLT3 scFv was poor, the maximum concentration of the scFv was used. Labeled α-FLT3-A192 was dosed at 150 μl/20g BW of 360 μM rhodamine (220 μM total protein concentration) and α-FLT3 scFv was dosed at 150 μl/20g BW of 30 μM rhodamine (110 μM total protein concentration). Blood samples (10–30 μl of blood) were collected by venipuncture and mixed with 80 μl of heparinized cold PBS (1,000 U/ml). Nanoparticle plasma samples were obtained at 3 min, 30 min, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h, 36 h, 48 h, and 72 h post injection. scFv alone plasma samples were obtained at 3 min, 15 min, 30 min, 45 min, 1 h, 2 h, 4 h, 6 h, 10 h, 24 h, 48 h, and 72 h post injection. Samples were centrifuged at 2,000 g for 10 minutes to collect diluted plasma. The diluted plasma was transferred to a 96-well plate, and a Biotek Synergy H1 Hybrid Multi-Mode Reader (Biotek, VT, USA), which was employed to measure the fluorescence (Excitation/Emission: 540/580 nm). Standard curves were generated from the rhodamine concentrations of 2, 5, 14, 41, 123, 370, 1111, 3333, and 10000 nM for α-FLT3-A192 Nanoparticles. For α-FLT3 scFv, standard curve was plotted with the fluorescence measurements of 12, 37, 111, 333, and 1000 nM of rhodamine-labeled α-FLT3 scFv. After obtaining the rhodamine-labeled protein concentration in the plasma over time, the collected sera were run on an SDS-PAGE gel, and the gel was imaged with a ChemiDoc Touch Image System. All fluorescent proteins were visualized on the gel, and the fraction of the protein of interest was calculated using ImageJ at each time point (Fig. S4 and Table S3, S4, S5, S6). The fraction was applied to the measured concentration of α-FLT3 scFv and α-FLT3-A192, and the concentration over time was plotted to obtain PK curves.
Relative accumulation of α-FLT3-A192 nanoparticles using whole-tissue fluorescence imaging
To analyze the accumulation of rhodamine-labeled α-FLT3-A192, the liver, kidneys, and spleen were collected from NSG mice (n=5) after 96 hours post injection, and the organs were imaged with In Vivo Imaging System (IVIS) Lumina Series III (PerkinElmer, MA, USA). The exposure time for the imaging was 1 second. Similarly, to analyze the accumulation of rhodamine-labeled αFLT3 scFv, the liver, kidneys, and spleen were collected from NSG mice (n=5) after 96 hours post injection, and the organs were imaged with iBright FL1000 Imaging System (Thermo Fisher, MA, USA). The exposure time for the imaging was set to 1 second. To compare the relative accumulation of α-FLT3-A192 and α-FLT3 scFv in the liver, kidneys, and spleen, organ images collected from IVIS and iBright FL1000 Imaging System were analyzed with ImageJ, and % mean fluorescence was calculated in those organs by using the following equation:
| (6) |
where the total mean fluorescence is the sum of mean fluorescence of the liver, kidneys, and spleen. Mean fluorescence from each kidney was averaged to measure the accumulation to the kidneys.
Cell culture
MV4–11 cells were purchased from ATCC. MOLM-13 and U937 cells, were kindly provided by Dr. Wendy Stock’s lab. All the cell lines were authenticated at the University of Arizona Cell Authentication Core. All cell lines were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium (Thermo Fisher, MA, USA) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin (Thermo Fisher, MA, USA).
Viability assays
Cell viability experiments were performed by incubating 0.5×105 cells with either control-A192 or α-FLT3-A192 (1,10, 25 and 50 μM) for 30 minutes on ice and then seeding the cells at a concentration of 5×105/mL in a 12 well plate. The number of live cells at 72 hours was counted using trypan blue (cat.no:15250–061, Life Technologies, Carlsbad, USA), and cell viability was determined by the ratio of the number of live cells in treated samples to that in untreated cells. Experiments were performed three times. To determine the IC50 for MOLM-13, MV4–11 and U937 cells 5×104 cells treated with α-FLT3-A192 (0.01, 0.1, 0.5, 1, 10, 25, 50, 75 and 100 μM) were seeded in triplicate wells in a 96 well plate. Viability was then measured using alamar blue assay (cat no: DAL 1100, Invitrogen, Carlsbad, USA). Briefly, 72 hours after seeding cells, 10 μl of Alamar blue was added per well, and incubated for four hours at 37 °C, after which fluorescence was measured on a synergy H1 microplate reader. Change in viability was calculated by normalizing the fluorescence of treated cells to that of untreated cells. IC50 was calculated based on non-linear regression.
Flow cytometry analysis
The binding of A192 or α-FLT3-A192 to cell-surface FLT3 was assessed by flow cytometry. Briefly, 0.5×105 FLT3-ITD+ MOLM-13, MV4–11 cells or the FLT3null U937 cells were incubated with either rhodamine-labeled A192 or α-FLT3-A192 (1, 10, and 25 μM) for 30 minutes on ice. Cells were then washed to remove unbound antibody. Bound rhodamine-labeled A192 and α-FLT3-A192 were measured by assessing the shift in the mean fluorescence intensity (MFI) by flow cytometry. Data were analyzed by normalizing MFI to unstained cells. For in vitro apoptosis assays, cells were stained using the Annexin V and PI APC kit according to the manufactures protocol (Invitrogen, Cat no:88–8007-72). The percentage of APC+ cells were compared among groups. For in vivo experiments, engraftments were measured using cellsurface expression of human CD45 (huCD45: cat.no:25–0459-41, eBioscience, CA, USA). Cells were stained with PE-Cy7-A conjugated anti-huCD45. Mean florescence intensity (MFI) of PE-Cy7-A was used to quantify data. Data were analyzed using the LSRII BD Fortessa X20 flow cytometer and processed using FloJo software (BD, Franklin Lakes, NJ, USA).
Immunoblotting
Immunoblotting was performed by lysing cells in Pierce-Protease lysis buffer (cat.no:8788, Thermo Fisher, MA, USA), supplemented with a protease inhibitor mix (cat.no: A32959, Thermo Fisher, MA, USA). Protein concentrations were measured using the Bicinchoninic acid (BCA) protein assay reagent (Pierce, Thermo Scientific, MA, USA). 30 μg of total cellular lysates were added to each lane of SDS-PAGE gels. Lysates were resolved by electrophoresis and transferred overnight to a polyvinylidene fluoride membrane. Membranes were blocked with 5% non-fat milk or bovine serum albumin and probed with indicated antibodies: ERK (Cell Signaling, cat.no.9102), P-ERK (Cell Signaling, cat.no.9101), STAT5 (Cell Signaling, cat.no.9351), STAT5 (Santa Cruz, cat.no.sc-835), GAPDH (Santa Cruz, cat.no. sc-32233. Horseradish peroxidase (HRP)conjugated secondary antibodies (Santa Cruz Biotechnology, TX, USA) were used for detection. Immunodetection was achieved with the SuperSignal Chemiluminescent Substrate (Thermo Fisher) and detected by a Bio-Rad Chemidoc Gel Imaging machine.
In vivo efficacy studies
All animal protocols were approved by the Institution for Animal Care and Use Committee of the University of Southern California. For xenograft experiments, male and female 4- to 6-week-old NOD-scid /Il2rg−/− (NSG) were purchased from Jackson Laboratory (Bar Harbor, ME). Approximately 2.5×106 MOLM-13 cells were administered via tail vein injection. Data were obtained and summarized from two independent experiments. For the first experiment male mice were engrafted and treated with A192 (n=5), α-FLT3-A192 (n=5) or midostaurin (n=3). For the second experiment female mice were engrafted with MOLM-13 cells were treated with A192 (n=3) or α-FLT3-A192 (n=3). Based on IC50 of α-FLT3-A192, it was decided to use the maximum possible concentration of α-FLT3-A192 and so that the plasma concentration can be close to the measured IC50 in MOLM-13 cells. In both experiments, mice were treated with 200 μL of 220 μM A192 or α-FLT3-A192 via tail vein on day 7, 10, 13 and 16 post engraftments. 100mg/kg midostaurin was administered daily by oral gavage on days 7 to 11 based on previous reports [24, 25]. Mice were euthanized on day 17 and organs (spleen, bone marrow and blood) were collected for further analyses. Isolated cells from bone marrow and peripheral blood, tissues were stained for huCD45 and analyzed using flow cytometry. For survival analysis, female mice engrafted with MOLM-13 cells were treated with 200 μL of 220 μM of A192 (n=7) or α-FLT3-A192 (n=7) on days 7, 10, 13 and 16 post leukemia engraftments. Mice were euthanized when they displayed signs of distress including: lethargy, severe weight loss, hair loss, and/or hunched appearance.
Statistical Analysis
A global one-way ANOVA followed by the Tukey post-hoc test was used to determine whether mean values are significantly different between groups for confocal and IVIS image analysis. All the data is presented as mean ± standard deviation (SD). Student t test was used to determine if the difference in means between two groups was statistically significant for viability, apoptosis, and engraftment analysis. For the mice survival analysis, Kaplan Meier survival analysis was performed based on Log-rank (Mantel Cox). p < 0.05 was considered significant.
Results
Characterization of α-FLT3-A192 nanoparticles
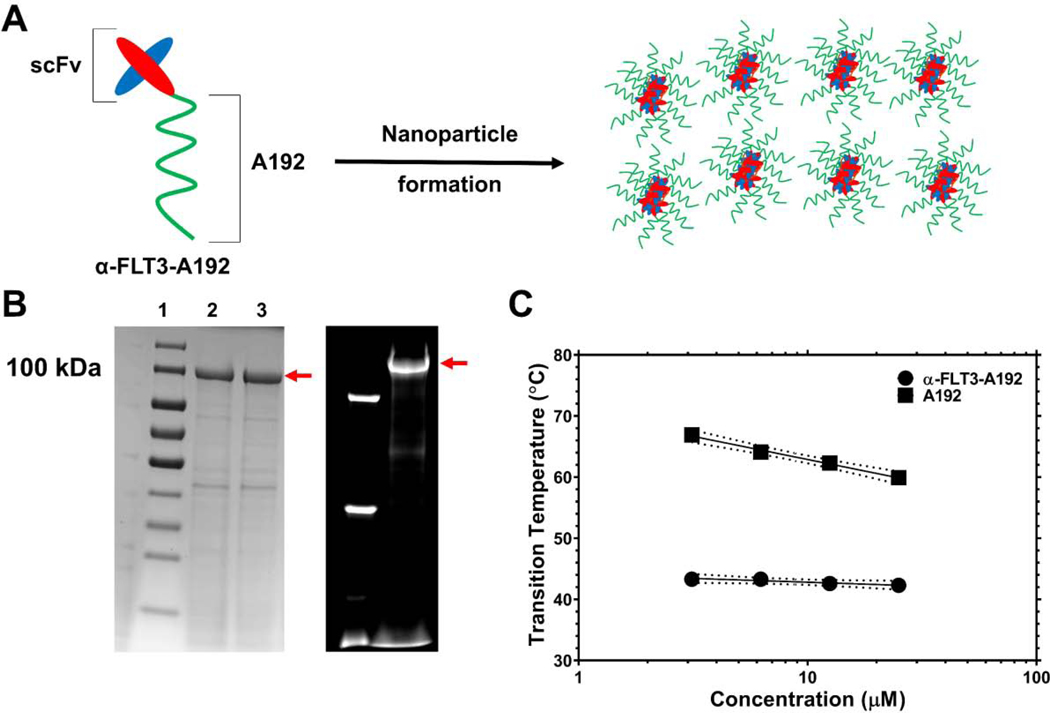
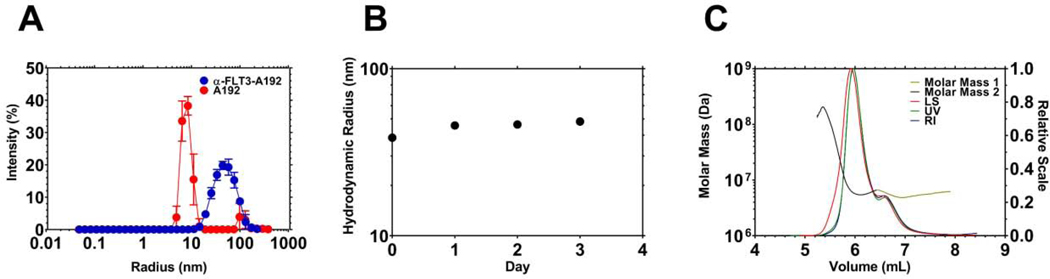
The fusion protein, α-FLT3-A192, was expressed in Clearcoli BL21, and the produced fusion protein was soluble in PBS. Both ELPs (Table 1) undergo reversible phase separation above a transition temperature (Tt) [18]. Through induction of this phase separation, α-FLT3-A192 was purified at a level of 77 mg/L culture with an apparent purity of ~82 % (Fig. 1B). The purity measured on fluorescent imaging of an SDS-PAGE gel with NHS-rhodamine labelled α-FLT3-A192 was ~92 %. The fusion of α-FLT3 scFv to A192 substantially lowered Tt compared to A192. As shown in Fig. 1C, α-FLT3-A192 phase separates above 42.3 °C while A192 phase separates above 59.9 °C (25 μM in PBS). Using the chromogenic endotoxin testing kit, Pyrochrome®, the endotoxin level of A192 and α-FLT3-A192 was measured to be 0.06 EU/nmol and 23.9 EU/nmol respectively. Since the α-FLT3-A192 was produced in ClearColi expression system, these nanoparticles cannot activate human TLR-4 [26]. In addition, the ClearColi expression system significantly lowered the detected LAL endotoxin level of α-FLT3-A192. When the fusion protein was expressed in Shuffle® T7 cells (New England Biolabs, MA, USA), we observed significantly higher endotoxin level (3400 EU/nmol) compared with that in the ClearColi cells (Table S1). DLS analysis shows that α-FLT3-A192 forms nanoparticles at 37 °C (Fig. 2A, Table S2). The hydrodynamic radius of α-FLT3-A192 was monitored for 72 hours, which demonstrated that α-FLT3-A192 nanoparticles are stable colloids at 37 °C in PBS (Fig. 2B). The average of absolute mass of nanoparticles was analyzed with SEC-MALS and indicated that approximately 60 α-FLT3-A192 molecules form a nanoparticle. SEC resolved a minor peak and major peak with an absolute mass of 5.6×106 Da and 6.4×106 Da, respectively (Fig. 2C). The Rg/Rh ratio = 1.1 for the major peak, which is consistent with an extended rod-like nanostructure (Table 1).
Table 1.
Biophysical characterization of elastin-like polypeptides evaluated in this manuscript
| ELP | Amino acid sequence | M.W. (kDa)b | Tt (°C)c |
Rh (nm)d |
Rg (nm)d |
Rg/Rh | Shapee |
|---|---|---|---|---|---|---|---|
| A192 | MG(VPGAG)192Y | 73.6 | 59.9 | 7.5 (0.2) |
N/A | N/A | N/A |
| a-FLT3- A192 |
a-FLT3a - G(VPGAG)192Y | 100.1 | 42.3 | 38.6 (0.7) |
42.6 (0.7) | 1.1 | extended |
the amino acid sequence is: MEVQLVQSGAEVKKPGASVKVSCKASGYTFTSYYMHWVRQAPGQGLEWMGIINPSGGSTSYAQKFQGRVTMTRDTSTSTVYMELSSLRSEDTAVYYCARGVGAHDAFDIWGQGTTVTVSSGGGGSGGGGSGGGGSDVVMTQSPLSLPVTPGEPASISCRSSQSLLHSNGNNYLDWYLQKPGQSPQLLIYLGSNRASGVPDRFSGSGSDTDFTLQISRVEAEDVGVYYCMQGTHPAISFGQGTRLEIK-LVPRGS
expected molecular weight based on the amino acid sequence and confirmed by SDS-PAGE (Fig. 1B)
transition temperature defined as the maximum first derivative of the optical density at 350 nm for 25 μM ELP in PBS (Fig. 1C)
values indicate the mean (standard deviation) of n=5
the Rg/Rh ratio of α-FLT3-A192 is consistent with a ‘rod-like’ nanoparticle of an extended aspect ratio as estimated using DLS and SEC-MALS
Figure 1. Characterization of an elastin-like polypeptide (ELP) fusion protein targeting the FLT3 receptor tyrosine kinase.
A) A single-chain variable fragment (scFv) targeting the FLT3 receptor tyrosine kinase was genetically fused to the amino-terminus of a high molecular weight ELP, A192 and expressed in E. coli. Dynamic light scattering (DLS) and size exclusion chromatography with multi-angle light scattering (SEC-MALS) suggest that α-FLT3-A192 forms nanoparticles. B) The recombinant fusion protein was purified using ELP-mediated phase separation, which was induced by increasing temperature and sodium chloride concentration. Using cycles of ‘cold’ and ‘hot’ centrifugation, recombinant fusion protein was obtained at high purity as demonstrated by SDS-PAGE stained by Coomassie. The red arrow indicates the major band for α-FLT3-A192. Lanes 2, 3 show the sample after 2, 3 purification cycles respectively. The purity of the major band in Lane 3 was estimated at 81.8%. On the right side, the fluorescent imaging of an SDS-PAGE gel shows the results of rhodamine labeling of α-FLT3-A192 used for PK study. The purity of the major band observed in the fluorescent imaging was estimated at 92.3 %. C) Optical density (350 nm) was used to evaluate ELP phase behavior over a range of temperatures and concentrations. While attachment of the scFv reduces the phase diagram curve with respect to free A192, at physiological salt concentrations (PBS) these ELP nanoparticles are expected to remain soluble during circulation at 37 °C.
Figure 2. Recombinant α-FLT3-A192 fusions form stable nanoparticles.
A) The hydrodynamic radius (Rh) of recombinant α-FLT3-A192 was measured using dynamic light scattering (DLS) at 37 °C. While A192 has a size consistent with the molecular weight of a free polymer, α-FLT3-A192 formed a larger and more polydisperse population of nanoparticles. B) Nanoparticle stability at 37 °C was measured with DLS over three days. The Rh did not change substantially, which suggests the nanoparticles remain stable colloids. C) Size exclusion chromatography multi-angle light scattering (SEC-MALS) was employed to measure the absolute molar mass and the radius of gyration of the nanoparticles. Two peaks were observed, and their average molecular weights were 6.4×106 Da and 5.6×106 Da for major peak 1, minor peak 2 respectively. Based on the expected molecular weight of the expressed fusion protein, these nanoparticles are composed of ~sixty FLT3-A192 molecules. These nanoparticles have Rg/Rh ratio = 1.1, which is consistent with an extended nanoworm shape.
α-FLT3-A192 binds specifically to FLT3 surface receptor in vitro
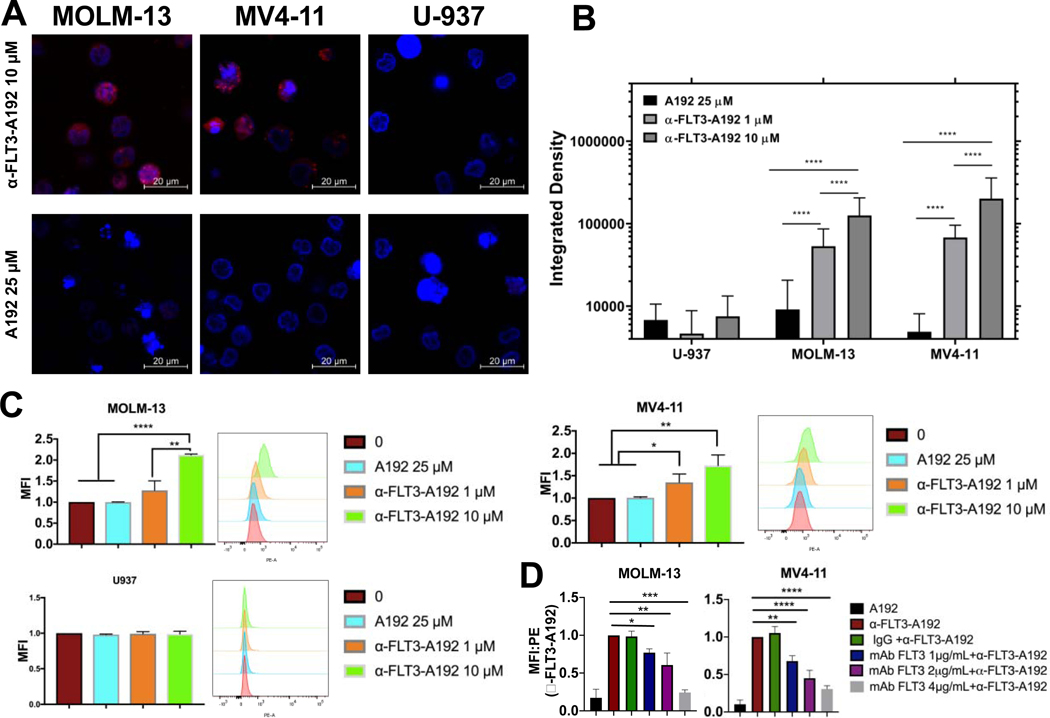
To assess the binding affinity and specificity of α-FLT3-A192 to the FLT3 receptor, MOLM-13 and MV4–11 cells (FLT3-ITD+ AML cell lines) or U937 cells (FLT3− AML cell line) were incubated with 1 μM or 10 μM of rhodamine-labeled α-FLT3-A192 or 25 μM of rhodamine labeled A192 for 30 minutes. Confocal microscopy images showed dose dependent increase in the fluorescence signals obtained from MOLM-13 and MV4–11 cells treated with α-FLT3-A192 compared with negative signal in A192 treated cells, which is consistent with positive binding. In addition, FLT3 negative U937 cells did not show any binding of α-FLT3-A192, which suggests that the fusion protein is specific to FLT3 receptors. A192 did not result in fluorescence signals in any of the cell lines, suggesting lack of binding. (Fig. 3A and 3B).
Figure 3. Recombinant α-FLT3-A192 binds to FLT3 receptor tyrosine kinase.
FLT3 ITD+ AML cell lines MOLM-13 and MV4–11 and FLT3 negative U937 cells (0.5 × 106 cells each) were treated with 1 μM or 10 μM of rhodamine labeled α-FLT3-A192 or 25 μM of rhodamine labeled A192 for 30 minutes on ice. Binding was only observed in MOLM-13 and MV4–11 via A) Laser scanning confocal microscopy. B) Cell fluorescence was quantified with ImageJ by analyzing images obtained from laser scanning confocal microscopy. Higher fluorescence signal was observed in cells treated with 10 μM α-FLT3-A192 compared with 1 μM. (U-937, n=35, 56, 20; MOLM-13, n=203, 70, 84; MV4–11, n=76, 62, 24) C) Binding of rhodamine labeled α-FLT3-A192 to MV4–11 and MOLM-13 cells was confirmed via flow cytometry by measuring peak shift in rhodamine to bounds cells and quantifying based on mean fluorescence imaging. No binding was observed in U937 cells (FLT3 negative). A rhodamine-labeled A192 control failed to bind to any of the cell lines. D) Competitive binding assay was performed in MOLM-13 and MV4–11 cells by pre-treating cells with either anti-FLT3mAb (1, 2, 4 μg/mL) or IgG and then observing binding with rhodamine α-FLT3-A192 (10 μM). Binding was measured via Flow cytometry by measuring the peak shift in rhodamine to bound cell and quantifying based on mean fluorescence intensity. MFI was normalized to cells treated with α-FLT3-A192 alone. Data represented as mean ± SD, n=3. * p≤0.05 ** p≤0.01*** p≤0.001**** p≤0.0001
The binding of rhodamine labelled α-FLT3-A192 to cell surface FLT3 was also assessed using flow cytometry. In MOLM-13 cells a significant increase in binding was observed in cells treated with α-FLT3-A192 10 μM compared to A192 (normalized MFI 1.0 vs. 2.0; p<0.0001). In MV4–11, a significant increase in binding was observed between cells treated with A192 vs 1 μM of α-FLT3-A192 (normalized MFI 1.0 vs. 1.36; p=0.03) and in α-FLT3-A192 10 μM (normalized MFI 1.0 vs. 1.7; p=0.006). On the contrary, α-FLT3-A192 showed minimal binding in FLT3 negative U937 cells (Fig. 3C).
To further establish the specificity of α-FLT3-A192 binding to FLT3, a competitive binding assay was performed. MOLM-13 and MV4–11 cells were pre-treated with anti-FLT3mAb and were then washed and treated with rhodamine-labeled α-FLT3-A192. After normalizing to untreated cells, MFI of samples pre-treated with anti-FLT3mAb were normalized to cells treated with α-FLT3-A192 alone. In both MOLM-13 and MV4–11 cells pre-treated with anti-FLT3mAb (0.1, 0.2 and 0.4 μg) had a significant decrease in binding to α-FLT3-A192. Pre-treatment with an irrelevant IgG did not affect the binding of α-FLT3-A192 to surface FLT3. No binding was observed in cells treated with rhodamine-labeled A192 (Fig 3D).
Antileukemic activity of α-FLT3-A192 in AML cells
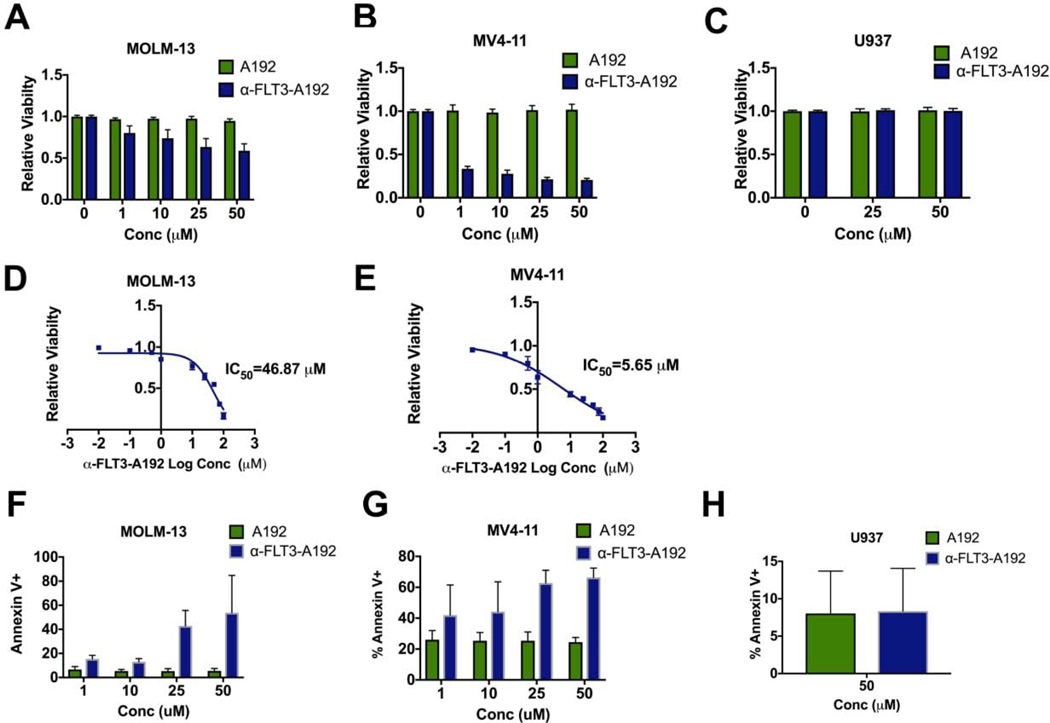
Next, to determine the antileukemia effects of α-FLT3-A192, FLT3-ITD+ cells (MOLM-13 and MV4–11 cells) were treated with α-FLT3-A192 at 1, 10, 25 and 50 μM, or the same concentrations of A192. Viable cells were counted using the trypan blue assay. The percentage of viable cells was significantly reduced when MOLM-13 cells were treated with α-FLT3-A192 compared with A192 at 1 μM (p=0.0009, 17% decrease), 10 μM (p=0.0002, 23% decrease), 25 μM (p<0.0001, 34% decrease) and 50 μM (p<0.0001, 36% decrease) (Fig. 4A). Similarly, in MV4–11 cells α-FLT3-A192 significantly reduced cell viability at 1 μM (p<0.0001, 67% decrease), 10 μM (p<0.0001, 71% decrease), 25 μM (p<0.0001, 80% decrease) and 50 μM (p<0.0001, 83% decrease) (Fig. 4B). No effect was seen on FLT3 negative U937 cells (Fig. 4C). We also performed viability assay using the α-FLT3 scFv that was generated using the same sequence but in mammalian cells and found strong anti-leukemic activity (Fig. S3). IC50 values of α-FLT3A192 for MOLM-13 and MV4–11 cells were determined as 46.87 μM and 5.65 μM, respectively (Fig. 4D and 4E respectively). We also assessed whether the decrease in cell viability was due to an enhanced apoptosis in α-FLT3-A192 treated cells. Apoptosis measured by Annexin V staining was significantly increased at 72 hours for MOLM-13 cells treated with α-FLT3-A192 (25 μM: p=0.007, 5.27% vs. 42.76%; and 50 μM: p=0.05, 5.58% vs. 53.83%; Fig. 4F). Similarly, MV411 cells treated with α-FLT3-A192 showed higher levels of apoptosis compared with A192 treated cells (25 μM: p=0.003, 25.36% vs 62.77%; and 50 μM: p=0.0004, 24.43% vs 66.36%; Fig. 4G). No effect was observed in FLT3 negative U937 cells (Fig. 4H).
Figure 4: α-FLT3-A192 has anti-leukemic activity in AML cells.
A-C) A trypan blue viability assay was performed in MOLM-13, MV4–11 (FLT3 ITD+) and U937 (FLT3 negative) cells treated with α-FLT3-A192 or an A192 control for 72 hours. The number of live cells was normalized to untreated cells. Data represented as mean ± SD, n=6. D-E) An IC50 of α-FLT3-A192 was measured using alamar blue staining in MOLM-13 and MV4–11 cells at 72 hours post treatment with the increasing concentration of α-FLT3-A192 and plotted based on non-linear regression. F-H) Apoptosis was measured by flow cytometry in MOLM-13, MV4–11 (FLT3 ITD+) and U937 (FLT3 negative) cells at 72 hours post treatment with α-FLT3-A192 or control A192 using APC conjugated Annexin V stain and normalized to untreated cells. Data represented as mean ± SD, n=3.
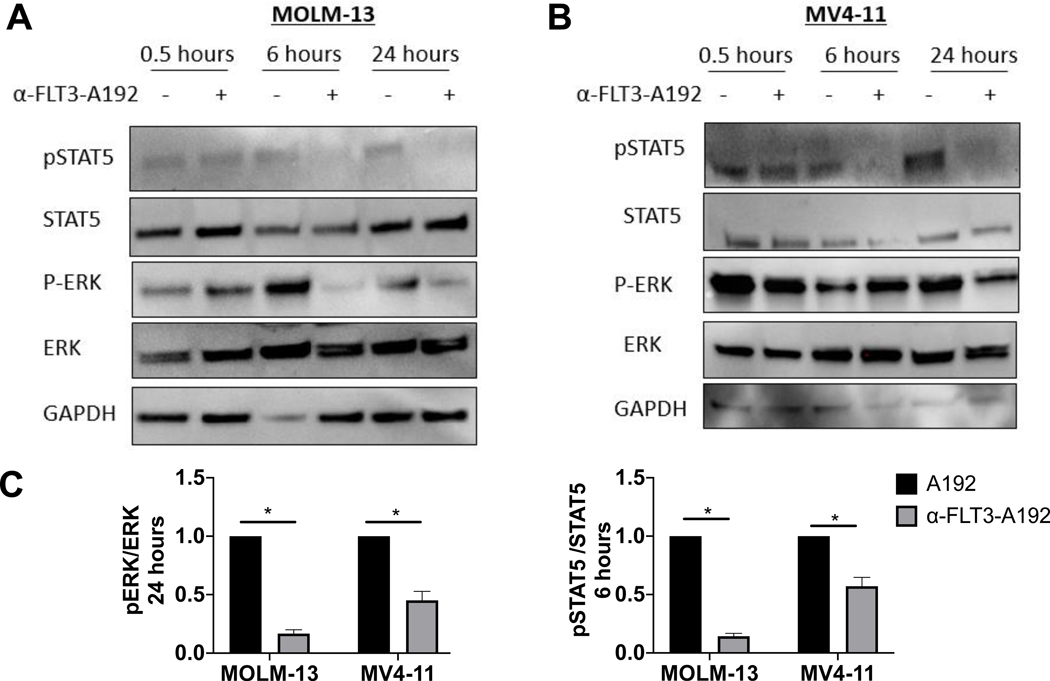
α-FLT3-A192 inhibits the FLT3 signaling pathway
FLT3-ITD mutations cause a constitutive activation of FLT3 kinase and subsequent activation of its downstream proliferative signaling pathways, including the Ras/MAPK kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway and PI3K/Akt pathway [27]. In contrast to wild-type FLT3 signaling, FLT3-ITD also activates the STAT5 pathway [28–30]. To determine the effect of α-FLT3-A192 nanoparticles on FLT3-ITD downstream signaling, FLT3ITD+ MOLM-13 and MV4–11 cells were treated with α-FLT3-A192 (25 μM) for 1, 6 and 24 hours. FLT3 targets phospho-STAT5 and phospho-ERK were assessed by western blot. There was a significant decrease in both phospho-STAT5 and phospho-ERK at 6 and 24 hours post treatment with α-FLT3-A192 compared with cells treated with control-A192 (25 μM). (MOLM-13 cells, Fig. 5A; MV4–11 cells, Fig. 5B)
Figure 5: α-FLT3-A192 inhibits FLT3 downstream signaling.
A) MOLM-13 and B) MV4–11 cells were treated with A192 or α-FLT3-A192 (25 μM). Cell lysates were collected at 0.5, 6 and 24 hours and prepared for western blot analysis; immunoblots were probed for STAT5 and ERK pathway. C) Quantified analysis for pERK/ERK and pSTAT5/STAT4 in MOLM-13 and MV4–11 cells. Experiment was performed in duplicates.
PK parameters of α-FLT3-A192 outperform α-FLT3 scFv
PK study of α-FLT3-A192 and α-FLT3 scFv showed that both proteins fit a biexponential decay model using the following equation:
| (7) |
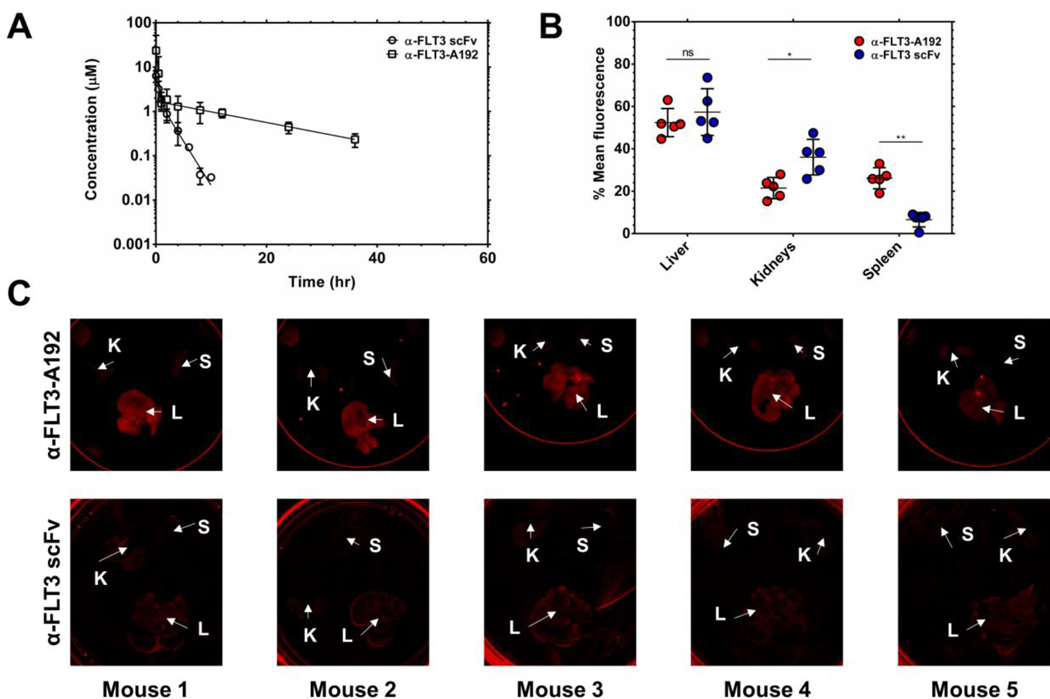
Where A, B, α, and β are ‘macroconstants’ that can be fit with nonlinear regression. Using a noncompartmental and two-compartmental analysis, the PK parameters of α-FLT3-A192 and α-FLT3 scFv were obtained. The terminal half-life of α-FLT3 scFv was measured to be 2.34 hours and mean residence time was 1.74 hours. The clearance of the protein was calculated to be 0.03 ml/hr·g, and the steady-state volume of distribution 0.06 mL/g. On the other hand, the terminal half-life of the fusion protein was determined to be 14.7 hours, and mean residence time was 18.7 hours. The clearance of the fusion protein was determined to be 0.07 mL/hr·g, and the steady-state volume of distribution was calculated to be 1.50 mL/g (Fig. 6A, Table 2, and Table 3). Images of spleen, kidneys, and liver showed that both α-FLT3 scFv and α-FLT3-A192 nanoparticles still resided in those organs after 96 hours from the injection (Fig. 6B and 6C). The whole-tissue fluorescence imaging analysis indicated that the relative accumulation of α-FLT3-A192 in the kidneys was significantly lower than that of α-FLT3 scFv (p≤0.05). On the other hand, the relative liver accumulation of α-FLT3-A192 was not statistically different from that of α-FLT3 scFv (p>0.05). The spleen accumulation of α-FLT3-A192 was significantly higher than that of α-FLT3 scFv (p≤0.01).
Figure 6. The pharmacokinetic terminal half-life of α-FLT3-A192 nanoparticles exceed that of a control α-FLT3 scFv:
220 μM of rhodamine-labeled α-FLT3-A192 and 110 μM of α-FLT3 scFv in PBS was injected via the tail vein to mice (n=5) in 150 μl/25g B.W. Plasma was collected and measured for fluorescence. Rhodamine-labeled α-FLT3-A192 and α-FLT3 scFv concentrations were calculated using a standard curve. As quality control, plasma samples were all analyzed by SDS-PAGE to determine what fraction of total fluorescence remained associated with the intact, labeled protein (Fig. S4). A) The plasma concentrations were fit to Eq. 7 to obtain the PK curve, and the generated curve shows rhodamine-labeled α-FLT3-A192 and α-FLT3 scFv concentration in the plasma over time. After 96 hours from the injection, spleen, kidneys, and liver were collected from mice treated with rhodamine-labeled α-FLT3-A192. B-C) IVIS images (α-FLT3-A192) and iBright images (α-FLT scFv) of organs were obtained 96 hours post-injection and quantified based on the mean fluorescence measured from images using ImageJ. The fraction of total mean fluorescence (the sum of mean fluorescence of the liver, kidneys, and spleen) was calculated for each organ (Eq. 6), and the % mean fluorescence was plotted to compare the relative accumulation in major clearance organs was plotted. The accumulation of α-FLT3-A192 in the liver was not significantly different from that of α-FLT3 scFv (p>0.05). On the other hand, the accumulation of α-FLT3-A192 in the kidneys was significantly lower than that of α-FLT3 scFv (p≤0.05). In contrast, accumulation of α-FLT3-A192 in the spleen was significantly higher than that of α-FLT3 scFv (p≤0.01).
Table 2.
Pharmacokinetic Parameters of α-FLT3-scFv and α-FLT3-A192 nanoparticles following IV administration using a non-compartmental analysis
| Parameter (Unit) | α-FLT3 scFva (n=5) | α-FLT3-A192a (n=5) | p value |
|---|---|---|---|
| CL/g BW (ml/h·g) | 0.03 (0.01) | 0.07 (0.02) | 0.009 |
| AUC (yM*h) | 7.14 (2.14) | 39.7 (14.9) | 0.001 |
| AUMC (μM*h2) | 11.7 (1.70) | 665 (238) | 0.0003 |
| MRT (h) | 1.74 (0.46) | 18.7 (7.8) | 0.001 |
| Vd/g BW (ml/g) | 0.03 (0.01) | 0.17 (0.09) | 0.008 |
| Vss/g (ml/g) | 0.06 (0.04) | 1.50 (0.76) | 0.003 |
values indicate the mean (standard deviation) of n=5 following a dose of 2.7 nanomoles of rhodamine-labeled α-FLT3-A192 (1.7 nanomoles of the total dose of α-FLT3-A192) per gram of mouse body weight or a dose of 0.2 nanomoles of rhodamine-labeled α-FLT3 scFv (0.8 nanomoles of the total dose of α-FLT3 scFv) alone.
Table 3.
Pharmacokinetic Parameters of α-FLT3-scFv and α-FLT3-A192 nanoparticles following IV administration using a two-compartment model
| Parameter (Unit) | α-FLT3 scFva (n=5) | α-FLT3-A192a (n=5) | p value |
|---|---|---|---|
| dose of total protein (nmol) | 20.6 (1.7) | 31.1 (5.7) | 0.004 |
| dose of rhodamine (nmol) | 5.52 (0.45) | 50.9 (9.4) | <0.0001 |
| Body weight (g) | 25.0 (2.0) | 18.8 (3.5) | 0.009 |
| A (μM) | 5.45 (1.80) | 31.6 (43.7) | 0.21 |
| B (μM) | 2.30 (1.37) | 1.83 (1.27) | 0.59 |
| α (h−1) | 12.1 (18.9) | 4.56 (1.63) | 0.40 |
| β (h−1) | 0.44 (0.19) | 0.056 (0.030) | 0.002 |
| t1/2α (h) | 0.26 (0.21) | 0.17 (0.05) | 0.36 |
| t1/2β (h) | 2.34 (2.20) | 14.7 (5.6) | 0.002 |
| C0 (μM) | 7.75 (2.31) | 33.4 (44.9) | 0.24 |
| AUC (μmol*h) | 6.73 (2.05) | 38.8 (14.6) | 0.001 |
| CL/g BW (ml/h·g) | 0.04 (0.01) | 0.08 (0.02) | 0.009 |
| V1/g BW (ml/g) | 0.03 (0.01) | 0.17 (0.09) | 0.008 |
| kelimination (h−1) | 1.17 (0.27) | 0.69 (0.62) | 0.15 |
| kplasma→tissue (h−1) | 5.18 (8.19) | 3.47 (1.52) | 0.66 |
| ktissue→plasma (h−1) | 6.18 (10.9) | 0.46 (0.28) | 0.27 |
values indicate the mean (standard deviation) of n=5 following a dose of 2.7 nanomoles of rhodamine-labeled α-FLT3-A192 (1.7 nanomoles of the total dose per gram BW of α-FLT3-A192) per gram of mouse body weight or a dose of 0.2 nanomoles of rhodamine-labeled α-FLT3 scFv (0.8 nanomoles per gram BW of the total dose of α-FLT3 scFv) alone. The average mouse body weight across these studies is 22 g.
Antileukemia activity of α-FLT3-A192 in FLT3-ITD xenograft murine model
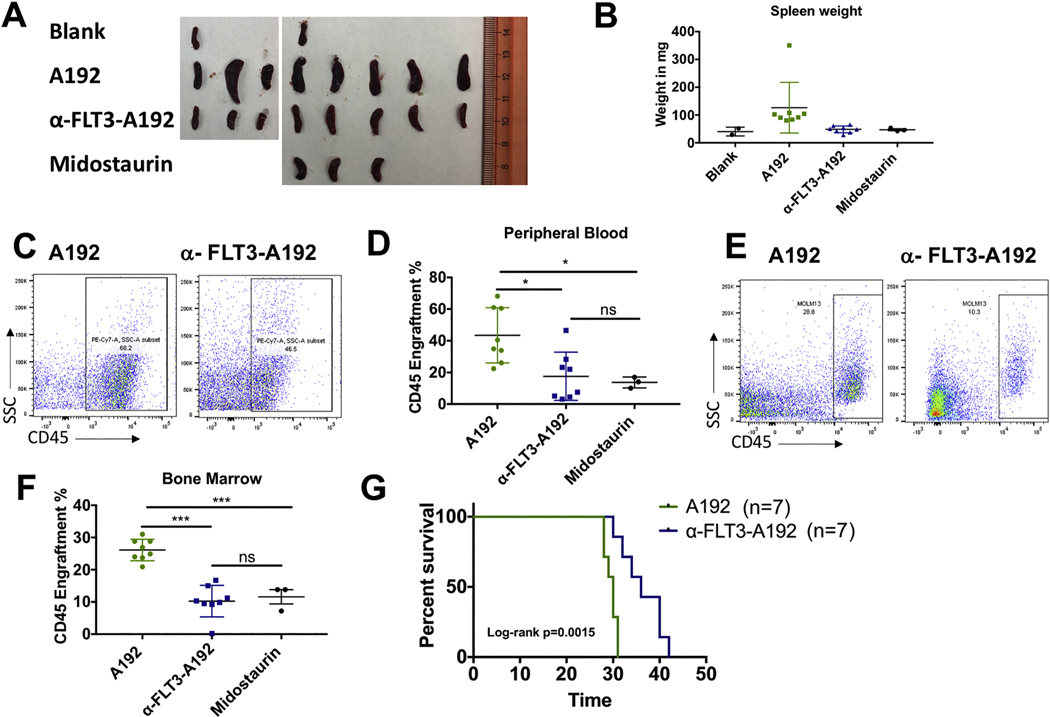
To assess the in vivo therapeutic potential of α-FLT3-A192 nanoparticles, the MOLM-13 leukemia murine model was used. NSG mice were engrafted with 2.5×106 MOLM-13 cells. Mice were then randomized into two treatment groups. Mice were treated with 176 mg/kg/mouse with either A192 (n=8) or α-FLT3-A192 (n=8) on day 7, 10, 13 and 16 or daily 100mg/kg midostaurin (n=3) on days 7–11 post leukemia engraftments. Mice were euthanized on day 17 and organs were evaluated for leukemia burden. Mice treated with α-FLT3-A192 had smaller spleens that weighed significantly less compared with the A192 treated mice (48 vs. 126 mg, p=0.03) (Fig. 7A). Flow cytometry analysis of huCD45 for leukemia engraftment revealed that α-FLT3-A192 mice had significantly less engraftment compared with the A192 mice in the peripheral blood (%huCD45: 17.5 vs. 43.44, p=0.01; Fig. 7C-D) and the bone marrow (%huCD45: 10.2 vs. 26.1%, p<0.0001; Fig. 7E-F). To demonstrate that this model is responsive to a kinase inhibitor of FLT3, a therapeutic regimen of midostaurin was used a positive control. Midostaurin resulted in smaller spleens as compared with A192 treated mice (46.83 vs. 126 mg). Analysis of huCD45 for leukemia engraftment revealed that midostaurin significantly reduced engraftment compared with the A192 mice in the peripheral blood (%huCD45: 13.73 vs. 43.44, p=0.03; Fig. 7C-D) and the bone marrow (%huCD45: 11.59 vs. 26.1%, p=0.0003; Fig. 7E-F). There was no difference in the engraftment of mice treated with midostaurin or α-FLT3-A192.
Figure 7.
Anti-leukemia activity of α-FLT3-A192 in FLT3-ITD murine model. Fig. A-F: 2.5 × 106 cells MOLM-13 cells/mouse were engrafted in NSG mice. Mice were treated with 200 μL of 220 μM of A192 (n = 8) or α-FLT3-A192 (n = 8) on day 7, 10, 13 and 16 post engraftment and euthanized on day 17. 100 mg/kg Midostaurin (N = 3) on day 7–11 post leukemia engraftments. A) Images of mice spleens show decreased in spleen size in mice treated with α-FLT3-A192 compared with A192 treated mice. Enlargement of spleen is consistent with progression of this AML model in A192 control mice. B) Spleen’s weights were measured, and spleens of mice treated with α-FLT3-A192 weighed significantly less compared with the A192 treated mice (0.48 vs. 126 mg, p = .03). C-D) Leukemia engraftment was measured using human CD45 (huCD45) antibody by flow cytometry in the peripheral blood (%huCD45- α-FLT3-A192: 17.5 vs. A192: 43.44, p =0 .01) and (%huCD45- midostaurin: 13.73 vs. A192: 43.44, p = .03) E-F) Similarly, CD45 engraftment was reduced by α-FLT3-A192 treatment in the bone marrow (%huCD45- α-FLT3-A192: 10.2 vs. A192: 26.1%, p < .0001) and (%huCD45- midostaurin: 11.59 vs. A192: 26.1%, p = .0003). There was no difference in the engraftment of mice treated with midostaurin or α-FLT3-A192. G) 2.5 × 106 MOLM-13 cells/mouse were engrafted in NSG mice and treated with 200 μL of 220 μM A192 (n = 7) or α-FLT3-A192 on day 7, 10, 13 and 16 post engraftment and survival was recorded. Kaplan Meier survival analysis showed that mice in the α-FLT3-A192 treatment group survived significantly longer than mice in the A192 group (median survival: 36 vs. 30 days, p = .0015).
Next, the effect of α-FLT3-A192 was observed on the survival of MOLM-13 engrafted mice. For survival analysis, 2.5×106 MOLM-13 cells were engrafted in NSG mice. Mice were treated with four doses 176 mg/kg/mouse of either A192 (n=7) or α-FLT3-A192 (n=7) on day 7, 10, 13 and 16. Mice in the α-FLT3-A192 treatment group survived significantly longer than mice in the A192 group (median survival: 36 vs. 30 days, p=0.0015).
Discussion
The FLT3 receptor is a key therapeutic target for AML because it is frequently overexpressed or mutated in leukemic blasts. Also, the presence of the ITD mutation is associated with a poor prognosis [31–34]. Targeted therapies against FLT3-ITD have long been investigated, including small-molecule FLT3 tyrosine kinase inhibitors (TKIs) and FLT3 antibodies, which showed promising results in clinical trials. Recently two FLT3 inhibitors received FDA approval for treating FLT3-ITD+ patients. Midostaurin is approved for the treatment of pre-transplant patients in combination with standard therapy. Despite progress, half of these patients die of their disease within 4 years. Gilteritinib is approved for relapsed and refractory AML in patients with FLT3-ITD. Yet, the Phase III ADMIRAL Trial showed that only 37% of treated patients survived longer than one year [2, 6]. In addition, the potential resistance emerging from the selective oncogene inhibition by a potent and highly specific kinase inhibitor like gilteritinib is a major clinical challenge. The presence of wild-type FLT3, which is sensitive to FLT3 ligand and relatively resistant to FLT3 inhibitors, may also contribute to resistance. In addition, the high level of FLT3 ligand found in the bone marrow microenvironment leads to persistent activity of the FLT3/MAPK pathway and provides survival signals to leukemic blasts. Activation of pathways downstream of FLT3 and FLT3-ITD, such as MAPK and STAT5, also contribute to inherent resistance to FLT3 inhibitors [10, 35, 36]. Furthermore, due to the limited specificity of first-generation TKIs there is increased toxicity associated with their high dose clinical use [37–39]. Altogether, these data challenge us to design and develop better therapeutic approaches for patients with FLT3-ITD. Antibodies against FLT3 were developed in an attempt to address the limitations associated with TKIs. Previously, two human anti-FLT3 mAbs, IMC-EB10 and IMC-NC7 were shown to have promising antileukemia activity both in vitro and in FLT3-ITD murine models [40]. However, phase 1 clinical trial (NCT00887926) to determine the safety of IMC-EB10 (LY3012218) in patients with relapsed or refractory AML was terminated due to lack of efficacy, even though the drug was safe [41]. It remains unclear why this antibody failed clinical testing. However, limitations associated with monoclonal antibody development may have contributed to this outcome. Factors related to the lack of therapeutic concentrations of the antibody on the target cells and limited clustering on the target receptors are critical pharmacological challenges of antibody therapies. Our study presents the development and validation of a new antibody-based FLT3 nanoparticle, which is composed of α-FLT3 single-chain variable fragment (scFv) linked to A192, an elastin-like polypeptide (ELP), derived from human tropoelastin.
Many studies regarding scFvs produced from E. coli are reported to forms aggregates, known as inclusion bodies; furthermore, these aggregates require complicated strategies to recover insoluble scFvs [42–44]. Interestingly, the fusion of A192 to α-FLT3 scFv generated a high yield of fusion protein (77 mg/L) in the soluble fraction after bacterial lysis. ELPs phase separate above tunable transition temperatures, and this characteristic facilitates the chromatography-free purification of the fusion protein. By using hot and cold centrifugation steps during protein purification, high purity of α-FLT3-A192 (82, 92 % purity by Coomassie, Rhodamine-labeling respectively) was obtained without attaching any additional purification tags, which can hinder protein activity [45, 46]. As humanized biomaterials, ELPs are biocompatible and biodegradable [18]. Conjugating A192 to α-FLT3 scFv increases its molecular weight and stabilizes these antibodies into nanoparticles. Although A192 consists of repeats containing aliphatic amino acids, their solubility is enhanced due to hydration of a predominantly random-coil peptide backbone. Conversely, most scFvs tend to form inclusion bodies when produced in E. coli as well as precipitation during protein refolding, which represents their tendency to oligomerize into aggregates. In this light, the most likely mechanism of assembly is that fusion of A192 blocks formation of inclusion bodies of α-FLT3 scFv, favoring the formation of stable colloids. Neither A192 nor α-FLT3-A192 nanoparticles undergo ELP-mediated coacervation until temperatures much higher than 37 °C. Meanwhile, the nanoparticles are observed even down to 10 °C. Thus, the tendency of scFv to precipitate and the high solubility of A192 suggests the nanoparticle core forms through oligomerization of the scFv protein domains.
Based on DLS and SEC-MALS, Rg/Rh ratio suggests that α-FLT3-A192 forms nanoworms, but there is a limitation of using the ratio as the fusion protein consists of two completely different proteins. To verify the fusion protein forms nanoworms, Cryo-TEM imaging can be employed in future. The stability of scFv fragments has always been a major challenge for developing scFvs as therapeutic antibodies [47–49]. However, the engineered α-FLT3-A192 nanoparticles proved stable for three days at 37 °C. This supports the feasibility for moving forward with this approach into further clinical development. The endotoxin levels of α-FLT3-A192 were significantly reduced by expressing the fusion protein in ClearColi cells instead of the Shuffle T7 cells. Lipopolysaccharide is the major pyrogen of concern for recombinant protein therapeutics; however, traditional anion exchange ‘polymixin’ chromatography is ineffective at removing endotoxin signal from these nanoparticles. As an alternative, these fusions have been expressed in Clear Coli bacteria, which produce a LipidA1 variant that is unable to activate human TLR4 and lacks the extended polysaccharide chain [26]. This strategy also reduced observed endotoxin levels on the chromogenic LAL test; however, levels remain higher than for free A192 or for FDA recommended guidelines. Thus while in vitro studies show the nanoparticles directly apoptose human cell lines and that free endotoxin is unable to do so, we cannot rule out TLR4-mediated recruitment of the innate immune system that remains in NOD/SCID mice.
The PK and pharmacodynamic testing of α-FLT3-A192 suggested excellent activity of this compound. In vitro, α-FLT3-A192 nanoparticles inhibit STAT5 and ERK phosphorylation in FLT3ITD+ cells (Fig. 5C), pathways that are known to drive cell survival and proliferation induced by FLT3-ITD mutations [50, 51]. This mechanistically corroborates that the α-FLT3-A192 nanoparticles indeed act on the FLT3 signaling pathway. This is consistent with a previous study showing that an anti-FLT3 antibody, IMC-EB10 that targets the same domain as α-FLT3-A192 (domain 4 of FLT3) inhibited the FLT3-mediated activation of downstream signaling kinases [22]. We also observed one magnitude difference in IC50 between MOLM-13 and MV4–11 cells. It is important to note that MV4–11 carries homozygous FLT3-ITD mutation while the MOLM-13 is heterozygous. This suggest that the MV4–11 cells are more dependent on the FLT3 signaling pathway compared with the MOLM-13 cells. This may potentially explain the higher sensitivity observed in the MV4–11 cells.
In the NSG murine model treatment reduced the leukemia burden and improved survival of MOLM-13 engrafted mice. Due to their small molecular weight, scFvs are susceptible to rapid renal clearance. The PK for a sample of scFvs administered to mice were assessed in brief review of the literature, which is detailed in the supplemental materials. Datapoints were extracted from the published plots to calculate half-lives and mean residence time (MRT) of several scFvs. The half-lives of scFvs were short, averaging about 2.2 ± 1.5 hours (mean ± 95% Cl, n=7). The MRT of scFvs was also short, averaging 2.4 ± 2.0 hours (mean ± 95% Cl, n=7) (Table S7) [11, 52–57]. In addition, α-FLT3 scFv showed an MRT and terminal half-life (Table 2 and Table 3) that are consistent with these prior reports. In contrast, the terminal half-life of α-FLT3-A192 nanoparticles was 14.7 hours and the MRT was 18.7 hours. Both of these are significantly greater than values expected for free scFvs. While additional studies are required to understand their pharmacology and PK in other species, the nearly day-long MRT of α-FLT3-A192 nanoparticles in mice likely contributed to its activity in the MOLM-13 model of FLT3-ITD AML. In addition, we also evaluated the activity of α-FLT3 scFv that was generated using the same scFv sequence but expressed in mammalian cells. Although the fusion of α-FLT3 scFv to A192 significantly increased the half-life, lower anti-FLT3 activity was observed compared to α-FLT3 scFv in vitro (Fig. S3B). If the scFv forms the core of the nanoparticles, then the otherwise soluble A192 may cause steric hindrance to receptor binding, which reduced the anti-FLT3 activity of α-FLT3-A192. In addition, as α-FLT3 scFv was purified from modified HEK293 cells, differences in post-translational modification and correct protein folding could further enhance the anti-FLT3 activity of α-FLT3 scFv compared to α-FLT3-A192. Therefore, future experiments that compare α-FLT3-A192 with the cleaved form generated with the same system α-FLT3 scFv is needed. This leukemia mouse model is aggressive due to rapid expansion of leukemic cells, so it is possible that the FLT3 receptor concentration in the model could be much higher than the anti-FLT3-A192 concentration. However, the PK study was done in healthy NOD-SCID mice where target saturation is not expected; therefore, we cannot exclude the possibility of target-mediated drug disposition (TMDD) effect on the PK.
Formulation of nanoparticles targeting FLT3 with a long half-life can bring advantages for AML treatment over anti-FLT3 mAbs. Targeting leukemic cells in both the systemic circulation and the bone marrow microenvironment is essential for an effective treatment for AML treatment. For mAbs to access the bone marrow, they need to be internalized through the reticuloendothelial uptake across the sinusoidal endothelial barrier [58]. The size of monoclonal antibodies is on the order of about 10 nm [59], which allows monoclonal antibodies to extravasate to many normal tissues [60, 61]. In general, mAbs do not specifically target the bone marrow microenvironment. Unlike a monoclonal antibody, the nanoparticulate nature α-FLT3-A192 appears to target two organs (liver, spleen) of the reticular endothelial system (Fig. 6B), which suggest they may also be substrates for reticuloendothelial uptake and retention in the bone marrow. Further, the average diameter of α-FLT3-A192 nanoparticles is 77.2 nm, which will restrict their extravasation to normal tissues in comparison with a mAb without impacting their retention in the bone marrow vasculature [62]. Further, the vasculature is leakier in patients with AML than in healthy people [58, 63], which may promote additional accumulation of α-FLT3-A192 in the bone marrow than for a monoclonal antibody. Therefore, in terms of targeting both peripheral blood and bone marrow, α-FLT3-A192 nanoparticles have the potential to be superior to anti-FLT3 mAbs. The above conjecture must be experimentally evaluated in future studies. With growing efforts to target FLT3 and a high need to overcome current limitations, we have developed a novel therapeutic approach to target FLT3 in AML using α-FLT3-A192. These fusion proteins assemble stable nanoparticles with high specificity and efficacy in vitro and effective therapeutic and PK properties in a mouse xenograft of AML. This report was designed to demonstrates that α-FLT3A192 nanoparticles obtained from bacterial fermentation have therapeutic efficacy and potential as a platform technology. This manuscript shows that nanoparticle pharmacokinetics are significantly prolonged compared to single-chain antibody control. To definitively evaluate their potential in future studies, nanoparticle efficacy and toxicology must be compared with traditional single chain and monoclonal antibodies as a function of dose and dose frequency.
Supplementary Material
Acknowledgements
This work was made possible by University of Southern California (USC), the Gavin S. Herbert Professorship, the National Institutes of Health R01 GM114839 to JM, the USC Ming Hsieh Institute, P30 CA014089 to the USC Norris Comprehensive Cancer Center, P30 EY029220 to the USC Ophthalmology Center Core Grant for Vision Research, Center of Excellence in Nanobiophysics at USC, and the Translational Research Laboratory at USC School of Pharmacy.
Footnotes
Declaration of interest statement
JAM, MP, VPV, and HA are inventors on pending provisional patent application related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lowenberg B, Acute myeloid leukemia: the challenge of capturing disease variety, Hematology Am Soc Hematol Educ Program, (2008) 1–11. [DOI] [PubMed] [Google Scholar]
- [2].Daver N, Schlenk RF, Russell NH, Levis MJ, Targeting FLT3 mutations in AML: review of current knowledge and evidence, Leukemia, 33 (2019) 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, Asou N, Kuriyama K, Jinnai I, Shimazaki C, Akiyama H, Saito K, Oh H, Motoji T, Omoto E, Saito H, Ohno R, Ueda R, Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia, Blood, 93 (1999) 3074–3080. [PubMed] [Google Scholar]
- [4].Takahashi S, Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications, J Hematol Oncol, 4 (2011) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ravandi F, Kantarjian H, Faderl S, Garcia-Manero G, O’Brien S, Koller C, Pierce S, Brandt M, Kennedy D, Cortes J, Beran M, Outcome of patients with FLT3-mutated acute myeloid leukemia in first relapse, Leuk Res, 34 (2010) 752–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Dohner K, Marcucci G, Lo-Coco F, Klisovic RB, Wei A, Sierra J, Sanz MA, Brandwein JM, de Witte T, Niederwieser D, Appelbaum FR, Medeiros BC, Tallman MS, Krauter J, Schlenk RF, Ganser A, Serve H, Ehninger G, Amadori S, Larson RA, Dohner H, Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation, N Engl J Med, 377 (2017) 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chappell G, Geer M, Gatza E, Braun T, Churay T, Brisson J, Bixby D, Marini B, Perissinotti A, Frame D, Parkin B, Reddy P, Magenau J, Choi SW, Maintenance sorafenib in FLT3-ITD AML following allogeneic HCT favorably impacts relapse and overall survival, Bone Marrow Transplant, 54 (2019) 1518–1520. [DOI] [PubMed] [Google Scholar]
- [8].Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, Montesinos P, Baer MR, Larson RA, Ustun C, Fabbiano F, Erba HP, Di Stasi A, Stuart R, Olin R, Kasner M, Ciceri F, Chou WC, Podoltsev N, Recher C, Yokoyama H, Hosono N, Yoon SS, Lee JH, Pardee T, Fathi AT, Liu C, Hasabou N, Liu X, Bahceci E, Levis MJ, Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML, N. Engl. J. Med, 381 (2019) 17281740. [DOI] [PubMed] [Google Scholar]
- [9].Daver N, Cortes J, Ravandi F, Patel KP, Burger JA, Konopleva M, Kantarjian H, Secondary mutations as mediators of resistance to targeted therapy in leukemia, in: Blood, 2015, pp. 3236–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Williams AB, Nguyen B, Li L, Brown P, Levis M, Leahy D, Small D, Mutations of FLT3/ITD confer resistance to multiple tyrosine kinase inhibitors, Leukemia, 27 (2013) 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hutt M, Farber-Schwarz A, Unverdorben F, Richter F, Kontermann RE, Plasma half-life extension of small recombinant antibodies by fusion to immunoglobulin-binding domains, J. Biol. Chem, 287 (2012) 4462–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hu X, O’Hara L, White S, Magner E, Kane M, Wall JG, Optimisation of production of a domoic acid-binding scFv antibody fragment in Escherichia coli using molecular chaperones and functional immobilisation on a mesoporous silicate support, Protein Expr. Purif, 52 (2007) 194–201. [DOI] [PubMed] [Google Scholar]
- [13].Skerra A, Pluckthun A, Assembly of a functional immunoglobulin Fv fragment in Escherichia coli, Science, 240 (1988) 1038–1041. [DOI] [PubMed] [Google Scholar]
- [14].Hayhurst A, Harris WJ, Escherichia coliskp chaperone coexpression improves solubility and phage display of single-chain antibody fragments, Protein Expr. Purif, 15 (1999) 336–343. [DOI] [PubMed] [Google Scholar]
- [15].Schein CH, Production of Soluble Recombinant Proteins in Bacteria, Bio-Technology, 7 (1989) 1141–1147. [Google Scholar]
- [16].Lavallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM, A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm, Biotechnology. (N. Y.), 11 (1993) 187. [DOI] [PubMed] [Google Scholar]
- [17].Thomson CA, Olson M, Jackson LM, Schrader JW, A simplified method for the efficient refolding and purification of recombinant human GM-CSF, PLoS One, 7 (2012) e49891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Despanie J, Dhandhukia JP, Hamm-Alvarez SF, MacKay JA, Elastin-like polypeptides: Therapeutic applications for an emerging class of nanomedicines, J. Control. Release, 240 (2016) 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yeboah A, Cohen RI, Rabolli C, Yarmush ML, Berthiaume F, Elastin-like polypeptides: A strategic fusion partner for biologics, Biotechnol. Bioeng, 113 (2016) 1617–1627. [DOI] [PubMed] [Google Scholar]
- [20].Christensen T, Amiram M, Dagher S, Trabbic-Carlson K, Shamji MF, Setton LA, Chilkoti A, Fusion order controls expression level and activity of elastin-like polypeptide fusion proteins, Protein Sci, 18 (2009) 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Aluri SR, Shi P, Gustafson JA, Wang W, Lin YA, Cui H, Liu S, Conti PS, Li Z, Hu P, Epstein AL, MacKay JA, A hybrid protein-polymer nanoworm potentiates apoptosis better than a monoclonal antibody, ACS Nano, 8 (2014) 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li Y, Li H, Wang MN, Lu D, Bassi R, Wu Y, Zhang H, Balderes P, Ludwig DL, Pytowski B, Kussie P, Piloto O, Small D, Bohlen P, Witte L, Zhu Z, Hicklin DJ, Suppression of leukemia expressing wild-type or ITD-mutant FLT3 receptor by a fully human anti-FLT3 neutralizing antibody, Blood, 104 (2004) 1137–1144. [DOI] [PubMed] [Google Scholar]
- [23].Pace CN, Vajdos F, Fee L, Grimsley G, Gray T, How to Measure and Predict the Molar Absorption-Coefficient of a Protein, Protein Sci, 4 (1995) 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Weisberg E, Boulton C, Kelly LM, Manley P, Fabbro D, Meyer T, Gilliland DG, Griffin JD, Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412, Cancer Cell, 1 (2002) 433–443. [DOI] [PubMed] [Google Scholar]
- [25].Weisberg E, Meng C, Case AE, Tiv HL, Gokhale PC, Buhrlage SJ, Yang J, Liu X, Wang J, Gray N, Adamia S, Sattler M, Stone R, Griffin JD, Effects of the multi-kinase inhibitor midostaurin in combination with chemotherapy in models of acute myeloid leukaemia, J Cell Mol Med, 24 (2020) 2968–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mamat U, Wilke K, Bramhill D, Schromm AB, Lindner B, Kohl TA, Corchero JL, Villaverde A, Schaffer L, Head SR, Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins, Microb Cell Fact., 14 (2015) 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gilliland DG, Griffin JD, The roles of FLT3 in hematopoiesis and leukemia, Blood, 100 (2002) 1532–1542. [DOI] [PubMed] [Google Scholar]
- [28].Choudhary C, Schwable J, Brandts C, Tickenbrock L, Sargin B, Kindler T, Fischer T, Berdel WE, Muller-Tidow C, Serve H, AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations, Blood, 106 (2005) 265–273. [DOI] [PubMed] [Google Scholar]
- [29].Mizuki M, Schwable J, Steur C, Choudhary C, Agrawal S, Sargin B, Steffen B, Matsumura I, Kanakura Y, Bohmer FD, Muller-Tidow C, Berdel WE, Serve H, Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations, Blood, 101 (2003) 3164–3173. [DOI] [PubMed] [Google Scholar]
- [30].Grundler R, Miething C, Thiede C, Peschel C, Duyster J, FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model, Blood, 105 (2005) 4792–4799. [DOI] [PubMed] [Google Scholar]
- [31].Drexler HG, Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells, Leukemia, 10 (1996) 588–599. [PubMed] [Google Scholar]
- [32].Hawley TS, T.T.H. From the Oncology Gene Therapy Program, and the Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada, U.o.W.r. the Institute of Pathology, Würzburg, Germany, S. and Immunex Corp, WA., A.Z.C. Fong, T.T.H. From the Oncology Gene Therapy Program, and the Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada, U.o.W.r. the Institute of Pathology, Würzburg, Germany, S. and Immunex Corp, WA., H. Griesser, T.T.H. From the Oncology Gene Therapy Program, and the Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada, U.o.W.r. the Institute of Pathology, Würzburg, Germany, S. and Immunex Corp, WA., S.D. Lyman, T.T.H. From the Oncology Gene Therapy Program, and the Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada, U.o.W.r. the Institute of Pathology, Würzburg, Germany, S. and Immunex Corp, WA., R.G. Hawley, T.T.H. From the Oncology Gene Therapy Program, and the Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada, U.o.W.r. the Institute of Pathology, Würzburg, Germany, S. and Immunex Corp, WA., Leukemic Predisposition of Mice Transplanted With Gene-Modified Hematopoietic Precursors Expressing flt3 Ligand, Blood, 92 (2019) 2003–2011. [Google Scholar]
- [33].Abu-Duhier FM, Goodeve AC, Wilson GA, Gari MA, Peake IR, Rees DC, Vandenberghe EA, Winship PR, Reilly JT, FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group, Br J Haematol, 111 (2000) 190–195. [DOI] [PubMed] [Google Scholar]
- [34].Zheng R, Levis M, Piloto O, Brown P, Baldwin BR, Gorin NC, Beran M, Zhu Z, Ludwig D, Hicklin D, Witte L, Li Y, Small D, FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells, Blood, 103 (2004) 267–274. [DOI] [PubMed] [Google Scholar]
- [35].Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, Brandts C, Serve H, Roesel J, Giles F, Feldman E, Ehninger G, Schiller GJ, Nimer S, Stone RM, Wang Y, Kindler T, Cohen PS, Huber C, Fischer T, Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain, Blood, 107 (2006) 293–300. [DOI] [PubMed] [Google Scholar]
- [36].von Bubnoff N, Engh RA, Aberg E, Sanger J, Peschel C, Duyster J, FMS-like tyrosine kinase 3-internal tandem duplication tyrosine kinase inhibitors display a nonoverlapping profile of resistance mutations in vitro, Cancer Res, 69 (2009) 3032–3041. [DOI] [PubMed] [Google Scholar]
- [37].Serve H, Krug U, Wagner R, Sauerland MC, Heinecke A, Brunnberg U, Schaich M, Ottmann O, Duyster J, Wandt H, Fischer T, Giagounidis A, Neubauer A, Reichle A, Aulitzky W, Noppeney R, Blau I, Kunzmann V, Stuhlmann R, Kramer A, Kreuzer KA, Brandts C, Steffen B, Thiede C, Muller-Tidow C, Ehninger G, Berdel WE, Sorafenib in combination with intensive chemotherapy in elderly patients with acute myeloid leukemia: results from a randomized, placebo-controlled trial, J Clin Oncol, 31 (2013) 3110–3118. [DOI] [PubMed] [Google Scholar]
- [38].Fischer T, Stone RM, Deangelo DJ, Galinsky I, Estey E, Lanza C, Fox E, Ehninger G, Feldman EJ, Schiller GJ, Klimek VM, Nimer SD, Gilliland DG, Dutreix C, HuntsmanLabed A, Virkus J, Giles FJ, Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3, J Clin Oncol, 28 (2010) 4339–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Levis M, Ravandi F, Wang ES, Baer MR, Perl A, Coutre S, Erba H, Stuart RK, Baccarani M, Cripe LD, Tallman MS, Meloni G, Godley LA, Langston AA, Amadori S, Lewis ID, Nagler A, Stone R, Yee K, Advani A, Douer D, Wiktor-Jedrzejczak W, Juliusson G, Litzow MR, Petersdorf S, Sanz M, Kantarjian HM, Sato T, Tremmel L, Bensen-Kennedy DM, Small D, Smith BD, Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse, Blood, 117 (2011) 3294–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Piloto O, Levis M, Huso D, Li Y, Li H, Wang M-N, Bassi R, Balderes P, Ludwig DL, Witte L, Zhu Z, Hicklin DJ, Small D, Inhibitory Anti-FLT3 Antibodies Are Capable of Mediating Antibody-Dependent Cell-Mediated Cytotoxicity and Reducing Engraftment of Acute Myelogenous Leukemia Blasts in Nonobese Diabetic/Severe Combined Immunodeficient Mice, (2005). [DOI] [PubMed] [Google Scholar]
- [41].Sanford D, Blum WG, Ravandi F, Klisovic RB, Borthakur G, Walker AR, Garcia-Manero G, Marcucci G, Wierda WG, Whitman SP, Kantarjian HM, Cortes JE, Efficacy and safety of an anti-FLT3 antibody (LY3012218) in patients with relapsed acute myeloid leukemia, Journal of Clinical Oncology, 33 (2015) 7059–7059. [Google Scholar]
- [42].Sanchez L, Ayala M, Freyre F, Pedroso I, Bell H, Falcon V, Gavilondo JV, High cytoplasmic expression in E. coli, purification, and in vitro refolding of a single chain Fv antibody fragment against the hepatitis B surface antigen, J. Biotechnol, 72 (1999) 13–20. [DOI] [PubMed] [Google Scholar]
- [43].Kipriyanov SM, Moldenhauer G, Little M, High level production of soluble single chain antibodies in small-scale Escherichia coli cultures, J. Immunol. Methods, 200 (1997) 69–77. [DOI] [PubMed] [Google Scholar]
- [44].Heo MA, Kim SH, Kim SY, Kim YJ, Chung J, Oh MK, Lee SG, Functional expression of single-chain variable fragment antibody against c-Met in the cytoplasm of Escherichia coli, Protein Expr. Purif, 47 (2006) 203–209. [DOI] [PubMed] [Google Scholar]
- [45].Sabaty M, Grosse S, Adryanczyk G, Boiry S, Biaso F, Arnoux P, Pignol D, Detrimental effect of the 6 His C-terminal tag on YedY enzymatic activity and influence of the TAT signal sequence on YedY synthesis, BMC Biochem, 14 (2013) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goel A, Colcher D, Koo JS, Booth BJ, Pavlinkova G, Batra SK, Relative position of the hexahistidine tag effects binding properties of a tumor-associated single-chain Fv construct, Biochim. Biophys. Acta, 1523 (2000) 13–20. [DOI] [PubMed] [Google Scholar]
- [47].Worn A, Pluckthun A, Different equilibrium stability behavior of ScFv fragments: identification, classification, and improvement by protein engineering, Biochemistry, 38 (1999) 8739–8750. [DOI] [PubMed] [Google Scholar]
- [48].Brockmann EC, Cooper M, Stromsten N, Vehniainen M, Saviranta P, Selecting for antibody scFv fragments with improved stability using phage display with denaturation under reducing conditions, J. Immunol. Methods, 296 (2005) 159–170. [DOI] [PubMed] [Google Scholar]
- [49].Worn A, Pluckthun A, Stability engineering of antibody single-chain Fv fragments, J. Mol. Biol, 305 (2001) 989–1010. [DOI] [PubMed] [Google Scholar]
- [50].Wingelhofer B, Maurer B, Heyes EC, Cumaraswamy AA, Berger-Becvar A, de Araujo ED, Orlova A, Freund P, Ruge F, Park J, Tin G, Ahmar S, Lardeau CH, Sadovnik I, Bajusz D, Keserű GM, Grebien F, Kubicek S, Valent P, Gunning PT, Moriggl R, Pharmacologic inhibition of STAT5 in acute myeloid leukemia, in: Leukemia, 2018, pp. 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Benekli M, Baumann H, Wetzler M, Targeting Signal Transducer and Activator of Transcription Signaling Pathway in Leukemias, in: J Clin Oncol, 2009, pp. 4422–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pavlinkova G, Beresford GW, Booth BJ, Batra SK, Colcher D, Pharmacokinetics and biodistribution of engineered single-chain antibody constructs of MAb CC49 in colon carcinoma xenografts, J. Nucl. Med, 40 (1999) 1536–1546. [PubMed] [Google Scholar]
- [53].Pavlinkova G, Beresford G, Booth BJ, Batra SK, Colcher D, Charge-modified single chain antibody constructs of monoclonal antibody CC49: generation, characterization, pharmacokinetics, and biodistribution analysis, Nucl. Med. Biol, 26 (1999) 27–34. [DOI] [PubMed] [Google Scholar]
- [54].Rios X, Compte M, Gomez-Vallejo V, Cossio U, Baz Z, Morcillo MA, Ramos-Cabrer P, Alvarez-Vallina L, Llop J, Immuno-PET Imaging and Pharmacokinetics of an Anti-CEA scFvbased Trimerbody and Its Monomeric Counterpart in Human Gastric Carcinoma-Bearing Mice, Mol. Pharm, 16 (2019) 1025–1035. [DOI] [PubMed] [Google Scholar]
- [55].Muller D, Karle A, Meissburger B, Hofig I, Stork R, Kontermann RE, Improved pharmacokinetics of recombinant bispecific antibody molecules by fusion to human serum albumin, J. Biol. Chem, 282 (2007) 12650–12660. [DOI] [PubMed] [Google Scholar]
- [56].Yang K, Basu A, Wang M, Chintala R, Hsieh MC, Liu S, Hua J, Zhang Z, Zhou J, Li M, Phyu H, Petti G, Mendez M, Janjua H, Peng P, Longley C, Borowski V, Mehlig M, Filpula D, Tailoring structure-function and pharmacokinetic properties of single-chain Fv proteins by site-specific PEGylation, Protein Eng, 16 (2003) 761–770. [DOI] [PubMed] [Google Scholar]
- [57].Li Q, Hudson W, Wang D, Berven E, Uckun FM, Kersey JH, Pharmacokinetics and biodistribution of radioimmunoconjugates of anti-CD19 antibody and single-chain Fv for treatment of human B-cell malignancy, Cancer Immunol. Immunother, 47 (1998) 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mu CF, Shen J, Liang J, Zheng HS, Xiong Y, Wei YH, Li F, Targeted drug delivery for tumor therapy inside the bone marrow, Biomaterials, 155 (2018) 191–202. [DOI] [PubMed] [Google Scholar]
- [59].Reth M, Matching cellular dimensions with molecular sizes, Nat. Immunol, 14 (2013) 765–767. [DOI] [PubMed] [Google Scholar]
- [60].Stylianopoulos T, EPR-effect: utilizing size-dependent nanoparticle delivery to solid tumors, Ther. Deliv, 4 (2013) 421–423. [DOI] [PubMed] [Google Scholar]
- [61].Sarin H, Physiologic upper limits of pore size of different blood capillary types and another perspective on the dual pore theory of microvascular permeability, J. Angiogenes. Res., 2 (2010) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bergqvist L, Sundberg R, Ryden S, Strand SE, The Critical Colloid Dose in Studies of Reticuloendothelial Function, J. Nucl. Med, 28 (1987) 1424–1429. [PubMed] [Google Scholar]
- [63].Passaro D, Di Tullio A, Abarrategi A, Rouault-Pierre K, Foster K, Ariza-McNaughton L, Montaner B, Chakravarty P, Bhaw L, Diana G, Lassailly F, Gribben J, Bonnet D, Increased Vascular Permeability in the Bone Marrow Microenvironment Contributes to Disease Progression and Drug Response in Acute Myeloid Leukemia, Cancer Cell, 32 (2017) 324–341 e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.